Abstract
BACKGROUND & AIMS
In healthy individuals, interactions between intestinal epithelial cells and lamina propria lymphocytes give rise to a population of CD8+ T cells with suppressor functions (Ts cells). Disruption of Ts cell activities can lead to mucosal inflammation. We investigated what factors were required for expansion of the Ts cell population or loss of their activity in patients with Crohn’s disease (CD).
METHODS
We developed a method to generate Ts cell lines from freshly isolated lamina propria lymphocytes from patients with ulcerative colitis (UC), patients with CD, or healthy individuals (controls). Cells were stimulated with a monoclonal antibody against CD3, interleukin (IL)-7, and IL-15. After 14 days in culture, CD8+T cells were purified and cultured with IL-7 and IL-15. The resulting Ts cells were analyzed for suppressor activity, expression of surface markers, and cytokine secretion profiles. RNA was isolated from the 3 groups of Ts cells and used in microarray analyses.
RESULTS
Ts cells from patients with UC and controls suppressed proliferation of CD4+ T cells; the suppression required cell contact. In contrast, Ts cells from patients with CD had a reduced capacity to suppress CD4+ T-cell proliferation. The difference in suppressive ability was not associated with surface or intracytoplasmic markers or secretion of cytokines. Microarray analysis identified changes in expression of genes regulated by transforming growth factor (TGF)-β that were associated with the suppressive abilities of Ts cells. We found that TGF-β or supernatants from Ts cells of patients with CD reduced the suppressor activity of control Ts cells.
CONCLUSIONS
Ts cells isolated from patients with CD have a reduced ability to suppress proliferation of CD4+T cells compared with control Ts cells. TGF-β signaling reduces the suppressor activity of Ts cells.
Keywords: Regulatory T Cells, Treg Cells, Immune Regulation, Inflammatory Bowel Disease
Regulatory T (Treg) cells or suppressor T (Ts) cells are known to be critically important T-cell subsets for homeostasis of the immune system and are primarily responsible for dampening immune responses. Dysfunction of any of these cell subsets may lead to a number of immune/inflammatory disorders, leading to autoimmunity, tumors, transplant rejection, and so on. Controlled inflammation in the gastrointestinal tract is maintained partially by regulatory lymphocytes. Inflammatory bowel disease (IBD), including Crohn’s disease (CD) and ulcerative colitis (UC), reflects a loss of mucosal homeostasis. CD4+-expressing FoxP3+ CD25+ Treg cells have been extensively studied in the context of IBD and shown to suppress proliferation of T cells and to protect from the induction of experimental colitis by CD4+ CD45RBhigh-expressing cells transferred into SCID mice.1–3 Yet, CD4+ Treg cells have also been shown to be up-regulated in the mucosa of both patients with UC and patients with CD and to have intact suppressor activity (suppressing the proliferation of effector CD4+ CD25− T cells4,5). Therefore, there is a question as to whether CD4+ Treg cells are the critical cell type mediating mucosal homeostasis.
CD8+ Ts cells are present in the systemic and mucosal immune system. In fact, there may be many distinct subsets. A role for CD8+ Ts cells has been implicated in several IBD animal models. CD8+ Ts cells have in vivo suppressor activity and are able to prevent the development of colitis in a CD45RBhi CD4+ T-cell transfer model and spontaneous ileitis in TNFΔARE mice.6–8
We have previously shown that human CD8+ Ts cells are activated and expanded when cultured with isolated human intestinal epithelial cells (IECs) through a complex of the nonclassic class I molecule CD1d with glycoprotein gp180, a member of the carcinoembryonic antigen subfamily.9–11 Defective expression of this complex in patients with IBD is associated with the inability of IBD IECs to activate CD8+ Ts cells, specifically T cells that had biased expression of BV5.1. In this report, we show a more global defect in CD8+ lamina propria (LP) Ts cells in patients with CD and investigate the nature of these differences compared with controls. Using a high-profiling approach, we have identified molecular differences between CD8+ Ts lines that have high and low suppressor activity. We have correlated the expression of multiple genes associated with transforming growth factor (TGF)-β signaling with suppressor activity. Furthermore, we have shown that TGF-β is increased in tissue derived from patients with CD (compared with controls) and that the presence of TGF-β inhibits the suppressor activity of CD8+ Ts cells.
Materials and Methods
Patients and Tissues
Surgical specimens from patients undergoing bowel resection for cancer or IBD at Mount Sinai Medical Center were used as the source for lamina propria lymphocytes (LPLs). “Normals” (NLs) consisted of patients undergoing bowel resection for colon cancer, tubulovillous adenoma, or diverticulitis. Within this group, cells were always isolated from normal tissue >10 cm from the tumor (except diverticulitis) and the samples in this group were derived from noninflamed tissues. UC and CD patient samples were all isolated from areas containing moderate to severe inflammation. Patients with UC and patients with CD shared common medications (corticosteroids, infliximab, azathioprine, mesalamine). This study was approved by the Mount Sinai Institutional Review Board.
Cell Purification
LPLs were isolated according to an established protocol.12 Peripheral blood mononuclear cells (PBMCs) were isolated from heparinized venous blood on a Ficoll-Hypaque density gradient (Amersham Biosciences, Piscataway, NJ) according to standard procedures.
Lines of CD8+ Ts Cells
Whole LPLs were stimulated with the soluble humanized non–FcR-binding αCD3 monoclonal antibody visilizumab (100 ng/mL; PDL, Biopharma Inc, Incline Village, NV). Five days later, cells were treated with interleukin (IL)-7 (10 ng/mL), IL-15 (20 ng/mL) (R&D Systems, Minneapolis, MN), and feeder cells (irradiated allogeneic PBMCs, 3000 rads at a 1:1 ratio). Fourteen days later, CD8+ T cells were positively selected using a CD8+ T-cell selection kit (CD8+ Selection Kit; Stemcell Technologies, Vancouver, British Columbia, Canada). Lines were tested and were shown to be >96% CD8αβ. Cells were maintained in complete medium (RPMI 1640 supplemented with 10% fetal bovine serum, penicillin, streptomycin, glutamine, amphotericin B), IL-7, and IL-15 and expanded for 3 to 6 months.
Tissue Explant Cultures
Explant cultures from surgical specimens were performed following a modification as previously described.13 Briefly, intestinal mucosa (2-cm segments) was washed in phosphate-buffered saline, standardized by weight, and cultured in 24-well culture plates in serum-free RPMI 1640 medium supplemented with penicillin, streptomycin, glutamine, amphotericin B, a protease inhibitor cocktail (Thermo Scientific, Rockford, IL), and a phosphatase inhibitor cocktail (Thermo Scientific). The volume of the media was adjusted to the tissue weight. After 24 hours, supernatants were collected and stored at −20°C.
In Vitro Suppression Assay
We studied the effect of CD8+ Ts lines on CD3/CD28-stimulated PBMCs. PBMCs were labeled with carboxyfluorescein succinimidyl ester (CFSE) (Invitrogen, Carlsbad, CA) and either unstimulated or stimulated with αCD3/CD28 beads (Invitrogen) at a 1:1 ratio. The CD8+ T cells were washed and added to PBMCs at a ratio of 1:1 to 1:16 (1 CD8+ Ts line to 16 PBMC). Proliferation of CD4+ T cells was analyzed by flow cytometry. Percent suppression was calculated based on the following equation: .
Recombinant human TGF-β (BioLegend, San Diego, CA) was added to the suppressor assay as noted. Tissue supernatants were added to the suppressor assay in a 1:100 dilution. αTGF-β neutralizing antibody was used (10 μg/mL; Abcam, Cambridge, MA). Transwell cultures (6.5-mm diameter inserts, 3.0-mm pore size; Corning, Lowell, MA) were used to physically separate the responders from the suppressor cells. A total of 1 × 106 cells were placed in each chamber of the Transwell. αCD3/CD28 beads were placed in both chambers, and thus both responder and suppressor cells were activated.
