SUMMARY
Death Inducer Obliterator 3 (Dido3) is implicated in the maintenance of stem cell genomic stability and tumorigenesis. Here, we show that Dido3 regulates the expression of stemness genes in embryonic stem cells through its plant homeodomain (PHD) finger. Binding of Dido3 PHD to histone H3K4me3 is disrupted by threonine phosphorylation that triggers Dido3 translocation from chromatin to the mitotic spindle. The crystal structure of Dido3 PHD in complex with H3K4me3 reveals an atypical aromatic-cage-like binding site that contains a histidine residue. Biochemical, structural, and mutational analyses of the binding mechanism identified the determinants of specificity and affinity and explained the inability of homologous PHF3 to bind H3K4me3. Together, our findings reveal a link between the transcriptional control in embryonic development and regulation of cell division.
INTRODUCTION
Misregulation of the Death Inducer Obliterator (Dido) gene is linked to an increased incidence of myeloid cancers, melanoma, and infertility (Braig and Bosserhoff, 2013; Douet-Guilbert et al., 2008; Fütterer et al., 2005; Prieto et al., 2009). Dido codes for three proteins, termed Dido1, Dido2, and Dido3, which are generated through alternative splicing (Fütterer et al., 2005; Trachana et al., 2007). The largest isoform, Dido3, is found ubiquitously expressed in all human tissues, whereas the two smaller splice variants can be detected only transiently. Recent studies have located Dido3 in the cell nucleus, the mitotic spindle, and the synaptonemal complex (Fütterer et al., 2005; Ozlü et al., 2010; Prieto et al., 2009; Trachana et al., 2007). In agreement with its wide range of subcellular localizations, Dido3 mutations cause blockage of stem cell differentiation, chromosome segregation defects, and meiosis prophase alterations (Fütterer et al., 2005; Ozlü et al., 2010; Prieto et al., 2009; Trachana et al., 2007). The multiple subcellular locations and effects, however, have precluded identification of a molecular function of Dido3 until now.
Bioinformatics analysis of Dido3 composition suggests a role in the maintenance of genomic stability (Rojas et al., 2005). Dido3 comprises a plant homeodomain (PHD) finger, a transcription elongation factor S-II subunit M (TFSIIM) domain, a spen paralog and ortholog (SPOC) module, and a long C-terminal region (CT) of unknown homology (Figures 1A and S1A). Although precise functions of the TFSIIM and SPOC modules of Dido3 have not been determined, the TFSIIM domain of the transcription elongation factor (TFIIS) binds to and facilitates activity of RNA polymerase II (Kettenberger et al., 2003), and the SPOC module is proposed to mediate protein-protein interactions. The PHD domain belongs to a large family of zinc fingers that recognize posttranslationally modified histone tails, including methylated lysine residues (Musselman and Kutateladze, 2011; Musselman et al., 2012); however, its role in the diverse activities of Dido3 remains unclear. Elucidating the mechanisms that govern these activities may shed light on the importance of Dido3 mutations in promoting tumorigenesis.
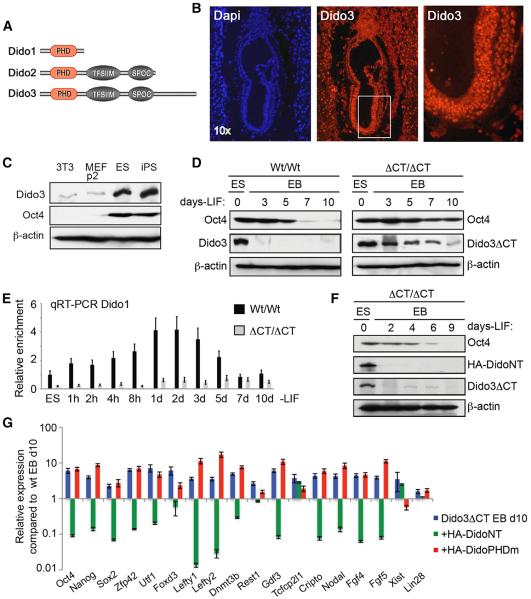
Figure 1. Dido3 Is Necessary for Early Embryonic Stem Cell Development.
(A) The three isoforms encoded by the Dido3 locus (Dido1, Dido2, and Dido3).
(B) Expression of Dido3 was analyzed in Wt/Wt embryos at E7.5 by immunofluorescence.
(C) Expression of Dido3 and Oct4 was analyzed by western blot in 3T3 and primary MEF somatic fibroblasts and in pluripotent embryonic and induced stem cells; β-actin was used as loading control.
(D) Wt/Wt and ΔCT/ΔCT ESCs were aggregated into EBs and maintained without LIF in untreated bacterial plates for the time indicated to induce differentiation. In western blot, we analyzed Oct4 and endogenous Dido3 expression in WT ESCs and their EBs, as well as Oct4 and Dido3ΔCT in mutant ESCs and EBs.
(E) In contrast to Dido3, which shows no significant differences (data not shown), Dido1 is upregulated at the onset of differentiation in Wt/Wt cells but not in ΔCT/ΔCT cells. Quantitative RT-PCR was used to determine Dido1 expression; values were normalized to actin.
(F) ΔCT/ΔCT ESCs expressing HA-DidoNT were treated as in (D) and analyzed by western blot for expression of Oct4, endogenous Dido3ΔCT, as well as for overexpressed HA-DidoNT.
(G) Quantitative RT-PCR was used to determine expression of selected stemness markers in d10 ΔCT/ΔCT EBs (blue bars) overexpressing HA-DidoNT (green) or HA-DidoPHDm (red).
Data show mean ± SD (n = 3) for one representative experiment of two performed, indicating expression of the markers analyzed in cells transfected with HA-DidoNT or HA-DidoPHDm, relative to d10 Wt/Wt EBs (baseline).
See also Figure S1.
