In Lotus japonicus, the LHK1 cytokinin receptor performs an essential function but also works partially redundantly with LHK1A and LHK3 to mediate nodule primordium formation within the root cortex. LHK1 is also expressed in the root epidermis, where it likely participates in signaling to restrict rhizobial infection.
Abstract
Previous analysis of the Lotus histidine kinase1 (Lhk1) cytokinin receptor gene has shown that it is required and also sufficient for nodule formation in Lotus japonicus. The L. japonicus mutant carrying the loss-of-function lhk1-1 allele is hyperinfected by its symbiotic partner, Mesorhizobium loti, in the initial absence of nodule organogenesis. At a later time point following bacterial infection, lhk1-1 develops a limited number of nodules, suggesting the presence of an Lhk1-independent mechanism. We have tested a hypothesis that other cytokinin receptors function in at least a partially redundant manner with LHK1 to mediate nodule organogenesis in L. japonicus. We show here that L. japonicus contains a small family of four cytokinin receptor genes, which all respond to M. loti infection. We show that within the root cortex, LHK1 performs an essential role but also works partially redundantly with LHK1A and LHK3 to mediate cell divisions for nodule primordium formation. The LHK1 receptor is also presumed to partake in mediating a feedback mechanism that negatively regulates bacterial infections at the root epidermis. Interestingly, the Arabidopsis thaliana AHK4 receptor gene can functionally replace Lhk1 in mediating nodule organogenesis, indicating that the ability to perform this developmental process is not determined by unique, legume-specific properties of LHK1.
INTRODUCTION
Intracellular accommodation of nitrogen-fixing rhizobia is supported by many leguminous plants as well as a single nonlegume genus, Parasponia, of the Ulmaceae family (Doyle, 2011; Op den Camp et al., 2011; Santi et al., 2013). In most cases, a host plant builds organs, called root nodules, which provide lodging and optimal conditions for symbiotic bacteria to perform nitrogen fixation (Sprent and James, 2007; Desbrosses and Stougaard, 2011).
In the majority of legume-Rhizobium systems that have been analyzed, the main stimulus that initiates nodule formation comes from a compatible bacterium in the form of chemically decorated lipochitin oligosaccharide molecules, known as nodulation or Nod factors (NFs) (Lerouge et al., 1990; Bek et al., 2010). Perception of NFs by the host plant LysM motif kinase receptors (Madsen et al., 2003; Radutoiu et al., 2003) activates an ancient root response pathway, called the common symbiosis pathway (Duc et al., 1989; Kistner et al., 2005). Together with other signaling events, this mediates and coordinates bacterial entry inside the root with the formation of nodule structures (Madsen et al., 2010; Held et al., 2010; Hossain et al., 2012). As a result, functional nitrogen-fixing root nodules are developed, rendering the growth of the host plant independent from soil nitrogen.
Entry into the root or even the physical presence of bacteria is not required, as the application of NFs is sufficient to induce cell divisions for nodule structure formation (Truchet et al., 1991). Central to this process is the ability of NFs to incite the functioning of the common symbiosis pathway, which evokes and then interprets intracellular Ca2+ signaling (Wais et al., 2000; Walker and Downie, 2000; Sieberer et al., 2012). This leads to the activation of downstream effectors for rhizobial infection and nodule formation, including several key transcription regulators such as NODULATION SIGNALING PATHWAY1 (NSP1), NSP2, NODULE INCEPTION (NIN), and NUCLEAR FACTOR Y (NF-Y) (Schauser et al., 1999; Kaló et al., 2005; Smit et al., 2005; Combier et al., 2006, 2008; Heckmann et al., 2006; Murakami et al., 2006; Marsh et al., 2007; Middleton et al., 2007; Laloum et al., 2013; Sayano et al., 2013). CALCIUM- AND CALMODULIN-DEPENDENT KINASE (CCaMK), an element of the common symbiosis pathway, is presumed to be the key interpreter of NF-dependent Ca2+ signaling (Hayashi et al., 2010; Liao et al., 2012; Singh and Parniske, 2012). Deleterious mutations in the CCaMK gene prevent the transcriptional reprograming of plant cells for symbiosis, effectively blocking bacterial infection and also abolishing nodule primordium formation in the root cortex (Lévy et al., 2004; Mitra et al., 2004). Conversely, gain-of-function, autoactivated CCaMK molecules, such as CCaMKT265I and CCaMKT265D, induce spontaneous nodule formation in the absence of rhizobia or NFs, indicating that the activation of CCaMK is sufficient for nodule structure formation (Gleason et al., 2006; Tirichine et al., 2006a; Hayashi et al., 2010).
Early research suggested the involvement of the plant hormone auxin in nodule formation (Thimann, 1936), and this has been confirmed by subsequent studies (Hirsch et al., 1989; Grunewald et al., 2009). In both Lotus japonicus, which develops determinate nodules that lack permanent meristem, and Medicago truncatula indeterminate nodules, auxin signaling is thought to be generated downstream from NF perception and cytokinin signaling to participate in the regulation of cell divisions that build a nodule primordium (Mathesius et al., 2000; Plet et al., 2011; Suzaki et al., 2012).
An elegant study by Cooper and Long (1994) demonstrated that the cytokinin trans-zeatin synthesis and secretion system engineered into Sinorhizobium meliloti functionally replaced NF in its ability to incite nodule primordium formation in M. truncatula. A similar effect can be obtained without bacteria by the external application of a small amount of cytokinin to legume roots, including L. japonicus (Bauer et al., 1996; Heckmann et al., 2011).
To date, no direct evidence exists to support the involvement of bacterially produced cytokinin in nodule formation. To the contrary, the identification of spontaneous nodule formation phenotypes in tetraploid alfalfa (Medicago sativa) (Truchet et al., 1989) and in mutants of diploid L. japonicus (Tirichine et al., 2006b) indicates the presence of an inherent plant signaling process for nodule formation. Current data are most consistent with a model in which the NF-dependent activation of CCaMK leads to local accumulation of cytokinin, which, in turn, stimulates root cortical cell divisions for nodule primordium formation (Frugier et al., 2008). Such an interpretation is in agreement with the observation that L. japonicus plants carrying the spontaneous nodule formation1 (snf1) gain-of-function allele of CCaMK do not produce spontaneous nodules in the absence of the functional L. japonicus HISTIDINE KINASE1 (LHK1) cytokinin receptor (Madsen et al., 2010). Conversely, the L. japonicus snf2 gain-of-function allele of Lhk1 induces spontaneous nodule formation independent of CCaMK, supporting an epistatic relationship in which Lhk1 acts downstream from CCaMK (Tirichine et al., 2007).
Using a combined bioinformatics and functional genomics approach, Ariel et al. (2012) demonstrated that NSP2, one of the earliest acting transcriptional switches for symbiotic differentiation of the host cells (Kaló et al., 2005; Heckmann et al., 2006; Murakami et al., 2006), was among numerous genes that contain the M. truncatula nodule-associated Response Regulator1 binding site and are rapidly upregulated by ectopic cytokinin. Previous data have documented that, in addition to NSP2, other genes that encode nodule-associated transcriptional regulators, such as NSP1, ERN1, NIN, and NF-YA1 (formerly HAP2A), respond to ectopic cytokinin, although this response necessitates the presence of active protein biosynthesis (Murray et al., 2007; Heckmann et al., 2011; Plet et al., 2011). Cytokinin receptors, LHK1 in L. japonicus and CRE1 in M. truncatula, were shown to be essential in this context (Gonzalez-Rizzo et al., 2006; Murray et al., 2007; Heckmann et al., 2011; Plet et al., 2011), confirming and extending the genetic data demonstrating that the snf2-dependent spontaneous nodule formation requires NSP2 and NIN (Tirichine et al., 2007).
Functional analyses of loss-of-function and gain-of-function alleles of LHK1 defined this cytokinin receptor as being required and also sufficient for nodule organogenesis in L. japonicus (Murray et al., 2007; Tirichine et al., 2007; Heckmann et al., 2011). Unlike M. truncatula cre1, the L. japonicus mutant carrying a loss-of function lhk1-1 allele (formerly known as hit1; Murray et al., 2006) is hyperinfected by Mesorhizobium loti, with infection threads heavily present in segments of the root epidermis and cortex in the initial absence of nodule organogenesis (Murray et al., 2007). lhk1-1 develops a limited number of nodules at a later time point upon inoculation by M. loti (Murray et al., 2007), suggesting the presence of an LHK1-independent signaling mechanism for nodule formation. Therefore, we tested the hypothesis that other cytokinin receptors function in at least a partially redundant manner with LHK1 to mediate nodule organogenesis in L. japonicus. Our data demonstrate that LHK1 exerts a unique function in the root epidermis but works partially redundantly with LHK1A and LHK3 within the root cortex to mediate nodule formation.
RESULTS
The L. japonicus Cytokinin Receptor Gene Family Comprises at Least Four Members
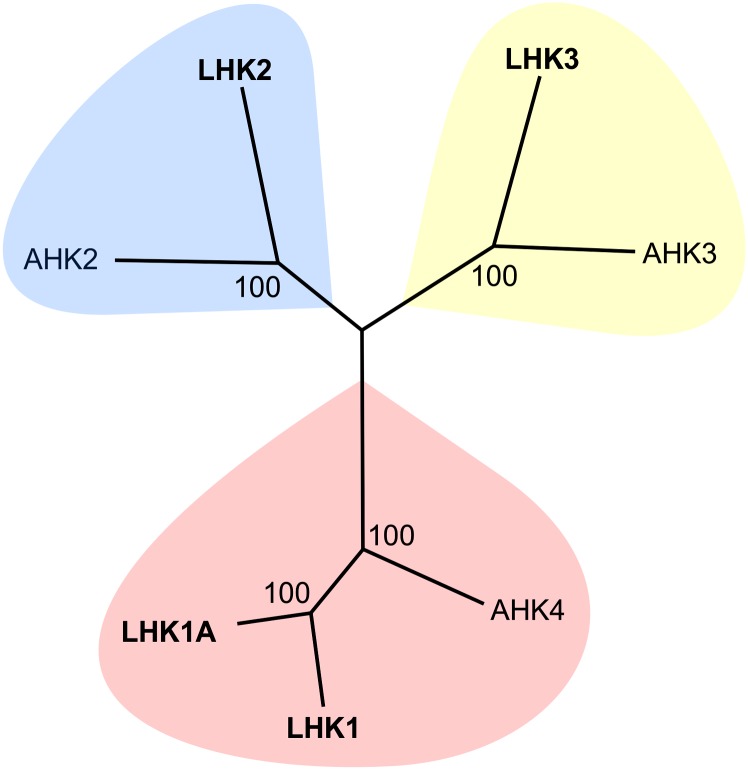
A search of the L. japonicus genome and cDNA sequences resulted in the prediction of a small family of four Lhk cytokinin receptor genes, including the previously described Lhk1 and Lhk2 (Murray et al.., 2007). Based on the clustering pattern of the corresponding LHK proteins with Arabidopsis thaliana cytokinin receptors (Figure 1), the Lhk2 gene was renamed Lhk1A, while the two putative cytokinin receptor genes described in this study were named Lhk2 and Lhk3. The corresponding L. japonicus proteins are referred to as LHK1, LHK1A, LHK2, and LHK3 (Figure 1).
Figure 1.
The L. japonicus LHK Protein Family.
This unrooted relationship tree is based on an amino acid alignment of full-length sequences from L. japonicus (LHK1, LHK1A [formerly LHK2; Murray et al., 2007], LHK2, and LHK3) and Arabidopsis (AHK4, AHK2, and AHK3). Protein sequences were aligned with Clustal Omega using the default settings (Supplemental Data Set 1), and the MEGA 6.0.5 phylogeny tool was used to portray the relationships between proteins. The LHK3 variant 1 protein (Supplemental Figure 4) was used for the alignment.
Full-length transcripts were obtained for three Lhk mRNAs (i.e., Lhk1A, Lhk2, and Lhk3; see Methods), and these were used to decipher the corresponding gene structures. Like Lhk1, the Lhk3 gene has 11 exons, while 12 and 14 exons are predicted for the Lhk1A and Lhk2 genes, respectively. The length of the predicted open reading frames was found to be 2991, 3654, and 2958 bp for Lhk1A, Lhk2, and Lhk3, respectively; these reflect the corresponding proteins of 997, 1218, and 986 amino acids.
LHK Proteins Contain Domains Characteristic of Known Cytokinin Receptors
To test the prediction that, like LHK1, the LHK1A, LHK2, and LHK3 proteins constitute bona fide cytokinin receptors, their amino acid sequences were analyzed. Subsequently, functional assays in heterologous yeast and Escherichia coli systems were performed (see below).
