Abstract
The association between xeroderma pigmentosum complementation group D (XPD) Asp312Asn and Lys751Gln gene polymorphisms and breast cancer risk has been widely reported, but the results were inconsistent. In order to derive a more precise estimation of the relationship, a meta-analysis was performed. A comprehensive search strategy was conducted towards the electronic databases including Medline, PubMed, Web of Science, Embase, and Chinese Biomedical Literature Database (Chinese). The association between the XPD polymorphism and breast cancer risk was conducted by odds ratios (ORs) and 95 % confidence intervals (95 % CIs). A total of 22 studies with 18,136 cases and 18,351 controls were included in our meta-analysis. Among these, 12 studies with 7,667 cases and 7,480 controls for Asp312Asn polymorphism and 20 studies with 10,469 cases and 10,871 controls for Lys751Gln polymorphism. With regard to Asp312Asn polymorphism, no significantly associated was found with breast cancer risk. However, significant association was found between Lys751Gln polymorphism and breast cancer risk under all genetic models in overall populations (C vs. A—OR = 1.10, 95 % CI = 1.04–1.17, P = 0.002; CC vs. AA—OR = 1.17, 95 % CI = 1.06–1.30, P = 0.003; AC vs. AA—OR = 1.06, 95 % CI = 1.01–1.12, P = 0.032; CC vs. AC/AA—OR = 1.17, 95 % CI = 1.04–1.32, P = 0.009; CC/AC vs. AA—OR = 1.07, 95 % CI = 1.02–1.12, P = 0.005). In subgroup analysis base on ethnicity, significance was found in Caucasians and mix. The results suggest that XPD Asp312Asn polymorphism was not associated with breast cancer. The XPD Lys751Gln polymorphism significantly increased breast cancer risk, especially for Caucasian and mix.
Keywords: XPD, Polymorphism, Breast cancer, Meta-analysis
Introduction
Breast cancer is the most frequently diagnosed and most prevalent invasive cancers and the leading cause of cancer-related death among women, accounting for 23 % of total cancer cases and 14 % of cancer deaths and 30 % of the new malignant tumors in females over the worldwide, which has become a major public health challenge [1–3]. In fact, the incidence of breast cancer is significantly higher in developed countries than in developing ones, which was the first malignant disease to pose a significant threat to women [4]. But unfortunately, the pathogenesis and progression of breast cancer are still not fully understood. Many studies have concluded that breast cancer is the cumulative result of multiple environmental factors and genetic alterations [5, 6]. Previous studies have suggested that the stimulation of estrogen [7], high weight of birth [8], obesity [9], and family history of breast cancer [10, 11] were associated with increased risk of breast cancer especially in postmenopausal women. But not all people exposed to these risk factors are suffering from breast cancer, which indicated that genetic plays an important role in the development of breast cancer.
In recent years, several common low-penetrant genes have been identified as potential breast cancer susceptibility genes [12]. Xeroderma pigmentosum complementation group D (XPD) or called excision repair cross-complimentary group 2 (ERCC2) is one of the most important low-penetrant gene, which is locating at chromosome 19q13.3 and involving in the nucleotide excision repair (NER) pathway, removes certain DNA cross-links, ultraviolet photolesions, and bulky chemical adducts [13, 14]. The Asp312Asn and Lys751Gln have been identified as the two most common polymorphisms in the coding region of XPD and the most extensively studied [15, 16]. The XPD Asp312Asn polymorphism (rs1799793) at position 312 in exon 10 is characterized by a G to A substitution resulting in aspartic acid (Asp [D]) to asparagine (Asn [N]) amino acid, whereas the Lys751Gln polymorphism (rs13181) is at position 751 in exon 23 and characterized by an A to C substitution causing a lysine (Lys [K]) to glutamine [Gln (Q)]) amino acid exchange [15, 16]. This polymorphism and the association of breast cancer risk has been a research focus in the scientific community and have drawn increasing attention. There were a number of studies reporting the role of XPD Asp312Asn and Lys751Gln polymorphisms in breast cancer risk [17–38], but the results are inconclusive, this may partially be because of the possible small effect of the polymorphism on breast cancer risk and the relatively small sample size in each of the published studies. In order to derive a more precise conclusion, we performed this meta-analysis.
Materials and methods
Search strategy
A comprehensive search strategy was conducted towards the electronic databases including Medline, PubMed, Web of Science, Embase, and Chinese Biomedical Literature Database (Chinese), with keywords “breast cancer”, “breast neoplasm”, “XPD”, “ERCC2”, “polymorphism”, and “variant” for all studies; there were no limitations to the language of publications. Reference lists of the selected papers were screened by hand for potentially relevant articles; review articles were also examined to find additional eligible studies.
Inclusion and exclusion criteria
Studies were selected if they satisfy the following inclusion criteria: (a) case–control design; (b) evaluation of the XPD polymorphism and breast cancer risk; (c) the publications must offer the sample size, distribution of alleles, genotypes, or others information for estimating the odds ratio (OR) and 95 % confidence interval (CI); (d) when multiple publications reported on the same or overlapping data, we used the most recent or largest population. The exclusion criteria were as follows: (a) not a case–control study, (b) no usable data reported, (c) studies contained duplicate data, and (d) case reports or reviews.
Data extraction
Information was carefully extracted from all eligible publications independently by two investigators according to the inclusion criteria mentioned above. If the conflicting evaluations are encountered, an agreement was reached following a discussion; if an agreement could not be reached, then a third author was consulted to resolve the debate The following information were extracted: the name of first author, year of publication, country of origin, ethnicity, genotyping methods, source of the control group, and the distribution of genotypes in case and control groups. We also evaluated whether the genotype distributions were in Hardy–Weinberg equilibrium.
