Abstract
Chondrocyte mechanotransduction is the process by which cartilage cells transduce mechanical loads into biochemical and biological signals. Previous studies have identified several pathways by which chondrocytes transduce mechanical loads, yet a general understanding of which signals are activated and in what order remains elusive. This study was performed to identify candidate mediators of chondrocyte mechanotransduction using SW1353 chondrocytes embedded in physiologically stiff agarose. Dynamic compression was applied to cell-seeded constructs for 0–30 minutes, followed immediately by whole-cell metabolite extraction. Metabolites were detected via LC-MS, and compounds of interest were identified via database searches. We found several metabolites which were statistically different between the experimental groups, and we report the detection of 5 molecules which are not found in metabolite databases of known compounds indicating potential novel molecules. Targeted studies to quantify the response of central energy metabolites to compression found a transient increase in the ratio of NADP+ to NADPH and a continual decrease in the ratio of GDP to GTP, suggesting a flux of energy into the TCA cycle. These data are consistent with the remodeling of cytoskeletal components by mechanically induced signaling, and add substantial new data to a complex picture of how chondrocytes transduce mechanical loads.
Introduction
The field of cellular mechanotransduction seeks to identify mechanisms by which cells respond to their mechanical loading environments. Mammalian cells have the ability to respond to a variety of loads by altering signaling pathways in a diverse set of cells and tissues [1,2,3,4]. These and other studies demonstrate the ability of mammalian cells to respond to exogenous mechanical loading. However, knowledge of the mechanisms by which cells sense and respond to loading remains incomplete.
Articular cartilage is the smooth tissue lining the surfaces of articulating joints (e.g. knee) which deforms during physiological activity [5,6]. Articular chondrocytes, the cells of articular cartilage, respond to applied loading via multiple pathways, including activation of GTPase signaling via Rho-A and ROCK [7,8]. Osteoarthritis (OA) is a major medical problem that involves deterioration of articular cartilage [9], and osteoarthritic chondrocytes demonstrate differences in mechanotransduction compared with healthy chondrocytes. For example, cyclical strain reduces AKT phosphorylation in OA chondrocytes [10], and OA chondrocytes fail to produce sulfated glycosaminoglycans (sGAG) in response to load whereas normal chondrocytes exhibit loading-induced increases in sGAG production [11]. In the present study, we provide high-dimensional data regarding changes in expression levels and flux of thousands of metabolites (i.e. cytosolic molecules smaller than ~1000 Da) in response to highly controlled compression of SW1353 chondrocytes [12].
Chondrocytes within articular cartilage are surrounded by a pericellular matrix (PCM) which is composed primarily of Type VI collagen and other proteins [13,14,15]. The chondrocyte PCM has a stiffness of ~25–200 kPa [16] which is diminished in OA [17]. Previous studies of chondrocyte mechanotransduction have utilized various three dimensional culture methods [18,19,20,21,22] most with stiffness values of 5–10 kPa or less, which are markedly lower than those present in the human pericellular matrix [16,17]. The present study sought to build on previous methodology [23,24] by using high concentrations of agarose to support the chondrocytes and form a high-stiffness gel capable of applying physiological deformation to chondrocytes.
Previous research indicates that central energy metabolism is altered both in inflammation and OA, including the balance between glycolysis and oxidative phosphorylation [25,26]. Energy metabolism may be affected by loading because activation of AMP-activated protein kinase can prevent catabolism induced by mechanical injury [27]. Based on these and other data, we hypothesized that dynamic compression within the physiological range will increase glycolytic metabolism to maintain the environment of the PCM. As a first step in evaluating this hypothesis, we conducted the present study to develop and demonstrate methods for targeted quantification of metabolites associated with the central metabolism of SW1353 chondrosarcoma chondrocytes in response to applied dynamic compression in the physiological range. To our knowledge, this is the first application of either targeted or untargeted metabolomics studying chondrocyte mechanotransduction.
The objective of this study was to use targeted and untargeted metabolomics to identify candidate mediators of chondrocyte mechanotransduction. We identified loading-induce changes in ~4000 metabolites in untargeted studies and measured quantitative changes in 36 targeted metabolites relevant to central metabolism and protein production. From this untargeted metabolomics detection, 54 novel mediators of chondrocyte mechanotransduction were identified. These data define the functional response of chondrocytes to applied loading. Future studies to build on these results will aim to develop a more detailed systems understanding of chondrocyte mechanotransduction.
Materials and Methods
Chondrocyte Culture and Encapsulation
Human SW1353 chondrosarcoma cells were cultured in 5% CO2 in DMEM with 10% fetal bovine serum and antibiotics (10,000 I.U./mL penicillin and 10,000 μg/mL streptomycin). For encapsulation, cells were trypsinized, counted, and resuspended in media at 11X. Agarose/PBS solution was prepared using low-gelling-temperature agarose (Sigma: Type VII-A A0701) at 1.1X of desired final concentration and placed into a water bath at 40°C. The cell-suspension was added to the agarose with vortexing to distribute the cells throughout the liquid hydrogel. Gels were subsequently cast in an anodized aluminum mold for 5 minutes at 23°C with diameter of 7mm and height of 12.7 mm [28]. Cell-seeded agarose constructs were removed from the molds and cultured in antibiotic free media for 72 hours at 37°C under 5% CO2. These methods have been shown to provide uniform compressive deformations [29] to chondrocytes by modeling the stiffness of the pericellular matrix [17]. The stiffness of the pericellular matrix provides in vivo deformations to the relatively less-stiff chondrocytes [30], and the advantage of this approach is that it provides observable cellular deformations while providing homogeneous, uniaxial unconfined compression as the defined mechanical stimulus[29].
