Abstract
Genome-wide molecular approaches have substantially elucidated molecular alterations and pathways involved in the oncogenesis of brain tumors. In gliomas, several molecular biomarkers including IDH mutation, 1p/19q co-deletion, and MGMT promotor methylation status have been introduced into neuropathological practice. Recently, mutations of the ATRX gene have been found in various subtypes and grades of gliomas and were shown to refine the prognosis of malignant gliomas in combination with IDH and 1p/19q status. Mutations of ATRX are associated with loss of nuclear ATRX protein expression, detectable by a commercially available antibody, thus turning ATRX into a promising prognostic candidate biomarker in the routine neuropathological setting.
Keywords: gliomas, genomic aberrations, molecular biomarker, ATRX, IDH, 1p/19q
Molecular biomarkers in gliomas
In the last years, genome- and transcriptome-wide molecular approaches have deciphered molecular alterations and pathways involved in the oncogenesis of various brain tumors, especially in medulloblastomas and gliomas. This led to the introduction of molecular diagnostic, prognostic, and predictive biomarkers in neuro-oncology practice (for review see [1]). In gliomas, IDH1 mutation, 1p/19q co-deletion, and MGMT promoter methylation can be routinely assessed and the results of these analyses help to refine the diagnosis and prognosis, and improve the treatment of the affected patients (for review see [2]).
Recently, mutations in the ATRX (α-thalassemia/mental retardation syndrome X-linked) gene have been detected in gliomas of various subtypes and grades [3, 4, 5, 6, 7, 8, 9, 10]. In independent studies, a prognostic impact of ATRX mutations in the context of other molecular markers could be demonstrated [4, 11], thus turning ATRX into a new candidate biomarker for routine clinical practice.
ATRX
The ATRX gene is located on chromosome Xq21.1 and encodes a protein that belongs to the H3.3-ATRX-DAXX chromatin-remodeling pathway.
Mutations in ATRX give rise to characteristic developmental abnormalities including severe mental retardation, facial dysmorphism, urogenital abnormalities and α-thalassemia. ATRX is required for the incorporation of the histone variant H3.3 at pericentric heterochromatin and at telomeres, as well as at several transcription factor binding sites [12]. Perturbation of ATRX has been associated with a wide range of effects: altered patterns of DNA methylation (at subtelomeres, heterochromatic repeats, and ribosomal DNA), aberrant chromosome congression in mitosis, and segregation in meiosis, as well as telomere dysfunction [13].
ATRX mutations in gliomas
In 2011, mutations in the ATRX gene were described for the first time in a small fraction of adult and pediatric glioblastomas (GBM), as well as oligodendrogliomas (OG), and a significant correlation with alternative lengthening of telomeres (ALT), a presumed precursor to genomic instability, was demonstrated [3]. In 2012, mutations in the H3F3A and ATRX genes were detected in 30 – 40% of pediatric glioblastomas and in a smaller percentage of adult glioblastomas [9]. Analyses of ATRX in a series of 363 gliomas of various subtypes and grades revealed mutations in 25.6% of all tumors [4]. They occurred frequently in grade II astrocytomas (67%), grade III astrocytomas (73%), secondary GBMs (57%), and in tumors of mixed astrocytic and oligodendrocytic lineage (68%), whereas they were rare in primary GBMs (4%) and uncommon in pediatric GBMs (20%) and pure oligodendroglial tumors (14%). Furthermore, the correlation between ATRX, p53, and IDH1/2 mutations, the status of chromosome arms 1p and 19q, as well as mutations of the CIC and FUBP1 genes (candidate genes in 1p/19q co-deleted tumors) revealed a significant correlation between ATRX, IDH1, and p53 mutations [4]. In contrast, ATRX mutation and combined 1p/19q co-deletion rarely co-occurred. Thus, based on the combination of these molecular changes, the authors defined three different groups of gliomas. 1) “group I-CF”(IDH-CIC/FUBP1): tumors with IDH1/2 mutation and 1p/19q codeletion, CIC or FUBP1 mutation, 2) “group I-A”: tumors with IDH1/2 and ATRX mutation, 3) “group I-X”: tumors that did not fall into group 1 or 2. Correlations of these three groups with morphology, patient age and outcome revealed that astrocytic gliomas primarily had a I-A (32%) or I-X (66%) genetic signature, whereas oligodendroglial tumors were primarily I-CF (77%), but a significant subset harbored the I-A (16%) or I-X (7%) genetic signatures. Gliomas with I-A tumors were significantly younger than patients with I-CF tumors and patients with I-X tumors (mean ages 34, 44, and 54 years, respectively). Among all grade II – IV glioma patients, those whose tumors bore the I-A and I-CF signatures survived significantly longer than those with I-X tumors (median 51, 96, and 13 months, respectively), whereas in grade II glioma or grade II – III oligodendroglioma patients, no significant differences in median survival between the three groups could be observed.
