Abstract
The momentum of genomic science will carry it far into the future and into the heart of research on typical and atypical behavioral development. The purpose of this paper is to focus on a few implications and applications of these advances for understanding behavioral development. Quantitative genetics is genomic and will chart the course for molecular genomic research now that these two worlds of genetics are merging in the search for many genes of small effect. Although current attempts to identify specific genes have had limited success, known as the missing heritability problem, whole-genome sequencing will improve this situation by identifying all DNA sequence variation including rare variants. Because the heritability of complex traits is caused by many DNA variants of small effect in the population, polygenic scores that are composites of hundreds or thousands of DNA variants will be used by developmentalists to predict children’s genetic risk and resilience. The most far-reaching advance will be the widespread availability of whole-genome sequence for children, which means that developmentalists would no longer need to obtain DNA or to genotype children in order to use genomic information in research or in the clinic.
Keywords: quantitative genetics, molecular genetics, DNA, missing heritability, whole-genome sequencing
Introduction
In 2011, when the editor of this journal invited us to write this paper, he encouraged us: “… to set out the future agenda for research, translation, and policy in the decades ahead… I want your paper to be forward-thinking and visionary in ambition. I am not asking you to provide a systematic review article; rather, I am seeking a contribution that is conceptual, opinion-driven and innovative—one that provides a blueprint for future empirical research, translational research, and policy implications” (D. Cicchetti, personal communication, June 15, 2011).
We have written this paper for behavioral researchers and clinicians interested in development rather than for experts in genomics. Following the editor’s advice, we have tried to make the paper conceptual and opinion-driven, rather than providing a systematic review. Our goal was to discuss a few issues that will shape the future of developmental research and eventually translate to the clinic. No crystal ball is needed to see some of the future of genomics because the breath-taking momentum of current advances will sweep the field into the next decade. However, genomics is moving so fast that we can confidently predict that some important advances will be completely new and unanticipated.
We waited until 2013 for three reasons. First, we wanted to contribute to this 25th anniversary issue of the journal, which has promoted genetic research in development over the decades by publishing more than 100 papers on genetics, even early on when genetics was not as popular as it is now. Second, we wanted to wait until the publication in November 2012 of this journal’s special issue, ‘Genomic sciences for developmentalists: The current state of affairs’ (Grigorenko & Cicchetti, 2012). This special issue provides reviews of research in genetics and genomics and shows the great extent to which developmental psychopathologists have increasingly incorporated genomics in their research. The special issue frees us to focus on the future because it covers background issues to which we refer along the way; more details about these background issues can be found elsewhere (Plomin, 2013; Plomin, DeFries, Knopik, & Neiderhiser, 2013). We also provide a glossary at the end of this paper so that we can avoid definitions in the text; we denote words included in the glossary with bold-face type, beginning with genetics and genomics. Finally, we wanted to wait until the next big thing, whole-genome sequencing, began to materialize through the dust settling from the recent explosion of research on genome-wide association studies using DNA arrays.
What is inherited is DNA sequence variation – everything else is a phenotype
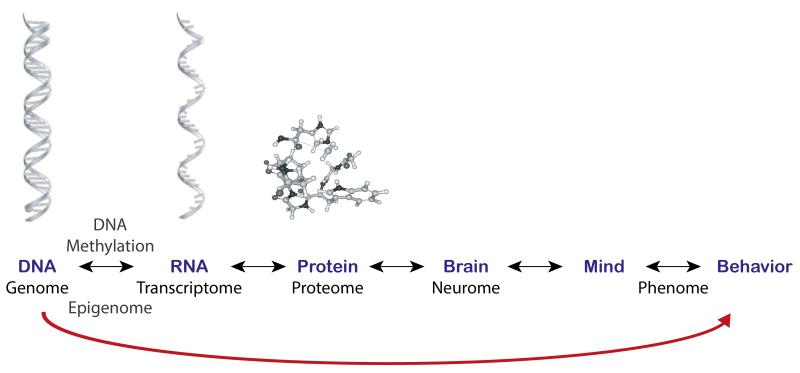
We begin with a general conceptual issue about DNA that is critical for understanding the future role of genomics in developmental research and translation. In this paper, we focus on DNA -- genetics and genomics -- because that is what is inherited. Specifically, we focus on less than 1 percent of the 3 billion base pairs of DNA that differs from person to person. This DNA sequence variation is responsible for all inherited physical, physiological and psychological differences between individuals. Although the 99 percent of DNA that does not differ between individuals is vitally important for development, we focus on what makes individuals differ. All of the other –omics in between genomics and behavior are important endophenotypes for understanding pathways between genes and individual differences in outcomes, but they are not inherited from parent to offspring. (See Figure 1.) For example, many mechanisms affect the extent to which DNA is transcribed into RNA in response to internal and external environments, such as adding methyl groups to DNA (epigenomics) or other mechanisms of gene expression (transcriptomics). Several papers in the special genomics issue of this journal report research on epigenetics and epigenomics (Bick et al., 2012; Knopik, Maccani, Francazio, & McGeary, 2012; Monk, Spicer, & Champagne, 2012) and transcriptomics (Carlyle et al., 2012; Naumova et al., 2012). After DNA is transcribed into RNA, some RNA is translated into amino acid sequences that produce our entire complement of proteins (proteomics). One of the most exciting findings in recent years is about the ‘dark matter’ of DNA, which refers to the more than 98 percent of the genome that is not translated into amino acid sequences and had been thought to be ‘junk’. We now know that more than half of this DNA can be expressed in the sense that it is transcribed into RNA although it is not translated into amino acid sequences; the RNA itself can be functional in the sense of regulating the expression of other genes (Pennisi, 2012).
Figure 1. All the ‘-omic’ pathways between the genome and behavior are important for understanding individual differences.
However, DNA sequence variation in the genome is ultimately responsible for hereditary influences on behavior. Correlations between DNA sequence variation and behavior are causal, as indicated by the single-headed arrow from the genome to behavior. Although behavior is often referred to as the observable ‘phenome’, all levels between the genome and behavior can be considered as phenotypes, which we show as connected by double-headed arrows signifying possible two-way directions of effect. The genetic and environmental origins of individual differences in these phenotypes need to be assessed rather than assumed.
Although the ultimate goal is to understand all the mechanisms by which DNA sequence variation contributes to individual differences in behavioral development, all of these processes downstream from DNA can be viewed as phenotypes. For example, individual differences in the amount of methylation across the genome (epigenomics) or individual differences in the number of RNA transcripts for all transcribed DNA in the genome (transcriptomics) are phenotypes that are age-specific, state-specific, and tissue-specific. The genetic and environmental origins of individual differences in these processes need to be assessed rather than assumed, like any other phenotype. Often discussed as a possible exception to this rule -- that all that is inherited is DNA sequence variation -- is the inheritance of epigenetic markings from parent to offspring (Slatkin, 2009). For example, some methylation markings that silence genes in mothers may be transmitted to their offspring (Hackett et al., 2013). However, the extent of transmitted methylation seems limited because as sperm and eggs develop, markings from the parent are erased and a further clean-up job happens when the egg is fertilized. Only a few markings fail to be erased and those that slip through might not matter. Empirically, twin studies suggest that individual differences in DNA methylation appear to be largely environmental in origin rather than inherited (Kaminsky et al., 2009; Wong et al., 2010; Wong et al., 2013). Epigenetics may be an especially good biomarker of early developmental adversity (Bell & Spector, 2011; Gordon et al., 2012; Heijmans & Mill, 2012; Knopik et al., 2012; Monk et al., 2012). Again, we emphasize that all of these processes between genes and behavior are important in their own right. However, all that is inherited is DNA sequence variation – everything else is a phenotype.
For this reason, DNA sequence variation is in a causal class of its own in the sense that there is no direction of effects issue when it comes to correlations between genes and behavior. That is, correlations between DNA sequence variation and behavior are ultimately causal from genes to behavior because our behavior and experiences do not change DNA sequence variation. Other correlations between behavior and biology, including all the –omics and the brain, are questionable concerning the direction of effects – whether the correlation is caused by the effects of behavior on biology or vice versa. As a result, one of the great strengths of DNA sequence variation is that it can be used to predict problems long before they appear, even prenatally. This predictive ability will enable research on interventions to prevent disorders rather than trying to reverse a disorder after it appears and causes collateral damage. The ultimate hope is for personalized genomics, individualized gene-based diagnoses and treatment programs (Collins, 2010).
The future includes quantitative genetics
Quantitative genetics might seem to belong to the past rather than the future. Quantitative genetics refers to designs such as family, twin and adoption studies in humans and inbred strains and selection studies in nonhuman animals that estimate the overall extent to which the genome and the environome account for individual differences in a trait without knowledge of specific genetic or environmental factors. These designs are called quantitative genetic not because they are limited to quantitative dimensions – these methods can also be applied to qualitative disorders – but because they assume that many genes are responsible for genetic influence (polygenic). Quantitative genetics is often contrasted with molecular genetics, a term that has been used broadly to refer to research using DNA. Until the last two decades, these two worlds had been growing apart since their births a century ago. In relation to inheritance, the goal of molecular genetics was to identify and understand genes, and the field focused on single-gene mutations of very large effect. The genes responsible for most of the thousands of monogenic disorders have been identified, and molecular geneticists have turned to complex traits and common disorders that are highly polygenic. Meanwhile, quantitative geneticists had not attempted to identify the genes responsible for genetic influence on complex traits because single-gene research strategies seemed inappropriate for what was assumed to be highly polygenic traits. A momentous shift is underway: These two worlds of genetics are merging as genomic technology makes it possible to identify the polygenic origins of individual differences in complex traits and common disorders.
Nearly all that we know with certainty about the genetics of the development of complex disorders and dimensions comes from quantitative genetic research designs such as family, twin and adoption studies. We predict that quantitative genetic studies will become increasingly important in the era of molecular genetics as these two worlds of genetics merge (Haworth & Plomin, 2010), a prediction supported by the fact that the number of twin studies around the world has doubled during the past decade (Hur & Craig, 2013). Quantitative genetic research has gone well beyond the rudimentary discovery of the ubiquitous and powerful influence of genes. Although it is an important first step to ask whether and how much genes affect behavioral dimensions and disorders (heritability), the key developmental question is how – how genotypes become phenotypes. For example, longitudinal studies have revealed genetic change as well as continuity in development, multivariate studies have found a surprising degree of genetic overlap as well as specificity between traits, and studies of gene-environment interplay have demonstrated the importance of gene-environment correlation as well as interaction (Plomin, DeFries, et al., 2013). One major direction for growth is the application of quantitative genetic research designs, primarily twin studies, to individual differences at all the –omic levels between genes and behavior (van Dongen, Slagboom, Draisma, Martin, & Boomsma, 2012). The twin design is uniquely powerful for understanding not only the genetic and environmental etiologies of the variance at each level but, importantly, the covariance between levels. The effort in launching such large twin studies insures that most of them continue longitudinally, making them especially valuable for developmentalists. Although the ‘and environmental’ phrase has often been ignored except as the residual variance not explained by genetics, more and more research is taking advantage of the power of the twin method to investigate the environment. For instance, developmental studies increasingly use genetically sensitive designs such as the twin method to identify causal environmental effects free of genetic mediation (Harold, Elam, Lewis, Rice, & Thapar, 2012; Jaffee & Price, 2012), and epigenomic studies use discordant MZ twins to identify biomarkers of non-shared environmental influences that make genetically identical co-twins different, as indicated earlier.