The identity and similarity of LHK protein sequences within the predicted L. japonicus cytokinin receptor family and to representatives from Arabidopsis are summarized in Supplemental Table 1. Briefly, LHK1 and LHK1A are the most similar, sharing 80% identity at the amino acid level. Conservation between these two proteins and the Arabidopsis Histidine Kinase 4 (AHK4) cytokinin receptor is also high at 68 and 69%, respectively. In contrast, LHK1 and LHK1A share only ∼50% identity with LHK2 and LHK3. Amino acid sequence conservation is greater between LHK2 and LHK3 and also to their presumed orthologs from Arabidopsis (AHK2 and AHK3, respectively) than between these two proteins and LHK1 or LHK1A (Supplemental Table 1).
The N-terminal portions of all four LHKs contain a predicted cyclase/histidine kinase–associated sensory extracellular (CHASE) domain (Heyl et al., 2007, 2012), which is highly conserved within the LHK family and also between LHKs and CHASE domains of other known cytokinin receptors, such as Arabidopsis AHK4 (Supplemental Figure 1).
As expected, the predicted cytosolic portion of the LHK proteins contains the highly conserved kinase domain (Hwang et al., 2012) with the canonical H, N, G1, F, and G2 consensus motifs and a highly conserved His (H) residue (Supplemental Figure 2). Downstream, a C-terminal receiver or output domain is also present in all predicted LHK receptors. This domain is known to participate in the phosphotransfer from the kinase domain to downstream signaling elements, such as His phosphotransfer proteins (Ferreira and Kieber, 2005). The functional receiver domain carries three characteristic motifs named the DD, D, and K motifs for their conserved amino acid residues (Ueguchi et al.., 2001). All of these conserved motifs are present in the predicted LHK receptors, including an absolutely invariant Asp residue in the D motif (Supplemental Figure 3).
Lhk Transcripts Are Present in Different L. japonicus Tissues
A survey of L. japonicus gene atlas data (http://ljgea.noble.org/v2/; Verdier et al., 2013) shows that the Lhk transcripts are present in all organs analyzed, including roots, stems, leaves, and nodules. Two variants of the Lhk3 mRNA (variants 1 and 2) were identified through 5′ rapid amplification of cDNA ends experiments (Supplemental Figure 4A). These variants differ in length, with Lhk3 mRNA variant 2 being longer by 228 bp in comparison with variant 1. Transcript-specific primers were designed against each predicted Lhk3 mRNA variant (see Methods), and the presence of both mRNA species was confirmed via RT-PCR of L. japonicus nodule total RNA (Supplemental Figure 4B).
Identification of Cytokinin Receptor Mutant Alleles
The targeted induced localized lesions in genomes (TILLING) approach was employed to identify mutations in the Lhk1A, Lhk2, and Lhk3 loci. A 1-kb region within the highly conserved kinase domain was targeted (see Methods). Several L. japonicus lines carrying single-nucleotide substitutions were identified in the Lhk1A, Lhk2, and Lhk3 genes (Supplemental Table 2). For Lhk1A, a mutant line carrying the Gly4439-to-Ala transition (named lhk1a-1) was chosen for detailed analyses, as this was predicted to change a Trp residue in the kinase domain to a premature stop codon (Trp-565 to stop; Supplemental Table 2). For the Lhk2 and Lhk3 loci, mutant lines carrying the lhk2-5 and lhk3-1 alleles were selected, as the corresponding mutations were predicted to change invariant residues within the conserved G1 (Gly-605 to Arg) and N (Arg-561 to Gln) box motifs of the kinase domain, respectively (Supplemental Figure 2). The original TILLING lines were backcrossed to wild-type L. japonicus ecotype Gifu, and homozygote lhk1a-1, lhk2-5, and lhk3-1 single mutant individuals were selected from among segregating F2 individuals. Their progeny were used in subsequent analyses (see Methods).
Lhk1A Confers Cytokinin-Responsive Growth to the sln1Δ Mutant of Saccharomyces cerevisiae
Wild-type and mutant alleles were used in parallel to functionally evaluate different LHKs in the sln1Δ mutant of S. cerevisiae (Maeda et al., 1994). We previously used this yeast strain to demonstrate the cytokinin-responsive function of the LHK1 receptor and the deleterious nature of the lhk1-1 mutation (Murray et al., 2007).
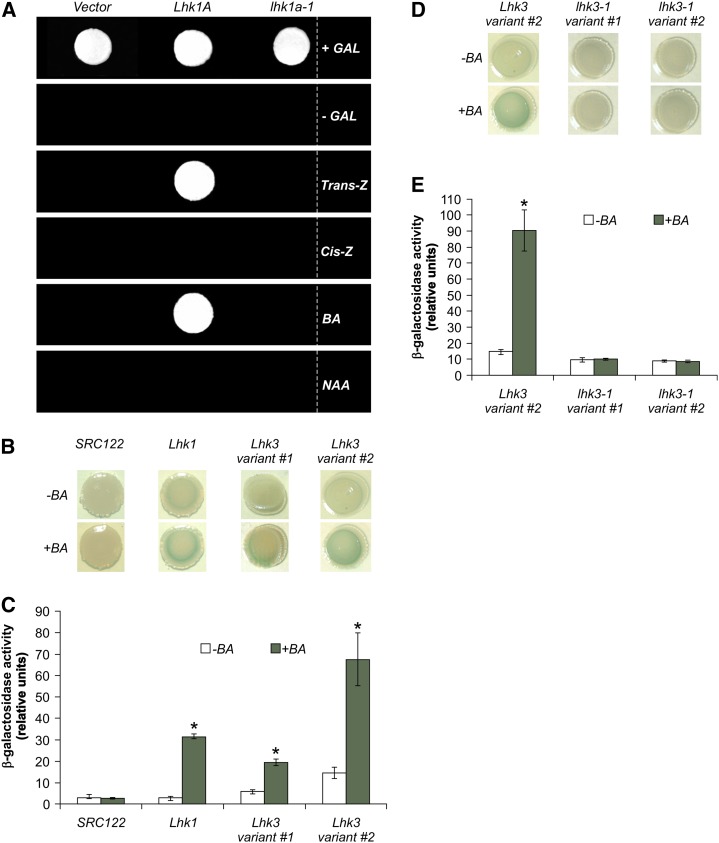
Like Lhk1, the wild-type Lhk1A cDNA restored the viability of the sln1Δ strain in a cytokinin-dependent fashion. In contrast, the lhk1a-1 cDNA failed to do so, demonstrating the deleterious nature of the lhk1a-1 mutation (Figure 2A).
Figure 2.
Lhk1A and Lhk3 Encode Functional Cytokinin Receptors.
(A) In the absence of Gal (-GAL), the wild-type Lhk1A cDNA confers cytokinin (Trans-Z and BA)-dependent growth of the sln1Δ mutant of S. cerevisiae. The lhk1a-1 mutant cDNA is unable to do so. +Gal, 2% Gal supplement; Trans-Z, trans-zeatin; Cis-Z, cis-zeatin; NAA, 1-naphthaleneacetic acid.
(B) to (E) Wild-type ([B] and [C]) and lhk3-1 mutant ([D] and [E]) cDNAs corresponding to the two Lhk3 mRNA variants (variants 1 and 2) were cloned into the pSTV28 expression vector and transformed into the sensor-negative E. coli SRC122 strain. The Lhk1 cDNA was used as a positive control for the experiments shown in (B) and (C). Upon application of BA, a noticeable increase in the β-galactosidase reporter activity (blue color) can be observed in the presence of Lhk1 and both wild-type variants of Lhk3 cDNA ([B] and [D]). Quantification of the β-galactosidase activity as driven by different Lhk cDNAs in the presence or absence of BA is shown in (C) and (E). SRC122 is the sensor-negative E. coli strain used as a negative control. The lhk3-1 mutation abolishes the cytokinin-responsive function of the LHK3 receptor ([D] and [E]). The wild-type Lhk3 variant 2 was used as a positive control in (D) and (E). In all cases, values represent means ± 95% confidence interval (n = 3). Asterisks denote significant differences (Student’s t test, P < 0.05).
Repeated attempts to perform a similar functional study with Lhk2 and the two variants of the Lhk3 cDNA have failed due to an apparent toxicity of the products of these cDNAs in the yeast cells. Therefore, an alternative approach based on an E. coli two-component phosphorelay assay (Yamada et al., 2001; Tirichine et al., 2007) was used.
Lhk3 Confers Cytokinin Responsiveness to the Sensor-Negative SRC122 E. coli Mutant Strain
The introduction of a functional His kinase receptor and an appropriate ligand to the SRC122 E. coli strain results in activation of the cps:LacZ reporter fusion (Yamada et al., 2001). Application of cytokinin to the SRC122 E. coli strain carrying wild-type copies of either of the two splice variants of Lhk3 cDNA significantly induced the β-galactosidase activity above the control level of the untreated samples, thus confirming their cytokinin-responsive function (Figures 2B and 2C). The lhk3-1 mutation completely abolished the cytokinin responsiveness, irrespective of the cDNA variant used, which indicates that the lhk3-1 mutant form is nonfunctional (Figures 2D and 2E).
When transformed into SRC122, the Lhk2 cDNA–containing replicon was highly unstable, such that no intact receptor sequence could be recovered. This outcome was not entirely unexpected, given the reports of similar problems with the Arabidopsis AHK2 gene (Yamada et al., 2001). Thus, the LHK2 receptor remains functionally undefined by this work and is considered hereafter as a presumed cytokinin receptor.
Single and Double Mutants of lhk1a-1, lhk2-5, and lhk3-1 Do Not Affect Nodule Formation
Early events that characterize the epidermal program for symbiosis, such as the formation of bacterial microcolonies trapped within curled root hairs and the subsequent development of infection threads, were evaluated 7 d after inoculation (DAI) with an M. loti strain carrying the hemA:LacZ reporter gene fusion. Unlike the hyperinfected root phenotype of lhk1-1 (Murray et al., 2007), lhk1a-1, lhk2-5, and lhk3-1 single mutants displayed a wild-type number of infection events (Supplemental Figure 5A). Furthermore, the number of nodule primordia and nodules were at wild-type levels in lhk1a-1 and lhk3-1 single mutants (Supplemental Figure 5B). The lhk2-5 mutant formed slightly but significantly fewer nodules than the wild type (Supplemental Figure 5B). However, this is likely an indirect effect, as the overall growth of the lhk2 mutant, including root elongation (Supplemental Figure 6B), was also significantly affected. Under the same growth conditions, lhk1-1 formed a strongly reduced number of nodules, confirming our previous data (Murray et al., 2007). All double receptor mutants also had wild-type or close to wild-type (in the case of plants carrying the lhk2-5 allele; see above) nodulation phenotypes, except for those carrying the lhk1-1 allele, where a greatly reduced number of nodules was apparent (Supplemental Figure 5B).
LHK1 Is the Main Sensor of Exogenous Cytokinin
In wild-type L. japonicus plants, root elongation is significantly inhibited by the external application of cytokinin (Murray et al., 2007). Deleterious mutations in the Lhk1 receptor gene, such as lhk1-1, render mutant roots insensitive to exogenous 6-benzylaminopurine (BA) up to 10−7 M (Murray et al., 2007). This indicates that, in addition to its significant role during nodule organogenesis, the LHK1 receptor mediates root responses to external signals, such as cytokinin. To analyze whether other L. japonicus cytokinin receptors partake of this physiological response, all L. japonicus lhk mutant lines, including lhk1-1, were subjected to the root elongation assay in the presence or absence of BA. In contrast with lhk1-1, which was insensitive to external cytokinin, lhk1a-1, lhk2-5, and lhk3-1 mutants responded to the exogenous BA by reducing root growth in a manner similar to the wild-type L. japonicus (Supplemental Figure 6A). It is worth noting here that the growth of untreated lhk1-1, lhk1a-1, and lhk3-1 mutant roots was not significantly different from that of the wild type. The root length of lhk2-5, however, was slightly yet significantly affected (Supplemental Figure 6B).
