Abstract
G protein coupled receptors (GPCRs) are key players in signal recognition and cellular communication making them important therapeutic targets. Large-scale production of these membrane proteins in their native form is crucial for understanding their mechanism of action and target-based drug design. Here we report the overexpression system for a GPCR, the cannabinoid receptor subtype 2 (CB2), in Escherichia coli C43(DE3) facilitated by two fusion partners: Mistic, an integral membrane protein expression enhancer at the N-terminal, and TarCF, a C-terminal fragment of the bacterial chemosensory transducer Tar at the C-terminal of the CB2 open reading frame region. Multiple histidine tags were added on both ends of the fusion protein to facilitate purification. Using individual and combined fusion partners, we found that CB2 fusion protein expression was maximized only when both partners were used. Variable growth and induction conditions were conducted to determine and optimize protein expression. More importantly, this fusion protein Mistic-CB2-TarCF can localize into the E. coli membrane and exhibit functional binding activities with known CB2 ligands including CP55,940, WIN55,212-2 and SR144528. These results indicate that this novel expression and purification system provides us with a promising strategy for the preparation of biologically active GPCRs, as well as general application for the preparation of membrane-bound proteins using the two new fusion partners described.
Keywords: Mistic, TarCF, fusion partner, E. coli expression, cannabinoid receptor 2
Introduction
The physiological effects of endogenous and synthetic cannabinoid ligands are mediated by two cell surface receptors, belonging to the Rhodopsin family of G protein coupled receptors (GPCRs) [1]. These two receptors, cannabinoid receptor subtype 1 (CB1) expressing abundantly in the brain and subtype 2 (CB2) expressing mainly in the immune system, share 68% similarity in their transmembrane domains and 44% similarity in their overall receptor sequences [2–5]. After stimulation, the CB2 receptor couples to Gαi to negatively regulate cyclic AMP levels by inhibiting adenylase cyclase activity [6–7], and to the Gβγ domain to enhance MAPK and PI3K activation, ceramide production and downstream gene expression [8–10]. Clinically, modulation of the CB2 signaling exhibits great potential for the treatment of inflammatory and autoimmune diseases, cancer, heart and bone disorders as well as neurodegenerative disorders [11–15]. In addition, CB2 activation has also shown to have neuroprotective and analgesic effects in animals via unclear mechanisms [16–17]. CB1 is highly expressed in the brain and therapeutic modulations of this receptor have resulted in adverse psychotropic side effects [18–19]. Selective modulation of CB2, however, would be able to achieve the desired therapeutic effect without such psychotropic side effects due to no or very low expression of CB2 in the central nervous system (CNS). Therefore, the CB2 receptor is a significant and desirable target for therapeutic intervention requiring more in-depth information regarding the receptor structure and function to design highly selective ligands. However, expression levels of CB2 are very low in native tissues, and structure determination of CB2 has been impeded due to the inability to produce sufficient amounts of the receptor proteins with high homogeneity and natural ligand binding activity.
Different hosts have been employed to improve the expression levels of GPCRs. Baculovirus-infected insect cell lines have been used to produce GPCRs including the cannabinoid receptor 2 [20], beta 2-adrenergic receptor [21–23], chemokine receptor [24] and the A2a adenosine receptor [25–26]; most of which have been structurally modified to facilitate receptor stability and crystallization. Yeast cells also provide eukaryotic environment for post-translational modification of the exogenous GPCRs [27–28]. However, compared to mammalian cells, they differ in membrane composition and posttranslational modification [29]. While lacking post-translational modifications, the bacterial system offers several unbeatable advantages for the expression of exogenous proteins: fast, homogeneity in protein production, low cost and ability to isotopically label the protein of interest for subsequent NMR studies [30]. Previously, E. coli was used in our lab to express CB2 receptor fragments by directing the fragment expression to inclusion bodies using the Trp LE leader sequence [31–32]. The CB2 receptor fragment produced in E. coli and reconstituted in Brij 58 showed > 75% preservation of the alpha helical structure [33]. However, the methodology developed in these studies may not be applied to the intact receptor without substantial modifications.
To heterologously express eukaryotic membrane proteins, fusion protein technology in E. coli has been successfully applied for the neurotensin receptor, an integral membrane protein for which the expression level was enhanced 40-fold when neurotensin was fused to maltose binding protein (MBP) at the at the N terminus and the signal peptide sequence Endotoxin B at the C terminus [34]. Related methods have also been used for the production of the rat neurokinin A receptor [35] and human adenosine A2a receptors [36]. In addition, expression of the CB2 receptor by using MBP as an N-terminal fusion partner and Thioredoxin as a C-terminal fusion partner has also been reported [37–39].
Determining the correct fusion partner(s) to optimize GPCR expression is not empirical but largely depends on the receptor in question. Mistic is an unusual B. subtilis membrane protein [40–41]; TarCF is the C-terminal fragment of bacterial aspartate chemosensory transducer Tar [42–43]. While Mistic and TarCF have been used as fusion partners to enhance protein expression and stabilization their effects on the expression and stabilization of GPCRs are obscure and remain unexplored. In the present study, we have evaluated the roles of several fusion partners including Mistic, TarCF and TrxA, alone or in combination, to drive the functional expression of the CB2 receptor in E. coli. To facilitate the fusion protein release and purification, Factor Xa/TEV sequences and multi-His tags were introduced into the expression construct. Culture conditions were optimized to determine the conditions for maximum fusion protein yield.
Materials and Methods
Expression bacteria strain and reagents
The expression bacteria strain E. coli C43(DE3) was purchased from Lucigen (Middleton, WI). Strain C43(DE3) contains no intrinsic plasmids and expresses the T7 polymerase from the lacUV5 promoter upon IPTG induction. In addition, C43(DE3) shows no proteolytic activity towards exogenously overexpressed proteins [44]. 3H-CP55,940 (specific activity: 88.3 Ci/mmol), CP55,940, WIN55,212-2 and SR144,528 were obtained from RTI International (Research Triangle Park, NC). Isopropyl-β-D-thiogalactoside (IPTG), Benzonase nuclease and lysozyme were purchased from EMD Chemicals (Gibbstown, NJ). Protease inhibitor cocktail was purchased from Sigma (St. Louis, MO). All restriction and DNA modifying enzymes were purchased from New England Biolabs (Ipswich, MA).
