Abstract
Small-molecule inhibitors and microRNAs (miRNAs) are two newly emerging classes of tools for optimizing induced pluripotent stem cell (iPSC) generation. We report here that sodium butyrate (NaB), a small-molecule inhibitor of histone deacetylases (HDACs), upregulates transcriptional levels of the miR-302/367 cluster by enhancing Oct4 transcriptional activity at the miR-302/367 cluster promoter. NaB does not affect the OCT4 DNA-binding domain; instead it enhances transactivity of the OCT4 transactivation domains. We elucidate that OCT4 transcriptional activity is usually dampened by its associated HDACs in cells and can be derepressed by NaB by impairing the interaction between Oct4 and HDACs, which leads to an elevated expression of the miR-302/367 cluster. Our new findings suggest a novel molecular mechanism for NaB in promoting somatic cell reprogramming via the miR-302/367 cluster.
Introduction
Induced pluripotent stem cells (iPSCs) are generated from somatic cells by forced expression of defined transcription factor (TF) combinations (i.e., Oct4, Sox2, Klf4, and c-Myc, or Oct4, Sox2, Nanog, and Lin28), closely resembling embryonic stem cells (ESCs). iPSCs hold great potential in basic stem cell biology and regenerative medicine (Lowry et al., 2008; Meissner et al., 2007; Park et al., 2008; Takahashi et al., 2007; Yu et al., 2007). Because of the oncogenic features of c-Myc and Klf4 and low efficiency with slow kinetics for iPSC generation, many strategies have been developed to reduce tumorigenesis or enhance reprogramming efficiency (Liao et al., 2011; Mali et al., 2010).
microRNAs (miRNAs) are 18- to 24-nucleotide single-stranded RNAs that can degrade target mRNAs or repress the translation of encoded proteins by binding to partially complementary target sites (Bartel, 2004). Due to specific modulation mechanisms, one miRNA may target hundreds of target genes, resulting in the formation of complex regulatory feedback networks. Recently, miRNAs have been found to enhance reprogramming efficiency (Anokye-Danso et al., 2011; Miyoshi et al., 2011). Among these miRNAs, the miRNA-302/367 cluster not only enhances reprogramming efficiency and substitutes c-Myc in mouse embryonic fibroblasts (MEFs), but also directly reprograms human somatic cells into iPSCs (Anokye-Danso et al., 2011; Lin et al., 2011; Lipchina et al., 2012; Miyoshi et al., 2011). The miR-302/367 cluster is a polycistronic miRNA that consists of five mature miRNAs (i.e., miR-302b/c/a/d and miR-367), and they are expressed primarily in pluripotent cell populations.
Small molecules such as the histone deacetylase (HDAC) inhibitor sodium butyrate (NaB), transforming growth factor-β (TGF-β signaling pathway inhibitor SB431542, and MAP/extracellular signal–regulated kinase (ERK) (MEK) signaling pathway inhibitor PD0325901 can enhance reprogramming with faster kinetics (Liang et al., 2010; Lin et al., 2009; Mali et al., 2010; Zhang et al., 2011). Recently, we reported a simple approach for generating human iPSCs by using a single polycistronic retroviral vector expressing four TFs, combined with a cocktail consisting of these three small molecules (3-SM) (Zhang et al., 2011). The 3-SM cocktail selectively promotes generation of fully reprogramed human iPSCs with higher efficiency and faster kinetics (Zhang et al., 2011). Interestingly, a previous report showed that NaB reverts human ESCs and mouse epiblast stem cells (EPiSCs) to an earlier developmental stage and slows down differentiation of human (h) ESCs cultured in conditioned medium (CM), as evidenced by a slower decline in the miR-302/367 family (Ware et al., 2009). These reports implicate a potential relationship between NaB and the miR-302/367 cluster in maintaining the ESC pluripotency and reprogramming process. Indeed, we recently found that a 3-SM cocktail or NaB alone upregulates the expression of the miR-302/367 cluster and enhances generation of iPSCs from human fibroblasts. However, the molecular mechanisms of how NaB regulates expression of the miR-302/367 cluster remain largely unknown.
Here, we report that the 3-SM cocktail or NaB upregulated the transcriptional level of the miR-302/367 cluster by enhancing hOCT4 transcriptional activity in the miR-302/367 promoter. We dissected three functional domains of hOCT4 and identified the domains required for NaB or 3-SM to enhance hOCT4 transactivity. In addition, we elucidated the molecular mechanisms by which NaB augments transactivation activity of hOCT4.
Materials and Methods
Cell culture
The primary human foreskin fibroblasts (HFFs) were purchased from Millipore and cultured in FibroGRO™-LS Complete Media. 293T cells were maintained in Dulbecco's Modified Eagle Medium (DMEM) (high glucose) containing 10% fetal bovine serum (FBS). hiPSCs or hESCs were cultured on MEF feeder cells (or BD gel) in conventional human ESC culture medium [DMEM/F12, 20% knockout serum replacement, 1% GlutaMAX, 1% nonessential amino acids, 1% penicillin/streptomycin, 0.1 mM β-mercaptoethanol, and 20 ng/mL basic fibroblast growth factor (bFGF)]. All cell culture products were purchased from Invitrogen, except where mentioned.
iPSC generation using a retrovirus-mediated gene delivery approach
The 293T cells were plated at 4×105 cells per well of a six-well plate, cultured overnight, and then transfected with a mixture of DNA containing 2.5 μg of pMig-OKSM (Zhang et al., 2011) and 1.5 μg of pCL-Ampho (IMGENEX) by FuGENE HD (Promega), according to the manufacturer's instructions. The supernatant of the transfected cells was collected 24 h posttransfection and filtered through a 0.45- μm pore-size filter. HFFs were seeded in a 12-well plate at 1×104 cells per well of 12-well plate at 1 day before transduction and incubated with retrovirus-containing supernatant supplemented with 5 μg/mL Polybrene (Sigma), followed by centrifugation at 1000×g for 30 min. Four days postinfection, infected cells were split using 0.025% trypsin-EDTA and plated on MEF feeders cultured in fibroblast medium. After 24 h, the medium was switched to conventional human ESC medium as described above and treated with a small-molecule cocktail containing different combinations of 0.5 mM NaB (Sigma), 2 μM SB431542 (Stemgent), and 0.5 μM PD0325901 (Stemgent). Medium was changed every other day until induced colonies were picked up based on human ESC colony morphology at day 21 or were stained by alkaline phosphatase in situ.
