Expression of BRAFV600E in the mouse lung epithelium elicits benign tumors that fail to progress to cancer due to an apparent senescence-like proliferative arrest. Juan et al. demonstrate that nuclear β-catenin → c-MYC signaling is essential for early stage proliferation of BRAFV600E-induced lung tumors and is inactivated in the subsequent senescence-like state. Inhibition of Porcupine, an acyl-transferase essential for WNT ligand secretion, also inhibited BRAFV600E-initiated lung tumorigenesis. These data indicate that inactivation of WNT → β-catenin → c-MYC signaling triggers the senescence-like proliferative arrest that constrains the malignant progression of BRAFV600E-initiated lung tumors.
Keywords: β-catenin, BRAF, non-small-cell lung cancer, c-MYC
Abstract
Oncogene-induced senescence (OIS) is proposed as a cellular defense mechanism that restrains malignant progression of oncogene-expressing, initiated tumor cells. Consistent with this, expression of BRAFV600E in the mouse lung epithelium elicits benign tumors that fail to progress to cancer due to an apparent senescence-like proliferative arrest. Here we demonstrate that nuclear β-catenin → c-MYC signaling is essential for early stage proliferation of BRAFV600E-induced lung tumors and is inactivated in the subsequent senescence-like state. Furthermore, either β-catenin silencing or pharmacological blockade of Porcupine, an acyl-transferase essential for WNT ligand secretion and activity, significantly inhibited BRAFV600E-initiated lung tumorigenesis. Conversely, sustained activity of β-catenin or c-MYC significantly enhanced BRAFV600E-induced lung tumorigenesis and rescued the anti-tumor effects of Porcupine blockade. These data indicate that early stage BRAFV600E-induced lung tumors are WNT-dependent and suggest that inactivation of WNT → β-catenin → c-MYC signaling is a trigger for the senescence-like proliferative arrest that constrains the expansion and malignant progression of BRAFV600E-initiated lung tumors. Moreover, these data further suggest that the trigger for OIS in initiated BRAFV600E-expressing lung tumor cells is not simply a surfeit of signals from oncogenic BRAF but an insufficiency of WNT → β-catenin → c-MYC signaling. These data have implications for understanding how genetic abnormalities cooperate to initiate and promote lung carcinogenesis.
Non-small-cell lung cancer (NSCLC) is a leading cause of cancer mortality, with adenocarcinoma its most common subtype (Herbst et al. 2008). Driver mutations in proto-oncogenes have been identified in NSCLC, providing a rational strategy for the successful clinical deployment of pathway targeted therapies targeting oncoproteins such as EGF receptor, ROS1, or AML4-ALK in genetically defined subsets of lung cancer patients (Shepherd et al. 2005; Corcoran et al. 2012; Heist and Engelman 2012; Gainor et al. 2013).
Mutationally activated KRAS is the most common driver oncoprotein in NSCLC (≥25%) but also the most pharmacologically intractable (Heist and Engelman 2012). However, certain KRAS effectors are directly implicated as bona fide NSCLC oncogenes, such as BRAF, which is mutated in ∼8% of NSCLC, with one-quarter of such mutations encoding the constitutively active BRAFV600E oncoprotein (Heist and Engelman 2012; The Cancer Genome Atlas Research Network, in prep.). While agents that specifically inhibit BRAFV600E have been developed, BRAF mutation has proven to be an unreliable predictive marker for the clinical effectiveness of such agents. Indeed, whereas vemurafenib is successful in treating BRAF-mutated melanoma, it displays limited efficacy against BRAF-mutated colorectal or thyroid cancer (Flaherty et al. 2009; Chapman et al. 2011; Prahallad et al. 2012; Montero-Conde et al. 2013). Although BRAF-mutated NSCLC cell lines are MEK inhibitor-sensitive in vitro, it remains to be seen whether patients with BRAF-mutated lung cancer will be clinically responsive to agents that target BRAFV600E → MEK → ERK signaling (Pratilas et al. 2008; Trejo et al. 2012).
To explore mechanisms of lung carcinogenesis in detail, we generated mice (BrafCA) carrying a conditional allele of BRaf in which expression of normal BRAF is converted to BRAFV600E following the action of Cre recombinase (Supplemental Fig. S1A; Dankort et al. 2007, 2009; Charles et al. 2011; Collisson et al. 2012). Expression of BRAFV600E in the lung epithelium elicits multifocal, MEK-dependent lung tumors with short latency and 100% penetrance (Dankort et al. 2007; Trejo et al. 2012, 2013). However, these tumors remain benign and rarely progress to NSCLC unless combined with additional genetic events (Dankort et al. 2007; Trejo et al. 2013).
Key unresolved questions in this model are as follows: (1) What promotes an initiated BRAFV600E-expressing lung epithelial cell to become a benign lung tumor? (2) What triggers the senescence-like proliferative arrest of BRAFV600E-induced benign tumors? (3) Does bypass of senescence inevitably result in malignant progression to NSCLC? To address these questions, we explored the potential importance of WNT signaling, a pathway implicated in lung development, homeostasis, and cancer, in BRAFV600E-induced lung tumorigenesis (Maretto et al. 2003; Zhang et al. 2008; Morrisey and Hogan 2010; Beers and Morrisey 2011; Pacheco-Pinedo et al. 2011; Stewart 2014). Our results indicate that WNT → β-catenin → c-MYC signaling is essential for BRAFV600E-induced lung tumorigenesis. Moreover, they indicate a striking linearity of the role of this pathway in sustaining BRAFV600E-driven lung tumor growth. However, whereas coexpression of BRAFV600E with mutated β-catenin led to malignant NSCLC, coexpression of BRAFV600E with c-MYC did not. Taken together, these data emphasize the key role of cooperating signaling pathways in the initiation of lung tumorigenesis, the maintenance of tumor cell proliferation, and the acquisition of malignant characteristics.
Results
β-Catenin activity correlates with proliferation of BRAFV600E-initiated lung tumors
To test for a role for WNT signaling in BRAFV600E-induced lung tumorigenesis, we generated mice carrying a Cre-activated BRafCA allele and a transgenic reporter for nuclear β-catenin/TCF activity (BAT-GAL) (Supplemental Fig. S1B; Maretto et al. 2003). BRAFV600E expression was initiated in the lung epithelium of compound BRafCA; BAT-GAL mice using adenovirus-Cre (Ad-Cre; 107 plaque-forming units [pfu]), with mice euthanized for analysis at 6 or 12 wk post-initiation. Six weeks post-initiation, BRAFV600E-induced lung tumors expressed BAT-GAL activity (Fig. 1A) and remained proliferative, as assessed by Ki67 expression in surfactant protein C-expressing (SPC+) tumor cells (Fig. 1A,B) or by bromodeoxyuridine (BrdU) incorporation (data not shown). In contrast, 12 wk post-initiation, BAT-GAL activity and proliferation in BRAFV600E-induced lung tumor cells were greatly diminished (Fig. 1B), consistent with the onset of senescence-like proliferative arrest. As a control, we assessed BAT-GAL activity in BRAFV600E/TP53Null lung tumors emerging in similarly initiated BRafCA; Trp53f/f; BAT-GAL mice. These tumors sustain their proliferation, do not undergo cell cycle arrest, and also retain BAT-GAL activity, providing evidence of ongoing β-catenin signaling (Fig. 1A,B). These data indicate that BAT-GAL activity correlates with the proliferation status of BRAFV600E-induced lung tumor cells.
Figure 1.
β-Catenin is required for BRAFV600E-induced lung tumorigenesis. Lungs of mice of the indicated genotypes were infected with Ad-Cre and monitored for 6 or 12 wk as indicated, at which time they were euthanized and frozen, or FFPE lung sections were processed for hematoxylin and eosin (H&E), lacZ activity, or immunofluorescence. (A) Representative lacZ-stained tissue sections from BRafCA; BAT-GAL mice euthanized at 6 wk (top left) or 12 wk (top middle) and from BRafCA; Trp53f/f; BAT-GAL mice euthanized at 12 wk (top right). Immunofluorescence analysis of histological sections from BRafCA; BAT-GAL mice euthanized at 6 wk (bottom left) or 12 wk (bottom middle) post-initiation and from BRafCA; Trp53f/f; BAT-GAL mice euthanized at 12 wk (bottom right). DAPI is in blue, SPC is in green, and Ki67 is in red. All bars, 50 μm. (B) Quantification of the percentage of LacZ+ tumors and the percentage of Ki67+/SPC+ cells from BRafCA; BAT-GAL mice euthanized at 6 or 12 wk post-initiation and from BRafCA; Trp53f/f; BAT-GAL mice euthanized at 12 wk. (C) Representative H&E-stained tissue sections from BRafCA (left) or BRafCA; Ctnnb1f/f (right) mice euthanized 12 wk post-initiation. All bars, 4 mm. Immunofluorescence analysis of histological sections from BRafCA; Ctnnb1f/f mice euthanized 12 wk post-initiation. DAPI is in blue, and β-catenin is in green. All bars, 50 μm. (D) Quantification of tumor number, size, and burden in BRafCA; Ctnnb1f/+ and BRafCA; Ctnnb f/f mice euthanized 12 wk post-initiation.
