Alternatively spliced transcripts often have different fates depending on their cellular locations. This article presents an easily applicable method using molecular beacons and standard laser scanning microscopy to monitor endogenous transcripts within a plant cell. The results show that intron retention transcripts, which are insensitive to nonsensemediated decay, are localized to the nucleus.
Abstract
Alternative splicing (AS) is an important regulatory process that leads to the creation of multiple RNA transcripts from a single gene. Alternative transcripts often carry premature termination codons (PTCs), which trigger nonsense-mediated decay (NMD), a cytoplasmic RNA degradation pathway. However, intron retention, the most prevalent AS event in plants, often leads to PTC-carrying splice variants that are insensitive to NMD; this led us to question the fate of these special RNA variants. Here, we present an innovative approach to monitor and characterize endogenous mRNA splice variants within living plant cells. This method combines standard confocal laser scanning microscopy for molecular beacon detection with a robust statistical pipeline for sample comparison. We demonstrate this technique on the localization of NMD-insensitive splice variants of two Arabidopsis thaliana genes, RS2Z33 and the SEF factor. The experiments reveal that these intron-containing splice variants remain within the nucleus, which allows them to escape the NMD machinery. Moreover, fluorescence recovery after photobleaching experiments in the nucleoplasm show a decreased mobility of intron-retained mRNAs compared with fully spliced RNAs. In addition, differences in mobility were observed for an mRNA dependent on its origin from an intron-free or an intron-containing gene.
INTRODUCTION
Alternative splicing (AS) enlarges the protein-coding potential of higher eukaryotes and may thereby increase their complexity. The increased number of protein isoforms per gene can be achieved by combinatorial removal of intervening sequences during splicing. However, some AS events lead to the production of transcripts that contain a premature termination codon (PTC) or other features that can be recognized by the nonsense-mediated decay (NMD) pathway, a regulatory RNA degradation pathway within the cytoplasm (reviewed in Nicholson and Mühlemann, 2010). Regulated AS with concomitant destruction of the transcript by NMD, therefore, functions as an important regulator of gene expression. AS occurs frequently in mammals; more than 90% of intron-containing genes show AS in various cell types or under certain environmental conditions. In contrast, AS in plants was grossly underestimated. Recent analyses (Marquez et al., 2012) showed that at least 61% of all Arabidopsis thaliana intron-containing genes undergo AS under normal growth conditions. The refined study of a selected set of AS genes using a high-throughput RT-PCR panel revealed that AS/NMD regulates around 13 to 18% of intron-containing genes; most of the sensitive transcripts exhibit features characteristic for NMD targets, including the presence of a PTC upstream of an exon splice junction, long 3′ untranslated regions (UTRs), and certain upstream open reading frames within the 5′ UTRs (Simpson et al., 2008; Kalyna et al., 2012). Unlike in animals, the most common AS event in plants is intron retention (IR), which constitutes around 40% of all AS events (Filichkin et al., 2010; Marquez et al., 2012). However, the AS/NMD study (Kalyna et al., 2012) also revealed that most transcripts with IR events are resistant to the plant NMD degradation pathway, even though they possess features that should make them clear targets. This was surprising, as plant-specific IR variants were previously detected on ribosomes (Ner-Gaon et al., 2004; Jiao and Meyerowitz, 2010); this is a known prerequisite for NMD, since it requires a first round of translation. In addition, data from other organisms also suggest that transcripts with retained introns containing PTCs are subject to NMD (Sayani et al., 2008; de Lima Morais and Harrison, 2010). These results led us to question the cellular fate of these special transcripts, as nuclear localization would render them insensitive to NMD.
We were interested to establish a method to monitor the distribution of distinct AS transcripts in living plant cells. However, the number of in vivo techniques allowing the sequence-specific detection of RNA on the subcellular level remains limited (Christensen et al., 2010). Genetically encoded reporter proteins such as MS2 coat protein (Stockley et al., 1995; Fusco et al., 2003), λN22 (Daigle and Ellenberg, 2007) peptide, or Pumilio-HD (a bimolecular fluorescence complementation approach; Ozawa et al., 2007) bind with high specificity to distinctively tagged and, therefore, altered RNA molecules. However, large fluorescent marker proteins may influence the spatiotemporal features of RNAs by altering the naturally occurring secondary and tertiary structures. Moreover, many experimental approaches use direct injection of labeled RNA transcripts. Such a strategy may prevent these transcripts from proceeding toward their natural processing steps (e.g., cotranscriptional splicing, polyadenylation, capping, export, etc.) and hence alter their localization and protein complex formation. Therefore, in order to investigate RNA in its living context, it is important to image endogenous, unaltered RNA transcripts. One elegant approach for RNA imaging is the use of short fluorescent hybridization-sensitive DNA oligonucleotides, named molecular beacons (MBs), which are fluorescently quenched in their unbound state (Tyagi and Kramer, 1996). MBs have already proven their applicability in mammalian cells (Grünwald and Singer, 2010), providing insights into RNA transcription, maturation, subnuclear molecule movement, and transport (Tyagi and Kramer, 1996; Bratu et al., 2003; Vargas et al., 2005; Santangelo et al., 2009).
In this article, we describe a method that uses MBs conjugated with single fluorophores targeted to an unmodified endogenous transcript; this allows us to monitor their subcellular localization in living Arabidopsis thaliana cells. We use a standard confocal laser scanning microscope and our statistical analysis pipeline in order to characterize a population made of individual cells. Our approach combines in vivo imaging of RNA via MBs and a standard biochemical cell fractionation protocol with RT-PCR analysis of the transcript variants. We demonstrate this technique on the splice variants of two genes, RS2Z33 and the SEF factor. In both cases, NMD-resistant splice variants with IR are localized in the nucleus of plant cells. Furthermore, we show that MBs can be used for RNA fluorescence recovery after photobleaching (FRAP) experiments, yielding information about the movement of the targeted molecules through the nucleoplasm. Interestingly, we show that the nucleus-retained IR transcripts move slower through the nucleoplasm than their fully spliced (FS) version.
RESULTS
Experimental Setup for Monitoring Transcripts in Living Plant Cells
As the subcellular localization of AS transcripts may explain why certain transcripts are not targeted by NMD, we used MBs to monitor such splice variants in vivo (reviewed in Reddy et al., 2012). We first had to determine whether MBs are sufficiently sensitive to be used as reporters in living plant suspension cells. AS transcripts tend to be lower in abundance than the FS variants; therefore, we established an inducible cell suspension system of the Ser/Arg-rich domain splicing factor RS2Z33. We used a stably transformed cell line carrying a β-estradiol–inducible promoter fused to an intron-free RS2Z33 gene and examined cells either treated or not treated with β-estradiol. RS2Z33 is a plant-specific Ser/Arg-rich domain protein and possesses AS events, which are upregulated by overexpression of RS2Z33 (Lopato et al., 2002; Kalyna et al., 2003). After the induction of protein expression with β-estradiol, the overexpressed protein upregulates the alternatively spliced endogenous transcript containing intron 3 (Supplemental Figure 1).
MBs were designed according to published considerations about their hybridization dynamics (Sokol et al., 1998). To make MB binding kinetics as comparable as possible for all probes, we accounted for secondary structure (avoiding extended stems and internal base pairs), duplex, mono, dimer, and target-opening energies. Binding of an MB to the RNA transcript creates an RNA-DNA hybrid that may be hydrolyzed by RNase H, as was shown in animal cell experiments (Walder and Walder, 1988). To test whether MBs cause an altered degradation of their bound transcript, we tested RNA MBs with 2′-O-methylated backbone modifications, which are not targets for RNase H (Bratu et al., 2003). Cells transfected with 2′-O-methylated RNA MB showed similar binding specificity to a DNA MB (Supplemental Figure 2I). We observed similar average intensities (ratio of 1.08), and the signal stayed stable for at least 24 h. Since RNA MBs exhibited a similar distribution to DNA MBs, but with the disadvantage of accumulating within the nucleus (and especially the nucleolus) in stressed cells, and also since we showed that degradation of the target does not seem to be an issue, we decided to use DNA MBs instead of 2′-O-methylated RNA MBs.