RNA Extraction and Processing for Microarray Analysis
RNA was isolated from 72-hour CD3/CD28-stimulated CD8+ Ts cells using the RNeasy Micro Kit (Qiagen, Valencia, CA). A total of 200 ng of total RNA was amplified using the Illumina TotalPrep-96 RNA Amplification Kit (Applied Biosystems/Invitrogen, Grand Island, NY). A total of 750 ng of amplified antisense complementary RNA targets was hybridized to Illumina Human HT-12 V4 BeadChip Arrays (Illumina, San Diego, CA) and incubated at 58°C for 17 hours. The arrays were washed, stained, and then scanned on an Illumina Beadstation 500 (Illumina). Signal intensity was captured for each probe on the array using BeadArray Reader software (Illumina).
Microarray Data Analysis
After background subtraction, intensity values were scaled to the median average intensity of the entire sample set using the average normalization function available in GenomeStudio V2009.1 software (Illumina). GeneSpring GX version 7.3.1 (Agilent Technologies, Santa Clara, CA) was used for all downstream analyses. Intensity values were floored to 10, and the intensity of each probe in each sample was normalized to the median intensity of this probe across all samples.
Scoring Scheme to Determine Statistical Enrichment for Protein Kinases and Transcription Factors Given an Input List of Differentially Expressed and Background Networks
Given an input list of differentially expressed genes, the enrichment P values for transcription factors listed in a database developed from chromatin immunoprecipitation (ChIP)-ChIP and ChIP-seq studies14 were analyzed using the Fisher exact test. The top 10 transcription factors were used as seed nodes to construct a protein-protein interaction network, utilizing a merged database of protein-protein interactions, using the shortest path algorithm.15 Comparisons were made by statistical enrichment for protein kinases using the Fisher exact test and a database of kinase-substrate interactions.16
Statistical Analysis
All statistical analyses (other than the microarray analysis) were performed with Prism software (GraphPad, La Jolla, CA). Statistical significance was determined by one-way analysis of variance or t test when appropriate. P < .05 was considered statistically significant.
Results
CD CD8+ Ts Lines Display Reduced Suppressor Activity When Compared With Control Derived Lines
It was previously shown that CD8+ T cells derived from the LP of healthy controls have suppressor activity.11 In the present study, we sought to study the phenotype, function, and characteristics of CD8+ Ts cells from patients with and without IBD. We established an ex vivo expansion protocol that allowed us to preferentially expand CD8+ Ts cells to have enough cells to perform functional and phenotypic characterizations. The expanded CD8+ Ts cells were maintained as a cell line in culture for approximately 3 to 6 months. We assessed different expansion protocols to define the best strategy to selectively expand LP CD8+ Ts cells. The unique effects of visilizumab on CD8+ T cells were first described by Yu et al in 2008.17 They showed that visilizumab induced apoptosis in activated CD4+ but not in CD8+ T cells. We expanded CD8+ Ts cells from the LP using visilizumab and a combination of cytokines that are produced by gut IECs and were shown to play an important role in homeostasis of CD8+ T cells. We found that the combination of IL-7 and IL-15 (not individually) was required for outgrowth of these CD8+ LPL lines for an extended time. Freshly isolated NL LPLs were plated using different experimental conditions, and the percentages of CD8+ T cells were measured 15 days after plating (gating on CD8α). As shown in Supplementary Table 1, the percentage of CD8+ T cells increased dramatically on stimulation with visilizumab (6.19% CD8+ T cells with feeder cells alone, 30.43% CD8+ T cells with feeder cells and visilizumab). We then assessed the effects of IL-7 and IL-15 on the expansion of CD8+ T cells. Five days after plating the cells (stimulated with visilizumab or not), feeder cells and cytokines were added, as noted in Supplementary Table 1A. We observed a significant increase in the percentage of CD8+ T cells in the presence of IL-15 (30.43% without IL-15 and 56.27% with IL-15). In contrast, IL-7 did not have the same effect on CD8+ T-cell expansion (56.27% with IL-15 and 48.6% with IL-15 and IL-7), but the lines were maintained for a longer time. Eliminating the IL-15 resulted in a reduction in Ts activity.
Fourteen days after plating the cells, we isolated the CD8+ T cells (by magnetic bead separation) and maintained the cells in culture in the presence of IL-7 and IL-15 for 3 to 6 months. The expanded CD8+ T cells had suppressor function, and their suppressor activity was further characterized (Figure 1A). Moreover, we tested the function of visilizumab on the suppressor activity by generating matching lines with and without visilizumab stimulation. Visilizumab increased the suppressor activity of the lines (Supplementary Figure 1A). This may indicate the ability of visilizumab to expand global CD8+ Ts cell subsets and is in contrast to our previous data using IECs (presumably antigen driven) to stimulate CD8+ Treg cells.
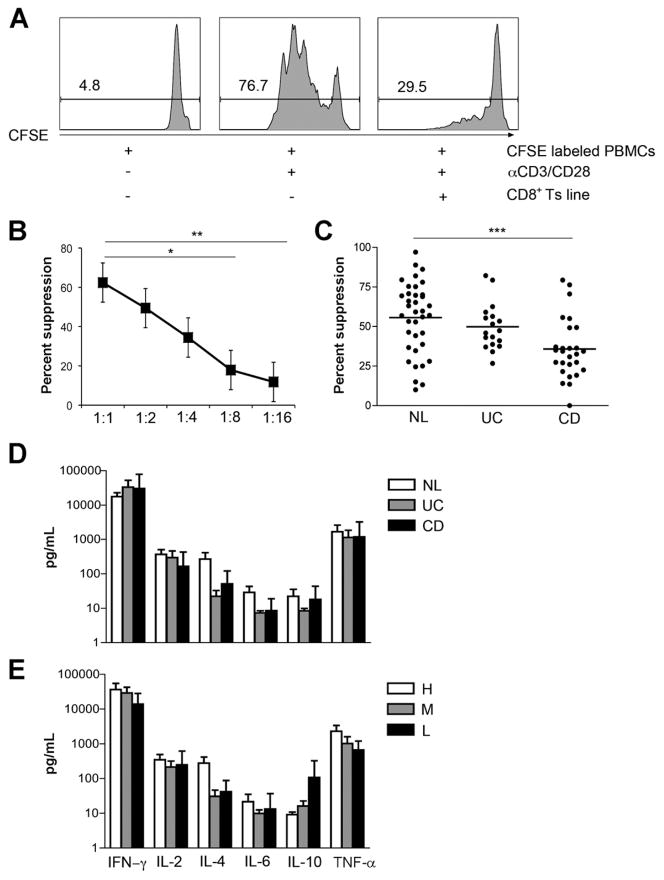
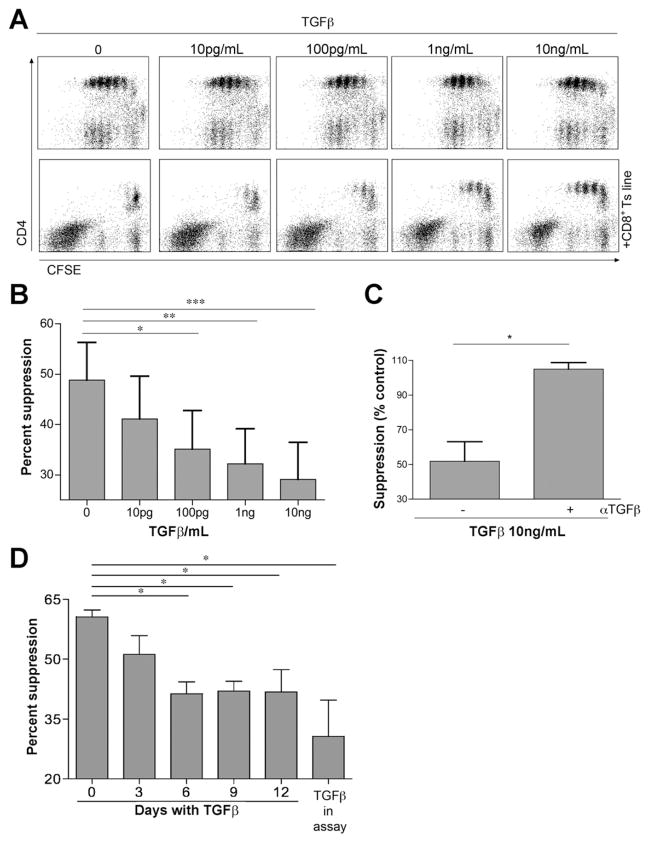
Figure 1.