Statistical analysis
The possible association between the XPD Asp312Asn polymorphism and breast cancer risk was evaluated by OR and 95%CI according to allele contrast (A vs. G), homozygote (AA vs. GG), heterozygote (GA vs. GG), recessive (AA vs. GA/GG), and dominant (AA/GA vs. GG) models. While the strength of association between the XPD Lys751Gln polymorphism and breast cancer risk was assessed by OR and 95% CI according to allele contrast (C vs. A), homozygote (CC vs. AA), heterozygote (CA vs. AA), dominant model (CC/AC vs. AA), and recessive model (CC vs. AC/AA), respectively. Subgroup analyses were assessed according to ethnicity. Heterogeneity among studies was checked by a chi-square-based Q statistic test. The effect of heterogeneity was quantified by using a P value as well as I 2 value [39]. An I 2 value of <50 % or P > 0.10 suggest no heterogeneity was existed among studies; ORs were pooled by fixed-effects model (the Mantel–Haenszel method) [40]. Otherwise, the random-effects model (DerSimonian and Laird method) [41] was used.
The publication bias was assessed both by Egger's test (P < 0.05 was considered representative of statistically significant publication bias) [42] and visual observation of funnel plot [43]. A professional web-based program (http://ihg2.helmholtz-muenchen.de/cgibin/hw/hwa1.pl) was conducted to assess the Hardy–Weinberg equilibrium [44] of controls. At P > 0.05, it suggests that controls followed the Hardy–Weinberg equilibrium (HWE) balance. Sensitivity analysis was performed to evaluate the stability of the results. A single study involved in the meta-analysis was omitted each time to reflect the influence of the individual dataset to the pooled ORs [45]. When the Hardy–Weinberg equilibrium disequilibrium existed (P < 0.05), the sensitivity analysis was also conducted. All statistical tests were performed with STATA Software (version 9.2, Stata Corp).
Results
Search results and study characteristics
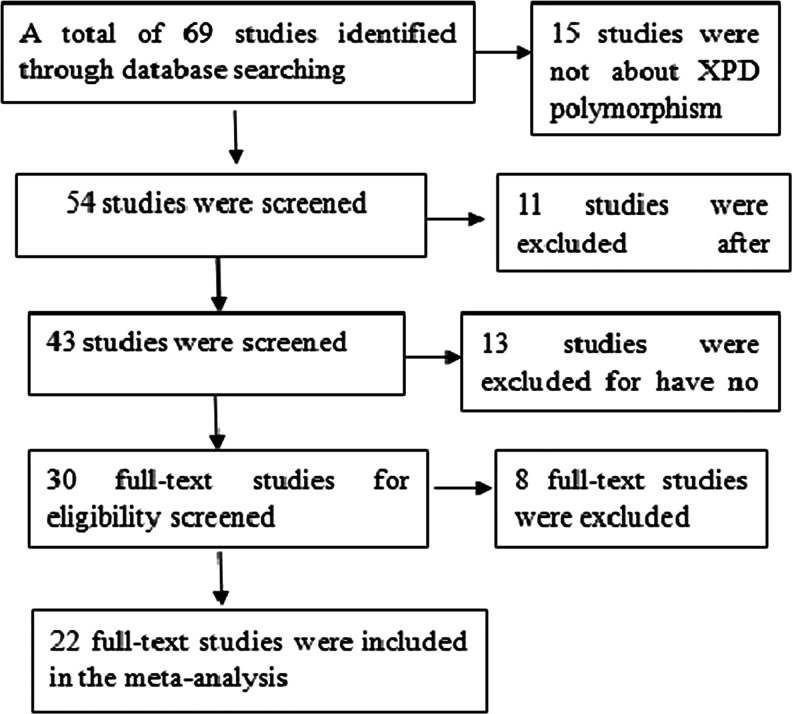
After being examined carefully according to the inclusion criteria (Fig. 1), a total of 22 studies [17–38] with 18,136 cases and 18,351 controls were included in our meta-analysis. Among these, 12 studies with 7,667 cases and 7,480 controls for Asp312Asn polymorphism (Table 1) were included in our meta-analysis and 20 studies with 10,469 cases and 10,871 controls for Lys751Gln polymorphism (Table 2) were included in our meta-analysis. The genotype distribution in the controls of all studies was consistent with HWE (all P > 0.05).
Fig. 1.
Flowchart of selection of studies for inclusion in the meta-analysis
Table 1.
Characteristics of case–control studies included in XPD Asp312Asn (G312A) polymorphism and breast cancer risk
| First author | Year | Country | Ethnicity | Genotyping methods | Source of Control | Cases | Controls | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| GG | GA | AA | GG | GA | AA | ||||||
| Hussien | 2012 | Egypt | Caucasian | ARMS-PCR | HB | 12 | 45 | 43 | 25 | 50 | 25 |
| Jelonek | 2010 | Poland | Caucasian | PCR-RFLP | PB | 41 | 59 | 21 | 85 | 123 | 23 |
| Wang | 2010 | China | Asian | PCR-RFLP | PB | 624 | 388 | 220 | 925 | 315 | 193 |
| Crew | 2007 | USA | Mix | Taqman | PB | 415 | 478 | 138 | 490 | 454 | 139 |
| Shen | 2006 | USA | Mix | Taqman | PB | 60 | 80 | 16 | 59 | 64 | 30 |
| Jorgensen | 2007 | USA | Mix | Taqman | PB | 110 | 128 | 22 | 102 | 142 | 29 |
| Debniak | 2006 | Poland | Caucasian | PCR-RFLP | PB | 672 | 785 | 269 | 492 | 597 | 173 |
| Mechanic(a) | 2006 | USA | Caucasian | PCR-RFLP | PB | 543 | 589 | 130 | 489 | 516 | 128 |
| Mechanic(b) | 2006 | USA | African-American | PCR-RFLP | PB | 564 | 181 | 15 | 517 | 145 | 13 |
| Brauch | 2004 | Germany | Caucasian | PCR-RFLP | PB | 347 | 173 | 47 | 276 | 255 | 79 |
| Shi | 2004 | USA | Caucasian | PCR-RFLP | PB | 29 | 32 | 8 | 46 | 27 | 6 |
| Tang | 2002 | USA | Mix | PCR-RFLP | PB | 52 | 31 | 7 | 74 | 28 | 10 |
PCR-RFLP PCR-restriction fragment length polymorphism, HB hospital based, PB population based
Table 2.