Previous studies repeatedly reported that DNA MBs and 2′-O-methylated MBs are actively transported into mammalian and vertebrate nuclei (Tyagi and Alsmadi, 2004; Chen et al., 2007, 2008). However, in living plant suspension cells, we found that both entities were evenly distributed within the cytoplasmic and nuclear compartments (Supplemental Figure 2; for more details on the distribution of DNA MBs, see Supplemental Figure 5), making DNA MBs suitable for RNA imaging in plant suspension cells.
To begin testing MB sensitivity in Arabidopsis cell culture, it was first important to examine binding dynamics and to establish controls. Our specifically binding MBs are DNA oligonucleotides carrying spectrally distinct dyes on their 5′ and 3′ ends, namely Atto550 (fluorophore) and BHQ2 (quencher). As an internal control, an MB labeled with FAM and BHQ1 was designed that had no target in the Arabidopsis genome or transcriptome. We refer to this MB as the “scrambled” MB, since it has a similar GC content to all specifically binding MBs. The scrambled MB was always cotransfected, with all MBs specifically targeting the splice variants of interest to correct for background. The background signal is a complex composite of the interaction between MB and the cell organelles, proteins, DNA, and RNA as well as MB degradation and changes in microenvironments (pH, ion strength, etc.). All these factors contribute to unspecific signal either by changing the quenching efficiency of the BHQ quencher or by the release of single fluorophores. The scrambled MB was used to estimate this background.
Okabe et al. (2011) reported that MBs bind up to 70 times slower when compared with linear probes measured in mammalian COS-7 cells due to the self-complementary stem. Therefore, we tested equilibration times for the MBs in plant suspension cells. It takes an expected time period of 6 to 14 h after transfection until the MB signal equilibrates and remains constant for at least another 24 h.
Next, we acquired a two-color image with a confocal laser microscope, which was used to generate a ratiometric map of the cell; the focal plane was set to intersect the nucleus. The cells, cotransfected with two MBs, were imaged with laser lines 488 and 561 nm. In order to compare the multicolor images, direct ratios of the images (pixel by pixel) were created. The generation of the ratiometric image required corrections for the quenching efficiency, excitation efficiency, and detection efficiency. The correction factors were derived from several preliminary control experiments and are described in more detail in Methods. The image ratios between the specific and unspecific channels directly correspond to the abundance of mRNA and can be used to compare the mRNA distribution in subcellular compartments and cell populations. The ratios were determined by average analysis in a manually defined area of interest (i.e., cellular compartment). In order to prevent biases due to subjective measures, we decided to always analyze the entire compartment and, subsequently, average the ratio for that compartment. Finally, the splice variant ratios of all analyzed cells are represented in a plot of ratios versus ratio frequencies (Figure 1). We compared the nuclear and cytoplasmic intensity ratios of induced and uninduced cell populations (i.e., cells overexpressing the RS2Z33 protein versus cells bearing the endogenous [wild-type] levels of RS2Z33). Since the overexpression of this splicing factor leads to an altered ratio of the investigated splice variants, an observable shift of the intensity histograms in the respective compartments was expected. For the characterization of the experimental data, we chose a parameter-free analysis. In order to characterize the changes between the induced and uninduced cell populations, we compared fitted histograms of the data. For analysis, two nonparametrical statistical tests were performed, a two-sample Kolmogorow-Smirnow (KS) test and a Mann-Whitney (MW) test. The acquired intensity ratio distributions gave us insights into the abundance and subcellular localizations of particular mRNA splice variants of the RS2Z33 gene (Figure 2B).
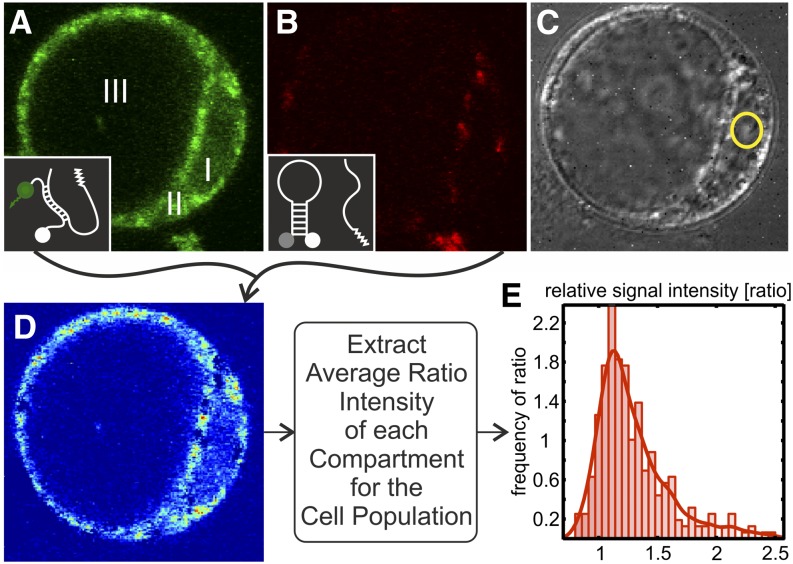
Figure 1.
Protoplasts from Arabidopsis Suspension Cells Transfected with MBs via Electroporation.
(A) Cell with specific MBs (EJ6) labeled with Atto550 and BHQ2. I, nuclear region; II, cytoplasm; III, vacuole.
(B) The same cell cotransfected with a scrambled MB labeled with FAM and BHQ1.
(C) Transmission image (with nucleolus in the yellow circle).
(D) Image processing. After correction for quantum yield, quenching efficiency, and laser power, a ratiometric image was created.
(E) The average intensity of the nuclear and cytoplasmic regions was plotted into a histogram (parameter-free fitting).
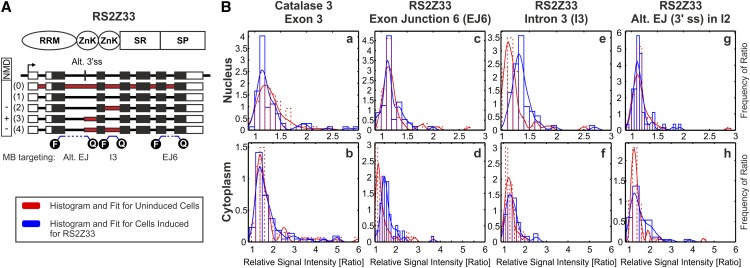
Figure 2.
MBs Detect Splice Variants in Living Plant Cells.
(A) At the top, the protein domains of the plant-specific SR protein RS2Z33 are shown. Below are the gene structure and alternative splice variants of RS2Z33. White boxes, UTRs; red boxes, retained intronic regions; blue lines, MBs targeting mRNA regions. Transcripts are numbered according to Figure 3. Alt 3′ss, alternative 3′ss; F, fluorophore; I, intron; Q, quencher; RRM, RNA recognition motif; SP, Ser/Pro-rich domain; SR, Ser/Arg-rich domain; ZnK, zinc knuckle. − and + indicate the sensitivity to NMD.
