This study uses a quantitative, transient approach in Nicotiana benthamiana to measure the complementarity between plant miRNAs and their targets required to achieve measurable target repression at both the mRNA and protein levels.
Abstract
MicroRNAs (miRNAs) guide RNA-induced silencing complexes to target RNAs based on miRNA-target complementarity. Using a dual-luciferase based sensor system in Nicotiana benthamiana, we quantitatively assessed the relationship between miRNA-target complementarity and silencing efficacy measured at both the RNA and protein levels, using several conserved miRNAs and their known target sites from Arabidopsis thaliana. We found that naturally occurring sites have variable efficacies attributable to their complementarity patterns. We also observed that sites with a few mismatches to the miRNA 3′ regions, which are common in plants, are often equally effective and sometimes more effective than perfectly matched sites. By contrast, mismatches to the miRNA 5′ regions strongly reduce or eliminate repression efficacy but are nonetheless present in several natural sites, suggesting that in some cases, suboptimal miRNA efficacies are either tolerated or perhaps selected for. Central mismatches fully abolished repression efficacy in our system, but such sites then became effective miRNA target mimics. Complementarity patterns that are functional in animals (seed sites, 3′-supplementary sites, and centered sites) did not reliably confer repression, regardless of context (3′-untranslated region or open reading frame) or measurement type (RNA or protein levels). Overall, these data provide a robust and empirical foundation for understanding, predicting, and designing functional miRNA target sites in plants.
INTRODUCTION
MicroRNAs (miRNAs) are small regulatory RNAs found in both animals and plants. These ∼21-nucleotide RNAs are derived from single-stranded hairpin precursors (Axtell et al., 2011). In complex with Argonaute (AGO) effector proteins, they recognize target mRNAs based on sequence complementarity and function as negative regulators involved in multiple gene regulatory circuits in both plants and animals (Bartel, 2009; Chen, 2009).
In animals, miRNA target sites are most often found within the 3′-untranslated region (UTR) of target mRNAs (Bartel, 2009). 5′-UTR and open reading frame (ORF) targeting are also functional, but less so, and occur much less frequently (Grimson et al., 2007; Lytle et al., 2007). Many animal miRNA target sites form seven consecutive base pairs from position two through eight (numbering from the 5′ end of the aligned miRNA); this is the “seed.” Both perfectly and imperfectly paired seed sites are occasionally supplemented by additional pairings in the miRNA 3′ region, resulting in enhanced target recognition (Grimson et al., 2007). In fewer cases, centered sites, comprising pairing at positions 4 to 15, also mediate target repression (Shin et al., 2010). By contrast, most experimentally verified miRNA target sites in plants are single sites located in ORFs, with a few exceptions in 5′-UTRs, 3′-UTRs, or in noncoding RNAs (Allen et al., 2005; Addo-Quaye et al., 2008; German et al., 2008). Also, in contrast with animal miRNA target sites, extensive complementarity (typically ≤5 mismatches) is the hallmark of all functionally verified miRNA target sites in plants to date. Base pairing at the miRNA 5′ region (from positions 2 to 13) is critical for plant miRNA-mediated target repression with pairing in the vicinity of the AGO-catalyzed slicing site (positions 9 to 11) being especially important (Mallory et al., 2004b; Parizotto et al., 2004; Schwab et al., 2005). Mismatches at the miRNA 3′ region are much less destructive than those at the 5′ or central region (Mallory et al., 2004b; Parizotto et al., 2004; Schwab et al., 2005; Lin et al., 2009).
Modes of miRNA-initiated repression are increasingly recognized as being rather similar in plants and animals; in both cases, targets are typically repressed by a combination of mRNA degradation and translational repression (Brodersen et al., 2008; Guo et al., 2010; Bazzini et al., 2012; S. Li et al., 2013). In animals, miRNA-mediated silencing typically results in the depletion of target mRNAs (Hendrickson et al., 2009; Guo et al., 2010) through a slicing-independent pathway (Bagga et al., 2005; Jing et al., 2005; Valencia-Sanchez et al., 2006). Target slicing is only favored upon forming a highly paired duplex (Yekta et al., 2004). Translational repression is also widely seen in animals and involves inhibition of translational initiation (Humphreys et al., 2005; Pillai et al., 2005; Mathonnet et al., 2007), which is followed by subsequent deadenylation and decapping of the mRNA (Wakiyama et al., 2007; Iwasaki et al., 2009). In plants, with many target sites being subject to endonucleolytic cleavage by AGOs, it has also been suggested that miRNA targets with lower complementarity are funneled exclusively toward translational repression without detectable mRNA destabilization (Dugas and Bartel, 2008). However, nearly all documented cases of translationally repressed plant miRNA targets involve canonical, highly complementary target sites (Aukerman and Sakai, 2003; Chen, 2004; Gandikota et al., 2007; Brodersen et al., 2008; Yang et al., 2012; J.-F. Li et al., 2013; S. Li et al., 2013). In an in vitro system, translational repression can be observed for perfectly matched sites only when the AGO residues required to catalyze target cleavage are mutated or for sites with central mismatches specifically located in the 5′-UTR; animal-like seed-only sites are not effective (Iwakawa and Tomari, 2013). However, the efficacies of potential target sites with more than five mismatches have not been systematically documented in vivo in a plant system to date. The ease of both predicting (Rhoades et al., 2002) and validating (German et al., 2008; Addo-Quaye et al., 2008; Llave et al., 2002) canonical, highly complementary plant miRNA target sites could conceivably have led to the neglect of a wider constellation of less complementary sites in plants.
Furthermore, most previous studies that explicitly examined complementarity requirements for plant miRNA function in vivo have relied heavily or exclusively on mRNA accumulation data (Mallory et al., 2004b; Parizotto et al., 2004; Schwab et al., 2005), or visual phenotypes (Lin et al., 2009). In this study, we used a dual-luciferase-based miRNA sensor system to quantitatively evaluate plant miRNA complementarity requirements at both the mRNA and protein levels. We tested a total of 60 different potential target sites for five different miRNAs and examined several key areas, including comparing the efficacies of naturally occurring miRNA target sites to perfectly paired sites, position-specific effects of mismatches on miRNA efficacy, function of animal-like patterns of complementarity, comparison of ORF versus 3′-UTR target site locations, and the pairing patterns required for effective miRNA target mimicry.
RESULTS
Perfectly Paired Sites Can Behave Differently in ORF versus 3′-UTR
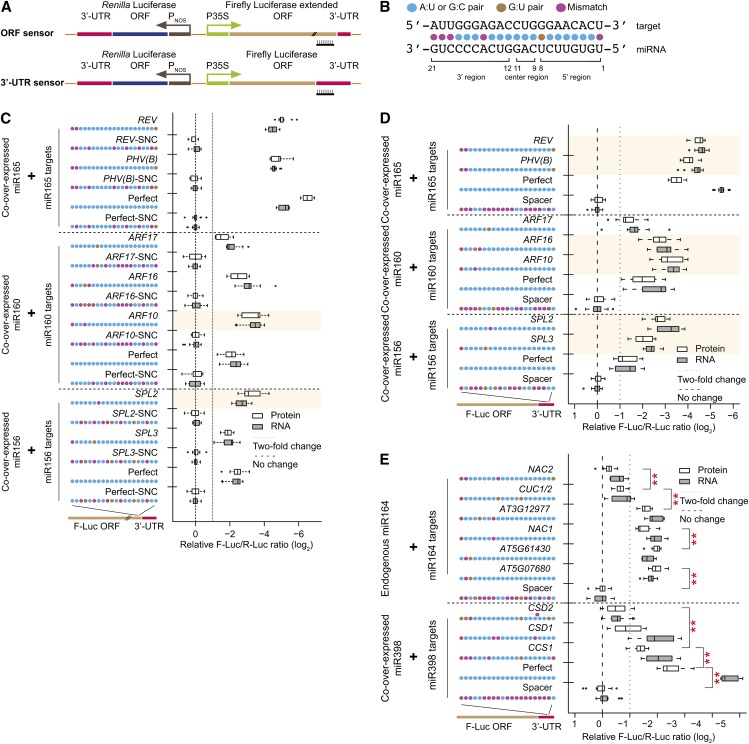
Agrobacterium tumefaciens–mediated transient expression assays in Nicotiana benthamiana leaves (agroinfiltrations) have been widely used to study plant miRNA–target interactions (Llave et al., 2002; Allen et al., 2005; Franco-Zorrilla et al., 2007). We designed a dual-luciferase-based reporter system to quantitatively assess target site efficacies through agroinfiltration (Figure 1A). miRNA target sites were inserted into either the ORF (ORF sensor) or the 3′-UTR (3′-UTR sensor) of firefly luciferase (F-Luc) (Figure 1B). Relative accumulation of the F-Luc sensor was evaluated at both the mRNA (via quantitative RT-PCR [qRT-PCR] with primers flanking the target site) and protein levels (by quantification of luciferase activities) using Renilla luciferase (R-Luc), expressed from the same T-DNA, as an internal control. For ORF sensors, each tested site changes the amino acid sequence of the extended F-Luc protein; thus, each requires a distinct synonymous negative control, which is a target site with maximally disrupted complementarity without changing the amino acid sequence (Figure 1C; Supplemental Table 1). For 3′-UTR sensors, a 21-nucleotide spacer of random sequence served as a common negative control (Figure 1D). Use of the F-Luc/R-Luc ratio allowed internal normalization that corrects for variation in transformation efficiencies between experiments.
Figure 1.
Naturally Occurring Imperfect Target Sites Can Be More or Less Effective Than Perfectly Complementary Sites.