Construction of recombinant CB2 receptor expression vectors
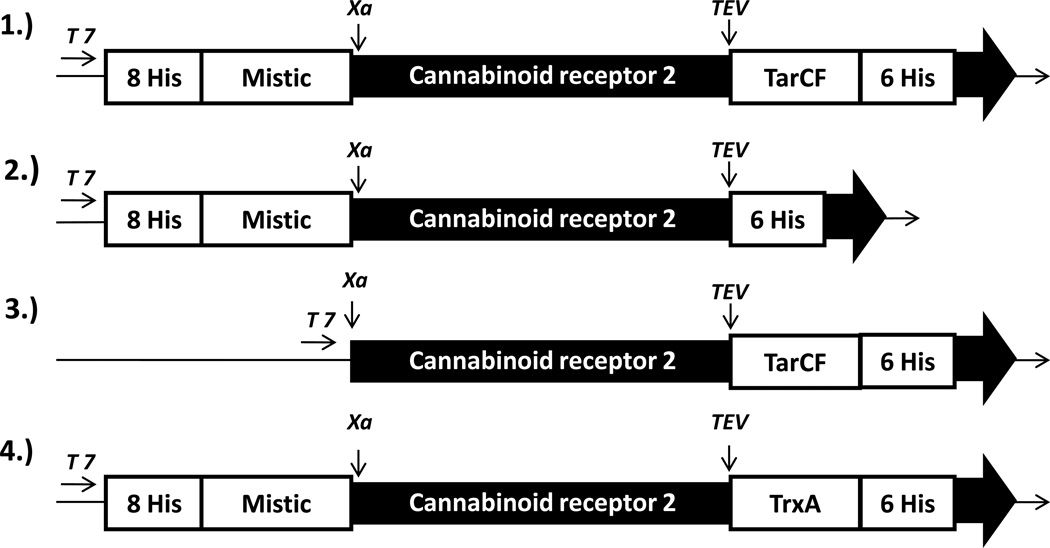
The constructs used in the present study are shown in Figure 1. All expression vectors were based upon the pET-21a vector backbone. Gene fragment encoding octahistidine tagged Mistic (8His-Mistic) was derived from the pMIS3.0E vector via polymerase chain reaction using specific primers (For: 5’-atatacatatgaaacaccaccacc-3’; Rev: 5’- aagcttaccactcaggatcatgtaat-3’). The forward and reverse primers included the restriction sites NdeI and HindIII respectively for subsequent cloning. The Human cannabinoid receptor 2 (CNR2) gene with the Factor Xa sequence (5’-attgagggacgc-3’) fused at its 5’ terminal end (Xa-CB2) was extracted from the pMMHb-TrpLE-Xa-CB2 vector using HindIII and BamHI sites. The pET-21a-TarCF construct was used as a template. The 8His-Mistic-Xa-CB2 encoding sequence was cloned upstream of the TarCF gene on the pET-21a-TarCF template using NdeI and BamHI sites. A Tobacco Etch Virus (TEV) (sequence 5’-gaaaacctatacttccaagga-3’) protease recognition site was introduced between TarCF and CB2 encoding sequences on the expression plasmid pET-21a for higher efficiency and specificity of protein cleavage. Similarly, the 8His-Mistic encoding sequence was subcloned into the pET-21a-CB2-TrxA template using NdeI and HindIII sites to create the construct (2). Constructs (3) and (4) were created by removing either the TarCF sequence (using HindIII and XhoI sites) or the 8His-Mistic sequence (using NdeI and AvaI sites) from construct (1), followed by subsequent Klenow treatments (or a subsequent Klenow treatment) and intramolecular ligation reaction. Double digestion of the constructs with AvaI and HindIII released the CB2 gene fragment confirming successful cloning. All construct sequences were verified by automated DNA sequencing at the University of Pittsburgh Genomics core facility.
Figure 1.
Schematic diagram of human CB2 fusion protein constructs. All expression plasmid vectors (1–4) were constructed on the pET-21a vector backbone under the control of the T7 promoter. Mistic, the N-terminal fusion tag, was separated from CB2 by the Factor Xa sequence while TarCF or TrxA, C-terminal fusion tag, were separated from the CB2 receptor by the TEV sequence. The boxes shown are not drawn to scale. TarCF, C-terminal fragment of bacterial aspartate chemosensory transducer Tar; TrxA, thioredoxin; TEV, tobacco etch virus sequence; His, Histidine residues.
Culture of E. coli C43(DE3) for protein expression
Minicultures were inoculated with single colonies from an LB-Ampicillin plate containing freshly transformed E. coli C43(DE3). The bacterial cultures were grown overnight in presence of Ampicillin (100µg/ml) in a shaker (at 250 rpm) at 37°C. Bacterial maxicultures (1 L) were inoculated with the minicultures and shaken at 250 rpm, 37°C until the culture reached an OD600 of 0.6. Expression of the recombinant CB2 protein was induced with 0.5 mM isopropyl-β-D-thiogalactoside (IPTG), followed by continuous shaking for another 4 h at 37°C. Cells were harvested by centrifugation. After a 50 mM Tris–HCl (pH 8.0) wash, the pellets were stored at −80°C for further experiments. Optimization of culture conditions and IPTG concentration were performed for maximum expression of Mistic-CB2-TarCF. Briefly, E. coli C43(DE3) cultures were grown to OD600 0.6, induced with 0.5 mM or 1 mM IPTG and then maintained at 25°C or 30°C for 8, 22, 32, 48 and 72 h after IPTG induction.
Preparation of bacterial membrane fractions
The harvested bacterial pellet was washed twice with 0.1 M Tris-HCI (pH 8.0) buffer and resuspended in the same buffer containing 20% (w/v) sucrose. The OD600 of the cell suspension was adjusted to 10.0. The suspended pellet was incubated at 37°C for 25 minutes in the presence of the protease Inhibitor Cocktail (PIC) (430 µg/ml) and lysozyme (0.5 µl/g ) followed by immediate addition of EDTA to a final concentration 10 mM. After a 0.1 M Tris-HCI wash containing 20% sucrose, the pellet was then subjected to osmotic lysis by suspension in cold water and sonicated on ice. This suspension was incubated for 1 h with PIC, Benzonase nuclease and MgCl2 (10 mM). After a low speed centrifugation (4500 × g, 10 mins), the supernatant was subjected to a high speed spin (100,000 × g, 90 mins) at 4°C. The membrane pellet obtained was dissolved in Tris-HCI buffer with 20% sucrose and PIC. This was flash frozen and the aliquots were stored at −80°C for subsequent use.