(B) Distribution fits of ratios between the specific and unspecific channels of cells transfected with MBs. The red graphs represent the intensity ratio distribution of the uninduced RS2Z33 cells, and the blue graphs represent the intensity ratio distribution after the induction with estrogen, yielding RS2Z33-overexpressing cells. a and b show histograms and corresponding distribution fits of the nuclear and cytoplasmic fractions of cells transfected with MBs targeting exon 3 of catalase3 mRNA. The nuclear fraction shows a nonsignificant shift toward higher values in the distribution in the induced fraction (P = 0.19 by the KS test). No shift can be observed in the cytoplasmic fraction (P = 0.78 by the KS test). c and d show histograms and corresponding distribution fits of the nuclear and cytoplasmic fractions of cells transfected with MBs targeting EJ6 of all RS2Z33 mRNA variants. Within the nucleus, no shift between the induced and uninduced populations can be observed (P = 0.9 by the KS test). The cytoplasmic fraction shows a significant signal increase in the induced cells (P ∼ 10e−9 by the KS test). e and f show histograms and corresponding distribution fits of the nuclear and cytoplasmic fractions of cells transfected with MB targeting intron 3 of RS2Z33 mRNA variants. The nuclear fraction shows a very significant increase in RNA abundance of IR3-containing variants within the induced population (P ∼ 10e−10 by the KS test). In the cytoplasm, no difference between the two populations was detected (P = 0.9 by the KS test). g and h show histograms and corresponding distribution fits of the nuclear and cytoplasmic fractions of cells transfected with MB targeting the alternative EJ (Alt. EJ) in intron 2 of RS2Z33 mRNA variants. The nuclear fraction exhibits no difference between the induced and uninduced populations (P = 0.99 by the KS test). The cytoplasmic intensity ratio distribution shows an increase within the induced cell population (P = 0.0487 by the KS test). Each data set includes ∼70 cells from different biological replicates. The significance level α for all tests is 0.05.
MBs Can Be Used to Detect Endogenous mRNA Splice Variants in Living Cells
At the beginning of these experiments, it was uncertain whether MBs can be used to detect endogenous mRNAs within living plant cells. Therefore, in an initial experiment, we chose to investigate a gene with highly abundant and well-described transcripts, catalase3 (Mhamdi et al., 2012). We designed an MB targeting exon 3, which should detect all known transcripts of catalase3. The corresponding fitted histograms for MBs targeting catalase3 transcripts are shown in Figure 2B (a and b). The median values of the distribution are 1.5 and 1.22 for cytoplasm and nucleus, respectively. However, as expected, we did not observe a signal enhancement of the induced cell population versus the uninduced cell population (high distribution similarity within the nucleus and cytoplasm: P = 0.19 and 0.78, respectively, tested with the KS test and P = 0.174 and 0.589, respectively, tested with the MW test). This result was reassuring, since catalase3 is not expected to be upregulated upon RS2Z33 protein overexpression. Nevertheless, the determined ratios for catalase3 greater than 1 demonstrate that specific targeting of mRNAs via MBs is possible in plant cells.
In order to distinguish alternative transcripts, it was important to show that MBs can specifically bind to exon junctions (EJ). The corresponding fitted histograms for MBs targeting RS2Z33 transcripts that contain EJ6 are shown in Figure 2B (c and d). Again, we obtained ratios greater than 1 (i.e., that MBs can specifically target an EJ in living plant suspension cells; the median values of the distribution are 1.32 and 1.17 for cytoplasm and nucleus, respectively). The intensity ratios of cells transfected with MBs targeting the EJ6 exhibit a strong increase within the cytoplasm upon induction. According to the applied KS test resulting in a P value of ∼10e−9 (0.0 by the MW test), these distributions are clearly different from each other (i.e., transcripts carrying the EJ6 are accumulating in the cytoplasm of RS2Z33-overexpressing cells). The nuclear intensity ratio distribution of the induced population, however, is not shifted upon overexpression of RS2Z33 (KS test P value is ∼0.9 [0.79 by the MW test]; Figure 2B, c and d). In conclusion, the overexpression of the intron-free gene from the inducible construct results in the accumulation of mRNA in the cytoplasm.
Most importantly, we wanted to examine whether we can distinguish the various AS transcripts of RS2Z33 and determine their localizations. Therefore, we designed an MB targeting intron 3 of RS2Z33, which is retained in two splice variants, both of which have been shown to be insensitive to NMD (Kalyna et al., 2012; Supplemental Figure 3D). Due to the upregulation of AS transcripts upon RS2Z33 overexpression, an intensity increase of MB signals in the induced cell population should be detectable. Indeed, the intensity ratio distribution for transcripts carrying intron 3 indicates a significant expression increase within the nucleus upon induction (KS test P value of ∼10e−10 [0.0 by the MW test]). In the cytoplasm, no increase in specifically binding MBs targeting the IR events was observed (KS test P value of ∼0.9 [0.52 by the MW test]; Figure 2B, e and f). These results show an accumulation of RS2Z33 splice variants carrying IR3 within the nucleus. This suggests a potential function and specific evasion of the regular NMD machinery through nuclear retention, as NMD targets typically need an initial round of translation in the cytoplasm.
Another AS transcript of RS2Z33 was tested via an MB targeting the alternative 3′ splice site (3′ss) in intron 2 (Figure 2A, Alt. EJ). The splice variant with just this alternative 3′ss event is sensitive to the NMD pathway and, therefore, is predicted to be present in the cytoplasm. The second transcript targeted by this MB is a combination of AS at the 3′ss of intron 2 and retention of intron 3. The intensity ratio distribution of the MB displays only a weak increase in the signal; thus, the KS test P values are 0.0487 and 0.99 for the cytoplasm and nuclear fractions, respectively (0.039 and 0.78 by the MW test; Figure 2B, g and h). The majority of cells within this population showed a ratio of ∼1; only a small subpopulation showed a significantly shifted ratio, especially in the cytoplasmic fraction. Because of the low signal intensity and rare occurrence of cells exhibiting ratios above 1, we chose to abandon further investigation of this transcript.
In order to confirm the results with an alternative method, a cell fractionation experiment with subsequent RT-PCR analysis of the RS2Z33 transcripts was performed (Figure 3A). The results clearly show that the AS transcripts with retention of intron 3 are retained in the nuclear fraction, whereas the AS transcript with the AS 3′ss in intron 2 is cytoplasmic. This is consistent with the results obtained via in vivo imaging using MBs and shows that MBs can indeed be used as indicators for detecting AS variant transcripts in living plant cells.
Figure 3.
Splice Variants Carrying IR Events Are Retained within the Nucleus.
(A) Gel separation of RT-PCR products of RS2Z33 (At2g37340) splice variants in fractionated cell extracts. Since splice variants 3 and 4 are very low abundance within the cell suspension culture (Supplemental Figure 1A), two different primer (P) sets (A+B and C+B) were used to identify the localization of the splice variants of RS2Z33: the splice variant carrying the alternative 3′ss in intron 2, the splice variant carrying intron 3 (IR3), the splice variant carrying the combination of these splicing events (IR3 and Alt. 3′ss in I2), and the FS variant. Asterisks indicate unknown bands, and stop signs indicate the positions of the PTCs. T, total RNA; C, cytoplasmic RNA; N, nuclear RNA. − and + indicate the sensitivity to NMD.
(B) Gel separation of RT-PCR products of the SEF factor splice variants in fractionated cell extracts. Primers A and B were used to identify the localization of the splice variant retaining intron 2, the splice variant retaining introns 1 and 2, the FS variant, as well as the unspliced mRNA (US).
(C) PAGE gel separation of protein extracts from the same cell fractionation in Figure 3 and Supplemental Figures 1 and 5 with subsequent immunoblotting using fraction-specific antibodies. The cytoplasmic fraction is free of histone 3, and the nuclear fraction is free of the cytoplasmic isoform of fructose-1,6-bisphosphatase.
MBs Can Detect Endogenous AS Variants in Wild-Type Cells
Overexpression of RS2Z33 and the subsequent upregulation of splicing variants were used to show that with MBs, AS variants can indeed be distinguished. However, our main objective was to use MBs to monitor undisturbed endogenous mRNA transcripts in untransformed wild-type cells. We chose the well-characterized SEF factor (At5g37055, also known as Arabidopsis SWC6, a homolog of the yeast SWC6 protein; Deal et al., 2007; Choi et al., 2011), as it features a gene structure (Figure 4A) with two AS variants with either an IR event of intron 1 or IR of intron 1 and intron 2. These AS transcripts consist of features that should trigger NMD (PTC) but have been shown to be insensitive to this degradation pathway (Kalyna et al., 2012; Supplemental Figure 3D).
