Abstract
Genomes contain both a genetic code specifying amino acids, and a regulatory code specifying transcription factor (TF) recognition sequences. We used genomic DNaseI footprinting to map nucleotide resolution TF occupancy across the human exome in 81 diverse cell types. We find that ~15% of human codons are dual-use codons (`duons') that simultaneously specify both amino acids and TF recognition sites. Duons are highly conserved and have shaped protein evolution, and TF-imposed constraint appears to be a major driver of codon usage bias. Conversely, the regulatory code has been selectively depleted of TFs that recognize stop codons. >17% of single nucleotide variants within duons directly alter TF binding. Pervasive dual encoding of amino acid and regulatory information appears to be a fundamental feature of genome evolution.
The genetic code, common to all organisms, contains extensive redundancy, wherein most amino acids can be specified by 2–6 synonymous codons. The observed ratios of synonymous codons are highly non-random, and codon usage biases are fixtures of both prokaryotic and eukaryotic genomes (1). In organisms with short life spans and large effective population sizes codon biases have been linked to translation efficiency and mRNA stability (2–7). However, these mechanisms explain only a small fraction of observed codon preferences in mammalian genomes (7–11), which appear to be under selection (12),.
Genomes also contain a parallel regulatory code specifying recognition sequences for transcription factors (TFs) (13), and the genetic and regulatory codes have been assumed to operate independently of one another, and to be segregated physically into the coding and non-coding genomic compartments. However the potential for some coding exons to accommodate transcriptional enhancers or splicing signals has long been recognized (14–18).
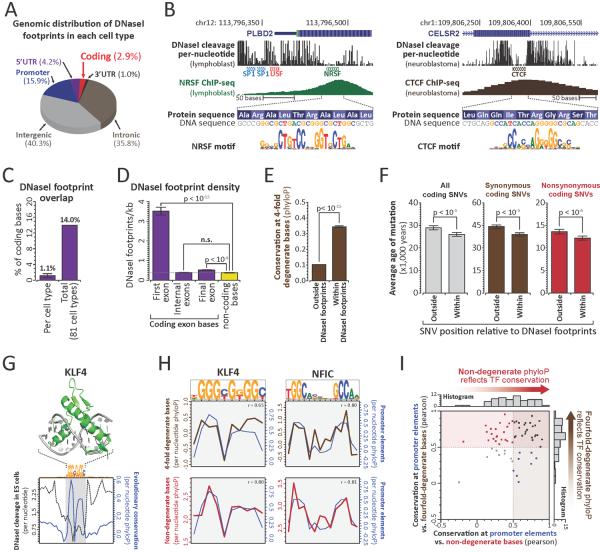
To define intersections between the regulatory and genetic codes, we generated nucleotide-resolution maps of transcription factor occupancy in 81 diverse human cell types using genomic DNaseI footprinting (19). Collectively, we defined 11,598,043 distinct 6–40bp footprints genome-wide (~1,018,514 per cell-type), 216,304 of which localized completely within protein-coding exons (~24,842 per cell-type) (Fig. 1A–B, S1A, Table S1). ~14% of all human coding bases contact a TF in at least one cell type (avg. 1.1% per cell type; Figs. 1C, S1B) and 86.9% of genes contained coding TF footprints (avg. 33% per cell type) (Figs. S1C–D).
Figure 1. TFs densely populate and evolutionarily constrain protein-coding exons.
(A) Distribution of DNaseI footprints. (B) Per-nucleotide DNaseI cleavage and ChIP-seq signal for coding CTCF (left) and NRSF (right) binding elements. (C) Proportion of coding bases within DNaseI footprints in each of 81 cell types (left), or any cell type (right). (D) Average footprint density within first, internal, or final coding exons (mean +/− SEM; p-value, paired t-test, n.s.: p-value> 0.1). (E) PhyloP conservation at 4FDBs within and outside footprints. (F) Estimated mutational age at all (grey), synonymous (brown) and nonsynonymous (red) coding SNVs (European) within and outside footprints (p-values per (21)) (G) Structure of DNA-bound KLF4 vs. average per-nucleotide DNaseI cleavage and evolutionary constraint at KLF4 footprints. (H) Average per-nucleotide conservation at 4FDBs (brown) and NDBs (red) overlapping KLF4 (left) and NFIC (right) footprints. (r = Pearson correlation, conservation at promoter bases vs. 4FDBs (top) or NDBs (bottom)). (I) Evolutionary constraint imparted by 63 TFs at promoter elements, 4FDBs and NDBs (Pearson correlations).