Detection of CB2 fusion protein expression in E. coli
Transformed E. coli cell pellets or membrane fractions were analyzed for CB2 expression by Coomassie blue staining and Western blot. Cell pellets or membrane fractions were lysed in buffer (10 mM Tris-HCl, 10 mM MgCl2, 10% SDS, 430 µg/ml PIC) and sonicated briefly. The lysate supernatants were subjected to SDS-PAGE, followed by Coomassie blue staining. For Western blot analysis, the lysate supernantant (30 µg) was heat-denatured, subjected to 12 % SDS-PAGE, and transferred to polyvinylidene fluoride membrane. Following, histidine tagged CB2 receptor were probed with anti His monoclonal (1:1000, Sigma) and anti CB2 polyclonal (1:1000, Cayman Chemicals) primary antibodies, the protein bands were detected using Amersham Enhanced Chemiluminescence-Western blotting detection reagents (GE Healthcare, Piscataway, NJ).
Saturation binding assay of the fusion protein
The saturation binding of 3H-CP55,940 to the membrane proteins was performed as described previously [45]. Briefly, the membrane fractions (20 µg) were incubated with increasing concentrations of 3H-CP55,940 (0.01–5 nM) in 96-well plates at 30°C with slow shaking for 1 h. The incubation buffer was composed of 50 mM Tris-HCl (pH 7.4), 5 mM MgCl2, 2.5 mM EGTA and 0.1% (w/v) fatty acid free BSA. Ligand was diluted in incubation buffer supplemented with 10% dimethyl sulfoxide and 0.4% methyl cellulose. Non-specific binding was determined in the presence of 1:1000 unlabeled CP55,940 (5000 nM) in excess. The reaction was terminated by rapid filtration through Unifilter GF/C filter plates using a Unifilter Cell Harvester (PerkinElmer). After the plate was allowed to dry overnight, 30 µl MicroScint-20 cocktail (PerkinElmer) was added to each well and the radioactivity was counted by using a PerkinElmer TopCounter. Data from these assays were analysed using GraphPad Prism 5.0 Software. The difference between total and nonspecific binding equals the receptor specific binding. Non-linear regression analysis revealed the receptor density (Bmax) and the equilibrium dissociation constant (Kd) values of 3H-CP55,940 for the CB2 receptor.
Competitive ligand displacement assay
CB2 receptor ligand displacement assay was performed as described previously [45]. The known CB2 ligands CP55,940 (unlabelled), WIN55,212-2 and SR144528 were used in this displacement assay to test whether the fusion proteins expressed in E. coli C43(DE3) exhibited receptor-ligand binding properties. Briefly, non-radioactive (or cold) ligands were diluted ( 10−2–103 nM ) in binding buffer [50 mM Tris-HCl (pH 7.4), 5 mM MgCl2, 2.5 mM EGTA and 0.1% (w/v) fatty acid free BSA], supplemented with 10% dimethyl sulfoxide and 0.4% methyl cellulose. Each assay plate well contained a total of 200 µl of reaction mixture comprised of 20 µg of membrane protein, labeled 3H-CP55,940 ligand at a final concentration of 4 nM and the unlabeled ligand at its varying dilutions as stated above. Plates were incubated at 30°C for 1 h with gentle shaking. Reactions were terminated and read as described in the previous section. All assays were performed in triplicate (n=3) and data points represented as mean±S.E.M. Bound radioactivity was analyzed for Ki values using non-linear regression analysis by GraphPad Prism 5.0 software.
Results and Discussion
Recombinant CB2 receptor produced in E. coli does not have the ability to translocate to the membrane and is devoid of membrane environment. This phenomenon has been proposed to be toxic towards the host and lead to misfolded protein aggregation, requiring the isolated protein to undergo refolding [46]. To enhance membrane protein expression and solubility with correct folding, as well as membrane localization of the recombinant GPCRs, researchers have employed several approaches including the identification of fusion partners linked with GPCRs in E. coli [47–48]. Previous studies have shown that MBP or thioredoxin (Trx) can stabilize and improve the expression and solubility of foreign fusion proteins in E. coli [49]. Furthermore, Trx fusion proteins can be folded correctly and express complete biological activity [50]. Mistic, a bacterial membrane-associating protein, has been found to enhance expression of eukaryotic membrane proteins at the bacterial membrane [40–41, 51]. The chemosensory aspartate receptor, Tar, is a resident membrane protein of the bacterial host which is expected to facilitate membrane protein expression [52]. Combining different fusion partners at both ends of a target gene has emerged as a promising strategy to facilitate expression and improve the solubility of recombinant proteins [39]. However, application of the fusion tags Mistic and TarCF for the expression of GPCRs in E. coli has not been investigated previously. For the first time, we report in this study the use of different fusion partner combinations (Mistic, TarCF and TrxA) for the functional expression of the recombinant CB2 receptor in E. coli C43(DE3). We show here, that the fusion protein Mistic-CB2-TarCF is overexpressed by E. coli and localized to the bacterial membrane with ligand binding properties comparable to those on mammalian cells.
Expression of cannabinoid receptor 2 fusion protein in E. coli
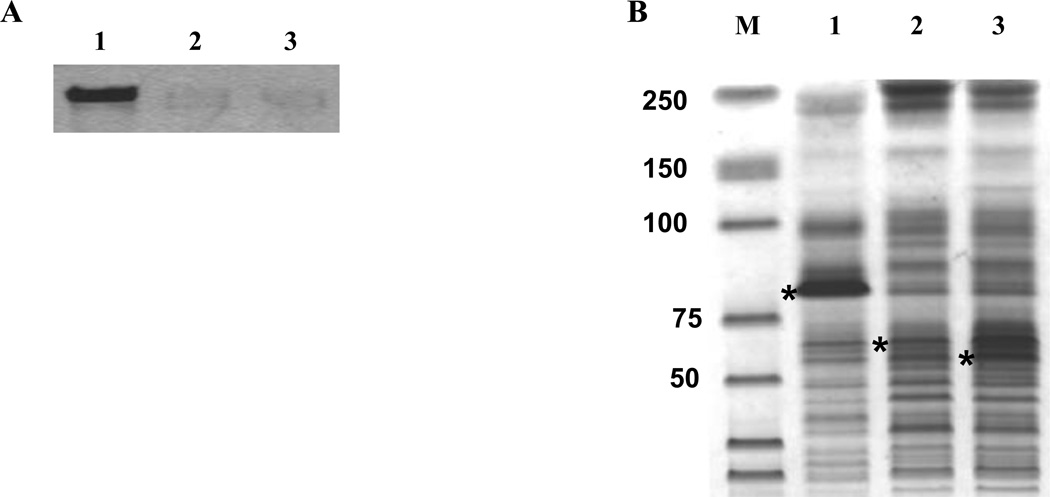
E. coli C43(DE3) cells were transformed respectively with the fusion constructs shown in Figure 1. Expression of the recombinant fusion proteins were detected by Western blot or Commassie Blue staining. Since all constructs contain a multi-histidine tag, we used either anti-His or anti-CB2 antibody to detect expression of the CB2 fusion protein. As shown in Figure 2A, Mistic and TarCF alone failed to boost the CB2 gene expression. Only when both partners were linked to CB2 in the proper order did the fusion protein expression increase dramatically. In addition, fusion protein expression was also observed with the Mistic-CB2-TrxA construct at a comparable expression level with that of the Mistic-CB2-TarCF (data not shown). However, the construct Mistic-CB2- TarCF was used for further optimization due to its novel combination of fusion partners and prominent expression levels of recombinant fusion protein.
