Abstract
The development of multicellular organisms involves the partitioning of the organism into territories of cells of specific structure and function. The information for spatial patterning processes is directly encoded in the genome. The genome determines its own usage depending on stage and position, by means of interactions that constitute gene regulatory networks (GRNs). The GRN driving endomesoderm development in sea urchin embryos illustrates different regulatory strategies by which developmental programs are initiated, orchestrated, stabilized or excluded to define the pattern of specified territories in the developing embryo.
Keywords: gene regulatory network, cis-regulatory module, network subcircuit, sea urchin, development
Introduction
The genome encodes the programs required to build the organism from the fertilized egg. Genomic information determines the sequence in which genes functionally interact to drive development. Gene regulatory networks (GRNs) make explicit this information, since they define precisely when, where, and why each gene is employed in development. The specialized cells of an animal are distinguished by the sets of genes they express, in order to accomplish the functions of the territory or structure to which they belong. Their specific patterns of gene expression are determined by the combination of transcriptional regulators that are being expressed at any given time, the sum of which is termed the regulatory state. But why are these regulators expressed in the right cells and at the right time? And how is this process encoded in the static DNA sequence? The presence of transcriptional regulators in the cell nucleus is the direct consequence of the previous regulatory states in each cell lineage. Transitions from one regulatory state to the next are precisely encoded by the cis-regulatory modules controlling the expression of the regulatory genes themselves; and the products of these genes, transcriptional regulatory factors, target other regulatory genes.
Transcription factors regulating the expression of other regulatory genes thus function as main drivers of developmental processes (Levine and Davidson, 2005; Davidson, 2006). The regulatory interactions between these transcription factors have the form of GRNs, because each gene is controlled by multiple regulatory inputs and each transcription factor regulates the expression of several target genes. The intrinsic logic of a GRN determines the sequence of regulatory states in every given developmental process.
The cis-regulatory modules of the genomic DNA respond in specific ways, according to their design, to the regulatory states they encounter in each cell. These functional control sequences, usually a few hundred bp in length, include clusters of transcription factor binding sites, and they control the expression of genes according to whether their respective regulatory factors are present and bound by them. The total cis-regulatory apparatus of many individual genes, and probably of all regulatory genes, consists of a number of distinct modules, each controlling expression in a particular temporal or spatial window (Kirchhamer et al., 1996; Davidson, 2001; Levine and Tjian, 2003; Davidson, 2006). Cis-regulatory elements serve as the computational units of GRNs. They gather regulatory input information and process it in order to output new regulatory information. The most important overall consequence is that the architecture of the GRN is directly determined by the genomic sequence.
Models of GRNs can also be regarded as logic maps. Once the regulatory interactions are solved, they reveal the molecular cause explaining when and where a specific process is taking place. GRN models summarize a dynamic cascade of events, i.e., the flow of regulatory information, which changes in time and space. At the upstream periphery of a GRN are an initial set of regulatory proteins which are interpreted by the control apparatus so as to cause the transcription of a specific set of regulatory genes. The furthest downstream outputs of GRNs are the instructions by which differentiation genes are activated. Unlike the internal genes of the network, which encode transcription factors, differentiation genes do not directly affect the expression of other genes in the network. Genes encoding signaling molecules, which transmit regulatory information to neighboring cells, are additional important outputs of GRNs, and such genes also provide inputs into them.
Despite the explanatory power of developmental GRNs, there have so far only been a few described at a satisfactory level (Stathopoulos et al., 2002; Loose and Patient, 2004; Stathopoulos and Levine, 2005; Davidson, 2006). The GRN controlling endomesoderm development before gastrulation in embryos of the purple sea urchin Strongylocentrotus purpuratus is characterized in relatively great detail (Davidson et al., 2002; Oliveri and Davidson, 2004; Ben-Tabou de-Leon and Davidson, 2007). Using examples from the endomesoderm GRN, we here discuss strategies used by the genome to initiate and stabilize specification programs, and to link the development of different cell lineages so as to ensure a fixed cascade of events that result in the formation of the embryo.
Gene regulatory network analysis in sea urchin embryos
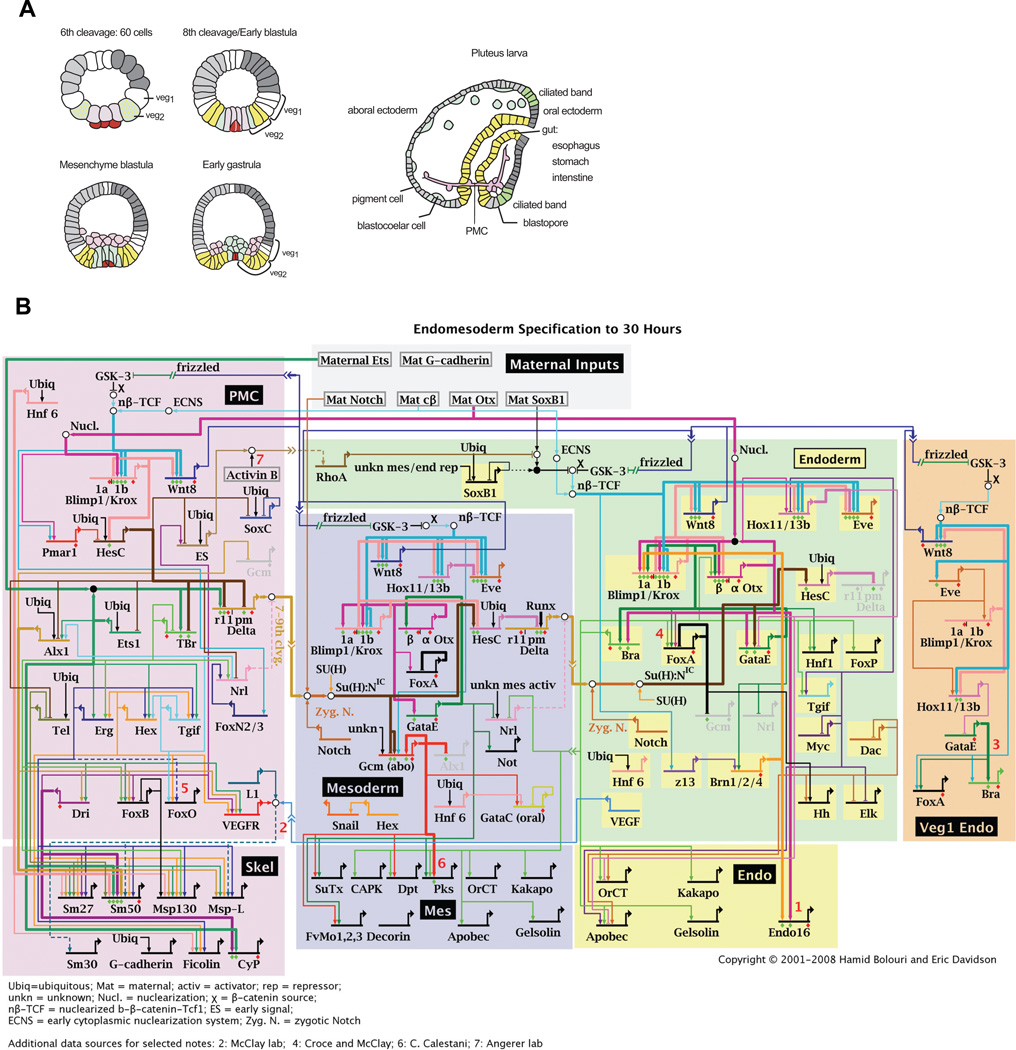
The advantage of using sea urchin embryos for regulatory network analysis lies in part in the biological simplicity of the system, and in part in the (unpredicted) natural advantages of this experimental material, e.g., the ease of gene transfer and of cis-regulatory analysis using DNA injection into eggs. Only a few territories are established in pre-gastrula stage embryos. GRN analysis at early developmental stages allows the use of systemic perturbation approaches, which are not applicable in a more complex system or at later stages, where secondary effects are more likely to occur. The endomesoderm in sea urchin embryos derives from the vegetal plate, more precisely from the skeletogenic micromeres and macromeres which are formed at the fourth cleavage. All micromere progeny will develop into skeletogenic mesenchyme (SM) cells (Fig. 1A, light purple). The specification program which occurs in macromere-derived cells is more complex, since they give rise to mesoderm, endoderm, and some ectodermal cells as well. At first, macromeres are divided to veg2 and veg1 cells. Veg2 cells are the precursors of non-skeletogenic mesenchyme cells (NSMs, Fig. 1A, blue) and endoderm (Fig. 1A, yellow), whereas veg1 cells give rise to endoderm and ectoderm (Fig. 1A, grey).