(A) Schematic diagram of dual-luciferase sensors. Top, ORF sensor; bottom, 3′-UTR sensor. P35S, promoter sequence of the cauliflower mosaic virus 35S gene; PNOS, promoter sequence of the nopaline synthase gene.
(B) Conventions for miRNA target site display and nomenclature.
(C) Naturally occurring miR156, miR160, and miR165 target sites in ORF sensors. Box plots summarize results from 15 independent replicates and show the median (thick line), extent of the 1st to 3rd quartile range (box), values extending to 1.5 times the interquartile range (whiskers), and outliers (black circles). Shaded area indicates that target protein downregulation is significantly stronger than the corresponding perfect site (P < 0.05, n = 15, ANOVA-Tukey honestly significantly different [HSD]). Synonymous negative controls (SNC) are negative controls with maximal disruption of their cognate sites while maintaining identical amino acid sequences. Target site sequences are shown in Supplemental Table 1.
(D) As in (C), except for 3′-UTR sensors.
(E) Naturally occurring miR164 and miR398 sites in 3′-UTR sensors. ORF sensor data are in Supplemental Figure 1. Data display conventions as in (C). Double asterisks indicate significant differences at the protein level between two adjacent groups (P < 0.05, n = 15, ANOVA-Tukey HSD).
We first tested the efficacies of naturally occurring, experimentally validated miRNA target sites from Arabidopsis thaliana: miR165a (based on REVOLUTA, PHAVOLUTA, and PHABULOSA) (Mallory et al., 2004b), miR160a (based on AUXIN RESPONSE FACTOR17 [ARF17], ARF16, and ARF10) (Mallory et al., 2005), and miR156a (based on SQUAMOSA PROMOTER BINDING PROTEIN LIKE2 [SPL2] and SPL3) (Schwab et al., 2005; Gandikota et al., 2007). Perfectly paired sites served as positive controls. These miRNAs and target sites were chosen because they are members of deeply conserved miRNA families, and nearly all mismatches in these target sites are located in the 3′ regions of the miRNAs (Figure 1C). N. benthamiana small RNA sequencing (small RNA-seq) from leaves revealed a single miR156a and miR160a-related species whose sequences exactly matched their Arabidopsis counterparts and two predominant miR165a isoforms with zero and one mismatch, respectively, relative to Arabidopsis miR165a (Supplemental Table 2). In all cases, we co-overexpressed the relevant Arabidopsis miRNAs. The previously validated miRNA targets were repressed significantly at both the mRNA and protein levels in our system, which demonstrated that our assay reflects the bona fide effects of miRNA–target interaction in vivo. With sites placed in the ORF, efficacies for these targets were largely comparable to that of a perfectly complementary site (Figure 1C), which is consistent with a previous study using artificially designed miRNA (Park et al., 2009). By contrast, we found that many native sites acted stronger than perfect sites when being tested in the 3′-UTR sensors (Figure 1D). For example, ARF16- and ARF10-like miR160 target sites located in the 3′-UTR were significantly more effective than a perfect site, despite the presence of two or three mismatches (Figure 1D). Further investigation suggested that the differences between ORF and 3′-UTR efficacies were due to decreased performance of perfect sites when placed in the 3′-UTR. The efficacies of two perfect sites (miR156 and miR165), especially at the protein level, dropped in 3′-UTR sensors compared to ORF sensors (Figures 1C and 1D; ∼2-fold for miR156, P = 8.01E-07; ∼4-fold for miR165, P = 9.62E-08; not significant for miR160; Mann-Whitney U-test). Diminished function of perfect sites when placed in the 3′-UTR has also been observed using an in vitro miRNA target system (Iwakawa and Tomari, 2013).
5′ Mismatches Are Strong Indicators of Suboptimal Efficacy for Naturally Occurring miRNA Target Sites
Next, we compared efficacies among naturally occurring target sites for Arabidopsis miR164 and miR398. Target site designs were based on the Arabidopsis miR164a and miR398b mature sequences. N. benthamiana small RNA-seq from leaves revealed a single miR164-related species whose sequence exactly matches Arabidopsis miR164a and two predominant miR398 isoforms with one and two mismatches, respectively, relative to Arabidopsis miR398b (Supplemental Table 2). We thus co-overexpressed Arabidopsis miR398b in experiments with miR398 sensors. The frequency and position of mismatches present in these naturally occurring sites varies greatly among different targets of the same miRNA. Seven genes are experimentally verified miR164 targets in Arabidopsis: NAM/ATAF/CUC1 (NAC1), NAC2, CUP-SHAPED COTYLEDON1 (CUC1), CUC2, AT3G12977, AT5G07680, and AT5G61430 (Mallory et al., 2004a; Baker et al., 2005; Guo et al., 2005; Addo-Quaye et al., 2008). Analysis using 3′-UTR sensors and endogenous N. benthamiana miR164 showed that these target sites had variable efficacies (Figure 1E). The least effective natural target sites (NAC2 and CUC1/2) had mismatches or G-U wobbles in the central and 5′ regions of the target site (Figure 1E). By contrast, for the most effective sites (AT5G61430 and AT5G07680), non Watson-Crick pairs were limited to the 3′ region (Figure 1E). Three miR398 natural target sites from Arabidopsis (COPPER/ZINC SUPEROXIDE DISMUTASE1 [CSD1], CSD2, and COPPER CHAPERONE FOR SOD1 [CCS1]) (Dugas and Bartel, 2008; Beauclair et al., 2010) and a perfectly matched miR398 site were also repressed at different levels, with the perfect site showing the strongest activity. The CSD2 site, which had the most disrupted 5′ region, was the least effective target site (Figure 1E). This was consistent using both 3′-UTR (Figure 1E) and ORF sensors (Supplemental Figure 1). These data indicate that the efficacy of plant native miRNA target sites can vary among different targets of the same miRNA and that some are less than maximally effective. The amount of unpaired bases near the miRNA 5′ region seems to be a strong indicator of impaired regulatory efficiency.
The 5′ and 3′ Regions of Target Sites Are Differentially Tolerant of Consecutive Mismatches
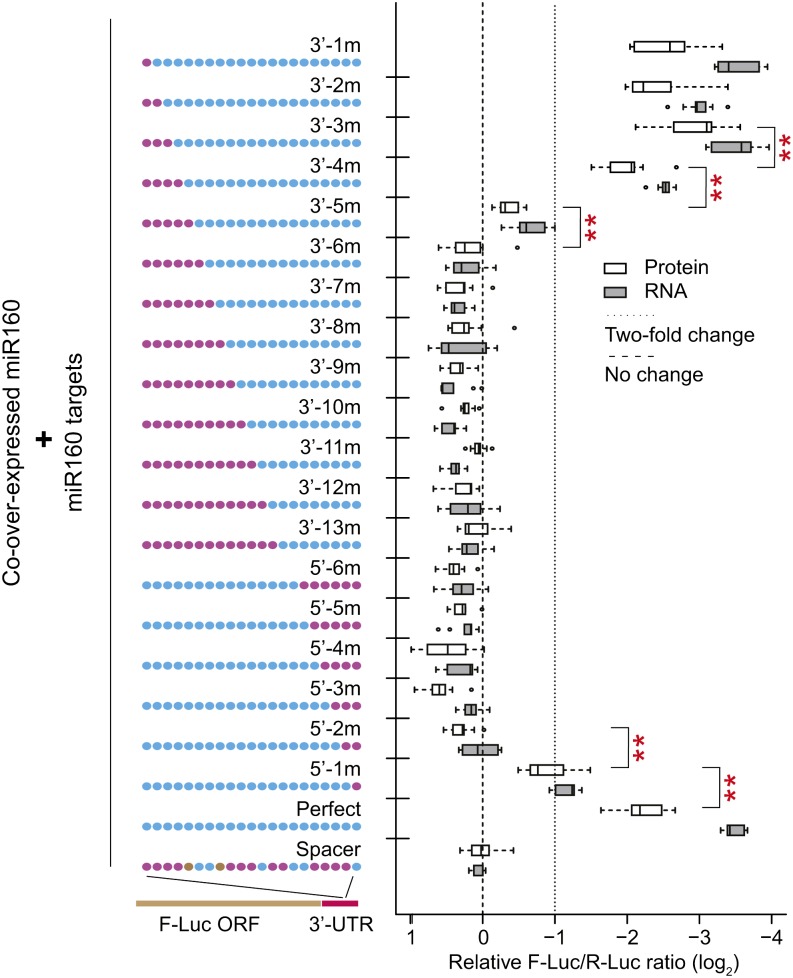
To systematically study target site sensitivity to mismatches on both ends of a target site, a series of miR160 targets with 0 to 13, or 0 to 6 consecutive mismatches relative to the miRNA 3′ and 5′ ends, respectively, were tested. Up to three consecutive mismatches at the miRNA 3′ end were tolerated without affecting site efficacy relative to the perfectly matched control (Figure 2). Target site efficacy was significantly diminished when four or five mismatches were introduced at the miRNA 3′ end, and no efficacy whatsoever was observed for sites with between 6 and 13 consecutive mismatches at the miRNA 3′ end (Figure 2). By contrast, a single mismatch at the miRNA 5′ end significantly diminished target site efficacy, and two or more consecutive mismatches at the miRNA 5′ end fully abolished it (Figure 2). The reduction in efficacy caused by single mismatch at the miRNA 5′ end (∼4-fold at the mRNA level and 2-fold at the protein level) is much stronger than data shown before based on an in vitro slicing assay (Mallory et al., 2004b). We attribute the difference in magnitude to the different systems being employed. In particular, we measured in vivo intact mRNA and protein levels as opposed to the in vitro accumulation of cleaved RNA fragments that were measured by Mallory et al. (2004b). Overall, these data are consistent with the canonical viewpoint that mismatches at the miRNA 5′ region are much more deleterious to target site function than those at the miRNA 3′ region (Mallory et al., 2004b; Schwab et al., 2005; Lin et al., 2009). Importantly, our results demonstrate that this is quantitatively true when measuring efficacy at the level of protein accumulation. Finally, we note that all of the sites with between 6 and 13 consecutive mismatches at the 3′ end would be considered functional animal target sites (Bartel, 2009), as they all contain intact seed pairing in a 3′-UTR context; there was no evidence for efficacy for any of these target sites at either the mRNA or protein levels.