In this study, we show that Dido3 regulates stemness genes in embryonic stem cells (ESCs) and that the PHD finger is required for this regulation. We found that phosphorylation of H3T3 or H3T6 abrogates binding of the PHD finger to histone H3K4me3 and releases Dido3 from chromatin during mitosis, thus imparting a cell-cycle-dependent molecular switch that drives relocation of the protein to the mitotic spindle. Structural and biochemical analysis of the PHD-H3K4me3 complex elucidated the molecular mechanism of H3K4me3 recognition and the repulsive effect of threonine phosphorylation. Together, our data demonstrate a link between transcriptional regulation in embryonic development and cell division and support the idea that chromatin condensation and spindle formation are coordinated through a phosphorylation-methylation crosstalk.
RESULTS
Dido3 Is Necessary for Early Embryonic Stem Cell Development
Epiblast integrity is critical to early embryonic development. Epiblast cells are pluripotent cells that give rise to all three germ layers during gastrulation. As shown in Figure 1B, the epiblast expresses very high levels of Dido3. Furthermore, Dido3 was highly expressed in both ESCs and induced pluripotent stem (iPS) cells; however, its expression level was noticeably reduced in somatic cells, indicating that Dido might be a stemness gene (Figure 1C). Because elimination of the Dido gene causes early embryonic lethality (Fütterer et al., 2012), to study the role of Dido3 in embryonic development, we generated a Dido mouse mutant in which the CT region of Dido3 (Dido3ΔCT) is replaced with a red fluorescent protein (RFP) tag.
Epiblast cell differentiation is triggered by repression of the transcription factors needed to establish and maintain stem cell pluripotency and self-renewal, such as Oct4. We monitored Dido3 and Oct4 levels in Wt/Wt and ΔCT/ΔCT ESCs and embryonic bodies (EBs). Whereas withdrawal of LIF (leukemia inhibitory factor) resulted in normal Oct4 downregulation in Wt/Wt EBs after 5 days, this stemness marker was detected in Dido3ΔCT EB cells even after 10 days (Figure 1D). Dido3 was downregulated even earlier than Oct4 in Wt/Wt EBs, indicating possible upstream control of Oct4 silencing, whereas persistent Dido3ΔCT protein expression appeared to be linked to Oct4 persistence. These data indicate that the ΔCT/ΔCT EB cells are unable to differentiate and thus can be used as a model for delayed stem cell differentiation.
Displacement of Dido3 by Dido1 Is Required for Repression of Stemness Genes
Unexpectedly, we found that Dido1 is transiently upregulated at the onset of differentiation in Wt/Wt cells, but not in ΔCT/ΔCT (Figure 1E). We therefore generated a constitutively active form of Dido1 (DidoNT), which contains the PHD finger but lacks a short Dido1-specific nuclear export signal at the C terminus. To examine the role of this Dido1 mimetic in differentiation, we ectopically expressed DidoNT constructs bearing either a wild-type PHD finger (HA-DidoNT) or a mutant that lacks two cysteines needed for domain stability (HA-DidoPHDm) in Dido3ΔCT ESCs. HA-DidoNT was sufficient to overcome the differentiation defect, as shown by Oct4 and Dido3ΔCT downregulation (Figure 1F). The PHD mutant HA-DidoPHDm neither induced differentiation nor silenced Oct4 or Dido3ΔCT, although the protein was expressed efficiently (Figure S1B). The wild-type PHD finger in the context of full-length HA-Dido3 also restored the ability of these cells to downregulate Oct4 (Figure S1B).
Quantitative RT-PCR assays of d10 Dido3ΔCT EB cells showed that most common stemness markers were also repressed by HA-DidoNT overexpression, but not by overexpression of the mutant HA-DidoPHDm (Figure 1G). Together, these data identify Dido3 as a putative stemness gene, displacement of which is necessary for silencing of a set of stemness genes and promoting differentiation, and suggest that the PHD finger plays a critical role in this process.
Dido3 PHD Interacts with H3K4me3 In Vivo and In Vitro
Expression or repression of stemness genes in early embryonic development depends on epigenetic changes, such as trimethylation of H3K4 and H3K9. The Drosophila ortholog of Dido3, PPS, or dDido has the same domain architecture as the human protein with a PHD finger being the only candidate for histone interaction. Interestingly, dDido associates with the first exon of the sxl gene in Drosophila independently of transcription factor recruitment (Johnson et al., 2010), and its binding overlaps with H3K4me3 but not with repressive marks, including H3K9me3, suggesting that Dido3 PHD recognizes trimethylated Lys4 (Figure 2A). To corroborate the interaction of Dido3 with this epigenetic mark, colocalization of the two in mouse embryonic fibroblasts (MEFs) was examined by fluorescence microscopy (Figure 2B). Dido3 colocalized with H3K4me3 in euchromatin but was absent from H3K9me3-positive pericentric heterochromatin, suggesting that the protein associates with H3K4me3 in vivo. To confirm the role of the PHD finger, copurification of native histone complexes by wild-type and mutant DidoNT was assayed in pull-down experiments (Figure 2C). Whereas pull-down of wild-type DidoNT allowed for copurification of histones, the PHD mutant, unable to promote downregulation of stemness genes, did not. Restoration of zinc coordination, through an exchange between histidine and cysteine, again enabled binding of DidoNT to histones.
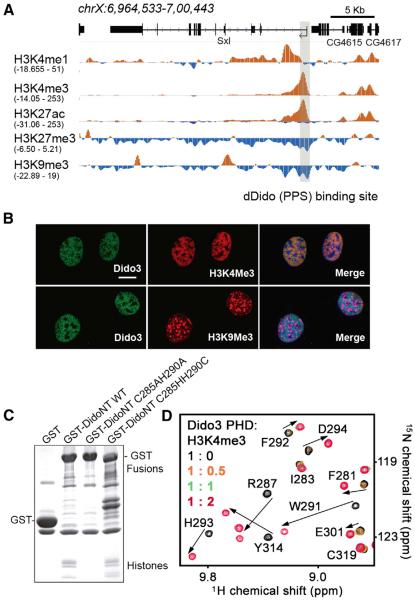
Figure 2. The Dido3 PHD Finger Binds H3K4me3.
(A) Snapshot of a 35 kb genomic region containing the Sxl gene. The dDido (PPS) binding site is shown in gray. Individual tracks for H3K4me1, H3K4me3, H3K27ac, and H3K9me3 chromatin immunoprecipitation sequencing (ChIP-seq) experiments were obtained from the modENCODE consortium (modENCODE Consortium et al., 2010). The range of the ChIP-seq reads is provided for each track.