Figure 2.
Enhanced expression of the CB2 receptor fusion protein (A) Representative immunoblot of His-tagged CB2 fusion protein detected in E. coli C43(DE3) membrane fractions using anti-His. Membrane fractions loaded from left are Lane 1: Mistic-CB2- TarCF; Lane 2: Mistic-CB2; Lane 3: CB2-TarCF. (B) Coomassie Brilliant Blue staining on the SDS-PAGE of extracted membrane fractions. The expected MW for the respective fusion proteins are as follows – Lane 1: Mistic-CB2-TarCF (86 kDa); Lane 2: Mistic-CB2 (71 kDa); Lane 3: CB2-TarCF (55 kDa). Asterisks show the corresponding fusion protein expression. M: protein marker. Care was taken to normalize the amount of E. coli C43(DE3) membrane fraction sample loaded on the gel.
Next, we investigated whether the expressed CB2 fusion protein possessed membrane affinity or localized to the E. coli membrane. Coomassie staining of the membrane enriched fractions revealed a prominent band at MW ~ 86 KDa for the fusion protein Mistic-CB2-TarCF while no bands were detected for the fusion proteins that carried either the Mistic or TarCF tag individually (Fig. 2B). Importantly, the membrane enriched fractions exhibited the same expression pattern as the whole E. coli cell lysates, indicating that all or the majority of CB2 fused protein driven by the two partners could localize or integrate into the E. coli membrane. Our data suggests that combination of the two tags (Mistic and TarCF) may contribute synergistic effects on the CB2 protein expression compared to either tag used alone. Our data also show that membrane fractions contain concentrated CB2 fusion protein compared to the whole cell lysate, indicating that most of the fusion protein is localized within the bacterial membrane. This is in accordance with previous studies where the effects of Mistic and other bacterial membrane resident protein to stabilize GPCR expression has been demonstrated [41, 53–54].
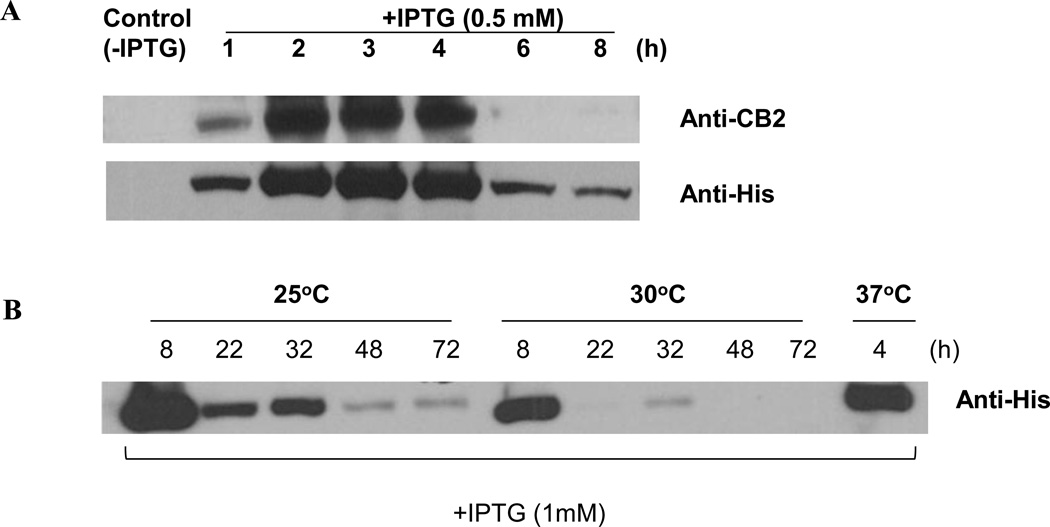
In absence of induction with IPTG, there was no expression of the fusion protein. However, after induction with IPTG, the fusion protein level increased significantly in a time-dependent manner (Fig. 3), suggesting that the expression cassette is under the tight control of the lac operon and T7/lac promoter. Also the pET21a carries the lacI gene together these elements explain why the expression is tightly controlled.
Figure 3.
Optimization of conditions for fusion protein expression in E. coli. (A) Cells transformed with Mistic-CB2-TarCF were grown for the indicated hours after induction with IPTG (0.5 mM). Expression levels of fusion protein Mistic-CB2-TarCF tagged with poly-histidine were detected by Western blot with anti-CB2 or anti-His antibody. Control group (0 hours) represented no IPTG induction. (B) Optimization of Mistic-CB2-TarCF fusion protein production in E coli. Different combinations of the parameters (IPTG, culture temperature and time) were tested and one representative setting was shown.
Control of recombinant protein expression under the tight regulation is necessary to avoid toxicity of protein expression to the host and ensure sufficient biomass of viable E. coli that would be available for membrane protein expression after induction.
To determine the time point of maximum receptor production, IPTG-induced cells were harvested at different time intervals from 1–8 hours. Whole cell lysates were analyzed by Western blot using mouse anti-His (1:1000 dilution) and rabbit anti-CB2 antibodies (1:500 dilution). As shown in Figure 3A, CB2 fusion protein expression levels steadily increased and reached maxima at 3–4 hours, followed by significantly reduced expression. Thus, from this experiment we can conclude that the expression level of the fusion protein peaked during culture at 37°C for 3–4 hours after IPTG induction. Since IPTG induction at lower temperature was previously reported to improve the exogenous protein production and correct folding [54], we optimized the culture conditions by combining different IPTG concentrations (0.5 mM and 1 mM), culture temperature and time. We found that the expression levels of the fusion protein Mistic-CB2-TarCF are weakly detected during culture period (2–48 h) at 22°C (data not shown). The fusion protein expression at 30°C was not distinct from the regular 37°C culture condition (Fig. 3B). However, once the transformed cultured underwent IPTG induction (1 mM) at 25°C for 8 h, the fusion protein levels were significantly increased 2-fold of that of regular conditions (Fig. 3B). Overall, 0.5 mM IPTG used for inducing protein expression resulted in lower amounts of fusion protein than 1 mM—especially for the conditions of culture temperatures at 25 or 30°C (data not shown).
