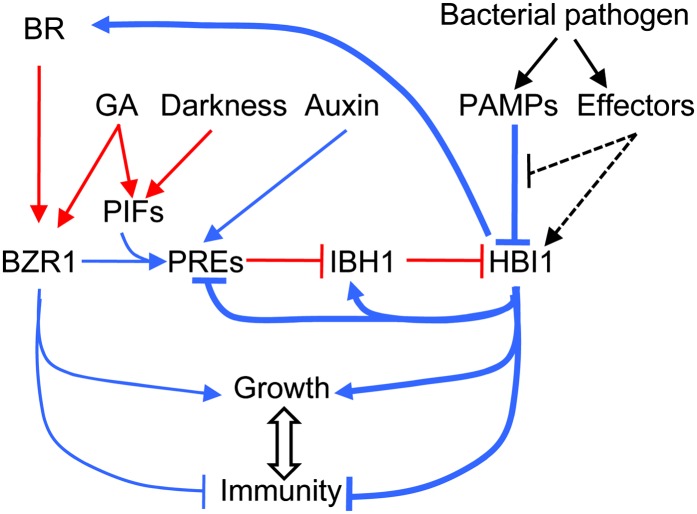
HBI1 is activated posttranscriptionally by growth-promoting hormonal and environmental signals through the triple-HLH/bHLH cascade but is repressed transcriptionally by pathogen-associated molecular pattern signals. HBI1 both activates growth and inhibits immunity, thereby acting as a crosstalk node that mediates the trade-off between growth and immunity.
Abstract
The trade-off between growth and immunity is crucial for survival in plants. However, the mechanism underlying growth-immunity balance has remained elusive. The PRE-IBH1-HBI1 tripartite helix-loop-helix/basic helix-loop-helix module is part of a central transcription network that mediates growth regulation by several hormonal and environmental signals. Here, genome-wide analyses of HBI1 target genes show that HBI1 regulates both overlapping and unique targets compared with other DNA binding components of the network in Arabidopsis thaliana, supporting a role in specifying network outputs and fine-tuning feedback regulation. Furthermore, HBI1 negatively regulates a subset of genes involved in immunity, and pathogen-associated molecular pattern (PAMP) signals repress HBI1 transcription. Constitutive overexpression and loss-of-function experiments show that HBI1 inhibits PAMP-induced growth arrest, defense gene expression, reactive oxygen species production, and resistance to pathogen. These results show that HBI1, as a component of the central growth regulation circuit, functions as a major node of crosstalk that mediates a trade-off between growth and immunity in plants.
INTRODUCTION
The trade-off between growth and immunity is crucial for optimal survival of plants in nature and is also important for agricultural productivity of crops. This trade-off is believed to require complex interactions between signal transduction pathways activated by growth signals and pathogen-generated signals (Robert-Seilaniantz et al., 2011). Plant growth is regulated by a wide range of signals, including endogenous hormones and environmental cues, such as light, temperature, and the presence of pathogens. These hormonal and environmental signals act through distinct signal transduction pathways, which have been studied extensively. Interactions between these pathways have also been observed at the molecular level (Depuydt and Hardtke, 2011), but the key molecular junctions regulated by both hormone and defense signaling pathways has remained elusive.
Brassinosteroids (BRs) are a group of growth-promoting hormones that regulate many developmental responses and also modulate immunity. BRs act through a well-defined signal transduction pathway (Kim and Wang, 2010; Wang et al., 2012). BRs directly interact with the extracellular domains of the receptor kinases BRASSINOSTEROID-INSENSITIVE1 (BRI1) and SOMATIC EMBRYOGENESIS RECEPTOR KINASE1 to induce their dimerization and trans-phosphorylation (Li et al., 2002; Nam and Li, 2002; Wang et al., 2008; Santiago et al., 2013). Activated BRI1 phosphorylates downstream receptor-like cytoplasmic kinase proteins BR-SIGNALING KINASE (BSK) and CONSTITUTIVE DIFFERENTIAL GROWTH1, which in turn phosphorylate and activate members of the BRI1 SUPPRESSOR1 (BSU1) family of phosphatases (Tang et al., 2008; Kim et al., 2009, 2011). BSU1 inactivates the GSK3-like kinase BRASSINOSTEROID-INSENSITIVE2 (BIN2) through Tyr dephosphorylation (Kim et al., 2009), leading to activation of BIN2’s substrates BRASSINAZOLE-RESISTANT1 (BZR1) and BZR2 (also named BRI1-EMS-SUPPRESSOR1 [BES1]) by PP2A-mediated dephosphorylation (Tang et al., 2011). Dephosphorylated BZR1 and BZR2 translocate into the nucleus to regulate the expression of BR response genes (Sun et al., 2010; Yu et al., 2011).
The transcriptional activity of BZR1 depends on its interactions with proteins regulated by other signaling pathways, including the PHYTOCHROME-INTERACTING FACTORs (PIFs) regulated by light, temperature, and circadian clock, and the DELLA proteins regulated by gibberellin (GA) (Bai et al., 2012b; Oh et al., 2012). BZR1 interacts with PIFs, which accumulate when plants are in the dark or shade, at promoters of common target genes to activate gene expression and promote cell elongation (Oh et al., 2012). The DELLA proteins, which accumulate when GA levels are low (Sun, 2011), inhibit the DNA binding activities of both BZR1 and PIFs (de Lucas et al., 2008; Feng et al., 2008; Bai et al., 2012b; Gallego-Bartolomé et al., 2012; Li et al., 2012). Therefore, the direct interactions among BZR1, PIF4, and DELLA integrate BR, phytochrome, and GA signals to regulate plant growth (Bai et al., 2012b; Oh et al., 2012; Wang et al., 2012).
The promotion of cell elongation by the BZR1-PIF4 module requires the PRE-IBH1-HBI1 tripartite helix-loop-helix/basic helix-loop-helix (HLH/bHLH) module. BZR1-PIF4 transcriptionally activates members of the PACLOBUTRAZOL-RESISTANT (PRE) family of non-DNA binding HLH factors, which sequester several HLH factors that otherwise inhibit DNA binding bHLH factors (Bai et al., 2012a; Oh et al., 2012). For example, PRE1 and PRE6/KIDARI interact with HFR1 and PAR1, which both inhibit PIF4 DNA binding, forming positive feedback loops (Hyun and Lee, 2006; Hornitschek et al., 2009; Hao et al., 2012). PRE1 also binds to IBH1 to prevent its inhibition of the bHLH factors HBI1 and ACTIVATORS FOR CELL ELONGATION (ACE1 to ACE3) (Bai et al., 2012a; Ikeda et al., 2012). Together, the BZR-PIF4 and HLH/bHLH modules form a central growth regulation transcription network that integrates hormonal and environmental signals. While genetic evidence supports an important role for HBI1 in promoting cell elongation (Bai et al., 2012a), the molecular functions of HBI1 in integrating signals into the network and specifying output remain unclear.
Pathogen-associated molecular pattern (PAMP) signals and growth-promoting hormones are known to antagonize each other to mediate the trade-off between growth and immunity, which is important for plant survival (Robert-Seilaniantz et al., 2011). The growth-promoting hormones BR and auxin inhibit PAMP-triggered immunity (PTI), and PAMPs (e.g., flagellin and elongation factor peptides flg22 and elf18, respectively) inhibit plant growth. Interactions between the BR and flagellin pathways have been studied extensively (Chinchilla et al., 2007; Kemmerling et al., 2007; Albrecht et al., 2012; Belkhadir et al., 2012; Lin et al., 2013; Shi et al., 2013). The BR receptor kinase BRI1 and the flagellin receptor kinase FLS2 share the coreceptor BAK1 and the substrates BSK1 and BOTRYTIS-INDUCED KINASE1 (BIK1) (Chinchilla et al., 2007; Kemmerling et al., 2007; Lin et al., 2013; Shi et al., 2013). BIK1 can be phosphorylated by both FLS2-BAK1 and BRI1, and positively regulates flagellin signaling but negatively regulates BR signaling (Lu et al., 2010; Lin et al., 2013). The functional importance of these shared components in the communication between the two pathways remains controversial. While BRI1 was shown to modulate FLS2 signaling through both BAK1-dependent and BAK1-independent mechanisms, BR does not affect the formation of the flg22-triggered FLS2-BAK1 complex (Albrecht et al., 2012; Belkhadir et al., 2012; Lozano-Duran et al., 2013). Furthermore, the weak effects of BR on flg22-induced BIK1 phosphorylation suggested that major crosstalk between the BR and FLS2 pathways occurs downstream of the membrane-bound kinases (Albrecht et al., 2012). A recent study reported that the activated BZR1 associates with WRKY40 to mediate repression of immune responses (Lozano-Duran et al., 2013). However, BES1/BZR2 was shown to be unaffected by PAMP signals (Albrecht et al., 2012), although whether BZR1 is affected by PAMPs remains unclear. Therefore, the major junction regulated by both PAMP and BR pathways might be downstream of BZR1.
Here, we performed chromatin immunoprecipitation followed by sequencing (ChIP-Seq) and RNA sequencing (RNA-Seq) experiments to identify the target genes of HBI1 in the Arabidopsis thaliana genome. The results show that HBI1 has overlapping functions with PIFs in activating genes involved in cell elongation but distinct functions in feedback regulation of the HLH/bHLH network and in regulating chloroplast function and immune responses. The expression of HBI1 and several homologs is repressed by PAMP signals, constitutive overexpression of HBI1 reduces PAMP-induced immune responses, and knockdown of HBI1 expression increases the resistance to bacterial infection. This study demonstrates that HBI1, as a key node of the central growth regulation network, mediates the integration of hormonal, environmental, and pathogen signals and plays a key role in the trade-off between immunity and growth.