(B) 3T3 cells were grown on coverslips and labeled with antibodies against Dido3 and antibodies against H3K4me3 (upper row) or H3K9me3 (lower row). Confocal fluorescence microscopy showed that Dido3 colocalized with H3K4me3 in euchromatin but was absent from H3K9me3-positive heterochromatin. Scale bar, 10 μm.
(C) The intact PHD finger is necessary to pull down native chromatin. DidoNT bearing a WT or mutated PHD domain was expressed in HEK293 cells and purified by affinity chromatography. GST alone was used as negative control. Whereas isolation of WT DidoNT allowed for copurification of histone complexes, destabilization of the PHD domain (C285AH290A) resulted in loss of interaction. Restoration of the PHD domain (C285HH290C) reestablished histone binding.
(D) Superimposed 1H, 15N HSQC spectra of Dido3 PHD collected upon titration with the H3K4me3 peptide. Spectra are color coded according to the protein:peptide molar ratio.
See also Figure S2.
The direct interaction between the Dido3 PHD finger and H3K4me3 was substantiated by NMR titration experiments. 1H,15N heteronuclear single quantum coherence (HSQC) spectra of uniformly 15N-labeled Dido3 PHD were recorded while unlabeled H3K4me3 peptide (amino acids [aa] 1–12 of H3) was gradually added to the NMR sample (Figure 2D). Extensive chemical-shift changes were observed in approximately one-third of the protein residues. A number of resonances corresponding to the unbound state of PHD decreased in intensity upon titration of the peptide, disappearing completely at a protein:peptide ratio of 1:1 (Figure 2D, green). Concomitantly, another set of resonances that corresponds to the bound state of PHD gradually appeared, fully replacing the ligand-free set at a ratio of 1:2 (Figure 2D, red). This pattern of chemical-shift changes is characteristic of the slow exchange regime on the NMR timescale and indicates robust interaction between Dido3 PHD and H3K4me3. Altogether, these data imply that the Dido3 PHD domain binds H3K4me3 in vitro and in vivo.
The Structure of Dido3 PHD in Complex with H3K4me3
To elucidate the molecular mechanism for recognition of H3K4me3, we determined a 1.35 Å resolution crystal structure of the Dido3 PHD finger bound to the H3K4me3 peptide (Figure 3; Table 1). The PHD finger folds into a short antiparallel β sheet, followed by a 310-helical turn, an α helix, and another α-helical turn at the far C terminus (Figure 3A). The characteristic C4HC3 motif of PHD fingers coordinates two zinc ions. The H3K4me3 peptide adopts an extended conformation and forms a third antiparallel β strand, pairing with β1 of the protein. The intermolecular hydrogen bonds are seen between the backbone carbonyl groups of Arg2 and Lys4me3 of the peptide and the backbone amino groups of C284 and M282 of the protein, respectively. The backbone amino group of Lys4me3 donates a hydrogen bond to the backbone carbonyl group of M282.
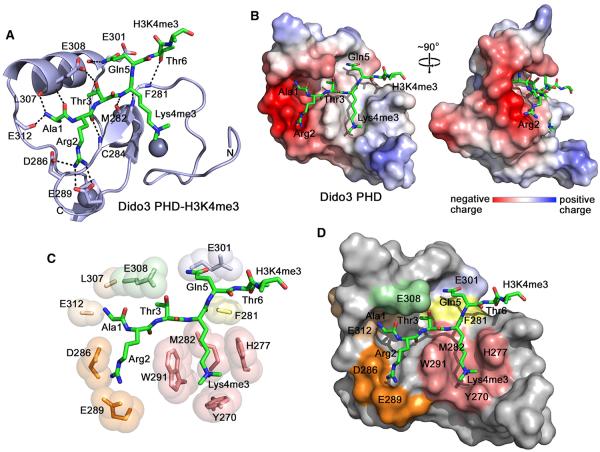
Figure 3. The Crystal Structure of the PHD Finger of Dido3 in Complex with the H3K4me3 Peptide.
(A) The ribbon diagram of the PHD finger. The H3K4me3 peptide is shown in a stick model. Dashed lines represent intermolecular hydrogen bonds. For clarity, here and throughout the text, individual residues of the PHD finger are denoted using a single-letter code, whereas individual residues of the histone peptide are denoted using a three-letter code.
(B) The electrostatic surface potential of Dido3 PHD is colored blue and red for positive and negative charges, respectively.
(C) A zoom-in view of the H3K4me3 binding pocket.
(D) The Dido3 PHD finger is depicted as a solid surface with the peptide shown in a stick model. The protein residues involved in the interactions with Ala1, Arg2, Thr3, Lys4me3, Gln5, and Thr6 of the peptide are colored wheat, orange, light green, salmon, light blue, and yellow, respectively.
See also Figure S3.
Table 1.
