Abstract
Most breast cancers expressing the estrogen receptor α (ERα) are treated successfully with the receptor antagonist tamoxifen (TAM), but many of these tumors recur. Elevated expression of the homeodomain transcription factor HOXB13 correlates with TAM-resistance in ERα-positive (ER+) breast cancer, but little is known regarding the underlying mechanism. Our comprehensive evaluation of HOX gene expression using tiling microarrays, with validation, showed that distant metastases from TAM-resistant patients also displayed high HOXB13 expression, suggesting a role for HOXB13 in tumor dissemination and survival. Here we show that HOXB13 confers TAM resistance by directly downregulating ERα transcription and protein expression. HOXB13 elevation promoted cell proliferation in vitro and growth of tumor xenografts in vivo. Mechanistic investigations showed that HOXB13 transcriptionally upregulated interleukin (IL)-6, activating the mTOR pathway via STAT3 phosphorylation to promote cell proliferation and fibroblast recruitment. Accordingly, mTOR inhibition suppressed fibroblast recruitment and proliferation of HOXB13-expressing ER+ breast cancer cells and tumor xenografts, alone or in combination with TAM. Taken together, our results establish a function for HOXB13 in TAM resistance through direct suppression of ERα and they identify the IL-6 pathways as mediator of disease progression and recurrence.
Introduction
Approximately 60% to 70% of breast cancers express estrogen receptor α (ERα) and/or progesterone receptor and merit the use of hormone therapies, such as the estrogen receptor antagonist tamoxifen (TAM; ref. 1). Use of TAM has improved outcomes for patients with these tumors, but in 30% to 40% of these patients the cancers recur, usually as distant metastasis, and the majority of these patients will die of their cancer. High expression of the gene HOXB13 and concomitant low expression of IL17BR have been found in multiple studies to predict TAM-R and disease recurrence in patients with ER-positive (ER+) breast cancer (2–5).
The HOX genes encode a family of highly conserved transcription factors that normally regulate temporospatial development of the extremities and organs. Aberrant expression of these genes in different tissues have been associated with tumorigenesis (6). In breast cancer, low expression of HOXA5 and HOXA10 is associated with decreased p53 expression (7, 8), whereas high expression of HOXB7 or HOXB13 has been associated with aggressive disease (3, 9). However, the mechanism by which HOXB13 promotes aggressive disease and TAM-R in ER+ breast cancer has not been defined. Without understanding these mechanisms, therapeutic alternatives for patients with TAM-R, HOXB13-expressing ER+ breast cancer cannot be rationally devised for greatest efficacy and minimal toxicity.
A comprehensive HOX cluster expression tiling array analysis of primary ER+ breast tumors and distant metastases (10) supported the involvement of HOXB13 in dissemination of disease following resistance to hormonal therapy. Here, we validate these findings and provide insights into the mechanism whereby HOXB13 mediates TAM-R and metastasis. HOXB13 promotes TAM-R by transcriptionally downregulating ERα expression. HOXB13 drives cell and tumor proliferation by inducing expression of interleukin (IL)-6 in the cancer cells, leading to activation of the AKT and mTOR pathways and also stimulating stromal recruitment. We also show that targeting these pathways with the mTOR inhibitor, rapamycin, suppresses the growth of TAM-R, HOXB13-expressing tumors.
Materials and Methods
Human tissue specimens
Normal breast epithelial preparations (organoids), primary breast tumors, and distant metastases were accessed with approval from the Johns Hopkins University Institutional Review Board, and RNA extracted as previously described (10). Detailed methods are presented in the Supplementary Methods section.
Reverse transcription-quantitative PCR validation of gene expression
A total of 200 ng of RNA from primary tissue samples, or 1 µg of RNA from cell lines, were reverse-transcribed using Superscript III (Invitrogen), per manufacturer protocol; 1 µL of yield was used per PCR. Taqman Gene Expression Assays for HOXB13 (Hs00197189_m1) and GAPDH (Hs99999905_m1) were used as primers and gene-specific fluorescent probes for PCR, using RampTaq polymerase (Denville Scientific) and supplied buffer. qPCR was conducted per manufacturer protocol, using the Applied Biosystem 7500 Real-Time PCR System for 40 cycles. A detection threshold of 0.01 was set for determination of Ct for each reaction. For each sample, qPCR was conducted to measure HOXB13 and GAPDH expression; each sample was tested in triplicate. The ΔΔCt method (GAPDH used for normalization) was used to determine the expression of HOXB13 in each reaction separately, using average lowest expression in organoid tissue as baseline. Relative expression was calculated as 2^(−ΔΔCt), and the 3 expression values averaged to determine HOXB13 expression in each sample. Primer compositions are presented in Supplementary Table S1.
Cell culture, plasmids, and cell line constructs
The breast cell linesMCF10A, MCF7, T47D, and BT474 were provided by NCI (IBC-45 panel) through the American Type Culture Collection; the fibroblast cell line NIH3T3 was obtained from lab stocks. MCF10A cells were grown in DMEM/F12 media (Mediatech) supplemented with 5% horse serum, 20 ng/mL EGF, 0.5 mg/mL hydrocortisone, 100 ng/mL cholera toxin, 10 µg/mL insulin, 50 IU/mL penicillin, and 50 µg/mL streptomycin sulfate. MCF7 cells were grown in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% FBS (Gemini Bio-Products), and T47D and BT474 cells were grown in RPMI media with 10% FBS. NIH3T3 cell lines were grown in DMEM with 10% normal calf serum.
Plasmids containing the full length cDNA of human HOXB13 in the pLPCX retroviral vector (pHOXB13), the empty pLPCX vector (Clontech), 2 short hairpin RNA (shRNA) lentiviral constructs targeting HOXB13 mRNA (shHOXB13), and scramble shRNA construct (PLKO.1/Thermo Scientific) were used to generate viral supernatant for overexpression or knock-down of HOXB13 in cell lines. Generation of HOXB13-modulated cell lines by retroviral infection is described in the Supplementary Methods.
Matrigel invasion assays
Invasion assays were conducted in BD Biocoat Matrigel (24-well format) Invasion Chambers per manufacturer protocol. Experiments were conducted in triplicate.
Promoter-luciferase reporter assay
MCF7 cells were transiently transfected with LipofectA-MINE 2000/DNA complexes (Invitrogen) of p-HOXB13, promoter-luciferase construct (pGL2; Promega), and β-galactosidase (βGAL) plasmid, and incubated for 24 hours. Luciferase and βGAL activity were measured per protocol (Promega). Assays were conducted in triplicate in a single experiment, and then as 3 independent experiments.
Western blots
Western blots were conducted as previously described (7); full methods and antibodies used can be found in the Supplementary Experimental Methods.
Drug in vitro cell survival
A total of 2.5 × 103 cells/well were plated in 96-well plates, in triplicate, with 4OH-TAM (Sigma-Aldrich) or rapamycin (Sell-eck Chem) at stated concentrations, a combination, or vehicle in 200 µL media. Media was changed every 2 days. MTT assays were conducted. Absorbance at 560 nm was measured, with background at 670 nm subtracted. Each sample was analyzed in triplicate, and percentage calculated of survival of drug-treated cells versus vehicle-treated cells. Experiments were conducted in triplicate. Antibody information is provided in Supplementary Table S2.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was conducted with the EZChip kit (Millipore) according to protocol; full details can be found in the Supplementary Methods, including primers for PCR (Supplementary Table S1).