β-Catenin is required for BRAFV600E-initiated lung tumorigenesis
To determine whether β-catenin is required for lung tumorigenesis, BRAFV600E expression was initiated in the lungs of BRafCA mice that were either heterozygous or homozygous for a conditional null allele of β-catenin (Ctnnb1f) (Supplemental Fig. S1C), with tumorigenesis assessed 12 wk post-initiation (Brault et al. 2001; Dankort et al. 2007; Trejo et al. 2012). The lungs of BRafCA; Ctnnb1f/+ mice exhibited tumorigenesis similar to that observed in control BRafCA mice (Fig. 1C). In contrast, BRafCA; Ctnnb1f/f mice displayed a significant reduction in tumor burden (29% vs. 4%) (Fig. 1C,D, P < 0.001). Moreover, immunofluorescence analysis of lung tumors that formed in BRafCA; Ctnnb1f/f mice revealed them to retain β-catenin expression, most likely due to the failure of Cre recombinase to silence both copies of Ctnnb1f (Fig. 1C). This striking reduction of lung tumorigenesis combined with the retention of β-catenin expression in those tumors that formed in BRafCA; Ctnnb1f/f mice provides compelling evidence for its importance in early stage BRAFV600E-induced lung tumorigenesis.
Stabilized β-catenin expression cooperates to promote BRAFV600E-induced lung carcinogenesis
We next tested whether a stabilized form of β-catenin would cooperate with BRAFV600E to sustain tumor growth and promote malignant progression to NSCLC. To do so, we used a Cre-activated β-catenin allele (Ctnnb1ex3(f)) (Supplemental Fig. S1D) in which exon 3 of Ctnnb1 is flanked by loxP sites such that Cre recombinase excises sequences required for APC-mediated β-catenin destruction, leading to expression of a stabilized form of β-catenin (CTNNB1*) (Harada et al. 1999). Consistent with previous data, CTNNB1* expression in mouse lungs failed to elicit any overt lung pathology at any time point analyzed (Pacheco-Pinedo et al. 2011; data not shown). However, BRAFV600E and CTNNB1* coexpression led to a dramatic increase in lung tumor burden whether measured at 4 or 10–12 wk post-initiation (Fig. 2A). This increased tumor burden led to a striking reduction in mouse survival, with 100% of BRafCA; Ctnnb1ex3(f)/+ mice reaching end-stage by 13 wk post-initiation In contrast, 100% of the control BRafCA or Ctnnb1ex3(f)/+ mice remained alive 16 wk post-initiation. As reported previously, we also observed cooperation between oncogenic KRASG12D and CTNNB1* in lung tumorigenesis (Supplemental Fig. S3; Pacheco-Pinedo et al. 2011). Ki67 staining revealed that BRAFV600E/CTNNB1*-coexpressing tumors displayed an elevated proliferative index compared with tumors expressing BRAFV600E alone at either early (4 wk) or late (12 wk) time points post-initiation (Fig. 2C,D). Most notably, CTNNB1* coexpression prevented the decrease in cell proliferation observed in lung tumors 12 wk after BRAFV600E expression. Immunofluorescence staining revealed that BRAFV600E/CTNNB1*-coexpressing tumors sustained c-MYC expression, a known β-catenin/TCF target gene, in comparison with tumors with BRAFV600E expression alone (Fig. 2C; He et al. 1998). However, despite the fact that Cyclin D1 is coregulated by β-catenin and ERK1/2 MAP kinase signaling in colorectal cancer cells (Tetsu and McCormick 1999), there was no difference in Cyclin D1 between the two groups of tumors, similar to previous reports (Fig. 2C,D; Sansom et al. 2005). These results suggest that, although it elicits no lung pathology on its own, activated β-catenin is a potent promoter of the proliferation of BRAFV600E-induced lung tumors.
Figure 2.
BRAFV600E cooperates with gain-of-function CTNNB1* to promote lung tumorigenesis. (A) Lungs of mice of the indicated genotypes were infected with Ad-Cre and monitored for 4 or 10–12 wk as indicated, at which time they were euthanized, and their lungs were processed for H&E staining. Representative H&E-stained tissue sections of BRAFV600E- or BRAFV600E/CTNNB1*-expressing lung tumors at 4 or 10–12 wk are presented. Bar, 4 mm. (B) BRafCA ;Ctnnb1ex3(f)/+ and BRafCA;Ctnnb1ex3(f)/+ mice were infected with Ad-Cre and monitored prospectively over ∼120 d. Mice were euthanized on development of end-stage disease per University of California at San Francisco Institutional Animal Care and Use Committee (IACUC) regulations, and a Kaplan-Meier survival curve was plotted. (C) Immunofluorescence analysis of histological sections from BRAFV600E- or BRAFV600E/CTNNB1*-expressing lung tumors from mice euthanized either 4 or 12 wk post-initiation, with DNA (DAPI) in blue, c-MYC in green, Ki67 and Cyclin D1 in red as indicated. Bar, 50 μm. (D) Quantification of the percentage of Ki67-, Cyclin D1-, or c-MYC-positive BRAFV600E or BRAFV600E/CTNNB1* lung tumor cells (marked by SPC expression) either 4 or 12 wk post-initiation as indicated.
Given the ability of CTNNB1* expression to promote BRAFV600E-induced lung tumorigenesis, we assessed whether BRAFV600E/CTNNB1*-coexpressing tumors displayed malignant progression using previously established grading criteria (Nikitin et al. 2004). Oncogene expression was initiated in either BRafCA or BRafCA; Ctnnb1ex3(f)/+ mice, with tumor grade assessed at 10–12 wk post-initiation (Fig. 3B; Supplemental Fig. S2). BRAFV600E-induced lung lesions were comprised almost exclusively of hyperplasias (15.31%) and grade I benign adenomas (83.37%), and only rarely (<2%) did we detect larger grade II benign adenomas (Fig. 3B). Indeed, even 16–24 wk post-initiation, BRAFV600E-induced lung tumors remained low-grade and benign (Fig. 6A, below; Dankort et al. 2007; Trejo et al. 2012, 2013). In contrast, BRAFV600E/CTNNB1*-coexpressing tumors displayed clear evidence of malignant progression as early as 10 wk post-initiation, at which time 34% of the tumors were grade III advanced adenomas and 7% were grade IV lung adenocarcinomas (Fig. 3B). Indeed, this degree of cancer progression is similar to that observed in BRAFV600E/TP53Null lung tumors at the same time point (Dankort et al. 2007). These data indicate that sustained activity of β-catenin, a genetic lesion that has no effect alone on lung epithelial cells, drives progression of BRAFV600E-initiated benign adenomas toward adenocarcinoma.
Figure 3.
BRAFV600E and CTNNB1* cooperate to promote malignant lung carcinogenesis. (A) BRAFV600E- or BRAFV600E/CTNNB1*-expressing lung tumors were initiated in mice of the appropriate genotype, with mice monitored for 4 wk (left) or 10–12 wk (right), at which time they were euthanized, and their lungs were processed for H&E staining. Bar, 50 µm. (B) The grade of BRAFV600E- or BRAFV600E/CTNNB1*-expressing lung tumors was assessed on a 5-point scale based on grading criteria described by Nikitin et al. (2004) and is described in more depth in Supplemental Figure 2. (C,D) Immunofluorescence analysis of histological sections of BRAFV600E- or BRAFV600E/CTNNB1*-expressing lung tumors either 4–8 or 10–12 wk post-initiation as indicated. DNA (DAPI) is in blue, SPC is in pink (top) and green (bottom), CC10 is in green (top) and pink (bottom), AQP5 and NKX2.1 are in pink, and NKX2.1.