Alkaline phosphatase and immunofluorescence staining
iPSCs were fixed with 4% formaldehyde in phosphate-buffered saline (PBS) for 10 min and stained using the Alkaline Phosphatase Staining Kit (Stemgent), according to the manufacturer's instructions. For immunostaining, the transfected cells were treated with NaB for 24 h and then fixed in 4% paraformaldehyde for 20 min at room temperature, washed three times with PBS, and blocked for 30 min with 5% FBS containing 0.05% Triton X-100, followed by primary and secondary antibody incubation. Antibodies were diluted in 1% FBS containing 0.05% Triton X-100. Anti-Oct4 and anti-Flag antibodies were purchased from Stemgent and Thermo Fisher Scientific Inc., respectively.
Gene expression analysis by qPCR
Total RNAs were extracted using Quick-RNA™ MicroPrep Kit (Zymo Research). For real-time qPCR analysis of individual mature miR-302/367 miRNAs, 500 ng of total RNA was reversed-transcribed using the NCode™ VILO™ miRNA cDNA Synthesis Kit (Invitrogen) and then followed by qPCR analysis. Primers for qPCR analysis are included in the Table S1. (Supplementary Data are available at www.liebertpub.com/cell/).
Plasmid and plasmid constructs
Plasmid pGL3-miR-302/367-Luc was constructed by PCR cloning. An approximately 1-kb region of the miR-302/367 promoter was inserted into KpnI and HindIII sites of the pGL3-basic vector (Promega) by using standard molecular techniques. To generate pGL3-miR-302/367(ΔOct4-RE)-Luc, the Oct4-binding site in miR302/367 cluster promoter was deleted by PCR in pGL3-miR-302/367-Luc. A pGL3-(Oct4-RE)3-TATA-Luc plasmid was generated by cloning the fragment consisting of the TATA-box and three copies of Oct4-binding sites into XhoI and HindIII sites of the pGL3-basic vector. Similarly, a Sox2-specific reporter (Sox2-RE)3-TATA-Luc was generated by cloning the TATA-box and three copies of Sox2 binding sites from the miR-302/367 cluster promoter into pGL3-basic vector. To generate pVP16-Oct4 DBD, a DNA fragment containing the OCT4 DNA-binding domain was fused in-frame to the carboxyl terminus of the VP16 activation domain in the pVP16 vector. Gal4-Oct4, Gal4-Oct4ND, Gal4-Oct4CD, and Gal4-Oct4NCD plasmids were constructed by in-frame fusing of full-length Oct4, the amino-terminal domain of Oct4, the carboxy-terminal domain of OCT4, and both amino- and carboxy-terminal domains to the carboxyl terminus of the Gal4 DNA-binding domain in pM2 vector. A pFR-Luc reporter plasmid that contains five Gal4 binding sites was purchased from Stratagene Inc. (La Jolla, CA, USA). Plasmids pMXs-Oct4, pMXs-Klf4, pMXs-Sox2, and pMXs-c-Myc were obtained from Addgene (Boston, MA, USA).
Transient transfection and reporter assay
293T cells and HFFs were transfected by FuGENE HD (Promega) and GeneIn (GlobalStem, Rockville, MD, USA), respectively, according to their manufacturer's protocols. Briefly, 5×104 cells were seeded in 24-well plates and cultured overnight. Cells at ∼80% confluence were transfected by mixing the indicated amount of DNA described in the figure legends. At 24 h after transfection, small molecules (as indicated in figures) were added to the cell culture medium, and transfected cells were harvested 48 h later by adding 200 μL of Passive Lysis Buffer (Promega). Transfections and reporter assays were performed in triplicate and were repeated independently twice. Luciferase activities of reporter vectors were assayed with a Bright-Glo luciferase assay kit (Promega) according to the provided instruction and were normalized by β-galactosidase activity from the same cells that were co-transfected with pCMV-LacZ as an internal control. Luciferase activities were expressed as relative luciferase/LacZ activities, normalized to those of control transfections in each experiment. For immunofluorescence staining, U2OS cells were plated on cover slides in six-well plates, cultured overnight, and then transfected with 1 μg of pMig-Oct4 and 1.0 μg of HDAC1/2-Flag using FuGENE HD (Promega), according to the manufacturer's instructions.
Co-immunoprecipitation and western blotting
293T cells were co-transfected with plasmids pMig-Oct4 and pCMV-HDAC1/2-Flag using FuGENE HD (Promega). At 12 h after transfection, cells were treated with NaB and then lysed in cell lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 10% glycerol with protease inhibitor mixture) for 30 min. Whole-cell extracts were collected and incubated with anti-Oct4 or anti-Flag antibodies to pull down Oct4 and HDAC1/2, respectively. Protein complex by immunoprecipitation was eluted and analyzed by western blot analysis using anti-Flag and anti-Oct4 antibodies, respectively.