Receptor saturation binding assay
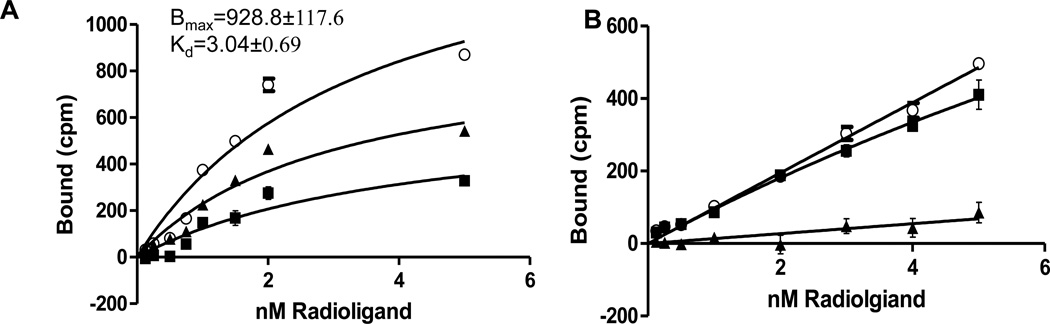
pET-21a-Mistic-CB2-TarCF transformed E. coli membranes were subjected to a saturation binding assay to determine receptor saturation with increasing concentrations of 3H-CP55,940. pET-21a-TarCF transformed E. coli membranes were used as the negative control. For the membrane proteins derived from Mistic-CB2-TarCF transformed E. coli, the maximal receptor density (Bmax) and dissociation constant (Kd) of 3H-CP55,940 for specific binding sites were 928.8 ± 117.6 fmol/mg protein and 3.04 ± 0.69 nM, respectively (Figure 4A). Membrane fractions clearly showed CB2 receptor binding characterization by the abundance of binding sites recognized by agonist 3H-CP55,940. For the negative control, however, no difference was observed between specific and nonspecific binding (Figure 4B), indicating that the overwhelming majority of the total binding was contributed by the nonspecific binding. This confirms the absence of CB2 receptor on pET-21a-TarCF transformed E. coli membranes.
Figure 4.
Saturation binding assay of the membrane fractions. Total (ο) and non-specific (■) binding was measured and the deduced specific binding saturation isotherm (▲) was obtained as the difference between total and nonspecific binding. (A) Mistic-CB2-TarCF; (B) pET 21-TarCF (negative control). Assay was performed in triplicate (n=3). Data presented as mean ±SEM.
Competitive ligand displacement assays
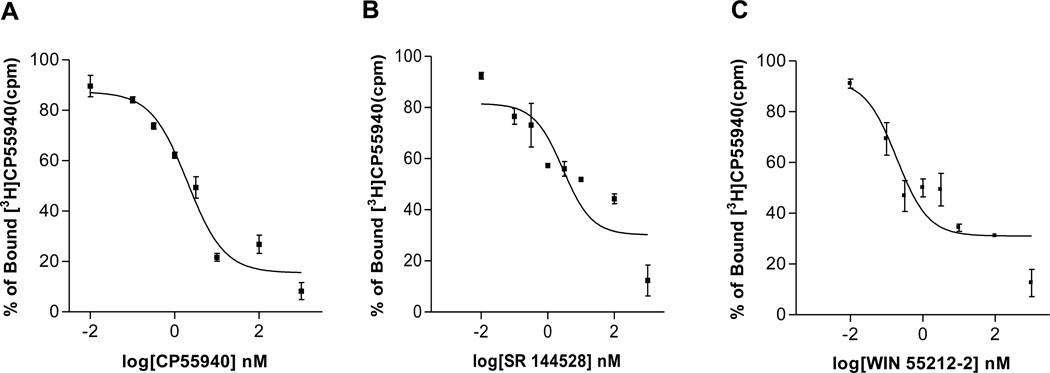
The conformational state of a receptor protein determines the functional state of a receptor. High affinity binding between a ligand and its receptors is often physiologically important when a portion of the binding energy can be used to cause a conformational change in the receptor, resulting in altered downstream signaling pathways. In the present study, to confirm whether the expressed fusion proteins from the E. coli exhibit functional binding activity, we used the well-known CB2 ligands to probe the interactions of these ligands with their cognate binding sites on the CB2 enriched membrane fractions (competitive binding assay), by quantifying the equilibrium dissociation constant (Ki). By using 10 µg of membrane fractions of Mistic-CB2-TarCF fusion protein in the binding assay, the Ki values for these ligands were well consistent with previous reports using the CB2 from mammalian cells: CP 55,940 (Ki= 1.43 nM), SR 144528 (Ki=−2.02 nM)m and WIN 55212-2 (Ki= 0.13 nM). These results indicate that the ligand binding domain of the CB2 receptor in the fusion protein is not perturbed by the physical presence of its neighboring fusion partners Mistic and TarCF.
