Abstract
Polarized kinesin-driven transport is crucial for development and maintenance of neuronal polarity. Kinesins are thought to recognize biochemical differences between axonal and dendritic microtubules in order to deliver their cargoes to the appropriate domain. To identify kinesins that mediate polarized transport, we prepared constitutively active versions of all the kinesins implicated in vesicle transport and expressed them in cultured hippocampal neurons. Seven kinesins translocated preferentially to axons and five translocated into both axons and dendrites. None translocated selectively to dendrites. Highly homologous members of the same subfamily displayed distinctly different translocation preferences and were differentially regulated during development. By expressing chimeric kinesins, we identified two microtubule-binding elements within the motor domain that are important for selective translocation. We also discovered elements in the dimerization domain of Kinesin-2 motors that contribute to their selective translocation. These observations indicate that selective interactions between kinesin motor domains and microtubules can account for polarized transport to the axon, but not for selective dendritic transport.
Keywords: kinesin, hippocampal neuron, polarity
INTRODUCTION
Neurons are highly polarized cells and nearly every aspect of neuronal function depends on the accurate trafficking of proteins to either the axonal or somatodendritic domain. By tracking the movements of individual vesicles that contain GFP-tagged axonal or dendritic membrane proteins, it has been established that selective microtubule-based transport plays a critical role in the maintenance of neuronal polarity (1, 2). Vesicles containing dendritic proteins are transported efficiently into the dendrites but they do not enter the axon beyond the initial segment; vesicles containing axonal membrane proteins are not entirely excluded from dendrites, but their transport is biased toward the axon.
In neurons, long-range anterograde transport is carried out principally by kinesins, members of a large superfamily of motor proteins that use ATP hydrolysis to power their translocation along microtubule tracks. All kinesins share a similar structure. They contain a globular “motor domain”, which binds to microtubules and hydrolyzes ATP in a microtubule-dependent manner, and an extended “tail domain”, which interacts with cargoes. Most kinesins are dimers and the coordinated binding and release of the two motors domains as they step along microtubules enables them to walk processively, taking many steps without detaching from the microtubule. Members of the Kinesin-1, −2, −3, and −4 sub-families, which all translocate toward the plus-ends of microtubules, are thought to function principally as motors for organelle transport (3). Individual neurons are thought to express many different kinesins from these subfamilies. For example, hippocampal pyramidal neurons express mRNAs encoding 14 kinesins whose levels and patterns of expression indicate they likely mediate organelle transport (4). Understanding the role these different kinesins play is a key issue in neuronal protein trafficking.
To account for the differential transport of proteins to axons or dendrites, Burack et al. (1) proposed that some kinesins are “smart”— that they can distinguish between axonal and dendritic microtubules and transport their vesicular cargoes preferentially to one or the other domain. Microtubules in axons and dendrites are biochemically distinct—they contain different populations of microtubule-associated proteins and are subject to different posttranslational modifications of tubulin, which modulate kinesin binding in vitro (5–9). Implicit in the smart motor hypothesis is the idea that interaction between microtubules and the kinesin motor domain is the primary determinant of a kinesin’s transport destination, although this may be modified by interactions with different cargoes (10).
Expressing fluorescently tagged, constitutively active kinesins in cultured neurons offers a convenient method to assess of the selectivity of kinesin motor domain translocation. Because 90% of axonal microtubules are oriented with their plus ends directed distally (11), and because kinesins are highly processive, constitutively active kinesins rapidly accumulate at the tip of the axon. Dendritic microtubules are of mixed polarity orientation but in distal dendrites plus-end out microtubules predominate (12), so that expressed truncated kinesins can also accumulate at dendritic tips (13). Previous studies established that KIF1A, a member of the Kinesin-3 subfamily, accumulates at the tips of both axons and dendrites in this assay; in contrast, members of the Kinesin-1 family accumulate selectively at axonal tips (13, 14). These results show that some kinesin motor domains translocate preferentially on different microtubule populations, as predicted by the smart motor hypothesis, but they leave many questions unanswered. In particular, we do not know which of the many kinesins expressed by neurons are capable of selective translocation and whether they translocate preferentially to axons or dendrites.
As a first step toward elucidating the role of different kinesins in neuronal organelle trafficking, we prepared constitutively active constructs of all of the relevant kinesins and expressed them in hippocampal neurons in order to assess the selectivity of their transport. Specifically, we sought to address the following questions: Which kinesins mediate selective transport? In addition to axon-preferring kinesins, are there dendrite-selective kinesins? Which structural elements within the kinesin motor domain enable recognition of different microtubule populations? Using this approach, we identified seven kinesins that accumulate selectively in axons and five that translocate into all neurites. None of the kinesins we examined translocates selectively to dendrites. Highly homologous members of the same kinesin subfamily displayed distinctly different selectivity patterns, suggesting that subtle differences in motor structure can lead to different patterns of translocation. Based on the expression of chimeric constructs of axon-preferring and nonselective kinesins, we demonstrated that loop 8 and loop 12 are the most important elements within the motor domain for selective translocation. Unexpectedly, we also discovered that elements within the dimerization domain of Kinesin-2 motors contribute to their translocation preferences, which suggests that regions outside the kinesin motor domain also interact with microtubules and influence the selectivity of transport. These observations indicate that the smart motor hypothesis can account for the preferential transport of vesicles to the axon, but by itself is unlikely to serve as the basis for selective dendritic transport.
RESULTS
Accumulation patterns of constitutively active homodimeric kinesins in cultured hippocampal neurons
Truncated kinesins that lack the auto-inhibitory tail domain exhibit enhanced, microtubule-dependent ATPase activity in biochemical assays and translocate processively toward microtubule plus ends in motility assays (15), (16). When expressed in cultured neurons, such constitutively active kinesins accumulate at the tips of the neurites (Fig. S1). The accumulation of a truncated kinesin is dependent on its dimer formation and ATPase activity (Fig S1, B and C). The selectivity of a given kinesin’s accumulation in this assay is thought to reflect the selectivity with which it transports its endogenous neuronal cargoes (13, 14).
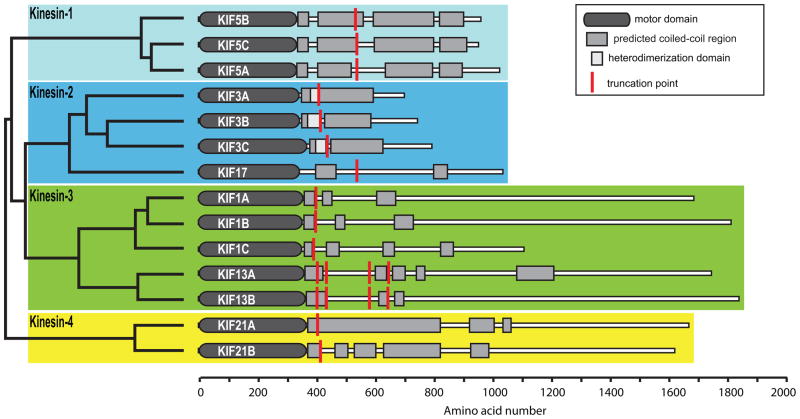
A previous study identified the kinesins that likely serve as organelle motors in hippocampal neurons (4). In order to determine which of these kinesins translocate selectively in axons or dendrites, we prepared truncation constructs that could be used in the neuronal accumulation assay and expressed them in hippocampal cultures. (Fig. 1; see Materials and Methods for details).