Tumor xenograft studies
Six- to 8-week-old female athymic nude mice were used, and study approved by Johns Hopkins Animal Use Committee. E2 pellets were implanted subcutaneously on day −3 in 6 mice. On day 0, 3 × 106 of the indicated tumor cells were injected subcutaneously into both flanks of each mouse. In a second experiment, mice were implanted with E2 pellets on day −3. On day 0, the indicated cancer cells were injected into the 4th mammary fat pads (mfp; 1.5 × 106 cells) or subcutaneously (3 × 106 cells) suspended in 0.1 mL of 1:1 PBS/Matrigel (BD Biosciences) in separate sets of mice. After 3 weeks, 6mice each with mfp or subcutaneous tumors were treated with either (i) TAM implants subcutaneously; (ii) rapamycin intraperitoneally with a loading dose of 9 mg/kg on day 21, then 3 mg/kg every other day, (iii) TAM implants plus rapamycin, or (iv) no additional treatment. Tumor growth was measured weekly. Mice were euthanized after 6 weeks and tumors harvested. Tumor volume was estimated by the calculation V = (length × width × height × 0.5236) mm3. Details are presented in the Supplementary Methods section.
Statistical analyses
All statistical analyses, except the analysis of the tiling arrays and the NKI dataset, were conducted using GraphPad Prism version 5.04 (GraphPad Software, Inc.). The NKI dataset (288 patients with ER+ breast cancer) uses a custom Rosetta oligonucleotide, on which HOXB13 and IL-6 are each represented by 2 probes. Spearman rank-based correlation coefficients were used to measure association, assessing significance using a permutation test.
For all reverse transcription-quantitative PCR (RT-qPCR) analyses and the invasion assays, one-way ANOVA analysis using the Kruskal– Wallis test was conducted to assess significant difference among groups, as well as two-tailed Student t test to compare specific pairs. For survival analyses, the logrank/Mantel–Cox test was conducted to calculate significant difference in survival. For sensitivity of cell lines to rapamycin, two-way ANOVA testing (cell line and drug concentration) was conducted, with Bonferroni posttest to compare cell lines. For tumor xenograft studies, tumor growth inhibition was analyzed using a mixed effects model by assuming an exchangeable covariance structure to account for correlation among measurements taken on the same animal. Tukey's procedure was used to adjust for multiple comparisons. For all other assays, two-tailed Student t tests with unpaired values were conducted.
Results
HOXB13 is overexpressed in distant metastases and associated with disease recurrence and death
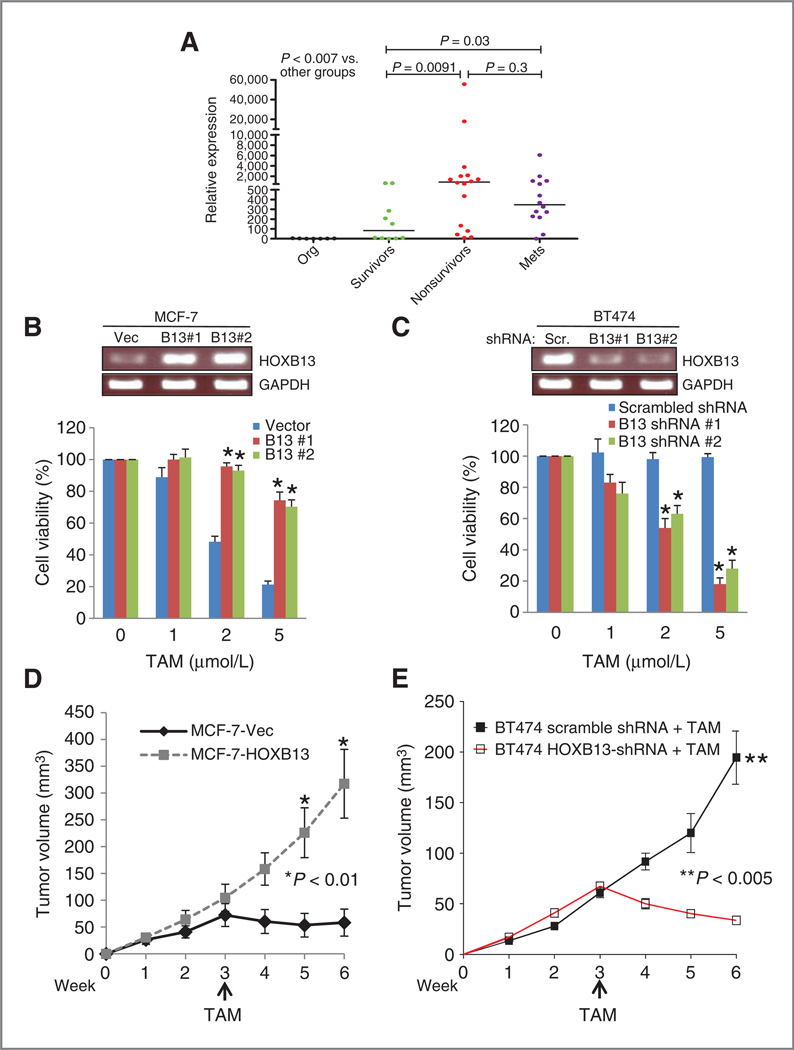
We evaluated HOX gene expression in ER+ breast cancer and distant metastatic disease through analysis of HOX tiling arrays (10) on normal breast organoids, primary tumors, and distant metastases. HOXB13 was overexpressed in the majority of tumor samples, including 5 of 6 metastases, compared with normal organoids (10). This finding was validated by RT-qPCR using a panel of ER+ primary tumors from long-term breast cancer survivors and nonsurvivors, and 14 metastases from multiple sites from 11 breast cancer patients who had undergone rapid autopsy for tissue donation (Fig. 1A). Compared with normal organoids, each group of tumors had significantly higher expression of HOXB13 (P < 0.007), and tumors from nonsurvivors and metastases had significantly higher expression than those from survivors (P = 0.009).
Figure 1.
HOXB13 is overexpressed in primary ER+ breast cancers and distant metastases and promotes TAM resistance. A, RT-qPCR validation of HOXB13 expression in normal breast organoids (org), ER+ primary breast tumors from long-term survivors or nonsurvivors of disease, and metastases (mets). Median expression is marked. B, RT-PCR analysis of HOXB13 expression in MCF7-HOXB13 cells (pools 1 and 2) compared with vector control; cell viability after treatment with 1, 2, or 5 µmol/L 4-OH TAM. C, RT-PCR analysis of BT474-HOXB13 shRNA cells (pools 1 and 2) compared with vector control; cell viability after treatment with 1, 2, or 5 µmol/L 4-OH TAM. D, tumor growth of MCF7-vector (black) and MCF7-HOXB13 cell subcutaneous xenografts in athymic mice with E2 supplementation, then treated with subcutaneously TAM implant (arrow; *,P < 0.01). E, tumor growth of BT474-scrambled shRNA (closed square) and BT474-HOXB13 shRNA (open square) cell subcutaneously xenografts in athymic mice with E2 supplementation, then treated with subcutaneously TAM implant (**, P < 0.005)
HOXB13 expression correlates with proliferation, resistance to TAM, and estrogen-independent growth
To study the mechanism by which HOXB13 mediates TAMR, we examined HOXB13 expression in breast cell lines. We showed that the immortalized normal breast cell line MCF10A had HOXB13 expression comparable to levels seen in primary normal breast organoid preparations (P = 0.9). Consistent with HOXB13 expression in primary tumors from survivors, expression in ER+, TAM-sensitive (TAM-S) cell lines, MCF7 and T47D, were 4- to 8-fold higher than MCF10A. In contrast, HOXB13 expression in an ER+, TAM-R cell line, BT474, was 400- to 500-fold higher than MCF10A, consistent with the level of expression in primary tumors from nonsurvivors (Supplementary Fig. S1A). In MCF7 and T47D, forced expression of HOXB13 raised levels of HOXB13 to within the range of expression seen in tumor samples from nonsurvivors (Supplementary Fig. S1B). In BT474 cells, lentiviral infection of 2 shRNA constructs resulted in reduced HOXB13 expression to 10% (shHOXB13 #1) and 5% (shHOXB13 #2) of the vector control of BT474 (shScramble) and below the median HOXB13 expression seen in tumors from survivors (Supplementary Fig. S1B).
