Abstract
The consumption of insects by apes has previously been reported based on direct observations and/or trail signs in feces. However, DNA-based diet analyses may have the potential to reveal trophic links for these wild species. Herein, we analyzed the insect-diet diversity of 9 feces obtained from three species of African great apes, gorilla (Gorilla gorilla gorilla), chimpanzee (Pan troglodytes) and bonobo (Pan paniscus), using two mitochondrial amplifications for arthropods. A total of 1056 clones were sequenced for Cyt-b and COI gene libraries, which contained 50 and 56 operational taxonomic units (OTUs), respectively. BLAST research revealed that the OTUs belonged to 32 families from 5 orders (Diptera, Isoptera, Lepidoptera, Coleoptera, and Orthoptera). While ants were not detected by this method, the consumption of flies, beetles, moths, mosquitoes and termites was evident in these samples. Our findings indicate that molecular techniques can be used to analyze insect food items in wild animals.
Investigating the diets of primate populations elucidates their behavioral ecology and evolution and clarifies dietary differences among the same species in different habitats and among different species in shared habitats1. The consumption of insects is widespread among nonhuman primates, and their entomophagy depends on their body size. Small primates such as galagos (Galago crassicaudatus and Galago senegalensis), pottos (Perodicticus potto), and tarsiers (Tarsius spectrum) are obligate insect eaters; their diet is composed mainly of insects belonging to the orders Lepidoptera, Orthoptera and Hymenoptera2,3,4. The diets of medium-bodied primates such as red colobus monkeys (Procolobus tephrosceles) and blue monkey (Cercopithecus mitis) consist mainly of young leaves, flowers, and unripe fruit; they are also known to eat insects less frequently than small primates5,6. Although the large-bodied great apes have a high frugivory7,8,9,10,11 and folivory component to their diet12,13, there is also an insectivorous component in the diet of gorilla, chimpanzee, bonobo, orangutan and gibbon populations14,15,16,17,18,19,20.
Insects comprise 0.1–1% of the fresh weight of the daily food ingested by both gorillas and chimpanzees21, but they may provide disproportionate nutritional benefits due to their role as protein sources. Insects are particularly valuable because they provide certain amino acids, vitamins (such as B12) and minerals (including iron (Fe) and manganese) that may be absent in plant foods17,22,23,24.
The insect prey items vary among gorilla species living in different habitats. In mountain gorillas (Gorilla beringei beringei), insectivory is uncommon18,25,26, but they feed on ants (Dorillus spp.) to a variable degree18,26. In contrast, Grauer's gorillas (Gorilla beringei graueri) consume ants more regularly than mountain gorillas, perhaps because the insects are more prevalent27. Furthermore, western gorillas (Gorilla gorilla gorilla) feed on insects, indicating that insects are an important aspect of the diet of these gorillas10,14,16,17,28,29.
There is considerable temporal variation in insect consumption among different chimpanzee populations. Both forest-inhabiting chimpanzees in central Africa and savannah-dwelling chimpanzees in western Africa consume insects, such as termites, throughout the year21,30,31,32. In East Africa and at Mt. Assirik in Senegal, chimpanzees feed on termites only seasonally33. There is not much variation in insect consumption by different chimpanzee groups. Generally, chimpanzees eat ants, termites, bees, wasps, caterpillars and beetle grubs29,34,35. In contrast, arthropods comprise a small fraction of the diet of wild bonobos. The most important non-plant food sources for bonobos are invertebrates, including larvae, termites, ants, bees, earthworms and millipedes36,37,38,39. Insectivory is also reported for orangutans; especially during times of fruit scarcity40, orangutans start to prey on insects including termites, ants, bees, gall wasps, crickets, caterpillars and bush crickets19,41,42.Wild Sumatran orangutans (Pongo pygmaeus abelii) make and use tools (twigs, sticks and branches) for extractive insect foraging41. Very little data is available on the diet of gibbons, although fruit is their main food source, which is supplemented with some arthropods including caterpillars and termites20,43.
The studies cited above are based either on direct observations of feeding apes or morphological classification of insect remains in ape feces. Another avenue for investigation of insectivory is the study of feeding remains left by apes (e.g. to identify insect species eaten from a recently disturbed ant nest or encountered termite mound). Although these classical methods can provide direct records for both qualitative and quantitative behavior (i.e. insect prey choice and frequency of insect consumption) and they are inexpensive. However, direct observations of insects consumed by apes are limited in each individual trial by difficulties in tracking apes in their native habitat; this approach can be time-consuming and therefore impractical for studying insectivory in non-human primates44. Furthermore, the damage to insect bodies and distortion of their morphology by the mechanical action of the digestive tract creates major challenges for direct analyses of insects in animal feces44. Alternative molecular methods may provide better identification and global detection of insects in primates feces compared with these classical methods and may also elucidate the feeding behavior of primates45,46.
The first PCR-based method for detecting animal prey in primate feces was used by Hofreiter et al.46, who detected fragments of vertebrate mitochondrial DNA in fecal samples of gorillas and bonobos. In a metagenomic study with primers targeting the arthropod cytochrome b (Cyt-b) gene, Pickett et al.45 investigated the insect diets of six sympatric New World monkeys living in a western Amazon rainforest. To date, molecular analyses of great ape insectivores using high throughput sequencing have not been reported. However, using this technique to target the cytochrome c oxidase I (COI) fragment is effective for studying the arthropod diet of bats and, thus, could be applicable to other insectivores47,48.
In this study, we evaluated the insect-diet diversity of three African great apes gorilla, chimpanzee and bonobo by analyzing fecal samples (N = 9) with DNA-barcoding primers targeting the Cyt-b49 and COI50 genes in arthropod mitochondrial genomes.