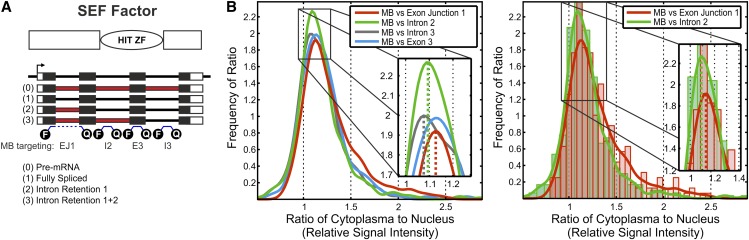
Figure 4.
MBs Detect Endogenous Splice Variants in Living Wild-Type Cells.
(A) Protein domains and gene structure as well as the four alternative splice variants of SEF factor (At5g37055). White boxes, UTRs; red boxes, retained intronic regions; blue lines, MBs. E, exon; F, fluorophore; HIT ZF, HIT-type zinc finger; I, intron; Q, quencher.
(B) Distribution fits between the ratios of the cytoplasm and the ratios of the nuclear regions (specific and unspecific signals of transfected MBs). The left panel represents the distribution of all MBs, and the right panel represents the MB against EJ1 and intron 2 with respective histograms. Plots display significant differences between the FS (MB versus EJ1; red) and the splice variants carrying intron 2 (MB versus intron 2; green) with a P value of 2.1086e−04 (by the KS test). Cells transfected with an MB targeting intron 3 (i.e., the pre-mRNA; dark gray) exhibit a significant difference in comparison with the FS variant (P value of 9.0444e−04 by the KS test). The significance level α for all tests is 0.05.
To demonstrate the distribution of these AS variants in living cells, we designed four different MB probes (Figure 4A). We monitored the total RNA with an MB targeted against exon 3 of the SEF factor, whereas we used an MB against the first EJ to measure the FS variant. In addition, an MB against intron 3 quantifies the level of pre-mRNA and an MB against intron 2 quantifies the abundance of AS variants with retained intron 2. With the overexpression system, it was possible to use the uninduced population as a reference in order to compare the intensities of different MBs in the cytoplasm or nucleus. For an analysis of two RNA populations within the same cell, the relative cytoplasmic and nuclear abundance of a certain RNA species has to be compared. Consequently, a comparison method for the analysis of endogenous RNA populations was established under the assumption of similar binding kinetics for all MBs. The intensity ratio distribution for the FS variant (monitored by an MB spanning EJ1) is significantly different from the intensity ratio distribution for the splice variants carrying intron 2 (monitored by an MB targeting intron 2), with a P value of 2.1086e−04 using the KS test (9.7158e−06 by the MW test, two sided, α of 5%; Figure 4B). Also, the intensity ratio distribution for the pre-mRNA (monitored by an MB targeting the constitutively spliced intron 3) is significantly smaller than the one for the FS mRNA (P value of 9.0444e−04 by the KS test [2.9541e−05 by the MW test]). It follows that the IR transcripts are localized to the nucleus, whereas the FS is in the cytoplasm.
Supporting these experiments, we also found AS transcripts with IR events in the nucleus by cell fractionation with subsequent RT-PCR experiments for the SEF factor (Figure 3B). With this method, we tested four other IR transcripts with features for NMD targets (Supplemental Figures 3A to 3D), which had been shown to be insensitive to NMD (Kalyna et al., 2012). Only NMD-sensitive transcripts were found in the cytoplasm, whereas NMD-insensitive transcripts were exclusively found in the nuclear fraction, indicating that in plant cells, IR mainly targets the transcript to the nucleus.
Together, these results indicate that SEF transcripts are retained within the nucleus if they possess an IR event. We also report that using the presented workflow and statistical pipeline, it is indeed possible to observe endogenous mRNA splice variants in wild-type cells.
Dynamics of Endogenous Splice Variants of RS2Z33
Since MBs bind to distinct AS transcripts of RS2Z33 in the living cell, we wanted to investigate their mobility. It was of interest to examine whether the exogenous transcripts (expressed from an intron-free RS2Z33 construct) move differently from transcripts derived from the endogenous intron-containing gene. Again, we used MBs against intron 3 (AS transcripts) and EJ6 (all transcripts of RS2Z33) to target the AS variants of RS2Z33 (Figure 2A) and performed an RNA FRAP experiment with the induced and uninduced populations (for details, see Methods).
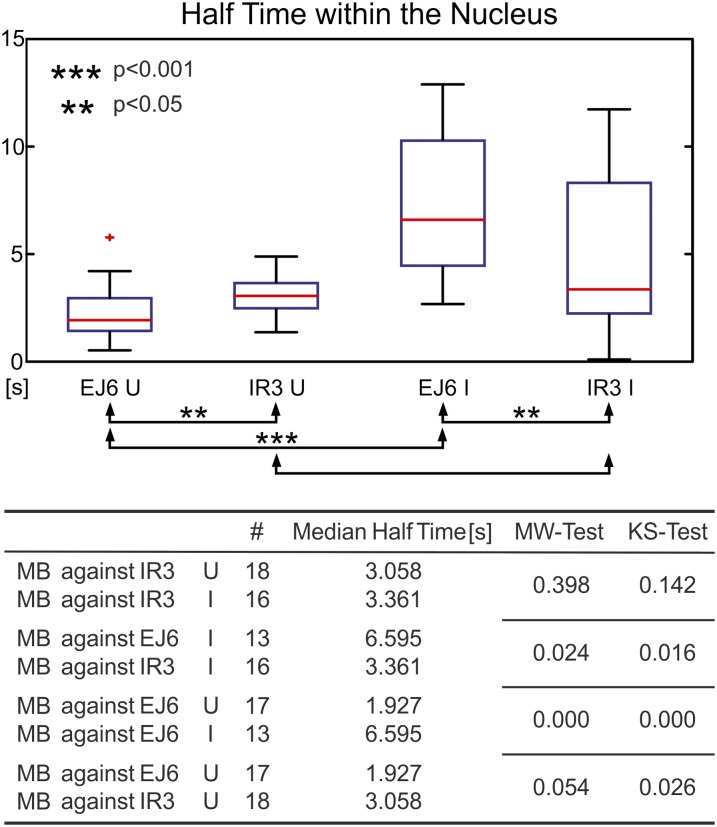
From the FRAP curves obtained by bleaching of the nucleoplasm of the uninduced cells, we determined that transcripts monitored with an MB against EJ6 move significantly faster than transcripts monitored with an MB for intron 3 (Figure 5). The MB against EJ6 monitors a mixture of all transcripts, including the variant with IR3 (i.e., the half-time of EJ6 is slightly overestimated due to the splice variant containing intron 3). This effect may be negligible, since the FS transcript is more abundant (Supplemental Figure 1). This suggests a different messenger ribonucleoprotein (mRNP) composition of the intron 3 containing transcripts. Interestingly, when we compared the mobility of MBs against EJ6 in the induced and uninduced cell populations, it became evident that the transcript within the induced cells moved much slower. As the majority of the transcripts within the induced cells come from the induction of the intron-free RS2Z33 construct, this finding suggests a different protein composition on this mRNP compared with the mRNP created by the intron-containing gene through the splicing process. As expected, the mobility of MBs targeting intron 3 in the induced and uninduced populations is not significantly different (Figure 5). This control was necessary to show that in induced cells, transcripts do not slow down their movement due to stress. Therefore, the transcript originating from the intron-free gene is moving slower than the endogenous FS version. In conclusion, we were able to use MBs for an RNA FRAP experiment monitoring the mobility of transcripts of RS2Z33.
Figure 5.