If we could identify all of the genes responsible for heritability, there would be no more need for quantitative genetic research. However, quantitative genetic research will continue to make important contributions because we are a long way from identifying all of the genes responsible for the heritability of any complex trait. This is the ‘missing heritability’ problem of genome-wide association studies, discussed later, in which heritability is defined by quantitative genetics. Quantitative genetics will continue to be an important part of the future of developmental research for three reasons. The most general reason is that quantitative genetics is genomic. That is, quantitative genetic designs appraise the net effect on traits of genes throughout the genome regardless of the types of genes (e.g., structural variants or noncoding genes in addition to traditional coding genes), the number of genes, the frequency of their alleles, the size of their effects, or the complications of their interactions with other genes or environments – these are the culprits thought to be responsible for missing heritability. The second reason is that quantitative genetics is as much about the environment as it is about genetics. Because heritability is always substantially less than 100 percent, quantitative genetic research provides the best available evidence for the importance of the environment, while controlling for the environment. Because heritability is substantial, environmental studies can exploit genetically sensitive designs to investigate the environment while controlling for possible confounding genetic influence and to explore the developmental interplay between the environment and genes.
Quantitative genetics using DNA alone
The third reason why quantitative genetics will continue to be important to developmentalists is a new technique that estimates the net effect of genetic influence using DNA alone in unrelated individuals rather than using familial resemblance in groups of special family members such as MZ and DZ twins who differ in genetic relatedness (Zaitlen & Kraft, 2012). The novelty and importance of this technique for future quantitative and molecular genetic research leads us to describe this advance in some detail.
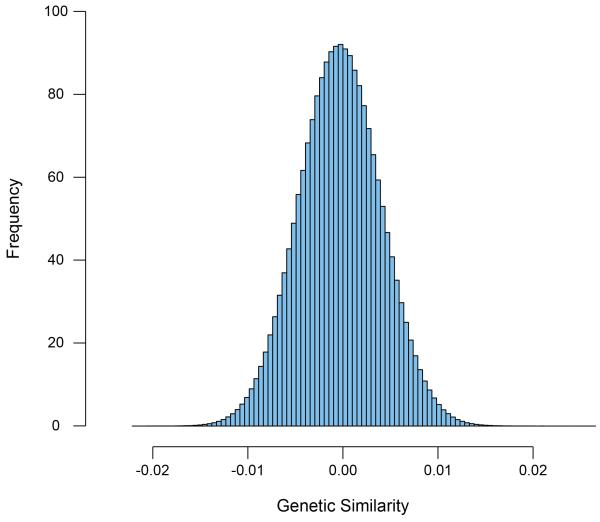
Like other quantitative genetic techniques, these new DNA-based methods use genetic similarity to predict phenotypic similarity. However, instead of using genetic similarity from groups differing markedly in genetic similarity, such as MZ and DZ twins, DNA-based methods use genetic similarity for each pair of unrelated individuals based on that pair‘s overall similarity across hundreds of thousands of single nucleotide polymorphisms (SNPs); each pair’s genetic similarity is then used to predict their phenotypic similarity. Even remotely related pairs of individuals are excluded – up to fifth-degree relatives whose genetic similarity is 3% -- so that chance genetic similarity is used as a random effect in a linear mixed model. Unlike MZ and DZ twins who are 100% and 50% similar genetically, the chance DNA similarity between pairs of unrelated individuals only varies from −2% to +2%. (See Figure 2.) Nonetheless, overall genetic influence can be estimated by the extent to which this random DNA similarity can predict phenotypic similarity.
Figure 2. The starting point for Genome-wide Complex Trait Analysis (GCTA) is a matrix of chance genetic similarity across hundreds of thousands of SNPs pair by pair for large samples of unrelated individuals.
(Adapted from Plomin, Haworth, et al., 2013.)
In contrast to the twin method which only requires a few hundred pairs of twins to estimate moderate heritability and does not need DNA, the demands of the DNA-based method are daunting: It requires not only DNA but also genotyping hundreds of thousands of SNPs for many thousands of individuals. Samples of many thousands are required because the method attempts to extract a slight signal of genetic similarity from the massive noise of hundreds of thousands of SNPs. The power of the method comes from comparing, not just two groups like MZ and DZ twins, but millions of pairs of individuals. For example, a sample of 3000 individuals provides nearly 5 million pair-by-pair comparisons. The first application of this approach was included in a software package with the clever acronym GCTA, Genome-wide Complex Trait Analysis (Yang et al., 2010; Yang, Lee, Goddard, & Visscher, 2011). We will refer to this DNA-based quantitative genetic method as GCTA, although other names have been proposed such as Linear Mixed Model and Genomic-Relatedness-Matrix Restricted Maximum Likelihood; other methods (So, Li, & Sham, 2011) and modifications (Speed, Hemani, Johnson, & Balding, 2012) are also emerging (Zaitlen & Kraft, 2012; Zhou, Carbonetto, & Stephens, 2013).
Despite the daunting demands of GCTA for hundreds of thousands of SNPs genotyped on many thousands of individuals, hundreds of datasets already exist that meet these requirements because these are the same requirements for genome-wide association studies, and these datasets are now being used to conduct GCTA. GCTA research across the life sciences generally yields evidence for significant genetic influence, confirming the results of twin studies. These include GCTA reports on common medical diseases (Lee et al., 2013), psychopathology (Lee, DeCandia, et al., 2012; Lee, Wray, Goddard, & Visscher, 2011; Lubke et al., 2012), cognitive abilities (Benyamin et al., 2013; Davies et al., 2011; Deary et al., 2012; Plomin, Haworth, et al., 2013), personality (Vinkhuyzen et al., 2012) and economic and political preferences (Benjamin et al., 2012). However, the first GCTA report for children’s behavior problems as rated by their parents, teachers and themselves found no significant genetic influence (Trzaskowski, Dale & Plomin, in press).
Like early twin studies, GCTA has primarily been used at first to demonstrate heritability but, like twin studies, the technique can be used to go beyond the estimation of heritability. For example, GCTA has confirmed one of the most interesting and puzzling findings about cognitive development: The heritability of general cognitive ability increases during development (Trzaskowski, Yang, Visscher, & Plomin, 2013). One interesting feature of GCTA is that, because it relies on comparisons between unrelated individuals, GCTA is entirely a between-family analysis, in contrast to traditional quantitative genetic methods like the twin method which are within-family analyses, essentially comparing differences within pairs of twins in a family to differences between families in the population. For developmentalists interested in gene-environment interplay, this feature of GCTA is important because it opens up the possibility of investigating genetic influence on family-wide environmental measures rather than being constrained to investigating child-specific environmental factors that differ for children in a family.
However, GCTA estimates of genetic influence are generally only about half the heritabilities estimated by twin studies. Although this could be because twin studies have overestimated heritability, it is clear that GCTA underestimates heritability for two reasons (Yang et al., 2011). First, GCTA only detects genetic effects that can be detected by the common SNPs (allele frequencies greater than 1%) that happen to be incorporated in the DNA microarrays used in genome-wide association studies. Second, GCTA is limited to detecting the additive effects of SNPs; it does not include gene-gene (or gene-environment) interaction. However, for these same reasons, GCTA provides critical clues for solving the missing heritability problem because GCTA gauges the extent to which the additive effects of common SNPs on current DNA arrays can potentially close the missing heritability gap if samples are sufficiently large. The answer is generally about 50%. The rest of the missing heritability might be found using rare DNA variants, other types of DNA variants, or non-additive (gene-gene and gene-environment) interactions (Plomin, 2013). When whole-genome sequence is available for large samples of individuals, GCTA will be able to include all types of DNA variants.
The value of GCTA has been greatly increased by extending it beyond the univariate analysis of the variance of a single trait to the bivariate analysis of the covariance between two traits (Lee, Yang, Goddard, Visscher, & Wray, 2012). The first application of this approach was to longitudinal IQ data from age 11 to age 70 (Deary et al., 2012), in which GCTA bivariate results showed that the remarkable phenotypic stability of IQ across 60 years from childhood to later life (phenotypic r = 0.63) is largely driven by genetic stability (genetic correlation = 0.62). Bivariate GCTA has also confirmed important twin study findings about cognitive development such as the extensive overlap (pleiotropy) of genetic effects on diverse cognitive abilities (Plomin, Haworth, et al., 2013; Trzaskowski, Davis et al., 2013; Trzaskowski, Shakeshaft, & Plomin, in press). An important feature of bivariate GCTA analysis is that its estimates of genetic correlations are similar to twin study estimates because GCTA estimates of genetic correlations are unbiased even though GCTA estimates of genetic variance and covariance underestimate twin study estimates (Trzaskowski, Yang et al., 2013). Extending GCTA from bivariate analysis to truly multivariate analysis will be especially valuable.
In the future, although traditional quantitative genetic designs such as the twin design will continue to make major contributions to developmental research, GCTA will provide a new approach that brings the potential of quantitative genetic analysis to any large sample of unrelated individuals for whom genome-wide genotypes are available. The promise of GCTA will be more fully realized as it is used to go beyond simply confirming the heritability of traits to investigate developmental, multivariate and gene-environment questions.
Despite the importance of having this completely new tool of GCTA in the armamentarium of quantitative genetics, nothing will advance the future of genomics research for developmentalists more than identifying the specific G, C, T, and A sequence variation responsible for heritability. In the next section, we briefly describe progress in this direction.
Finding the missing heritability
In summary, quantitative genetic research, which identifies genetic influence of any kind, finds substantial heritability for most traits, and can go beyond the rudimentary question of heritability to address developmental, multivariate, and gene-environment issues. GCTA, which is currently limited to detecting additive effects of common variants on commercially available DNA arrays, predicts that such effects can account for about half of the heritability. In this section, we consider the implications for future attempts to identify the DNA variants responsible for heritability. It would be wonderful if we were able to say that genomic research has served up a menu of genes to be chosen by developmentalists for use in their research, but this is not yet the case. However, progress is being made and it is important that developmental researchers be ready to take advantage of this genomics research, which will eventually make genomics relevant for all developmental researchers, as described later.
In this section, we focus on the implications of findings from genome-wide association (GWA) studies, which have dominated genomic research throughout the life sciences during the past five years. We will skip thousands of reports on linkage and candidate gene association. Linkage is systematic, using a few hundred DNA markers across the genome, to identify the chromosomal location of major gene effects by examining the co-segregation within family pedigrees between a DNA marker and a disorder. However, linkage is not powerful in detecting smaller genetic effects. Candidate gene association is powerful but not systematic. Association is a correlation in the population between an allele and a trait. For example, for late-onset Alzheimer disease, the frequency of a particular allele for the apolipoprotein E gene on chromosome 19 is about 40 percent in individuals with Alzheimer disease and about 15 percent in the rest of the population, one of the largest effect sizes found for a complex trait. The weakness of allelic association is that an association can only be detected if a DNA marker is itself the functional gene (called direct association) or very close to it (called indirect association or linkage disequilibrium). As a result, hundreds of thousands of DNA markers need to be genotyped to scan the genome thoroughly. For this reason, until very recently, allelic association has been used primarily to investigate associations with genes thought to be candidates for association.