Suppressor activity of CD8+ Ts lines was observed in LPLs derived from the intestinal mucosa of normal controls. (A) An in vitro suppression assay with CFSE-labeled PBMCs cultured with αCD3/CD28 beads in the presence or absence of an NL-derived CD8+ Ts line at a 1:1 ratio. The proliferation of responder CD4+ T cells assessed by CFSE dilution was analyzed by flow cytometry on day 4. The numbers above the lines indicate the percent proliferating CD4+ T cells. (B) Dose-response curve of CD8+ Ts cells. Suppression of responder CD4+ T-cell proliferation in the presence of NL CD8+ Ts lines incubated at the indicated effector/Treg ratios (n = 5, error bars show SE). *P < .05. **P < .01. (C) Suppressor activity analysis of CD8+ Ts lines in NLs (56.37% ± 22.38%, n = 34), patients with UC (45.35% ± 10.07%, n = 15), and patients with CD (37.39% ± 12.6%, n = 21). ***P < .001 compared with NL lines using a nonparametric test, Kruskal–Wallis one-way analysis of variance, and Dunn’s posttest. (D) CD CD8+ Ts lines (n = 8) exhibit cytokine secretion profiles similar to both normal (n = 11) and UC-derived CD8+ Ts lines (n = 8). Cytokine levels in supernatants were measured 72 hours after αCD3/CD28 stimulation by Cytometric Bead Array. There were no significant differences among the 3 groups. Error bars show the SD. (E) Low suppressor activity CD8+ Ts lines (n = 7) exhibit cytokine secretion profiles similar to both medium suppressor activity (n = 12) and high suppressor activity CD8+ Ts lines (n = 8). Cytokine levels in supernatants were measured 72 hours after αCD3/CD28 stimulation. There were no significant differences among the 3 groups. Error bars show the SD.
To determine whether the activity of the CD8+ Ts lines was stable, we measured the suppressor activity (n = 10) at weekly intervals. Suppressor activity did not change over time (Supplementary Figure 1B) and did not vary in relation to the site of the intestine from which the cells were isolated (data not shown). Suppression was dose dependent (Figure 1B), with significant suppression obtained at a ratio of 1:4 (35%). In contrast to the IEC-activated CD8+ T cells, these CD8+ T lines did not show TcR restriction (data not shown) probably due to the initial or more global nature of the stimulus (visilizumab).
CD8+ Ts lines were generated from the LP of 3 groups of patients: NL, UC, and CD. NL CD8+ Ts lines were derived from mostly noninflamed tissues (33 of 34 lines), whereas both UC and CD CD8+ Ts lines were derived from moderate to actively inflamed tissues. Consistent with our previous observation,11 CD8+ Ts lines generated from patients with CD showed significantly lower suppressor activity when compared with CD8+ Ts lines generated from NLs (P = .0005) (Figure 1C). Interestingly, CD8+ Ts lines derived from patients with UC, despite similar degrees of inflammation, did not show a significant reduction in suppressor activity. UC (as well as CD) IECs were shown18 to be unable to activate a pauciclonal BV5.1 CD8+ Treg subset. In this study, we showed that the CD8+ Ts cells isolated from the LP of patients with UC have intact suppressor activity. Multiple CD8+ Ts subsets may be participating in the homeostasis of the healthy bowel and are likely controlled by distinct mechanisms.
We sought to determine the differences between those CD8+ T cells derived and expanded from the LP of patients with CD versus NL and UC lines, phenotypically and functionally (no difference; see the following text). There was variability across groups in terms of the degrees of suppression. To account for these differences, we also compared the lines with high (56.7%–100%), medium (31.6%–56.7%), and low (0 –31.6%) suppressor activity. The high suppressor lines were enriched in NLs, whereas the low suppressor lines were enriched in patients with CD.
CD8+ Ts Lines Secrete Interferon γ and Tumor Necrosis Factor α on Stimulation With CD3/CD28
Given the differences in suppressor activity in the LP from NLs, patients with UC, and patients with CD, we assessed the cytokine secretion profile. We found that CD8+ Ts lines predominantly secreted interferon (IFN)-γ and tumor necrosis factor α 72 hours after stimulation with αCD3/CD28 beads. CD8+ Ts lines show only minimal secretion of IL-2, IL-4, IL-6, or IL-10 (Figure 1D) by Cytometric Bead Array (BD Bioscience, San Diego, CA). In addition, we were not able to detect any secretion of IL-17 or bioactive TGF-β (data not shown). We did not find any significant differences in cytokine secretion between NL-, UC-, or CD-derived CD8+ Ts lines or in the high, medium, and low suppressor activity groups (Figure 1E). Moreover, we measured cytokine secretion of individual lines at 1, 2, and 3 months in culture and found a similar cytokine secretion profile over time (Supplementary Figure 2). No correlation was found between IL-10 or IFN-γ and suppressor activity.
Suppression Mediated by CD8+ Ts Lines Is Contact Dependent
Because we could not detect any bioactive TGF-β and very low levels of IL-10 production by the CD8+ Ts lines, we focused on the contact-dependent nature of these cells. A Transwell culture was used to physically separate the responder PBMCs from the CD8+ Ts cells. The presence of a physical barrier between the responder and the suppressor cells significantly reduced the suppressor activity of CD8+ Ts cells (CD3/CD28 beads on both sides of the Transwell; Figure 2). Furthermore, supernatants collected from CD3/CD28-stimulated CD8+ Ts lines did not suppress responder cell proliferation (data not shown). Thus, it was unlikely that CD8+ Ts lines were secreting regulatory factors. Next, we tested the effects of exogenous IL-2 in the suppressor assay to test the possibility that CD8+ Ts cells were serving as a cytokine sink. IL-2 was added to the suppressor assay (as well as to a control containing only responder CFSE-labeled PBMCs); at the doses used (2, 20, and 200 U/mL), it did not reverse the suppressor activity of the NL CD8+ lines (Supplementary Figure 1C).
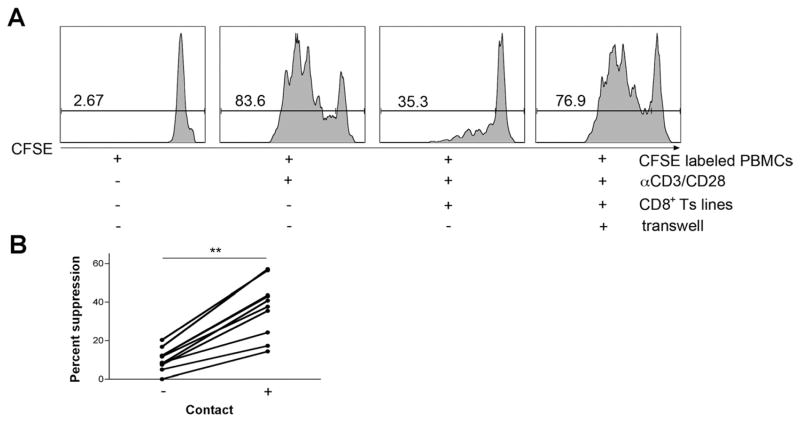
Figure 2.