Results
NaB increases the transcriptional level of miR-302/367 miRNAs and positively regulates its promoter
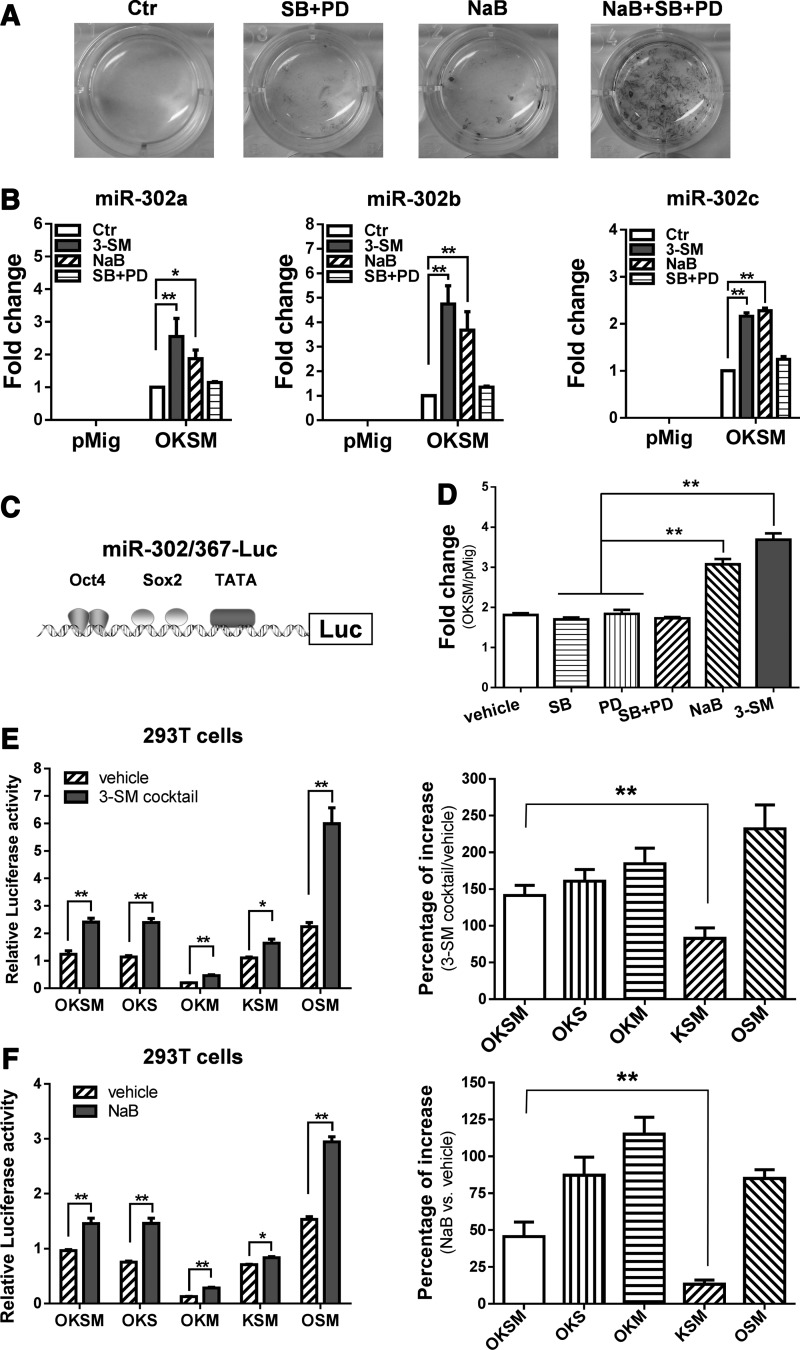
Recently, we found that a small molecule cocktail consisting of NaB, PD0325901, and SB431542 (3-SM cocktail) promotes generation of fully reprogrammed iPSCs (Zhang et al., 2011). As shown in Figure 1A, alkaline phosphatase (AP)-positive cell colonies only appeared noticeably in the groups treated with butyrate alone, a combination of PD0325901 and SB431542, or butyrate together with PD0325901 and SB431542, but not in groups without treatment. These data suggest that NaB has a unique ability to enhance reprogramming processes. By qPCR-based array analysis, we recently found that miRNAs from the miR-302/367 cluster were induced by more than five-fold with the addition of a 3-SM cocktail (Zhang and Wu, 2013). By qPCR analysis, we further found that NaB alone is sufficient to increase expression of mature miRNA members (miR-302a, b, and c) from the miR-302/367 cluster (Fig. 1B). These mature miRNAs were not detectable in the control group transduced with a pMig vector, even after treatment with the 3-SM cocktail, suggesting that action of NaB on the expression of these mature miRNAs is largely dependent on TFs. Because human ESCs highly express four TFs, we examined whether NaB induces expression of the miR-302/367 cluster in these cells. Our qPCR analysis showed that NaB treatment upregulates expression of five members of the miR-302/367 cluster in a time-dependent manner (Fig. S1).
FIG. 1.
NaB and the small-molecule cocktail enhances expression of the miR-302/367 cluster and regulates its promoter in a TF-dependent manner. (A) AP staining of cell cultures 21 days after treatment with small molecules. Primary HFFs were transduced with polycistronic retroviruses containing four TFs (pMig-OKSM), seeded at a density of 15,000 transduced cells per well of 12-well plate, and then treated with different combinations of small molecules (NaB, 0.5 mM; SB431542, 2 μM; PD0325901, 0.5 μM). Emerging colonies were stained by an AP staining kit. Ctr, control; SB, SB431542; PD, PD0325901; pMig, retroviral vector. (B) qPCR analysis of mature miRNA members of the miR-302/367 cluster in HFFs after treatment with different combinations of small molecules. HFFs were transduced with retroviruses containing pMig-OKSM or pMig only, and then treated with indicated small molecules for 72 h. miRNA members (miR-302a, b, c) were quantified by qPCR analysis. (*) p<0.05, (**) p<0.01. (C) The miR-302/367 promoter-driven luciferase reporter (miR-302/367-Luc). The miR-302/367 promoter contains a TATA-box, two separate Sox2-binding sites, and a binary response-element of Oct4. (D) The effects of the small molecules SB431542 and PD0325901 on upregulation of the pri-miR-302/367 promoter. 293T cells were transfected with 100 ng of miR-302/367-Luc reporter, together with 350 ng of pMig-OKSM (or pMig vector) and 50 ng of pCMV-LacZ and treated with or without individual small molecules (SB, PD, and NaB) and cocktails containing two small molecules (SB and PD) or three small molecules (SB, PD, and NaB), respectively. pCMV-LacZ was included in each transfection as an internal control. Relative luciferase activity was assayed 48 h after treatment and normalized by LacZ activity. Fold change in luciferase activity of miR-302/367-Luc reporter was calculated to compare the effects of small molecule treatment with the no treatment groups (right panel). (**) p<0.01. (E) The requirement of individual TFs for the 3-SM cocktail in upregulating the pri-miR-302/367 promoter. 293T cells were transfected with 400 ng of different combinations of indicated plasmids (pMig-Oct4/Klf4/Sox2/c-Myc), 100 ng of miR-302/367-Luc reporter, and 50 ng of pCMV-LacZ. Transfected cells were then treated with or without 3-SM cocktail for 48 h. Relative luciferase activity (left panel) and percentage increased in luciferase activity (right panel) of the miR-302/367-Luc reporter (3-SM cocktail treatment vs. without treatment) were calculated. O, Oct4; K, Klf4; S, Sox2; M, c-Myc. (*) p<0.05, (**) p<0.01. (F) The requirement of individual TFs for NaB in upregulating the pri-miR-302/367 promoter. 293T cells were transfected with a similar combination of plasmids as described in E, and then treated with or without NaB for 48 h. Relative luciferase activity (left panel) and percentage increased in luciferase activity (right panel) of the miR-302/367-Luc reporter (NaB cocktail treatment vs. without treatment) were calculated the same as E. (*) p<0.05, (**) p<0.01.