A problem with the candidate gene approach is that we often do not have strong hypotheses as to which genes are candidate genes. Indeed, any of the thousands of genes expressed in the brain could be considered as candidate genes for most behavioral traits. Moreover, candidate gene studies are limited to the 2 percent of the DNA that lies in coding regions. The biggest practical problem is that reports of candidate gene associations have been difficult to replicate (Tabor, Risch, & Myers, 2002), nor have they have they shown up in genome-wide association studies (Siontis, Patsopoulos, & Ioannidis, 2010). For example, a recent study of nearly 10,000 individuals was not able to replicate associations for ten of the most frequently reported candidate gene associations for general cognitive ability (Chabris et al., 2012). Nonetheless, as discussed later, candidate gene studies will continue, especially as they move beyond a single SNP to gene-based and network-based analyses, and as they are nominated as candidates on the basis of adequately powered empirical evidence.
In contrast, as explained elsewhere specifically for behavioral developmentalists (Plomin, 2013; Vrieze, Iacono, & McGue, 2012), GWA uses DNA arrays with hundreds of thousands of common SNPs that cover the entire genome in a systematic and a theoretical attempt to identify specific DNA variants additively associated with a trait. GWA has been successful in identifying hundreds of genes for hundreds of common diseases and quantitative traits (Sullivan, 2012; Visscher, Brown, McCarthy, & Yang, 2012).
The largest effects are very small
Despite the success of GWA, an important lesson from these studies is that the largest effect sizes for the additive effects of common SNPs on current DNA arrays are astonishingly small in the population. Although there are thousands of single-gene disorders with huge effects on affected individuals, these disorders are rare and thus do not individually account for detectable variance in the population.
GWA results from individual studies have been combined in meta-analyses to reach the huge sample sizes needed to detect such small effects after correcting for multiple testing of hundreds of thousands of SNPs. GWA analyses with the largest sample sizes have been reported for the quantitative traits of height and weight. Weight has yielded the largest GWA effect size so far for a quantitative trait. The FTO gene accounts for almost 1 percent of the variance of weight in independent samples (Speliotes et al., 2010). However, in GWAS analyses of 250,000 individuals, the effect sizes for the next largest associations were closer to 0.1 percent, comparable to a correlation of 0.03, which is similar to the largest effect sizes found for height in a meta-analysis of 183,000 individuals (Lango Allen et al., 2010). A meta-analytic GW study of structural MRI measures of volumes of brain regions for nearly 20,000 individuals found that the largest effect sizes accounted for 0.2% of the variance for hippocampal volume, 0.3% for intracranial volume, and 0.2% for total brain volume (Stein et al., 2012).
The largest effect sizes for behavioral traits are also tiny. For example, the largest study to date for a behavioral trait has recently been reported for the complex trait of total years of education (Rietveld et al., in press). In a GWA meta-analysis of more than 100,000 individuals, three SNPs reached genome-wide significance and were replicated in a sample of 25,000, but the largest association accounted for only .02 percent of the variance, that is, a correlation of 0.014. In a GWA meta-analysis of IQ for nearly 18,000 children, the largest effect size accounted for 0.2 percent of the variance, although even this association did not reach genome-wide significance in the discovery sample (Benyamin et al., 2013).
Such small effect sizes are not limited to quantitative traits such as height, weight and IQ. For example, large samples of cases have been amassed for psychiatric disorders. The largest effect size found for schizophrenia yielded an odds ratio of 1.1 in a GWA meta-analysis of more than 9000 cases (The Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium, 2011). Similarly small effect sizes have been reported for the largest meta-analytic GWA studies of bipolar disorder with more than 7000 cases (Psychiatric GWAS Consortium Bipolar Disorder Working Group, 2011), major depressive disorder with more than 9000 cases (Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium, 2012), and attention-deficit/hyperactivity disorder with 3000 cases (Neale et al., 2010). Associations for psychiatric disorders are smaller than those found for medical disorders, although even here the largest effect sizes involve odds ratios of less than 1.2 (Manolio, 2010; Visscher et al., 2012; The Wellcome Trust Case Control Consortium, 2007).
Although the power of GWA to detect such tiny effects is limited, GWA studies have overwhelming power to prove that there are no large effects in the population. For example, a study of 20,000 individuals has 99.9 percent power to detect an association that accounts for 1 percent of the variance (i.e., a correlation of 0.10). GWA research implies that if the largest effects are so small, the smallest effects are likely to be infinitesimal. Incredibly large samples are needed to detect such small effects. For example, nearly 40,000 individuals are needed to detect an effect size of 0.1 percent (i.e., a correlation of 0.01) that reaches genome-wide significance with 80 percent power.
Missing heritability from GWA research using common SNPs on current DNA arrays
Even though the effects sizes of individual SNPs are very small, their effects can be aggregated to estimate the size of the missing heritability gap. Genome-wide significance may be too strict a criterion for selecting DNA variants from GWA studies because the conventional genome-wide significance threshold of p<5×10−8 requires samples of many hundreds of thousands to reach 80% power to detect such miniscule effect sizes. Selecting DNA variants using less stringent criteria generally explains more variance in independent samples (The International Schizophrenia Consortium, 2009). That is, although some false positive ‘items’ will be included, on balance, using relaxed criteria helps more than it hurts. For example, for weight, 32 replicated SNPs explain about 2 percent of the variance, but thousands of SNPs explain about 5 percent of the variance (Speliotes et al., 2010). For height, 180 SNPs accounts for 10% of the variance, which increases to 13% when more SNPs are added (Lango Allen et al., 2010). For total years of education, the GWA meta-analysis mentioned earlier with more than 100,000 individuals explained about 1 percent of the variance using 3500 SNPs and 2.5 percent of the variance using all 2.5 million SNPs (Rietveld et al., in press). A similar pattern of results was reported for childhood IQ (Benyamin et al., 2013).
For medical disorders, where GWA consortia have been large enough to detect many reliable associations, specific SNPs in total account for up to about 8% of the liability (Lee et al., 2013). One recent example is coronary artery disease for which 150 SNPs accounted for about 5% of the total variance of liability (Deloukas et al., 2013). For schizophrenia, significant SNPs accounted for about 1% of the liability and all SNPs accounted for almost 6% of the liability (The Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium, 2011). For bipolar disorder, SNPs account for 1% to 3% of the liability (Psychiatric GWAS Consortium Bipolar Disorder Working Group, 2011).
Even in aggregate, the relatively small amount of variance explained by specific SNPs highlights the magnitude of the missing heritability problem. However, as indicated earlier, GCTA suggests that more heritability can be explained by additive effects of the common variants genotyped on current DNA arrays. As larger samples are amassed, the common SNPs on current DNA arrays will account for more of the missing heritability. The biggest question now in the genomic sciences is where the rest of the missing heritability can be found.
Beyond common SNPs on current DNA arrays
The most obvious direction for finding missing heritability gap is to consider less common DNA variants, including less common SNPs (Gibson, 2012). Common SNPs on currently available commercial DNA arrays have frequencies greater than one percent in the population. Many more SNPs are rarer, with frequencies that go down to ‘private mutations’ unique to an individual. More than 10 million SNPs have been validated in populations around the world; only about 2 million have frequencies greater than one percent in the population studied.
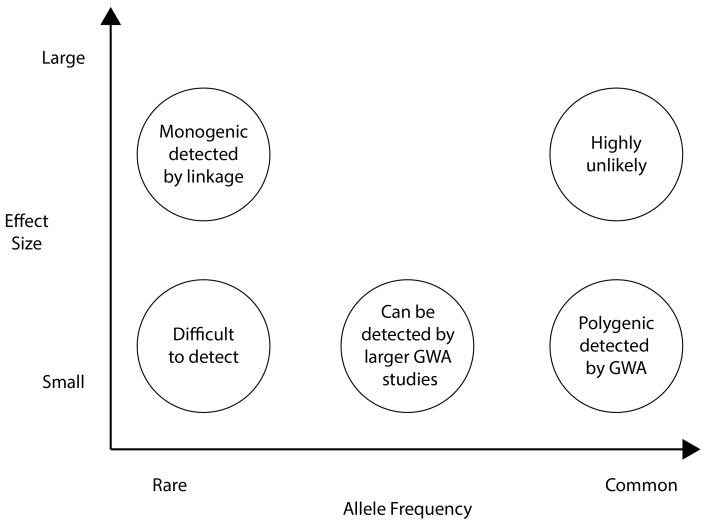
Figure 3 frames the issues by plotting allele frequency (from rare to common) against effect size (from small to large). Missing heritability could be found in all five circles in Figure 3, but the big question is the relative contributions of the five circles. So far, GWA has been limited to the lower right-hand corner (common SNPs of modest effect size); GCTA indicates that more of the missing heritability will be found here. As indicated in the upper left-hand corner, linkage analysis can detect rare alleles with large effects, which is why there has been a return to family-based designs (Ott, Kamatani, & Lathrop, 2011). Associations in the upper right-hand corner (common alleles with large effect) are highly unlikely based on results so far. The most daunting prospect is the lower left-hand corner: very rare alleles of small effect, which will be extremely difficult to detect. In between these extremes, in the middle of Figure 3, is a promising area for finding some of the missing heritability: less common alleles (< 1%) not well tagged by current microarrays yet common enough to show modest effects in the population.
Figure 3. Missing heritability: The relation between effect size and allele frequency.
(Adapted from McCarthy et al., 2008.)
The importance of rare variants has been highlighted by whole-exome sequencing which takes the first step towards whole-genome sequencing by sequencing the 2% of DNA that is transcribed into RNA and then translated into amino acid sequences. An early finding is changing the traditional view of gene-disrupting mutations as rare and very damaging, even though that is the case for thousands of well-established monogenic diseases. It appears that each of us inherits about 100 rare loss-of-function variants that lead to about 20 genes that are completely inactivated but most of which have not been clearly associated with disease (MacArthur et al., 2012). Most of these are recessive alleles so that in the heterozygous state the other normal allele is able to do the gene’s business, but even in the homozygous state, some of these gene-disrupting variants have no discernible phenotypic effect, suggesting a much more complicated system that tolerates gene disruption. Although these rare variants are not associated with known disorders, together they might well contribute to missing heritability even though individually their effects are small. A new DNA array called the Exome Chip includes most of the nonsynonymous SNPs in exons as rare as .001 (http://genome.sph.umich.edu/wiki/Exome_Chip_Design). The first paper using the Exome Chip has been published (Huyghe et al., 2013); many more such reports are in the pipeline. Although exomes are the location of most single-gene disorders, rare variants in the ‘dark matter’ outside these traditional gene regions are also likely to contribute to heritability, as discussed below.
Even more surprisingly, whole-exome sequencing has also revealed that we each have several dozen noninherited (de novo) gene-disrupting mutations, some of which have been shown to contribute to sporadic cases of neurodevelopmental disorders including intellectual disability, autism and schizophrenia (Gratten, Visscher, Mowry, & Wray, 2013; Stein, Parikshak, & Geschwind, 2013; Veltman & Brunner, 2012). These de novo mutations may be more damaging than inherited mutations because they have not been subjected to generations of selective pressure. Again, although the effects of these rare mutations are small in the population, collectively they could contribute to missing heritability if the mutations occur in the germline. It is possible that common disorders might be collections of such rare variants in many genes that are different for different individuals (Mitchell, 2012).
