Abstract
Synthetic polymers are ubiquitous in the development of drug and polynucleotide delivery vehicles, offering promise for personalized medicine. However, the polymer structure plays a central yet elusive role in dictating the efficacy, safety, mechanisms, and kinetics of therapeutic transport in a spatial and temporal manner. Here, we decipher the intracellular evolutionary pathways pertaining to shape, size, location, and mechanism of four structurally-divergent polymer vehicles (Tr455, Tr477, jetPEI™ and Glycofect™) that create colloidal nanoparticles (polyplexes) when complexed with fluorescently-labeled plasmid DNA (pDNA). Multiple high resolution tomographic images of whole HeLa (human cervical adenocarcinoma) cells were captured via confocal microscopy at 4, 8, 12 and 24 hours. The images were reconstructed to visualize and quantify trends in situ in a four-dimensional spatio-temporal manner. The data revealed heretofore-unseen images of polyplexes in situ and structure-function relationships, i.e., Glycofect™ polyplexes are trafficked as the smallest polyplex complexes and Tr455 polyplexes have expedited translocation to the perinuclear region. Also, all of the polyplex types appeared to be preferentially internalized and trafficked via early endosomes affiliated with caveolae, a Rab-5-dependent pathway, actin, and microtubules.
Keywords: Intracellular, trafficking, nucleic acid, polyplex, carbohydrate, 3D imaging, confocal
Targeted polymeric nanomedicines have shown success in the clinic to treat solid tumors in patients with polymer vehicles loaded with either chemotherapeutic drugs (i.e. docetaxel) or nucleic acids such as small interfering RNA (siRNA).1-3 These discoveries herald promise for personalized medicine with lower toxic side effects. To this end, the greatest challenge in this field remains the creative design and detailed understanding of vehicles that promote efficacious, stable, and reproducible delivery to targeted biological and intracellular sites. Concurrent with the advancements in the field of nanoparticles in drug and nucleic acid delivery, there has been an increase in efforts to better understand their trafficking both in vivo and in vitro to facilitate development of efficacious delivery vehicles.4-10 Indeed, understanding vehicle interaction with cell surfaces, mechanisms of internalization, and trafficking kinetics in a spatial and temporal manner are key to refining the structure, function, and efficacy of the materials used for delivery.
Our group and others have extensively studied the factors affecting the endocytosis of nanocomplexes based on polymers and polynucleotides (polyplexes) using a combination of inhibitor studies and 2D microscopy techniques. On the cellular level, nanomedicine transport involves trafficking across the plasma membrane and entry into cells,6 where intracellular barriers are encountered.4 For example, McLendon et al. discovered that endocytosis of glycopolymer-based nucleic acid vehicles exploit glycosaminoglycans on the cell surface to trigger internalization. Glycopolyplexes anchor to the cell surface, which appears to be a function of both charge and chemical interaction between the polymeric vehicles and cell surface glycosaminoglycans such as hyaluronate, heparan sulfate, etc.7 Thereafter glycopolyplexes appear to enter the cell via both clathrin and caveolae-mediated pathways but the caveolae pathway promotes active cellular trafficking, nuclear delivery, and transgene expression.6 Valuable insight into nanocomplex colocalization with cellular components such as endocytic vesicles, the Golgi, endoplasmic reticulum, mitochondria, nucleus, actin, and microtubules has also been gained through extensive 2D widefield and confocal microscopy studies.8, 10 Fichter et al. have recently discovered that the structure of the polymers comprising these delivery vehicles play an important role in their intracellular trafficking mechanisms. While both clathrin and caveolae mechanisms appear to contribute to cell internalization, the caveolae-mediated pathway appears to actively route glycopolyplexes though inter-organelle trafficking mechanisms en route to the nucleus, in a similar manner to viruses.8 Interestingly, in addition to the Golgi, these vehicles appear to be trafficked to the endoplasmic reticulum (ER) possibly via retrograde transport from the Golgi to ER via COP I (coat protein complex I) vesicles.8 In a related study, the importance of the structure of polymer was further demonstrated in a study by Reilly et al., which reported that PEI modified with histone H3 (a nuclear localization signal peptide) promoted endocytosis via caveolae and then trafficking to Golgi. From the Golgi these vehicles also appear to traffic to ER via the retrograde transport between these two organelles.15 Furthermore, in that study it was shown that polymeric vehicles comprised of only polyethyleneimine (PEI) appear to traffic via a different endocytic route.8, 16 It has also been shown in several studies that endosomes can also fuse with acidic lysosomes where the cargo could be degraded.17-19 The mechanism of polyplex entry into the nucleus has also been examined by Grandinetti et al.9 That study found that polycations such as PEI may cause permeabilization of the nuclear membrane, allowing physical penetration and nanomedicine entry into the nucleus, however, this effect is certainly reliant on the polymer-based vehicle chemical structure and molecular weight.9
Indeed, obtaining these data most often relies upon extrapolating three-dimensional trends from two-dimensional data, which can present difficulties in data interpretation and challenges in discerning structure-function trends. Traditional 2D cellular imaging methods and other techniques such as super resolution light microscopy could benefit from 3D image reconstruction and techniques to temporally-analyze large data sets in multiple cells. Studies utilizing three-dimensional imaging techniques have been very limited, yet when performed, provides significant information not available with 2D images. For example, a study by Unterstab et al. reported colocalization of polyomavirus BK agnoprotein with lipid droplets in Vero cells and studies by Duleh et al. examined the asymmetric distribution of WASH proteins (a nucleation promoting factor required for the proper cellular assembly of actin) with endosomes in COS7 cells.11, 12 Imaging cytometry has also been used to examine cellular internalization kinetics of nanoparticles (such as CdTe/ZnS core/shell quantum dots) and their intracellular distribution with human osteosarcoma cell populations, however, the resolution limits of this technique restricted spatio-data analysis in 3D.13 Tracking distributions of an adeno-associated viral vector in cells has also been reported in 3D to understand kinetics only.14
Studying nanoparticle vehicles at high resolution in cells is tremendously important to furthering the fundamental understanding in this field of nanomedicine trafficking. The capacity to resolve and reconstruct microscopy images in 3 dimensions as a function of time and polymer vehicle type significantly enhances the capacity to detect transport, volume, and other trends of nanomedicine trafficking. In this study, we focus on understanding cellular entry via endocytosis and intracellular trafficking of polyplexes en route towards the nucleus of a cell via four-dimensional data analysis. We have quantitatively analyzed and compared several parameters of polyplexes as a function of time and polymer vehicle structure. This has allowed us to distinguish intracellular trends for pDNA trafficking in four dimensions, facilitating a detailed understanding of nanocomplex size, shape, colocalization, and biological kinetics in situ as a function of polymer structure. Four different polymer types were complexed with FITC-labeled plasmid DNA (pDNA) and images of the colloidal complexes (polyplexes) in HeLa cells were obtained over time.
Specifically, the aim of the study was to obtain quantitative information on the following parameters as a function of time and polymer vehicle type: (i) polyplex volumes, (ii) distance between polyplexes within the cell, (iii) distances between each polyplex and the nucleus, (iv) colocalization with early and late endosomes, and (v) colocalization with the nucleus, and vi) intracellular mechanism of delivery (Figure 1, Movie S1). In addition, the unusual morphologies and variety of sizes characteristic of these nanocomplexes are observed for the first time in situ in the cellular environment. This allowed us to compare these data with endocytosis inhibition studies to investigate in detail the cellular internalization and trafficking pathways for polymeric nucleic acid delivery vehicles. To the best of our knowledge, this is the first report to utilize 4D image analysis to quantitatively examine cellular trafficking of polyplex-based nanomedicines.
Figure 1.
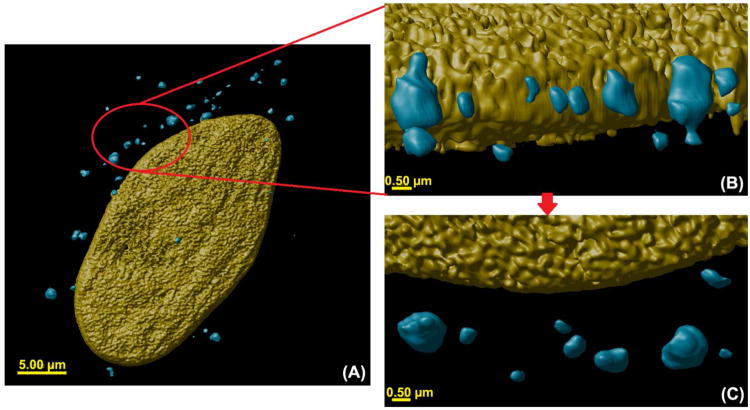
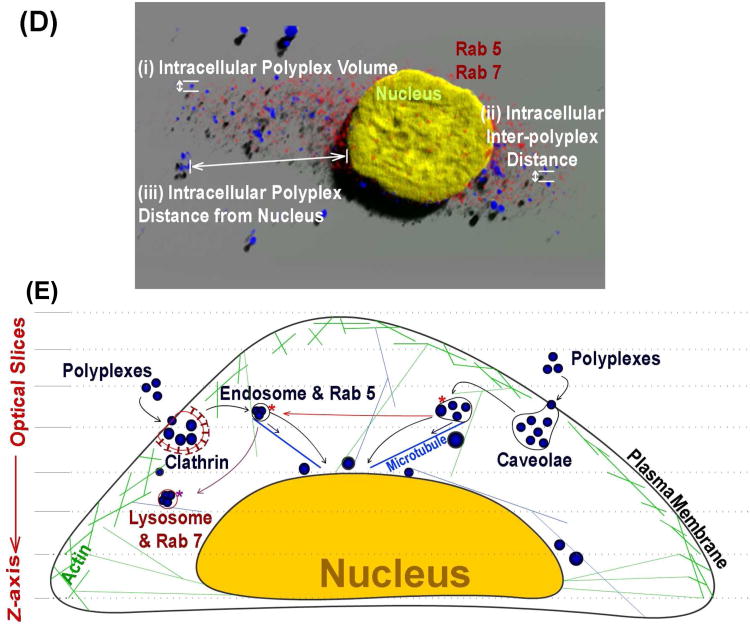
Three-dimensional (3D) volume rendering of polyplexes comprised of Tr455 polymer complexed with FITC-pDNA fluorescence (pseudo colored in blue) and a HeLa cell nucleus labeled with DAPI (pseudo colored in yellow) at 24 hours post transfection. (A) Zoomed out view of a single cell showing the nucleus (pseudo colored yellow) and polyplexes (pseudo colored blue). (B) A close-up view of the indicated part of the cell from ‘A’. Various globular morphologies of the polyplexes can be observed near the nucleus. (C) A ‘top view’ of the polyplexes shown in ‘C’. (D) Illustration of the polyplex confocal imaging parameters studied herein with the 4D imaging techniques. (E) The intracellular mechanisms studied herein with the polyplexes: (i) intracellular polyplex volume; (ii) intracellular inter-polyplex distance and (iii) intracellular polyplex distance from the nucleus. Also, analysis was performed to study 3D colocalization of polyplexes with intracellular organelles such as (i) endosomes (Rab 5), (ii) lysosomes (Rab 7) and the nucleus. In addition, endocytosis inhibition studies were performed to understand and compare polyplex internalization and trafficking.