Data Collection and Refinement Statistics
| Data Collection | Dido3 PHD-H3K4me3 (SAD) |
|---|---|
| Wavelength(Å) | 1 |
| Space group | P41212 |
| Resolution (Å) | 40.82-1.35 |
| Cell dimensions (Å) | a = b = 40.82 c = 80.62 α = β = γ = 90° |
| Number of measured reflections | 350737 |
| Number of unique reflections | 15723 |
| Completeness (%) | 100 (100) |
| Anomalous completeness (%) | 100 (100) |
| Redundancy | 22.3 (22.3) |
| Anomalous redundancy | 12.1 (11.7) |
| I/σ(I) | 24.4 (6.4) |
| Rmerge (%) | 6.2 (48.2) |
|
| |
| Refinement | |
|
| |
| Resolution (Å) | 28.68-1.35 (1.40-1.35) |
| Number of reflections | 28593 |
| R-factor (%) | 13.24 |
| R-free (%) | 14.22 |
| Number of protein atoms | 987 |
| Number of heterogen atoms | 75 |
| Number of zinc atoms | 2 |
| Number of water molecules | 73 |
| Figures of merit (FOM) | 0.92 |
| Rmsd from ideal values: Bond lengths (Å) | 0.007 |
| Rmsd from ideal values: Bond angles (°) | 1.169 |
| Average B values (Å2): Protein chain A | 22.9 |
| Average B values (Å2): Peptide chain U | 24.3 |
| Average B values (Å2): Water | 36.3 |
| Average B values (Å2): Zinc | 15.6 |
| Ramachandran plot analysis: Residues in most favored regions (%) |
92.50 |
| Ramachandran plot analysis: Residues in additional allowed regions (%) |
5.70 |
| Ramachandran plot analysis: Residues in generously allowed regions (%) |
1.90 |
| Ramachandran plot analysis: Residues in disallowed regions |
0 |
The binding interface is extensive. The first six residues of the H3K4me3 peptide are in direct contact with 16 surface residues in the PHD finger, which form a deep, branched channel. Whereas Ala1, Arg2, and Thr3 are bound in a highly acidic half of the channel, Lys4me3, Gln5, and Thr6 lay in a neutral or slightly basic half (Figure 3B). Three hydrogen bonds restrain Ala1. The two are formed between the N-terminal amino group of Ala1 and the backbone carbonyl groups of L307 and E312, and another is seen between the carbonyl group of Ala1 and the backbone amino group of E308. The fully extended side chain of Arg2 is bound in an elongated groove. The guanidino moiety of Arg2 is involved in the hydrogen bonding and ionic contacts with the carboxyl groups of D286 and E289. Thr3 of the peptide is buried in a narrow cleft flanked by W291 and E308. The hydroxyl group of Thr3 donates a hydrogen bond to the carboxyl group of E308, the side chain of which hangs over Thr3, making it largely solvent inaccessible. W291 also separates the Arg2- and Lys4me3-binding sites. The aromatic side chains of Y270, H277, and W291 are positioned almost perpendicular to each other and to the protein surface and together with M282, which lays at the bottom, create a cage around the trimethylammonium group of Lys4 (Figures 3C and 3D). The amide group of Gln5 occupies a shallow pocket and is stabilized through a hydrogen bond with the backbone carbonyl group of E301. The hydroxyl group of Thr6 is hydrogen bonded to the backbone carbonyl group of F281.
Dido3 PHD Is Specific for Histone H3K4me3
To determine whether Dido3 selects for the trimethylated state of lysine, we compared binding of the PHD finger to mono-, di-, tri-, and nonmethylated peptides (Figure 4). We assigned backbone and side chain 1H, 13C, and 15N chemical shifts of Dido3 PHD using a set of three-dimensional triple resonance experiments and analyzed perturbations in the 1H, 15N HSQC spectra of the protein upon binding to H3K4me3 (Figure 2D). Plotting chemical-shift changes for each backbone amide allowed us to identify the residues of Dido3 most affected due to the interaction (Figure 4B). In agreement with the crystal structure of the complex, in solution the protein residues located in the binding interface, particularly the aromatic cage residues Y270 and W291, were perturbed to the highest degree (Figure 4D). A very similar pattern of chemical-shift changes in the PHD finger was seen upon addition of H3K4me2 or H3K4me1 (aa 1–12 of H3), implying that these peptides occupy the same binding pocket; however, the intermediate exchange regime for the association with H3K4me1 pointed to a weaker interaction. This was confirmed through measuring equilibrium dissociation constants Kds using tryptophan fluorescence and NMR. We found that the Dido3 PHD finger exhibits a 1 μM affinity for H3K4me3, whereas it binds to H3K4me2 and H3K4me1 6- and 85-fold, respectively, weaker (Figures 4C and S2). Unmodified H3K4me0 peptide (aa 1–19 of H3) induced almost negligible changes, which indicated that the Dido3 PHD finger does not recognize nonmethylated H3 (Kd > 5 mM) (Figures 4A and 4C).
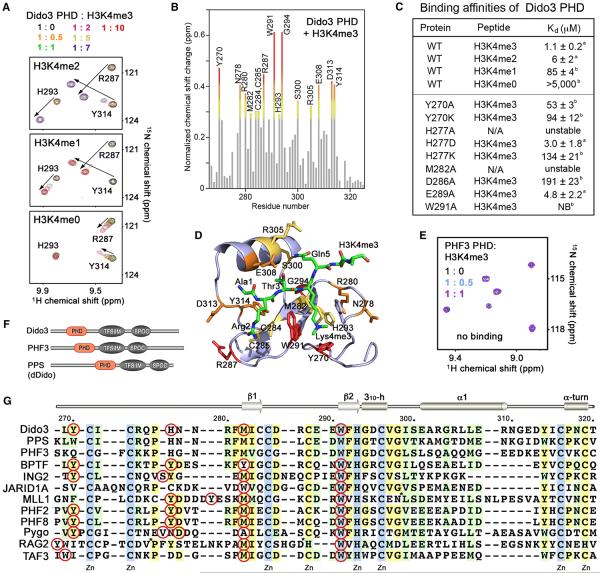
Figure 4. The Molecular Basis for H3K4me3 Recognition by Dido3 PHD.
(A) Superimposed 1H, 15N HSQC spectra of Dido3 PHD collected upon titration with H3K4me2 and H3K4me1 peptides (residues 1–12 of H3), as well as H3K4me0 (residues 1–19 of H3). Spectra are color coded according to the protein:peptide molar ratio.
(B) The normalized chemical-shift changes observed in 1H, 15N HSQC spectra of the PHD finger (depicted in Figure 2C) as a function of residue. Residues showing large chemical-shift differences are labeled. Differences greater than the average plus one and one-half SD, the average plus one SD, and the average plus one-half SD are shown in red, orange, and yellow, respectively.
(C) Binding affinities of wild-type (WT) and mutated Dido3 PHD for the indicated histone peptides measured by tryptophan fluorescence (A) or NMR (B). NB, no binding.
(D) The residues of the Dido3 PHD finger, most perturbed due to the interaction with H3K4me3, are labeled and colored as in (B).
(E) Superimposed 1H, 15N HSQC spectra of PHF3 PHD recorded as H3K4me3 peptide was titrated in.
(F) Architecture of the Dido/PHF3/PPS family.
