Abstract
While nucleic acids such as small interfering RNA (siRNA) and plasmid DNA (pDNA) are promising research tools and therapeutic modalities, their potential in medical applications is limited by a fundamental mechanistic understanding and inadequate efficiency. Herein, two series of carbohydrate-based polycations were synthesized and examined that varied in the degree of polymerization (n)—one containing trehalose [Tr4(n) series: Tr4(23), Tr4(55), Tr4(77)] and the other containing beta-cyclodextrin [CD4(n) series: CD4(10), CD4(26), CD4(39), CD4(143), CD4(239)]. In addition, two monosaccharide models were examined for comparison that contain tartaramidoamine (T4) and galactaramidoamine (G4 or Glycofect) repeats. Delivery profiles for pDNA were compared with those obtained for siRNA delivery and reveal that efficacy differs significantly as a function of carbohydrate type, nucleic acid type and dose, polymer length, and presence of excess polymer in the formulation. The Tr4 polymers yielded higher efficacy for pDNA delivery, yet, the CD4 polymers achieved higher siRNA delivery and gene down regulation. The T4 and Glycofect derivatives, while efficient for pDNA delivery, were completely ineffective for siRNA delivery. A strong polymer length and dose dependence on target gene knockdown was observed for all polymers tested. Also, free polymer in solution (uncomplexed) was demonstrated to be a key factor in promoting siRNA uptake and gene down regulation.
Keywords: RNAi, siRNA delivery, pDNA, polyplex, glycopolymers
INTRODUCTION
The discovery of RNA interference by Fire and Mello1 has been a milestone on the revolutionary journey of modern therapeutics. Dramatic attention has been drawn to the investigation and development of small interfering RNA (siRNA)-related gene silencing techniques, both as research tools and potential therapeutic strategies.2, 3 Currently, the major challenge to the translation of this discovery into novel therapeutics is the inadequate delivery of siRNA to target sites of therapeutic interest.4 Once in the cytoplasm, siRNA is able to assemble with the components of the RNA-induced silencing complex (RISC) to initiate the cleavage and degradation of its corresponding mRNA. In this manner, the production of the corresponding protein is diminished.2 However, the anionic nature of siRNA and its vulnerability to enzymatic degradation (when unmodified) limits the cellular internalization of ‘naked’ siRNA.4, 5 Therefore, development of safe and effective systems for siRNA delivery has received extraordinary attention.
There are many different approaches currently being taken to the development of delivery vehicles for siRNA, including polymers, lipids, viruses, gold nanoparticles, carbon nanotubes, and other nanosystems.4, 6-12 In particular, polymer-based delivery vehicles offer tunable structures and versatile combinations of different functionalities that allow achievement of desirable properties, including particle size, surface charge (zeta potential), and targeting ligand incorporation.12 Cationic polymers can bind with anionic siRNA to form polyplexes via electrostatic interactions and hydrogen bonding, thereby neutralizing the siRNA negative charge and protecting it from degradation. An early example of an efficient nucleic acid delivery polymer is polyethylenimine (PEI), which has had limited use in the clinic due, in large part, to its high toxicity at effective doses.13,14 Currently, there are some polymer-based candidate siRNA formulations at different stages of clinical trials.4 The first targeted delivery of siRNA in humans, reported in 2009, is based upon the use of a cyclodextrin-containing cationic polymer.15
Previously, we have developed several series of carbohydrate-based cationic polymers (Figures 1 and 2) that successfully deliver pDNA to various cell types in vitro.18-23 By alternating the carbohydrate type (both mono-, di-, and oligo-saccharides) and oligoethyleneamine length (between 1-6 in the repeat unit) within the polymer backbone, efficient pDNA delivery has been achieved with generally low concomitant cytotoxicity.16-21 The optimal structures for these series of polymers have mainly contained the pentaethylenetetramine moieties in the repeat unit. The structure-activity relationships have been extensively studied and shown to be strongly dependent on oligoethyleneamine length and the type of carbohydrate for pDNA delivery and polymer structures containing four sugar types generally revealed high efficiency for delivery of pDNA (tartarate, galatarate, trehalose, and beta-cyclodextrin).18-23 As noted in those previous publications, the sugars were incorporated to break up the charge density of the materials, lower toxicity, and offer interesting properties. For example, trehalose is known to promote stability against aggregation and beta-cyclodextrin provides a unique cup-like structure to non-covalently link in groups that could help stabilize polyplexes from aggregation and promote targeting. When considering preliminary studies with cationic beta-cyclodextrin and trehalose click polymers (Figure 1A and 1B), the delivery efficiency of pDNA and toxicity profile has been shown to depend on the molecular weight (typically, longer polymers reveal higher delivery efficiency and toxicity); however this is highly dependent on cell and polymer type.19, 20 Also, the trehalose analogs (Figure 1A) form stable polyplexes that have been found to remain mostly as discrete polyplexes in cell culture media containing fetal bovine serum, which could aid in cellular internalization and trafficking, promoting efficacy.18 While many delivery vehicles have been examined for pDNA delivery and assessed for structure-activity relationships (SAR) in vitro for gene expression, results examining siRNA SAR will likely be divergent. Because of the significant differences between siRNA and pDNA, including molecular weight and subcellular target site, previously-derived structure-activity relationships cannot be directly translated from pDNA delivery to siRNA delivery.24-26 In a recent study by Salcher et. al., a series of branched four-armed oligomers with either terminal cysteine or alanine groups affected polyplex stabilization and delivery efficiency of siRNA and pDNA.25 Structures with terminal cysteines appeared to stabilize the complexes and promote high delivery of both nucleic acid types. However, the type of oligoethyleneamine in the branches yielded clear differences in efficacy for siRNA (tetraethylenepentamine was best for gene knockdown) versus pDNA (pentaethylenehexamine was best to promote transgene expression).27 In addition, many new delivery systems are being developed and examined specifically for siRNA delivery. A related study by Frohlich et. al. has also reported the structure-activity relationships of a new family of oligo (ethane amino) amides with lipid modifications, which had different molecular shapes (described as ‘i’, ‘T’, and ‘U’ –shaped oligos); the oligo shape affected siRNA delivery and gene knockdown where C18 lipid modification appeared to play the largest role in increasing gene knockdown.28
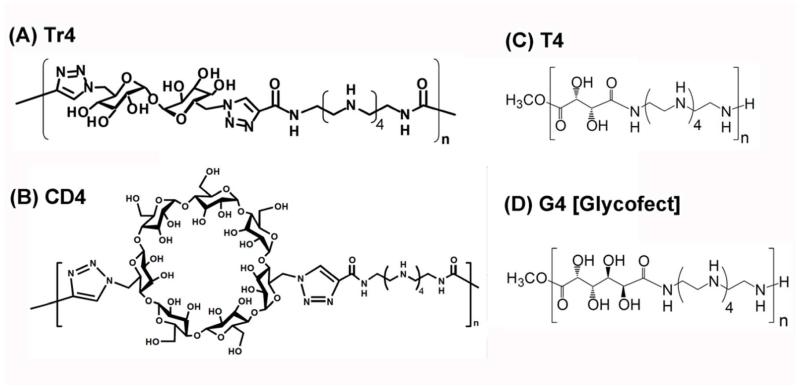
Figure 1.
Schematic structures of the carbohydrate-containing cationic polymers [Tr4,20 CD4,19 T4,21 and G4 (Glycofect)21] examined in this study, which span a large range in carbohydrate size (small to large).
Figure 2.
Schematic showing the different delivery destination of siRNA (cytoplasm) and pDNA (nucleus). In this study we also examine the role of free polymer, which also affects cellular entry and intracellular delivery.
Herein, a systematic evaluation of siRNA-mediated gene knockdown has been examined for a series of carbohydrate-containing ‘click’ polymers. The series of polymers were synthesized to contain either a trehalose [TR4(n)] or beta-cyclodextrin [CD4(n)] moiety copolymerized with a oligoethyleneamine monomer via copper-catalyzed azide/alkyne cycloaddition. In addition, models containing galactaramidoamine (Glycofect) or tartaramidoamine (T4) repeats were also created for comparison, thus spanning a large range in saccharide size (small to large, Figure 1). The effects of saccharide type, polymer length, and nucleic acid type (pDNA and siRNA), and presence of free polymer on polyplex formation and cellular delivery were examined. We reveal that all of these parameters all play a large role in the formation of polyplexes and delivery efficacy of both pDNA and siRNA in cultured U-87 (glioblastoma) cells largely due to chemical differences in the vehicles (Figure 1) and the inherent differences in mechanism/site of action for pDNA versus siRNA (Figure 2).