Materials and Methods
Materials
The four polymers used in the study were jetPEI™ (Polyplus-transfection Inc., New York, NY), Glycofect™ (Techulon Inc., Blacksburg, VA), Tr455 and Tr477 (both synthesized in the Reineke laboratory as previously described).20, 21 It should be noted that Glycofect has been denoted as polymer “G4” in our previous publications.22, 24, 25 The nucleic acid used for the study was fluorescein-labeled, 2.7 kb, double stranded, circular plasmid DNA (FITC-pDNA) (Mirus Bio LLC, Madison, WI). The primary antibodies for Rab 5 (specificity: detects Rab 5A, Rab 5B and Rab 5C) and Rab 7 (specificity: detects Rab 7a) were purchased from Santa Cruz Biotechnology, Inc., Santa Cruz, CA. The Alexa Fluor 555 goat anti-rabbit IgG secondary fluorescent antibody was purchased from Life Technologies, Carlsbad, CA. The clathrin, rabbit polyclonal antibody against clathrin heavy chain and caveolin, rabbit polyclonal antibody to caveolin-1 was purchased from Abcam (Cambridge, MA, USA). HeLa (human cervical adenocarcinoma) cells were obtained from American Type Culture Collection (ATCC, Manassas, VA). All the cell culture products were purchased from Life Technologies, Carlsbad, CA unless otherwise stated. Cell culture flasks and plates were purchased from Corning Inc., Lowell MA. The glass coverslips were purchased from Fisher Scientific, PA.
Cell Culture
HeLa cells were grown in 75 cm2 flasks in Advanced Dulbecco's Modified Eagle's Medium (DMEM) containing GlutaMAX™. This media was supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS) and 1% (v/v) antibiotic-antimycotic. For confocal, HeLa cells were cultured for 24 hours prior to transfection. The glass coverslips (15 CIR -2) used for the study were acid-washed prior to cell plating. The cells were cultured on these glass coverslips in a 12-well cell culture. The cell density at plating was 15,000 cells/well; 1 mL of cell suspension was added to each well.
Polyplex Preparation and Transfection
The polyplexes were formed based on N/P (nitrogen/phosphate) ratio, where ‘N’ corresponds to the number of secondary amines on polymer chain repeat unit and ‘P’ corresponds to the number of phosphate groups on the nucleic acid.34 The ratios used were 5 N/P for jetPEI™, 20 N/P for Glycofect™, and 7 N/P for Tr455 and Tr477. The N/P ratio of 20 was chosen for Glycofect™ polymer based on earlier studies by our group, which have indicated this N/P ratio promotes high delivery efficiency without toxicity.22, 24 The N/P ratio of 7 utilized for the trehalose polymers (for Tr453 and Tr475) were chosen based on the previous study by Srinivasachari et al.20, 21 Stock solutions of the polymers and FITC-pDNA were made in DNase/RNase-free H2O. The polymer solution (50 μL/well) was added to pDNA solution (50 μL/well at 0.02 μg/μL) and incubated at room temperature for 1 hour to form the polyplexes.
For observation of the cells after transfection via confocal microscopy, 1 mL/well of OptiMEM reduced-serum medium was added, followed by 100 μL/well of polyplex solution (containing 50 μL of pDNA at 0.02 μg/μL). The cells were then incubated at 37 °C and 5% CO2 for 4 hours. After 4 hours, the cells were washed with phosphate buffered saline (PBS, pH 7.4) to remove any unattached polyplexes or pDNA. In the case of the specimens observed and analyzed at the 4 hour time point, the cells were fixed immediately after this wash. In the case of the specimens observed and analyzed at the later time points at 8, 12, and 24 hours, each well was washed with 1 mL of PBS after 4 hours of incubation with the polyplex solutions. After this wash, 1 mL of DMEM containing 10% serum was added to each well and the cells were further incubated until the indicated time points.
Confocal Microscopy
The microscopy slides were prepared according to the methods outlined in the Supplementary Materials and Methods. The confocal microscopy was performed on fixed cells using the LSM 510 Meta, Carl Zeiss MicroImaging, Inc., Thornwood. The images were acquired using Zen software. The imaging was performed using a Plan-Apochromat 63×/1.4 Oil DIC objective. The lasers used for excitation were UV (364 nm) to excite DAPI (4′,6-diamidino-2-phenylindole) (excitation/emission maxima: 358/461 nm), 543 nm laser to excite Alexa Fluor 555 (excitation/emission maxima: 555/565 nm), argon (488 nm) to excite FITC (fluorescein isothiocyanate) (excitation/emission maxima: 495/518 nm) The z-stacks were sampled in the z-direction based on Nyquist Frequency, so as to ensure better deconvolution. The laser intensity, master gain and digital gain were held constant for all the channels, except for the DIC, which was varied for a few images. All the confocal microscope settings are listed in Table S9. The imaging was performed at room temperature. The controls were cells only, FITC only, DAPI only, Rab 5 primary antibody only, Rab 7 primary antibody only and secondary antibody only.
Stereology
Applying rule of stereology is essential to minimize bias in quantitative measurements of parameters.26 In the current study, the population of interest was the population of the polyplexes inside a cell. Therefore, the rule of stereology was considered to be applicable to the population of polyplexes, rather than the population of cells. The following steps were performed to minimize bias towards counting the number of polyplexes within a cell as follows: (1) X-Y plane of image: All of the polyplexes in a cell were kept in the field of view of 60x objective. (2) Z-slices: To satisfy the ‘rule of stereology’, a minimum number of slices along the z-axis is required to get a realistic estimate of the population studied. In the current study, up to approximately 40 slices/cell were taken along the z-axis of complete cell height such that each slice overlapped with its neighboring slice. (3) Finally, a total of 6 cells were imaged for each polymer for all experiments performed in this study (with the exception of three cells for polyplex colocalization with Rab 5 and Rab 7). (4) The cells were chosen for imaging at random from the coverslip. The imaging was limited to a total of 6 cells because it gave a substantial number of over 100 polyplexes to represent the intracellular polyplex-population for each polymer type. The total image data thus generated was ∼25 Gigabytes, which was computationally intensive and a limiting factor for analysis.
Confocal Image Analysis
The deconvolution of 3D image z-stacks was performed using a theoretical point spread function and the Classical Maximum Likelihood Estimation (CMLE) algorithm that is built into the software ‘Huygens Essential’, Scientific Volume Imaging SVI, Hilversum, The Netherlands. The theoretical point spread function (PSF) used for this deconvolution was generated by the software based on the confocal microscope parameters. The ‘Object Analyzer’ plugin was used to analyze the z-stacks. The thresholding values were as follows: for polyplexes, the threshold and seed was 24% and 11%; for nucleus, the threshold and seed was 4 - 7% and 52 - 87%, respectively; for Rab 7, the threshold and seed was 6% and 4%, respectively; and for Rab 5, the threshold and seed was 13% and 4%, respectively. The threshold for each of the channels was kept constant so as to be able to compare the results from different images. The threshold levels were chosen such that they best represented the image details. The area of interest for all the analysis, with the exception of the colocalization data with the nucleus, consisted of the region outside the nucleus and inside the cell. It should be noted that since the plasma membrane of the cell was not labeled, the DIC (differential interference contrast) image was used to locate the boundaries of each cell. The region of interest was set to the space inside the cell boundary. The polyplexes inside the cell boundary were considered to be inside the region of interest and hence inside the cell. Care was taken during slide preparation, to extensively wash off excessive polyplexes that were not attached to the cell surface. The polyplexes outside the region of interest were manually deleted by visualizing and comparing the polyplexes in each DIC image at different z-depths with the surface rendered 3D image using the Object Analyzer plugin in Huygens software. After deconvolution, the z-stacks were rendered to generate the 3D volume using the Object Analyzer plugin in Huygens Essential. The data from parameters measured such as volume, distance and colocalization was imported in Microsoft Excel for plotting and statistical analysis. Details of the statistical analysis methods are available in the Supplementary Materials and Methods.