RESULTS
Genome-Wide Identification of HBI1 Binding and Regulated Genes
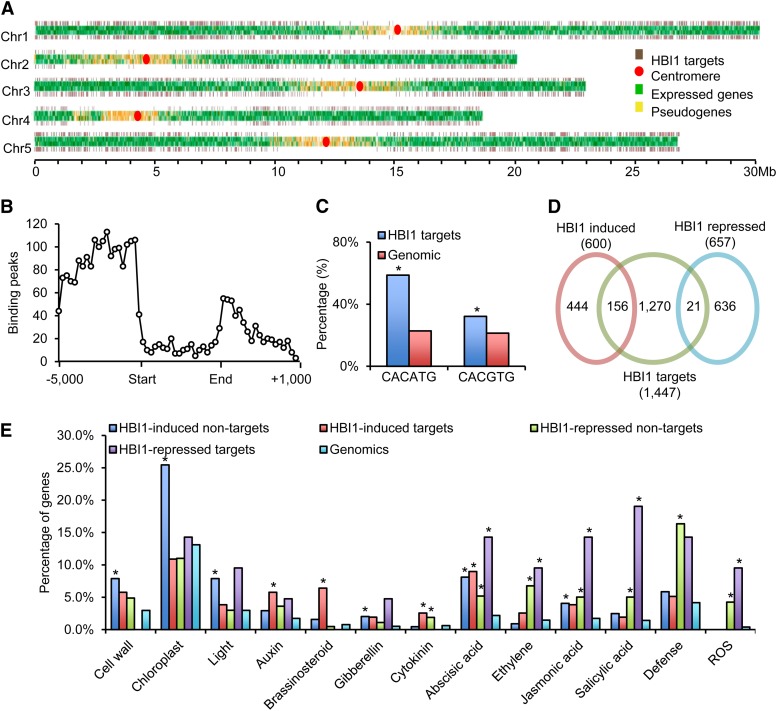
To understand the functions of HBI1, we mapped HBI1’s genomic binding sites using ChIP-Seq. Transgenic Arabidopsis plants expressing the HBI1 and yellow fluorescent protein (YFP) fusion protein driven by the native HBI1 promoter were used to carry out ChIP-Seq experiments with an anti-YFP antibody. A 35S:YFP transgenic line was used as a negative control. Analysis of the ChIP-Seq data with the statistical software CisGenome and PRI-CAT identified 1477 and 1851 HBI1 binding peaks, respectively. Among them, 1103 peaks were identified by both statistical methods and thus considered high-confidence HBI1 binding peaks and used for further analysis. The 1103 HBI1 binding peaks were linked to 1447 neighbor genes that were considered high-confidence HBI1 target genes (Figure 1A; Supplemental Data Set 1). The HBI1 target genes are distributed throughout the genome but are rare in the centromere regions (Figure 1A). Most of the HBI1 binding peaks are in the promoter regions (Figure 1B), consistent with HBI1’s molecular function as a transcription factor. A motif analysis showed that CACATG, the hormone up at dawn element (Michael et al., 2008), was the most enriched cis-element in the HBI1 binding sites, and the G-box motif (CACGTG) was also enriched but to a lesser degree (Figure 1C).
Figure 1.
Genome-wide Identification of HBI1 Binding and Regulated Genes.
(A) Distribution of HBI1 binding sites along the five chromosomes of Arabidopsis. The HBI1 target genes are depicted by brown bars, normal expressed genes are depicted by green bars, and pseudogenes are depicted by yellow bars. A red circle indicates the location of the centromere.
(B) Distribution of HBI1 binding peaks (frequency) relative to gene structure (−5 kb to +1 kb downstream of 3′ end).
(C) Frequency of shown cis-elements around HBI1 binding regions. Asterisk indicates significant difference from random genome (Fisher’s exact test; *P < 0.05).
(D) Venn diagram showing the overlap between the HBI1-regulated genes and HBI1 direct target genes.
(E) Gene Ontology analyses of HBI1 directly and indirectly regulated genes. Numbers indicate the percentages of genes belonging to each Gene Ontology category. Asterisk indicates significant difference from random genome (Fisher’s exact test; *P < 0.05).
To further define the HBI1-regulated genes, we performed RNA-Seq analysis of the transcriptomes of wild-type and transgenic Arabidopsis plants overexpressing HBI1 (HBI1-Ox). Plants were grown on half-strength Murashige and Skoog (MS) medium with 1% Suc under constant light for 5 d. RNA-Seq analysis identified 1257 genes that were affected >1.5-fold in HBI1-Ox compared with the wild type (Figure 1D; Supplemental Data Set 2). Quantitative RT-PCR analyses of 14 genes confirmed the gene expression changes identified by RNA-Seq (Supplemental Table 1). Among the HBI1-regulated genes, 156 out of 600 (26%) HBI1-induced genes and 21 out of 657 (3.2%) HBI1-repressed genes are HBI1 binding targets identified in the ChIP-Seq experiment, suggesting that HBI1 is a transcription activator for most of its target genes. These 177 HBI1-regulated genes were considered functional targets of HBI1 (Figure 1D; Supplemental Data Set 2). The overlap between HBI1-affected genes and the HBI1 binding target genes seems small and may be due to different tissues used in the experiments; however, similar levels of overlap has been reported for other transcription factors (Yu et al., 2011).
Functional classification of the HBI1 binding (i.e., target) and/or HBI1-regulated (i.e., induced or repressed) genes based on Gene Ontology categories showed that HBI1 directly and indirectly regulates a range of biological processes and cellular activities (Figure 1E; Supplemental Figure 1). For example, the genes involved in cell growth and chloroplast function were highly enriched in HBI1-induced, non-HBI1 targets. The genes involved in light response were enriched in HBI1-induced, HBI1 target, and nontarget genes. The genes involved in response to growth-promoting hormones such as BR and GA were enriched in HBI1-activated HBI1 target genes. By contrast, the genes involved in responses to stress hormones, including ethylene, jasmonic acid, and salicylic acid, were enriched in HBI1-repressed HBI1 target genes. The genes involved in auxin and abscisic acid response were enriched in both HBI1-activated and HBI1-repressed genes. In addition, the genes involved in defense and reactive oxygen species (ROS) production were highly enriched in HBI1-repressed, HBI1 targets, and nontarget genes, suggesting that HBI1 plays a role in repressing the plant defense response (Figure 1E; Supplemental Figure 1).
HBI1 activates, mostly directly, the growth-inhibiting HLH factors, such as IBH1, AIF1, AIF2, AIF3, AIF4, UPB1, PAR1, PAR2, HFR1, and three additional IBH1 homologs (At4G30180, At5G57780, and At4G30410) (Supplemental Figures 1, 2A, and 2B and Supplemental Data Set 2). A number of DNA binding bHLH factors are repressed by HBI1-Ox. Such extensive upregulation of the inhibitory HLH factors, many of which bind to and inactivate HBI1 and/or PIFs, suggests that a general feedback mechanism is built into the HLH/bHLH network to provide buffering function to the system.
Several gene families, many involved in defense and redox regulation, are consistently repressed by HBI1 overexpression (Supplemental Figure 1). These include about 31 cytochrome P450 genes, seven WRKY transcription factors, four VQ motif-containing proteins, three U-box proteins, four calmodulin-like proteins, nine FAD binding Berberine family proteins, 11 glutathione S-transferases, and nine members of the 2-oxoglutarate and Fe(II)-dependent oxygenase superfamily (Supplemental Figure 1). In addition, HBI1 activates four (GASA3, 4, 6, and 7) and represses one (GASA5) of the 14 members of the GASA gene family (Roxrud et al., 2007), three of them are direct target genes of HBI1 (Supplemental Figure 1 and Supplemental Data Set 2). The GASA genes encode secreted Cys-rich proteins that have been implicated in development, stress response, and inhibiting ROS (Ko et al., 2007; Rubinovich and Weiss, 2010; Nahirñak et al., 2012; Sun et al., 2013).
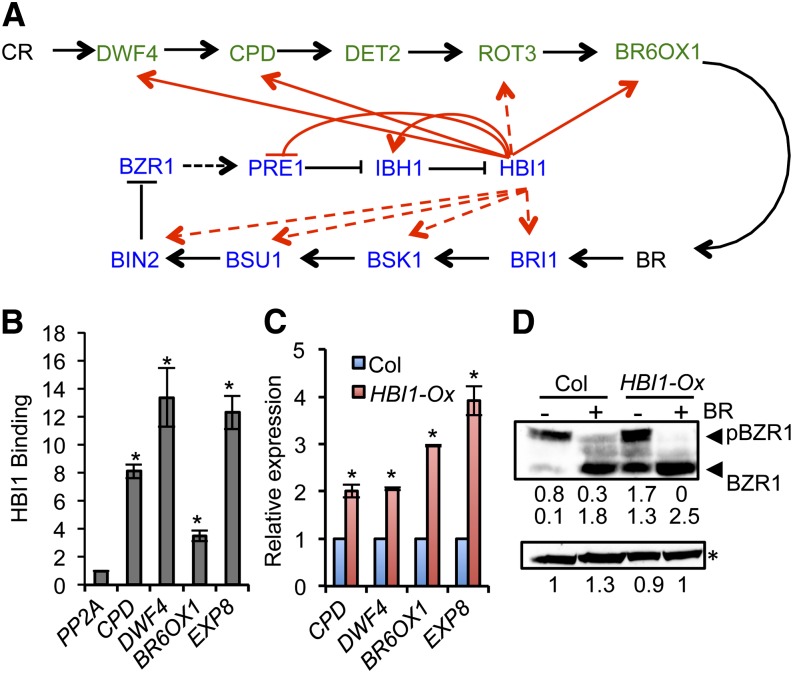
A previous study showed that HBI1 is a positive regulator of BR responses (Bai et al., 2012a). The ChIP-Seq results showed that many genes encoding BR biosynthetic and signaling components are HBI1 targets (Figure 2A). Quantitative ChIP-PCR and RT-PCR confirmed that HBI1 directly activates the expression of the BR biosynthetic genes CPD, DWF4, and BR6OX1 (Figures 2B and 2C), suggesting HBI1 positively regulates BR biosynthesis. Consistent with the positive effects of HBI1-Ox on BR biosynthesis and signaling, the transgenic plants overexpressing HBI1 showed increased BZR1 accumulation and dephosphorylation (Figure 2D).