Mechanical Stimulation
Cell-seeded agarose gels were subjected to cyclic compression via a custom made bioreactor for 0, 15, and 30 minutes. Dynamic unconfined compression was applied between impermeable platens in culture media at a frequency of 1.1 Hz [31] with an average compressive strain of 5% and an amplitude of 1.9% based on the initially measured height of 12.7 ± 0.1 mm. This sampling interval is based on previous observations of changes in central energy metabolism within a 30 minute timescale [32]. Physiological conditions (culture media at 37°C, 5% CO2) were maintained through the duration of the tests. To assess specificity of the mechanobiological response, unloaded control samples were placed in the bioreactor without deformational loading and analyzed for each timepoint.
Metabolite Extraction
After each time point samples were removed from the bioreactor, immediately wrapped in sterile foil and frozen in liquid nitrogen. Samples were then placed inside individual wells of a custom-made frozen aluminum mold for pulverization [28]. From here each sample was crushed using a sterilized stainless steel platen and a ballpeen hammer. Crushed gel particulate was then collected into 2 mL microcentrifuge tubes. Metabolites were extracted by adding 1 mL of a 70:30 solution of Methanol:Acetone and vortexing every 4–5 minutes for twenty minutes. Samples were extracted further at −20°C overnight. Solid content was pelleted by centrifugation at 13,000 rpm for 10 minutes at 4°C. The supernatant was placed into new 1.6 mL microcentrifuge tubes where solvent was removed via speedvac for six hours. The dried samples were then resuspended in 100 μL of a 50:50 Water:Acetonitrile solution for metabolomics analysis.
Untargeted and Targeted LC-MS
Detection of metabolites was performed via HPLC separation with ESI-MS (electrospray mass spectrometry) detection in the Montana State University Mass Spectrometry Core Facility [33,34,35]. HPLC was performed with an aqueous normal-phase, hydrophilic interaction chromatography (ANP/HILIC) HPLC column. A Cogent Diamond Hydride Type-C column with 4 μm particles and dimensions of 150 mm length and 2.1 mm diameter was used with an Agilent 1290 HPLC system. The column was maintained at 50 °C with a flow rate of 600 μL/min. Chromatography was as follows: solvent consisted of H2O with 0.1% (v/v) formic acid for channel “A” and acetonitrile with 0.1% formic acid for channel “B”. Following column equilibration at 95% B, the sample was injected via autosampler, and the column was flushed for 2.0 min to waste. From 2.0 min to the end of the run, the column eluant was directed to the MS source. From 2.0 min to 12.5 min, the gradient was linearly ramped from 95% to 25% B. From 12.5 to 13.5 min the column was held isocratically at 25% B, and from 13.5 to 15 minutes the column was re-equilibrated with 95% B. Blank solvent samples were run following each sample.
The mass spectrometer used was an Agilent 6538 Q-TOF with dual-ESI source. Resolution is ~20,000 and accuracy is ~5 ppm. Source parameters were: drying gas 12 L/min, nebulizer 60 psi, capillary voltage 3500 V, capillary exit 120 V. Spectra were collected in positive mode from 50 to 1000 m/z at a rate of 1 Hz. Quality control testing including mass accuracy calibration of all instruments is performed regularly and documented by the Mass Spectrometry Core Facility staff as part of routine operation.
Metabolites known to be involved in central metabolism [32,36] were targeted for LC-MS analysis. Using the isotopic distributions of these targeted masses (Agilent Technologies), a list of H+ and Na+ adducts was used to create 20 ppm mass windows for each ion, and pilot data was scanned to determine the range of retention times for each ion based on data from analytical standards (Biolog, Hayward, CA) run by the mass spectrometry core facility for 15 of the 36 targeted metabolites. These standard metabolites were run by Core staff under controlled conditions, and target validation ensured retention times within 0.1 min of core values for samples run on identical instruments under identical conditions. Targeted metabolite intensity was defined as the sum of the intensities of the isotopes of the ions and adducts associated with each metabolite as determined by the Quantitative Analysis package within MassHunter Workstation B.04.00 (Agilent Technologies).
Data Processing
Data analysis involved multiple software packages used to process the raw LC-MS data for feature identification, quantification, and metabolite identification (Figure S1). Raw LC-MS scans were converted to mzXML files using Agilent MassHunter and processed in MZmine2 [37] for the untargeted analysis. Unrefined lists of detected metabolites (i.e. detected mass values) with corresponding intensities were generated by aligning all LC-MS scans. These unrefined lists resulted in the identification of approximately 25K independent m/z values for positive mode and 18K m/z values for negative mode scans.