Results
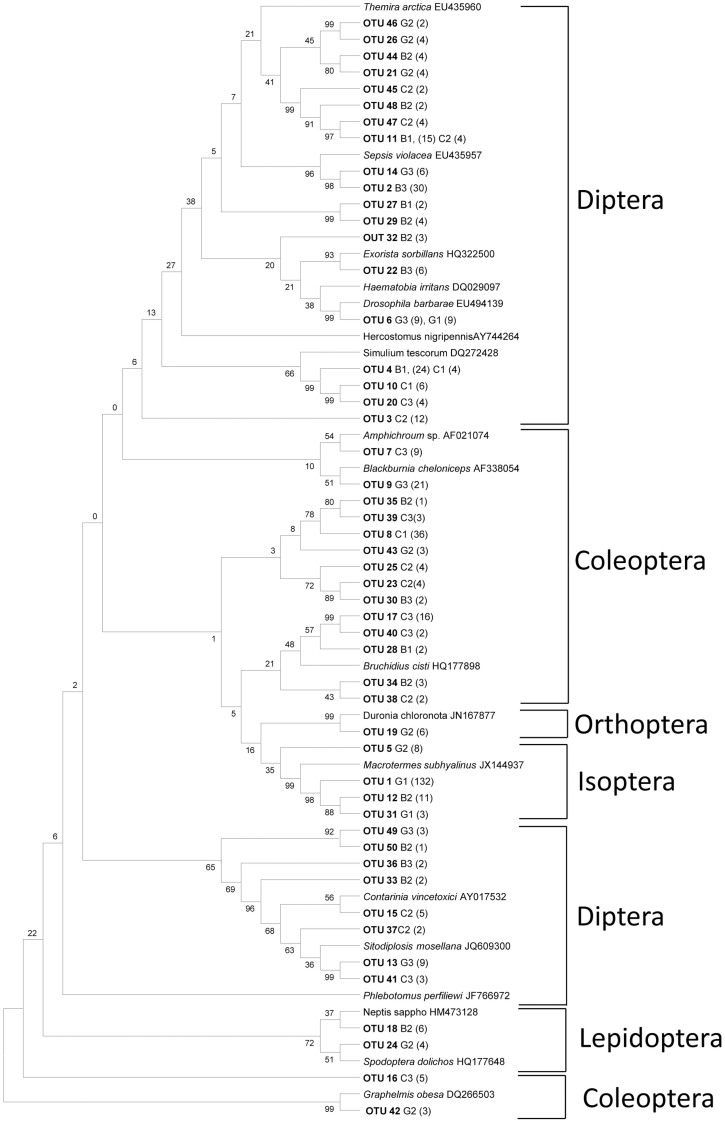
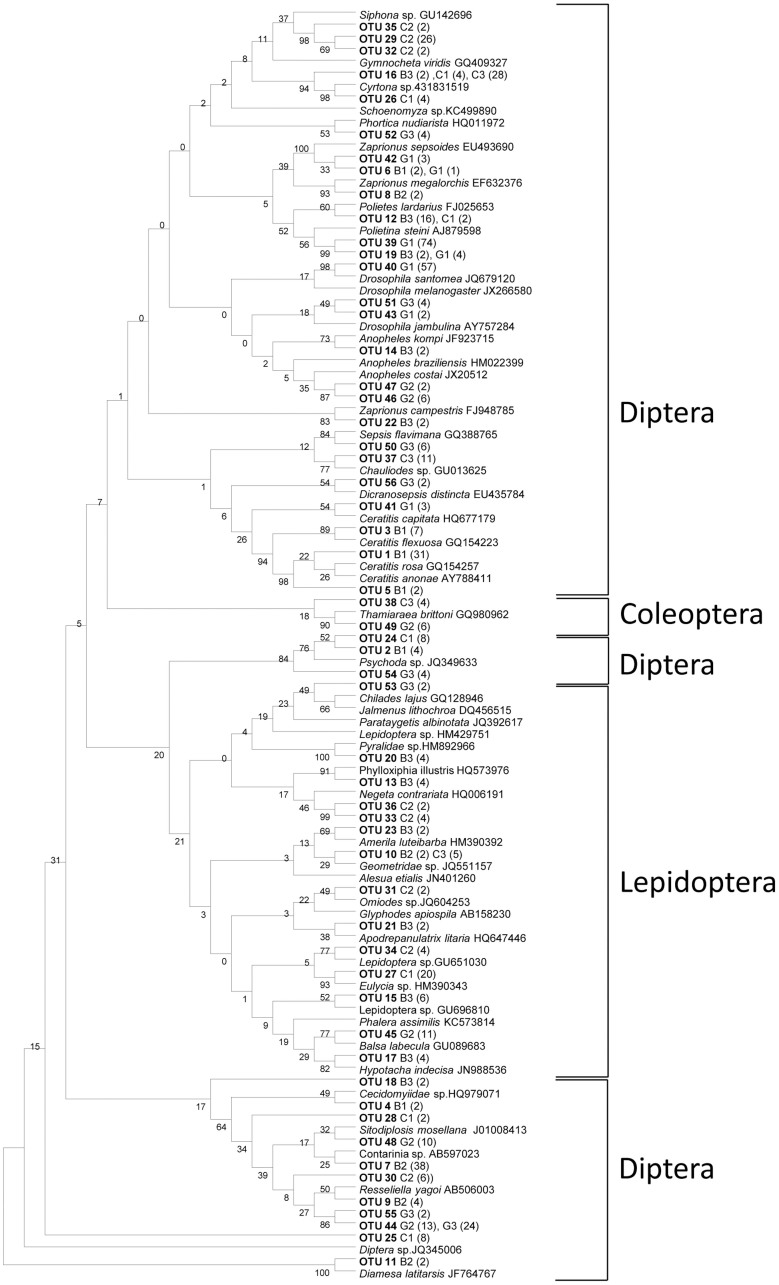
A total of 1056 clones were collected from the cloning libraries generated by using both primer sets. First, 24 clones per sample and per amplification were sequenced and analyzed. No additional operational taxonomic units (OTUs)51 were found when we sequenced 24 additional clones from each sample. Moreover, in one sample (G1), sequencing 144 clones did not increase the number of OTUs that were obtained from the first 24 sequenced clones. Finally, after examining the 1056 clones, 1006 clones presented good quality sequences that did not contain termination codons within the translated sequences. Analyses of these 1006 clones resulted in the detection of 106 different insect OTUs in the 9 fecal samples from bonobos (B1, B2 and B3), chimpanzees (C1, C2 and C3) and gorillas (G1, G2 and G3) with both Cyt-b and COI genes (Fig. 1 and 2).
Figure 1. Insect OTUs detected in African great ape fecal samples using CB3/CB4 primers targeting the Cyt-b gene.
The neighbor-joining tree was constructed using MEGA 5 and supported by 500 bootstrap replicates. The insect species listed with accession numbers are the closest matching sequences from GenBank. G1, G2 and G3: fecal samples from gorillas; C1, C2 and C3: chimpanzees; B1, B2 and B3: bonobos.
Figure 2. Insect OTUs detected in African great fecal samples using ZBJ-ArtF1c/ZBJ-ArtR2c primers targeting the COI gene.
The neighbor-joining tree was constructed using MEGA 5 and supported by 500 bootstrap replicates. The insect species listed with accession numbers are the closest matching sequences from GenBank. G1, G2 and G3: the fecal samples from gorillas; C1, C2 and C3: chimpanzees; B1, B2 and B3: bonobos.
Analysis of the Cyt-b gene revealed 18 OTUs from insects belonging to 12 families in 4 orders (Isoptera, Diptera, Lepidoptera and Coleoptera) in the fecal samples from bonobos (Fig. 1 and Table 1). Nineteen OTUs belonging to 10 families from Diptera and Coleoptera and 16 OTUs belonging to 11 families from 5 orders (Diptera, Isoptera, Lepidoptera, Coleoptera and Orthoptera) were identified in the feces of chimpanzees and gorillas, respectively, with the same primer set (Fig. 1 and Table 1).
