Summary
In a recent issue of Molecular Cell, Zheng et al. (2008) demonstrate that human DNA2, originally identified in yeast as a nuclear DNA replication and repair factor, functions exclusively in mammalian mitochondria in the recently discovered long-patch base excision repair pathway.
The longstanding dogma that mitochondria possess a single mode of DNA repair appears to be changing. The mitochondrial base excision repair (BER) pathway processes mtDNA that has been damaged by the reactive oxygen species produced during oxidative phosphorylation. In BER, a damaged DNA base is removed by a specific glycosylase, followed by AP endonuclease cleaving the DNA strand 5′ to the abasic site to generate a one nucleotide gap that also contains a 5′deoxyribose phosphate (dRP) group. Single nucleotide BER (SN-BER) in mitochondria involves DNA polymerase γ (Polγ) filling this gap and cleaving the 5′dRP moiety prior to ligation (Longley et al., 1998) (Figure 1A). Unlike nuclear BER, which can be accomplished through either the SN-BER pathway or a long-patch BER (LP-BER) pathway, mitochondria have long been considered to utilize exclusively the SN-BER pathway. This view has changed within the last year following several reports describing LP-BER activity in mitochondrial extracts and the identification of proteins required for LP-BER in mitochondria (Akbari et al., 2008; Liu et al., 2008; Szczesny et al., 2008).
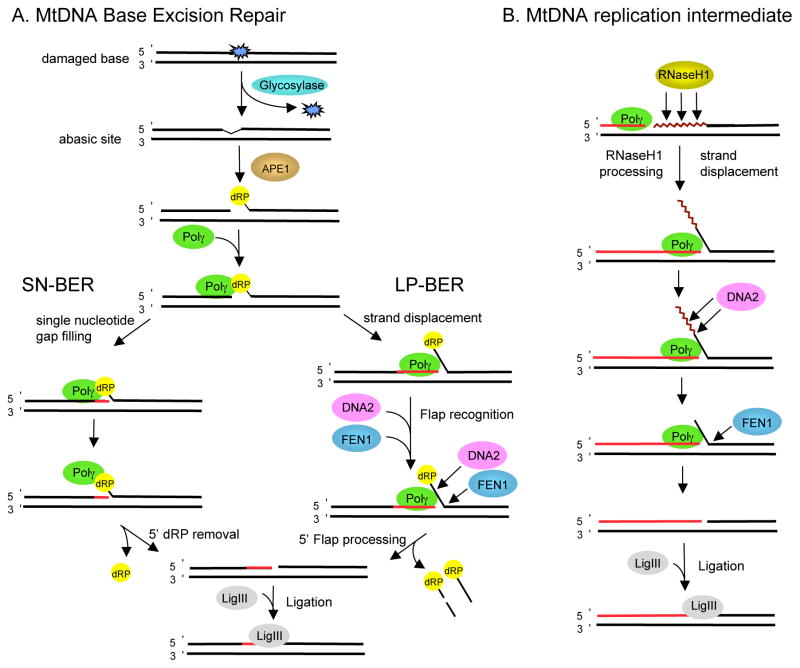
Figure 1. Proposed roles for DNA2 in mtDNA transactions.
A. Base excision repair pathways in mammalian mitochondria. An oxidized or damaged base is excised by a specific glycosylase, leaving an abasic site after which APE1 generates a strand break 5′ to the lesion and a 5′deoxyribose phosphate group on the downstream DNA. Repair can proceed via the SN-BER pathway (left) or the LP-BER pathway (right). In SN-BER, the single nucleotide gap is filled by Polγ and the 5′dRP moiety is removed by Polγ’s dRP lyase activity to make a ligatable substrate. In LP-BER, DNA synthesis by Polγ displaces the downstream strand to produce a 5′flap structure, which can be processed by DNA2 and FEN1. B. Possible involvement of DNA2 during mtDNA replication. Polγ is expected to encounter leftover RNA primers when terminating replication of circular mtDNA (continuous synthesis) or during Okazaki fragment maturation (discontinuous synthesis). If this RNA is not removed by RNaseH1, strand displacement DNA synthesis by Polγ generates an RNA-containing flap structure that is removed by DNA2 and processed further by FEN1 prior to ligation.
Utilizing mitochondrial extracts from Hela and HaCaT cells that were painstakingly prepared to be free from contaminating nuclear and cytosolic proteins, Akbari et al. (2008) observed both the generation and removal of 5′ protruding “flaps” that are hallmarks of LP-BER, although they did not identify specific factors that process these DNA intermediates. Liu et al. (2008) and Szczesny et al. (2008) exploited the fact that oxidation of dRP blocks the action of dRP lyase, essentially stopping progress through SN-BER and forcing repair of these intermediates through strand displacement DNA synthesis by Polγ in the more complex LP-BER pathway (Figure 1A). Utilizing such substrates and mitochondrial lysates carefully prepared from human lymphoblast cells, Liu et. al (2008) observed robust LP-BER activity in vitro and convincingly demonstrated participation of the flap endonuclease FEN1, an enzyme previously thought to function exclusively in the nucleus. Similarly, FEN1 in mitochondrial extracts from mouse tissues and human cell culture was shown to contribute in LP-BER, and our observation that immunoprecipitated complexes lacking FEN1 were nonetheless competent to perform LP-BER strongly suggested involvement of an unidentified 5′exonuclease/endonuclease (Szczesny et al., 2008). Residual LP-BER activity in extracts following immunodepletion of FEN1 also suggested the existence of an additional flap removing enzyme (Liu et al., 2008; Szczesny et al., 2008).