The prognostic impact of ATRX was corroborated in a second independent study, performed on a cohort of 133 malignant gliomas, treated within the German NOA-04 trial (NCT00717210) [11]. In this study, IDH mutations, MGMT promotor methylation, immunohistochemical loss of nuclear ATRX expression, which has been shown to correlate with ATRX mutations [3], and status of chromosome arms 1p and 19q were analyzed. Loss of nuclear ATRX expression was detected in 33% of all tumors and was significantly higher in anaplastic astrocytomas (AA, 45%) than in anaplastic oligoastrocytomas (AOA, 27%) and low in anaplastic oligodendrogliomas (AO, 10%). It occurred almost exclusively in tumors harboring IDH mutations (42/98 IDH mutated tumors). Nuclear ATRX loss and 1p/19q co-deletion were almost mutually exclusive. Furthermore, no association between MGMT promoter methylation and ATRX loss was observed. Similar to the study of Jiao et al, [4] patients with IDH mutation and ATRX loss were younger than patients with IDH mutation and ATRX expression, or IDH wild-type patients (mean age 35.7, 46.8, and 54.1 years, respectively).
In a multivariate Cox regression model only ATRX loss and 1p/19q status were significantly associated with prognosis, whereas the histological subtype, which was significantly correlated with survival in univariate analysis, lost its significance. Based on these results, the authors classified the tumors into three groups 1) “molecular astrocytomas”: tumors with IDH mutation and without 1p/19q co-deletion, including mixed AOA with ATRX loss and AA with and without ATRX loss; 2) “molecular oligodendrogliomas”: tumors with IDH mutation and ATRX expression including AOA harboring 1p/19q co-deletion and AO with and without 1p/19q co-deletion; 3) “molecular glioblastomas”: tumors with IDH wild-type. Outcome analyses showed that the group of molecular oligodendrogliomas had the best prognosis, followed by the molecular group of astrocytomas, whereas the molecular glioblastoma group had the poorest outcome. Importantly, in the group of molecular astrocytoma, patients with ATRX loss had a significantly better outcome than patients without ATRX loss, thus providing evidence that the molecular profile helps to refine the prognosis in malignant glioma patients.
In contrast to gliomas of adult patients, ATRX mutations are very rare in pediatric anaplastic gliomas and low-grade gliomas and have to date never been found in pilocytic astrocytomas [8, 9, 14].