Fig. 1. Schematic representation of sea urchin endomesoderm development and the underlying gene regulatory network (GRN).
(A) Schematic sections of embryos at different stages of development. At 6th cleavage stage, endomesoderm precursor cells include the macromere-derived veg2 (green) and veg1 cells, large micromere derivatives (light purple) and the small micromere lineage (dark purple). Mesomeres are depicted in grey and are the precursors of ectoderm. Both, veg2 and veg1 cells will give rise to two different territories, depending on their location relative to the vegetal pole. Proximal veg2 cells are the progenitors of NSMs (blue), whereas the remaining veg2-derived cells contribute to endoderm (yellow), as shown in the scheme for 8th cleavage stage embryos. Veg1-derived cells give rise to endoderm and ectoderm. By the mesenchyme blastula stage, the SMs have ingressed into the blastocoel. NSM precursors start to ingress at beginning of gastrulation and give rise to pigment cells and blastocoelar cells. At pluteus larva stage, SMs have fused and formed the embryonic skeleton. (B) Model for the GRN driving endomesoderm development. Color codes match the embryonic territories depicted in (A). Network nodes represent regulatory genes and connections represent regulatory interactions. Regulatory genes are grouped based on the process they are involved in: SM, NSM or endoderm specification. Updates of the endomesoderm GRN are available at: http://sugp.caltech.edu/e ndomes (Davidson et al., 2002).
The recently sequenced S. purpuratus genome opened the door to network analysis at a level which approaches completion. In an extensive gene expression study, the spatial and temporal embryonic expression patterns of all recognizable transcription factor genes were determined (Howard-Ashby et al., 2006b, a; Materna et al., 2006; Rizzo et al., 2006; Tu et al., 2006). Knowledge of the regulatory repertoire of each of the embryonic regions enables the regulatory relationships between them to be elaborated. In order to identify the target genes of regulatory proteins, expression of each regulatory protein is knocked down and resulting changes of transcript levels of all other regulatory genes in a network are determined. Since each of the regulatory inputs in a GRN model predicts the binding of a transcription factor to its respective binding site, we can directly verify the GRN structure by cis-regulatory analysis. Many of the nodes in the GRN driving sea urchin endomesoderm development indeed have been authenticated at the DNA level, by showing that the mutation of predicted binding sites leads to a change in gene expression either in terms of levels or spatial distributions.
The current version of the endomesoderm network is shown in Fig. 1B (current updates are available at http://sugp.caltech.edu/ endomes/). For didactic reasons, genes are grouped based on the processes they are involved in (SM, NSM and endoderm specification), although the regulatory interactions might occur also in cells of other embryonic territories.
Specification of the skeletogenic micromere lineages
The first cell lineage in sea urchin embryos showing a specific expression pattern is the micromere lineage, which gives rise to SM cells (Fig. 1, light purple). As their name suggests, these cells are also the first cells to ingress singly into the blastocoel, before the onset of gastrulation. Once within the blastocoel, SMs expressing biomineralization genes produce the larval skeleton. The SM lineage is unique in many respects. As the first endomesodermal cell lineage to embark on a specification program, it is expected not to be dependent on the developmental state of its neighboring cells. Indeed, micromere-derived cells are the only sea urchin cell lineage which shows autonomous specification. Micromeres isolated at fourth cleavage stage and cultured in vitro still carry out skeletogenesis, and on the normal schedule (Okazaki, 1975). However, removing the micromeres at fourth cleavage stage significantly affects the development of the other endomesoderm cells, which indicates that the micromeres provide signals required for normal endomesodermal specification (Hoerstadius, 1939; Ransick and Davidson, 1995). Transplantation of micromeres to the animal pole is sufficient to respecify adjacent prospective ectoderm cells into endomesodermal precursor cells (Ransick and Davidson, 1993). The presence of micromeres not only induces the expression of vegetal plate marker genes in animal pole cells, but also leads to the formation of a second gut.
Taken together, there are three main tasks to be accomplished by the SMs: 1) initiation of a specification program; 2) expression of signaling molecules which play an essential role in the specification of mesoderm and endoderm; and 3) expression of biomineralization genes. All these functions have to be encoded in the GRN which underlies SM development. The SM GRN has been analyzed in great detail (Oliveri et al., 2008) and it makes explicit the molecular cause of all these functions.
In the formation of territories within an embryo in general each territory is initially dependent either on signals from other territories or on maternally supplied, spatially localized cues. The initial regulatory information for SM specification comes is of maternal origin. Within the unfertilized egg, the protein Dishevelled is localized at the future vegetal pole of the embryo and determines its animal-vegetal (A/V) polarity (Logan et al., 1999; Weitzel et al., 2004; Leonard and Ettensohn, 2007). Dishevelled takes part in the regulation of the canonical Wnt signaling pathway and induces the stabilization and nuclearization of β-catenin (Weitzel et al., 2004). SM precursor cells are located at the vegetal pole and contain the highest levels of nuclear β- catenin within the 16 cell stage embryo. Nuclear β-catenin interacts with the transcription factor Tcf and acts as an activating cofactor. In the absence of β-catenin, Tcf is bound by Groucho, a transcriptional repressor (Range et al., 2005). Genes which are responsive to Tcf are therefore transcribed in cells with high nuclear levels of β-catenin and repressed in cells lacking the activating cofactor. This toggle-switch mechanism of gene regulation is observed also for other transcription factors downstream of signaling pathways and assures that target genes are silenced in the absence of the signaling ligand.