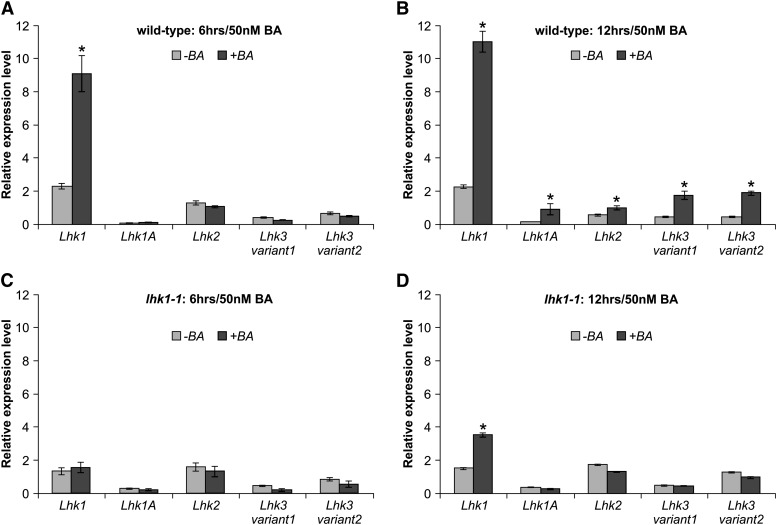
To further understand the unique role of LHK1 during the response of L. japonicus roots to ectopic cytokinin, we quantified steady state levels of the Lhk1 mRNA upon application of 50 nM BA. The four remaining cytokinin receptor mRNAs, including two variants of Lhk3, were also included in this analysis. The level of the Lhk1 mRNA was strongly and significantly upregulated within 3 h of BA treatment (Supplemental Figure 7), and this increase was further enhanced at 6 and 12 h (Figures 3A and 3B). By contrast, the steady state levels of the Lhk1A, Lhk2, and both variants of Lhk3 mRNA were significantly upregulated only after prolonged (12 h) treatment with BA (Figure 3B). Importantly, this increase in the levels of Lhk1A, Lhk2, and Lhk3 mRNAs in response to BA was not observed in lhk1-1 roots (Figures 3C and 3D), while the lhk1-1 mRNA still responded by a slight but significant increase at 12 h of the BA treatment (Figure 3D).
Figure 3.
Ectopic Cytokinin Increases the Steady State Level of Cytokinin Receptor mRNAs.
The relative steady state levels of Lhk1, Lhk1A, Lhk2, and Lhk3 transcripts in untreated (−BA) and cytokinin-treated (+BA) L. japonicus roots are given. Values (means ± se of three biological replicates) for L. japonicus wild-type ([A] and [B]) and lhk1-1 mutant ([C] and [D]) roots that were incubated in the absence or presence of 50 nM BA for 6 h ([A] and [C]) and 12 h ([B] and [D]) are shown.
The Transcriptional Output of Cytokinin Signaling during Nodule Formation
The results described above suggested a unique role for the LHK1 receptor in mediating nodule organogenesis and also in root responses to exogenous cytokinin. To explore this further, stable L. japonicus transgenic lines carrying the cytokinin two-component output sensor (TCS):GUS reporter (Müller and Sheen, 2008) or one of the four Lhk promoters transcriptionally fused to the GUS reporter gene (Lhkpro:GUS) were analyzed to map their expression domains (see Methods).
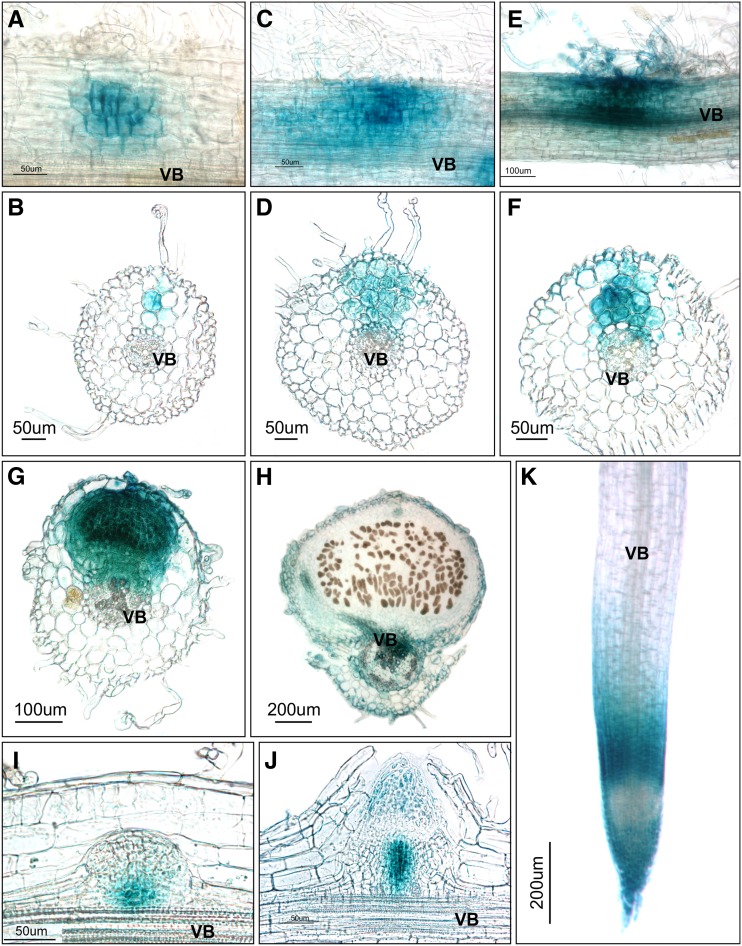
Following M. loti inoculation, the TCS-mediated GUS activity was initially detectable in the subepidermal layer and the second and third cortical cell layers, with the latter two showing most of the activity (Figures 4A and 4B). At this early stage in nodule development, GUS staining was observed only intermittently in the root epidermis, and if present, it was weak. At a slightly later developmental stage, GUS staining was present in cell layers encompassing the entire region from the root epidermis to the pericycle (Figures 4C and 4D) and continued to intensify across all cell layers that were actively engaged in nodule formation and also in the associated epidermis, including root hairs (Figures 4E and 4F). GUS activity was present in most cells of young nodules that had just emerged from the root epidermis (Figure 4G), but it was undetectable in fully mature nodules, except for a weak activity in the nodule parenchyma and vascular bundles (Figure 4H).
Figure 4.
Cytokinin Responses during Nodule and Root Development.
The TCS:GUS cytokinin output reporter activities are depicted (blue color) as associated with various stages of developing nodules ([A] to [H]), lateral roots ([I] and [J]), and the apical region of the main root (K). All images represent specimens collected at 7 or 14 DAI with M. loti. (A), (C), (E), and (K) represent whole mounts, and the other panels show ∼35-μm sections. VB, vascular bundles.
In comparison, during lateral root development, the TCS-driven GUS activity maxima were associated with a subset of proliferating cells at the base of developing primordia (Figure 4I) and, later on, also with the root apex (Figure 4J), consistent with the previous report (Lohar et al., 2004). In mature roots, GUS staining was mostly confined to the root apex and the root transition zone but was much weaker within the root proximal meristem (Figure 4K).
Expression of Lhk1, Lhk1A, Lhk2, and Lhk3 in Uninoculated L. japonicus Roots
Lhk1pro:GUS plants showed strong histochemical staining along the entire roots (Supplemental Figure 8A), including the root apex and a portion that is defined as a susceptible zone for M. loti infection, which spans the region positioned between 2 and 5 mm above the root tip (Supplemental Figure 8C). Longitudinal sectioning of several independent samples of Lhk1pro:GUS roots that were stained at room temperature (see Methods) showed GUS activity in all root cell layers, including the root epidermis; however, GUS staining was much weaker or entirely absent from cells of the proximal meristem (Supplemental Figure 8B) and in root cells located in a more proximal part of the root, above the root transition zone.
None of the other three Lhk promoters paralleled the activity of the Lhk1 promoter. As driven by the Lhk1A promoter, GUS activity was barely detectable in root cap cells (Supplemental Figure 8D) and in pericycle cells associated with lateral root emergence (see Figure 6D) but not within the susceptible zone (Supplemental Figure 8E). Lhk2 was active in the root vasculature (Supplemental Figure 8G), but GUS staining was undetectable in the root tip region (Supplemental Figure 8F), including the susceptible zone. Finally, Lhk3 was active in the proximal root meristem (Supplemental Figure 8H), but its activity was undetectable in the meristem proper or the main root vasculature (Supplemental Figures 8H and 8I).
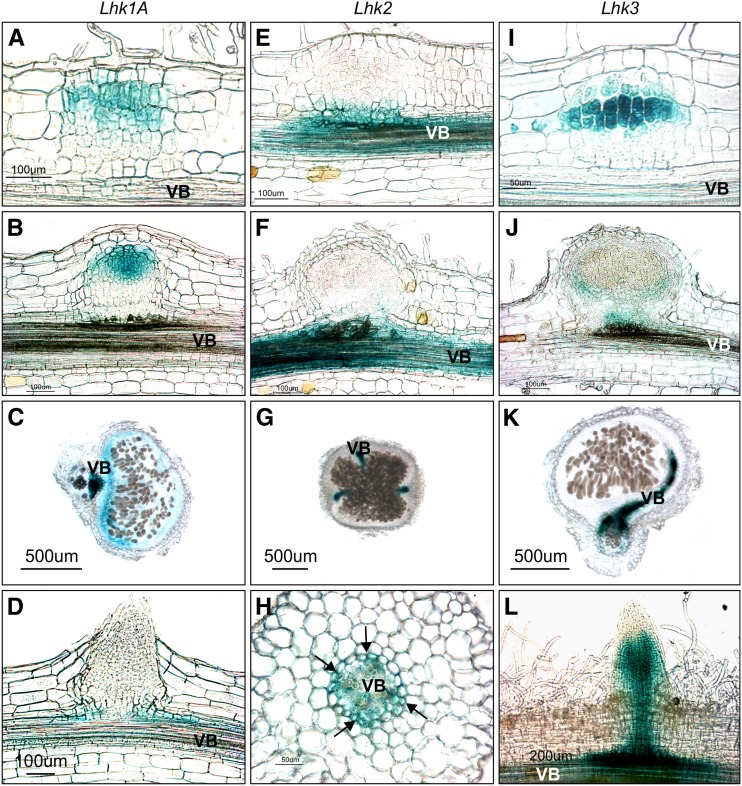
Figure 6.
Activities of the Lhk1A, Lhk2, and Lhk3 Promoters during Nodule and Lateral Root Development.
(A) to (D) Lhk1Apro:GUS reporter activity (blue) associated with nodule ([A] to [C]) and lateral root (D) development.
(E) to (H) Lhk2pro:GUS reporter activity associated with nodule ([E] to [G]) and lateral root (H) development. Note the specific GUS staining in root pericycle cells positioned opposite protoxylem poles (arrows in [H]).
(I) to (L) Lhk3pro:GUS reporter activity associated with nodule ([I] to [K]) and lateral root (L) development.
(L) represents a whole-mount image, and the other panels show 35-μm sections. All images represent specimens collected at 7 or 14 DAI with M. loti.
Activity of the Lhk1 Promoter during Nodule Formation
In order to monitor the activity of the Lhk1 promoter during nodule development, the temperature under which GUS staining was performed was reduced from 37°C to room temperature (see Methods). This decreased the overall intensity of root staining, making it possible to detect the GUS activity maxima.
At early stages of nodule primordium formation, where only very limited cortical cell divisions are present, GUS activity was detectable in a few subepidermal cells and the second and third cortical layers (Figures 5A and 5B). GUS staining was also detectable, albeit rather weakly and intermittently, in the associated root epidermis. With the advancement of cell divisions, the intensity of GUS staining increased across all cell layers and was now clearly detectable in the root epidermis, including root hairs (Figures 5C and 5D). This pattern of GUS staining persisted to the point of nodule emergence from the root epidermis (Figure 5E). In mature nodules, however, GUS activity was restricted to the nodule parenchyma and nodule vasculature, but it was undetectable in centrally located infected cells (Figure 5F).
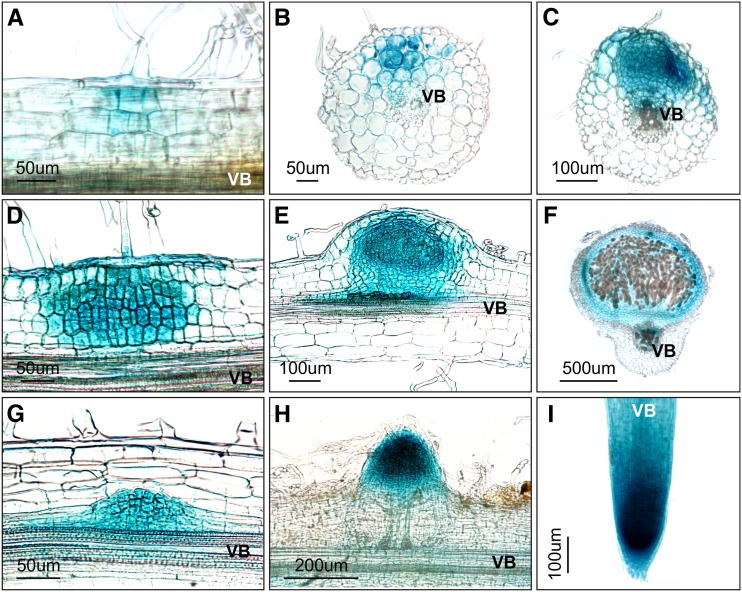
Figure 5.