Characteristics of case–control studies included in XPD Lys751Gln (A429C) polymorphisms and breast cancer risk
| First author | Year | Country | Ethnicity | Genotyping methods | Source of Control | Cases | Controls | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AC | CC | AA | AC | CC | ||||||
| Samson | 2010 | India | Asian | TaqMan | PB | 107 | 102 | 41 | 235 | 214 | 51 |
| Jelonek | 2010 | Poland | Caucasian | PCR-RFLP | PB | 54 | 47 | 22 | 116 | 128 | 29 |
| Wang | 2010 | China | Asian | PCR-RFLP | PB | 1136 | 81 | 15 | 1316 | 96 | 21 |
| Syamala | 2009 | India | Asian | PCR-RFLP | HB | 148 | 161 | 50 | 247 | 98 | 22 |
| Synowiec | 2008 | Poland | Caucasian | PCR-RFLP | PB | 15 | 24 | 4 | 30 | 17 | 1 |
| Rajaraman | 2008 | USA | Caucasian | TaqMan | PB | 342 | 377 | 120 | 428 | 494 | 158 |
| Makowska | 2007 | Poland | Caucasian | PCR-RFLP | PB | 16 | 19 | 57 | 26 | 52 | 32 |
| Kipikasova | 2008 | Slovak | Caucasian | PCR-RFLP | PB | 43 | 53 | 18 | 46 | 50 | 17 |
| Shen | 2006 | USA | Mix | Taqman | PB | 63 | 66 | 25 | 74 | 57 | 22 |
| Costa | 2007 | Portugal | Caucasian | PCR-RFLP | PB | 127 | 125 | 30 | 331 | 260 | 69 |
| Jorgensen | 2007 | USA | Mix | Taqman | PB | 30 | 175 | 104 | 34 | 159 | 125 |
| Debniak | 2006 | Poland | Caucasian | PCR-RFLP | PB | 703 | 850 | 277 | 432 | 547 | 162 |
| Onay | 2006 | Canada | Caucasian | Taqman | PB | 146 | 194 | 58 | 165 | 167 | 40 |
| Mechanic(a) | 2006 | USA | Caucasian | PCR-RFLP | PB | 525 | 590 | 158 | 445 | 538 | 150 |
| Mechanic(b) | 2006 | USA | African-American | PCR-RFLP | PB | 415 | 295 | 51 | 393 | 246 | 40 |
| Metsola | 2005 | Finland | Caucasian | PCR-RFLP | PB | 147 | 238 | 96 | 155 | 237 | 88 |
| Brauch | 2004 | Germany | Caucasian | PCR-RFLP | PB | 224 | 265 | 97 | 264 | 292 | 87 |
| Terry | 2004 | USA | Mix | Taqman | PB | 387 | 513 | 153 | 453 | 498 | 151 |
| Shi | 2004 | USA | Caucasian | PCR-RFLP | PB | 30 | 31 | 8 | 38 | 35 | 6 |
| Tang | 2002 | USA | Mix | PCR-RFLP | PB | 45 | 42 | 16 | 54 | 46 | 21 |
PCR-RFLP PCR-restriction fragment length polymorphism, HB hospital based, PB population based
Meta-analysis results
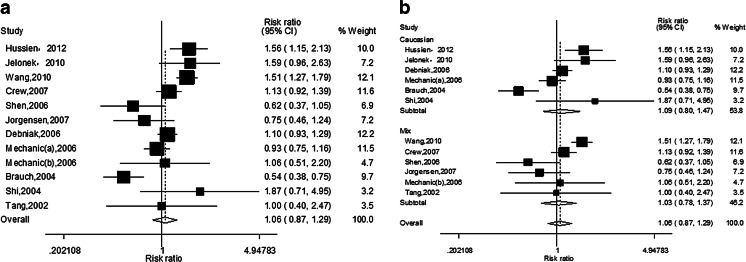
The main results of this meta-analysis and the heterogeneity test were shown in Tables 3 and 4. With regard to Asp312Asn polymorphism, no significant association was found with breast cancer risk in overall populations (A vs. G—OR = 1.06, 95 % CI = 0.95–1.18, P = 0.325; AA vs. GG—OR = 1.06, 95 % CI = 0.87–1.29, P = 0.591 (Fig. 2a); GA vs. GG—OR = 1.07, 95 % CI = 0.96–1.20, P = 0.223; AA vs. GA/GG—OR = 1.04, 95 % CI = 0.87–1.24, P = 0.654; AA/GA vs. GG—OR = 1.06, 95 % CI = 0.96–1.16, P = 0.273). Similarly, there was no association found with breast cancer risk in subgroup analysis base on ethnicity (Table 3; homozygote model is only shown in Fig. 2b).
Table 3.
Results of meta-analysis for XPD Asp312Asn (G312A) polymorphism and breast cancer risk
| Comparison | Population | N | Test of association | Model | Test of heterogeneity | |||
|---|---|---|---|---|---|---|---|---|
| OR | 95 % CI | P | P | I 2 | ||||
| A vs. G | Overall | 12 | 1.06 | 0.95–1.18 | 0.325 | R | 0 | 88.5 |
| Others | 6 | 1.08 | 0.93–1.25 | 0.341 | R | 0 | 85.3 | |
| Caucasians | 6 | 1.04 | 0.89–1.21 | 0.645 | R | 0 | 89.0 | |
| AA vs. GG | Overall | 12 | 1.06 | 0.87–1.29 | 0.591 | R | 0 | 77.3 |
| Others | 6 | 1.03 | 0.78–1.37 | 0.820 | R | 0 | 70.0 | |
| Caucasians | 6 | 1.09 | 0.80–1.47 | 0.584 | R | 0 | 82.2 | |
| GA vs. GG | Overall | 12 | 1.07 | 0.96–1.20 | 0.223 | R | 0 | 85.7 |
| Others | 6 | 1.16 | 0.99–1.36 | 0.067 | R | 0 | 82.0 | |
| Caucasians | 6 | 0.99 | 0.88–1.13 | 0.836 | R | 0 | 82.2 | |
| AA vs. GA / GG | Overall | 12 | 1.04 | 0.87–1.24 | 0.654 | R | 0.001 | 66.5 |
| Others | 6 | 0.97 | 0.75–1.26 | 0.821 | R | 0.025 | 61.0 | |
| Caucasians | 6 | 1.12 | 0.85–1.49 | 0.423 | R | 0.001 | 74.6 | |
| AA / GA vs. GG | Overall | 12 | 1.06 | 0.96–1.16 | 0.273 | R | 0 | 87.9 |
| Others | 6 | 1.11 | 0.97–1.28 | 0.142 | R | 0 | 84.6 | |
| Caucasians | 6 | 1.00 | 0.89–1.14 | 0.962 | R | 0 | 87.1 | |
OR odds ratio, CI confidence interval, F fixed effects model, R random effects model
Table 4.
