Abstract
IgASE1, a C18 Δ9-specific polyunsaturated fatty acid elongase from the marine microalga Isochrysis galbana, is able to convert linoleic acid and α-linolenic acid to eicosadienoic acid and eicosatrienoic acid in Arabidopsis. Eicosadienoic acid and eicosatrienoic acid are precursors of arachidonic acid, eicosapentaenoic acid, and docosahexaenoic acid, which are synthesized via the Δ8 desaturation biosynthetic pathways. This study shows that the IgASE1-expressing transgenic Arabidopsis exhibited altered morphology (decreased leaf area and biomass) and enhanced drought resistance compared to wild-type plants. The transgenic Arabidopsis were hypersensitive to abscisic acid (ABA) during seed germination, post-germination growth, and seedling development. They had elevated leaf ABA levels under well-watered and dehydrated conditions and their stomata were more sensitive to ABA. Exogenous application of eicosadienoic acid and eicosatrienoic acid can mimic ABA and drought responses in the wild type plants, similar to that found in the transgenic ones. The transcript levels of genes involved in the biosynthesis of ABA (NCED3, ABA1, AAO3) as well as other stress-related genes were upregulated in this transgenic line upon osmotic stress (300mM mannitol). Taken together, these results indicate that these two eicosapolyenoic acids or their derived metabolites can mitigate the effects of drought in transgenic Arabidopsis, at least in part, through the action of ABA.
Key words: ABA, abscisic acid, Arabidopsis, drought resistance, eicosapolyenoic acids, IgASE1
Introduction
Very-long-chain polyunsaturated fatty acids (VLCPUFAs), such as arachidonic acid (ARA; 20:4 Δ5,8,11,14) and eicosapentaenoic acid (20:5 Δ5,8,11,14,17), are key molecules that participate in various biological processes in the cell. In animals these VLCPUFAs affect health and nutrition by regulating the expression of genes through changes in the rate of transcription or post-transcriptional modifications (Schroeder et al., 2008; Dimri et al., 2010; Barnes et al., 2012). These include neonatal retinal and brain development (Lauritzen et al., 2001; Fleith and Clandinin, 2005; Leinster et al., 2010) and cardiovascular health and disease prevention (Breslow, 2006; Von Schacky and Harris, 2007; Serini et al., 2011). Since no higher plant can synthesize these VLCPUFAs naturally, genetic modification of plants to produce these health beneficial fatty acids has been the subject of intensive research in recent years. Consequently, various VCLPUFA-producing transgenic plants have been generated, including Arabidopsis (Qi et al., 2004), linseed (Abbadi et al., 2004), soybean (Kinney et al., 2004), and Brassica juncea (Wu et al., 2005; Cheng et al., 2010; Venegas-Calerón et al., 2010).
Although these VLCPUFAs are not commonly found in higher plants, they are abundant in lipids of pathogens seriously affecting crop yield, including Phytophthora species and related oomycetes (Sun et al., 2013). These fatty acids are released into plant tissue from spores during early stages of infection (Choi et al., 1992). Plants also respond to these fatty acids either by exogenous application or via pathogens containing them during infection, triggering the coordinated activation of defence-related responses (Bostock et al., 1986). Studies show that ARA and eicosapentaenoic acid are potent elicitors of programmed cell death and defence responses in Solanaceous plants (Bostock et al., 1981; García-Pineda et al., 2004). Thus, these eicosapolyenoic acids can function as signalling molecules in various organisms (Rozhnova et al., 2003; Savchenko et al., 2010).
Taking advantage of eicosapolyenoic acid-producing transgenic Arabidopsis (Qi et al., 2004), another study further validated the roles of ARA as a conserved signalling molecule in the regulation of biotic stresses in plants (Savchenko et al., 2010). The expression of jasmonic acid-biosynthetic genes in the transgenic plants was upregulated, thus increasing their resistance to pathogenic microorganisms (Savchenko et al., 2010). Therefore, eicosapolyenoic acids, such as ARA, play a regulatory role as novel pathogen-associated molecular patterns (or PAMPs) in oomycete-plant defence signalling networks (Bostock et al., 2011).
In plants, it is well recognized that C16 and C18 polyunsaturated fatty acids such as hexadecatrienoic acid (16:3 Δ7,10,13) and α-linolenic acid (ALA; 18:3 Δ9,12,15) as well as their derived metabolites can modulate signal transduction pathways evoked by abiotic stress (Zhang et al., 2005; Torres-Franklin et al., 2009). This is because these fatty acids could reduce the structural and/or functional damages of cellular membranes caused by physiological stresses (Somerville and Browse, 1991; Allakhverdiev et al., 2001; Iba, 2002; Upchurch, 2008). However, currently there are no reports on the roles that eicosapolyenoic acids, or indeed any other C20+ VLCPUFAs, play in abiotic stress responses in plants. Therefore, there is an urgent need to investigate the effect of these novel fatty acids on crop physiology and adaptation to the environment before introducing VLCPUFA-producing transgenic crops into the field for commercial production.
We generated transgenic Arabidopsis plant lines that produced appreciable amounts of the eicosapolyenoic acids eicosadienoic acid (EDA; 20:2 Δ11,14) and eicosatrienoic acid (ETrA; 20:3 Δ11,14,17) (Fraser et al., 2004). This was achieved by constitutive expression of a Δ9 elongase gene, IgASE1, from the docosahexaenoic acid-producing marine microalga Isochrysis galbana (Qi et al., 2002, 2003). Analysis of the fatty acids in the leaf glycerolipids revealed that these two novel fatty acids were particularly rich in membrane phospholipids such as phosphatidylcholine, phosphatidylethanolamine, phosphatidate, and phosphatidylinositol (Fraser et al., 2004). In this study, we isolated a higher EDA- and ETrA-producing transgenic line where these two fatty acids constituted approximately 25 mol% of total leaf fatty acids. In contrast to the previous lower-producing (15%) line, it exhibited some abnormal morphology coupled with enhanced drought resistance. As a stress signal, abscisic acid (ABA) plays important roles in regulating drought response in plants (Ashraf, 2010). Therefore, we monitored the transcript levels of genes involved in the biosynthesis of ABA as well as other stress-related genes in this transgenic line and found that the drought resistance of the transgenic Arabidopsis was both ABA-dependent and -independent.
Materials and methods
Plant material and growth conditions
Arabidopsis thaliana ecotype Columbia 4 (Col-4) and 35S:IgASE1 transgenic plants in the Col-4 background (Qi et al., 2004) were used for this study. Seed germination and plant growth conditions were as according to Pandey et al. (2006). Briefly, seed batches that were produced, harvested, and stored under identical conditions for 2 months were used. About 60 seeds from the wild-type (WT) and transgenic plants were surface sterilized and plated on the same petri dish containing 0.8% agar, ½ Murashige and Skoog (MS) salts, and 1% (w/v) sucrose. They were either chilled at 4 °C in the dark for 48h (stratified) or moved directly (non-stratified) to an environmental controlled growth room to germinate under a 16/8h light/dark regime at 21±1 °C. For soil-grown plants, 7-day-old plate-grown seedlings were transferred to 4×4cm pots filled with Levington F2 compost and cultivated under the same long day conditions. After 4 weeks the leaf areas of four individual plants were measured. For drought-stress assays, plants were grown in 6×6cm pots under 10/14h light/dark at 21±1 °C. After 3 weeks, water was withheld for 22 days and then pots were subsequently drenched with water.
For ABA and glycerol treatments, indicated concentrations of ABA and 2% glycerol were included in the ½ MS medium. To study the effect of EDA and ETrA, seeds were germinated on the same media combination that also contained the indicated amount of EDA, ETrA, or EDA+ETrA (sodium salt; Nu-Chek Prep, Elysian, MN, USA). Both EDA and ETrA were dissolved in 1% NP40 (Sigma-Aldrich, St Louis, MO, USA) to make a stock solution of 2.5mM. Germination is defined here as an emergence of the radicle through the seed coat. To study the response of dehydration by mannitol and glycerol, 3-day-old ½ MS-grown seedlings were transferred to fresh ½ MS, ½ MS containing 300mM mannitol, or 3% (v/v) glycerol and grown for further 7 days. The experiments were repeated three times. The data shown are means ± SE of all three experiments, unless stated otherwise.
Fatty acid analysis
Fatty acids from leaves were extracted and converted to their fatty acid methyl esters as described by Browse et al. (1986). The fatty acid methyl esters were analysed by gas chromatography on a 30 m × 0.25mm DB-23 column (J&W Scientific, Folsom, CA, USA) using heptadecanoic acid (17:0) as an internal standard and quantified by flame ionization detection (Fraser et al., 2004).
Stomatal aperture assay
Stomatal aperture was measured according to Pei et al. (1997) with slight modifications. Briefly, 10 of the first fully expanded rosette leaves were carefully removed from 4-week-old soil-grown plants cultivated under long days as described above. They were floated on buffer containing 10mM 2-(N-morpholine)-ethanesulphonic acid (MES), 50mM KCl, and 0.1mM CaCl2 (pH 6.15) and exposed to cool white light (120 µm·m−2·s−l) at 21 °C for 2h to induce stomatal opening. They were then transferred to the same buffer, with or without 10 μM ABA, and incubated for a further 2h. Abaxial epidermal peels were carefully removed and observed in the same buffers with an Olympus BX51 light microscope using a 20× objective lens. Images were captured using an Olympus DP71 U-TV0.5XC-3 digital camera attached to the microscope. Widths and lengths of approximately 60 stomata were measured that had guard cells with an inner edge of 16–22 µm (Pei et al., 1997), and estimated using Image J software. Data are the means of three experiments.