Figure 2.
Differential Sensitivity of the 5′ and 3′ Target Site Regions to Consecutive Mismatches.
Engineered miR160 target sites in 3′-UTR sensors. Data display conventions as in Figure 1. Double asterisks indicate significant difference at the protein level between two adjacent groups (P < 0.05, n = 9, ANOVA-Tukey HSD).
Complementarity Is a Major Determinant for Site Efficacy
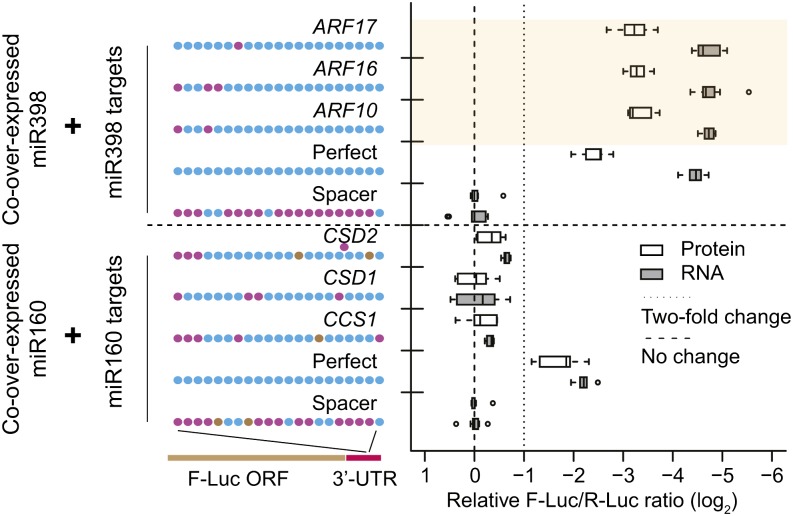
We next examined whether the differences in target site efficacies could be attributed to inherent functional differences between different miRNAs, instead of the complementarity patterns. To test this, we performed target swap experiments where the patterns of the natural miR398 targets were engineered for miR160 and vice versa. Target site patterns that were highly effective for miR160 (ARF10, ARF16, and ARF17; Figures 1C and 1D) were also highly effective for miR398 (Figure 3). By contrast, efficacies of targets with functional miR398 target patterns (CSD1, CSD2, and CCS1; Figure 1E; Supplemental Figure 1) were barely detectable, with only miR160-CSD2 showing marginal repression at the protein level (P = 4.8E-04, Mann-Whitney U-test; Figure 3). These data suggest that target site complementarity patterns are a major factor in determining target site efficacies.
Figure 3.
Complementarity Patterns Play a Major Role in Target Site Efficacy.
Target base pairing pattern swap between miR160 and miR398 in 3′-UTR sensors. Data display conventions as in Figure 1. Shaded area indicates targets with protein level downregulation significantly stronger than the perfect site (P < 0.05, n = 9, ANOVA-Tukey HSD).
Animal-Like miRNA Target Sites Are Ineffective
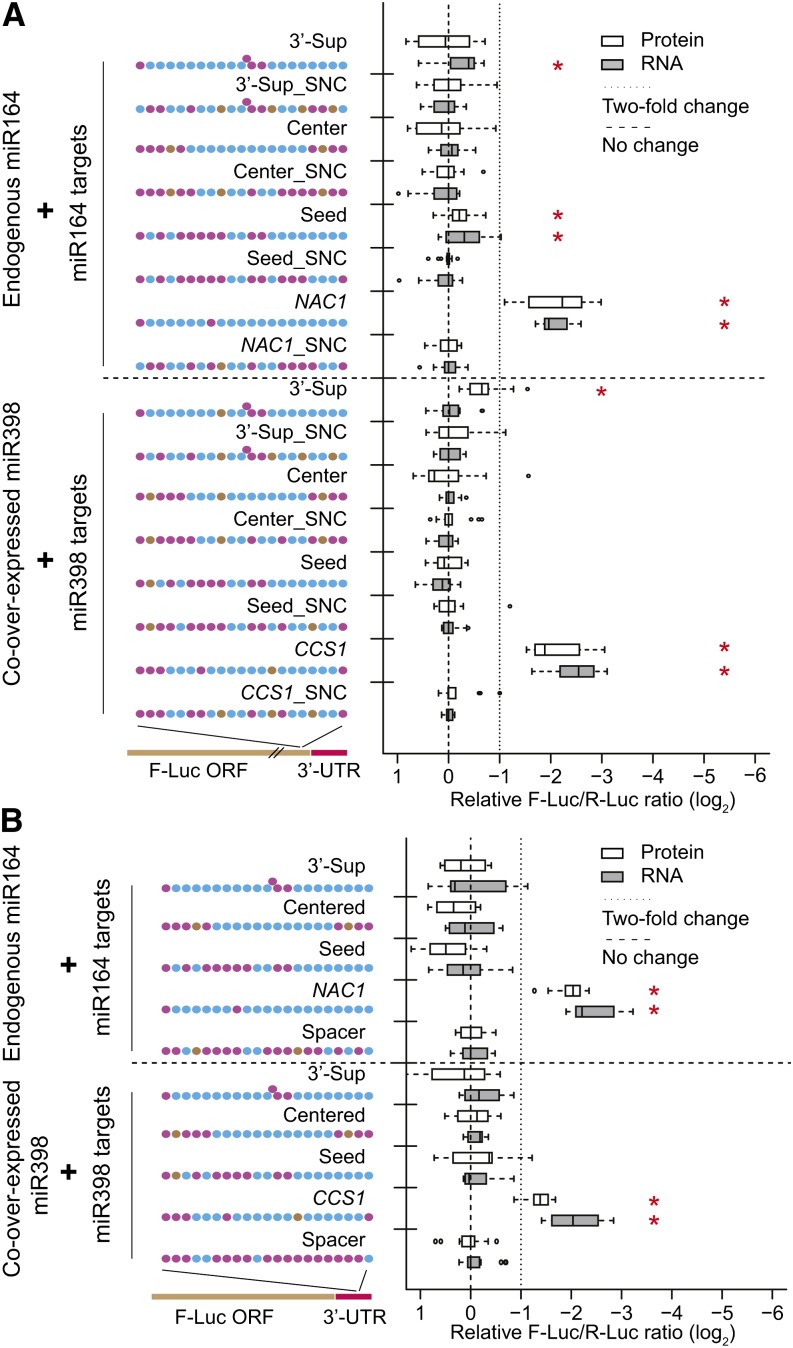
Since seed sites, the most typical pattern of animal miRNA targets, were not effective in our experiments (Figure 2), we then tested the functionality of two other target site patterns known to be functional in animals: centered sites (Shin et al., 2010) and 3′-supplemental sites (Reinhart et al., 2000), both of which have a relatively high degree of complementarity. Positive controls were based on known Arabidopsis miR164 and miR398 targets (NAC1 and CCS1, respectively; Mallory et al., 2004a; Beauclair et al., 2010). We examined the efficacies of various miR164 and miR398 animal-like target sites in both ORF (Figure 4A) and 3′-UTR sensors (Figure 4B). While the NAC1 and CCS1 positive controls were highly effective in both contexts, significant downregulation was rarely detected for the miR164 or miR398 animal-like target sites regardless of ORF versus 3′-UTR location or mRNA versus protein measurements (Figures 4A and 4B). Among the animal-like sites tested, only the seed site for miR164 and the 3′-supplemental site for miR398 resulted in a statistically significant amount of repression at the protein level and only in the ORF sensor (Figure 4A). The magnitudes of these effects were very small (<2-fold), not consistent for both miRNAs tested (Figure 4A), nor consistent in both the ORF and 3′-UTR contexts.
Figure 4.
Little to No Evidence for Functionality of Animal-Like Target Sites.
(A) Animal-like sites in ORF sensors. Data display conventions as in Figure 1. Synonymous negative controls (SNC) are negative controls with maximal disruption of their cognate sites while maintaining identical amino acid sequences. Extruded dots represent single nucleotide bulges on targets. Red asterisks indicate statistically significant downregulation (P < 0.05, Mann-Whitney U-test).
(B) Animal-like sites in 3′-UTR sensors. A 21-nucleotide spacer serves as common negative control for 3′-UTR–located target sites. Data display conventions as in Figure 1.