Results of meta-analysis for XPD Lys751Gln (A751C) polymorphisms and breast cancer risk
| Comparison | Population | N | Test of association | Model | Test of heterogeneity | |||
|---|---|---|---|---|---|---|---|---|
| OR | 95 % CI | P | P | I 2 | ||||
| C vs. A | Overall | 20 | 1.10 | 1.04–1.17 | 0.002 | R | 0 | 76.2 |
| Asian | 3 | 1.28 | 0.87–1.87 | 0.211 | R | 0 | 92.2 | |
| Caucasians | 12 | 1.07 | 1.01–1.14 | 0.020 | R | 0.002 | 63.0 | |
| Mix | 4 | 1.04 | 0.98–1.10 | 0.166 | F | 0.207 | 34,3 | |
| African | 1 | 1.09 | 0.96–1.23 | 0.200 | F | 0 | 0 | |
| CC vs. AA | Overall | 20 | 1.17 | 1.06–1.30 | 0.003 | R | 0.001 | 56.8 |
| Asian | 3 | 1.64 | 0.85–3.25 | 0.140 | R | 0.004 | 81.9 | |
| Caucasians | 12 | 1.09 | 1.00–1.17 | 0.045 | F | 0.119 | 33.9 | |
| Mix | 4 | 1.07 | 0.95–1.21 | 0.242 | F | 0.451 | 0 | |
| African | 1 | 1.19 | 0.80–1.76 | 0.398 | F | 0 | 0 | |
| AC vs. AA | Overall | 20 | 1.06 | 1.01–1.12 | 0.032 | R | 0 | 63.8 |
| Asian | 3 | 1.23 | 0.82–1.86 | 0.322 | R | 0 | 91.1 | |
| Caucasians | 12 | 1.01 | 0.97–1.04 | 0.751 | F | 0.187 | 26.2 | |
| Mix | 4 | 1.08 | 1.02–1.15 | 0.015 | F | 0.705 | 0 | |
| African | 1 | 1.08 | 0.95–1.23 | 0.254 | F | 0 | 0 | |
| CC vs. AC/AA | Overall | 20 | 1.17 | 1.04–1.32 | 0.009 | R | 0.001 | 58.0 |
| Asian | 3 | 1.53 | 0.93–2.52 | 0.097 | R | 0.047 | 67.3 | |
| Caucasians | 12 | 1.19 | 1.03–1.38 | 0.021 | R | 0.009 | 56.0 | |
| Mix | 4 | 0.97 | 0.85–1.12 | 0.715 | F | 0.476 | 0 | |
| African | 1 | 1.14 | 0.76–1.70 | 0.528 | F | 0 | 0 | |
| CC/AC vs. AA | Overall | 20 | 1.07 | 1.02–1.12 | 0.005 | R | 0 | 70.1 |
| Asian | 3 | 1.24 | 0.85–1.80 | 0.273 | R | 0 | 92.3 | |
| Caucasians | 12 | 1.02 | 0.99–1.05 | 0.285 | F | 0.120 | 33.7 | |
| Mix | 4 | 1.06 | 1.01–1.11 | 0.018 | F | 0.280 | 21.8 | |
| African | 1 | 1.08 | 0.96–1.21 | 0.203 | F | 0 | 0 | |
OR odds ratio, CI confidence interval, F fixed effects model, R random effects model
Fig. 2.
a The forest plot describing the meta-analysis under homozygous model for the association between XPD Asp312Asn polymorphism and the risk of breast cancer in overall population (AA vs. GG). b The forest plot describing the meta-analysis under homozygous model for the association between XPD Asp312Asn polymorphism and the risk of breast cancer in subgroup analysis base on ethnicity (AA vs. GG)
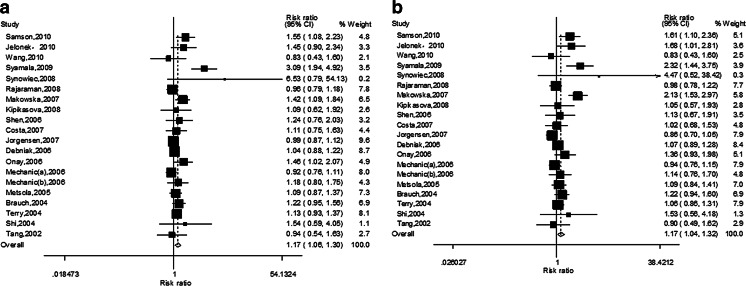
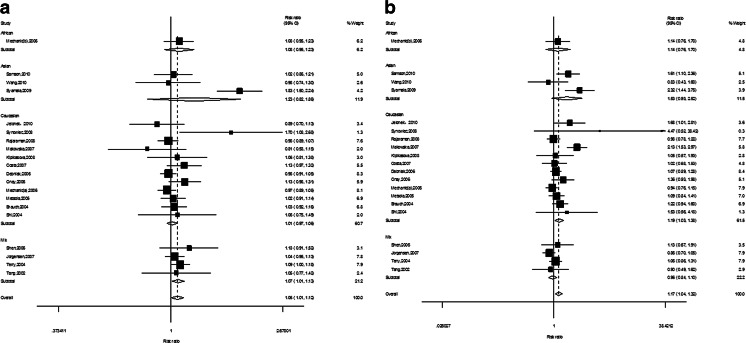
However, significant association was found between Lys751Gln polymorphism and breast cancer risk under all genetic models in overall populations (C vs. A—OR = 1.10, 95 % CI = 1.04–1.17, P = 0.002; CC vs. AA—OR = 1.17, 95 % CI = 1.06–1.30, P = 0.003 (Fig. 3a); AC vs. AA—OR = 1.06, 95 % CI = 1.01–1.12, P = 0.032; CC vs. AC/AA—OR = 1.17, 95 % CI = 1.04–1.32, P = 0.009 (Fig. 3b); CC/AC vs. AA—OR = 1.07, 95 % CI = 1.02–1.12, P = 0.005). In subgroup analysis base on ethnicity, significance was found in Caucasians (C vs. A—OR = 1.07, 95 % CI = 1.01–1.14, P = 0.020; CC vs. AA—OR = 1.09, 95 % CI = 1.00–1.17, P = 0.045; CC vs. AC/AA—OR = 1.19, 95 % CI = 1.03–1.38, P = 0.021 (Fig. 4b)) and mix (AC vs. AA—OR = 1.08, 95 % CI = 1.02–1.15, P = 0.015 (Fig.4a); CC/AC vs. AA—OR = 1.06, 95 % CI = 1.01–1.11, P = 0.018).