ABA assay
Leaves were harvested from 4-week-old WT and transgenic plants. Both fresh and dehydrated leaves (air-dried for 1h) were immediately frozen in liquid nitrogen. They were ground to a fine powder and homogenized in 90% (v/v) methanol containing 200 mg·l−1 diethydithiocarbamic acid sodium salt. The extracts were then incubated in a capped glass tube in darkness at 4 °C for 16h, followed by low-speed centrifugation at 2000rpm for 5min. The methanolic supernatant was recovered, dried, and the residue dissolved using methanolic Tris buffer (10% methanol/50mM Tris, pH 8.0/1mM MgCl2/150mM NaCl). An ABA ELISA quantitation kit was used to determine ABA content following the manufacturer’s instructions (Agdia, Elkhart, IN, USA; http://www.agdia.com).
Water loss from detached leaves
Five leaves of similar developmental stages from 4-week-old soil-grown plants were detached and weighed immediately. They were placed on a laboratory bench and the weight of individual leaves recorded every hour for 7h. The relative fresh weight at each time was calculated as the percentage of the initial fresh weight to indicate the rate of water loss from the leaves. The experiment was repeated three times.
RNA isolation and real-time PCR
Seven-day-old seedlings were transferred to liquid MS medium supplemented with 100 μM ABA, 300mM mannitol, or solvent control and incubated for 3h with gentle shaking. After this treatment seedlings were frozen in liquid nitrogen and ground into a fine powder. Total RNA was isolated from the frozen tissue using Trizol reagent (Transgen Biotech, Beijing, China) following the manufacturer’s instructions. RNA was further purified using the RNAeasy mini kit (Qiagen, Valencia, CA, USA) and 1 μg was used for the synthesis of first-strand cDNA using the SuperScript First-Strand Synthesis System (Life Technologies, Carlsbad, CA, USA). Real-time quantitative PCR was performed using gene-specific primers (Supplementary Table S1 available at JXB online) and TranStart Green qPCR SuperMix (Transgen Biotech). Actin2 gene (Yoo et al., 2010) was used as an internal normalization control. Fold change in gene expression was calculated using ΔCt values according to Schmittgen et al. (2008).
Statistical analysis
All data presented are from at least three replicate experiments. Mean values and standard errors of the means were calculated, and the significance of differences was evaluated by Student’s t test. One-way analysis of variance was used to evaluate significant differences between multiple treatments.
Results
Identification and characterization of the IgASE1 elongase gene in Arabidopsis
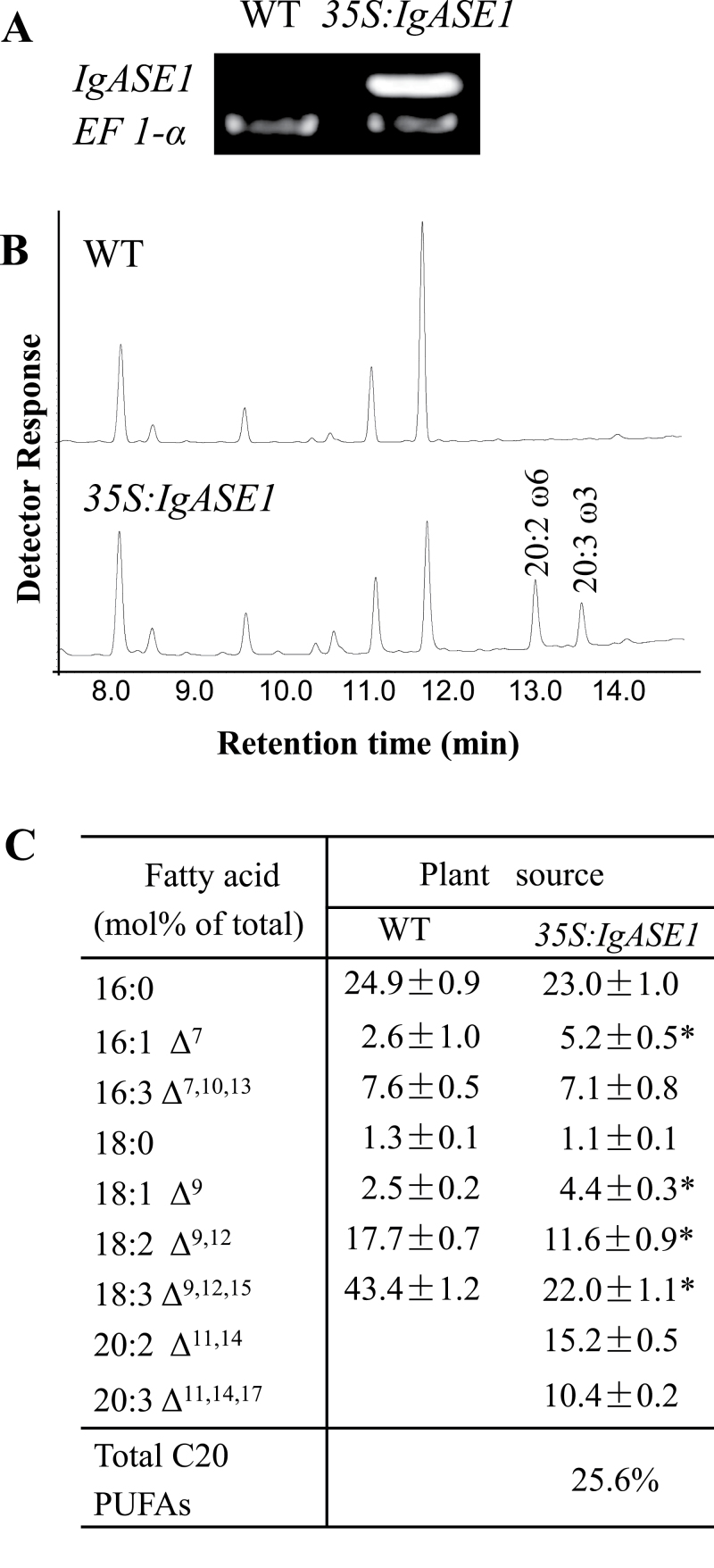
Homozygous single-copy transgenic Arabidopsis plants expressing IgASE1 were identified based on herbicide resistance. RT-PCR was used to detect the transcript level of IgASE1 from rosette leaves of 4-week-old plants and to show that IgASE1 was indeed expressed in these transgenic plants (Fig. 1A). Total fatty acid content in leaf tissue was measured by gas chromatography. Transgenic plants contained two additional fatty acids compared to the WT (Fig. 1B). These were previously identified as EDA and ETrA and considered to be the elongation products of the IgASE1 elongase component (Fraser et al., 2004). In one of the highest producers, these two fatty acids accumulated up to 15.2 and 10.4 mol% of total fatty acid, representing conversions of 56.7 and 32.1% of their precursor C18 substrates linoleic acid (LA) and ALA, respectively. This coincided with a significant decrease in LA and ALA and a small increase in 16:1 and 18:1 (Fig. 1C).
Fig. 1.
Expression of IgASE1 in Arabidopsis leads to production of EDA and ETrA. (A) RT-PCR analysis shows expression of the IgASE1 in transgenic Arabidopsis. PCR reactions were performed to amplify full-length transcript of IgASE1; EF1-1-α was used as a control. Samples were taken for analysis after 28 PCR cycles. (B) GC profiles of A. thaliana leaf fatty acid methyl esters. (C) Fatty acid composition. Total fatty acids were extracted from leaves of WT or transgenic plants. *P < 0.05 in a Student’s t test.
35S:IgASE1 transgenic Arabidopsis exhibited altered morphology
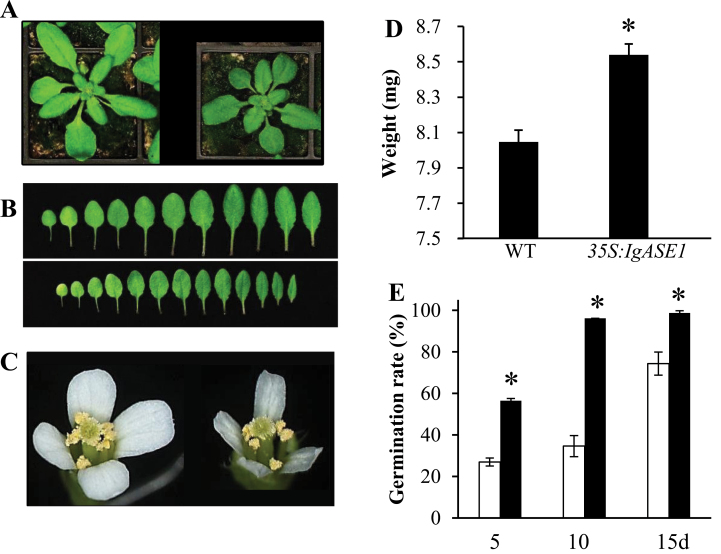
Although there were no significant phenotypic differences between WT plants and a previously identified transgenic Arabidopsis plant line in which the sum of the two fatty acid products accounted for about 15 mol% of the total fatty acids (Fraser et al., 2004), the newly identified high-EDA- and -ETrA-producing transgenic plant line exhibited some phenotypic differences compared to the WT Arabidopsis. At the rosette stage, transgenic leaves were smaller, rounder, and a darker green than WT leaves. Total rosette leaf area of the transgenic lines was 4.3±0.5cm2 (n = 4) compared to 6.9±0.6cm2 (n = 4) for WT plants, leading to an overall smaller plant stature (Fig. 2A, B). The flowers of the transgenic plants were smaller with smaller petals that appeared translucent and rarely opened fully during the early flowering stages (Fig. 2C). Although no significant differences in seed phenotype were apparent, the mean weight of 500 seeds from the transgenic line was higher, at 8.54mg, than that of WT seeds (8.1mg) (Fig. 2D).