Conclusion
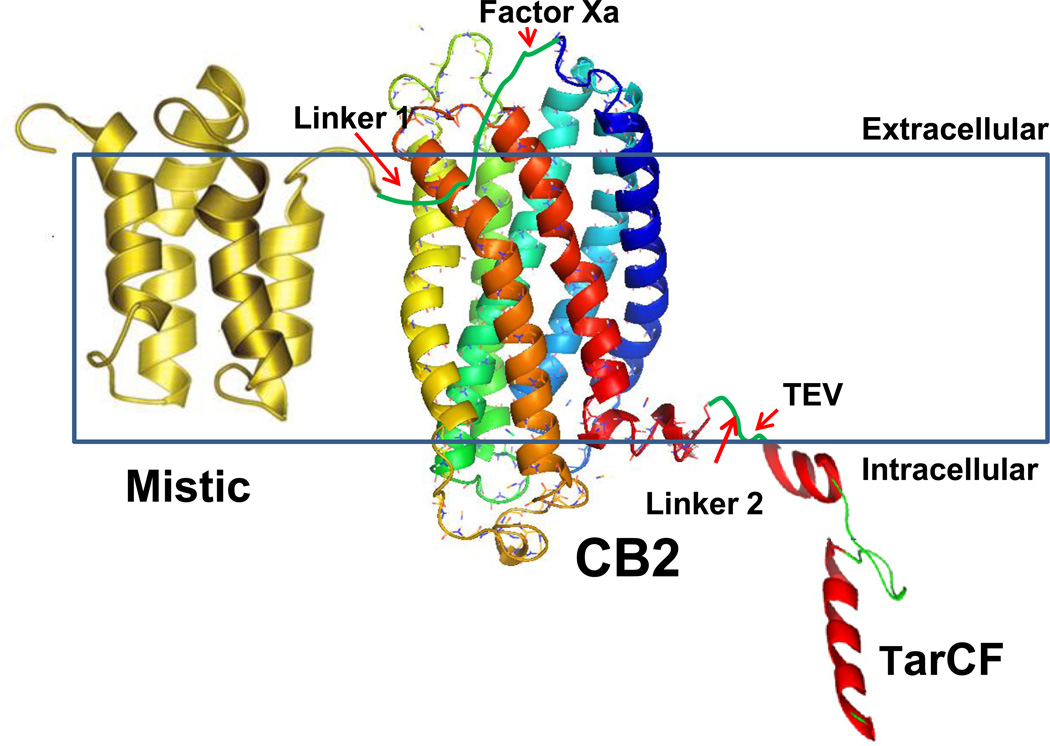
In summary, our data has demonstrated that the Mistic-CB2-TarCF construct can successfully express the CB2 receptor protein in E. coli C43(DE3). The obtained fusion proteins can localize at the bacterial membrane. Importantly, the Mistic-CB2-TarCF fusion proteins show effective binding activity with the known CB2 ligands. This suggests that the conformational state of the native CB2 receptor, used for specific ligand binding, is retained in the presence of fusion partners. Also, we found that the fusion partners – Mistic and TarCF – in combination, are more effective for enhancing protein expression in E. coli, than their use alone. Overall findings from this present study suggest that the targeting of fusion partners to the bacterial membrane is critical to the conformational stability of the expressed CB2 protein. The possible role of the fusion partners for the overexpression and stabilization the CB2 protein is illustrated schematically (Fig. 6) for easy comprehension. In this putative model, the CB2 receptor structure was adapted from the 3D CB2 model reported previously by Xie et. al [5], while the structure of Mistic and Tsr (structurally related to Tar) were determined by NMR (PDB:1YGM) [40] and cryo-electron microscopy [55] studies, respectively. However, confirming the putative model will be subject to further biophysical studies. Currently, we are using the entire fusion protein and cleaved receptor in parallel to carry out 2-dimensional crystal growth and analysis by cryo-electron microscopy. The trials for 2D crystal generation will be favorably facilitated by the increased molecular weight of the fusion protein complex [56].
Figure 6.
Putative schematic diagram illustrating the plausible structure and membrane bound state of the fusion protein construct. Mistic (golden) [PDB ID:1YGM] is joined to the N-terminal of the CB2 receptor (structure from homology model,[5]) (rainbow) via linker 1 containing Factor Xa sequence. The CB2 receptor is linked to TarCF (red) via linker 2 containing the TEV sequence. Mistic and TarCF contain 8 and 6 histidine residues in their N- and C- terminal domains respectively.
Our preliminary studies of detergent-screening in small-scale (data now shown) indicated the feasibility of rapid affinity purification for the fusion protein in the presence of detergents. These results strongly encourage us to optimize the extraction and purification conditions for large-scale production of human CB2 receptor to study its functional aspects. If necessary, we may use the refolding mechanism already demonstrated for the CB1 receptor [46]. Overall, our studies show new fusion partners for the functional expression of the cannabinoid receptor 2 in the bacterial membrane. We anticipate this approach will produce enough protein to conduct further biophysical studies.
Figure 5.
Competitive displacement of the 3H-CP55,940 was obtained by using an increased amount of cold ligands. Binding profile of (A) CP55,940 (unlabelled), Ki=1.43 nM; (B) SR144,528, Ki=2.02 nM; (C) WIN55,212-2, Ki=0.13 nM. Assay was performed in triplicate (n=3). Data presented as mean ±SEM.
Highlights.
In the present study we have designed and characterized a novel expression system for production of the GPCR-Cannabinoid Receptor 2 in the Escherichia coli. This novel combination strategy with fusion partners can be generally applicable to other GPCRs.
The expressed membrane receptor CB2 as fusion protein (Mistic-CB2-TarCF) can localize to the E. coli membrane.
Recombinant CB2 receptor fusion protein produced using this system shows the same binding properties as that of native CB2 receptor derived from mammalian cells.
Acknowledgements
The authors thank Dr. Peijun Zhang (Department of Structural Biology, University of Pittsburgh) for the pET-21a-TarCF vector and planned CryoEM studies; Dr. Senyon Choe (Department of Structural Biology, Salk Institute) for the pMIS3.0E vector. The work was supported by NIH RO1 DA025612.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Attwood TK, Findlay JB. Fingerprinting G-protein-coupled receptors. Protein Eng. 1994;7:195–203. doi: 10.1093/protein/7.2.195. [DOI] [PubMed] [Google Scholar]
- 2.Galiegue S, Mary S, Marchand J, Dussossoy D, Carriere D, Carayon P, Bouaboula M, Shire D, Le Fur G, Casellas P. Expression of central and peripheral cannabinoid receptors in human immune tissues and leukocyte subpopulations. Eur J Biochem. 1995;232:54–61. doi: 10.1111/j.1432-1033.1995.tb20780.x. [DOI] [PubMed] [Google Scholar]
- 3.Howlett AC, Barth F, Bonner TI, Cabral G, Casellas P, Devane WA, Felder CC, Herkenham M, Mackie K, Martin BR, Mechoulam R, Pertwee RG. International Union of Pharmacology. XXVII. Classification of cannabinoid receptors. Pharmacol Rev. 2002;54:161–202. doi: 10.1124/pr.54.2.161. [DOI] [PubMed] [Google Scholar]
- 4.Munro S, Thomas KL, Abu-Shaar M. Molecular characterization of a peripheral receptor for cannabinoids. Nature. 1993;365:61–65. doi: 10.1038/365061a0. [DOI] [PubMed] [Google Scholar]
- 5.Xie XQ, Chen JZ, Billings EM. 3D structural model of the G-protein-coupled cannabinoid CB2 receptor. Proteins. 2003;53:307–319. doi: 10.1002/prot.10511. [DOI] [PubMed] [Google Scholar]
- 6.Bayewitch M, Avidor-Reiss T, Levy R, Barg J, Mechoulam R, Vogel Z. The peripheral cannabinoid receptor: adenylate cyclase inhibition and G protein coupling. FEBS Lett. 1995;375:143–147. doi: 10.1016/0014-5793(95)01207-u. [DOI] [PubMed] [Google Scholar]
- 7.Gonsiorek W, Lunn C, Fan X, Narula S, Lundell D, Hipkin RW. Endocannabinoid 2-arachidonyl glycerol is a full agonist through human type 2 cannabinoid receptor: antagonism by anandamide. Mol Pharmacol. 2000;57:1045–1050. [PubMed] [Google Scholar]