Fig. 3.
a The forest plot describing the meta-analysis under homozygous model for the association between XPD Lys751Gln polymorphism and the risk of breast cancer in overall population (CC vs. AA). b The forest plot describing the meta-analysis under recessive model for the association between XPD Lys751Gln polymorphism and the risk of breast cancer in overall population (CC vs. CA/AA)
Fig. 4.
a The forest plot describing the meta-analysis under heterozygote model for the association between XPD Lys751Gln polymorphism and the risk of breast cancer in subgroup analysis base on ethnicity (CA vs. AA). b The forest plot describing the meta-analysis under recessive model for the association between XPD Lys751Gln polymorphism and the risk of breast cancer in subgroup analysis base on ethnicity (CC vs. CA/AA)
Sensitive analysis
Sensitivity analyses were conducted to determine whether modification of the inclusion criteria of the meta-analysis affected the final results. The statistical significance of the results was not change when any single study was omitted, indicating the stability of our results (data not shown). So, results of the sensitivity analysis suggest that the data in our meta-analysis are relatively stable and credible.
Publication bias
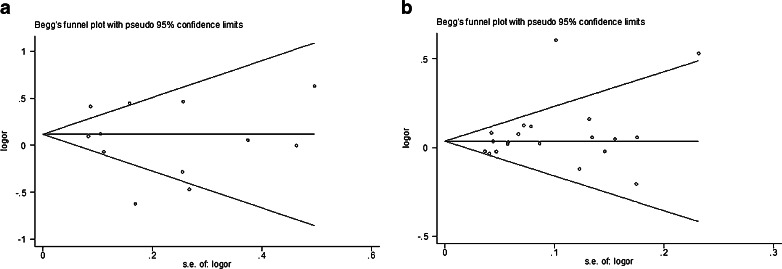
Both Funnel plot and Egger's test were performed to access the publication bias of our meta-analysis. Funnel plot is relatively straightforward to observe whether the publication bias is presence, and Egger's test was used to provide statistical evidence of symmetries of the plots. As shown in Fig. 5a (Asp312Asn polymorphism) and Fig. 5b (Lys751Gln polymorphism), the shape of the funnel plot did not show obvious asymmetry. Similarly, the results of Egger's test shows no publication bias was found too (all P > 0.05, data not shown).
Fig. 5.
a Begg funnel plot for publication bias test for the association between XPD Asp312Asn polymorphism and the risk of breast cancer under homozygous model (AA vs. GG). Each point represents a separate study for the indicated association. Log [OR], natural logarithm of OR. Horizontal line means effect size. b Begg funnel plot for publication bias test for the association between XPD Lys751Gln polymorphism and the risk of breast cancer under heterozygote model (CA vs. AA). Each point represents a separate study for the indicated association. Log [OR], natural logarithm of OR. Horizontal line means effect size
Heterogeneity analysis
There was a significant heterogeneity found in both Asp312Asn and Lys751Gln polymorphisms. To examine the source of heterogeneity, we analyze the dominant model by ethnicity, source of control (hospital or population based), genotyping methods (PCR-RFLP or TaqMan or ARMS-PCR), and sample size (≤400 subjects or >400 subjects). As a result, ethnicity (P = 0.012) but not sample size (P > 0.05), genotyping methods (P > 0.05) or source of control (P > 0.05) was found to contribute to substantial heterogeneity.
Discussion
Breast cancer is one of the most common malignant tumors and the leading causes of cancer-related death among females in the world and it is a threat to women's health. Many candidate genes have been reported to be involved in breast cancer susceptibility, such as CYP19 [46], CASP8 [47], GSM1 [48], hOGG1 [49], and so on. XPD is a DNA-dependent ATPase/helicase that is associated with the TFIIH transcription factor complex and plays an important role in NER pathway. XPD participates in the opening of the DNA helix to allow the excision of the DNA fragment containing the damaged base [50–52]. To date, a number of epidemiological studies have been conducted to evaluate the role of polymorphism Asp312Asn and Lys751Gln on breast cancer susceptibility, but the results remained controversial.
In order to derive a more precise estimation of the relationship, we performed this meta-analysis of 22 studies including 18,136 cases and 18,351 controls. Our results suggest that the XPD Asp312Asn polymorphism is not associated with breast cancer development, but strong association between XPD Lys751Gln polymorphism and breast cancer risk was found in overall populations; in the subgroup analysis base on ethnicity, a significant association was also found in Caucasians and mix suggesting a possible role of ethnic differences in genetic backgrounds and the environment they lived in.