Fig. 2.
Phenotypic comparison of WT and 35S:IgASE1 transgenic Arabidopsis plants. (A) Three-week-old WT (left) and transgenic (right) plants. (B) Comparison of rosette leaves from WT (top) and transgenic (bottom) plants. (C) Flowers from WT (left) and transgenic (right) plants. (D) Dry weight of 500 seeds of WT and transgenic Arabidopsis. The experiment was repeated three times and the data were averaged; values are means ± SE; n = 500 for each experiment. (E) Percentage germination of seeds of WT (white columns) and transgenic (black columns) plants that were dried for 5, 10, and 15 days after harvest; 56 seeds were germinated on ½ MS. Values are means ± SE from three repeats. *P < 0.05 in a Student’s t test. This figure is available in colour at JXB online.
Seeds of IgASE1 transgenic plants were less dormant than the WT
Seeds from the transgenic and WT plants were harvested and allowed to dry on a laboratory bench for 5, 10, and 15 days. They were then germinated on ½ MS agar plates without stratification and the percentage of seeds germinated after 48h were recorded. The germination rate of the transgenic seeds was 57, 98, and 100% compared to 25, 35, and 78%, respectively, for the WT seeds (Fig. 2E). Near 100% germination was found for stratified seeds of both WT and transgenic plants at all times (data not shown). Therefore, the germination rate of the transgenic seeds was higher than that of the WT seeds, implying that seeds from the IgASE1-expressing plants were less dormant and germinated sooner than the WT seeds.
35S:IgASE1 transgenic Arabidopsis was hypersensitive to ABA during germination and post-germination growth
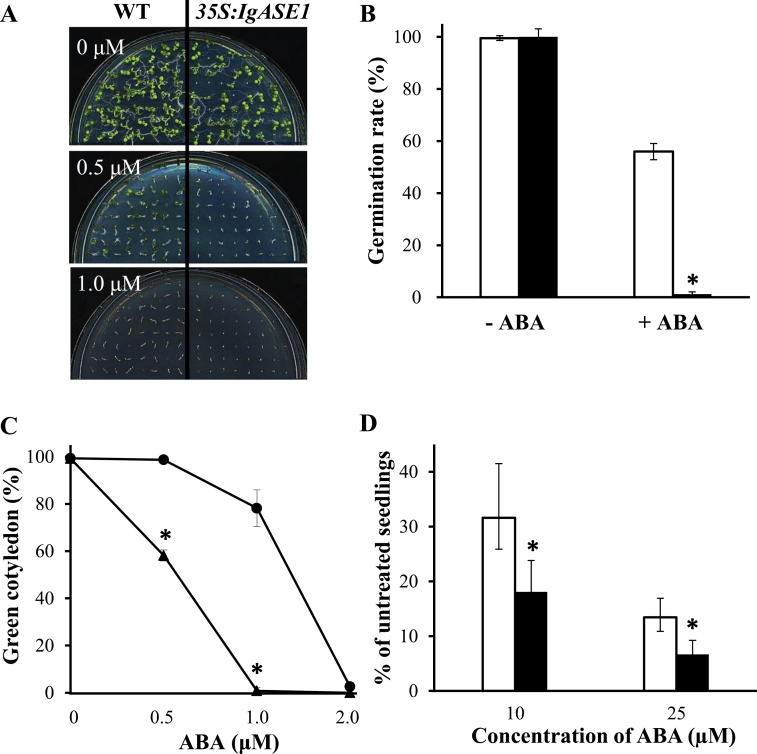
To test the sensitivity of transgenic Arabidopsis to ABA, germination and early seedling growth of the WT and the transgenic Arabidopsis plants were observed on ½ MS medium without ABA, or supplemented with different concentrations of ABA between 0 and 2 μM. Both germination and seedling growth of the WT and the transgenics were markedly inhibited as the ABA concentration increased (Fig. 3A). In the medium with 0.1 μM ABA, only about 1.2% of the transgenic seeds germinated whereas 56% of the WT seeds germinated after 48h (Fig. 3B). Thus germination of 35S:IgASE1 transgenic seeds was significantly more inhibited by ABA than for WT seeds, indicating the transgenic seeds were hypersensitive to ABA during germination.
Fig. 3.
Effect of exogenous ABA on seed germination and early seedling development. (A) Effect of ABA on early seedling growth. Sterilized and stratified seeds were grown on ½ MS medium without or with 0.5 and 1 μM ABA for 8 days. (B) Percentage seed germination after 48h incubation of WT (white columns) and transgenic Arabidopsis (black columns) on ½ MS medium with or without 0.1 μM ABA. Values are means ± SE from three repeats; n = 56 for both WT and transgenic plants. (C) Percentage of seedlings that developed green cotyledons 12 days after sowing on media in the presence of different concentrations of ABA; WT, black circles; transgenic, black triangles. Values are the mean percentage ± SE (n = 52) from three repeats. *P < 0.05 in a Student’s t test. (D) Percentage fresh weight of the shoots of WT (white columns) and transgenic (black columns) plants relative to the untreated seedlings grown on ½ MS medium for 18 days without or with 10 and 25 μM ABA. Values are mean weight ± SE (n = 15) from three repeats. *P < 0.05 in a Student’s t test. This figure is available in colour at JXB online.
The effect of ABA on post-germination growth of seedlings was analysed by scoring the percentage of germinated seeds that could form green cotyledons in the presence of different concentrations of ABA (Chen and Jones, 2004). Cotyledon opening and greening of both WT and transgenic plants were significantly inhibited as the ABA concentration increased (Fig. 3C). At 1 μM ABA almost all the transgenic cotyledons failed to turn green (as compared with those grown without ABA) while 78% of WT seedlings developed green cotyledons. Thus IgASE1-expressing seedlings were hypersensitive to ABA during post-germination growth.
The same trend was also observed for growth inhibition of seedlings during early development (Fig. 3D, Supplementary Fig. S1 available at JXB online). Five-day-old ½ MS-grown seedlings were transferred to fresh medium supplemented with 10 and 25 μM ABA (controls were transferred to fresh ½ MS medium without ABA). After 18 days, growth of both the transgenic and WT seedlings was much reduced by ABA compared to their untreated controls. However, ABA had a much greater effect on the transgenic than on the WT seedlings because while only 18.1 and 6.8% of the weights were retained in the 10 and 25 μM ABA-treated transgenic seedlings, that of the WT was nearly double that of the transgenic plants at 31.6 and 13.4% under the same ABA concentrations. Therefore, the growth of transgenic seedlings was reduced much more by ABA than that of WT seedlings, indicating that the IgASE1-expressing seedlings were hypersensitive to ABA during early seedling development.
Restricted transpiration of transgenic plants was associated with increased foliar ABA levels
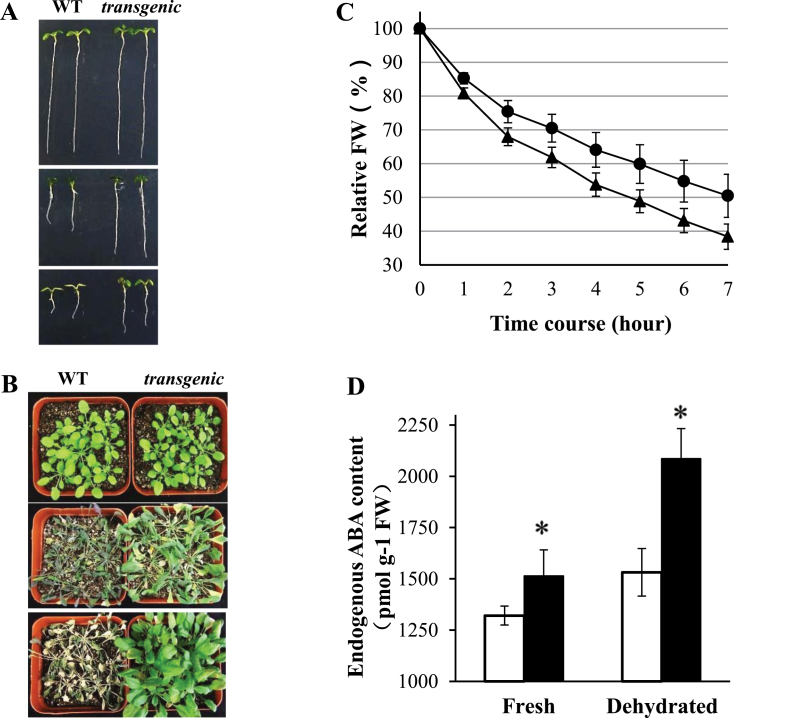
Since IgASE1-expressing transgenic Arabidopsis were hypersensitive to ABA treatment during seed germination and early seedling development, drought was mimicked by treating 3-day-old WT and transgenic seedlings with glycerol or mannitol for 7 days. Both mannitol and glycerol inhibited root growth of both the WT and transgenic plants, but root growth was significantly more inhibited in WT than transgenic seedlings (Fig. 4A). These results indicate that the transgenic Arabidopsis exhibited greater osmotic tolerance.
Fig. 4.