Previous studies have already shown that the miR-302/367 cluster is regulated by the ESC core TFs Oct4 and Sox2 (Card et al., 2008). Thus, it is possible that NaB or the 3-SM cocktail upregulates the transcriptional activity of the miR-302/367 cluster promoter. To test this possibility, we constructed a luciferase reporter driven by the proximal promoter region of the miR-302/367 cluster (Fig. 1C) and performed a luciferase reporter assay by transfecting this reporter (miR-302/367-Luc) into 293T cells and then treating them with small molecules. Our data showed that NaB, not SB431542 or PD0325901 or both SB431542 and PD0325901, enhances luciferase activity of the miR-302/367-Luc reporter (Fig. 1D). These data are consistent with the qPCR results (Fig. 1B).
Oct4 is required and sufficient for NaB to upregulate the transcriptional activity of the miR-302/367 cluster promoter
According to previous studies, the miR-302/367 cluster is a target of Oct4 and Sox2, and its promoter occupies responsive elements for both Oct4 and Sox2 (Card et al., 2008). Therefore, we asked whether Oct4 or Sox2 or both TFs are essential for NaB or the 3-SM cocktail to upregulate the miR-302/367 promoter. To do so, we transfected 293T cells with a miR-302/367-Luc reporter and different combinations of four TFs, followed by treatment with the 3-SM cocktail (Fig. 1E), NaB (Fig. 1F), or no treatment. By analysis of relative luciferase activity (Fig. 1E and 1F, left panels) and its percentage of increase (Fig. 1E and F, right panels), we found that elimination of Sox2 from the transfected plasmid mixture dramatically decreased the baseline activity of the miR302/367 cluster reporter (Fig. 1E, F), but did not abolish the response of this reporter to NaB treatment (Figure 1E, F, right panels). In contrast, exclusion of Oct4 from the transfected plasmid combinations did not significantly alter the baseline activity of the miR302/367 cluster reporter, but impaired the ability of NaB (Fig. 1E, F, right panel) or the 3-SM cocktail (Fig. 1E, F, right panels) in enhancing the transcriptional activity of the miR-302/367 cluster promoter. Together, these data suggest that Oct4 is essential for NaB and 3-SM to upregulate transcriptional level of miR-302/367 cluster.
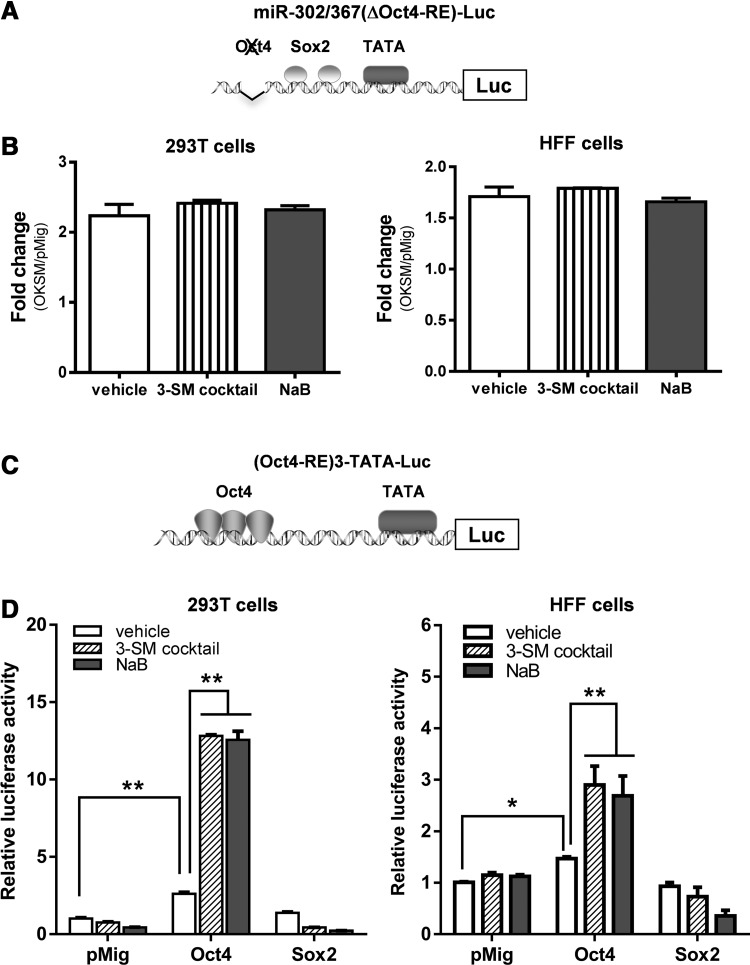
Because it has been reported that Oct4 directly transactivates the promoter of miR-302/367 cluster (Card et al., 2008), we asked whether the deletion of Oct4-responsive element (RE) in the promoter (Fig. 2A) would impair the ability of the 3-SM cocktail or NaB in upregulating this promoter. To address this question, we deleted Oct4-RE in the promoter, and the resulting reporter was designated as miR-302/367(ΔOct4-RE)-Luc. We transfected this new reporter with pMig-OKSM or pMig in 293T cells or HFFs, and then treated them with 3-SM cocktail or NaB. In contrast to the result in Figure 1D, our data showed that deletion of Oct4-RE abolished the ability of 3-SM cocktail or NaB in enhancing transcriptional activity of the miR-302/367 cluster promoter in the cells (Fig. 2B). Thus, our data suggest that Oct4 is a critical mediator for the action of the 3-SM cocktail and NaB on transcriptional regulation of the miR-302/367 cluster (Fig. 2B).
FIG. 2.
