CULLIN4-based E3 ubiquitin ligases and their associated substrate receptors, DDB1-CUL4–associated factors (DCAFs), regulate the selective ubiquitination of proteins, which ultimately leads to their degradation. This study reports that a DCAF, ABD1 (for ABA-hypersensitive DCAF1), directly binds to and affects the stability of ABI5, a central transcription factor in abscisic acid signaling.
Abstract
Members of the DDB1-CUL4–associated factors (DCAFs) family directly bind to DAMAGED DNA BINDING PROTEIN1 (DDB1) and function as the substrate receptors in CULLIN4-based E3 (CUL4) ubiquitin ligases, which regulate the selective ubiquitination of proteins. Here, we describe a DCAF protein, ABD1 (for ABA-hypersensitive DCAF1), that negatively regulates abscisic acid (ABA) signaling in Arabidopsis thaliana. ABD1 interacts with DDB1 in vitro and in vivo, indicating that it likely functions as a CUL4 E3 ligase substrate receptor. ABD1 expression is induced by ABA, and mutations in ABD1 result in ABA- and NaCl-hypersensitive phenotypes. Loss of ABD1 leads to hyperinduction of ABA-responsive genes and higher accumulation of the ABA-responsive transcription factor ABA INSENSITIVE5 (ABI5), hypersensitivity to ABA during seed germination and seedling growth, enhanced stomatal closure, reduced water loss, and, ultimately, increased drought tolerance. ABD1 directly interacts with ABI5 in yeast two-hybrid assays and associates with ABI5 in vivo by coimmunoprecipitation, and the interaction was found in the nucleus by bimolecular fluorescence complementation. Furthermore, loss of ABD1 results in a retardation of ABI5 degradation by the 26S proteasome. Taken together, these data suggest that the DCAF-CUL4 E3 ubiquitin ligase assembled with ABD1 is a negative regulator of ABA responses by directly binding to and affecting the stability of ABI5 in the nucleus.
INTRODUCTION
Due to their sessile nature, plants have evolved an extraordinary ability to alter their physiology, morphology, and development to adapt to changes in the environment. Environmental stress, particularly water-related stresses including drought and high salinity, presents an increasingly important challenge in agriculture as it reduces the potential yields as much as 70% in crop plants (Agarwal et al., 2006). Plant hormones play a central role in environmental adaptation by inducing many biochemical and physiological alterations to overcome abiotic stress such as drought, salinity, wounding, UV, and temperature change and biotic stress such as pathogen infection. The phytohormone abscisic acid (ABA) is an important regulator of plant growth and development that plays a key role in adaptive stress responses. ABA, at its fundamental level, mediates multiple physiological processes such as stomatal movement, seed development, embryo morphogenesis, seed dormancy, and synthesis of storage proteins and lipids. However, in response to abiotic and biotic stress, ABA levels rise substantially and play a pivotal role as a messenger in adaptive stress responses.
ABA regulates a much larger subset of genes compared with other plant hrmones, with just over 10% of protein-coding genes that are likely regulated by ABA in Arabidopsis thaliana plants (Nemhauser et al., 2006; Goda et al., 2008; Mizuno and Yamashino, 2008). In fact, a number of important components of this signaling network have been identified and characterized by a combination of biochemical, pharmacological, structural, and genetic approaches. In the presence of ABA, the ABA receptors PYR/PYL/RCAR (Pyrabactin resistance 1/Pyrabactin resistance 1-like/Regulatory Component of ABA Receptor) function at the apex of a negative regulatory pathway to directly bind to and negatively regulate type 2C Ser/Thr protein phosphatases (Ma et al., 2009; Park et al., 2009; Santiago et al., 2009; Nishimura et al., 2010). This allows for the activation of SnRK2 kinases (sucrose nonfermenting-1-related protein kinase class 2), which subsequently phosphorylate ABFs (ABA-responsive element binding factors) (Cutler et al., 2010). ABFs include members of the ABF/AREB/ABI5 clade of bZIP transcription factors (TFs), which represent the major TF family that regulates ABA-mediated gene expression under stress conditions. TFs in this family recognize the ABA response elements (ABREs), a G-box that is involved in ABA-regulated gene expression (Uno et al., 2000), and upregulate the expression of ABA-responsive genes such as RESPONSIVE TO DESSICATION 29A (RD29A) and RD29B (Nakashima et al., 2006). In addition to ABRE motifs, MYC and MYB recognition sites also have important roles in ABA signaling. The TFs MYC2 and MYB2 recognize these motifs, which are synthesized de novo in response to ABA and cooperatively activate stress inducible genes such as RD22 (Abe et al., 2003).
Adaptation to abiotic stress is also achieved through the ubiquitination and degradation of components specific to these stress-signaling pathways by the ubiquitin proteasome system (UPS). In fact, ABA INSENSITIVE5 (ABI5) is rapidly degraded after seed germination or removal of ABA, and its stability is regulated by ubiquitin-mediated degradation (Lopez-Molina et al., 2001; Lopez-Molina et al., 2003). The first evidence that ABI5 was regulated by the proteasome was with the identification of ABI FIVE BINDING PROTEIN1 (AFP1), which is a member of a small plant-specific protein family. AFP1 directly binds to and forms a high molecular weight complex with ABI5 and facilitates ubiquitin-mediated proteolysis of ABI5 (Lopez-Molina et al., 2003; Garcia et al., 2008). Moreover, when AFP1 and ABI5 are expressed together, the proteins become colocalized in nuclear bodies and may represent sites of ABI5 degradation (Lopez-Molina et al., 2003). More recently, two different types of E3 ligases have been identified as negative regulators of ABI5. KEEP ON GOING (KEG), a RING-ANK E3 ligase, is required for the regulation of ABI5 abundance. In vivo studies have shown that in the absence of ABA, KEG directly ubiquitinates ABI5, which causes the proteasomal degradation of ABI5. However, the presence of ABA triggers the self-ubiquitination and proteasomal degradation of KEG itself, leading to increased ABI5 stability (Stone et al., 2006; Liu and Stone, 2010). DWD hypersensitive to ABA1 (DWA1) and DWA2 are substrate adaptors for the CULLIN4-based E3 ligases that directly interact with each other and act together to mediate the degradation of ABI5 by the UPS (Lee et al., 2010).
The UPS polyubiquitinates target proteins for degradation by the 26S proteasome, which is accomplished by consecutive activities of three enzymes, ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3) (Hotton and Callis, 2008). The substrate specificity of the pathway is determined by the E3 ligase, which binds to a specific target protein and stimulates the conjugation of ubiquitin to the target protein. The largest E3 ligase family in Arabidopsis thaliana is the cullin-RING ubiquitin ligase (CRL) superfamily, with the potential to assemble many hundreds to a thousand distinct CRLs, which can be connected to almost all stages of plant physiology, growth, development, and stress response (Smalle and Vierstra, 2004; Hua and Vierstra, 2011). CRLs are assembled on a cullin protein, which provides scaffolding for two essential modules: a small RING-box domain protein (RBX1) on its C terminus and adaptors that are specific for each cullin protein, which are associated with substrate receptors that specifically recognize their corresponding target proteins on its N terminus (Hotton and Callis, 2008).
One of the cullin proteins, CULLIN4 (CUL4), the most recently identified cullin protein, has been described in greater detail in mammals recently due to its role in many key biological pathways. Unlike other cullin proteins, CUL4 assembles with DAMAGED DNA BINDING PROTEIN1 (DDB1) and a WD40 protein as its adaptor (Pintard et al., 2004; He et al., 2006). DDB1 consists of three WD40 β-propeller domains (BPA, BPB, and BPC) and a C-terminal helical domain. The BPB propeller mediates the interaction with CUL4, while the BPA and BPC propellers arrange themselves as a clam shape pocket, allowing for the association of DDB1 interacting partners, the substrate receptors for CUL4-RING ubiquitin ligases (CRL4s) (Angers et al., 2006; Li et al., 2006; Scrima et al., 2008; Bernhardt et al., 2010). DDB1-interacting partners contain a conserved domain called the DDB1 binding WD40 protein (DWD) box, which is a 16–amino acid motif that is found within WD40 proteins and is conserved in many eukaryotes (He et al., 2006). Furthermore, a majority of DDB1-interacting WD40 proteins contain one to two copies of a shorter four-amino motif called the WDxR, with Asp and Arg as essential residues for the interaction with DDB1 (Angers et al., 2006; Higa et al., 2006; Jin et al., 2006). This subgroup of WD40 proteins is interchangeably referred to as DDB1-CUL4–associated factor (DCAF) proteins, DWD proteins, WDxR proteins, or CUL4- and DDB1-associated WD40-repeat proteins (hereafter referred to as DCAF).
Compared with mammalian CRL4s, Arabidopsis uses similar machinery for its CRL4s. For example, DDB1 is encoded by two genes, DDB1a and DDB1b, compared with the human genome, which only encodes one (Schroeder et al., 2002; Bernhardt et al., 2010). Furthermore, 85 proteins in Arabidopsis and 78 proteins in rice (Oryza sativa) were found to contain at least one DWD motif, while the human genome contains upwards of 90 WD40 proteins that contain a DWD motif (He et al., 2006; Lee et al., 2008). Moreover, similar to mammalian studies, WD40 proteins with the WDxR motif are also able to interact with DDB1, with 119 out of 297 Arabidopsis WD40 proteins and 110 out of 223 rice WD40 proteins containing at least one copy of the motif (Zhang et al., 2008).
Initially, CRL4s were described as regulators of damaged DNA, with the major function of DDB1 described as participating in the recognition of damaged DNA and initiation of nucleotide excision repair processes (Keeney et al., 1993; Groisman et al., 2003; Zhong et al., 2003; Biedermann and Hellmann, 2010). Recently, other regulatory functions have been assigned to the CRL4 E3 ligases, such as photomorphogenesis, cell cycle regulation, flower timing and development, UV-B–induced photomorphogenesis and stress acclimation, and ABA stress response, but a majority of these E3 ligases still have no assigned function (Zhang et al., 2003; Chen et al., 2010; Gruber et al., 2010; Lee et al., 2010; Dumbliauskas et al., 2011; Lee et al., 2011; Pazhouhandeh et al., 2011; Nezames et al., 2012; Huang et al., 2013). Since it is proposed that upwards of 10% of coding genes are ABA regulated, it is likely that there are additional substrate receptors for CRL4 ubiquitin ligases, besides DWA1, DWA2, and DWA3 that participate in ABA signaling (Dreher and Callis, 2007; Lee et al., 2010, 2011). Moreover, DCAF proteins that only possess a WDxR motif (not an entire DWD domain) for ABA signaling have yet to be confirmed. Here, we demonstrate that ABD1, a DCAF protein that contains a WDxR motif, plays a critical role as a negative regulator in ABA signaling. We show that abd1 loss-of-function mutants display ABA hypersensitivity phenotypes in seed germination and postgermination growth. ABD1 functions as a substrate receptor in the CUL4-DDB1 E3 ligase machinery and directly interacts with ABI5 in vitro and in vivo, which modulates the stability of ABI5. Our findings also indicate that ABD1 functions separately from the DWA proteins (Lee et al., 2010, 2011). Taken together, ABD1 is a negative regulator of ABA signaling, acting as a substrate receptor in the CUL4-DDB1 E3 ligase by directly binding to ABI5 in the nucleus and negatively affecting the stability of ABI5.