EXPERIMENTAL SECTION
Materials and Methods
General
U-87 MG-luc2 (U-87_luc2) cells, human glioblastoma cells genetically engineered to constitutively express luciferase, were obtained from Caliper LifeSciences, Inc. (Mountain View, CA). Luc2 siRNA, which targets luc2 luciferase, and a Cy5-labeled Luc2 siRNA were obtained from Integrated DNA Technologies, Inc. (Coralville, IA). The sequence of the sense strand of the Luc2 siRNA is 5′-GGACGAGGACGAGCACUUCUU-3′, and the antisense strand sequence is 3′-UUCCUGCUCCUGCUCGUGAAG-5′. The Cy5 fluorophore within the Cy5-labeled Luc2 siRNA was conjugated to the 3′ terminus of the sense strand (5′-GGACGAGGACGAGCACUUCUU-Cy5-3′). Scrambled siRNA (siCon) was purchased from Dharmacon, Inc (Lafayette, CO). DMEM+GlutaMAX™-I (DMEM), Opti-MEM I+GlutaMAX™-I (Opti-MEM), UltraPure™ Agarose-1000, MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide), propidium iodide (1.0 mg/mL solution in water), PBS pH=7.4, trypsin, antibiotic-antimycotic and Lipofectamine™2000 (Lipo) were obtained from Invitrogen, Inc. (Carlsbad, CA). DEPC-treated water for the RNA work was obtained from Fisher Scientific (Pittsburgh, PA). INTERFERin™ was a gift from Polyplus-Transfection (Strasbourg, France). jetPEI™ was purchased from Polyplus-Transfection (above). The Luciferase Assay System was obtained from Promega Corporation (San Luis Obispo, CA). Bio-Rad DC Protein Assay Reagent A, Reagent B, and Reagent S were obtained from Bio-Rad Laboratories, Inc. (Hercules, CA). CellScrub™ Buffer was obtained from Genlantin, Inc. (San Diego, CA). Bovine albumin and all chemicals used in polymer synthesis were purchased from Sigma-Aldrich (St. Louis, MO). Spectra/Por® dialysis membranes (MWCO: 6,000-8,000 and MWCO: 3,500) were obtained from Spectrum Laboratories, Inc. (Rancho Dominguez, CA). Glycofect Transfection Reagent™, having a degree of polymerization of approximately 11 was donated by Techulon, Inc. (Blacksburg, VA). Polymer T4 (nw=12) was synthesized by our previously published method. 17, 23
Synthesis and characterization of the carbohydrate-containing monomers and polymers
The synthetic routes (see Supporting Information) for the monomers and polymers have been reported in our previous publications,18-20 with some slight modifications. In brief, the dialkyne-pentaethylenetetramine monomers were synthesized from pentaethylenehexamine by protecting the primary amines with trifluoroacetyl groups and secondary amines with tert-butoxylcarbonyls (Boc). The product was recrystallized twice from ethanol, followed by the deprotection of the primary amines by refluxing in aqueous methanol with potassium carbonate. The primary amines were then coupled to propiolic acid using dicyclohexyl carbodiimide (DCC). The dialkyne-pentaethylenetetramine monomer was purified by chromatography [silica gel, 5% (v:v) ethanol in dichloromethane (DCM)] and further recrystallized from ethyl acetate/hexane (1:3 v:v). The total % yield from starting material to final dialkyneoligoethylene amine monomer product was 34%.
2,3,4,2′,3′,4′-hexa-O-acetyl-6,6′-diazido-6,6′-dideoxyl-D-trehalose was also synthesized according to a previously-published procedure.18 The final product was purified on a silica gel column using 10% (v:v) diethyl ether in DCM. Fractions containing the product were collected, the solvent was evaporated, and the products were further recrystallized from 10% (v:v) ethyl acetate in diethyl ether to yield fine white crystals. The total % yield from starting materials to final product was 33%.
The protected (OAc) diazide-β-cyclodextrin monomers were synthesized and purified exactly according to our previously published procedure.19 The total % yield from starting materials to final product was 10%. Generally, the polymerization of the trehalose and β-cyclodextrin monomers with the dialkyne-pentaethylenetetramine monomer was performed in a 1:1 (v:v) solution of tert-butyl alcohol and water under catalysis of copper(I), generated in situ from CuSO4 and sodium ascorbate, at 50-70°C to yield the protected (OAc and Boc) polymers. The product was dried under vacuum, followed by the deprotection of the acetal groups with sodium methoxide in methanol and deprotection of Boc with trifluoroacetic acid in dichoromethane. The final products were purified by dialysis against ultrapure water, then lyophilized and analyzed by NMR and GPC (Table 1). The total % yields from polymerization to deprotection of the trehalose polymers and cyclodextrin polymers were 23% and 18% respectively. Characterization data for these polymers are available in the Supporting Information document.
Table 1.
GPC characterization of the click polymers. Mw: weight-averaged molecular weight. Mn: number-averaged molecular weight. Ð (Mw/Mn): polydispersity index. nw: weight-averaged number of repeat units. The degree of polymerization (n) for Glycofect is ~11 according to the manufacturer.
| Polymer | Mw | Mn | Ð (Mw/Mn) | nw |
|---|---|---|---|---|
| T4(12) | 4.3kDa | 3.9kDa | 1.1 | 12 |
| Tr4(23) | 17.2 kDa | 10.2 kDa | 1.7 | 23 |
| Tr4(55) | 40.5 kDa | 18.4 kDa | 2.2 | 55 |
| Tr4(77) | 56.1 kDa | 37.6 kDa | 1.5 | 77 |
| CD4(10) | 15.7 kDa | 12.6 kDa | 1.2 | 10 |
| CD4(26) | 39.0 kDa | 26.8 kDa | 1.5 | 26 |
| CD4(39) | 58.7 kDa | 34.2 kDa | 1.7 | 39 |
| CD4(143) | 217.6 kDa | 105.9 kDa | 2.0 | 143 |
| CD4(239) | 363.5 kDa | 177.5 kDa | 2.0 | 239 |
Polymer siRNA binding studies via gel electrophoresis shift assays
Agarose gels [2% (w/v)] were prepared by dissolving 1 g agarose in 50 mL TAE buffer (40 mM Tris-acetate, 1 mM EDTA) while heating. Immediately before the gel solution was poured in the gel electrophoresis chamber and cooled to ambient temperature, 4 μL of ethidium bromide (10 mg/mL) was added. The polyplexes were formed by adding 10 μL of each polymer solution at various concentrations to 10 μL of 2 μM siRNA solution in RNase-free (DEPC-treated) water. The N/P ratio indicates the polymer/nucleic acid ratio for polyplex formulation (the number of secondary amines on polymer backbones to the number of phosphate groups on the nucleic acid backbones to form the polyplex solutions) as previously described.18-20 The polyplexes were incubated at room temperature for 30 min. 2 μL of BlueJuice™ loading buffer (Invitrogen) was added to each polyplex solution shortly before loading onto the gel. Electrophoresis was performed at 65 V for 45 min. The complexation of siRNA by the polymer was indicated by the lack of gel migration (or shift) of the siRNA-containing bands.
Polyplex formation and analysis via dynamic light scattering (DLS)
Hydrodynamic diameters of the polymer-siRNA polyplex formulations were determined by dynamic light scattering (DLS) using a ZetaSizer (Nano ZS) instrument from Malvern, Inc. (Worcestershire, United Kingdom) equipped with a 633-nm laser. For each size measurement, polyplexes were formulated by addition of 33 μL of polymer solution in RNase-free water to 33 μL of 2 μM siRNA solution in RNase-free water at room temperature forming complexes at various N/P ratios. The formation of polyplexes was monitored by performing DLS measurements at 0, 5, 10, 15, 30 and 60 min after solution mixing. Each polyplex solution was then diluted to a final volume of 750 μL with RNase-free water for zeta potential measurements of each sample in triplicate.
The stability of the polyplexes in transfection media was studied by adding Opti-MEM (70 μL) or serum-containing DMEM (70 μL) to polyplex solutions (66 μL) in RNase-free water 30 min after mixing of the polymer and siRNA solutions. DLS measurements were then performed at the indicated time points (up to 24 h).
Cell culture
In case of siRNA, luciferase-expressing glioblastoma cells (U-87_luc2) were used for target gene (luciferase) down-regulation efficiency experiments, cellular uptake studies, and MTT assays for cell viability. The scrambled siRNA was included in the experiments to exclude the false positive (off target) data. Because in some circumstances, the low expression of target gene is due to the cytotoxicity of the delivery vehicle or low cellular metabolism rate, which is not sequence sensitive, and thus gene knockdown would be not be due to RNA interference but other factors. In addition non-luciferase expressing glioblastoma cells (U87MG) were used in up-regulation efficiency experiments and cellular uptake with plasmid DNA. The cells were grown in complete DMEM (supplemented with 10% (v:v) fetal bovine serum (FBS) and 1% antibiotic-antimycotic solution (containing penicillin, streptomycin, and amphotericin B) at 37°C and 5% CO2.
Luciferase assay and protein assay
Luciferase-expressing glioblastoma cells (U-87_luc2) were seeded at 50,000 cells/well in 24-well plates 24 h prior to transfection. In general, anti-luciferase (Luc2) siRNA, control (siCon) siRNA, and polymer stock solutions were diluted with RNase-free water, and the polyplexes were formed by the addition of 33 μL of polymer solution to 33 μL of siRNA solution, followed by incubation for 30 min at room temperature. The resulting polyplex solutions were then added to pre-warmed Opti-MEM or DMEM to yield transfection solutions. Cells were washed with PBS before the addition of 200 μL of the transfection solution.
The formation of siRNA-containing lipoplexes using Lipofectamine™ 2000 and complexes with INTEFERin™ were performed according to the manufacturer’s protocol. Polyplexes with the carbohydrate polymers were formulated at various N/P ratios as we previously described.19, 21 The cells were incubated with polyplex/lipoplex solutions for 4 h before complete DMEM was added. Forty eight hours later, the cells were washed with 500 μL PBS and treated with 1× cell lysis buffer (Promega, Madison, WI) for 15 min at room temperature. Aliquots (5 μL) of cell lysate were examined on 96-well plates with a luminometer (GENios Pro, TECAN US, Research Triangle Park, NC) for luciferase activity over 10 s. For each well, 100 μL of luciferase assay substrate (Promega, Madison, WI) was added. The average of duplicate measurements on each individual replicate was utilized for calculation. The amount of protein (mg) in cell lysates was calculated using a standard curve generated with bovine serum albumin by following the protocol included in Bio-Rad DC protein assay kit. The relative light unit (RLU)/mg protein was then calculated and averaged across replicate wells. The protein and luciferase levels of non-transfected cells were used for normalizing the data and calculating the extent of gene knockdown. Each treatment was tested in triplicate in 24-well plates.