It is important to note that the images captured from the confocal laser scanning microscope represent the diffracted light from polyplex/pDNA complexes. The resolution of these images is limited by the theoretical diffraction limit of light (x, y: approximately 148 nm and z: approximately 370 nm).27 The effect of this diffraction limit is more prominent in z-axis direction of a 3D reconstructed image. Therefore, a spherical object may appear elongated. This elongated appearance is due to the elliptical shape (football-like) of the point spread function (PSF) in z-axis direction. In this study, a mathematical deconvolution approach (CMLE) was used to correct the image using a theoretical PSF. The theoretical PSF was generated mathematically based on the type of microscope and the optics used. This deconvolution process can improve resolution of an image. Unfortunately, the images cannot be resolved beyond the diffraction limit of light. Therefore particles sizes below this limit were undetectable. The z-axis resolution may manifest itself towards part of the elongated appearance of 3D renderings of polyplex/pDNA complexes in z-direction. However, at any given location inside a cell, there is likely more than one polyplex and/or pDNA associated together in a group. Hence, they are referred to as polyplex/pDNA complexes in this manuscript. Also, because the polyplexes are likely together, it is possible they do so to form a complex of not only spherical shape but also a variety of other shapes (Figure 1).
In addition, the software Fiji (ImageJ, National Institutes of Health) was used to calculate the Manders coefficient of 2D confocal images showing colocalization with clathrin and caveolae.
Flow Cytometry
The experiment was performed as previously reported.6 Briefly, HeLa cells were plated in 6-well plates at a density of 150,000 cells/well at 24 hours prior to transfection. Polyplexes were formulated at 1 hour prior to transfection. The cells were pre-treated with inhibitors in OptiMEM a large variety of inhibitors to examine the uptake and trafficking routes of these polyplexes: filipin (1 μg/mL for 1 hour) which inhibits caveolae mediated endocytosis, chlorpromazine (5 μg/mL for 30 minutes) which inhibits clathrin mediated endocytosis, dimethylamiloride (100 μM for 5 minutes) which inhibits macropinocytosis, cytochalasin D (2 μg/mL for 15 minutes)6 which inhibits actin polymerization and nocodazole (0.006 μg/μL for 30 minutes) which inhibits microtubule polymerization.
Dynamic Light Scattering
Polyplexes were formulated as described before with unlabeled pDNA. The polyplex solution was transferred to a dust free glass tube for measurements. The data were recorded every 15 minutes for a total time of 4 hours on a Brookhaven Instruments (Holtsville, NY) dynamic light scattering instrument (DLS). The red laser source was Mini L-30 Laser (Brookhaven Instruments) at wavelength (λ) of 637 nm. The transient scattering intensity was detected by BI-APD avalanche photo detector. The signal was then processed by a BI-90000AT digital correlator. The optics were aligned on a BI-200SM goniometer. The refractive index (n) of water and OptiMEM that was used in the analysis was 1.33. The viscosity (η) of water and OptiMEM was 0.89 and 0.935 cP, respectively. The OptiMEM was filtered with a 0.20 μm syringe filter prior to diluting the polyplexes formed in DNase RNase water. Each time point represents the average for the scans for duration of 10 minutes (first delay: 1.0 μm; last delay: 100 μm; scan time: 10 minutes; angle (θ) 90°). The temperature was maintained at 37 °C. The autocorrelation function C(τ) was computed according to the equations in the Supplementary Materials and Methods in the Supporting Information.
Statistical Analysis
The distribution analysis and Power Law fit was performed in MS Excel software. A total of six cells (three cells stained for Rab 5 and three cells stained for Rab 7) were analyzed for polyplex distributions for each polymer for each of timepoint. The analysis of the distribution of polyplex volume, distance of polyplex from surface of the nucleus and inter-polyplex distance was performed. The raw data was first binned for the number of polyplexes in all six cells with the following increments: 0.01 μm3 for volume, 1 μm for distance from nucleus, and 0.5 μm for interpolyplex distance.
Two summed totals for the polyplexes in six cells were calculated as follows: (1) the total number of polyplexes in each individual bin range (or “total number of events per bin range”) and (2) the total number of all polyplexes from in allbins (or “total number of events”) (Table S3). Then to calculate probability for each bin range, the number of polyplexes in each bin range (1) was divided by the total number of polyplexes (2). Thus, the ‘events’ divided by ‘total number of events’ resulted in the probability value. This probability was the chance that the given number of polyplexes would exist at the given volume or distance range at the given time point. The probability value, thus obtained was plotted as the y-axis of the scatter plot, where the x-axis was the bin or range, (such as volume or distance) (Table S4).
For the volume distribution of the polyplexes, the Power Law model was fitted to the data to obtain the equation and correlation coefficient (R2-value). It should be noted that the fits were performed up to the bin range, which did not contain a zero number. The resulting equation was used to calculate the extrapolated probability values. These values were plotted with the probability on the y-axis and time on the x-axis for varying bin range.
For the distribution of polyplex distance from the surface of the nucleus data, the Power Law model was fitted as described before. For the data of the distribution of inter-polyplex distance (i.e. the distance of each polyplex from its nearest neighboring polyplex) the Power Law fit was performed on bin ranges from ‘>1 to 1.5’ μm and above; these ranges were analyzed because the three bin ranges below this did not appear to follow a specific trend.
Along with the correlation coefficient R2-value, the Kolmogorov-Smirnov test was used to test the goodness of fit. The normality of the data was analyzed by the Shapiro-Wilk W Test. Further, to calculate the significant differences, raw values were analyzed using JMP software (SAS Institute Inc., Cary, NC). First, an analysis of variance (ANOVA) test was performed. This was followed by post-hoc analysis using a Tukey's HSD (Honestly Significant Difference) test. Also, a student t-test was used to calculate statistically significant differences.
Results & Discussion
Intracellular Polyplex Volume Distribution
As shown in Figure 2, the intracellular polyplex size data revealed that a range of polyplex sizes from ∼267 to ∼724 nm in diameter are found within the cells.
Figure 2.
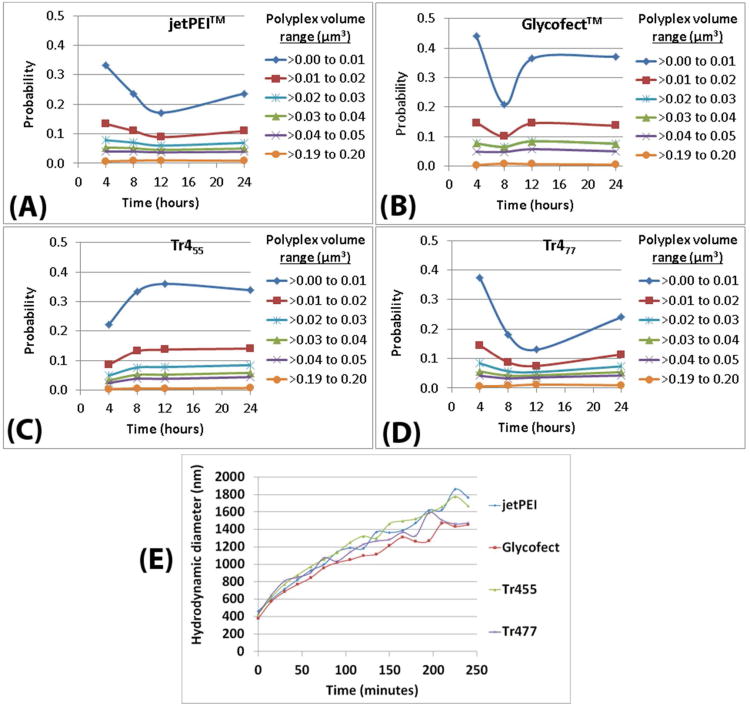
Polyplex volume data in HeLa cells as a function of time, polymer type, and condition (A-D). The size ranges of polyplexes in 3D in a cell was determined as a function of polyplex volume and time. The compiled data has been displayed as the extrapolated probability values for the number of polyplexes to exist in the cell within the indicated volumes at ∼0.01, ∼0.02, ∼0.03, ∼0.04, ∼0.05, and ∼0.20 μm3, as shown in Table S7A. The probability was calculated by dividing the total number of polyplexes in each volume-range (for each polymer; at each time point; within six separate cells) by the total number of polyplexes (for each polymer; at each time point; within six separate cells). (E) Hydrodynamic diameter (y-axis on left) at different time points for all four polymers in reduced serum media OptiMEM at 37 °C with pH ∼7.3.
The maximum amount of polyplexes (max probability) were observed in the lowest volume range (0.01 μm3; ∼267 nm diameter), which is consistent with the expected sizes of polyplexes that would be taken up by the cell.20, 28 The larger polyplex volume ranges beyond 0.02 μm3 (>336 nm diameter) had much lower probability values. This observation suggested that smaller polyplexes (up to ∼336 nm diameter) were more prevalent in the intracellular environment as compared to larger polyplexes (> ∼336 nm diameter). It should be noted that polyplexes below ∼267 nm in diameter, if present, could not be resolved because this small size is below the diffraction limit of light (and could not be analyzed). In support of observing this larger polyplex size in cells, Godbey et al. have reported that in the initial stages of endocytosis, groups of polyplexes can be endocytosed in “clumps”.29 Therefore, after such a group of polyplexes entered a cell, they would exist in a range of volume subgroups as we have observed (Figures 2 and S2).
Interestingly, as shown in Figure 2E, the average polyplex size data in cells were in agreement with the dynamic light scattering (DLS) analysis of polyplexes in the cell culture media only (Figure 2E). When aqueous polyplex solutions were immediately exposed to the culture media (OptiMEM), the polyplexes were found to measure about 400 nm in diameter, and with time, grew steadily in size to 800 nm, likely due to aggregation (calculated spherical volume for 400 nm diameter is ∼0.03 and for 800nm is ∼3.0 μm3). It should be noted that the polyplex sizes observed in OptiMEM represent the actual sizes of the polyplexes that the cell first encounters during the cellular uptake and transfection process, which likely plays a large role in the cellular uptake pathway (vide infra). These data suggested that polyplexes first encounter cells with a variance in size ranges depending on the time course they come into contact with the cell membrane. Interestingly, large polyplexes (or groups of polyplexes) of volume 3 to 5 μm3 (calculated spherical diameter of 0.89 to 1.06 μm) were also imaged within the cells, but such large polyplexes had an extremely low probability (∼0.001) to exist within the cells observed.