Activity of the Lhk1 Promoter during Nodule and Lateral Root Development.
Whole mounts ([A] and [I]) and 35-μm-thick root/nodule sections ([B] to [H]) are shown.
(A) to (F) Lhk1pro:GUS reporter activity (blue color) associated with the progressive stages in nodule development. Note that (B) shows a transverse section through a root region where nodule primordium formation has been initiated. (D) depicts the presence of GUS activity in the associated root hair.
(G) to (I) Lhk1pro:GUS activity during lateral root formation. (I) depicts the apical portion of a fully emerged lateral root.
All images represent specimens collected at 7 or 14 DAI with M. loti. VB, vascular bundles.
In comparison with nodules, GUS activity associated with lateral root development was observed in a discrete region of the root pericycle and was associated with all cells of the developing primordium (Figure 5G) before localizing to the apex in the emerging lateral root (Figure 5H). In fully emerged, growing lateral roots, the pattern of the Lhk1-dependent GUS activity was identical to that in the main root, showing strong staining within the meristematic region but also in other parts of the root (Figure 5I).
The Lhk1A and Lhk3 Promoters Show Partially Overlapping Activities with Lhk1 during Nodule Primordium Formation
As observed for Lhk1, the activities of the Lhk1A and Lhk3 promoters were clearly induced upon rhizobial infection (Figure 6). GUS staining was specifically associated with dividing cortical cells of young nodule primordia but was absent from surrounding root cortical cells (Figures 6A, 6B, and 6I). GUS activity could not be detected in the epidermal cells associated with the developing nodule primordium, even at later stages. Mature nodules showed GUS staining in the parenchyma and vasculature (Figure 6C) or only in the vasculature (Figures 6J and 6K) for Lhk1A or Lhk3, respectively.
Both promoters rendered rather different GUS activity profiles during lateral root development, with Lhk1A conferring a barely detectable histochemical signal in subtending pericycle cells (Figure 6D) and Lhk3:GUS plants showing rather strong GUS staining along the root vasculature and within the proximal meristem (Figure 6L).
Similar to uninoculated plants, the Lhk2:GUS reporter construct showed activity in the vasculature of roots and also nodules but not in dividing cortical cells of nodule primordia or emerging nodules (Figures 6E to 6G). Transverse sections of roots showed specific localization of the Lhk2-driven GUS activity in the pericycle cells that were positioned opposite root protoxylem poles (Figure 6H).
All Four Lhk Promoters Respond to M. loti Inoculation
In order to support the histochemical data, the steady state level of Lhk1, Lhk1A, Lhk2, and Lhk3 transcripts was quantified in uninoculated control roots and roots collected 2, 3, and 7 DAI with M. loti. In uninoculated wild-type L. japonicus roots, the level of the Lhk1 mRNA was the highest and that of Lhk1A was the lowest (∼13 to 33 times lower than the Lhk1 mRNA, depending on the specific time point), while steady state levels of Lhk2 and Lhk3 mRNA were intermediate (Supplemental Figure 9). Consistent with the histochemical observations, the steady state levels of Lhk1, Lhk1A, and Lhk3 transcripts were elevated upon inoculation. Quantitative PCR results also revealed that the steady state level of the Lhk2 transcript was also upregulated upon M. loti infection (Supplemental Figure 9).
Significant changes were observed in the lhk1-1 mutant. The steady state level of Lhk1 mRNA was markedly lower than that in wild-type roots, regardless of whether inoculated or uninoculated root samples were analyzed. Lhk1 remained responsive to M. loti inoculation; however, at 2 and 3 DAI, its mRNA reached only ∼50% of the levels in the corresponding wild-type roots (Supplemental Figure 9). Both Lhk2 and Lhk3 were rendered unresponsive to the infection, but the level of the Lhk2 mRNA in uninoculated lhk1-1 roots was elevated in comparison with wild-type roots. As in the wild type, the Lhk1A mRNA remained a relatively minor component and was still able to respond to M. loti infection (Supplemental Figure 9).
Bacterial Entry inside the Root Cortex Is Required for Nodule Formation in lhk1-1
Previously, we showed that initial colonization of the root cortex by M. loti occurs in lhk1-1 without concomitant nodule primordium formation but that a few nodules are eventually formed (Murray et al., 2007). Therefore, we tested the hypothesis that a prior colonization of the root cortex by M. loti is required for the nodule primordium inception in the lhk1-1 mutant. In the absence of functional LHK1, this was presumed to be mediated by bacterial signaling from within the root cortex through a partially redundant function of LHK1A and LHK3. Two previously characterized L. japonicus mutations, namely symRK-14 (Kosuta et al., 2011) and arpc1 (Hossain et al., 2012), which abort root hair–dependent bacterial entry inside the root while leaving the nodule organogenesis intact or even enhanced, were used to test this hypothesis. We reasoned that if bacterial entry is indeed needed to form nodules in the lhk1-1 background, combining these mutations with lhk1-1 should prevent infection of the root cortex, resulting in a nonnodulating (Nod−) phenotype. In agreement with this prediction, both symRK-14 and arpc1 mutations entirely aborted bacterial entry inside lhk1-1 roots when examined 21 DAI. While all three single mutants, lhk1-1 (x¯ = 6.3 ± 0.85; n = 25), symRK-14 (x¯ = 12.5 ± 3.2; n = 15), and arpc1 (x¯ = 28.5 ± 2.11; n = 15), formed nodules, the lhk1-1 symRK-14 (n = 15) and lhk1-1 arpc1 (n = 15) double mutants did not develop any nodules when analyzed 21 DAI with M. loti.
The lhk1-1 lhk1a-1 lhk3-1 Cytokinin Receptor Triple Mutant Does Not Form Nodules
We additionally tested the hypothesis that the LHK1, LHK1A, and LHK3 receptors work partially redundantly to mediate nodule formation by constructing and then analyzing the phenotype of the triple mutant line that combined mutations in these cytokinin receptor genes.
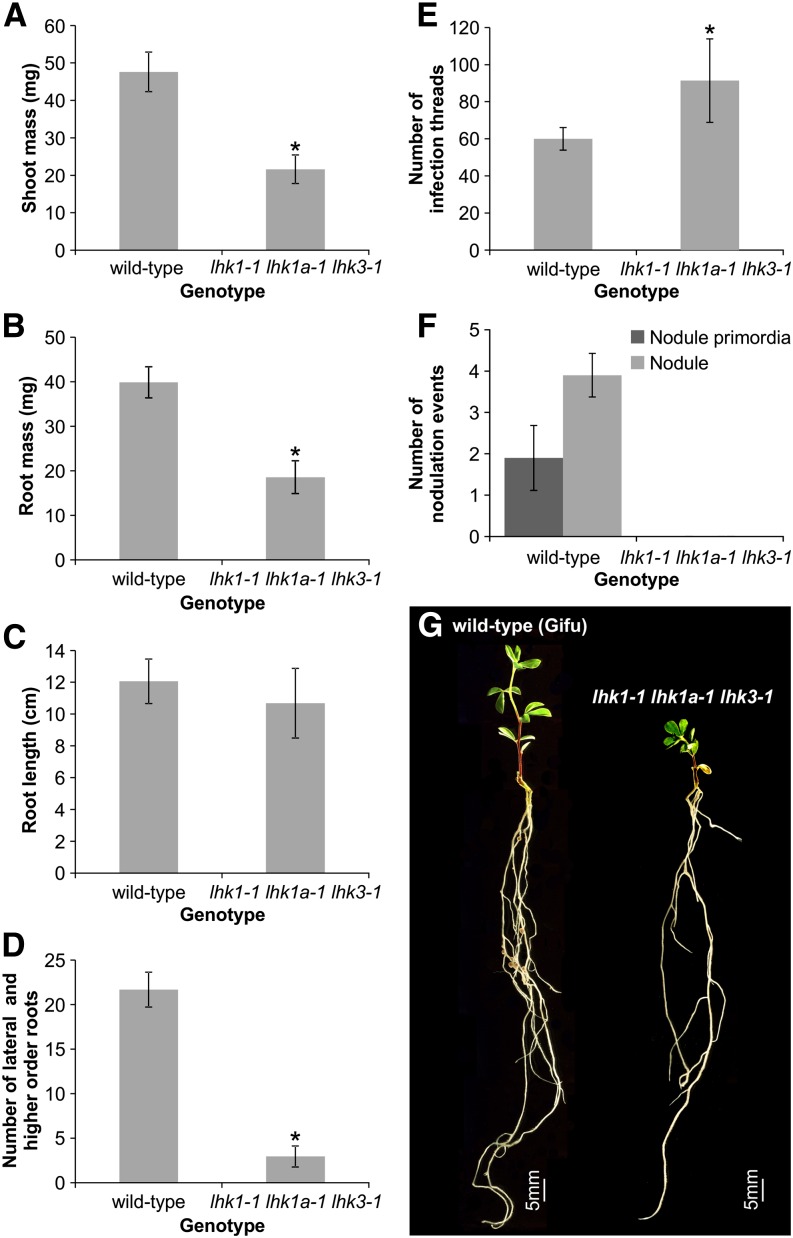
Although viable and relatively healthy, the growth of the lhk1-1 lhk1a-1 lhk3-1 triple mutant was significantly affected compared with wild-type L. japonicus. The average shoot and root mass of uninoculated triple mutant plants grown in the presence of KNO3 (see Methods) was only approximately half of the corresponding wild-type values when analyzed 28 d after sowing (Figures 7A and 7B). The length of the main root remained wild type (Figure 7C), but the number of lateral and higher order roots was at least five times lower in the triple mutant (Figure 7D). When analyzed 7 DAI with M. loti, the triple mutant formed significantly more infection threads than wild-type L. japonicus Gifu of the same age (Figure 7E), but no nodule primordia or nodules were present in the mutant roots (Figure 7F). As overall growth of the triple mutant is slower, we additionally tested its nodulation phenotype at 21 and 35 DAI. While wild-type L. japonicus formed on average 9 ± 0.74 (n = 32) and 24 ± 3.55 (n = 20) fully developed nodules, respectively, the triple mutant (n = 19 to 35) did not develop any nodules or nodule primordia.
Figure 7.
The lhk1-1 lhk1a-1 lhk3-1 Triple Mutant Does Not Form Nodules.
(A) to (D) Shoot (A) and root (B) masses, as well as the root length (C) and the number of lateral and higher order roots (D), were scored in 28-d-old uninoculated L. japonicus (Gifu) wild-type and lhk1-1 lhk1a-1 lhk3-1 triple receptor mutant plants grown in the presence of KNO3.
(E) and (F) Scores of infection threads (E) and nodulation events (i.e., nodule primordia and nodules) (F) are given for the L. japonicus wild type (Gifu) and the triple receptor mutant.
All values reported represent means ± 95% confidence interval (n = 10 to 35) as measured 28 d after sowing ([A] to [D]) or 7 DAI with M. loti ([E] and [F]) for nodulation counts. Asterisks denote significant differences (Student’s t test, P < 0.05).
(G) Phenotypes of the wild type and the triple mutant are shown 21 DAI.
Other Receptors Can Substitute for LHK1 during Nodule Formation
Given the prominent role of LHK1, we asked whether other cytokinin receptors can substitute for its essential function in mediating nodule organogenesis. One important aspect of this was to test whether a nonlegume cytokinin receptor could function in the nodulation pathway. If confirmed, this would indicate that the ability to respond to rhizobial signaling and/or ectopic cytokinin by inciting nodule formation is not due to neofunctionalization of cytokinin receptors in legumes.