To determine if HOXB13 expression correlates with TAM-R, cells were treated with 1 to 5 µmol/L 4-hydroxytamoxifen (4-OH TAM, the active metabolite of TAM) in vitro, for 48 hours. Both the MCF7-HOXB13 cell pools with high HOXB13 expression had significantly higher survival at 2 and 5 µmol/L 4OH-TAM compared with vector control (Fig. 1B). Conversely, BT474-shHOXB13 cell pools, with low HOXB13 expression, had lower survival (Fig. 1C). Growth phenotypes of these cells were also tested in vivo. ER+ cell lines require estradiol supplementation (E2) to grow as xenografts and are TAM-S, which causes growth arrest and tumor regression. MCF7-HOXB13 cells injected subcutaneously into athymic nude mice formed tumors that proliferated without E2, whereas the MCF7-vector control cells did not (Supplementary Fig. S1C), suggesting that HOXB13-expressing cells had lost dependence on E2 for growth. To test response of the tumors to TAM, E2 pellets were implanted into mice 3 days before xenograft implantation; after 3 weeks the pellets were replaced with TAM pellets. MCF7-HOXB13 xenografts grew faster and continued to grow despite TAM, whereas MCF-vector xenograft growth was suppressed by TAM (Fig. 1D). Conversely, xenografts of TAM-R, BT474-vector control cells continued to grow whereas BT474-shHOXB13-2 xenografts regressed upon TAM treatment (Fig. 1F). Thus, the phenotype of both the HOXB13-overexpressing and -depleted breast cancer cells reflect the TAM-R phenotype associated with HOXB13 overexpression in vitro and in clinical studies.
HOXB13 directly suppresses ERα transcription
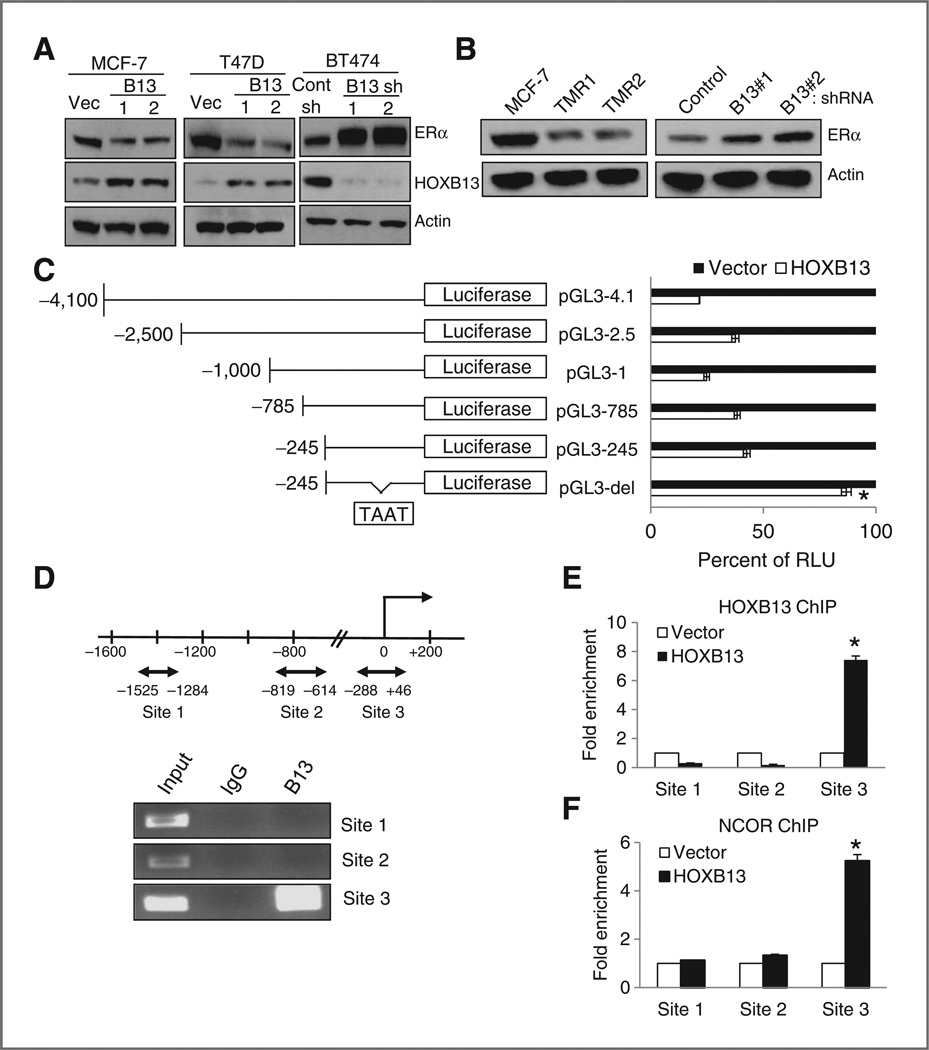
Despite extensive evidence for a strong correlation between HOXB13 and TAM-R, few studies have addressed the mechanisms linking the two factors. ERα activation with E2 in cultured breast cancer cells decreased HOXB13 mRNA levels (11). Conversely, ERα suppression with TAM in breast cells resulted in increased levels of HOXB13 mRNA(11). In ovarian cancer cells, a reverse phenomenon was reported—HOXB13 expression enhanced expression of ERα protein and ERE-linked luciferase activity (12). We investigated the correlation between HOXB13 and ERα in our cell line models. By Western blot analysis, we observed that both T47D-HOXB13 and MCF7-HOXB13 pools showed reduced expression of ERα (Fig. 2A). Conversely, two pools of BT474-shHOXB13 cells showed increased expression of ERα (Fig. 2A). In MCF7-TMR1 and MCF7-TMR2 cells (grown for more than 6 months in 0.1 or 1 µmol/L 4-OH TAM, respectively), ERα mRNA expression (Supplementary Fig. S2A) and protein expression (Fig. 2B) was significantly lower compared with parental MCF7 cells. HOXB13 caused this downregulation, as shRNA-mediated depletion of HOXB13 expression in MCF7-TMR1 cells resulted in increased ERα protein (Fig. 2B) and mRNA (Supplementary Fig. S2B) expression. These changes are reflected in the sensitivity of these cells to TAM. Treatment of MCF7-TMR1 cells with 1 to 5 µmol/L 4-OH TAM for 48 hours had little effect on cell viability (Supplementary Fig. S2C). However, the MCF7-TMR1 shHOXB13#1 and #2 cells regained sensitivity to 4-OH TAM (Supplementary Fig. S2C). Thus, HOXB13 overexpression and TAM-Rare closely linked; reducing HOXB13 levels in the cells confers TAM sensitivity.