ATRX in neuropathological practice
ATRX expression
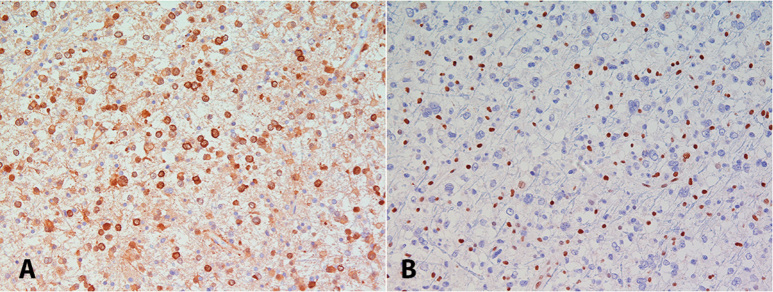
Loss of nuclear ATRX seems to be a good surrogate marker for ATRX mutations [3] and ATRX expression can – similarly to mutant IDH1 protein (Figure 1A) – be easily assessed in the routine neuropathological setting with a commercially available antibody (HPA001906, Sigma-Aldrich, St. Louis, MO, USA). Physiologically, ATRX protein is ubiquitously expressed in cell nuclei. Mutations in the ATRX gene result in a loss of nuclear protein expression in tumor cells, but retained expression in non-tumor cells (e.g., endothelial cells, pre-existing glial cells), which serve as a positive internal control (Figure 1B), analogously to the expression of INI1/SMARCB1 protein in atypical teratoid/rhabdoid tumors. Yet, interpretation of the immunohistochemical staining results might be difficult in diffusely infiltrating tumors with very low tumor cell content and areas with squeezing artifacts which are usually not well stained.
Figure 1. Diffuse astrocytoma with expression of mutant IDH1 protein (A) and loss of nuclear ATRX expression (B).
Diagnostic value of ATRX
As ATRX mutations have been to date not detected in pilocytic astrocytomas, ATRX expression analysis might be helpful in diagnostically challenging small biopsies, especially with regard to the differential diagnosis of pilocytic versus diffuse astrocytoma.
Prognostic value of ATRX
ATRX is a prognostic candidate biomarker in adult patients with malignant gliomas, where it might help to define a group of anaplastic astrocytomas with a better prognosis. The currently available literature suggests that anaplastic astrocytomas, oligodendrogliomas, and mixed oligoastrocytomas can be divided based on IDH and ATRX mutations and 1p/19q status in three prognostically different groups:
Tumors with IDH mutations, no ATRX mutation and 1p/19q co-deletion. These tumors are most frequently oligodendrogliomas and oligoastrocytomas and have the best prognosis.
Tumors with IDH mutations and ATRX mutation and without 1p/19q co-deletion. These tumors are most frequently astrocytomas or oligoastrocytomas and show an intermediate but significantly better prognosis than group 3 tumors.
Tumors without IDH mutation. These tumors have the poorest prognosis and seem to behave clinically like glioblastomas.
Open questions
The impact of ATRX as prognostic marker in low-grade gliomas is to date still unclear.
A fraction of gliomas (e.g., IDH-mutated tumors with or without both ATRX mutation and 1p/19 co-deletion) cannot be classified according to the proposed algorithm.
The association between ATRX mutations and loss of nuclear expression should be confirmed in larger series.
As the histopathological diagnosis of glioma subtypes is prone to interobserver variability and molecular markers seem to improve the prediction of biological behavior, it will be a matter of discussion whether and how IDH, 1p/19q status, and ATRX can be used to complement and refine the diagnosis of glioma subtypes.