Figure 2.
HBI1 Positively Regulates Components of the BR Pathway.
(A) A diagram of BR pathway. The BR biosynthetic enzymes are shown in green, and the BR signaling components are in blue. Black arrows and bar ends show activation and inhibition at the protein level. Red arrows show HBI1 binding to the promoter of these genes, with solid lines indicating HBI1 activation and dashed lines indicating no evidence for HBI1 regulation according to our RNA-Seq data.
(B) Quantitative ChIP-PCR analysis of HBI1 binding to the promoter of selected genes. The chromatin of pHBI1:HBI1-YFP and 35S:YFP transgenic plants was immunoprecipitated with anti-YFP antibody, and the precipitated DNA was quantified by quantitative PCR. Enrichment of DNA was calculated as the ratio between pHBI1:HBI1-YFP and 35S:YFP, normalized to that of the PP2A coding region. Error bars indicate standard deviation of three biological repeats. Asterisk indicates significant difference from control gene PP2A (t test; *P < 0.05).
(C) qRT-PCR analysis of the expression of BR biosynthetic genes in the wild-type (Columbia [Col]) and HBI1-Ox. Error bars indicate standard deviation of three biological repeats. Asterisk indicates significant difference from wild-type control (t test; *P < 0.05).
(D) Anti-BZR1 immunoblot analysis of BZR1 phosphorylation status in 4-week-old plants. The relative band intensity was quantified by ImageJ software and labeled under the gel. Asterisk indicates the nonspecific bands to show equal loading. Experiment was repeated four times with similar results.
HBI1 and PIF Have Overlapping and Distinct Functions
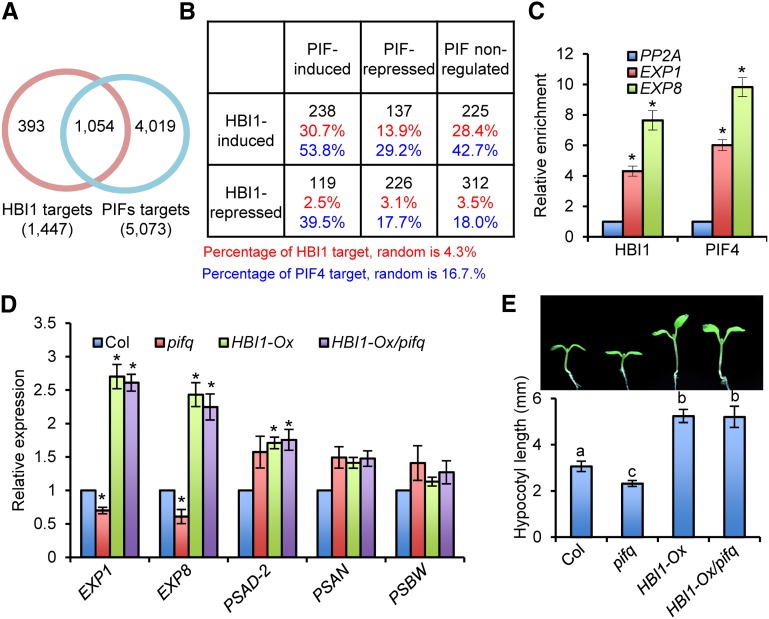
Previous studies showed that HBI1 and PIFs promote cell elongation and bind to E-box and G-box elements (Bai et al., 2012a; Oh et al., 2012). To understand the relationship between these transcription factors, we compared the genomic targets of HBI1 to those of PIF1, PIF3, PIF4, or PIF5 (Oh et al., 2009, 2012; Hornitschek et al., 2012; Zhang et al., 2013). Of the 1477 HBI1 target genes, 1054 (71.4%) were also targets of at least one of the four PIFs (Figure 3A). A comparison between the HBI1-regulated genes and the PIF-regulated genes identified based on differential expression in the pifq mutant (pif1 pif3 pif4 pif5) (Oh et al., 2012) showed that HBI1 and PIFs coregulate 720 genes, which include 464 genes (64.4%) regulated in the same direction and 256 (35.6%) in opposite direction (Figure 3B).
Figure 3.
HBI1 and PIF Have Overlapping and Distinct Functions.
(A) Venn diagram showing the overlap between HBI1 target genes and PIF target genes. The PIF target genes include the genes associated with PIF1, PIF3, PIF4, or PIF5.
(B) The table shows the overlap of HBI1-regulated genes and PIF-regulated genes and the percentage of HBI1 target genes or PIFs target genes among the gene sets that HBI1 and/or PIFs regulate. The top black numbers are the numbers of genes regulated by HBI1 and/or PIFs, the middle red numbers are the percentage of HBI1 targets, and the bottom blue numbers are the percentage of PIF targets.
(C) ChIP-qPCR analysis of HBI1 and PIF4 binding to the promoters of selected genes. Chromatin immunoprecipitation was performed with anti-YFP antibody or anti-Myc antibody using pHBI1:HBI1-YFP, 35S:YFP, pPIF4:PIF4-myc/pifq, and pifq plants grown in the dark for 5 d. Enrichment of DNA was calculated as the ratio between pHBI1:HBI1-YFP and 35S:YFP, or pPIF4:PIF4-myc/pifq and pifq, normalized to that of the PP2A coding region as the internal control. Error bars indicate standard deviation of three biological repeats. Asterisks indicate significant difference from control gene PP2A (t test; *P < 0.05).
(D) qRT-PCR analyses of EXP1, EXP8, PSAD-2, PSAN, and PSBW mRNA levels in wild-type (Col), pifq, and HBI1-Ox/pifq plants. PP2A was used as the internal control. Error bars indicate standard deviation from three biological repeats. Asterisks indicate significant difference from the wild type (t test; *P < 0.05).
(E) Overexpression of HBI1 rescues the dwarf phenotype of pifq. The top picture shows Columbia, pifq, HBI1-Ox, and HBI1-Ox/pifq grown for 7 d under constant light. Bottom graph shows the quantification of hypocotyl lengths. Error bars indicate standard deviation from 20 biological repeats. Different letters indicate statistically significant differences between the samples (t test, P < 0.05).
The genes activated by both HBI1 and PIFs include a high percentage of direct targets of HBI1 (30.7%) and PIFs (53.8%), whereas genes repressed by both HBI1 and PIFs showed the random probability of being direct targets, consistent with both HBI1 and PIFs acting mostly as transcription activators (Figure 3B). A significant portion of the genes activated by HBI1 but not affected by PIFs were also targets of HBI1 (28.4%) and PIFs (42.7%) (Figure 3B). These genes might be regulated by HBI1 and PIFs in an additive or redundant manner and thus are not affected in the pifq mutant.
Gene Ontology analysis showed that the genes activated by both HBI1 and PIFs are highly enriched with genes involved in BR and GA responses and cell elongation, which is consistent with HBI1’s function in promoting cell elongation downstream of these hormone pathways (Supplemental Figure 3A). Chromatin immunoprecipitation–quantitative PCR (ChIP-qPCR) assays confirmed that both HBI1 and PIF4 bind to the promoters of EXP1 and EXP8 (Figure 3C). Quantitative RT-PCR analysis showed that the expression levels of EXP1 and EXP8 were reduced in the pifq mutant but increased by overexpression of HBI1 (Figure 3D). Consistent with the levels of expansin gene expression, the pifq mutant had shorter hypocotyls, but this defect was more than rescued by overexpression of HBI1 (Figure 3E). Overexpression of HBI1 also rescued additional phenotypes of the pifq mutant, including dwarfism and deetiolation in the dark (Supplemental Figure 3B). These results suggest that HBI1 and PIFs have interchangeable biochemical activities in regulating plant growth and photomorphogenesis.
On the other hand, the 226 genes regulated in opposite ways by HBI1 and PIFs suggest that HBI1 also has unique functions in a subset of responses (Supplemental Data Set 2). For example, the genes involved in light response and chloroplast/photosynthesis, such as PSAD-2, PSAN, and PSBW, are enriched in the HBI1-induced but PIF-repressed gene class (Figure 3D; Supplemental Figure 1). Genes involved in defense, salicylic acid–dependent responses, and ethylene-dependent responses are enriched in the gene sets that are repressed by HBI1 but unaffected or affected by PIFs in complex ways (Supplemental Figure 3A).