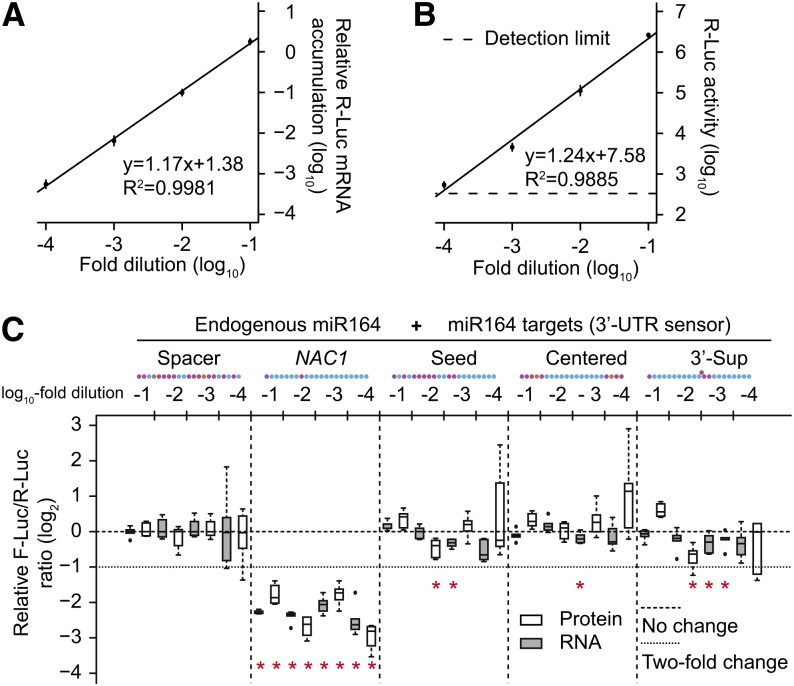
We considered the possibility that animal-like target sites could have been inactive only because the miRNA-AGO complexes were saturated with the overwhelmingly expressed sensors. This was a particular concern for the miR164 experiments, which relied solely upon endogenous miR164. We addressed this question by performing a series of agroinfiltrations using the miR164 3′-UTR sensors where the concentration of Agrobacterium inoculum was diluted in 10-fold increments. Analysis of R-Luc (which lacks engineered miRNA target sites) mRNA (Figure 5A) and protein (Figure 5B) accumulation showed that sensor expression responded to inoculum dilution in a linear fashion. The positive control NAC1 site had strong efficacy across the dilution series (Figure 5C). By contrast, sensors bearing animal-like sites remained ineffective across the dilution series, with only a few cases showing significant repression, and none more than 2-fold (Figure 5C). These data argue against the hypothesis that lack of efficacy is due to saturation brought on by overexpression. Overall, we do not find compelling evidence suggesting that animal-like target sites can enable reliable target repression, regardless of location (ORF versus 3′-UTR) or of measurement (mRNA versus protein).
Figure 5.
Animal-Like Sites Remain Nonfunctional Regardless of Sensor Expression Level.
(A) Relative R-Luc mRNA accumulation levels across a 10-fold dilution series of Agrobacterium concentrations. Mean (n = 24) relative R-Luc mRNA levels ±sd are shown, along with the linear regression.
(B) As in (A) except for R-Luc luciferase activities.
(C) Relative F-Luc target mRNA accumulation and luciferase activity of 3′-UTR sensors across a dilution series of Agrobacterium concentrations. Data display conventions as in Figure 1. Results from two independent replicates are shown.
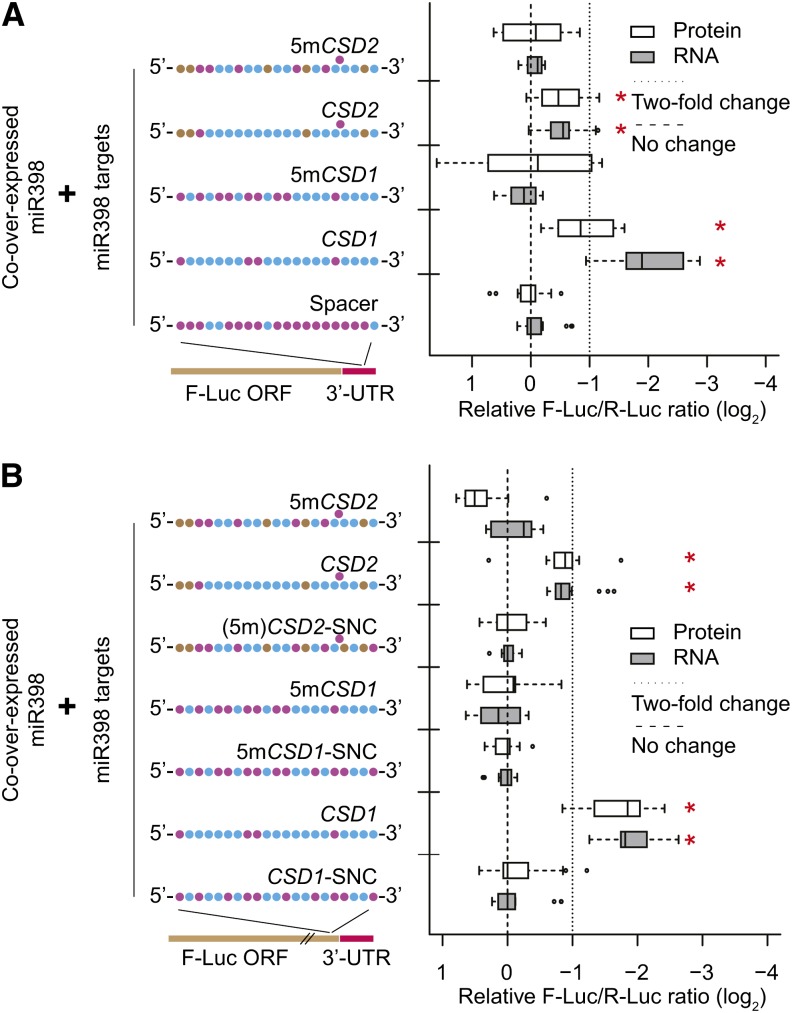
5mCSD1 and 5mCSD2 Are Not Functional miR398 Targets
A previous study showed that two Arabidopsis miR398 targets, CSD1 and CSD2, remained sensitive to miR398 induction even after the miRNA-target complementarity was severely disrupted (Dugas and Bartel, 2008). The disrupted variants, 5mCSD1 and 5mCSD2, lost regulation at the mRNA level but retained miR398-mediated repression of protein accumulation, suggesting that miRNA-induced translational repression in plants required a lower complementarity threshold than miRNA-induced reductions in mRNA levels. However, when placed in our F-Luc sensor, 5mCSD1 and 5mCSD2 sites were ineffective at both the mRNA and protein levels in both the 3′-UTR and ORF contexts (Figures 6A and 6B). Although it is possible that these sites require their native mRNA flanking sequence contexts to function, the simplest conclusion is that the severely disrupted 5mCSD1 and 5mCSD2 sites themselves are nonfunctional. The previously reported miR398-dependent repression of 5mCSD1 and 5mCSD2 protein levels could instead have been due to indirect posttranslational mechanisms. The fact that the CCS1 mRNA, which encodes a chaperone required for the stability of the CSD1 and CSD2 proteins, is also a miR398 target (Beauclair et al., 2010) provides a basis for this hypothesis. This result highlights a potential pitfall of assessing miRNA function via native target protein accumulation data alone: Posttranslational effects caused by other targets of the miRNA in question could confound such assays.
Figure 6.
No Evidence for Functionality of 5mCSD1 or 5mCSD2 Sites.
(A) Results from 3′-UTR sensors. Data display conventions as in Figure 1. 5mCSD1 and 5mCSD2 sites are miR398 targets with five mutations compared with CSD1 and CSD2 sites, respectively, as described by Dugas and Bartel (2008).
(B) Results from ORF sensors. Data display conventions as in Figure 1.
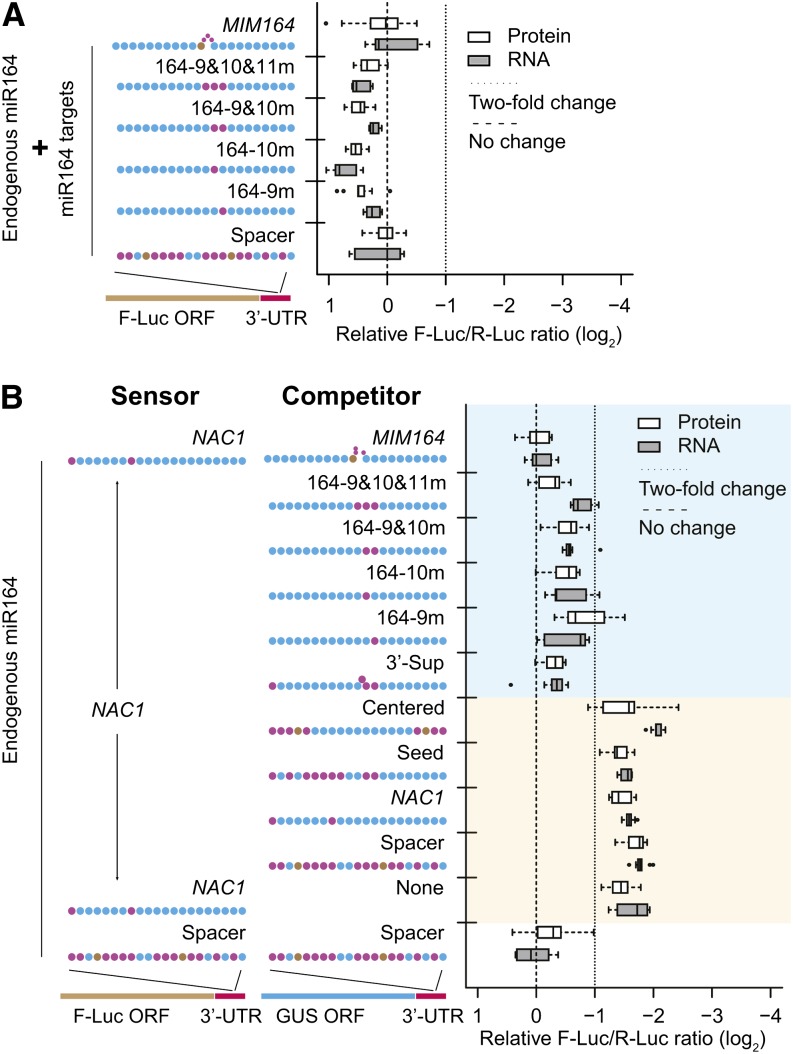
Multiple Pairing Patterns with Central Mismatches Are Effective miRNA Target Mimics
Based largely on measurements of mRNA accumulation, pairing at the central positions, 9 to 11, is thought to be especially critical for plant miRNA function (Schwab et al., 2005; Franco-Zorrilla et al., 2007; Todesco et al., 2010). Consistent with this hypothesis, we found that single nucleotide mismatches at positions 9 and 10, as well as combinations of mismatches at positions 9, 10, and 11, showed complete elimination of the responsiveness of a miR164-targeted 3′-UTR sensor (Figure 7A). Importantly, these results held true regardless of whether efficacy was measured at the mRNA or protein level (Figure 7A). Notably, Arabidopsis miR173 is known to direct AGO-catalyzed slicing of several noncoding RNA targets despite mismatches at positions 9 (TAS1a, TAS1c, and TAS2) or at both positions 9 and 10 (TAS1b) (Allen et al., 2005). However, the miRNA-programmed RNA-induced silencing complex in this case is likely to have unique properties because miR173 is a member of ancient superfamily of plant miRNAs that, instead of simply repressing target accumulation, are instead specialized to recruit RNA-dependent RNA polymerases to their targets (Allen et al., 2005; Chen et al., 2010; Cuperus et al., 2010; Xia et al., 2013).