RESULTS
Identification of a DCAF Protein Involved in ABA Response
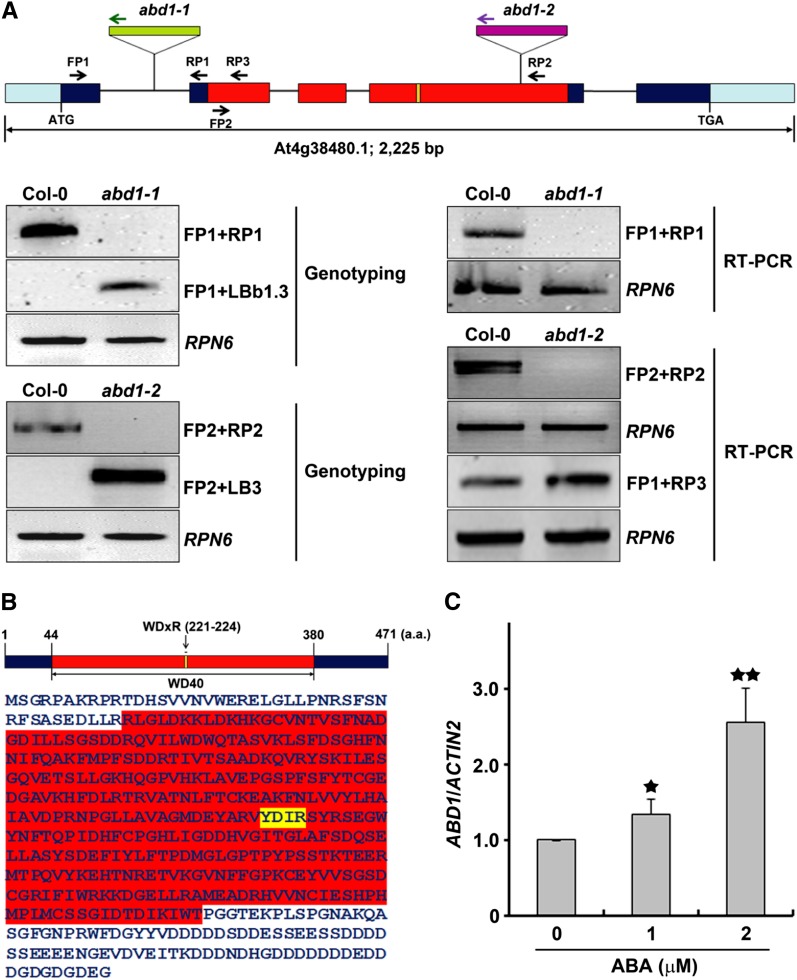
To investigate the possible involvement of DCAFs with a WDxR motif during the ABA response, we analyzed their transcriptional levels in response to ABA or NaCl using microarray data from the AtGenExpress Visualization Tool (Kilian et al., 2007), since salt stress and ABA responses are largely overlapped with each other (Zhu, 2002). One At4g38480 named ABD1 (for ABA-hypersensitive DCAF1) was selected since its transcripts were upregulated 1.9 times in seeds after treatment with 3 μM ABA for 24 h and 2.3 times in roots after treatment with 150 mM NaCl for 6 h based on AtGenExpress Visualization Tool data. To check if the loss of ABD1 leads to altered sensitivity in response to ABA, two independent T-DNA insertion lines, abd1-1 and abd1-2, were obtained, representing insertions in the first intron and fifth exon, respectively, which were analyzed for ABD1 expression (Figure 1A). RT-PCR analysis revealed that both T-DNA insertion lines abolish ABD1 expression (Figure 1A). Since the T-DNA insertion in abd1-2 occurs at the 3′ end of ABD1, we further tested for the expression of a partial ABD1 transcript, upstream of the T-DNA insertion in abd1-2. RT-PCR analysis revealed that abd1-2 expresses a truncated ABD1 transcript (Figure 1A). In our initial screenings, abd1-1 and abd1-2 displayed hypersensitivity to ABA and NaCl; therefore, they were selected for characterization.
Figure 1.
Schematic Structure of ABD1 and Isolation of abd1-1 and abd1-2.
(A) The genomic structure and T-DNA insertions in Arabidopsis ABD1. Exons are depicted as colored boxes, introns are represented by lines, and pale-blue boxes represent the 5′- and 3′-untranslated regions. The T-DNA insertion sites, abd1-1 and abd1-2, are represented by open inverted triangles. Genotyping and transcript analysis of abd1-1 and abd1-2. RPN6 primers were used as an internal control for the both the genotyping and transcript analyses. RT-PCR analysis of the ABD1 transcript was performed in wild-type, abd1-1, and abd1-2 mutant seedlings.
(B) Protein structure of ABD1 depicting the predicted WD40 region in red and the WDxR motif in yellow.
(C) Quantitative PCR analysis of ABD1 expression in 7-d-old Col-0 wild-type seedlings in the absence or presence of 1 or 2 μM ABA. Values are means ± sd (n = 3). Significant difference was determined by a Student’s t test; single or double stars indicate a P value of <0.05 or <0.01, respectively.
ABD1 has a 1416-bp-long coding region, which encodes a 471–amino acid protein (Figures 1A and 1B). Sequence analysis indicated that ABD1 has seven predicted WD40 repeats between amino acids 44 to 380 and one WDxR motif that is found within the fourth WD40 repeat between amino acids 221 to 224 (Figure 1B). From the GenBank reference protein database, we identified homologs of ABD1 from several other representative model organisms, including rice, mouse (Mus musculus), human (Homo sapiens), budding yeast (Saccharomyces cerevisiae), fission yeast (Schizosaccharomyces pombe), methane-producing Archaebacterium (Methanosarcina barkei and Methanosaeta harundinacea), and cyanobacterium. Together, this suggests that ABD1 has been evolutionarily conserved among all species, possibly with cyanobacterial ancestry. Arabidopsis ABD1 also has 42 and 43% sequence similarity to human and mouse DCAF8, respectively. In all three homologs, the WDxR motif is conserved, indicating that this motif is likely important for the function of these proteins (Supplemental Figure 1). Furthermore, ABD1 transcripts are induced upon exposure to increasing concentrations of ABA (Figure 1C), suggesting that ABD1 plays a role in ABA signaling in Arabidopsis.
ABD1 Regulates ABA-Mediated Seed Germination and Postgermination Growth
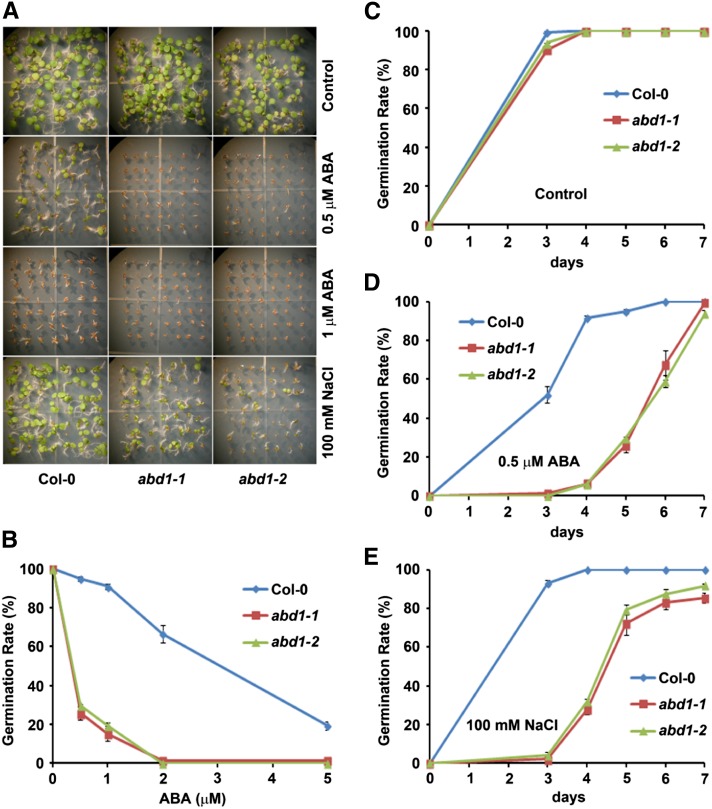
To further characterize the role of ABD1 in the ABA signaling pathway, abd1-1 and abd1-2 seeds were germinated on increasing concentrations of ABA as well as NaCl. In the presence of ABA and NaCl, abd1-1 and abd1-2 seedlings exhibited significantly reduced seed germination (Figure 2A). A dose–response analysis using increasing concentrations of ABA was performed to compare the germination of Columbia-0 (Col-0) wild type, abd1-1, and abd1-2 after 5 d. Treatment with increasing concentrations of ABA resulted in a decrease in seed germination in concentrations as low as 0.5 μM ABA (Figure 2B). To confirm that the reduced germination rate in abd1-1 and abd1-2 was not the result of a developmental defect in seeds, Col-0 wild-type, abd1-1, and abd1-2 seeds were germinated in the absence of ABA. As shown in Figure 2C, both abd1-1 and abd1-2 have a slightly delayed germination, but reach 100% germination after 4 d. When germinated on medium containing 0.5 μM ABA, abd1-1 and abd1-2 seeds germinated much later than in the wild type but were able to eventually germinate to levels similar to the wild type after 7 d (Figure 2D). Furthermore, abd1-1 and abd1-2 displayed delayed germination rate to 100 mM NaCl compared with the wild type (Figure 2E).
Figure 2.
ABD1 Is Required for Normal Seed Germination and Postgerminative Growth in Response to ABA and NaCl.
(A) Visual comparison of Col-0, abd1-1, and abd1-2 seed germination and postgerminative growth after 7 d in the absence or presence of 0.5 μM ABA, 1 μM ABA, or 100 mM NaCl.
(B) Col-0 wild type, abd1-1, and abd1-2 were grown for 5 d on an increasing concentration of ABA, after which the germination rate was determined. Mean ± sd values were determined from three replicates (n = 144).
(C) to (E) Germination rate of Col-0 wild type, abd1-1, and abd1-2 over 7 d in the absence (C) or presence (D) of 0.5 μM ABA or 100 mM NaCl (E). Mean ± sd values were determined from three replicates (n = 144).
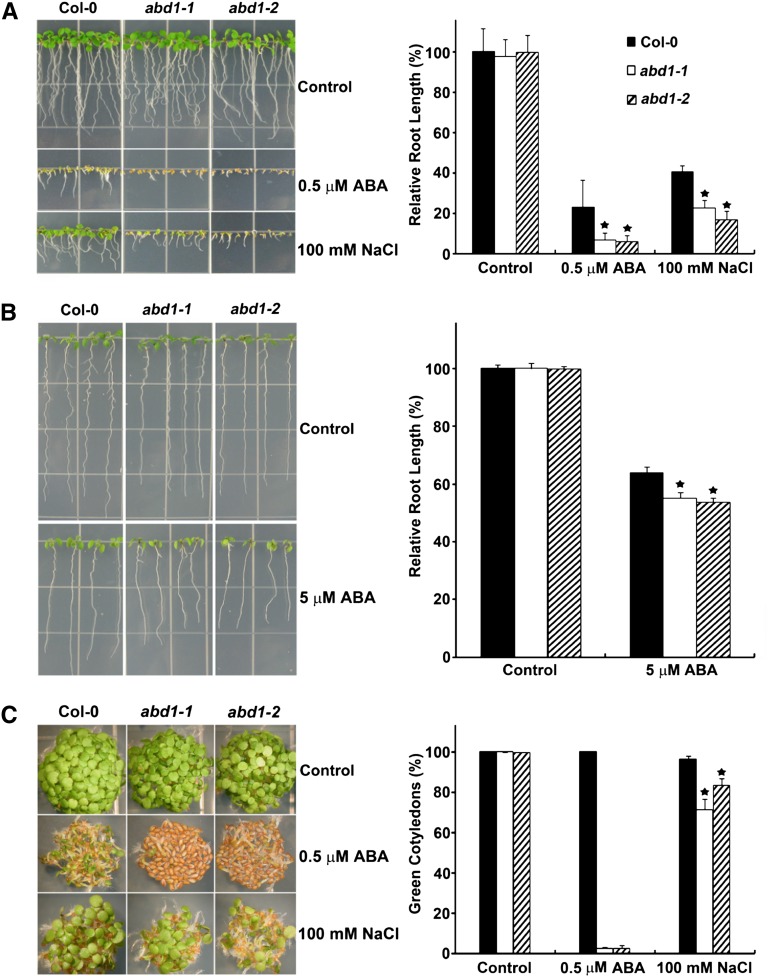
When abd1-1 and abd1-2 were germinated and grown in the presence of low levels of ABA and NaCl, both mutants displayed hypersensitivity to both treatments (Figure 3A). To determine if the reduced root growth in the abd1 mutants was solely due to a defect in seed germination and not the inhibition of primary root growth, the seeds of abd1-1 and abd1-2 were allowed to germinate and were then transferred to media containing a high level of ABA. In contrast with the dwa mutants (Lee et al., 2008, 2010), there was a significant reduction in primary root growth of abd1-1 and abd1-2 when compared with the wild type (Figure 3B). Furthermore, when cotyledon greening was observed, both abd1-1 and abd1-2 had reduced cotyledon greening compared with the wild type (Figure 3C). Together, these results demonstrate that ABD1 acts as a negative regulator of ABA signaling during both seed germination and postgermination seedling growth.