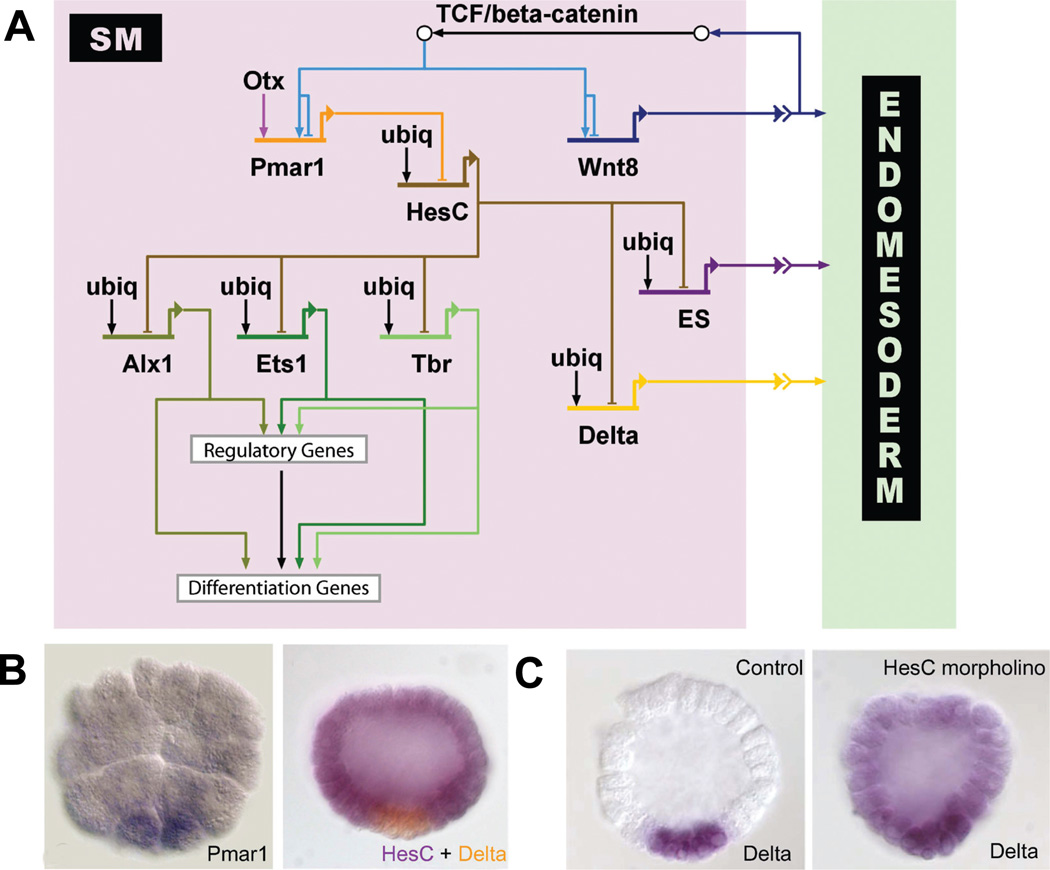
The crucial Tcf target gene in SM precursors is pmar1, as shown in the network diagram in Fig. 2A. In addition to Tcf, maternal Otx contributes to the regulation of pmar1 gene expression. Pmar1 encodes a repressor protein which is exclusively expressed in the micromere lineage, as shown in Fig. 2B, and is sufficient to drive SM specification (Oliveri et al., 2002; Oliveri et al., 2003). Micromere-derived cells, in which the nuclearization of β-catenin has been inhibited by injection of D-cadherin mRNA, are not able to develop into SMs or to form an embryonic skeleton. However, this phenotype is rescued by co-injection of pmar1 mRNA. Normal embryos in which 4th cleavage micromeres have been removed and replaced by micromeres expressing D-cadherin and pmar1 show normal development of SMs and embryonic skeleton. These results indicate that the necessary and sufficient function of nuclear β-catenin in SM precursors is the activation of pmar1 expression, and that pmar1 controls the activation of the SM GRN.
Fig. 2. Expression patterns of genes involved in skeletogenic mesenchyme (SM) specification and SM-gene retulatory network (GRN) model.
(A) Model of the GRN driving SM development. The double repression by Pmar1 and HesC gives rise to a micromere- specific expression of alx1, ets1 and t-brain (tbr). All three transcription factors further activate other regulatory genes as well as the differentiation gene battery. The SM GRN drives the expression of three genes encoding signaling molecules which are required for endomesoderm specification: wnt8, the early signal and delta. (B) Left: Whole-mount in situ hybridization (WMISH) of 5th cleavage stage embryos detecting pmar1 gene expression in micromeres. (Reprinted from Oliveri et al., 2002; with permission from Elsevier). Right: Two color WMISH monitoring hesC (violet) and delta (orange) gene expression in 7th cleavage stage embryos. (Reprinted from Revilla-i-Domingo et al., 2007; with permission from the National Academy of Sciences, USA). Whereas delta is expressed exclusively in the micromeres, hesC expression is ubiquitous except for repression in micromere derivatives by Pmar1. (C) Expression of delta was detected by WMISH in micromere-derived cells of control embryos (left) and ubiquitously in embryos in which HesC expression has been blocked by morpholinos (right). The results show that delta expression is activated by ubiquitous transcription factors and repressed by HesC in all cells except the micromere derivatives, where HesC is not present. (Reprinted from Revilla-i-Domingo et al., 2007; with permission from the National Academy of Sciences, USA).
The function of the Pmar1 repressor is to prevent expression in the SM lineage of the hesC gene, which encodes another repressor protein (Revilla-i-Domingo et al., 2007). The hesC gene is expressed in a pattern inverse to pmar1 at early stages. It is transcribed throughout the embryo, except in micromere-derived cells, where it is repressed by Pmar1 (Fig. 2B). A number of genes which are specifically expressed in the SM-precursor lineage are regulated by HesC. The HesC target genes include the genes encoding transcription factors Alx1, Ets1, Tel, and T-brain as well as the Notch ligand Delta. When expression of HesC is sharply diminished by injection of a gene-specific morpholino, these target genes are expressed uniformly in the whole embryo, as shown for delta in Fig. 2C (Revilla-i-Domingo et al., 2007). The activating inputs into the HesC target genes are therefore present ubiquitously, but their expression is only allowed in the SM lineage, where they are not exposed to the HesC repressor.
The network subcircuit responsible for the onset of the SM GRN is termed a double-negative gate, in which two negatively acting genes are coupled in order to cause expression of a specific set of regulatory genes (Fig. 2A). This network structure relies on the cell type-specific expression of a single repressor in order to activate the downstream GRN in a cell type-restricted manner. At the same time, it assures the exclusion of regulatory proteins involved in the SM GRN from cell lineages other than the SM precursors.
The second important function of the micromere lineage is to provide the signaling inputs into the specification programs of adjacent endomesoderm cells. As the SM GRN model depicted in Fig. 2A indicates, there are at least three signaling molecules expressed in micromere-derived cells. The early signal (ES) must be expressed immediately after formation of the micromeres at fourth cleavage, also apparently under the control of HesC. At the same time, under the control of β-catenin/Tcf, wnt8 expression becomes apparent in SM precursors (Minokawa et al., 2005). This signaling system is employed in a way which puts the regulation of the signaling ligand (Wnt8) under the control of the downstream effector of the signaling pathway (β-catenin/Tcf) itself. The implications of this network subcircuit are discussed below. The third signal is mediated by the Delta/Notch signaling pathway. By 7th cleavage delta is expressed in the SM precursors (Sweet et al., 2002). Cis-regulatory analyses of delta have led to the identification of a cis-regulatory module which drives specific expression of a GFP reporter gene in the micromere lineage, comparable to endogenous delta expression (Revilla-i-Domingo et al., 2004). Consistent with the results for delta expression, this cis-regulatory module is responsive to the pmar1-hesC subcircuit and co-injection of the reporter construct with either pmar1 mRNA or hesC morpholino strongly induces ectopic GFP expression (Revilla-i-Domingo, 2007). In addition a functional HesC site is present in a proximal cis-regulatory module of the delta gene (Smith and Davidson, 2008). These results confirm the position of delta in the SM GRN downstream of the double-negative gate (Fig. 2A).
The three HesC target genes Alx1, Ets1, and T-brain activate the expression of a number of other transcription factors in the micromere lineage, which in turn control the expression of additional regulatory genes (Fig. 1B). Most of the transcription factors expressed downstream of HesC contribute directly to the transcriptional control of the differentiation genes involved in biomineralization (Oliveri et al., 2002; Livingston et al., 2006; Oliveri et al., 2008).
Re-deployment of the skeletogenic mesenchyme gene regulatory network (SM GRN)
Although the double-negative gate is required for the induction of the SM GRN in micromere-derived cells, it has recently been shown that ectopic expression of the HesC target gene alx1 in macromere daughter cells is sufficient to induce the skeletogenic GRN at an ectopic location in postgastrular embryos of the sea urchin Lytechinus variegatus (Ettensohn et al., 2007). The ectopic expression of alx1 and t-brain is induced by the absence of properly specified SMs, which results in respecification of other types of mesodermal cells. This implies that the SMs of the normal late embryo must emit a signal which represses the skeletogenic cell fate, including alx1 and tbr expression in the other mesodermal cell lineages.