The exonic TF footprints we observed likely underestimate the true fraction of protein-coding bases that contact TFs since (i) TF footprint detection increases substantially with sequencing depth (13), and (ii) the 81 cell types sampled, though extensive, is far from complete, as we saw little evidence of saturation of coding TF footprint discovery (Fig. S2).
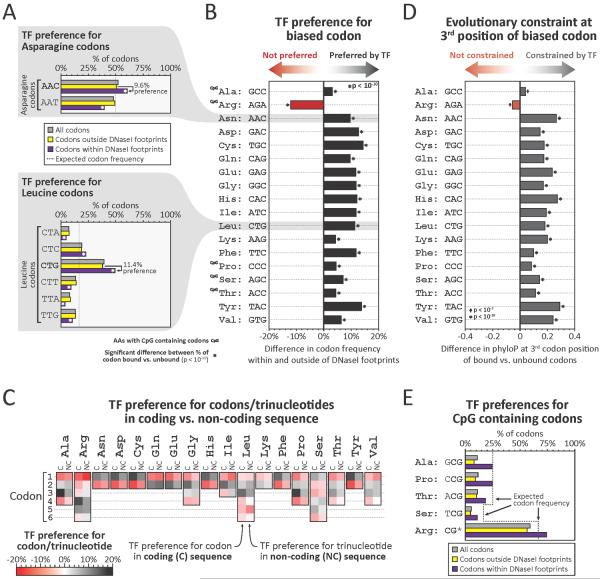
Figure 2. Transcription factors modulate global codon biases.
(A) Proportions of all codons (grey), or codons outside of (yellow), or within (purple) footprints, that encode asparagine (top) or leucine (bottom). Note that codons with bias (AAC for asparagine and CTG for leucine) preferentially localize within footprints. (B) Preferential footprinting of biased codons, calculated as in (A) (p-values, Pearson's chi-squared test). (C) Preferential footprinting of each codon trinucleotide in coding vs non-coding regions (C = coding, NC = non-coding). (D) Difference in average evolutionary constraint at 3rd positions of biased codons outside vs. within footprints (p-values, Mann-Whitney test). (E) Proportions of amino acids encoded by CpG-containing codons among all codons (grey), codons outside footprints (yellow), or codons within footprints (purple)
To ascertain coding footprints more completely, we developed an approach for targeted exonic footprinting via solution-phase capture of DNaseI-seq libraries using RNA probes complementary to human exons (19). Targeted capture footprinting of exons from abdominal skin and mammary stromal fibroblasts yielded ~10-fold increases in DNaseI cleavage, equivalent to sequencing >4 billion reads per sample using conventional genomic footprinting (Fig. S3A), quantitatively exposing many additional TF footprints (Fig. S3B–D). Overall, we identified an average of ~175,000 coding footprints per cell type (Fig. S1E), 7-12-fold more than conventional footprinting.
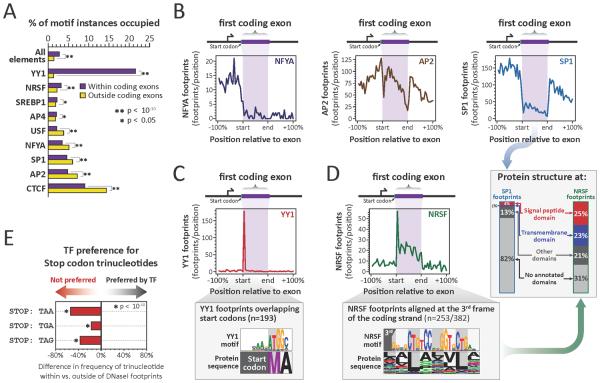
Figure 3. TFs exploit and avoid specific coding features.