Figure 3.
ABA and NaCl Responses of Col-0 Wild-Type, abd1-1, and abd1-2 Plants in Terms of Seedling Growth.
(A) ABA and NaCl hypersensitivity of abd1-1 and abd1-2. Root growth of Col-0 wild type, abd1-1, and abd1-2 that were vertically grown on MS medium in the absence or presence of 0.5 µM ABA or 100 mM NaCl for 7 d. Values are mean ± sd (n = 12). Significant difference was determined by a Student’s t test; a star indicates a P value of <0.0001.
(B) Root growth inhibition of seedlings in the presence ABA. Seedlings were grown for 3 d on MS plates and then transferred to 0 or 5 μM ABA. After 5 d of growth, photographs were taken and root lengths were measured. Values are means ± sd (n = 10). Significant difference was determined by a Student’s t test; a star indicates a P value of <0.0001.
(C) Cotyledon greening of Col-0 wild type, abd1-1, and abd1-2. Seedlings were germinated and grown on MS plates in the absence or presence of 0.5 µM ABA or 100 mM NaCl for 7 d, after which photographs were taken. Cotyledon greening percentage was determined from an average of >100 seeds with three independent experiments. Values are means ± sd (n = 144). Significant difference was determined by a Student’s t test; a star indicates a P value of <0.0001.
ABD1 Regulates ABA-Mediated Stomatal Closure and Plant Drought Tolerance
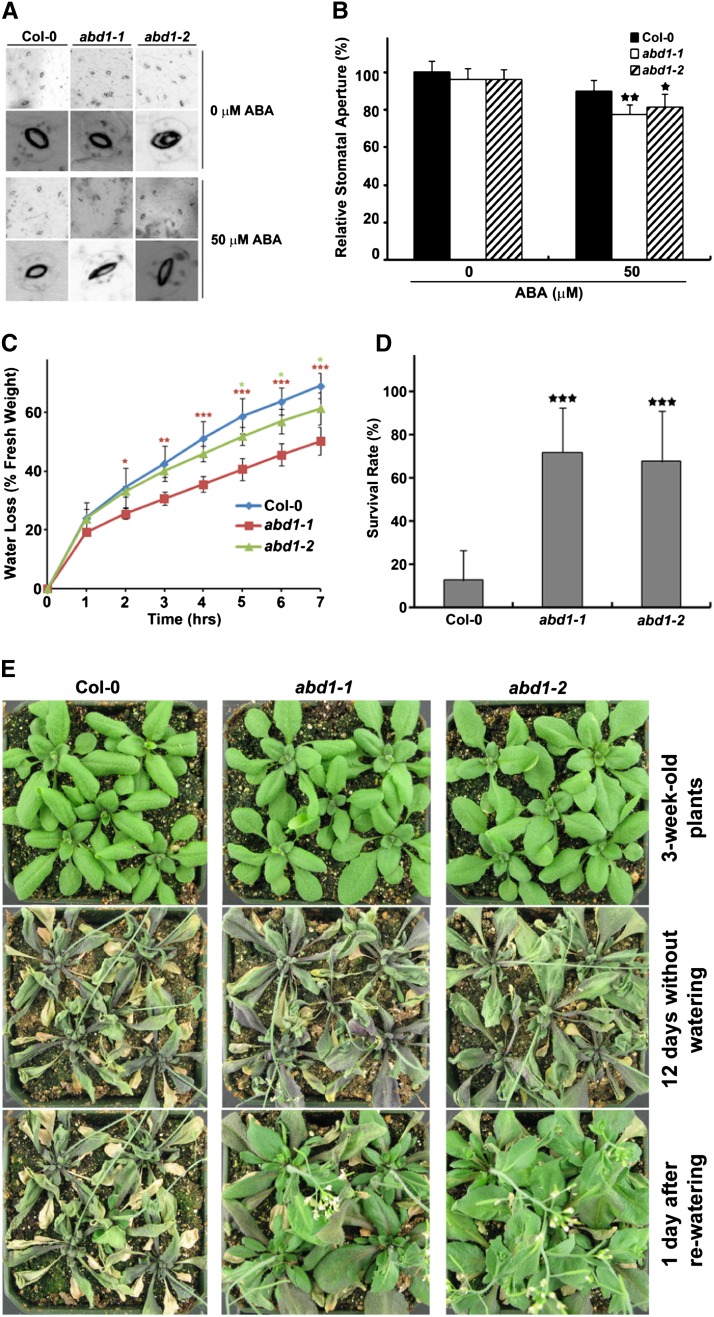
Since loss of ABD1 confers hypersensitivity to ABA during both seed germination and postgermination growth, it was of interest to determine if loss of ABD1 results in other ABA-associated phenotypes, such as enhanced ABA-induced stomatal closing and enhanced drought tolerance. ABA-mediated stomatal closure is one of the ABA-regulated processes that determine the rate of transpiration under drought conditions. For this experiment, epidermal peels from both wild-type and abd1 mutants were used to analyze the stomatal response to ABA. As shown in Figure 4, treatment of wild-type plants with 50 μM ABA for 2.5 h led to stomatal apertures of the wild type being reduced by 10%, whereas the abd1 mutants had stomatal apertures reduced by 20% (Figures 4A and 4B).
Figure 4.
Loss of ABD1 Confers Increased Drought Tolerance.
(A) ABA-induced stomatal closure of Col-0 wild type, abd1-1, and abd1-2. Epidermal peels from Col-0 wild type, abd1-1, and abd1-2 were measured for stomatal aperture in response to ABA as described by Li et al. (2011).
(B) Relative stomatal aperture compared with that on ABA-free medium. Results are from three replicates, and values represent means ± sd (n = 30). Significant difference was determined by a Student’s t test; single and double stars indicate a P value 0.01 ≤ P < 0.05 and P < 0.01, respectively.
(C) Water loss assay of Col-0 wild type, abd1-1, and abd1-2 detached leaves. Results are from three replicates, and values represent means ± sd (n = 3). Statistically significant difference was determined by a Student’s t test; single, double, and triple stars indicate P values of <0.05, <0.005, and <0.001, respectively.
(D) Drought tolerance assay of 3-week-old Col-0 wild-type, abd1-1, and abd1-2 plants was performed by withholding water for 12 d and subsequently rewatering and examining after 1 d. Values represent means ± sd (n = 132). Significant difference was determined by a Student’s t test; triple stars indicate a P value of <0.0001.
(E) Representative plants from drought tolerance assay described in (D).
Since ABD1 loss-of-function mutants displayed enhanced stomatal closing in response to ABA treatment, it was likely that ABD1 may play a role in regulating plant response to drought. We assessed whether the enhanced stomatal response of abd1 mutants altered their transpiration rates and ultimately contributes to their better survival under drought stress. As shown in Figure 4C, the water loss rate of abd1-1 and abd1-2 detached leaves exhibited slower water loss than wild-type plants. Consistent with the stomatal response to ABA (Figure 4B), abd1-2 displayed little effect on leaf water loss, suggesting that abd1-2 is a weaker allele, compared with abd1-1.
To confirm that loss of ABD1 results in increased drought tolerance, 3-week-old wild-type and abd1-1 and abd1-2 plants were assayed for survival after drought tolerance. As shown in Figure 4D, after withholding water for 12 d in 3-week-old plants and then allowing them to recover after rewatering for 1 d, over 70% of the abd1 mutants survived, while just over 10% of Col-0 wild-type plants survived. In support of the enhanced survival, in Figure 4E, after withholding water for 12 d, all of the tested plants appeared withered, whereas upon rewatering, both loss-of-function mutants, abd1-1 and abd1-2, survived the drought stress, while wild-type plants remained withered.
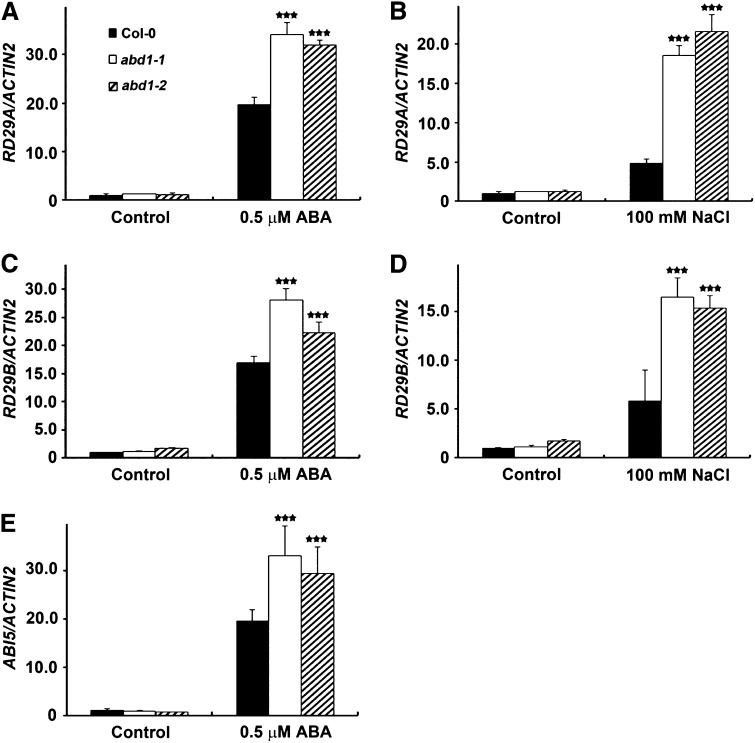
ABD1 Affects Expression Patterns of ABA-Responsive Genes
ABI5 and MYC2 activate a variety of ABA-inducible genes in response to ABA and salt treatment, such as RD29A, RD29B, and RD22. It has been previously reported RD29A and RD29B have ABREs in their promoters and are transactivated by ABI5, whereas RD22 is transactivated by MYC2 (Abe et al., 2003; Nakashima et al., 2006). We tested the expression of these ABA-inducible genes, as well as ABI5, in Col-0 wild type and abd1 mutants in response to 0.5 μM ABA and 100 mM NaCl (Figure 5; Supplemental Figure 2). RD29A, RD29B, and ABI5 were all hyperinduced in abd1-1 and abd1-2 compared with the wild type (Figure 5). We further tested to see if ABI3 induction due to loss of ABD1 was causing the overexpression of ABI5. As shown in Supplemental Figure 2E, we did not find any significant change in the transcript levels between Col-0 wild type and abd1 before and after treatment with ABA, indicating that ABI5 overexpression in abd1 is not a result of ABI3 overexpression due to loss of ABD1.
Figure 5.
abd1-1 and abd1-2 Seedlings Have Increased Expression of Abiotic Stress–Responsive Genes after ABA and NaCl Treatment.
Seven-day-old Col-0 wild-type, abd1-1, and abd1-2 seeds were grown in the absence or presence of 0.5 μM ABA or 100 mM NaCl. mRNA levels were determined by quantitative real-time PCR analysis. Relative amounts of transcripts were normalized to the levels of ACTIN2 within the same sample. Results are from three biological replicates and values represent means ± sd (n = 9). Statically significant difference was determined by a Student’s t test; triple black stars indicate a significant difference between the wild type and the abd1 mutants (P < 0.0001).
(A) RD29A after ABA treatment.
(B) RD29A after NaCl treatment.
(C) RD29B after ABA treatment.
(D) RD29B after NaCl treatment.
(E) ABI5 after ABA treatment.
Furthermore, RD22 expression level did not show a significant hyperinduction when compared with the wild type, suggesting that MYC2 activated pathway is not compromised in ABD1 loss-of-function plants. Moreover, the expression of the drought- and salt-inducible gene RD20 was also investigated. Loss of ABD1 also did not result in a significant hyperinduction of RD20 when compared with the wild type (Supplemental Figure 2).