The acquisition of a specialized cell lineage for the production of embryonic skeleton is an evolutionary invention of modern sea urchins. The only other Class of echinoderms which shows an embryonic skeleton, the ophiuroids, does not form a micromere cell lineage (Primus, 2005). The GRN which drives the specification of a skeletogenic cell lineage, however, was presumably assembled long before its use in embryonic skeleton formation (Gao and Davidson, 2008). Most of the transcription factors in the SM GRN downstream of the pmar1-hesC double repression system are expressed in the skeletonization centers of sea urchin larvae, where the adult skeleton is being formed. Very similar results have also been obtained in the distantly related sea star Asterina miniata (Gao and Davidson, 2008). Despite 500 Mya divergent evolution, both species presumably employ a similar regulatory toolkit for the specification of the adult skeletogenic cell lineages. In sea urchin embryos, this preexisting network might have been recruited to the micromere cell lineage by bringing it under the control of a simple regulatory switch system, the double negative gate.
The onset of the non-skeletogenic mesenchyme gene regulatory network depends on inputs provided by the skeletogenic gene regulatory network
The successful initiation of the SM specification process leads to the production of different signaling molecules which are required inputs into GRNs driving equivalent processes in other endomesoderm lineages. At the early blastula stage, there are 32 veg2 macromere-derived cells surrounding the SM precursor cells. These cells give rise to the mesoderm lineages, which constitute pigment cells, blastocoelar cells, muscle cells and coelomic pouches, and to the endoderm territory, which will form the larval gut. However at this early stage there is a common endomesodermal regulatory state expressed in all veg2-derived cells, waiting for a signal which initiates either mesoderm or endoderm developmental programs. The most important input for the specification of NSMs, discriminating them from the endoderm cell fate, is the Delta/Notch signaling pathway (Sherwood and McClay, 1999; Sweet et al., 1999; Sweet et al., 2002). The property of this signaling system requires that cells expressing Delta ligand are in physical contact with the cells they signal to. In this way, this signaling system can transmit spatial information, ensuring that only adjacent cells expressing Notch receptor can respond to the signal. Delta expression in micromere-derived cells is controlled by the pmar1-hesC subcircuit, whereas the Notch receptor is provided maternally and is present ubiquitously at this stage (Sherwood and McClay, 1997; Sweet et al., 2002). Only the veg2-derived cells which are located adjacent to the SM precursors respond to the Delta/Notch signal and induce the NSM specification program. Activation of the Notch receptor by Delta leads to cleavage of Notch, which allows the intracellular part (NICD) to enter the nucleus and interact with the transcription factor Suppressor of Hairless (Su(H)) to activate target gene expression. In the absence of Notch signaling, Su(H) is associated with Groucho and this complex represses Delta/Notch target genes. Interfering with the expression of either Delta or Notch in sea urchin embryos severely affects the development of NSMs, resulting in albino pluteus larvae, which lack NSM-derived pigment cells (Sweet et al., 2002). Injection of delta mRNA into L. variegatus embryos on the other hand gives rise to pluteus larvae showing an increased number of NSM-derived pigment cells, blastocoelar cells, and muscle fibers.
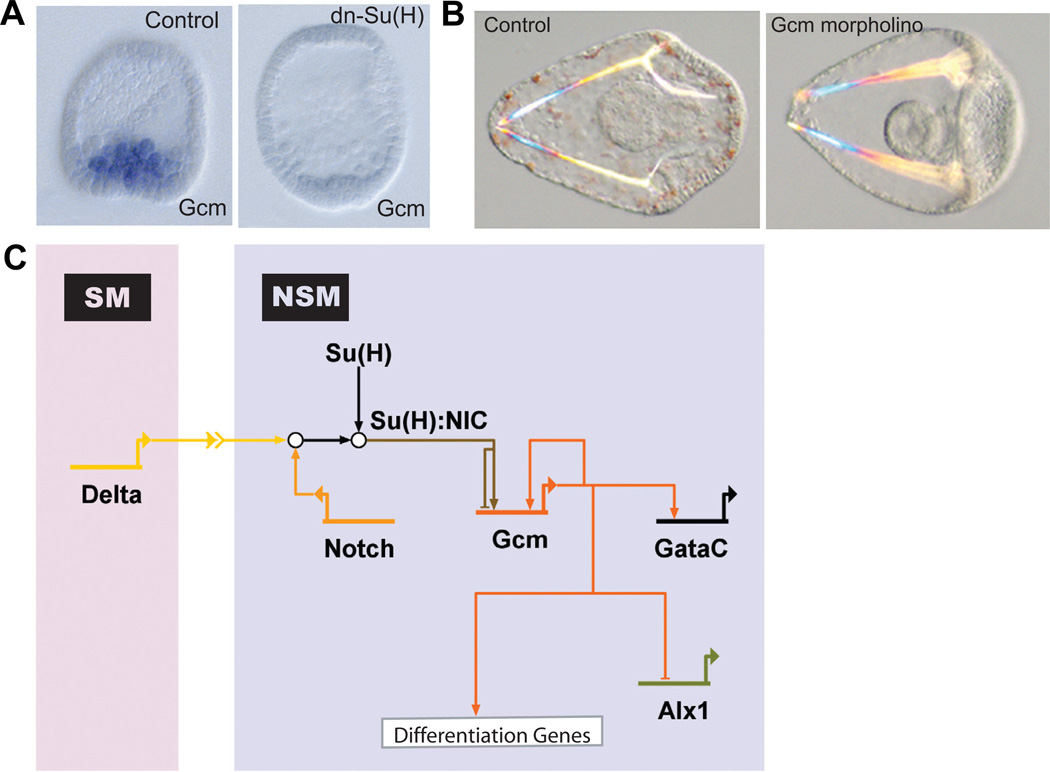
Shortly after the onset of delta expression in micromeres, adjacent NSM precursor cells start to express glial cell missing (gcm) (Ransick and Davidson, 2006). As expected for a gene responsive to the Delta/Notch signaling pathway, gcm expression drops significantly if the Notch signaling pathway is blocked by over expression of a dominant negative form of Su(H) (dn-Su(H)), as shown in Fig. 3A. If gcm is indeed a target of the Notch signaling pathway, it must contain binding sites for the transcription factor Su(H) within its regulatory apparatus. This prediction is directly testable by analyzing the cis-regulatory sequences controlling gcm expression. Putative gcm cis- regulatory modules have been identified recently, which when placed upstream of a GFP reporter construct, induce GFP expression mainly in the NSM domain (Ransick and Davidson, 2006). One of these modules contains several candidate binding sites for Su(H). Mutation of these Su(H) binding sites led to a decrease in NSM-specific GFP expression, but also to an increase in ectopic gene expression in prospective endoderm and ectoderm cells. These results reflect the toggle-switch function for Su(H), repressing gcm in the absence of Notch signaling and acting as a transcriptional activator in the context of NICD.
Fig. 3. Role of gcm in the nonskeletogenic mesenchyme gene regulatory network (NSM GRN).
(A) WMISH for gcm on control embryos (left) and embryos injected with mRNA encoding a dominant-negative (dn) form of the transcription factor Su(H) (dn-Su(H)) (right). Su(H) mediates a toggle-switch mechanism by interacting either with the repressor protein Groucho or, in the presence of Delta/Notch signaling, with the intracellular part of Notch (NIC). Gcm is expressed in NSM precursors at blastula stage in control embryos, but does not express significantly in embryos injected with dn-Su(H), indicating that gcm is a target gene of Su(H). (Reprinted from Ransick et al., 2006; with permission from Elsevier). (B) Control pluteus larva (left) and pluteus larva that developed after injection of gcm-specific morpholinos (right). Note the lack of pigment cells as a result of interfering with gcm expression, showing that Gcm is required for pigment cell formation. (Reprinted from Ransick et al., 2006; with permission from Elsevier). (C) Model for the GRN underlying NSM specification. Delta expression in micromere-derived cells induces the cleavage of Notch receptor in adjacent NSM precursor cells. NIC interacts with Su(H) leading to activation of gcm expression. Gcm expression is maintained by an autoregulatory feedback loop. Gcm also provides activating inputs into gataC and the differentiation genes. To exclude alternative cell fates, Gcm represses alx1.