Figure 6.
c-MYC fails to promote malignant cancer progression of BRAFV600E-initiated lung tumors. (A) BRAFV600E-, BRAFV600E/c-MYCWT-, and BRAFV600E/c-MYCT58A-expressing lung tumors were initiated in mice of the appropriate genotype, with mice monitored for 4–8 wk (left), 10–14 wk (middle), or 16+ weeks (right), at which time they were euthanized, and their lungs were processed for H&E staining. Bar, 50 µm. (B) The grade of BRAFV600E/c-MYCT58A-expressing lung tumors was assessed as described in Figure 3A and Supplemental Figure 1 based on criteria established for the classification of proliferative pulmonary lesions of the mouse (Nikitin et al. 2004). (C,D) Immunofluorescence analysis of histological sections BRAFV600E/c-MYCT58A-expressing lung tumors either 4–8 or 10–12 wk post-initiation as indicated. DNA (DAPI) is in blue, SPC is in pink (top) and green (bottom), CC10 is in green (top) and pink (bottom), AQP5 and NKX2.1 are in pink, and NKX2.1. Bar, 50 μm. (E) Lung tumorigenesis was initiated in mice of the indicated genotypes, with mice monitored for 12 wk, at which time they were euthanized, and their lungs were processed for H&E staining. Representative H&E-stained tissue sections from BRafCA; Ctnnb1f/+; R26+/+, BRafCA; Ctnnb1f/fR26+/+, BRafCA; Ctnnb1f/+; R26MYC(T58A)/+, and BRafCA; Ctnnb1f/f;R26MYC(T58A)/+ mice euthanized at 12 wk. Bar, 50 µm. (F) Immunofluorescence analysis of histological sections of lung tumors from BRafCA; Ctnnb1f/+; R26MYC(T58A)/+ and BRafCA; Ctnnb1f/f; R26MYC(T58A)/+ mice euthanized 12 wk after Ad-Cre infection as indicated. DNA (DAPI) is in blue, SPC is in green, and β-catenin is in pink as indicated. Bar , 50 μm. Quantification of β-catenin-positive, mixed β-catenin-negative/positive, and β-catenin-negative lung tumors in BRafCA, BRafCA;Ctnnb1f/f, and BRafCA;Ctnnb1f/f;R26MYC(T58A)/+ mice euthanized 12 wk after Ad-Cre infection.
To determine whether coexpression of CTNNB1* would influence tumor cell differentiation, tissue sections of BRAFV600E- or BRAFV600E/CTNNB1*-expressing lung tumors at either early (4–8 wk) or late (12 wk) times post-initiation were analyzed by immunostaining (Fig. 3C,D). As observed previously, BRAFV600E-induced benign lung tumors expressed the NKX2.1 transcription factor, SPC, and Aquaporin 5 (AQP5) but failed to express Clara cell antigen (CC10) (Trejo et al. 2012). To our surprise, BRAFV600E/CTNNB1*-expressing lung tumors displayed the same pattern of marker expression at both early and late time points. We noted that occasional late stage BRAFV600E/CTNNB1*-expressing lung tumors displayed reduced NKX2.1 and SPC expression with evidence of elevated N-cadherin and α-smooth muscle actin expression (Fig. 3D, bottom panel as indicated; data not shown), but the significance of this remains unclear. Consequently, despite its ability to promote malignant progression, expression of CTNNB1* did not appear to have a major influence on the differentiation status of the majority of BRAFV600E-induced lung tumor cells.
Tumor cell proliferation is under the dual control of both BRAFV600E and CTNNB1* signaling through effects on c-MYC
Nuclear β-catenin regulates expression of RNAs involved in processes central to the aberrant behavior of cancer cells (Clevers and Nusse 2012). Two well-characterized target genes are Cyclin D1 and c-MYC (He et al. 1998; Tetsu and McCormick 1999). As reported previously, we noted that c-MYC, but not Cyclin D1, expression correlated with diminished β-catenin activity in BRAFV600E-induced benign lung tumors (Fig. 2D; Sansom et al. 2005). To further explore this, we generated BRAFV600E/CTNNB1*/TP53Null (BCT) lung cancer-derived cells from an end-stage BRafCA; Ctnnb1ex3(f)/+; Trp53f/f mouse (Fig. 4A,B; Materials and Methods). Untreated BCT cells display detectable phospho-ERK1/2 (p-ERK1/2) and express normal and activated CTNNB1* (upper and lower bands, respectively) as well as c-MYC, Cyclin D1, and CDK4 (Fig. 4A). Treatment of BCT cells with either of two different siRNAs against CTNNB1 (siβ-cat1/2) extinguished expression of both normal and mutationally activated CTNNB1 and inhibited c-MYC expression (>50%) but had no effect on either Cyclin D1 or CDK4 expression (Fig. 4A). Interestingly, either inhibition of CTNNB1* expression or pharmacological inhibition of BRAFV600E → MEK → ERK signaling (PD325901) had inhibitory effects on c-MYC mRNA and protein abundance, but combined inhibition of both pathways largely extinguished c-MYC expression (Fig. 4B,C; Ohren et al. 2004). These data suggest that c-MYC expression is under the dual control of both β-catenin and BRAFV600E and is consistent with previous data indicating an important role for these pathways in the regulation of c-MYC in various settings, including lung tumorigenesis (He et al. 1998; Aziz et al. 1999; Soucek et al. 2013).
Figure 4.
Dual regulation of c-MYC expression by CTNNB1* and BRAFV600E. (A–D) BCT lung cancer-derived cells were transfected with siRNAs against β-catenin (siβ-cat1 or siβ-cat2), MEK inhibitor (1 μM PD325901), or a combination of siβ-cat1 and MEK inhibitor as described in the Materials and Methods. (A,B) Cell extracts were analyzed by immunoblotting as indicated. (C) RT–PCR was used to measure c-MYC mRNA levels in BCT cells treated as indicated. (D) Cell cycle status of BCT cells treated as indicated was assessed by staining with propidium iodide. (E,F) BCT cell proliferation was assessed over 7 d, with viable cell number quantified by using Trypan blue staining in conjunction with a Countess automated cell counter as indicated in the Materials and Methods (shown in E). (F) Extracts of BCT cells were analyzed by immunoblotting as indicated.
Treatment with either siβ-cat1 or PD325901 led to a striking reduction in the percentage of BCT cells in S phase with a concomitant increase of cells in G1, indicating that coordinate BRAFV600E and β-catenin signaling is required for BCT cell cycle progression (Fig. 4D). In separate experiments, we showed that the anti-proliferative effects of MEK1/2 inhibition in BRAFV600E/TP53Null lung cancer cells could be overcome by c-MYC:ER activation (Eilers et al. 1989; Littlewood et al. 1995; data not shown). To test whether the effects of β-catenin silencing could also be overcome by sustained c-MYC activity, BCT cells were engineered to express c-MYC:ER. As expected, treatment of BCT cells with siβ-cat1 suppressed their proliferation by ∼50% (Fig. 4E). Although activation of c-MYC:ER (with 4-HT) had no effect on the baseline proliferation of BCT cells, it was able to substantially restore the proliferation of cells treated with siβ-cat1 (Fig. 4E). Importantly, expression/activation of c-MYC:ER had no effect on BRAFV600E → MEK → ERK activity in BCT cells (Fig. 4F). Hence, the growth inhibitory effects of β-catenin silencing can be substantially overcome by ectopic c-MYC activity.
Sustained c-MYC expression promotes BRAFV600E-induced benign lung tumorigenesis
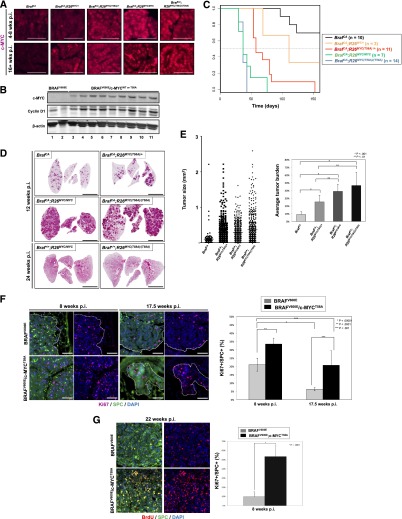
Given the importance of c-MYC in the cooperation of BRAFV600E and CTNNB1* for lung tumorigenesis, we tested the ability of sustained c-MYC expression to promote BRAFV600E-induced benign lung tumorigenesis using mouse strains (RFSMYC) in which cDNAs encoding either wild-type or a phospho-site mutant (T58A) of c-MYC were recombined into the Rosa26 locus downstream from a floxed STOP element (Supplemental Fig. S1E; Sears et al. 2000; Ashton et al. 2010; Wang et al. 2011). Hence, Ad-Cre treatment of BRafCA; RFSMYC mice leads to coexpression of both BRAFV600E and either wild-type or T58A c-MYC in the lung epithelium.