For the experiments examining pDNA delivery, non-luciferase expressing U87MG cells were plated 24 hours prior to transfection at a cell density of 50,0000 cells/well in 24-well plates in DMEM culture media containing 10% FBS. Polyplexes were formulated with luciferase plasmid DNA (gWiz-luc) at respective N/P ratios and incubated at room temperature for 1 h prior to transfection. The cells were then transfected in the presence of either (1) serum-free a OptiMEM media or (2) DMEM media containing 10% FBS.
Cellular uptake measurement by flow cytometry
Flow cytometry was performed to examine the cellular uptake of Cy5-labeled siRNA with various formulations at 3 h post-transfection. In general, U-87_luc2 glioblastoma cells were seeded at 300,000/well in 6-well plates 24 h prior to transfection. To transfect, 33 μL of each polymer solution was added to 33 μL of 2 μM Cy5-labeled siRNA solution. After 30 min incubation at room temperature (to form the polyplexes), each polyplex solution was pipetted into 1584 μL of pre-warmed Opti-MEM to yield the final transfection solution. Each well was treated with 500 μL of the obtained transfection solution. After 3 h, the media was removed and cells were washed with 500 μL/well CellScrub™ Buffer for 15 min at room temperature. The CellScrub™ Buffer was then aspirated and cells were exposed to trypsin (0.05% (w/v), 500 μL/well) for 3 min to provide detachment from the plate, then complete DMEM (500 μL/well) was applied to inhibit trypsin. The cell suspension was collected and centrifuged at 1000 rpm for 10 min at 4°C. The supernatant was removed and cells were twice washed with 0.5 mL PBS and centrifuged to remove the extracellular polyplexes. Finally, 1 mL PBS was added and the suspensions were kept on ice prior to flow cytometry analysis. Propidium iodide (2.5 μL) was added prior to the analysis. The flow cytometer (FACSCalibur and FACSVerse, Becton Dickenson, San Jose, CA) equipped with a helium-neon laser to excite Cy5 at 633 nm was used to count twenty thousand events for each sample. The threshold fluorescence level was defined by manually adjusting the positive region such that <1% of negative control cells were positive for fluorescence. Each treatment was performed in triplicate. For pDNA delivery experiments, non-luciferase expressing cells (U87MG) were transfected with Cy5 labeled plasmid DNA and analyzed for uptake via flow cytometry analysis.
MTT assay
MTT reagent (3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide) was used to estimate the cytotoxicity of the formulations. Typically, U-87_luc2 glioblastoma cells were seeded at 50,000 cells/well in 24-well plates 24 h prior to transfection. The polyplexes were formed following the procedure described above. 200 μL of transfection media was added to each well; 4 h later, complete DMEM was added at 1 mL/well. Then, 24 h later, the media was aspirated and the cells were washed with PBS (500 μL/well). 1 mL of serum-containing DMEM with 0.5 mg/mL of MTT was added to each well and cells were incubated for 1 h. The media was then replaced with 600 μL of DMSO for 15 min at room temperature. A 200 μL aliquot of the media was transferred to a well of a 96-well plate for analysis by colorimeter with wavelength at 570 nm. Samples of non-transfected cells were used for normalization.
Statistical Analysis
Statistical analysis was performed at alpha level of 0.05 using JMP software (SAS Campus Drive, Cary, NC). The Tukey-Kramer HSD (honestly significant difference) was used to test significant differences among all means. In addition, a Student’s t-test was used to further find significant differences in cases where the differences were apparent but not detectable by Tukey’s test. All significant differences are denoted by asterisk (*) mark.
RESULTS
Preparation and characterization of polymer vehicles
We have previously published the main synthetic procedures for all of the model polymers examined in this study. T4 and Glycofect (G4) were examined at one length (degree of polymerization, n ~ 12) due to difficulties in the synthesis of these models at longer lengths.18, 20, 27 For the click polymer models, we obtained two series of polymers that vary in the carbohydrate type (trehalose or beta-cyclodextrin) in the repeat unit and the polymer length (Table 1), which was obtained by varying the reaction conditions as previously described.16, 17,18, 29 Each novel polymer prepared for this study is referred to by a name indicating the carbohydrate (i.e. ‘Tr’ for trehalose, ‘CD’ for β-cyclodextrin, “T” for tartarate), number of secondary amines (4), and the weight-averaged number of repeat units (nw).
Gel electrophoresis study of polymer-siRNA complexation
Formulations of siRNA at various N/P ratios (polymer-amines / siRNA-phosphates) were prepared with each of the ten different cationic polymers. Three trehalose-containing click polymers (Tr4(23), Tr4(55), and Tr4(77)) and five β-cyclodextrin-containing click polymers (CD4(10), CD4(26), CD4(39), CD4(143), and CD4(239)) were examined. In addition, the results here in were compared to two polymers described previously; the poly(glycoamidoamine) structures containing four secondary amines per repeat group and either L-tartarate (T4) or meso-galactarate (G4 or “Glycofect”).18,20 These formulations were subjected to electrophoretic analysis in an agarose gel to assess polymer-siRNA interaction (Figure 3 and S1).
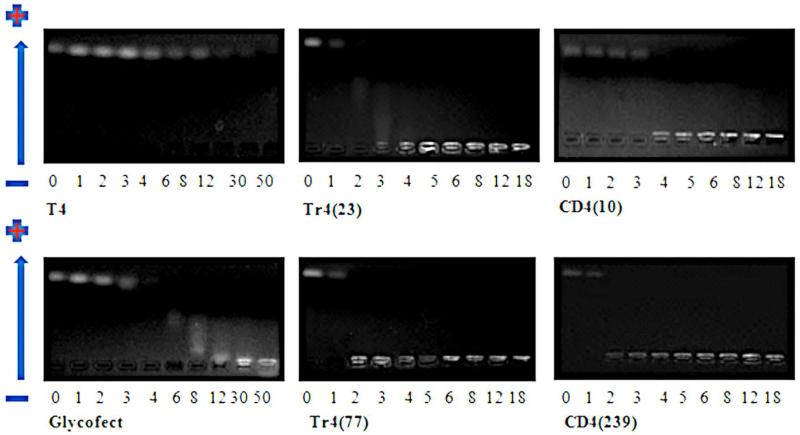
Figure 3.
The binding of polymers with siRNA examined by agarose gel electrophoresis shift assays. Photographs of gels for T4, Glycofect, Tr4(23), Tr4(77), CD4(10) and CD4(239) at various N/P ratios (charge ratio of “number of positive secondary amines (N) on polymer” to the “number of negative phosphate groups (P) on the nucleic acid”). The wells where the gel was loaded is at the bottom of each gel photograph (near the − electrode) and the pDNA, if uncomplexed, migrates to the top of the gel photograph (towards the + electrode). Gels for Tr4(55), CD4(26), CD4(39) and CD4(143) have similar results to their analogs shown here and can be found in the Supporting Information Figure S1.
Polymer T4 containing 12 repeat units has been successfully used to complex pDNA at a low N/P ratio of 1 and effectively delivers pDNA to various cell lines at N/P ratios of 5-30; 17; however, surprisingly, evidence of free siRNA in the gel is apparent for all N/P ratios tested that were at or below 30. At some N/Ps, blurred areas can be seen, which indicates some interaction between polymer and siRNA, but full complexation was not achieved even at the highest tested N/P ratio of 50. A similar affect was noticed with Glycofect (complexes pDNA at an N/P ratio of 2),22 however, retardation of siRNA migration was noticed at a high N/P ratio of 30 and 50 (yet blurring of the pDNA band was still noticed). Binding of siRNA by all analogs of the trehalose and β-cyclodextrin polymers was noticed at significantly lower N/P ratios, with N/P=4 being sufficient to ensure the complete retardation of siRNA migration (thus indicating binding and polyplex formation) for even the shortest versions of these polymers [Tr4(23) and CD4(10)]. This indicates that carbohydrate type plays a role in complex formation since the T4, Glycofect, CD4(10), and Tr4(23) polymers all had similar degrees of polymerization yet very different N/P ratios of binding siRNA. Comparing the binding ability among polymers of different length within the same series (Figure 3), no clear trend was observed; however, it should be noted that longer Tr4 and CD4 polymers tend to retard the migration of siRNA at somewhat lower N/P ratios than their shorter analogues. Because the siRNA-polyplexes with T4 and Glycofect did not appear to be stably complexed (Figure 3), the DLS data for these polymers was not reported in the subsequent section.
Dynamic light scattering (DLS) study
Particle size and zeta potential (surface charge) are two physical parameters that affect the performance of polyplexes both in vitro and in vivo for nucleic acid delivery. To study the complexation process, dynamic light scattering (DLS) measurements of polyplex hydrodynamic diameter were performed at 0, 5, 10, 15, 30 and 60 min time points after the addition of a polymer solution to siRNA solution (that were diluted with RNase free water). After 60 min, size measurement was made and the zeta potential of each formulation was also measured. The polyplexes were formulated at an N/P ratio of 50 as that was found to yield optimal gene down regulation in culture experiments (vide infra).