As shown in Figure S2, we also computed a percentage value, which directly analyzed the number of polyplexes in each volume range divided by the total number of polyplexes, and then averaged these percentages (for each polymer averaging the data in 6 cells at each timepoint). For the volume distribution, we observed that the percentage of polyplexes at the lower volume range ∼0.01 μm3 (∼267 nm) at 4 hours was significantly higher (p < 0.01) than polyplexes in the larger volume ranges (∼0.02 μm3, ∼0.03 μm3, ∼0.04 μm3 and ∼0.05 μm3) for jetPEI™, Glycofect™ and Tr455. Yet, a different trend was observed for Tr477 polyplexes. The percentage of polyplexes in the lowest volume range ∼0.01 μm3 (at 4 hours) was significantly (p < 0.01) higher than that observed in the volume ranges of ∼0.03 μm3 to ∼0.05 μm3, however it was not significantly different than the percentage found in the ∼0.02 μm3 (calculated spherical diameter: of 336 nm) volume range. In addition, at later time points, the Tr477 polyplexes showed no significant differences in sizes/volumes across the entire volume range (Figure S2). This indicated that the Tr477 polyplexes had a broader range of sizes in its intracellular population. The percentage of Glycofect™ polyplexes of volume ∼0.01 μm3 at 4 hours was significantly higher when compared to the other time points for this system and the other three polyplex variants. This indicates that a greater number of smaller Glycofect™ polyplexes (∼267 nm diameter) are being endocytosed during the first 4 hours of transfection. Because the molecular weight of Glycofect™ (formerly reported as G4, Mw = 4.6 kD30) is much lower than that of the other polymers tested (40.5 kDa for Tr455, 56.1 kD for Tr477, and about 25 kDa for PEI15, similar to jetPEI™), these data may be indicative of accelerated cellular entry with polymeric vehicles having smaller sizes (which could be indirectly linked to polymer molecular weight).
It should be noted that, while deconvolution was performed with the Huygens software, certain parameters such as region of interest (ROI) and thresholding for a channel, etc., had to be defined manually. This thresholding of the volume rendered image could affect the shape and size of the objects. Because there was no rule to set the value for thresholding, a criterion was set. This criterion was to first achieve the same shape, size and location of the objects in the image in x-y direction as the original image and then the thresholding in the z-direction was adjusted. Once these values were set, all the volume rendered images were then processed using the exact same value for all images (with the exception of a few of the images, where the thresholding had to be changed slightly and this variation could be due to change in the intensity of light recorded). It should also be noted that it has been previously reported that labeling a plasmid DNA with a fluorophore may affect intracellular trafficking.31 Also, different methods of labeling plasmid DNA may also alter results.31 In the current study, the same FITC-pDNA batch was used to eliminate variability induced due to different labeling methods. At the same time, although trafficking of naked-pDNA could be different than FITC-pDNA, the addition of a fluorophore was required for detection on confocal microscope.
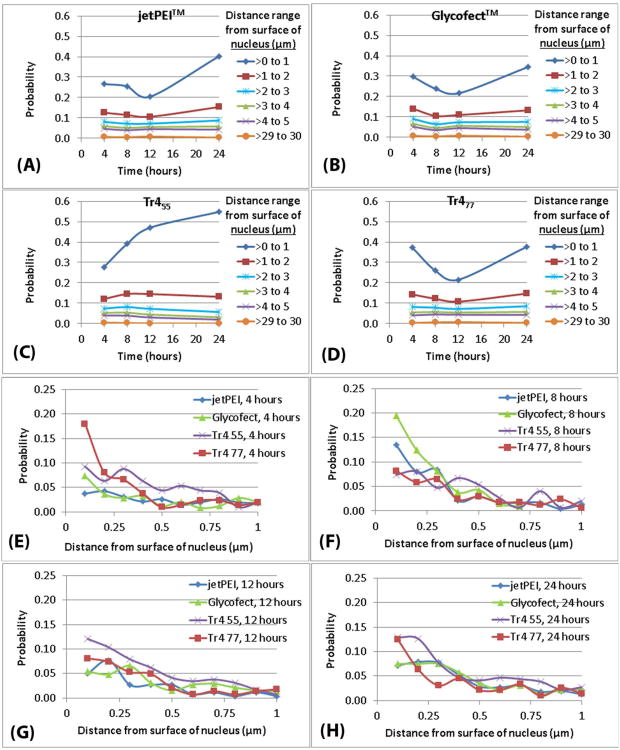
Polyplex distance from the nucleus and concentric-nuclear zones
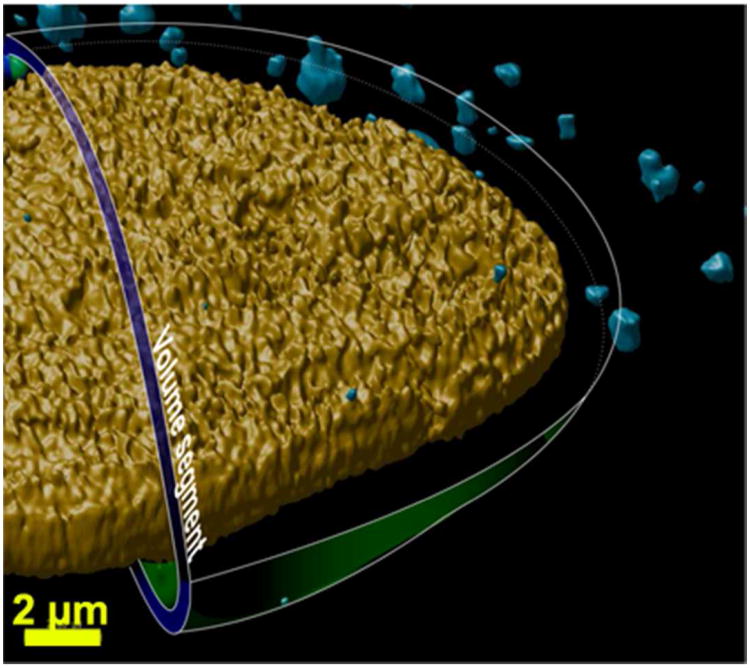
The distribution of polyplexes around the nucleus in the intracellular space in three dimensions was also examined in 3D space as a function of time (Figures 3, 4 and 5). The polyplex distance from the surface of the nucleus data was categorized into ‘intracellular 3D concentric distance-based nuclear zones’ inside a cell (Figure 4). The concentric distances of less than 5 μm were denoted as ‘perinuclear zones’. Interestingly, the probability of polyplexes to exist beyond 5 μm up to the farthest zone at a distance of ∼30 μm was rare (probability ∼0.008) when compared to the higher probability of polyplexes in the perinuclear zone (probability ≥ 0.01) at all timepoints. The low probability at large distances for all timepoints, implied that if a polyplex enters a cell at a longer distance, its presence in that zone is transient. Alternatively, this could imply that a polyplex would rarely enter a cell at these longer distances. Interestingly, the probability values increased as the distance from the nucleus decreased from 5 to 1 μm and below (Figure 5). This relationship further suggests that the polyplexes are rapidly trafficked toward the nucleus in membrane-bound endocytic vesicles (likely along microtubules and actin, vide infra) and have an increased tendency to reside in the closest perinuclear zones. Based on this observation, we propose a new theory that if a polyplex enters the cell at any point on the plasma membrane via an endocytic route (independent of the distance from the nucleus in 3D space), it is trafficked in this vesicle rapidly toward the inner perinuclear zone (∼ 5 μm and below) (Figure 5). Thereafter, when the polyplexes reach the inner zone, they may be trafficked at a slower speed (resulting in a higher probability to exist in the inner zones). As reported by Kulkarni et al., such trafficking towards the perinuclear region may involve a combination of both direction-oriented and random trafficking steps.32 This could be attributed to the physical state of polyplexes that could be either (i) a polyplex in the process of releasing nucleic acid, and/or (ii) the released nucleic acid in the process of entering the nucleus. In addition, this may be due to localization with organelles and other cellular components near the nucleus (such as the ER, as previously mentioned).8 Polyplex colocalization with the nucleus was also analyzed (Figure S5). Significant colocalization of all polyplex types within the nucleus was observed for all time points. However, significant trends or differences in these data were not observed when comparing the polyplex types at all timepoints.
Figure 3.