Figure 1. The truncated kinesins examined in this study.
The dendrogram (left) shows the phylogenetic relationship of these kinesins, based on sequence homologies within the motor domains. The schematics (right) indicate the domain structure of each kinesin. The site where each kinesin was truncated is indicated by a red line.
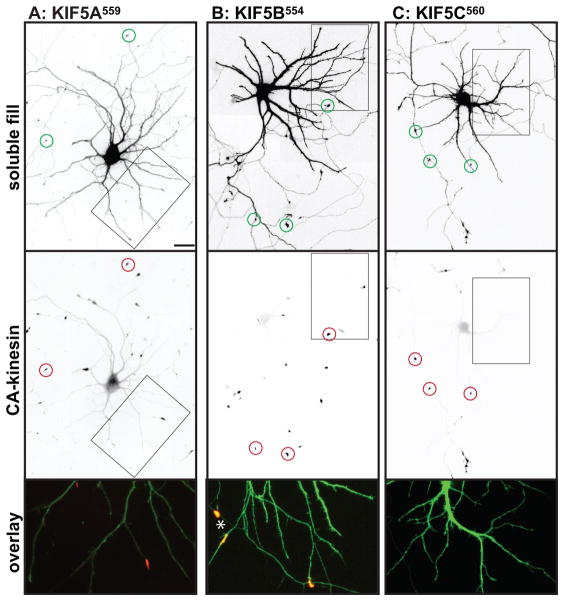
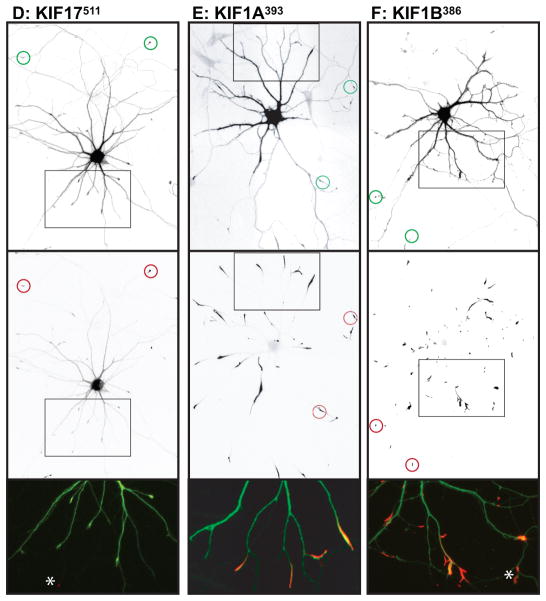
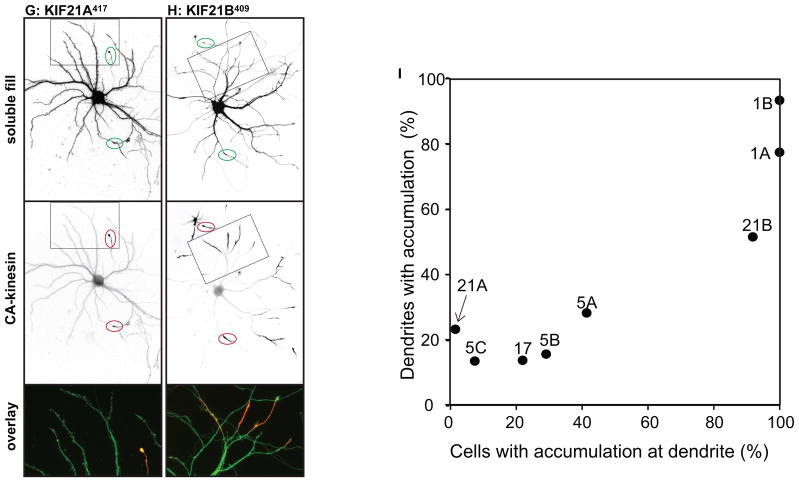
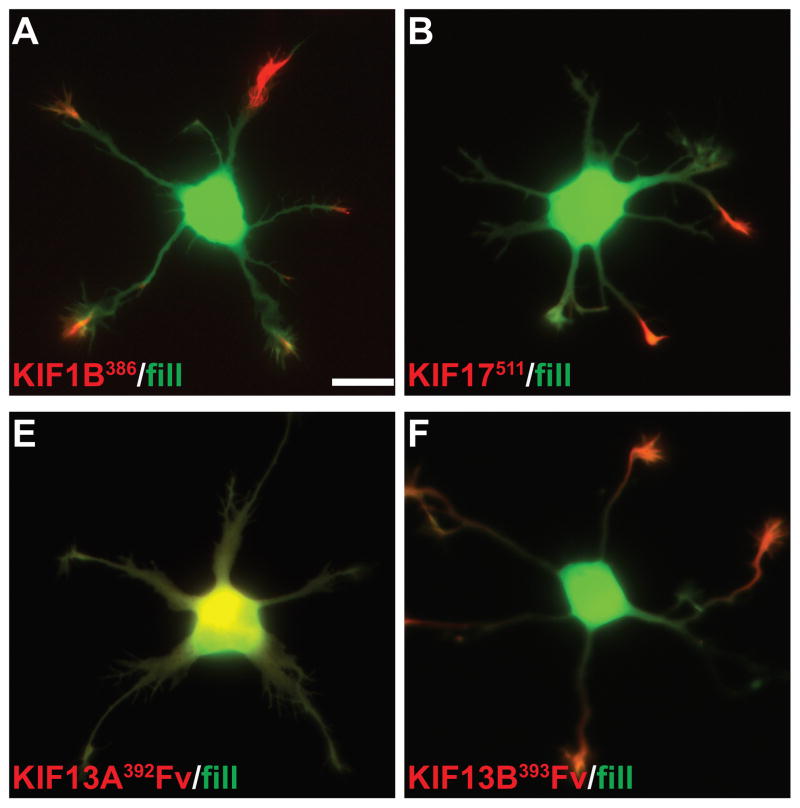
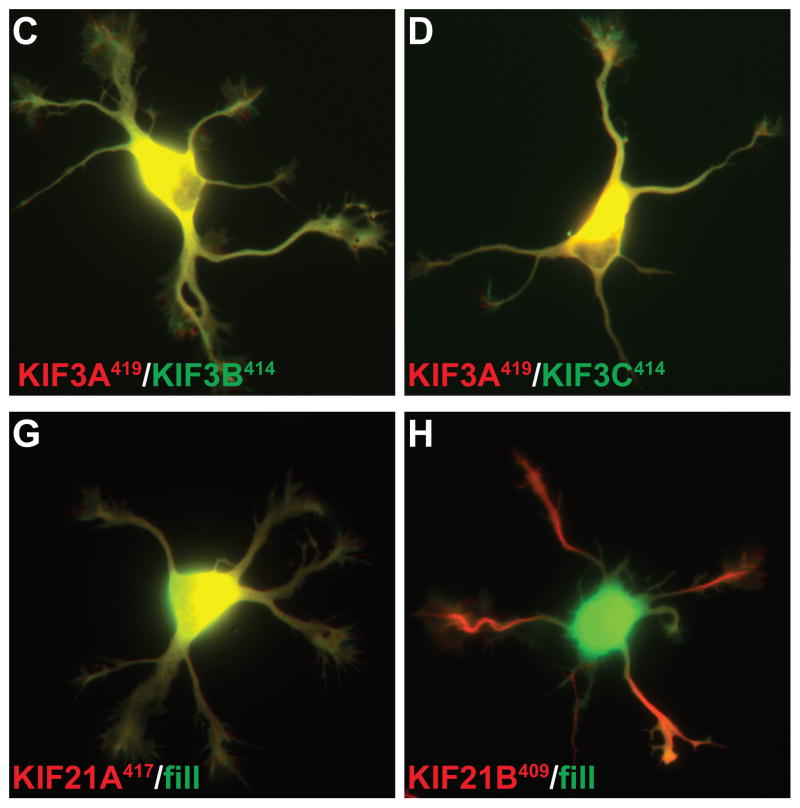
We examined the localization of these constructs in neurons at stage 4 of development (17), when cells have well-developed axonal and dendritic arbors (Fig. 2); cell morphology was visualized by co-expressing soluble GFP or an appropriate color variant. The dendrites, which are thick at their origin and taper as they extend outward, usually terminate within 150–250 μm from the cell body (boxed regions in Fig. 2A). The axon is much longer, extending far beyond the field shown, but gives off many branches, including some that terminate near the cell body (Fig. 2A, circles indicate the tips of axons). Representative examples of the accumulation patterns of several of these kinesins are shown in Fig. 2. All of the constitutively active kinesins illustrated accumulated at axonal tips (red circles in low magnification images), including tips of the many distal axon branches that extend beyond the fields shown. In contrast, there were striking differences among the different kinesins in the extent to which they accumulated at dendritic tips. Three kinesins showed prominent accumulation at dendrite tips: Kinesin-3 family members KIF1A393 and KIF1B386, and Kinesin-4 family member KIF21B409 (Fig. 2E, F, H). Kinesins that accumulated preferentially at axon tips included all three members of the Kinesin-1 family, the sole Kinesin-2 family member that forms homodimers, KIF17, and Kinesin-4 family member, KIF21A417 (Fig. 2A, B, C, D, G). A diffusely distributed pool of KIF17511 and KIF21A417 was present throughout the dendritic arbor (Fig. 2D, G), but this was readily distinguished from an active accumulation in higher magnification images (e.g., compare with Fig. 2E, F, H). The truncation constructs of three Kinesin-3 family members, KIF13A, KIF13B, and KIF1C, failed to accumulate at any neurite tips in this assay. Further studies of these three kinesins are described in the following section.
Figure 2. Different homodimeric kinesins exhibit different translocation selectivites in cultured hippocampal neurons.
(A–H) To evaluate the translocation preferences of members of the Kinesin-1, −2, −3, and −4 families, truncated kinesins tagged at their C-termini were expressed in stage 4 neurons. Several kinesins accumulated exclusively at axon tips whereas others accumulated at dendritic tips as well. The upper panels illustrate cell morphology, based on signals (γ-value adjusted) from co-expressed soluble fluorescent protein, and the middle panels show the distribution of truncated kinesins. In the lower panels, which show enlargements of the boxed regions that contain multiple dendritic tips, the kinesin is shown in red and the soluble fill in green. Axonal tips are indicated by circles in the upper panels and asterisks in the enlargements. (I) Quantification of the selectivity of each kinesin construct (based on the analysis of 29–81 cells). The X-axis indicates the percentage of transfected cells with kinesin labeling of at least one dendritic tip. For those cells with dendritic labeling, the percentage of dendritic tips that were labeled is shown on the Y-axis. Scale bars: 20μm. Images shown in panels A, B, D, and E are composites of 4 to 9 microscopic fields assembled into a single image as described in Materials and Methods.
These eight homodimeric kinesins fell into two distinct groups based on their translocation preferences: three kinesins--KIF1A, KIF1B, and KIF21B--displayed no selectivity. More than 90% of cells expressing these kinesins exhibited dendritic labeling and the great majority of dendritic tips were labeled. The remaining kinesins--KIF5A, 5B, 5C, KIF17, KIF21A--translocated selectively to axonal tips. In most cells, no dendritic accumulation was detected; in the few cells that did exhibit dendritic accumulation, only one or two tips were labeled. That said, there were subtle but significant variations in the selectivity of axon-preferring kinesins that could be detected by quantitative analysis. As shown in Fig. 2I, we determined the percentage of cells in which at least one dendritic tip was labeled (X-axis) and for such cells we determined the fraction of dendritic tips with an accumulation of kinesin (Y-axis). Kinesins that lie near the lower left corner in this graph translocate into axons with a high degree of selectivity whereas those that lie near the upper right corner accumulate robustly in dendrites as well as axons. Quantitative comparison of axon-selective kinesins revealed that less than 10% of cells expressing KIF5C559 had one or more labeled dendrites compared with 30% of cells expressing KIF5B555 and 42% of cells expressing KIF5A559 (t-test, KIF5C vs KIF5B, p< 0.05; KIF5C vs KIF5A, p<0.001).
As noted above, a significant pool of diffusely distributed, unaccumulated protein was observed following expression of one Kinesin-1 (KIF5A559), a Kinesin-2 (KIF17511), and the twoKinesin-4s (Fig. 2C, D, G, H). The presence of this diffusely distributed pool may indicate that these kinesins translocate less efficiently. To estimate the amount of accumulated versus diffusely distributed kinesins, we compared the fluorescence intensity in the cell body and in axonal tips of the same cell (Fig. S2). The tip to soma ratio among different kinesins varied more than twenty-fold. Importantly, the efficiency of accumulation did not correlate with translocation preference. Some axon-selective kinesins had high tip-to-soma ratios (>20 for KIF5B and 5C), others had low ratios (<3 for KIF5A, KIF17 and KIF21).
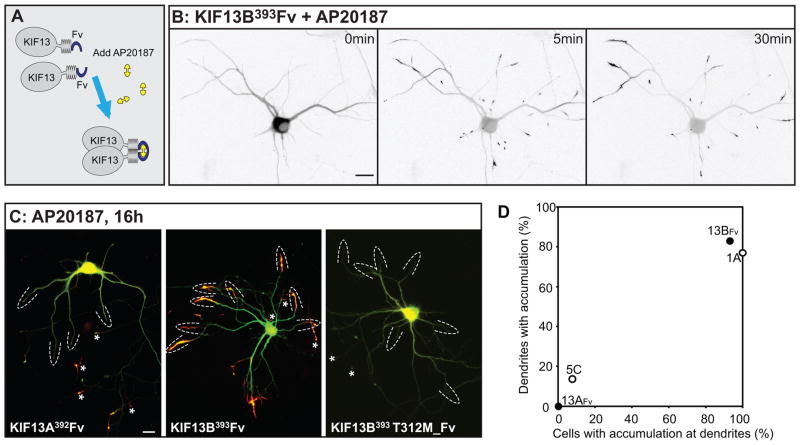
Chemically induced dimerization of KIF13A and KIF13B reveals their accumulation preferences
Truncation constructs of two Kinesin-3 family members, KIF13A and 13B, failed to accumulate when expressed in hippocampal neurons, even when extended stretches of coiled-coil domain were included (see Fig. 1). Huckaba et al. (18) noted that Kinesin-73, the Drosophila homolog of mammalian KIF13s, does not dimerize efficiently, but can be induced to do so by inclusion of a leucine zipper dimerization domain (see also (19)). We prepared truncation constructs of KIF13A and 13B that included a variant of the dimerization domain from FK506 binding protein (referred to as FV) that can be induced to homodimerize by adding AP20187, a homolog of FK1012 (Fig. 3A). In the absence of the dimerizing drug, neither GFP-tagged KIF13A392FV nor KIF13B393Fv accumulated at neurite tips (Fig. 3B and data not shown). Within 5 min of adding AP20187, KIF13B393 Fv became enriched in distal neurites and by 30 minutes had accumulated at axonal and dendritic tips (Fig. 3B). KIF13A392 Fv also accumulated at neurite tips in the presence of AP20187, but with a slower time course. A non-processive KIF13B mutant (KIF13B393 Fv T311M) failed to accumulate (Fig. 3C), confirming that the accumulation of FV- tagged truncated kinesins depends on their microtubule-induced ATPase activity. Remarkably, the accumulation patterns of these highly similar kinesins were distinct (Fig. 3B and C). KIF13A accumulated selectively in axons, whereas KIF13B accumulated to dendrites and axons both. In KIF13B393 Fv -expressing cells that showed dendritic accumulation after AP20187 treatment (N=11), 83% of the dendritic tips (N=117) were labeled. We also prepared a construct of KIF1C that included the Fv dimerization domain (KIF1C389 Fv), but this construct did not accumulate at neurite tips, even after overnight exposure to the dimerizing reagent (data not shown). Therefore, its accumulation preference could not be ascertained.