Response of WT and transgenic Arabidopsis plants to dehydration stress. (A) Response of seedlings to dehydration by mannitol and glycerol. Three-day-old ½ MS-grown seedlings were transferred to fresh ½ MS (top), ½ MS containing 300mM mannitol (middle), or 3% (v/v) glycerol (bottom) and grown for a further 7 days. (B) Drought treatment of soil-grown plants. 3-week-old plants (top) had water withheld for 22 days (middle) and were subsequently re-watered (bottom). (C) Water loss of detached leaves. Relative fresh weight (FW) is the percentage fresh weight of a leaf at a dry time point relative to its fresh weight at time 0. WT, black triangles; transgenics, black circles. The experiment was repeated three times and the data were averaged; values are means ± SE from five leaves for each experiment. (D) Endogenous ABA levels of the WT (white columns) and 35S:IgASE1 transgenic plants (black columns) estimated from untreated (Fresh) and 1-h air-dried (dehydrated) leaves. ABA concentration is expressed as pmol per gram of fresh weight. *P < 0.05 in a Student’s t test. This figure is available in colour at JXB online.
Furthermore, 3-week-old plants were subjected to drought stress by withholding water for 22 days. Both WT and transgenic plants were withered at the end of the treatment. However, when these plants were drenched with water most of the transgenic plants quickly recovered and continued to grow to maturity while none of the WT plants survived under these conditions (Fig. 4B), indicating that the transgenic plants are more drought-resistant than the WT ones.
To test whether these responses could result, at least in part, from lower transpiration rates of transgenic plants, detached leaf water loss was measured over time. Relative fresh weight of the detached leaves of IgASE1-expressing transgenics was significantly higher than that of the WT leaves throughout the experiment. After 7h the transgenic leaves retained 50.5% of their original weight whereas only 38.4% of the original weight of the WT leaves was retained (Fig. 4C).
Endogenous foliar ABA levels in both fresh leaves (immediately detached) and leaves dehydrated for 1h were higher in transgenic plants compared to WT plants (Fig. 4D). This suggests that ABA production was elevated in the transgenic plants in both normal and dehydration conditions.
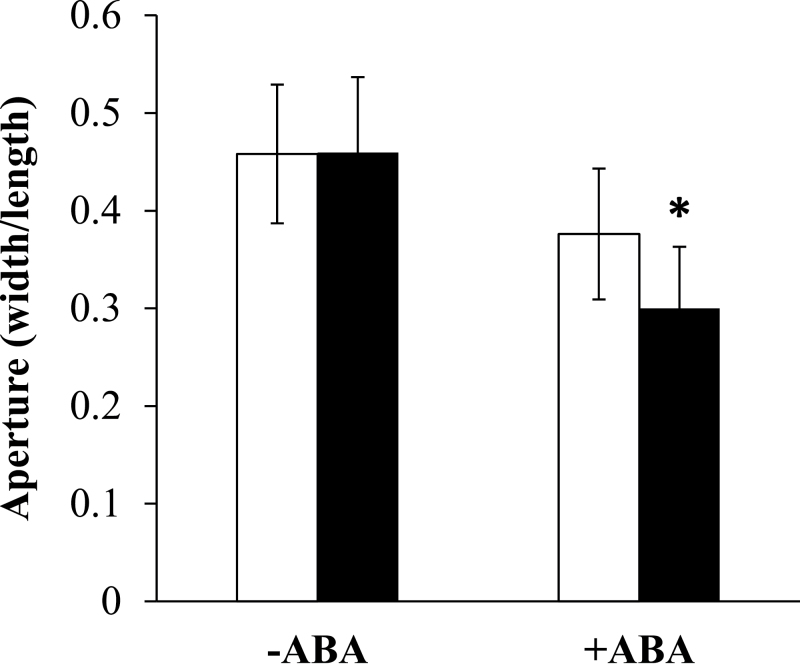
Because water loss is regulated via stomata, the stomatal apertures of transgenic and WT plants were compared in the presence and absence of ABA. While both the 35S:IgASE1 transgenic leaves and the WT leaves had similar stomatal apertures without ABA treatment (7.7±0.6 µm in width and 16.8±1.4 µm in length for WT, 8.2±0.7 µm width and 18.0±0.5 µm length for the transgenics), the aperture of the stomata of the transgenic plants was smaller than that of the WT plants when ABA was present (6.0±0.7 µm width and 15.9±1.4 µm length for WT, 4.8±0.3 µm width and 15.9±1.3 µm length for the transgenic plants; Fig. 5), suggesting that 35S:IgASE1 transgenic leaves were more sensitive to ABA-induced stomatal closure than the WT leaves.
Fig. 5.
ABA-induced stomatal closure in WT (white columns) and 35S:IgASE1 transgenic plants (black columns). Data represent the mean ± SE from three independent experiments (n = 50–60 per experiment). *P < 0.05 in a Student’s t test.
These combined data indicate that the drought resistance of IgASE1-expressing plants is regulated via the ABA-mediated stress-response pathway.
Exogenous application of EDA and ETrA can mimic ABA and drought responses in WT plants, similar to that found in transgenic plants
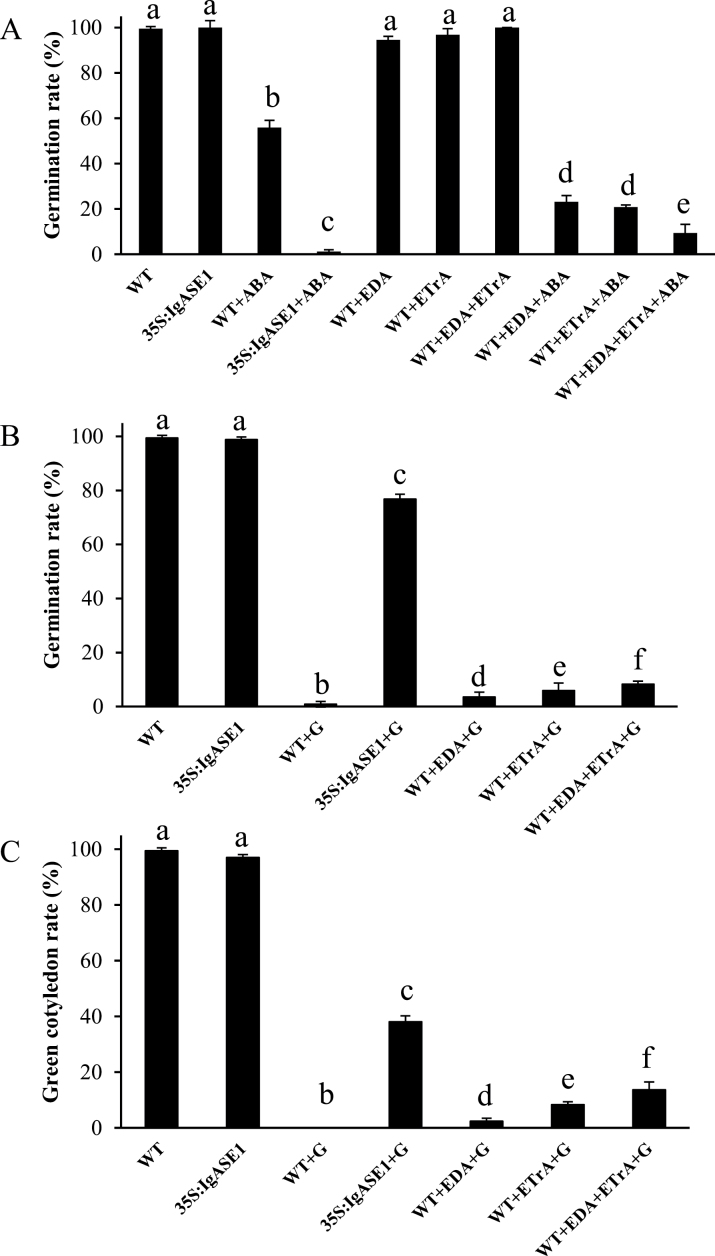
To determine whether the hypersensitivity to ABA in the transgenic plants was caused by the newly synthesized C20 VLCPUFAs EDA and ETrA in these plants, exogenous application treatments with these two fatty acids were performed on WT plants during germination. Germination rates of both WT and transgenic seeds were close to 100% and were not significantly altered by exogenous application of EDA, ETrA, or EDA+ETrA to the germination media (Fig. 6A). However, in the presence of 0.1 μM ABA the germination rate of WT seeds was reduced to nearly 56%. This inhibitory effect was enhanced by exogenous supplementation of EDA, ETrA, or EDA+ETrA, under which treatments only 23.2, 20.8, and 9.5% of the seeds germinated, respectively (Fig. 6A). Next, different amounts of EDA+ETrA were added to the germination media. Although 92% of the untreated seeds germinated after 48h, only 47% germinated when 0.5 µM ABA was present (Supplementary Fig. S2 available at JXB online). This reduction in seed germination by ABA was significantly enhanced by adding 2.5, 5, and 10 μM of each of EDA and ETrA in a dose-dependent manner, where only 32.5, 7.6, and 6.0% of the seeds germinated, respectively (Supplementary Fig. S2 available at JXB online). Although this was higher than the ABA-treated transgenic seeds, of which only 1.2% germinated, it suggests that the increased sensitivity to ABA of IgASE1-expressing plants is most likely due to the endogenous EDA and ETrA or their derived metabolites in the transgenic plants.
Fig. 6.
The effect of exogenous application of EDA and ETrA on seed germination and seedling establishment. (A) Germination of WT and 35S:IgASE1 transgenic Arabidopsis on ½ MS media with or without ABA (0.1 μM), EDA (10 µM), ETrA (10 µM), or EDA+ETrA (10 µM each) after 48h. (B) Germination of WT and 35S:IgASE1 transgenic plants on ½ MS media without or with glycerol (G; 2%), EDA (10 µM), ETrA (10 µM), or both (10 µM each) after 48h. (C) Quantitative analysis of the percentage of greening cotyledons on WT and transgenic plants grown on media without or with glycerol (2%), EDA (10 µM), ETrA (10 µM), or both (10 µM each) after 7 days. Different letters indicate statistically different values after one-way ANOVA.