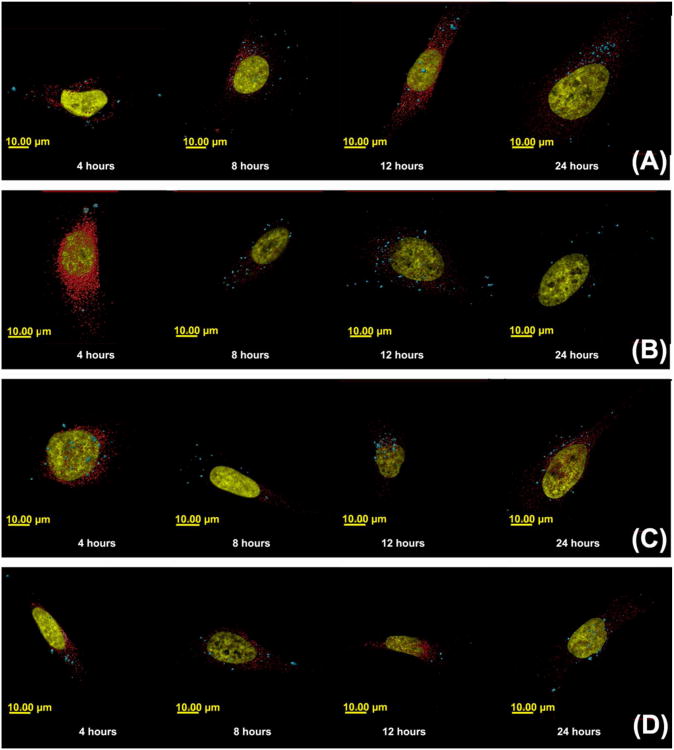
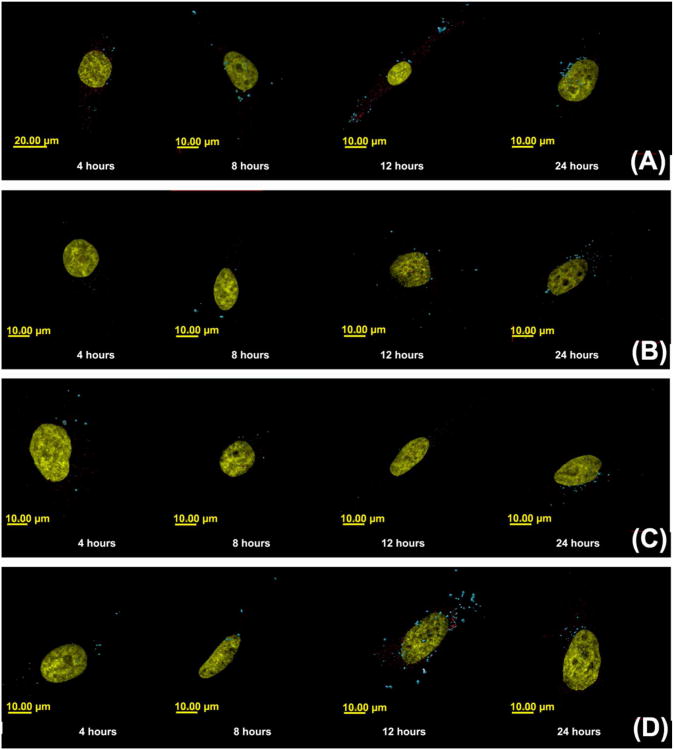
Three-dimensional reconstructed images of cells transfected with polyplexes formulated with Cy5-pDNA and (A) jetPEI™, (B) Glycofect™, (C) Tr455 and (D) Tr477. With each polymer type, the cells were fixed after 4, 8, 12, and 24 hours. The nucleus is shown in gold and the polyplexes (detected by imaging Cy5-labeled plasmid DNA) are shown in blue. Note: In the time series A-D, the nucleus was set as transparent to view colocalizing polyplexes. The polyplexes colocalizing with the nucleus appear ‘light blue/white’ in color. The controls were cells only, FITC only, DAPI only, Rab 5 primary antibody only, Rab 7 primary antibody only and secondary antibody only. The parameters measured were: (1) volume (μm3) of polyplexes and was determined by the volume of a voxel multiplied by the number of voxels in each physical volume; (2) distance of the polyplex/pDNA complexes from the surface of the nucleus (μm).
Figure 4.
Proposed concept of a concentric-nuclear zone traversed by the polyplexes in the cell. The image was generated by compiling the two-dimensional confocal microscopy images and three dimensionally rendering the data allowing visualization of the nucleus (yellow) and polyplexes (blue). This image also shows an illustration of an intracellular three-dimensional concentric-nuclear zone (in blue-green) in the perinuclear space to help visualize how the “polyplex distance from the nucleus” was determined.
Figure 5.
(A-D) Probability values for the number of polyplexes to exist at the indicated distance range from the surface of the nucleus. (E-H) Probability values for the number of polyplexes to exist at a distance below 1 μm from the surface of the nucleus. The raw data to a distance of 1 μm was analyzed at 0.1 μm increments. All probability values in A-H were calculated by dividing the total number of polyplexes in each bin or distance-range (for each polymer; at each time point; in six cells) by the total number of polyplexes (for each polymer; at each time point; in six cells) and were extrapolated from the power law fit equations (Table S7B).
Power law model was also fit to the probability values for the polyplex distance from the surface of nucleus (Figure 5; Table S7B). The fit was performed for all polyplex variants at each time point and categorized into zones. Further analysis of these data revealed that the polyplexes primarily existed in the zones at ∼1 μm to ∼2 μm (observed probability greater than 0.10) for all polymers at all timepoints, Figure 5. The data delineating polyplex distance from the nucleus was further analyzed in the perinuclear zones from 1 μm to the surface of the nucleus, Figure 5. The distance of concentric-nuclear zones in this step of analysis was set at an increment of 0.1 μm. Interestingly, as shown in Figure 3 and the quantitative results in Figure 5, the trehalose-containing polyplexes (Tr455 and Tr477) showed a higher probability for the polyplexes to exist closest to the nucleus (at ∼0.1 μm) as early as 4 hours post transfection and even at 12 and 24 hours when compared to the other polyplex types. However at the 8 hour timepoint, Glycofect™ polyplexes have a higher probability to exist at ∼0.1 μm distance from the nucleus than polyplexes formed with the trehalose polymers. This may be attributed to a slower trafficking speed of Glycofect™, polyplexes which appeared to have taken 8 hours to reach the perinuclear zone of 0.1 μm. This is particularly evident visually in Figure 3C for Tr455 polyplexes. These results suggested that the trafficking to the perinuclear region with the Tr455 and Tr477 polyplexes occurred more rapidly than with jetPEI™ or Glycofect™ polyplexes. We speculate that the larger molecular weights of these polymers and/or the repeated trehalose structure may possibly contribute to this accelerated intracellular trafficking toward the nucleus.
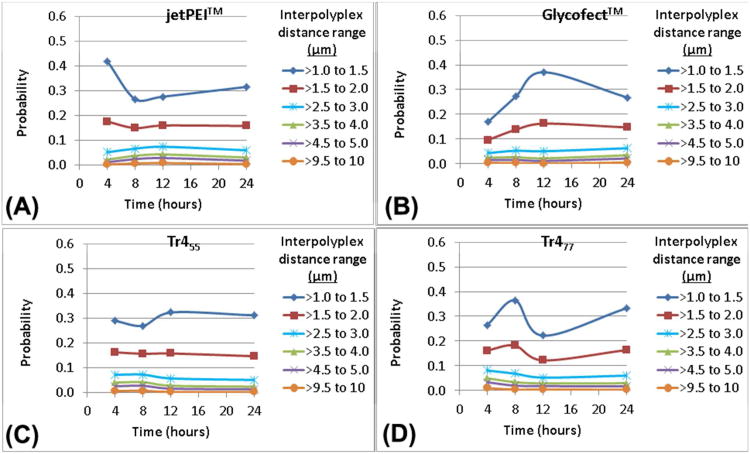
Inter-Polyplex Distance Distribution
The distance between nearest-neighboring polyplexes was analyzed at an increment of 0.5 μm (Figure S4). This distance varied from ∼0.5 μm to ∼5.0 μm (probabilities between 0.4 to 0.01; Figure 6). When analyzing inter-polyplex distances greater than 5 μm, the probability reduced dramatically (less than 0.001). These data reveal that all polyplex variants have a higher probability to exist in a close proximity to each other ∼1.5 μm as compared to larger distances (∼5-10 μm) at all timepoints. This suggests that polyplexes rapidly traffic to similar regions of the cell despite the location of initial internalization. Interestingly, these data also revealed that the polyplexes maintained a proximity of ∼1.5 μm distance from its neighbors at all timepoints as shown in Figure 6. Also, from these data, it was concluded that once endocytosed, the polyplexes have only a negligible tendency to form large intracellular aggregates (of the order of >724 nm spherical diameter i.e. >0.20 μm3 spherical volume). To form a large aggregate from two or more neighboring polyplexes, their inter-polyplex distance would need to be at least equal to or less than the sum of the distance between their centers. The results indicated that the most prominent inter-polyplex distance that existed inside a cell was ∼1.5 μm (probability up to 0.4). This strongly suggested that intracellular aggregation of individual polyplexes to form larger complexes (of diameter 1.5 μm or calculated spherical volume 1.7 μm3) may not favorable. It is important to note that the data for inter-polyplex distances from ∼0.5 μm up to ∼1.0 μm, did not follow any specific trend.
Figure 6.
(A-D) Probability values for the polyplexes formed with pDNA and either jetPEI™, Glycofect™, Tr455 or Tr477 to exist in various inter-polyplex distance ranges from 1-10 μm. The probability values were calculated by dividing the total number of polyplexes in each distance-range by the total number of polyplexes (for each polymer; at each time point; for six cells). The probability values were extrapolated from the power law fit equations (Table S7C).