Figure 3. KIF13A and KIF13B, closely related members of the Kinesin-3 family, have different translocation preferences.
(A) Addition of an Fv domain to the C-termini of KIF13A or 13B enabled induction of homodimerization by addition of AP20187. (B) Addition of AP20187 to a cell expressing KIF13B393Fv caused the kinesin to accumulate at neurite tips. Just before drug addition (0 min), GFP-KIF13B393Fv was distributed diffusely throughout the cell. Within 5 min of adding AP20187 (10 nM), GFP-KIF13B393Fv began to accumulate at neurite tips. The contrast was inverted so that fluorescent signals appear dark on a light background. (C) Distribution of GFP-KIF13A392Fv, GFP-KIF13B393Fv, and GFP-KIF13B393T311M_Fv after incubation with 10nM AP20187. KIF13A392Fv accumulated in axonal but not dendritic tips whereas KIF13B393Fv accumulated in both axonal and dendritic tips. KIF13B393Fv, with a mutation that disrupts microtubule-stimulated ATPase activity, failed to accumulate at either axonal or dendritic tips. Fluorescent signal from the kinesins is shown in red and soluble mRFP in green. Dashed arcs indicate dendritic tips and asterisks indicate axonal tips. Scale bars= 20μm. (D) Translocation preferences of KIF13A392Fv and KIF13B393Fv constructs, based on the analysis of 18 or 15 cells quantified as in Fig. 2. Data points for KIF1A393 and KIF5C559 were taken from the experiment shown in Figure 2I. The image of GFP-KIF13B393T311M_Fv was assembled from 4 microscopic fields as described in Materials and Methods.
Translocation preferences of heterodimeric members of the Kinesin-2 family
Kinesin-2 family members KIF3A, KIF3B, and KIF3C normally form heterodimeric complexes: KIF3A dimerizes with either KIF3B or KIF3C. When expressed singly in Stage 4 hippocampal neurons (data not shown), KIF3A419 and KIF3C441 were diffusely distributed with no evidence of accumulation at neurite tips; KIF3B414 did accumulate at axons tips, although there was also a pool of diffusely distributed protein. In vitro studies suggest that KIF3B, unlike other KIF3s, may be capable of forming homodimers (20), which suggests that KIF3B414 forms homodimers that translocate selectively to axon tips.
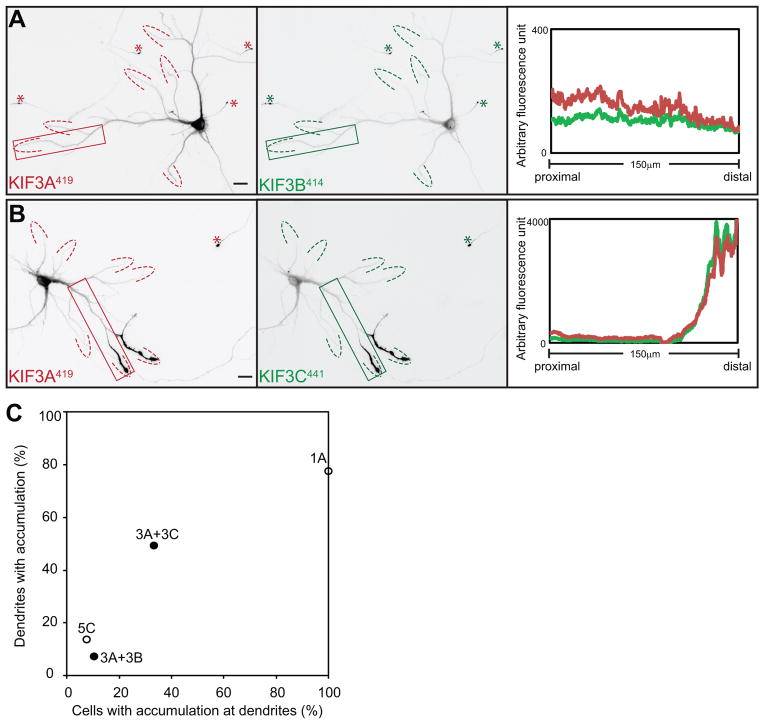
We next investigated the translocation preferences of KIF3 heterodimers. The domains that mediate heterodimer formation by KIF3s have not been clearly established. Some evidence points to the highly charged region following the neck-linker (20) whereas other studies indicate that coiled-coil regions near the C-terminus are crucial (21). In C. elegans, the C-terminal region initiates heterodimerization of Kinesin- 2 family members KLP11 and KLP20 in vitro, but these proteins lack the highly charged region following the neck linker that is characteristic of mammalian KIF3s (22). We prepared constructs truncated just beyond the highly charged region and co-expressed them in pairs in rat embryo fibroblasts to determine whether they dimerized. We found that KIF3A419 co-immunoprecipitated with KIF3B414 and with KIF3C441 (Fig. S3), demonstrating that the highly charged region following the neck-linker is sufficient for heterodimer formation of truncated KIF3s. For simplicity, we refer to this region as the heterodimerization domain, but this is not meant to imply that other regions do not contribute to dimerization in the full-length protein.
When expressed in hippocampal neurons, the two pairs of truncated KIF3s showed a clear difference in their patterns of accumulation (Fig. 4). When KIF3A419 and KIF3B414 were co-expressed, the motors accumulated almost exclusively at axon tips (Fig. 4A). Dendritic labeling was detected in only 10% of cells (n=67), and in those cells only 7 % of dendritic tips were labeled. When KIF3A419 and KIF3C441 were expressed together, 33% of cells (n=51) exhibited dendritic labeling and in these cells 49% of the tips were labeled (Fig. 4B). Both of these differences were highly significant (t-test, p<0.0001). The accumulation of co-expressed KIF3A419 and KIF3C441 at dendritic tips did not correlate to the expression level of the constructs. This raises the possibility that the translocation preference of this kinesin is regulated by some physiological process that differs from cell to cell.
Figure 4. Kinesin-2 heterodimers exhibit different translocation preferences.
Distribution of truncated KIF3s following co-expression with their heterodimer-forming partners. (A) When expressed together, both KIF3A419-tdTomato and KIF3B414-YFP accumulated at axonal tips; (B) co-expression of KIF3A419-tdTomato and KIF3C441-GFP resulted in labeling at axonal tips and some dendritic tips. Line scans of the boxed regions confirm strong enrichment of KIF3A/KIF3C heterodimers at dendritic tips. Scale bars= 20μm. (C) Translocation preferences of truncated KIF3A/3B and KIF3A/3C heterodimers, based on the analysis of 67 or 51 cells quantified as in Fig. 2. Data points for KIF1A393 and KIF5C559 were taken from the experiment shown in Figure 2I. Images in A and B were assembled from four microscope fields as described in Materials and Methods.
Accumulation of truncated kinesins at early stages of neuronal development
Early in the development of hippocampal neurons, before the axon has formed (developmental Stage 2), KIF5C accumulates selectively at the tips of only one or two neurites and its accumulation is dynamic—it can disappear from one neurite and appear in another within tens of minutes (14). Coincident with axon specification, truncated KIF5C accumulates only in the emerging axon and no longer appears in any other neurite. It is thought that these changes in kinesin accumulation mirror dynamic molecular changes in microtubules, including posttranslational modifications of tubulin (23). We wondered whether other axon-selective kinesins undergo similar dynamic changes in their accumulation early in development. We expressed truncated kinesin constructs by electroporation prior to cell plating and examined the pattern of kinesin accumulation in Stage 2 neurons (Fig. 5). Like the axon-selective Kinesin-1 family members, KIF17511 exhibited dynamic changes in its accumulation (illustrated in (24)). Surprisingly, other axon-selective kinesins failed to accumulate in Stage 2 cells. Moreover, highly homologous kinesins differed in their ability to accumulate at this stage of development. In the Kinesin-4 sub-family, KIF21A417 failed to accumulate whereas KIF21B409 accumulated at the tips of most minor neurites (Fig. 5C, D). The Kinesin-3 family member KIF13B393Fv accumulated in most neurites but the closely related KIF13A392 Fv failed to accumulate at all (Fig. 5E, F). The heterodimeric Kinesin-2s also failed to accumulate at the tips of Stage 2 neurites (Fig. 5G, H). Thus at Stage 2 some kinesins accumulate at the tips of most neurites, some accumulate only in a subset of neurites, and others fail to accumulate at any neurite. It is surprising that some of these kinesins fail to accumulate, given that Stage 2 neurites contain stable, plus-end out microtubules (11, 12, 25) and that extensive anterograde vesicle transport is observed in these neurites (4).
Figure 5. Truncated kinesins exhibit different patterns of accumulation early in neuronal development.
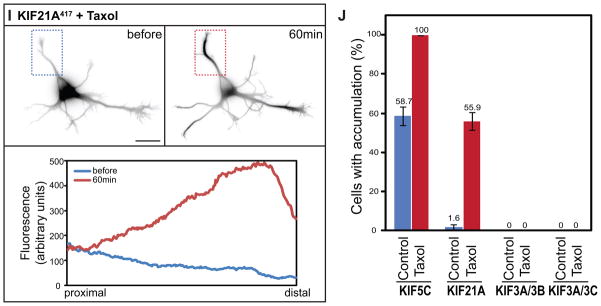
Truncated KIF1B (A) and KIF21B (D) accumulated at all neurite tips whereas truncated KIF17 (B) only accumulated at a subset of neurite tips. The remaining kinesins failed to accumulate at any tips. For KIF13-Fv constructs, cells were incubated with 10nM AP21087 for 4–6h before imaging. (I) KIF21A,, which is unable to accumulate in immature neurons, translocated to neurite tips following Taxol treatment (10 nM) for 1 h. Line scans of the average intensity along the neurite in the boxed region (below) confirm the redistribution of KIF21A417-GFP. (J) The accumulation of KIF21A and KIF5C but not Kinesin-2s in Stage 2 cells was enhanced by 10nM Taxol treatment for 4h. The figure shows the percentage of cells with truncated kinesin accumulation in at least one neurite; values are means and SEMs from 3 independent experiments. Dissociated hippocampal neurons were co-electroporated with tdTomato and a truncated kinesin or with a pair of truncated KIF3s. Scale bar: 20μm.