Before whole-exome sequencing uncovered so many single-nucleotide mutations, noninherited de novo variants were found that involved deletions and duplications of hundreds to millions of base pairs of DNA, called structural variants or copy number variants (CNVs) (Conrad et al., 2010; Weischenfeldt, Symmons, Spitz, & Korbel, 2013). Rare, large CNVs involving hundreds of thousands or even millions of base pairs have been shown to be risk factors for several common diseases (The Wellcome Trust Case Control Consortium, 2010), including autism, attention-deficit/hyperactivity disorder, and schizophrenia (Rucker & McGuffin, 2012; Williams et al., 2012) and learning disabilities (Cooper et al., 2011; Gill, 2012; Topper, Ober, & Das, 2011). The effects of such rare and large CNVs appear to be nonspecific, showing effects on multiple neurocognitive disorders (Malhotra & Sebat, 2012), and the effects are compounded for children with more than one such CNV (Girirajan et al., 2012). Because many of these CNVs are not inherited in that they appear in children but not their parents (i.e., de novo), they are especially sought in sporadic cases, that is, in cases whose families have no prior history of the disorder. Nonetheless, these CNVs can contribute to missing heritability because they can occur during the formation of gametes and thus will be shared by identical twins but are unlikely to be shared by fraternal twins. In addition, more than 10,000 smaller common CNVs have been identified with a frequency of at least 5 percent in the population (Conrad et al., 2010), although these are largely tagged by SNP arrays, which suggests that they will not make a major contribution to missing heritability (The Wellcome Trust Case Control Consortium, 2010). We all have CNVs peppered throughout our genome without obvious effect, despite all the extra or missing segments of DNA that include whole genes. However, these CNVs might have more subtle effects that contribute to the heritability of neurodevelopmental problems in the population (Geschwind, 2011; Sahoo et al., 2011).
Variation in DNA sequence can be seen as a continuum from single nucleotide base pairs to small CNVs to larger CNVs to whole chromosomes. In addition, individuals differ in the number of certain repeating nucleotide base pairs, such as the triplet repeat CAG responsible for Huntington disease. Repeat sequence variants consist of two, three, or four base pairs that are repeated up to a hundred times, creating multiple alleles, and have been found at as many as 50,000 loci throughout the genome. The value of whole-genome sequencing is that it will reveal variants of any kind.
Methods are also being developed to bring bioinformatics to bear on detecting small effects in the population rather than just relying on the blind brute force of larger samples. For instance, because a particular gene can have mutations at several locations, gene-based analyses can aggregate multiple DNA variants in a gene, including rare variants, which also reduces the multiple-testing problem (Bacanu, 2012; Liu et al., 2010). As an extreme example, hundreds of different mutations have been found in the gene responsible for phenylketonuria, and some of these different mutations have different effects (Scriver, 2007). At a broader level, it is possible to group genes in terms of pathways networks of related functions (Ramanan, Shen, Moore, & Saykin, 2012), including recent empirically based pathway discovery (Lehne & Schlitt, 2012). Another general example is to reduce the multiple testing problem by focusing on DNA variants known to be functional, such as those known to affect gene expression (Pickrell et al., 2010). Another example is to aggregate rare variants because many are likely to be deleterious (Li & Leal, 2008; Morris & Zeggini, 2010).
These various strategies are likely to help find some of the missing heritability exposed by GWA studies relying on common SNPs. A worrying issue is the reliance of GWA research on additive effects. If heritability is substantially due to nonadditive gene-gene or gene-environment interactions, it will be extremely difficult to identify these effects because power to detect two-way interactions is diminished (Vukcevic, Hechter, Spencer & Donnelly, 2011) and declines sharply with each additional interaction. We can only hope that quantitative genetic research is correct in its conclusion that most genetic variance is additive (Flint, DeFries, & Henderson, 2004; Plomin, DeFries, et al., 2013). There are also evolutionary reasons to expect that most genetic variance is additive (Hill, Goddard, & Visscher, 2008).
Much more attention should also be paid to better assessment of phenomes, not just genomes. The need for careful behavioral measurement, for including assessments of the environment, and for a developmental perspective are well covered in this journal’s special issue (McGrath, Weill, Robinson, Macrae, & Smoller, 2012; Vrieze et al., 2012).
Polygenic scores
How will this wealth of genomic data be incorporated in developmental research? As discussed above, the main message from GWA and GCTA research is that many DNA variants of small effect size are responsible for most of the heritability of complex traits, and that DNA variants other than common SNPs will be part of the solution to the missing heritability problem. In the future, developmental research will use aggregate genotypic scores consisting of hundreds or thousands of DNA variants to predict children’s genetic risk and resilience because these composite genotypic scores can be used with feasible sample sizes, even though huge samples are required to identify each association of small effect size. For example, if a polygenic predictor accounts for 5% of the variance (i.e., 10% of the heritable variance for a trait with 50% heritability), a sample size of 150 would have 80% power to detect its effect (p = .05, one-tailed). These polygenic scores consisting of thousands of DNA variants will replace studies of a few SNPs in a candidate gene. With custom DNA arrays (see below), it will soon be possible to genotype hundreds of thousands of DNA variants for the current cost of genotyping a few candidate genes. Eventually, for a similar cost, it will be possible to sequence the entire genome (see below).
At least a dozen labels have been used to denote aggregated genotypic scores such as polygenic susceptibility scores (Pharoah et al., 2002), genomic profiles (Khoury, Yang, Gwinn, Little, & Flanders, 2004), SNP sets (Harlaar et al., 2005), and aggregate risk scores (The International Schizophrenia Consortium, 2009). Although most of these labels involve the notion of risk, we prefer the term polygenic score because it makes more sense for quantitative traits with positive as well as negative poles. As described, later, this semantic distinction has some important implications. Some of the labels include SNP but the generic word polygenic is better because DNA variants other than SNPs, such as CNVs, will be included in aggregate genotypic scores. Unlike GCTA, which is a quantitative genetic technique for estimating genetic influence on average in a population without knowing the specific DNA variants responsible for heritability, polygenic scores are based on specific DNA variants associated with a trait.
DNA variants shown to be associated with a trait can simply be summed like items on a test. For example, 100 associations that each account for .1% of the variance of a trait on average would together account for 10% of the variance because DNA variants are uncorrelated unless they are very close together on a chromosome. Like positive and negative items on a questionnaire, the genotype scores for each DNA variant must be added in the correct direction. For SNPs, which have only two alleles (A1 and A2), there are three genotypes (A1A1, A1A2, A2A2) that can be assigned additive genotypic scores of 0, 1, and 2, respectively, to reflect increasing dosage of the A2 allele. This is an additive polygenic score, which is typically used for polygenic scores because heritability appears to be largely due to additive genetic factors, although nonadditivity at a locus (dominance) or between loci (epistasis) can also be incorporated in polygenic scores. Thus, if 100 SNPs were included in a polygenic score in this way, scores could vary from 0 to 200. Like a factor score for items on a test, when SNPs are summed to create a polygenic score, they are often weighted by the strength of their association with the phenotype (Dudbridge, 2013).
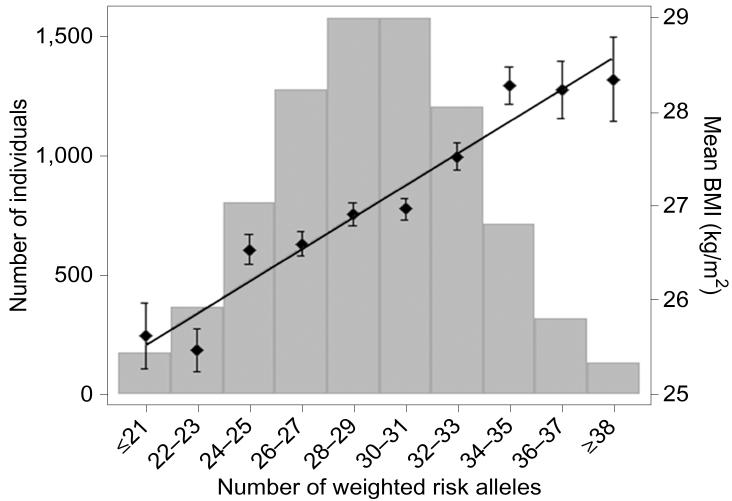
For example, as indicated earlier, a polygenic score for weight in adulthood created from 32 SNPs identified in GWA studies correlates 0.12 with weight in independent samples, which means that it accounts for 1.5 percent of the variance in weight or about 2 percent of the heritability of 70 percent (Speliotes et al., 2010). Even with such a modest correlation, the top and bottom 2% of the weight distribution differ by about 8 kg. (See Figure 4.) This opens up the possibility of genotypic selection of individuals with low versus high polygenic scores from large samples for whom genome-wide genotypes are available.
Figure 4. A polygenic score based on 32 SNPs associated with weight corrected for height (body mass index, BMI) correlates 0.12 with BMI.
Although the polygenic score only accounts for 1.5 percent of the total variance of BMI, individuals with the lowest and highest 2 percent polygenic scores differ by 8 kg, suggesting the possibility of reasonably sized research samples selected solely on the basis of polygenic scores (genotypic selection). Figure used with permission from Speliotes et al. (2010, Figure 2.1, p.940).
Polygenic scores can be used in the same way that candidate genes have been used. A neuroscientist might not find a polygenic score useful for investigating molecular pathways between genes and behavior through the brain, except perhaps to emphasize the need for a network approach governed by pleiotropy (each gene affects many traits) and polygenicity (each trait is affected by many genes). However, developmental researchers can use polygenic scores to investigate developmental, multivariate, and gene-environment issues as genetic predictors.
For example, a polygenic score constructed from SNPs associated with adult BMI has been shown to be just as strongly associated with BMI in childhood, suggesting substantial genetic stability for BMI between childhood and adulthood (Belsky et al., 2012). Other interesting developmental results from this study include finding that the polygenic BMI score was associated with weight gain from birth to age 3 and with adiposity rebound at age 6, but not with birth weight.
Two conceptual implications of polygenic scores will affect future developmental research. First, the genetic liabilities for common disorders are normally distributed, which implies that from a genetic perspective there are no common disorders, just the extremes of quantitative liabilities. Second, polygenic scores will draw attention to the positive end of the normal distribution of polygenic scores, which has been called positive genetics (Plomin, Haworth, & Davis, 2009). Using Figure 4 as an example, who are the children with low polygenic scores for obesity risk? Do they merely have the absence of risk factors or are there different mechanisms involved such as being especially resistant to the temptations of a fast-food nation? The positive end of polygenic scores will stimulate research on how children flourish rather than fail and about resilience rather than vulnerability. However, it is also possible that being at very low risk for obesity has its own problems such as being a finicky eater more prone to eating disorders. Perhaps the positive end of polygenic score distributions is not actually positive -- ‘all things in moderation’ might apply to polygenic scores as well.
In the near future, custom DNA arrays will become available that will assess hundreds of thousands of DNA variants relevant to particular traits at a cost less than the current cost of genotyping a few candidate genes. For example, custom arrays are available for several hundred thousand DNA variants related to immunology (ImmunoChip), cardiovascular functioning (CardioChip), and metabolic functioning (Metabochip). DNA arrays could be customized to produce polygenic scores for any and all aspects of behavioral development, including age-specific associations and associations involved in gene-environment interactions and correlations.