Cell-to-cell contact is required for mediating suppression. (A) CFSE-labeled PBMCs were cultured with αCD3/CD28 activation beads, and CD8+ Ts lines were either placed in the same well with responder cells or separated from responder cells by a Transwell. CD8+ Ts lines were not able to mediate suppression when separated from the responder cells (both sides of the Transwell were stimulated with anti-CD3/CD28 beads; data are representative of 10 NL/UC CD8+ Ts lines). (B) Summary figure: 10 lines were tested for their ability to mediate suppression in a Transwell; in all, suppression was significantly reduced. **P < .001 using a nonparametric Wilcoxon matched pairs t test.
CD8+ Ts Lines Show Phenotypic Characteristics of Known CD8+ Ts Cells
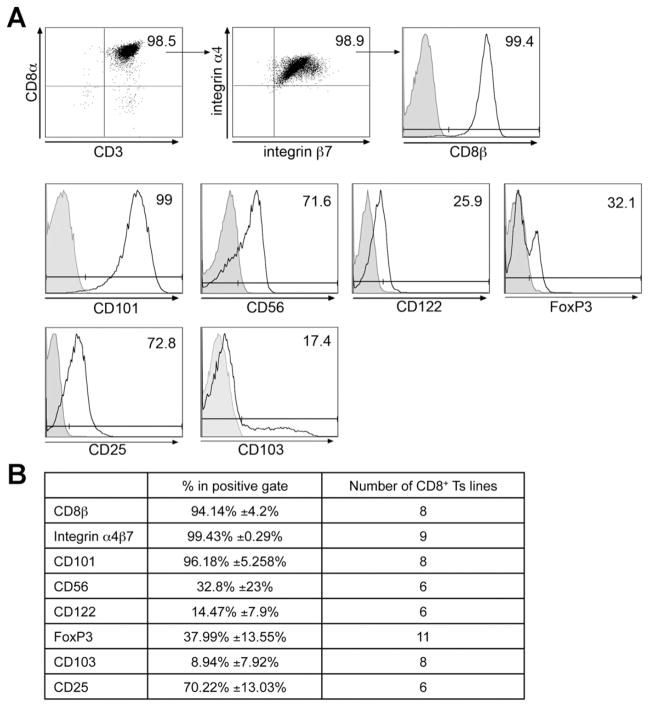
One trivial difference to account for the lack of suppression in the cell lines from patients with CD versus NLs/patients with UC could have been the differences in input cell populations (baseline characteristics of the expanded cell populations). As shown in Supplementary Table 1B, we saw no differences in the percentage of CD8α cells expressing CD45RAhi CD62L between cells derived from the mucosa of healthy individuals and patients with CD. In fact, the baseline characteristics (CD4/ CD8, CD3, FoxP3, and so on) were no different among the various groups (data not shown). Given the contact dependency, we analyzed whether surface molecules were involved in mediating suppression. We assessed the expression of surface markers known to be associated with CD4+ Treg cells or CD8+ Ts cells (at 4 weeks in culture). We found that CD8+ Ts lines were CD8αβ positive and expressed high levels of the integrin α4β7 (imprinted for gut homing). Furthermore, CD8+ Ts lines expressed high levels of CD101, shown to be expressed on both CD8+ TrE cells9 and CD4+-expressing FoxP3 cells.19 CD8+ Ts lines also shared phenotypic features of previously described CD8+ Ts cells, expressing CD56 and low levels of CD122,20–22 but expressed low levels of FoxP3 and CD103, both markers that are expressed by CD4+ Treg cells.23,24 Because FoxP3 has been used as a marker for CD4+ Treg cells, we looked for a correlation of FoxP3 expression levels with suppressor activity but found no such correlation. CD8+ Ts lines did express CD25, which is associated with CD4+ Treg cells but can also be a measure of activation (Figure 3). The expression levels of each surface or intracellular marker were found to be comparable among the 3 groups (Supplementary Table 2) and did not correlate with any suppressor cell function. Moreover, CD8+ Ts lines did not express CTLA4, ICOS, or CD28, as well as perforin, granzyme B, or CD16, known to be associated with cytotoxic T lymphocytes or natural killer T cells (Supplementary Figure 3).
Figure 3.
CD8+ Ts lines display some phenotypic similarities with known CD8+ Ts cells. (A) Surface staining of CD8+ Ts lines at 4 weeks in culture with IL-7 and IL-15. Surface expression of CD8α β, CD3, α4β7, CD101, CD56, CD122, FoxP3, CD25, and CD103. Open histograms indicate specific staining (antibodies to markers below the plots), and filled histograms indicate isotype-matched control antibodies. Numbers adjacent to the outlined areas and above lines indicate the percentage of cells in the gate. No differences were found between NL-, UC-, and CD-derived lines. Data are representative of at least 6 independent experiments. (B) Summary table indicating the number of cell lines stained for a specific cell marker as well as the percentage of cells in the positive gate.
Transcript Profiling and Gene List Enrichment Analysis
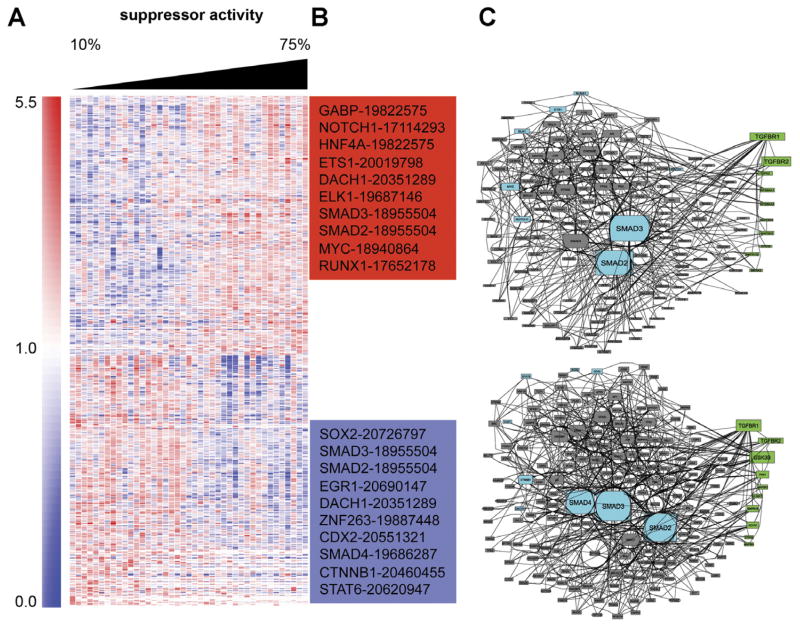
To further analyze the cellular differences between CD8+ lines with high and low suppressor activity, we used a genome-wide profiling approach. Forty-one CD8+ T-cell lines were collected. For each, suppressor activity was measured and RNA was isolated. CD8+ T-cell lines were stimulated with αCD3/CD28 beads for 3 days before RNA isolation. The lines were collected from all 3 groups (CD, UC, NL) and were only ranked based on suppressor activity. Lines with low suppressor activity were enriched for CD lines, and lines with high suppressor activity were enriched for NL lines. We looked for genes displaying differential expression levels that were either positively or negatively correlated with suppressor activity, that is, genes that showed low expression levels in the low suppressor lines and high expression levels in the high suppressor lines (positive correlation) or genes that showed high expression levels in the low suppressor lines and low expression levels in the high suppressor lines (negative correlation). A set of 24,977 probes with a detection precision P value<.01 in at least one sample was selected (detection P value is determined by calculating precision of gene beads compared with control beads, background on the chip). Next, the levels of expression of those transcripts were correlated with suppressor activity (Spearman correlation). The top 600 transcripts with significant positive or negative correlations were selected (in each list several genes were represented by more than one probe; hence, only 531 and 511 genes are listed, respectively; Supplementary Table 3). A heat map of the selected genes was created by hierarchical clustering (Figure 4A). To explore the functional theme of the genes that correlated with suppressor activity, we performed gene list enrichment analysis using the software Lists2Networks,15 which uses many gene list libraries of prior biological knowledge such as pathways, gene ontology, mouse phenotype, microRNA/messenger RNA, and disease gene libraries.