Gcm itself encodes for a transcriptional regulatory factor. In the absence of Gcm, the NSM specification program is disturbed and embryos show a similar albino phenotype, exactly as in the absence of Notch signaling (Fig. 3B). Consistent with its function as a main integrator of Notch signaling, gcm occupies a central role in the GRN driving NSM specification, as shown by the network model in Fig. 3C. Later, when SMs start to enter the blastocoel, they are no longer in direct contact with prospective NSMs to provide the Delta signaling ligand. The disappearance of the Notch input into gcm is compensated by a positive feedback-loop, in which Gcm activates its own gene expression (Fig. 3C; A. Ransick and E.H. Davidson, unpublished results). Like many other regulatory proteins in the GRN, Gcm is also involved in the transcriptional control of other transcription factor genes as well as of differentiation genes. Since SM and NSM precursor cells have been physically separated for only a short time at this early stage, their regulatory states are still quite similar. The GRNs underlying both specification programs in fact include mechanisms to avoid the acquisition of a wrong cell fate. Gcm apparently plays a dual role, acting both as effector and as target of this cell fate decision process. Thus in SMs, gcm expression is repressed by the HesC target gene alx1 (P. Oliveri and E.H. Davidson, unpublished results). And in turn, gcm interferes with the SM GRN by repressing the expression of alx1 (Fig. 3C; S. Damle and E.H. Davidson, unpublished results). Such exclusion of alternative cell fates is a very commonly observed feature of specification GRNs in many animal systems (Oliveri and Davidson, 2007).
We see that one of the outputs of the SM GRN, delta expression, determines directly the onset of the NSM GRN. The coupling of GRNs ensures a unidirectional process, coordinating the specification programs which take place in different cell types. In this system, the Delta/Notch signaling pathway effectively transmits the information about when and where the NSM specification process should take place. The essential input into the NSM GRN is primarily encoded by two cis-regulatory sequences. The first one integrates the inputs from the early transcription factors in the SM GRN to induce the expression of delta in micromere-derived cells. The second cis-regulatory region, driving the expression of gcm, contains the binding site for the transcription factor mediating Delta/Notch signaling, Su(H). The information underlying both giving and receiving the signal which induces NSM specification is therefore directly encoded in the regulatory genome.
Conservation of an endoderm subcircuit
Cells of the veg2 macromere lineage which are located further away from SM precursors will develop into endodermal cell lineages. The regulatory state in the veg2 lineage at blastula stage consists predominantly of factors activated by TCF/β-catenin, viz. Blimp1, Eve and Hox11/13b, and Otx (Yuh et al., 2002; Arenas-Mena et al., 2006; Smith et al., 2008). In the absence of an early Notch signaling input, these broadly expressed endomesodermal transcription factors initiate the endoderm regulatory program (Fig. 1B). Their immediate target genes are the brachyury, foxa and gatae genes, the latter two of which have a widely conserved role in endoderm formation. Brachyury and GataE are thereupon responsible for inducing the expression of the transcription factors Hnf1, Tgif and FoxP shortly before the onset of gastrulation (I. Peter and E.H. Davidson, unpublished results). The regulatory state at this stage defines a pre-endoderm specification state, which will shortly thereafter diverge to initiate the different regulatory programs underlying the formation of the fore-, mid- and hindgut. Before gastrulation, however, transcription factors which are later restricted to a specific gut compartment are expressed in the same cells.
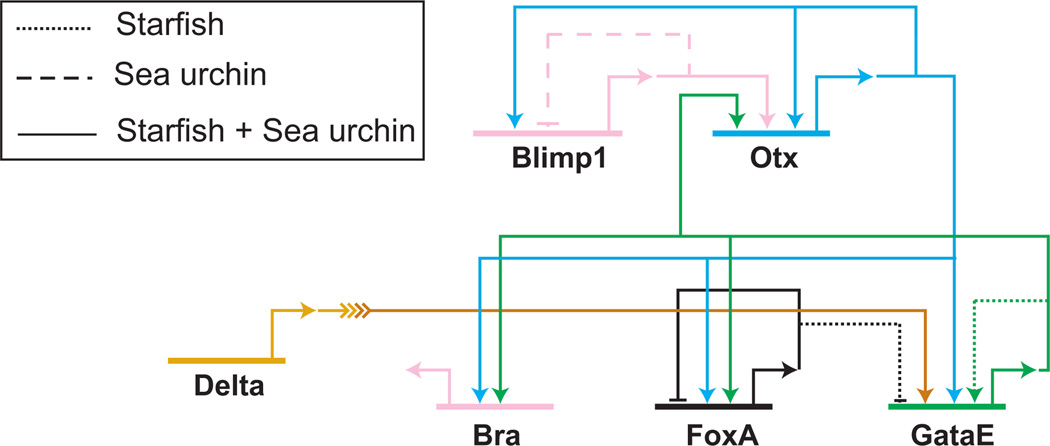
The initial subcircuit of the endoderm GRN involves a highly wired regulatory subcircuit of five genes. The five nodes in this subcircuit are blimp1, otx, gatae, foxA and brachyury, as shown in Fig. 4. The blimp1 gene, which is expressed under the control of TCF/β-catenin and Otx, produces an activating input into otxβ1/2, an alternative transcript of the otx gene which is exclusively zygotically expressed (Yuh et al., 2002). Otx expression is further on stabilized by additional positive feedback loops. The first feedback is provided by the activating autoregulation of otx. In addition to this, there are two Otx target genes encoding the transcription factors GataE and Brachyury, which both feed back to control the expression of otx ((Yuh et al., 2002); I. Peter and E.H. Davidson, unpublished results). The binding sites of three postulated inputs, Blimp1, Otx and GataE, have been identified in a cis-regulatory module driving otxβ expression (Yuh et al., 2004). Positive feedback loops are frequently observed motifs in GRNs since they assure stable gene expression after the initial input has disappeared. And indeed, otxβ and gatae expression are maintained long after blimp1 expression has been turned off in veg2-derived cells. Since gatae requires not only an activating input provided by Otx, but is also responsive to Delta/Notch signaling, its expression is at first restricted to the prospective NSMs and is turned on in endoderm precursor cells at the early mesenchyme blastula stage, when NSM precursor cells start to express delta. One of the gatae cis-regulatory modules has been shown to reflect the early expression pattern of the endogenous gene, when driving the expression of a GFP reporter gene (Lee et al., 2007). Interfering with the activity of either Otx or Su(H) only moderately affects reporter gene expression, whereas simultaneously blocking both regulatory inputs strongly reduces GFP expression. Otx and GataE together regulate the expression of two other regulatory genes in this subciruit, foxA and brachyury.
Fig. 4. Evolutionary conservation of a network subcircuit.
The regulatory linkages between blimp1, otx, bra, foxA and gataE have been analyzed in sea urchin and starfish embryos. Most of the regulatory interactions between these genes are identical and are shown by solid lines. Otx expression is first induced by Blimp1 and is later on maintained by two positive feedback loops: Otx and one of its target genes, GataE, provide an input into otx. Otx together with GataE control the expression of bra and foxA. Interactions that are unique in one of the species are shown as dashed (sea urchin) or dotted (starfish) lines. Not conserved regulatory linkages include the auto-repression of blimp1 in sea urchin and the autoregulation of gataE in starfish. (Modified from Hinmann et al., 2003).
The endoderm subcircuit contains all the information required for initializing a developmental program. Its activation is linked to the Wnt signaling input from the SM GRN via blimp1. It is also dependent on the NSM GRN, which provides the Delta/Notch signaling input into gataE. Furthermore, this subcircuit contains a stabilizing function in the form of direct positive feedback loops into otx mediated by cis-regulatory target sites. Two of its components, namely GataE and Brachyury are responsible for the activation of later endoderm regulatory factors (I. Peter and E.H. Davidson, unpublished results). And finally, FoxA prevents the activation of the NSM GRN in the endoderm by repressing gcm (Oliveri et al., 2006). It is not surprising that each of these five regulatory proteins is absolutely required for endoderm development: interfering with the expression of each of them inhibits the formation of a gut.