Experiments then determined whether the drought resistance in the transgenic Arabidopsis was caused by endogenous EDA and ETrA, by germinating WT seeds in media containing these two fatty acids coupled with 2% glycerol to mimic dehydration conditions. When glycerol was present, fewer than 1% of the WT seeds germinated whereas 77% of the transgenic seeds germinated. The inhibitory effect of glycerol on WT seed germination was alleviated by supplementing the media with EDA and ETrA, for which 3.6, 6.0, and 8.3% germinated with EDA, ETrA, or EDA+ETrA present, respectively (Fig. 6B).
The percentage of seedlings that developed green cotyledons on glycerol-containing media was 38% for the 35S:IgASE1 transgenic plants, while no WT seedlings were recovered. However, when EDA, ETrA, or both EDA and ETrA were supplemented in the presence of glycerol, the numbers of WT seedlings bearing green cotyledons increased to 2.4, 8.3, and 13.7% for EDA, ETrA or EDA+ETrA, respectively (Fig. 6C). Thus EDA and ETrA can partially alleviate the inhibitory effects of glycerol on seed germination and early seedling development in the WT. Thus responses of 35S:IgASE1 transgenic Arabidopsis plants are due, at least in part, to these two newly synthesized C20 VLCPUFAs or their derived metabolites.
Transcript levels of ABA biosynthetic and catabolic genes were affected in transgenic Arabidopsis under stress conditions
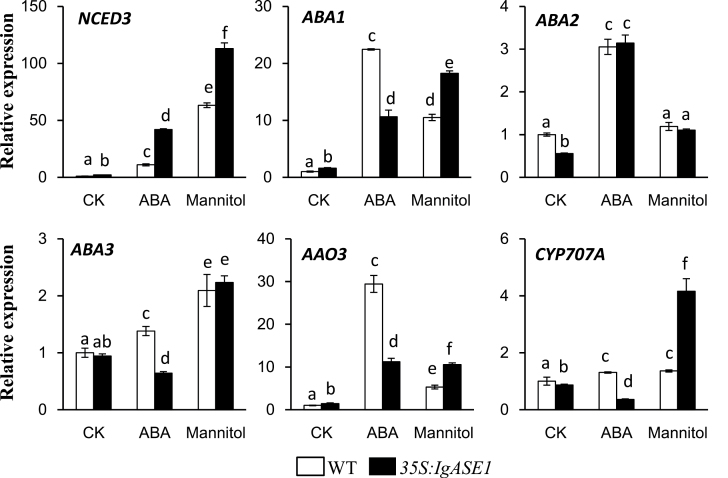
To determine whether foliar ABA accumulation of 35S:IgASE1 transgenics could be explained by changes in gene expression, the expression profiles of some ABA biosynthetic genes, such as 9-cis-epoxycarotenoid dioxygenase 3 (NCED3), ABA1, ABA2, ABA3, and abscisic aldehyde oxidase 3 (AAO3), as well as the ABA catabolic gene CYP707A, were determined by quantitative RT-PCR (Fig. 7). Samples were analysed following 3h ABA (100 µM) or mannitol (300mM) treatment in 7-day-old plate-grown seedlings.
Fig. 7.
Transcript levels of genes involved in ABA metabolic pathways. Transcript levels were measured by quantitative RT-PCR from untreated solvent control (CK), ABA (100 µM), and mannitol (300mM)-treated 7-day-old seedlings. Experiments were performed three times. Each bar represents the mean ± SE (n = 3). Different letters indicate statistically different values after one-way ANOVA.
In the untreated control seedlings, mRNA levels of NCED3 and ABA1 were much higher while those of ABA2 and CYP707A were much lower in the transgenic plants than in the WT, although those of ABA3 and AAO3 were similar. Upon treatment with mannitol, NCED3, ABA1, ABA3, and AAO3 transcripts were upregulated in both the WT and transgenic plants and the expression levels of NCED3 (1.8-fold), ABA1 (1.7-fold), ABA2 (2.0-fold), and AAO3 (2.0-fold) were much higher in the 35S:IgASE1 transgenic seedlings than in the WT, while expression of ABA3 was similar between WT and transgenic plants (Fig. 7). However, expression of CYP707A in the 35S:IgASE1 transgenic Arabidopsis was also 3.1-fold higher than in WT plants. These combined results indicate that genes regulating ABA turnover were more active in the transgenic Arabidopsis under dehydration stress.
In ABA-treated seedlings, mRNA levels of NCED3, ABA1, ABA2, and AAO3 were increased in both the WT and the transgenic plants. However, among them only NCED3 was expressed at much higher levels in the transgenics than in the WT. Levels of ABA1, ABA3, and AAO3 mRNAs in the transgenic plants were lower than those in the WT while ABA2 mRNA levels were similar in both sets of plants. Interestingly, ABA inhibited the expression of CYP707A in the transgenic plants (Fig. 7). This suggests that the inhibitory effect of exogenous ABA during seedling growth of the IgASE1 transgenic plants (Fig. 3C, D) may be enhanced by the upregulation of NCED3 coupled with less ABA being degraded by the less-active CYP707A, leading to an overall higher level of endogenous ABA being produced in the transgenic compared to the WT Arabidopsis.
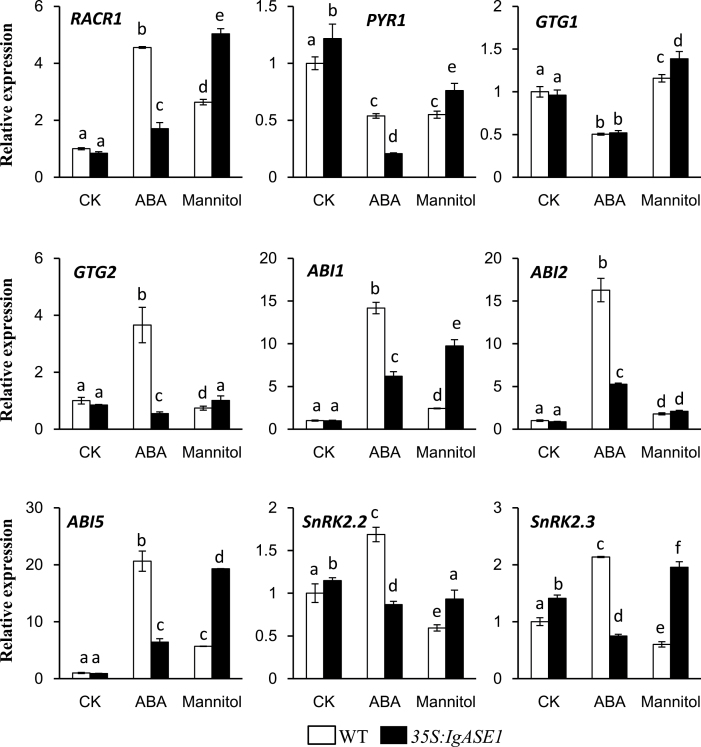
Expression levels of ABA receptor and signalling genes were altered in the transgenic Arabidopsis
The ABA receptor genes RCAR1, PYR1, GTG1, and GTG2 as well as signalling-related genes (ABI1, ABI2, ABI5, SnRK2.2, and SnRK2.3) were also monitored in the untreated and ABA- or mannitol-treated seedlings. mRNA levels of most of the tested genes were similar in the control seedlings apart from PYR and SnRK2.3, which were slightly upregulated in the transgenic plants compared to the WT (Fig. 8). However, mannitol strongly upregulated the expression of RACR1, ABI1, and ABI5 in both the WT and the transgenic seedlings; this was more so in the transgenic than in the WT seedlings. Of the four ABA receptor genes studied here, only RACR1 was significantly upregulated by mannitol and its expression level was 1.9-fold higher in the transgenic plants than in the WT, while the expression of PYR1 was decreased, GTG2 was similar, and GTG1 was only modestly upregulated. The signalling-related genes ABI1 and ABI5 were also significantly upregulated while ABI2 was only slightly upregultaed in both the WT and transgenic seedlings and the mRNA levels of ABI1 and ABI5 were 4.0- and 3.4-fold higher in the transgenic plants than in WT, respectively. SnRK2.2 and SnRK2.3 were decreased in the WT while SnRK2.2 was similarly expressed and SnRK2.3 was slightly upregulated in the transgenic compared to the WT plants (Fig. 8). These results imply that the drought resistance of the 35S:IgASE1 transgenic plants was at least partly due to the upregulation of genes in the ABA-mediated drought stress-response pathway. ABA, on the other hand, seems to have less effect on the transcript levels of these genes in the transgenic plants than in the WT (Fig. 8).
Fig. 8.
Expression levels of genes involved in ABA signalling pathways. Transcript levels were measured by quantitative RT-PCR from untreated solvent control (CK), ABA (100 µM), and mannitol (300mM)-treated 7-day old seedlings. Experiments were performed three times. Each bar represents the mean ± SE (n = 3). Different letters indicate statistically different values after one-way ANOVA.