Using established methods [38], filtered datasets were generated as follows. Chromatograms were built using centroidal mass detection with a minimum signal level of 1000, minimum timespan of 0.02 seconds, minimum peak height of 1000, and an m/z tolerance of 0.05. Peak deconvolution was performed with a chromatographic threshold of 85%, search minimum in retention time of 0.03 seconds, minimum relative height of 5%, minimum absolute height of 10000, and minimum ratio of top/edge of 1.0. The chromatograms were then normalized by retention times with a retention time tolerance of 0.25 min and a minimum standard intensity of 1000. Chromatograms were aligned and a duplicate peak filter was applied with an m/z tolerance of 0.1 and a retention time difference maximum of 1 minute. These refined lists were used for statistical analysis and candidate metabolite identification. Intensity was quantified via peak height in the total ion intensity chromatogram.
Data Analysis and Candidate Selection
The experimental procedures were repeated for n = 5 independent samples. To assess the biological effects of physiological loading, compressed samples were compared to unloaded controls. The cell-seeded agarose gels that received no mechanical stimulation (0 minute time point extraction) acted as the unloaded controls (UC), while cell-seeded agarose gels that received either 15 or 30 minutes of mechanical stimulation made up the dynamically compressed groups, DC15 and DC30 respectively. Data analysis started with the unrefined lists generated in MZmine2. We defined detected masses as those present in the majority of samples (i.e. ≥ 3 samples). To minimize false positives associated with multiple comparisons, conservative statistical methods were used to make comparisons between groups. Comparisons were performed using t-tests [39] and p-value corrections using a standard false discovery rate (FDR) calculation [39]. For metabolites detected in one group (e.g. DC15) but not in another, statistical comparisons were enabled by the use of small random values for the non-detected intensities. These small values were < 2% of the minimally detected value and <0.04% of the median value, and were required to calculate the appropriate FDR. Two comparisons were performed: UC vs. DC15 and UC vs. DC30.
Ordination techniques were subsequently used to help identify candidate metabolites from the untargeted data for which statistical significance was not achieved. The point of this analysis was to generate a list of potentially important metabolites for future studies. Data matrices were generated with data binned according to masses and time points. Only masses that were detected in at least three of five time point replicates were analyzed. Principal components analysis was performed using the rda function of the Vegan package in R statistical software [40]. All masses were standardized to unit variance and displayed using the biplot function (scaling=−1). This approach allowed us to visually identify candidate mediators with relatively large magnitudes that were responsible for differentiating between 0, 15, and 30 minutes of loading.
In order to assess metabolite flux over the 30 minute experimental timecourse, Pearson’s correlation coefficients were determined using loading time (0, 15, or 30 minutes) as the independent variable and the intensity of each detected metabolite as the dependent variable. Significant positive correlations indicated metabolite accumulation, and negative correlations indicated metabolite consumption. Candidate mediators were defined as the twenty largest (i.e. accumulated) and twenty smallest (i.e. depleted) statistically significant correlations. Targeted metabolite profiles were analyzed by cluster and correlation analyses. Additionally, the median ratios of ATP:ADP, NADP+:NADPH, NAD+:NADH, and GDP:GTP were calculated as a function of time to assess relative changes in energy metabolism.
Compound Identification
Putative compound identifications were made by comparing the metabolite mass to charge (m/z) ratio to previous results using the METLIN and HMDB databases, which contain over 80,000 identifiable metabolites [41,42] using a mass tolerance of 20 ppm. METLIN parameters were charged masses and adducts of either +1H+ or +1Na+. Compounds with LipidMAPS identifications [43] were designated as human unless detected in a non-human species at the time of database search.
Results
These data provide an initial systems-level view of the cytosolic metabolite profile of human chondrocytes in response to applied compression. We defined 54 novel compounds as candidate mediators (Figure 1B, Table S1) of chondrocyte mechanotransduction, 40 from correlation analysis of untargeted metabolites and 14 from ordination analysis of targeted metabolites. No identification was possible for 5 of the 54 (11%) mediator masses based on database searches, indicating that these may be novel compounds. The remaining 49 (89%) masses map to a total of 180 metabolites due to isobaric redundancy and structural isomers (Table S1).
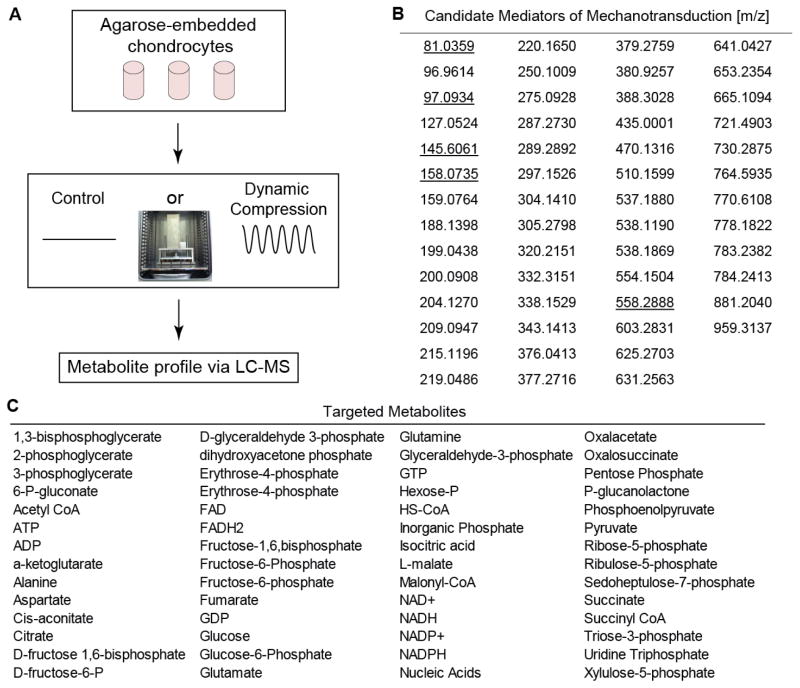
Figure 1.