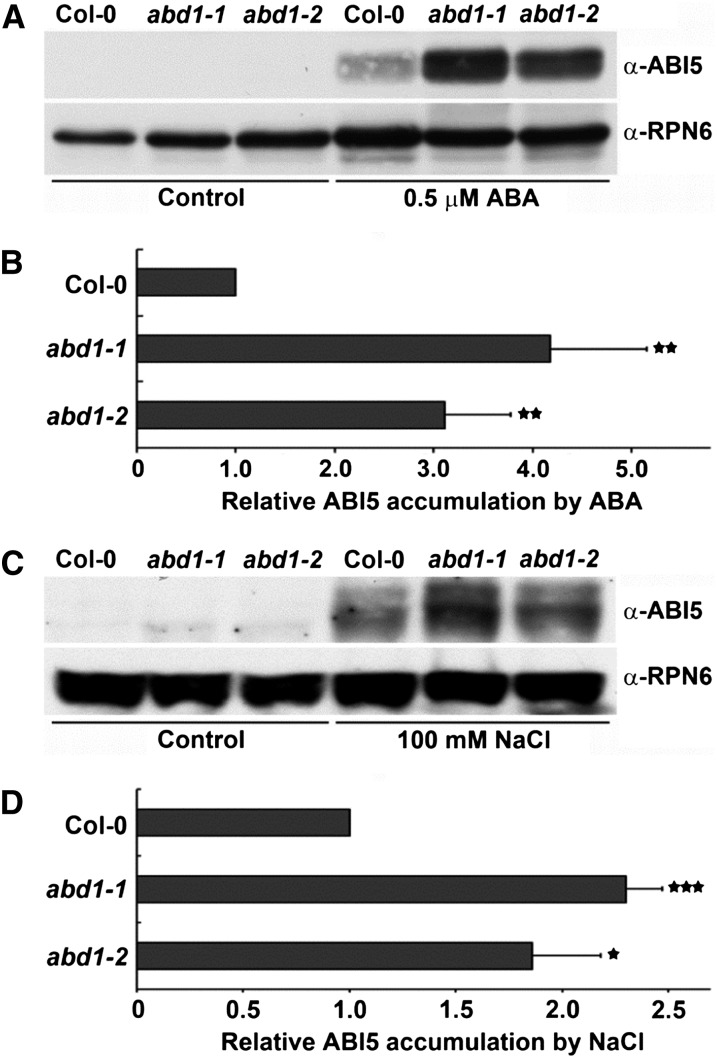
Since abd1 mutants exhibited ABA hypersensitivity and ABI5-regulated genes are hyperinduced in abd1 mutants, we checked the protein levels of ABI5. The wild type and abd1 mutants were germinated and grown on Murashige and Skoog (MS) plates supplemented with 0.5 μM ABA or 100 mM NaCl. After 7 d of growth, protein level of ABI5 was examined. In immunoblot assays, ABI5 appears as three different isoforms (52.5, 51, and 50 kD) using the ABI5 antibody, with the 52.5- and 50-kD bands predominating in the blot (Stone et al., 2006). Similar to what has been previously reported, we also detected the two predominate ABI5 bands in our immunoblot assays in response to ABA treatment. As shown in Figure 6, abd1 mutants hyperaccumulate ABI5 in response to ABA, about 4-fold more ABI5 protein compared with the wild-type plants (Figures 6A and 6B). Furthermore, abd1 mutants also hyperaccumulate ABI5 in response to salt, about 2-fold more ABI5 protein compared with the wild-type plants (Figures 6C and 6D). Although, we found that loss of ABD1 affects the transcript levels of ABI5 only upon ABA treatment (Figure 5E), the hyperaccumulation of ABI5 in abd1 by ABA cannot be explained by only transcriptional regulation since the increased fold of ABI5 by loss of ABD1 is much lower than that of ABI5 protein by loss of ABD1. Moreover, previous reports describe that ABI5 is necessary for ABI5 expression and that ABI5 hyperaccumulation results in increased ABI5 transcript abundance due to ABI5 autoregulation (Brocard et al., 2002). This suggests that ABD1 is required for the appropriate degradation of ABI5, likely by acting as a substrate receptor in the CUL4-DDB1–based E3 ubiquitin ligase machinery.
Figure 6.
ABD1 Represses the Accumulation of ABI5 Protein.
(A) Col-0 wild type, abd1-1, and abd1-2 were grown on MS plates in the absence or presence of 0.5 µM ABA for 7 d. Proteins were extracted and ABI5 protein level was determined by immunoblot assay. The ABI5 bands represent the two predominant forms of ABI5.
(B) Quantification of immunoblot assay. Relative amounts of ABI5 protein were normalized to the levels of RPN6 within the same sample.
(C) Col-0 wild type, abd1-1, and abd1-2 were grown on MS plates in the absence or presence of 100 mM NaCl for 7 d. Proteins were extracted and ABI5 protein level was determined by immunoblot assay.
(D) Quantification of immunoblot assay. Relative amounts of ABI5 protein were normalized to the levels of RPN6 within the same sample. Values are means ± sd (n = 3). Significant difference was determined by a Student’s t test; single, double, or triple stars indicate a P value of <0.05, <0.009, and <0.0001, respectively.
ABD1 Interacts With DDB1
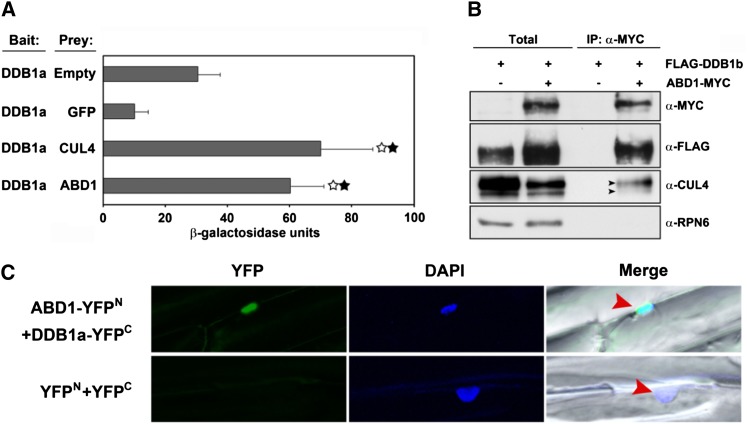
To investigate the possibility that ABD1 is acting as a substrate receptor in the CUL4-DDB1–based E3 ubiquitin ligase machinery, full-length ABD1 and DDB1a were cotransformed into yeast for a two-hybrid assay. As expected, increased β-galactosidase activity was observed, indicating that ABD1 interacts with DDB1a in yeast (Figure 7A). β-Galactosidase activity was at least 2-fold greater than the empty vector control and more than 6-fold greater than the green fluorescent protein (GFP) negative control. The interaction also showed a similar β-galactosidase activity when compared with the positive control, CUL4. To confirm that ABD1 interacts with DDB1 and forms a CUL4 E3 ligase complex in planta, MYC-tagged ABD1 was introduced into FLAG-DDB1b/ddb1a lines. In vivo coimmunoprecipitation (co-IP) assays were then performed to test for interactions between DDB1, CUL4, and ABD1. As shown in Figure 7B, when MYC-tagged proteins were immunoprecipitated from plant extracts using anti-MYC antibody, ABD1-MYC was detected together with FLAG-tagged DDB1b as well as CUL4. Finally, to elucidate the cellular compartment where ABD1 and DDB1a directly interact, we performed bimolecular fluorescence complementation (BiFC). ABD1 and DDB1a interact with each other specifically in the nucleus (Figure 7C). These results demonstrate that ABD1 interacts with DDB1 in Arabidopsis in the nucleus and likely participates as a substrate receptor in the CUL4-based E3 ligase machinery.
Figure 7.
ABD1 Directly Interacts with DDB1.
(A) Interaction between ABD1 and DDB1a by yeast two-hybrid assays. Assays were performed with ABD1 protein as prey and DDB1a as bait for monitoring their interactions. CUL4 was used as positive control, while empty vector and GFP were used as negative controls. β-Galactosidase activities were quantified after growing yeast strains in liquid culture using o-nitrophenyl-β-d-galactopyranoside as a substrate. Values are means ± sd (n = 3). Statistically significant difference was determined by a Student’s t test; white stars indicate a significant difference between the empty vector as prey (P < 0.02), and black stars indicate a significant difference between GFP as prey (P < 0.005).
(B) In vivo co-IP of ABD1 and DDB1b. Transgenic Arabidopsis plants overexpressing FLAG-tagged DDB1b with or without MYC-tagged ABD1 were used to detect the interaction between ABD1 and DDB1b. An α-MYC affinity matrix was used for immunoprecipitation, and α-FLAG and α-MYC antibodies were used for immunoblotting. The immunoblot used RPN6 as an internal control. Total: 5 μg of total proteins from FLAG-DDB1b and FLAG-DDB1b; ABD1-MYC transgenic lines were loaded in each lane and were used as a control for the corresponding co-IP assays.
(C) BiFC assay showing ABD1 directly interacts with DDB1a in the nucleus. Onion (Allium cepa) epidermal cells coexpressing ABD1-YFPN and DDB1a-YFPC fusion proteins through cobombardment. The nucleus, depicted in blue, is stained with 4′,6-diamidino-2-phenylindole (DAPI). The arrows indicate the nucleus in the merged image.
ABD1 Directly Interacts with and Influences the Stability of ABI5
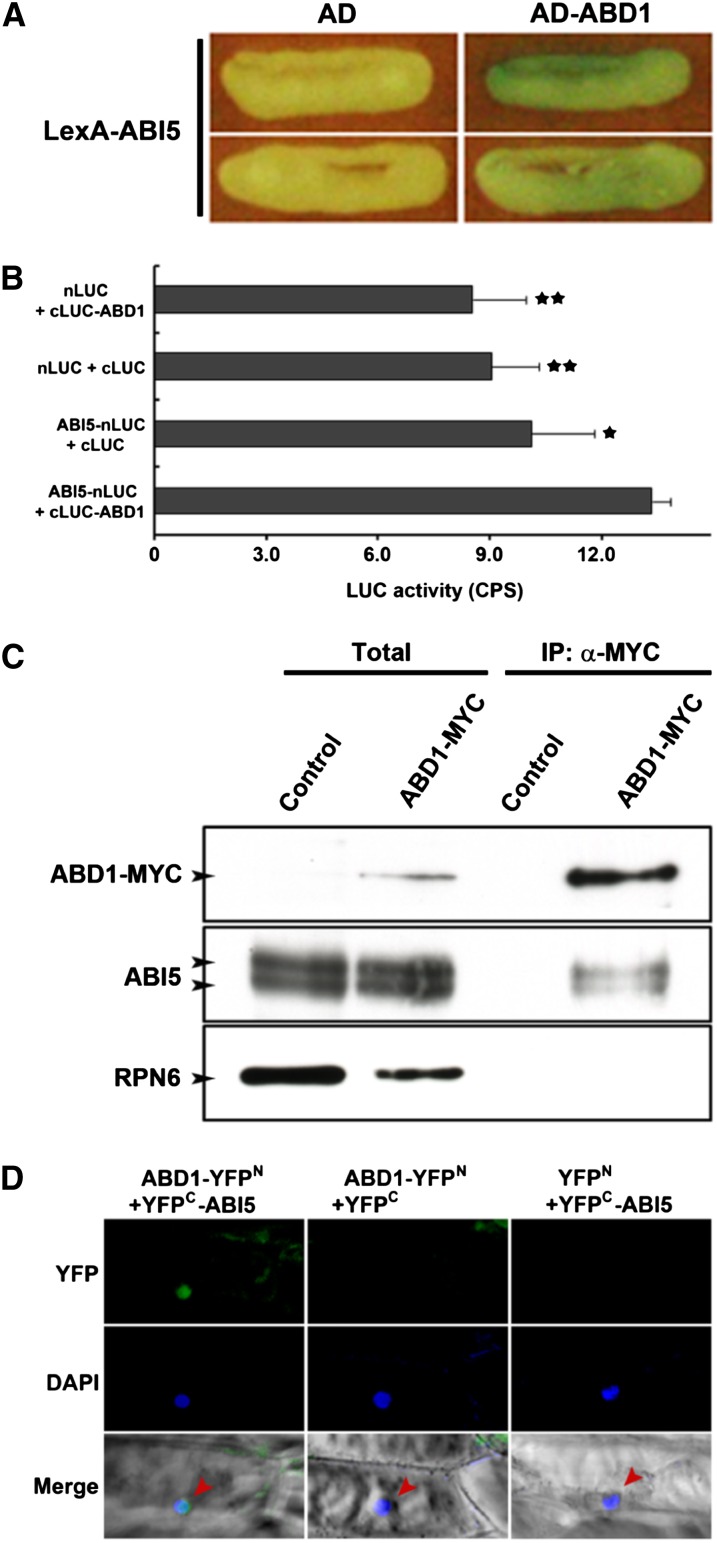
Since loss ABD1 results in hyperaccumulation of ABI5, we checked for a direct interaction between ABD1 and ABI5. In yeast two-hybrid analyses, we found that ABD1 and ABI5 directly interact (Figure 8A). Furthermore, since ABI5 is a member of the bZIP/ABRE family, we tested the five closest related TFs in this family to ABI5 (Kim et al., 2002), which included ABSCISIC ACID RESPONSIVE ELEMENT BINDING FACTOR1 (ABF1), DC3 PROMOTER BINDING FACTOR2 (DPBF2), ABSCISIC ACID-RESPONSIVE ELEMENT BINDING PROTEIN1 (AREB1), AREB2, and AREB3, for an interaction with ABD1. No obvious interaction was observed between ABD1 and ABF1 or DPBF2, the two that display the highest protein homology to ABI5, or any of the other members tested, suggesting that ABD1 may act specifically in the ABI5 pathway (Supplemental Figure 3). Since it was previously reported that two other DCAF proteins, DWA1 and DWA2, directly interact with each other to mediate ABI5 degradation and three DCAF proteins, DWA1, DWA2, and DWA3, display similar ABA hypersensitivity (Lee et al., 2010, 2011), we tested for ABD1’s interaction with DWA1, DWA2, and DWA3. No obvious interaction was observed between ABD1 and the other reported DWA proteins in yeast (Supplemental Figure 3), indicating that ABD1 likely acts apart from the other DWA proteins. Additionally, we checked for a direct interaction between ABI3 and ABD1 since ABI5 and ABI3 appear to act in combination to regulate seed sensitivity to ABA. When we checked the interaction between ABD1 and ABI3 in yeast, there was no binding activity between the two (Supplemental Figure 3).