(G) Alignment of PHD finger sequences: absolutely, moderately, and weakly conserved residues are colored light blue, pale yellow, and light green, respectively. The aromatic cage residues are indicated by red circles. Each tenth residue of Dido3 is labeled. Secondary structure elements of Dido3 PHD are shown on the top. For simplicity, eight residues in MLL1 and seven residues in RAG2 are deleted, as indicated by asterisks.
Molecular Mechanism of the H3K4me3 Recognition by Dido3
Alignment of the amino acid sequences of Dido3 PHD and the PHD fingers from other proteins known to interact with H3K4me3 reveals some similarities but also highlights a uniqueness of the aromatic cage of Dido3 (Figure 4G). The cage contains an invariable tryptophan (W291) two residues preceding the zinc-coordinating histidine, a highly conserved methionine (M282) three residues preceding the third zinc-coordinating cysteine, and a moderately conserved tyrosine (Y270) preceding the first zinc-coordinating cysteine. Unexpectedly, we found that Dido3 uses a histidine (H277) as the fourth residue in the cage. Its aromatic moiety is oriented parallel to the aromatic ring of W291 and orthogonal to the aromatic ring of Y270 (Figure 3C). The three aromatic residues are perfectly positioned to make cation-π interactions with the trimethylammonium group of Lys4. The distance between the aromatic groups of W291, Y270, and H277 and the nitrogen atom of Lys4me3 is ~4.3, 4.2, and 4.6 Å, respectively. The hydrophobic M282 is lining the bottom side of the cage, with the polarizable lone electron pair of its sulfur atom likely offering additional stabilization.
The importance of the aromatic cage residues was confirmed through mutagenesis. Substitution of W291 with an alanine eliminated binding of Dido3 to H3K4me3 but did not compromise the protein structure (Figures 4C and S2). Replacement of M282 in the hydrophobic PHD core, however, caused unfolding, implying that M282 is required for structural stability. Mutations of either Y270 or H277 decreased binding, though the structures of the mutants, except for H277A, remained intact (Figures 4C and S2). Interestingly, the PHD finger of PHF3, a paralog of Dido3, contains all but one conserved residues of the aromatic cage, including a tryptophan, a methionine, and a histidine, but has a glutamine instead of a tyrosine (Y270 in Dido3) (Figures 4F and 4G). We produced 15N-labeled PHF3 PHD and tested its binding to H3K4me3 by NMR (Figure 4E). A lack of any changes in 1H, 15N HSQC spectra of the PHF3 PHD finger upon titration with the H3K4me3 peptide indicated that PHF3 does not recognize this epigenetic mark and that other factors besides the aromatic cage can influence this interaction. The Drosophila ortholog of Dido3, PPS, contains a tryptophan residue in place of Y270 and thus most likely binds to H3K4me3 (Figure 4G).
In the Dido3 PHD-H3K4me3 complex, the guanidino group of Arg2 of the peptide is fixed through the interactions with D286 and E289 of the protein. We found that binding of the D286A mutant to H3K4me3 was decreased ~200-fold, and binding of E289A was reduced ~4-fold (Figure 4C). A similar reduction of binding to H3K4me3 was observed when the corresponding Asp and Glu residues in the ING2 PHD finger were mutated (Peña et al., 2006) (Figure S3). Thus, restraining of Arg2 by Dido3 favorably contributes to the complex formation, and the fact that the PHF3 PHD finger has a glycine residue in place of D286 (Dido3 numbering) may explain its full inactivity toward H3K4me3 (Figure 4G).
Phylogenetic analysis of the PHD fingers reveals that Dido3 PHD is closely related to the PHD modules of PHF2 and PHF8. We overlaid the structures of the PHD fingers of Dido3, PHF2, and PHF8 in complex with H3K4me3 (Horton et al., 2010; Wen et al., 2010) (Figure S3). On the whole, the histone peptide and the aromatic cage residues are superimposed well; however, the Arg2-binding groove in PHF2/8 is less acidic, with only one glutamic acid contacting Arg2. More importantly, the side chains of Thr3, Gln5, and Thr6 of the peptide in the PHF2/8 complexes are not restrained, and the Thr3-Thr6 sequence is notably solvent exposed (Figure S3). In contrast, in the Dido3 complex the side chains of Thr3, Gln5, and Thr6 are fixed, and Thr3 is significantly less solvent accessible, which has implications for the Dido3 biological activity (see below).
Phosphorylation of H3T3 or H3T6 Abolishes Binding to H3K4me3
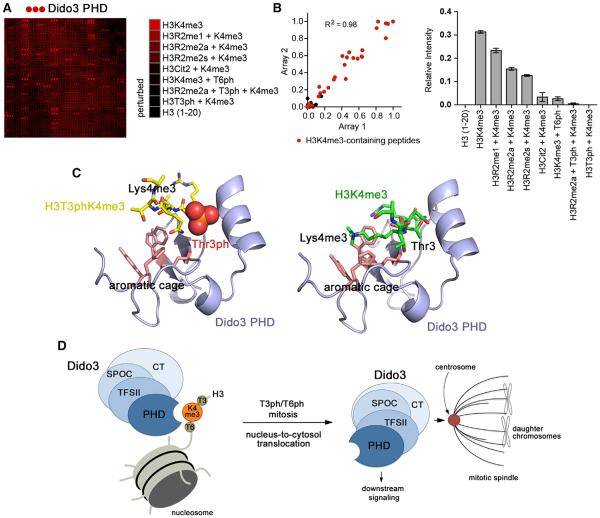
We next examined the effect of posttranslational modifications (PTMs) in the vicinity of Lys4 on the interaction of the Dido3 PHD finger with H3K4me3 by a high-throughput peptide microarray (Figure 5). In the microarray, a library of ~200 biotinylated peptides corresponding to the tails of histones H3, H4, H2A, and H2B were immobilized on streptavidin-coated glass slides. The peptides contained single or multiple covalently modified residues, including acetylated and mono-, di-, and trimethylated lysine, monomethylated, and symmetrically and asymmetrically dimethylated arginine, citrulline, and phosphorylated serine and threonine residues (Table S1). Screening GST-tagged Dido3 PHD against the library revealed that the protein recognizes various H3K4me3-containing peptides but does not associate with PTMs on other histones. We found that PTMs on Arg2, Thr3, and Thr6 of histone H3 substantially affect the interaction with H3K4me3 (Figures 5A and 5B). Phosphorylation of Thr3 (H3T3ph) completely abrogated this interaction and phosphorylation of Thr6 (H3T6ph) strongly inhibited it. As expected, methylation of Arg (H3R2me) or citrullination of Arg2 (H3Cit2) also reduced binding.