Lung tumorigenesis was initiated in five cohorts of mice—(1) BRafCA, (2) BRafCA; RFSMYC(WT)/+, (3) BRafCA; RFSMYC(WT)/(WT), (4) BRafCA; RFSMYC(T58A)/+, and (5) BRafCA; RFSMYC(T58A)/(T58A)—with mice euthanized for analysis at 4–8 or >16 wk post-initiation. As expected, expression of endogenous c-MYC was detected by immunofluorescence in BRAFV600E-induced lung tumors at the early but not the later time point (Fig. 5A). In contrast, c-MYC was readily detected in tumors derived from all of the other genotypes at both early and late time points, indicating coactivation of the BRafCA and RFSMYC alleles in lung tumors (Fig. 5A), a result confirmed by immunoblotting of tumor-derived lysates from BRafCA (Fig. 5B, lanes 1,2), BRafCA; RFSMYC(WT or T58A)/+ (Fig. 5B, lanes 3–6), or BRafCA; RFSMYC(WT)/(WT) (Fig. 5B, lanes 7–11) mice (Fig. 5B).
Figure 5.
Sustained c-MYC expression promotes BRAFV600E-induced lung tumorigenesis but fails to promote malignant lung cancer progression. (A) Immunofluorescence analysis of histological sections from (1) BRafCA, (2) BRafCA; RFSMYC(WT)/+, (3) BRafCA; RFSMYC(WT)/(WT), (4) BRafCA; RFSMYC(T58A)/+, and (5) BRafCA; RFSMYC(T58A)/(T58A) mice euthanized 4–8 or 16+ wk post-initiation as indicated with c-MYC expression (in red). Bar, 50 μm. (B) Expression of c-MYC was assessed by immunoblot analysis of lysates of BRAFV600E- or BRAFV600E/c-MYC-expressing lung tumors isolated directly from mice 6–9 wk post-initiation. (Lanes 1,2) BRafCA mice. (lanes 3–6) BRafCA; RFSMYC(WT)/+ or BRafCA; RFSMYC(T58A)/+ mice. (Lanes 7–11) BRafCA; RFSMYC(WT)/(WT) or BRafCA; RFSMYC(T58A)/(T58A) mice. (C) Lung tumorigenesis was initiated in mice of the various indicated BRafCA and RFSMYC genotypes, which were infected with Ad-Cre and monitored prospectively over ∼180 d. Mice were euthanized upon development of end-stage disease per University of California at San Francisco IACUC regulations, and a Kaplan-Meier survival curve was plotted. (D) Lung tumorigenesis was initiated in mice of the various indicated BRafCA and RFSMYC genotypes, with mice monitored for 12 or 24 wk as indicated, at which time they were euthanized, and their lungs were processed for H&E staining. Representative H&E-stained lung sections from various indicated BRafCA and RFSMYC genotypes were stained with H&E. (E) Quantification of lung tumor number, size, and burden in mice of the various indicated BRafCA and RFSMYC genotypes at 12 wk post-initiation. (F,G) Immunofluorescence analysis of histological sections from BRafCA and BRafCA; R26MYC(T58A)/+ mice euthanized 8, 17.5, or 22 wk post-initiation as indicated with DNA (DAPI) in blue, SPC in green, Ki67 in pink, and BrdU in red as indicated. Bar, 50 μm. Quantification of the percentage of Ki67+/SPCs and the percentage of BrdU+/SPC+ cells in BRAFV600E or BRAFV600E/c-MYCT58A tumors.
Kaplan-Meier analysis indicated that expression c-MYC (wild type or T58A) in the lung had no effect on mouse survival (data not shown). In contrast, BRafCA mice either heterozygous or homozygous for the wild-type or T58A RFSMYC alleles displayed significantly reduced survival compared with BRafCA controls (P < 0.001 vs. RFSMYC/+ and P < 0.0001 vs. RFSMYC/MYC) (Fig. 5C). Moreover, there appeared to be an activity and gene dosage effect such that BRafCA mice heterozygous for RFSMYC(T58A) reached end-stage more rapidly than BRafCA mice heterozygous for RFSMYC(WT) (P < 0.001). In addition, BRafCA mice homozygous for either RFSMYC allele succumbed to end-stage disease more rapidly than their heterozygous littermates (P < 0.0001). In this case, there was no statistically significant difference between the c-MYC alleles used (Fig. 5D).
Consistent with previous reports, expression of c-MYC alone failed to elicit any overt pathology in the lung even at 24 wk post-initiation (Murphy et al. 2008). However, expression of a single RFSMYC allele with BRAFV600E led to a significant increase in tumor size and burden (Fig. 5D,E). Consistent with their further diminished survival, BRafCA mice homozygous for either RFSMYC allele displayed a significant increase in tumor burden compared with relevant controls (Fig. 5E). Hence, like CTNNB1*, c-MYC significantly enhanced BRAFV600E-induced lung tumorigenesis without having an overt pathological effect itself on the mouse lung epithelium.
To assess the effects of c-MYC on the proliferation of BRAFV600E-induced tumors, oncogene expression was initiated in a second group of mice at lower-dose Ad-Cre (106 pfu) to induce fewer tumors and allow the mice to live longer. Using Ki67 expression as an indicator, BRAFV600E/c-MYC-coexpressing tumors displayed both elevated and sustained proliferation (Fig. 5F,G) at both early (8 wk post-initiation, 35% vs. 20%) and late (17.5 wk post-initiation, 20% vs. 5%) time points (Fig. 5F) compared with control BRAFV600E-induced lung tumors. Furthermore, when transit through S phase was assessed by long-term BrdU labeling (7 d), >50% of SPC+ BRAFV600E/c-MYCT58A-expressing tumor cells incorporated BrdU, whereas <10% of BRAFV600E-expressing tumor cells were BrdU-positive (Fig. 5G). These results support the hypothesis that an insufficiency of β-catenin → c-MYC signaling is responsible for the proliferative arrest of late stage BRAFV600E-initiated lung tumor cells.
c-MYC expression fails to promote malignant progression of BRAFV600E-induced lung tumors
c-MYC’s ability to promote BRAFV600E-induced lung tumorigenesis prompted us to test whether BRAFV600E/c-MYC coexpression would promote lung carcinogenesis. To that end, tumorigenesis was initiated with a low dose (106 pfu) of Ad-Cre in each of the BRafCA; RFSMYC cohorts described above, with mice euthanized at 4–8, 10–14, or 16+ weeks post-initiation to assess lung tumor grade (Fig. 6; Supplemental Fig. S2; Nikitin et al. 2004). As before, BRAFV600E-induced lung lesions were largely hyperplasias or benign grade I adenomas no matter the time of analysis (Fig. 6A). Although >70% of BRAFV600E/c-MYC-induced tumors displayed progression to grade II large benign adenomas, none of these lesions were grade III high-grade adenomas or grade IV adenocarcinomas (Fig. 6A). This was true for all combinations of BRAFV600E and c-MYC (wild type or T58A) and regardless of the time point analyzed (data not shown). Hence, despite c-MYC’s ability to promote early stage BRAFV600E-induced tumorigenesis, it was unable to cooperate with BRAFV600E for malignant transformation of cells, in contrast to other genetic alterations such as CTNNB1*, PIK3CAH1047R, or silencing of TP53 or INK4A/ARF (Dankort et al. 2007; Finch et al. 2009; Trejo et al. 2013). These data are consistent with those of others using a similar system to analyze the effects of sustained MYC activity on BRAFV600E-induced lung tumorigenesis (V Tabor, M Bocci, N Alikhani, R Kuiper, and LG Larsson, in prep.). Finally, BRAFV600E/c-MYCT58A-coexpressing tumors expressed NKX2.1, SPC, and AQP5 at all time points, indicating that c-MYC had not altered the differentiation status of BRAFV600E-initiated lung tumor cells (Fig. 6C,D).
c-MYC rescues the inhibitory effects of β-catenin silencing on BRAFV600E-induced lung tumorigenesis
Although c-MYC is regarded as a potentially important β-catenin effector, there are numerous targets of β-catenin that may play an important role in BRAFV600E-induced lung tumorigenesis (Sansom et al. 2005; Finch et al. 2009; Clevers and Nusse 2012). Since β-catenin silencing inhibited BRAFV600E-induced lung tumorigenesis, we tested the ability of c-MYC to rescue that effect. To do so, lung tumorigenesis was initiated in BRafCA mice either heterozygous or homozygous for Ctnnb1f/f and either with or without one RFSMYC(T58A) allele, with mice euthanized for analysis 12 wk post-initiation (Fig. 6E). As before, β-catenin silencing inhibited BRAFV600E-induced lung tumorigenesis (Figs. 2, 6E,F). However, when c-MYCT58A was coexpressed with BRAFV600E, we observed substantial rescue of lung tumorigenesis in BRafCA mice homozygous for the Ctnnb1f/f allele (Fig. 6E,F). These data suggest that c-MYC expression rescued the proliferation of BRAFV600E/CTNNB1Null-initiated lung tumor cells.