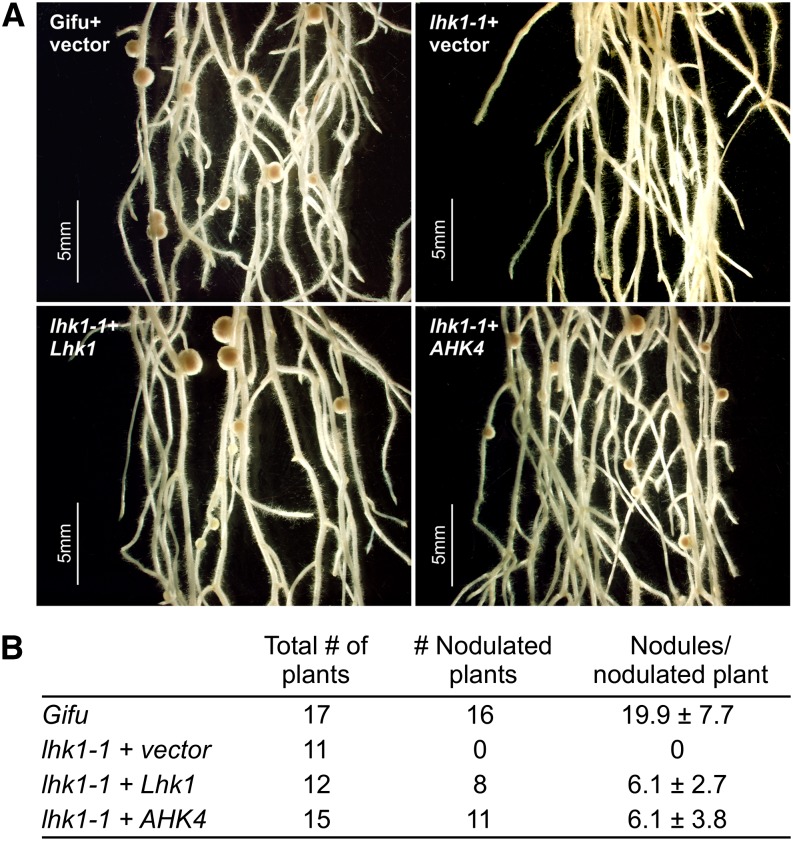
Hairy roots induced on lhk1-1 mutant shoots via Agrobacterium rhizogenes–mediated transformation exaggerate the mutant nodulation phenotype, which results in a Nod− or almost Nod− phenotype (Murray et al., 2007). We demonstrated previously that nodulation can be restored to hairy roots formed on lhk1-1 shoots by expressing the wild-type Lhk1 gene (Murray et al., 2007). A similar complementation result was obtained in this study with the Lhk1 cDNA expressed under the control of a constitutive cauliflower mosaic virus 35S promoter (Supplemental Figures 10A and 10B). Nodulation was also restored to lhk1-1 hairy roots by expressing either of the two Lhk3 cDNA variants; however, Lhk1A and Lhk2 cDNAs failed to complement the lhk1-1 mutant phenotype (Supplemental Figures 10A and 10B). We then tested whether the Arabidopsis AHK4 gene, including its own promoter and terminator, could restore nodule formation to lhk1-1 transgenic hairy roots, and indeed, this was the case (Figures 8A and 8B).
Figure 8.
The Arabidopsis Cytokinin Receptor Mediates Nodule Organogenesis.
(A) Nodulation phenotypes of hairy roots formed on wild-type and lhk1-1 mutant shoots after transformation with the A. rhizogenes strain AR12 containing empty vector (+vector), the entire Lhk1 gene, including 5′ and 3′ untranslated regions (+Lhk1), or a vector containing the Arabidopsis AHK4 gene, including its cognate promoter and terminator (+AHK4) (see Methods).
(B) The nodulation phenotypes were scored 21 DAI with M. loti. Numbers of plants and nodules in each transformation category are given. Wild-type L. japonicus ecotype Gifu transformed with A. rhizogenes AR12 was used as a positive control. In all cases, values represent means ± 95% confidence interval.
DISCUSSION
We show here that L. japonicus contains a small family of four cytokinin receptor genes, including Lhk2, encoding a presumed cytokinin receptor, which all respond to M. loti infection. While further highlighting the prominent role of LHK1 (Murray et al., 2007; Tirichine et al., 2007; Heckmann et al., 2011), our data also demonstrate the involvement of other cytokinin receptors during L. japonicus nodule formation. We show that only Lhk1 is expressed in the root epidermis but is also essential within the root cortex, where it mediates cell divisions for nodule primordium formation in a partially redundant manner with Lhk1A and Lhk3. Lhk2 is not expressed in the root cortex and therefore is unlikely to be involved in the stimulation of cortical cell divisions, as mediated by LHK1, LHK1a, and LHK3. Consistent with this, the presence of Lhk2 is not sufficient to induce nodule primordium formation in the lhk1-1 lhk1a-1 lhk3-1 triple mutant.
Arabidopsis AHK4 Mediates Nodule Organogenesis
The loss-of-function allele, lhk1-1, had roots that were insensitive to growth inhibition by applied cytokinin; none of the mutations in other cytokinin receptor genes had this effect. In this respect, LHK1 resembles its closest homolog in Arabidopsis, AHK4, which is the main sensor of external cytokinin in that species (Inoue et al., 2001; Nishimura et al., 2004). The similarity in the functions of the two genes extends to their promoters, both of which are active almost ubiquitously in roots (Nishimura et al., 2004). We show, however, that Lhk1 is less active in the L. japonicus root proximal meristem. Unlike Lhk1, the expression of Lhk2 and Lhk3 in L. japonicus uninoculated roots was detectable only in the vasculature and the proximal meristem, respectively. This is different from the expression patterns described for their presumed Arabidopsis counterparts, AHK2 and AHK3 (Nishimura et al., 2004).
Our data show that AKH4, a cytokinin receptor gene from a nonlegume species that does not form nodular symbiosis with rhizobia, can functionally substitute for Lhk1 in mediating nodule organogenesis in L. japonicus upon M. loti infection. Lhk3 also rescued the lhk1-1 nodulation defect in hairy root experiments, but this effect could not be achieved with Lhk1A or Lhk2 cDNAs. The lack of complementation by Lhk1A, which is the closest L. japonicus homolog of Lhk1, is puzzling and may reflect a need for additional regulatory sequences beyond the cDNA used herein. Regardless, the ability of AHK4 to complement nodulation suggests that the evolution of signaling for nodule primordium formation involved the recruitment of a cytokinin receptor that has not been subjected to major, legume-specific modifications (Szczyglowski and Amyot, 2003). Consequently, the specificity of cytokinin signaling during nodule formation must be exerted downstream from the cytokinin perception. However, we cannot rule out the possibility that the capacity for nodule formation is orchestrated by other cues that provide a unique legume-specific context to cytokinin signaling.
Unique Properties of Lhk1
A distinct role for LHK1 is supported by several additional observations. Lhk1, but not other Lhk promoters, directs GUS activity to the root epidermis. External application of cytokinin to L. japonicus roots induced an increase in the steady state level of the Lhk1 mRNA within the first 3 h, a response not mimicked by other Lhk mRNAs. This increase appears to be largely the consequence of an autoregulatory mechanism, because it requires a functional copy of the Lhk1 gene (Figure 3). The steady state level of Lhk1 mRNA was lower in untreated lhk1-1 mutant roots as compared with wild-type roots, which further supports the existence of an inherent autoregulatory feedback mechanism.
Upregulation of Lhk1A, Lhk2, and Lhk3 mRNA levels after a prolonged (12 h) treatment with BA was also Lhk1 dependent. The level of Lhk1 mRNA in the lhk1-1 mutant roots was still somewhat increased under these conditions (i.e., 12 h at 50 nM BA) (Figure 3D), perhaps due to the effects of other cytokinin receptors.
When grown in soil in the presence or absence of M. loti, the existence of a regulatory relationship between different cytokinin receptors was also discernible. The steady state level of Lhk1 was significantly diminished in the lhk1-1 mutant. Lhk2 and Lhk3 genes became unresponsive to, while the Lhk1 mRNA was elevated by, M. loti infection, although its level remained at or below that of the uninoculated wild-type roots (Supplemental Figure 9).
Taken together, these data demonstrate the prominent role of LHK1 during the perception and response of L. japonicus roots to both applied cytokinin and M. loti infection. They also suggest some degree of regulatory capacity, where LHK1 appears to exert a dominant role over its own expression but also over those of other cytokinin receptors. The underlying mechanism is unknown; nevertheless, given the rather distinct expression domains for Lhk genes, a functional link might be required to maintain homeostasis during root development.
LHK1 Mediates Signaling without Bacterial Entry into Roots
Unlike wild-type L. japonicus, the lhk1-1 mutant is unable to form nodules upon the application of BA (Heckmann et al., 2011) but it does so in response to M. loti inoculation, albeit with a noticeable delay and significant reduction in nodule number (Murray et al., 2007). Thus, bacterial infection must be exerting an effect beyond what is being mimicked by ectopic cytokinin.
Consistent with this prediction, we show that the formation of lhk1-1 nodules is prevented by blocking bacterial entry inside roots with either the symRK-14 or arpc1-1 mutation. This is unlike in L. japonicus plants carrying the wild-type Lhk1 gene, where empty or initially empty nodules readily develop in the presence of various mutations that affect infections (Karas et al., 2005; Yokota et al., 2009; Groth et al., 2010), including symRK-14 and arpc1-1 (Kosuta et al., 2011; Hossain et al., 2012).
The simplest explanation for these observations is to assume that LHK1 participates, either directly or indirectly, in transducing a signal from the root epidermis to the subtending root cortex, regardless of whether the initial stimulus is ectopic cytokinin or M. loti NF. In the lhk1-1 mutant, therefore, applied cytokinin would be expected to fail in inducing nodule formation, which indeed is the case (Heckmann et al., 2011). Bacterial infection, however, could bypass the epidermis and initiate signaling for cell divisions from within the root cortex via redundantly acting LHK1A and/or LHK3 receptors. Although we cannot entirely rule out this explanation, our histochemical data point to a more complex signaling circuit during the response to bacterial infection.
LHK1, the Master of Symbiotic Events
It was proposed by Heckmann et al. (2011) that the induction of nodule primordia organogenesis in L. japonicus is regulated by a cytokinin-dependent mechanism that operates in the root cortex. Consistent with this, M. loti infection-dependent cytokinin signaling (as monitored by the TCS:GUS histochemical activity) was initially localized to the second and/or third cortical layers. This is where cell divisions for nodule primordium formation are first initiated in L. japonicus (van Spronsen et al., 2001). This expression pattern was paralleled, at least to some extent, by GUS activity driven by the Lhk1 promoter, which also peaked initially within the middle cortex, although by default the Lhk1 promoter is also active in the root epidermis.
A similar, inner cortex–localized primary cytokinin response was documented in M. truncatula roots responding to S. meliloti infection (Plet et al., 2011). It is likely, therefore, that another NF-dependent mechanism generates a cell nonautonomous signaling event that originates in the root epidermis and is rapidly translocated to incite the initial peak accumulation of bioactive cytokinin in the inner cortex. This could explain why a much longer time is needed to induce nodule formation in L. japonicus upon external application of cytokinin than upon bacterial infection (Heckmann et al., 2011).
We postulate that the initial cytokinin burst, as perceived by LHK1 within the root cortex, leads to an LHK1-dependent autostimulation (Figure 9A). This is manifested by a local increase in the level of the Lhk1 mRNA. Our results show that, in addition to Lhk1, the activities of Lhk1A, Lhk2, and Lhk3 promoters are also enhanced by M. loti infection. We have interpreted these events as leading to a local increase in cell sensitivity to cytokinin, a mechanism that is primarily reliant on LHK1 and by which a threshold is reached sufficient to initiate first cortical cell divisions for nodule primordium formation.
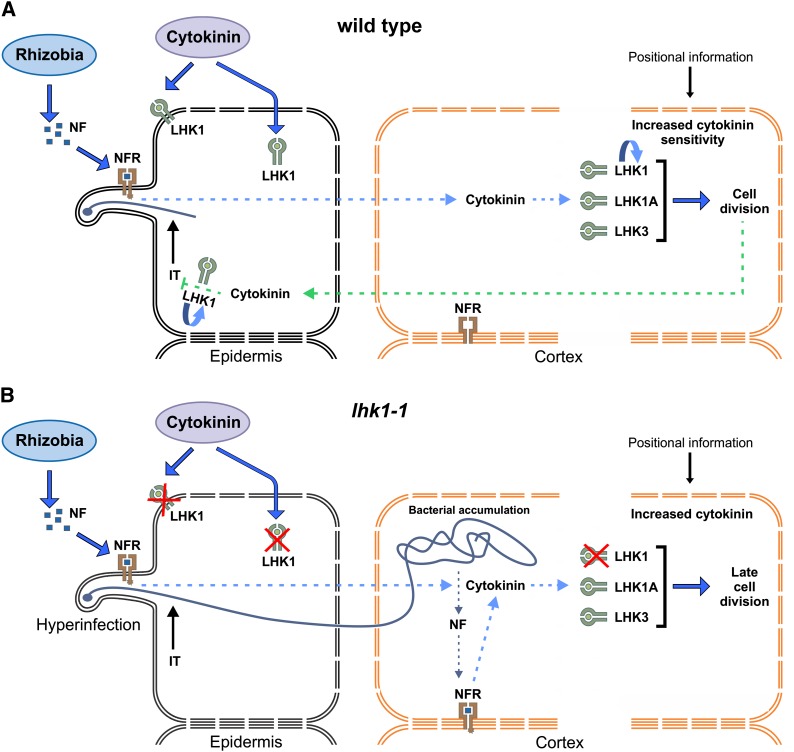
Figure 9.