The important role of Oct4 for NaB in the regulation of the miR-302/367 promoter. (A) The miR-302/367 promoter-driven luciferase reporter [miR-302/367 (ΔOct4)-Luc] with a deletion of the Oct-binding site. (B) The requirement of the Oct4-binding site in the regulation of the miR-302/367 cluster promoter. 293T cells (left panel) or HFFs (right panel) were transfected with 100 ng of miR-302/367(ΔOct4)-Luc, 350 ng of pMig-OKSM (or pMig vector), and 50 ng of pCMV-LacZ. Transfected cell were then treated with or without the 3-SM cocktail or NaB, respectively, at 12 h after transfection. pCMV-LacZ was included in each transfection as an internal control. Relative luciferase activity was assayed 48 h after treatment and normalized by LacZ activity. Fold change in luciferase activity were calculated (treatment vs. without treatment). (C) An artificial luciferase reporter with a TATA-box and three copies of Oct4 binding sites, designated as (Oct4-RE)3-TATA-Luc. (D) Effect of small molecules on transcriptional activity of Oct4. A 100-ng amount of (Oct4-RE)3-TATA-Luc reporter plasmid was transfected together with 350 ng of Oct4 or Sox2 expression plasmids (or pMig vector) and 50 ng of pCMV-LacZ into 293T cells (left panel) or HFFs (right panel). Transfected cells were treated with or without the 3-SM cocktail or NaB for 48 h before measurement of luciferase and normalization by lacZ activity. Relative luciferase activity was calculated (treatment vs. without treatment). (*) p<0.05, (**) p<0.01.
To further prove that Oct4 is a key mediator for 3-SM cocktail and NaB, we generated a Oct4-specific luciferase reporter (Oct4-RE)3-TATA-Luc that contains a TATA-box and three copies of Oct4-RE into a pGL3-basic vector (Fig. 2C). We transfected 293T cells or HFFs with this reporter and Oct4 or Sox2 expression plasmids, and then treated the cells with or without the 3-SM cocktail or NaB (Fig. 2D). As expected, our data showed that expression of Oct4 but not Sox2 alone increases the transcriptional activity of the (Oct4-RE)3-TATALuc reporter (Fig. 2D). The addition of NaB and 3-SM cocktail enhanced Oct4 transactivity by six-fold and three-fold in 293T and HFFs, respectively (Fig. 2D). Furthermore, we created a Sox2-specific luciferase reporter (Sox2-RE)3-TATA-Luc comprising a TATA-box and three copies of Sox2-RE (Fig. S2A). By reporter assays, we showed that Sox2 but not Oct4 could transactivate the (Sox2-RE)3-TATA-Luc reporter by 50-fold, but failed to further enhance luciferase activity of this reporter after the treatment with NaB or the 3-SM cocktail (Fig. S2B).
Taken together, our data demonstrated that NaB or the 3-SM cocktail positively regulates transcriptional activity of the miRNA-302/367 cluster promoter through enhancing transactivity of Oct4 but not Sox2.
Oct4 transactivation domains but not DNA-binding domains are required for butyrate to upregulate the miR-302/367 cluster promoter
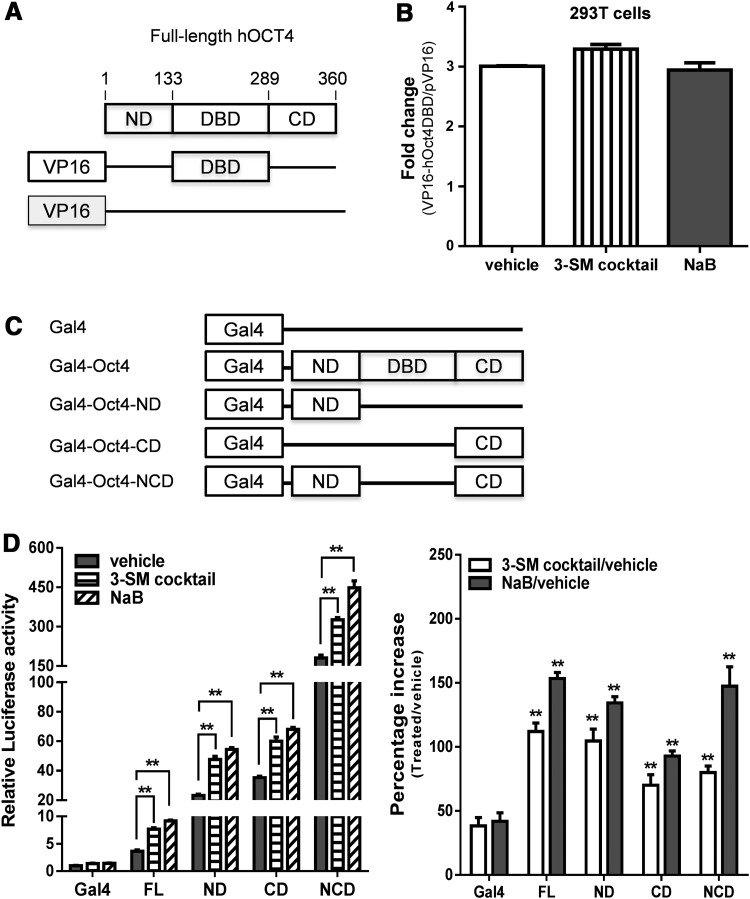
Our results indicated that Oct4 TF is essential for butyrate or the 3-SM cocktail to upregulate the miR-302/367 promoter (Figs. 1 and 2). Next, we asked whether specific OCT4 domains are essential for butyrate or the 3-SM cocktail to increase the transcriptional activity of miR-302/367 cluster promoter. On the basis of previous studies (Lim et al., 2009), human OCT4 (hOCT4) protein consists of an amino-terminal domain [ND, amino acid (aa) 1–133], a POU DNA-binding domain (DBD, aa 134–289), and a carboxy-terminal domain (CD, aa 290–360) (Fig. 3A). It was shown that hOCT4 ND and CD are the transactivation domains, separated by a DBD (Brehm et al., 1997; Lim et al., 2009). To examine whether NaB or the 3-SM cocktail increases DNA-binding activity of hOCT4, we first fused the hOCT4 DBD to the VP16 transactivation domain, designated as pVP16-hOCT4 DBD (Fig. 3A). By performing a luciferase reporter assay using (Oct4-RE)3-TATA-Luc reporter and pVP16-hOCT4 DBD or pVP16 (vector), we found that NaB or the 3-SM cocktail does not affect the transcriptional activity of VP16-hOCT4 DBD on this reporter (Fig. 3B). Thus, it is unlikely that NaB or the 3-SM cocktail increases the DNA-binding activity of hOCT4. To examine if the two transactivation domains (ND and CD) of hOCT4 are important for the action of NaB and 3-SM cocktail, we fused either one or both hOCT4 ND and CD to the carboxyl terminus of the Gal4 DNA-binding domain (Gal4 DBD, aa 1–147) to generate an array of constructs with the same Gal4 DBD, but different hOCT4 domains (Fig. 3C).