(A) Schematic of Experimental Methods. SW1353 Chondrocytes were encapsulated in 4.5% agarose (stiffness ~35 kPa), cultured for 72 hours, and exposed to 0, 15, or 30 minutes of dynamic compression. Metabolites were extracted and analyzed via LC-MS. Inset shows loading apparatus for applying unconfined compression. (B) 59 metabolites were identified as candidate mediators of chondrocyte mechanotransduction in the untargeted studies. These were detected by either significant correlation with time of loading or ordination based on principal components analysis. Underlined values were not detected in database searches and may indicate novel metabolites. Note compound identifications available in Table S1.
Untargeted Metabolomics
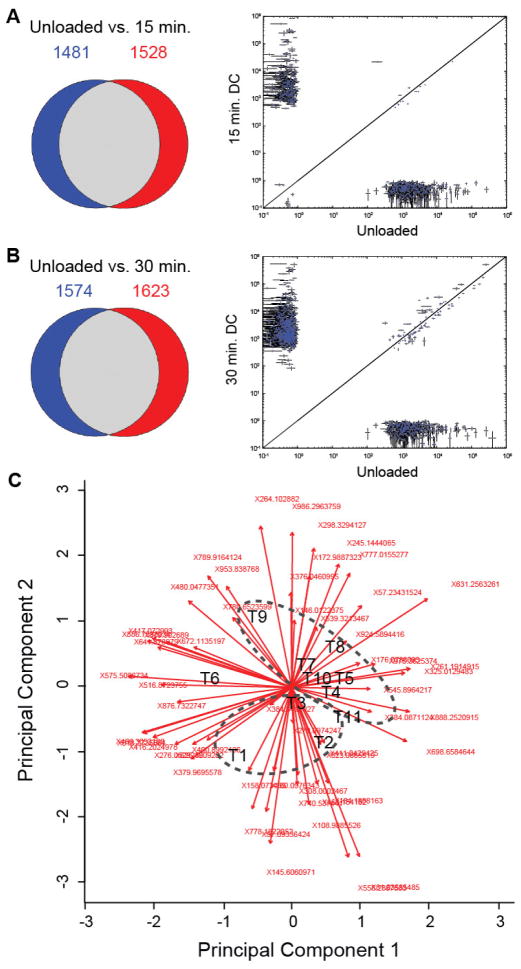
Untargeted studies detected 2438 to 3211 individual metabolites per sample. 1481 metabolites were detected in unloaded samples that were not detected following 15 minutes of loading, whereas 1528 unique metabolites were detected following 15 minutes of loading that were not present in unloaded control samples (Figure 2A). 1574 unique metabolites were detected in unloaded samples that were not detected following 30 minutes of loading, whereas 1623 metabolites were detected in samples subjected to 30 minutes of dynamic compression that were not detected in unloaded samples (Figure 2B).
Figure 2.
Loading-specific differences in untargeted metabolite expression. Mechanical loading resulted in statistically significant changes in expression of targeted and untargeted metabolites. Left shows Venn diagrams of unique metabolites detected in each group. Right shows quantitative comparisons for metabolites detected in both groups. (A) Unloaded controls versus 15 minutes of dynamic compression (DC). (B) Unloaded controls versus 30 minutes of DC. (C) Principal Components Analysis used for ordination analysis. The first two principal components contained 37.8% of the overall variance for this dataset (3-3-5). The timepoints are represented as follows: unloaded control: T1–3, DC15: T4–6, and DC30: T7–11. Ellipses represent unloaded controls (T1, T2, and T3) and dynamically compressed samples (≥T4). Principal Component 1 and Principal Component 2 were associated with 22.3% and 15.5% of the total variance among masses.
Many of the metabolites were significantly regulated in response to dynamic compression. Fifteen minutes of dynamic compression resulted in significant changes in 255 metabolites with 223 metabolites having increased intensity and 32 metabolites having decreased intensity compared with unloaded control samples (Table S2). Thirty minutes of dynamic compression resulted in significant changes in 787 metabolites with 689 metabolites having increased intensity and 98 metabolites having decreased intensity compared with unloaded control samples (Table S2). LC-MS analysis of naïve agarose control samples without chondrocytes found <15 metabolites (data not shown), which also served as solvent, reagent, and process controls.
Untargeted metabolite intensity was significantly correlated with time of loading for 254 metabolites (Figure 3 and Table S3). Of the significantly correlated metabolites, 157 were negatively correlated with time indicating cellular consumption and 97 were positively correlated with time indicating accumulation. Identification of these metabolites found peptides, lipids, substrates, and products.