Table 1. Insect OTUs detected in 9 fecal samples from wild African great apes using Cyt-b targeting primers.
| Ape | Sample | OTU (No. of Clones) | Order | Family | Common name |
|---|---|---|---|---|---|
| Bonobo | B1 | 4 (24) | Diptera | Simuliidae | Flies (Black flies) |
| Bonobo | B1 | 11 (15) | Diptera | Sepsidae | Flies (Ensign flies) |
| Bonobo | B1 | 27 (2) | Diptera | Tephritidae | Flies (Fruit flies) |
| Bonobo | B1 | 28 (2) | Coleoptera | Chrysomelidae | Beetles (Leaf beetles) |
| Bonobo | B2 | 12 (11) | Isoptera | Termitidae | Termite |
| Bonobo | B2 | 18 (11) | Lepidoptera | Nymphalidae | Butterflies (Brush-footed butterflies) |
| Bonobo | B2 | 29 (4) | Diptera | Tephritidae | Flies (Fruit flies) |
| Bonobo | B2 | 32 (3) | Diptera | Drosophilidae | Flies (Fruit flies) |
| Bonobo | B2 | 44 (4) | Diptera | Muscidae | Flies (House flies) |
| Bonobo | B2 | 48 (2) | Diptera | Sepsidae | Flies (Ensign flies) |
| Bonobo | B2 | 50 (1) | Diptera | Fanniidae | Flies (True Flies) |
| Bonobo | B2 | 33 (2) | Diptera | Cecidomyiidae | Flies (Gall gnats) |
| Bonobo | B2 | 34 (3) | Coleoptera | Chrysomelidae | Beetles (Leaf beetles) |
| Bonobo | B2 | 35 (1) | Coleoptera | Carabidae | Beetles |
| Bonobo | B3 | 2 (30) | Diptera | Sepsidae | Flies (Ensign flies) |
| Bonobo | B3 | 22 (6) | Diptera | Muscidae | Flies (House flies) |
| Bonobo | B3 | 36 (2) | Diptera | Cecidomyiidae | Flies (Gall gnats) |
| Bonobo | B3 | 30 (2) | Coleoptera | Staphylinidae | Beetles (Rove beetles) |
| Chimpanzee | C1 | 4 (4) | Diptera | Simuliidae | Flies (Black flies) |
| Chimpanzee | C1 | 10 (6) | Diptera | Chironomidae | Flies (nonbiting midges) |
| Chimpanzee | C1 | 8 (36) | Coleoptera | Scarabaeidae | Beetles (Scarab beetles) |
| Chimpanzee | C2 | 3 (12) | Diptera | Tachinidae | Flies (True flies) |
| Chimpanzee | C2 | 11 (4) | Diptera | Sepsidae | Flies (Ensign flies) |
| Chimpanzee | C2 | 15 (5) | Diptera | Cecidomyiidae | Flies (Gall gnats) |
| Chimpanzee | C2 | 37 (2) | Diptera | Cecidomyiidae | Flies (Gall gnats) |
| Chimpanzee | C2 | 45 (2) | Diptera | Anthomyiidae | Flies |
| Chimpanzee | C2 | 47 (4) | Diptera | Anthomyiidae | Flies (House flies) |
| Chimpanzee | C2 | 23 (4) | Coleoptera | Scarabaeidae | Beetles (Scarab beetles) |
| Chimpanzee | C2 | 25 (4) | Coleoptera | Chrysomelidae | Beetles (Leaf beetles) |
| Chimpanzee | C2 | 38 (2) | Coleoptera | Chrysomelidae | Beetles (Leaf beetles) |
| Chimpanzee | C3 | 7 (9) | Coleoptera | Staphylinidae | Beetles (Rove beetles) |
| Chimpanzee | C3 | 16 (5) | Coleoptera | Scarabaeidae | Beetles (Scarab beetles) |
| Chimpanzee | C3 | 17 (16) | Coleoptera | Dytiscidae | Beetles (Water beetles) |
| Chimpanzee | C3 | 39 (3) | Coleoptera | Scarabaeidae | Beetles (Scarab beetles) |
| Chimpanzee | C3 | 40 (2) | Coleoptera | Dytiscidae | Beetles (Water beetles) |
| Chimpanzee | C3 | 20 (4) | Diptera | Chironomidae | Flies (nonbiting midges) |
| Chimpanzee | C3 | 41 (3) | Diptera | Cecidomyiidae | Flies (Gall gnats) |
| Gorilla | G1 | 1 (132) | Isoptera | Termitidae | Termite |
| Gorilla | G1 | 31 (3) | Isoptera | Termitidae | Termite |
| Gorilla | G1 | 6 (9) | Diptera | Drosophilidae | Flies (Fruit flies) |
| Gorilla | G2 | 42 (3) | Coleoptera | Carabidae | Beetles |
| Gorilla | G2 | 43 (3) | Coleoptera | Chrysomelidae | Beetles (Leaf beetles) |
| Gorilla | G2 | 19 (6) | Orthoptera | Acrididae | Grasshoppers |
| Gorilla | G2 | 21 (4) | Diptera | Sepsidae | Flies (Ensign flies) |
| Gorilla | G2 | 26 (4) | Diptera | Muscidae | Flies (House flies) |
| Gorilla | G2 | 46 (2) | Diptera | Muscidae | Flies (House flies) |
| Gorilla | G2 | 24 (4) | Lepidoptera | Arctiidae | Moths (tiger moths) |
| Gorilla | G2 | 5 (8) | Isoptera | Rhinotermitidae | Termite |
| Gorilla | G3 | 6 (9) | Diptera | Drosophilidae | Flies (Fruit flies) |
| Gorilla | G3 | 13 (9) | Diptera | Cecidomyiidae | Flies (Gall gnats) |
| Gorilla | G3 | 14 (6) | Diptera | Sepsidae | Flies (Ensign flies) |
| Gorilla | G3 | 49 (3) | Diptera | Fanniidae | Flies (True flies) |
| Gorilla | G3 | 9 (21) | Coleoptera | Carabidae | Beetles |
G1, G2 and G3: fecal samples from gorillas; C1, C2 and C3: chimpanzees; B1, B2 and B3: bonobos.
The COI bonobo clone libraries yielded 23 OTUs belonging to 12 families within the orders Diptera and Lepidoptera (Fig. 2 and Table 2). Similarly, 19 and 21 OTUs from 12 and 10 families within the orders Diptera, Lepidoptera and Coleoptera were retrieved from the COI chimpanzee and gorilla clone libraries, respectively (Fig. 2 and Table 2).
Table 2. Insect OTUs detected in 9 fecal samples from wild African great apes using COI targeting primers.