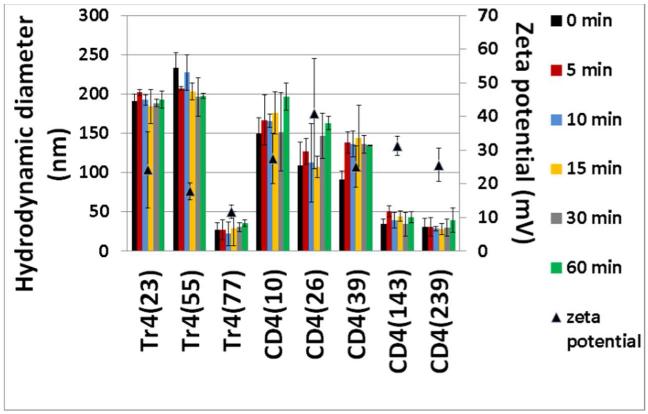
By monitoring the formation of polyplexes over 60 min (Figure 4), we did not observe any significant change in size over time. That is, the hydrodynamic diameter measured immediately after mixing the siRNA and polymer solutions (0 min) is essentially the same throughout 60 min of incubation, indicating that the polyplex formation is extremely rapid and that, once formed, the complexes do not change in size. For the Tr4 series of polymers, Tr4(23) and Tr4(55) form polyplexes with hydrodynamic diameters of around 200 nm; Tr4(77), however, yields significantly smaller (36 nm) polyplexes. The same phenomenon was observed for the CD4 series: while shorter polymers (10, 26 and 39 repeat units) form polyplexes with siRNA of about 130-180 nm, the longer structures (143 and 239 repeat units) yielded polyplexes with hydrodynamic diameters around 40 nm. It is interesting to note that T4 and Glycofect “complexes” at N/P = 50 were also analyzed via DLS, however, these polymers did not form polyplexes with siRNA according to DLS measurements (also in line with the gel shift assay data).
Figure 4.
Dynamic light scattering study to monitor complexation of siRNA and each of the polymers at N/P=50 in RNase-free water at 25 °C. Nanoparticle size (hydrodynamic diameter) was measured at the indicated timepoints from 0 min to 60 min. Zeta potential (triangles) was measured after the 60 min timepoint. All the experiments were completed in triplicate. It should be noted here that both T4 and Glycofect did not form polyplexes with siRNA according to DLS.
All of the Tr4 and CD4 series of polymers tested form polyplexes with siRNA that have positive ζ-potentials. For the Tr4 series, a trend was observed where it was noticed that the ζ-potential decreased with increasing polymer length. The ζ-potential drops from +24 mV for polyplexes formed with Tr4(23) to +12mV for Tr4(77)-containing polyplexes. Surprisingly, a similar trend was not observed for the CD4 series. All of the CD4 polymers, with an exception of CD4(26), formed polyplexes with ζ-potential of +25 to +30 mV. Because of the high N/P ratio (50) used for all formulations tested, there is expected to be a significant amount of “free” polymer in solution (that is, polymer not complexed within the polyplexes), which may contribute to these results.
Effect of polyplex formulation on siRNA delivery to cultured glioblastoma cells
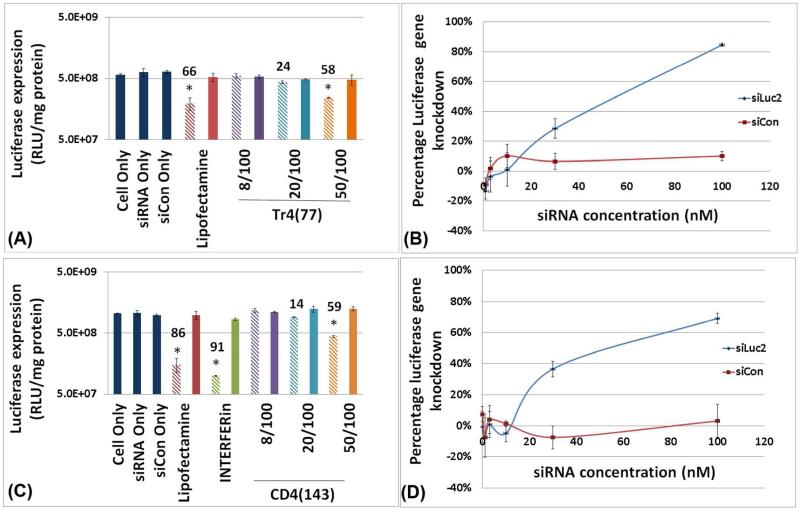
The N/P ratio and siRNA concentration were systematically examined in evaluation of siRNA sequence-specific target (luciferase) down-regulation (Figure 5). The siRNA concentrations were varied from 1 to 100 nM and N/P ratios from 8 to 50. As shown in Figure 5 for both polymer vehicles tested [Tr4(77) and CD4(143)], the most potent target gene down-regulation was observed at a siRNA concentration of 100 nM and N/P=50. Both polyplex types exhibited a dependence of luciferase down-regulation on siRNA concentration; an inhibitory effect was observed at 30 nM, although to a lesser extent than at 100 nM, while no inhibitory effect was seen for siRNA concentrations of 10 nM and below. Similarly, dose dependence with respect to the amount of polymer (N/P ratio) was noticed for both polymers. The luciferase down-regulation was strongest at N/P=50 and was close to 60% for both vehicles. At an N/P ratio of 20, the down regulation was slightly reduced, and a biological effect was completely absent for the polyplexes at an N/P ratio of 8. Finally, all samples in which luciferase expression was reduced showed the expected siRNA sequence specificity, for example, the same polyplex siRNA concentration and N/P ratio, substitution of siCon for siLuc2 abrogated target down-regulation, which indicates that the gene knockdown was not due to off target effects or toxicity of the polymer vehicle. T4 and G4 were also examined for siRNA delivery (vide infra) and were completely ineffective for siRNA delivery (promotion of gene down regulation).
Figure 5.
Influence of siRNA concentration and N/P ratio on polyplex-induced target gene (luciferase) down-regulation in U-87_luc2 glioblastoma cells. Notation is as follows: hashed bar (lighter color) indicates treatment with polyplexes containing siRNA and the solid bar (darker color) indicates treatment with polyplexes containing siCon. The numbers above the bars indicate percentage of gene down-regulation. The notation Tr4(77) 50/100 indicates that polyplexes were formulated with siRNA and Tr4(77) at an N/P ratio of 50 and with 100 nM siRNA concentration. “siLuc2” indicates anti-luciferase siRNA; “siCon” indicates scrambled siRNA negative control. (A) Knockdown efficiency response to polyplexes formed with Tr4(77) with 100 nM siRNA at differing N/P ratio, (B) Extent of knockdown response to Tr4(77) polyplexes formulated at an N/P ratio of 50 at siRNA concentrations from 1 nM to 100 nM. (C) Knockdown efficiency response to polyplexes formed with CD4(143) with 100 nM siRNA at differing N/P ratios, (D) Extent of knockdown response to CD4(143) formulated at an N/P ratio of 50 at siRNA concentrations from 1 nM to 100 nM. [Note: The “*” symbol indicates significantly different values as compared to ‘cells only’ control (p<0.05)].
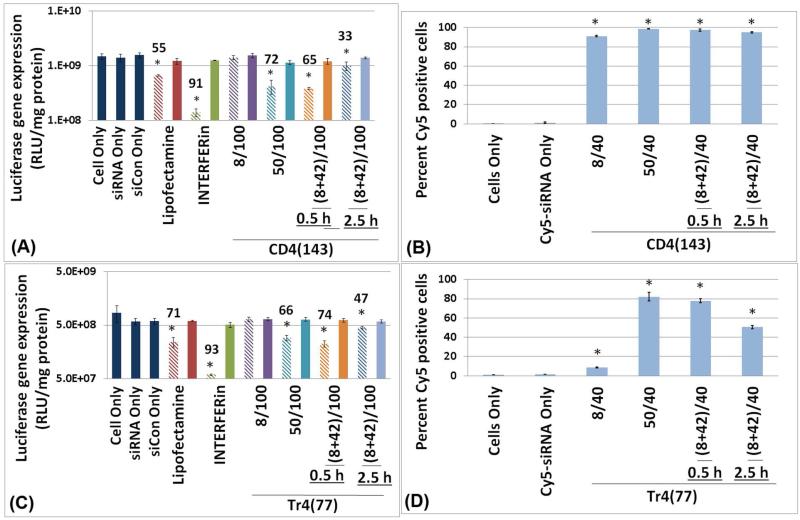
Promotion of siRNA-mediated gene down regulation by addition of free polymers
To investigate the effect of free polymer, we treated U-87_luc2 glioblastoma cells with free polymer 0.5 h or 2.5 h after initial exposure of the cells to CD4(143)/siRNA or Tr4(77)/siRNA polyplexes that were formulated at an N/P ratio of 8 (Figure 6). The amount of free polymer added was such that the total amount of polymer to which cells were exposed (sum of polymer in polyplexes plus subsequently-added free polymer) was equivalent to N/P=50 (Figure 6). Addition of free polymers at the earlier time point (0.5 h after initiation of polyplex exposure), significantly increased siRNA internalization and gene down regulation for the Tr4(77) formulations but not as dramatically for the CD4(143) formulation, which was found to be very high at low N/P. There was close to a 10-fold increase (from 8% to 77%) in the percentage of Cy5 positive cells for Tr4(77) polyplexes with addition of free polymer, however, for CD4(143) polyplexes, a smaller increase from 91% to 97% was noticed. Likewise, when comparing the extent of target gene down-regulation for these formulations, siRNA-mediated gene knockdown increased from 5% for CD4(143) and 25% for Tr4(77) at N/P of 8 to 65% and 74% in cells with free polymer 0.5 h after transfection for CD4(143) and Tr4(77) respectively, achieving an effect comparable to that obtained with polyplexes formulated at N/P=50. Addition of free polymer at a later time point (2.5 h after original polyplex exposure) was shown to promote cellular internalization of siRNA to a lesser degree for Tr4(77) from 8% to 50%, and for CD4(143) from 91% to 94%. The increase in luciferase gene knockdown was also lower for this time point for polyplexes formed with both Tr4(77) (increased from 25% to 47%) and for CD4(143) (increased from 5% to 33%). Overall, these results indicate that addition of free polymer during polyplex exposure increases uptake and potency, suggesting that free polymer within polyplexes formed at high N/P ratios contributes significantly to their efficacy and likely endosomal escape.