Immunofluorescence analysis of β-catenin expression revealed that ∼100% of tumors that formed in either BRafCA or BRafCA; Ctnnb1f/f mice expressed β-catenin (Fig. 6F). In contrast, >75% of tumors forming in BRafCA; Ctnnb1f/f; RFSMYC(T58A) mice were either completely negative for CTNNB1 expression or displayed a mixture of CTNNB1-positive and CYNNB1-negative tumor cells (Fig. 6F). Hence, the ability of c-MYC to rescue the anti-tumor effects observed in BRAFV600E/CTNNB1Null cells strongly suggests that c-MYC expression is a crucial target downstream from β-catenin required for BRAFV600E-induced benign lung tumor cell proliferation.
BRAFV600E-induced lung tumorigenesis is dependent on WNT → β-catenin → c-MYC signaling
β-Catenin plays an important role in the assembly and function of adherens junctions and is also a downstream mediator of canonical WNT signaling (Clevers and Nusse 2012). Hence, we tested whether the initiator of nuclear β-catenin signaling required for BRAFV600E-induced lung tumorigenesis might be WNT ligands in the microenvironment of incipient BRAFV600E-initated tumor cells. Mice and humans express 19 WNT ligands such that conventional knockout experiments are logistically challenging. However, WNT acylation, which is essential for secretion and activity, is critically dependent on a palmitoyl transferase known as Porcupine (PORCN), against which pharmacological inhibitors have been developed (Siegfried et al. 1994; Hofmann 2000; Lum and Clevers 2012). One such agent, LGK974, shows specificity and selectivity for PORCN over other similar enzymes (Lum and Clevers 2012; Jiang et al. 2013).
To test the WNT dependency of BRAFV600E-induced tumorigenesis, lung tumors were initiated in BRafCA mice that were then treated with either vehicle or LGK974 (5 mg/kg once per day, 5 d per week) starting 1 wk post-initiation, with mice euthanized for analysis 5 wk later. Similar to β-catenin silencing, LGK974 inhibited BRAFV600E-induced tumorigenesis by >75% (Fig. 7A,B). Moreover, analysis of tumor cell proliferation (Ki67) revealed that the small tumors that formed in LGK974-treated mice displayed diminished proliferation (Fig. 7C). To test the specificity of the anti-tumor effects of LGK974, we initiated tumorigenesis in BRafCA mice heterozygous for either the Cre-activated Ctnnb1ex3f/+ or RFSMYC(T58A) alleles to determine whether tumor cell-autonomous maintenance of β-catenin → c-MYC signaling would rescue the inhibitory effects of LGK974 on lung tumorigenesis. Remarkably, expression of either CTNNB1* or c-MYCT58A completely rescued the inhibitory effects of LGK974 on BRAFV600E-induced tumorigenesis (Fig. 7A) whether assessed by overall tumor number, size, or total burden (Fig. 7B). These data confirm the key inhibitory specificity of LGK974 for the WNT → β-catenin → c-MYC signaling pathway in this model system. Moreover, notwithstanding important differences in lung cancer progression, these data demonstrate the critical dependency of BRAFV600E-induced lung tumorigenesis on the WNT → β-catenin →c-MYC signaling axis.
Figure 7.
BRAFV600E-induced lung tumorigenesis is WNT ligand-dependent. (A) BRAFV600E-, BRAFV600E/CTNNB1*-, or BRAFV600E/c-MYCT58A-expressing lung tumors were initiated in mice of the appropriate genotype. One week later, mice were dosed for 5 wk with vehicle control or LGK974 as described in the Materials and Methods, at which time mice were euthanized, and their lungs were prepared for H&E staining. All bars, 4 μm. (B) Quantification of the effects of LGK974 on lung tumor number, size, and overall burden in mice bearing BRAFV600E-, BRAFV600E/CTNNB1*-, or BRAFV600E/c-MYCT58A-expressing lung tumors as described in the Materials and Methods. (C) Immunofluorescence analysis of BRAFV600E tumors dosed for 5 wk with vehicle control or LGK974 and euthanized 6 wk post-initiation as indicated. DNA (DAPI) is in blue, Ki67 is in pink, and SPC is in green. Quantification of the percentage of Ki67+/SPC+ cells from BRAFV600E-induced tumors dosed for 5 wk with vehicle control or LGK974 and euthanized at 6-wk after Ad-Cre infection. Bar, 50 μm.
Discussion
BRAF is mutated in 6%–8% of human lung cancers, but in genetically engineered mouse (GEM) models, expression of BRAFV600E in the lung epithelium results only in benign lung tumors that rarely progress to cancer due to a senescence-like proliferative arrest with some similarity to oncogene-induced senescence (OIS) (Dankort et al. 2007; Haigis et al. 2007; The Cancer Genome Atlas Research Network, in prep.). In cultured cells, sustained RAF → MEK → ERK signaling elicits an immediate proliferative arrest followed by OIS without any initial phase of cell proliferation (Woods et al. 1997; Zhu et al. 1998). In contrast, expression of BRAFV600E in lung epithelial cells elicits clonally derived tumors that undergo ∼15–20 population doublings prior to the onset of senescence-like growth arrest (Dankort et al. 2007; Trejo et al. 2013). Here we demonstrate that WNT → β-catenin signaling, a pathway essential for normal lung development and homeostasis, is critically required for early stage BRAFV600E-induced lung tumorigenesis. Furthermore, the senescence-like growth arrest observed in late stage BRAFV600E-expressing tumors appears to be due to an insufficiency of β-catenin activity, leading to reduced c-MYC expression. While it remains unclear by what mechanisms WNT ligand signaling becomes limiting in this situation, one possibility is that BRAFV600E-expressing tumor cells are responding to a stromal source of WNT ligands (for example, from macrophages) such that as the tumors reach a certain size, there is insufficient exposure to WNT, thereby triggering cell cycle arrest through diminished β-catenin → c-MYC signaling (Boulter et al. 2012). An alternative would be that sustained BRAFV600E → MEK → ERK activity generates a negative feedback circuit to WNT receptors or other components of WNT signaling that restricts the cells’ ability to respond to WNT ligands in the tumor microenvironment, as suggested in OIS triggering in other circumstances (Courtois-Cox et al. 2006). An obvious question is whether a requirement for β-catenin signaling is unique to BRAFV600E-initiated lung tumorigenesis or is a general requirement for lung cancers driven by other oncogenes (Herbst et al. 2008; Heist and Engelman 2012). In this regard, we and others have demonstrated that expression of CTNNB1* accelerated KRASG12D-induced lung carcinogenesis in GEM models, suggesting a more general dependency on WNT signaling for lung carcinogenesis (Supplemental Fig. S3; Pacheco-Pinedo et al. 2011; Stewart 2014). Finally, the role of β-catenin in BRAFV600E-induced neoplasia is not restricted to lung tumorigenesis, as both primary tumor growth and subsequent metastatic dissemination of BRAFV600E/PTENNull melanomas is diminished in the absence of β-catenin (Damsky et al. 2011).
As indicated by analysis of cultured BCT cells, expression of c-MYC appears under the dual control of both oncogenic BRAFV600E and WNT → β-catenin signaling such that loss of either signal has potent anti-tumor effects through effects on c-MYC (He et al. 1998; Aziz et al. 1999; Soucek et al. 2013). This bears a similarity to the situation in KRAS-mutated colorectal cancer (CRC) cells, where Cyclin D1 is under the dual control of the same two pathways (Tetsu and McCormick 1999). However, in CRC, both pathways are mutationally altered through KRAS mutation in combination with either APC silencing or mutation of β-catenin (Powell et al. 1992). However, during early stage lung tumorigenesis, the signal for β-catenin → c-MYC signaling does not require mutational activation but instead relies on the presence of WNT ligands, most likely from the tumor microenvironment. The importance of c-MYC as a downstream effector of oncogenic KRASG12D or BRAFV600E is emphasized by the anti-tumor effects of an inhibitor of c-MYC (Omo-MYC) that suppresses KRASG12D-induced lung tumorigenesis (Soucek et al. 2008, 2013). Moreover, BRAFV600E cooperates with other deregulated signaling pathways, such as the PI3′-kinase pathway, to sustain c-MYC expression in either lung or prostate tumorigenesis (Wang et al. 2012; Trejo et al. 2013). However, the fact that expression/activation of c-MYC failed to elicit any pathology in the lung emphasizes that c-MYC is necessary but not sufficient to initiate lung tumorigenesis (Fig. 5; Murphy et al. 2008). Finally, in contrast to previous reports, we obtained no evidence that c-MYC promoted invasion or metastasis of BRAFV600E-induced lung tumors (Rapp et al. 2009).