Neuronal microtubules undergo profound developmental changes in posttranslational modification, in their population of microtubule-associated proteins, and in tubulin isoform composition (26, 27). The developmental changes in kinesin translocation may reflect changes in the biochemical properties of microtubules that make them efficient kinesin substrates. Taxol treatment of neurons has been shown to rapidly enhance posttranslational modifications of tubulin and allows constitutively active KIF5C to accumulate continuously at all neurite tips (28). Thus we investigated whether Taxol treatment could enable accumulation of non-accumulating kinesins in Stage 2 cells. We compared the pattern of kinesin accumulation in cells from control and Taxol-treated cultures and used time-lapse microscopy to follow Taxol-induced changes in the distribution of truncated kinesins in individual cells. As shown in Fig. 5I, Taxol treatment reduced the amount of diffusely distributed KIF21A417 and enabled it to accumulate at multiple neurite tips. Analysis of cells fixed 1 hr after addition of Taxol showed that the accumulation of truncated KIF5C and KIF21A but not heterodimeric KIF3s could be enhanced by Taxol treatment (Fig. 5J). These results suggest that different kinesins require different microtubule specializations in order to walk efficiently in cultured neurons and that microtubules that allow efficient translocation of some kinesins fail to support the translocation of others.
Motor domain sequences that influence the selectivity of kinesin accumulation
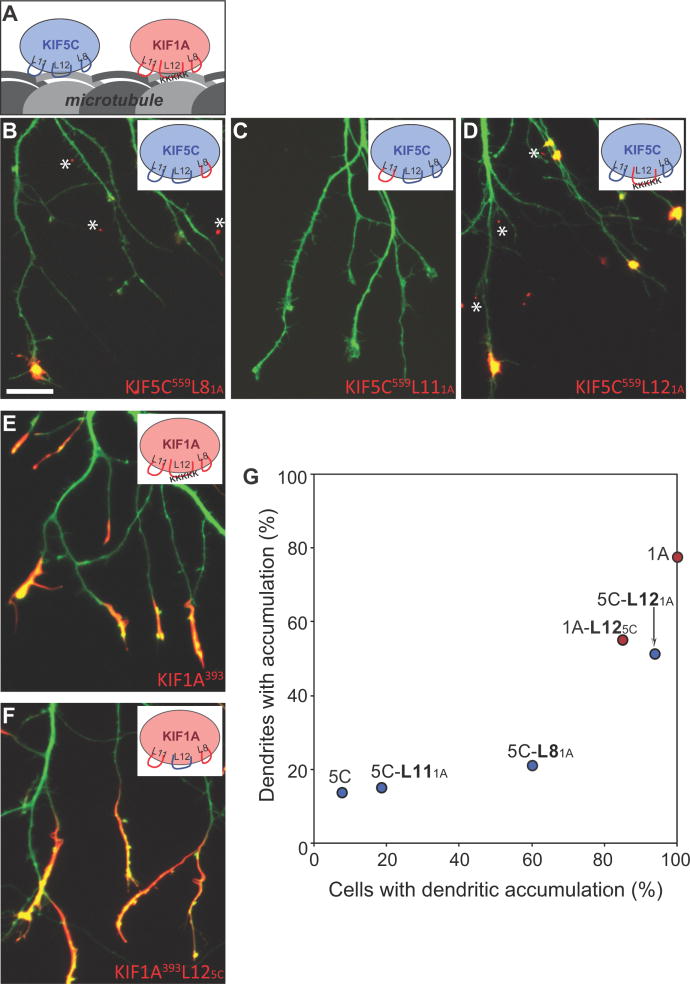
It is thought that the kinesins that accumulate preferentially in the neuronal expression assay recognize molecular differences between axonal and dendritic microtubules (14). What elements within these kinesins enable them to accumulate preferentially in axons? To address this question, we began by comparing KIF5C and KIF1A, two well-characterized motors (29–32) that are polar opposites in terms of their patterns of accumulation. KIF5C559 is highly axon-selective (it labeled fewer than 2% of dendritic tips) whereas KIF1A393 accumulated non-selectively (more than 75% of dendritic tips were labeled). Three sequence elements that interact with microtubules within the motor domains have been identified: β5-loop 8, loop 11-β7, and α4-loop 12-α5 (33) (Fig. 6A). To identify the elements within KIF5C that contribute to its selectivity, we prepared chimeric constructs in which each of its three microtubule-interacting domains was replaced with the corresponding domain from KIF1A (Fig. 6B, C, D). Replacement of the loop 12 domain had the greatest effect. A KIF5C construct with the loop 12 domain from KIF1A (KIF5C559L121A) lost its axonal selectivity. It accumulated at dendritic tips in more than 75% of cells and almost 50% of dendritic tips were labeled; both values are significantly different than for KIF5C559 (p<0.001, t-test). Replacement of the β5-loop 8 region also increased the number of cells with dendritic labeling (from 8% to 60%; p<0.001, t-test) and the percentage of labeled dendrites (from 2% to 15%), but the effects were less pronounced than those following replacement of loop 12 domain. Replacing the loop 11 region of KIF5C with the corresponding region from KIF1A had only a small effect. These results differ somewhat from a recent report by Konishi and Setou (9), who reported that replacing a short sequence within the β5 region with the corresponding sequence from KIF1A redirects KIF5B to dendrites. In our hands, KIF5C559SKLA failed to accumulate at dendritic tips, but exhibited a large component of diffusely distributed motor that extended throughout the somatodendritic domain (Fig. S4), similar to KIF5C mutants with impaired microtubule binding (34). Inspection of fig 1E in Konishi and Setou (9) also shows a large diffusely distributed dendritic pool with minimal accumulation at dendritic tips.
Figure 6. Two loops in the microtubule binding domain of KIF5C determine its selective translocation to axons.
(A) Cartoon illustration of the three MT-binding loops (L8, L11, and L12) of KIF5C and KIF1A. (B–D) Localization of chimeric kinesins in which each of the microtubule binding regions of KIF5C559 was exchanged with the corresponding region of KIF1A. Replacement of L12 caused KIF5C to accumulate at nearly all dendritic tips; replacement of L8 also increased dendritic labeling, but to a lesser extent. When KIF5C559 constructs translocated to dendrites, they accumulated in tiny spots (asterisks) rather than elongated segments like KIF1A. (E, F) Exchanging the L12 of KIF1A with that of KIF5C did not alter the localization of truncated KIF1A393. (G) Translocation preferences of KIF5C559 and KIF1A393 chimeric constructs, based on the analysis of 29–82 cells quantified as in Fig. 2. Data for wild type KIF1A and KIF5C were taken from the experiment shown in Figure 2I. Scale bar: 20μm.
We also attempted to identify elements within KIF5C that were sufficient to confer axon selectivity on KIF1A. A KIF1A construct with the loop 12 region from KIF5C (KIF1A393L125C,) accumulated at both axon and dendrite tips (Fig. 6F), but the percentage of dendrite tips with accumulation was reduced (from 78% to 47%; p<0.001, t-test). Constructs of KIF1A that included the loop 8 region from KIF5C or both the loop 11 and loop 12 regions failed to accumulate, probably because these substitutions disrupted the overall motor domain structure. Taken together, the results of expressing chimeric constructs of KIF5C/KIF1A motor domains demonstrate that loop 12 and to a lesser extent loop 8 play a critical role in determining the selectivity of kinesin translocation in mature hippocampal neurons. In this series of experiments, we also noted that constructs of KIF5C that were redirected to dendrites displayed an unusual pattern of labeling (Fig. 6B, D). Only the most distal tips of dendrites were labeled and tiny accumulations of the motor also occurred at the tips of filopodia-like projections along dendritic shafts. This unique accumulation pattern could be traced to sequences between residues 389 and 559 of KIF5C (Fig. S4).
Sequences within dimerization domains can also influence the selectivity of kinesin accumulation
Do differences in β5-loop 8 and loop 12 regions account for differences in the selectivity of other kinesins? Comparison of pairs of kinesins (KIF13A versus KIF13B, KIF21A versus 21B) did not reveal obvious differences in their microtubule binding domains that could account for their different translocation preferences. We did note that the loop 11 region of KIF3C contains a 25-residue insert which is enriched in glycines and serines that is not present in other kinesins. When we expressed a chimeric construct of KIF3C and the loop 11 region from KIF3B (KIF3C441L113B ) together with KIF3A, it accumulated in dendritic tips in 63% of the cells (data not shown), slightly more than KIF3C441. Thus the loop 11 region of KIF3C is not required for dendritic accumulation of KIF3A/3C heterodimers.
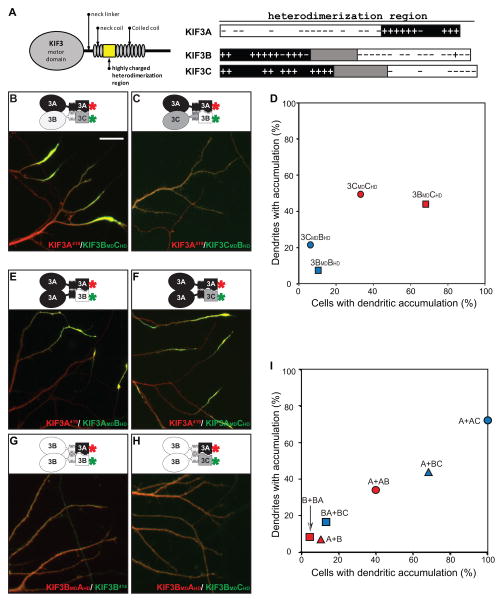
The KIF3 kinesins contain a unique region of charged amino acids just C-terminal to the neck coil (Fig. 7A). In KIF3A, this domain consists of a region highly enriched in acidic amino acids followed by a region highly enriched in basic amino acids; in KIF3B and 3C, the organization of charged residues is reversed. These regions are proposed to govern partner recognition of KIF3 motors (35, 36) and our results show that this region is sufficient for heterodimer formation (Fig. 4). Interestingly, there are significant differences between KIF3B and KIF3C in this region; the positively charged region is shorter and the negatively charged region is longer in KIF3B. Studies of Kinesin-1 indicate that residues C-terminal to the motor domain can influence kinesin interactions with microtubules (37). We therefore examined the possibility that differences in the heterodimerization domain of KIF3B and KIF3C contribute to their different translocation preferences.
Figure 7. The heterodimerization domains of KIF3s contribute to their translocation preferences.