Figure 6.
Enhancement of siRNA delivery by polymer vehicles Tr4(77) and CD4(143) upon the addition of free polymer to solution. To understand the role of free polymer in gene down regulation, polyplexes formed at N/P = 8 were added to cells, then at time points of 0.5 h or 2.5 h after initial transfection, a solution containing free polymer equivalent to N/P=42 was added to the wells (brining effective polymer-siRNA ratio to N/P=50 after a time delay). For luciferase gene down regulation (graphs A and C), the cells were transfected with 200 μL/well of siRNA or siCon (at a final RNA concentration of 100 nM/well). For Cy5-siRNA cellular uptake (graphs B and D), the cells were transfected with 500 μL/well of Cy5-siRNA (at a final concentration 40 nM/well). (A) Luciferase gene down-regulation 48 h after transfection with CD4(143)-siRNA polyplexes. (B) Cellular uptake of Cy5-siRNA in the different polyplex formulations with CD4(143) 3 h after transfection. Data is reported as the percentage of live cells positive for Cy5. (C) Luciferase gene down-regulation 48 h after transfection with Tr4(77)-siRNA polyplexes. (D) Cellular uptake of Cy5-siRNA in the different polyplex formulations with Tr4(77) 3 h after transfection. Data is reported as the percentage of live cells positive for Cy5. Notation is as follows: hashed bar (lighter color) indicates treatment with polyplexes containing siRNA and the solid bar (darker color) indicates treatment with polyplexes containing siCon negative control. The numbers above the bars indicate percentage of gene down-regulation. [Note the * symbol indicates significantly different values as compared to the cells only control (p<0.05)].
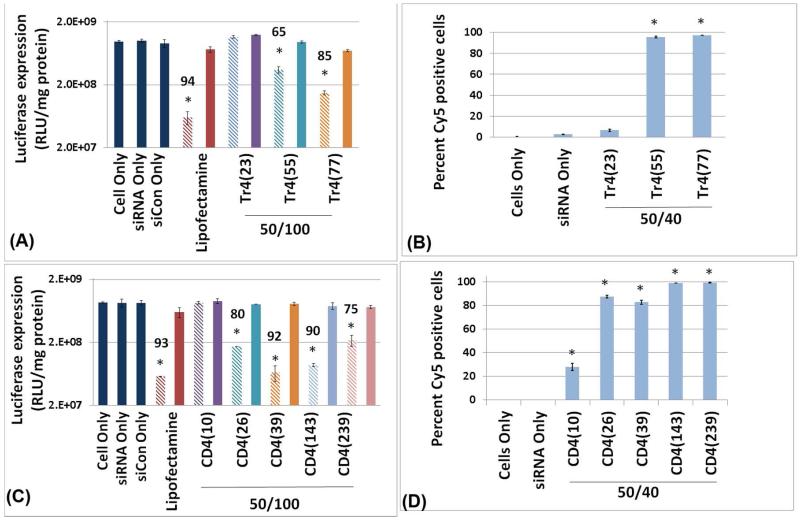
Influence of polymer length on polyplex uptake and efficacy
The molecular weight of polymeric biomaterials may play a significant role in their properties. We investigated the influence of the molecular weight (polymer length) on the siRNA delivery efficiency and target gene down-regulation of polyplexes made with Tr4 or CD4 polymers (Figure 7). For the Tr4 series, an increase in polymer length improves the cell internalization (denoted by an increase in the percentage of cells positive for Cy5-siRNA Figure 7B and 7D) and the extent of luciferase down-regulation (Figure 7A and 7C). Interestingly, polyplexes made with Tr4(23) yield virtually no detectable siRNA internalization. However, Tr4(55) and Tr4(77) polyplexes deliver siRNA into 90%+ of the cells. The CD4 series also demonstrated a similar trend. Polyplexes created from the shortest polymer, CD4(10), were only internalized by about 30% of cells and did not reveal gene down regulation. Indeed, longer polymers were needed to achieve maximal siRNA uptake and target gene down-regulation up to a point, and then the highest molecular weight analog was not as effective. Interestingly, polyplexes formed with CD4(26) and CD4(39) were found to be internalized by about 80% of the cells and yielded significant gene knockdown (80% and 92% respectively). This result was surprising as Tr4(23) was completely inactive and Tr4(55) also was less potent than CD4(26). A similar result was found for CD4(143) were polyplexes were internalized by close to 100% of cells and 90% gene down regulation was found. Yet, while polyplexes formed with the longest system, CD4(239), were internalized by close to 100% of cells, gene down regulation decreased to 75%. It should be noted that for all polymer-containing formulations examined in this study, no apparent cytotoxicity was observed 24 h after transfection according to MTT assays (for polyplexes formulated at an N/P=50 and siRNA concentration of 100 nM, as shown in Figure S6, Supporting Information).
Figure 7.
siRNA-mediated gene down regulation with U87-luc2 cells as a function of polyplex type (formed with either Tr4 or CD4 polymers) and polymer degree of polymerization. (A) Luciferase gene down-regulation facilitated by polyplexes formulated with polymer Tr4 (nw =23, 55, 77) at an N/P = 50 using 100 nM siRNA (50/100). (B) Uptake of Cy5-labeled siRNA within Tr4-containing polyplexes formulated at an N/P = 50 with 40 nM of siRNA (50/40) as measured by flow cytometry 3 h after transfection. (C) Luciferase gene down-regulation facilitated by polyplexes formulated with polymer CD4 (nw =10, 26, 39, 143, 239), at an N/P = 50 using 100 nM siRNA (50/100). (D) Uptake of Cy5-labeled siRNA within CD4-containing polyplexes formulated at an N/P = 50 with 40 nM of siRNA (50/40) as measured by flow cytometry 3 h after transfection. In graphs A and C, the hashed bars indicate data for cells treated with siRNA-containing polyplexes and the solid bars indicate data for cells treated with siCon-containing polyplexes (control). The numbers above the bars indicate percentage of gene down-regulation. Note the * symbol indicates significantly different values as compared to the cells only sample (p<0.05).
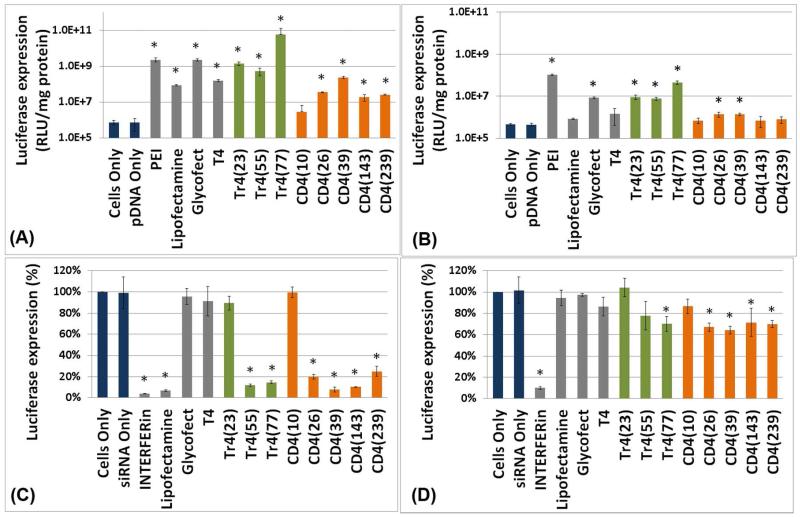
Comparison of pDNA and siRNA delivery profiles
As indicated in Figure 8A, polyplexes created from pDNA and all polymers in the Tr4 and CD4 series (and controls), delivered pDNA to U87 glioblastoma cells as significant luciferase activity is observed in cells transfected in OptiMEM. For cells transfected in DMEM supplemented with 10% fetal bovine serum (FBS), the luciferase activity was lower than in OptiMEM (Figure 8C). As published previously, both Glycofect and T4 yield high efficiency for pDNA delivery as indicated by high luciferase gene expression in HeLa cells.21, 30 Also, initial work has shown the Tr4 and CD4 polymer vehicles were very active for pDNA delivery with these cell types and in some cases a trend was noticed that gene expression increased with increasing polymer molecular weight.19, 20 However, these polymers have not been directly assessed for siRNA delivery and these data not directly compared for pDNA delivery. For the CD4 series, luciferase expression increased with polymer length from nw=10 to nw=39, but this trend did not continue for the high molecular weight versions nw=143 or nw=239. For the Tr4 series, the longest polymer tested (nw=77) yielded the highest transgene expression (Figure 8A) out of any polymer and control in serum-free OptiMEM and second only to PEI in DMEM. When comparing siRNA delivery results, as shown in Figure 8C, polymers T4, Glycofect, Tr4(23) and CD4(10) failed to effectively deliver anti-luciferase siRNA in U87_luc2 glioblastoma cells as a significant reduction in cellular bioluminescence was not observed. In contrast, for both the Tr4 and CD4 series, polymers of larger molecular weight—Tr4(55), Tr4(77), CD4(26), CD4(39), CD4(143) and CD4(239), all yielded significant luciferase down-regulation. The results in serum-free OptiMEM were dramatic, where 80-90% gene down regulation was found. Polymer CD4(39) yielded the highest gene down regulation of all the glycopolymers. In DMEM (contains 10% serum), the siRNA-mediated gene down regulation results were less dramatic (Figure 8D), where 60% gene knockdown was observed. Similar to the pDNA delivery results, maximum delivery efficacy was obtained for both pDNA and siRNA for both the Tr4(77) and CD4(39) polymers. Interestingly, while T4 and Glycofect are potent pDNA delivery vehicles, they are completely inactive for siRNA delivery. In addition, at similar N/P ratios, the Tr4 polymer series yielded higher delivery efficacy for pDNA (than the CD4 series); however, the opposite was true with siRNA, the CD4 series yielded the highest efficacy (Figures 7 and 8).