Figure 4.
Heat map of expression values of the 1042 transcripts differentially expressed and correlated with suppression activity. (A) Samples (columns) were arranged by increasing suppressor activity (left to right). Genes (rows) were grouped by hierarchical clustering based on similarity in expression profiles. Intensity values are normalized to the median calculated across all samples. Values that are greater than the median are represented in red and lower than the median are in blue. (B) Two lists of positively and negatively correlated genes (with suppressor activity) were used to determine the top 10 transcription factors that are most likely to be the upstream regulators of the differentially expressed genes. The numbers next to the transcription factors are the PubMed IDs of the ChIP-seq/ChIP articles used to extract the interactions with the transcription factor. (C) Using the 2 lists of the top 10 transcription factors, a protein-protein interaction subnetwork that “connected” the factors using additional protein-protein interactions collected from 12 databases was constructed and displayed. The protein kinases enriched in substrates in each subnetwork were identified using a database of kinase substrate interactions. Cyan nodes are identified transcription factors, gray nodes are intermediate proteins from known protein-protein interaction networks, and green nodes are the inferred protein kinases. The size of the nodes is determined based on the degree of connectivity. Transcription factors and protein kinases are ordered based on their enrichment scores.
Genes that were positively correlated with suppressor activity revealed intriguing biological pathways. We found enrichment for 9 genes that, when knocked out in mice, display abnormal intestine physiology (P = .044). The negatively correlated gene list (with suppressor activity) indicated that members of the TGF-β signaling pathway were enriched (P = .0068).25 See Table 1 for additional enriched biological categories.
Table 1.
Enrichment Analysis With Lists2Networks
| Category | Term | P value | Genes |
|---|---|---|---|
| Positively correlated genes | |||
| BioCarta pathways | TNFR1PATHWAY | .0069466925 | CASP2, DFFA, MAP3K7, PRKDC |
| BioCarta pathways | RHOPATHWAY | .0088238497 | ACTR2, ARHGAP4, ARPC1B, PIP5K1A |
| KEGG pathways | HSA00030_PENTOSE_PHOSPHATE_PATHWAY | .0046581186 | PFKM, PGM1, PRPS2, RPIA |
| KEGG pathways | HSA00071_FATTY_ACID_METABOLISM | .0080360017 | ACADSB, ACSL3, ADH5, DCI, GCDH |
| Chromosome location | chr6q13 | .0088 | BCKDHB, HDDC2, OGFRL1, RWDD1 |
| MGI mouse phenotype | MP0010155_abnormal_intestine_physiology | .044 | CUL4A, LIG4, MAP3K7, PCID2, PMS2, PRKDC, SAV1, TLR5, VDR |
| MicroRNAs | MIR-433 | .0061 | AZIN1, CLYBL, CRLF3, CTBP1, CUL4A, GRB2, HNRNPR, LBR, MAPK1IP1L, PAPD5, PCCB, SRFBP1, TMEM33, TOP1, WAC, YWHAZ |
| Negatively correlated genes | |||
| KEGG pathways | HSA05120_EPITHELIAL_CELL_SIGNALING_IN_HELICOBACTER_PYLORI_INFECTION | .0017853991 | ATP6V0C, ATP6V1C1, CCL5, GIT1, JUN, MAPK11, PLCG2 |
| KEGG pathways | HSA04060_CYTOKINE_CYTOKINE_RECEPTOR_INTERACTION | .0071917045 | ACVR1, CCL15, CCL18, CCL5, CXCL16, FASLG, IL12RB2, IL18RAP, IL1R2, IL2RB, LEPR, TNFRSF1A, TNFRSF6B, TNFSF14 |
| KEGG pathways | HSA00562_INOSITOL_PHOSPHATE_METABOLISM | .0096720277 | ITPK1, PI4KA, PIP5K1C, PLCG2, SYNJ2 |
| WikiPathways | Hs_TGF_Beta_Signaling_Pathway_WP560_21858 | .0068 | JUN, SMAD7, STAT3, TGIF1, ZEB2 |
| PPI hubs | HDAC3 | .0063818991 | BCOR, JUN, MAPK11, NCOR2, PPP4R1, STAT3 |
| GO biological process | proton transport GO_0015992 | .0004597864 | ATP6, V0CATP6V1C1NOX1UCP2UCP3 |
| GO molecular function | activin binding GO_0048185 | .0008475625 | ACVR1, ENG, SMAD7 |
NOTE. Summary of the enrichment analysis of the positively and negatively correlated gene list (with suppression). The table shows what types of categories were enriched, what genes derived this enrichment, by what criteria, and its P value.
The 2 gene lists of negatively and positively correlated genes were subjected to promoter analysis. We used the software ChEA14 to determine the top 10 transcription factors that were most likely to regulate the expression of the genes in each list (Figure 4B). For both lists, SMAD2 and SMAD3 transcription factors were identified as enriched. We next used protein-protein interactions to construct a subnetwork that “connects” the top 10 predicted transcription factors using additional proteins that are known to interact with those factors (Supplementary Table 4). Interestingly, SMAD2 and SMAD3 were highly connected central hubs (Figure 4A and Supplementary Figure 4). Finally, we used KEA software16 to identify the protein kinases that have many substrates in the protein interaction networks centered around these transcription factors (Supplementary Table 4). Both TGFBR1 and TGFBR2 were highly enriched, using KEA software, further implicating the SMAD pathway.
TGF-β Blocks Suppressor Activity Mediated by CD8+ Ts Lines
The downstream kinases and transcription factors of TGF-β signaling pathways were highlighted by several analysis patterns; members of the TGF-β signaling pathway correlated with suppressor activity and SMAD2 and SMAD3, which are downstream of the TGF-β receptor, were predicted to be central in regulating gene expression correlating with suppressor activity. SMAD signaling was shown to positively and negatively regulate gene transcription by recruiting co-activators or corepressors to the SMAD complex.26 Because TGF-β signaling was central for both up-regulation and down-regulation of target genes, we tested the effect of recombinant human TGF-β on the suppressor activity of CD8+ Ts lines. TGF-β was added to the suppressor assay (as well as to a control containing only responder CFSE-labeled PBMCs), and at the doses used (0.01–10 ng/mL) it did not reduce CD4+ T-cell proliferation but rather significantly reduced suppressor activity (in a dose-dependent fashion; Figure 5A and B). TGF-β–mediated FoxP3 expression and Treg cell differentiation in naïve CD4+ T cells occurs after 5 days.27 In our experimental setting, the suppressor assay cell activity was measured at 3 to 4 days, not enough time to induce CD4+ Treg cells that possibly could adversely affect the CD8+ Treg cells. Addition of αTGF-β neutralizing antibody to the TGF-β–supplemented suppressor assay reversed the effect (Figure 5C). This is the first report implicating TGF-β as an inhibitor of suppressor cells.