In a recent issue of Molecular Cell, Zheng et al. (2008) identify DNA2 as the missing flap endonuclease in human mitochondrial LP-BER. DNA2 was originally identified in yeast as a nuclear DNA helicase with an endonuclease activity well suited for removing part of an RNA or DNA flap structure, and yeast DNA2 has been known for some time to function in the nucleus along with FEN1 to process 5′ flaps (Budd and Campbell, 1997). However, Zheng et al. (2008) surprisingly found no evidence of human DNA2 in the nucleus; instead they found hDNA2 to be exclusively localized to the mitochondria. Recognizing the complexities of intracellular protein trafficking and the limitations of bioinformatics to predict mitochondrial targeting sequences, these investigators resolved this apparent contradiction through immunofluorescence studies and an elegant deletion analysis to identify an internal targeting peptide that is both necessary and sufficient to direct hDNA2 to mitochondria. Together with an immunoblot confirmation of known markers for various sub-cellular compartments, this study provides compelling evidence that hDNA2 is localized to mitochondria and is not a cytosolic or nuclear contaminant (Zheng et al., 2008). Human DNA2 significantly stimulated primer extension by Polγ, and co-precipitation experiments indicated a direct physical interaction between these two enzymes. Zheng et al. (2008) also demonstrated that hDNA2 and FEN1 function synergistically to define the end of a repair patch by efficiently cleaving flaps in advance of DNA ligase III (Figure 1A). Importantly, siRNA knockdown of hDNA2 during oxidative stress caused a significant accumulation of oxidized lesions specifically in mtDNA, suggesting selective repair of mtDNA by hDNA2 in vivo.
Why does DNA2 have a nuclear function in yeast and a mitochondrial function in mammalian cells? Does DNA2 actually function in both compartments in yeast? One hint that DNA2 might indeed function in yeast mitochondria came from a recent study in which dna2Δ in a pif1-m2 background generated a high petite frequency, whereas the pif1-m2 single mutant did not display this mitochondrial phenotype (Budd et al., 2006). Interaction between these loci is likely complex because Pif1 is a 5′-3′ DNA helicase functioning in multiple pathways in both the mitochondria and nucleus. Nonetheless, the current work implies that mammals have evolved to utilize FEN1 as the only nuclear flap endonuclease, whereas both FEN1 and DNA2 appear to function together in mitochondria.
The identification of hDNA2 as a mitochondrial LP-BER enzyme also contributes to the discussion of the mechanisms underlying mammalian mtDNA replication. Current models are incomplete, and competing mechanisms include an asynchronous, strand displacement model and a strand coupled, bidirectional replication model (Bowmaker et al., 2003; Brown et al., 2005). The helicase activity of hDNA2 and its ability to remove RNA primers from flap structures resembling replication intermediates are directly relevant to both models, and the observation that hDNA2 binds Polγ and stimulates strand-displacement DNA synthesis is a crucial piece to this puzzle (Figure 1B).
Many questions about the biological consequences of LP-BER in mitochondria persist, including questions regarding oxidation of 5′dRP moieties in vivo, FEN1 compartmentalization and trafficking, and factors regulating the switch between SN-BER and LP-BER. Currently, strand displacement DNA synthesis during SN-BER appears to be the deciding factor that commits a repair event to the LP-BER pathway (Figure 1A). Given the weak dRP lyase activity of Polγ and the strong strand displacement activity of Polγ in vitro, LP-BER might be the predominant mode for repairing damaged bases in vivo. As mutations in genes whose exclusive function is to maintain mtDNA integrity are commonly associated with heritable neuromuscular disorders (Copeland, 2008), the participation of hDNA2 in mitochondrial LP-BER clearly makes DNA2 a potential locus for mitochondrial disease. Subsequent research promises to be fruitful in identifying all the players in mitochondrial SN-BER and LP-BER as well as the role of hDNA2 in mtDNA replication.
References
- Akbari M, Visnes T, Krokan HE, Otterlei M. DNA Repair (Amst) 2008;7:605–616. doi: 10.1016/j.dnarep.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Bowmaker M, Yang MY, Yasukawa T, Reyes A, Jacobs HT, Huberman JA, Holt IJ. J Biol Chem. 2003;278:50961–50969. doi: 10.1074/jbc.M308028200. [DOI] [PubMed] [Google Scholar]
- Brown TA, Cecconi C, Tkachuk AN, Bustamante C, Clayton DA. Genes & Development. 2005;19:2466–2476. doi: 10.1101/gad.1352105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd ME, Campbell JL. Mol Cell Biol. 1997;17:2136–2142. doi: 10.1128/mcb.17.4.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd ME, Reis CC, Smith S, Myung K, Campbell JL. Mol Cell Biol. 2006;26:2490–2500. doi: 10.1128/MCB.26.7.2490-2500.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland WC. Annu Rev Med. 2008;59:131–146. doi: 10.1146/annurev.med.59.053006.104646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Qian L, Sung JS, de Souza-Pinto NC, Zheng L, Bogenhagen DF, Bohr VA, Wilson DM, 3rd, Shen B, Demple B. Mol Cell Biol. 2008;28:4975–4987. doi: 10.1128/MCB.00457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longley MJ, Prasad R, Srivastava DK, Wilson SH, Copeland WC. Proc Natl Acad Sci U S A. 1998;95:12244–12248. doi: 10.1073/pnas.95.21.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesny B, Tann AW, Longley MJ, Copeland WC, Mitra S. J Biol Chem. 2008;283:26349–26356. doi: 10.1074/jbc.M803491200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Zhou M, Guo Z, Lu H, Qian L, Dai H, Qiu J, Yakubovskaya E, Bogenhagen DF, Demple B, Shen B. Mol Cell. 2008:31. doi: 10.1016/j.molcel.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]