FIG. 3.
Butyrate enhances transcriptional activity of Oct4 via transactivation domains. (A) Schematic structure of full-length Oct4 and VP16-tagged Oct4 DNA-binding domain. Human Oct4 consists of 360 amino acids. Amino-terminal domain (ND), DNA-binding domain (DBD), and carboxy-terminal domain (CD) are located in amino-terminal, middle, and carboxy-terminal region of Oct4, respectively. Numbers indicate those of amino acid residues from the amino-terminal portion. The Oct4 DBD was fused in-frame with VP16 (activation domain) by PCR, and the resulting construct was designated as pVP16-Oct4DBD. Numbers indicate amino acid positions of Oct4. (B) Effect of NaB on the DNA-binding activity of Oct4. A 100-ng amount of the (Oct4-RE)3-TATA-Luc reporter plasmid was transfected together with 350 ng of the pVP16-Oct4 DBD plasmid (or pVP16 vector) and 50 ng of pCMV-LacZ into 293T cells. Transfected cells were treated with or without 3-SM cocktail or NaB for 48 h before luciferase activity was measured and normalized by LacZ activity. Fold change in luciferase activity (right panel) was calculated (treatment vs. without treatment). (C) Schematic maps of Oct4 mutants fused to Gal4 DBD. Full-length Oct4 or Oct4 mutants containing ND, CD, or both of ND and CD were fused in-frame to the carboxyl terminus of the Gal4 DBD in the pM2 vector and designated as Gal4-Oct4, Gal4-Oct4-ND, Gal4-Oct4-CD, and Gal4-NCD, respectively. (D) Effect of small molecules on the transcriptional activity of Oct4 mutants. 293T cells were transfected with 350 ng of Gal4-Oct4 mutants (or pM2 vector), 100 ng of pFR-Luc consisting of five copies of Gal4-binding sites, and 50 ng of pCMV-LacZ. pCMV-LacZ was included in each transfection as an internal control. Relative luciferase activity was assayed 48 h after transfection and normalized by LacZ activity (left panel). Percentage increases in luciferase activity (right panel) were calculated by comparing relative luciferase activity for full-length Oct4 or each Oct4 mutant (treatment vs. without treatment). (*) p<0.05, (**) p<0.01.
These chimeric TFs should transactivate the pFR-Luc reporter plasmid because it contains five copies of Gal4-binding sites in the upstream region of the transcription start. After confirming the expression of different Gal4 DBD-hOCT4 fusion proteins by these constructs (data not shown), we co-transfected their encoding plasmids with a pFR-Luc reporter plasmid into 293T cells, then treated them with NaB, 3-SM cocktail, or vehicle (control). Consistent with previous reports (Brehm et al., 1997; Lim et al., 2009), our data show that either hOCT4 ND or CD was sufficient to activate transcription of the reporter whereas both domains lead to a synergistic transactivation activity (Fig. 3D, left panel). After treatment with NaB or the 3-SM cocktail, transcriptional activity of the reporter in the transfection groups with full-length or mutant hOCT4 was significantly increased (Fig. 3D, right panel). Interestingly, the mutants showed a much higher transactivation activity than full-length hOCT4 (Fig. 3D, left panel), which is in agreement with the previous report (Brehm et al., 1997; Lim et al., 2009). Our data suggested that hOCT4 DBD has an inherent inhibitory effect on transactivation activity of hOCT4 and that NaB or the 3-SM cocktail could enhance hOCT4 transactivity through its ND and CD. Collectively, our data indicated that the ND and CD of hOCT4 are essential for NaB and 3-SM cocktail to activate the transcriptional activity of the miR-302/367 cluster promoter.
NaB impairs the association and co-localization between hOCT4 and HDACs
It is well known that histone acetylation influences the accessibility of DNA to the transcriptional machinery for gene expression. Histone deacetylation is generally associated with a closed chromatin state, and inhibitors of HDACs such as NaB are employed to enhance nuclear reprogramming (Liang et al., 2010; Mali et al., 2010). Our data showed that NaB could enhance hOCT4 transcription activity in the promoter of the miR-302/367 cluster. Thus, it is possible that HDACs associate with HDACs and thereby hinder the transcriptional activity of hOCT4. To test this possibility, we co-transfected expression plasmids for hOCT4 and Flag-tagged HDAC1/2 into 293T cells and then treated them with NaB before performing co-immunoprecipitation (co-IP). Our result showed that immunoprecipitation using anti-hOCT4 antibody could indeed pull down HDAC1/2 (Fig. 4A, upper panel); and reciprocally, HDAC1/2 also co-precipitates with OCT4 (Fig. 4A, lower panel). Significantly, treatment with NaB impairs the association between OCT4 and HDAC1/2 (Fig. 4A, lower panel). Next, we examined the localization of hOCT4 and HDAC1/2 by co-transfecting their expression plasmids into cells. Our data showed that both hOCT4 and HDAC1/2 are localized in the cell nucleus and they co-localized in cells when without NaB treatment. Interestingly, HDAC1/2 was mainly relocated to the cell nucleus membrane after NaB treatment and failed to co-localize with hOCT4 in the center area of the cell nucleus (Fig. 4B). Furthermore, we transfected expression plasmids for HDAC1//2 and four reprogramming factors into cells, which were then cultured in hESC medium for 7 days. We examined the localization of hOCT4 and HDAC1/2 and found that our data are similar with the result in Figure 4B (Fig. S3). These results strongly suggest that the presence of NaB interferes with the interaction of HDACs with hOCT4, therefore releasing full transcriptional activity of hOCT4 in the promoter of the miR-302/367 cluster and thus enhances the transcriptional level of miR-302/367 (Fig. 4C).
FIG. 4.