Figure 8.
Comparison of pDNA and siRNA delivery efficiency for luciferase expression and down regulation respectively, in the absence and presence of serum. (A, B) Delivery of pDNA encoding the firefly luciferase reporter gene into U87 glioblastoma cells with the carbohydrate-containing polymers and controls in (A) OptiMEM and (B) DMEM. Trehalose (Tr4) or β-cyclodextrin (CD4)-containing polyplexes were formulated with pDNA at N/P=7, while pDNA complexes with T4 or G4 were formulated at N/P=20 (maximum gene expression as previously published).17, 22 (C, D) Delivery of anti-luciferase siRNA into U87-luc2 glioblastoma cells with the carbohydrate-containing polymers and controls in (C) OptiMEM and (D) DMEM. All of the carbohydrate-containing polymers were formulated at a siRNA concentration of 100 nM and N/P=50. Note the asterisk “*”, symbol indicates significantly different values as compared to the ‘cells only’ sample (p<0.05).
DISCUSSION
In an effort to optimize the siRNA delivery performance with glycopolymer-based polyplexes and understand the polymer structure – siRNA delivery efficiency relationships, we assessed and compared how polyplex formation and nucleic acid delivery (pDNA versus siRNA) differed with a series of glycopolycation vehicles developed in our laboratory. The chemical structure of polymeric vehicles has a direct influence on its ability to deliver pDNA or siRNA.25 Numerous studies have been published in the literature that are devoted to understanding the structure-activity relationships for pDNA delivery. For example, with PEI, both branched and linear versions of this polymer have been found to effectively deliver pDNA.31 However, when considering siRNA delivery, that study also revealed that branched PEI was much more effective at siRNA-mediated gene down regulation (60% gene knockdown) when compared to linear PEI, which proved completely inactive.31 It was hypothesized that the branched structure of PEI was able to form more stable polyplexes with siRNA.31 The current study herein was undertaken to investigate the influence of carbohydrate type, polymer length, dose response, and the presence of excess polymer on polyplex formation, and nucleic acid delivery efficiency with siRNA. With this family of polymer vehicles (Figure 1), we also sought to compare siRNA delivery to efficiency of pDNA delivery, which have much different intracellular locations and mechanisms of action (Figure 2). The polymer-siRNA binding was examined by gel electrophoresis (Figure 3 and Figure S1). We observed that Glycofect can minimally retard siRNA migration at N/P ratios below 10, however, full binding of siRNA was still not observed until an extremely high N/P ratio of 50. Also, with polymer T4, there is no retardation of siRNA migration at N/P values below 30. These results are in stark contrast to what has been previously published by our group for pDNA delivery (where these polymers fully bind pDNA and retard migration on the gel at low N/P=2).22, 32 These data indicate that these short oligomeric polymers (n ≈12) containing monosaccharides have very low relatively binding affinity to siRNA (but much stronger to pDNA). Molecular weight of both the polymer and the nucleic acid play a large role in polyplex formation. It should be noted that CD4(10), which has the same degree of polymerization as Glycofect and T4, exhibited a significant improvement in siRNA binding affinity (fully bound at N/P = 4, Figure 3) according to the gel shift assays. Despite having the same number of repeats, these polymers differ significantly in molecular weight due to the large difference in sizes of their carbohydrate units (and the presence of the triazole groups). Interestingly, this indicates that the carbohydrate size plays a role in polyplex formation and binding; previous studies from our group have shown that this is likely promoted through hydrogen bonding between the nucleic acid and the hydroxyl units in the polymerized carbohydrate monomer.33, 34 When comparing the CD4 and Tr4 polymer series, there was little to no difference in siRNA binding among the polymers of different lengths as all polymer analogs bound siRNA at low N/P ratios (between 2-4), which is similar to that previously published by our group for pDNA.19, 20, 29 There is a possibility that such a difference could be observed for even shorter (nw<10) polymers within these series; however, such shorter polymers are likely to be poor candidates for siRNA delivery, as can be seen from the trends observed in the gene knockdown experiments (vide infra).
To examine the polymer-siRNA complexation process in aqueous conditions, we monitored the sizes of species present over 60 min after mixing siRNA and polymer solutions. All of the Tr4 and CD4 polymers can readily form polyplexes (Figure 4). As observed with the gel experiments, T4 and Glycofect (G4) did not form well-defined polyplexes (not able to observe polyplex formation in DLS) likely due to their low molecular weight and low binding affinity. Again, this is in contrast to the ability of these polymers to form well defined polyplexes with pDNA.16-21,32 Similar to that observed with pDNA, rapid siRNA complexation was observed for all of the Tr4 and CD4 polymer series, and the dimensions of the polyplexes did not change significantly over 60 min incubation in water at room temperature. The size of the polyplexes formed with the Tr4 or CD4 polymers decreased with increasing polymer length; the shorter Tr4 and CD4 polymers yielded complexes having diameters of ~100 nm or above, while polymers Tr4(77), CD4(143), and CD4(239) yielded complexes of <50 nm (Figure 4). It is also interesting to compare the siRNA polyplexes formed with the longer cyclodextrin polymers of ~50 nm diameter (N/P=50; CD4(143), CD4(239)) to that previously reported for pDNA, where polyplexes of ~100 nm diameter were formed with comparable polymers of similar length (N/P=10; time = 45 min; CD4(93) and CD4(200)).19 On the contrary, the smaller cyclodextrin polymers formed larger polyplexes with siRNA of ~150 to 200 nm diameter (CD4(26), CD4(39)), which is similar to the reported a size of ~70 to 100 nm for pDNA polyplexes (N/P=10; time = 45 min; CD4(27) and CD4(47)).19
These data suggest that the mechanism of polyplex formulation with long/flexible pDNA and short/rigid siRNA may be different. The plasmid DNA used in this study is several kilobase pairs, which ranges in size from 30 to 100 nm in solution. However, siRNA is typically 21-27 bp and is quite small, ranging in size from 2 to 7 nm.25 Thus, the number of negative charges on each siRNA for electrostatic attraction of cationic polymers is substantially less than pDNA.25 The short polymers nw=23 to 55 do not seem to be optimal for wrapping siRNA strands into a compact polyplex. A small increase in the polymer length to nw=77 seems to offer enough length for stable siRNA polyplex formation. A similar trend was observed with the CD4 polymer series. When considering these data along with that previously published for these polymers for pDNA, it appears that polymers with larger molecular weight are needed to serve as a “template” for stable siRNA polyplex formation. Yet, stable and compact polyplexes are able to be formed with short oligos/polymers and large plasmids, which may serve as the template.18-20 Nucleic acid type (DNA versus RNA) could also drive differences in binding due to variances in helicity and the ribose sugar, which could affect complexation and H-bonding. Collectively, when considering the size, zeta potential, cellular uptake, and gene knockdown efficiency of different polyplex formulations, correlations relating size and Zeta potential to delivery efficiency were not observed. For example, polymeric vehicles Tr4(23) and CD4(10) failed to achieve significant siRNA delivery efficiency in vitro, despite having similar polyplex size and Zeta potential to Tr4(55) and CD4(39), respectively, both of which demonstrated markedly improved cellular uptake and gene knockdown in U-87 glioblastoma cells (Figure 7).