HBI1 Is a Negative Regulator for Flagellin-Regulated Gene Expression
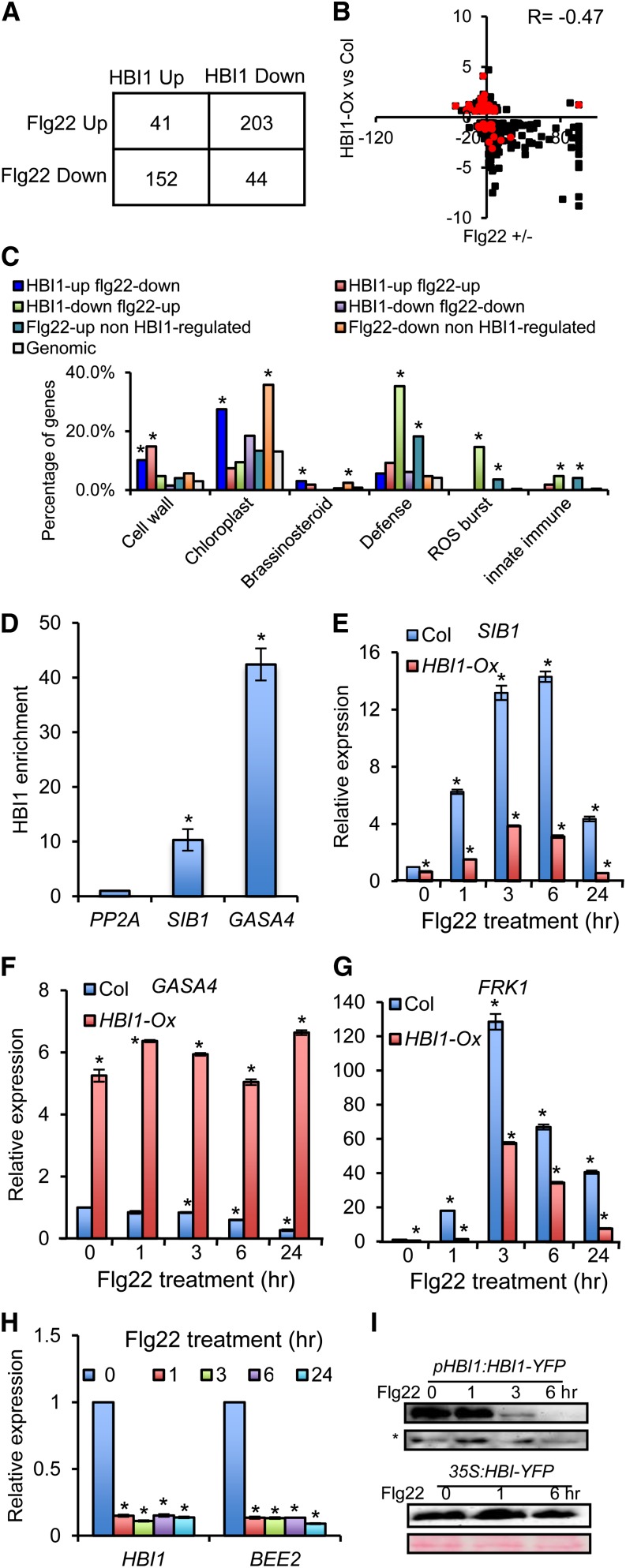
The HBI1 inhibition of genes involved in defense responses prompted us to further analyze the relationship between HBI1 and immunity pathways. A comparison of the HBI1-Ox RNA-Seq data with the previously identified microarray data sets of genes affected by flg22 (Denoux et al., 2008) showed that 440 (35%) of the HBI1-regulated genes were affected by flg22, of which 355 genes (80.7%) were regulated by HBI1 and flg22 in opposite directions, with a correlation coefficient R = −0.47 (Figures 4A and 4B). Similar negative correlation was found between HBI1-mediated gene expression changes and those caused by another PAMP signal, elf18 (Tintor et al., 2013) (Supplemental Figures 4A and 4B). These data suggest that HBI1 negatively regulates a subset of PAMP-induced defense genes.
Figure 4.
HBI1 and Flg22 Oppositely Regulate Gene Expression.
(A) Overlaps between gene sets regulated by HBI1 and flg22.
(B) Scatterplot of log2 fold change values of the genes coregulated by HBI1 and flg22. Red color indicates the HBI1 target genes. Col, Columbia.
(C) GO analyses of gene sets regulated by HBI1 and/or flg22. Numbers indicate the percentage of genes belonging to each GO category. Asterisk indicates significant difference from random genome (Fisher’s exact test; *P < 0.05).
(D) Quantitative ChIP-PCR analysis of the HBI1 enrichment in the promoter of selected genes. The chromatin of pHBI1:HBI1-YFP and 35S:YFP transgenic plants was immunoprecipitated with anti-YFP antibody, and the precipitated DNA was quantified by quantitative PCR. Enrichment of DNA was calculated as the ratio between pHBI1:HBI1-YFP and 35S:YFP, normalized to that of the PP2A coding region. Error bars indicate standard deviation from three biological repeats. Asterisk indicates significant difference from control gene PP2A (t test; *P < 0.05).
(E) to (G) qRT-PCR analysis of flg22-regulated gene expression in wild-type (Col) and HBI1-Ox seedlings. Error bars indicate standard deviation from three biological repeats. Asterisk indicates significant difference from the wild type with mock treatment (t test; *P < 0.05).
(H) qRT-PCR analyses of flg22 effect on the expression of HBI1 and BEE2. PP2A was used as the internal control. Error bars indicate standard deviation from three biological repeats. Asterisk indicates significant difference from the wild type with mock treatment (t test; *P < 0.05).
(I) Flg22 treatment reduces the HBI1-YFP protein accumulation in the pHBI1:HBI1-YFP but not 35S:HBI1-YFP plants. The immunoblots were analyzed using anti-YFP antibody. The nonspecific band (asterisk) and Ponceau S staining were used to show the equal loading.
Gene Ontology analyses showed that the subset of HBI1-activated but flg22-repressed genes is enriched with cell wall– and chloroplast-related functions (Figure 4C). These data suggest that flg22 may repress cell elongation and photosynthetic functions partly through an HBI1-dependent mechanism. Strikingly, many of the defense-related genes activated by flg22 are repressed by HBI1 (Figure 4C; Supplemental Figure 1). These include many WRKY transcription factors and their interacting partner VQ-motif proteins, the plant U-box proteins, calmodulin-like proteins, and NBS-LRR proteins (Supplemental Figure 1). In addition, genes involved in ROS metabolism were dramatically enriched in the HBI1-repressed but flg22-induced genes (Figure 4C; Supplemental Figure 1). On the other hand, three of the four HBI1-induced GASA genes were repressed by flg22 (Supplemental Figure 1). These observations suggest that HBI1 is a negative regulator of PAMP-induced responses.
To confirm the RNA-Seq data, we performed quantitative RT-PCR (qRT-PCR) on selected genes. Among the HBI1-regulated target genes (Supplemental Data Sets 1 and 2), SIB1 and GASA4 have been recently reported to play important roles in immunity. Both loss- and gain-of-function genetic analysis demonstrated that SIB1 is a positive regulator for the expression of several defense genes and resistance to bacterial pathogen Pseudomonas syringae pv tomato DC3000 (Pst DC3000) and necrotrophic fungal pathogen Botrytis cinerea (Xie et al., 2010; Lai et al., 2011). ChIP-qPCR analysis showed that HBI1 binds to the SIB1 promoter (Figure 4D), and qRT-PCR analysis showed that the flg22-induced SIB1 expression was dramatically repressed in the HBI1-Ox plants (Figure 4E). By contrast, the expression level of GASA4, which represses ROS production (Rubinovich and Weiss, 2010), is repressed by flg22 but directly activated by HBI1 (Figures 4D and 4F). The flg22 induction of SIB1 is diminished and repression of GASA4 is abolished in the HBI1-Ox plants (Figures 4E and 4F). qRT-PCR analyses of additional pathogen response genes showed that the expression levels of FRK1, At2g17740, PR1, JAZ6, and MPK11 were increased by flg22 application in wild-type plants, but their induction was significantly reduced in the HBI1-Ox plants, and the expression levels of FRK1, At2g17740, PR1, and MPK11 were also much lower in HBI1-Ox plants compared with the wild type without flg22 treatment (Figure 4G; Supplemental Figures 5A to 5D). Flg22 activation of MPK3/6 seemed normal or slightly enhanced in the HBI1-Ox plants (Supplemental Figure 5E), suggesting that reduced MPK11 expression has no effect on PAMP activation of other mitogen-activated protein kinases, consistent with that observed in the mpk11 mutant (Bethke et al., 2012). These results demonstrate that flg22 repression of HBI1 mediates the regulation of a subset of defense response genes.
The opposite effects of HBI1 and PAMP signals on gene expression suggest that PAMPs may inhibit HBI1 activity and/or HBI1 may repress a branch of PAMP-elicited signaling pathways. Based on microarray data (Denoux et al., 2008; Tintor et al., 2013), both flg22 and elf18 inhibit the expression of HBI1. Our qRT-PCR assays verified that flg22 treatment decreases the transcript levels of HBI1 and some of its homologs (i.e., BEE1, BEE2, BEE3, and CIB1) (Figure 4H; Supplemental Figure 6). The level of HBI1-YFP protein expressed from the endogenous HBI1 promoter decreased rapidly after flg22 treatment, whereas the HBI1-YFP protein expressed from the constitutive 35S promoter did not change dramatically upon flg22 treatment (Figure 4I), indicating that flg22 mainly regulates HBI1 at the transcriptional level. Flg22 repression of HBI1 expression and the inhibitory effects of HBI1 overexpression on many flg22-induced genes indicate that HBI1 mediates flagellin regulation of a subset of genes.
HBI1 Negatively Regulates PAMP Responses
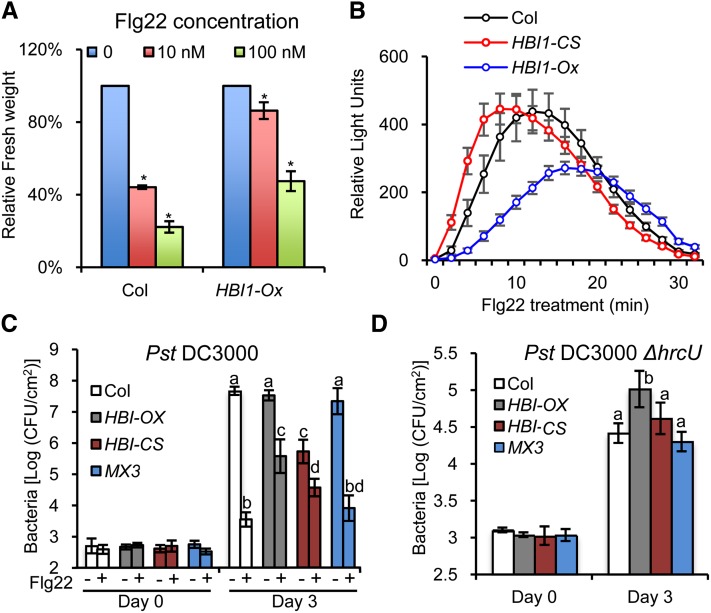
Next, we tested if the repression of HBI1 expression contributes to the flg22-triggered growth inhibition and immunity responses. Indeed, the growth of HBI1-Ox was much less inhibited by flg22 and elf18 than that of wild-type plants (Figure 5A; Supplemental Figures 4D and 4E), indicating that normal PAMP inhibition of plant growth requires repression of HBI1 expression.