(A) Percentage of TF motifs occupied in coding vs. non-coding regions (p-values, paired t-test). (B) Density of NFYA (left), AP2 (middle) and SP1 (right) footprints relative to translated region of first coding exons. (C) (top) Density of YY1 footprints across first coding exons. (bottom) YY1 recognition sequence and corresponding amino acid sequence within YY1 footprints overlapping start codons. (D) (top left and bottom) For NRSF as per (C). (right, arrow) Protein domain annotation of first exon third-frame NRSF footprints vs. SP1 footprints. (E) TF preference (avoidance) of stop codon trinucleotides within vs. outside footprints in non-coding regions (p-values, Pearson's chi-squared test).
While coding sequences are densely occupied by TFs in vivo, the density of TF footprints at different genic positions varied widely, with many genes exhibiting sharply increased density in the translated portion of their first coding exon (Figs. 1D, S4A). By contrast, internal coding exons were as likely as flanking intronic sequences to harbor TF footprints (Fig.1D). The total number of coding DNaseI footprints within a gene was related both to the length of the gene, and to its expression level (Fig. S4B–D).
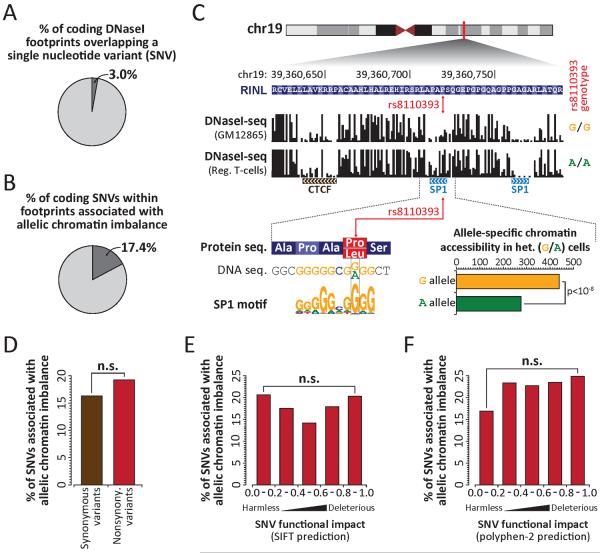
Figure 4. Genetic variation in duons frequently alters TF occupancy.
(A) Proportion of coding footprints overlapping a SNV in any of 81 cell-types. (B) Proportion of SNVs in duons that allelically alter TF occupancy. (C) (top) Per-nucleotide DNaseI cleavage at common nonsynonymous G→A SNV (rs8110393) in G/G and A/A homozygous cells. (bottom) Allelic SP1 occupancy in heterozygous (G/A) cells. (D) Proportion of synonymous and nonsynonymous variants in duons that allelically alter TF occupancy. (E–F) Proportion of nonsynonymous variants from (D) grouped by predicted impact of coding variant on protein function using (E) SIFT or (F) Polyphen-2. Note that none of the bins are significantly different (Fisher's exact test; n.s. indicates p-value > 0.1).
Given their abundance, we sought to determine whether exonic TF binding elements were under evolutionary selection. 4-fold degenerate coding bases are frequently used as a model of neutral (or nearly neutral) evolution (20), but may exhibit constraint when a functional signal impinges on coding sequence (11). Across the coding compartment, 4-fold degenerate bases (4FDBs) within TF footprints show significantly greater evolutionary constraint vs. non-footprinted 4FDBs (Figs. 1E, S5A–B), indicating that TF-DNA recognition constrains the third codon position.
To test for evolutionary constraint at coding footprints in modern human populations, we quantified the age of mutations arising within or outside of coding footprints using exome sequencing data from 4,298 individuals of European ancestry (Fig. S5C) and 2,217 individuals of African American ancestry (Fig. S5D) (21). This analysis revealed that mutations within coding footprints were on average 10.2% younger than those outside of footprints (Figs. 1F, S5E), signaling influence of coding TF elements on human fitness.