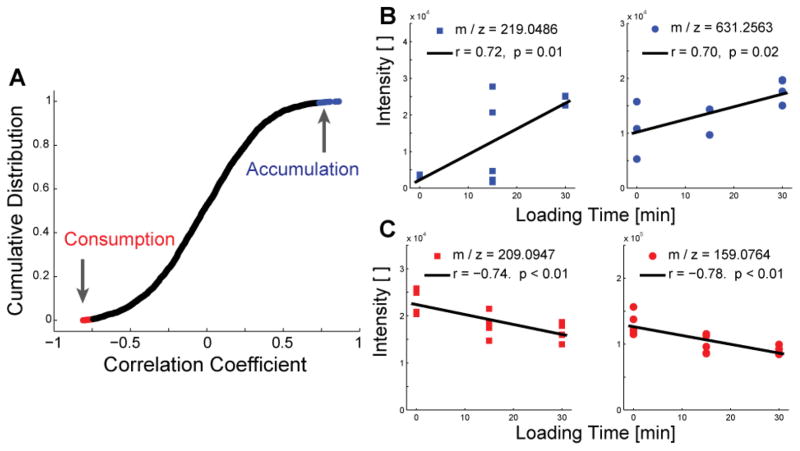
Figure 3.
Dynamic compression results in both accumulation and depletion of untargeted metabolites. (A) The cumulative distribution of correlation coefficients of untargeted metabolites were used to identify accumulating (blue) and consumed (red) metabolites. The top and bottom twenty correlations were identified as candidate mediators of chondrocyte mechanotransduction. (B) Accumulation of selected candidate mediators of mechanotransduction. Metabolites were database-matched as gulonic acid (left) and harderoporphyrin (right). (C) Depletion of selected candidate mediators of mechanotransduction, which were database-matched as kynurenine and 4-methylene-L-glutamine.
Targeted Metabolomics
Targeted metabolites related to central energy metabolism exhibited distinct expression patterns with loading time (Figure 4A). Clustering of targeted data resulted in groups of metabolites with increasing intensities with respect to time, intensities peaking at 15 minutes of compression, and decreasing intensities with respect to time. Guanosine tri-phosphate (GTP) was positively correlated with time (Figure 4B, p = 0.042). The ratio of NADP+ to NADPH peaked at 15 minutes of loading, as did the ratio of ATP to ADP to a lesser extent (Figure 4C). There were small changes in the ratio of NAD+ to NADH, and the ratio of GDP to GTP declined substantially with respect to loading time, consistent with the accumulation of GTP (Figure 4B).
Figure 4.
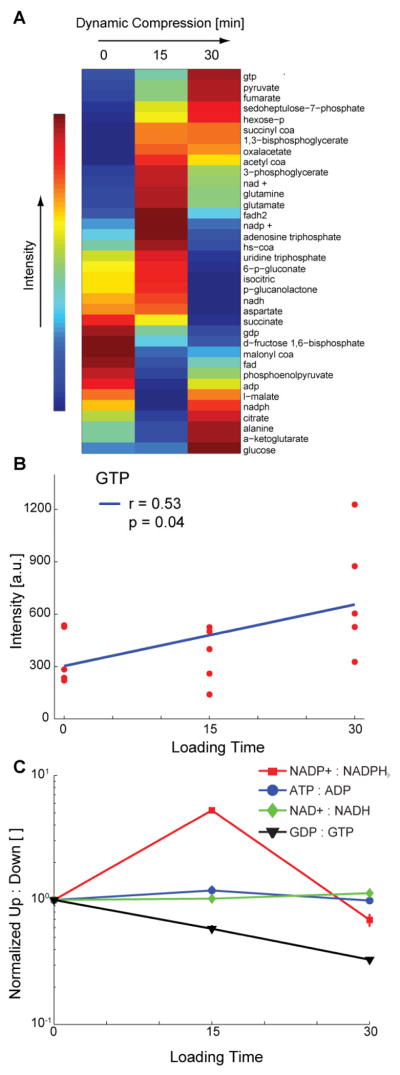
Changes in expression of targeted central-energy-related metabolites over from 0–30 minutes of applied compression. Metabolites associated with central energy metabolism and protein production (e.g. amino acids) were targeted for detailed analysis via detection of multiple ionized adducts and ions. (A) Clustered heatmap of co-expressed metabolites. (B) Significant accumulation (correlation) of GTP and marginally-significant depletion of GDP in response to applied compression. (C) Ratios of upstream to downstream mediators of central energy metabolism. The peak in NADP+ to NADPH indicates decreased energy flow toward the pentose phosphate pathway, and the continual decrease in GDP to GPT indicates increased flux through the TCA cycle.
Discussion
The objective of this study was to identify mediators of chondrocyte mechanotransduction using targeted and untargeted metabolomics. SW1353 chondrocytes were embedded in agarose with stiffness in the physiologic range of the human PCM. By providing quantitative measures of presence, absence, and relative abundance of cytosolic metabolites (i.e. reactant and product levels for biochemical reactions involved in multiple pathways) these data provide a systems-level view of how the functional chondrocyte fingerprint changes in response to mechanical compression. The significance of this study is that to our knowledge this is the first report of mechanically induced changes in the cellular metabolome. Furthermore, by encapsulating chondrocytes in physiologically-stiff agarose capable of inducing cellular deformations, we were able to identify 54 metabolites mediating chondrocyte mechanotransduction.
The untargeted metabolome is a high-dimensional measure of the functional state of the cell as defined by the individual metabolite levels. The targeted metabolome of central-energy-related metabolites can be used for rigorous hypothesis testing via systems biology. Future studies can utilize these data to map the full mechanisms of chondrocyte mechanotransduction. Below we discuss specific pathways motivated by this initial analysis.