Figure 5.
TGF-β reduces CD8+ Ts suppressor activity. (A) TGF-β was added to the suppressor assay in increasing amounts. Interestingly, CD4+ T-cell proliferation was not inhibited. In contrast, TGF-β reduced the ability of CD8+ Ts cells to mediate suppression. (B) Summary figure. The effect of TGF-β was tested in 10 different NL, UC, and CD CD8+ Ts lines with medium or high suppressor activity using one-way analysis of variance and Bonferroni posttest. TGF-β suppressed the Ts suppressor function in all cases. (C) Addition of αTGF-β neutralizing antibody (105 ± 3.68, n = 2) reversed the effect of TGF-β on the suppressor assay (51.98% ± 11.2%, n = 2). A 100% suppression is equivalent to the amount of suppression in the absence of TGF-β and αTGF-β antibody using an unpaired t test. (D) CD8+ NL Ts lines were pretreated with TGF-β for 12, 9, 6, and 3 days or not treated. Suppressor activity was reduced with the amount of time that lines were treated with TGF-β. TGF-β was added to the un-pretreated cells and reduced suppressor activity as well. n = 3; data are representative of at least 3 independent experiments. Error bars represent SEM. *P < .05, **P < .01, ***P < .001.
We looked at TGFBR expression levels in low and high suppressor lines and found the expression levels of TGFBR1 and TGFBR2 to be comparable (Supplementary Figure 5), suggesting that the difference between the responses to TGF-β is not the result of hyporesponsiveness or hyperresponsiveness to TGF-β in the low suppressor lines. To determine whether tissue-derived TGF-β was responsible for the reduced suppressor activity in CD, we collected supernatants from tissue explant cultures. Supernatants from explants derived from patients with CD had significantly higher amounts of bioactive TGF-β when compared with controls (Figure 6A). Next we measured the effect of these supernatants on suppressor activity. Supernatants derived from patients with CD significantly reduced the suppression in NL CD8+ T-cell lines whereas the control supernatants did not (Figure 6B). To support the idea that TGF-β, and not other factors present in the supernatants, were capable of mediating the reduction in suppressor activity, we added an αTGF-β neutralizing antibody to the assay. This antibody was able to reverse the inhibition of suppression mediated by CD-derived supernatants, showing that TGF-β is the cytokine responsible for this inhibition (Figure 6C). We wanted to determine if acute exposure to TGF-β was sufficient to affect the suppressor cell activity. NL CD8+ Ts lines were treated with 5 ng/mL TGF-β for 12, 9, 6, and 3 days and washed before placing them in the suppressor assay. Suppressor activity was reduced when cells were treated for 6 days or longer, suggesting a preconditioned response (Figure 5D). However, the effect of TGF-β in the suppressor assay was more pronounced, indicating that other factors in the CD microenvironment may contribute to the defect found in CD-derived CD8+ Ts lines.
Figure 6.
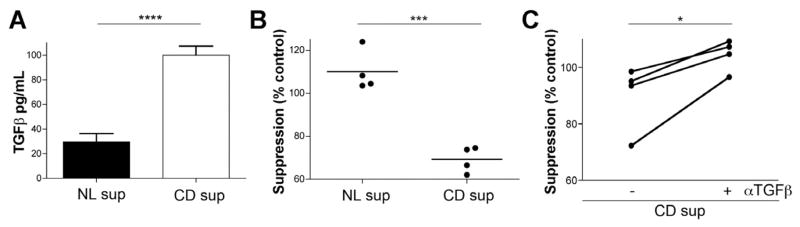
(A) Bioactive levels of TGF-β in tissue explants derived from NLs and patients with CD. Error bars represent SD, using an unpaired t test with Welch’s correction (n = 3). (B) Change in suppressor activity in response to supernatants derived from tissue explants. CD, but not NL, supernatants reversed Ts activity in all cases. A 100% suppression is equivalent to the amount of suppression in the absence of supernatants using an unpaired t test with Welch’s correction. (C) Addition of αTGF-β neutralizing antibody reversed the effect of CD supernatants on the suppressor assay. Four different CD supernatants were added to the suppressor assay. Data are representative of at least 3 independent experiments using an unpaired t test. *P < .05; **P < .001; ****P < .0001.
Discussion
Defects in the induction of regulatory CD8+ Ts cells by IBD epithelial cells led us to investigate the characteristics of CD8+ Ts cells derived from the LP of healthy controls and patients with IBD. Here we show that functional CD8+ Ts cells are present in the LP of controls and are defective in the LP of patients with CD although not in the LP of patients with UC despite similar levels of inflammation and similar medications. We have established a protocol to expand such cells, enabling us to maintain CD8+ Ts lines in culture for an extended time. Anti-CD3 antibodies have been used in the treatment of patients with autoimmune diseases, IBD, and allograft rejection and have been shown to promote immune tolerance by both depleting or inactivating effector T cells as well as increasing the numbers of CD8+ Treg cells. Thus, we used a humanized FcR nonbinding anti-CD3 monoclonal antibody, visilizumab, in our cultures. CD8+ Ts cells were shown to express CD25, CD56, CD122, CD103, and CD101, which were previously shown to be expressed on CD8+ Ts cells.9,15,16,28 The cells did not express CTLA4, which is reported to be associated with CD4+ Treg cells.29,30 The cells express low levels of FoxP3, which was not associated with suppressor activity, because lines that lacked FoxP3 expression had intact suppressor activity. Moreover, we found that CD8+ Ts lines have no TcR restriction, suggesting that the suppression is not constrained to T cells recognizing a specific antigen, as opposed to our previously reported IEC-activated CD8 Treg cells. These cells were not shown to secrete IL-10 or bioactive TGF-β, known to be associated with suppression. Rather, they were inhibited by cell contact.
Gene expression analysis was used to shed light on the molecular differences between lines with high and low suppressor activity (enriched in NL vs CD lines, respectively). The results point toward the TGF-β signaling pathway, a main candidate in controlling the suppressor activity of CD8+ Ts cells. This was evident by enrichment analysis of gene lists with both positive and negative correlations with suppressor activity. TGF-β is a multi-functional cytokine with a crucial role in many cellular pathways, including cell growth, apoptosis, fibrosis, differentiation, and immune responses. TGF-β inhibits immune responses through multiple mechanisms such as the induction and maintenance of FoxP3-expressing CD4+ Treg cells and inhibition of T-cell proliferation. Nevertheless, TGF-β also has proinflammatory effects, inducing the differentiation of Th17 cells stimulating expression of retinoic acid receptor–related orphan nuclear receptor γt. In the presence or absence of IL-6, TGF-β induces the development of Th17 versus Treg cells, respectively.31 Intestinal fibrosis and stricture formation are often part of the natural course of CD. TGF-β1 has been implicated in fibrosis through its ability to promote extracellular matrix synthesis and fibroblast contraction. Both TGF-β and its receptors are overexpressed in the intestine of patients with CD32,33 and are consistent with our findings of higher TGF-β levels produced by mucosal samples derived from patients with CD compared with healthy controls. It is intriguing to note that despite this overproduction, CD4+ T-cell immune responses are not inhibited. The existence of TGF-β in CD tissue can explain both the high levels of Th1734–36 and the defect in CD8+ Ts cells in patients with CD. We have shown how the presence of TGF-β reduces the suppressor activity of CD8+ Ts cells in vitro. The effects of TGF-β on CD8+ Ts cells and on regulatory CD4+ FoxP3+ cells (and TGF-β secreting) are clearly dichotomous. The effect of TGF-β in CD tissue may act to suppress the CD8+ Ts cells while promote active inflammation (in combination with IL-1β and IL-6).