Figure 5. Phosphorylation of H3T3 or H3T6 Abolishes Binding to H3K4me3.
(A) GST-Dido3 PHD was hybridized to a histone peptide microarray. Red spots indicate binding of Dido3 PHD to various H3K4me3 peptides. The complete list of singly and multiply modified peptides used in the microarray is shown in Table S1.
(B) Left: scatterplot of two arrays probed with Dido3 PHD. Red dots indicate H3K4me3-containing peptides. The correlation coefficient was calculated by linear regression analysis using GraphPad Prism v5. Right: relative average signal intensities for peptide microarrays probed with Dido3 PHD. Error bars represent SEM from pooled averages of two independent array experiments.
(C) Modeling of the Dido3 PHD-H3T3phK4me3 and Dido3 PHD-H3K4me3 complexes using HADDOCK. The lowest energy structures from the top clusters are shown.
(D) A model for Dido3 translocation from chromatin to the mitotic spindle in mitosis.
See also Figure S4.
A model of Dido3 PHD in complex with H3T3phK4me3 and H3K4me3, which was generated using the docking algorithm HADDOCK, helped to explain the inhibitory effect of phosphorylation (Figure 5C). Phosphorylation of Thr3 is sterically and electrostatically unfavorable because this residue is bound in a narrow cleft and is sheltered from solvent. A bulky phosphate group on Thr3 prevents insertion of K4me3 into the aromatic cage and eliminates the hydrogen bond to the carboxylic group of E308. Likewise, phosphorylation of Thr6 disrupts the hydrogen bond with F281 and may also be sterically prohibitive.
Cell-Cycle-Dependent Localization of Dido3
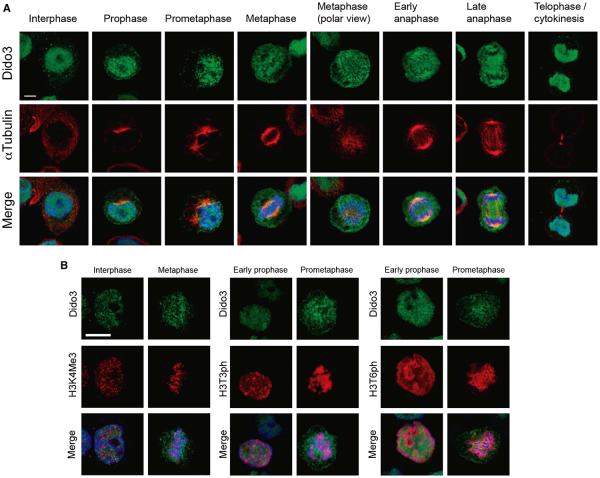
Phosphorylation of Thr3 and Thr6 are PTMs closely linked to chromatin condensation during mitosis (Sawicka and Seiser, 2012). In mitosis, Dido3 is bound to the mitotic spindle, where it controls microtubule organization (Trachana et al., 2007). We tested the idea that Dido3 is released from chromatin to associate with microtubules in a cell-cycle-dependent manner. HT29 colon cancer cells were labeled with anti-Dido3 and anti-Tubulin antibodies and studied by fluorescence microscopy (Figure 6A). Whereas Dido3 associated with chromatin in the interphase nucleus, it was found directly surrounding condensed chromosomes when cells entered mitosis. At this stage, the labeling pattern followed the nucleus outline, indicating that Dido3 disengages from condensed chromosomes in prophase before nuclear envelope breakdown. Later in mitosis, Dido3 was found more diffusely distributed in the cytosol as chromosomes migrated (prometaphase), and subsequently enriched on microtubules until chromosomes decondensed (metaphase to telophase). In conclusion, Dido3 exclusion from chromatin closely follows chromosome condensation in mitosis.
Figure 6. Dido3 Is Excluded from Chromatin in Mitosis.
(A) HT29 cells were seeded on coverslips, labeled with antibodies against Dido3 and αTubulin, and visualized by confocal fluorescence microscopy. Mitotic phases represented are indicated on the left. Dido3 exclusion from the chromatin is first observed in prophase, is maintained while chromosomes are condensed, and finally reverts in telophase.
(B) HT29 cells were seeded on coverslips, labeled with antibodies against Dido3 and antibodies against H3K4me3, H3T3ph, or H3T6ph, and subsequently studied by confocal fluorescence microscopy. Dido3 loses affinity for condensed chromosomes, even though the H3K4me3 mark is maintained (left). Dido3is excluded from H3T3ph- and H3T6-positive chromatin (middle and right) in early prophase, before chromatin condensation is evident, and maintained in fully condensed chromosomes. Scale bars, 10 μm.
See also Figures S5 and S6.
H3K4me3 persists in a stable manner when somatic cells pass through mitosis. To assess the role of H3K4me3 recognition by Dido, we monitored localization of Dido3 in the interphase and metaphase. As shown in Figure 6B, left panel, Dido3 is released from mitotic chromosomes even though trimethylation is preserved, suggesting that other PTMs control Dido3 release in mitosis. Because H3T3ph and H3T6ph, responsible for the loss of H3K4me3 binding, appear during early prophase (Polioudaki et al., 2004; Sawicka and Seiser, 2012), these modifications most likely account for cell-cycle-dependent disengagement of Dido3 from chromatin. To confirm this, HT29 cells were double-labeled with anti-Dido3 and anti-H3T3ph or anti-H3T6ph antibodies. Histone H3 phosphorylation was first observed early in prophase before chromosome condensation became evident (Figure 6B, middle and right panels). Dido3 was excluded from chromatin regions positive for either H3T3ph or H3T6ph, illustrating the importance of histone H3 phosphorylation for Dido3 translocation. Although T3ph and T6ph had a similar effect on Dido3 release, their individual distribution revealed striking differences. Because Lys4 trimethylation partially inhibits Thr3 phosphorylation (Karimi-Ashtiyani and Houben, 2013), Thr6 phosphorylation could represent a complementary mechanism of prophase chromatin signaling. Similar results were observed using wild-type and Dido3ΔCT ESCs (Figure S4). Dido3 colocalized with H3K4me3-enriched chromatin in the interphase but was excluded in mitosis. Taken together, these results demonstrate that histone H3 phosphorylation is a key determinant of Dido3 redistribution in early mitosis, suggesting coordination between chromatin condensation and mitotic spindle formation.