Dynamics of MB-Labeled Splice Variants within the Nucleoplasm Measured by FRAP Experiments.
Box-plot representation of the half-time of recovery after bleaching a defined spot within the nucleoplasm. Results are shown for MBs against EJ6 of RS2Z33 in induced/uninduced cells as well as MBs against IR3 in induced and uninduced cells. FS variants move faster than intron 3–containing variants. mRNA derived from a cDNA gene moves slower than naturally processed FS mRNA. The median and outcome of the statistical tests are depicted in the table at bottom. I, induced; U, uninduced; #, number of cells.
[See online article for color version of this figure.]
DISCUSSION
Confocal imaging of MB localization in combination with cell fractionation experiments and subsequent RT-PCR analysis has proven to be a powerful system to detect and distinguish endogenous mRNA splice variants at the subcellular level in living plant cells. To show that discrimination between particular splice variants can be achieved, fitted histograms of image ratios were compared between subcellular compartments. Using MBs targeting the respective introns of the RS2Z33 gene and the SEF factor, we were able to show the nuclear localization of certain splice variants that carry IR and how by that they avoid degradation by NMD. The fact that these IR transcripts are not upregulated in NMD mutants or by cycloheximide treatment (Kalyna et al., 2012; Supplemental Figure 3D) indicates that they are completely retained in the nucleus and not partially distributed to the cytoplasm. One explanation for their nuclear localization could be that IR transcripts are recognized by certain proteins (e.g., UA-rich binding proteins; Kim et al., 2009), eventually obstructing the export through the nuclear pores to the cytoplasm. However, this concept has not been fully clarified, since genome-wide studies showed the association of certain IR transcripts with ribosomes (Ner-Gaon et al., 2004; Jiao and Meyerowitz, 2010). Interestingly, 7 out of 11 genes exhibiting NMD-insensitive IR events identified by Kalyna et al. (2012) are listed as being involved in stress responses when comparing the Gene Ontology biological process classification, and 3 of them do not yet have a class (Supplemental Table 1). Leviatan et al. (2013) showed that most of the investigated, cold-regulated IR events in Arabidopsis are putative NMD targets; however, they are not degraded by this mechanism. The authors of that study argue that these alternative transcripts may generate truncated proteins that competitively inhibit full-length proteins; however, our results suggest that such transcripts are retained within the nucleus. Another idea comes from observations in animal cells exhibiting paraspeckles. These nuclear granules are capable of specifically immobilizing certain transcripts due to the presence of a hairpin structure, which is recognized by the structural components of the paraspeckle. Upon certain stress stimuli, the retained transcripts are posttranscriptionally spliced and released into the cytoplasm (reviewed in Bond and Fox, 2009). Thus, the nuclear retention of certain transcripts could serve as a readily recruitable transcript pool in stress situations. Neither of these hypotheses have been experimentally addressed in plants. However, a recent interesting study was able to demonstrate the presence of IR transcripts in the dry microspore of the fern Marsilea vestita, which the authors convincingly showed to be posttranscriptionally spliced in spermatogenesis and used for translation in a development-dependent manner (Boothby et al., 2013). This suggests that IR events, which are common in plants, contribute to the regulation of gene expression during development.
The MB targeting intron 3 of RS2Z33 monitored the distribution of splice variants containing intron 3, including the pre-mRNA. Pre-mRNA, which also contains intron 3, is only present at a low level and can be disregarded for this purpose (Supplemental Figure 3E). These results, therefore, lead to the conclusion that a major part of the observed shift of the distribution fits for MB targeting intron 3 is due to the retention of these splice variants within the cell nucleus. To apply the method to a second endogenous gene in an uninduced system, we transfected wild-type cells with MBs targeting splice variants of the SEF factor. The analysis method only needed a minor adjustment within the reference system to become applicable for wild-type cells. Also in this case, the transcripts with retained introns showed nuclear localization.
It is unknown whether an MB bound to mRNA is dislocated by the translating ribosome or if it stalls the ribosome (Tyagi and Alsmadi, 2004). We observed a stable fluorescent signal for 24 h in the cytoplasm corresponding to a steady state level of the transcript. Only if the MB-bound transcript were partially degraded, leaving the hybrid intact, would an increased signal over time be expected.
Another important question is whether MBs change the subnuclear localization and export of bound transcripts. At least for export, it has been shown that even RNA molecules labeled with much larger tags such as the MS2-Coat protein still can be exported (Grünwald and Singer, 2010). It is important to consider whether large tags lead to increased dwelling times, altered diffusion coefficients, or distorted cellular and subcellular distributions. In comparison, MBs represent a very small label, in fact one of the smallest labels available for imaging endogenous mRNA (Okamoto, 2011). However, it has been shown that for the export function, a complex interplay between mRNP proteins is necessary, especially the fully assembled EJ complex (Schmidt et al., 2009). The EJ complex is located ∼20 nucleotides upstream of the splice junction (Mishler et al., 2008), and the target regions of the MBs used in this study are spatially distinct from these binding sites. Also in our study, comparison of the biochemical and the imaging approaches led to the same results. These findings suggest that the binding of the MB to its targeted EJ is not impairing export functionality.
The mobility of an RNA molecule is an important property in its life cycle and is dependent on the mRNP’s protein composition. Since MBs represent a tool to specifically target RNA splice variants in living cells, we became interested in their dynamics. Are there differences in speed and/or transport between the various transcripts? Since MBs can be detected with a confocal laser scanning setup, we decided to perform FRAP in the nucleoplasm. Interestingly, the FS RNA recovered faster than the AS variants with an IR event, although the intron is only 140 nucleotides long. This finding indicates that retained AS transcripts (IR3) move slower through the nucleoplasm than FS transcripts. Also, the half-time of recovery was faster than expected within a viscous volume but not as fast as free fluorophores are expected to diffuse (Kühn et al., 2011). Studies from animal cells provide evidence for intranuclear dynamics by monitoring bulky MS2- or array-labeled RNA molecules. These studies suggest a passive diffusion of mRNPs, with an average diffusion coefficient of 0.04 to 0.7 µm2/s (Politz et al., 1999; Shav-Tal et al., 2004; Vargas et al., 2005; Shav-Tal and Gruenbaum, 2009; Mor et al., 2010). Through using single MBs, our estimated average diffusion constant (1.6 µm2/s, measured in the uninduced cell population) suggests that endogenous mRNA molecules in living plant cells are moving faster. These differences may be caused by the alternative detection system used (single MB versus MB RNA arrays versus tagged RNA and protein binding). However, further investigations of the intranuclear dynamics of mRNPs monitoring endogenous, unaltered RNA in animal cells are necessary to evaluate the differences in diffusion coefficients in plant cells. The control experiments of the FRAP study also showed an unexpected result, as the FS mRNAs displayed different mobility depending on their origins. The mRNA from the intron-free gene construct moved much slower than the mRNA created through a cotranscriptional splicing event. As the sequence of both mRNAs is identical, the difference in mobility is probably due to the different fates (splicing versus nonsplicing, presence of UTRs, different promoters) they have gone through, which may result in different mRNP packaging and, hence, different mobility.
In conclusion, in vivo RNA imaging and RT-PCR analysis show that the IR-containing and NMD-insensitive transcripts of RS2Z33 and the SEF factor are retained within the nucleus, thus avoiding the NMD machinery. This phenomenon may hold true for other IR events in plants as long as there are no other typical NMD features present that may overrule this decision. The presented method is a simple and easily applicable way of gaining reliable qualitative and quantitative data on the subcellular localization of endogenous mRNA in living plant cells. It creates the possibility of measuring the distribution of splice variants in a multitude of different conditions (e.g., upon drug treatment, cell cycle synchronization, pathogen influence, DNA damage, etc.) and, therefore, represents a powerful tool for future localization, single-particle tracking, and superresolution experiments.