Figure 2.
Direct transcriptional downregulation of ERα by HOXB13. A, immunoblot analysis of ERα and HOXB13 expression in MCF7-HOXB13, T47D-HOXB13, and BT474shHOXB13 cells compared with vector control cells. B, immunoblot analysis of ERα in MCF7-TMR1 and TMR2 cells compared with parental MCF7 cells (left). shRNA-mediated HOXB13 knockdown in MCF7-TMR1 cells results in increased ERα expression (right). C, ERα promoter-luciferase construct maps (left) and luciferase activity of each, cotransfected with vector or HOXB13 plasmid. A region 245 bp upstream of the TSS, including a HOX binding sequence, is critical to HOXB13-directed suppression (*, P < 0.01). D, diagram of regions in the ERα promoter with HOXB13 binding sites (double arrow, top). ChIP on MCF7-HOXB13 cells with anti-HOXB13 antibody or control IgG shows Site 3 is enriched with HOXB13 expression (bottom). E, qPCR analysis of putative HOXB13 binding sites in the ERα promoter after HOXB13 ChIP assay (*,P < 0.01) F, qPCR analysis of the putative HOXB13 binding sites after NCOR ChIP assay (*, P < 0.01).
The negative correlation between HOXB13 and ERα expression led us to investigate if HOXB13 acts directly upon the ERα promoter to repress expression. ERα promoter-luciferase constructs were made, from −4,100 to −245 bp of the transcriptional start site (TSS) of ERα (Fig. 2C). Cotransfection experiments showed that HOXB13 significantly suppressed ERα promoter-luciferase activity in all constructs as compared with empty vector, and identified a consensus HOX-binding sequence, TAAT, within −245 bp of the TSS. Deletion of 10 base pairs inclusive of the putative HOX-binding site resulted in loss of the suppressive effect of HOXB13 on ERα promoter (Fig. 3C). To show direct interaction between HOXB13 and the ERα promoter, we conducted ChIP for HOXB13 in the MCF7-HOXB13 cell line. Three regions of the ERα promoter with putative HOX-binding sequences (Fig. 2D) were tested; HOXB13 specifically bound to a region extending from −288 to +46 bp. Compared withMCF7-vector cells, ChIP using an anti-HOXB13 antibody in MCF7-HOXB13 cells showed specific enrichment for binding to this region, in contrast to other regions of the promoter (Fig. 2D). Consistent with the presence of a repressive complex, ChIP for the corepressor, NCOR, showed enriched binding to this region of the ERα promoter in the MCF7-HOXB13 cells (Fig. 2F).
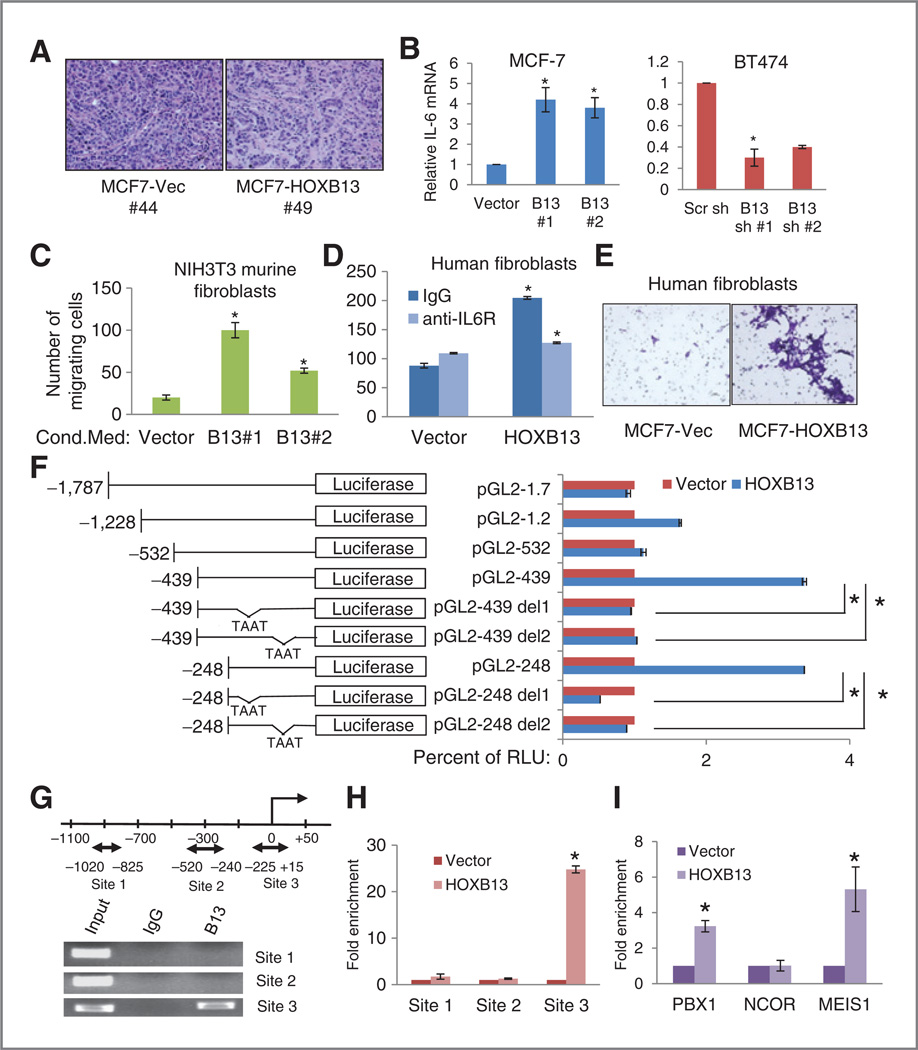
Figure 3.
HOXB13 promotes stromal recruitment through upregulation of IL-6. A, hematoxylin and eosin-stained sections ofMCF7 xenografts.MCF7-vector tumors (left) grow as uniform sheets of cancer cells, with little stromal component. In contrast, MCF7-HOXB13 tumors (right) have significantly more stromal infiltration. B, RT-qPCR analysis of IL6 expression in MCF7-HOXB13 (left) and BT474-shHOXB13 (right) cells compared with the control cells. C and D, cell invasion/chemotaxis analysis using MCF7-HOXB13 conditioned media as the chemoattractant for untreated NIH3T3 fibroblasts (C) or primary human fibroblasts pretreated with IgG or anti-IL-6R antibody (D). E, representative photomicrographs (× 10) of crystal violet-stained invading fibroblasts on the bottom of the insert membrane, attracted to conditioned media from MCF7-vector or MCF7-HOXB13 cells. F, IL-6 promoter-luciferase construct maps (left) and luciferase activity of each construct, cotransfected with vector (red) or pHOXB13 (blue), identifying a region 248 bp upstream of the TSS, including 2HOX binding sequences, as critical to HOXB13-directed enhancement (*, P < 0.01).G, diagram of regions in the IL6 promoter with HOXB13 binding sites (double arrow). ChIP on MCF7-HOXB13 cells with either anti-HOXB13 antibody or control IgG shows Site 3 is enriched with HOXB13 expression (bottom). H, qPCR analysis of the 3 putative HOXB13 binding sites in IL6 promoter after HOXB13 ChIP assay. I, qPCR analysis of HOXB13 binding Site 3 after PBX1, NCOR, or MEIS1 ChIP assay (*, P < 0.01).