Strengths of DNA arrays include their speed, accuracy and low cost. However, the problem with DNA arrays is that as new DNA variants relevant to a trait continue to be identified and need to be added to a polygenic score, it could be necessary to genotype the sample again using another DNA array. As discussed later, the huge advantage of whole genome sequencing is that, once a genome is sequenced, there is no more genotyping to be done. A tipping point will come in the next few years as the plummeting cost of whole genome sequencing approaches the cost of DNA arrays.
Pleiotropic polygenic scores
An important finding from quantitative genetics is that genetic effects tend to be general across phenotypes rather than specific to a single phenotype. In genetics, pleiotropy denotes the manifold effects of genes; that is, each gene affects many traits. Pleiotropy might drive polygenicity: If each gene affects many traits then many traits will be influenced by many genes. Pleiotropy has been highlighted in multivariate genetic research in the domain of cognitive and learning abilities and disabilities, where genetic correlations are as high as 0.80 between diverse cognitive processes such as verbal and non-verbal cognitive abilities and reading and mathematics (Plomin & Kovas, 2005), findings that have been confirmed using bivariate GCTA, as noted earlier. Substantial pleiotropy has also been found for childhood psychopathology (Lichtenstein, Carlstrom, Rastam, Gillberg, & Anckarsater, 2010) and adult psychosis (Lichtenstein et al., 2009), which suggests a genetic architecture that differs greatly from current diagnostic classifications based on symptoms. For example, even though schizophrenia and bipolar disorder are the first branching point in psychiatric diagnosis, the genetic correlation between them is 0.60 (Lichtenstein et al., 2009). GWA studies have come face to face with the reality of this quantitative genetic evidence for genetic overlap because most SNPs and CNVs associated with schizophrenia have turned out to be associated with bipolar disorder and vice versa (Girirajan et al., 2012; Huang et al., 2010; Liu et al., 2011; Malhotra & Sebat, 2012; Psychiatric GWAS Consortium Bipolar Disorder Working Group, 2011; Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium, 2011). The first polygenic score created for schizophrenia was found to be almost as highly correlated with liability for bipolar disorder (The International Schizophrenia Consortium, 2009). Extensive overlap of DNA variants has also been found for other medical disorders, especially autoimmune diseases (Cotsapas et al., 2011; Zhernakova et al., 2011).
Rather than attempting to identify genes associated with one trait and then asking whether the genes are also associated with other traits, one direction for research is multivariate GWA studies that focus on genetic associations in common across traits. A multivariate GWA analysis of five major psychiatric disorders, including autism spectrum disorder and attention deficit-hyperactivity disorder, found three SNPs that were associated with all five disorders in a meta-analysis of more than 30,000 cases (Cross-Disorder Group of the Psychiatric Genomics Consortium, 2013). This approach is multivariate in the sense that, although it begins with separate GWA analyses for each disorder, it looks for SNPs that are most associated across disorders regardless of whether the SNPs are among the most highly significant for any particular disorder. If a SNP is associated with two traits, it contributes to the phenotypic covariance and to the genetic correlation between the traits.
However, a more truly multivariate GWA analysis would focus on the covariance per se between traits rather than the variance of each trait. This is easier to see for quantitative traits: Rather than beginning with univariate GWA analyses of each trait and then asking which SNPs show associations across traits, it would be more direct and more powerful to conduct GWA analyses of the covariance between traits. For example, two traits (X, Y) or more could be included in a multiple regression predicting SNP genotypes: The betas for X and Y would index trait-specific associations, and the multiple R would index the association due to covariance between X and Y. Software for such multivariate genetic analyses is available (O’Reilly et al., 2012).
Finding DNA variation that accounts for this pervasive pleiotropy could have far-reaching implications for assessing and understanding typical and atypical development. Pleiotropic polygenic scores could be created that predict what is in common across broad domains such as psychopathology and cognitive abilities and disabilities. These pleiotropic polygenic scores could be used to look at the DNA-based architecture of psychopathology and cognition free from a century of phenotypic diagnoses and definitions. Future research can explore the psychopathological and cognitive traits that best characterize these pleiotropic polygenic factors and the mechanisms that underlie them, chart their developmental course, and consider their interplay with the environment. Trait-specific polygenic scores that remove the considerable genetic variance spread across traits are also likely to provide cleaner targets for research on typical and atypical development.
The end of genotyping: Whole genome sequencing
Rather than genotyping particular DNA variants on a DNA array, whole genome sequencing has two huge advantages: It reveals all variants in the entire DNA sequence and it only needs to be done once. The cost of whole genome sequencing is dropping so quickly that it will soon become competitive with the cost of DNA arrays. Before DNA arrays became available, it cost about $1 to genotype one SNP for one individual; in other words, genotyping a million SNPs for just one individual would cost $1 million, which is why GWA studies were not feasible other than pooling DNA for low and high groups (e.g., Plomin et al., 2001). When commercially available DNA arrays became available in the early 2000s, it cost about $1000 for a DNA array that genotyped 10,000 SNPs for one individual (i.e., $0.10 per genotype) but in a couple of years it cost less for a DNA array that genotyped 100,000 SNPs (< $0.01 per genotype). Now it costs about $500 for an array that genotypes a million SNPs (~ $0.0001 per genotype). Despite the low cost per genotype, the average GWA sample size of 2000 will cost $1 million, and, as discussed earlier, we now know that a sample size of 2000 is much too small to detect the expected small effect sizes.
The cost of sequencing the entire genome of 3 billion nucleotide base pairs of DNA has also plummeted. The first human genome sequence was announced in 2003 and took 2000 researchers 5 years at a cost of some $2 billion (http://www.genome.gov/11006943). Sequencing the entire genome of an individual now takes one day and costs about $5000. A completely new technology is predicted to bring the cost down to under $1000 (Maitra, Kim, & Dunbar, 2012).
Because sequencing yields all DNA sequence variation, it means that no more genotyping is required. This has far-reaching implications for developmentalists: If an individual’s DNA sequence is known, there is no need to do any further genotyping for that individual – indeed there is no need to obtain DNA from that individual. Although storage, quality control, and analysis of such massive amounts of data is currently challenging (Altmann et al., 2012), as whole genome sequencing becomes routine, so too will the necessary data management and analytic tools, as has happened for analysis of DNA array data.
As the cost of sequencing continues to fall, many individuals will pay to have their DNA sequenced, just as hundreds of thousands of people have already paid to have their DNA genotyped on DNA arrays by direct-to-consumer companies such as 23&Me to detect 250 single-gene diseases and determine ancestry (https://www.23andme.com/). There are of course many concerns with direct-to-consumer genotyping (Guttmacher, McGuire, Ponder, & Stefansson, 2010), and these concerns will be greatly magnified with direct-to-consumer sequencing, which is already on offer. Parents have also begun to pay to have their children’s DNA sequenced (Rochman, 2012). Indeed, it is likely that in some countries, all children will have their DNA sequenced. In an excellent book on the potential of the genomics revolution for personalized medicine, Francis Collins, former director of the Human Genome Project and currently director of the U.S. National Institutes of Health, after noting some caveats and cautions, makes this prediction: “I am almost certain that complete genome sequencing will become part of newborn screening in the next few years… It is likely that within a few decades people will look back on our current circumstance with a sense of disbelief that we screened for so few conditions” (Collins, 2010, p.50).
If this prediction is correct, it will be a game-changer for developmentalists. If DNA sequence data were widely available for children -- whether through direct-to-consumer testing or through systematic neonatal screening -- developmentalists would no longer need to collect DNA, to genotype it, or to sequence it. Our long-term prediction for the future of genomics for developmentalists is the routine use of polygenic scores derived from DNA sequence data. Developmental clinicians can use polygenic scores to provide diagnostic information based on etiology rather than symptomatology, suggest personalized interventions, and enable early prediction that will lead to prevention. Developmental researchers can use polygenic scores to predict genetic propensities for children, to trace how those genetic dispositions develop longitudinally, how they overlap with other traits, and how they interact and correlate with the environment. Polygenic scores associated with behavioral traits will lead to research attempting to understand links between the genome, epigenome, transcriptome, proteome, neurome, and behavior. The grandest implication for science is that genomics will serve as a common denominator integrating all the life sciences.
Acknowledgments
RP is supported by a Medical Research Council Research Professorship award [G19/2] and a European Advanced Investigator award [295366].
Glossary
- additive genetic variance
Individual differences caused by the independent effects of alleles or loci that “add up.” In contrast to nonadditive genetic variance, in which the effects of alleles or loci interact.
- allele
An alternative form of a gene at a locus, for example, A1 versus A2.
- allelic association
An association between allelic frequencies and a phenotype. For example, the frequency of allele 4 of the gene that encodes apolipoprotein E is about 40 percent for individuals with Alzheimer disease and 15 percent for control individuals who do not have the disorder.
- allelic frequency
Population frequency of an alternate form of a gene. For example, the frequency of the PKU allele is about 1 percent. (In contrast, see genotypic frequency.)
- base pair (bp)
One step in the spiral staircase of the double helix of DNA, consisting of adenine bonded to thymine, or cytosine bonded to guanine.
- bioinformatics
Techniques and resources to study the genome, transcriptome, and proteome, such as DNA sequences and functions, gene expression maps, and protein structures.
- candidate gene
A gene whose function suggests that it might be associated with a trait. For example, dopamine genes are considered as candidate genes for hyperactivity because the drug most commonly used to treat hyperactivity, methylphenidate, acts on the dopamine system.
- coding region
The portion of a gene’s DNA composed of exons that code for proteins.
- copy number variant (CNV)
A structural variation that involves duplication or deletion of long stretches of DNA (one thousand to many thousands of base pairs in length), often encompassing protein-coding genes as well as noncoding genes. CNVs account for more than 10% of the human genome.
- direct association
An association between a trait and a DNA marker that is the functional polymorphism that causes the association. In contrast to indirect association, in which the DNA marker is not the functional polymorphism.
- DNA (deoxyribonucleic acid)
The double-stranded molecule that encodes genetic information. The two strands are held together by hydrogen bonds between two of the four bases, with adenine bonded to thymine, and cytosine bonded to guanine.
- DNA array
A collection of DNA probes (short DNA sequences) attached to a solid surface used to genotype hundreds of thousands of DNA sequence variants. Also called DNA microarray or DNA chip.
- DNA marker
A polymorphism in DNA itself, such as a single-nucleotide polymorphism (SNP) or copy number variant (CNV).
- DNA sequence
The order of base pairs on a single chain of the DNA double helix.
- dominance
The effect of one allele depends on that of another. A dominant allele produces the same phenotype in an individual regardless of whether one or two copies are present. (Compare with epistasis, which refers to nonadditive effects between genes at different loci.)
- effect size
The proportion of individual differences for the trait in the population accounted for by a particular factor. For example, heritability estimates the effect size of genetic differences among individuals.
- endophenotype
An “inside” or intermediate phenotype that does not involve overt behavior.
- environome
All environmental factors that affect a phenotype.
- epigenetics
DNA methylation or histone modifications that affect gene expression without changing DNA sequence. Can be involved in long-term developmental changes in gene expression.