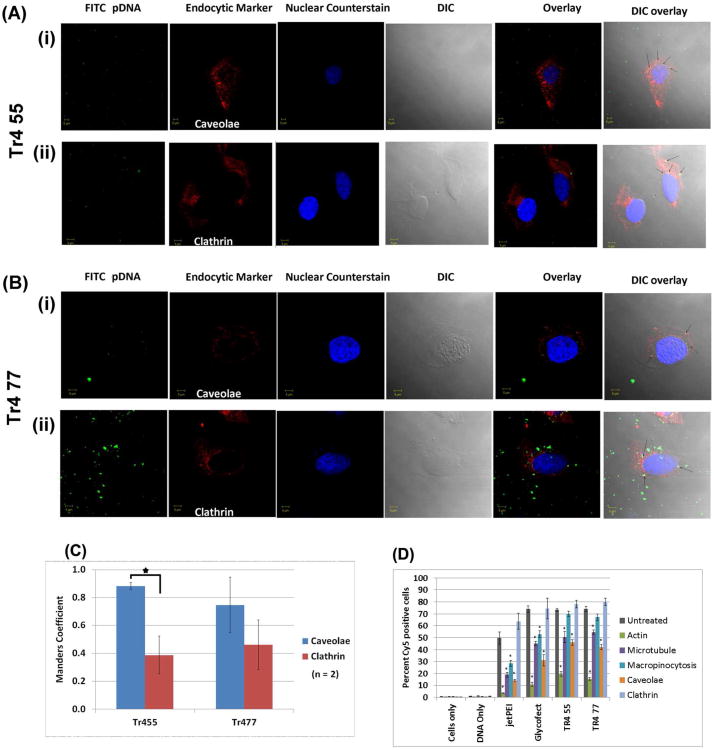
Pharmacological inhibition (Figure 7D) and confocal microscopy experiments (Figure 7A-B) further showed that cellular internalization via endocytosis was highly influenced by the polymer vehicle type. Interestingly, jetPEI™, Glycofect™, Tr455, and Tr477 appear to be internalized into HeLa cells primarily via the caveolae pathway (Figure 7). Supporting these data, significant polyplex colocalization with Rab-5, a marker for early endosomes, including caveolae, was found for all polyplex types and timepoints (Figure 8).
Figure 7.
(A-B) Confocal images showing colocalization of Tr455 and Tr477 polymer containing polyplexes with clathrin and caveolae in HeLa cells. The experiment was performed as reported previously for Glycofect™.6 (C) Manders coefficient for colocalization of polyplexes (pseudo colored ‘green’) with clathrin (marker for clathrin heavy chain) and caveolae (marker for caveolin-1) (pseudo colored ‘red’) using ImageJ (n = 2 cells). The higher Manders coefficient of 0.8 indicates higher colocalization of polyplexes with either clathrin or caveolae; whereas the lower values of 0.4 indicate partial colocalization; (D) Pharmacological inhibition of polyplex internalization pathway (polyplexes were formed with Cy5-pDNA and the indicated polymer). The values on the y-axis denote the percent Cy5 positive cells. Actin was inhibited by cytochalasin D6 (green bars; 2 μg/mL for 15 minutes), microtubules were depolymerized with nocodazole (violet bars; 0.006 μg/μL for 30 minutes), macropinocytosis was inhibited by dimethylamiloride (turquoise bars; 100 μM for 5 minutes), caveolae was inhibited by filipin III (orange bars; 1 μg/mL for 1 hour), and clathrin was inhibited by chlorpromazine (light blue bars; 5 μg/mL for 30 minutes). Note: The ‘untreated’ (grey bars) controls represent an average of values from two different experiments. Note: ‘*’ indicates significant differences as compared to cells only at an alpha value of 0.05.
Figure 8.
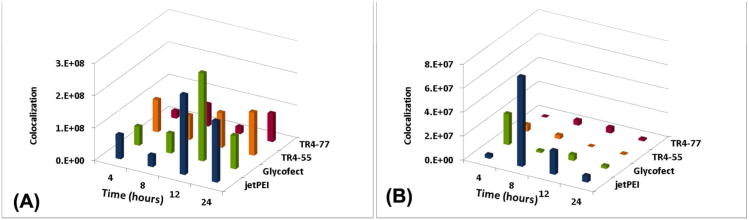
Colocalization of polyplexes with Rab 5 proteins, a marker of early endosomes, at time points of 4, 8, 12, and 24 hours. Images A-D show confocal images of HeLa cells showing polyplex colocalization (monitored by imaging Cy5-pDNA) with Rab 5 protein for early endosomes at 4, 8, 12 and 24 hour timepoints for polyplexes formed with Cy5-pDNA and (A) jetPEI™, (B) Glycofect™, (C) Tr455 and (D) Tr477. The nucleus is pseudo colored yellow, the polyplexes (monitored by imaging Cy5-pDNA) are pseudo colored blue, and Rab 5 is pseudo colored in red. The z-axis is directed into the plane of image, which is shown in the x-y plane. The parameter measured was 3D colocalization of the polyplex/pDNA with Rab 5 (early endosome marker in arbitrary units), which was determined by the total of overlapping volumes, rendered from the fluorescence, between the two channels.
As shown in Figure 7 A-C, the microscopy data revealed that during intracellular trafficking, Tr455 and Tr477 polyplexes colocalized with markers for both clathrin and caveolae, which was corroborated by a previous study from our group that showed that both Glycofect™ and jetPEI™ also colocalized with both endocytic markers6. However, Figure 7D showed that for all polyplex types, cellular internalization was primarily via caveolae. When the cells were treated with filipin to deplete cholesterol domains on the cell surface, the resulting disruption of caveolae decreased or inhibited uptake by ∼40 to 70%. This suggested that caveolae-mediated endocytosis was a major internalization pathway. In contrast, the treatment of cells with chlorpromazine to disrupt formation of clathrin coated pits, did not appear to decrease polyplex internalization, which suggests that it may not be a major route of endocytosis. Similarly, Gabrielson et al. also reported that the caveolae mediated endocytosis was a primary route for cellular uptake that lead to transgene expression for PEI-based polyplexes in HeLa cells.33 The inhibitory effect of dimethylamiloride (for macropinocytosis; ∼10 to 50%) is less than the effect due to inhibition of actin, microtubules, and caveolae, which suggests that macropinosomes may not be a primary route of polyplex internalization into the cell, particularly for the polyplexes formed with Tr455 and Tr477. These pharmacological inhibition data revealed that polyplex internalization occurs primarily via caveolae. This is further supported by the DLS and 3D imaging data, which indicated that the polyplexes are actually relatively large in size (> ∼300 nm) both in the cell and in culture media. It is important to note here that such large polyplexes could be internalized via caveolae, which concurs with previously reported values where Fluoresbrite® fluorescent polystyrene latex microspheres of 500 nm were preferentially endocytosed via caveolae in B16-F10 cells.46
In addition, to understand the endocytic pathways, the roles of both actin and microtubules were investigated in polyplex internalization into cells. Actin inhibition with cytochalasin D reduced polyplex uptake to the largest degree (∼70 to 90%). This may be because the actin cytoskeleton provides mechanical scaffolding for the formation of endocytic vesicles on the inner side of the plasma membrane.34 Therefore, inhibiting actin may disrupt vesicle formation and hence its subsequent endocytosis.34 Further, microtubules are the primary intracellular trafficking highways of the cell,35 and therefore we also investigated the role of microtubules by its depolymerization with nocodazole. Depolymerization of microtubules did have a large effect on inhibiting polyplex internalization (∼35 to 70% knockdown), however, this effect was less than the inhibitory effect observed by knocking down actin (Figure 7D). This suggests that actin is primarily responsible for the early stages of polyplex endocytosis. The cytoskeletal framework of actin has also been reported to be involved in the formation of endocytic vesicles.36 Once the polyplexes enclosed in such a vesicle enter a cell, the subsequent trafficking towards the nucleus may be largely via microtubules, which are the intracellular highways. Overall, the entire process of internalization and trafficking of polyplexes is mostly active transport.6
To further examine these intracellular trafficking pathways with our 3D imaging technique, we examined whether the polyplexes colocalized with specific endocytic trafficking markers such as Rab 5 and Rab 7 (Figure 8 and 9). Rab 5 has been reported to primarily participate in early endosome fusion, trafficking between -coated vesicles and early endosomes, and sorting of endosomes.37 Rab 5 vesicles have also been reported to be associated with the caveolin-1 protein (and thus caveolae vesicles).38 Rab 7 has been reported to participate in trafficking of late endosomes and lysosomes.37,39 Indeed, these data suggested that all four polyplex types colocalize with both endocytic markers. It is important to note that in this experiment, the Rab 7 was stained with the same antibody concentration as Rab 5, which resulted in a much dimmer staining for Rab 7. Therefore, we can only qualitatively conclude that polyplexes may colocalize with Rab 7 vesicles to a lesser degree as compared to Rab 5 vesicles because of the staining differences. Also, Figures 8, 9, and 10 showed that the overall levels of early endosomes (Rab 5) in a cell was higher than levels of late endosomes/lysosomes (Rab 7 vesicles) at the time points of study. These results suggested that the polyplexes appeared to have a low occurrence of trafficking to acidic lysosomes (causes degradation), with the exception of jetPEI™ polyplexes, which did appear to localize to Rab 7 vesicles (late endosomes) to a higher degree (particularly at 8 hours) than the other polyplex types. Therefore, these data taken together, indicated that the four polyplex types appear to be internalized and trafficked primarily via the early endosomal pathway mostly involving caveolae, which could be related to the size of the polyplexes and structure of polymers that were encountered by the cell surface. These data also suggested that to some extent, the clathrin-mediated pathway was also used by all four polymers and jetPEI™ polyplexes appear to be trafficked via the late endosomal pathway to the greatest extent than the other polyplex types.
Figure 9.
Colocalization of polyplexes with Rab-7 proteins, a marker for late endosomes. Images A-D show confocal images of HeLa cells monitoring colocalization of polyplexes with Rab 7 proteins for late endosomes at 4, 8, 12 and 24 hour timepoints for polyplexes formed with Cy5-pDNA and (A) jetPEI™, (B) Glycofect™, (C) Tr455 and (D) Tr477. Note: The nuclei are pseudo colored in yellow, the polyplexes are pseudo colored in blue, and Rab 7 is pseudo colored in red. The z-axis is directed into the plane of image which shown in the x-y plane. The parameter measured was 3D colocalization of the polyplex/pDNA with Rab 7 (late endosome marker) in arbitrary units), which was determined by the total of overlapping volumes, rendered from the fluorescence, between the two channels.