Strikingly, both synonymous and nonsynonymous mutations within coding footprints were significantly younger than those outside of footprints (Figs. 1F, S5E), indicating that coding TF binding constrains both codon and amino acid evolution. The genome-wide recognition sequence landscape of each TF has evolved to fit the molecular topography of its protein-DNA binding interface (13) (Fig. 1G). To study how specific TFs influence codon and amino acid choice at their recognition sites, we compared the per-nucleotide evolutionary conservation profiles of TF recognition sequences at non-coding, 4FDBs and non-degenerate coding bases (NDBs). For example, the conservation profiles at 4FBDs and NDBs at KLF4 and NFIC recognition sites closely mirror those of recognition sites in non-coding regions (promoter; Fig. 1H). As such, these TFs constrain both codon choice (via constraint on 4FDBs), and amino acid choice (via NDBs) encoded at their recognition sites. Analysis of conservation profiles for 63 TFs with prevalent occupancy within coding regions (19) showed that 73% constrain 4FDBs, and 51% constrain NDBs (Figs. 1I, S6, S7). Thus, individual TFs may influence both codon and amino acid choice.
To examine how TF binding relates to codon usage patterns, we examined -binding at preferred (biased) vs. non-preferred codons. For example, across all human proteins Asparagine is encoded by the AAC codon 52% of the time (vs. AAT, 48%), indicating a generalized 4% bias in favor of this codon. However, genome-wide, 60.4% of Asn codons within footprints are AAC, vs. only 50.8% outside of footprints (i.e., a 9.6% occupancy bias towards the preferred codon) (Fig. 2A). Strikingly, apart from Arginine (see below), for all amino acids encoded by two or more codons, the codon that is preferentially utilized genome-wide is also preferentially occupied by TFs (Fig. 2B, Table S2).
To determine whether preferential occupancy of biased codons is inherent to TF recognition sequences, we compared trinucleotide frequencies within coding vs. non-coding footprints. Trinucleotide combinations favored by TFs within coding sequence were equivalent to those favored in non-coding sequence (Fig. 3C), indicating that global TF binding preferences are directly reflected in the frequency of different codons. Notably, baseline trinucleotide frequencies within coding and non-coding sequence are largely independent of one another (Table S2). The fact that the third position of preferred codons overlapping footprints is under excess evolutionary constraint (Fig. 2D, Table S2) supports a general role for TFs in potentiating codon usage biases through the selective preservation of preferred codons.
While nearly all codon biases parallel TF recognition preferences genome-wide, Arginine, one of the 5 amino acids encoded by codons containing CpGs (4 out of 6 codons), was a notable exception. CpGs frequently occur in regulatory DNA (Table S2), yet have an elevated mutational rate (22). Consequently, although TFs may favor CpG-containing codons (Fig. 2E), and impart excess constraint thereto (Table S2), the higher mutational rate at such codons is likely incompatible with preferential utilization.
We note that codons outside footprints still exhibit usage biases (Fig. 2A and Table S2); however, it is likely that these biases also reflect the actions of TFs. Firstly, our conclusions above are drawn from a conservative and incomplete annotation of duons. Secondly, because TF trinucleotide preferences and codon biases have not changed substantially since the divergence of humans and mice (Fig. S8), preferences at any given codon may result from a TF binding element extant in some ancestral species to human. Third, codon usage bias can be exaggerated due to mutual reinforcement with other cellular factors such as tRNA abundances (23, 24). Indeed, such mechanisms could be linked to codon biases created by exonic TF occupancy through a feedback mechanism that potentiates intrinsic TF-imposed biases, resulting in both abundant and rare codons and associated tRNAs, differences in which could in turn affect protein synthesis and stability (25–27).
To analyze positional occupancy patterns of specific TFs within coding sequence, we systematically matched TF recognition sequences with footprints, providing an accurate measure of a TF's in vivo occupancy (13, 28). This analysis revealed that a subset of TFs selectively avoid coding sequences (Fig. 3A). Intriguingly, TFs involved in positioning the transcriptional pre-initiation complex, such as NFYA and SP1 (29), preferentially avoid the translated region of the first coding exon (Fig. 3A), and typically occupy elements immediately upstream of the methionine start codon (Figs. 3B, S9A). Conversely, TFs involved in modulating promoter activity, such as YY1 and NRSF, preferentially occupy the translated region of the first coding exon (30, 31) (Fig. 3A,C). These findings indicate that that the translated portion of the first coding exon may serve functionally as an extension of the canonical promoter.