Figure 7.
Multiple Pairing Patterns Are Effective miRNA Target Mimics.
(A) Results from central mismatch–containing sites in 3′-UTR sensors. Data display conventions as Figure 1.
(B) Results from a target mimicry assay. Data display conventions as in Figure 1. NAC1 3′-UTR site activity was measured in the presence of coinfiltrated GUS-based competitors. Blue shaded area indicates effective competitors with median values of relative F-Luc repression <2-fold. Peach shaded area indicates nonfunctional competitors with median relative F-Luc repression stronger than 2-fold.
Central bulges are also a hallmark of miRNA target mimics in plants, which have frequently been based on the At4/IPS1 family of noncoding RNAs and contain a central 3-nucleotide asymmetric bulge (Franco-Zorrilla et al., 2007; Todesco et al., 2010). Sites with two consecutive central mismatches (not bulges) at positions 10 and 11 have also been demonstrated to perform as intermediate-strength target mimics, and all types of target mimicry sites appear ineffective at conferring repression when embedded in the 3′-UTRs of protein-coding transcripts (Ivashuta et al., 2011). Consistent with expectations, we observed that a canonical miRNA target mimic site for miR164 (MIM164) was unable to confer any repression in our system (Figure 7A). To test our series of central mismatch sites for mimicry activity, we took advantage of the fact that target mimic sites are effective decoys when embedded into the 3′-UTR of β-glucuronidase (GUS) (Ivashuta et al., 2011). We thus designed a series of GUS-based competitors with various miR164 target sites in the 3′-UTRs and coagroinfiltrated them with a functional NAC1 dual-luciferase 3′-UTR sensor. As a negative control, GUS with a spacer site had no effect on miR164-mediated repression (Figure 7B). By contrast, the positive control (GUS with a MIM164 site in its 3′-UTR) had a strong decoy effect, fully preventing detection of miR164-mediated repression (Figure 7B). All variants with central mismatches (9m, 10m, 9mand10m, and 9mand10mand11m) were moderately effective target mimics, though none were as strong as MIM164 (Figure 7B). These results are largely consistent with those of Ivashuta et al. (2011), except that, in our hands, a single mismatch at position 10 was a moderately effective mimic instead of a noneffective one. The discrepancy may be due to increased sensitivity in our assay or perhaps to intrinsic differences between the miRNAs that were tested. We also found that the animal-like 3′-supplementary site pattern, based on a classic let-7 target site from Caenorhabditis elegans lin-41 (Reinhart et al., 2000), was a moderately effective target mimic (Figure 7B). This was predictable because the pattern of let-7 site is similar to canonical plant target mimicry sites. By contrast, centered and seed sites were ineffective target mimics (Figure 7B). Finally, the NAC1 site, itself a highly potent target site in terms of repression (Figure 1E), was ineffective as a miRNA target mimic (Figure 7B). We conclude that many complementarity patterns involving central mismatches flanked by two highly complementary regions, beyond the classic At4/IPS1-like pattern and the mismatch 10-11 pattern, are functional miRNA target mimics in plants. Our data from target sites within 3′-UTRs reinforce the conclusion that effective target mimicry is mutually exclusive with effective repression (Ivashuta et al., 2011): All of the sites that function well as target mimics failed to confer repression, and a site that confers robust repression failed to act as a target mimic. However, we do not rule out the possibility that target mimic sites in other mRNA contexts may function differently. For instance, single sites with central mismatches mediate translational repression with catalytically active AGO1 in vitro only when placed in the 5′-UTR (Iwakawa and Tomari, 2013).
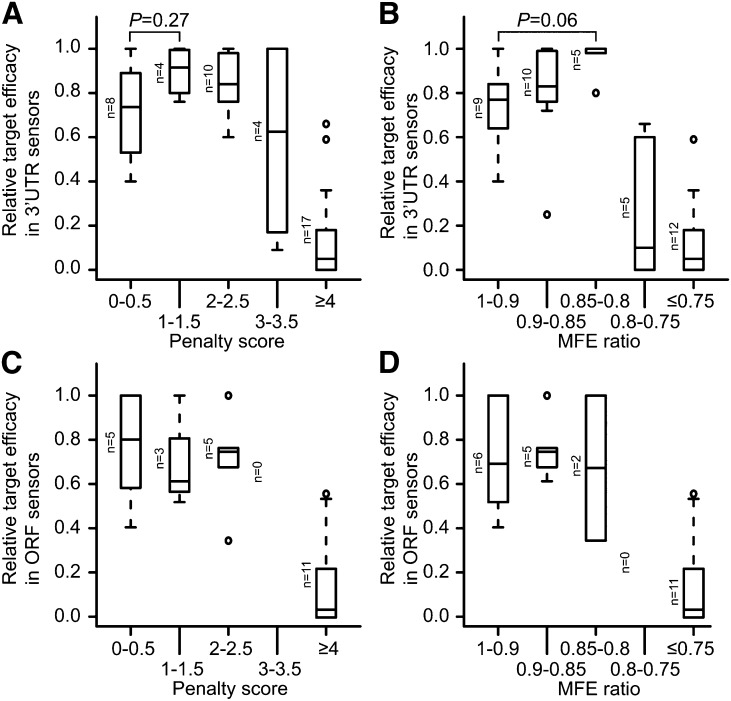
Summary of Target Site Properties
Many studies have reported scoring schemes for computational predictions of plant miRNA targets (Rhoades et al., 2002; Allen et al., 2005; Schwab et al., 2005; Alves et al., 2009). However, there is limited knowledge on the correlation between target prediction scores and target site efficacies, especially at the protein level. We thus scored the target sites used in our studies with respect to two different scoring schemes: the position-specific scoring matrix of Allen et al. (2005) and the minimum free energy (MFE) ratio between the site and a perfectly complementary site. We also split the data into ORF and 3′-UTR categories to further reveal the positional effects on target efficacies. At extreme values, both methods in both sensors identified sharp decreases in efficacy after a critical threshold was reached; an Allen et al. score of four or higher (Figures 8A and 8C) or an MFE ratio of <0.75 was associated with marginal or noneffective sites (Figures 8B and 8D). When focusing only on 3′-UTR sensors, these scoring methods seemed inversely correlated with efficacy for cases above the threshold: The maximum median efficacies were observed for sites with Allen et al. scores of 1 to 1.5 (Figure 8A) and MFE ratios between 0.8 and 0.85 (Figure 8B). Although the differences were small and the P values relatively large (due both to small sample size and variability within categories), this observation suggests that maximally effective target sites in plants are not necessarily perfectly complementary. However, using ORF sensors, we did not observe an inverse correlation (although the number of observations was extremely low). Instead, in the ORF, perfect sites had a similar distribution of efficacies compared with sites with slightly higher scores/lower MFE ratios. Nevertheless, the data imply that, for computational prediction of miRNA targets, it should not be assumed that the target site scoring regime necessarily predicts the quantitative efficacy of the target site since perfect sites are often equivalent to or even sometimes less effective than sites with a few 3′ mismatches.
Figure 8.
Correlation of Target Site Scores with Protein-Level Efficacies.
(A) Box plots of relative target efficacy in 3′-UTR sensors at the protein level with respect to target penalty score. Target penalty score was calculated according to Allen et al. (2005) (P = 0.27, Mann-Whitney U-test).
(B) As in (A) except for the MFE ratio between the sites and a perfectly complementary site (P = 0.06, Mann-Whitney U-test).
(C) As in (A) except for ORF sensors.
(D) As in (B) except for ORF sensors.