HOXB13 expression is associated with increased stroma in MCF7 xenografts and increased fibroblast recruitment in vitro, and acts by directly promoting IL-6 expression
Because the cell lines with higher HOXB13 expression grow faster as xenografts, and display TAM-R and E2-independent proliferation, HOXB13 is likely utilizing ER-independent pathways to potentiate these phenotypes. We also noted a striking difference in the histology of the tumor xenografts-MCF7-vector control xenografts grow with little stroma, whereas tumors of MCF7-HOXB13 cells had significantly larger stromal component interspersed among the cancer cells (Fig. 3A). Based on the vast literature of stromal recruitment and participation in breast cancer aggressiveness (13–15), we entertained the notion that the factors that drive proliferation in HOXB13-expressing tumors may include those that affect their microenvironment.
Genes implicated in different aspects of lymphocyte, macrophage, and stromal recruitment and cellular invasion include IL6, IL8, CXCL12, CXCR4, and MMP1 (16–18). RT-qPCR was conducted on RNA from the MCF7-HOXB13 and BT474-HOXB13 shRNA cells and their vector controls, to analyze mRNA expression levels of these genes (Supplementary Fig. S3A and S3B). Of those tested, IL6 showed consistent patterns of direct correlation of expression with HOXB13 across both sets of cell lines (Fig. 3B, Supplementary Fig. S3C and S3D), and MCF7-TMR cell lines (Supplementary Fig. S3E and S3F).
To evaluate the ability of HOXB13-expressing cancer cells to attract fibroblasts and to define the role of IL-6 in this function, we conducted cell invasion/chemotaxis assays. NIH3T3 mouse fibroblasts respond to chemotactic stimuli in a manner similar to human tumor stroma-derived fibroblasts (19, 20). Media conditioned by MCF7-HOXB13 cells was used as the chemoattractant in Matrigel invasion assays, using NIH3T3 fibroblasts (Fig. 3C) or primary human fibroblasts (Fig. 3D and E) as invading cells. In both cases, conditioned medium from MCF7-HOXB13 cells attracted significantly more fibroblasts than MCF7-vector (Fig. 3D and E, Supplementary Fig. S3G). Furthermore, pretreatment of the fibroblasts with an anti-IL-6R antibody abrogated this effect (Fig. 3D), suggesting that the major stromal attractant was IL-6.
To evaluate if HOXB13 was directly driving IL6 expression, deletion constructs of IL6 promoter-luciferase plasmids (−248 to −1787 bp of the TSS) were constructed. Significantly increased luciferase activity was observed with constructs −439 bp or less (Fig. 3F) in MCF7 cells transiently cotransfected with pHOXB13, compared with empty vector. Deletion of 10 base pairs that included either of 2 distinct putative HOX-binding sites from the −439 or −248 bp promoter-luciferase constructs resulted in loss of the HOXB13 effect (Fig. 3F). These results suggested that HOXB13 binds directly to the IL6 promoter and upregulates its expression. ChIP analysis for HOXB13 identified a region −225 to +15 bp of the TSS that was enriched for binding in MCF7-HOXB13 cells (Fig. 3G) as compared with vector control cells (Fig. 3H). Also, ChIP analysis showed enrichment at the IL6 promoter of 2 HOX cofactors, PBX1 and MEIS1, but not of the corepressor NCOR (Fig. 3I). These findings support the hypothesis that HOXB13 binds the IL6 promoter directly and promotes IL6 transcription.
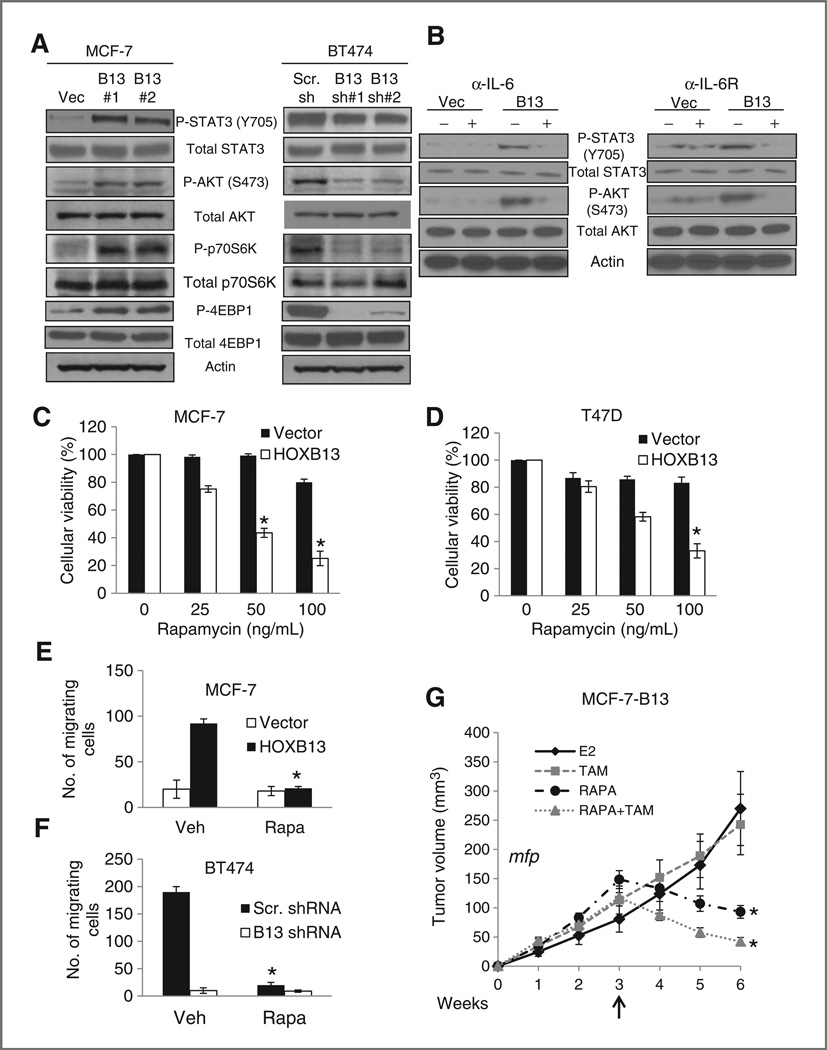
HOXB13-driven IL-6 expression activates AKT and mTOR, and this activation can be blocked by rapamycin
IL-6 promotes tumorigenesis through multiple mechanisms, including cell proliferation and stromal recruitment (21–23), and causes these effects, in part, through phosphorylation of AKT and activation of mTOR-mediated pathways (21, 24). Consistent with high IL-6 activity, Western blot analysis of the MCF7 and BT474 cell line sets showed that, in MCF7-HOXB13 and BT474 cells, STAT3, AKT, and downstream targets of mTOR were phosphorylated (Fig. 4A, left). Conversely, knockdown of HOXB13 in BT474 cells resulted in slight decrease in p-STAT-3 but a marked decrease in p-AKT, pp70S6K, and p-4EBP1, the downstream effectors of the mTOR pathway (Fig. 4A, right). To determine if the stromal recruitment effects are largely mediated by IL-6, human fibroblasts were grown in conditioned media from MCF7-vector or MCF7-HOXB13 cells after treatment with either anti-IL-6 or IL-6R blocking antibody, followed by immunoblot analysis of p-STAT3 (Y705) and p-AKT (S473). We found phosphorylation of STAT3 and AKT was blocked despite treatment with medium from MCF7-HOXB13 cells (Fig. 4B).