Further evidence for the significance of this subcircuit comes from evolutionary studies using the starfish Asterina miniata (Hinman et al., 2003). The surprising result was that the architecture of the regulatory linkages between the five network nodes described above is identical in sea urchin and starfish, including the all-important gatae-otx feedback loop, despite 500 million years of independent evolution. Most other parts of the GRN are however unique to each of these organisms, as shown in Fig. 4 (Hinman et al., 2003; Hinman and Davidson, 2007). For example, blimp1 gene expression is turned off by an autoregulatory loop in sea urchin but not in starfish. In starfish, on the other hand, the positive feedback loop between otxβ and gatae is further amplified by an autoregulatory activity of GataE that is lacking in the sea urchin. As this example shows, studying the evolution of network structure provides an approach to the modularity of GRNs.
A module driving dynamic gene expression patterns
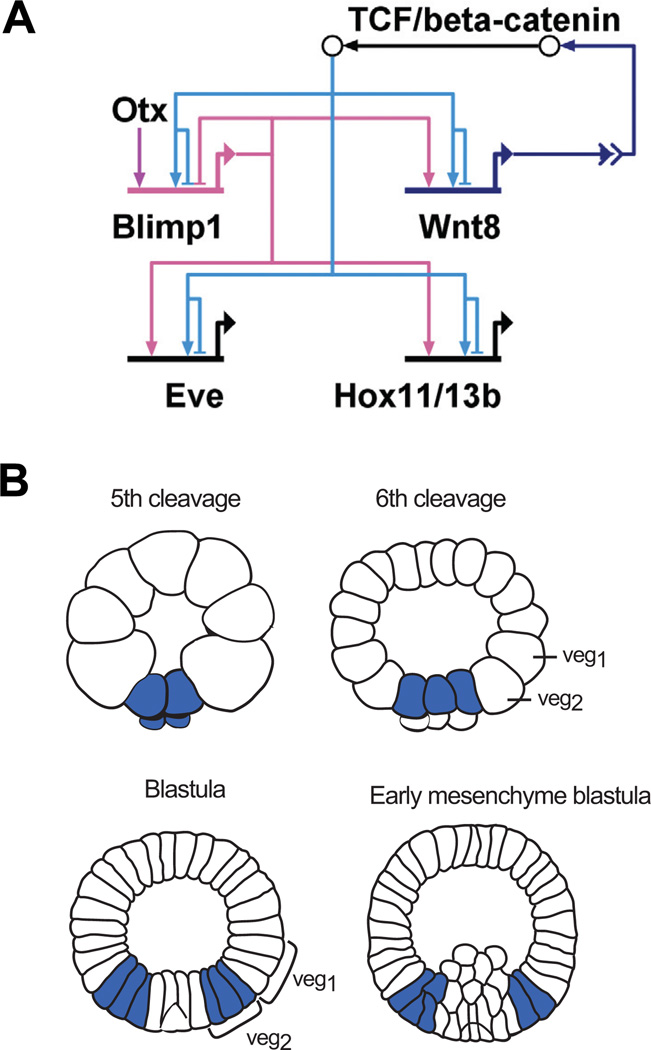
The sea urchin GRN also contains a subcircuit which is active in all endomesoderm cell lineages, albeit at different time points. This subcircuit, which is shown in Fig. 5A, starts with the activation of blimp1 and wnt8 gene expression by β-catenin/Tcf. Cis-regulatory analyses have shown that blimp1 expression requires the activating input of two transcription factors, Otx and Tcf, and that these factors function in an AND logic gate (Smith et al., 2007). Blimp1 further activates the expression of wnt8. The binding sites for both Blimp1 and Tcf have been identified in wnt8 cis-regulatory modules and functionality confirmed by mutational analysis (Minokawa et al., 2005). The expression of wnt8 is therefore controlled by two indirect positive feedback loops. The first feedback loop consists of β- catenin/Tcf activating wnt8 expression. This in turn leads to the secretion of Wnt8, the activation of the Wnt signaling pathway and further nuclearization of β-catenin (β- catenin/Tcf-wnt8). Since the second step in this loop is the result of a signal transduction pathway which involves many other molecules, we refer to this circuit structure as an indirect feedback loop. The second positive feedback loop consists of β-catenin/Tcf-wnt8-blimp1, including blimp1 as a target of β-catenin/Tcf and an activator of wnt8 expression.
Fig. 5. Structure and output of a subcircuit driving dynamic gene expression.
(A) Model of a subcircuit which results in a dynamic gene expression pattern. Two positive feedback loops induce the activation of this subcircuit in adjacent cells: β- catenin/TCF activates the expression of wnt8 and blimp1. Blimp1 provides an additional input into wnt8. Reception of the Wnt signal leads to nuclearization of β-catenin and activation of this subcircuit in neighboring cells. Blimp1 and β-catenin/TCF are both required to induce the expression of eve and hox11/13b. The expression of blimp1 is turned off by its own gene product, leading to inactivation of the whole subcircuit. (B) Schematic presentation of wnt8 expression pattern. Wnt8 is expressed in the micromere lineage at 5th cleavage, from where it expands to veg2-and veg1-derived cells. Wnt8 is no longer expressed in the SM precursors at blastula stage and in NSM precursors at early mesenchyme blastula. (Modified from Smith et al., 2007).
The secretion of signaling ligands can have two regulatory effects: It can result in the synchronization of regulatory states within cells of the same domain (“community effect”) or provide a regulatory input to cells of an adjacent territory (induction). In fact, it has been shown that the nuclearization of β-catenin and the expression of wnt8 and blimp1 are first induced in the micromere lineage and later expand to the other lineages of the endomeso-derm, as shown for wnt8 in Fig. 5B (Logan et al., 1999; Wikramanayake et al., 2004; Minokawa et al., 2005; Livi and Davidson, 2006). However, only a few hours after being activated, this subcircuit is turned off in each cell lineage, again first in the micromere lineage and then in NSM precursors. The mechanism has been shown to depend on the autoregulatory activity of blimp1. Thus Blimp1 binding sites within the blimp1 cis-regulatory region are responsible for the down regulation of blimp1 expression in mesoderm precursor cells (Smith et al., 2007). Deletion of these Blimp1 binding sites in a construct consisting of blimp1 cis-regulatory sequences driving blimp1 cDNA resulted in sustained expression not only of blimp1 but also of wnt8 in prospective mesenchyme cells. Furthermore, Blimp1 and β-catenin/Tcf together regulate the expression of target genes including eve and hox11/13b (Fig. 5A; (Smith et al., 2008)). The binding sites for both Blimp1 and Tcf have been identified in eve cis-regulatory sequences and shown to introduce an AND logic function, since mutation of the binding sites for either of these factors abolishes reporter gene expression.
The special architecture of these regulatory linkages leads to the activation of this subcircuit in successive tiers of cells, and its inactivation where it had been active earlier, resulting in an expanding torus of gene expression. The genomic cis-regulatory sequences thus directly encode a modular network subcircuit which gives rise to dynamic gene expression patterns. The most important ultimate consequence of this subcircuit is that it controls successive domains of Notch signaling (Smith and Davidson, 2008): following delta expression in the SM and Notch signaling in the NSM as discussed above, the delta gene is activated in the NSM and now sends its signal to the endoderm. The mechanism is that the blimp1-wnt8 subcircuit also controls hesC expression, and when hesC is made to turn off in the NSM the delta gene is allowed to go on. In addition the same subcircuit controls transcription of the notch gene itself. There are a number of consequences of these relations in addition to direct determination of the progressive spatial domains of Notch signaling (Smith and Davidson, 2008). For one thing they ensure that as development proceeds, wherever in the endomesoderm wnt8 is expressed, delta is not, and vice versa; for another, wherever Notch signaling is occurring, the delta gene is silent. All of these spatial signaling functions are directly encoded in the regulatory genome, and all therefore contribute to the precise activation of the NSM and endodermal GRNs in space and time.