Regulation of ABA-dependent and -independent drought-response genes in transgenic plants
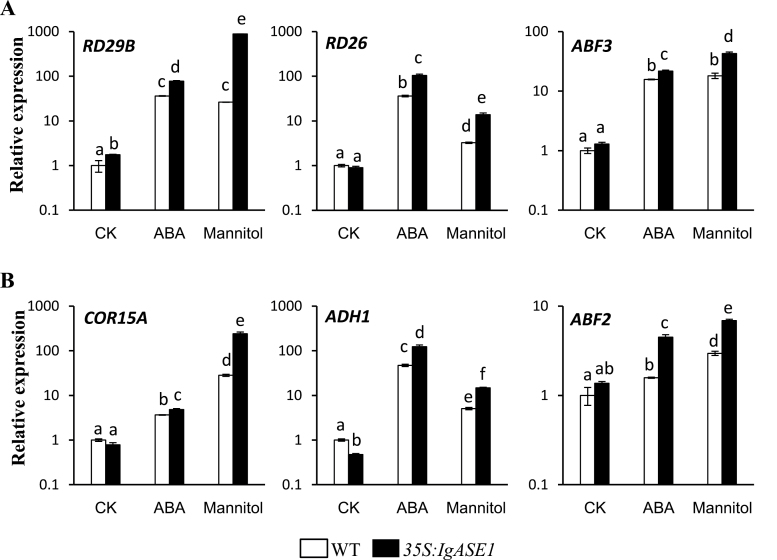
To determine the involvement of EDA and ETrA in the drought response, and whether its role was limited to the ABA-regulated pathway, expression levels of several drought-inducible genes that display different sensitivity to exogenously applied ABA (Ding et al., 2011) were assayed. The ABA-dependent genes RD29B, RD26, and ABF3 were strongly induced by both exogenous ABA and mannitol (Fig. 9A) in both IgASE1-expressing and WT plants. However, their expression levels were much higher in the 35S:IgASE1 transgenic plants than in the WT.
Fig. 9.
Expression levels of genes involved in the ABA-dependent and ABA-independent pathways. Transcript levels were measured by quantitative RT-PCR from untreated solvent control (CK), ABA (100 µM), and mannitol (300mM)-treated 7-day old seedlings. (A) Transcript levels of the three ABA-dependent genes. (B) Transcript levels of the three ABA-independent genes. Experiments were performed three times. Each bar represents the mean ± SE (n = 3). Different letters indicate statistically different values after one-way ANOVA.
We next monitored the expression of three ABA-independent genes, COR15A, ADH1, and ABF2. The transcript levels of COR15A and ABF2 were similar between the WT and transgenic plants while ADH1 was much lower in the transgenic than the WT plants in the untreated control samples. Followed ABA treatment the transcript levels of COR15A, ABF2, and ADH1 were increased by 3.6- and 6.2-, 47.2- and 261.5-, and 1.6- and 3.3-fold for the WT and transgenic plants, respectively. Mannitol had similar effect where the transcript levels were increased by 28.1- and 307.2-fold for COR15A, 5.1- and 31.4-fold for ADH1, and 3.0- and 5.0-fold for ABF2 in WT and transgenic plants, respectively. Therefore, both ABA and mannitol can strongly induce the expression of all three genes and their transcript levels were much higher in the 35S:IgASE1 transgenic plants than in the WT (Fig. 9B).
Discussion
Fatty acids and their derived metabolites are not only major structural components of the cell membrane, but also function as modulators of a multitude of signal transduction pathways evoked by environmental and developmental changes. Therefore, remodelling the fatty acid composition of an organism can alter metabolism as well as the complex interaction array of signalling cascades that affect a range of physiological responses (Savchenko et al., 2010). While studies show that the VLCPUFAs, such as ARA and other eicosapolyenoic acids, play important roles during biotic stress in plants (Bostock et al., 2011), hitherto there have been no reports of their effects on abiotic stress in plants.
We utilized a transgenic Arabidopsis line expressing a fatty acid elongase gene from I. galbana that was specific for the two C18 Δ9 long-chain unsaturated fatty acids LA and ALA (Qi et al., 2002). This transgenic line can produce EDA and ETrA to 25 mol% of total leaf fatty acids. Effects of these transgene products on plant growth and development and the modulation of ABA in relation to seed germination, seedling establishment, and drought resistance in young vegetative plants were assayed. Phenotypic differences between this transgenic line and WT Arabidopsis were detected in leaves, flowers, and seed size (Fig. 2). While a previous study with a different transgenic line that contained less EDA and ETrA (15%) reported that the transgenics were morphologically indistinguishable from the WT plants (Fraser et al., 2004), differences in morphology between the two transgenic lines could be the direct result of different amounts of EDA and ETrA accumulated. Arabidopsis plants that overexpress Fatty Acid Elongase 1 (FAE1) under the control of the CaMV35S promoter showed dramatic phenotypic alterations (Millar et al., 1998). FAE1 is a seed-specific fatty acid elongase that catalyses the production of C20+ very-long-chain saturated and mono-unsaturated fatty acids (VLCFAs) in Arabidopsis seed oil (O’Neill et al., 2003). WT Arabidopsis leaves contain less than 1% of these VLCFAs, which are mainly present in the epidermal cuticular waxes (Millar and Kunst, 1997). However, the CaMV35S:FAE1 transgenic Arabidopsis plants can accumulate high levels (>30%) of VLCFAs in their leaf membrane lipids. Although the transgenic plants with relatively low levels of VLCFAs appeared to be like the WT, the transgenic plants with high levels of VLCFAs exhibited a wide range of morphological changes and some even failed to survive (Millar et al., 1998; Bach et al., 2008). Recent studies show that VLCFAs have a wide range of physiological and structural functions and are crucial for many biological processes such as cell division and expansion, cell proliferation, or differentiation (Bach et al., 2011). High levels of VLCPUFAs also had similar, although reduced, effects on plant growth and development (Fig. 2), which suggests that they may function in the same pathways as VLCFAs.
Seeds of the transgenic plants displayed lower levels of dormancy than the WT (Fig. 2E). When first harvested, seeds are usually highly dormant and this dormancy is gradually reduced during dry storage to allow germination. It is well established that the plant hormone ABA plays an essential role in the induction of seed dormancy and inhibition of germination (Carrera et al., 2008; Finkelstein et al., 2008). In line with this, germination of the transgenic seeds was strongly inhibited by ABA (Fig. 3A). ABA also inhibited early seedling establishment and growth (Fig. 3A–C), indicating that the accumulation or perception of ABA might be different in the transgenics compared to the WT during seed maturity, germination and post-germination periods. The fact that these effects could be altered by exogenous application of EDA and ETrA to the WT (Fig. 6, Supplementary Fig. S2 available at JXB online) indicates the direct involvement of these two eicosapolyenoic acids in the regulation of ABA-mediated seed germination and early seedling development. Further, and perhaps most importantly, adult 35S:IgASE1 transgenic plants were apparently more drought resistant than the WT (Fig. 4). This is likely due to their smaller leaf area and lower transpiration rate of the transgenic plants (Figs 2A, B and 4C). They contained more endogenous ABA than the WT and their stomata more readily closed upon ABA treatment (Figs 4D and 5). Since inhibitory effects of mannitol and glycerol on seed germination and seedling growth were reduced by EDA and ETrA, it suggests that these two fatty acids played positive roles in regulating drought response in the transgenic Arabidopsis (Fig. 6).
ABA biosynthesis and degradation contribute to ABA homeostasis during plant development and stress conditions. To determine whether the overall production of ABA in the transgenic plants was different from the WT the endogenous ABA levels were measured. This revealed that the transgenic plants contained more ABA in their leaves; this was more so under dehydration conditions (Fig. 4D). This indicates that the drought resistance in the 35S:IgASE1 transgenic Arabidopsis is involved in the accumulation and/or perception of ABA. Indeed, the transcript levels of ABA biosynthesis and signalling genes were very different in the control untreated as well as osmotic stress-/ABA-treated transgenic seedlings compared to WT (Figs 7 and 8). Although ABA2, ABA3, and AAO3 transcript levels were lower and similar respectively in the transgenics compared to the WT, the expression of ABA1 and NCED3 were significantly higher in the 35S:IgASE1 transgenic seedlings (Fig. 7). Further, the transcript level of the major enzyme for ABA catabolism, CYP707A, was much lower in the 35S:IgASE1 transgenic plants than in WT. This suggests that the higher foliar ABA level in the transgenic plants was due to the combined action of higher activity of ABA1 and NCED3 for biosynthesis and lower CYP707A activity for ABA degradation, resulting in overall higher ABA content in the transgenics than in the WT in untreated plants. Upon treatment with mannitol the transcript levels of four out of the five ABA biosynthetic genes (apart from ABA2 in the WT) as well as the catabolic gene CYP707A were all significantly upregulated, more so in the transgenics than in the WT (Fig. 7). The increase in expression of CYP707A during osmotic stress can aid the plants to rapidly degrade the excess ABA generated; therefore, the homeostatic ABA levels can be maintained (Kushiro et al., 2004).