Heterogeneity plays an important role when performing meta-analysis; so, finding the source of heterogeneity is very important for the final result of the meta-analysis. In current study, an obvious heterogeneity between the study was found in the overall population. Heterogeneity cannot be explained by several possible source of heterogeneity such as source of control (hospital or population based), genotyping methods (PCR-RFLP or TaqMan or MassARRAY) or sample size (≤400 subjects or >400 subjects). By conducting meta-regression, we found that ethnicity was the major source of high heterogeneity in our meta-analysis, which could be explained by the race-specific effect of XPD Asp312Asn polymorphism and Lys751Gln polymorphism on the susceptibility to breast cancer because different countries may have different genetic backgrounds and life styles. However, ethnicity did not explain all heterogeneity in this meta-analysis and other sources need further investigating. It is possible that other limitations of the recruited studies may partially contribute to the observed heterogeneity. For this reason, we conducted analyses using the random effects model. Publication bias is another important aspect which may have a negative effect on meta-analysis. In our meta-analysis, both Funnel plot and Egger's test were used to test the publication bias of the included studies. As a result, both the shape of the funnel plot and statistical results show no obvious publication bias, this suggests that the publication bias have little effect on the results in our study and the results of our meta-analysis are relatively stable.
Although comprehensive analysis was performed to show the association between XPD Asp312Asn and Lys751Gln polymorphisms and breast cancer risk, there are still some limitations that should be pointed out. Firstly, the number of studies and the number of samples included in the meta-analysis were relatively small. Secondly, the controls were not uniformly defined. Some studies used controls that were population-based, while others used hospital-based controls, which may not be representative of the general population. Thirdly, in the subgroup analysis, the number of Africans was relatively small, thus not having enough statistical power to explore the real association. Finally, our results were based on unadjusted estimates, while a more precise analysis should be conducted if individual data were available, which would allow for the adjustment by other covariants including age, menopausal status, obesity, environmental factors, and lifestyle.
Despite the limitations above, our meta-analysis also had several advantages. Firstly, a meta-analysis of the association of XPD polymorphism on breast cancer risk is statistically more powerful than any other single study. Secondly, the quality of case–control studies included in the meta-analysis was met our inclusion criteria and was satisfactory; the sensitivity analysis and publication bias analysis indicated the results of our meta-analysis are stability, credibility, and convincing. Thirdly, strict searching strategy, which combines computer-assisted with manual search, makes the eligible studies included as much as possible.
In summary, the results suggest that XPD Asp312Asn was not associated with breast cancer. While XPD Lys751Gln polymorphism significantly increased breast cancer risk, especially for Caucasian and mix. Considering the limited sample size and ethnicities included in the meta-analysis, further larger-scaled and well-designed studies are needed to confirm our results. Moreover, gene–gene and gene–environment interactions should also be considered in future analysis.
Footnotes
Yulan Yan, Hongjie Liang, and Morning Light contributed equally to this work so that they should be considered as the co-fist authors.
Contributor Information
Yulan Yan, Email: xueying201120521@163.com.
Hongjie Liang, Email: lianghongjie2004@163.com.
Morning Light, Email: morninglight2008@hotmail.com.
Taijie Li, Email: 715085562@qq.com.
Yan Deng, Email: 2609025345@qq.com.
Meng Li, Email: 1825725213@qq.com.
Shan Li, Email: panyan1002@163.com.
Xue Qin, Email: xijunshi56234@163.com.
References
- 1.Smigal C, Jemal A, Ward E, Cokkinides V, Smith R, Howe HL, et al. Trends in breast cancer by race and ethnicity: update 2006. CA Cancer J Clin. 2006;56:168–83. doi: 10.3322/canjclin.56.3.168. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 4.Sturgeon SR, Schairer C, Grauman D, El Ghormli L, Devesa S. Trends in breast cancer mortality rates by region of the united states, 1950–1999. Cancer Causes Control. 2004;15:987–95. doi: 10.1007/s10552-004-1092-2. [DOI] [PubMed] [Google Scholar]
- 5.Pei J, Li F, Wang B: Single nucleotide polymorphism 6q25.1 rs2046210 and increased risk of breast cancer. Tumour Biol 2013 [DOI] [PubMed]
- 6.Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, et al. Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 7.Cheung KL. Endocrine therapy for breast cancer: an overview. Breast. 2007;16:327–43. doi: 10.1016/j.breast.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Silva Idos S, De Stavola B, McCormack V. Birth size and breast cancer risk: re-analysis of individual participant data from 32 studies. PLoS Med. 2008;5:e193. doi: 10.1371/journal.pmed.0050193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaman K, Bodmer A, Pralong F, Castiglione-Gertsch M. Breast cancer and obesity, a dangerous relation. Rev Med Suisse. 2012;8:1101–4. [PubMed] [Google Scholar]