Figure 5.
HBI1 Negatively Regulates PTI Signaling.
(A) HBI1-Ox plants show reduced sensitivity to Flg22-induced growth inhibition. Growth is represented relative to untreated plants. Error bars indicate standard deviation from three biological repeats. Asterisk indicates significant difference from mock treatment (t test; *P < 0.05).
(B) Oxidative burst triggered after flg22 treatment (100 nM) in wild-type, HBI1-CS, and HBI1-Ox plants. Error bars indicate standard deviation from three biological repeats.
(C) Pst DC3000 growth in Columbia (Col) wild-type, HBI-Ox, HBI1-CS, and pBZR1:bzr1-1D-CFP (MX3) plants pretreated with 1 µM flg22 or water. Leaves were inoculated with105 colony-forming units (CFU)/mL of bacteria. Bacterial growth was quantified at 0 and 3 d after inoculation. Data points represent mean log (colony-forming units/cm2). Error bars indicate standard deviation from 12 biological repeats. Different letters above the day 3 bars indicate statistically significant differences between the samples (one-way analysis of variance and Tukey's honestly significant difference test, P < 0.05).
(D) Pst DC3000ΔhrcU growth in Columbia wild-type, HBI-Ox, HBI1-CS, and MX3 plants. Leaves were inoculated with 2 × 105 colony-forming units/mL of bacteria. Bacterial growth was quantified at 0 and 3 d after inoculation. Data points represent mean log (colony-forming units/cm2). Error bars indicate standard deviation from 12 biological repeats. Different letters above day 3 bars indicate statistically significant differences between the samples (one-way analysis of variance and Tukey's honestly significant difference test, P < 0.05).
The effects of HBI1 overexpression on PAMP-induced defense gene expression suggested that HBI1 might also modulate immunity. As part of the defense mechanism, ROS production is induced by flg22 or elf18 treatment in wild-type plants, but the ROS response was slower and reached a lower magnitude in the HBI1-Ox plants (Figure 5B; Supplemental Figure 4F). By contrast, the flg22-elicited ROS response occurred earlier in the HBI1 cosuppressing plants (HBI1-CS) compared with the wild type (Figure 5B), confirming a role of HBI1 in inhibiting ROS production.
We next tested the susceptibility of plants to infection with the virulent hemibiotrophic bacterium Pst DC3000. Without flg22 treatment, the HBI1-Ox plants exhibited a similar level of susceptibility as wild-type plants. Pretreatment of wild-type plants with flg22 markedly reduced bacterial growth due to activation of PTI (Figure 5C), but this effect of flg22 on bacterial growth was significantly reduced in the HBI1-Ox plants (Figure 5C), indicating that repression of HBI1 expression is required for flagellin to fully induce immunity. Furthermore, the HBI1-CS plants were more resistant to Pst DC3000 than the wild type without flg22 pretreatment, demonstrating that HBI1 is an essential negative regulator of immunity and that reducing HBI1 expression is sufficient to partly turn on immune responses. Interestingly, the HBI1-CS plants were less resistant to Pst DC3000 than wild-type plants after flg22 pretreatment. One possibility is that the HBI1-CS plants, due to the feedback regulation of HLH factors by HBI1, have an elevated level/activity of homologous bHLH factors that are not repressed by flg22. Alternatively, HBI1 might be involved in other feedback desensitization mechanisms, and long-term reduction of HBI1 level leads to decreased flg22 elicitation of immunity. In contrast with HBI1, the dominant active bzr1-1D and cyan fluorescent protein (CFP) fusion protein expressed from the native BZR1 promoter had little effect on the susceptibility to Pst DC3000.
The Pst DC3000 ∆hrcU mutant is defective in effector delivery and lacks the effector-mediated dampening of host PTI and thus shows reduced growth in wild-type leaves compared with Pst DC3000 (Figure 5D). Pst DC3000 ∆hrcU grew to higher titers in the HBI1-Ox plants than in wild-type plants (Figure 5D), consistent with the observation that HBI1 suppresses flg22-induced resistance (Figure 5C). Pst DC3000 ∆hrcU grew normally in the HBI1-CS plants compared with its growth in wild-type and bzr1-1D-CFP plants (Figure 5D). This is also consistent with the weak response of HBI1-CS plants to flg22 elicitation (Figure 5C). One possibility suggested by this observation is that the wild-type HBI1 gene is not functional in regulating immunity against Pst DC3000 ∆hrcU because HBI1 is fully inactivated by PAMP signaling triggered by this nonpathogenic strain. Taken together, these results demonstrate that the growth regulator HBI1 is a negative regulator of immunity, and it is effectively switched off upon PAMP signaling to enhance immunity and suppress growth at the same time.
DISCUSSION
We previously identified the PRE-IBH1-HBI1 tripartite HLH/bHLH cascade as an important module downstream of the BZR1-PIF-DELLA module in a central transcriptional network that mediates growth regulation by multiple hormonal and abiotic signals (Bai et al., 2012a, 2012b; Oh et al., 2012) (Figure 6). While HBI1 was shown to promote cell elongation similar to other DNA binding components of this network, its specific function in the network has remained unclear. Detailed characterization of HBI1 in this study, at both the genome-wide and molecular levels, reveals major functions of HBI1 in specifying the output of the network, feedback coordinating other components within the network, and integration of additional signals into the network. Importantly, HBI1 is activated by hormones but repressed by PAMP signals, and HBI1 both activates growth and represses immunity. Therefore, HBI1 is a key node for the crosstalk between growth hormones and immune signals (Figure 6).
Figure 6.
Diagram of the Signaling Network Integrating Hormonal, Biotic, and Abiotic Signals.
HBI1 is activated posttranscriptionally by growth-promoting hormonal and environmental signals through the PRE-IBH1-HBI1 cascade but is repressed transcriptionally by PAMP signals. HBI1 both activates growth and inhibits immunity, thereby acting as a crosstalk node that mediates the trade-off between growth and immunity. Arrows show activation, bar-ended lines show inhibition, red lines show regulation by protein–protein interactions, blue lines show transcriptional regulation, and the dashed lines show hypothetical mechanisms. The mechanisms elucidated in this study are marked by thick lines.
HBI1 Promotes Cell Elongation and Feedback Regulates the Hormone Network
Our previous studies showed an essential role of the PRE-IBH1-HBI1 module in the regulation of cell elongation by several hormones, light, and temperature. HBI1 is the DNA binding bHLH factor that specifies the transcriptional output of the module (Bai et al., 2012a). Among the 1257 HBI1-regulated genes (including 177 directly regulated by HBI1 binding), HBI1 activates large numbers of genes encoding cell wall–related proteins, consistent with its role in promoting cell elongation (Bai et al., 2012b). Many of the HBI1-activated genes are also activated by PIF4, and this is likely due to overlapping function with PIF4 on shared target promoters and/or HBI1 activation of hormones. HBI1 overexpression activated genes involved in BR biosynthesis and signal transduction and increased dephosphorylation of BZR1, suggesting a function in positive feedback regulation of upstream signaling. Such positive feedback might be important for maintaining the growth condition of young organs, as inactivation of HBI1 by elevated IBH1 expression in mature tissues (Zhang et al., 2009) would be expected to downregulate hormone synthesis and signaling. These molecular functions are consistent with HBI1’s essential role in promoting cell elongation.
HBI1 also modulates the HLH/bHLH network via a feedback mechanism by transcriptionally activating IBH1 and several other HLH factors that act in parallel to IBH1 as negative regulators of growth, including PAR1, PAR2, HFR1, AIF1, AIF2, AIF3, AIF4, UPB1, and three homologs of IBH1 (Supplemental Figures 1 and 2A). Activation of these inhibitory HLH factors would inhibit not only HBI1 but also other growth-promoting bHLH factors, such as PIFs and ACEs, with which they also interact (Hornitschek et al., 2009; Bai et al., 2012a; Ikeda et al., 2012). As such, this feedback regulation would allow a change of HBI1 activity to indirectly modulate the relative activities of other positive bHLH factors. For example, when HBI1 is repressed (e.g., by initial PAMP exposure or cosuppression in HBI1-CS plants), expression of IBH1 would decrease, which would lead to subsequent derepression of other bHLH factors, such as PIFs and ACEs. High levels of these non-PAMP-regulated bHLH factors relative to HBI1 would reduce the ability of plants to respond to PAMP elicitation, as observed in the HBI1-CS plants. Therefore, the feedback mechanism can potentially not only help maintain the overall activity level of the positive bHLH factors, but also fine-tune the sensitivity and output of the network. Therefore, both protein–protein interaction and feedback transcriptional regulation provide buffering mechanisms that are important for the functional homeostasis of the tripartite HLH/bHLH system (Figure 6).
HBI1 Is a Negative Regulator of Immunity
This study helps uncover a major function of HBI1 in cross regulation between growth and immunity. HBI1 regulates a set of genes in an opposite way to flg22 and elf18 signals, apparently due to PAMP-mediated repression of HBI1 and a number of its homologs. The oppositely regulated genes include not only genes involved in cell wall expansion and growth, but also defense-related genes. Many HBI1 regulated genes have been shown to function in immunity. For example, HBI1 suppresses the expression of five VQ-domain proteins (VQ1, VQ10, VQ23/SIB1, VQ28, and VQ32); three of these are induced by flg22. VQ23/SIB1 has been shown by both loss-of-function and overexpression experiments to be a positive regulator of the defense pathways and to act both as a chloroplast transcription regulator and as a nuclear cofactor that enhance WRKY33 DNA binding (Xie et al., 2010; Lai et al., 2011). VQ10 also interacts with WRKY transcription factors to mediate immunity (Cheng et al., 2012). In addition, a large number of PAMP-induced WRKY factors are also repressed by HBI1, including WRKY33, which interacts with VQ1, VQ10, and VQ23/SIB1 (Cheng et al., 2012) (Supplemental Figure 1 and Supplemental Data Set 2). HBI1 suppression of both interacting partners is likely to have a strong effect on the activity of the VQ-WRKY transcription complex. Additional genes repressed by HBI1-Ox, induced by PAMPs, and shown to be implicated in plant–pathogen interaction include MPK11, calmodulin-like proteins CML42 and CML43 (Chiasson et al., 2005), the plant U-box proteins PUB22, PUB23, and PUB24 (PUB triplet) (Trujillo et al., 2008), and many cytochrome P450 genes. MPK11 is one of the four mitogen-activated protein kinases activated by PAMPs (Bethke et al., 2012). There is evidence that the CMLs function as positive regulators of PAMP early signaling (Chiasson et al., 2005), whereas the PUB triplet has been shown to be negative regulators of PAMP signaling and mediate the attenuation of PAMP signaling after the initial burst of response (Trujillo et al., 2008). Among the 21 HBI1-repressed cytochrome P450 genes, CYP79B2, CYP79B3, and CYP71A12 have been shown to be induced by flg22 and in turn convert Trp to camalexin and glucosinolate, which are required for Arabidopsis immunity (Hiruma et al., 2013; Møldrup et al., 2013). HBI1 regulation of these genes apparently leads to inhibition of immune responses.