TGF-β was shown to negate the effects of IL-15 by down-regulating the expression of CD122 and inducing apoptosis (and not cell survival) in cytolytic CD8+ T cells.37,38 TGF-β did not affect CD8+ Ts cells in the same fashion. The antagonistic effects of IL-15 and TGF-β should be further investigated because both cytokines are present in the intestinal mucosa of patients with CD and their effects can play a role in the defect seen in patients with CD.
The mechanism of suppression used by CD4+ Treg cells is poorly understood. Direct cell contact was shown to be crucial in mediating suppression,39,40 yet definition of a surface molecule has been elusive. Membrane-bound TGF-β has been recognized to have a role in CD4+ FoxP3 Treg cells, but there are conflicting reports. However, TGF-β does not mediate CD8+ Ts cell suppression; in contrast, its presence abolishes their suppressor activity. It would be of interest to study the mechanism of action of these CD8+ Ts cells by investigating the molecular consequences of TGF-β–treated CD8+ Ts cells. A negative relationship has been proposed between TGF-β and SMAD7 in effector T cells.41 It will be of interest if similar responses are observed in suppressor CD8+ Ts cells.
Along those lines, several genes, identified by microarray analysis, bring up interesting possibilities. Endoglin (ENG) is a type III TGF-β receptor that is overexpressed in CD stricture fibroblasts. ENG is a negative regulator of TGF-β signaling. We have found that ENG expression in CD8+ Ts cells is negatively correlated with suppressor activity (Supplementary Table 1). ENG messenger RNA is up-regulated in CD8+ Ts cells from patients with CD compared with controls and is up-regulated in NL- or UC-derived lines (with high suppressor activity) on treatment with TGF-β (Supplementary Figure 6).42 Moreover, we have found JUN and TG-interacting factor to be up-regulated in CD-derived CD8+ Ts lines and in response to treatment with TGF-β. It has been reported that the interaction between c-JUN and TG-interacting factor can suppress SMAD2 transcriptional activity.43 IL18RAP was also negatively correlated with suppressor activity and is up-regulated in CD-derived CD8+ Ts lines and in response to treatment with TGF-β in NL and UC lines. Interestingly, IL18RAP has been identified in genome-wide association studies to be associated with CD.44,45 Further analysis of the levels of these proteins and other components of the TGF-β signaling pathway should shed light on the mechanism of CD8+ suppressor activity.
In summary, we have shown that there is a defect in CD8+ Ts cells derived from the LP of patients with CD that can be accounted for by excess TGF-β in the tissue (patients with UC, an inflammatory control, do not have this defect and do not have excess TGF-β; Supplementary Figure 7). This lack of suppression may account for the unopposed proinflammatory (Th17) and profibrogenic (TGF-β) activities seen in patients with CD.
Supplementary Material
Acknowledgments
Funding
Supported by National Institutes of Health grants AI044236, AI084952, DK072201, and DK086605.
Abbreviations used in this paper
- CFSE
carboxyfluorescein succinimidyl ester
- ChIP
chromatin immunoprecipitation
- ENG
endoglin
- IEC
intestinal epithelial cell
- IFN
interferon
- IL
interleukin
- LP
lamina propria
- LPL
lamina propria lymphocyte
- NL
normal
- PBMC
peripheral blood mononuclear cell
- TGF
transforming growth factor
- Treg cell
regulatory T cell
- Ts cell
suppressor T cell
Footnotes
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at http://dx.doi:10.1053/j.gastro.2012.12.001.
Conflicts of interest
The authors disclose no conflicts.
References
- 1.Morrissey PJ, Charrier K, Braddy S, et al. CD4+ T cells that express high levels of CD45RB induce wasting disease when transferred into congenic severe combined immunodeficient mice. Disease development is prevented by cotransfer of purified CD4+ T cells. J Exp Med. 1993;178:237–244. doi: 10.1084/jem.178.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Powrie F, Correa-Oliveira R, Mauze S, et al. Regulatory interactions between CD45RBhigh and CD45RBlow CD4+ T cells are important for the balance between protective and pathogenic cell-mediated immunity. J Exp Med. 1994;179:589–600. doi: 10.1084/jem.179.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Powrie F, Leach MW, Mauze S, et al. Phenotypically distinct subsets of CD4+ T cells induce or protect from chronic intestinal inflammation in C. B-17 scid mice. Int Immunol. 1993;5:1461–1471. doi: 10.1093/intimm/5.11.1461. [DOI] [PubMed] [Google Scholar]
- 4.Yu QT, Saruta M, Avanesyan A, et al. Expression and functional characterization of FOXP3+ CD4+ regulatory T cells in ulcerative colitis. Inflamm Bowel Dis. 2007;13:191–199. doi: 10.1002/ibd.20053. [DOI] [PubMed] [Google Scholar]
- 5.Saruta M, Yu QT, Fleshner PR, et al. Characterization of FOXP3+CD4+ regulatory T cells in Crohn’s disease. Clin Immunol. 2007;125:281–290. doi: 10.1016/j.clim.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Menager-Marcq I, Pomie C, Romagnoli P, et al. CD8+CD28- regulatory T lymphocytes prevent experimental inflammatory bowel disease in mice. Gastroenterology. 2006;131:1775–1785. doi: 10.1053/j.gastro.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho J, Kurtz CC, Naganuma M, et al. A CD8+/CD103high T cell subset regulates TNF-mediated chronic murine ileitis. J Immunol. 2008;180:2573–2580. doi: 10.4049/jimmunol.180.4.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujiwara D, Chen L, Wei B, et al. Small intestine CD11c+ CD8+ T cells suppress CD4+ T cell-induced immune colitis. Am J Physiol Gastrointest Liver Physiol. 2011;300:G939–G947. doi: 10.1152/ajpgi.00032.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allez M, Brimnes J, Shao L, et al. Activation of a unique population of CD8(+) T cells by intestinal epithelial cells. Ann N Y Acad Sci. 2004;1029:22–35. doi: 10.1196/annals.1309.004. [DOI] [PubMed] [Google Scholar]
- 10.Allez M, Brimnes J, Dotan I, et al. Expansion of CD8+ T cells with regulatory function after interaction with intestinal epithelial cells. Gastroenterology. 2002;123:1516–1526. doi: 10.1053/gast.2002.36588. [DOI] [PubMed] [Google Scholar]
- 11.Brimnes J, Allez M, Dotan I, et al. Defects in CD8+ regulatory T cells in the lamina propria of patients with inflammatory bowel disease. J Immunol. 2005;174:5814–5822. doi: 10.4049/jimmunol.174.9.5814. [DOI] [PubMed] [Google Scholar]
- 12.Allez M, Brimnes J, Dotan I, et al. Expansion of CD8+ T cells with regulatory function after interaction with intestinal epithelial cells. Gastroenterology. 2002;123:1516–1526. doi: 10.1053/gast.2002.36588. [DOI] [PubMed] [Google Scholar]