Untargeted experiments found hundreds of metabolites that were statistically different between loading time points (Figure 2, Table S2). Among these molecules were 54 mediators that have not been previously associated with chondrocyte mechanotransduction. Database searching of these candidates resulted in the identification of 180 compounds from 49 of the masses: such a non-unique expansion of targets is inevitable due to structural isomers with identical formulae and masses. Molecular mass is a linear Diophantine function of elemental composition, and multiple different molecular formulas can produce the same overall mass [44]. Future work will employ MS/MS analyses to examine these candidate molecules.
These data suggest several specific signaling pathways, many of which have not previously been associated with mechanotransduction. Accumulated molecules of m/z values ~219 and ~631 may include gulonic acid and harderoporphyrin, respectively (Table S2). The accumulation of gulonic acid may indicate decreased pentose phosphate pathway-related energy metabolism [45]. The accumulation of harderoporphyrin, an intermediate in heme biosynthesis [46], may indicate compression-induced expression of oxidative phosphorylation consistent with the targeted results (see below), although chondrocytes likely reside in a hypoxic environment and have not been demonstrated to produce hemoglobin [47]. Compounds identified from depleted metabolites include kynurenine and 4-methylene-L-glutamine (Figure 3C). Consistent with the presently observed changes in GDP to GTP ratio (Figure 4C) and the known metabolism of pyruvate [45,48], the depletion of kynurenine and 4-methylene-L-glutamine may represent mechanically-induced re-direction of cellular energy via increased TCA cycle flux.
Human central energy metabolism of glucose involves the pentose phosphate pathway, glycolysis, and the tri-carboxylic acid cycle [36]. The ratio of upstream to downstream metabolites for a particular pathway (e.g. glycolysis) can be used as a surrogate measure of energy flux, under the assumptions of constant enzyme concentration and activity which are reasonable for the short timeframe employed by these experiments. For example, energy flow to the pentose phosphate pathway can be inferred by the ratio of NADP to NADPH with larger ratios indicating decreased flux. Our data show a transient increase in NADP:NADPH at 15 minutes (Figure 4C) which suggests decreased energy flow to the pentose phosphate pathway, which is typically used for nucleic acid production [32]. The ratio of GDP to GTP may represent energy flow to the tri-carboxylic acid cycle. We found a continual decrease in the ratio of GDP:GTP suggesting increased flux through the tri-carboxylic acid cycle, which may represent a re-direction of cellular energy in response to mechanical loading.
GTP, guanosine tri-phosphate, is a substrate required for activation of GTPase signaling pathways. In this study, GTP was significantly upregulated following 30 minutes of dynamic compression (Results and Figure 4B). Previous studies have found early-time (i.e. 15 minutes and shorter) activation of the GTPase RhoA [8] which is involved in activation of ROCK to stimulate cytoskeletal remodeling [8] and can also activate the master chondrogenic transcription factor of Sox9 [49]. These data add to a complex picture of chondrocyte mechanotransduction involving directed changes in energy metabolism to maintain and remodel the cytoskeleton. Future studies will build on this work to elucidate both mechanisms and consequences of modifying energy metabolism in response to mechanical loading.
While this study found several candidate mediators of chondrocyte mechanotransduction using targeted and untargeted metabolomics some limitations apply. First, the temporal resolution of the metabolome was limited to sampling at 15 minute intervals in these initial experiments, and flux inferences assume constant enzyme concentration and activity. Future studies may build on these results by using shorter time intervals, measuring enzyme concentrations and activities, and extending the loading duration. Second, this study quantified and compared the levels of thousands of metabolites. Although we used previously published LC-MS data analysis procedures, the possibility of both false positive and false negative results remains. We controlled the risk of false positive comparisons using a false discovery rate of 0.05 [39]. Third, these studies were performed using SW1353 chondrocytes encapsulated in 4.5% agarose. While the high concentration of agarose modeled the stiffness of the human chondrocyte pericellular environment [16,17], the in vivo chondrocyte pericellular matrix likely contains substantial additional signals for regulating cellular behavior. Additionally, dynamic loading of cartilage can result in a diverse set of mechanical signals including streaming potentials and fluid flow [50,51] which have been observed previously for lower concentrations of agarose, but have not been measured in the high stiffness in vitro system used in this study. Therefore, the pathways and mechanisms reported herein likely represent a subset of mechanically-activated pathways, which represent cell-intrinsic mechanisms of mechanotransduction. Finally, although putative compound identifications were made, a general limitation of metabolomics is the challenge of unique identification, which future studies may address using NMR and MS/MS. Despite these limitations, to our knowledge, this is the first report of the mechanically-induced metabolome for human chondrocytes, which provides substantial new data describing how chondrocytes respond to mechanical loads.
Conclusions
This study demonstrates the power and challenges associated with high-dimensional data for systems biology and provides both a methodological and data-based foundation for future studies. In untargeted experiments, we found hundreds of statistically-significant changes in metabolites induced by mechanical loading and using advanced statistical methods identified 54 candidate mediators of chondrocyte mechanotransduction. In targeted experiments, we found that mechanical loading induces significant changes in metabolites associated with cellular energy usage. Future studies will identify the mechanisms of these changes and address the cell-type specificity of these responses
Supplementary Material
SW1353 chondrocytes were mechanically compressed in a physiologically stiff 3D environment.