Figure 4.
mTOR pathway is activated in HOXB13-expressing cells via IL-6. A, immunoblot analysis of p-STAT3 (Y705), p-AKT (S473), p-p70S6K, and p-4EBP1 in MCF7-HOXB13 and BT474-shHOXB13 cells compared with control cells. B, immunoblot analysis of p-STAT3 (Y705) and p-AKT (S473) in human fibroblasts after treatment with conditioned media from MCF7-vector or MC7-HOXB13, with and without treatment of either anti-IL-6 (left) or IL-6R blocking (right) antibody. C and D, cell viability analysis following treatment with 25, 50, and 100 ng/mL rapamycin in MCF7-HOXB13 (C) and T47DHOXB13 (D) cells compared with vector control cells (*, P < 0.005). E and F, cell invasion/chemotaxis analysis of NIH3T3 cells pretreated with vehicle or 50 ng/mL rapamycin using conditioned medium from MCF7-HOXB13 or control (E), or BT474-HOXB13shRNA or control as the chemoattractant (F). G, growth of MCF7-HOXB13 xenografts implanted in athymic mouse mammary fat pad (mfp). Xenografts grew in the presence of E2 for 3 weeks; mice were then treated with TAM (square), rapamycin (circle), or rapamycin and TAM (triangle) for 3 weeks. Tumor volume was significantly smaller in rapamycin and rapamycin + TAM treated groups compared with either vehicle or tamoxifen alone (*, P < 0.005).
Rapamycin, an inhibitor of mTOR, has been shown to suppress IL-6–driven pathways (21, 25). If so, rapamycin would suppress phenotypes that are associated with high expression of HOXB13. Indeed, both MCF7-HOXB13 and T47D-HOXB13 cells were sensitive to rapamycin, with significantly decreased viability compared with vector-control cells with low HOXB13 expression (Fig. 4C and D). Next, we tested if rapamycin could block fibroblast recruitment by the cancer cells in culture. Cancer cell lines were grown in complete medium with and without 50 nmol/L rapamycin for 24 hours; this media was then used as the chemoattractant for NIH3T3 cells in the matrigel invasion assay. Although MCF7-HOXB13 and BT474-conditioned media successfully increased fibroblast invasion, rapamycin-treated conditioned media failed to attract fibroblasts more than media from control cells (Fig. 4E and F), suggesting that blockade of mTOR-mediated pathways successfully suppressed that phenotype.
Rapamycin can inhibit proliferation of HOXB13-expressing ER+ breast cancer cells in vivo
Given the suppressive effect of rapamycin on HOXB13-expressing ER+ cells in vitro, we tested its effect in vivo. Because HOXB13-expressing cells still expressed low levels of ERα, we also evaluated if there would be a combinatorial effect of using rapamycin with TAM. MCF7-HOXB13 xenografts were grown for 3 weeks. Then mice were treated with either rapamycin, TAM, or rapamycin and TAM for 3 additional weeks. In MCF7-HOXB13 xenografts grown in the mammary fat pad (Fig. 4G) or subcutaneously (Supplementary Fig. S4A), treatment with rapamycin alone, or together with TAM caused significant tumor regression, and was significantly more effective than TAM alone (weeks 5 and 6, P < 0.003; Supplementary Tables S3–S6).
HOXB13 expression correlates directly with IL6 expression and inversely with ERα (ESR1) expression in primary tumor samples and is associated with poor prognosis
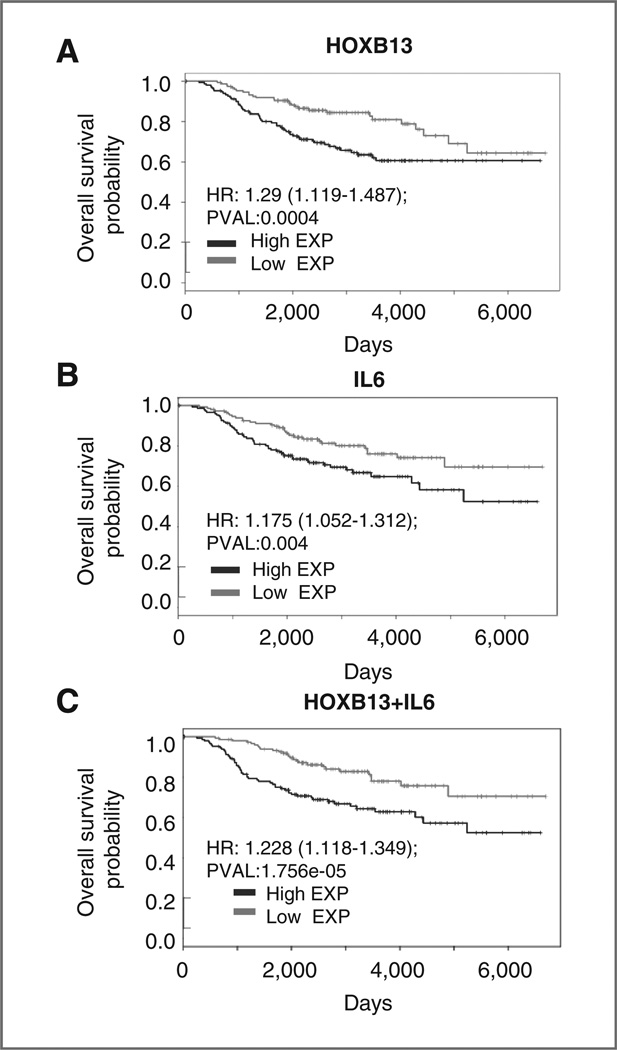
To study the correlation between HOXB13 and IL6 expression and whether a combination of these markers might provide better predictive and prognostic significance compared to either one alone, we analyzed an independent dataset of tumors from 288 patients with ER+ breast cancer (26). As supported by multiple publications (3–5), HOXB13 expression was significantly prognostic of 5-year overall survival (OS; Fig. 5A; HR = 1.29, P = 4 × 10−4). Patients whose primary tumors had high HOXB13 expression had a significantly higher risk of disease recurrence [HR = 3.55; 95% confidence interval (CI), 1.068–11.84] and death from disease (HR = 3.438; 95% CI, 1.036–11.41) compared with those with low HOXB13 expression. Next, we evaluated the role of IL6 expression, in relation to HOXB13 expression, as a clinical biomarker. High IL6 expression was prognostic of 5-year OS (Fig. 5B; HR = 1.175, P = 0.0041). Furthermore, a combination of IL6 and HOXB13 expression was significantly prognostic of OS and more powerful than either gene alone (Fig. 5C; HR = 1.228, P = 1.75 × 10−5). These findings all support the role of upregulation of HOXB13 and IL6 in disease aggression and TAM-R, ER+ breast cancer.