Figure 8.
ABD1 Directly Interacts with ABI5.
(A) Interaction between ABD1 and ABI5 by yeast two-hybrid assays. Assays were performed with ABD1 protein as prey and ABI5 as bait for monitoring their interactions. Empty vector was used as a negative control. Yeast was grown in the presence of X-Gal for 26 h, after which images were taken.
(B) LCI assays showing that ABD1 and ABI5 interact in N. benthamiana leaf cells. Values are means ± sd (n = 3). Significant difference was determined by a Student’s t test; single or double stars indicate P values of <0.01 or <0.0007, respectively.
(C) In vivo co-IP of ABD1 and ABI5. Transgenic Arabidopsis plants overexpressing MYC-tagged ABD1 were used to detect the interaction between ABD1 and ABI5. An α-MYC affinity matrix was used for immunoprecipitation. α-MYC and α-ABI5 antibodies were used for immunoblotting. The immunoblot used RPN6 as an internal control. Control, Col-0 wild type was used as a negative control; Total, 5 μg of total proteins from Col-0 wild type and the ABD1-MYC transgenic line were loaded in each lane and were used as a control for the corresponding co-IP assays.
(D) BiFC assay showing ABD1 directly interacts with ABI5 in the nucleus. Onion epidermal cells coexpressing ABD1-YFPN and YFPC-ABI5 fusion proteins through cobombardment. The nucleus, depicted in blue, is stained with 4′,6-diamidino-2-phenylindole (DAPI). The arrows indicate the nucleus in the merged image.
To confirm that ABD1 and ABI5 interact in planta, luciferase complementation imaging (LCI) assays (Chen et al., 2008) and in vivo co-IP assays were performed. The LCI assays indicated that cLUC-ABD1 interacted with ABI5-nLUC (Figure 8B). In vivo co-IP assays were performed using MYC-tagged ABD1 introduced into wild-type Arabidopsis plants and showed that ABD1-MYC was coimmunoprecipitated with ABI5 but not with RPN6 in Arabidopsis (Figure 8C). Unlike previous co-IP experiments with DCAF proteins (Lee et al., 2010), ABD1 was able to pull down all of the ABI5 isoforms (Figure 8C). To elucidate the cellular compartment where ABD1 and ABI5 directly interact, we performed BiFC. ABD1 and ABI5 interact with each other specifically in the nucleus (Figure 8D), which further supports the interaction between ABD1 and ABI5 in vivo and indicates that ABI5 is likely the substrate for ABD1-DDB1-CUL4 E3 ligase complexes.
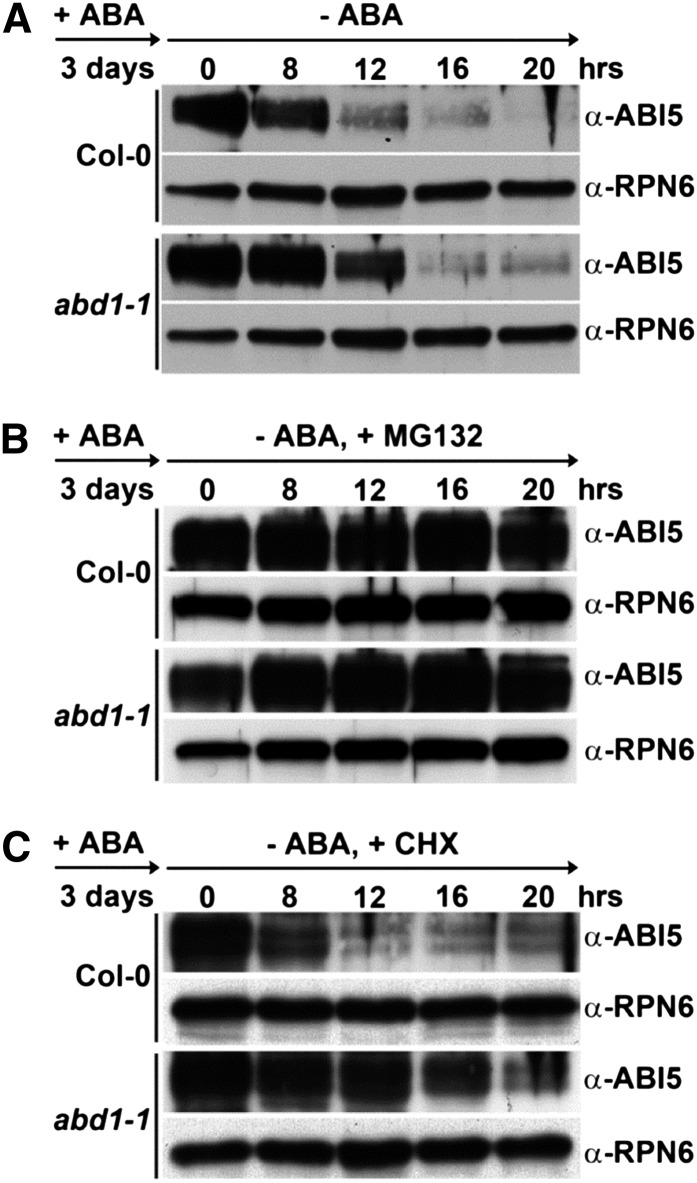
Previous reports have shown that ABI5 is stabilized in the presence of ABA and is then rapidly degraded via the 26S proteasome when the ABA treatment is removed (Lopez-Molina et al., 2001). To determine how ABD1 functions in this process and whether ABD1 perturbs ABI5 degradation, we treated Col-0 wild-type and abd1-1 seeds with 5 μM ABA for 3 d, washed out the ABA, and grew the seeds on MS liquid medium without ABA. We harvested the samples at various incubation time points (0, 8, 12, 16, and 20 h) for protein extraction. We found that after removal of ABA, ABI5 was more abundant in abd1-1 (Figure 9A; Supplemental Figure 4).
Figure 9.
ABI5 Protein Degradation after ABA Removal Requires ABD1.
Immunoblot assays of ABI5 protein in Col-0 wild-type and abd1-1 seeds that were treated with 5 µM ABA in white light for 3 d and then harvested at the indicated times after the removal of ABA by either being washed out with liquid medium (A) or liquid medium supplemented with the proteasome inhibitor MG132 (50 µM) (B) or the protein synthesis inhibitor CHX (100 µM) (C). A total of 10 µg was used in each lane. RPN6 was used as a loading control. Experiments were repeated three times with similar results.
We then treated the samples with the proteasome inhibitor MG132 or the protein synthesis inhibitor cycloheximide (CHX) during the ABA washout and then monitored ABI5 protein levels in both Col-0 wild type and abd1-1. As shown in Figure 9B and Supplemental Figure 4, ABI5 degradation in both Col-0 wild type and abd1-1 was markedly delayed under the presence of MG132, indicating that ABI5 degradation shown in Figure 9A is dependent on the 26S proteasome. To exclude the possibility that the result from Figure 9A was due to increased ABI5 transcripts in abd1-1 compared with Col-0 wild type, CHX treatment was added during the ABA washout. After this treatment, the degradation of ABI5 is significantly delayed compared with the wild type, confirming that the high accumulation of ABI5 in abd1-1 results from the decreased degradation rate of ABI5 due to the loss of ABD1 (Figure 9C; Supplemental Figure 4). Together, these data support that ABD1 is required for posttranscriptional regulation of ABI5 in response to reduced ABA levels and ABD1 is responsible for the degradation of ABI5.
DISCUSSION
For plants, as sessile organisms, the UPS is an essential system that allows the selective removal of short-lived regulatory proteins, which allows plants to rapidly adapt to their environment and to redirect growth and development (Biedermann and Hellmann, 2011). Furthermore, plant hormones such as ABA, auxin, ethylene, gibberellin, brassinosteroids, and jasmonic acid also play a key role in this adaptation. The crosstalk between these two systems is essential since most known target proteins of the UPS are key transcriptional activators or repressors, and modifying their half-lives seems to establish a crucial regulatory point in hormone signaling (Smalle and Vierstra, 2004). Although there have been several recent reports that describe the roles of the UPS in hormonal signal transduction, it is still unclear how many E3 ligases participate in ABA signaling and what their role is in this pathway. Moreover, the function of DCAFs, the substrate receptors for CRL4s, in ABA signal transduction is still largely unknown.
To further understand the relationship between CRL4s and ABA, we report the role of ABD1, a DCAF protein, which plays a negative role regulating ABA signaling and stress responses. ABD1 contains a WDxR motif, which is a truncated DDB1 interacting motif. We identified two T-DNA insertion lines, abd1-1 and abd1-2. Loss of ABD1 results in ABA-associated phenotypes such as ABA and salt hypersensitivity in seed germination (Figure 2) and postgerminative growth (Figure 3B), enhanced stomatal closure (Figures 4A and 4B), and reduced leaf water loss (Figure 4C), which ultimately leads to enhanced drought tolerance (Figures 4D and 4E). Furthermore, ABD1 transcripts are also induced by ABA treatment (Figure 1C). Taken together, these data suggest that ABD1 is a negative regulator of ABA signaling.
Interestingly, abd1-1 and abd1-2 were found to behave differently, which we believe is due to the location of the T-DNA insertion mutation, which results in different phenotypes. The abd1-1 mutation occurs at the 5′ end of ABD1, while the abd1-2 mutation occurs at the 3′ end of ABD1. Although we have shown by RT-PCR that both abd1-1 and abd1-2 fail to produce full-length ABD1 transcript, in Figure 1A, we found that abd1-2 produces a truncated ABD1 transcript. It is likely that abd1-2 produces a partially functional protein, and this results in the altered phenotypes observed in Figures 2 to 4 and 6 and Supplemental Figure 2.