METHODS
Plasmids and Cell Lines
Plasmids for the expression of hemagglutinin (HA)-tagged RS2Z33 in plant protoplasts under the control of a β-estradiol–inducible promoter (Zuo et al., 2000) were created by cloning corresponding coding sequences of cDNA (i.e., without introns or UTRs) into a pER8-derived vector named pMDC7-RS2Z33-HA (hygromycin resistance). Protoplasts of an Arabidopsis thaliana suspension culture from 10-d-old seedlings (accession Columbia-0) have been stably transformed using this construct before (polyethylene glycol transformation according to Lorković et al. [2004]) and selected using 100 µg/mL hygromycin.
Isolation of Arabidopsis Protoplasts
Arabidopsis cell suspension protoplasts (derived from 10-d-old seedlings) were prepared as described by Lorković et al. (2004). Protoplasts were collected 24 h after induction with 20 µM β-estradiol (dissolved in DMSO) or as a control with only DMSO. Two hours after isolation, cells were used for electroporation and for the preparation of fractionated RNA and protein extracts.
Induced cells overexpressed an HA-tagged RS2Z33 protein measurable by immunoblotting using an antibody against HA. Furthermore, in induced cells, the AS variant carrying IR3 was ∼1.49 times more highly expressed than in uninduced cells.
Transfection
Protoplast cells (105) were washed with 0.275 M calcium-nitrate buffer and pelleted in a microcentrifuge (150g for 2 min). For electroporation, 0.7 M mannitol buffer was used mixed with a final concentration of 500 nM specific MB and 500 nM scrambled MB. A final volume of 800 μL in a 4-mm cuvette (Biozym Scientific) was pulsed with the following settings of the device (Easyject Optima; EquiBio): 240 V, 75 µF, two pulses with a duration of ∼7.5 ms and an interval of 20 s. Electroporation conditions for cells used in the FRAP experiments were 280 V, 1050 µF, one pulse with a duration of ∼13.5 ms. Cells were electroporated in a final volume of 800 μL of EP buffer (5 mM CaCl2, 150 mM NaCl, 10 mM HEPES, pH 7.9, and 200 mM d-mannitol) and placed on ice for 4 min. Subsequently, cells were fed with 0.34 M GM buffer (3163 mg/L Gamborg B5 powder including vitamins [Duchefa], 170 mM d-glucose, 170 mM d-mannitol, and 1 mg/L 2,4-D, pH 5.5, adjusted with KOH) and incubated at room temperature, in the dark, for 24 h. Shortly before measurement, cells were washed twice with GM buffer. During measurements, cells were tested for viability using the membrane-impermeable dye To-pro-3-iodide (Invitrogen).
RT-PCR Analysis of Fractionated Cells
Protoplast cells (106) were washed with 0.275 M calcium-nitrate buffer and pelleted in a microcentrifuge (900g for 2 min). The pellet was resuspended in 9 volumes of nuclear isolation buffer (10 mM MES/KOH, pH 5.5, 200 mM Suc, 2.5 mM EDTA, pH 8.0, 2.5 mM DTT, 0.1 mM spermine, 10 mM NaCl, 10 mM KCl, 0.15% Triton X-100, and 5 μ/mL RNasin] and subsequently passed through a 27G syringe until the total disappearance of intact protoplasts was visible by stereo light microscopy. An aliquot was taken resembling the total fraction. Eventually, the residual lysate was spun in a microcentrifuge at 1500g for 10 min at 4°C. The supernatant was recovered and spun again at 15,000g for 15 min at 4°C. The resulting supernatant represents the cytoplasmic fraction and was frozen in liquid nitrogen. The pellet from the first centrifugation step was subject to RNasin and washed three times in nuclear isolation buffer at 4°C before freezing in liquid nitrogen. The resulting pellet represents the nuclear fraction. All steps were performed on ice or at 4°C, if not stated otherwise.
RNA was isolated from the total, cytoplasmic, and nuclear samples via RNA Trizol purification, treated with Turbo-free DNase (Zymo Research), and subsequently purified using the DNA-free RNA purification system (Zymo Research).
Synthesis of cDNA and RT-PCR Analysis of Splice Variants
Total, cytoplasmic, and nuclear RNA was transcribed into cDNA using the RS2Z33-specific primer RS2Z33-exon 5 rev (RT-PCR system; Promega). Subsequently, cDNA was amplified using additional oligonucleotides listed in Supplemental Table 2. Proteins were separated on a 12% SDS-PAGE gel. Detection was performed using a polyclonal antibody against cytosolic fructose-1,6-biphosphatase raised in rabbit (1:5000; Agrisera), a polyclonal antibody against histone-3 raised in rabbit (1:5000; Agrisera), and a polyclonal goat anti-rabbit IgG antibody conjugated to horseradish peroxidase (1:10,000; Agrisera).
MB Design and Optimization
The sequence length was kept constant at 35 to 36 nucleotides to reduce the quenching effects in an open state. The self-complementary stem sequence of seven nucleotides in length had a high GC percentage. Moreover, the stems of the specific MBs differed from the scrambled sequence to prevent cross-linking between the stems.
Since DNA/RNA hybrids are more stable than DNA double-stranded sequences, MBs prefer to bind to target RNA. However, heavy secondary structures of the target RNA can influence the MB hybridization efficiency.
To minimize locus effects, the web-based program RNAup (http://rna.tbi.univie.ac.at/cgi-bin/RNAup.cgi) was used to predict the accessibility of the target region. Since there is no possibility to adjust for ion strength or temperature, this test is a mere estimate for accessibility. The aim was to minimize the opening energy for all MBs. For simulating MB dimer and monomer secondary structures and energies, the DINAmelt web server was used (mfold.rna.albany.edu/?q=DINAMelt). The MB homodimers (low probability) and monomers were designed to build stable structures within the cellular environment. The dimer/monomer energies (monomer binding energy > homodimer binding energy) were kept significantly lower than duplex energies in order to favor MB binding. Furthermore, the secondary structure was adapted to have minimal internal base pairing, and extended stems were avoided. In that way, a closed conformation within the cellular environment should be favored.
The introduced MBs are spectrally separated. For the specific probes, we used a 5′-Atto550 fluorophore and a 3′-BHQ2 quencher, and for the scrambled sequence, we used a 5′-FAM and a 3′-BHQ1. All MBs were ordered from Microsynth Austria. The observed fluorescent signals in the cells were constant over time (up to 24 h). Such behavior indicates an equilibrium state between the monomer/dimer MB (dark states) and RNA-bound structures (fluorescence), which benefits the duplex binding (RNA binding).
The sequences of the MBs can be found in Supplemental Table 2.
Imaging and Analysis
The transfected MBs were imaged with a Zeiss LSM 510 meta microscope equipped with laser light sources (for the analysis of wild-type cells, a Zeiss LSM 710 microscope was used). For our analysis, three laser lines (488, 561, and 633 nm) were used. Wild-type cells did not exhibit any considerable autofluorescence in these channels (Supplemental Figure 4). The distribution of MBs in closed conformation was tested via a dual-labeled MB (FAM and Atto550). When MBs are closed, a FRET signal can be observed. Supplemental Figure 5 demonstrates that MBs in closed conformation are nearly evenly distributed within the nucleus and cytoplasm. To allow a direct comparability between the images, different quantum efficiencies, environmental effects, excitation efficiencies, and laser intensities were taken into account, before a ratio of the images was made. In order to generate a direct ratio image, the fluorescence of the specific channel was corrected for differences in laser intensity and excitation efficiency for the dyes used. The correction of the detection efficiency considers the properties of the optical filters and the detector sensitivity at a particular wavelength. Moreover, we also investigated the influence of microenvironmental changes on fluorophore emission (e.g., quencher efficiencies and fluorescent lifetime changes). The power correction for the LSM 510 microscope was determined via phycoerythrin-doped fluorescent beads (the number of fluorophores is known; BD Biosciences). Phycoerythrin can be excited by both wavelengths, 488 and 561 nm, and has been used to correct for different excitation powers. In additional experiments, the cells were transfected with two MBs identical in sequence (targeting 33mRNA_EJ6) and labeled with either the fluorophore/quencher pair FAM/BHQ1 or Atto550/BHQ-2. The MBs provided information about the differences in dye intensities due to quenching effects and quantum yields. The FAM/BHQ1-labeled probe showed approximately 18% lower intensity in comparison with the Atto550/BHQ2 probe (Supplemental Figure 6). The experiment with MBs targeting EJ6 confirmed as well the difference in excitation efficiency (in total, 58% lower signal in blue than in green). For the LSM 710 microscope, a similar experiment was performed, and a difference of 32% in the detection between the Atto550 and FAM MB signals were determined. The 32% difference includes all parameters: the different excitation power, quantum yield, and quenching efficiency. A crosstalk of the laser excitation between the two channels was not observed.