The decrease of ROS production in HBI1-Ox is likely due to a combination of HBI1’s effect on both PAMP signaling components and genes involved in redox regulation and/or metabolism. HBI1 directly activates three GASA genes, which are repressed by flg22 in wild-type plants but not in HBI1-Ox. GASA genes encode secreted Cys-rich proteins that have been implicated in hormone crosstalk and redox homeostasis (Nahirñak et al., 2012). GASA4 has been shown to display redox activity and overexpression of GASA4 and GASA14 suppressed ROS accumulation (Rubinovich and Weiss, 2010; Sun et al., 2013). The high-level and constitutive GASA4 expression in HBI1-Ox is likely to contribute to the reduced ROS production. It will be interesting to test in the future whether direct activation of GASAs by HBI1 leads to indirect repression of immunity and of the redox enzymes such as the FAD binding Berberine family proteins, glutathione S-transferases, and 2-oxoglutarate and Fe(II)-dependent oxygenases (Supplemental Data Set 2 and Supplemental Figure 1).
HBI1 may also inhibit immunity through regulation of photosynthesis. Recent studies have uncovered molecular crosstalk between PAMP-triggered immunity and photosynthesis (Xie et al., 2010; Göhre et al., 2012). PAMP signaling is known to inhibit the expression of many genes encoding chloroplast proteins and to alter photosynthetic activities. In particular, PAMP exposure leads to a rapid decrease in nonphotochemical quenching, and impairment of nonphotochemical quenching by mutation of PsbS/NPQ4 enhances PAMP-triggered ROS production and defense gene expression (Göhre et al., 2012). Our data show that HBI1 overexpression increases the expression of PsbS/NPQ4 together with 20 other photosynthetic genes; half of these genes are repressed by flg22. In addition, SIB1 is localized in both the nucleus and plastid and is involved in immune response (Xie et al., 2010; Göhre et al., 2012).
Taken together, our results demonstrate a central role of HBI1 in modulating a trade-off between growth and immunity. HBI1 activates growth-related genes but represses defense genes, providing opposite output for growth and defense; its activity level is posttranslationally increased by growth hormones through interaction of PREs and IBH1 but transcriptionally repressed by PAMP signaling, thereby mediating antagonistic interactions between growth and immune pathways.
Previous studies have demonstrated that the BR- and flagellin-signaling pathways share several upstream components, including the coreceptor BAK1 and the substrates BSK1 and BIK1 shared by BRI1 and FLS2 (Chinchilla et al., 2007; Kemmerling et al., 2007; Albrecht et al., 2012; Belkhadir et al., 2012; Lin et al., 2013; Shi et al., 2013). The functions of these molecular interactions in the antagonistic interaction between the two pathways remain unclear (Albrecht et al., 2012; Belkhadir et al., 2012; Lozano-Duran et al., 2013). The lack of obvious effects of BR treatment on flg22-induced BIK1 phosphorylation and of flg22 treatment on BES1 phosphorylation suggested that the sharing of upstream components does not play an important role in the crosstalk (Albrecht et al., 2012). Recently, Lozano-Duran et al. (2013) reported evidence that BZR1 mediates repression of immunity by interacting with WRKY40 and directly activating other WRKY factors that inhibit immune responses. However, we observed that bzr1-1D has a much weaker effect on pathogen resistance than HBI1-Ox or active BZR1 defective in binding to the 14-3-3 proteins, which are phosphopeptide binding proteins that prevent phospho-BZR1 from accumulating in the nucleus (Gampala et al., 2007; Lozano-Duran et al., 2013). Our latter findings may be due to lower expression of the BZR1 mutant using the native BZR1 promoter or a requirement of specifically abolishing the interaction with the 14-3-3 proteins. Nevertheless, both BZR1 and HBI1 directly target genes involved immunity, and they regulate distinct sets of WRKY genes, suggesting that BZR1 and HBI1 contribute to inhibition of immunity through distinct transcription responses. Since HBI1 can activate BZR1 by activating the BR biosynthetic and signaling genes, and BZR1 can activate HBI1 through increasing PRE1 expression, it is clear that HBI1 and BZR1 are parts of a positive feedback loop. Increased susceptibility caused by activation of either BZR1 or HBI1 could be partly due to indirect activation of either protein.
Unlike BZR1, which appears to be unaffected by PAMPs, HBI1 is not only activated by growth-promoting hormones but also effectively inhibited by PAMP signals. In the absence of pathogen, a high level of HBI1 activates BR synthesis and promotes growth while suppressing immunity pathways. Upon pathogen infection, PAMP-triggered repression of HBI1 contributes to both growth inhibition and activation of immune responses. Therefore, while many molecular mechanisms may have evolved to ensure optimal balance between BR-promoted growth and PAMP-triggered immunity, our results support that HBI1 is a key node of crosstalk mediating the trade-off.
As a component of the central growth transcription network, HBI1 may mediate crosstalk between additional growth signals and PAMP signals. Previous studies have shown antagonism of PAMP signaling with auxin and GA responses. Flg22 inhibits auxin response through microRNA-mediated suppression of auxin receptors as well as a salicylic acid–dependent mechanism to promote resistance against biotrophic pathogens (Navarro et al., 2006; Wang et al., 2007). Defense signaling also stabilizes the GA-signaling repressor DELLA proteins, which contribute to both growth inhibition and defense (Navarro et al., 2008; Yang et al., 2012). The mechanisms by which auxin and GA inhibit immunity have remained unclear. Our study suggests that auxin and GA may inhibit immunity through HBI1, as both auxin and GA activate expression of PREs, which activate HBI1 by sequestering IBH1 (Bai et al., 2012a). We propose that the trade-off between growth and PAMP-induced immunity in Arabidopsis is mainly mediated by the central growth regulation transcription network, in which HBI1 functions as a key junction between the growth and immunity pathways (Figure 6).
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana Columbia-0 ecotype was used as wild-type control for phenotype comparison and for generating the transgenic plants. The HBI1-Ox and HBI1-CS lines have been described previously (Bai et al., 2012a). Seeds were either surface sterilized and plated on half-strength MS basal salt medium (Phyto-Technology Laboratories) or grown directly in soil.
Seedling Growth Inhibition Assay
Seedling growth inhibition was assessed as previously described (Belkhadir et al., 2012). Seedlings were grown in half-strength MS medium containing 1% Suc under constant light for 5 d, then transferred to liquid half-strength MS medium containing 1% Suc supplemented with the indicated concentration of flg22 or elf18 peptides. Seedlings were weighted 8 d after treatment.
ROS Assay
ROS assay was performed as described previously (Kunze et al., 2004). Each data point consists of at least 12 replicates.
Bacterial Growth Assays
Arabidopsis plants were grown in pots in Pro-Mix soil (Premier Horticulture) in a growth chamber (22°C, 80% RH, 125 µE·m−2·s−1 fluorescent illumination) on a 10-h-light/14-h-dark cycle. Pst DC3000 and Pst DC3000 hrcU:Tn3gus (Pst DC3000 ΔhrcU) mutant strain were grown on nutrient yeast glycerol agar with appropriate antibiotics (100 µg/mL rifampicin and/or 50 µg/mL kanamycin) at 28°C (Mudgett and Staskawicz, 1999). To monitor Pst DC3000 or Pst DC3000 ΔhrcU growth in plants, leaves of 4- to 5-week-old plants were hand-infiltrated with a 1 or 2 × 105 colony-forming units/mL suspension of bacteria in 1 mM MgCl2 using a needless syringe. For flg22 pretreatment, the leaves were hand-infiltrated with water or 1 µM flg22 a day before the bacterial inoculation. Leaf discs per treatment per time point were collected and ground in 1 mM MgCl2 and then spotted on nutrient yeast glycerol agar plates in triplicate to determine the bacterial titer. Each replicate includes four leaf discs and 12 biological replicates were used, and the experiment was repeated at least three times with similar results. The average bacterial titer ±sd is reported.
Protein Gel Blot Analysis
Total protein samples were extracted from 10-d-old seedlings or 4-week-old plants using 2× SDS sample buffer, separated on SDS-PAGE gels, transferred to a nitrocellulose membrane, and probed with a polyclonal anti-BZR1 antibody (custom-made, 1:1000 dilution) or anti-phosphop44/42 mitogen-activated protein kinase (Erk1/2) (Thr202/Tyr204) antibody (Cell Signaling; 1:1000 dilution).