The observation that gene knockdown was enhanced at higher N/P ratios despite being able to retard siRNA migration in the gel shift assays at lower N/P ratios, led us to hypothesize that the polyplex solutions contain significant free polymer, which may strongly influence delivery efficiency. To investigate the effect of addition of free polymer, transfection was performed with the timed addition of free polymer to the culture medium. As can be seen from Figure 5, transfection of U87_luc2 glioblastoma cells with CD4(143)-siRNA complexes at a low N/P=8 followed by the addition of free polymer into the transfection medium had a dramatic impact on the transfection efficiency. The addition of free polymer (up to a total polymer amount equivalent to N/P=50) at an early time point (0.5 h after the initiation of transfection) substantially improved the target gene (luciferase) down-regulation; the extent of down-regulation increased 7-fold compared to the polyplexes samples only formulated at N/P=8 (without free polymer addition). When free polymer was added at a later time point (2.5 h after the initiation of transfection), the efficiency improved to a lesser extent (2-fold increase in luciferase down-regulation). With the CD(143) delivery system, free polymer did not have a substantial effect on internalization as polyplex uptake was found to be above 80% at N/P ratios of 8, 50, and in the experiments with the timed polymer addition. A similar study was performed with Tr4(77) polymer and the results follow a similar trend as compared to CD4(143) polymer with some key differences. Polyplexes formulated at N/P = 8 did not show significant gene down regulation (Figure 6C). Significant luciferase knockdown was observed at N/P=50 (Figure 6C). Similar to the CD4(143) polyplexes, addition of free polymer (up to a total polymer amount equivalent to N/P=50), increased the gene knockdown significantly (~ 74 %) for the earlier addition time point (0.5 h post transfection) and to a lower degree for the later time point. However, with Tr4(77) internalization was much lower at N/P=8 (~10% cells) but the cells positive for polyplex internalization did significantly improve at N/P= 50 and after free polymer addition, again indicating that the CD4 series yields more effective siRNA delivery. The results further indicate that for the free polymer to enhance gene knockdown, the free polymers needed to be present near the beginning of the transfection experiment (a longer duration time of 3.5 h incubation). The shorter incubation time of 1.5 h (for 2.5 h post transfection time point) showed lower (~ 47 %) gene knockdown (Figure 6C). This may be explained by the reduced uptake for Tr4(77) polyplexes at the 2.5 h post transfection time point. Taken together, these data validate the hypothesis that free polymer is necessary to achieve maximal siRNA delivery efficiency. Others have also shown this to be true, for example, Boeckle et al. revealed the effects of free polymer addition during transfection of CT 26 (fibroblast cells derived from colon of BALB/c mouse) cells with non-purified (contained free polymer) and purified PEI (did not contain free polymer) polyplexes at 1 h and 4 with varying concentrations of pDNA. They found that free polymer increased the luciferase expression by many fold as compared to purified PEI polyplexes.35 When considering the data for CD4(143), although 90% of cells were positive for siRNA internalization at N/P 8, target gene down regulation was minimal at this low N/P ratio. However, when free polymer (equivalent to N/P = 42) was added to the culture media 0.5 hours after transfection at N/P 8, gene down regulation was equivalent to that observed for polyplexes formulated at N/P =50. This was not the case for Tr4(77) at N/P 8, both internalization and gene down regulation was minimal. These results could indicate that free polymer aids in endosomal escape of siRNA formulations, which intern, aids gene knockdown and was particularly noticed with the cyclodextrin analogs. We speculate that the known affinity for cyclodextrin to extract cholesterol from cell membranes36-39 could play a role in aiding endosomal disruption and polyplex release into the cytoplasm with the CD4 polymer series.
Despite the ability of Tr4(23) and CD4(26) to bind siRNA, Tr4(23) was unable to effectively deliver siRNA into glioblastoma cells, but CD4(26) was effective for siRNA internalization to a large fraction (80%) of U87 cells. Interestingly, at equivalent N/P ratios, the cyclodextrin polymers were found to be more potent than the analogous Tr4 polymer in terms of both cellular uptake and gene down regulation (Figures 6-8). These findings suggest that cyclodextrin, a larger carbohydrate unit, seems to be more suitable than trehalose for designing polymer vehicles that are active for siRNA delivery, in terms of both uptake and target gene down-regulation and could be related to more stable complex formation, enhanced endosomal release, and greater cytoplasmic delivery.
When the polyplexes were collectively examined for transfection efficiency (Figure 8), the shortest polymers [including T4(10), Glycofect, CD4(10), as well as Tr4(23)] were completely inactive towards siRNA-mediated luciferase gene down regulation with U87_Luc cells, which is likely due, in part, to their inability to form stable complexes (Figure 3). Yet, the longer Tr4 and CD4 polymers achieved siRNA-mediated gene knockdown and a polymer length dependence on delivery efficacy was evident (Figure 7 and 8). Importantly, all but the short Tr4 and CD4 polymer analogs were effective at promoting internalization of Cy5-labeled siRNA in 80%+ of U87 cells. In addition, while toxicity was not revealed according to MTT assays, the decrease in gene down regulation with increasing molecular weight could point towards an off target effect or decreased siRNA release.
Previously, we have shown with pDNA that the trehalose polymers reveal a distinct trend of an increase in gene expression with an increase in polymer length (nw=35, 53, 75, 100) with HeLa cells.20 However with H9c2(2-1) cells, such a trend was not observed, as the gene expression levels plateaued with polymer lengths greater than nw=35.20 In the current study with U87MG cells, the observed gene expression was equivalent for pDNA being carried with Tr4 polymers nw=23 to 55; however it then increased at nw=77 (which is seen in both Opti-MEM and DMEM). Interestingly, for siRNA polyplexes formed with Tr4 polymers, gene down regulation was insignificant for Tr4(23) but drastically increased to ~90% for Tr4(55) and Tr4(77) in the presence of Opti-MEM. In the presence of DMEM containing 10% serum, a similar result was found but gene down regulation was dampened (~30% knockdown). Thus, the length of the trehalose and cyclodextrin polymers appears to be an important variable in cellular delivery of siRNA and pDNA. Many other reports have shown that varying polymer length affects delivery; for example Stand et. al. showed that the most efficient gene silencing was achieved using fully de-N-acetylated chitosans with intermediate chain lengths (DPn 100–300).40
When directly comparing the efficacy of the T4, Glycofect, Tr4, and CD4 polymers for pDNA versus siRNA delivery, clear differences and trends in the combined data were observed (Figure 8). As we have previously published, both T4 and Glycofect are effective pDNA delivery vehicles. Among all the polymers tested, Tr4(77) obtained the highest transgene expression in U87 glioblastoma cells, yielding over two orders of magnitude higher transgene expression for than CD4(39), the most potent cyclodextrin polymer for pDNA delivery. Also, Tr4(77) revealed over an order of magnitude higher gene expression than Glycofect. In contrast to their high potency for siRNA delivery, the CD4 polymers were shown to be less effective in the delivery of pDNA than the other polymers examined (Figure 8). A comparison of these carbohydrate-containing polymers for pDNA delivery revealed a slight correlation to the polymer length for both the Tr4 and CD4 series. As shown, the longest trehalose polymer revealed the highest gene expression, also, there was some correlation to the polymer length for the cyclodextrin polymers, where pDNA expression increased as the polymer molecular weight increased up to CD4(39), then the gene expression slightly decreased. In the presence of serum, however, gene expression was dampened. This is likely due to the presence of serum proteins in this media that may bind to the polyplexes. Because the CD4 series does promote nucleic acid delivery to a large amount of cells, the low gene expression efficacy could be due to two possible combined factors: i) the CD4 polymer series could promote higher delivery of nucleic acids to the cytoplasm than the Tr4 series (wrong destination for pDNA delivery), and ii) the CD4 polymers could bind very tightly to pDNA, discouraging release.
The dependence of potency on polymer type and length was more dramatic for siRNA delivery. We were surprised to discover that, despite the high potency of T4 and Glycofect for pDNA delivery, both of these polymers were completely ineffective at forming polyplexes with and delivering siRNA in vitro (Figure 3 and Figure 8D). Also, a dramatic dependence on molecular weight was observed with the trehalose and cyclodextrin polymers. Both the low molecular weight versions of these systems [Tr4(23) and CD4(10)] revealed no activity for siRNA delivery. However, interestingly, as the degree of polymerization was further increased, the cyclodextrin polymers CD(26) and CD(39) were very active for siRNA delivery (80%, and 90% gene down regulation respectively), more so than the Tr4 polymer series. However, the two highest molecular weight analogs of the CD4 series were less effective for delivery. Collectively, these data reveal two very important trends: i) length of the polymer has an effect on gene expression and gene knockdown and ii) the carbohydrate type/size plays a clear role in siRNA binding, compaction, and delivery. The collective data for the CD4 series is particularly intriguing; the CD4 polymers clearly promote effective binding and encapsulation of both pDNA and siRNA, polyplex internalization into 90% of cells even at low N/P ratio, low gene expression but high siRNA-mediated gene knockdown. This points toward the direct role of the sugar in mediating both binding and delivery of nucleic acids. Cyclodextrin clearly promotes higher binding affinity, likely due to H-bonding via the hydroxyl groups and the polymerized nucleotides. In addition, cyclodextrin likely plays a role in promoting endosomal release of siRNA (as Tr4 analogs had lower affinity) possibly due to the known affinity of the hydrophobic cyclodextrin cup to extracting cholesterol out of the membrane, which could promote leaky endosomes. This was also clearly noticed when free polymer was added, which also clearly plays a role in promoting endosomal release of polyplexes carrying siRNA. Lastly, the efficacy was significantly reduced in serum, possibly due to the affinity of the cyclodextrin to binding serum proteins, which likely coat the polyplexes.
CONCLUSION
The observations with these polymers illustrate that, because of their structural differences and the difference in intracellular sites of action, indeed, knowledge based upon pDNA delivery cannot necessarily predict the performance of polymers for siRNA delivery. Indeed, efficacy differs significantly as a function of carbohydrate type, nucleic acid type and dose, polymer length, and presence of excess polymer in the polyplex formulation. The trehalose polymers (Tr4) were more effective for pDNA delivery, yet, the cyclodextrin-containing polymers (CD4) offered higher siRNA delivery and gene down regulation. Interestingly, the systems containing the smaller carbohydrate analogs (T4 and Glycofect) were completely ineffective for siRNA delivery despite having high pDNA delivery and gene expression values. A strong polymer length and dose dependence on target gene knockdown was found and free polymer in solution (uncomplexed) was demonstrated to be a key factor in promoting siRNA uptake and gene down regulation. These findings also highlight the potential of utilizing the Tr4 and CD4 series of polymers as therapeutic pDNA and siRNA delivery agents and demonstrate the subtle effects of chemical structure on the activity of pDNA and siRNA delivery.