Recent studies identified and confirmed the PYR/RCAR proteins as ABA receptors that interact with the protein phosphatase 2Cs (PP2Cs), such as ABI1 and ABI2, which function as negative regulators of ABA signalling (Ma et al., 2009; Park et al., 2009). In contrast, SnRK2s act as positive regulators of downstream signalling (Vlad et al., 2009). Thus, in the absence of ABA, the PP2Cs are active and repress SnRK2 activity and downstream signalling. In the presence of ABA, PYR/RCARs interact with PP2Cs and inhibit phosphatase activity, allowing SnRK2 activation and phosphorylation of target proteins (Hubbard et al., 2010). Enhancing ABA signalling through PYR/PYR1-LIKE (PYL)/RCAR ABA receptors has been shown to improve plant drought resistance. For example, transgenic Arabidopsis plants overexpressing ABA receptor PYL4, particularly its mutated variant PYL4 A194T, exhibited an enhanced response to ABA and drought resistance via ABA-dependent inhibition of PP2Cs (Pizzio et al., 2013). The 35S:IgASE1 transgenic Arabidopsis were hypersensitive to ABA during seed germination and early seedling development (Fig. 3). This could be explained by the higher transcript levels of PYR1, SnRK2.2, and SnRK2.3 in the transgenics than WT (Fig. 8). Upon treatment with mannitol only RCAR1 and GTG1 of the four ABA receptor genes assayed were significantly upregulated. The signalling-related genes, such as ABI1, ABI2, and ABI5, were also significantly upregulated in both WT and 35S:IgASE1 transgenic plants, and higher levels of expression of these genes were found in the transgenics compared to WT (Fig. 8). Therefore, our quantitative RT-PCR data demonstrated that ABA signalling was also modified by upregulating receptor genes and signalling-related genes in the transgenic plants under osmotic stress condition (Fig. 8). However, exogenous application of ABA had the opposite effect on gene expression of RACR1, ABI1, ABI2, ABI5, SnRK2.2, and SnRK2.3 where lower levels of transcripts were found in transgenic seedlings compared to WT (Fig. 8). In addition, we also found that the ABA-dependent (RD29B, RD26, and ABF3) as well as the ABA-independent (COR15A, ADH1, and ABF2) drought-inducible genes displayed increased transcript levels in the transgenics following both ABA and osmotic stress (Fig. 9A, B), implying that stress adaptation in the EDA- and ETrA-producing transgenic Arabidopsis was complex, involving both ABA-dependent and -independent pathways.
It is thought that plant drought tolerance is largely dependent on the inherent level of fatty acid unsaturation and/or the ability to maintain or adjust fatty acid unsaturation (Mikami and Murata 2003). For example, water deficit decreased ALA (18:3) and LA (18:2) concentrations in Brassica napus (Dakhma et al., 1995). Relatively higher proportions and levels of unsaturated fatty acids, particularly 18:3 and 18:2, are associated with leaf dehydration tolerance and post-drought rehydration in Kentucky bluegrass (Xu et al., 2011). Overexpression of the Arabidopsis FAD3 and FAD8 ω-3 desaturases in tobacco cells and plants increases the ratio of 18:3 to 18:2, and increases cellular resistance to osmotic stress and whole-plant drought tolerance (Zhang et al., 2005). The major polyunsaturated fatty acids in Arabidopsis leaves are 18:3 (43.4%) and 18:2 (17.7%) (Fig. 1B, C). In the 35S:IgASE1 transgenic Arabidopsis, although the amounts of both 18:2 and 18:3 are significantly reduced due to the production of the two C20 polyunsaturated fatty acids 20:2 and 20:3 (Fig. 1B, C), the plant exhibited enhanced drought and osmotic resistance (Fig. 4). This suggests that the two newly synthesized C20 polyunsaturated fatty acids EDA and ETrA play a positive role in the drought stress response that is similar to 18:2 and 18:3; hence, the transgenic plants were not compromised by the replacement of the two C18 polyunsaturated fatty acids with EDA and ETrA. Instead, the enrichment of EDA and ETrA in the membrane phospholipids phosphatidylcholine, phosphatidylethanolamine, phosphatidate, and phosphatidylinositol (Fraser et al., 2004) may have helped to maintain membrane structure and fluidity. This is important for membrane integrity and the functionality of integral membrane proteins, including ABA-biosynthetic and signalling machinery proteins.
Although the exact mechanism of the direct involvement of the two eicosapolyenoic acids EDA and ETrA in ABA metabolism and signalling is unknown, we nevertheless provide evidence that they can stimulate the expression of ABA biosynthesis genes under osmotic stress. They also control the expression of stress regulation and ABA-responsive genes. Through these combined actions, they may enhance the metabolism and responsiveness to ABA of the plant cell, therefore helping to maintain water balance by decreasing water loss through stomatal closure and hence improving drought resistance in these transgenic plants.
Here we show that eicosapolyenoic acids such as EDA and ETrA are involved in a range of activities during the life cycle of Arabidopsis as well as in the adaptation of plants to abiotic stress such as drought, by regulating ABA-dependent and -independent pathways.
Supplementary material
Supplementary material is available at JXB online.
Table S1. Sequences of primers used in this study.
Figure S1. IgASE1-expressing seedlings (35:IgASE1) were more sensitive to ABA during early seedling development. WT and transgenic seeds were germinated on ½ MS medium for 5 days. They were then transferred to ½ MS medium supplemented without (0 µM) or with 10 and 25 µM ABA. They were scanned after 18 days culture in an environmental controlled growth room under long-day-length conditions.
Figure S2. Effect of different amounts of exogenously supplied fatty acids to percentage of germinated seeds of the WT Arabidopsis in the presence of 0.5 μM ABA. Seeds were germinated for 48h. FA1, 2.5 μM of each EDA and ETrA; FA2, 5 μM of each EDA and ETrA; FA3, 10 μM of each EDA and ETrA. Different letters indicate statistically different values after one-way analysis of variance.
Acknowledgements
We thank Dr James Doughty, University of Bath, UK for critical reading of the manuscript. We also thank Dr Ian Dodd, Lancaster University, UK and 3 anonymous reviewers for their constructive comments. This work was supported by grants awarded to BQ from the National Natural Science Foundation of China (30970222 and 31170233) and National Genetically Modified Organisms Breeding Major Project of China (009ZX08005-024B).
Glossary
Abbreviations:
- ABA
abscisic acid
- ALA
α-linolenic acid
- ARA
arachidonic acid
- EDA
eicosadienoic acid
- EtrA
eicosatrienoic acid
- LA
linoleic acid
- MS
Murashige and Skoog
- PP2C
protein phosphatase 2C
- VLCFA
very-long-chain saturated and mono-unsaturated fatty acid
- VLCPUFA
very-long-chain polyunsaturated fatty acid
- WT
wild-type.
References
- Abbadi A, Domergue F, Bauer J, Napier JA, Welti R, Zahringer U, Cirpus P, Heinz E. 2004. Biosynthesis of very-long-chain polyunsaturated fatty acids in transgenic oilseeds: constraints on their accumulation. The Plant Cell 16, 2734–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allakhverdiev SI, Kinoshita M, Inaba M, Suzuki I, Murata N. 2001. Unsaturated fatty acids in membrane lipids protect the photosynthetic machinery against salt-induced damage in Synechococcus. Plant Physiology 125, 1842–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf M. 2010. Inducing drought tolerance in plants: recent advances. Biotechnology Advances 28, 169–183 [DOI] [PubMed] [Google Scholar]
- Bach L, Gissot L, Marion J, Tellier F, Moreau P, Satiat-Jeunemaitre B, Palauqui JC, Napier JA, Faure JD. 2011. Very-long-chain fatty acids are required for cell plate formation during cytokinesis in Arabidopsis thaliana . Journal of Cell Science 124, 3223–3234 [DOI] [PubMed] [Google Scholar]
- Bach L, Michaelson LV, Haslam R, et al. 2008. The very-long-chain hydroxy fatty acyl-CoA dehydratase PASTICCINO2 is essential and limiting for plant development. Proceedings of the National Academy of Sciences, USA 105, 14727–14731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes CM, Prox D, Christison-Lagay EA, et al. 2012. Inhibition of neuroblastoma cell proliferation with omega-3 fatty acids and treatment of a murine model of human neuroblastoma using a diet enriched with omega-3 fatty acids in combination with sunitinib. Pediatric Research 71, 168–178 [DOI] [PubMed] [Google Scholar]
- Bostock RM, Kuc JA, Laine RA. 1981. Eicosapentaenoic and arachidonic acids from Phytophthora infestans elicit fungitoxic sesquiterpenes in the potato. Science 212, 67–69 [DOI] [PubMed] [Google Scholar]
- Bostock RM, Savchenko T, Lazarus C, Dehesh K. 2011. Eicosapolyenoic acids: novel MAMPs with reciprocal effect on oomycete-plant defense signaling networks. Plant Signaling and Behavior 6, 531–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostock R, Schaeffer D, Hammerschmidt R. 1986. Comparison of elicitor activities of arachidonic acid, fatty acids and glucans from Phytophthora infestans in hypersensitivity expression in potato tuber. Physiological and Molecular Plant Pathology 29, 349–360 [Google Scholar]
- Breslow JL. 2006. n-3 fatty acids and cardiovascular disease. American Journal of Clinical Nutrition 83, 1477S–1482S [DOI] [PubMed] [Google Scholar]
- Browse J, McCourt PJ, Somerville CR. 1986. Fatty acid composition of leaf lipids determined after combined digestion and fatty acid methyl ester formation from fresh tissue. Analytical Biochemistry 152, 141–145 [DOI] [PubMed] [Google Scholar]
- Carrera E, Holman T, Medhurst A, Dietrich D, Footitt S, Theodoulou FL, Holdsworth MJ. 2008. Seed after-ripening is a discrete developmental pathway associated with specific gene networks in Arabidopsis. The Plant Journal 53, 214–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Jones AM. 2004. AtRGS1 function in Arabidopsis thaliana. Methods in Enzymology 389, 338–350 [DOI] [PubMed] [Google Scholar]