Following compression, untargeted and targeted metabolomics studies were performed.
This study identified candidate mediators of cellular mechanostransduction.
This research implicates central metabolism as a mechanosensitive pathway.
Acknowledgments
We thank Drs. Ross Carlson, Brian Bothner, and Edward Dratz, MSU, for critical insight provided during discussions. The SW1353 cell line was kindly donated by Martin Lotz. Funding was provided by NIH P20GM10339405S1, Montana State University, and the Murdock Charitable Trust.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ward CW, Prosser BL, Lederer WJ. Mechanical stretch induced activation of ROS/RNS signaling in striated muscle. Antioxid Redox Signal. 2013 doi: 10.1089/ars.2013.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ko FC, Dragomir C, Plumb DA, Goldring SR, Wright TM, Goldring MB, van der Meulen MC. In vivo cyclic compression causes cartilage degeneration and subchondral bone changes in mouse tibiae. Arthritis Rheum. 2013;65:1569–1578. doi: 10.1002/art.37906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grygorczyk R, Furuya K, Sokabe M. Imaging and characterization of stretch-induced ATP release from alveolar A549 cells. J Physiol. 2013;591:1195–1215. doi: 10.1113/jphysiol.2012.244145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neu CP, Khalafi A, Komvopoulos K, Schmid TM, Reddi AH. Mechanotransduction of bovine articular cartilage superficial zone protein by transforming growth factor beta signaling. Arthritis Rheum. 2007;56:3706–3714. doi: 10.1002/art.23024. [DOI] [PubMed] [Google Scholar]
- 5.Niehoff A, Muller M, Bruggemann L, Savage T, Zaucke F, Eckstein F, Muller-Lung U, Bruggemann GP. Deformational behaviour of knee cartilage and changes in serum cartilage oligomeric matrix protein (COMP) after running and drop landing. Osteoarthritis Cartilage. 2011;19:1003–1010. doi: 10.1016/j.joca.2011.04.012. [DOI] [PubMed] [Google Scholar]
- 6.Coleman JL, Widmyer MR, Leddy HA, Utturkar GM, Spritzer CE, Moorman CT, 3rd, Guilak F, DeFrate LE. Diurnal variations in articular cartilage thickness and strain in the human knee. J Biomech. 2013;46:541–547. doi: 10.1016/j.jbiomech.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haudenschild DR, D’Lima DD, Lotz MK. Dynamic compression of chondrocytes induces a Rho kinase-dependent reorganization of the actin cytoskeleton. Biorheology. 2008;45:219–228. [PubMed] [Google Scholar]
- 8.Haudenschild DR, Nguyen B, Chen J, D’Lima DD, Lotz MK. Rho kinase-dependent CCL20 induced by dynamic compression of human chondrocytes. Arthritis Rheum. 2008;58:2735–2742. doi: 10.1002/art.23797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adatia A, Rainsford KD, Kean WF. Osteoarthritis of the knee and hip. Part I: aetiology and pathogenesis as a basis for pharmacotherapy. J Pharm Pharmacol. 2012;64:617–625. doi: 10.1111/j.2042-7158.2012.01458.x. [DOI] [PubMed] [Google Scholar]
- 10.Kawakita K, Nishiyama T, Fujishiro T, Hayashi S, Kanzaki N, Hashimoto S, Takebe K, Iwasa K, Sakata S, Nishida K, Kuroda R, Kurosaka M. Akt phosphorylation in human chondrocytes is regulated by p53R2 in response to mechanical stress. Osteoarthritis Cartilage. 2012;20:1603–1609. doi: 10.1016/j.joca.2012.08.022. [DOI] [PubMed] [Google Scholar]
- 11.Holledge MM, Millward-Sadler SJ, Nuki G, Salter DM. Mechanical regulation of proteoglycan synthesis in normal and osteoarthritic human articular chondrocytes--roles for alpha5 and alphaVbeta5 integrins. Biorheology. 2008;45:275–288. [PubMed] [Google Scholar]
- 12.Fogh J. Human tumor lines for cancer research. Cancer Invest. 1986;4:157–184. doi: 10.3109/07357908609038260. [DOI] [PubMed] [Google Scholar]
- 13.Mansfield J, Yu J, Attenburrow D, Moger J, Tirlapur U, Urban J, Cui Z, Winlove P. The elastin network: its relationship with collagen and cells in articular cartilage as visualized by multiphoton microscopy. J Anat. 2009;215:682–691. doi: 10.1111/j.1469-7580.2009.01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen U, Allen JM, White R, Moscibrocki C, Bruckner P, Bateman JF, Fitzgerald J. WARP interacts with collagen VI-containing microfibrils in the pericellular matrix of human chondrocytes. PLoS One. 2012;7:e52793. doi: 10.1371/journal.pone.0052793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macdonald DW, Squires RS, Avery SA, Adams J, Baker M, Cunningham CR, Heimann NB, Kooyman DL, Seegmiller RE. Structural variations in articular cartilage matrix are associated with early-onset osteoarthritis in the spondyloepiphyseal dysplasia congenita (sedc) mouse. Int J Mol Sci. 2013;14:16515–16531. doi: 10.3390/ijms140816515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darling EM, Wilusz RE, Bolognesi MP, Zauscher S, Guilak F. Spatial mapping of the biomechanical properties of the pericellular matrix of articular cartilage measured in situ via atomic force microscopy. Biophys J. 2010;98:2848–2856. doi: 10.1016/j.bpj.2010.