| Ape | Sample | OTU (No. of clones) | Order | Family | Common name |
|---|---|---|---|---|---|
| Bonobo | B1 | 1 (31) | Diptera | Tephritidae | Flies (Fruit flies) |
| Bonobo | B1 | 2 (4) | Diptera | Psychodidae | Flies (drain flies) |
| Bonobo | B1 | 3 (7) | Diptera | Tephritidae | Flies (Fruit flies) |
| Bonobo | B1 | 4 (2) | Diptera | Cecidomyiidae | Flies (Gall gnats) |
| Bonobo | B1 | 5 (2) | Diptera | Tephritidae | Flies (Fruit flies) |
| Bonobo | B1 | 6 (2) | Diptera | Drosophilidae | Flies (Fruit flies) |
| Bonobo | B2 | 7 (38) | Diptera | Cecidomyiidae | Flies (Gall gnats) |
| Bonobo | B2 | 8 (2) | Diptera | Drosophilidae | Flies (Fruit flies) |
| Bonobo | B2 | 9 (4) | Diptera | Cecidomyiidae | Flies (Gall gnats) |
| Bonobo | B2 | 11 (2) | Diptera | Chironomidae | Flies (nonbiting midges) |
| Bonobo | B2 | 10 (2) | Lepidoptera | Noctuidae | Moths (Owlet moths) |
| Bonobo | B3 | 12 (16) | Diptera | Muscidae | Flies (House flies) |
| Bonobo | B3 | 14 (2) | Diptera | Culicidae | Mosquitoes |
| Bonobo | B3 | 16 (2) | Diptera | Curtonotidae | Flies (Quasimodo flies) |
| Bonobo | B3 | 18 (2) | Diptera | Cecidomyiidae | Flies (Gall gnats) |
| Bonobo | B3 | 19 (2) | Diptera | Muscidae | Flies (House flies) |
| Bonobo | B3 | 22 (2) | Diptera | Drosophilidae | Flies (Fruit flies) |
| Bonobo | B3 | 13 (4) | Lepidoptera | Sphingidae | Moths (Hawk moths) |
| Bonobo | B3 | 15 (6) | Lepidoptera | Nymphalidae | Butterflies (Brush-footed butterflies) |
| Bonobo | B3 | 17 (4) | Lepidoptera | Noctuidae | Moths (Owlet moths) |
| Bonobo | B3 | 20 (4) | Lepidoptera | Pyralidae | Moths (snout moths) |
| Bonobo | B3 | 21 (2) | Lepidoptera | Noctuidae | Moths (Owlet moths) |
| Bonobo | B3 | 23 (2) | Lepidoptera | Noctuidae | Moths (Owlet moths) |
| Chimpanzee | C1 | 12 (2) | Diptera | Muscidae | Flies (House flies) |
| Chimpanzee | C1 | 16 (4) | Diptera | Curtonotidae | Flies (Quasimodo flies) |
| Chimpanzee | C1 | 24 (8) | Diptera | Psychodidae | Flies (drain flies) |
| Chimpanzee | C1 | 25 (8) | Diptera | Sciaridae | Flies (Dark-winged fungus gnats) |
| Chimpanzee | C1 | 26 (4) | Diptera | Curtonotidae | Flies (Quasimodo flies) |
| Chimpanzee | C1 | 28 (2) | Diptera | Cecidomyiidae | Flies (Gall gnats) |
| Chimpanzee | C1 | 27 (20) | Lepidoptera | Geometridae | Moths (geometer moths) |
| Chimpanzee | C2 | 29 (26) | Diptera | Tachinidae | Flies (True flies) |
| Chimpanzee | C2 | 30 (6) | Diptera | Cecidomyiidae | Flies (Gall gnats) |
| Chimpanzee | C2 | 32 (2) | Diptera | Tachinidae | Flies (True flies) |
| Chimpanzee | C2 | 35 (2) | Diptera | Tachinidae | Flies (True flies) |
| Chimpanzee | C2 | 31 (2) | Lepidoptera | Geometridae | Moths (geometer moths) |
| Chimpanzee | C2 | 33 (4) | Lepidoptera | Cosmopterigidae | Moths (Cosmet moths) |
| Chimpanzee | C2 | 34 (4) | Lepidoptera | Erebidae | Moths |
| Chimpanzee | C2 | 36 (2) | Lepidoptera | Cosmopterigidae | Moths (Cosmet moths) |
| Chimpanzee | C3 | 10 (5) | Lepidoptera | Noctuidae | Moths (Owlet moths) |
| Chimpanzee | C3 | 16 (28) | Diptera | Curtonotidae | Flies (Quasimodo flies) |
| Chimpanzee | C3 | 37 (11) | Diptera | Simuliidae | Flies (Black flies) |
| Chimpanzee | C3 | 38 (4) | Coleoptera | Carabidae | Beetles |
| Gorilla | G1 | 6 (1) | Diptera | Muscidae | Flies (House flies) |
| Gorilla | G1 | 19 (4) | Diptera | Muscidae | Flies (House flies) |
| Gorilla | G1 | 39 (74) | Diptera | Muscidae | Flies (House flies) |
| Gorilla | G1 | 40 (57) | Diptera | Drosophilidae | Flies (Fruit flies) |
| Gorilla | G1 | 41 (3) | Diptera | Drosophilidae | Flies (Fruit flies) |
| Gorilla | G1 | 42 (3) | Diptera | Drosophilidae | Flies (Fruit flies) |
| Gorilla | G1 | 43 (2) | Diptera | Drosophilidae | Flies (Fruit flies) |
| Gorilla | G2 | 44 (13) | Diptera | Cecidomyiidae | Flies (Gall gnats) |
| Gorilla | G2 | 46 (6) | Diptera | Drosophilidae | Flies (Fruit flies) |
| Gorilla | G2 | 47 (2) | Diptera | Drosophilidae | Flies (Fruit flies) |
| Gorilla | G2 | 48 (10) | Diptera | Cecidomyiidae | Flies (Gall gnats) |
| Gorilla | G2 | 49 (6) | Coleoptera | Staphylinidae | Beetles (Rove beetles) |
| Gorilla | G2 | 45 (11) | Lepidoptera | Noctuidae | Moths (Owlet moths) |
| Gorilla | G3 | 44 (24) | Diptera | Cecidomyiidae | Flies (Gall gnats) |
| Gorilla | G3 | 50 (6) | Diptera | Sepsidae | Flies (Ensign flies) |
| Gorilla | G3 | 51 (4) | Diptera | Drosophilidae | Flies (Fruit flies) |
| Gorilla | G3 | 52 (4) | Diptera | Syrphidae | Flies (Hoverflies) |
| Gorilla | G3 | 54 (4) | Diptera | Psychodidae | Flies (drain flies) |
| Gorilla | G3 | 55 (2), | Diptera | Cecidomyiidae | Flies (Gall gnats) |
| Gorilla | G3 | 56 (2) | Diptera | Tephritidae | Flies (Fruit flies) |
| Gorilla | G3 | 53 (2) | Lepidoptera | Riodinidae | Butterflies (metalmarks) |
G1, G2 and G3: fecal samples from gorillas; C1, C2 and C3: chimpanzees; B1, B2 and B3: bonobos.