Supplementary Material
ACKNOWLEDGMENTS
A special thanks to Dr. Karina Kizjakina and Dr. Antons Sizovs for discussion on some of the synthetic procedures. We gratefully acknowledge our funding agencies: Grant # DP2OD006669-01 (NIH Director’s New Innovator Award Program).
Footnotes
SUPPORTING INFORMATION AVAILABLE
Supporting information includes initial binding study of siRNA with polymer Tr4(55), CD4(26), CD4(39), CD4(143). The 1H-NMR and SEC data of polymer Tr4(77) and CD4(26). This information is available free of charge via the Internet at http://pubs/acs.org/
COI Statement: TMR has stock options in Techulon, Inc. Techulon has licensed and markets Glycofect Transfection Reagent, which was developed in the Reineke Lab.
REFERENCES
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Dykxhoorn DM, Novina CD, Sharp PA. Nat. Rev. Mol. Cell Biol. 2003;4(6):457–467. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- 3.Hammond SM, Caudy AA, Hannon GJ. Nat. Rev. Genet. 2001;2(2):110–119. doi: 10.1038/35052556. [DOI] [PubMed] [Google Scholar]
- 4.Whitehead KA, Langer R, Anderson DG. Nat. Rev. Drug Discovery. 2009;8(2):129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reischl D, Zimmer A. Nanomedicine. 2009;5(1):8–20. doi: 10.1016/j.nano.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 6.Tseng Y-C, Mozumdar S, Huang L. Lipid-based systemic delivery of siRNA. Adv. Drug Delivery Rev. 2009;61(9):721–731. doi: 10.1016/j.addr.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herrero MA, Toma FM, Al-Jamal KT, Kostarelos K, Bianco A, Da Ros T, Bano F, Casalis L, Scoles G, Prato M. J. Am. Chem. Soc. 2009;131(28):9843–9848. doi: 10.1021/ja903316z. [DOI] [PubMed] [Google Scholar]
- 8.de Fougerolles A, Vornlocher H-P, Maraganore J, Lieberman J. Nat. Rev. Drug Discovery. 2007;6(6):443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding W, Hattori Y, Qi X, Kitamoto D, Maitani Y. Chem. Pharm. Bull. 2009;57(2):138–143. doi: 10.1248/cpb.57.138. [DOI] [PubMed] [Google Scholar]
- 10.Gao K, Huang L. Mol. Pharmaceutics. 2008;6(3):651–658. doi: 10.1021/mp800134q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang H-L, Kwon J-T, Kim E-M, Kim Y-K, Arote R, Jere D, Jeong H-J, Jang M-K, Nah J-W, Xu C-X, Park I-K, Cho M-H, Cho C-S. J. Controlled Release. 2008;131(2):150–157. doi: 10.1016/j.jconrel.2008.07.029. [DOI] [PubMed] [Google Scholar]
- 12.Mintzer MA, Simanek EE. Chem. Rev. 2008;109(2):259–302. doi: 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]
- 13.Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. Proc. Natl. Acad. Sci. U. S. A. 1995;92(16):7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Werth S, Urban-Klein B, Dai L, Höbel S, Grzelinski M, Bakowsky U, Czubayko F, Aigner A. J. Controlled Release. 2006;112(2):257–270. doi: 10.1016/j.jconrel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 15.Davis ME. Mol. Pharmaceutics. 2009;6(3):659–668. doi: 10.1021/mp900015y. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y, Reineke TM. Bioconjugate Chem. 2005;17(1):101–108. doi: 10.1021/bc050275+. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, Wenning L, Lynch M, Reineke Theresa M. Polymeric Drug Delivery I. Vol. 923. American Chemical Society; 2006. Gene Delivery with Novel Poly(l-tartaramidoamine)s; pp. 217–227. [Google Scholar]
- 18.Srinivasachari S, Liu Y, Zhang G, Prevette L, Reineke TM. J. Am. Chem. Soc. 2006;128(25):8176–8184. doi: 10.1021/ja0585580. [DOI] [PubMed] [Google Scholar]
- 19.Srinivasachari S, Reineke TM. Biomaterials. 2009;30(5):928–938. doi: 10.1016/j.biomaterials.2008.09.067. [DOI] [PubMed] [Google Scholar]
- 20.Srinivasachari S, Liu Y, Prevette LE, Reineke TM. Biomaterials. 2007;28(18):2885–2898. doi: 10.1016/j.biomaterials.2007.01.037. [DOI] [PubMed] [Google Scholar]
- 21.Lee C-C, Liu Y, Reineke TM. Bioconjugate Chem. 2008;19(2):428–440. doi: 10.1021/bc7001659. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, Reineke TM. J. Am. Chem. Soc. 2005;127(9):3004–3015. doi: 10.1021/ja0436446. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, Wenning L, Lynch M, Reineke TM. J. Am. Chem. Soc. 2004;126(24):7422–7423. doi: 10.1021/ja049831l. [DOI] [PubMed] [Google Scholar]
- 24.Smith AE, Sizovs A, Grandinetti G, Xue L, Reineke TM. Biomacromolecules. 2011;12(8):3015–3022. doi: 10.1021/bm200643c. [DOI] [PubMed] [Google Scholar]
- 25.Scholz C, Wagner E. J. Controlled Release. 2012;161(2):554–565. doi: 10.1016/j.jconrel.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 26.Schallon A, Synatschke CV, Jérôme V, Müller AHE, Freitag R. Biomacromolecules. 2012;13(11):3463–3474. doi: 10.1021/bm3012055. [DOI] [PubMed] [Google Scholar]
- 27.Salcher EE, Kos P, Fröhlich T, Badgujar N, Scheible M, Wagner E. J. Controlled Release. 2012;164(3):380–386. doi: 10.1016/j.jconrel.2012.06.023. [DOI] [PubMed] [Google Scholar]
- 28.Fröhlich T, Edinger D, Kläger R, Troiber C, Salcher E, Badgujar N, Martin I, Schaffert D, Cengizeroglu A, Hadwiger P, Vornlocher H-P, Wagner E. J. Controlled Release. 2012;160(3):532–541. doi: 10.1016/j.jconrel.2012.03.018. [DOI] [PubMed] [Google Scholar]
- 29.Srinivasachari S, Fichter KM, Reineke TM. J. Am. Chem. Soc. 2008;130:4618–4627. doi: 10.1021/ja074597v. [DOI] [PubMed] [Google Scholar]
- 30.Grandinetti G, Reineke TM. Mol. Pharmaceutics. 2012;9(8):2256–2267. doi: 10.1021/mp300142d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwok A, Hart SL. Nanomedicine. 2011;7(2):210–219. doi: 10.1016/j.nano.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 32.Reineke TM, Davis ME. 9.26 - Nucleic Acid Delivery via Polymer Vehicles. In: Krzysztof M, Martin M, editors. Polymer Science: A Comprehensive Reference. Elsevier; Amsterdam: 2012. pp. 497–527. [Google Scholar]
- 33.Prevette LE, Kodger TE, Reineke TM, Lynch ML. Langmuir. 2007;23(19):9773–9784. doi: 10.1021/la7009995. [DOI] [PubMed] [Google Scholar]
- 34.Prevette LE, Lynch ML, Kizjakina K, Reineke TM. Langmuir. 2008;24(15):8090–8101. doi: 10.1021/la800120q. [DOI] [PubMed] [Google Scholar]
- 35.Boeckle S, von Gersdorff K, van der Piepen S, Culmsee C, Wagner E, Ogris M. J. Gene Med. 2004;6(10):1102–1111. doi: 10.1002/jgm.598. [DOI] [PubMed] [Google Scholar]
- 36.Yancey PG, Rodrigueza WV, Kilsdonk EPC, Stoudt GW, Johnson WJ, Phillips MC, Rothblat GH. J. Biol. Chem. 1996;271(27):16026–16034. doi: 10.1074/jbc.271.27.16026. [DOI] [PubMed] [Google Scholar]
- 37.Rodal SK, Skretting G, Garred Ø, Vilhardt F, van Deurs B, Sandvig K. Mol. Biol. Cell. 1999;10(4):961–974. doi: 10.1091/mbc.10.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Aa MAEM, Huth US, Häfele SY, Schubert R, Oosting RS, Mastrobattista E, Hennink WE, Peschka-Süss R, Koning GA, Crommelin DJA. Pharm. Res. 2007;24(8):1590–1598. doi: 10.1007/s11095-007-9287-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kline MA, O’Connor Butler ES, Hinzey A, Sliman S, Kotha SR, Marsh CB, Uppu RM, Parinandi NL. A Simple Method for Effective and Safe Removal of Membrane Cholesterol from Lipid Rafts in Vascular Endothelial Cells: Implications in Oxidant-Mediated Lipid Signaling. In: Uppu RM, Murthy SN, Pryor WA, Parinandi NL, editors. Free Radicals and Antioxidant Protocols. Vol. 610. Humana Press; Hertfordshire: 2010. pp. 201–211. [DOI] [PubMed] [Google Scholar]
- 40.Malmo J, Sørgård H, Vårum KM, Strand SP. J. Controlled Release. 2012;158(2):261–268. doi: 10.1016/j.jconrel.2011.11.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.