Working Models for LHK1-Dependent and LHK1-Independent Signaling for L. japonicus Nodule Formation.
(A) In wild-type L. japonicus, M. loti infection generates a presumed cell nonautonomous signaling event (dotted blue arrow), which triggers the accumulation of bioactive cytokinin and the subsequent stimulation of cortical cell divisions for nodule primordium formation prior to bacterial entry into the root. A feedback loop is induced downstream from advancing cell divisions, which results in the accumulation of cytokinin in the root epidermis; this is presumed to locally block subsequent infections (IT) in an LHK1-dependent manner.
(B) In the lhk1-1 mutant, bacterial entry and accumulation inside the root are required for the initiation of cortical cell divisions (leading to late cell divisions). Lack of the LHK1-dependent feedback loop results in the hyperinfection of root epidermis and cortex.
The “u-turn” arrows in (A) denote autostimulation of LHK1 expression. Ectopic cytokinin can be perceived either at the plasma membrane or upon movement inside the cell by the endoplasmic reticulum–localized cytokinin receptors (Wulfetange et al., 2011). “Positional information” refers to as yet undefined spatial information that dictates the position of the first cortical cell division for nodule primordium formation. NFR, nodulation factor receptor complex. For further details, see text.
In the absence of functional LHK1, cell divisions for nodule primordium formation are initiated only upon bacterial entry (Murray et al., 2007), and we show that the presence of Lhk1A or Lhk3 is critical in this context (Figure 9B). Lack of functional LHK1 leads to significantly reduced levels of Lhk1, which could explain why the required threshold for the stimulation of cortical cell divisions is not reached during the initial bacterial signaling, prior to their entry within the root cortex. Following heavy colonization of lhk1-1 roots by M. loti, direct perception of NFs within the root cortex might contribute to overcoming this limitation, which eventually leads to cell divisions (Figure 9B).
The evidence for NF perception in the root cortex was provided in the context of transcellular cortical infection threads, as formed by the L. japonicus nfr1 nfr5 snf1 symrk-3 quadruple mutant (Madsen et al., 2010). The same mechanism likely accounts for delayed nodulation in the lhk1-1 mutant. In many legumes, rhizobia are able to bypass the root epidermis, entering the root by a crack-entry mechanism (Sprent and James, 2007). We and others have shown that a crack-entry–like mechanism also operates in L. japonicus (Karas et al., 2005; Madsen et al., 2010; Kosuta et al., 2011), and signaling for nodule formation from within the root cortex in lhk1-1 likely reflects this latent ability.
Rapid expansion to and/or intensification of cytokinin signaling in the root epidermis, including root hairs, were clearly observable early on, following some initial advancement in nodule primordium formation. This was true for the cytokinin response as well as the activity of the Lhk1 promoter. Unlike M. truncatula cre1 (Gonzalez-Rizzo et al., 2006), the lhk1-1 mutant is hyperinfected by M. loti (Murray et al., 2007), and we are currently testing the hypothesis that increased cytokinin activity in the root epidermis as mediated by LHK1 provides a mechanism by which L. japonicus locally restricts subsequent infection events, thus preventing hyperinfection (Figure 9B). Hence, LHK1, in addition to being the master of cortical events, may also prove to be the master of epidermal infections.
METHODS
Plant Growth Conditions
Seeds of wild-type and mutant Lotus japonicus ecotype Gifu plants were surface-sterilized and grown on wet filter paper for a period of 7 d and planted as described (Szczyglowski et al., 1998). The extent of root growth was assessed for various genotypes using the protocol described by Wopereis et al. (2000). Briefly, 2-d-old seedlings were transferred to vertical plates containing half-strength B5 with minimal organics, 2.5 mM MES, 4.5% Suc, and 0.8% phytagel. Roots were allowed to elongate for 7 d at room temperature in total darkness. Root elongation was scored and averaged for 10 to 20 roots per genotype. Where appropriate, BA was added (10−8, 10−7, 10−6, and 10−5 M).
Assessment of Symbiotic Phenotypes
For nodulation assays, 7-d-old seedlings were transferred (under sterile conditions) to pots containing a mixture of vermiculite and sand (6:1) and watered with 1× B&D nutrient solution (Broughton and Dilworth, 1971) supplemented with 0.5 mM KNO3. Standard growth conditions of 18 h of light at 23°C and 6 h of dark at 18°C were used. Seven days after planting, the seedlings were inoculated with either the wild-type Mesorhizobium loti strain NZP 2355 or M. loti containing the hemA:LacZ reporter cassette for visualization of bacterial infection (Wopereis et al., 2000).
Characterization of Cytokinin Receptor Transcripts
Total mRNA was isolated from different L. japonicus (Gifu) tissues using the RNeasy Plant Mini kit (Qiagen). Rapid amplification of cDNA ends (both 5′ and 3′) was performed using the FirstChoice RLM-RACE kit (Ambion). For RT-PCR, cDNA was synthesized using the Thermoscript RT-PCR system (Invitrogen) from uninoculated root tissue, nodules, and leaves of wild-type Gifu plants. Lhk transcripts were routinely amplified from these tissues using High Fidelity Platinum Taq (Invitrogen). The PCR conditions used were as follows: 94°C for 5 min, followed by 30 cycles of 94°C for 30 s, 58°C for 30 s, and 68°C for 30 s, with a final extension of 7 min at 68°C. Gene-specific primers designed to amplify products encompassing the final exon and 3′ untranslated region were as follows: Lhk1A F, 5′-ATGGACGGATTTGAAGCAAC-3′; Lhk1A R, 5′-CAAGATCTCTTTCGGTCTGC-3′; Lhk2 F, 5′-CACTCATTGCAGGAGAAGAGG-3′; Lhk2 R, 5′-TTTTCCATCTTAGCCCCTCA-3′; Lhk3 F, 5′-TGGAACACAATGTGAACAGAGA-3′; Lhk3 R, 5′-CCCATTTCTCCCATCCTTCT-3′. Lhk1 RT-PCR primers were the same as those used by Murray et al. (2007).
Alternative Splicing at the Lhk3 Locus
Primers were designed for PCR-based detection and expression-based studies of the two alternative splice variants produced by the Lhk3 locus (named Lhk3 variants 1 and 2). For the comparative analysis of these two alternatively spliced transcripts, 5′ end products were amplified from cDNA templates using a common reverse primer and transcript-specific forward primers (see below). The PCR cycling program was as follows: 5 min at 94°C, followed by 30 cycles of 94°C for 30 s, 58°C for 30 s, and 68°C for 1 min, followed by 7 min at 68°C. The primers were as follows: Lhk3 variant 1 F, 5′-CTTATATGAAGGGTGGTTTTGG-3′; Lhk3 variant 2 F, 5′-GGTTGGTTACTGTTGTGGATGA-3′; common reverse primer, 5′-CTTTCCAGAAAGCACGTCAAC-3′.
The Lhk Mutants
The lhk1-1 mutant has been described (Murray et al., 2007). TILLING was utilized for the identification of mutant alleles at the Lhk1A, Lhk2, and Lhk3 loci.
The following primers were used to generate amplicons for TILLING: LHK1A forward, 5′-TGTCCATGCTTCGAGCCCAATGAGTCC-3′; LHK1A reverse, 5′-AAACCCACCCAGAATGGAAAATATGTC-3′; LHK2 forward, 5′-ACAGTTGGCTGTTTATGCATCT-3′; LHK2 reverse, 5′-TTTTTAGGCAGCGCTCTAATGCCAATG-3′; LHK3 forward, 5′-TGATGTACGGGCAATTCTGGATGATGT-3′; LHK3 reverse, 5′-GCTTACCCATCCATTTCTGGCATTTGA-3′.
Lhk2 TILLING was performed using an ABI3730 capillary sequencer following the method of Le Signor et al. (2009), whereas Lhk1A and Lhk3 TILLING was performed on a LI-COR instrument according to Horst et al. (2007).
All selected TILLING mutant lines were backcrossed once to wild-type L. japonicus Gifu before extensive phenotypic analyses were conducted. Double mutants were developed by genetic crosses between primary homozygous single mutants for each of the two loci being analyzed. The F1 plants were allowed to self-fertilize and produce F2 segregating populations, where the desired double mutant was selected for by using a combination of sequence analysis and cleaved-amplified polymorphic sequence or derived cleaved-amplified polymorphic sequence markers, depending on the specific line. The F3 progeny from confirmed, homozygous double mutants were utilized for phenotypic evaluation.
The lhk1-1 lhk1a-1 lhk3-1 cytokinin receptor triple mutant was selected from the F2 population of a cross between homozygous lhk1-1 lhk3-1 and lhk1a-1. For the analysis of nonsymbiotic phenotypes in uninoculated plants, including shoot and root mass, root length, and lateral root number, 7-d-old wild-type and triple mutant seedlings were transferred to pots containing a mixture of vermiculite and coarse sand and were supplemented with Hoagland nutrient solution containing 6 mM KNO3 (Hoagland and Arnon, 1950). Plants were harvested and analyzed 28 d after sowing. The nodulation phenotype of the triple mutant was assessed as described above, except that an additional time point at 35 DAI with M. loti was also included in this analysis.
Cytokinin-Responsive Assay in Saccharomyces cerevisiae
All Lhk cDNAs were directionally cloned into the multicloning site of a yeast expression vector (P415CYC; Mumberg et al., 1995). The previously analyzed Lhk1 cDNA was used as a positive control (Murray et al., 2007). The resultant constructs were transformed into the sln1Δ mutant of S. cerevisiae (a kind gift from Tatsuo Kakimoto, Osaka University, Japan) and analyzed for their responses to treatments with different plant hormones, including BA, trans-zeatin, cis-zeatin, and the nonspecific ligand 1-naphthaleneacetic acid, as described by Murray et al. (2007). Putative loss-of-function mutant cDNAs were also analyzed using this assay.
Cytokinin-Responsive Assay in Escherichia coli
The mutant and wild-type Lhk cDNAs were cloned into the E. coli expression vector pSTV28 (Tirichine et al., 2007) using either SacI and SalI (for Lhk1 and both variants of the Lhk3 mRNA) or EcoRI and SalI (for Lhk2). The resultant constructs were transformed into the sensor-negative E. coli SRC122 strain (a kind gift from Takafumi Yamashino, Nagoya University, Japan). Following transformation, colonies were grown on Luria-Bertani plates or in liquid Luria-Bertani medium containing 40 mM sodium phosphate buffer and 20 mM Glc with or without the addition of 200 μM BA. For analysis of β-galactosidase activity, a standard assay was used as described (Tirichine et al., 2007).
Stable Transgenics and GUS Staining
To develop promoter-GUS fusions for Lhk1, Lhk1A, Lhk2, and Lhk3, ∼4-kb promoter fragments were first amplified and cloned into the promoterless pKGWFS7,0 destination vector using the Gateway technology (Invitrogen). After validation of the insert by sequencing, the corresponding vectors were transferred to Agrobacterium tumefaciens LBA4404. Standard transformation protocols (Lombari et al., 2005) were used to regenerate fully transgenic plants from hypocotyl segments of wild-type (Gifu) L. japonicus plants. At least seven independent transgenic plants were used for the analyses of promoter expression.
A synthetic cytokinin TCS was used to follow the presence of bioactive cytokinin. TCS harbors the concatemerized B-type Arabidopsis thaliana response regulator binding motifs and a minimal 35S promoter, followed by the tobacco mosaic virus Ω translational enhancer sequence, as described (Müller and Sheen 2008). The TCS promoter was amplified from the TCS min35S-ΩeGFP ER vector (pCB302; a kind gift from Bruno Müller, University of Zürich, Switzerland) and was cloned into the pKGWFS7,0 destination vector using the Gateway technology (Invitrogen).
Detection of the GUS reporter activity was routinely conducted using a staining solution that contained 0.1 M potassium phosphate buffer, 5 mM EDTA, 0.5 mM potassium ferric cyanide and ferrous cyanide, and 0.5 mg/mL 5-bromo-4-chloro-3-indolyl glucuronide cyclohexylammonium salt (Fermentas). All tissues were vacuum-infiltrated for 15 min, stained overnight at room temperature or 37°C, and cleared as described previously (Wopereis et al., 2000).
Primers used to amplify TCS were as follows: F, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTGAAGCTTATGCTAGCAAAATCT-3′; and R, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTGTTATATCTCCTTGGATCGAT-3′.