Figure 5.
High HOXB13 and IL6 expression in primary tumors from the NKI dataset (288 patients with ER+ breast cancer; ref. 26) are associated with poor overall survival. A, Kaplan–Meier plot displaying overall survival probability of ER+ breast cancer patients with tumors expressing high or low levels of HOXB13. B, Kaplan–Meier plot displaying overall survival probability of ER+ breast cancer patients with tumors expressing high or low levels of IL6. C, Kaplan–Meier plot displaying overall survival probability of ER+ breast cancer patients stratified by high or low sum expression of HOXB13 and IL6.
Discussion
Numerous studies have showed the predictive value of HOXB13 expression in ER+ breast cancer (2, 3, 11), establishing it as a potentially useful clinical biomarker. Prior work had found HOXB13 mRNA expression correlates inversely with ESR1 expression in breast cancer and, in ER+ TAM-S cell lines, E2 suppresses HOXB13 expression (11). However, no studies have examined the mechanisms by which HOXB13 contributes to disease aggression, specifically in TAM-R and metastatic recurrence. An evaluation of HOXB13 function in TAM-R breast cancer is critical to identifying new therapeutic targets. We conducted the first such examination of HOXB13 in breast cancer and showed its role in aggressive disease, consistent with clinical studies.
For the first time, we have assessed HOXB13 in distant metastatic tumors. We confirmed tumors from nonsurvivors had significantly higher mRNA levels of HOXB13 compared with those from survivors, and HOXB13 expression significantly predicts disease recurrence and death from disease. The metastatic samples used were from a variety of body sites, including liver, lung, lymph nodes, and ovaries. Subset analysis using only one body site was not possible while maintaining statistical power, but we are now evaluating the role of HOXB13 in metastasis to specific anatomic sites.
We showed that HOXB13 suppressed but did not fully abrogate ERα expression, and it also promoted proliferation and TAM-R in vivo, consistent with the association of HOXB13 expression with metastatic disease recurrence in patients despite treatment with TAM. The experimental evidence and the increased stromal component in HOXB13 tumor xenografts suggested HOXB13 promoted tumor formation and disease recurrence also by promoting other pathways, including proliferation and stromal recruitment.
An extensive body of literature is emerging that supports cross-talk and cooperation between stromal cells and tumor cells at multiple levels such as—the soluble factors elicited by tumor cells and those in response by the stromal cells, changes in the extracellular matrix, and alterations in cell adhesion, which collectively lead to a microenvironment conducive to tumor progression (27, 28). A major cell type that alters the tumor milieu is the stromal fibroblast. To study the notable stromal recruitment visible in HOXB13 overexpressing tumors, and to decipher the role of IL-6–mediated pathways in this function, we modeled the in vivo conditions in tissue culture. We showed the ability of normal murine and human fibroblasts to migrate toward conditioned medium from HOXB13 overexpressing tumor cells, but not from vector control cells; and that the action was blunted by pretreating the fibroblasts with anti-IL-6 antibodies (Fig. 3). Although this approach replicated the recruitment of naïve fibroblasts to the tumor site by IL-6 secreted by tumor cells, it would also be important to study, in the future, the molecular and biological differences between cancer-associated-fibroblasts (CAF) existing in the tumors and in normal fibroblasts in the breast, and how CAF action relates to hormone resistance mediated by HOXB13.
In this study, we confirmed HOXB13-directed transcriptional activation of IL6, a common mediator of stromal recruitment IL6 is an activator of the AKT pathway, and this activation is suppressed by mTOR inhibitors (21, 24, 25). We specifically showed that the mTOR inhibitor rapamycin can suppress the proliferation of ER+, HOXB13-expressing cells in vitro and in vivo, alone or in combination with TAM. This work led us to conclude that HOXB13 expression in ER+ breast cancer cells not only allows for estrogen-independent proliferation and tumorigenesis but makes the cells dependent on mTOR-mediated pathways for this proliferation. These pathways need to be more fully defined to identify potential mechanisms of multidrug resistance. Nonetheless, we show that the use of rapamycin with TAM suppresses ER+, HOXB13-expressing breast cancer growth better than either drug alone and can even cause tumor regression in vivo.
In the past year, two major clinical trials showed the efficacy of everolimus, a second generation mTOR inhibitor, in combination with anti-estrogen therapy in recurrent ER+, anti-endocrine-resistant breast cancer (29, 30). Our findings provide a mechanistic explanation for this benefit. There are currently 3 clinically available tests, including the HOXB13/IL17BR RT-qPCR/MGI test known as the Breast Cancer Index (Biotheranostics), that have been well-validated as predictive of TAM-resistance in ER+ tumors. However, no treatment option has been previously suggested for those patients for whom TAM alone would be an unsuccessful treatment. Our work provides a novel and logical mechanistic basis for the use of everolimus with TAM for treatment of ER+ breast tumors with high HOXB13 expression at initial diagnosis. This combination is particularly attractive as everolimus is well studied and well tolerated in humans. Our work provides a rationale for clinical studies to evaluate the use of the Breast Cancer Index to recommend combined use of an mTOR inhibitor and TAM at diagnosis to prevent disease recurrence. These studies should also inform the rapid development of new therapies for TAM-R, HOXB13-expressing ER+ breast cancers.
Supplementary Material
Acknowledgments
The NCI IB45 kits are gratefully acknowledged. The authors thank A. Rein for critically reviewing the manuscript.
Grant Support
This work was supported by grants from the National Institutes of Health (SPORE Program in Breast Cancer, P50 CA 88843) and the Department of Defense COE: W81XWH-04-1-0595 to S. Sukumar, Susan G. Komen Foundation Postdoctoral fellowship KG101506 to K. Jin, and the Hartwell Foundation Biomedical Research Fellowship No.105408 to N. Shah.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
S. Sukumar is a consultant/advisory board member of CBCRF. No potential conflicts of interest were disclosed by the other authors.