(A) A drawing of the N-terminal portion of a KIF3 kinesin illustrating the motor domain and regions thought to contribute to dimerization. The enlarged view (right) compares the distribution of charged residues in the putative heterodimerization domains of KIF3A, 3B, and 3C. (B, C) Co-expression of KIF3A together with chimeras of KIF3B and 3C shows that the dimerization domain of KIF3B enhances axon-selective translocation. Constructs with red asterisks were tagged with tdTomato and pseudocolored red and those labeled with green asterisks were tagged with eGFP and pseudocolored green. (D) Quantification of cells expressing KIF3A together with different KIF3B and 3C combinations confirms that constructs which include the dimerization domain of KIF3B (red symbols) are far more axon-selective than those corresponding constructs with the 3C dimerization domain (blue symbols). (E–H) Translocation preferences of truncated kinesins that include two copies of the KIF3A motor domain (E, F) or two copies of the KIF3B motor domain (G, H). Kinesins with two copies of the B motor domain were far more axon-selective than those with two copies of the A motor domain, regardless of whether they contained dimerization domains from KIF3A and 3B (E, G) or from KIF3A and 3C (F, H). (I) Quantification of kinesin localization based on the analysis of 23–67 cells expressing truncated kinesins with two copies of the 3A motor domain (circles), two copies of the 3B motor domain (squares), or one copy of each (triangles). Red symbols correspond to kinesin dimers with dimerization domains from KIF3A and 3B, blue symbols to kinesin dimers with dimerization domains from KIF3A and 3C. Single letters (A, B) refer to wild-type KIF3A and 3B. Pairs of letters (e.g., AB) refer to chimeras, with the first letter identifying the source of the motor domain (A) and the second letter the source of the dimerization domain (B). Data for wild-type truncated KIF3A/3B or KIF3A/3C heterodimers were taken from Fig 4C. Scale bar=20μm.
We prepared chimeric constructs consisting of the motor domain (MD) of KIF3B or KIF3C with the heterodimerization domain (HD) of the other and examined the translocation preference of each construct when co-expressed with truncated KIF3A (Fig. 7B-D). When KIF3BMD3CHD was co-expressed with KIF3A419, the resulting heterodimer accumulated at both axonal and dendritic tips in nearly 70% of cells, far higher than the percentage of cells that showed a dendritic accumulation of KIF3A419/ KIF3B414. In contrast, when KIF3CMD3BHD was co-expressed with KIF3A419, a dendritic accumulation was observed in only 6% of cells, far less than the percentage of cells that exhibit dendritic labeling following co-expression of wild-type KIF3A and 3C. Thus heterodimers that contain the dimerization domain from KIF3B are far more axon-selective than heterodimers that contain the dimerization domain from 3C, regardless of whether they contain the motor domain from KIF3B or from KIF3C. These results demonstrate that sequences outside the motor domains of KIF3s play a key role in determining translocation selectivity.
We further utilized this approach to assess the translocation preferences of dimers containing two copies of the same motor domain. Dimers with two copies of the KIF3A motor domain accumulated in both axons and dendrites regardless of whether they assembled with dimerization domains from KIF3A and 3B (Fig. 7E) or KIF3A and KIF3C (Fig. 7F). Truncated kinesins that contained two copies of the 3B motor domain accumulated rather selectively in axons (Fig. 7G, H). These results indicate that the intrinsic translocation preference of KIF3A is to both axon and dendrite while that of KIF3B is axon selective. Motors with two copies of KIF3C motor domain failed to accumulate in this assay.
Quantification of these two experiments revealed the relative contributions of the motor and heterodimerization domains to the truncation preferences of the KIF3 kinesins (Fig. 7I). When assembled with dimerization domains from KIF3A and KIF3C, truncated kinesins that contained two copies of the 3B motor domain accumulated rather selectively in axons, kinesins that contained two copies of the 3A motor domain accumulated in both dendrites and axons, and kinesins with one copy of each displayed an intermediate distribution. Parallel results were obtained when assembly was mediated by dimerization domains from KIF3A and 3B; constructs with two copies of the 3B motor domain were most axon-selective and those with two copies of the 3A motor domain were the least selective. Taken together these results demonstrate that sequences within the motor domains and the heterodimerization domains both contribute to the translocation preference of KIF3 kinesins. The motor and dimerization domains from KIF3B both contain sequences that promote selective transport to the axon. The KIF3A motor domain and the KIF3C heterodimerization contain elements that promote the dendritic translocation of KIF3 dimers.
DISCUSSION
The principal goal of this study was to systematically compare the translocation preferences of all of the kinesins relevant to organelle transport in a single, cell-based assay. We identified seven axon-selective kinesins, but found no kinesins that translocate preferentially to dendrites. Highly homologous kinesins exhibited different patterns of accumulation and also differed in their ability to translocate efficiently in immature neurons. These findings have important implications concerning the factors that contribute to selective kinesin translocation and the role these kinesins play in the development and maintenance of neuronal polarity.
Translocation preferences of different kinesins and their possible roles in polarized transport
The principal reason for undertaking this study was to evaluate the translocation preferences of the kinesins expressed in hippocampal neurons as a first step toward understanding the distinctive role each plays in neuronal protein trafficking. As summarized in Table I, with a single exception, the twelve constitutively active kinesins we examined fell into one of two distinct groups: seven accumulated selectively at axon terminals whereas four others accumulated at both dendritic and axonal tips. Only the heterodimeric Kinesin-2 consisting of KIF3A and KIF3C motor domains displayed an intermediate pattern. This kinesin accumulated at dendritic tips in only about 35% of cells, but in that subpopulation it labeled the great majority of dendritic tips (comparable to kinesins in the non-selective group). This raises the possibility that the translocation preference of this kinesin is regulated by some physiological process that differs from cell to cell.
Table I.
Translocation preferences of kinesin motor domains in cultured hippocampal neurons
| Stage 4 | Stage 2 | ||
|---|---|---|---|
| Kinesin-1 | KIF5A | axonal | subset of neurites |
| KIF5B | axonal | subset of neurites | |
| KIF5C | axonal | subset of neurites | |
|
| |||
| Kinesin-2 | KIF3A/3B | axonal | fails to accumulate |
| KIF3A/3C | weakly axonal | fails to accumulate | |
| KIF17 | axonal | subset of neurites | |
|
| |||
| Kinesin-3 | KIF1A | non-selective | most neurites |
| KIF1B | non-selective | most neurites | |
| KIF13A | axonal | fails to accumulate | |
| KIF13B | non-selective | most neurites | |
|
| |||
| Kinesin-4 | KIF21A | axonal | fails to accumulate |
| KIF21B | non-selective | most neurites | |
Perhaps most important, we found no kinesin motor domains that were dendrite-selective in the neuronal expression assay. One of the key features of neuronal protein trafficking is that the vesicles which carry dendritically polarized membrane proteins do not enter the axonal domain. Our results indicate that the exclusion of dendritic vesicles from the axon is not solely due to properties of the motor domains of the kinesins responsible for their transport, as predicted by the smart motor hypothesis. Instead the inherent translocation preferences of kinesin motor domains may be modified by binding to dendritic cargoes, as proposed in the cargo steering model of Setou et al (10), or that the translocation of dendritic cargoes is selectively inhibited as they approach the axon initial segment (38). It is also possible that dyneins or myosins mediate the selective dendritic transport of some cargoes (39, 40). That said, the five kinesins that accumulate at dendritic as well as axonal tips seem the most likely candidates to mediate selective transport.
Vesicles that carry axonal proteins are preferentially transported to the axon and the seven axon-selective kinesins we identified in the expression assay are likely candidates to mediate their transport. The Kinesin-1s and the KIF3A/3B heterodimer of Kinesin-2 have already been implicated as motors for axonal transport based on their association with axonal cargoes, their accumulation in ligated axons, and on the phenotypes observed in genetically null mice (41–45). The finding that KIF17 is axon-selective in the neuronal expression assay was unexpected. Previous studies implicated KIF17 as a motor that mediates dendritic transport (40, 46, 47), although this has never been directly demonstrated by a live imaging assay of endogenous cargo. We identified two additional axon-selective kinesins, a Kinesin-3 (KIF13A) and a Kinesin-4 (KIF21A). The role that these two kinesins may play in axonal transport has received little attention. Some of the non-selective kinesins identified in this report, including the Kinesin-3 family members KIF1A and KIF1Bβ, also mediate axonal transport based on biochemical, genetic, and imaging studies (42, 43, 48–50). It would be of interest to know whether there are differences in the efficiency of axonal transport mediated by axon-preferring versus non-selective kinesins.
The structural elements within kinesins that determine their translocation preferences
Our results show that kinesins with only subtle differences in sequence can have very different translocation selectivities. For example, KIF13A and KIF21A accumulate selectively at axon tips, whereas the highly homologous family members KIF13B and KIF21B accumulate nonselectively. By comparing the translocation preferences of chimeric constructs of the axon-selective KIF5C and the non-selective KIF1A, we established that Loop 12 and to a lesser extent Loop 8 are particularly important in conferring axonal selectivity in stage 4 hippocampal neurons. In structural models of kinesin-microtubule interactions, these two regions interact with the C-terminal of β-tubulin, which is the site for posttranslational modifications that can influence kinesin selectivity. Previous studies established that the positively charged insert in loop 12 of KIF1A, referred to as the K-loop, strongly enhances the affinity of this kinesin for microtubules in the weak binding state and increases its processivity in vitro (51). Adding the KIF1A Loop 12 to KIF5C disrupts its axonal selectivity, but replacing the K-loop from KIF1A with the corresponding region of KIF5C does not confer axon selectivity. KIF1B, another non-selective member of the Kinesin-3 family, also has a K-loop with 5 lysine residues, but the presence or absence of a K-loop alone cannot account for translocation preferences of other kinesins. For example, KIF3A (non-selective) and 3B (axonal) differ markedly in translocation preference, but both have a single lysine in loop 12; the axon-selective KIF13A and the non-selective KIF13B both have 3 lysines in loop 12. Future experiments will be needed to identify the kinesin domains that determine the translocation selectivity in the case of these kinesins. Nakata et al. also recently analyzed the translocation preferences of KIF1A/KIF5C chimeras early in neural development, at stage 2 or stage 3 (52). For the most part our results are similar except that in their experiments, replacing the loop11 region of KIF5C with the corresponding region from KIF1A resulted in non-selective accumulation. Future experiments will be required to determine whether this difference is due to the different developmental stages examined or to differences in the truncation constructs used (which included amino acids 1–381 in Nakata et al. (52) versus 1–559 in our study).
Our analysis of the heterodimeric Kinesin-2 motors demonstrates that sequences outside the motor domain also regulate the selectivity of the kinesin translocation. The dimerization domain of KIF3B confers axon selectivity whereas heterodimers that contain the KIF3C dimerization domain are non-selective. Although there is precedent for interactions between microtubules and sequences outside the motor domain--for example, a sequence in the neck coil of Kinesin-1 interacts with microtubules and increases the processivity of truncated Kinesin-1 in vitro (37)-- it is quite remarkable that sequences outside the motor domain play such a critical role in enabling kinesins to distinguish different populations of microtubules. Scanning other kinesins for additional microtubule-interacting segments may reveal more elements that contribute to selective translocation.
Developmental regulation of kinesin translocation
Perhaps the most unexpected finding from the present study concerns the differences among kinesins seen at early stages of neuronal development (summarized in Table I). From previous work (14), it was known that KIF1A accumulates at the tips of most neurites whereas KIF5C accumulates selectively at the tips of only one or two Stage 2 neurites. We identified several additional kinesins in each category. Surprisingly, we also identified several kinesins that fail to accumulate at any neurite tips in immature neurons, even though they accumulate robustly in mature neurons. This group of kinesins included KIF13A, KIF21A, and the heterodimeric kinesins KIF3A/3B and KIF3A/3C. The failure of these kinesins to accumulate is remarkable, given that Stage 2 neurites contain stable, plus-end out microtubules (11, 12, 25) and that extensive anterograde vesicle transport is observed in these neurites (4). Among the kinesins that fail to accumulate in immature neurons, some are axon-selective and some are non-selective when expressed in mature neurons. The inability of KIF13A and KIF21A to accumulate is particularly surprising since the closely related kinesins KIF13B and KIF21B accumulate in most or all Stage 2 neurites. The creation of motor domain chimeras between these pairs of kinesins could be used to determine the features required for efficient translocation in immature neurons.