- epigenome
Epigenetic events throughout the genome.
- epistasis
Nonadditive interaction between genes at different loci. The effect of one gene depends on that of another. (Compare with dominance, which refers to nonadditive effects between alleles at the same locus.)
- exome
All exons in the genome.
- exon
The 2% of DNA transcribed into messenger RNA and translated into protein.
- gamete
Mature reproductive cell (sperm or ovum) that contains a haploid (half) set of chromosomes.
- gene
The basic unit of heredity. A sequence of DNA bases that codes for a particular product. Includes DNA sequences that regulate transcription. (See also allele; locus.)
- gene expression
Transcription of DNA into mRNA.
- genetics
Methods and theory related to inheritance. Includes both quantitative genetic and molecular genetic approaches.
- genome
All the DNA sequences of an organism. The human genome contains about 3 billion DNA base pairs across 23 pairs of chromosomes.
- genome-wide association
An association (correlation) between DNA sequence variation and a phenotype found in a systematic search throughout the genome.
- genome-wide complex trait analysis (GCTA)
A technique to estimate the extent to which phenotypic variance for a trait can potentially be explained by all genotyped DNA variants. For a sample of thousands of individuals, overall genotypic similarity pair by pair is used to predict phenotypic similarity. Does not identify specific allelic associations.
- genomics
Theory and methods that investigates DNA sequence variation throughout the genome.
- genotype
An individual’s combination of alleles at a particular locus.
- heritability
The proportion of phenotypic differences among individuals that can be attributed to genetic differences in a particular population. Broad-sense heritability involves all additive and nonadditive sources of genetic variance, whereas narrow-sense heritability is limited to additive genetic variance.
- heterozygosity
The presence of different alleles at a given locus on both members of a chromosome pair.
- homozygosity
The presence of the same allele at a given locus on both members of a chromosome pair.
- indirect association
An association between a trait and a DNA marker that is not itself the functional polymorphism that causes the association. In contrast to direct association, in which the DNA marker itself is the functional polymorphism.
- linkage
Loci that are close together on a chromosome. Linkage is an exception to Mendel’s second law of independent assortment, because closely linked loci are not inherited independently within families.
- linkage disequilibrium
A violation of Mendel’s law of independent assortment in which genes are uncorrelated. It is most frequently used to describe how close DNA markers are together on a chromosome; linkage disequilibrium of 1.0 means that the alleles of the DNA markers are perfectly correlated; 0.0 means that there is complete non-random association (linkage equilibrium).
- locus (plural, loci)
The site of a specific gene on a chromosome. Latin for “place.”
- methylation
An epigenetic process by which gene expression is inactivated by adding a methyl group to a chromosome region.
- missing heritability
The difference between the total variance accounted for by known genomewide associations and heritability estimates from quantitative genetic studies.
- molecular genetics
The investigation of the effects of specific genes at the DNA level. In contrast to quantitative genetics, which partitions phenotypic variances and covariances into genetic and environmental components.
- multivariate genetic analysis
Quantitative genetic analysis of the covariance between traits.
- nonadditive genetic variance
Individual differences due to nonlinear interactions between alleles at the same (dominance) or different (epistasis) loci. (In contrast, see additive genetic variance.)
- non-coding RNA (ncRNA)
DNA that is transcribed into RNA but not translated into amino acid sequences. Examples include introns and microRNA.
- nonsynonymous SNP
A SNP that alters the amino acid sequence of a protein and is thus likely to make a functional difference. In contrast, synonymous SNPs do not alter the amino acid sequence of proteins.
- odds ratio
An effect size statistic for association calculated as the odds of an allele in cases divided by the odds of the allele in controls. An odds ratio of 1.0 means that there is no difference in allele frequency between cases and controls.
- phenotype
An observed characteristic of an individual that results from the combined effects of genotype and environment.
- pleiotropy
Multiple effects of a gene.
- polygenic trait
A trait influenced by many genes.
- polymorphism
A locus with two or more alleles. Greek for “multiple forms.”
- proteome
All the proteins translated from RNA (transcriptome).
- quantitative genetics
Theory and methods (such as the twin and GCTA methods for human analysis) to estimate overall genetic and environmental contributions to phenotypic variance and covariance in a population.
- repeat sequence variant
Short sequences of DNA—two, three, or four nucleotide bases of DNA—that repeat a few times to a few dozen times. Used as DNA markers.
- single-nucleotide polymorphism (SNP)
The most common type of DNA polymorphism which involves a mutation in a single nucleotide. SNPs (pronounced “snips”) can produce a change in an amino acid sequence (called nonsynonymous, i.e., not synonymous).
- transcription
The synthesis of an RNA molecule from DNA in the cell nucleus.
- transcriptome
RNA transcribed from all the DNA in the genome.
- translation
Assembly of amino acids into peptide chains on the basis of information encoded in messenger RNA. Occurs on ribosomes in the cell cytoplasm.
- whole-exome sequence
Determining the complete sequence of DNA nucleotide base pairs for all exons in the genome.
- whole-genome sequencing
Determining the complete sequence of DNA nucleotide base pairs for a genome.
References
- Altmann A, Weber P, Bader D, Preuss M, Binder EB, Muller-Myhsok B. A beginners guide to SNP calling from high-throughput DNA-sequencing data. Human Genetics. 2012;131:1541–1554. doi: 10.1007/s00439-012-1213-z. doi: 10.1007/s00439-012-1213-z. [DOI] [PubMed] [Google Scholar]
- Bacanu S-A. On optimal gene-based analysis of genome scans. Genetic Epidemiology. 2012;36:333–339. doi: 10.1002/gepi.21625. doi: 10.1002/gepi.21625. [DOI] [PubMed] [Google Scholar]
- Bell JT, Spector TD. A twin approach to unraveling epigenetics. Trends in Genetics. 2011;27:116–125. doi: 10.1016/j.tig.2010.12.005. doi: 10.1016/j.tig.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsky DW, Moffitt TE, Houts R, Bennett GG, Biddle AK, Blumenthal JA, Caspi A. Polygenic risk, rapid childhood growth, and the development of obesity evidence from a 4-decade longitudinal study. Archives of Pediatrics and Adolescent Medicine. 2012;166:515–521. doi: 10.1001/archpediatrics.2012.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin DJ, Cesarini D, van der Loos MJHM, Dawes CT, Koellinger PD, Magnusson PKE, Visscher PM. The genetic architecture of economic and political preferences. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:8026–8031. doi: 10.1073/pnas.1120666109. doi: 10.1073/pnas.1120666109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyamin B, Pourcain B, Davis OS, Davies G, Hansell NK, Brion MJ, Visscher PM. Childhood intelligence is heritable, highly polygenic and associated with FNBP1L. Molecular Psychiatry. 2014;19:253–258. doi: 10.1038/mp.2012.184. doi: 10.1038/mp.2012.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bick J, Naumova O, Hunter S, Barbot B, Lee M, Luthar SS, Grigorenko EL. Childhood adversity and DNA methylation of genes involved in the hypothalamus–pituitary–adrenal axis and immune system: Whole-genome and candidate-gene associations. Development and Psychopathology. 2012;24:1417–1425. doi: 10.1017/S0954579412000806. doi: 10.1017/S0954579412000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlyle BC, Duque A, Kitchen RR, Bordner KA, Coman D, Doolittle E, Simen AA. Maternal separation with early weaning: A rodent model providing novel insights into neglect associated developmental deficits. Development and Psychopathology. 2012;24:1401–1416. doi: 10.1017/S095457941200079X. doi: 10.1017/S095457941200079X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabris CF, Hebert BM, Benjamin DJ, Beauchamp J, Cesarini D, van der Loos M, Laibson D. Most reported genetic associations with general intelligence are probably false positives. Psychological Science. 2012;23:1314–1323. doi: 10.1177/0956797611435528. doi: 10.1177/0956797611435528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS. The language of life: DNA and the revolution in personalised medicine. Harper Collins; New York: 2010. [Google Scholar]
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang YJ, Hurles ME. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, Eichler EE. A copy number variation morbidity map of developmental delay. Nature Genetics. 2011;43:838–844. doi: 10.1038/ng.909. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotsapas C, Voight BF, Rossin E, Lage K, Neale BM, Wallace C, Daly MJ. Pervasive sharing of genetic effects in autoimmune disease. PLoS Genetics. 2011;7:e1002254. doi: 10.1371/journal.pgen.1002254. doi: 10.1371/journal.pgen.1002254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G, Tenesa A, Payton A, Yang J, Harris SE, Liewald D, Deary IJ. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Molecular Psychiatry. 2011;16:996–1005. doi: 10.1038/mp.2011.85. doi: 10.1038/mp.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deary IJ, Yang J, Davies G, Harris SE, Tenesa A, Liewald D, Visscher PM. Genetic contributions to stability and change in intelligence from childhood to old age. Nature. 2012;482:212–215. doi: 10.1038/nature10781. doi: 10.1038/nature10781. [DOI] [PubMed] [Google Scholar]
- Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes T, Thompson J, Samani N. Large-scale association analysis identifies new risk loci for coronary artery disease. Nature Genetics. 2013;45:25–33. doi: 10.1038/ng.2480. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genetics. 2013;9:e1003348. doi: 10.1371/journal.pgen.1003348. doi: 10.1371/journal.pgen.1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J, DeFries JC, Henderson ND. Little epistasis for anxiety-related measures in the DeFries strains of laboratory mice. Mammalian Genome. 2004;15:77–82. doi: 10.1007/s00335-003-3033-x. doi: 10.1007/s00335-003-3033-x. [DOI] [PubMed] [Google Scholar]
- Geschwind DH. Genetics of autism spectrum disorders. Trends in Cognitive Sciences. 2011;15:409–416. doi: 10.1016/j.tics.2011.07.003. doi: 10.1016/j.tics.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G. Rare and common variants: Twenty arguments. Nature Reviews Genetics. 2012;13:135–145. doi: 10.1038/nrg3118. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill M. Developmental psychopathology: The role of structural variation in the genome. Development and Psychopathology. 2012;24:1319–1334. doi: 10.1017/S0954579412000739. doi: 10.1017/S0954579412000739. [DOI] [PubMed] [Google Scholar]