Authors' Contributions
Conception and design: N. Shah, K. Jin, S. Park, S. Sukumar
Development of methodology: N. Shah, K. Jin, S. Park, H.Y. Chang, S. Sukumar
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): N. Shah, L.-A. Cruz, H. Sadik, H. Nakshatri, R. Gupta, H.Y. Chang, A. Cimino-Mathews, C. Umbricht
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): N. Shah, K. Jin, S. Cho, C.P. Goswami, Z. Zhang, A. Cimino-Mathews, L. Cope, S. Sukumar
Writing, review, and/or revision of the manuscript: N. Shah, K. Jin, S. Park, S. Cho, C.P. Goswami, Z. Zhang, A. Cimino-Mathews, L. Cope, S. Sukumar
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): N. Shah, H. Nakshatri, C. Umbricht, S. Sukumar
Study supervision: N. Shah, K. Jin, S. Sukumar
References
- 1.EBCTCG. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 2.Ma XJ, Wang Z, Ryan PD, Isakoff SJ, Barmettler A, Fuller A, et al. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5:607–616. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 3.Jansen MP, Sieuwerts AM, Look MP, Ritstier K, Meijer-van Gelder ME, van Staveren IL, et al. HOXB13-to-IL17BR expression ratio is related with tumor aggressiveness and response to tamoxifen of recurrent breast cancer: a retrospective study. J Clin Oncol. 2007;25:662–668. doi: 10.1200/JCO.2006.07.3676. [DOI] [PubMed] [Google Scholar]
- 4.Jerevall PL, Ma XJ, Li H, Salunga R, Kesty NC, Erlander MG, et al. Prognostic utility of HOXB13:IL17BR and molecular grade index in early-stage breast cancer patients from the Stockholm trial. Br J Cancer. 2011;104:1762–1769. doi: 10.1038/bjc.2011.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma XJ, Salunga R, Tuggle JT, Gaudet J, Enright E, McQuary P, et al. Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci U S A. 2003;100:5974–5979. doi: 10.1073/pnas.0931261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10:361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 7.Raman V, Martensen SA, Reisman D, Evron E, Odenwald WF, Jaffee E, et al. CompromisedHOXA5 function can limit p53 expression in human breast tumours. Nature. 2000;405:974–978. doi: 10.1038/35016125. [DOI] [PubMed] [Google Scholar]
- 8.Chu MC, Selam FB, Taylor HS. HOXA10 regulates p53 expression and matrigel invasion in human breast cancer cells. Cancer Biol Ther. 2004;3:568–572. doi: 10.4161/cbt.3.6.848. [DOI] [PubMed] [Google Scholar]
- 9.Wu X, Chen H, Parker B, Rubin E, Zhu T, Lee JS, et al. HOXB7, a homeodomain protein, is overexpressed in breast cancer and confers epithelial-mesenchymal transition. Cancer Res. 2006;66:9527–9534. doi: 10.1158/0008-5472.CAN-05-4470. [DOI] [PubMed] [Google Scholar]
- 10.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z, Dahiya S, Provencher H, Muir B, Carney E, Coser K, et al. The prognostic biomarkers HOXB13, IL17BR, and CHDH are regulated by estrogen in breast cancer. Clin Cancer Res. 2007;13:6327–6334. doi: 10.1158/1078-0432.CCR-07-0310. [DOI] [PubMed] [Google Scholar]
- 12.Miao J, Wang Z, Provencher H, Muir B, Dahiya S, Carney E, et al. HOXB13 promotes ovarian cancer progression. Proc Natl Acad Sci U S A. 2007;104:17093–17098. doi: 10.1073/pnas.0707938104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qian BZ, Li J, Zhang H, Kitamura T, Zhang J, Campion LR, et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475:222–225. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liao D, Luo Y, Markowitz D, Xiang R, Reisfeld RA. Cancer associated fibroblasts promote tumor growth and metastasis by modulating the tumor immune microenvironment in a 4T1 murine breast cancer model. PLoS One. 2009;4(11):e7965. doi: 10.1371/journal.pone.0007965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Acharyya S, Oskarsson T, Vanharanta S, Malladi S, Kim J, Morris PG, et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell. 2012;150:165–178. doi: 10.1016/j.cell.2012.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arihiro K, Oda H, Kaneko M, Inai K. Cytokines facilitate chemotactic motility of breast carcinoma cells. Breast Cancer. 2000;7:221–230. doi: 10.1007/BF02967464. [DOI] [PubMed] [Google Scholar]
- 17.Nutt JE, Lunec J. Induction of metalloproteinase (MMP1) expression by epidermal growth factor (EGF) receptor stimulation and serum deprivation in human breast tumour cells. Eur J Cancer. 1996;32A:2127–2135. doi: 10.1016/s0959-8049(96)00261-4. [DOI] [PubMed] [Google Scholar]
- 18.Rundqvist H, Johnson RS. Hypoxia and metastasis in breast cancer. Curr Top Microbiol Immunol. 2010;345:121–139. doi: 10.1007/82_2010_77. [DOI] [PubMed] [Google Scholar]
- 19.Dong J, Grunstein J, Tejada M, Peale F, Frantz G, Liang WC, et al. VEGF-null cells require PDGFR alpha signaling-mediated stromal fibroblast recruitment for tumorigenesis. EMBO J. 2004;23:2800–2810. doi: 10.1038/sj.emboj.7600289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edin ML, Howe AK, Juliano RL. Inhibition of PKA blocks fibroblast migration in response to growth factors. Exp Cell Res. 2001;270:214–222. doi: 10.1006/excr.2001.5345. [DOI] [PubMed] [Google Scholar]
- 21.Hashimoto I, Koizumi K, Tatematsu M, Minami T, Cho S, Takeno N, et al. Blocking on the CXCR4/mTOR signalling pathway induces the anti-metastatic properties and autophagic cell death in peritoneal disseminated gastric cancer cells. Eur J Cancer. 2008;44:1022–1029. doi: 10.1016/j.ejca.2008.02.043. [DOI] [PubMed] [Google Scholar]
- 22.Silzle T, Kreutz M, Dobler MA, Brockhoff G, Knuechel R, Kunz-Schughart LA. Tumor-associated fibroblasts recruit blood monocytes into tumor tissue. Eur J Immunol. 2003;33:1311–1320. doi: 10.1002/eji.200323057. [DOI] [PubMed] [Google Scholar]
- 23.Katoh H, Hosono K, Ito Y, Suzuki T, Ogawa Y, Kubo H, et al. COX-2 and prostaglandin EP3/EP4 signaling regulate the tumor stromal proangiogenic microenvironment via CXCL12-CXCR4 chemokine systems. Am J Pathol. 2010;176:1469–1483. doi: 10.2353/ajpath.2010.090607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sahara N, Takeshita A, Ono T, Sugimoto Y, Kobayashi M, Shigeno K, et al. Role for interleukin-6 and insulin-like growth factor-I via PI3-K/Akt pathway in the proliferation of CD56- and CD56þ multiple myeloma cells. Exp Hematol. 2006;34:736–744. doi: 10.1016/j.exphem.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 25.Kim JH, Kim JE, Liu HY, Cao W, Chen J. Regulation of interleukin-6-induced hepatic insulin resistance by mammalian target of rapamycin through the STAT3-SOCS3 pathway. J Biol Chem. 2008;283:708–715. doi: 10.1074/jbc.M708568200. [DOI] [PubMed] [Google Scholar]
- 26.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, et al. Agene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 27.Mao Y, Keller ET, Garfield DH, Shen K, Wang J. Stromal cells in tumor microenvironment and breast cancer. Cancer Metastasis Rev. 2013;32:303–315. doi: 10.1007/s10555-012-9415-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Place AE, Jin Huh S, Polyak K. The microenvironment in breast cancer progression: biology and implications for treatment. Breast Cancer Res. 2011;13:227. doi: 10.1186/bcr2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baselga J, Campone M, Piccart M, Burris HA, 3rd, Rugo HS, Sahmoud T, et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med. 2012;366:520–529. doi: 10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bachelot T, Bourgier C, Cropet C, Ray-Coquard I, Ferrero JM, Freyer G, et al. Randomized phase II trial of everolimus in combination with tamoxifen in patients with hormone receptor-positive, human epidermal growth factor receptor 2-negative metastatic breast cancer with prior exposure to aromatase inhibitors: a GINECO study. J Clin Oncol. 2012;30:2718–2724. doi: 10.1200/JCO.2011.39.0708. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.