It seems likely that the developmental changes in kinesin translocation reflect changes in the biochemical properties of microtubules that make them efficient kinesin substrates. Neuronal microtubules undergo profound developmental changes in posttranslational modification, in their population of microtubule-associated proteins, and in tubulin isoform composition (26, 27). Treatment of immature neurons with Taxol facilitated the accumulation of some kinesins, which is also consistent with the idea that microtubule properties are primarily responsible for developmental changes in kinesin translocation.
Verhey and Gaertig suggest that the heterogeneity of posttranslational modifications of tubulin can provide a ‘tubulin code’ for directing events along the microtubule polymer, analogous to the ‘histone code’ that spatially regulates transcriptional events along chromatin (53). Our results do not speak to the biochemical nature of the tubulin code, but they do indicate that the code is more complicated than previously appreciated. It is possible to distinguish four different groups of kinesins, based on their behavior in neurons at developmental Stage 2: some kinesin motor domains accumulate at nearly all neurite tips, some accumulate at only a subset of tips, some do not accumulate at any tips but can be induced to do so by Taxol treatment, and some do not accumulate at all. These findings imply that there must be at least four different biochemical features of microtubules that influence the efficiency of kinesin translocation. Identifying the microtubule features that govern kinesin translocation is a key challenge for future work.
MATERIALS AND METHODS
Truncated kinesin constructs
Table II provides details of the constructs used in this study. The constitutively active truncation constructs of KIF5B, KIF5C, and KIF17 are based on those described in previous reports (13, 14, 54). We prepared a KIF5A construct by truncating at a homologous site. The KIF3’s, also members of the Kinesin-2 family, are thought to function as heterodimers consisting of KIF3A/KIF3B or KIF3A/KIF3C. Based on in vitro interactions of synthetic peptide, Chana et al (20) proposed that the charged region immediate following the neck coil mediates heterodimer formation (20). We therefore created constructs of KIF3A, 3B, and 3C by truncating just after this charged region. Kinesin-3 family members (KIF1A, KIF1B, KIF1C, KIF13A, and KIF13B) all share a similar domain structure, including a predicted coiled-coil region just after the neck linker. This coiled-coil region is sufficient for dimerization of KIF1A (16). We therefore created constructs of KIF1B, 1C, 13A, and 13B kinesin that were truncated at a homologous position. Members of the Kinesin-4 family are thought to function as dimers, but the domains required for their dimerization have not been established experimentally. We prepared truncation constructs of KIF21A and 21B that included the first coiled-coil domain, which ends around amino acid 400. All of these truncation constructs were tagged at their C-termini with GFP or a color variant.
Table II.
Kinesin Constructs
| a. Wild- type truncation constructs:
| |||
|---|---|---|---|
| Kinesin | Amino acids | FP tag | Reference |
| KIF5A | 1–559 | eGFP | NP 032473 |
| KIF5B | 1–555 | YFP | NP 032474 |
| KIF5C | 1–388, 1–559 | eGFP | NP 001101200 |
| KIF3A | 1–419 | tdTomato | NP 032469 |
| KIF3B | 1–414 | YFP | NP 032470 |
| KIF3C | 1–441 | eGFP | NP 032471 |
| KIF17 | 1–511 | YFP | NP 034753 |
| KIF1A | 1–393 | tdTomato | XP 343631 |
| KIF1B | 1–386 | tagRFP | NP 476548 |
| KIF1C | 1–389 | eGFP | NP 006603 |
| KIF13A | 1–392, 1–435, 1–565, 1–639 | eGFP | NP 034747 |
| KIF13B | 1–393, 1–436, 1–568, 1–648 | eGFP | NP 001074646 |
| KIF21A | 1–417 | eGFP | NP 001102511 |
| KIF21B | 1–409 | eGFP | NP 001034561 |
| b. Chimeras with interchanged microtubule-binding regions
| |||
|---|---|---|---|
| Name | Deleted region | Inserted region | FP tag |
| KIF5C559L81A | KIF5C142–176 | KIF1A153–187 | eGFP |
| KIF5C559L111A | KIF5C238–253 | KIF1A254–269 | eGFP |
| KIF5C559L121A | KIF5C272–275 | KIF1A288–302 | eGFP |
| KIF1A393L85C | KIF1A153–187 | KIF5C142–176 | tdTomato |
| KIF1A393L115C | KIF1A254–269 | KIF5C238–253 | tdTomato |
| KIF1A393L125C | KIF1A288–302 | KIF5C272–275 | tdTomato |
| KIF3B414L113C | KIF3B253–262 | KIF3C254–289 | YFP |
| KIF3C441L113B | KIF3C254–289 | KIF3B253–262 | eGFP |
| c. Chimeras with interchanged dimerization domains
| |||
|---|---|---|---|
| Name | N-terminal (motor domain) | C-terminal (dime- rization domain) | FP tag |
| KIF3AMD3BHD | KIF3A1–361 | KIF3B357–414 | YFP |
| KIF3AMD3CHD | KIF3A1–357 | KIF3C380–441 | eGFP |
| KIF3BMD3AHD | KIF3B1–356 | KIF3A362–419 | tdTomato |
| KIF3BMD3CHD | KIF3B1–352 | KIF3C380–441 | eGFP |
| KIF3CMD3AHD | KIF3C1–379 | KIF3A358–419 | tdTomato |
| KIF3CMD3BHD | KIF3C1–379 | KIF3B353–414 | YFP |
| KIF1AMD5C559 | KIF1A1–352 | KIF5C326–559 | tdTomato |
cDNAs encoding KIF1A, KIF21A, and KIF21B were generously provided by Dr Bruce Schnapp and Dr. Lawrence Goldstein. The remaining truncated kinesins were generated using polymerase chain reaction from a mouse brain cDNA library (BioChain, Hayward, CA). Fragments were subcloned into plasmids for in-frame fusion to various fluorescent proteins and protein expression was driven by the CAG β-actin promoter (55). For the KIF13 dimerization assay, an ARGENT™ Regulated Homodimerization Kit (Fv) was obtained from ARIAD Pharmaceuticals (Cambridge, MA). Kinesins were tagged with eGFP, YFP, tagRFP2 (Evrogen, Moscow, Russia), tdTomato or mCherry (kindly provided by Dr. Roger Tsien). Unless otherwise indicated, tags were placed at the C-terminus. All constructs were verified by DNA sequencing.
Cell Culture and Transfection
Primary hippocampal cultures were prepared from embryonic day 18 rats and maintained in Minimal Essential Medium with N2 supplements, as described previously (56). To examine the translocation preferences of truncated kinesins in Stage 4 hippocampal neurons, constructs were transfected into 9–11 day-old neurons using Lipofectamine 2000 (Invitrogen). For most experiments, cultures were fixed 6–24 hrs after transfection and mounted in elvanol, as previously described (57). To examine the accumulation of truncated kinesins in the minor neurites of Stage2 neurons, truncated kinesin constructs were electroporated into dissociated hippocampal neurons prior to plating using a nucleofection protocol (Lonza) and fixed 22 to 26 hrs later. For live cell imaging, cells were maintained at 32–34°C in a heated chamber (Warner Instruments, Hamden, CT) containing imaging medium (Hanks BSS with Ca2+ and Mg2+, 0.6% Glucose, and 10mM HEPES) or Hibernate E without phenol red (BrainBits, Springfield, IL). When individual kinesins were expressed, cells were co-transfected with a soluble fluorescent protein in a complementary color.
Image Acquistion and Analysis
Images were acquired with a Zeiss Observer Z1 microscope and AxioVision software (Carl Zeiss, Gottingen, Germany) using LCI Plan-Neofluar 25×/ 0.8 or LCI Plan-Apochromat 40×/1.3 objectives and an Axiocam MRm camera or with a Leica DM RXA micfoscope (Leica Microsystems, Wetzlar, Germany) using MetaMorph software (Molecular Devices, Sunnyvale, CA) with a HCX PL APO 40×/ 1.25 and a Micromax CCD camera (Princeton Instruments, Trenton, NJ). Images illustrating the pattern of axonal and dendritic arborization (based on expression of soluble fluorescent proteins) were obtained by adjusting the gamma-values of soluble fill images in Photoshop to better illustrate the finest caliber neurites. Composite images were acquired using the MosaiX function of AxioVision and aligned using MetaMorph (montage function) or Photoshop.
Images were analyzed using MetaMorph software (Molecular Devices, Sunnyvale, CA). Background intensity values were subtracted from each image before quantification. Neurites were scored as positive for truncated kinesin accumulation if the fluorescence intensity at the tip was threefold higher than the fluorescence intensity of the shaft. To quantify the fluorescence intensity ratio of axon tips vs cell body, image thresholds were set to exclude pixels that dkid not fall over the cell body or neurites, then the average fluorescence intensity of each region was obtained. For each cell, the average intensity value from 4 axon tips was compared to the intensity value from the cell body
Supplementary Material
Figure S1. The accumulation of truncated KIF1A at neurite tips depends on its dimerization and microtubule-stimulated ATPase activity (A) KIF1A393 tdTomato accumulated at the tips of axons and dendrites, which were visualized by co-expression of GFP. Higher magnification images of the boxed regions reveal that the KIF1A393 tdTomato (red) is highly enriched at dendritic tips, compared with soluble GFP (green). Axonal tips are indicated by small circles; unlabeled tips are dendritic. (B) KIF1A393T312M-tdTomato, a construct with point mutation that alters MT-binding and ATPase cycles, does not accumulate at neurite tips. It is diffusely distributed throughout the cell. (C) KIF1A366-GFP, which does not dimerize, also fails to accumulate at neurite tips. Boxes enclose dendritic regions and circles indicate axonal tips. Scale bars: 20μm. The image shown in C is a montage of four microscopic fields prepared as described in Materials and Methods
Figure S2: Kinesins differ in the efficiency of their accumulation at neurite tips. The ratio of fluorescence intensity at axon tips versus the cell body was measured to assess the accumulation efficiency of each truncated kinesin construct shown in Fig.2. For soluble GFP, which is evenly distributed throughout the cell, the tip-to-soma ratio averaged 0.05 (reflecting the much greater thickness of the cell body compared with the axon tip). KIF5B, 5C, 1A, and 1B accumulated with high efficiency (>200 times greater than that of soluble fill), whereas KIF5A, KIF17, KIF21A and 21B were comparatively less efficient (with axonal tip concentrations 20 to 60 times greater than that of GFP). Images of neurons expressing each truncated kinesin were analyzed as described in Materials and Methods. Between 10 and 22 cells were analyzed for each construct. Error bars show SEM.