Figure 10.
3D volume colocalization of polyplexes with intracellular trafficking proteins: (A) Rab 5, a marker for early endosomes and (B) Rab 7, a marker for lysosomes. The colocalization is sum of intensity of overlapping voxels (in arbitrary units). The reported values within each plot were not significant at an alpha value of 0.05.
All polyplex types colocalized with Rab 5 vesicles (associated with early endosomes including caveolae) to the largest degree, and to a much lesser degree, localized with Rab-7 vesicles (not normally associated with caveolae); these results collectively supports studies of others in this field.40 Supporting our results, several other reports have shown that cellular internalization of larger particles greater than 200 nm (specifically, 500 nm microspheres) primarily occurred via the caveolae pathway.41, 42 For example, Godbey et al. and Rejman et al. have found that endocytosis may be size dependent and that smaller polyplexes are endocytosed faster and in larger numbers than larger particles.29, 41 These reports indicated that smaller particles (fluorescent latex beads up to 200 nm in diameter) were preferably endocytosed via a clathrin-mediated pathway in a murine melanoma cell line B16-F10, whereas the larger particles (fluorescent latex beads 200 to 500 nm in diameter) are internalized through the caveolae pathway.41 Interestingly, Rejman et al. observed that 1 μm particles were not internalized by B16-F10 cells, which also suggests that both particle size and cell type play a role in nanomedicine internalization (and mechanism).41 In addition, Gratton et al. observed shape and size dependent behavior of nanoparticle endocytosis, where larger cylindrical particles of 1 μm (length) made via lithography (PRINT, Particle Replication In Non-wetting Templates) were able to be internalized by HeLa cells, and were endocytosed preferentially via a clathrin mediated pathway, however, 150 nm - 200 nm (length) cylindrical particles were endocytosed via a caveolae mediated pathway in in the same cell type.43 Grosse et al. reported that <100 nm PEI polyplexes are endocytosed via caveolae in human tracheal epithelial ΣCFTE29o- cells.44 They further observed that in addition to size, the polymer structure also largely affected the mode of endocytosis.44 Gersdorff et al. reported that the in addition to the size and polymer type, endocytosis also depended on the cell type when comparing results in HUH-7, COS-7 and HeLa cells.45 They concluded that polyplexes of initial size ∼100 nm were endocytosed via both clathrin and caveolae mediated pathway in HUH-7 and HeLa cells and that particles larger than 1 μm were internalized by clathrin and caveolae independent pathways.45
In the current study, our results indicate that the intracellular distribution of nanoparticle polyplexes was in the range ∼267 to ∼724 nm, largely between ∼267 to ∼424 nm, which appeared to be primarily endocytosed via a caveolae-mediated pathway in HeLa cells. These results support findings by Rejman et al. suggesting that larger polyplexes are able to be endocytosed via a caveolae-mediated pathway.46 Finally, in our study, to correlate the endocytosis and intracellular trafficking, a luciferase assay was performed with the four polyplex types. The results indicated that the longer chain Tr477 polymer showed significantly (p-value < 0.05) higher gene expression as compared to Glycofect™ and Tr455 (Figure S6) in HeLa cells. In view of the above findings, it is clear that the polyplex endocytosis pathway may be governed by a combination of five key factors such as: (1) cell type, (2) nanocomplex shape, (3) chemical and structural features of the nanomedicine complex, (4) size distribution, and (5) medium in which nanoparticles approach a cell (such as transfection media).
Conclusions
In this work, 4D spatio-temporal cellular imaging and statistical data analysis have yielded heretofore unrealized quantitative evidence that the structural characteristics of polymer-based delivery vehicles play an important role in determining the intracellular polyplex size, trafficking kinetics, and delivery mechanisms. Collectively, this study reveals several interesting trends important for understanding the cellular behavior of nanomedicines created from polymers. (i) At 4 hours post transfection, the smaller size particles (volume of < 0.02 μm3; diameter of 336 nm) formed the majority of the intracellular polyplex population for all polymer vehicle types when compared to larger polyplexes (∼ 2 μm3 volume; diameter of 1562 nm). Thus, the smaller low molecular weight Glycofect™ polyplexes (diameter ∼ 267 nm) dominate the intracellular population as compared to Tr477 polyplexes, which have a broader size range(ii) When comparing within a specific class of polymer vehicle, the smaller size polyplexes trafficked the fastest and reached the perinuclear zones at the earlier time points than the larger polyplexes. For all of the polymer-pDNA complexes studied, the percentage of polyplexes in the perinuclear zone (0 to 1 μm) was significantly (p < 0.05) higher than all other zones (at distances above 1 μm) at the 24 hour timepoint. By 4 hours, it was the trehalose-containing polymers that trafficked faster in the cell to reach the perinuclear region the earliest and they appeared to maintain this state up to 24 hours. For Glycofect™ polyplexes, although they entered the cell first and in greater numbers, they appear to be trafficked slower to the perinuclear region. (iii) For all polymer vehicles and time points, the inter-polyplex distance was primarily between 1.0 to 1.5 μm, suggesting that polyplex nanosystems are rapidly trafficked to similar regions within the cell despite the location of initial internalization. However, the polyplexes do not appear to be collected as aggregates in the cell, which could indicate that they are generally not sorted into late endosomes. (iv) There was no significant difference in the degree of pDNA colocalization with the nucleus for all polyplex types at all timepoints. (v) The polyplexes appear to be internalized by a caveolae/Rab-5-dependent pathway and, for the most part, have a low occurrence of trafficking to late endosomes associated with Rab-7 (with the exception of jetPEI™ polyplexes, where some colocalization with late endosomes was noticed at 8 and 12 hours). Lastly, successful intracellular trafficking to the perinuclear region and nuclear delivery was corroborated via gene expression experiments. In particular, pDNA delivered by polymeric vehicles comprising of the longer chain trehalose polymer Tr477 showed higher gene expression, which supports the data that the Tr4-series of polymers have more rapid trafficking to the nucleus.
Collectively, we conclude that the nature of the chemical backbone in the polymer plays a vital role in uptake, trafficking, and kinetics. The analysis techniques utilized herein can be employed to further understand intracellular trafficking of nanomedicines and colocalization of nanomedicines with organelles, which is beyond the scope of the current work. Indeed, the resulting data can be used in the de novo design of materials to enhance the biological properties of nanoparticle complexes and can be broadly applied to study, in 4D, the behavior of all biological systems able to be observed via light microscopy in a spatio-temporal manner.
Supplementary Material
Acknowledgments
The authors wish to thank the National Institutes of Health (NIH) Director's New Innovator Award Program (DP2OD006669-01) for financial support of this project. We thank Techulon, Inc. for the gift of Glycofect™ transfection reagent. We wish to thank Professor T. Lodge and the Polymer Group at University of Minnesota for use of the dynamic light scattering equipment.
Funding Sources: National Institutes of Health (NIH).
Abbreviations
- FITC
fluorescein isothiocyanate
Footnotes
Competing Financial Interests: TMR has stock options in Techulon, Inc., which sells Glycofect Transfection Reagent.