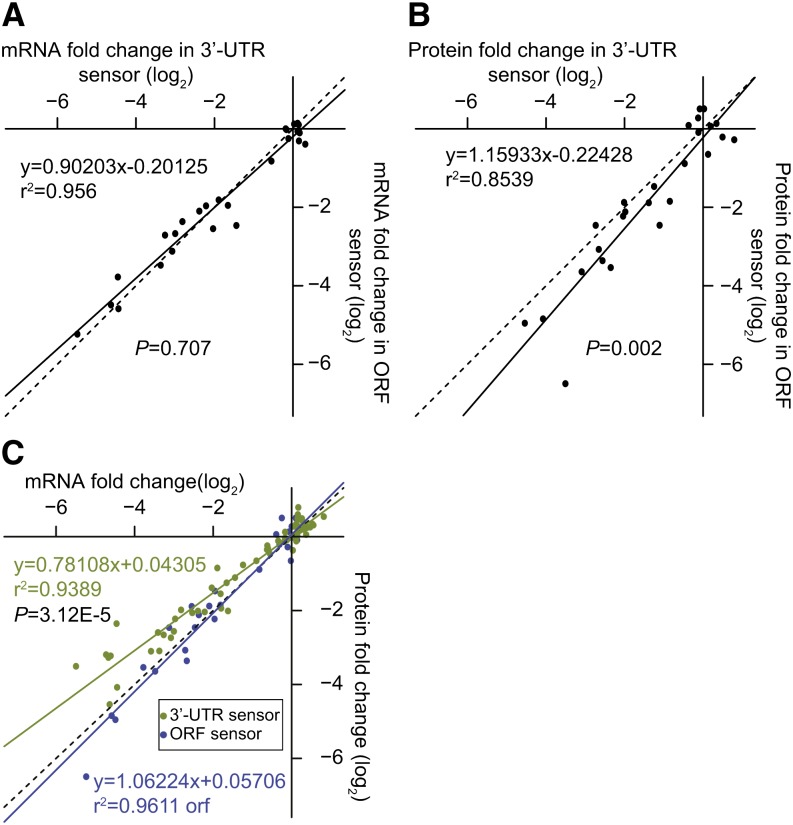
Next, we compared overall efficacies between the ORF and 3′-UTR. ORF-located and 3′-UTR–located site efficacies were correlated in a one-to-one manner at the mRNA level (Figure 9A). However, target repression at the protein level was more effective in ORF sensors than in 3′-UTR sensors (Figure 9B; P < 0.05, analysis of covariance [ANCOVA]). Our data also allowed us to compare magnitudes of repression at the mRNA versus protein levels. For 3′-UTR–located sites, we found that the magnitude of repression measured at the mRNA level was frequently stronger than that measured at the protein level (Figure 9C; P < 0.05, ANCOVA), while for ORF sites, mRNA and protein-based efficacies were correlated in a one-to-one manner (Figure 9C). We did not generally see stronger levels of repression at the protein level compared with the mRNA level (Figure 9C). However, this does not necessarily indicate that translational repression is not contributing to miRNA-mediated repression in our system. In the absence of specific measurements of both the translational efficiencies of the F-Luc mRNAs and the F-Luc protein turnover rates, mechanistic interpretations of these data are not feasible.
Figure 9.
Protein-Level Repression Is More Effective for ORF Sites Than for 3′-UTR Sites.
(A) Comparison of target repression at the mRNA level between 3′-UTR sensors and ORF sensors. Each dot represents the median target repression at the mRNA level in 3′-UTR (x value) and ORF (y value) sensors (n = 26). The linear regression is shown (solid line), as is a reference line with slope of 1 (dotted). P value represents the significance of difference between slope of the experimental regression line and the reference line (ANCOVA).
(B) As in (A), except for repression efficacies measured at the protein level (n = 26).
(C) Correlation between target repression at mRNA levels and protein levels. Each dot represents the median mRNA (x value) and protein (y value) change of a unique target site. Data are pooled from miR164, miR398, miR156, miR160, and miR165 experiments (n = 113). The 3′-UTR targets and ORF targets are plotted separately in green and blue, respectively. The corresponding linear regression is shown (solid line), as is the reference line with slope of 1 (dotted). P value represents the significance of difference between slope of the UTR regression and ORF regression (ANCOVA).
Changes in Target Site Secondary Structures Are Unlikely to Have Affected Our Measurements
Target site accessibility may be important for miRNA function in plants: GC-poor codons tend to flank known target sites (Gu et al., 2012) and transcriptome-wide RNA structure analysis indicates a slight increase in target site accessibility relative to flanking bases (Li et al., 2012). In our experiments, we wished to minimize any effects of site accessibility by testing all sites in the same two flanking contexts (ORF and 3′-UTR sensor). However, it remained possible that the differing sequences of the sites themselves affected site accessibilities in our various sensors. Therefore, we tested whether inadvertent changes in F-Luc secondary structures involving the target sites might have affected our results. Predicted RNA secondary structures surrounding sensor target sites were generally very weak when compared against a control set of known functional structures (MIRNA hairpins from plants; Supplemental Figure 2A). No correlation was observed between target efficacies and the MFE ratios of experimental versus control target sites (Supplemental Figure 2B). Additionally, in terms of absolute target site occlusion (the number of predicted intramolecular paired nucleotides within a target site), there were essentially no differences among all the sensors we tested (Supplemental Figure 2C). We conclude that inadvertent changes in sensor site accessibilities are unlikely to have had a strong influence on our results and that our data primarily reflect the inherent efficacies of the sites themselves.
DISCUSSION
Our data demonstrate that, instead of being “all or nothing” regulators, naturally occurring target sites in plants have a range of inherent efficacies. This implies that targets are fine-tuned by miRNAs to acquire desired biological properties. Consistent with this idea, the variation of spike density among different barley cultivars (Hordeum vulgare) is a direct consequence of the varied efficacies of miR172 target sites due to slight variations in target site complementarities (Houston et al., 2013).
We found that the presence of mismatches near miRNA 3′ ends maintained optimal efficacy when placed in the 3′-UTR, in contrast with perfectly paired sites, whose performance decreased in the 3′-UTR relative to the ORF. The differential performance of perfect sites in the ORF and 3′-UTR is consistent with prior observations suggesting that target sites located in the 5′ regions of mRNAs are often more effective (Iwakawa and Tomari 2013; J.-F. Li et al., 2013). Our data suggest that this position-specific effect can be lessened when the target sites contain 3′ mismatches. Indeed, the fact that 3′ end mismatches enhance small interfering RNA efficacy in human cells (De et al., 2013) argues that this is a conserved feature of AGO-bound small RNAs between plants and animals. This observation may also explain why many known target sites in plants possess mismatches at the 3′ end of the site as opposed to being perfectly complementary. One hypothesis to explain the effect of 3′-mismatches is that following AGO1-mediated endonucleolytic cleavage, the RNA-induced silencing complex disassociates more slowly from perfectly paired target sites and thus delays the process of product release and subsequent turnover of the cleaved mRNA fragments. With perfect sites in 3′-UTR sensors, the protein coding region of the target is entirely contained within the upstream cleavage fragment. Therefore, the coding region remains intact after AGO-catalyzed cleavage and because of the delay in product release remains translatable for an extended period resulting in less efficient repression. This hypothesis could also account for the stronger effects upon mRNA levels versus protein levels seen in the 3′-UTR sensors; our qRT-PCR assay for mRNA levels spans the cleavage site, so that cleaved sensor mRNA will not be detected, even if the upstream fragment remains stable and translatable for some time. We also observed other natural miRNA target sites with more extensive mismatches in the central and/or 5′ regions, and correspondingly suboptimal efficacies, such as the miR398 targets (Figure 1E; Supplemental Figure 1). This suggests that natural selection either tolerates, or perhaps has even selected for, suboptimal miRNA efficacies for some targets. Consistent with this idea, a range of different target site efficacies was also observed for miR396 targets (Debernardi et al., 2012).
Single animal-like target sites, including seed, 3′-supplementary, and centered sites, were ineffective in our assay. Importantly, these results are true when quantitatively measured at both the mRNA and protein levels. It should be noted that in animals, effects observed for most target sites are generally subtle, and miRNA target sites frequently act synergistically (Grimson et al., 2007; Baek et al., 2008; Guo et al., 2010). These data support the canonical view that plant miRNAs require a higher complementarity threshold to exert repression on targets than do animal miRNAs. Recently, it has been shown that in the green alga Chlamydomonas reinhardtii miRNA seed sites can confer ∼20% repression (Yamasaki et al., 2013). Thus, the ineffectiveness of seed sites may be a trait unique to the land plants. The mechanistic reasons why animals, plants, and green algae have differing complementarity thresholds for miRNA function are currently unclear.
Effective miRNA-directed target repression at either the protein or mRNA levels in our assay requires pairing at the central nucleotides and minimal mismatches in the 5′ region. By contrast, an expanded set of target site patterns, united by the presence of central mismatches flanked by highly paired regions on both sides, are effective target mimics in plants. This suggests that searches for new naturally occurring target mimics, which have already identified a large number of candidates (Ivashuta et al., 2011; Wu et al., 2013), should be expanded to include a wider variety of complementarity patterns.
The dual-luciferase sensor system allows rapid, quantitative testing of a large number of target sites. It also has the advantage of not being confounded by redundant systems of miRNA-induced posttranslational control that can affect native target protein accumulation levels. In addition, sensor assays can ensure that the expression domains of the target and miRNA have 100% overlap, avoiding artifacts due to distinct cell- or tissue-type-specific expression of native miRNAs and targets. However, the assay is based upon ectopic overexpression, in the context of a massive Agrobacterium infection. Thus, the system is potentially subject to artifacts arising both from saturation by overexpressed targets and from the modulation of small RNA pathways by Agrobacterium infection (Dunoyer et al., 2006; Wang et al., 2011). Our titration assays indicate that saturation does not appear to be an issue (Figure 5), but nonetheless we cannot rule out entirely such artifacts. Therefore, it is theoretically possible that some properties of this system do not entirely match those that occur under normal cellular conditions. For instance, it is possible that the apparent lack of effectiveness for animal-like targets is caused by low sensitivity or some other artifact particular to agroinfiltration. However, in this regard, we point out that despite more than a decade of intense study of miRNAs, there are no reports showing positive data in favor of the hypothesis that animal-like target sites are functional in plants.