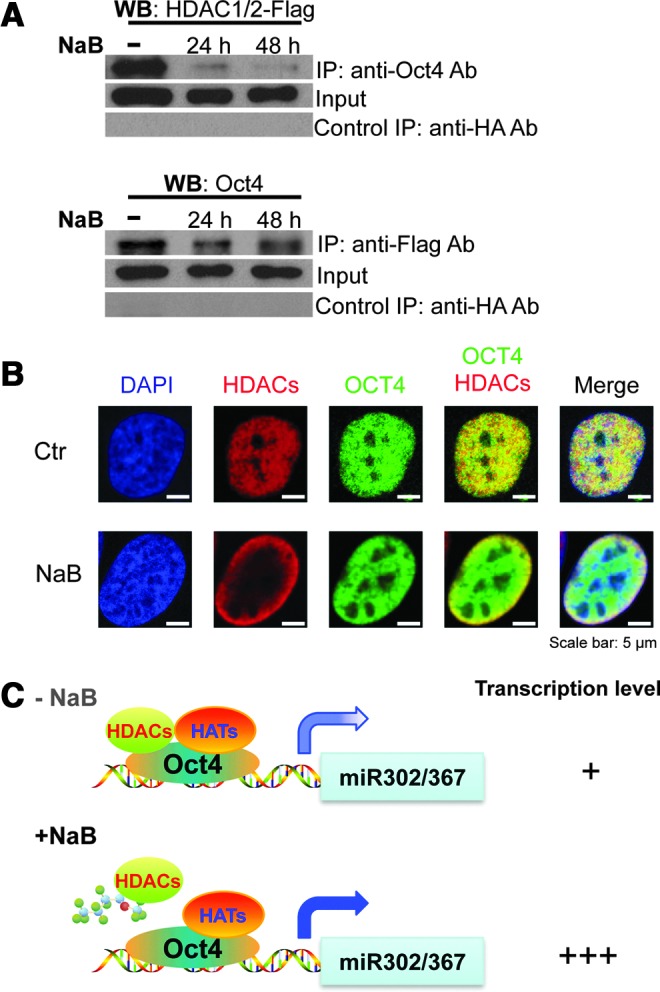
NaB impairs the interaction between Oct4 and HDAC1/2. (A) Co-immunoprecipitation of Oct4 and HDAC1/2. 293T cells were co-transfected with Oct4 and HDAC1/2-Flag expression constructs, cultured overnight, and then treated with NaB for 24 h or 48 h. Cells were lysed, subjected to mmunoprecipitation, and analyzed by western blotting analysis with indicated antibodies. (B) Co-localization of Oct4 with HDAC1/2 in cells. U2OS cells were co-transfected with Oct4 and HDAC1/2-Flag expression plasmids, cultured overnight, and then treated with NaB for 24 h. Cells were fixed and processed for immunofluorescence with anti-Oct4 (green) and anti-Flag (red) and were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Scale bar, 5 μm. (C) Model for NaB's action in the regulation of the miR-302/367 cluster promoter. In the absence of NaB, HDACs and HATs co-associate with Oct4 within the Oct4-binding site in the miR-302/367 cluster promoter, therefore suppressing its transcription. When NaB is present, it will bind to HDACs and therefore result in dissociating HDACs from the Oct4–HATs complex, leading to elevated transcription of the miR-302/367 cluster.
Discussion
Recent evidence has shown that NaB maintains self-renewal of mouse and human ESCs without the need for feeder conditioning or recombinant growth factors (Ware et al., 2009). Also, butyrate greatly enhances human iPSC generation by promoting epigenetic remodeling and the expression of pluripotency-associated genes (Liang et al., 2010; Mali et al., 2010). These studies focused primarily on the HDAC inhibitor's feature of butyrate in the hESC self-renewal and reprogramming process, but did not provide complete insight into understanding the roles of NaB. Ware et al. reported that butyrate exposure slows down the decline of miR-302 family miRNAs in hESCs (Ware et al., 2009). However, there are no studies showing that butyrate can enhance the transcriptional level of the miR-302/367 cluster in reprogramming process. This shows that butyrate upregulates the transcriptional level of the miR-302/367 cluster by relieving HDACs from hOCT4, thus it presents a novel finding. The miR-302/367 cluster is a target of Oct4 and Sox2, and its proximal promoter region occupies binary consensus binding motifs for Oct4 and two separate binding sites for Sox2 (Card et al., 2008).
Our current findings indicate that the 3-SM cocktail and NaB upregulate expression of the miR-302/367 cluster by enhancing hOCT4 transcriptional activity in its promoter, suggesting that Oct4, but not Sox2, is a key regulator of the miR-302/367 cluster. Notably, iPSCs have been generated without Sox2 from melanocytes (Utikal et al., 2009) and with Oct4 alone from adult neural stem cells (Kim et al., 2009). Ding's group has generated human iPSCs from epidermal keratinocytes and cell types other than HFFs using Oct4 plus SM cocktail consisting of four to six inhibitors (Zhu et al., 2010). So far, there are no reports showing that an approach omitting Oct4 TF by replacing it with other TFs or small molecules can generate iPSCs. This further suggests that Oct4 is a key regulator in cellular reprogramming.
As a key regulator for reprogramming and hESC self-renewal, hOCT4 contains two transactivation domains and a DNA-binding domain (Lim et al., 2009). The amino-terminal domain of Oct4 is a constitutive activation domain and the carboxy-terminal domain mediates cell-type specific transcriptional activation (Brehm et al., 1997; Lim et al., 2009). Our findings showing the synergistic activity of the two transactivation domains and amino-terminal domain as the main target of butyrate further support that the amino-terminal domain is indeed the critical element (Brehm et al., 1997). Interestingly, recent studies show that the Oct4-centered protein interaction network plays critical roles in regulating self-renewal and pluripotency of ESCs (van den Berg et al., 2010), and HDACs are among these Oct4-associated proteins that form NuRD (nucleosome remodeling deacetylase) or NODE (Nanog and Oct4-associated deacetylase) complexes in ESCs (Kaji et al., 2006; Liang et al., 2008). Although these studies show that Oct4 associates with HDACs, whether HDAC inhibitors such as butyrate affect the interaction of Oct4 with HDACs has not been documented. Thus, our current findings that NaB impairs the interaction of Oct4 and HDACs present a novel mechanism for butyrate in cellular reprogramming.
In summary, we report here that butyrate increases the transcriptional level of the miR-302/367 cluster via enhancing transactivation activity of hOCT4 ND and CD in the promoter of this cluster. These novel findings will contribute to a broader understanding of molecular mechanisms and functions of butyrate with regard to cellular reprogramming processes and self-renewal of pluripotent stem cells.
Supplementary Material
Acknowledgments
We thank members of the Wu laboratory for discussion and support and Fransisca Heriyanto for her technical support. Parts of the study were supported by an National Institute of Child Health and Human Development (NICHD)/National Institutes of Health (NIH) grant (5R21HD061777) and the Jordan family's endowment fund. Z.Z. was supported by a California Institute for Regenerative Medicine (CIRM) Berkeley scholarship (CIRM training grant TG2-01164).