Typically, it is challenging to identify the substrate receptors for E3 ligases, since there are thousands of potential targets. There has been evidence that most known E3 ligase targets are key transcriptional activators or repressors. We set out to identify the target of ABD1 and tested the expression level of ABA-responsive genes RD29A, RD29B, RD22, and RD20 (Figure 5; Supplemental Figure 2). Since loss of ABD1 resulted in misregulation of RD29A and RD29B but not RD22 and RD20, we tested key TFs that are known to regulate ABA-responsive genes RD29A and RD29B but not RD22 or RD20. Although the direct transcriptional regulation of RD20 has not been investigated, it has been suggested MYC2 activates RD20 transcription due the presence of MYC2 cis-regulatory elements in the RD20 promoter and hyperactivation of RD20 in MYC2-overexpressing lines (Abe et al., 2003; Aubert et al., 2010). Furthermore, it was previously reported that RD22 is transactivated by MYC2 (Abe et al., 2003). RD29A and RD29B are both positively regulated by ABI5, indicating that ABI5 may be the target substrate for ABD1. Several lines of evidence also suggest that ABI5 is the target substrate for ABD1. We found that loss of ABD1 is highly associated with hyperaccumulation of ABI5 at the transcript and protein level (Figures 5E and 6) and can specifically bind to ABI5 but not other closely related members of the bZIP transcription family, such as ABF1 and DPBF2 (Figure 8; Supplemental Figure 3). We found that ABD1 directly binds to all isoforms of ABI5, and this interaction occurs in the nucleus (Figure 8). Furthermore, we found that ABI5 degradation is dependent on the 26S proteasome, the increased stability of ABI5 observed in abd1-1 results from the decreased degradation rate of ABI5 in abd1-1, and ABD1 is responsible for the degradation of ABI5 (Figure 9). These observations suggest that the ABA hypersensitivity of ABD1 loss-of-function mutants is due to the reduced proteolysis of ABI5 caused by the hyperaccumulation of ABI5, resulting in upregulation of downstream ABA-responsive gene expression.
We cannot exclude the possibility that a portion of ABI5 protein overaccumulation observed in abd1 mutants upon ABA treatment was due to ABI5 autoregulation (Brocard et al., 2002). Alternatively, the increase/decrease of other positive/negative regulators in ABA signaling, which are located upstream of ABI5, caused from the loss of ABD1 might lead to the accumulation of ABI5. We tested the transcript levels of ABI3, a TF known to trigger and act upstream of ABI5 after ABA treatment (Lopez-Molina et al., 2002). In Supplemental Figure 2E, we did not find any significant change in the transcript levels between Col-0 wild type and abd1 before and after the treatment with ABA, indicating that ABI5 overexpression in abd1 is not a result of ABI3 overexpression due to loss of ABD1. Additionally, when we checked the interaction between ABD1 and ABI3 in yeast (Supplemental Figure 3C), there was no binding activity between the two, which implies that ABD1 does not seem to directly modulate the stability of ABI3 protein.
Loss of ABD1 also results in drought tolerance in vegetative tissue, and this observation is similar to the expression pattern of ABI5 in vegetative tissue. ABI5 expression is not seed specific, and ABI5 function is required for the full induction of some LATE EMBRYOGENESIS ABUNDANT genes expressed at low levels in ABA-treated vegetative tissue. Furthermore, the ABA hypersensitivity caused by the ectopic expression of ABI3, a TF that acts upstream of ABI5, in vegetative tissues appears to be partially facilitated by the hyperinduction of ABI5 expression (Finkelstein and Lynch, 2000; Lopez-Molina et al., 2002). Moreover, histochemical studies of the ABI5 promoter activity revealed that ABI5 is expressed in vegetative tissues (Brocard et al., 2002). In addition, ABI5 is regulated by sugar and stress in 6-d-old seedlings, displaying that the ABI5 protein induction in response to ABA is not restricted to a narrow developmental window and that ABI5 also plays a role in regulating stress response (Brocard et al., 2002; Arroyo et al., 2003). In support of these studies, we also found that loss of ABD1 results in hyperaccumulation of ABI5 protein in 7-d-old seedlings in response to NaCl stress (Figures 6C and 6D). Although we did not monitor ABI5 levels under drought stress in vegetative tissues, it is possible that ABD1 could affect ABI5 levels under these conditions as well, since loss of ABD1 results in drought tolerance. Understanding how ABI5 levels are affected in this condition could further help elucidate to the role of ABD1 in vegetative tissues.
Unlike other DCAF proteins described to date, ABD1 appears to act in a pathway apart from the MYC2 ABA-responsive pathway. DWA1, DWA2, and DWA3 all show overlapping functions with other ABA-responsive pathways since loss of any of the DWA proteins causes an overexpression of RD22 and hyperaccumulation of MYC2 after ABA treatment (Lee et al., 2010, 2011). It has also been speculated that DWA1 and DWA2 are involved in the degradation of other positive regulators in ABA signaling pathway, such as other AREB/ABF/DPBF bZIP proteins (Lee et al., 2010). The incomplete match of DWA1/DWA2 and ABI5 expression profiles suggests that ABI5 degradation in vivo may require other pathways as well. There are several examples of differential utilization of E3 ligases to modify one substrate depending on the physiological context (Mazzucotelli et al., 2006). In support of this, ABD1 does not directly interact with DWA1, DWA2, or DWA3 and has other characteristic differences (Figures 3B, 5E, and 8C; Supplemental Figure 2A), suggesting that ABD1 acts in a pathway apart from the DWA proteins.
In addition to DWA1 and DWA2, KEG was also reported as a negative regulator of ABI5 and is a single-subunit E3 ligase (Stone et al., 2006; Liu and Stone, 2010). KEG is a multidomain E3 ligase that consists of a RING E3 ligase and kinase domain with a series of ankyrin and HERC2-like repeats. Loss of KEG results in hypersensitivity to ABA, hyperaccumulation of ABI5, and growth arrest immediately after germination, which ultimately results in seedling lethality (Stone et al., 2006). Moreover, it was shown that KEG ubiquitinates ABI5 in vitro and regulates its stability. However, the introduction of the abi5-1 mutation into keg seedlings resulted in only a partial rescue of the early grown arrest phenotype by keg-1, suggesting that KEG has other functions. Furthermore, both the loss of function and the overexpression of ABI5 resulted in phenotypes that are relatively mild, contrasting with the severe phenotype of keg-1 (Lopez-Molina et al., 2001, 2003; Brocard et al., 2002). Recently, the subcellular localization of KEG was described. KEG is localized to the trans-Golgi network/early endosome and does not appear to be localized to the nucleus in either Nicotiana benthamiana or Arabidopsis protoplasts or stabile transgenic Arabidopsis lines (Gu and Innes, 2011, 2012). KEG appears to be a key component in regulating multiple post-Golgi trafficking events in plants, including vacuole biogenesis, targeting of membrane-associated proteins to the vacuole, and secretion of apoplastic proteins (Gu and Innes, 2012). This is a surprising finding since ABI5 is constitutively localized to the nucleus (Lopez-Molina et al., 2002, 2003). A recent study suggests that ABI5 is continually shuttled between the nucleus and the cytoplasm and that ABI5 and KEG directly interact in the cytoplasm. The authors hypothesize that the KEG-dependent turnover of ABI5 within the cytoplasm occurs in the absence of ABA. In response to ABA, KEG self-ubiquitinated and is degraded, while ABI5 is activated by phosphorylation. When ABI5 is no longer required, ABI5 is then degraded by other E3s (Liu and Stone, 2013). It is likely that ABD1, DWA1, DWA2, and KEG work in concert to modulate ABI5 stability.
Interestingly, ABD1 interacts strongly with the two predominate isoforms of ABI5 in in vivo co-IP experiments. Moreover, previous in vivo co-IP experiments reveal that CUL4 is also able to interact strongly with the two predominate isoforms of ABI5 (Lee et al., 2010). By contrast, DWA1 and DWA2 only interact with the slower migrating form of ABI5 in vivo (Lee et al., 2010). It is unclear what these isoforms represent, since ABI5 undergoes multiple posttranslational modifications and to date the nature of this banding pattern in immunoblot analyses is uncertain (Lopez-Molina et al., 2001, 2002; Stone et al., 2006; Miura et al., 2009). It was previously reported that the banding pattern of ABI5 represented different phosphorylation statuses, but there are conflicting reports stating that treatment with protein phosphatase modifies the mobility of ABI5 (Lopez-Molina et al., 2001; Stone et al., 2006; Piskurewicz et al., 2008). The multiple isoforms of ABI5 are only observed during the expression of native ABI5, while overexpression lines with HA-tagged ABI5 only display one isoform whose size can be modified by phosphorylation/dephosphorylation (Piskurewicz et al., 2008). Furthermore, immunoblot analyses of FyPP1 and FyPP2, two type 2C protein phosphatases that play a key role in promoting dephosphorylation and subsequent degradation of ABI5, reveal no difference in ABI5 banding patterns in immunoblot assays in either overexpression or loss-of-function lines of FyPP1 and FyPP2 (Dai et al., 2013). Why plants have evolved two sets of E3 ligases to target different forms of ABI5 is also unclear. Based on the binding difference of DWA1/2 and ABD1 with ABI5 (Lee et al., 2010; Figures 8A and 8C), it seems that ABD1 influences the stability of all ABI5 isoforms, while DWA1/2 only affect the slow migrating form of ABI5.
Overall, our work raises several questions for future investigation. Several regulators, such as KEG, DWA1, DWA2, and ABD1, are responsible for the regulation of ABI5 stability. Further examination of the co-IP pattern of ABI5 with CUL4 in specific genetic backgrounds, such as dwa1, dwa2, abd1, and keg, could help elucidate a detailed mode of action of each regulator in ABI5-mediated ABA signaling. Furthermore, how many E3 ligases influence the stability of ABI5? Further investigation of these E3 ligases will likely provide novel insights into how CRL4’s negatively regulate the ABI5 signaling pathway. In summary, ABD1 is involved in regulating ABA and stress responses during multiple stages of plant growth and development. In our study, we propose that when ABI5 is no longer needed or its levels need to be reduced, such as after stress or ABA removal, ABI5, including phosphorylated ABI5, is recognized by the ABD1-DDB1-CUL4 E3 ligase in the nucleus, which targets ABI5 for degradation. This event indirectly causes reduced transcription of ABI5 due to the lower amounts of ABI5 available to activate ABI5 transcription.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Col-0 was used in this study. The abd1-1 (SALK_051074) and abd1-2 (SAIL_648_G02) T-DNA insertion mutants were obtained from the ABRC (http://www.arabidopsis.org/). Oligonucleotide sequences of the primer pairs used for genotyping analysis of T-DNA insertion mutants are shown in Supplemental Table 1. FLAG-DDB1b transgenic lines were the same ones as those from Lee et al. (2008). Arabidopsis seedlings were grown as described previously (Lee et al., 2008, 2010). For the assays shown in Figures 3A and 3B, all seedlings were germinated and grown vertically on 1× MS plates with or without 0.5 μM ABA (Sigma-Aldrich) or 100 mM NaCl for 7 d after stratification under continuous white light in a controlled-environment chamber at 22°C or grown for 3 d under continuous white light and then the seedlings were transferred onto fresh MS plates with or without 5 μM ABA and grown for an additional 5 d under continuous white light. For adult plants, 2-week-old seedlings grown on MS plates were transferred to soil and kept under long-day conditions (16 h light/8 h dark) in a controlled environment chamber at 22°C. For the assays shown in Figure 9, Col-0 wild-type and abd1-1 seeds were grown on MS plates with 5 µM ABA in white light for 3 d. Seeds were washed two times with liquid MS medium and transferred to either liquid MS medium, liquid MS medium supplemented with 50 µM MG132 (Sigma-Aldrich), or liquid MS medium supplemented with 100 µM CHX (Sigma-Aldrich) and harvested at the indicated times after the removal of ABA. To minimize the effect of harvesting time on seed germination, all seed batches compared in this study were harvested on the same day from plants grown in the same growth chamber with identical environmental conditions.