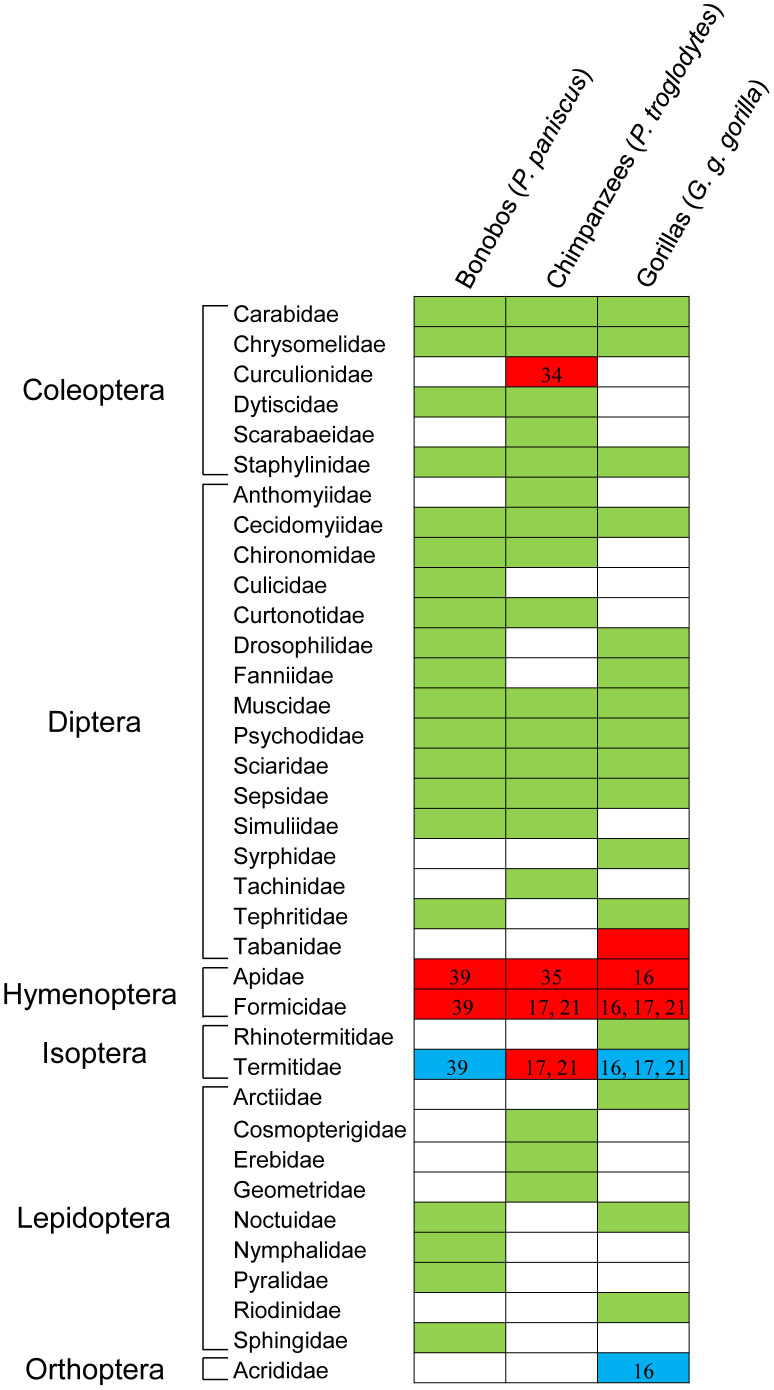
Taking the PCR results together, we detected a variety of arthropod OTUs in the fecal samples from these three African great apes, including fruit flies, moths, beetles, butterflies, mosquitoes and termites (Fig. 1 and 2 and Tables 1 and 2). A total of 32 families from 5 orders were present in at least one of the African great apes studied (Fig. 3). Eight of these families (Carabidae, Chrysomelidae and Staphylinidae from the order Coleoptera and Cecidomyiidae, Muscidae, Psychodidae, Sciaridae and Sepsidae from the order Diptera) were commonly detected in feces from the 3 African great apes species (Fig. 3). Three types of OTUs belonging to the order Isoptera (termites) were only identified in gorilla and bonobo feces (G1, G2 and B2), as shown in Fig. 1 and 3. However, no OTUs assigned to the families Formicidae, Apidae and Tabanidae were identified in our samples (Fig. 3).
Figure 3. Summary of insect consumption by three species of African great apes.
Orders and families of known insect prey identified through DNA analysis, direct observation or morphological analysis of prey remains in feces. Arthropod families obtained from Cyt-b and COI sequences and from different individuals of the same species were pooled together. Green cells indicate insect families that were identified as diet items in this study. Red cells indicate insect families that were not detected in this study. Blue cells indicate insect families that were detected in this study by DNA and in other studies through classical methods.
Discussion
Investigating the diets of animals by applying molecular methods to fecal samples is useful for the study of wild animals that are difficult to observe52,53,54. Very few studies have examined the insect-diet diversity eaten by primates through molecular approaches. Hofreiter et al.46 investigated the presence of vertebrate prey DNA in bonobo and gorilla feces through PCR-based methods, and more recently, Pickett et al.45 evaluated insect diversity in fecal samples from new world monkeys using a single arthropod primer set. Thus, our study is the first to analyze insect diversity in fecal samples from African great apes using two primers (targeting the Cyt-b and COI genes in the mitochondrial genome of arthropods). These primers have been used to successfully analyze the arthropod prey of tiger beetles and bats49,50. In this study, we detected 106 insect OTUs (41, 38 and 37 for bonobos, chimpanzees and gorillas, respectively) using these primers (Fig. 1 and 2, Tables 1 and 2). Compared with behavioral observations and/or analysis of trail signs in ape feces, we found many previously unknown insect families that are consumed by African great apes (Fig. 3). Many insects, such as species in the orders Coleoptera and Lepidoptera or caterpillars detected in this study (Tables 1 and 2) have strong associations with plants55,56. Some of these insect species, such as member of family Chrysomelidae, feed on different plant parts57. Consequently, they could be eaten incidentally (secondary predation) when African great apes feed on plants. Thus, one advantage of using a molecular approach to examine insects consumed is the inclusion of those consumed via secondary predation. These species may not have been otherwise detected through classical approaches, but they are still components of the diet that have nutritional value. These indirectly eaten insects may also contribute to an understanding of the feeding ecology and foraging strategy of a species. Interestingly, different OTUs from termites were identified in both gorilla and bonobo feces (Fig. 1 and Table 1). Termites are commonly preyed on by African great apes21,33 (Fig. 3). Macrotermes species, which are the largest termites in Africa and live in different habitat types, are an important insect meal for both western gorillas and chimpanzees residing in southeastern Cameroon21 (Fig. 3). Despite the wide global distribution of some of the insect species found in our work, such as those belonging to Drosophilidae, no data on the occurrence of the remaining species in Cameroon and DRC are available.
We were unable to amplify any of Formicidae species in this study (Fig. 3 and Tables 1 and 2) using both arthropod primers, and the macroscopic examination of fecal samples did not detect any visible hard parts (e.g. chitinous exoskeleton or jointed appendages) of these insects. Their absence could be due to degradation of insect DNA during its journey through the digestive tube, the small sample size or insufficient mitochondrial DNA caused by bias in the DNA extraction, amplification and cloning process.
While DNA-based fecal analyses are useful for evaluating the diets of wild animals, several challenges and limitations should be noted when employing this method. Although, no hard insect parts were visible in feces prior to analysis, fecal sample contamination is possible and, unfortunately, difficult to investigate especially for insects that burrow into feces. However, we minimized this limitation by using the inner part of the fecal mass for DNA extraction, which reduced contamination with soil organisms, flies and their eggs deposition.