Author Disclosure Statement
The authors declare that no conflicting financial interests exist. Z.Z. and D.X. performed the experiments and evaluated the data. Z.Z. and W.S.W wrote the manuscript.
References
- Anokye-Danso F., Trivedi C.M., Juhr D., Gupta M., Cui Z., Tian Y., Zhang Y., Yang W., Gruber P.J., Epstein J.A., and Morrisey E.E. (2011). Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 8, 376–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- Brehm A., Ohbo K., and Scholer H. (1997). The carboxy-terminal transactivation domain of Oct-4 acquires cell specificity through the POU domain. Mol. Cell. Biol. 17, 154–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card D.A., Hebbar P.B., Li L., Trotter K.W., Komatsu Y., Mishina Y., and Archer T.K. (2008). Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol. Cell. Biol. 28, 6426–6438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji K., Caballero I.M., MacLeod R., Nichols J., Wilson V.A., and Hendrich B. (2006). The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat. Cell Biol. 8, 285–292 [DOI] [PubMed] [Google Scholar]
- Kim J.B., Sebastiano V., Wu G., Arauzo-Bravo M.J., Sasse P., Gentile L., Ko K., Ruau D., Ehrich M., van den Boom D., Meyer J., Hübner K., Bernemann C., Ortmeier C., Zenke M., Fleischmann B.K., Zaehres H., and Schöler H.R. (2009). Oct4-induced pluripotency in adult neural stem cells. Cell 136, 411–419 [DOI] [PubMed] [Google Scholar]
- Liang G., Taranova O., Xia K., and Zhang Y. (2010). Butyrate promotes induced pluripotent stem cell generation. J. Biol. Chem. 285, 25516–25521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J., Wan M., Zhang Y., Gu P., Xin H., Jung S.Y., Qin J., Wong J., Cooney A.J., Liu D., and Songyang Z. (2008). Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat. Cell Biol. 10, 731–739 [DOI] [PubMed] [Google Scholar]
- Liao B., Bao X., Liu L., Feng S., Zovoilis A., Liu W., Xue Y., Cai J., Guo X., Qin B., Zhang R., Wu J., Lai L., Teng M., Niu L., Zhang B., Esteban M.A., and Pei D. (2011). MicroRNA cluster 302-367 enhances somatic cell reprogramming by accelerating a mesenchymal-to-epithelial transition. J. Biol. Chem. 286, 17359–17364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H.Y., Do H.J., Lee W.Y., Kim D.K., Seo H.G., Chung H.J., Park J.K., Chang W.K., Kim J.H., and Kim J.H. (2009). Implication of human OCT4 transactivation domains for self-regulatory transcription. Biochem. Biophys. Res. Commun. 385, 148–153 [DOI] [PubMed] [Google Scholar]
- Lin S.L., Chang D.C., Lin C.H., Ying S.Y., Leu D., and Wu D.T. (2011). Regulation of somatic cell reprogramming through inducible mir-302 expression. Nucleic Acids Res. 39, 1054–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T., Ambasudhan R., Yuan X., Li W., Hilcove S., Abujarour R., Lin X., Hahm H.S., Hao E., Hayek A., and Ding S. (2009). A chemical platform for improved induction of human iPSCs. Nat. Methods 6, 805–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipchina I., Studer L., and Betel D. (2012). The expanding role of miR-302-367 in pluripotency and reprogramming. Cell Cycle 11, 1517–1523 [DOI] [PubMed] [Google Scholar]
- Lowry W.E., Richter L., Yachechko R., Pyle A.D., Tchieu J., Sridharan R., Clark A.T., and Plath K. (2008). Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc. Natl. Acad. Sci. USA 105, 2883–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Chou B.K., Yen J., Ye Z., Zou J., Dowey S., Brodsky R.A., Ohm J.E., Yu W., Baylin S.B., Yusa K., Bradley A., Meyers D.J., Mukherjee C., Cole P.A., and Cheng L. (2010). Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells 28, 713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A., Wernig M., and Jaenisch R. (2007). Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat. Biotechnol. 25, 1177–1181 [DOI] [PubMed] [Google Scholar]
- Miyoshi N., Ishii H., Nagano H., Haraguchi N., Dewi D.L., Kano Y., Nishikawa S., Tanemura M., Mimori K., Tanaka F., Saito T., Nishimura J., Takemasa I., Mizushima T., Ikeda M., Yamamoto H., Sekimoto M., Doki Y., and Mori M. (2011). Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell 8, 633–638 [DOI] [PubMed] [Google Scholar]
- Park I.H., Zhao R., West J.A., Yabuuchi A., Huo H., Ince T.A., Lerou P.H., Lensch M.W., and Daley G.Q. (2008). Reprogramming of human somatic cells to pluripotency with defined factors. Nature 451, 141–146 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., and Yamanaka S. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 [DOI] [PubMed] [Google Scholar]
- Utikal J., Maherali N., Kulalert W., and Hochedlinger K. (2009). Sox2 is dispensable for the reprogramming of melanocytes and melanoma cells into induced pluripotent stem cells. J. Cell Sci. 122, 3502–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg D.L., Snoek T., Mullin N.P., Yates A., Bezstarosti K., Demmers J., Chambers I., and Poot R.A. (2010). An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell 6, 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware C.B., Wang L., Mecham B.H., Shen L., Nelson A.M., Bar M., Lamba D.A., Dauphin D.S., Buckingham B., Askari B., Lim R., Tewari M., Gartler S.M., Issa J.P., Pavlidis P., Duan Z., and Blau C.A. (2009). Histone deacetylase inhibition elicits an evolutionarily conserved self-renewal program in embryonic stem cells. Cell Stem Cell 4, 359–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R., and Slukvin I.I., and Thomson J.A. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Gao Y., Gordon A., Wang Z.Z., Qian Z., and Wu W.S. (2011). Efficient generation of fully reprogrammed human iPS cells via polycistronic retroviral vector and a new cocktail of chemical compounds. PloS One 6, e26592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., and Wu W.S. (2013). Sodium butyrate promotes generation of Human iPS cells through induction of the miR302/367 cluster. Stem Cells Dev. 22, 2268–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Li W., Zhou H., Wei W., Ambasudhan R., Lin T., Kim J., Zhang K., and Ding S. (2010). Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell 7, 651–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.