Subsequently, a ratiometrical display of the cell was created by division of the corrected images from specific and unspecific color channels. The local intensities in the nucleus and cytoplasm were determined by average intensity analysis of the image with a custom-made MatLab-based tool (Supplemental Data Set 1). Moreover, in order to equally level the images, the ratios were background corrected. The background intensity was analyzed inside the vacuole, which did not contain any MBs. The background outside the cells showed higher intensity fluctuations due to varying protoplast washing efficiency.
The analysis of the localization of alternative splice variants in wild-type cells is based on a comparison of intensity ratio distributions between the cytoplasm and nucleus. Such distributions express the relative RNA abundance within the cell. Moreover, the intensities of the nuclear/cytoplasmic fractions are normalized by the signal of a cotransfected scrambled MB. Due to the constant unspecific binding probability of this MB, we corrected for background intensity heterogeneities within the cytoplasm and nucleus. All intensities used in the analysis were background corrected, and the background signal was determined from the vacuole. Effectively, the splice variant distributions are represented by an intensity ratio distribution of normalized RNA abundance within the nuclear or cytoplasmic compartment.
For comparison of the inducible cell system, two populations (induced and uninduced) were compared. For comparison, all individual cytoplasmic and nuclear ratios were displayed in a fitted histogram. The distributions were compared using a parameter-free MW test and a two-sample KS test. The data were analyzed for distribution similarity (null hypothesis) at the 5% significance level. Correction of the images for fluorophore and setup properties and creating a ratio image were conducted with a pixel-by-pixel method, and leveling of the dark count/signal correction and determination of intensity ratios from cell compartments were conducted by an average signal/area method.
MB Kinetics
As expected, the binding kinetics of MBs was slow until a stable equilibrium could be reached. Also, possible energy barriers created by secondary RNA structures can be disregarded, since the incubation time interval is long enough to reach binding equilibrium. An exception to this is the presence of stable structures (e.g., tetraloops). The probability of targeting such regions can be minimized by choosing highly accessible sequence regions via analysis with programs such as RNAup. Since the splicing and processing of RNA is a relatively rapid process (Hoskins et al., 2011a, 2011b), the slow binding kinetics of MBs measures the steady state level of transcript abundance; thus, MB interference with RNA processing can be disregarded. We observed around a 10-fold increase of fluorescent signal in dead cells over living cells (due to DNase activity). We observed a 10.14 ± 1.9 higher average signal obtained from dead cells than from living cells transfected with a specific MB (five dead and five living cells). This observation and the in vivo distribution of a dual-labeled MB led us to the conclusion (Supplemental Figure 5) that unbound MBs within the cell are highly abundant and not rate limiting.
FRAP Analysis
For FRAP analysis, three methods were used for fitting. A monoexponential and biexponential as well as a biexponential Phair fit were applied to the data, and the data were always prenormalized according to Phair et al. (2004). To estimate τ and diffusion coefficients, the best fit result was chosen.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: RS2Z33 (At2g37340), catalase3 (AT1G20620), and SEF factor (At5g37055).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Splice Variants Carrying IR3 Are Upregulated in the Induced RS2Z33 Cell Culture.
Supplemental Figure 2. Localization of 2′-O-Methylated MBs in Comparison with a Scrambled MB.
Supplemental Figure 3. Splice Variants Carrying NMD-Negative but PTC-Positive IR Events Are Present within the Nuclear Fraction.
Supplemental Figure 4. The Autofluorescence of Wild-Type Cells in the Relevant Channels.
Supplemental Figure 5. Distribution of MBs within Transfected Cells.
Supplemental Figure 6. Correction for Quenching Efficiency and Quantum Yields.
Supplemental Table 1. Gene Ontology Biological Process Classification of Genes with NMD-Insensitive IR Events.
Supplemental Table 2. Sequences of the Oligonucleotides and MBs.
Supplemental Data Set 1. MatLab Script for Power Correction.
Supplementary Material
Acknowledgments
We thank Nick Fulcher for helpful comments and for carefully editing the manuscript, Olga Bannikova for advice on plant tissue culture, and the members of the laboratory for fruitful discussions. This work was supported by the Austrian Science Fund (Grants DK W1207 and SFB RNA-REG F43-P10 to A.B.), the Austria Genomic Program, and the European Union FP6 Programme Network of Excellence on Alternative Splicing (Grant LSHG-CT-2005-518238).
AUTHOR CONTRIBUTIONS
J.G. and J.J. designed and performed the research and analyzed data. J.J. contributed new computational tools. A.B. guided the project. All authors wrote the article.
Glossary
- AS
alternative splicing
- PTC
premature termination codon
- NMD
nonsense-mediated decay
- UTR
untranslated region
- IR
intron retention
- MB
molecular beacon
- FRAP
fluorescence recovery after photobleaching
- FS
fully spliced
- KS
Kolmogorow-Smirnow
- MW
Mann-Whitney
- 3′ss
3′ splice site
- mRNP
messenger ribonucleoprotein
- EJ
exon junction
- HA
hemagglutinin
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Bond C.S., Fox A.H. (2009). Paraspeckles: Nuclear bodies built on long noncoding RNA. J. Cell Biol. 186: 637–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boothby T.C., Zipper R.S., van der Weele C.M., Wolniak S.M. (2013). Removal of retained introns regulates translation in the rapidly developing gametophyte of Marsilea vestita. Dev. Cell 24: 517–529. [DOI] [PubMed] [Google Scholar]
- Bratu D.P., Cha B.J., Mhlanga M.M., Kramer F.R., Tyagi S. (2003). Visualizing the distribution and transport of mRNAs in living cells. Proc. Natl. Acad. Sci. USA 100: 13308–13313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A.K., Behlke M.A., Tsourkas A. (2007). Avoiding false-positive signals with nuclease-vulnerable molecular beacons in single living cells. Nucleic Acids Res. 35: e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A.K., Behlke M.A., Tsourkas A. (2008). Efficient cytosolic delivery of molecular beacon conjugates and flow cytometric analysis of target RNA. Nucleic Acids Res. 36: e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K., Kim J., Hwang H.J., Kim S., Park C., Kim S.Y., Lee I. (2011). The FRIGIDA complex activates transcription of FLC, a strong flowering repressor in Arabidopsis, by recruiting chromatin modification factors. Plant Cell 23: 289–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen N.M., Oparka K.J., Tilsner J. (2010). Advances in imaging RNA in plants. Trends Plant Sci. 15: 196–203. [DOI] [PubMed] [Google Scholar]
- Daigle N., Ellenberg J. (2007). LambdaN-GFP: An RNA reporter system for live-cell imaging. Nat. Methods 4: 633–636. [DOI] [PubMed] [Google Scholar]