- 10.Berclaz G, Li S, Price KN, Coates AS, Castiglione-Gertsch M, Rudenstam CM, et al. Body mass index as a prognostic feature in operable breast cancer: the International Breast Cancer Study group experience. Ann Oncol. 2004;15:875–84. doi: 10.1093/annonc/mdh222. [DOI] [PubMed] [Google Scholar]
- 11.Hankinson SE. Circulating levels of sex steroids and prolactin in premenopausal women and risk of breast cancer. Adv Exp Med Biol. 2008;617:161–9. doi: 10.1007/978-0-387-69080-3_15. [DOI] [PubMed] [Google Scholar]
- 12.Qiu LX, Yao L, Zhang J, Zhu XD, Zhao XM, Xue K, et al. Xpd lys751gln polymorphism and breast cancer susceptibility: a meta-analysis involving 28,709 subjects. Breast Cancer Res Treat. 2010;124:229–35. doi: 10.1007/s10549-010-0813-3. [DOI] [PubMed] [Google Scholar]
- 13.Flejter WL, McDaniel LD, Johns D, Friedberg EC, Schultz RA. Correction of xeroderma pigmentosum complementation group D mutant cell phenotypes by chromosome and gene transfer: involvement of the human ercc2 DNA repair gene. Proc Natl Acad Sci U S A. 1992;89:261–5. doi: 10.1073/pnas.89.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coin F, Marinoni JC, Rodolfo C, Fribourg S, Pedrini AM, Egly JM. Mutations in the XPD helicase gene result in XP and TTD phenotypes, preventing interaction between XPD and the p44 subunit of TFIIH. Nat Genet. 1998;20:184–8. doi: 10.1038/2491. [DOI] [PubMed] [Google Scholar]
- 15.Shen MR, Jones IM, Mohrenweiser H. Nonconservative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humans. Cancer Res. 1998;58:604–8. [PubMed] [Google Scholar]
- 16.Pabalan N, Francisco-Pabalan O, Sung L, Jarjanazi H, Ozcelik H. Meta-analysis of two ERCC2 (XPD) polymorphisms, asp312asn and lys751gln, in breast cancer. Breast Cancer Res Treat. 2010;124:531–41. doi: 10.1007/s10549-010-0863-6. [DOI] [PubMed] [Google Scholar]
- 17.Hussien YM, Gharib AF, Awad HA, Karam RA, Elsawy WH. Impact of DNA repair genes polymorphism (XPD and XRCC1) on the risk of breast cancer in Egyptian female patients. Mol Biol Rep. 2012;39:1895–901. doi: 10.1007/s11033-011-0935-7. [DOI] [PubMed] [Google Scholar]
- 18.Samson M, Singh SS, Rama R, Sridevi V, Rajkumar T. Xpd lys751gln increases the risk of breast cancer. Oncol Lett. 2011;2:155–9. doi: 10.3892/ol.2010.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jelonek K, Gdowicz-Klosok A, Pietrowska M, Borkowska M, Korfanty J, Rzeszowska-Wolny J, et al. Association between single-nucleotide polymorphisms of selected genes involved in the response to DNA damage and risk of colon, head and neck, and breast cancers in a Polish population. J Appl Genet. 2010;51:343–52. doi: 10.1007/BF03208865. [DOI] [PubMed] [Google Scholar]
- 20.Wang HC, Liu CS, Wang CH, Tsai RY, Tsai CW, Wang RF, et al. Significant association of XPD asp312asn polymorphism with breast cancer in Taiwanese patients. Chin J Physiol. 2010;53:130–5. doi: 10.4077/CJP.2010.AMK005. [DOI] [PubMed] [Google Scholar]
- 21.Syamala VS, Syamala V, Sreedharan H, Raveendran PB, Kuttan R, Ankathil R. Contribution of XPD (lys751gln) and XRCC1 (arg399gln) polymorphisms in familial and sporadic breast cancer predisposition and survival: an Indian report. Pathol Oncol Res. 2009;15:389–97. doi: 10.1007/s12253-008-9135-8. [DOI] [PubMed] [Google Scholar]
- 22.Synowiec E, Stefanska J, Morawiec Z, Blasiak J, Wozniak K. Association between DNA damage, DNA repair genes variability and clinical characteristics in breast cancer patients. Mutat Res. 2008;648:65–72. doi: 10.1016/j.mrfmmm.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 23.Rajaraman P, Bhatti P, Doody MM, Simon SL, Weinstock RM, Linet MS, et al. Nucleotide excision repair polymorphisms may modify ionizing radiation-related breast cancer risk in US radiologic technologists. Int J Cancer. 2008;123:2713–6. doi: 10.1002/ijc.23779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Romanowicz-Makowska H, Sobczuk A, Smolarz B, Fiks T, Kulig A. Xpd lys751gln polymorphism analysis in women with sporadic breast cancer. Pol J Pathol. 2007;58:245–9. [PubMed] [Google Scholar]
- 25.Kipikasova L, Wolaschka T, Bohus P, Baumohlova H, Bober J, Blazejova J, et al. Polymorphisms of the XRCC1 and XPD genes and breast cancer risk: a case–control study. Pathol Oncol Res. 2008;14:131–5. doi: 10.1007/s12253-008-9034-z. [DOI] [PubMed] [Google Scholar]
- 26.Crew KD, Gammon MD, Terry MB, Zhang FF, Zablotska LB, Agrawal M, et al. Polymorphisms in nucleotide excision repair genes, polycyclic aromatic hydrocarbon-DNA adducts, and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2007;16:2033–41. doi: 10.1158/1055-9965.EPI-07-0096. [DOI] [PubMed] [Google Scholar]
- 27.Shen J, Desai M, Agrawal M, Kennedy DO, Senie RT, Santella RM, et al. Polymorphisms in nucleotide excision repair genes and DNA repair capacity phenotype in sisters discordant for breast cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:1614–9. doi: 10.1158/1055-9965.EPI-06-0218. [DOI] [PubMed] [Google Scholar]
- 28.Costa S, Pinto D, Pereira D, Rodrigues H, Cameselle-Teijeiro J, Medeiros R, et al. DNA repair polymorphisms might contribute differentially on familial and sporadic breast cancer susceptibility: a study on a Portuguese population. Breast Cancer Res Treat. 2007;103:209–17. doi: 10.1007/s10549-006-9364-z. [DOI] [PubMed] [Google Scholar]
- 29.Jorgensen TJ, Visvanathan K, Ruczinski I, Thuita L, Hoffman S, Helzlsouer KJ. Breast cancer risk is not associated with polymorphic forms of xeroderma pigmentosum genes in a cohort of women from Washington County, Maryland. Breast Cancer Res Treat. 2007;101:65–71. doi: 10.1007/s10549-006-9263-3. [DOI] [PubMed] [Google Scholar]