- 8.Bouaboula M, Poinot-Chazel C, Marchand J, Canat X, Bourrie B, Rinaldi-Carmona M, Calandra B, Le Fur G, Casellas P. Signaling pathway associated with stimulation of CB2 peripheral cannabinoid receptor. Involvement of both mitogen-activated protein kinase and induction of Krox-24 expression. Eur J Biochem. 1996;237:704–711. doi: 10.1111/j.1432-1033.1996.0704p.x. [DOI] [PubMed] [Google Scholar]
- 9.Bouaboula M, Desnoyer N, Carayon P, Combes T, Casellas P. Gi protein modulation induced by a selective inverse agonist for the peripheral cannabinoid receptor CB2: implication for intracellular signalization cross-regulation. Mol Pharmacol. 1999;55:473–480. [PubMed] [Google Scholar]
- 10.Bouaboula M, Dussossoy D, Casellas P. Regulation of peripheral cannabinoid receptor CB2 phosphorylation by the inverse agonist SR 144528. Implications for receptor biological responses. J Biol Chem. 1999;274:20397–20405. doi: 10.1074/jbc.274.29.20397. [DOI] [PubMed] [Google Scholar]
- 11.Alexander A, Smith PF, Rosengren RJ. Cannabinoids in the treatment of cancer. Cancer Lett. 2009;285:6–12. doi: 10.1016/j.canlet.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 12.Lozano-Ondoua AN, Wright C, Vardanyan A, King T, Largent-Milnes TM, Nelson M, Jimenez-Andrade JM, Mantyh PW, Vanderah TW. A cannabinoid 2 receptor agonist attenuates bone cancer-induced pain and bone loss. Life Sci. 2010;86:646–653. doi: 10.1016/j.lfs.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin-Moreno AM, Reigada D, Ramirez BG, Mechoulam R, Innamorato N, Cuadrado A, de Ceballos ML. Cannabidiol and other cannabinoids reduce microglial activation in vitro and in vivo: relevance to Alzheimers' disease. Mol Pharmacol. 2011 doi: 10.1124/mol.111.071290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pacher P, Mechoulam R. Is lipid signaling through cannabinoid 2 receptors part of a protective system? Prog Lipid Res. 2011;50:193–211. doi: 10.1016/j.plipres.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zajicek JP, Apostu VI. Role of cannabinoids in multiple sclerosis. CNS Drugs. 2011;25:187–201. doi: 10.2165/11539000-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 16.Cabral GA, Raborn ES, Griffin L, Dennis J, Marciano-Cabral F. CB2 receptors in the brain: role in central immune function. Br J Pharmacol. 2008;153:240–251. doi: 10.1038/sj.bjp.0707584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anand P, Whiteside G, Fowler CJ, Hohmann AG. Targeting CB2 receptors and the endocannabinoid system for the treatment of pain. Brain Res Rev. 2009;60:255–266. doi: 10.1016/j.brainresrev.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelly DL, Gorelick DA, Conley RR, Boggs DL, Linthicum J, Liu F, Feldman S, Ball MP, Wehring HJ, McMahon RP, Huestis MA, Heishman SJ, Warren KR, Buchanan RW. Effects of the cannabinoid-1 receptor antagonist rimonabant on psychiatric symptoms in overweight people with schizophrenia: a randomized, double-blind, pilot study. J Clin Psychopharmacol. 2011;31:86–91. doi: 10.1097/JCP.0b013e318204825b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cahill K, Ussher M. Cannabinoid type 1 receptor antagonists (rimonabant) for smoking cessation. Cochrane Database Syst Rev. 2007:CD005353. doi: 10.1002/14651858.CD005353.pub2. [DOI] [PubMed] [Google Scholar]
- 20.Nowell KW, Pettit DA, Cabral WA, Zimmerman HW, Jr., Abood ME, Cabral GA. High-level expression of the human CB2 cannabinoid receptor using a baculovirus system. Biochem Pharmacol. 1998;55:1893–1905. doi: 10.1016/s0006-2952(98)00081-1. [DOI] [PubMed] [Google Scholar]
- 21.Rasmussen SG, Choi HJ, Fung JJ, Pardon E, Casarosa P, Chae PS, Devree BT, Rosenbaum DM, Thian FS, Kobilka TS, Schnapp A, Konetzki I, Sunahara RK, Gellman SH, Pautsch A, Steyaert J, Weis WI, Kobilka BK. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, Schertler GF, Weis WI, Kobilka BK. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 23.Sarramegna V, Talmont F, Demange P, Milon A. Heterologous expression of G-protein-coupled receptors: comparison of expression systems fron the standpoint of large-scale production and purification. Cell Mol Life Sci. 2003;60:1529–1546. doi: 10.1007/s00018-003-3168-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EY, Lane JR, Ijzerman AP, Stevens RC. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naider F, Estephan R, Englander J, Suresh Babu VV, Arevalo E, Samples K, Becker JM. Sexual conjugation in yeast: A paradigm to study G-protein-coupled receptor domain structure. Biopolymers. 2004;76:119–128. doi: 10.1002/bip.10567. [DOI] [PubMed] [Google Scholar]
- 28.Kim TK, Zhang R, Feng W, Cai J, Pierce W, Song ZH. Expression and characterization of human CB1 cannabinoid receptor in methylotrophic yeast Pichia pastoris. Protein Expr Purif. 2005;40:60–70. doi: 10.1016/j.pep.2004.10.026. [DOI] [PubMed] [Google Scholar]
- 29.Sarramegna V, Talmont F, Demange P, Milon A. Heterologous expression of G-protein-coupled receptors: comparison of expression systems from the standpoint of large-scale production and purification. Cell Mol Life Sci. 2003;60:1529–1546. doi: 10.1007/s00018-003-3168-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hockney RC. Recent developments in heterologous protein production in Escherichia coli. Trends Biotechnol. 1994;12:456–463. doi: 10.1016/0167-7799(94)90021-3. [DOI] [PubMed] [Google Scholar]
- 31.Xie XQ, Zhao J, Zheng H. Expression, purification, and isotope labeling of cannabinoid CB2 receptor fragment, CB2(180-233) Protein Expr Purif. 2004;38:61–68. doi: 10.1016/j.pep.2004.07.020. [DOI] [PubMed] [Google Scholar]