Although CTNNB1 is mutated in ∼5% of human NSCLCs, there is insufficient evidence regarding coincident mutation with either KRAS or BRAF (Heist and Engelman 2012). Importantly, WNT → β-catenin signaling is reported to be active in NSCLC-derived cell lines through a multiplicity of mechanisms, including autocrine production of WNT ligands and diminished secretion of WNT antagonists such as Dickkopf (DKK1–4) or WNT inhibitory factor 1 (WIF1) (Mazieres et al. 2004, 2005; You et al. 2004; Soucek et al. 2013; Stewart 2014). The observation that BRAFV600E/TP53Null lung tumors retained BAT-GAL activity even at late time points is consistent with reports linking TP53 silencing to β-catenin through effects on miR34 expression (Cha et al. 2012). Consequently, the frequency of CTNNB1 mutation may underestimate the importance of WNT → β-catenin signaling in NSCLC (Stewart 2014). Given that there are now reliable target genes and reporters for nuclear β-catenin activity and inhibitors of WNT signaling, it should be possible to parse out the importance of this pathway in different cancer types, leading to predictive marker-guided clinical trials of agents that target this pathway.
Silencing of TP53 or INK4A/ARF or expression of PIK3CAH1047R can promote bypass of the senescence-like growth arrest in BRAFV600E-induced benign lung tumors, thereby promoting malignant lung cancer progression (Dankort et al. 2007; Trejo et al. 2013). This begged the question as to whether bypass of senescence is synonymous with malignant progression. Our results indicated that, whereas CTNNB1* promoted malignant progression of BRAFV600E-induced lung tumors, c-MYC did not. These data suggest that malignant progression requires more than simple bypass of the senescence-like proliferative arrest displayed by BRAFV600E-induced lung tumors and is consistent with previous data indicating that c-MYC does not fully rescue the effects of loss of β-catenin function in the gastrointestinal tract (Finch et al. 2009). Although β-catenin → c-MYC signaling is critically required for sustained proliferation of BRAFV600E-expressing lung tumors, there must be branch points downstream from CTNNB1*, independent of c-MYC, that influence malignant progression. Future experiments will be directed to those branch points using GEM models and human lung cancer cell lines.
GEM models of KRAS- or BRAF-initiated lung tumorigenesis have revealed the remarkable complexity of biochemistry and cell biology that occurs even in the earliest stages of transformation. Such tumors are regulated by both instructive (initiated by the oncogene) and permissive (regulated by the tumor microenvironment) signals required for tumor initiation and clonal expansion. Previous studies have emphasized the importance of tumor cell-autonomous pathways instructed by oncogenic KRAS or BRAF, such as the ERK MAP kinase, PI3′-kinase → AKT, Rac1 → NF-κB, NKX2.1 signaling, and others (Dankort et al. 2007; Ji et al. 2007; Kissil et al. 2007; Meylan et al. 2009; Winslow et al. 2011; Trejo et al. 2012, 2013). However, these data indicate the potential importance of growth factor signaling in providing a permissive tumor microenvironment that renders lung epithelial cells responsive to mutational activation of oncogenes. Moreover, it remains likely, as observed in other circumstances, that factors present in the tumor microenvironment will play an important role in the response of BRAF-mutated cancer cells to pathway targeted therapy (Straussman et al. 2012; Wilson et al. 2012).
Materials and methods
Mice and adenovirus delivery
All animal experiments were conducted in accordance with protocols approved by the University of California at San Francisco Institutional Animal Care and Use Committee. BRafCA (BRaftm1Mmcm), LSL-KRasG12D (Krastm4Tyj), RFSMYC(WT) or (T58A) [Gt(ROSA)26Sortm1(myc)Rcse, Gt(ROSA)26Sortm1(myc*T58A)Rcse], BAT-GAL [B6.Cg-Tg(BAT-lacZ)3Picc/J], Ctnnb1ex3(f) (Ctnnb1tm1Mmt), and Ctnnb1f (Ctnnb1tm2Kem) mice were bred and genotyped as previously described (Harada et al. 1999; Brault et al. 2001; Jackson et al. 2001; Maretto et al. 2003; Dankort et al. 2007; Wang et al. 2011). Stocks of adenovirus encoding Cre recombinase (Ad-CMV-Cre) were purchased from Viraquest and instilled into the lungs of adult mice as previously described (Fasbender et al. 1998; Dankort et al. 2007).
Histology and quantification of lung tumor burden
Lungs were processed for analysis as described previously (Trejo et al. 2013). Hematoxylin and eosin (H&E)-stained slides were scanned with an Aperio ScanScope scanner, with quantification performed using Aperio Spectrum ImageScope software. Tumor number and size were measured per lobe, and overall tumor burden was calculated as (area of lung lobe occupied by tumor)/(total area of lobe) in square micrometers (Trejo et al. 2013).
Treatment of mice with pathway targeted therapeutics
For treatment of mice, NVP-LGK974-AE (LGK974, Novartis) was formulated in 0.5% (w/v) methylcellulose (Fluka Analytical)/0.5% (v/v) Tween-80 (Sigma-Aldrich) and administered by oral gavage at 5 mg/kg per mouse once per day for 5 d per week.
Immunostaining of mouse lung tissue, LacZ detection, and immunoblotting
Mouse lungs were fixed in zinc-buffered formalin, processed, embedded in paraffin, cut into 5-μm sections, and mounted on glass slides. Citrate-mediated antigen retrieval was performed, and then the following antibodies were used for detection: anti-β-catenin (Cell Signaling Technology), anti-AQP5 (Calbiochem), anti-Ki67, (Abcam), anti-BrdU, anti-CC10, anti-Cyclin D1, anti-SPC, anti-NKX2.1 (also known as TTF-1) (Santa Cruz Biotechnology), and c-MYC (Epitomics).
β-Galactosidase activity driven by the BAT-GAL transgene was assessed in fresh-frozen sections incubated with 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) for 16 h as described previously (Maretto et al. 2003).
Fifty-microgram aliquots of cell proteins were probed with antisera against c-MYC (Epitomics), Cyclin D1, p-ERK1/2 (pT202/PY204), total ERK1/2 (Cell Signaling Technology), and anti-β-actin (Sigma). Immunoblots were visualized using the Odyssey Classic or Odyssey Fc, with data analyzed using either the Odyssey application software version 3.0.30 or Image Studio version 2.0 software (LI-COR Biosciences).
Cell proliferation assays
Cell proliferation was assessed by counting the percentage of surfactant protein-positive (SPC+) tumor cells that were also BrdU- or Ki67-positive by double-label immunofluorescence. In BRafCA mice, nine tumors were analyzed with nine grids each for a total of 5900 SPC+ cells evaluated. In BRafCA; R26MYC(T58A)/+ mice, 10 tumors were analyzed with 10 grids each for a total of 5718 SPC+ cells evaluated. Similar numbers of cells were evaluated for the presence of SPC/Ki67 double-positive cells in tumors from BRafCA, BRafCA; R26MYC(T58A)/+, BRafCA; Ctnnb1f/f, and BRafCA; Ctnnb1ex3(f)/+ mice.
Lung tumor cell isolation, culture, and analysis
Lungs of tumor-bearing mice were perfused with dispase (BD Bioscience), and individual lobes were minced and incubated with 2 mg/mL dispase/collagenase (Roche). Cell suspensions from BCT-induced lung cancers were then filtered and plated directly into culture in Hams-F12/Glutamax medium supplemented with 10% (v/v) fetal bovine serum. Following the outgrowth of a mixed population of lung cancer-derived cells, a number of single-cell-derived clones were isolated by limiting dilution and then expanded to create BCT lung tumor-derived cells.
Ecotropic virus was produced by transient transfection of pBabePuro-MYC:ER (gift of Dr. Gerard Evan) into Plat-E cells (Littlewood et al. 1995). BCT cells were infected and selected in DMEM containing 2 µg/mL puromycin. RNAi-mediated inhibition of β-catenin expression was performed using β-catenin siRNA 1 (si-βcat1, 5′-ATCAACTGGATAGTCAGCACC-3′) or 2 (si-βcat2, 5′-ATTCATAAAGGACTTGGGAGG-3′). A siRNA targeting GFP (AACCACTACCTGAGCACCCAG) was used as a negative control. BCT cells were transfected with 5 nM siRNA by RNAi-Max (Invitrogen) either alone or in the presence of a MEK inhibitor (1 μM PD0325901, Hansun Trading Co.) (Sebolt-Leopold 2004). BCT cells expressing c-MYC:ER were treated with either 100 nM ethanol or 100 nM 4-hydroxytamoxifen to activate the fusion protein. Cell cycle analysis was performed by incubating cells at 50% confluency with pathway targeted inhibitors for 24 h, with 10 µM BrdU (Becton-Dickinson) added for the final 3 h. Floating and adherent cells were collected, fixed in ethanol, and stained with anti-BrdU-FITC (Becton-Dickinson) and propidium iodide (Sigma).