DNA extraction, PCR amplification and cloning biases were also limitations in this study. The amount of feces used for DNA extractions represented only a small fraction of the total feces bulk. Low DNA concentration, species with low frequency in samples and the limited number of tested clones also may have prevented the detection of certain prey items. Moreover, the primers used may not have amplified the mitochondrial sequences of some insect species, so these sequences were lost. Another challenge with molecular diet analyses using DNA from feces is that one cannot determine whether the insects present in the guts of African great apes were the result of primary or secondary predation or contamination from the edible parts of plants. Although the primer sets we used amplified a wide range of arthropods, the incompleteness of insect sequence data in GenBank prevented us from assigning the recovered OTUs at genus or species level. For this reason, the taxonomic identifications were done at family level according to the E-values of BLAST results. Exhaustive studies using molecular techniques along with conventional morphological taxonomy are needed to enrich the public databases with complete arthropod mitochondrial genome sequences, which will identify insects at the species level. Finally, prey sequence counts recovered using traditional cloning/sequencing or pyrosequencing58 cannot quantify insect consumption or provide insights to insect choice by African great apes. Thus, combining molecular tools with classical observation methods provides a more complete picture of the complex diet of wild animals, including primates.
In conclusion, molecular fecal analysis is an emerging method that can provide valuable insight into the insectivory aspect of primate diet and can expand the list of insect foods ingested by a study population. The high number of insect OTUs detected in fecal samples using this strategy confirms the importance of molecular applications for analyzing direct and indirect insect consumption in African great apes. For detailed descriptions of the occurrence and abundance of insects in African great ape feces using next generation sequencing, additional studies using larger numbers of samples collected in different seasons are needed. This approach will clarify intra- and inter-species dietary variation in wild primates.
Methods
Source of the fecal samples
A total of 9 fecal samples were used in this study. Three fecal samples from wild gorillas (G. g. gorilla) were collected at a forest site near Messok (sample G1) and at a forest site near Mambele (samples G2, G3) (Messok and Mambele are two towns located southeast and east of Cameroon, respectively). Three fecal samples from chimpanzees (P. troglodytes) were collected at a forest site near Mambele (samples C1, C2) and at a forest site near Messok (sample C3), and three fecal samples (B1, B2 and B3) from bonobos (P. paniscus) were collected at the Lomako-Yokokala faunal reserve in the Democratic Republic of Congo (DRC). The sample collection protocol was described previously59. Briefly, samples were collected near night nests or feeding sites. For all samples, the time, date and GPS position were recorded, and the species of origin was determined according to prints, collection sites and morphological aspects of the samples as well as by amplifying a 386 bp mtDNA fragment spanning the 12SrRNA gene (using primers 12S-L1091 5′-AAAAAGCTTCAAACTGGGATTAGATACCCCACTAT-3′ and 12S-H1478 5′-TGACTGCAGAGGGTGACGGGCGGTGTGT-3′). The time between defection and collection was estimated at <24 h, according to the physical texture of the samples. Collected samples were saved at base camps at ambient temperature in RNAlater (Ambion, Austin, TX) for less than 3 weeks and transported to a central laboratory for storage at −80°C. No experimentation was conducted on these animals. The collection of fecal samples from the soil was approved by the Ministry of Scientific Research and Innovation of Cameroon and DRC. No other permit was required, as this research was non-invasive work, and the collection of the samples did not disrupt wild fauna.
DNA extraction
The outer layer of fecal bulk was peeled carefully with a sterile scalpel and polystyrene tweezers (approximately 1–2 mm was removed). The inner part of the fecal bulk was used for extraction to avoid a possible contamination with soil organisms and/or the risk of egg deposition by some flies, which would result in the amplification of non-prey organisms. DNA was extracted using a modified version of the Qiagen fecal procedure (QIAamp DNA Tissue Kit, Qiagen Inc., Germany)60. A 200-mg aliquot of each fecal sample was placed in a 2-ml tube containing 200 mg of a mixture of 0.1-, 0.5-, and 2-mm zirconium beads and 1.5 ml of ASL buffer (Qiagen). The sample was bead-beaten at 3200 rpm for 90 seconds, followed by heating at 95°C for 10 minutes. The final pellet was suspended in 180 μl of tissue lysis buffer and incubated with proteinase K for 2 hours at 55°C. The manufacturer's recommendations were followed for the purification and elution of DNA.
Primer selection and genomic amplification
To minimize primer bias, two different primers targeting Cyt-b and COI genes in the mitochondrial genome of arthropods were used49,50. The first PCR was previously used to study the prey of tiger beetles after whole DNA extraction from predatory specimens, which provided lower identity sequences compared with the GenBank database but sufficiently high scores to identify the order, family and occasionally the genus49. The second PCR was used to analyze arthropod prey in bat feces, enabling the authors to identify 37 prey taxa from 15 fecal samples (identification at the species level in 72% of the analyzed clones)50. The primer sets ZBJ-ArtF1c (AGATATTGGAACWTTATATTTTATTTTTGG)/ZBJ-ArtR2c (WACTAATCAATTWCCAAATCCTCC)49 and CB3 (GAGGAGCAACTGTAATTACTAA)/CB4 (AAAAGAAARTATCATTCAGGTTGAAT)50 amplified 157 and 410 bp segments of mitochondrial DNA, respectively. The PCR reaction mixture (final volume 50 μl) contained 5 μl of dNTPs (2 mM of each nucleotide), 5 μl of 10× DNA polymerase buffer (QIAGEN, Courtaboeuf, France), 1 μl of MgCl2 (25 mM), 0.25 μl of HotStarTaq DNA polymerase (5 U) (QIAGEN, Courtaboeuf, France), 1 μl of each primer (10 pmol/μl), and 5 μl of extracted DNA. The PCR was performed with an initial denaturation at 95°C for 15 minutes, followed by 40 cycles of 95°C for 45 seconds, an annealing temperature specific for the primers used (57°C for ZBJ-ArtF1c/ZBJ-ArtR2c and 46°C for CB3/CB4) for 30 seconds, 72°C for 1 minute, and a final extension at 72°C for 5 minutes. The PCR products were analyzed using agarose gel electrophoresis (1.5%) and visualized with ethidium bromide staining. Positive PCR products were subsequently purified using the NucleoFast® 96 PCR Kit (MACHEREY-NAGEL, Hoerdt, France) according to the manufacturer's instructions.
Cloning procedures and insert amplification
The purified PCR products were cloned with the pGEM®-T Easy Vector System 2 Kit (Promega, Madison, USA), as recommended by the manufacturer. All white colonies were analyzed by PCR with the M13d (5′-GTAAAACGACGGCCAG) and M13r (5′-CAGGAAACAGCTATGAC) primers, as described previously60.
Sequencing and data analysis
Correct sizes of purified PCR-M13 inserts were sequenced in both directions using the Big Dye® Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems, Courtaboeuf, France). The M13d and M13r primers were used for sequencing. The sequencing products were run on an ABI PRISM 3130 automated sequencer (Applied Biosystems, Foster city, CA, USA).
The sequences were corrected with the CodonCode Aligner software (4.0.1). Sequence similarity was determined through multiple alignment software using ClustalW2. Sequences with base-pair mismatches, insertions or deletions leading to stop codons in every possible reading frame were excluded from further analysis. A cloned sequence was designated an “operational taxonomic unit” (OTU)51 if it had more than 2% sequence divergence from all other recovered sequences across approximately 400 bp and 157 bp of Cyt-B and COI, respectively45. Phylogenetic analyses (neighbor-joining tree) were performed using MEGA 5, supported by 500 bootstrap replicates.