Generation of FLAG-DDB1b;ABD1-MYC Transgenic Arabidopsis Plants
ABD1 cDNA was amplified using cDNA synthesized from total mRNA from Arabidopsis leaves and was cloned into the SpeI site of the binary vector, pMYC-fused pJIM19 (gentamycin) (Lee et al., 2010). The resulting ABD1-MYC construct was introduced into FLAG-DDB1b;ddb1a plants by Agrobacterium tumefaciens strain GV3101–mediated transformation using the floral dip method (Clough and Bent, 1998).
Stomatal Aperture Analysis
Stomatal bioassay experiments were performed as previously described (Li et al., 2011). To study the promotion of stomatal closure by ABA, leaves from 4-week-old plants grown in the same conditions described above were harvested in darkness at the end of the night. Paradermal sections of abaxial epidermis obtained were incubated in stomatal opening solution containing 50 mM KCl, 10 mM CaCl2, and 10 mM MES-KOH, pH 6.15, at 22 to 25°C and blocked from light for 2.5 h. Subsequently, 50 μM ABA was added to the solution, and the samples were exposed to light for 2.5 h. After treatment, stomatal apertures (the length and width of the stomata) were measured with a LSM 510 Meta confocal laser scanning microscope (Carl Zeiss). The apertures of 30 stomata were measured in three independent experiments according to Roelfsema and Prins (1995).
Drought Stress
Seeds were germinated and grown on MS plates for 7 d under continuous white light in a controlled-environment growth chamber. The seedlings were then transferred into soil and grown in long-day (16 h light/8 h dark) conditions in a growth chamber side by side for 3 weeks with identical environmental conditions. Drought stress was initiated by withholding watering for 12 d, and then the plants were tested for survival 1 d after rewatering. All experiments were performed in triplicate and representative results are shown.
Determination of Water Loss
Analysis of water loss was performed as previously described (Cheong et al., 2007). Briefly, three rosette leaves per plant were detached from 3-week-old wild-type and abd1 mutant plants. The leaves were kept on the laboratory bench for a total of 7 h. After every hour, their fresh weights were measured. Water loss represents the percentage of weight loss at the indicated time versus initial fresh weight. To minimize variation, three independent experiments were performed, and similar results were obtained.
RNA Isolation, RT-PCR, and Quantitative PCR Analysis
Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen) and reverse transcribed via the SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. RT-PCR was performed as described by Lee et al. (2008). Quantitative PCR was performed using 50 ng of cDNA in each reaction with the SYBR Green PCR Master Mix (Applied Biosystems) according to the manufacturer’s instructions using the CFX96 real-time PCR detection system (Bio-Rad). Relative amounts of transcripts were calculated using the comparative cycle threshold method, which was normalized to ACTIN2 expression from the same sample. All quantitative PCR experiments were independently performed in triplicate. The primers for RT-PCR and quantitative PCR are shown in Supplemental Tables 1 and 2, respectively.
Protein Isolation and Immunoblot Analysis
Proteins were isolated with the extraction buffer containing 50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 1 mM PMSF, 1× Complete protease inhibitor cocktail (Roche), 5% glycerol, 1 mM EDTA, and 1 mM DTT. Yeast proteins were extracted using the Y-PER yeast protein extraction reagent (Thermo Scientific) according to the manufacturer’s instructions with 1× Complete protease inhibitor cocktail (Roche). Protein concentration was determined by the Bradford assay following the manufacturer’s instructions (Bio-Rad). The samples were mixed with 2× SDS sample buffer and boiled for 3 min and then separated on a 10% SDS protein gel. The membrane transfer and protein gel blot assays were performed as described previously (Lee et al., 2010). Subsequent immunoblot assays were performed with anti-ABI5 (Abcam) and anti-RPN6 (Chen et al., 2006) antibodies at dilutions of 1:4000 and 1:500, respectively. For Supplemental Figure 3, anti-LexA (Sigma-Aldrich) antibodies were used at a 1:3000 dilution.
In Vivo Pull-Down Assay
Arabidopsis tissue was homogenized in 50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 1 mM PMSF, 1× Complete protease inhibitor cocktail (Roche), 5% glycerol, 1 mM EDTA, and 1 mM DTT. The extracts were centrifuged twice at 13,000g for 10 min, and the protein concentration in the supernatant was determined by Bradford assay (Bio-Rad). For co-IP, 2 mg of total protein was incubated for 4 h at 4°C with 30 μL of anti-MYC monoclonal antibodies immobilized on Sepharose Fast Flow beads (Covance). The precipitated samples were washed at least four times with the protein extraction buffer and then eluted by the addition of 2× SDS protein loading buffer with boiling for 5 min. Subsequent immunoblot assays were performed with anti-FLAG (Sigma-Aldrich), anti-MYC (Cell Signaling), anti-CUL4 (Chen et al., 2006), anti-RPN6 (Chen et al., 2006), or anti-ABI5 (Abcam) antibodies at dilutions of 1:2000, 1:1000, 1:1000, 1:500, and 1:4000, respectively.
LCI
For LCI assays, full-length coding DNA sequence (CDS) of ABI5 and ABD1 were amplified by PCR (Supplemental Table 3) and inserted into the KpnI sites of pCAMBIA133-cLUC and nLUC vectors (Chen et al., 2008). The LCI assays were performed as previously described (Chen et al., 2008). Briefly, various nLUC and cLUC fusion vectors were transformed into the Agrobacterium strain GV2260. Young Nicotiana benthamiana leaves were infiltrated with equal amounts of the appropriate bacterial strains, mixed pairwise. After infiltration, the plants were incubated in constant light at room temperature for 3 d before analysis. The luciferase activity was then determined using a Xenogen IVIS Spectrum imaging system and quantified with Living Image software (Caliper).
Yeast Two-Hybrid Assay
Full-length CDS of ABD1 was cloned into the EcoRI site of pB42AD vector (Clontech). Full-length CDS of ABI5, DWA1, and DWA3 were cloned into the EcoRI site of pLexA vector (Clontech), CDS of AREB1 and AREB2 were cloned into EcoRI-NotI sites of pLexA vector, CDS of ABF1, DPBF2, AREB3, and ABI3 were cloned into BamHI and NotI sites of pLexA vector, and the CDS of DWA2 was cloned into the XhoI site of pLexA vector (Supplemental Table 4). pLexA-DDB1a, pB42AD-CUL4, and pB42AD-GFP were previously described (Lee et al., 2008). Yeast two-hybrid assay was performed according to the Matchmaker LexA Two-Hybrid System manual (Clontech). Briefly, all constructs were cotransformed into yeast strain EGY48 containing p80p-LacZ. Transformants were selected by growth on SD/-His/-Trp/-Ura plates prior to X-Gal selection. An interaction between two proteins was quantified by measuring β-galactosidase activity in liquid culture using ο-nitrophenyl-β-d-galactopyranoside (Sigma-Aldrich) as a substrate.
BiFC
Full-length CDS of ABD1, ABI5, and DDB1a were amplified by PCR (Supplemental Table 5) and subsequently cloned into the NotI site of pSY728 vector, the BamHI site of pSY735 vector, and the NotI site of pSY738 vector, respectively (Bracha-Drori et al., 2004). Pairwise combinations as indicated in Figures 7C and 8D were cobombarded into onion (Allium cepa) epidermal cells as previously described (Shen et al., 2009). Yellow fluorescent protein fluorescence was visualized by a LSM 510 Meta confocal laser scanning microscope (Carl Zeiss).
Protein Sequence Alignment
ABD1 homologs were identified from GenBank using the protein basic local alignment search tool (BLASTp) (http://www.ncbi.nlm.nih.gov/BLAST/). Sequence alignment was performed by the Clustal Omega method. Alignment shading was performed using GeneDoc 3.2.0 software (http://www.nrbsc.org/gfx/genedoc/index.html).
Accession Numbers
Sequence data from this study can be found in the Arabidopsis Genome Initiative database under the following accession numbers: At4g38480 (ABD1), At2g19430 (DWA1), At1g76260 (DWA2), At1g61210 (DWA3), At5g46210 (CUL4), At4g05420 (DDB1a), At4g21100 (DDB1b), At2g36270 (ABI5), At1g49720 (ABF1), At3g44460 (DPBF2), At1g45249 (AREB1), At3g19290 (AREB2), At3g56850 (AREB3), At3g24650 (ABI3), At5g52310 (RD29A), At5g52300 (RD29B), At5g25610 (RD22), At2g33380 (RD20), At3g18780 (ACTIN2), and At1g29150 (RPN6). Sequence data for the ABD1 homologs used in the phylogenetic analysis were obtained from the GenBank database with the following accession numbers: NP_056541 (human), NP_705783 (mouse), and NP_001065201 (rice).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Protein Alignment of ABD1 from Other Model Organisms.
Supplemental Figure 2. abd1-1 and abd1-2 Seedlings Have No Change in Expression of RD20 and RD22 after ABA and NaCl Treatment.
Supplemental Figure 3. ABD1 Does Not Directly Interact with DWA Proteins, ABI5 Homologs, or ABI3 by Yeast Two-Hybrid Assays.
Supplemental Figure 4. Quantification of ABI5 Protein Degradation after ABA Removal.
Supplemental Table 1. List of Primers Used for Genotyping and RT-PCR Analysis.
Supplemental Table 2. List of Primers Used in Quantitative Real-Time PCR Analysis.
Supplemental Table 3. List of Primers Used for LCI Analysis.
Supplemental Table 4. List of Primers Used for Yeast Two-Hybrid Analysis.
Supplemental Table 5. List of Primers Used for BiFC Assay.
Supplementary Material
Acknowledgments
We thank the Ohio State University Arabidopsis Biological Resource for providing seed stocks. This work was supported by a National Institutes of Health grant (GM47850) and a National Science Foundation 2010 Program grant (MCB0929100) to X.W.D. and by a grant from the Next-Generation BioGreen 21 Program (PJ00901002) and the National Academy of Agricultural Science agenda program (PJ00859802), Rural Development Administration, Republic of Korea to M.-O.B. This research was also supported by Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education, Science, and Technology (NRF-2012R1A1A1001564) and by iPET (Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry, and Fisheries), Ministry of Agriculture, Food, and Rural Affairs (112030-1; to J.-H.L.). C.D.N. is a Yale University Brown Postdoctoral Fellow.
AUTHOR CONTRIBUTIONS
K.-I.S. mainly performed the experiments for this research. J.-H.L. and X.W.D. designed the research. J.-H.L. carried out some experiments related to ABA sensitivity and germination rate. C.D.N., S.Z., and M.-O.B. carried out some experiments and expression analysis. E.S. investigated ABA inducibility of the ABD1 gene. K.-I.S., J.-H.L., and C.D.N. analyzed the data. C.D.N. and X.W.D. wrote the article.
Glossary
- ABA
abscisic acid
- TF
transcription factor
- ABRE
ABA response element
- UPS
ubiquitin proteasome system
- Col-0
Columbia-0
- MS
Murashige and Skoog
- co-IP
coimmunoprecipitation
- BiFC
bimolecular fluorescence complementation
- LCI
luciferase complementation imaging
- CHX
cycloheximide
- CDS
coding DNA sequence
Footnotes
Online version contains Web-only data.