Quantitative Real-Time RT-PCR Assay
Total RNA was extracted using the RNeasy Plant Mini kit (Qiagen) and was treated with DNase I. The concentration and purity of RNA were determined by measuring absorbance at 260/280 nm. cDNA was prepared from 2 µg of total RNA using the High Capacity cDNA Synthesis kit (Applied Biosystems) with random primers. Negative control reactions to which no reverse transcriptase was added were included for each RNA sample. Quantitative RT-PCR was performed in triplicate (i.e., three biological and three technical replicates) on the CFX-96 Real-Time PCR Detection System (Bio-Rad) using PerfeCTa SYBR Green FastMix (Quanta Biosciences). Three reference genes, UBC (ubiquitin-conjugating enzyme), PP2A (protein phosphatase 2A), and TB2C (tubulin β-chain), were used to normalize the results as described previously (Tirichine et al., 2007).
Primer sequences used to produce Lhk gene-specific amplicons were as follows: Lhk1 F, 5′-GTGCTTAAATTGTGGGATGGA-3′; Lhk1 R, 5′-ATTGATGCTGGGAGAAGTTGA-3′; Lhk1A F, 5′-TCAAAGCCATTTGAGGAACAG-3′; Lhk1A R, 5′-GCATAGTTTACCTGCAACATCTG-3′; Lhk2 F, 5′-ATGGATGGCTACGTGTCAAAG-3′; Lhk2 R, 5′-GCATACGTTGTTGATTGAATGC-3′; Lhk3-variant 1 F, 5′-CTTATATGAAGGGTGGTTTTGG-3′; Lhk3-variant 1 R, 5′-TTGTCTCTTCACTCCCTTGGA-3′; Lhk3-variant 2 F, 5′-TCAGCTGCAATTCACAAACTC-3′; Lhk3-variant 2 R, 5′-ACAACCCAGCAACATAGCACT-3′.
Complementation of the lhk1-1 Nodulation Defect
For hairy root complementation, the binary vector BIN19 containing the entire AHK4 gene, including its cognate promoter and terminator regions, was used (a kind gift from Chiharu Ueguchi, Nagoya University).
To overexpress different Lhk mRNAs in the lhk1-1 mutant background, Lhk cDNAs were cloned into the pEarleyGate100 destination vector using the Gateway technology and subsequently transformed into Agrobacterium rhizogenes strain AR1193. Standard A. rhizogenes–mediated transformation procedures were followed to induce the formation of hairy roots on lhk1-1 mutant shoots (Petit et al., 1987). The chimeric plants that developed hairy roots were transferred to soil, and at least 10 independent plants per genotype were assessed for the presence of nodules 21 DAI with M. loti.
Microscopy and Image Analysis
All microscopic observations were performed on a Nikon SMZ1500 dissecting or Zeiss Axioskop 2 compound light microscope. Both microscopes were integrated with a Nikon DXM1200 digital camera using the ACT1 media software (Nikon). All images captured were taken in a TIFF format at a resolution of 3840 × 3072. Longitudinal and cross sections of root and/or nodule segments were generated by embedding specimens in 3% (w/v) agar blocks and sectioning to 35 μm thickness using a VT 1000S vibratome (Leica Microsystems).
Statistical Analyses
In all cases, means were calculated from data ranges containing no fewer than 10 plants per genotype or per treatment. Pairwise comparisons were made using a Student’s t test assuming unequal variance.
Phylogenetic Analysis
All sequence alignments were generated in Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) using default settings. The phylogenetic tree (Figure 1) was constructed based on the alignment shown in Supplemental Data Set 1. The maximum likelihood method of the MEGA 6.0.5 package (http://www.megasoftware.net/mega.php) and the branch support test from 1000 bootstrap repetitions were used.
Computer Analyses
Databases were searched with standard protein BLAST (http://www.ncbi.nlm.nih.gov/), which also predicts putative conserved domains.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: Lhk1 (ABI48271), Lhk1A (DQ848998; formerly Lhk2 [Murray et al., 2007]), Lhk2 (KJ361851), Lhk3 (KJ361852), AHK4 (NP_565277.1), AHK2 (NP_568532.1), and AHK3 (NP_564276.1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Amino Acid Sequence Alignment of CHASE Domains, as Predicted for L. japonicus LHK Proteins and Arabidopsis AHK4.
Supplemental Figure 2. Amino Acid Sequence Alignment of Protein Kinase Domains, as Predicted for L. japonicus LHK Proteins and Compared with Arabidopsis AHK4.
Supplemental Figure 3. Amino Acid Sequence Alignment of Receiver Domains, as Predicted for L. japonicus LHK Proteins and Arabidopsis AHK4.
Supplemental Figure 4. Alternative Splicing of Lhk3.
Supplemental Figure 5. Bacterial Infection and Nodule Formation Are Unaffected by the lhk1a-1, lhk2-5, and lhk3-1 Mutations.
Supplemental Figure 6. Responses of the L. japonicus Wild-Type and lhk Mutant Roots to Exogenous Cytokinin.
Supplemental Figure 7. Ectopic Cytokinin Increases the Steady State Level of the Lhk1 mRNAs.
Supplemental Figure 8. Activities of Lhk Promoters in Uninoculated L. japonicus Roots.
Supplemental Figure 9. All Four Lhks Respond to M. loti Inoculation.
Supplemental Figure 10. Lhk3 Functionally Replaces Lhk1.
Supplemental Table 1. Amino Acid Conservation among L. japonicus LHK Proteins and Their Presumed Arabidopsis Counterparts.
Supplemental Table 2. A list of lhk1a, lhk2, and lhk3 Mutant Alleles as Identified by TILLING.
Supplemental Data Set 1. Protein alignment for Figure 1.
Supplementary Material
Acknowledgments
We thank Alex Molnar for his expert help in the preparation of the figures. This work was supported by the Agriculture and Agri-Food Canada Crop Genomics Initiative and the National Science and Engineering Research Council of Canada (Grant 3277A01 to K.S.), by a National Science and Engineering Research Council of Canada fellowship (Grant NSERC-PGS-D2 to M.H.), and by the Biotechnology and Biological Science Research Council of the United Kingdom (Grant BBS/B/02401 to Martin Parniske and T.L.W. and Grant BB/F010591/1 to T.L.W.).
AUTHOR CONTRIBUTIONS
K.S. designed the research. M.H., H.H., M.M., C.H., M.S.H., and L.R. performed most of the research. S.S. and S.T. identified candidate Lhk loci in the L. japonicus genome and provided relevant sequence information. J.P. and T.L.W. performed TILLING experiments. M.H., L.R., and K.S. wrote the article.
Glossary
- NF
nodulation factor
- CHASE
cyclase/histidine kinase–associated sensory extracellular
- TILLING
targeted induced localized lesions in genomes
- DAI
days after inoculation
- BA
6-benzylaminopurine
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Ariel F., et al. (2012). Two direct targets of cytokinin signaling regulate symbiotic nodulation in Medicago truncatula. Plant Cell 24: 3838–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer P., Ratet P., Crespi M.D., Schultze M., Kondorosi A. (1996). Nod factors and cytokinins induce similar cortical cell division, amyloplast deposition and MsEnod12A expression patterns in alfalfa roots. Plant J. 10: 91–105. [Google Scholar]
- Bek A.S., Sauer J., Thygesen M.B., Duus J.O., Petersen B.O., Thirup S., James E., Jensen K.J., Stougaard J., Radutoiu S. (2010). Improved characterization of nod factors and genetically based variation in LysM receptor domains identify amino acids expendable for nod factor recognition in Lotus spp. Mol. Plant Microbe Interact. 23: 58–66. [DOI] [PubMed] [Google Scholar]
- Broughton W.J., Dilworth M.J. (1971). Control of leghaemoglobin synthesis in snake beans. Biochem. J. 125: 1075–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combier J.P., de Billy F., Gamas P., Niebel A., Rivas S. (2008). Trans-regulation of the expression of the transcription factor MtHAP2-1 by a uORF controls root nodule development. Genes Dev. 22: 1549–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combier J.P., Frugier F., de Billy F., Boualem A., El-Yahyaoui F., Moreau S., Vernié T., Ott T., Gamas P., Crespi M., Niebel A. (2006). MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev. 20: 3084–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J.B., Long S.R. (1994). Morphogenetic rescue of Rhizobium meliloti nodulation mutants by trans-zeatin secretion. Plant Cell 6: 215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbrosses G.J., Stougaard J. (2011). Root nodulation: A paradigm for how plant-microbe symbiosis influences host developmental pathways. Cell Host Microbe 10: 348–358. [DOI] [PubMed] [Google Scholar]
- Doyle J.J. (2011). Phylogenetic perspectives on the origins of nodulation. Mol. Plant Microbe Interact. 24: 1289–1295. [DOI] [PubMed] [Google Scholar]
- Duc G., Trouvelot A., Gianinazzi-Pearson V., Gianinazzi S. (1989). First report of non-mycorrhizal plant mutants (Myc−) obtained in pea (Pisum sativum L.) and fababean (Vicia faba L.). Plant Sci. 60: 215–222. [Google Scholar]
- Ferreira F.J., Kieber J.J. (2005). Cytokinin signaling. Curr. Opin. Plant Biol. 8: 518–525. [DOI] [PubMed] [Google Scholar]
- Frugier F., Kosuta S., Murray J.D., Crespi M., Szczyglowski K. (2008). Cytokinin: Secret agent of symbiosis. Trends Plant Sci. 13: 115–120. [DOI] [PubMed] [Google Scholar]
- Gleason C., Chaudhuri S., Yang T., Muñoz A., Poovaiah B.W., Oldroyd G.E.D. (2006). Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 441: 1149–1152. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Rizzo S., Crespi M., Frugier F. (2006). The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell 18: 2680–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth M., Takeda N., Perry J., Uchida H., Dräxl S., Brachmann A., Sato S., Tabata S., Kawaguchi M., Wang T.L., Parniske M. (2010). NENA, a Lotus japonicus homolog of Sec13, is required for rhizodermal infection by arbuscular mycorrhiza fungi and rhizobia but dispensable for cortical endosymbiotic development. Plant Cell 22: 2509–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W., van Noorden G., Van Isterdael G., Beeckman T., Gheysen G., Mathesius U. (2009). Manipulation of auxin transport in plant roots during Rhizobium symbiosis and nematode parasitism. Plant Cell 21: 2553–2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Banba M., Shimoda Y., Kouchi H., Hayashi M., Imaizumi-Anraku H. (2010). A dominant function of CCaMK in intracellular accommodation of bacterial and fungal endosymbionts. Plant J. 63: 141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann A.B., Lombardo F., Miwa H., Perry J.A., Bunnewell S., Parniske M., Wang T.L., Downie J.A. (2006). Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol. 142: 1739–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann A.B., Sandal N., Bek A.S., Madsen L.H., Jurkiewicz A., Nielsen M.W., Tirichine L., Stougaard J. (2011). Cytokinin induction of root nodule primordia in Lotus japonicus is regulated by a mechanism operating in the root cortex. Mol. Plant Microbe Interact. 24: 1385–1395. [DOI] [PubMed] [Google Scholar]
- Held M., Hossain M.S., Yokota K., Bonfante P., Stougaard J., Szczyglowski K. (2010). Common and not so common symbiotic entry. Trends Plant Sci. 15: 540–545. [DOI] [PubMed] [Google Scholar]
- Heyl A., Riefler M., Romanov G.A., Schmülling T. (2012). Properties, functions and evolution of cytokinin receptors. Eur. J. Cell Biol. 91: 246–256. [DOI] [PubMed] [Google Scholar]
- Heyl A., Wulfetange K., Pils B., Nielsen N., Romanov G.A., Schmülling T. (2007). Evolutionary proteomics identifies amino acids essential for ligand-binding of the cytokinin receptor CHASE domain. BMC Evol. Biol. 7: 62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch A.M., Bhuvaneswari T.V., Torrey J.G., Bisseling T. (1989). Early nodulin genes are induced in alfalfa root outgrowths elicited by auxin transport inhibitors. Proc. Natl. Acad. Sci. USA 86: 1244–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland D.R., Arnon D.I. (1950). The water-culture method for growing plants without soil. Calif. Agric. Exper. Stat. Circ. 347: 1–32. [Google Scholar]
- Horst I., Welham T., Kelly S., Kaneko T., Sato S., Tabata S., Parniske M., Wang T.L. (2007). TILLING mutants of Lotus japonicus reveal that nitrogen assimilation and fixation can occur in the absence of nodule-enhanced sucrose synthase. Plant Physiol. 144: 806–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M.S., Liao J., James E.K., Sato S., Tabata S., Jurkiewicz A., Madsen L.H., Stougaard J., Ross L., Szczyglowski K. (2012). Lotus japonicus ARPC1 is required for rhizobial infection. Plant Physiol. 160: 917–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I., Sheen J., Müller B. (2012). Cytokinin signaling networks. Annu. Rev. Plant Biol. 63: 353–380. [DOI] [PubMed] [Google Scholar]