- Girirajan S, Rosenfeld JA, Coe BP, Parikh S, Friedman N, Goldstein A, Eichler EE. Phenotypic heterogeneity of genomic disorders and rare copy-number variants. New England Journal of Medicine. 2012;367:1321–1331. doi: 10.1056/NEJMoa1200395. doi: doi:10.1056/NEJMoa1200395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon L, Joo JE, Powell JE, Ollikainen M, Novakovic B, Li X, Saffery R. Neonatal DNA methylation profile in human twins is specified by a complex interplay between intrauterine environmental and genetic factors, subject to tissue-specific influence. Genome Research. 2012;22:1395–1406. doi: 10.1101/gr.136598.111. doi: 10.1101/gr.136598.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratten J, Visscher PM, Mowry BJ, Wray NR. Interpreting the role of de novo protein-coding mutations in neuropsychiatric disease. Nature Genetics. 2013;45:234–238. doi: 10.1038/ng.2555. doi: 10.1038/ng.2555. [DOI] [PubMed] [Google Scholar]
- Grigorenko EL, Cicchetti D. Genomic sciences for developmentalists: The current state of affairs. Development and Psychopathology. 2012;24:1157–1164. doi: 10.1017/S0954579412000612. doi: 10.1017/S0954579412000612. [DOI] [PubMed] [Google Scholar]
- Guttmacher AE, McGuire AL, Ponder B, Stefansson K. Personalized genomic information: preparing for the future of genetic medicine. Nature Reviews Genetics. 2010;11:161–165. doi: 10.1038/nrg2735. doi: 10.1038/nrg2735. [DOI] [PubMed] [Google Scholar]
- Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, Surani MA. Germline DNA demethylation dynamics and imprint erasure through 5-Hydroxymethylcytosine. Science. 2013;339:448–452. doi: 10.1126/science.1229277. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlaar N, Butcher LM, Meaburn E, Sham P, Craig IW, Plomin R. A behavioural genomic analysis of DNA markers associated with general cognitive ability in 7-year-olds. Journal of Child Psychology and Psychiatry. 2005;46:1097–1107. doi: 10.1111/j.1469-7610.2005.01515.x. doi: 10.1111/j.1469-7610.2005.01515.x. [DOI] [PubMed] [Google Scholar]
- Harold GT, Elam KK, Lewis G, Rice F, Thapar A. Interparental conflict, parent psychopathology, hostile parenting, and child antisocial behavior: Examining the role of maternal versus paternal influences using a novel genetically sensitive research design. Development and Psychopathology. 2012;24:1283–1295. doi: 10.1017/S0954579412000703. doi: 10.1017/S0954579412000703. [DOI] [PubMed] [Google Scholar]
- Haworth CMA, Plomin R. Quantitative genetics in the era of molecular genetics: Learning abilities and disabilities as an example. Journal of the American Academy of Child and Adolescent Psychiatry. 2010;49:783–793. doi: 10.1016/j.jaac.2010.01.026. doi: 10.1016/j.jaac.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijmans BT, Mill J. Commentary: The seven plagues of epigenetic epidemiology. International Journal of Epidemiology. 2012;41:74–78. doi: 10.1093/ije/dyr225. doi: 10.1093/ije/dyr225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Goddard ME, Visscher PM. Data and theory point to mainly additive genetic variance for complex traits. PLoS Genetics. 2008;4:e1000008. doi: 10.1371/journal.pgen.1000008. doi: 10.1371/journal.pgen.1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Perlis RH, Lee PH, Rush AJ, Fava M, Sachs GS, Smoller JW. Cross-disorder genomewide analysis of schizophrenia, bipolar disorder, and depression. American Journal of Psychiatry. 2010;167:1254–1263. doi: 10.1176/appi.ajp.2010.09091335. doi: 10.1176/appi.ajp.2010.09091335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur Y-M, Craig JM. Twin registries worldwide: An important resource for scientific research. Twin Research and Human Genetics. 2013;16:1–12. doi: 10.1017/thg.2012.147. doi: 10.1017/thg.2012.147. [DOI] [PubMed] [Google Scholar]
- Huyghe JR, Jackson AU, Fogarty MP, Buchkovich ML, Stancakova A, Stringham HM, Mohlke KL. Exome array analysis identifies new loci and low-frequency variants influencing insulin processing and secretion. Nature Genetics. 2013;45:197–201. doi: 10.1038/ng.2507. doi: 10.1038/ng.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffee SR, Price TS. The implications of genotype–environment correlation for establishing causal processes in psychopathology. Development and Psychopathology. 2012;24:1253–1264. doi: 10.1017/S0954579412000685. doi: 10.1017/S0954579412000685. [DOI] [PubMed] [Google Scholar]
- Kaminsky ZA, Tang T, Wang SC, Ptak C, Oh GHT, Wong AHC, Petronis A. DNA methylation profiles in monozygotic and dizygotic twins. Nature Genetics. 2009;41:240–245. doi: 10.1038/ng.286. doi: 10.1038/ng.286. [DOI] [PubMed] [Google Scholar]
- Khoury MJ, Yang QH, Gwinn M, Little JL, Flanders WD. An epidemiologic assessment of genomic profiling for measuring susceptibility to common diseases and targeting interventions. Genetics in Medicine. 2004;6:38–47. doi: 10.1097/01.gim.0000105751.71430.79. doi: 10.1097/01.GIM.0000105751.71430.79. [DOI] [PubMed] [Google Scholar]
- Knopik VS, Maccani MA, Francazio S, McGeary JE. The epigenetics of maternal cigarette smoking during pregnancy and effects on child development. Development and Psychopathology. 2012;24:1377–1390. doi: 10.1017/S0954579412000776. doi: 10.1017/S0954579412000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, Hirschhorn JN. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, DeCandia TR, Ripke S, Yang J, The Schizophrenia Psychiatric Genome-Wide Association Study Consortium (PGC-SCZ) The International Schizophrenia Consortium (ISC) Wray NR. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nature Genetics. 2012;44:247–250. doi: 10.1038/ng.1108. doi: 10.1038/ng.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Harold D, Nyholt DR, Goddard ME, Zondervan KT, Williams J, Visscher PM. Estimation and partitioning of polygenic variation captured by common SNPs for Alzheimer’s disease, multiple sclerosis and endometriosis. Human Molecular Genetics. 2013;22:832–841. doi: 10.1093/hmg/dds491. doi: 10.1093/hmg/dds491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. American Journal of Human Genetics. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Yang J, Goddard ME, Visscher PM, Wray NR. Estimation of pleiotropy between complex diseases using SNP-derived genomic relationships and restricted maximum likelihood. Bioinformatics. 2012;28:2540–2542. doi: 10.1093/bioinformatics/bts474. doi: 10.1093/bioinformatics/bts474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehne B, Schlitt T. Breaking free from the chains of pathway annotation: de novo pathway discovery for the analysis of disease processes. Pharmacogenomics. 2012;13:1967–1978. doi: 10.2217/pgs.12.170. doi: 10.2217/pgs.12.170. [DOI] [PubMed] [Google Scholar]
- Li BS, Leal SM. Methods for detecting associations with rare variants for common diseases: Application to analysis of sequence data. American Journal of Human Genetics. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein P, Carlstrom E, Rastam M, Gillberg C, Anckarsater H. The genetics of autism spectrum disorders and related neuropsychiatric disorders in childhood. American Journal of Psychiatry. 2010;167:1357–1363. doi: 10.1176/appi.ajp.2010.10020223. doi: 10.1176/appi.ajp.2010.10020223. [DOI] [PubMed] [Google Scholar]
- Lichtenstein P, Yip BH, Bjork C, Pawitan Y, Cannon TD, Sullivan PF, Hultman CM. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: A population-based study. Lancet. 2009;373:234–239. doi: 10.1016/S0140-6736(09)60072-6. doi: 10.1016/S0140-6736(09)60072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, Macgregor S. A versatile gene-based test for genome-wide association studies. American Journal of Human Genetics. 2010;87:139–145. doi: 10.1016/j.ajhg.2010.06.009. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Blackwood DH, Caesar S, de Geus EJC, Farmer A, Ferreira MAR, Wellcome Trust Case Control Consortium Meta-analysis of genome-wide association data of bipolar disorder and major depressive disorder. Molecular Psychiatry. 2011;16:2–4. doi: 10.1038/mp.2009.107. doi: 10.1038/mp.2009.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubke GH, Hottenga JJ, Walters R, Laurin C, de Geus EJ, Willemsen G, Boomsma DI. Estimating the genetic variance of major depressive disorder due to all single nucleotide polymorphisms. Biological Psychiatry. 2012;72:707–709. doi: 10.1016/j.biopsych.2012.03.011. doi: 10.1016/j.biopsych.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur DG, Balasubramanian S, Frankish A, Huang N, Morris J, Walter K, Tyler-Smith C. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra RD, Kim J, Dunbar WB. Recent advances in nanopore sequencing. Electrophoresis. 2012;33:3418–3428. doi: 10.1002/elps.201200272. doi: 10.1002/elps.201200272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium A mega-analysis of genome-wide association studies for major depressive disorder. Molecular Psychiatry. 2012;18:497–511. doi: 10.1038/mp.2012.21. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra D, Sebat J. CNVs: Harbingers of a rare variant revolution in psychiatric genetics. Cell. 2012;148:1223–1241. doi: 10.1016/j.cell.2012.02.039. doi: 10.1016/j.cell.2012.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA. Genomewide association studies and assessment of the risk of disease. New England Journal of Medicine. 2010;363:166–176. doi: 10.1056/NEJMra0905980. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JPA, Hirschhorn JN. Genome-wide association studies for complex traits: Consensus, uncertainty and challenges. Nature Reviews Genetics. 2008;9:356–369. doi: 10.1038/nrg2344. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- McGrath LM, Weill S, Robinson EB, Macrae R, Smoller JW. Bringing a developmental perspective to anxiety genetics. Development and Psychopathology. 2012;24:1179–1193. doi: 10.1017/S0954579412000636. doi: 10.1017/S0954579412000636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell KJ. What is complex about complex disorders? Genome Biology. 2012;13:237. doi: 10.1186/gb-2012-13-1-237. doi: 10.1186/gb-2012-13-1-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk C, Spicer J, Champagne FA. Linking prenatal maternal adversity to developmental outcomes in infants: The role of epigenetic pathways. Development and Psychopathology. 2012;24:1361–1376. doi: 10.1017/S0954579412000764. doi: 10.1017/S0954579412000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AP, Zeggini E. An evaluation of statistical approaches to rare variant analysis in genetic association studies. Genetic Epidemiology. 2010;34:188–193. doi: 10.1002/gepi.20450. doi: 10.1002/gepi.20450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumova OY, Palejev D, Vlasova NV, Lee M, Rychkov SY, Babich ON, Grigorenko EL. Age-related changes of gene expression in the neocortex: Preliminary data on RNA-Seq of the transcriptome in three functionally distinct cortical areas. Development and Psychopathology. 2012;24:1427–1442. doi: 10.1017/S0954579412000818. doi: 10.1017/S0954579412000818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Medland SE, Ripke S, Asherson P, Franke B, Lesch K-P, Nelson S. Meta-analysis of genome-wide association studies of attention-deficit/hyperactivity disorder. Journal of the American Academy of Child and Adolescent Psychiatry. 2010;49:884–897. doi: 10.1016/j.jaac.2010.06.008. doi: 10.1016/j.jaac.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly PF, Hoggart CJ, Pomyen Y, Calboli FC, Elliott P, Jarvelin MR, Coin LJ. MultiPhen: Joint model of multiple phenotypes can increase discovery in GWAS. PLoS ONE. 2012;7:e34861. doi: 10.1371/journal.pone.0034861. doi: 10.1371/journal.pone.0034861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J, Kamatani Y, Lathrop M. Family-based designs for genome-wide association studies. Nature Reviews Genetics. 2011;12:465–474. doi: 10.1038/nrg2989. doi: 10.1038/nrg2989. [DOI] [PubMed] [Google Scholar]