- 30.Debniak T, Scott RJ, Huzarski T, Byrski T, Masojc B, van de Wetering T, et al. XPD common variants and their association with melanoma and breast cancer risk. Breast Cancer Res Treat. 2006;98:209–15. doi: 10.1007/s10549-005-9151-2. [DOI] [PubMed] [Google Scholar]
- 31.Onay VU, Briollais L, Knight JA, Shi E, Wang Y, Wells S, et al. SNP–SNP interactions in breast cancer susceptibility. BMC Cancer. 2006;6:114. doi: 10.1186/1471-2407-6-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mechanic LE, Millikan RC, Player J, de Cotret AR, Winkel S, Worley K, et al. Polymorphisms in nucleotide excision repair genes, smoking and breast cancer in African Americans and whites: a population-based case–control study. Carcinogenesis. 2006;27:1377–85. doi: 10.1093/carcin/bgi330. [DOI] [PubMed] [Google Scholar]
- 33.Brewster AM, Jorgensen TJ, Ruczinski I, Huang HY, Hoffman S, Thuita L, et al. Polymorphisms of the DNA repair genes XPD (lys751gln) and XRCC1 (arg399gln and arg194trp): relationship to breast cancer risk and familial predisposition to breast cancer. Breast Cancer Res Treat. 2006;95:73–80. doi: 10.1007/s10549-005-9045-3. [DOI] [PubMed] [Google Scholar]
- 34.Metsola K, Kataja V, Sillanpaa P, Siivola P, Heikinheimo L, Eskelinen M, et al. XRCC1 and XPD genetic polymorphisms, smoking and breast cancer risk in a Finnish case–control study. Breast Cancer Res. 2005;7:R987–97. doi: 10.1186/bcr1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Justenhoven C, Hamann U, Pesch B, Harth V, Rabstein S, Baisch C, et al. ERCC2 genotypes and a corresponding haplotype are linked with breast cancer risk in a German population. Cancer Epidemiol Biomarkers Prev. 2004;13:2059–64. [PubMed] [Google Scholar]
- 36.Terry MB, Gammon MD, Zhang FF, Eng SM, Sagiv SK, Paykin AB, et al. Polymorphism in the DNA repair gene XPD, polycyclic aromatic hydrocarbon-DNA adducts, cigarette smoking, and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2004;13:2053–8. [PubMed] [Google Scholar]
- 37.Shi Q, Wang LE, Bondy ML, Brewster A, Singletary SE, Wei Q. Reduced DNA repair of benzo[a]pyrene diol epoxide-induced adducts and common xpd polymorphisms in breast cancer patients. Carcinogenesis. 2004;25:1695–700. doi: 10.1093/carcin/bgh167. [DOI] [PubMed] [Google Scholar]
- 38.Tang D, Cho S, Rundle A, Chen S, Phillips D, Zhou J, et al. Polymorphisms in the DNA repair enzyme xpd are associated with increased levels of PAH-DNA adducts in a case–control study of breast cancer. Breast Cancer Res Treat. 2002;75:159–66. doi: 10.1023/A:1019693504183. [DOI] [PubMed] [Google Scholar]
- 39.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–58. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 40.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–48. [PubMed] [Google Scholar]
- 41.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–88. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 42.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–101. doi: 10.2307/2533446. [DOI] [PubMed] [Google Scholar]
- 44.Zamora-Ros R, Rothwell JA, Scalbert A, Knaze V, Romieu I, Slimani N, Fagherazzi G, Perquier F, Touillaud M, Molina-Montes E, Huerta JM, Barricarte A, Amiano P, Menendez V, Tumino R, de Magistris MS, Palli D, Ricceri F, Sieri S, Crowe FL, Khaw KT, Wareham NJ, Grote V, Li K, Boeing H, Forster J, Trichopoulou A, Benetou V, Tsiotas K, Bueno-de-Mesquita HB, Ros M, Peeters PH, Tjonneland A, Halkjaer J, Overvad K, Ericson U, Wallstrom P, Johansson I, Landberg R, Weiderpass E, Engeset D, Skeie G, Wark P, Riboli E, Gonzalez CA: Dietary intakes and food sources of phenolic acids in the European Prospective Investigation into Cancer and Nutrition (epic) study. Br J Nutr 2013:1–12. [DOI] [PubMed]
- 45.Tobias A, Campbell MJ. Modelling influenza epidemics in the relation between black smoke and total mortality. A sensitivity analysis. J Epidemiol Community Health. 1999;53:583–4. doi: 10.1136/jech.53.9.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma X, Qi X, Chen C, Lin H, Xiong H, Li Y, et al. Association between cyp19 polymorphisms and breast cancer risk: results from 10,592 cases and 11,720 controls. Breast Cancer Res Treat. 2010;122:495–501. doi: 10.1007/s10549-009-0693-6. [DOI] [PubMed] [Google Scholar]
- 47.Sergentanis TN, Economopoulos KP. Association of two casp8 polymorphisms with breast cancer risk: a meta-analysis. Breast Cancer Res Treat. 2010;120:229–34. doi: 10.1007/s10549-009-0471-5. [DOI] [PubMed] [Google Scholar]
- 48.Qiu LX, Yuan H, Yu KD, Mao C, Chen B, Zhan P, et al. Glutathione S-transferase m1 polymorphism and breast cancer susceptibility: a meta-analysis involving 46,281 subjects. Breast Cancer Res Treat. 2010;121:703–8. doi: 10.1007/s10549-009-0636-2. [DOI] [PubMed] [Google Scholar]
- 49.Yuan W, Xu L, Feng Y, Yang Y, Chen W, Wang J, et al. The hOGG1 ser326cys polymorphism and breast cancer risk: a meta-analysis. Breast Cancer Res Treat. 2010;122:835–42. doi: 10.1007/s10549-009-0722-5. [DOI] [PubMed] [Google Scholar]
- 50.Benhamou S, Sarasin A. ERCC2/XPD gene polymorphisms and cancer risk. Mutagenesis. 2002;17:463–9. doi: 10.1093/mutage/17.6.463. [DOI] [PubMed] [Google Scholar]
- 51.Manuguerra M, Saletta F, Karagas MR, Berwick M, Veglia F, Vineis P, et al. XRCC3 and XPD/ERCC2 single nucleotide polymorphisms and the risk of cancer: a huge review. Am J Epidemiol. 2006;164:297–302. doi: 10.1093/aje/kwj189. [DOI] [PubMed] [Google Scholar]
- 52.Yin QH, Liu C, Hu JB, Meng RR, Li L, Wang YJ. Xpd lys751gln and asp312asn polymorphisms and gastric cancer susceptibility: a meta-analysis of case–control studies. Asian Pac J Cancer Prev. 2013;14:231–6. doi: 10.7314/APJCP.2013.14.1.231. [DOI] [PubMed] [Google Scholar]