ChIP Assay
To generate transgenic plants expressing HBI1-YFP from a native HBI1 promoter (pHBI1:HBI1-YFP), an HBI1 genomic fragment including 1.3 kb upstream of the transcription start site (TSS) and all coding sequence was cloned into pENTRY/SD/D-TOPO vectors (Invitrogen) and then recombined into destination vector pEG-TW (Kim et al., 2009). The pHBI1:HBI1-YFP binary vector was transformed into Agrobacterium tumefaciens strain GV3101 and then transformed into the wild-type Arabidopsis (Columbia-0) plants. A line that showed wild-type phenotype but expressed detectable HBI1-YFP protein was selected and used for the ChIP assay. The pHBI1:HBI1-YFP and 35S:YFP plants were grown in a greenhouse with a 16-h-light/8-h-dark cycle at 22-24°C for 4 weeks. ChIP assays were performed as described previously (Bai et al., 2012b), using an affinity-purified anti-YFP polyclonal antibody (custom-made, 10 μg for each reaction). The ChIP products were analyzed by quantitative real-time PCR (primer sequences are listed in Supplemental Table 2), and enrichment was calculated as the ratio between the transgenic samples expressing HBI1-YFP and the 35S:YFP control sample. The ChIP experiments were performed with three biological replicates, from which the means and standard deviations were calculated from three biologic repeats.
ChIP-Seq Analysis
For ChIP-Seq, a library was constructed from 10 ng of ChIP DNA, pooled from 12 biological repeats to reduce sample variation, with barcodes using a NEBNext ChIP-Seq Library Prep Reagent Set for Illumina kit (New England Biolabs). Two barcode libraries were pooled together and sequenced by Illumina HiSeq2000. Total reads were mapped to the Arabidopsis genome (TAIR10; www.arabidopsis.org) using TopHat software (Trapnell et al., 2009). The uniquely mapped reads were analyzed using CisGenome and PRI-CAT online software with default parameters (http://www.ab.wur.nl/pricat/) (Ji et al., 2008; Muiño et al., 2011). The data of the 35S:YFP sample were used as a negative control, and the HBI1 binding peaks were defined using fold change >1.5, false discovery rate–adjusted P value < 0.05. Two nearest neighbor genes flanking each binding site and genes that contain binding site within the transcribed region were defined as putative HBI1 binding target genes.
To discover the in vivo HBI1 binding motifs, DNA sequences of the binding peaks were analyzed by MEME-ChIP (Machanick and Bailey, 2011). The motifs identified by MEME-ChIP were further analyzed by comparing the frequencies of the motifs in the binding peaks to those in the Arabidopsis total genome (TAIR9; www.arabidopsis.org).
To determine the genomic distribution of HBI1 binding peaks relative to gene structure, we divided the genome into three regions: 5 kb upstream of the TSS to TSS, the TSS to the 3′ end of the gene, and the 3′ end of the gene to 1 kb downstream of the gene. We then calculated the frequency of binding peaks in these three regions. If a peak was located within 5 kb upstream of one gene and 1 kb downstream of another gene, the peak was counted in both regions. Peaks outside these regions were not included and peaks existing within 5 kb upstream of two different genes were counted twice.
qRT-PCR Analysis
Total RNA was extracted from Arabidopsis seedlings using the Spectrum Plant Total RNA kit (Sigma-Aldrich). The first-strand cDNA was synthesized using RevertAid reverse transcriptase (Fermentas) and used as RT-PCR templates. Quantitative PCR analyses were performed on a plate-based LightCycler 480 (Roche) using a SYBR Green reagent (Bio-Rad) with gene-specific primers (Supplemental Table 2). The conditions for PCR amplification were as follows: 98°C for 10 min; 45 cycles of 98°C for 30 s; 65°C for 45 s and 72°C for 30 s; one cycle of 72°C for 10 min. The relative expression was calculated as ratio between the transgenic plant and the wild type and then normalized by the PP2A (AT1G13320) gene, which is a constitutively expressed reference gene (Czechowski et al., 2005). The means and standard deviations were calculated from three biological repeats.
RNA-Seq Analysis
Plants were grown on half-strength MS medium for 5 d under constant light. Total RNA was extracted from two biological repeat samples with the Spectrum Plant Total RNA Kit (Sigma-Aldrich), and mRNA sequencing libraries were constructed with barcodes using the TrueSeq RNA Sample Preparation Kit (Illumina). Four barcoded libraries were pooled together and sequenced by Illumina HiSeq2000. Total reads were mapped to the Arabidopsis genome (TAIR10; www.arabidopsis.org) using the TopHat software (Trapnell et al., 2009). Read counts for every gene were generated using HTSeq with union mode. Differential expressed genes between samples were defined by DESeq using two separate models (Anders and Huber, 2010), based on fold change>1.5 and false discovery rate–adjusted P value < 0.05. Gene Ontology analysis was assisted by the VirtualPlant package (Katari et al., 2010), and the functional categories were determined using the annotation of the Arabidopsis genome (TAIR10; www.arabidopsis.org).
Accession Numbers
Sequence data for ChIP-Seq and RNA-Seq can be found in the Gene Expression Omnibus database under accession numbers GSE53099 and GSE53078.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Representative HBI1 Binding and Regulated Genes with Known Functions in Various Developmental and Cellular Processes.
Supplemental Figure 2. Growth-Promoting and Growth-Inhibiting HLH Factors Are Directly Repressed and Induced by HBI1, Respectively.
Supplemental Figure 3. HBI1 and PIF Have Overlapping and Distinct Functions.
Supplemental Figure 4. HBI1 Negatively Regulates elf18- and flg22-Mediated PTI Responses.
Supplemental Figure 5. Activation of HBI1 Results in the Suppression of PTI Marker Gene Expression but Not MAPK Activation.
Supplemental Figure 6. Quantitative RT-PCR Analyses of flg22 Effect on the Expression of HBI1 Homolog Genes.
Supplemental Table 1. Quantitative RT-PCR Validation of the RNA-Seq Data.
Supplemental Table 2. Oligonucleotide Sequences Used in This Study.
Supplemental Data Set 1. ChIP-Seq Analysis of HBI1 Binding Sites.
Supplemental Data Set 2. RNA-Seq Analysis of Genes Affected in HBI1-Ox Plants.
Supplementary Material
Acknowledgments
We thank the Stanford Center for Genomics and Personalized Medicine led by Michael Snyder and Arend Sidow for the sequencing service and Ziming Weng for carrying out the sequencing. This study was supported by grants from the National Institutes of Health (R01GM066258 to Z.-Y.W. and 2R01GM068886-06A1 to M.B.M.), a “Qilu Scholarship” from Shandong University of China (11200083963009 to M.-Y.B.), and the National Research Foundation of Korea Grant funded by the Korean Government (NRF-2011-220-C00059 to S.-K.K.). We declare no competing financial interests.
AUTHOR CONTRIBUTIONS
M.-Y.B. and Z.-Y.W. designed the experiments. M.-Y.B. performed ChIP-Seq, RNA-Seq, and together with E.O. analyzed sequencing data. M.F. together with T.W. and L.C. performed ROS burst assay, RT-qPCR, ChIP-qPCR, and statistical analysis of plant growth. J.-G.K. performed pathogen growth assays. J.-G.K. and M.B.M. helped with data interpretation and article editing. M.F. performed all other experiments. M.-Y.B. and Z.-Y.W. wrote the article.
Glossary
- BR
brassinosteroid
- GA
gibberellin
- HLH/bHLH
helix-loop-helix/basic helix-loop-helix
- PTI
PAMP-triggered immunity
- ChIP-Seq
chromatin immunoprecipitation followed by sequencing
- RNA-Seq
RNA sequencing
- MS
Murashige and Skoog
- ROS
reactive oxygen species
- ChIP-qPCR
ChIP–quantitative PCR
- qRT-PCR
quantitative RT-PCR
- Pst DC3000
Pseudomonas syringae pv tomato DC3000
- TSS
transcription start site
- PAMP
pathogen-associated molecular pattern (
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Albrecht C., Boutrot F., Segonzac C., Schwessinger B., Gimenez-Ibanez S., Chinchilla D., Rathjen J.P., de Vries S.C., Zipfel C. (2012). Brassinosteroids inhibit pathogen-associated molecular pattern-triggered immune signaling independent of the receptor kinase BAK1. Proc. Natl. Acad. Sci. USA 109: 303–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M.Y., Fan M., Oh E., Wang Z.Y. (2012a). A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell 24: 4917–4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M.Y., Shang J.X., Oh E., Fan M., Bai Y., Zentella R., Sun T.P., Wang Z.Y. (2012b). Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 14: 810–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir Y., Jaillais Y., Epple P., Balsemão-Pires E., Dangl J.L., Chory J. (2012). Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc. Natl. Acad. Sci. USA 109: 297–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke G., Pecher P., Eschen-Lippold L., Tsuda K., Katagiri F., Glazebrook J., Scheel D., Lee J. (2012). Activation of the Arabidopsis thaliana mitogen-activated protein kinase MPK11 by the flagellin-derived elicitor peptide, flg22. Mol. Plant Microbe Interact. 25: 471–480. [DOI] [PubMed] [Google Scholar]
- Cheng Y., Zhou Y., Yang Y., Chi Y.J., Zhou J., Chen J.Y., Wang F., Fan B., Shi K., Zhou Y.H., Yu J.Q., Chen Z. (2012). Structural and functional analysis of VQ motif-containing proteins in Arabidopsis as interacting proteins of WRKY transcription factors. Plant Physiol. 159: 810–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiasson D., Ekengren S.K., Martin G.B., Dobney S.L., Snedden W.A. (2005). Calmodulin-like proteins from Arabidopsis and tomato are involved in host defense against Pseudomonas syringae pv. tomato. Plant Mol. Biol. 58: 887–897. [DOI] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J.D., Felix G., Boller T. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500. [DOI] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M., Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484. [DOI] [PubMed] [Google Scholar]
- Denoux C., Galletti R., Mammarella N., Gopalan S., Werck D., De Lorenzo G., Ferrari S., Ausubel F.M., Dewdney J. (2008). Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol. Plant 1: 423–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depuydt S., Hardtke C.S. (2011). Hormone signalling crosstalk in plant growth regulation. Curr. Biol. 21: R365–R373. [DOI] [PubMed] [Google Scholar]