Figure S3: Truncated KIF3 constructs form heterodimers when expressed in rat embryo fibroblasts. Western blot analyses revealed that immunoprecipitation with antibodies that recognize KIF3B414-YFP or KIF3C441-GFP co-precipitated KIF3A419-tdTomato. REF52 cells were transfected with KIF3A419-tdTomato together with either KIF3B414-YFP, KIF3C441-GFP, or soluble GFP (as a control). Cell lysates were collected 48 h after trasnfection and subjected to immunoprecipitation using rabbit α-GFP antibody (Invitrogen) and Protein G Sepharose 4 Fast Flow (GE Health Care). Total lysates and immunoprcipitates were subjected to western blot analysis using chicken anti-GFP antibody (Aves Labs) and rabbit anti-RFP antibody (Rockland) and bands were detected using the Odyssey infrared fluorescent imaging system (LI-COR).
Figure S4: Sequences within both the motor and stalk domains influence the accumulation of KIF5C. (A) Replacing four amino acids within the loop 8 region of KIF5C with the corresponding residues from KIF1A markedly inhibited its ability to accumulate; most of the expressed motor was distributed diffusely throughout the cell. (B, C) Elements within the stalk region of KIF5C influence the pattern of accumulation of chimeric constructs that accumulate in dendrites. A construct consisting of the motor domain of KIF1A and the stalk of KIF5 (KIF1AMD5C326–559) labeled tiny spots at dendritic tips and the tips of filopodia rather than the dendritic shaft proper (compare with KIF1A393, Fig. S1A and Fig. 2E). A shorter KIF5C chimera that includes Loop 12 from KIF1A and ends at residue 388 (KIF5C388 L121A) labeled the distal dendritic shaft and failed to label filopodial tips. Truncation constructs of chimeras between KIF5C/KIF1A chimeras were expressed in stage 4 neurons. The upper panels illustrate cell morphology, based on signals from co-expressed soluble fluorescent protein, while the middle panels show the distribution of truncated kinesins. In the lower panels, which show enlargements of the boxed regions which contain multiple dendritic tips, the kinesin is shown in red and the soluble fill in green. Scale bar=20μm. Images shown in panels A and B are montages of four microscopic fields prepared as described in Materials and Methods.
Acknowledgments
This work was supported by NIH grant MH066179. Live-cell imaging was performed at the Advanced Light Microscopy Core @ The Jungers Center, which is supported in part by NIH P30-NS06180 (Aicher, P.I.). We thank Julie Luisi and Barbara Smoody for their outstanding technical assistance, Dr. Stefanie Kaech for her advice on imaging and image analysis, Dr. Bruce Schnapp for his comments on the manuscript, and members of the Banker lab for their helpful advice throughout this project.
References
- 1.Burack MA, Silverman MA, Banker G. The role of selective transport in neuronal protein sorting. Neuron. 2000;26(2):465–472. doi: 10.1016/s0896-6273(00)81178-2. [DOI] [PubMed] [Google Scholar]
- 2.Silverman MA, Kaech S, Jareb M, Burack MA, Vogt L, Sonderegger P, Banker G. Sorting and directed transport of membrane proteins during development of hippocampal neurons in culture. Proc Natl Acad Sci U S A. 2001;98(13):7051–7057. doi: 10.1073/pnas.111146198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirokawa N, Noda Y. Intracellular transport and kinesin superfamily proteins, KIFs: structure, function, and dynamics. Physiol Rev. 2008;88(3):1089–1118. doi: 10.1152/physrev.00023.2007. [DOI] [PubMed] [Google Scholar]
- 4.Silverman MA, Kaech S, Ramser EM, Lu X, Lasarev MR, Nagalla S, Banker G. Expression of kinesin superfamily genes in cultured hippocampal neurons. Cytoskeleton (Hoboken) 2010;67(12):784–795. doi: 10.1002/cm.20487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larcher JC, Boucher D, Lazereg S, Gros F, Denoulet P. Interaction of kinesin motor domains with alpha- and beta-tubulin subunits at a tau-independent binding site. Regulation by polyglutamylation. J Biol Chem. 1996;271(36):22117–22124. doi: 10.1074/jbc.271.36.22117. [DOI] [PubMed] [Google Scholar]
- 6.Liao G, Gundersen GG. Kinesin is a candidate for cross-bridging microtubules and intermediate filaments. Selective binding of kinesin to detyrosinated tubulin and vimentin. J Biol Chem. 1998;273(16):9797–9803. doi: 10.1074/jbc.273.16.9797. [DOI] [PubMed] [Google Scholar]
- 7.Reed NA, Cai D, Blasius TL, Jih GT, Meyhofer E, Gaertig J, Verhey KJ. Microtubule acetylation promotes kinesin-1 binding and transport. Curr Biol. 2006;16(21):2166–2172. doi: 10.1016/j.cub.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 8.Dunn S, Morrison EE, Liverpool TB, Molina-Paris C, Cross RA, Alonso MC, Peckham M. Differential trafficking of Kif5c on tyrosinated and detyrosinated microtubules in live cells. J Cell Sci. 2008;121(Pt 7):1085–1095. doi: 10.1242/jcs.026492. [DOI] [PubMed] [Google Scholar]
- 9.Konishi Y, Setou M. Tubulin tyrosination navigates the kinesin-1 motor domain to axons. Nat Neurosci. 2009;12(5):559–567. doi: 10.1038/nn.2314. [DOI] [PubMed] [Google Scholar]
- 10.Setou M, Seog DH, Tanaka Y, Kanai Y, Takei Y, Kawagishi M, Hirokawa N. Glutamate-receptor-interacting protein GRIP1 directly steers kinesin to dendrites. Nature. 2002;417(6884):83–87. doi: 10.1038/nature743. [DOI] [PubMed] [Google Scholar]
- 11.Baas PW, Deitch JS, Black MM, Banker GA. Polarity orientation of microtubules in hippocampal neurons: uniformity in the axon and nonuniformity in the dendrite. Proc Natl Acad Sci U S A. 1988;85(21):8335–8339. doi: 10.1073/pnas.85.21.8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baas PW, Black MM, Banker GA. Changes in microtubule polarity orientation during the development of hippocampal neurons in culture. J Cell Biol. 1989;109(6 Pt 1):3085–3094. doi: 10.1083/jcb.109.6.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakata T, Hirokawa N. Microtubules provide directional cues for polarized axonal transport through interaction with kinesin motor head. J Cell Biol. 2003;162(6):1045–1055. doi: 10.1083/jcb.200302175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobson C, Schnapp B, Banker GA. A change in the selective translocation of the Kinesin-1 motor domain marks the initial specification of the axon. Neuron. 2006;49(6):797–804. doi: 10.1016/j.neuron.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Friedman DS, Vale RD. Single-molecule analysis of kinesin motility reveals regulation by the cargo-binding tail domain. Nat Cell Biol. 1999;1(5):293–297. doi: 10.1038/13008. [DOI] [PubMed] [Google Scholar]
- 16.Hammond JW, Cai D, Blasius TL, Li Z, Jiang Y, Jih GT, Meyhofer E, Verhey KJ. Mammalian Kinesin-3 motors are dimeric in vivo and move by processive motility upon release of autoinhibition. PLoS Biol. 2009;7(3):e72. doi: 10.1371/journal.pbio.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dotti CG, Sullivan CA, Banker GA. The establishment of polarity by hippocampal neurons in culture. J Neurosci. 1988;8(4):1454–1468. doi: 10.1523/JNEUROSCI.08-04-01454.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huckaba TM, Gennerich A, Wilhelm JE, Chishti AH, Vale RD. Kinesin-73 Is a Processive Motor That Localizes to Rab5-containing Organelles. J Biol Chem. 2011;286(9):7457–7467. doi: 10.1074/jbc.M110.167023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomishige M, Klopfenstein DR, Vale RD. Conversion of Unc104/KIF1A kinesin into a processive motor after dimerization. Science. 2002;297(5590):2263–2267. doi: 10.1126/science.1073386. [DOI] [PubMed] [Google Scholar]
- 20.Chana MS, Tripet BP, Mant CT, Hodges R. Stability and specificity of heterodimer formation for the coiled-coil neck regions of the motor proteins Kif3A and Kif3B: the role of unstructured oppositely charged regions. J Pept Res. 2005;65(2):209–220. doi: 10.1111/j.1399-3011.2005.00210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Marco V, De Marco A, Goldie KN, Correia JJ, Hoenger A. Dimerization properties of a Xenopus laevis kinesin-II carboxy-terminal stalk fragment. EMBO Rep. 2003;4(7):717–722. doi: 10.1038/sj.embor.embor884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vukajlovic M, Dietz H, Schliwa M, Okten Z. How kinesin-2 forms a stalk. Mol Biol Cell. 2011;22(22):4279–4287. doi: 10.1091/mbc.E11-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammond JW, Huang CF, Kaech S, Jacobson C, Banker G, Verhey KJ. Posttranslational modifications of tubulin and the polarized transport of kinesin-1 in neurons. Mol Biol Cell. 2010;21(4):572–583. doi: 10.1091/mbc.E09-01-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaech S, Huang CF, Banker G. Live imaging of developing hippocampal neurons in culture. In: Wong R, Sharpe J, editors. Imaging in Developmental Biology: A Laboratory Manual. Woodbury, NY: Cold Spring Harbor Laboratory Press; 2011. pp. 449–467. [Google Scholar]
- 25.Dotti CG, Banker G. Intracellular organization of hippocampal neurons during the development of neuronal polarity. J Cell Sci Suppl. 1991;15:75–84. doi: 10.1242/jcs.1991.supplement_15.11. [DOI] [PubMed] [Google Scholar]
- 26.Janke C, Kneussel M. Tubulin post-translational modifications: encoding functions on the neuronal microtubule cytoskeleton. Trends Neurosci. 2010;33(8):362–372. doi: 10.1016/j.tins.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 27.Poulain FE, Sobel A. The microtubule network and neuronal morphogenesis: Dynamic and coordinated orchestration through multiple players. Mol Cell Neurosci. 2010;43(1):15–32. doi: 10.1016/j.mcn.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 28.Hammond JW, Blasius TL, Soppina V, Cai D, Verhey KJ. Autoinhibition of the kinesin-2 motor KIF17 via dual intramolecular mechanisms. J Cell Biol. 2010;189(6):1013–1025. doi: 10.1083/jcb.201001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kikkawa M, Okada Y, Hirokawa N. 15 A resolution model of the monomeric kinesin motor, KIF1A. Cell. 2000;100(2):241–252. doi: 10.1016/s0092-8674(00)81562-7. [DOI] [PubMed] [Google Scholar]
- 30.Sindelar CV, Budny MJ, Rice S, Naber N, Fletterick R, Cooke R. Two conformations in the human kinesin power stroke defined by X-ray crystallography and EPR spectroscopy. Nat Struct Biol. 2002;9(11):844–848. doi: 10.1038/nsb852. [DOI] [PubMed] [Google Scholar]