- Deal R.B., Topp C.N., McKinney E.C., Meagher R.B. (2007). Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z. Plant Cell 19: 74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lima Morais D.A., Harrison P.M. (2010). Large-scale evidence for conservation of NMD candidature across mammals. PLoS ONE 5: e11695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichkin S.A., Priest H.D., Givan S.A., Shen R., Bryant D.W., Fox S.E., Wong W.K., Mockler T.C. (2010). Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 20: 45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco D., Accornero N., Lavoie B., Shenoy S.M., Blanchard J.M., Singer R.H., Bertrand E. (2003). Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr. Biol. 13: 161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünwald D., Singer R.H. (2010). In vivo imaging of labelled endogenous β-actin mRNA during nucleocytoplasmic transport. Nature 467: 604–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins A.A., Friedman L.J., Gallagher S.S., Crawford D.J., Anderson E.G., Wombacher R., Ramirez N., Cornish V.W., Gelles J., Moore M.J. (2011b). Ordered and dynamic assembly of single spliceosomes. Science 331: 1289–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins A.A., Gelles J., Moore M.J. (2011a). New insights into the spliceosome by single molecule fluorescence microscopy. Curr. Opin. Chem. Biol. 15: 864–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Meyerowitz E.M. (2010). Cell-type specific analysis of translating RNAs in developing flowers reveals new levels of control. Mol. Syst. Biol. 6: 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyna M., Lopato S., Barta A. (2003). Ectopic expression of atRSZ33 reveals its function in splicing and causes pleiotropic changes in development. Mol. Biol. Cell 14: 3565–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyna M., et al. (2012). Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 40: 2454–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Koroleva O.A., Lewandowska D., Pendle A.F., Clark G.P., Simpson C.G., Shaw P.J., Brown J.W. (2009). Aberrant mRNA transcripts and the nonsense-mediated decay proteins UPF2 and UPF3 are enriched in the Arabidopsis nucleolus. Plant Cell 21: 2045–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn T., Ihalainen T.O., Hyväluoma J., Dross N., Willman S.F., Langowski J., Vihinen-Ranta M., Timonen J. (2011). Protein diffusion in mammalian cell cytoplasm. PLoS ONE 6: e22962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leviatan N., Alkan N., Leshkowitz D., Fluhr R. (2013). Genome-wide survey of cold stress regulated alternative splicing in Arabidopsis thaliana with tiling microarray. PLoS ONE 8: e66511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopato S., Forstner C., Kalyna M., Hilscher J., Langhammer U., Indrapichate K., Lorković Z.J., Barta A. (2002). Network of interactions of a novel plant-specific Arg/Ser-rich protein, atRSZ33, with atSC35-like splicing factors. J. Biol. Chem. 277: 39989–39998. [DOI] [PubMed] [Google Scholar]
- Lorković Z.J., Hilscher J., Barta A. (2004). Use of fluorescent protein tags to study nuclear organization of the spliceosomal machinery in transiently transformed living plant cells. Mol. Biol. Cell 15: 3233–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez Y., Brown J.W., Simpson C., Barta A., Kalyna M. (2012). Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 22: 1184–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhamdi A., Noctor G., Baker A. (2012). Plant catalases: Peroxisomal redox guardians. Arch. Biochem. Biophys. 525: 181–194. [DOI] [PubMed] [Google Scholar]
- Mishler D.M., Christ A.B., Steitz J.A. (2008). Flexibility in the site of exon junction complex deposition revealed by functional group and RNA secondary structure alterations in the splicing substrate. RNA 14: 2657–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor A., Suliman S., Ben-Yishay R., Yunger S., Brody Y., Shav-Tal Y. (2010). Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat. Cell Biol. 12: 543–552. [DOI] [PubMed] [Google Scholar]
- Ner-Gaon H., Halachmi R., Savaldi-Goldstein S., Rubin E., Ophir R., Fluhr R. (2004). Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J. 39: 877–885. [DOI] [PubMed] [Google Scholar]
- Nicholson P., Mühlemann O. (2010). Cutting the nonsense: The degradation of PTC-containing mRNAs. Biochem. Soc. Trans. 38: 1615–1620. [DOI] [PubMed] [Google Scholar]
- Okabe K., Harada Y., Zhang J., Tadakuma H., Tani T., Funatsu T. (2011). Real time monitoring of endogenous cytoplasmic mRNA using linear antisense 2′-O-methyl RNA probes in living cells. Nucleic Acids Res. 39: e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto A. (2011). ECHO probes: A concept of fluorescence control for practical nucleic acid sensing. Chem. Soc. Rev. 40: 5815–5828. [DOI] [PubMed] [Google Scholar]
- Ozawa T., Natori Y., Sato M., Umezawa Y. (2007). Imaging dynamics of endogenous mitochondrial RNA in single living cells. Nat. Methods 4: 413–419. [DOI] [PubMed] [Google Scholar]
- Phair R.D., Gorski S.A., Misteli T. (2004). Measurement of dynamic protein binding to chromatin in vivo, using photobleaching microscopy. Methods Enzymol. 375: 393–414. [DOI] [PubMed] [Google Scholar]
- Politz J.C., Tuft R.A., Pederson T., Singer R.H. (1999). Movement of nuclear poly(A) RNA throughout the interchromatin space in living cells. Curr. Biol. 9: 285–291. [DOI] [PubMed] [Google Scholar]
- Reddy A.S., Day I.S., Göhring J., Barta A. (2012). Localization and dynamics of nuclear speckles in plants. Plant Physiol. 158: 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo P.J., Lifland A.W., Curt P., Sasaki Y., Bassell G.J., Lindquist M.E., Crowe J.E., Jr (2009). Single molecule-sensitive probes for imaging RNA in live cells. Nat. Methods 6: 347–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayani S., Janis M., Lee C.Y., Toesca I., Chanfreau G.F. (2008). Widespread impact of nonsense-mediated mRNA decay on the yeast intronome. Mol. Cell 31: 360–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt U., Im K.B., Benzing C., Janjetovic S., Rippe K., Lichter P., Wachsmuth M. (2009). Assembly and mobility of exon-exon junction complexes in living cells. RNA 15: 862–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal Y., Darzacq X., Shenoy S.M., Fusco D., Janicki S.M., Spector D.L., Singer R.H. (2004). Dynamics of single mRNPs in nuclei of living cells. Science 304: 1797–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal Y., Gruenbaum Y. (2009). Single-molecule dynamics of nuclear mRNA. F1000 Biol Rep 1: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson C.G., Fuller J., Maronova M., Kalyna M., Davidson D., McNicol J., Barta A., Brown J.W. (2008). Monitoring changes in alternative precursor messenger RNA splicing in multiple gene transcripts. Plant J. 53: 1035–1048. [DOI] [PubMed] [Google Scholar]
- Sokol D.L., Zhang X., Lu P., Gewirtz A.M. (1998). Real time detection of DNA.RNA hybridization in living cells. Proc. Natl. Acad. Sci. USA 95: 11538–11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockley P.G., Stonehouse N.J., Murray J.B., Goodman S.T., Talbot S.J., Adams C.J., Liljas L., Valegård K. (1995). Probing sequence-specific RNA recognition by the bacteriophage MS2 coat protein. Nucleic Acids Res. 23: 2512–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi S., Alsmadi O. (2004). Imaging native beta-actin mRNA in motile fibroblasts. Biophys. J. 87: 4153–4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi S., Kramer F.R. (1996). Molecular beacons: Probes that fluoresce upon hybridization. Nat. Biotechnol. 14: 303–308. [DOI] [PubMed] [Google Scholar]
- Vargas D.Y., Raj A., Marras S.A., Kramer F.R., Tyagi S. (2005). Mechanism of mRNA transport in the nucleus. Proc. Natl. Acad. Sci. USA 102: 17008–17013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walder R.Y., Walder J.A. (1988). Role of RNase H in hybrid-arrested translation by antisense oligonucleotides. Proc. Natl. Acad. Sci. USA 85: 5011–5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J., Niu Q.W., Chua N.H. (2000). Technical advance. An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24: 265–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.