- 13.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, et al. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 14.Lachmann A, Xu H, Krishnan J, et al. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26:2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berger SI, Posner JM, Ma’ayan A. Genes2Networks: connecting lists of gene symbols using mammalian protein interactions databases. BMC Bioinformatics. 2007;8:372. doi: 10.1186/1471-2105-8-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lachmann A, Ma’ayan A. KEA: kinase enrichment analysis. Bioinformatics. 2009;25:684–686. doi: 10.1093/bioinformatics/btp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu QT, Saruta M, Papadakis KA. Visilizumab induces apoptosis of mucosal T lymphocytes in ulcerative colitis through activation of caspase 3 and 8 dependent pathways. Clin Immunol. 2008;127:322–329. doi: 10.1016/j.clim.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 18.Allez M, Brimnes J, Shao L, et al. Activation of a unique population of CD8(+) T cells by intestinal epithelial cells. Ann N Y Acad Sci. 2004;1029:22–35. doi: 10.1196/annals.1309.004. [DOI] [PubMed] [Google Scholar]
- 19.Fernandez I, Zeiser R, Karsunky H, et al. CD101 surface expression discriminates potency among murine FoxP3+ regulatory T cells. J Immunol. 2007;179:2808–2814. doi: 10.4049/jimmunol.179.5.2808. [DOI] [PubMed] [Google Scholar]
- 20.Endharti AT, Rifa’I M, Shi Z, et al. Cutting edge: CD8+CD122+ regulatory T cells produce IL-10 to suppress IFN-gamma production and proliferation of CD8+ T cells. J Immunol. 2005;175:7093–7097. doi: 10.4049/jimmunol.175.11.7093. [DOI] [PubMed] [Google Scholar]
- 21.Davila E, Kang YM, Park YW, et al. Cell-based immunotherapy with suppressor CD8+ T cells in rheumatoid arthritis. J Immunol. 2005;174:7292–7301. doi: 10.4049/jimmunol.174.11.7292. [DOI] [PubMed] [Google Scholar]
- 22.Rifa’i M, Kawamoto Y, Nakashima I, et al. Essential roles of CD8+CD122+ regulatory T cells in the maintenance of T cell homeostasis. J Exp Med. 2004;200:1123–1134. doi: 10.1084/jem.20040395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allakhverdi Z, Fitzpatrick D, Boisvert A, et al. Expression of CD103 identifies human regulatory T-cell subsets. J Allergy Clin Immunol. 2006;118:1342–1349. doi: 10.1016/j.jaci.2006.07.034. [DOI] [PubMed] [Google Scholar]
- 24.Leithauser F, Meinhardt-Krajina T, Fink K, et al. Foxp3-expressing CD103+ regulatory T cells accumulate in dendritic cell aggregates of the colonic mucosa in murine transfer colitis. Am J Pathol. 2006;168:1898–1909. doi: 10.2353/ajpath.2006.050228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith CL, Goldsmith CA, Eppig JT. The Mammalian Phenotype Ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome Biol. 2005;6:R7. doi: 10.1186/gb-2004-6-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wrana JL. Crossing Smads. Sci STKE. 2000;2000:re1. doi: 10.1126/stke.2000.23.re1. [DOI] [PubMed] [Google Scholar]
- 27.Zhou L, Lopes JE, Chong MM, et al. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453:236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rifa’i M, Kawamoto Y, Nakashima I, et al. Essential roles of CD8+CD122+ regulatory T cells in the maintenance of T cell homeostasis. J Exp Med. 2004;200:1123–1134. doi: 10.1084/jem.20040395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zemann B, Schwaerzler C, Griot-Wenk M, et al. Oral administration of specific antigens to allergy-prone infant dogs induces IL-10 and TGF-beta expression and prevents allergy in adult life. J Allergy Clin Immunol. 2003;111:1069–1075. doi: 10.1067/mai.2003.1411. [DOI] [PubMed] [Google Scholar]
- 30.Contractor N, Louten J, Kim L, et al. Cutting edge: Peyer’s patch plasmacytoid dendritic cells (pDCs) produce low levels of type I interferons: possible role for IL-10, TGFbeta, and prostaglandin E2 in conditioning a unique mucosal pDC phenotype. J Immunol. 2007;179:2690–2694. doi: 10.4049/jimmunol.179.5.2690. [DOI] [PubMed] [Google Scholar]
- 31.Yang XO, Nurieva R, Martinez GJ, et al. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 2008;29:44–56. doi: 10.1016/j.immuni.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Di Sabatino A, Jackson CL, Pickard KM, et al. Transforming growth factor beta signalling and matrix metalloproteinases in the mucosa overlying Crohn’s disease strictures. Gut. 2009;58:777–789. doi: 10.1136/gut.2008.149096. [DOI] [PubMed] [Google Scholar]
- 33.Burke JP, Mulsow JJ, O’Keane C, et al. Fibrogenesis in Crohn’s disease. Am J Gastroenterol. 2007;102:439–448. doi: 10.1111/j.1572-0241.2006.01010.x. [DOI] [PubMed] [Google Scholar]
- 34.Fujino S, Andoh A, Bamba S, et al. Increased expression of interleukin 17 in inflammatory bowel disease. Gut. 2003;52:65–70. doi: 10.1136/gut.52.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nielsen OH, Kirman I, Rudiger N, et al. Upregulation of interleukin-12 and -17 in active inflammatory bowel disease. Scand J Gastroenterol. 2003;38:180–185. doi: 10.1080/00365520310000672. [DOI] [PubMed] [Google Scholar]
- 36.Seiderer J, Elben I, Diegelmann J, et al. Role of the novel Th17 cytokine IL-17F in inflammatory bowel disease (IBD): upregulated colonic IL-17F expression in active Crohn’s disease and analysis of the IL17F p.His161Arg polymorphism in IBD. Inflamm Bowel Dis. 2008;14:437–445. doi: 10.1002/ibd.20339. [DOI] [PubMed] [Google Scholar]
- 37.Sanjabi S, Mosaheb MM, Flavell RA. Opposing effects of TGF-beta and IL-15 cytokines control the number of short-lived effector CD8+ T cells. Immunity. 2009;31:131–144. doi: 10.1016/j.immuni.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lucas PJ, Kim SJ, Mackall CL, et al. Dysregulation of IL-15-mediated T-cell homeostasis in TGF-beta dominant-negative receptor transgenic mice. Blood. 2006;108:2789–2795. doi: 10.1182/blood-2006-05-025676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dieckmann D, Bruett CH, Ploettner H, et al. Human CD4(+)CD25(+) regulatory, contact-dependent T cells induce interleukin 10-producing, contact-independent type 1-like regulatory T cells [corrected. J Exp Med. 2002;196:247–253. doi: 10.1084/jem.20020642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jonuleit H, Schmitt E, Kakirman H, et al. Infectious tolerance: human CD25(+) regulatory T cells convey suppressor activity to conventional CD4(+) T helper cells. J Exp Med. 2002;196:255–260. doi: 10.1084/jem.20020394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monteleone G, Kumberova A, Croft NM, et al. Blocking Smad7 restores TGF-beta1 signaling in chronic inflammatory bowel disease. J Clin Invest. 2001;108:601–609. doi: 10.1172/JCI12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burke JP, Watson RW, Mulsow JJ, et al. Endoglin negatively regulates transforming growth factor beta1-induced profibrotic responses in intestinal fibroblasts. Br J Surg. 2010;97:892–901. doi: 10.1002/bjs.6996. [DOI] [PubMed] [Google Scholar]
- 43.Pessah M, Prunier C, Marais J, et al. c-Jun interacts with the corepressor TG-interacting factor (TGIF) to suppress Smad2 transcriptional activity. Proc Natl Acad Sci U S A. 2001;98:6198–6203. doi: 10.1073/pnas.101579798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhernakova A, Festen EM, Franke L, et al. Genetic analysis of innate immunity in Crohn’s disease and ulcerative colitis identifies two susceptibility loci harboring CARD9 and IL18RAP. Am J Hum Genet. 2008;82:1202–1210. doi: 10.1016/j.ajhg.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Festen EA, Goyette P, Green T, et al. A meta-analysis of genome-wide association scans identifies IL18RAP, PTPN2, TAGAP, and PUS10 as shared risk loci for Crohn’s disease and celiac disease. PLoS Genet. 2011;7:e1001283. doi: 10.1371/journal.pgen.1001283. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.