References
- Abe H., Urao T., Ito T., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal P.K., Agarwal P., Reddy M.K., Sopory S.K. (2006). Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 25: 1263–1274. [DOI] [PubMed] [Google Scholar]
- Angers S., Li T., Yi X., MacCoss M.J., Moon R.T., Zheng N. (2006). Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 443: 590–593. [DOI] [PubMed] [Google Scholar]
- Arroyo A., Bossi F., Finkelstein R.R., Leon P. (2003). Three genes that affect sugar sensing (abscisic acid insensitive 4, abscisic acid insensitive 5, and constitutive triple response 1) are differentially regulated by glucose in Arabidopsis. Plant Physiol. 133: 231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert Y., Vile D., Pervent M., Aldon D., Ranty B., Simonneau T., Vavasseur A., Galaud J.P. (2010). RD20, a stress-inducible caleosin, participates in stomatal control, transpiration and drought tolerance in Arabidopsis thaliana. Plant Cell Physiol. 51: 1975–1987. [DOI] [PubMed] [Google Scholar]
- Bernhardt A., Mooney S., Hellmann H. (2010). Arabidopsis DDB1a and DDB1b are critical for embryo development. Planta 232: 555–566. [DOI] [PubMed] [Google Scholar]
- Biedermann S., Hellmann H. (2010). The DDB1a interacting proteins ATCSA-1 and DDB2 are critical factors for UV-B tolerance and genomic integrity in Arabidopsis thaliana. Plant J. 62: 404–415. [DOI] [PubMed] [Google Scholar]
- Biedermann S., Hellmann H. (2011). WD40 and CUL4-based E3 ligases: Lubricating all aspects of life. Trends Plant Sci. 16: 38–46. [DOI] [PubMed] [Google Scholar]
- Bracha-Drori K., Shichrur K., Katz A., Oliva M., Angelovici R., Yalovsky S., Ohad N. (2004). Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J. 40: 419–427. [DOI] [PubMed] [Google Scholar]
- Brocard I.M., Lynch T.J., Finkelstein R.R. (2002). Regulation and role of the Arabidopsis abscisic acid-insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiol. 129: 1533–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Huang X., Gusmaroli G., Terzaghi W., Lau O.S., Yanagawa Y., Zhang Y., Li J., Lee J.H., Zhu D., Deng X.W. (2010). Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell 22: 108–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Shen Y., Tang X., Yu L., Wang J., Guo L., Zhang Y., Zhang H., Feng S., Strickland E., Zheng N., Deng X.W. (2006). Arabidopsis CULLIN4 forms an E3 ubiquitin ligase with RBX1 and the CDD complex in mediating light control of development. Plant Cell 18: 1991–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.M. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong Y.H., Pandey G.K., Grant J.J., Batistic O., Li L., Kim B.G., Lee S.C., Kudla J., Luan S. (2007). Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J. 52: 223–239. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cutler S.R., Rodriguez P.L., Finkelstein R.R., Abrams S.R. (2010). Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 61: 651–679. [DOI] [PubMed] [Google Scholar]
- Dai M., Xue Q., McCray T., Margavage K., Chen F., Lee J.H., Nezames C.D., Guo L., Terzaghi W., Wan J., Deng X.W., Wang H. (2013). The PP6 phosphatase regulates ABI5 phosphorylation and abscisic acid signaling in Arabidopsis. Plant Cell 24: 517–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher K., Callis J. (2007). Ubiquitin, hormones and biotic stress in plants. Ann. Bot. (Lond.) 99: 787–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumbliauskas E., Lechner E., Jaciubek M., Berr A., Pazhouhandeh M., Alioua M., Cognat V., Brukhin V., Koncz C., Grossniklaus U., Molinier J., Genschik P. (2011). The Arabidopsis CUL4-DDB1 complex interacts with MSI1 and is required to maintain MEDEA parental imprinting. EMBO J. 30: 731–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R.R., Lynch T.J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M.E., Lynch T., Peeters J., Snowden C., Finkelstein R. (2008). A small plant-specific protein family of ABI five binding proteins (AFPs) regulates stress response in germinating Arabidopsis seeds and seedlings. Plant Mol. Biol. 67: 643–658. [DOI] [PubMed] [Google Scholar]
- Goda H., et al. (2008). The AtGenExpress hormone and chemical treatment data set: Experimental design, data evaluation, model data analysis and data access. Plant J. 55: 526–542. [DOI] [PubMed] [Google Scholar]
- Groisman R., Polanowska J., Kuraoka I., Sawada J., Saijo M., Drapkin R., Kisselev A.F., Tanaka K., Nakatani Y. (2003). The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113: 357–367. [DOI] [PubMed] [Google Scholar]
- Gruber H., Heijde M., Heller W., Albert A., Seidlitz H.K., Ulm R. (2010). Negative feedback regulation of UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. Proc. Natl. Acad. Sci. USA 107: 20132–20137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Innes R.W. (2011). The KEEP ON GOING protein of Arabidopsis recruits the ENHANCED DISEASE RESISTANCE1 protein to trans-Golgi network/early endosome vesicles. Plant Physiol. 155: 1827–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Innes R.W. (2012). The KEEP ON GOING protein of Arabidopsis regulates intracellular protein trafficking and is degraded during fungal infection. Plant Cell 24: 4717–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y.J., McCall C.M., Hu J., Zeng Y., Xiong Y. (2006). DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes Dev. 20: 2949–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa L.A., Wu M., Ye T., Kobayashi R., Sun H., Zhang H. (2006). CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat. Cell Biol. 8: 1277–1283. [DOI] [PubMed] [Google Scholar]
- Hotton S.K., Callis J. (2008). Regulation of cullin RING ligases. Annu. Rev. Plant Biol. 59: 467–489. [DOI] [PubMed] [Google Scholar]
- Hua Z., Vierstra R.D. (2011). The cullin-RING ubiquitin-protein ligases. Annu. Rev. Plant Biol. 62: 299–334. [DOI] [PubMed] [Google Scholar]
- Huang X., Ouyang X., Yang P., Lau O.S., Chen L., Wei N., Deng X.W. (2013). Conversion from CUL4-based COP1-SPA E3 apparatus to UVR8-COP1-SPA complexes underlies a distinct biochemical function of COP1 under UV-B. Proc. Natl. Acad. Sci. USA 110: 16669–16674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Arias E.E., Chen J., Harper J.W., Walter J.C. (2006). A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell 23: 709–721. [DOI] [PubMed] [Google Scholar]
- Keeney S., Chang G.J., Linn S. (1993). Characterization of a human DNA damage binding protein implicated in xeroderma pigmentosum E. J. Biol. Chem. 268: 21293–21300. [PubMed] [Google Scholar]
- Kilian J., Whitehead D., Horak J., Wanke D., Weinl S., Batistic O., D'Angelo C., Bornberg-Bauer E., Kudla J., Harter K. (2007). The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50: 347–363. [DOI] [PubMed] [Google Scholar]
- Kim S.Y., Ma J., Perret P., Li Z., Thomas T.L. (2002). Arabidopsis ABI5 subfamily members have distinct DNA-binding and transcriptional activities. Plant Physiol. 130: 688–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Terzaghi W., Deng X.W. (2011). DWA3, an Arabidopsis DWD protein, acts as a negative regulator in ABA signal transduction. Plant Sci. 180: 352–357. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Terzaghi W., Gusmaroli G., Charron J.B., Yoon H.J., Chen H., He Y.J., Xiong Y., Deng X.W. (2008). Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. Plant Cell 20: 152–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Yoon H.J., Terzaghi W., Martinez C., Dai M., Li J., Byun M.O., Deng X.W. (2010). DWA1 and DWA2, two Arabidopsis DWD protein components of CUL4-based E3 ligases, act together as negative regulators in ABA signal transduction. Plant Cell 22: 1716–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Jiang H., Bu Q., Zhao Q., Sun J., Xie Q., Li C. (2011). The Arabidopsis RING finger E3 ligase RHA2b acts additively with RHA2a in regulating abscisic acid signaling and drought response. Plant Physiol. 156: 550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Chen X., Garbutt K.C., Zhou P., Zheng N. (2006). Structure of DDB1 in complex with a paramyxovirus V protein: Viral hijack of a propeller cluster in ubiquitin ligase. Cell 124: 105–117. [DOI] [PubMed] [Google Scholar]
- Liu H., Stone S.L. (2010). Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 22: 2630–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Stone S.L. (2013). Cytoplasmic degradation of the Arabidopsis transcription factor ABSCISIC ACID INSENSITIVE 5 is mediated by the RING-type E3 ligase KEEP ON GOING. J. Biol. Chem. 288: 20267–20279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., Chua N.H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 98: 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., Kinoshita N., Chua N.H. (2003). AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev. 17: 410–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., McLachlin D.T., Chait B.T., Chua N.H. (2002). ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 32: 317–328. [DOI] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., Grill E. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068. [DOI] [PubMed] [Google Scholar]
- Mazzucotelli E., Belloni S., Marone D., De Leonardis A., Guerra D., Di Fonzo N., Cattivelli L., Mastrangelo A. (2006). The e3 ubiquitin ligase gene family in plants: Regulation by degradation. Curr. Genomics 7: 509–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Lee J., Jin J.B., Yoo C.Y., Miura T., Hasegawa P.M. (2009). Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc. Natl. Acad. Sci. USA 106: 5418–5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T., Yamashino T. (2008). Comparative transcriptome of diurnally oscillating genes and hormone-responsive genes in Arabidopsis thaliana: Insight into circadian clock-controlled daily responses to common ambient stresses in plants. Plant Cell Physiol. 49: 481–487. [DOI] [PubMed] [Google Scholar]
- Nakashima K., Fujita Y., Katsura K., Maruyama K., Narusaka Y., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2006). Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol. Biol. 60: 51–68. [DOI] [PubMed] [Google Scholar]
- Nemhauser J.L., Hong F., Chory J. (2006). Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475. [DOI] [PubMed] [Google Scholar]
- Nezames C.D., Sjogren C.A., Barajas J.F., Larsen P.B. (2012). The Arabidopsis cell cycle checkpoint regulators TANMEI/ALT2 and ATR mediate the active process of aluminum-dependent root growth inhibition. Plant Cell 24: 608–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N., Sarkeshik A., Nito K., Park S.Y., Wang A., Carvalho P.C., Lee S., Caddell D.F., Cutler S.R., Chory J., Yates J.R., Schroeder J.I. (2010). PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. 61: 290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazhouhandeh M., Molinier J., Berr A., Genschik P. (2011). MSI4/FVE interacts with CUL4-DDB1 and a PRC2-like complex to control epigenetic regulation of flowering time in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 3430–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintard L., Willems A., Peter M. (2004). Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family. EMBO J. 23: 1681–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U., Jikumaru Y., Kinoshita N., Nambara E., Kamiya Y., Lopez-Molina L. (2008). The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 20: 2729–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelfsema M.R.G., Prins H.B.A. (1995). Effect of abscisic acid on stomatal opening in isolated epidermal strips of abi mutants of Arabidopsis thaliana. Physiol. Plant. 95: 373–378. [Google Scholar]
- Santiago J., Rodrigues A., Saez A., Rubio S., Antoni R., Dupeux F., Park S.Y., Marquez J.A., Cutler S.R., Rodriguez P.L. (2009). Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 60: 575–588. [DOI] [PubMed] [Google Scholar]
- Schroeder D.F., Gahrtz M., Maxwell B.B., Cook R.K., Kan J.M., Alonso J.M., Ecker J.R., Chory J. (2002). De-etiolated 1 and damaged DNA binding protein 1 interact to regulate Arabidopsis photomorphogenesis. Curr. Biol. 12: 1462–1472. [DOI] [PubMed] [Google Scholar]
- Scrima A., Konickova R., Czyzewski B.K., Kawasaki Y., Jeffrey P.D., Groisman R., Nakatani Y., Iwai S., Pavletich N.P., Thoma N.H. (2008). Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell 135: 1213–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Zhou Z., Feng S., Li J., Tan-Wilson A., Qu L.J., Wang H., Deng X.W. (2009). Phytochrome A mediates rapid red light-induced phosphorylation of Arabidopsis FAR-RED ELONGATED HYPOCOTYL1 in a low fluence response. Plant Cell 21: 494–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J., Vierstra R.D. (2004). The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55: 555–590. [DOI] [PubMed] [Google Scholar]
- Stone S.L., Williams L.A., Farmer L.M., Vierstra R.D., Callis J. (2006). KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18: 3415–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno Y., Furihata T., Abe H., Yoshida R., Shinozaki K., Yamaguchi-Shinozaki K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 97: 11632–11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Ransom C., Ludwig P., van Nocker S. (2003). Genetic analysis of early flowering mutants in Arabidopsis defines a class of pleiotropic developmental regulator required for expression of the flowering-time switch flowering locus C. Genetics 164: 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Feng S., Chen F., Chen H., Wang J., McCall C., Xiong Y., Deng X.W. (2008). Arabidopsis DDB1-CUL4 ASSOCIATED FACTOR1 forms a nuclear E3 ubiquitin ligase with DDB1 and CUL4 that is involved in multiple plant developmental processes. Plant Cell 20: 1437–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W., Feng H., Santiago F.E., Kipreos E.T. (2003). CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature 423: 885–889. [DOI] [PubMed] [Google Scholar]
- Zhu J.K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53: 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.