To make taxonomy assignments, the OTUs were compared with a BLAST database of pre-assigned sequences in GenBank (available at the National Center for Biotechnology Information website: http://www.ncbi.nlm.nih.gov/), and the closest available matching hit with the lowest E-value was assigned as the taxonomic identification for each OTU. As many arthropod species and genera shared or had very close E-values, OTUs were assigned to the family level.
Nucleotide sequence accession numbers
All sequences generated from the primer set CB3/CB4 were deposited in the GenBank database with the accession numbers JX500757 to JX501234. The sequences obtained from the primer set ZBJ-ArtF1c/ZBJ-ArtR2c were not deposited because sequences shorter than 200 bp are not accepted in the GenBank database.
Author Contributions
D.R. and F.B. designed the experiments; I.H. conducted the experiments; I.H., E.D., D.R. and F.B. analyzed the results; I.H. and F.B. prepared the figures; I.H. and F.B. wrote the manuscript. All authors reviewed the manuscript.
Acknowledgments
Fadi Bittar was supported by a Chair of Excellence IRD provided by the Institut de Recherche pour le Développement. We are grateful to Dr. Martine Peeters for providing us the fecal samples.
References
- Tutin C. E. G. & Fernandez M. Composition of the diet of chimpanzees and comparisons with that of sympatric lowland gorillas in the lopé, reserve, gabon. Am. J. Primatol. 30, 195–211 (1993). [DOI] [PubMed] [Google Scholar]
- Gursky S. Tarsiiformes. In: Primates in Perspective, (eds. Campbell, C., Fuentes, A., Mackinnon, K., Bearder, S., Stump, R.) 79–90 (Oxford University Press, UK, 2001). [Google Scholar]
- Harcourt C. Seasonal variation in the diet of South African Galagos. Int. J. Primatol. 7, 491–506 (1986). [Google Scholar]
- Isbell L. A. & Young T. P. Interspecific and temporal variation of ant species within Acacia drepanolobium ant domatia, a staple food of patas monkeys (Erythrocebus patas) in Laikipia, Kenya. Am. J. Primatol. 69, 1387–1398 (2007). [DOI] [PubMed] [Google Scholar]
- Chapman C. A. et al. in Variation in the diets of Cercopithecus species: differences within forests, among forests, and across species (ed Cords, M. & Glenn, M. E.) 325–350 (Academic/Plenum, New York: Kluwer, 2002). [Google Scholar]
- Struhsaker T. The Red Colobus Monkeys: Variation in Demography, Behavior and Ecology of Endangered Species. (The University of Chicago Press, Chicago, 2010). [Google Scholar]
- Rogers M. E. et al. Western gorilla diet: a synthesis from six sites. Am. J. Primatol. 64, 173–192 (2004). [DOI] [PubMed] [Google Scholar]
- Yamagiwa J. & Mwanza N. Day-journey length and daily diet of solitary male gorillas in lowland and highland habitats. Int. J. Primatol. 15, 207–224 (1994). [Google Scholar]
- Remis M. J. Western lowland gorillas (Gorilla gorilla gorilla) as seasonal frugivores: use of variable resources. Am. J. Primatol. 43, 87–109 (1997). [DOI] [PubMed] [Google Scholar]
- Doran D. M. et al. Western lowland gorilla diet and resource availability: new evidence, cross-site comparisons, and reflections on indirect sampling methods. Am. J. Primatol. 58, 91–116 (2002). [DOI] [PubMed] [Google Scholar]
- Inogwabini B. I. & Matungila B. Bonobo food items, food availability and bonobo distribution in the Lake Tumba Swampy Forests, Democratic Republic of Congo. Open. Conservat. Bio. J. 3, 14–23 (2009). [Google Scholar]
- Schaller G. B. The mountain gorilla: ecology and behavior. (University of Chicago Press, 1963). [Google Scholar]
- Jones C. & Sabater P. J. Comparative Ecology of Gorilla gorilla & Pan troglodytes in Rio Muni West Africa. Bibl. Primatol. 13, 1–96 (1971). [Google Scholar]
- Cipolletta C. et al. Termite Feeding by Gorilla gorilla gorilla at Bai Hokou, Central African Republic. Int. J. Primatol. 28, 457–476 (2007). [Google Scholar]
- Deblauwe I. & Dekoninck W. Diversity and distribution of ground-dwelling ants in a lowland rainforest in southeast Cameroon. Insect. Soc. 54, 334–342 (2007). [Google Scholar]
- Deblauwe I., Dupain J., Nguenang G. M., Werdenich D. & Van Elsacker L. Insectivory by Gorilla gorilla gorilla in Southeast Cameroon. Int. J. Primatol. 24, 493–502 (2003). [Google Scholar]
- Deblauwe I. & Janssens G. P. New insights in insect prey choice by chimpanzees and gorillas in southeast Cameroon: the role of nutritional value. Am. J. Phys.Anthropol. 135, 42–55 (2008). [DOI] [PubMed] [Google Scholar]
- Ganas J. & Robbins M. M. Intrapopulation differences in ant eating in the mountain gorillas of Bwindi Impenetrable National Park, Uganda. Primates. 45, 275–278 (2004). [DOI] [PubMed] [Google Scholar]
- Galdikas B. F. Orangutan diet, range, and activity at Tanjung Puting, Central Borneo. Int. J. Primatol. 9, 1–35 (1988). [Google Scholar]
- Kim S., Lappan S. & Choe J. C. Diet and ranging behavior of the endangered Javan gibbon (Hylobates moloch) in a submontane tropical rainforest. Am. J. Primatol. 73, 270–280 (2011). [DOI] [PubMed] [Google Scholar]
- Deblauwe I. Temporal Variation in Insect-eating by Chimpanzees and Gorillas in Southeast Cameroon: Extension of Niche Differentiation. Int. J. Primatol. 30, 229–252 (2009). [Google Scholar]
- McGrew W. C. in Meat-eating and human evolution (eds Stanford, C. B. & Bunn, H. T.) 160–178 (Oxford University Press, 2001). [Google Scholar]
- Finke M. D. Complete nutrient composition of commercially raised invertebrates used as food for insectivores. Zoo.Bio. 21, 269–285 (2002). [Google Scholar]
- Barker D., Fitzpatrick M. P. & Dierenfeld E. S. Nutrient composition of selected whole invertebrates. Zoo. Bio. 17, 123–134 (1998). [Google Scholar]
- Harcourt A. H. & Harcourt S. A. Insectivory by gorillas. Folia Primatol. 43, 229–233 (1984). [Google Scholar]
- Watts D. P. Ant eating behaviour of mountain gorillas. Primates 30, 121–125 (1989). [Google Scholar]
- Yamagiwa J., Mwanza N., Yumoto T. & Maruhashi T. Ant eating by eastern lowland gorillas. Primates 32, 247–253 (1991). [Google Scholar]
- Calvert J. J. Food selection by western gorillas (G.g. gorilla) in relation to food chemistry. Oecol. 65, 236–246 (1985). [DOI] [PubMed] [Google Scholar]