While miRNA-target complementarity patterns are clearly pivotal for miRNA function, there are several other factors that may contribute to miRNA target site efficacy. Changes in nucleotide compositions flanking target sites might influence local mRNA secondary structure resulting in variations in target site accessibility. Experimentally determined structural occlusions are slightly depleted within miRNA target sites relative to flanking residues (Li et al., 2012), and GC-poor codons are enriched around target sites (Gu et al., 2012). Both observations suggest that selection has favored minimizing local mRNA secondary structure surrounding miRNA target sites. Target site accessibility could also be influenced by the positioning of various mRNA binding proteins, which at present are very hard to accurately predict a priori. The relative position on the target mRNA may also play a role in regulating target-site efficacy. We observed that perfectly matched sites were often less effective in the 3′-UTR relative to the ORF, and the same is true in an in vitro AGO1 targeting system (Iwakawa and Tomari, 2013). In a systematic analysis of multiple artificial miRNAs, all with similar or identical levels of very high target site complementarity, sites located toward the 5′ of the mRNA target tended to be more effective (J.-F. Li et al., 2013). Our development of a rapid and quantifiable system to measure miRNA target site function in vivo will facilitate further study of the role of mRNA secondary structure and target site positioning. Mechanisms of miRNA-initiated target repression can also differ between cell types (Grant-Downton et al., 2013), and of course the miRNA and the mRNA bearing a potential target site must accumulate in at least some of the same cell type(s) or tissues in order for a site to be measurably functional. Future studies in this area should focus on incorporating all of these parameters to produce a more refined model for the efficacies of miRNA target sites in plants.
METHODS
GUS-Based Competitors
GUS ORF was amplified by PCR using Phusion High-Fidelity DNA Polymerase (New England Biolabs). PCR products were double digested with XhoI and EcoRI and were ligated into pGreenII-0229 previously digested by the same enzymes. Cauliflower mosaic virus 35S promoter (5′-ApaI and 3′-XhoI) and terminator (5′-SpeI and 3′-NotI) were also PCR amplified and were placed upstream and downstream, respectively, from GUS. A 45-nucleotide artificial 3′-UTR sequence flanked by PstI and SpeI was synthesized (Integrated DNA Technologies) and placed in between the GUS ORF and the 35S terminator. Two unique restriction sites (AvrII and AgeI) were embedded in the artificial 3′-UTR sequence to facilitate the insertion of different target sites. Target site inserts were generated by hybridizing synthetic oligonucleotides flanked by AvrII and AgeI sites, ligated into the 3′-UTR of GUS previously digested by the same enzymes, and confirmed by sequencing. Supplemental Table 3 contains oligonucleotide sequences.
Dual-Luciferase Sensors
R-Luc ORF was amplified by PCR using Phusion High-Fidelity DNA Polymerase. PCR products were double digested with SalI and XbaI and ligated into NOS-cassette of pUC119 vector previously digested by the same enzymes. NOS-R-Luc cassette was inserted into pGreenII-0000 previously digested with HpaI through blunt-end ligation. F-Luc ORF with (3′-UTR sensor) or without (ORF sensor) stop codon were also PCR amplified and double digested with XhoI and EcoRI. The 35S-F-Luc cassette was created by replacing GUS ORF in pGreenII-0229 with F-Luc ORF. The whole cassette (P35S-F-Luc-3′UTR-ter) was cut by ApaI and NotI and ligated into pGreenII-0000 bearing NOS-R-Luc cassette. Target site inserts were generated by hybridizing synthetic oligonucleotides flanked by AvrII and AgeI sites, ligated into the 3′-UTR of F-Luc previously digested by the same enzymes, and confirmed by sequencing (see Supplemental Table 1 for target site sequence and Supplemental Table 3 for oligonucleotides used).
miRNA Overexpressers
All MIRNA sequences were PCR amplified from Columbia-0 genomic DNA using Phusion High-Fidelity DNA Polymerase. The complete hairpin structures and ∼50 nucleotides both upstream and downstream of the hairpin region were amplified. XhoI and EcoRI sites were incorporated into the 5′ and 3′ends of the PCR product followed by double digestion to replace the GUS ORF in GUS-based competitor. Supplemental Table 3 contains oligonucleotide sequences.
Agrobacterium tumefaciens–Mediated Transient Expression in Nicotiana benthamiana
Dual-luciferase sensors were transformed into Agrobacterium GV3101 (pMP90 pSoup). Positive transformants were selected on Luria-Bertani agar medium (25 μg/mL kanamycin, 25 μg/mL gentamycin, 50μg/mL rifampicin, and 5 μg/mL tetracycline). Target inserts were confirmed by sequencing the PCR products. N. benthamiana plants were grown at 22°C under 24-h-light conditions. One-month-old plants were used for infiltration. Two days before infiltration, 2-mL cultures of the Agrobacterium strains were inoculated from single colonies on plates and grown for 24 h at 28°C. The working cultures were inoculated from the starter culture at a 1:1000 ratio. Cells were harvested by centrifugation at 3000g, 22°C, for 5 min. Cell pellets were resuspended in infiltration medium (10 mM MgCl2, 10 mM MES, pH 5.7, and 150 μM acetosyringone) with OD600 adjusted to 0.5. Resuspended cell cultures were kept on a bench-top for 4 h before infiltration. Only one construct was infiltrated into each plant. Leaf discs were collected 48 h after infiltration using a hole puncher. Samples from the same plant were used for measurements at both mRNA level (qRT-PCR) and protein level (dual-luciferase assay).
RNA Extraction Followed by qRT-PCR
Around 50 mg of N. benthamiana leaf samples was ground to a fine powder in liquid nitrogen and homogenized in QIAzol lysis reagent (miRNeasy mini kit; Qiagen). Total RNAs were extracted per the manufacturer's instructions. cDNA was obtained with the QuantiTect reverse transcription kit (Qiagen), using 1 μg of total RNA. qRT-PCR, using oligos flanking the target site in the case of F-Luc, was performed using the QuantiTect SYBR Green RT-PCR kit (Qiagen) to derive F-Luc to R-Luc ratios. Supplemental Table 3 contains oligonucleotide sequences.
Dual-Luciferase Assay
The same leaf samples collected for RNA extraction were also used for dual-luciferase assay. Around 20 mg of leaf discs was ground to a fine powder in liquid nitrogen and homogenized in Passive Lysis Buffer (Promega). Nondissolved plant tissues were clarified by performing a quick spin at 12,000g, 4°C, for 30 s. Ten microliters of supernatant was used for dual-luciferase assay using the Dual-Luciferase Reporter Assay System (Promega) on a GloMax 96 Microplate Luminometer equipped with dual injectors (Promega).
Small RNA-Seq
Two replicate RNA samples from 1-month-old N. benthamiana leaves were extracted and used as input for TruSeq Small RNA sample preparation kit (Illumina), followed by 50-nucleotide single-end sequencing on a HiSeq2500 (Illumina) in rapid run mode. Data have been deposited at the National Center for Biotechnology Information Gene Expression Omnibus (accession number GSE48886). After adapter trimming, reads were aligned to Arabidopsis thaliana miRNAs from miRBase 19 using bowtie 1 (Langmead et al., 2009).
RNA Secondary Structure Analyses
Hybridization energy between miRNAs and corresponding targets was calculated using RNAcofold (from ViennaRNA; Lorenz et al., 2011) with default settings. Target site sequences were extended six nucleotides upstream and downstream to avoid dangling ends. MFE of the predicted secondary structure flanking target site was calculated using RNALfold (ViennaRNA; Lorenz et al., 2011), with default settings. Upstream 500-nucleotide and downstream 200-nucleotide sequences were added to the target sequence as input.
Accession Number
Sequence data from this article can be found in the National Center for Biotechnology Information Gene Expression Omnibus under accession number GSE48886.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Naturally Occurring miR398 Target Sites Are Less Effective Than a Perfectly Complementary Site.
Supplemental Figure 2. No Correlation between Predicted Target Accessibility and Protein-Level Target Efficacy.
Supplemental Table 1. miRNA-Target Site Alignments for Sensors Used in This Study.
Supplemental Table 2. Identification of N. benthamiana miRNAs.
Supplemental Table 3. Oligonucleotides.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation Award 1121438 to M.J.A.
AUTHOR CONTRIBUTIONS
Q.L. and M.J.A. designed the experiments. Q.L. and F.W. performed the experiments. Q.L. analyzed the data. M.J.A. did the bioinformatic analysis. Q.L. and M.J.A. wrote the article with input and editing from F.W.