The PHD-Finger Containing NT Region of Dido3 Is Essential in Cell Division
To determine the role of the PHD finger in the mitotic function of Dido3, we overexpressed wild-type Dido3 and the Dido3 mutant lacking the N-terminal region harboring the PHD finger (Dido3ΔNT) in MEFs. As shown in Figure S5, Dido3ΔNT and Dido3 accumulate well at centrosomes, indicating that the N-terminal region, and therefore the PHD finger, is not required for Dido3 localization to the mitotic spindle. However, the Dido3ΔNT mutant caused the amplification of centrosomes, implying that this region is necessary for proper cell division process (Figure S5).
DISCUSSION
Dido3 has been found alternatively in the nucleus and in complex with the mitotic apparatus (Fütterer et al., 2005; Ozlü et al., 2010; Prieto et al., 2009; Trachana et al., 2007). Although the different locations suggest divergent roles in gene expression and spindle checkpoint signaling and active protein targeting, the key determinants for Dido3 activities and redistribution remain unknown. Here, we show that in the nucleus, Dido3 associates with chromatin through binding of its PHD finger to H3K4me3, a histone mark generally associated with gene transcription. In stem cells, H3K4me3 is enriched at the promoters of actively transcribed stemness genes, including the master regulator Oct4. Because Dido3 does not have an enzymatic activity, to regulate stemness-relevant gene expression, it may interact with transcriptional activators and other regulatory complexes through the TFSIIM and SPOC domains.
Transcriptional silencing of Oct4 and other stemness genes is dependent on LIF withdrawal in vitro, and on developmentally regulated signals delivered at the onset of gastrulation in vivo. These signals trigger transient upregulation of Dido1, which can displace Dido3 at its targets, leading to Dido3 release and downregulation of a subset of stemness genes (Figure S6). The inability of ΔCT/ΔCT cells to upregulate Dido1 causes Dido3ΔCT to be constitutively active. It remains bound to H3K4me3 and maintains self-renewal conditions, thus impeding silencing of the stemness genes. Overexpression of the Dido1 mimetic, HA-DidoNT, which contains a wild-type PHD finger but lacks the TFSIIM and SPOC domains, releases Dido3ΔCT and triggers Oct4 downregulation. In contrast, the HA-DidoPHDm mutant that is unable to bind H3K4me3, does not displace the Dido3ΔCT protein, and maintains stem cells in self-renewal conditions, preventing differentiation.
In this context, we believe that through targeting H3K4me3, Dido3 maintains stemness-related genes in a “primed” state by recruiting the activator or other regulatory complexes to the target promoters. This mechanism permits pluripotent cells to rapidly integrate the molecular signals that trigger differentiation, including the transient upregulation of Dido1. Displacement of Dido3 from its H3K4me3-enriched targets provides a fine-tuned mechanism that could be vital during early embryogenesis, when rapid cell cycling reduces the time window for coordinated expression of genes that determine cell fate and embryonic patterning.
Prophase chromatin condensation is characterized by the phosphorylation of the N-terminal tail of histone H3. We found that phosphorylation of H3T3 and H3T6 provides a negative signal recognized by the Dido3 PHD finger, allowing for Dido3 translocation to microtubules and subsequent activation of its mitotic role (Figure 5D). Although the PHD finger is not required for Dido3 localization to the centrosome, the loss of the N-terminal region of Dido3 results in centrosome amplification abnormalities, suggesting its role in downstream signaling processes leading to cell division. In conclusion, our data reveal a phosphorylation-methylation crosstalk that modulates localization and activities of Dido3, linking transcriptional regulation, embryonic development, and cell division.
EXPERIMENTAL PROCEDURES
Mice and Embryonic Stem Cells
Dido3 knockout mice were generated by targeting the Dido3-specific exon 16 of the Dido3 locus and were handled in accordance with National and European Union legislation, as well as with approval of the Committee for Ethics in Animal Experimentation of the Centro Nacional de Biotecnología (Fütterer et al., 2012).
ESCs were maintained in DMEM medium (Gibco, Invitrogen) with 20% fetal calf serum (FCS), nonessential amino acids, 0.05 mM β-mercaptoethanol, 100 U/ml penicillin, 0.1 mg/ml streptomycin, and 1,000 U/ml murine LIF (Chemicon International) on a layer of irradiated or mitomycin-C-treated MEF. For EB formation, ESCs were cultured in LIF-free culture medium in uncoated Petri dishes in suspension and harvested at the indicated time points. For further details, please refer to Extended Experimental Procedures.
Protein Crystallization and Structure Determination by X-Ray Crystallography
Purified Dido3 PHD (aa 266–325, 12.5 mg/ml) was incubated overnight with the H3K4me3 (1–12) peptide at a 1:1.5 molar ratio prior to crystallization. Coarse screens were set up in 96-well sitting drop plates at 18°C, mixing 1 μl protein-peptide complex and 1 μl precipitant in the drop. The crystallization condition was optimized to 0.1 M HEPES (pH 8) and 30% PEG6K. Good-quality diffracting crystals were grown by hanging-drop vapor diffusion at 18°C. Data sets were collected at National Synchrotron Light Source X25 beamline. The structure of the protein-peptide complex was solved using Single Anomalous Dispersion method with Zn anomalous signal at 1.00 Å. Data set (1.35 Å) was processed using iMosflm and Scala, and the location of the Zn atoms was determined using PHENIX AutoSol (Adams et al., 2002). The initial model was built using PHENIX AutoBuild. Manual modeling was done in Coot, and the structure was refined using PHENIX.refine. The final structure was verified by PROCHECK.