- Cheng B, Wu G, Vrinten P, Falk K, Bauer J, Qiu X. 2010. Towards the production of high levels of eicosapentaenoic acid in transgenic plants: the effects of different host species, genes and promoters. Transgenic Research 19, 221–229 [DOI] [PubMed] [Google Scholar]
- Choi D, Ward BL, Bostock RM. 1992. Differential induction and suppression of potato 3-hydroxy-3-methylglutaryl coenzyme A reductase genes in response to Phytophthora infestans and to its elicitor arachidonic acid. The Plant Cell 4, 1333–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daklma WS, Zarrouk M, Cherif A.1995. Effects of drought stress on lipids in rape leaves. Phytochemistry 40, 1383–1386 [Google Scholar]
- Dimri M, Bommi PV, Sahasrabuddhe AA, Khandekar JD, Dimri GP. 2010. Dietary omega-3 polyunsaturated fatty acids suppress expression of EZH2 in breast cancer cells. Carcinogenesis 31, 489–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Avramova Z, Fromm M. 2011. The Arabidopsis trithorax-like factor ATX1 functions in dehydration stress responses via ABA-dependent and ABA-independent pathways. The Plant Journal 66, 735–744 [DOI] [PubMed] [Google Scholar]
- Finkelstein R, Reeves W, Ariizumi T, Steber C. 2008. Molecular aspects of seed dormancy. Annual Review of Plant Biology 59, 387–415 [DOI] [PubMed] [Google Scholar]
- Fleith M, Clandinin MT. 2005. Dietary PUFA for preterm and term infants: review of clinical studies. Critical Review in Food Science and Nutrition 45, 205–229 [DOI] [PubMed] [Google Scholar]
- Fraser TC, Qi B, Elhussein S, Chatrattanakunchai S, Stobart AK, Lazarus CM. 2004. Expression of the Isochrysis C18-delta9 polyunsaturated fatty acid specific elongase component alters Arabidopsis glycerolipid profiles. Plant Physiology 135, 859–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Pineda E, Castro-Mercado E, Lozoya-Gloria E. 2004. Gene expression and enzyme activity of pepper (Capsicum annuum L.) ascorbate oxidase during elicitor and wounding stress. Plant Science 166, 237–243 [Google Scholar]
- Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI. 2010. Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes and Development 24, 1695–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iba K. 2002. Acclimative response to temperature stress in higher plants: approaches of gene engineering for temperature tolerance. Annual Review of Plant Biology 53, 225–245 [DOI] [PubMed] [Google Scholar]
- Kinney AJ, Cahoon EB, Damude HG, Hitz WD, Kolar CW, Liu Z. 2004. Production of very long chain polyunsaturated fatty acids in oilseed plants. World Patent Application No. WO 71467, A2 [Google Scholar]
- Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, Asami T, Hirai N, Koshiba T, Kamiya Y, Nambara E. 2004. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: key enzymes in ABA catabolism. EMBO Journal 23, 1647–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauritzen L, Hansen HS, Jorgensen MH, Michaelsen KF. 2001. The essentiality of long chain n-3 fatty acids in relation to development and function of the brain and retina. Progress in Lipid Research 40, 1–94 [DOI] [PubMed] [Google Scholar]
- Leinster VH, Robson LG, Shortland PJ. 2010. Differential effects of riluzole on subpopulations of adult rat dorsal root ganglion neurons in vitro. Neuroscience 166, 942–951 [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. 2009. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068 [DOI] [PubMed] [Google Scholar]
- Mikami K, Murata N. 2003. Membrane fluidity and the perceptionof environmental signals in cyanobacteria andplants. Progress in Lipid Research 42, 527–543 [DOI] [PubMed] [Google Scholar]
- Millar AA, Kunst L. 1997. Very-long-chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. The Plant Journal 12, 121–131 [DOI] [PubMed] [Google Scholar]
- Millar AA, Wrischer M, Kunst L. 1998. Accumulation of very-long-chain fatty acids in membrane glycerolipids is associated with dramatic alterations in plant morphology. The Plant Cell 10, 1889–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill CM, Gill S, Hobbs D, Morgan C, Bancroft I. 2003. Natural variation for seed oil composition in Arabidopsis thaliana . Phytochemistry 64, 1077–1090 [DOI] [PubMed] [Google Scholar]
- Pandey S, Chen JG, Jones AM, Assmann SM. 2006. G-protein complex mutants are hypersensitive to abscisic acid regulation of germination and postgermination development. Plant Physiology 141, 243–256 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Park SY, Fung P, Nishimura N, et al. 2009. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei ZM, Kuchitsu K, Ward JM, Schwarz M, Schroeder JI. 1997. Differential abscisic acid regulation of guard cell slow anion channels in Arabidopsis wild-type and abi1 and abi2 mutants. The Plant Cell 9, 409–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzio GA, Rodriguez L, Antoni R, Gonzalez-Guzman M, Yunta C, Merilo E, Kollist H, Albert A, Rodriguez PL. 2013. The PYL4 A194T mutant uncovers a key role of PYR1-LIKE4/PROTEIN PHOSPHATASE 2CA interaction for abscisic acid signaling and plant drought resistance. Plant Physiology 163, 441–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi B, Beaudoin F, Fraser T, Stobart AK, Napier JA, Lazarus CM. 2002. Identification of a cDNA encoding a novel C18-Delta9 polyunsaturated fatty acid-specific elongating activity from the docosahexaenoic acid (DHA)-producing microalga, Isochrysis galbana. FEBS Letters 510, 159–165 [DOI] [PubMed] [Google Scholar]
- Qi B, Fraser TC, Bleakley CL, Shaw EM, Stobart AK, Lazarus CM. 2003. The variant ‘his-box’ of the C18-Delta9-PUFA-specific elongase IgASE1 from Isochrysis galbana is essential for optimum enzyme activity. FEBS Letters 547, 137–139 [DOI] [PubMed] [Google Scholar]
- Qi B, Fraser T, Mugford S, Dobson G, Sayanova O, Butler J, Napier JA, Stobart AK, Lazarus CM. 2004. Production of very long chain polyunsaturated omega-3 and omega-6 fatty acids in plants. Nature Biotechnology 22, 739–745 [DOI] [PubMed] [Google Scholar]
- Rozhnova N, Gerashchenkov G, Babosha A. 2003. The effect of arachidonic acid and viral infection on the phytohemagglutinin activity during the development of tobacco acquired resistance. Russian Journal of Plant Physiology 50, 661–665 [Google Scholar]
- Savchenko T, Walley JW, Chehab EW, et al. 2010. Arachidonic acid: an evolutionarily conserved signaling molecule modulates plant stress signaling networks. The Plant Cell 22, 3193–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen, Thomas D, Livak, Kenneth J. 2008. Analyzing real-time PCR data by the comparative CT method. Nature Protocols 3, 1101–1108 [DOI] [PubMed] [Google Scholar]
- Schroeder F, Petrescu AD, Huang H, et al. 2008. Role of fatty acid binding proteins and long chain fatty acids in modulating nuclear receptors and gene transcription. Lipids 43, 1–17 [DOI] [PubMed] [Google Scholar]
- Serini S, Fasano E, Piccioni E, Cittadini AR, Calviello G. 2011. Differential anti-cancer effects of purified EPA and DHA and possible mechanisms involved. Current Medicinal Chemistry 18, 4065–4075 [DOI] [PubMed] [Google Scholar]
- Somerville C, Browse J. 1991. Plant lipids: metabolism, mutants, and membranes. Science 252, 80–87 [DOI] [PubMed] [Google Scholar]
- Sun Q, Liu J, Zhang Q, Qing X, Dobson G, Li X, Qi B. 2013. Characterization of three novel desaturases involved in the delta-6 desaturation pathways for polyunsaturated fatty acid biosynthesis from Phytophthora infestans. Applied Microbiology and Biotechnology 97, 7689–7697 [DOI] [PubMed] [Google Scholar]
- Torres-Franklin ML, Repellin A, Huynh VB, D’Arcy-Lameta A, Zuily-Fodil Y, Pham-Thi AT. 2009. Omega-3 fatty acid desaturase (FAD3, FAD7, FAD8) gene expression and linolenic acid content in cowpea leaves submitted to drought and after rehydration. Environmental and Experimental Botany 65, 162–169 [Google Scholar]
- Upchurch RG. 2008. Fatty acid unsaturation, mobilization, and regulation in the response of plants to stress. Biotechnology Letters 30, 967–977 [DOI] [PubMed] [Google Scholar]
- Venegas-Calerón M, Sayanova O, Napier JA. 2010. An alternative to fish oils: metabolic engineering of oil-seed crops to produce omega-3 long chain polyunsaturated fatty acids. Progress in Lipid Research 49, 108–119 [DOI] [PubMed] [Google Scholar]
- Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, Leung J, Rodriguez PL, Lauriere C, Merlot S. 2009. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. The Plant Cell 21, 3170–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Schacky C, Harris WS. 2007. Cardiovascular benefits of omega-3 fatty acids.Cardiovascular Research 73, 310–315 [DOI] [PubMed] [Google Scholar]
- Wu G, Truksa M, Datla N, Vrinten P, Bauer J, Zank T, Cirpus P, Heinz E, Qiu X. 2005. Stepwise engineering to produce high yields of very long-chain polyunsaturated fatty acids in plants. Nature Biotechnology 23, 1013–1017 [DOI] [PubMed] [Google Scholar]
- Xu L, Han L, Huang B. 2011. Membrane fatty acid composition and saturation levels associated with leaf dehydration tolerance and post-drought rehydration in Kentucky bluegrass. Crop Science 51, 273–281 [Google Scholar]
- Yoo CY, Pence HE, Jin JB, Miura K, Gosney MJ, Hasegawa PM, Mickelbart MV. 2010. The Arabidopsis GTL1 transcription factor regulates water use efficiency and drought tolerance by modulating stomatal density via transrepression of SDD1. The Plant Cell 22, 4128–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Barg R, Yin M, Gueta-Dahan Y, Leikin-Frenkel A, Salts Y, Shabtai S, Ben-Hayyim G. 2005. Modulated fatty acid desaturation via overexpression of twodistinct ω-3 desaturases differentially alters tolerance tovarious abiotic stresses in transgenic tobacco cells and plants. The Plant Journal 44, 361–371 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.