Supporting Information Available: Materials, Calculations for Polyplex Distributions, Dynamic Light Scattering Information, Figures S1 to S6, Tables S1 to S8, Movie S1, Movie S1 Caption. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Shen H, Sun T, Ferrari M. Nanovector delivery of siRNA for cancer therapy. Cancer Gene Ther. 2012;19:367–373. doi: 10.1038/cgt.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis ME, Zuckerman JE, Choi CHJ, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A. Evidence of RNAi in Humans from Systemically Administered siRNA via Targeted Nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hrkach J, Von Hoff D, Ali MM, Andrianova E, Auer J, Campbell T, De Witt D, Figa M, Figueiredo M, Horhota A, et al. Preclinical Development and Clinical Translation of a PSMA-Targeted Docetaxel Nanoparticle with a Differentiated Pharmacological Profile. Sci Transl Med. 2012;4:1–11. doi: 10.1126/scitranslmed.3003651. [DOI] [PubMed] [Google Scholar]
- 4.Davis ME. Non-viral gene delivery systems. Curr Opin Biotechnol. 2002;13:128–131. doi: 10.1016/s0958-1669(02)00294-x. [DOI] [PubMed] [Google Scholar]
- 5.Davis ME. The First Targeted Delivery of siRNA in Humans via a Self-Assembling, Cyclodextrin Polymer-Based Nanoparticle: From Concept to Clinic. Mol Pharm. 2009;6:659–668. doi: 10.1021/mp900015y. [DOI] [PubMed] [Google Scholar]
- 6.McLendon PM, Fichter KM, Reineke TM. Poly(glycoamidoamine) Vehicles Promote pDNA Uptake through Multiple Routes and Efficient Gene Expression via Caveolae-Mediated Endocytosis. Mol Pharm. 2010;7:738–750. doi: 10.1021/mp900282e. [DOI] [PubMed] [Google Scholar]
- 7.McLendon PM, Buckwalter DJ, Davis EM, Reineke TM. Interaction of Poly(glycoamidoamine) DNA Delivery Vehicles with Cell-Surface Glycosaminoglycans Leads to Polyplex Internalization in a Manner Not Solely Dependent on Charge. Mol Pharm. 2010;7:1757–1768. doi: 10.1021/mp100135n. [DOI] [PubMed] [Google Scholar]
- 8.Fichter KM, Ingle NP, Mclendon PM, Reineke TM. Polymeric Nucleic Acid Vehicles Exploit Active Interorganelle Trafficking Mechanisms. ACS Nano. 2012 doi: 10.1021/nn304218q. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grandinetti G, Smith AE, Reineke TM. Membrane and Nuclear Permeabilization by Polymeric pDNA Vehicles: Efficient Method for Gene Delivery or Mechanism of Cytotoxicity? Mol Pharm. 2011;9:523–538. doi: 10.1021/mp200368p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grandinetti G, Ingle NP, Reineke TM. Interaction of Poly(ethylenimine)–DNA Polyplexes with Mitochondria: Implications for a Mechanism of Cytotoxicity. Mol Pharm. 2011;8:1709–1719. doi: 10.1021/mp200078n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Unterstab G, Gosert R, Leuenberger D, Lorentz P, Rinaldo CH, Hirsch HH. The polyomavirus BK agnoprotein co-localizes with lipid droplets. Virology. 2010;399:322–331. doi: 10.1016/j.virol.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 12.Duleh SN, Welch MD. WASH and the Arp2/3 complex regulate endosome shape and trafficking. Cytoskeleton. 2010;67:193–206. doi: 10.1002/cm.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Summers HD, Rees P, Holton MD, Rowan Brown M, Chappell SC, Smith PJ, Errington RJ. Statistical analysis of nanoparticle dosing in a dynamic cellular system. Nat Nanotechnol. 2011;6:170–174. doi: 10.1038/nnano.2010.277. [DOI] [PubMed] [Google Scholar]
- 14.Xiao PJ, Li C, Neumann A, Samulski RJ. Quantitative 3D Tracing of Gene-delivery Viral Vectors in Human Cells and Animal Tissues. Mol Ther. 2012;20:317–328. doi: 10.1038/mt.2011.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reilly MJ, Larsen JD, Sullivan MO. Polyplexes Traffic through Caveolae to the Golgi and Endoplasmic Reticulum en Route to the Nucleus. Mol Pharm. 2012;9:1280–1290. doi: 10.1021/mp200583d. [DOI] [PubMed] [Google Scholar]
- 16.Forrest ML, Pack DW. On the Kinetics of Polyplex Endocytic Trafficking: Implications for Gene Delivery Vector Design. Mol Ther. 2002;6:57–65. doi: 10.1006/mthe.2002.0631. [DOI] [PubMed] [Google Scholar]
- 17.Khalil IA, Kogure K, Akita H, Harashima H. Uptake Pathways and Subsequent Intracellular Trafficking in Nonviral Gene Delivery. Pharmacol Rev. 2006;58:32–45. doi: 10.1124/pr.58.1.8. [DOI] [PubMed] [Google Scholar]
- 18.van der Aa MAEM, huth US, Hafele SY, Schubert R, Oosting RS, et al. Cellular Uptake of Cationic Polymer-DNA Complexes Via Caveolae Plays a Pivotal Role in Gene Transfection in COS-7 Cells. Pharm Res. 2007;24:1590–1598. doi: 10.1007/s11095-007-9287-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dominska M, Dykxhoorn DM. Breaking down the barriers: siRNA delivery and endosome escape. J Cell Sci. 2010;123:1183–1189. doi: 10.1242/jcs.066399. [DOI] [PubMed] [Google Scholar]
- 20.Srinivasachari S, Liu Y, Prevette LE, Reineke TM. Effects of trehalose click polymer length on pDNA complex stability and delivery efficacy. Biomaterials. 2007;28:2885–2898. doi: 10.1016/j.biomaterials.2007.01.037. [DOI] [PubMed] [Google Scholar]
- 21.Srinivasachari S, Liu Y, Zhang G, Prevette L, Reineke TM. Trehalose Click Polymers Inhibit Nanoparticle Aggregation and Promote pDNA Delivery in Serum. J Am Chem Soc. 2006;128:8176–8184. doi: 10.1021/ja0585580. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, Reineke TM. Hydroxyl Stereochemistry and Amine Number within Poly(glycoamidoamine)s Affect Intracellular DNA Delivery. J Am Chem Soc. 2005:127, 3004–3015. doi: 10.1021/ja0436446. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, Reineke TM. Degradation of Poly(glycoamidoamine) DNA Delivery Vehicles: Polyamide Hydrolysis at Physiological Conditions Promotes DNA Release. Biomacromolecules. 2010;11:316–325. doi: 10.1021/bm9008233. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y, Wenning L, Lynch M, Reineke TM. New Poly(d-glucaramidoamine)s Induce DNA Nanoparticle Formation and Efficient Gene Delivery into Mammalian Cells. J Am Chem Soc. 2004;126:7422–7423. doi: 10.1021/ja049831l. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y, Reineke TM. Degradation of Poly(glycoamidoamine) DNA Delivery Vehicles: Polyamide Hydrolysis at Physiological Conditions Promotes DNA Release. Biomacromolecules. 2010;11:316–325. doi: 10.1021/bm9008233. [DOI] [PubMed] [Google Scholar]
- 26.Gundersen HJG, Østerby R. Optimizing sampling efficiency of stereological studies in biology: or ‘Do more less well!’. J Microsc. 1981;121:65–73. doi: 10.1111/j.1365-2818.1981.tb01199.x. [DOI] [PubMed] [Google Scholar]
- 27.Bolte S, Cordelieres FP. A Guided Tour into Subcellular Colocalization Analysis in Light Microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 28.Wan L, You Y, Zou Y, Oupický D, Mao G. DNA Release Dynamics from Bioreducible Poly(amido amine) Polyplexes. J Phys Chem B. 2009;113:13735–13741. doi: 10.1021/jp901835u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Godbey WT, Wu KK, Mikos AG. Tracking the Intracellular Path of Poly(ethylenimine)/DNA Complexes for Gene Delivery. Proc Natl Acad Sci U S A. 1999;96:5177–5181. doi: 10.1073/pnas.96.9.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prevette LE, Kodger TE, Reineke TM, Lynch ML. Deciphering the Role of Hydrogen Bonding in Enhancing pDNA-Polycation Interactions. Langmuir. 2007;23:9773–9784. doi: 10.1021/la7009995. [DOI] [PubMed] [Google Scholar]
- 31.Gasiorowski JZ, Dean DA. Postmitotic Nuclear Retention of Episomal Plasmids Is Altered by DNA Labeling and Detection Methods. Mol Ther. 2005;12:460–467. doi: 10.1016/j.ymthe.2005.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kulkarni RP, Wu DD, Davis ME, Fraser SE. Quantitating intracellular transport of polyplexes by spatio-temporal image correlation spectroscopy. Proc Natl Acad Sci U S A. 2005;102:7523–7528. doi: 10.1073/pnas.0501950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gabrielson NP, Pack DW. Efficient Polyethylenimine-Mediated Gene Delivery Proceeds via a Caveolar Pathway in HeLa Cells. J Control Release. 2009;136:54–61. doi: 10.1016/j.jconrel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 34.Qualmann B, Kessels MM, Kelly RB. Molecular Links between Endocytosis and the Actin Cytoskeleton. J Cell Biol. 2000;150:F111–F116. doi: 10.1083/jcb.150.5.f111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mundy DI, Machleidt T, Ying Ys, Anderson RGW, Bloom GS. Dual control of caveolar membrane traffic by microtubules and the actin cytoskeleton. J Cell Sci. 2002;115:4327–4339. doi: 10.1242/jcs.00117. [DOI] [PubMed] [Google Scholar]
- 36.Carreno S, Engqvist-Goldstein ÅE, Zhang CX, McDonald KL, Drubin DG. Actin dynamics coupled to clathrin-coated vesicle formation at the trans-Golgi network. J Cell Biol. 2004;165:781–788. doi: 10.1083/jcb.200403120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 38.Hagiwara M, Shirai Y, Nomura R, Sasaki M, Kobayashi KI, Tadokoro T, Yamamoto Y. Caveolin-1 activates Rab5 and enhances endocytosis through direct interaction. Biochem Biophys Res Commun. 2009;378:73–78. doi: 10.1016/j.bbrc.2008.10.172. [DOI] [PubMed] [Google Scholar]
- 39.Vonderheit A, Helenius A. Rab7 Associates with Early Endosomes to Mediate Sorting and Transport of Semliki Forest Virus to Late Endosomes. PLoS Biol. 2005;3:e233. doi: 10.1371/journal.pbio.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hayer A, Stoeber M, Ritz D, Engel S, Meyer HH, Helenius A. Caveolin-1 is ubiquitinated and targeted to intralumenal vesicles in endolysosomes for degradation. J Cell Biol. 2010;191:615–629. doi: 10.1083/jcb.201003086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rejman J, Oberle V, Zuhorn IS, Hoekstra D. Size-dependent internalization of particles via the pathways of clathrin- and caveolae-mediated endocytosis. Biochem J. 2004;377:159–169. doi: 10.1042/BJ20031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rejman J, Conese M, Hoekstra D. Gene Transfer by Means of Lipo- and Polyplexes: Role of Clathrin and Caveolae-Mediated Endocytosis. J Liposome Res. 2006;16:237–247. doi: 10.1080/08982100600848819. [DOI] [PubMed] [Google Scholar]
- 43.Gratton SEA, Ropp PA, Pohlhaus PD, Luft JC, Madden VJ, Napier ME, DeSimone JM. The effect of particle design on cellular internalization pathways. Proc Natl Acad Sci U S A. 2008;105:11613–11618. doi: 10.1073/pnas.0801763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grosse S, Aron Y, Thevenot G, Francois D, Monsigny M, Fajac I. Potocytosis and Cellular Exit of Complexes as Cellular Pathways for Gene Delivery by Polycations. J Gene Med. 2005;7:1275–1286. doi: 10.1002/jgm.772. [DOI] [PubMed] [Google Scholar]
- 45.von Gersdorff K, Sanders NN, Vandenbroucke R, De Smedt SC, Wagner E, Ogris M. The Internalization Route Resulting in Successful Gene Expression Depends on both Cell Line and Polyethylenimine Polyplex Type. Mol Ther. 2006;14:745–753. doi: 10.1016/j.ymthe.2006.07.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.