More broadly, the repressor NRSF preferentially occupies and evolutionarily constrains sequences coding for leucine-rich protein domains, such as signal peptide and transmembrane domains (Figs. 3D, S9B,C). Also, TFs such as CTCF and SREBP1 preferentially occupy and constrain splice sites (Fig. S10A–D), which are otherwise generally depleted of DNaseI footprints (Fig. S10E). The above results suggest that specific protein structural and splicing features may undergo exaptation for specific regulatory purposes.
We also found that the occupancy of specific TFs within coding sequence parallels the extent of CpG methylation at their binding site (Fig. S11). This raises the possibility that gene body methylation, which is paradoxically extensive at actively transcribed genes (32, 33), may provide a tunable mechanism for thwarting opportunistic TF occupancy within coding sequence during transcription.
If TFs, through selective recognition sequences, could impose changes in protein sequence, deleterious consequences could arise if such changes resulted in a nonsense substitution. We observed that TFs generally avoid stop codons (Fig. S10E). Surprisingly, this finding extends to non-coding regions, where stop codon trinucleotides (TAA, TAG and TGA) are selectively depleted within footprints. This indicates that the global TF repertoire has been selectively purged of DNA binding domains capable of recognizing, and thus preferentially stabilizing, nonsense codons (Fig. 3E and S10F).
The high sequencing coverage provided by genomic footprinting revealed 592,867 heterozygous single nucleotide variants (SNVs) across the 81 cell type samples, and 3% of coding footprints harbored heterozygous SNVs (Fig. 4A). Functional SNVs that disrupt TF occupancy quantitatively skew the allelic origins of DNaseI cleavage fragments (13), and 17.4% of all heterozygous coding SNVs within footprints showed this signature (Figs. 4B, S12), including both synonymous and nonsynonymous variant classes (Fig. 4C). The potential of a coding SNV to disrupt overlying TF occupancy was independent of the class of variant (Fig. 4D), or whether a nonsynonymous variant was predicted to be deleterious to protein function (Fig. 4E–F).
Notably, 13.5% of common disease- and trait-associated SNVs identified by genome-wide associated studies (GWAS) (19) fall within duons (Fig. S13A). GWAS SNPs in duons encompass both synonymous (12%) and nonsynonymous (88%) substitutions (Fig. S13A), and may directly affect pathogenetic mechanisms (Fig. S13B–F, Table S3). As such, disease-associated variants within duons may compromise both regulatory and/or protein-structural functions. These findings have substantial practical implications for the interpretation of genetic variation in coding regions.
In summary, our results indicate that simultaneous encoding of amino acid and regulatory information within exons is a major functional feature of complex genomes. The information architecture of the received genetic code is optimized for superimposition of additional information (34, 35), and this intrinsic flexibility has been extensively exploited by natural selection. While TF binding within exons may serve multiple functional roles, we note that our analyses above is agnostic to these roles, which may be complex (36).
Supplementary Material
ACKNOWLEDGMENTS
We thank many colleagues for their insightful comments and critical readings of the manuscript. We also thank many colleagues who provided individual cell samples for DNaseI analysis. We also thank Eric Rynes for his technical assistance. This work was supported by NIH grants U54HG004592, U54HG007010 and U01ES01156 to J.A.S.. A.B.S. was supported by grant FDK095678A from NIDDK. J.M.A. is a paid consultant for Glenview Capital. All data from this study are available through the ENCODE data repository at UCSC (http://www.encodeproject.org) and the Roadmap Epigenomics data repository at NCBI http://www.ncbi.nlm.nih.gov/epigenomics.