- Feng S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J., Minguet E.G., Grau-Enguix F., Abbas M., Locascio A., Thomas S.G., Alabadí D., Blázquez M.A. (2012). Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 13446–13451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gampala S.S., et al. (2007). An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell 13: 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göhre V., Jones A.M., Sklenář J., Robatzek S., Weber A.P. (2012). Molecular crosstalk between PAMP-triggered immunity and photosynthesis. Mol. Plant Microbe Interact. 25: 1083–1092. [DOI] [PubMed] [Google Scholar]
- Hao Y., Oh E., Choi G., Liang Z., Wang Z.Y. (2012). Interactions between HLH and bHLH factors modulate light-regulated plant development. Mol. Plant 5: 688–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiruma K., Fukunaga S., Bednarek P., Pislewska-Bednarek M., Watanabe S., Narusaka Y., Shirasu K., Takano Y. (2013). Glutathione and tryptophan metabolism are required for Arabidopsis immunity during the hypersensitive response to hemibiotrophs. Proc. Natl. Acad. Sci. USA 110: 9589–9594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P., Kohnen M.V., Lorrain S., Rougemont J., Ljung K., López-Vidriero I., Franco-Zorrilla J.M., Solano R., Trevisan M., Pradervand S., Xenarios I., Fankhauser C. (2012). Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 71: 699–711. [DOI] [PubMed] [Google Scholar]
- Hornitschek P., Lorrain S., Zoete V., Michielin O., Fankhauser C. (2009). Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 28: 3893–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Y., Lee I. (2006). KIDARI, encoding a non-DNA binding bHLH protein, represses light signal transduction in Arabidopsis thaliana. Plant Mol. Biol. 61: 283–296. [DOI] [PubMed] [Google Scholar]
- Ikeda M., Fujiwara S., Mitsuda N., Ohme-Takagi M. (2012). A triantagonistic basic helix-loop-helix system regulates cell elongation in Arabidopsis. Plant Cell 24: 4483–4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H., Jiang H., Ma W., Johnson D.S., Myers R.M., Wong W.H. (2008). An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol. 26: 1293–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katari M.S., Nowicki S.D., Aceituno F.F., Nero D., Kelfer J., Thompson L.P., Cabello J.M., Davidson R.S., Goldberg A.P., Shasha D.E., Coruzzi G.M., Gutiérrez R.A. (2010). VirtualPlant: A software platform to support systems biology research. Plant Physiol. 152: 500–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmerling B., et al. (2007). The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Curr. Biol. 17: 1116–1122. [DOI] [PubMed] [Google Scholar]
- Kim T.W., Guan S., Burlingame A.L., Wang Z.Y. (2011). The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol. Cell 43: 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Guan S., Sun Y., Deng Z., Tang W., Shang J.X., Sun Y., Burlingame A.L., Wang Z.Y. (2009). Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 11: 1254–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Wang Z.Y. (2010). Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu. Rev. Plant Biol. 61: 681–704. [DOI] [PubMed] [Google Scholar]
- Ko C.B., Woo Y.M., Lee D.J., Lee M.C., Kim C.S. (2007). Enhanced tolerance to heat stress in transgenic plants expressing the GASA4 gene. Plant Physiol. Biochem. 45: 722–728. [DOI] [PubMed] [Google Scholar]
- Kunze G., Zipfel C., Robatzek S., Niehaus K., Boller T., Felix G. (2004). The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16: 3496–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z., Li Y., Wang F., Cheng Y., Fan B., Yu J.Q., Chen Z. (2011). Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell 23: 3824–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wen J., Lease K.A., Doke J.T., Tax F.E., Walker J.C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222. [DOI] [PubMed] [Google Scholar]
- Li Q.-F., Wang C., Jiang L., Li S., Sun S.S., He J.X. (2012). An interaction between BZR1 and DELLAs mediates direct signaling crosstalk between brassinosteroids and gibberellins in Arabidopsis. Sci. Signal. 5: ra72. [DOI] [PubMed] [Google Scholar]
- Lin W., Lu D., Gao X., Jiang S., Ma X., Wang Z., Mengiste T., He P., Shan L. (2013). Inverse modulation of plant immune and brassinosteroid signaling pathways by the receptor-like cytoplasmic kinase BIK1. Proc. Natl. Acad. Sci. USA 110: 12114–12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Durán R., Macho A.P., Boutrot F., Segonzac C., Somssich I.E., Zipfel C. (2013). The transcriptional regulator BZR1 mediates trade-off between plant innate immunity and growth. Elife 2: e00983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Wu S., Gao X., Zhang Y., Shan L., He P. (2010). A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA 107: 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick P., Bailey T.L. (2011). MEME-ChIP: Motif analysis of large DNA datasets. Bioinformatics 27: 1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael T.P., Breton G., Hazen S.P., Priest H., Mockler T.C., Kay S.A., Chory J. (2008). A morning-specific phytohormone gene expression program underlying rhythmic plant growth. PLoS Biol. 6: e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møldrup M.E., Salomonsen B., Geu-Flores F., Olsen C.E., Halkier B.A. (2013). De novo genetic engineering of the camalexin biosynthetic pathway. J. Biotechnol. 167: 296–301. [DOI] [PubMed] [Google Scholar]
- Mudgett M.B., Staskawicz B.J. (1999). Characterization of the Pseudomonas syringae pv. tomato AvrRpt2 protein: demonstration of secretion and processing during bacterial pathogenesis. Mol. Microbiol. 32: 927–941. [DOI] [PubMed] [Google Scholar]
- Muiño J.M., Hoogstraat M., van Ham R.C.H.J., van Dijk A.D.J. (2011). PRI-CAT: A web-tool for the analysis, storage and visualization of plant ChIP-seq experiments. Nucleic Acids Res. 39: W524-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahirñak V., Almasia N.I., Hopp H.E., Vazquez-Rovere C. (2012). Snakin/GASA proteins: Involvement in hormone crosstalk and redox homeostasis. Plant Signal. Behav. 7: 1004–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam K.H., Li J. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212. [DOI] [PubMed] [Google Scholar]
- Navarro L., Bari R., Achard P., Lisón P., Nemri A., Harberd N.P., Jones J.D. (2008). DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr. Biol. 18: 650–655. [DOI] [PubMed] [Google Scholar]
- Navarro L., Dunoyer P., Jay F., Arnold B., Dharmasiri N., Estelle M., Voinnet O., Jones J.D.G. (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312: 436–439. [DOI] [PubMed] [Google Scholar]
- Oh E., Kang H., Yamaguchi S., Park J., Lee D., Kamiya Y., Choi G. (2009). Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell 21: 403–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Zhu J.Y., Wang Z.Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14: 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A., Grant M., Jones J.D. (2011). Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 49: 317–343. [DOI] [PubMed] [Google Scholar]
- Roxrud I., Lid S.E., Fletcher J.C., Schmidt E.D., Opsahl-Sorteberg H.G. (2007). GASA4, one of the 14-member Arabidopsis GASA family of small polypeptides, regulates flowering and seed development. Plant Cell Physiol. 48: 471–483. [DOI] [PubMed] [Google Scholar]
- Rubinovich L., Weiss D. (2010). The Arabidopsis cysteine-rich protein GASA4 promotes GA responses and exhibits redox activity in bacteria and in planta. Plant J. 64: 1018–1027. [DOI] [PubMed] [Google Scholar]
- Santiago J., Henzler C., Hothorn M. (2013). Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science 341: 889–892. [DOI] [PubMed] [Google Scholar]
- Shi H., Shen Q., Qi Y., Yan H., Nie H., Chen Y., Zhao T., Katagiri F., Tang D. (2013). BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell 25: 1143–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Wang H., Yu H., Zhong C., Zhang X., Peng J., Wang X. (2013). GASA14 regulates leaf expansion and abiotic stress resistance by modulating reactive oxygen species accumulation. J. Exp. Bot. 64: 1637–1647. [DOI] [PubMed] [Google Scholar]
- Sun T.P. (2011). The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr. Biol. 21: R338–R345. [DOI] [PubMed] [Google Scholar]
- Sun Y., et al. (2010). Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 19: 765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Kim T.W., Oses-Prieto J.A., Sun Y., Deng Z., Zhu S., Wang R., Burlingame A.L., Wang Z.Y. (2008). BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., et al. (2011). PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 13: 124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tintor N., Ross A., Kanehara K., Yamada K., Fan L., Kemmerling B., Nürnberger T., Tsuda K., Saijo Y. (2013). Layered pattern receptor signaling via ethylene and endogenous elicitor peptides during Arabidopsis immunity to bacterial infection. Proc. Natl. Acad. Sci. USA 110: 6211–6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. (2009). TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo M., Ichimura K., Casais C., Shirasu K. (2008). Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr. Biol. 18: 1396–1401. [DOI] [PubMed] [Google Scholar]
- Wang D., Pajerowska-Mukhtar K., Culler A.H., Dong X. (2007). Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 17: 1784–1790. [DOI] [PubMed] [Google Scholar]
- Wang X., Kota U., He K., Blackburn K., Li J., Goshe M.B., Huber S.C., Clouse S.D. (2008). Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev. Cell 15: 220–235. [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Bai M.Y., Oh E., Zhu J.Y. (2012). Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 46: 701–724. [DOI] [PubMed] [Google Scholar]
- Xie Y.D., Li W., Guo D., Dong J., Zhang Q., Fu Y., Ren D., Peng M., Xia Y. (2010). The Arabidopsis gene SIGMA FACTOR-BINDING PROTEIN 1 plays a role in the salicylate- and jasmonate-mediated defence responses. Plant Cell Environ. 33: 828–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D.L., et al. (2012). Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA 109: E1192–E1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Li L., Zola J., Aluru M., Ye H., Foudree A., Guo H., Anderson S., Aluru S., Liu P., Rodermel S., Yin Y. (2011). A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 65: 634–646. [DOI] [PubMed] [Google Scholar]
- Zhang L.Y., et al. (2009). Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21: 3767–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Mayba O., Pfeiffer A., Shi H., Tepperman J.M., Speed T.P., Quail P.H. (2013). A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 9: e1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.