- 32.Zheng H, Zhao J, Wang S, Lin CM, Chen T, Jones DH, Ma C, Opella S, Xie XQ. Biosynthesis and purification of a hydrophobic peptide from transmembrane domains of G-protein-coupled CB2 receptor. J Pept Res. 2005;65:450–458. doi: 10.1111/j.1399-3011.2005.00239.x. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Xie XQ. Biosynthesis, purification, and characterization of a cannabinoid receptor 2 fragment (CB2(271-326)) Protein Expr Purif. 2008;59:249–257. doi: 10.1016/j.pep.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grisshammer R, Duckworth R, Henderson R. Expression of a rat neurotensin receptor in Escherichia coli. Biochem J. 1993;295(Pt 2):571–576. doi: 10.1042/bj2950571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grisshammer R, Little J, Aharony D. Expression of rat NK-2 (neurokinin A) receptor in E. coli. Receptors Channels. 1994;2:295–302. [PubMed] [Google Scholar]
- 36.Weiss HM, Grisshammer R. Purification and characterization of the human adenosine A(2a) receptor functionally expressed in Escherichia coli. Eur J Biochem. 2002;269:82–92. doi: 10.1046/j.0014-2956.2002.02618.x. [DOI] [PubMed] [Google Scholar]
- 37.Berger C, Ho JT, Kimura T, Hess S, Gawrisch K, Yeliseev A. Preparation of stable isotope-labeled peripheral cannabinoid receptor CB2 by bacterial fermentation. Protein Expr Purif. 2010;70:236–247. doi: 10.1016/j.pep.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krepkiy D, Gawrisch K, Yeliseev A. Expression and purification of CB2 for NMR studies in micellar solution. Protein Pept Lett. 2007;14:1031–1037. doi: 10.2174/092986607782541051. [DOI] [PubMed] [Google Scholar]
- 39.Yeliseev AA, Wong KK, Soubias O, Gawrisch K. Expression of human peripheral cannabinoid receptor for structural studies. Protein Sci. 2005;14:2638–2653. doi: 10.1110/ps.051550305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roosild TP, Greenwald J, Vega M, Castronovo S, Riek R, Choe S. NMR structure of Mistic, a membrane-integrating protein for membrane protein expression. Science. 2005;307:1317–1321. doi: 10.1126/science.1106392. [DOI] [PubMed] [Google Scholar]
- 41.Kefala G, Kwiatkowski W, Esquivies L, Maslennikov I, Choe S. Application of Mistic to improving the expression and membrane integration of histidine kinase receptors from Escherichia coli. J Struct Funct Genomics. 2007;8:167–172. doi: 10.1007/s10969-007-9033-4. [DOI] [PubMed] [Google Scholar]
- 42.Krikos A, Conley MP, Boyd A, Berg HC, Simon MI. Chimeric chemosensory transducers of Escherichia coli. Proc Natl Acad Sci U S A. 1985;82:1326–1330. doi: 10.1073/pnas.82.5.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Antommattei FM, Munzner JB, Weis RM. Ligand-specific activation of Escherichia coli chemoreceptor transmethylation. J Bacteriol. 2004;186:7556–7563. doi: 10.1128/JB.186.22.7556-7563.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 45.Leifert WR, Bucco O, Abeywardena MY, Patten GS. Radioligand binding assays: application of (125)I]angiotensin II receptor binding. Methods Mol Biol. 2009;552:131–141. doi: 10.1007/978-1-60327-317-6_9. [DOI] [PubMed] [Google Scholar]
- 46.Michalke K, Huyghe C, Lichiere J, Graviere ME, Siponen M, Sciara G, Lepaul I, Wagner R, Magg C, Rudolph R, Cambillau C, Desmyter A. Mammalian G protein-coupled receptor expression in Escherichia coli: II. Refolding and biophysical characterization of mouse cannabinoid receptor 1 and human parathyroid hormone receptor 1. Anal Biochem. 2010;401:74–80. doi: 10.1016/j.ab.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 47.Zuo X, Li S, Hall J, Mattern MR, Tran H, Shoo J, Tan R, Weiss SR, Butt TR. Enhanced expression and purification of membrane proteins by SUMO fusion in Escherichia coli. J Struct Funct Genomics. 2005;6:103–111. doi: 10.1007/s10969-005-2664-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Korepanova A, Gao FP, Hua Y, Qin H, Nakamoto RK, Cross TA. Cloning and expression of multiple integral membrane proteins from Mycobacterium tuberculosis in Escherichia coli. Protein Sci. 2005;14:148–158. doi: 10.1110/ps.041022305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kapust RB, Waugh DS. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci. 1999;8:1668–1674. doi: 10.1110/ps.8.8.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.LaVallie ER, DiBlasio EA, Kovacic S, Grant KL, Schendel PF, McCoy JM. A thioredoxin gene fusion expression system that circumvents inclusion body formation in the E. coli cytoplasm. Biotechnology (N Y) 1993;11:187–193. doi: 10.1038/nbt0293-187. [DOI] [PubMed] [Google Scholar]
- 51.Blain KY, Kwiatkowski W, Choe S. The functionally active Mistic-fused histidine kinase receptor, EnvZ. Biochemistry. 2010;49:9089–9095. doi: 10.1021/bi1009248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meir Y, Jakovljevic V, Oleksiuk O, Sourjik V, Wingreen NS. Precision and kinetics of adaptation in bacterial chemotaxis. Biophys J. 2010;99:2766–2774. doi: 10.1016/j.bpj.2010.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baneyx F. Recombinant protein expression in Escherichia coli. Curr Opin Biotechnol. 1999;10:411–421. doi: 10.1016/s0958-1669(99)00003-8. [DOI] [PubMed] [Google Scholar]
- 54.Freigassner M, Pichler H, Glieder A. Tuning microbial hosts for membrane protein production. Microb Cell Fact. 2009;8:69. doi: 10.1186/1475-2859-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khursigara CM, Wu X, Zhang P, Lefman J, Subramaniam S. Role of HAMP domains in chemotaxis signaling by bacterial chemoreceptors. Proc Natl Acad Sci U S A. 2008;105:16555–16560. doi: 10.1073/pnas.0806401105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smyth DR, Mrozkiewicz MK, McGrath WJ, Listwan P, Kobe B. Crystal structures of fusion proteins with large-affinity tags. Protein Sci. 2003;12:1313–1322. doi: 10.1110/ps.0243403. [DOI] [PMC free article] [PubMed] [Google Scholar]