ACCESSION NUMBERS
Atomic coordinates for the structure have been deposited to the Protein Data Bank under accession number 4L7X.
Supplementary Material
ACKNOWLEDGMENTS
We thank Patricia Garcia-Arias Garcia-Morato for assistance with immunofluorescence and Annie Heroux and the mail-in data collection service at the National Synchrotron Light Source (NSLS) for synchrotron data collection. This research is supported by NIH grants GM096863 and GM101664 (to T.G.K.), GM068088 (to B.D.S.), and HL65440 (to M.G.) and Spanish government grants PS09/00572 (to K.H.M.v.W) and SAF2010-21295, PIB2010BZ-00564, and MITIC (S2010/BMD-2502) (to C.M.A.). K.H.M.v.W. is supported by a JAE-doc fellowship from the Spanish National Research Council (CSIC). S.B.R. is supported by the UNC Lineberger Comprehensive Cancer Center Basic Sciences Training Program (T32CA09156) and a Postdoctoral Fellowship from the American Cancer Society (PF-13-085-01-DMC). Financial support for NSLS comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy and from the National Center for Research Resources of the National Institutes of Health (NIH).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, six figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2013.06.014.
REFERENCES
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Braig S, Bosserhoff AK. Death inducer-obliterator 1 (Dido1) is a BMP target gene and promotes BMP-induced melanoma progression. Oncogene. 2013;32:837–848. doi: 10.1038/onc.2012.115. [DOI] [PubMed] [Google Scholar]
- Douet-Guilbert N, Laï JL, Basinko A, Gueganic N, Andrieux J, Pollet B, Plantier I, Delattre C, Crépin O, Corm S, et al. Fluorescence in situ hybridization characterization of ider(20q) in myelodysplastic syndrome. Br. J. Haematol. 2008;143:716–720. doi: 10.1111/j.1365-2141.2008.07436.x. [DOI] [PubMed] [Google Scholar]
- Fütterer A, Campanero MR, Leonardo E, Criado LM, Flores JM, Hernández JM, San Miguel JF, Martínez-A C. Dido gene expression alterations are implicated in the induction of hematological myeloid neoplasms. J. Clin. Invest. 2005;115:2351–2362. doi: 10.1172/JCI24177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fütterer A, Raya A, Llorente M, Izpisúa-Belmonte JC, de la Pompa JL, Klatt P, Martínez-A C. Ablation of Dido3 compromises lineage commitment of stem cells in vitro and during early embryonic development. Cell Death Differ. 2012;19:132–143. doi: 10.1038/cdd.2011.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JR, Upadhyay AK, Qi HH, Zhang X, Shi Y, Cheng X. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat. Struct. Mol. Biol. 2010;17:38–43. doi: 10.1038/nsmb.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ML, Nagengast AA, Salz HK. PPS, a large multido-main protein, functions with sex-lethal to regulate alternative splicing in Drosophila. PLoS Genet. 2010;6:e1000872. doi: 10.1371/journal.pgen.1000872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi-Ashtiyani R, Houben A. In vitro phosphorylation of histone h3 at threonine 3 by Arabidopsis haspin is strongly influenced by posttranslational modifications of adjacent amino acids. Mol. Plant. 2013;6:574–576. doi: 10.1093/mp/sss149. [DOI] [PubMed] [Google Scholar]
- Kettenberger H, Armache KJ, Cramer P. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell. 2003;114:347–357. doi: 10.1016/s0092-8674(03)00598-1. [DOI] [PubMed] [Google Scholar]
- modENCODE Consortium, E.C. Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman CA, Kutateladze TG. Handpicking epigenetic marks with PHD fingers. Nucleic Acids Res. 2011;39:9061–9071. doi: 10.1093/nar/gkr613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman CA, Lalonde ME, Côté J, Kutateladze TG. Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 2012;19:1218–1227. doi: 10.1038/nsmb.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozlü N, Monigatti F, Renard BY, Field CM, Steen H, Mitchison TJ, Steen JJ. Binding partner switching on microtubules and aurora-B in the mitosis to cytokinesis transition. Mol. Cell. Proteomics. 2010;9:336–350. doi: 10.1074/mcp.M900308-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polioudaki H, Markaki Y, Kourmouli N, Dialynas G, Theodoropoulos PA, Singh PB, Georgatos SD. Mitotic phosphorylation of histone H3 at threonine 3. FEBS Lett. 2004;560:39–44. doi: 10.1016/S0014-5793(04)00060-2. [DOI] [PubMed] [Google Scholar]
- Prieto I, Kouznetsova A, Fütterer A, Trachana V, Leonardo E, Alonso Guerrero A, Cano Gamero M, Pacios-Bras C, Leh H, Buckle M, et al. Synaptonemal complex assembly and H3K4Me3 demethylation determine DIDO3 localization in meiosis. Chromosoma. 2009;118:617–632. doi: 10.1007/s00412-009-0223-7. [DOI] [PubMed] [Google Scholar]
- Rojas AM, Sanchez-Pulido L, Fütterer A, van Wely KH, Martinez-A C, Valencia A. Death inducer obliterator protein 1 in the context of DNA regulation. Sequence analyses of distant homologues point to a novel functional role. FEBS J. 2005;272:3505–3511. doi: 10.1111/j.1742-4658.2005.04759.x. [DOI] [PubMed] [Google Scholar]
- Sawicka A, Seiser C. Histone H3 phosphorylation - a versatile chromatin modification for different occasions. Biochimie. 2012;94:2193–2201. doi: 10.1016/j.biochi.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachana V, van Wely KH, Guerrero AA, Fütterer A, Martínez-A C. Dido disruption leads to centrosome amplification and mitotic checkpoint defects compromising chromosome stability. Proc. Natl. Acad. Sci. USA. 2007;104:2691–2696. doi: 10.1073/pnas.0611132104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H, Li J, Song T, Lu M, Kan PY, Lee MG, Sha B, Shi X. Recognition of histone H3K4 trimethylation by the plant homeodomain of PHF2 modulates histone demethylation. J. Biol. Chem. 2010;285:9322–9326. doi: 10.1074/jbc.C109.097667. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.