In conclusion, ATRX seems to be a promising candidate biomarker in gliomas, which could help to refine, in combination with IDH and 1p/19q status, the prognosis of patients with malignant gliomas.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Berghoff AS Stefanits H Woehrer A Heinzl H Preusser M Hainfellner JA Clinical neuropathology practice guide 3-2013: levels of evidence and clinical utility of prognostic and predictive candidate brain tumor biomarkers. Clin Neuropathol. 2013; 32: 148–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weller M Pfister SM Wick W Hegi ME Reifenberger G Stupp R Molecular neuro-oncology in clinical practice: a new horizon. Lancet Oncol. 2013; 14: e370–379 [DOI] [PubMed] [Google Scholar]
- 3.Heaphy CM de Wilde RF Jiao Y Klein AP Edil BH Shi C Bettegowda C Rodriguez FJ Eberhart CG Hebbar S Offerhaus GJ McLendon R Rasheed BA He Y Yan H Bigner DD Oba-Shinjo SM Marie SK Riggins GJ Kinzler KW Vogelstein B Hruban RH Maitra A Papadopoulos N Meeker AK Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011; 333: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiao Y Killela PJ Reitman ZJ Rasheed AB Heaphy CM de Wilde RF Rodriguez FJ Rosemberg S Oba-Shinjo SM Nagahashi Marie SK Bettegowda C Agrawal N Lipp E Pirozzi C Lopez G He Y Friedman H Friedman AH Riggins GJ Holdhoff M Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget. 2012; 3: 709–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kannan K Inagaki A Silber J Gorovets D Zhang J Kastenhuber ER Heguy A Petrini JH Chan TA Huse JT Whole-exome sequencing identifies ATRX mutation as a key molecular determinant in lower-grade glioma. Oncotarget. 2012; 3: 1194–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khuong-Quang DA Buczkowicz P Rakopoulos P Liu XY Fontebasso AM Bouffet E Bartels U Albrecht S Schwartzentruber J Letourneau L Bourgey M Bourque G Montpetit A Bourret G Lepage P Fleming A Lichter P Kool M von Deimling A Sturm D K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol. 2012; 124: 439–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Killela PJ Pirozzi CJ Reitman ZJ Jones S Rasheed BA Lipp E Friedman H Friedman AH He Y McLendon RE Bigner DD Yan H The genetic landscape of anaplastic astrocytoma. Oncotarget. 2013; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu XY Gerges N Korshunov A Sabha N Khuong-Quang DA Fontebasso AM Fleming A Hadjadj D Schwartzentruber J Majewski J Dong Z Siegel P Albrecht S Croul S Jones DT Kool M Tonjes M Reifenberger G Faury D Zadeh G Frequent ATRX mutations and loss of expression in adult diffuse astrocytic tumors carrying IDH1/IDH2 and TP53 mutations. Acta Neuropathol. 2012; 124: 615–625 [DOI] [PubMed] [Google Scholar]
- 9.Schwartzentruber J Korshunov A Liu XY Jones DT Pfaff E Jacob K Sturm D Fontebasso AM Quang DA Tonjes M Hovestadt V Albrecht S Kool M Nantel A Konermann C Lindroth A Jager N Rausch T Ryzhova M Korbel JO Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012; 482: 226–231 [DOI] [PubMed] [Google Scholar]
- 10.Sturm D Witt H Hovestadt V Khuong-Quang DA Jones DT Konermann C Pfaff E Tonjes M Sill M Bender S Kool M Zapatka M Becker N Zucknick M Hielscher T Liu XY Fontebasso AM Ryzhova M Albrecht S Jacob K Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012; 22: 425–437 [DOI] [PubMed] [Google Scholar]
- 11.Wiestler B Capper D Holland-Letz T Korshunov A von Deimling A Pfister SM Platten M Weller M Wick W ATRX loss refines the classification of anaplastic gliomas and identifies a subgroup of IDH mutant astrocytic tumors with better prognosis. Acta Neuropathol. 2013; 126: 443–451 [DOI] [PubMed] [Google Scholar]
- 12.Clynes D Higgs DR Gibbons RJ The chromatin remodeller ATRX: a repeat offender in human disease. Trends Biochem Sci. 2013; 38: 461–466 [DOI] [PubMed] [Google Scholar]
- 13.Clynes D Gibbons RJ ATRX and the replication of structured DNA. Curr Opin Genet Dev. 2013; 23: 289–294 [DOI] [PubMed] [Google Scholar]
- 14.Zhang J Wu G Miller CP Tatevossian RG Dalton JD Tang B Orisme W Punchihewa C Parker M Qaddoumi I Boop FA Lu C Kandoth C Ding L Lee R Huether R Chen X Hedlund E Nagahawatte P Rusch M Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet. 2013; 45: 602–612 [DOI] [PMC free article] [PubMed] [Google Scholar]