- Tutin C. E. G. & Fernandez M. Insect-eating by sympatric Lowland gorillas (Gorilla g. gorilla) and chimpanzees (Pan t. troglodytes) in the lopé, Reserve, Gabon. Am. J. Primatol. 28, 29–40 (1992). [DOI] [PubMed] [Google Scholar]
- Suzuki S., Kuroda S. & Nishihara T. Tool-set for termite-fishing by chimpanzees in the Ndoki Forest, Congo. Behaviour 132, 219–235 (1995). [Google Scholar]
- Sanz C., Morgan D. & Gulick S. New insights into chimpanzees, tools, and termites from the Congo Basin. Am.Nat. 164, 567–581 (2004). [DOI] [PubMed] [Google Scholar]
- Bogart S. L. & Pruetz J. D. Insectivory of savanna chimpanzees (Pan troglodytes verus) at Fongoli, Senegal. Am.J.Phys.Anthropol. 145, 11–20 (2011). [DOI] [PubMed] [Google Scholar]
- McGrew W. C. & Collins D. A. Tool use by wild chimpanzees (Pan troglodytes) to obtain termites (Macrotermes herus) in the Mahale Mountains, Tanzania. Am. J. Primatol. 9, 47–62 (1985). [DOI] [PubMed] [Google Scholar]
- Sugiyama Y. Tool-use for catching ants by chimpanzees at Bossou and Monts Nimba, West Africa. Primates 36, 193–205 (1995). [Google Scholar]
- Deblauwe I. New evidence of honey-stick use by chimpanzees in southeast Cameroon. Pan. Africa. News 13, 4 (2006). [Google Scholar]
- Badrian N., Badrian A. & Susman R. Preliminary observations on the feeding behavior of Pan paniscus in the lomako forest of central Zaïre. Primates 22, 173–181 (1981). [Google Scholar]
- Susman R., Badrian N., Badrian A. & Handler N. Positional behavior and feeding ecology of the pygmy chimpanzee (Pan paniscus): First year results of the Lomako Forest Pygmy Chimpanzee Project. Nat. Geo. Res. 20, 725–739 (1985). [Google Scholar]
- Bermejo M., Illera G. & Sabater J. Animals and mushrooms consumed by bonobos (Pan paniscus): New records from lilungu (Ikela), Zaire. Int. J. Primatol. 16, 879–898 (1995). [Google Scholar]
- Sabater Pi J. V. J. Comparative inventory of foods consumed by the wild pygmy chimpanzee (Pan paniscus; mammalia) in the Lilungu-Lokofe region of the Republic of Zaïre. J. Afr. Zoo. 108, 381–396 (1994). [Google Scholar]
- Knott C. Changes in Orangutan Caloric Intake, Energy Balance, and Ketones in Response to Fluctuating Fruit Availability. Int. J. Primatol. 19, 1061–1079 (1998). [Google Scholar]
- Schaik C. P., Fox E. A. & Sitompul A. F. Manufacture and use of tools in wild Sumatran orangutans. Naturwissenschaften 83, 186–188 (1996). [DOI] [PubMed] [Google Scholar]
- Hardus M. et al. Behavioral, Ecological, and Evolutionary Aspects of Meat-Eating by Sumatran Orangutans (I). Int. J. Primatol. 33, 287–304 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappeler M. in The Lesser Apes: Evolutionary and Behavioural Biology (eds Preuschoft, H., Chivers, D., Brocklman, W. & Creel, N.) 209–218 (Edinburg University Press, Edinburg, 1984). [Google Scholar]
- Moreno-Black G. The use of scat samples in primate diet analysis. Primates 19, 215–221 (1978). [Google Scholar]
- Pickett S. B., Bergey C. M. & Di F. A. A metagenomic study of primate insect diet diversity. Am. J. Primatol. 74, 622–631 (2012). [DOI] [PubMed] [Google Scholar]
- Hofreiter M., Kreuz E., Eriksson J., Schubert G. & Hohmann G. Vertebrate DNA in fecal samples from bonobos and gorillas: evidence for meat consumption or artefact? PLoS One 5, 0009419 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmann K. et al. Molecular diet analysis of two african free-tailed bats (molossidae) using high throughput sequencing. PLoS One 6, e21441 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesterinen E. J., Lilley T., Laine V. N. & Wahlberg N. Next generation sequencing of fecal DNA reveals the dietary diversity of the widespread insectivorous predator Daubenton's Bat (Myotis daubentonii) in Southwestern Finland. PLoS One 8, e82168 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons J. DNA-based identification of preys from non-destructive, total DNA extractions of predators using arthropod universal primers. Mol. Ecol. Notes 6, 623–626 (2006). [Google Scholar]
- Zeale M. R., Butlin R. K., Barker G. L., Lees D. C. & Jones G. Taxon-specific PCR for DNA barcoding arthropod prey in bat faeces. Mol. Ecol. Resour. 11, 236–244 (2011). [DOI] [PubMed] [Google Scholar]
- Blaxter M. et al. Defining operational taxonomic units using DNA barcode data. Philos Trans. R. Soc. Lond. B. Biol. Sci. 360, 1935–1943 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley B. J. et al. Plant DNA sequences from feces: potential means for assessing diets of wild primates. Am. J. Primatol. 69, 699–705 (2007). [DOI] [PubMed] [Google Scholar]
- Clare E. L., Fraser E. E., Braid H. E., Fenton M. B. & Hebert P. D. Species on the menu of a generalist predator, the eastern red bat (Lasiurus borealis): using a molecular approach to detect arthropod prey. Mol. Ecol. 18, 2532–2542 (2009). [DOI] [PubMed] [Google Scholar]
- Clare E. L., Barber B. R., Sweeney B. W., Hebert P. D. & Fenton M. B. Eating local: influences of habitat on the diet of little brown bats (Myotis lucifugus). Mol. Ecol. 20, 1772–1780 (2011). [DOI] [PubMed] [Google Scholar]
- Jurado-Rivera J. A., Vogler A. P., Reid C. A., Petitpierre E. & Gomez-Zurita J. DNA barcoding insect-host plant associations. Proc. Biol. Sci. 276, 639–648 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce N. E. Predatory and parasitic Lepidoptera: Carnivores living on plants. J. Lepidopterists' Soc. 49, 412–453 (1995). [Google Scholar]
- Hawkeswood T. in Novel aspects of the biology of Chrysomelidae 50 (eds Jolivet Pierre, H., Cox, M. L. & Petitpierre, E.) 12, 191–204 (Springer Netherlands, 1994). [Google Scholar]
- Deagle B. E., Thomas A. C., Shaffer A. K., Trites A. W. & Jarman S. N. Quantifying sequence proportions in a DNA-based diet study using Ion Torrent amplicon sequencing: which counts count? Mol. Ecol. Resour. 13, 620–633 (2013). [DOI] [PubMed] [Google Scholar]
- Keita A. K. et al. Molecular evidence for the presence of Rickettsia Felis in the feces of wild-living African apes. PLoS One 8, e54679 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hamad I., Sokhna C., Raoult D. & Bittar F. Molecular Detection of Eukaryotes in a Single Human Stool Sample from Senegal. PLoS One 7, e40888 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]