Quantitative RT–PCR
RNAs were purified from cultured cells using RNeasy (Qiagen) and reverse-transcribed using the Advantage RT-for-PCR kit (Takara-Clontech). Real-time quantitative PCR was performed using SYBR Green Premix EX Taq (Takara) on an Applied Biosystems 7500 real-time PCR system (Applied Biosystems). Sense and antisense primers were β-actin (GTGCTTCTAGGCGGACTGTT and TGCGCAAGTTAGGTTTTGTCA) and c-MYC (GCCCAGTGAGGATATCTGGA and ATCGCAGATGAAGCTCTGGT). The following cycle parameters were used: denaturation for 10 sec at 95°C and annealing and elongation for 30 sec at 60°C.
Acknowledgments
We thank current and former members of the McMahon laboratory, especially David Dankort, Roch-Philippe Charles, Vicky Marsh, Christy Trejo, Anny Shai, Jillian Silva, and J. Edward van Veen, for training, advice, guidance, and support throughout this project. We thank Jane Gordon, Yunita Lim, and Cynthia Cowdrey from the University of California at San Francisco Laboratory for Cell Analysis and the Brain Tumor Research Center for assistance with microscopy, FACS analysis, and processing of formalin fixed samples, respectively. We thank Meredith West and Sandy DeVries from the University of California at San Francisco Radiation Therapy Oncology Group for use of the Aperio scanner; Rolf Kemler (Max Planck Institute, Freiburg) and Makoto Taketo (Kyoto University) for access to modified Ctnnb1 mice; Gerard Evan (Cambridge University) for c-MYC:ER; Shifeng Pan, Joseph Growney, and Margaret McLaughlin (Novartis, Inc.) for LGK974; and Lars-Gunnar Larsson (Karolinska Institute) for communicating unpublished results. J.J. is supported by a Genentech Foundation Graduate Student Fellowship, T.M. is supported by a fellowship from the Japanese Society for Promotion of Science, R.C.S. and M.M. are supported by the National Cancer Institute (CA129040 and CA100855 to R.C.S., and CA131261 to M.M.), and M.M. is supported by Uniting Against Lung Cancer.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.233627.113.
References
- Ashton GH, Morton JP, Myant K, Phesse TJ, Ridgway RA, Marsh V, Wilkins JA, Athineos D, Muncan V, Kemp R, et al. 2010. Focal adhesion kinase is required for intestinal regeneration and tumorigenesis downstream of Wnt/c-Myc signaling. Dev Cell 19: 259–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz N, Cherwinski H, McMahon M 1999. Complementation of defective colony-stimulating factor 1 receptor signaling and mitogenesis by Raf and v-Src. Mol Cell Biol 19: 1101–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beers MF, Morrisey EE 2011. The three R’s of lung health and disease: Repair, remodeling, and regeneration. J Clin Invest 121: 2065–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulter L, Govaere O, Bird TG, Radulescu S, Ramachandran P, Pellicoro A, Ridgway RA, Seo SS, Spee B, Van Rooijen N, et al. 2012. Macrophage-derived Wnt opposes Notch signaling to specify hepatic progenitor cell fate in chronic liver disease. Nat Med 18: 572–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, Sommer L, Boussadia O, Kemler R 2001. Inactivation of the β-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development 128: 1253–1264 [DOI] [PubMed] [Google Scholar]
- Cha YH, Kim NH, Park C, Lee I, Kim HS, Yook JI 2012. MiRNA-34 intrinsically links p53 tumor suppressor and Wnt signaling. Cell Cycle 11: 1273–1281 [DOI] [PubMed] [Google Scholar]
- Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, et al. 2011. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 364: 2507–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles RP, Iezza G, Amendola E, Dankort D, McMahon M 2011. Mutationally activated BRAF(V600E) elicits papillary thyroid cancer in the adult mouse. Cancer Res 71: 3863–3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H, Nusse R 2012. Wnt/β-catenin signaling and disease. Cell 149: 1192–1205 [DOI] [PubMed] [Google Scholar]
- Collisson EA, Trejo CL, Silva JM, Gu S, Korkola JE, Heiser LM, Charles RP, Rabinovich BA, Hann B, Dankort D, et al. 2012. A central role for RAF → MEK → ERK signaling in the genesis of pancreatic ductal adenocarcinoma. Cancer Discov 2: 685–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran RB, Cheng KA, Hata AN, Faber AC, Ebi H, Coffee EM, Greninger P, Brown RD, Godfrey JT, Cohoon TJ, et al. 2012. Synthetic lethal interaction of combined BCL-XL and MEK inhibition promotes tumor regressions in KRAS mutant cancer models. Cancer Cell 23: 121–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois-Cox S, Genther Williams SM, Reczek EE, Johnson BW, McGillicuddy LT, Johannessen CM, Hollstein PE, MacCollin M, Cichowski K 2006. A negative feedback signaling network underlies oncogene-induced senescence. Cancer Cell 10: 459–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damsky WE, Curley DP, Santhanakrishnan M, Rosenbaum LE, Platt JT, Gould Rothberg BE, Taketo MM, Dankort D, Rimm DL, McMahon M, et al. 2011. β-Catenin signaling controls metastasis in Braf-activated Pten-deficient melanomas. Cancer Cell 20: 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dankort D, Filenova E, Collado M, Serrano M, Jones K, McMahon M 2007. A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes Dev 21: 379–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE Jr, You MJ, DePinho RA, McMahon M, Bosenberg M 2009. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet 41: 544–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M, Picard D, Yamamoto KR, Bishop JM 1989. Chimaeras of myc oncoprotein and steroid receptors cause hormone-dependent transformation of cells. Nature 340: 66–68 [DOI] [PubMed] [Google Scholar]
- Fasbender A, Lee JH, Walters RW, Moninger TO, Zabner J, Welsh MJ 1998. Incorporation of adenovirus in calcium phosphate precipitates enhances gene transfer to airway epithelia in vitro and in vivo. J Clin Invest 102: 184–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch AJ, Soucek L, Junttila MR, Swigart LB, Evan GI 2009. Acute overexpression of Myc in intestinal epithelium recapitulates some but not all the changes elicited by Wnt/β-catenin pathway activation. Mol Cell Biol 29: 5306–5315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty K, Puzanov I, Sosman J, Kim K, Ribas A, McArthur G, Lee R, Grippo JF, Nolop K, Chapman P 2009. Phase I study of PLX4032: Proof of concept for V600E BRAF mutation as a therapeutic target in human cancer. J Clin Oncol 25: 1 [Google Scholar]
- Gainor JF, Varghese AM, Ou SH, Kabraji S, Awad MM, Katayama R, Pawlak A, Mino-Kenudson M, Yeap BY, Riely GJ, et al. 2013. ALK rearrangements are mutually exclusive with mutations in EGFR or KRAS: An analysis of 1683 patients with non-small cell lung cancer. Clin Cancer Res 19: 4273–4281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis KM, Wistuba II, Kurie JM 2007. Lung premalignancy induced by mutant B-Raf, what is thy fate? To senesce or not to senesce, that is the question. Genes Dev 21: 361–366 [DOI] [PubMed] [Google Scholar]
- Harada N, Tamai Y, Ishikawa T, Sauer B, Takaku K, Oshima M, Taketo MM 1999. Intestinal polyposis in mice with a dominant stable mutation of the β-catenin gene. EMBO J 18: 5931–5942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW 1998. Identification of c-MYC as a target of the APC pathway. Science 281: 1509–1512 [DOI] [PubMed] [Google Scholar]
- Heist RS, Engelman JA 2012. SnapShot: Non-small cell lung cancer. Cancer Cell 21: 448–448.e2 [DOI] [PubMed] [Google Scholar]
- Herbst RS, Heymach JV, Lippman SM 2008. Lung cancer. N Engl J Med 359: 1367–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K 2000. A superfamily of membrane-bound O-acyltransferases with implications for wnt signaling. Trends Biochem Sci 25: 111–112 [DOI] [PubMed] [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA 2001. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 15: 3243–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Wang Z, Perera SA, Li D, Liang MC, Zaghlul S, McNamara K, Chen L, Albert M, Sun Y, et al. 2007. Mutations in BRAF and KRAS converge on activation of the mitogen-activated protein kinase pathway in lung cancer mouse models. Cancer Res 67: 4933–4939 [DOI] [PubMed] [Google Scholar]