- 31.Kikkawa M, Hirokawa N. High-resolution cryo-EM maps show the nucleotide binding pocket of KIF1A in open and closed conformations. EMBO J. 2006;25(18):4187–4194. doi: 10.1038/sj.emboj.7601299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nitta R, Okada Y, Hirokawa N. Structural model for strain-dependent microtubule activation of Mg-ADP release from kinesin. Nat Struct Mol Biol. 2008;15(10):1067–1075. doi: 10.1038/nsmb.1487. [DOI] [PubMed] [Google Scholar]
- 33.Woehlke G, Ruby AK, Hart CL, Ly B, Hom-Booher N, Vale RD. Microtubule interaction site of the kinesin motor. Cell. 1997;90(2):207–216. doi: 10.1016/s0092-8674(00)80329-3. [DOI] [PubMed] [Google Scholar]
- 34.Morfini GA, You YM, Pollema SL, Kaminska A, Liu K, Yoshioka K, Bjorkblom B, Coffey ET, Bagnato C, Han D, Huang CF, Banker G, Pigino G, Brady ST. Pathogenic huntingtin inhibits fast axonal transport by activating JNK3 and phosphorylating kinesin. Nat Neurosci. 2009;12(7):864–871. doi: 10.1038/nn.2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chana M, Tripet BP, Mant CT, Hodges RS. The role of unstructured highly charged regions on the stability and specificity of dimerization of two-stranded alpha-helical coiled-coils: analysis of the neck-hinge region of the kinesin-like motor protein Kif3A. J Struct Biol. 2002;137(1–2):206–219. doi: 10.1006/jsbi.2002.4446. [DOI] [PubMed] [Google Scholar]
- 36.Rashid DJ, Wedaman KP, Scholey JM. Heterodimerization of the two motor subunits of the heterotrimeric kinesin, KRP85/95. J Mol Biol. 1995;252(2):157–162. doi: 10.1006/jmbi.1995.0484. [DOI] [PubMed] [Google Scholar]
- 37.Thorn KS, Ubersax JA, Vale RD. Engineering the processive run length of the kinesin motor. J Cell Biol. 2000;151(5):1093–1100. doi: 10.1083/jcb.151.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song AH, Wang D, Chen G, Li Y, Luo J, Duan S, Poo MM. A selective filter for cytoplasmic transport at the axon initial segment. Cell. 2009;136(6):1148–1160. doi: 10.1016/j.cell.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 39.Lewis TL, Jr, Mao T, Svoboda K, Arnold DB. Myosin-dependent targeting of transmembrane proteins to neuronal dendrites. Nat Neurosci. 2009;12(5):568–576. doi: 10.1038/nn.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kapitein LC, Schlager MA, Kuijpers M, Wulf PS, van Spronsen M, MacKintosh FC, Hoogenraad CC. Mixed microtubules steer dynein-driven cargo transport into dendrites. Curr Biol. 2010;20(4):290–299. doi: 10.1016/j.cub.2009.12.052. [DOI] [PubMed] [Google Scholar]
- 41.Hirokawa N, Takemura R. Molecular motors and mechanisms of directional transport in neurons. Nat Rev Neurosci. 2005;6(3):201–214. doi: 10.1038/nrn1624. [DOI] [PubMed] [Google Scholar]
- 42.Niwa S, Tanaka Y, Hirokawa N. KIF1Bbeta- and KIF1A-mediated axonal transport of presynaptic regulator Rab3 occurs in a GTP-dependent manner through DENN/MADD. Nat Cell Biol. 2008;10(11):1269–1279. doi: 10.1038/ncb1785. [DOI] [PubMed] [Google Scholar]
- 43.Okada Y, Yamazaki H, Sekine-Aizawa Y, Hirokawa N. The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell. 1995;81(5):769–780. doi: 10.1016/0092-8674(95)90538-3. [DOI] [PubMed] [Google Scholar]
- 44.Takeda S, Yamazaki H, Seog DH, Kanai Y, Terada S, Hirokawa N. Kinesin superfamily protein 3 (KIF3) motor transports fodrin-associating vesicles important for neurite building. J Cell Biol. 2000;148(6):1255–1265. doi: 10.1083/jcb.148.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xia CH, Roberts EA, Her LS, Liu X, Williams DS, Cleveland DW, Goldstein LS. Abnormal neurofilament transport caused by targeted disruption of neuronal kinesin heavy chain KIF5A. J Cell Biol. 2003;161(1):55–66. doi: 10.1083/jcb.200301026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chu PJ, Rivera JF, Arnold DB. A role for Kif17 in transport of Kv4.2. J Biol Chem. 2006;281(1):365–373. doi: 10.1074/jbc.M508897200. [DOI] [PubMed] [Google Scholar]
- 47.Setou M, Nakagawa T, Seog DH, Hirokawa N. Kinesin superfamily motor protein KIF17 and mLin-10 in NMDA receptor-containing vesicle transport. Science. 2000;288(5472):1796–1802. doi: 10.1126/science.288.5472.1796. [DOI] [PubMed] [Google Scholar]
- 48.Lee JR, Shin H, Ko J, Choi J, Lee H, Kim E. Characterization of the movement of the kinesin motor KIF1A in living cultured neurons. J Biol Chem. 2003;278(4):2624–2629. doi: 10.1074/jbc.M211152200. [DOI] [PubMed] [Google Scholar]
- 49.Wagner OI, Esposito A, Kohler B, Chen CW, Shen CP, Wu GH, Butkevich E, Mandalapu S, Wenzel D, Wouters FS, Klopfenstein DR. Synaptic scaffolding protein SYD-2 clusters and activates kinesin-3 UNC-104 in C. elegans. Proc Natl Acad Sci U S A. 2009;106(46):19605–19610. doi: 10.1073/pnas.0902949106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lo KY, Kuzmin A, Unger SM, Petersen JD, Silverman MA. KIF1A is the primary anterograde motor protein required for the axonal transport of dense-core vesicles in cultured hippocampal neurons. Neurosci Lett. 2011;491(3):168–173. doi: 10.1016/j.neulet.2011.01.018. [DOI] [PubMed] [Google Scholar]
- 51.Okada Y, Hirokawa N. Mechanism of the single-headed processivity: diffusional anchoring between the K-loop of kinesin and the C terminus of tubulin. Proc Natl Acad Sci U S A. 2000;97(2):640–645. doi: 10.1073/pnas.97.2.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakata T, Niwa S, Okada Y, Perez F, Hirokawa N. Preferential binding of a kinesin-1 motor to GTP-tubulin-rich microtubules underlies polarized vesicle transport. J Cell Biol. 2011;194(2):245–255. doi: 10.1083/jcb.201104034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verhey KJ, Gaertig J. The tubulin code. Cell Cycle. 2007;6(17):2152–2160. doi: 10.4161/cc.6.17.4633. [DOI] [PubMed] [Google Scholar]
- 54.Vale RD, Funatsu T, Pierce DW, Romberg L, Harada Y, Yanagida T. Direct observation of single kinesin molecules moving along microtubules. Nature. 1996;380(6573):451–453. doi: 10.1038/380451a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108(2):193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 56.Kaech S, Banker G. Culturing hippocampal neurons. Nat Protoc. 2006;1(5):2406–2415. doi: 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
- 57.Goslin K, Asmussen A, Banker G. Rat Hippocampal Neurons in Low-density Culture. In: Banker G, Goslin K, editors. Culturing Nerve cells. 2. Cambridge, MA: The MIT Press; 1998. pp. 339–370. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The accumulation of truncated KIF1A at neurite tips depends on its dimerization and microtubule-stimulated ATPase activity (A) KIF1A393 tdTomato accumulated at the tips of axons and dendrites, which were visualized by co-expression of GFP. Higher magnification images of the boxed regions reveal that the KIF1A393 tdTomato (red) is highly enriched at dendritic tips, compared with soluble GFP (green). Axonal tips are indicated by small circles; unlabeled tips are dendritic. (B) KIF1A393T312M-tdTomato, a construct with point mutation that alters MT-binding and ATPase cycles, does not accumulate at neurite tips. It is diffusely distributed throughout the cell. (C) KIF1A366-GFP, which does not dimerize, also fails to accumulate at neurite tips. Boxes enclose dendritic regions and circles indicate axonal tips. Scale bars: 20μm. The image shown in C is a montage of four microscopic fields prepared as described in Materials and Methods
Figure S2: Kinesins differ in the efficiency of their accumulation at neurite tips. The ratio of fluorescence intensity at axon tips versus the cell body was measured to assess the accumulation efficiency of each truncated kinesin construct shown in Fig.2. For soluble GFP, which is evenly distributed throughout the cell, the tip-to-soma ratio averaged 0.05 (reflecting the much greater thickness of the cell body compared with the axon tip). KIF5B, 5C, 1A, and 1B accumulated with high efficiency (>200 times greater than that of soluble fill), whereas KIF5A, KIF17, KIF21A and 21B were comparatively less efficient (with axonal tip concentrations 20 to 60 times greater than that of GFP). Images of neurons expressing each truncated kinesin were analyzed as described in Materials and Methods. Between 10 and 22 cells were analyzed for each construct. Error bars show SEM.
Figure S3: Truncated KIF3 constructs form heterodimers when expressed in rat embryo fibroblasts. Western blot analyses revealed that immunoprecipitation with antibodies that recognize KIF3B414-YFP or KIF3C441-GFP co-precipitated KIF3A419-tdTomato. REF52 cells were transfected with KIF3A419-tdTomato together with either KIF3B414-YFP, KIF3C441-GFP, or soluble GFP (as a control). Cell lysates were collected 48 h after trasnfection and subjected to immunoprecipitation using rabbit α-GFP antibody (Invitrogen) and Protein G Sepharose 4 Fast Flow (GE Health Care). Total lysates and immunoprcipitates were subjected to western blot analysis using chicken anti-GFP antibody (Aves Labs) and rabbit anti-RFP antibody (Rockland) and bands were detected using the Odyssey infrared fluorescent imaging system (LI-COR).
Figure S4: Sequences within both the motor and stalk domains influence the accumulation of KIF5C. (A) Replacing four amino acids within the loop 8 region of KIF5C with the corresponding residues from KIF1A markedly inhibited its ability to accumulate; most of the expressed motor was distributed diffusely throughout the cell. (B, C) Elements within the stalk region of KIF5C influence the pattern of accumulation of chimeric constructs that accumulate in dendrites. A construct consisting of the motor domain of KIF1A and the stalk of KIF5 (KIF1AMD5C326–559) labeled tiny spots at dendritic tips and the tips of filopodia rather than the dendritic shaft proper (compare with KIF1A393, Fig. S1A and Fig. 2E). A shorter KIF5C chimera that includes Loop 12 from KIF1A and ends at residue 388 (KIF5C388 L121A) labeled the distal dendritic shaft and failed to label filopodial tips. Truncation constructs of chimeras between KIF5C/KIF1A chimeras were expressed in stage 4 neurons. The upper panels illustrate cell morphology, based on signals from co-expressed soluble fluorescent protein, while the middle panels show the distribution of truncated kinesins. In the lower panels, which show enlargements of the boxed regions which contain multiple dendritic tips, the kinesin is shown in red and the soluble fill in green. Scale bar=20μm. Images shown in panels A and B are montages of four microscopic fields prepared as described in Materials and Methods.