REFERENCES AND NOTES
- 1.Grantham R, Gautier C, Gouy M, Mercier R, Pavé A. Nucleic acids research. 1980;8:r49–r62. doi: 10.1093/nar/8.1.197-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ikemura T. Journal of molecular biology. 1981;151:389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- 3.Grantham R, Gautier C, Gouy M, Jacobzone M, Mercier R. Nucleic acids research. 1981;9:r43–74. doi: 10.1093/nar/9.1.213-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gouy M, Gautier C. Nucleic acids research. 1982;10:7055–74. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eyre-Walker A, Bulmer M. Nucleic acids research. 1993;21:4599–603. doi: 10.1093/nar/21.19.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlini DB, Stephan W. Genetics. 2003;163:239–43. doi: 10.1093/genetics/163.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.dos Reis M, Savva R, Wernisch L. Nucleic acids research. 2004;32:5036–44. doi: 10.1093/nar/gkh834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parmley JL, Chamary JV, Hurst LD. Molecular biology and evolution. 2006;23:301–9. doi: 10.1093/molbev/msj035. [DOI] [PubMed] [Google Scholar]
- 9.Warnecke T, Weber CC, Hurst LD. Biochemical Society transactions. 2009;37:756–61. doi: 10.1042/BST0370756. [DOI] [PubMed] [Google Scholar]
- 10.Gu W, Zhou T, Wilke CO. PLoS computational biology. 2010;6:e1000664. doi: 10.1371/journal.pcbi.1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin MF, et al. Genome research. 2011;21:1916–28. doi: 10.1101/gr.108753.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Z, Nielsen R. Molecular biology and evolution. 2008;25:568–79. doi: 10.1093/molbev/msm284. [DOI] [PubMed] [Google Scholar]
- 13.Neph S, et al. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hyder SM, Nawaz Z, Chiappetta C, Yokoyama K, Stancel GM. The Journal of biological chemistry. 1995;270:8506–13. doi: 10.1074/jbc.270.15.8506. [DOI] [PubMed] [Google Scholar]
- 15.Lang G, Gombert WM, Gould HJ. Immunology. 2005;114:25–36. doi: 10.1111/j.1365-2567.2004.02073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ritter DI, Dong Z, Guo S, Chuang JH. PloS one. 2012;7:e35202. doi: 10.1371/journal.pone.0035202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khan AH, Lin A, Smith DJ. PloS one. 2012;7:e46098. doi: 10.1371/journal.pone.0046098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Birnbaum RY, et al. Genome research. 2012;22:1059–68. doi: 10.1101/gr.133546.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.See methods.
- 20.Li W-H. Molecular Evolution (Sinauer Associates, Incorporated. 1997:487. [Google Scholar]
- 21.Fu W, et al. Nature. 2013;493:216–20. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coulondre C, Miller JH, Farabaugh PJ, Gilbert W. Nature. 1978;274:775–80. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- 23.Bulmer M. Nature. 1987;325:728–30. doi: 10.1038/325728a0. [DOI] [PubMed] [Google Scholar]
- 24.Bulmer M. Genetics. 1991;129:897–907. doi: 10.1093/genetics/129.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duan J, et al. Human molecular genetics. 2003;12:205–16. doi: 10.1093/hmg/ddg055. [DOI] [PubMed] [Google Scholar]
- 26.zur Megede J, et al. Journal of virology. 2000;74:2628–35. doi: 10.1128/jvi.74.6.2628-2635.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coleman JR, et al. Science. 2008;320:1784–7. doi: 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samstein RM, et al. Cell. 2012;151:153–66. doi: 10.1016/j.cell.2012.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKnight S, Tjian R. Cell. 1986;46:795–805. doi: 10.1016/0092-8674(86)90061-9. [DOI] [PubMed] [Google Scholar]
- 30.Zhang C, et al. Nucleic acids research. 2006;34:2238–46. doi: 10.1093/nar/gkl248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xi H, et al. Genome research. 2007;17:798–806. doi: 10.1101/gr.5754707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hellman A, Chess A. Science. 2007;315:1141–3. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 33.Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Nature genetics. 2007;39:61–9. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 34.Itzkovitz S, Alon U. Genome research. 2007;17:405–12. doi: 10.1101/gr.5987307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Itzkovitz S, Hodis E, Segal E. Genome research. 2010;20:1582–9. doi: 10.1101/gr.105072.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mercer TR, et al. Nature Genetics. 2013 doi:10.1038/ng.2677. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.