Glossary
- miRNA
microRNA
- UTR
untranslated region
- ORF
open reading frame
- qRT-PCR
quantitative RT-PCR
- small RNA-seq
small RNA sequencing
- MFE
minimum free energy
- ANCOVA
analysis of covariance
- HSD
honestly significantly different
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Addo-Quaye C., Eshoo T.W., Bartel D.P., Axtell M.J. (2008). Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 18: 758–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E., Xie Z., Gustafson A.M., Carrington J.C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221. [DOI] [PubMed] [Google Scholar]
- Alves L., Jr, Niemeier S., Hauenschild A., Rehmsmeier M., Merkle T. (2009). Comprehensive prediction of novel microRNA targets in Arabidopsis thaliana. Nucleic Acids Res. 37: 4010–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman M.J., Sakai H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell M.J., Westholm J.O., Lai E.C. (2011). Vive la différence: Biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 12: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D., Villén J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P. (2008). The impact of microRNAs on protein output. Nature 455: 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E. (2005). Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122: 553–563. [DOI] [PubMed] [Google Scholar]
- Baker C.C., Sieber P., Wellmer F., Meyerowitz E.M. (2005). The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr. Biol. 15: 303–315. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. (2009). MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini A.A., Lee M.T., Giraldez A.J. (2012). Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 336: 233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauclair L., Yu A., Bouché N. (2010). MicroRNA-directed cleavage and translational repression of the copper chaperone for superoxide dismutase mRNA in Arabidopsis. Plant J. 62: 454–462. [DOI] [PubMed] [Google Scholar]
- Brodersen P., Sakvarelidze-Achard L., Bruun-Rasmussen M., Dunoyer P., Yamamoto Y.Y., Sieburth L., Voinnet O. (2008). Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190. [DOI] [PubMed] [Google Scholar]
- Chen H.-M., Chen L.-T., Patel K., Li Y.-H., Baulcombe D.C., Wu S.-H. (2010). 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci. USA 107: 15269–15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2009). Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol. 25: 21–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus J.T., Carbonell A., Fahlgren N., Garcia-Ruiz H., Burke R.T., Takeda A., Sullivan C.M., Gilbert S.D., Montgomery T.A., Carrington J.C. (2010). Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 17: 997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De N., Young L., Lau P.-W., Meisner N.-C., Morrissey D.V., MacRae I.J. (2013). Highly complementary target RNAs promote release of guide RNAs from human Argonaute2. Mol. Cell 50: 344–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debernardi J.M., Rodriguez R.E., Mecchia M.A., Palatnik J.F. (2012). Functional specialization of the plant miR396 regulatory network through distinct microRNA-target interactions. PLoS Genet. 8: e1002419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas D.V., Bartel B. (2008). Sucrose induction of Arabidopsis miR398 represses two Cu/Zn superoxide dismutases. Plant Mol. Biol. 67: 403–417. [DOI] [PubMed] [Google Scholar]
- Dunoyer P., Himber C., Voinnet O. (2006). Induction, suppression and requirement of RNA silencing pathways in virulent Agrobacterium tumefaciens infections. Nat. Genet. 38: 258–263. [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla J.M., Valli A., Todesco M., Mateos I., Puga M.I., Rubio-Somoza I., Leyva A., Weigel D., García J.A., Paz-Ares J. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39: 1033–1037. [DOI] [PubMed] [Google Scholar]
- Gandikota M., Birkenbihl R.P., Höhmann S., Cardon G.H., Saedler H., Huijser P. (2007). The miRNA156/157 recognition element in the 3' UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 49: 683–693. [DOI] [PubMed] [Google Scholar]
- German M.A., et al. (2008). Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 26: 941–946. [DOI] [PubMed] [Google Scholar]
- Grant-Downton R., Kourmpetli S., Hafidh S., Khatab H., Le Trionnaire G., Dickinson H., Twell D. (2013). Artificial microRNAs reveal cell-specific differences in small RNA activity in pollen. Curr. Biol. 23: R599–R601. [DOI] [PubMed] [Google Scholar]
- Grimson A., Farh K.K.-H., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. (2007). MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell 27: 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Wang X., Zhai C., Xie X., Zhou T. (2012). Selection on synonymous sites for increased accessibility around miRNA binding sites in plants. Mol. Biol. Evol. 29: 3037–3044. [DOI] [PubMed] [Google Scholar]
- Guo H., Ingolia N.T., Weissman J.S., Bartel D.P. (2010). Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H.-S., Xie Q., Fei J.F., Chua N.H. (2005). MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 17: 1376–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson D.G., Hogan D.J., McCullough H.L., Myers J.W., Herschlag D., Ferrell J.E., Brown P.O. (2009). Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 7: e1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston K., et al. (2013). Variation in the interaction between alleles of HvAPETALA2 and microRNA172 determines the density of grains on the barley inflorescence. Proc. Natl. Acad. Sci. USA 110: 16675–16680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys D.T., Westman B.J., Martin D.I., Preiss T. (2005). MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc. Natl. Acad. Sci. USA 102: 16961–16966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashuta S., Banks I.R., Wiggins B.E., Zhang Y., Ziegler T.E., Roberts J.K., Heck G.R. (2011). Regulation of gene expression in plants through miRNA inactivation. PLoS ONE 6: e21330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa H.O., Tomari Y. (2013). Molecular insights into microRNA-mediated translational repression in plants. Mol. Cell 52: 591–601. [DOI] [PubMed] [Google Scholar]
- Iwasaki S., Kawamata T., Tomari Y. (2009). Drosophila argonaute1 and argonaute2 employ distinct mechanisms for translational repression. Mol. Cell 34: 58–67. [DOI] [PubMed] [Google Scholar]
- Jing Q., Huang S., Guth S., Zarubin T., Motoyama A., Chen J., Di Padova F., Lin S.-C., Gram H., Han J. (2005). Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell 120: 623–634. [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Zheng Q., Vandivier L.E., Willmann M.R., Chen Y., Gregory B.D. (2012). Regulatory impact of RNA secondary structure across the Arabidopsis transcriptome. Plant Cell 24: 4346–4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.-F., Chung H.S., Niu Y., Bush J., McCormack M., Sheen J. (2013). Comprehensive protein-based artificial microRNA screens for effective gene silencing in plants. Plant Cell 25: 1507–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., et al. (2013). MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell 153: 562–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.-S., Wu H.-W., Elena S.F., Chen K.-C., Niu Q.-W., Yeh S.-D., Chen C.-C., Chua N.-H. (2009). Molecular evolution of a viral non-coding sequence under the selective pressure of amiRNA-mediated silencing. PLoS Pathog. 5: e1000312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C., Xie Z., Kasschau K.D., Carrington J.C. (2002). Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297: 2053–2056. [DOI] [PubMed] [Google Scholar]
- Lorenz R., Bernhart S.H., Höner Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L. (2011). ViennaRNA package 2.0. Algorithms Mol. Biol. 6: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytle J.R., Yario T.A., Steitz J.A. (2007). Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc. Natl. Acad. Sci. USA 104: 9667–9672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A.C., Bartel D.P., Bartel B. (2005). MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 17: 1360–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A.C., Dugas D.V., Bartel D.P., Bartel B. (2004a). MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr. Biol. 14: 1035–1046. [DOI] [PubMed] [Google Scholar]
- Mallory A.C., Reinhart B.J., Jones-Rhoades M.W., Tang G., Zamore P.D., Barton M.K., Bartel D.P. (2004b). MicroRNA control of PHABULOSA in leaf development: Importance of pairing to the microRNA 5′ region. EMBO J. 23: 3356–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathonnet G., et al. (2007). MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science 317: 1764–1767. [DOI] [PubMed] [Google Scholar]
- Parizotto E.A., Dunoyer P., Rahm N., Himber C., Voinnet O. (2004). In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev. 18: 2237–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W., Zhai J., Lee J.-Y. (2009). Highly efficient gene silencing using perfect complementary artificial miRNA targeting AP1 or heteromeric artificial miRNA targeting AP1 and CAL genes. Plant Cell Rep. 28: 469–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai R.S., Bhattacharyya S.N., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W. (2005). Inhibition of translational initiation by Let-7 microRNA in human cells. Science 309: 1573–1576. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G. (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906. [DOI] [PubMed] [Google Scholar]
- Rhoades M.W., Reinhart B.J., Lim L.P., Burge C.B., Bartel B., Bartel D.P. (2002). Prediction of plant microRNA targets. Cell 110: 513–520. [DOI] [PubMed] [Google Scholar]
- Schwab R., Palatnik J.F., Riester M., Schommer C., Schmid M., Weigel D. (2005). Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8: 517–527. [DOI] [PubMed] [Google Scholar]
- Shin C., Nam J.-W., Farh K.K.-H., Chiang H.R., Shkumatava A., Bartel D.P. (2010). Expanding the microRNA targeting code: Functional sites with centered pairing. Mol. Cell 38: 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco M., Rubio-Somoza I., Paz-Ares J., Weigel D. (2010). A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. 6: e1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia-Sanchez M.A., Liu J., Hannon G.J., Parker R. (2006). Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 20: 515–524. [DOI] [PubMed] [Google Scholar]
- Wakiyama M., Takimoto K., Ohara O., Yokoyama S. (2007). Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes Dev. 21: 1857–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Soyano T., Machida S., Yang J.-Y., Jung C., Chua N.-H., Yuan Y.A. (2011). Molecular insights into plant cell proliferation disturbance by Agrobacterium protein 6b. Genes Dev. 25: 64–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.-J., Wang Z.-M., Wang M., Wang X.-J. (2013). Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol. 161: 1875–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia R., Meyers B.C., Liu Z., Beers E.P., Ye S., Liu Z. (2013). MicroRNA superfamilies descended from miR390 and their roles in secondary small interfering RNA biogenesis in eudicots. Plant Cell 25: 1555–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki T., Voshall A., Kim E.-J., Moriyama E., Cerutti H., Ohama T. (2013). Complementarity to an miRNA seed region is sufficient to induce moderate repression of a target transcript in the unicellular green alga Chlamydomonas reinhardtii. Plant J. 76: 1045–1056. [DOI] [PubMed] [Google Scholar]
- Yang L., Wu G., Poethig R.S. (2012). Mutations in the GW-repeat protein SUO reveal a developmental function for microRNA-mediated translational repression in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 315–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta S., Shih I.H., Bartel D.P. (2004). MicroRNA-directed cleavage of HOXB8 mRNA. Science 304: 594–596. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.