- Pennisi E. ENCODE project writes eulogy for junk DNA. Science. 2012;337:1159–1161. doi: 10.1126/science.337.6099.1159. doi: 10.1126/science.337.6099.1159. [DOI] [PubMed] [Google Scholar]
- Pharoah PDP, Antoniou A, Bobrow M, Zimmern RL, Easton DF, Ponder BAJ. Polygenic susceptibility to breast cancer and implications for prevention. Nature Genetics. 2002;31:33–36. doi: 10.1038/ng853. doi: 10.1038/ng853. [DOI] [PubMed] [Google Scholar]
- Pickrell JK, Marioni JC, Pai AA, Degner JF, Engelhardt BE, Nkadori E, Pritchard JK. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomin R. Child development and molecular genetics: 14 years later. Child Development. 2013;84:104–120. doi: 10.1111/j.1467-8624.2012.01757.x. doi: 10.1111/j.1467-8624.2012.01757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomin R, DeFries JC, Knopik VS, Neiderhiser JM. Behavioral genetics. 6th ed. Worth Publishers; New York: 2013. [Google Scholar]
- Plomin R, Haworth CMA, Davis OSP. Common disorders are quantitative traits. Nature Reviews Genetics. 2009;10:872–878. doi: 10.1038/nrg2670. doi: 10.1038/nrg2670. [DOI] [PubMed] [Google Scholar]
- Plomin R, Haworth CMA, Meaburn EL, Price T, Wellcome Trust Case Control Consortium 2. Davis OSP. Common DNA markers can account for more than half of the genetic influence on cognitive abilities. Psychological Science. 2013;24:562–568. doi: 10.1177/0956797612457952. doi: 10.1177/0956797612457952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomin R, Hill L, Craig I, McGuffin P, Purcell S, Sham P, Owen MJ. A genome-wide scan of 1842 DNA markers for allelic associations with general cognitive ability: A five-stage design using DNA pooling and extreme selected groups. Behavior Genetics. 2001;31:497–509. doi: 10.1023/a:1013385125887. doi: 10.1023/A:1013385125887. [DOI] [PubMed] [Google Scholar]
- Plomin R, Kovas Y. Generalist genes and learning disabilities. Psychological Bulletin. 2005;131:592–617. doi: 10.1037/0033-2909.131.4.592. doi: 10.1037/0033-2909.131.4.592. [DOI] [PubMed] [Google Scholar]
- Psychiatric GWAS Consortium Bipolar Disorder Working Group Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nature Genetics. 2011;43:977–983. doi: 10.1038/ng.943. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan VK, Shen L, Moore JH, Saykin AJ. Pathway analysis of genomic data: concepts, methods, and prospects for future development. Trends in Genetics. 2012;28:323–332. doi: 10.1016/j.tig.2012.03.004. doi: 10.1016/j.tig.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld CA, Medland SE, Derringer J, Yang J, Esko T, Martin NW, Koellinger PD. Common genetic variants are associated with educational attainment. Science. in press. [Google Scholar]
- Rochman B. ‘Want to know my future’? Parents grapple with delving into their kids’ DNA. Time. 2012 Dec 13; [Google Scholar]
- Rucker JJH, McGuffin P. Genomic structural variation in psychiatric disorders. Development and Psychopathology. 2012;24:1335–1344. doi: 10.1017/S0954579412000740. doi: 10.1017/S0954579412000740. [DOI] [PubMed] [Google Scholar]
- Sahoo T, Theisen A, Rosenfeld JA, Lamb AN, Ravnan JB, Schultz RA, Shaffer LG. Copy number variants of schizophrenia susceptibility loci are associated with a spectrum of speech and developmental delays and behavior problems. Genetics in Medicine. 2011;13:868–880. doi: 10.1097/GIM.0b013e3182217a06. doi: 10.1097/GIM.0b013e3182217a06. [DOI] [PubMed] [Google Scholar]
- The Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium Genome-wide association study identifies five new schizophrenia loci. Nature Genetics. 2011;43:969–976. doi: 10.1038/ng.940. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scriver CR. The PAH gene, phenylketonuria, and a paradigm shift. Human Mutation. 2007;28:831–845. doi: 10.1002/humu.20526. doi: 10.1002/humu.20526. [DOI] [PubMed] [Google Scholar]
- Siontis KCM, Patsopoulos NA, Ioannidis JPA. Replication of past candidate loci for common diseases and phenotypes in 100 genome-wide association studies. European Journal of Human Genetics. 2010;18:832–837. doi: 10.1038/ejhg.2010.26. doi: 10.1038/ejhg.2010.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M. Epigenetic inheritance and the missing heritability problem. Genetics. 2009;182:845–850. doi: 10.1534/genetics.109.102798. doi: 10.1534/genetics.109.102798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So HC, Li MX, Sham PC. Uncovering the total heritability explained by all true susceptibility variants in a genome-wide association study. Genetic Epidemiology. 2011;35:447–456. doi: 10.1002/gepi.20593. doi: 10.1002/gepi.20593. [DOI] [PubMed] [Google Scholar]
- Speed D, Hemani G, Johnson MR, Balding DJ. Improved heritability estimation from genome-wide SNPs. American Journal of Human Genetics. 2012;91:1011–1021. doi: 10.1016/j.ajhg.2012.10.010. doi: 10.1016/j.ajhg.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Loos RJ. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nature Genetics. 2010;42:937–948. doi: 10.1038/ng.686. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, Enhancing Neuro Imaging Genetics through Meta-Analysis Consortium Identification of common variants associated with human hippocampal and intracranial volumes. Nature Genetics. 2012;44:552–561. doi: 10.1038/ng.2250. doi: 10.1038/ng.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JL, Parikshak NN, Geschwind DH. Rare inherited variation in autism: Beginning to see the forest and a few trees. Neuron. 2013;77:209–211. doi: 10.1016/j.neuron.2013.01.010. doi: 10.1016/j.neuron.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan P. Don’t give up on GWAS. Molecular Psychiatry. 2012;17:2–3. doi: 10.1038/mp.2011.94. doi: 10.1038/mp.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor HK, Risch NJ, Myers RM. Candidate-gene approaches for studying complex genetic traits: Practical considerations. Nature Reviews Genetics. 2002;3:391–397. doi: 10.1038/nrg796. doi: 10.1038/nrg796. [DOI] [PubMed] [Google Scholar]
- The International Schizophrenia Consortium Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topper S, Ober C, Das S. Exome sequencing and the genetics of intellectual disability. Clinical Genetics. 2011;80:117–126. doi: 10.1111/j.1399-0004.2011.01720.x. doi: 10.1111/j.1399-0004.2011.01720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzaskowski M, Dale PS, Plomin R. No genetic influence for childhood behavior problems from DNA analysis. Journal of the American Academy of Child and Adolescent Psychiatry. doi: 10.1016/j.jaac.2013.07.016. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzaskowski M, Davis OSP, DeFries JC, Yang J, Visscher PM, Plomin R. DNA evidence for strong genome-wide pleiotropy of cognitive and learning abilities. Behavior Genetics. 2013;43:267–273. doi: 10.1007/s10519-013-9594-x. doi: 10.1007/s10519-013-9594-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzaskowski M, Shakeshaft N, Plomin R. Intelligence indexes generalist genes for cognitive abilities. Intelligence. doi: 10.1016/j.intell.2013.07.011. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzaskowski M, Yang J, Visscher PM, Plomin R. DNA evidence for strong genetic stability and increasing heritability of intelligence from age 7 to 12. Molecular Psychiatry. 2014;19:380–384. doi: 10.1038/mp.2012.191. doi: 10.1038/mp.2012.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen J, Slagboom PE, Draisma HHM, Martin NG, Boomsma DI. The continuing value of twin studies in the omics era. Nature Reviews Genetics. 2012;13:640–653. doi: 10.1038/nrg3243. doi: 10.1038/nrg3243. [DOI] [PubMed] [Google Scholar]
- Veltman JA, Brunner HG. De novo mutations in human genetic disease. Nature Reviews Genetics. 2012;13:565–575. doi: 10.1038/nrg3241. doi: 10.1038/nrg3241. [DOI] [PubMed] [Google Scholar]
- Vinkhuyzen AAE, Pedersen NL, Yang J, Lee SH, Magnusson PKE, Iacono WG, Wray NR. Common SNPs explain some of the variation in the personality dimensions of neuroticism and extraversion. Translational Psychiatry. 2012;2:e102. doi: 10.1038/tp.2012.27. doi: 10.1038/tp.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. American Journal of Human Genetics. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze SI, Iacono WG, McGue M. Confluence of genes, environment, development, and behavior in a post genome-wide association study world. Development and Psychopathology. 2012;24:1195–1214. doi: 10.1017/S0954579412000648. doi: 10.1017/S0954579412000648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukcevic D, Hechter E, Spencer C, Donnelly P. Disease model distortion in association studies. Genetic Epidemiology. 2011;35:278–290. doi: 10.1002/gepi.20576. doi: 10.1002/gepi.20576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J, Symmons O, Spitz F, Korbel JO. Phenotypic impact of genomic structural variation: insights from and for human disease. Nature Reviews Genetics. 2013;14:125–138. doi: 10.1038/nrg3373. doi: 10.1038/nrg3373. [DOI] [PubMed] [Google Scholar]
- The Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Wellcome Trust Case Control Consortium Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010;464:713–720. doi: 10.1038/nature08979. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams NM, Franke B, Mick E, Anney RJL, Freitag CM, Gill M, Faraone SV. Genome-wide analysis of copy number variants in attention deficit hyperactivity disorder: The role of rare variants and duplications at 15q13.3. American Journal of Psychiatry. 2012;169:195–204. doi: 10.1176/appi.ajp.2011.11060822. doi: 10.1176/appi.ajp.2011.11060822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CCY, Caspi A, Williams B, Craig IW, Houts R, Ambler A, Mill J. A longitudinal study of epigenetic variation in twins. Epigenetics. 2010;5:516–526. doi: 10.4161/epi.5.6.12226. doi: 10.4161/epi.5.6.12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CCY, Meaburn EL, Ronald A, Price TS, Jeffries AR, Schalkwyk LC, Mill J. Methylomic analysis of monozygotic twins discordant for autism spectrum disorder (ASD) and related behavioural traits. Molecular Psychiatry. 2014;19:495–503. doi: 10.1038/mp.2013.41. doi: 10.1038/mp.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Visscher PM. Common SNPs explain a large proportion of the heritability for human height. Nature Genetics. 2010;42:565–569. doi: 10.1038/ng.608. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JA, Lee SH, Goddard ME, Visscher PM. GCTA: A tool for genome-wide complex trait analysis. American Journal of Human Genetics. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitlen N, Kraft P. Heritability in the genome-wide association era. Human Genetics. 2012;131:1655–1664. doi: 10.1007/s00439-012-1199-6. doi: 10.1007/s00439-012-1199-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhernakova A, Stahl EA, Trynka G, Raychaudhuri S, Festen EA, Franke L, Plenge RM. Meta-analysis of genome-wide association studies in celiac disease and rheumatoid arthritis identifies fourteen non-HLA shared loci. PLoS Genetics. 2011;7:e1002004. doi: 10.1371/journal.pgen.1002004. doi: 10.1371/journal.pgen.1002004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Carbonetto P, Stephens M. Polygenic modeling with bayesian sparse linear mixed models. PLoS Genetics. 2013;9:e1003264. doi: 10.1371/journal.pgen.1003264. doi: 10.1371/journal.pgen.1003264. [DOI] [PMC free article] [PubMed] [Google Scholar]