Conclusions
We have discussed here some of the subcircuits underlying the specification program of three different endomesoderm cell lineages in the sea urchin embryo. Although there are obvious differences, both at the level of cell lineage development as well as at the level of network structure, these subcircuits share some general principles.
Control of signaling
Signaling between adjacent cells deriving from the same progenitor cell introduces a switch function to enable closely related regulatory states to activate different GRNs. Signaling processes link the SM and NSM GRNs (delta-Su(H)- gcm); the NSM and endoderm GRNs (delta-Su(H)-gataE); and all three GRNs by the superimposed (β-catenin/Tcf-wnt8- blimp1) subcircuit just discussed.
Stabilization
The primary inputs are rapidly processed and positive feedback loops then stabilize the regulatory state and uncouple the specification process from the primary inputs. These are typically transient. These positive feedback loops are either direct, as in the feedback between erg, hex, and tgif in the SM GRN, and between otx and gatae in the pre-endoderm GRN; or they are intercellular and indirect as in the wnt8 – β-catenin loop.
Exclusion of alternative cell fates
Regulatory states are very similar in territories which have been physically separated only a few cell cleavages earlier. Therefore, some mechanisms have to be introduced to avoid the acquisition of alternative cell fates (Oliveri and Davidson, 2007). Network subcircuits frequently repress regulatory genes associated with different specification programs, in order to keep their activation mutually exclusive, and several examples are discussed above.
Expression of differentiation genes
At the downstream periphery of every GRN are the differentiation genes, which encode for proteins that will execute the functions of differentiated cells. Some of the regulatory factors contributing to a specification GRN directly feed into the regulation of the differentiation genes, irrespective of their position in the network.
Each of these features of GRNs is the result of regulatory interactions encoded in the regulatory genome and each contributes to the control of patterning. The list of network principles will certainly be extended in the future as more GRN architectures become deciphered.
Acknowledgments
We thank Andrew Ransick for helpul discussion and critical reading of the manuscript. Research was funded by NIH grant HD37105; I.P. was supported by a fellowship from the Swiss National Science Foundation.
Abbreviations used in this paper
- GRN
gene regulatory network
- NSM
non-skeletogenic mesenchyme
- QPCR
quantitative PCR
- SM
skeletogenic mesenchyme
- WMISH
whole-mount in situ hybridization
References
- Arenas-Mena C, Cameron RA, Davidson EH. Hindgut specification and cell-adhesion functions of sphox11/13b in the endoderm of the sea urchin embryo. Dev. Growth Differ. 2006;48:463–472. doi: 10.1111/j.1440-169X.2006.00883.x. [DOI] [PubMed] [Google Scholar]
- Ben-Tabou De-Leon S, Davidson EH. Gene regulation: Gene control network in development. Annu. Rev. Biophys. Biomol. Struct. 2007;36:191–212. doi: 10.1146/annurev.biophys.35.040405.102002. [DOI] [PubMed] [Google Scholar]
- Davidson EH. Genomic regulatory systems: Development and evolution. San Diego: Acdemic Press; 2001. [Google Scholar]
- Davidson EH. The regulatory genome. Gene regulatory networks in development and evolution. San Diego, CA.: Academic Press/Elsevier; 2006. [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, et al. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- Ettensohn CA, Kitazawa C, Cheers MS, Leonard JD, Sharma T. Gene regulatory networks and developmental plasticity in the early sea urchin embryo: Alternative deployment of the skeletogenic gene regulatory network. Development. 2007;134:3077–3087. doi: 10.1242/dev.009092. [DOI] [PubMed] [Google Scholar]
- Gao F, Davidson EH. Transfer of a large gene regulatory apparatus to a new developmental address in echinoid evolution. Proc. Natl. Acad. Sc.i U. S. A. 2008;105:6091–6096. doi: 10.1073/pnas.0801201105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman VF, Davidson EH. Evolutionary plasticity of developmental gene regulatory network architecture. Proc. Natl. Acad. Sci. U. S. A. 2007;104:19404–19409. doi: 10.1073/pnas.0709994104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman VF, Nguyen AT, Cameron RA, Davidson EH. Developmental gene regulatory network architecture across 500 million years of echinoderm evolution. Proc. Natl. Acad. Sci. U.S.A. 2003;100:13356–13361. doi: 10.1073/pnas.2235868100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoerstadius S. The mechanics of sea urchin development, studied by operative methods. Rev. Cambridge Phil. Soc. 1939;14:132–179. [Google Scholar]
- Howard-Ashby M, Materna SC, Brown CT, Chen L, Cameron RA, Davidson EH. Gene families encoding transcription factors expressed in early development of strongylocentrotus purpuratus. Dev. Biol. 2006a;300:90–107. doi: 10.1016/j.ydbio.2006.08.033. [DOI] [PubMed] [Google Scholar]
- Howard-Ashby M, Materna SC, Brown CT, Chen L, Cameron RA, Davidson EH. Identification and characterization of homeobox transcription factor genes in strongylocentrotus purpuratus, and their expression in embryonic development. Dev. Biol. 2006b;300:74–89. doi: 10.1016/j.ydbio.2006.08.039. [DOI] [PubMed] [Google Scholar]
- Kirchhamer CV, Yuh CH, Davidson EH. Modular cis-regulatory organization of developmentally expressed genes: Two genes transcribed territorially in the sea urchin embryo, and additional examples. Proc. Natl. Acad. Sci. U. S. A. 1996;93:9322–9328. doi: 10.1073/pnas.93.18.9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PY, Nam J, Davidson EH. Exclusive developmental functions of gatae cis- regulatory modules in the strongylocentrorus purpuratus embryo. Dev. Biol. 2007;307:434–445. doi: 10.1016/j.ydbio.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard JD, Ettensohn CA. Analysis of dishevelled localization and function in the early sea urchin embryo. Dev. Biol. 2007;306:50–65. doi: 10.1016/j.ydbio.2007.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M, Davidson EH. Gene regulatory networks for development. Proc. Natl. Acad. Sci. USA. 2005;102:4936–4942. doi: 10.1073/pnas.0408031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- Livi CB, Davidson EH. Expression and function of blimp1/krox, an alternatively transcribed regulatory gene of the sea urchin endomesoderm network. Dev. Biol. 2006;293:513–525. doi: 10.1016/j.ydbio.2006.02.021. [DOI] [PubMed] [Google Scholar]
- Livingston BT, Killian CE, Wilt F, Cameron A, Landrum MJ, Ermolaeva O, Sapojnikov V, Maglott DR, Buchanan AM, Ettensohn CA. A genome-wide analysis of biomineralization-related proteins in the sea urchin strongylocentrotus purpuratus. Dev. Biol. 2006;300:335–348. doi: 10.1016/j.ydbio.2006.07.047. [DOI] [PubMed] [Google Scholar]
- Logan CY, Miller JR, Ferkowicz MJ, Mcclay DR. Nuclear beta-catenin is required to specify vegetal cell fates in the sea urchin embryo. Development. 1999;126:345–357. doi: 10.1242/dev.126.2.345. [DOI] [PubMed] [Google Scholar]
- Loose M, Patient R. A genetic regulatory network for Xenopus mesendoderm formation. Dev. Biol. 2004;271:467–478. doi: 10.1016/j.ydbio.2004.04.014. [DOI] [PubMed] [Google Scholar]