- Jiang X, Hao HX, Growney JD, Woolfenden S, Bottiglio C, Ng N, Lu B, Hsieh MH, Bagdasarian L, Meyer R, et al. 2013. Inactivating mutations of RNF43 confer Wnt dependency in pancreatic ductal adenocarcinoma. Proc Natl Acad Sci 110: 12649–12654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissil JL, Walmsley MJ, Hanlon L, Haigis KM, Bender Kim CF, Sweet-Cordero A, Eckman MS, Tuveson DA, Capobianco AJ, Tybulewicz VL, et al. 2007. Requirement for Rac1 in a K-ras induced lung cancer in the mouse. Cancer Res 67: 8089–8094 [DOI] [PubMed] [Google Scholar]
- Littlewood TD, Hancock DC, Danielian PS, Parker MG, Evan GI 1995. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res 23: 1686–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum L, Clevers H 2012. Cell biology. The unusual case of Porcupine. Science 337: 922–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maretto S, Cordenonsi M, Dupont S, Braghetta P, Broccoli V, Hassan AB, Volpin D, Bressan GM, Piccolo S 2003. Mapping Wnt/β-catenin signaling during mouse development and in colorectal tumors. Proc Natl Acad Sci 100: 3299–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazieres J, He B, You L, Xu Z, Lee AY, Mikami I, Reguart N, Rosell R, McCormick F, Jablons DM 2004. Wnt inhibitory factor-1 is silenced by promoter hypermethylation in human lung cancer. Cancer Res 64: 4717–4720 [DOI] [PubMed] [Google Scholar]
- Mazieres J, He B, You L, Xu Z, Jablons DM 2005. Wnt signaling in lung cancer. Cancer Lett 222: 1–10 [DOI] [PubMed] [Google Scholar]
- Meylan E, Dooley AL, Feldser DM, Shen L, Turk E, Ouyang C, Jacks T 2009. Requirement for NF-κB signalling in a mouse model of lung adenocarcinoma. Nature 462: 104–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero-Conde C, Ruiz-Llorente S, Dominguez JM, Knauf JA, Viale A, Sherman EJ, Ryder M, Ghossein RA, Rosen N, Fagin JA 2013. Relief of feedback inhibition of HER3 transcription by RAF and MEK inhibitors attenuates their antitumor effects in BRAF-mutant thyroid carcinomas. Cancer Discov 3: 520–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrisey EE, Hogan BL 2010. Preparing for the first breath: Genetic and cellular mechanisms in lung development. Dev Cell 18: 8–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DJ, Junttila MR, Pouyet L, Karnezis A, Shchors K, Bui DA, Brown-Swigart L, Johnson L, Evan GI 2008. Distinct thresholds govern Myc’s biological output in vivo. Cancer Cell 14: 447–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitin AY, Alcaraz A, Anver MR, Bronson RT, Cardiff RD, Dixon D, Fraire AE, Gabrielson EW, Gunning WT, Haines DC, et al. 2004. Classification of proliferative pulmonary lesions of the mouse: Recommendations of the mouse models of human cancers consortium. Cancer Res 64: 2307–2316 [DOI] [PubMed] [Google Scholar]
- Ohren JF, Chen H, Pavlovsky A, Whitehead C, Zhang E, Kuffa P, Yan C, McConnell P, Spessard C, Banotai C, et al. 2004. Structures of human MAP kinase kinase 1 (MEK1) and MEK2 describe novel noncompetitive kinase inhibition. Nat Struct Mol Biol 11: 1192–1197 [DOI] [PubMed] [Google Scholar]
- Pacheco-Pinedo EC, Durham AC, Stewart KM, Goss AM, Lu MM, Demayo FJ, Morrisey EE 2011. Wnt/β-catenin signaling accelerates mouse lung tumorigenesis by imposing an embryonic distal progenitor phenotype on lung epithelium. J Clin Invest 121: 1935–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell SM, Zilz N, Beazer-Barclay Y, Bryan TM, Hamilton SR, Thibodeau SN, Vogelstein B, Kinzler KW 1992. APC mutations occur early during colorectal tumorigenesis. Nature 359: 235–237 [DOI] [PubMed] [Google Scholar]
- Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, Beijersbergen RL, Bardelli A, Bernards R 2012. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 483: 100–103 [DOI] [PubMed] [Google Scholar]
- Pratilas CA, Hanrahan AJ, Halilovic E, Persaud Y, Soh J, Chitale D, Shigematsu H, Yamamoto H, Sawai A, Janakiraman M, et al. 2008. Genetic predictors of MEK dependence in non-small cell lung cancer. Cancer Res 68: 9375–9383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp UR, Korn C, Ceteci F, Karreman C, Luetkenhaus K, Serafin V, Zanucco E, Castro I, Potapenko T 2009. MYC is a metastasis gene for non-small-cell lung cancer. PLoS ONE 4: e6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom OJ, Reed KR, van de Wetering M, Muncan V, Winton DJ, Clevers H, Clarke AR 2005. Cyclin D1 is not an immediate target of β-catenin following Apc loss in the intestine. J Biol Chem 280: 28463–28467 [DOI] [PubMed] [Google Scholar]
- Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR 2000. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev 14: 2501–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebolt-Leopold JS 2004. MEK inhibitors: A therapeutic approach to targeting the Ras–MAP kinase pathway in tumors. Curr Pharm Des 10: 1907–1914 [DOI] [PubMed] [Google Scholar]
- Shepherd FA, Rodrigues Pereira J, Ciuleanu T, Tan EH, Hirsh V, Thongprasert S, Campos D, Maoleekoonpiroj S, Smylie M, Martins R, et al. 2005. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med 353: 123–132 [DOI] [PubMed] [Google Scholar]
- Siegfried E, Wilder EL, Perrimon N 1994. Components of wingless signalling in Drosophila. Nature 367: 76–80 [DOI] [PubMed] [Google Scholar]
- Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, Karnezis AN, Swigart LB, Nasi S, Evan GI 2008. Modelling Myc inhibition as a cancer therapy. Nature 455: 679–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek L, Whitfield JR, Sodir NM, Masso-Valles D, Serrano E, Karnezis AN, Swigart LB, Evan GI 2013. Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes Dev 27: 504–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart DJ 2014. Wnt signaling pathway in non-small cell lung cancer. J Natl Cancer Inst 106: djt356. [DOI] [PubMed] [Google Scholar]
- Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, Davis A, Mongare MM, Gould J, Frederick DT, et al. 2012. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature 487: 500–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetsu O, McCormick F 1999. β-Catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398: 422–426 [DOI] [PubMed] [Google Scholar]
- Trejo CL, Juan J, Vicent S, Sweet-Cordero A, McMahon M 2012. MEK1/2 inhibition elicits regression of autochthonous lung tumors induced by KRASG12D or BRAFV600E. Cancer Res 72: 3048–3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trejo CL, Green S, Marsh V, Collisson EA, Iezza G, Phillips WA, McMahon M 2013. Mutationally activated PIK3CAH1047R cooperates with BRAFV600E to promote lung cancer progression. Cancer Res 73: 6448–6461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Cunningham M, Zhang X, Tokarz S, Laraway B, Troxell M, Sears RC 2011. Phosphorylation regulates c-Myc’s oncogenic activity in the mammary gland. Cancer Res 71: 925–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Kobayashi T, Floc’h N, Kinkade CW, Aytes A, Dankort D, Lefebvre C, Mitrofanova A, Cardiff RD, McMahon M, et al. 2012. B-Raf activation cooperates with PTEN loss to drive c-Myc expression in advanced prostate cancer. Cancer Res 72: 4765–4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, Peng J, Lin E, Wang Y, Sosman J, et al. 2012. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature 487: 505–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winslow MM, Dayton TL, Verhaak RG, Kim-Kiselak C, Snyder EL, Feldser DM, Hubbard DD, DuPage MJ, Whittaker CA, Hoersch S, et al. 2011. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature 473: 101–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods D, Parry D, Cherwinski H, Bosch E, Lees E, McMahon M 1997. Raf-induced proliferation or cell cycle arrest is determined by the level of Raf activity with arrest mediated by p21Cip1. Mol Cell Biol 17: 5598–5611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You L, He B, Xu Z, Uematsu K, Mazieres J, Mikami I, Reguart N, Moody TW, Kitajewski J, McCormick F, et al. 2004. Inhibition of Wnt-2-mediated signaling induces programmed cell death in non-small-cell lung cancer cells. Oncogene 23: 6170–6174 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Goss AM, Cohen ED, Kadzik R, Lepore JJ, Muthukumaraswamy K, Yang J, DeMayo FJ, Whitsett JA, Parmacek MS, et al. 2008. A Gata6-Wnt pathway required for epithelial stem cell development and airway regeneration. Nat Genet 40: 862–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Woods D, McMahon M, Bishop JM 1998. Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev 12: 2997–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]