- Inoue T., Higuchi M., Hashimoto Y., Seki M., Kobayashi M., Kato T., Tabata S., Shinozaki K., Kakimoto T. (2001). Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409: 1060–1063. [DOI] [PubMed] [Google Scholar]
- Kaló P., et al. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308: 1786–1789. [DOI] [PubMed] [Google Scholar]
- Karas B., Murray J., Gorzelak M., Smith A., Sato S., Tabata S., Szczyglowski K. (2005). Invasion of Lotus japonicus root hairless 1 by Mesorhizobium loti involves the nodulation factor-dependent induction of root hairs. Plant Physiol. 137: 1331–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistner C., Winzer T., Pitzschke A., Mulder L., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J., Webb K.J., Szczyglowski K., Parniske M. (2005). Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. Plant Cell 17: 2217–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuta S., Held M., Hossain M.S., Morieri G., Macgillivary A., Johansen C., Antolín-Llovera M., Parniske M., Oldroyd G.E., Downie A.J., Karas B., Szczyglowski K. (2011). Lotus japonicus symRK-14 uncouples the cortical and epidermal symbiotic program. Plant J. 67: 929–940. [DOI] [PubMed] [Google Scholar]
- Laloum T., De Mita S., Gamas P., Baudin M., Niebel A. (2013). CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci. 18: 157–166. [DOI] [PubMed] [Google Scholar]
- Lerouge P., Roche P., Faucher C., Maillet F., Truchet G., Promé J.C., Dénarié J. (1990). Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344: 781–784. [DOI] [PubMed] [Google Scholar]
- Le Signor C., Savois V., Aubert G., Verdier J., Nicolas M., Pagny G., Moussy F., Sanchez M., Baker D., Clarke J., Thompson R. (2009). Optimizing TILLING populations for reverse genetics in Medicago truncatula. Plant Biotechnol. J. 7: 430–441. [DOI] [PubMed] [Google Scholar]
- Lévy J., et al. (2004). A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303: 1361–1364. [DOI] [PubMed] [Google Scholar]
- Liao J., Singh S., Hossain M.S., Andersen S.U., Ross L., Bonetta D., Zhou Y., Sato S., Tabata S., Stougaard J., Szczyglowski K., Parniske M. (2012). Negative regulation of CCaMK is essential for symbiotic infection. Plant J. 72: 572–584. [DOI] [PubMed] [Google Scholar]
- Lohar D.P., Schaff J.E., Laskey J.G., Kieber J.J., Bilyeu K.D., Bird D.M. (2004). Cytokinins play opposite roles in lateral root formation, and nematode and rhizobial symbioses. Plant J. 38: 203–214. [DOI] [PubMed] [Google Scholar]
- Lombari, P., Ercolano, E., El Alaouri, E., and Chiurazzi, M. (2005). Agrobacterium-mediated in vitro transformation. In Lotus japonicus Handbook, A.J. Marquez, ed (Dordrecht, The Netherlands: Springer), pp. 87–95. [Google Scholar]
- Madsen E.B., Madsen L.H., Radutoiu S., Olbryt M., Rakwalska M., Szczyglowski K., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J. (2003). A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640. [DOI] [PubMed] [Google Scholar]
- Madsen L.H., Tirichine L., Jurkiewicz A., Sullivan J.T., Heckmann A.B., Bek A.S., Ronson C.W., James E.K., Stougaard J. (2010). The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat Commun 1: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T., Wurgler-Murphy S.M., Saito H. (1994). A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369: 242–245. [DOI] [PubMed] [Google Scholar]
- Marsh J.F., Rakocevic A., Mitra R.M., Brocard L., Sun J., Eschstruth A., Long S.R., Schultze M., Ratet P., Oldroyd G.E. (2007). Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 144: 324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathesius U., Charon C., Rolfe B.G., Kondorosi A., Crespi M. (2000). Temporal and spatial order of events during the induction of cortical cell divisions in white clover by Rhizobium leguminosarum bv. trifolii inoculation or localized cytokinin addition. Mol. Plant Microbe Interact. 13: 617–628. [DOI] [PubMed] [Google Scholar]
- Middleton P.H., et al. (2007). An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19: 1221–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra R.M., Gleason C.A., Edwards A., Hadfield J., Downie J.A., Oldroyd G.E., Long S.R. (2004). A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: Gene identification by transcript-based cloning. Proc. Natl. Acad. Sci. USA 101: 4701–4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B., Sheen J. (2008). Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453: 1094–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D., Müller R., Funk M. (1995). Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119–122. [DOI] [PubMed] [Google Scholar]
- Murakami Y., Miwa H., Imaizumi-Anraku H., Kouchi H., Downie J.A., Kawaguchi M., Kawasaki S. (2006). Positional cloning identifies Lotus japonicus NSP2, a putative transcription factor of the GRAS family, required for NIN and ENOD40 gene expression in nodule initiation. DNA Res. 13: 255–265. [DOI] [PubMed] [Google Scholar]
- Murray J., et al. (2006). Genetic suppressors of the Lotus japonicus har1-1 hypernodulation phenotype. Mol. Plant Microbe Interact. 19: 1082–1091. [DOI] [PubMed] [Google Scholar]
- Murray J.D., Karas B.J., Sato S., Tabata S., Amyot L., Szczyglowski K. (2007). A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315: 101–104. [DOI] [PubMed] [Google Scholar]
- Nishimura C., Ohashi Y., Sato S., Kato T., Tabata S., Ueguchi C. (2004). Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16: 1365–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Op den Camp R., Streng A., De Mita S., Cao Q., Polone E., Liu W., Ammiraju J.S., Kudrna D., Wing R., Untergasser A., Bisseling T., Geurts R. (2011). LysM-type mycorrhizal receptor recruited for rhizobium symbiosis in nonlegume Parasponia. Science 331: 909–912. [DOI] [PubMed] [Google Scholar]
- Petit A., Stougaard J., Kühle A., Marcker K.A., Tempé J. (1987). Transformation and regeneration of the legume Lotus corniculatus: A system for molecular studies of nitrogen fixation. Mol. Gen. Genet. 207: 245–250. [Google Scholar]
- Plet J., Wasson A., Ariel F., Le Signor C., Baker D., Mathesius U., Crespi M., Frugier F. (2011). MtCRE1-dependent cytokinin signaling integrates bacterial and plant cues to coordinate symbiotic nodule organogenesis in Medicago truncatula. Plant J. 65: 622–633. [DOI] [PubMed] [Google Scholar]
- Radutoiu S., Madsen L.H., Madsen E.B., Felle H.H., Umehara Y., Grønlund M., Sato S., Nakamura Y., Tabata S., Sandal N., Stougaard J. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592. [DOI] [PubMed] [Google Scholar]
- Santi C., Bogusz D., Franche C. (2013). Biological nitrogen fixation in non-legume plants. Ann. Bot. (Lond.) 111: 743–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayano T., Kouchi H., Hirota A., Hayashi M. (2013). Nodule inception directly targets NF-Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus. PLoS Genet. 9: e1003352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauser L., Roussis A., Stiller J., Stougaard J. (1999). A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195. [DOI] [PubMed] [Google Scholar]
- Sieberer B.J., Chabaud M., Fournier J., Timmers A.C., Barker D.G. (2012). A switch in Ca2+ spiking signature is concomitant with endosymbiotic microbe entry into cortical root cells of Medicago truncatula. Plant J. 69: 822–830. [DOI] [PubMed] [Google Scholar]
- Singh S., Parniske M. (2012). Activation of calcium- and calmodulin-dependent protein kinase (CCaMK), the central regulator of plant root endosymbiosis. Curr. Opin. Plant Biol. 15: 444–453. [DOI] [PubMed] [Google Scholar]
- Smit P., Raedts J., Portyanko V., Debellé F., Gough C., Bisseling T., Geurts R. (2005). NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308: 1789–1791. [DOI] [PubMed] [Google Scholar]
- Sprent J.I., James E.K. (2007). Legume evolution: Where do nodules and mycorrhizas fit in? Plant Physiol. 144: 575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki T., Yano K., Ito M., Umehara Y., Suganuma N., Kawaguchi M. (2012). Positive and negative regulation of cortical cell division during root nodule development in Lotus japonicus is accompanied by auxin response. Development 139: 3997–4006. [DOI] [PubMed] [Google Scholar]
- Szczyglowski K., Amyot L. (2003). Symbiosis, inventiveness by recruitment? Plant Physiol. 131: 935–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczyglowski K., Shaw S.R., Wopereis J., Hamburger D., Copeland S., Dazzo F.B., de Bruijn F.J. (1998). Nodule organogenesis and symbiotic mutants of the model legume Lotus japonicus. Mol. Plant Microbe Interact. 11: 684–697. [Google Scholar]
- Thimann K.V. (1936). On the physiology of the formation of nodules on legume roots. Proc. Natl. Acad. Sci. USA 22: 511–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirichine L., et al. (2006a). Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441: 1153–1156. [DOI] [PubMed] [Google Scholar]
- Tirichine L., James E.K., Sandal N., Stougaard J. (2006b). Spontaneous root-nodule formation in the model legume Lotus japonicus: A novel class of mutants nodulates in the absence of rhizobia. Mol. Plant Microbe Interact. 19: 373–382. [DOI] [PubMed] [Google Scholar]
- Tirichine L., Sandal N., Madsen L.H., Radutoiu S., Albrektsen A.S., Sato S., Asamizu E., Tabata S., Stougaard J. (2007). A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 315: 104–107. [DOI] [PubMed] [Google Scholar]
- Truchet G., Barker D.G., Camut S., de Billy V.J., Huguet T. (1989). Alfalfa nodulation in the absence of Rhizobium. Mol. Gen. Genet. 219: 65–68. [Google Scholar]
- Truchet G., Roche P., Lerouge P., Vasse J., Camut S., de Billy F., Promé J.-C., Dénarié J. (1991). Sulphated lipo-oligosaccharide signals of Rhizobium meliloti elicit root nodule organogenesis in alfalfa. Nature 351: 670–673. [Google Scholar]
- Ueguchi C., Koizumi H., Suzuki T., Mizuno T. (2001). Novel family of sensor histidine kinase genes in Arabidopsis thaliana. Plant Cell Physiol. 42: 231–235. [DOI] [PubMed] [Google Scholar]
- van Spronsen P.C., Grønlund M., Pacios Bras C., Spaink H.P., Kijne J.W. (2001). Cell biological changes of outer cortical root cells in early determinate nodulation. Mol. Plant Microbe Interact. 14: 839–847. [DOI] [PubMed] [Google Scholar]
- Verdier J., Torres-Jerez I., Wang M., Andriankaja A., Allen S.N., He J., Tang Y., Murray J.D., Udvardi M.K. (2013). Establishment of the Lotus japonicus Gene Expression Atlas (LjGEA) and its use to explore legume seed maturation. Plant J. 74: 351–362. [DOI] [PubMed] [Google Scholar]
- Wais R.J., Galera C., Oldroyd G., Catoira R., Penmetsa R.V., Cook D., Gough C., Denarié J., Long S.R. (2000). Genetic analysis of calcium spiking responses in nodulation mutants of Medicago truncatula. Proc. Natl. Acad. Sci. USA 97: 13407–13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker S.A., Downie J.A. (2000). Entry of Rhizobium leguminosarum bv. viciae into root hairs requires minimal Nod factor specificity, but subsequent infection thread growth requires nodO or nodE. Mol. Plant Microbe Interact. 13: 754–762. [DOI] [PubMed] [Google Scholar]
- Wopereis J., Pajuelo E., Dazzo F.B., Jiang Q., Gresshoff P.M., De Bruijn F.J., Stougaard J., Szczyglowski K. (2000). Short root mutant of Lotus japonicus with a dramatically altered symbiotic phenotype. Plant J. 23: 97–114. [DOI] [PubMed] [Google Scholar]
- Wulfetange K., Lomin S.N., Romanov G.A., Stolz A., Heyl A., Schmülling T. (2011). The cytokinin receptors of Arabidopsis are located mainly to the endoplasmic reticulum. Plant Physiol. 156: 1808–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H., Suzuki T., Terada K., Takei K., Ishikawa K., Miwa K., Yamashino T., Mizuno T. (2001). The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol. 42: 1017–1023. [DOI] [PubMed] [Google Scholar]
- Yokota K., et al. (2009). Rearrangement of actin cytoskeleton mediates invasion of Lotus japonicus roots by Mesorhizobium loti. Plant Cell 21: 267–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.