- Materna SC, Howard-Ashby M, Gray RF, Davidson EH. The c2h2 zinc finger genes of strongylocentrotus purpuratus and their expression in embryonic development. Dev. Biol. 2006;300:108–120. doi: 10.1016/j.ydbio.2006.08.032. [DOI] [PubMed] [Google Scholar]
- Minokawa T, Wikramanayake AH, Davidson EH. Cis-regulatory inputs of the wnt8 gene in the sea urchin endomesoderm network. Dev. Biol. 2005;288:545–558. doi: 10.1016/j.ydbio.2005.09.047. [DOI] [PubMed] [Google Scholar]
- Okazaki K. Spicule formation by isolated micromeres of the sea urchin embryo. Amer. Zool. 1975;15:567–581. [Google Scholar]
- Oliveri P, Carrick DM, Davidson EH. A regulatory gene network that directs micromere specification in the sea urchin embryo. Dev. Biol. 2002;246:209–228. doi: 10.1006/dbio.2002.0627. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Davidson EH. Gene regulatory network controlling embryonic specification in the sea urchin. Curr. Opin. Genet. Dev. 2004;14:351–360. doi: 10.1016/j.gde.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Davidson EH. Development. Built to run not fail. Science. 2007;315:1510–1511. doi: 10.1126/science.1140979. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Davidson EH, Mcclay DR. Activation of pmar1 controls specification of micromeres in the sea urchin embryo. Dev. Biol. 2003;258:32–43. doi: 10.1016/s0012-1606(03)00108-8. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Tu Q, Davidson EH. Global regulatory logic for specification of an embryonic cell lineage. Proc. Natl. Acad. Sci. U. S. A. 2008;105:5955–5962. doi: 10.1073/pnas.0711220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveri P, Walton KD, Davidson EH, Mcclay DR. Repression of mesodermal fate by foxa, a key endoderm regulator of the sea urchin embryo. Development. 2006;133:4173–4181. doi: 10.1242/dev.02577. [DOI] [PubMed] [Google Scholar]
- Primus AE. Regional specification in the early embryo of the brittle star ophiopholis aculeata. Dev. Biol. 2005;283:294–309. doi: 10.1016/j.ydbio.2005.04.022. [DOI] [PubMed] [Google Scholar]
- Range RC, Venuti JM, Mcclay DR. Lvgroucho and nuclear beta-catenin functionally compete for tcf binding to influence activation of the endomesoderm gene regulatory network in the sea urchin embryo. Dev. Biol. 2005;279:252–267. doi: 10.1016/j.ydbio.2004.12.023. [DOI] [PubMed] [Google Scholar]
- Ransick A, Davidson EH. A complete second gut induced by transplanted micromeres in the sea urchin embryo. Science. 1993;259:1134–1138. doi: 10.1126/science.8438164. [DOI] [PubMed] [Google Scholar]
- Ransick A, Davidson EH. Micromeres are required for normal vegetal plate specification in sea urchin embryos. Development. 1995;121:3215–3222. doi: 10.1242/dev.121.10.3215. [DOI] [PubMed] [Google Scholar]
- Ransick A, Davidson EH. Cis-regulatory processing of notch signaling input to the sea urchin glial cells missing gene during mesoderm specification. Dev. Biol. 2006;297:587–602. doi: 10.1016/j.ydbio.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Revilla-i-Domingo R, Minokawa T, Davidson EH. R11: A cis-regulatory node of the sea urchin embryo gene network that controls early expression of spdelta in micromeres. Dev. Biol. 2004;274:438–451. doi: 10.1016/j.ydbio.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Revilla-i-Domingo R, Oliveri P, Davidson EH. A missing link in the sea urchin embryo gene regulatory network: Hesc and the double-negative specification of micromeres. Proc. Natl. Acad. Sci. USA. 2007;104:12383–12388. doi: 10.1073/pnas.0705324104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo F, Fernandez-Serra M, Squarzoni P, Archimandritis A, Arnone MI. Identification and developmental expression of the ets gene family in the sea urchin (strongylocentrotus purpuratus) . Dev. Biol. 2006;300:35–48. doi: 10.1016/j.ydbio.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Sherwood DR, Mcclay DR. Identification and localization of a sea urchin notch homologue: Insights into vegetal plate regionalization and notch receptor regulation. Development. 1997;124:3363–3374. doi: 10.1242/dev.124.17.3363. [DOI] [PubMed] [Google Scholar]
- Sherwood DR, Mcclay DR. Lvnotch signaling mediates secondary mesenchyme specification in the sea urchin embryo. Development. 1999;126:1703–1713. doi: 10.1242/dev.126.8.1703. [DOI] [PubMed] [Google Scholar]
- Smith J, Davidson EH. Gene regulatory network subcircuit controlling a dynamic spatial pattern of signaling in the sea urchin embryo. Proc. Natl. Acad. Sci. USA. 2008;105:20089–20094. doi: 10.1073/pnas.0806442105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Kraemer E, Liu H, Theodoris C, Davidson E. A spatially dynamic cohort of regulatory genes in the endomesodermal gene network of the sea urchin embryo. Dev. Biol. 2008;313:863–875. doi: 10.1016/j.ydbio.2007.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Theodoris C, Davidson EH. A gene regulatory network subcircuit drives a dynamic pattern of gene expression. Science. 2007;318:794–797. doi: 10.1126/science.1146524. [DOI] [PubMed] [Google Scholar]
- Stathopoulos A, Levine M. Genomic regulatory networks and animal development. Dev. Cell. 2005;9:449–462. doi: 10.1016/j.devcel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Stathopoulos A, Van Drenth M, Erives A, Markstein M, Levine M. Whole-genome analysis of dorsal-ventral patterning in the drosophila embryo. Cell. 2002;111:687–701. doi: 10.1016/s0092-8674(02)01087-5. [DOI] [PubMed] [Google Scholar]
- Sweet HC, Gehring M, Ettensohn CA. Lvdelta is a mesoderm-inducing signal in the sea urchin embryo and can endow blastomeres with organizer-like properties. Development. 2002;129:1945–1955. doi: 10.1242/dev.129.8.1945. [DOI] [PubMed] [Google Scholar]
- Sweet HC, Hodor PG, Ettensohn CA. The role of micromere signaling in notch activation and mesoderm specification during sea urchin embryogenesis. Development. 1999;126:5255–5265. doi: 10.1242/dev.126.23.5255. [DOI] [PubMed] [Google Scholar]
- Tu Q, Brown CT, Davidson EH, Oliveri P. Sea urchin forkhead gene family: Phylogeny and embryonic expression. Dev. Biol. 2006;300:49–62. doi: 10.1016/j.ydbio.2006.09.031. [DOI] [PubMed] [Google Scholar]
- Weitzel HE, Illies MR, Byrum CA, Xu R, Wikramanayake AH, Ettensohn CA. Differential stability of beta-catenin along the animal-vegetal axis of the sea urchin embryo mediated by dishevelled. Development. 2004;131:2947–2956. doi: 10.1242/dev.01152. [DOI] [PubMed] [Google Scholar]
- Wikramanayake AH, Peterson R, Chen J, Huang L, Bince JM, Mcclay DR, Klein WH. Nuclear beta-catenin-dependent wnt8 signaling in vegetal cells of the early sea urchin embryo regulates gastrulation and differentiation of endoderm and mesodermal cell lineages. Genesis. 2004;39:194–205. doi: 10.1002/gene.20045. [DOI] [PubMed] [Google Scholar]
- Yuh CH, Brown CT, Livi CB, Rowen L, Clarke PJ, Davidson EH. Patchy interspecific sequence similarities efficiently identify positive cis-regulatory elements in the sea urchin. Dev. Biol. 2002;246:148–161. doi: 10.1006/dbio.2002.0618. [DOI] [PubMed] [Google Scholar]
- Yuh CH, Dorman ER, Howard ML, Davidson EH. An otx cis-regulatory module: A key node in the sea urchin endomesoderm gene regulatory network. Dev. Biol. 2004;269:536–551. doi: 10.1016/j.ydbio.2004.02.025. [DOI] [PubMed] [Google Scholar]