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilusz RE, Zauscher S, Guilak F. Osteoarthritis Cartilage. 2013. Micromechanical Mapping of Early Osteoarthritic Changes in the Pericellular Matrix of Human Articular Cartilage. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knight MM, Toyoda T, Lee DA, Bader DL. Mechanical compression and hydrostatic pressure induce reversible changes in actin cytoskeletal organisation in chondrocytes in agarose. J Biomech. 2006;39 doi: 10.1016/j.jbiomech.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 19.Kisiday JD, Lee JH, Siparsky PN, Frisbie DD, Flannery CR, Sandy JD, Grodzinsky AJ. Catabolic responses of chondrocyte-seeded peptide hydrogel to dynamic compression. Ann Biomed Eng. 2009;37:1368–1375. doi: 10.1007/s10439-009-9699-9. [DOI] [PubMed] [Google Scholar]
- 20.Vaughan NM, Grainger J, Bader DL, Knight MM. The potential of pulsed low intensity ultrasound to stimulate chondrocytes matrix synthesis in agarose and monolayer cultures. Med Biol Eng Comput. 2010;48:1215–1222. doi: 10.1007/s11517-010-0681-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haudenschild DR, Chen J, Pang N, Steklov N, Grogan SP, Lotz MK, D’Lima DD. Vimentin contributes to changes in chondrocyte stiffness in osteoarthritis. J Orthop Res. 2011;29:20–25. doi: 10.1002/jor.21198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farnsworth NL, Antunez LR, Bryant SJ. Dynamic compressive loading differentially regulates chondrocyte anabolic and catabolic activity with age. Biotechnol Bioeng. 2013 doi: 10.1002/bit.24860. [DOI] [PubMed] [Google Scholar]
- 23.Campbell JJ, Blain EJ, Chowdhury TT, Knight MM. Loading alters actin dynamics and up-regulates cofilin gene expression in chondrocytes. Biochem Biophys Res Commun. 2007;361:329–334. doi: 10.1016/j.bbrc.2007.06.185. [DOI] [PubMed] [Google Scholar]
- 24.Chen J, Irianto J, Inamdar S, Pravincumar P, Lee DA, Bader DL, Knight MM. Cell mechanics, structure, and function are regulated by the stiffness of the three-dimensional microenvironment. Biophys J. 2012;103:1188–1197. doi: 10.1016/j.bpj.2012.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishida T, Kubota S, Aoyama E, Takigawa M. Impaired glycolytic metabolism causes chondrocyte hypertrophy-like changes via promotion of phospho-Smad1/5/8 translocation into nucleus. Osteoarthritis Cartilage. 2013;21:700–709. doi: 10.1016/j.joca.2013.01.013. [DOI] [PubMed] [Google Scholar]
- 26.O’Neill LA, Hardie DG. Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature. 2013;493:346–355. doi: 10.1038/nature11862. [DOI] [PubMed] [Google Scholar]
- 27.Petursson F, Husa M, June R, Lotz M, Terkeltaub R, Liu-Bryan R. Linked decreases in liver kinase B1 and AMP-activated protein kinase activity modulate matrix catabolic responses to biomechanical injury in chondrocytes. Arthritis Res Ther. 2013;15:R77. doi: 10.1186/ar4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jutila AA. Development and Validation of a System for Studying Chondrocyte Mechanotransduction wtih Preliminary Metabolomic Results. Mechanical and Industrial Engineering, Montana State University; Bozeman, MT: 2013. p. 154. [Google Scholar]
- 29.Zignego DL, Jutila AA, Gelbke MK, Gannon DM, June RK. The mechanical microenvironment of high concentration agarose for applying deformation to primary chondrocytes. J Biomech. 2013 doi: 10.1016/j.jbiomech.2013.10.051. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Darling EM, Zauscher S, Guilak F. Viscoelastic properties of zonal articular chondrocytes measured by atomic force microscopy. Osteoarthritis Cartilage. 2006;14:571–579. doi: 10.1016/j.joca.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 31.Umberger BR, Martin PE. Mechanical power and efficiency of level walking with different stride rates. J Exp Biol. 2007;210:3255–3265. doi: 10.1242/jeb.000950. [DOI] [PubMed] [Google Scholar]
- 32.Munger J, Bennett BD, Parikh A, Feng XJ, McArdle J, Rabitz HA, Shenk T, Rabinowitz JD. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat Biotechnol. 2008;26:1179–1186. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heinemann J, Hamerly T, Maaty WS, Movahed N, Steffens JD, Reeves BD, Hilmer JK, Therien J, Grieco PA, Peters JW, Bothner B. Expanding the paradigm of thiol redox in the thermophilic root of life. Biochim Biophys Acta. 2013 doi: 10.1016/j.bbagen.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 34.Ortmann AC, Brumfield SK, Walther J, McInnerney K, Brouns SJ, van de Werken HJ, Bothner B, Douglas T, van de Oost J, Young MJ. Transcriptome analysis of infection of the archaeon Sulfolobus solfataricus with Sulfolobus turreted icosahedral virus. J Virol. 2008;82:4874–4883. doi: 10.1128/JVI.02583-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Secor PR, Jennings LK, James GA, Kirker KR, Pulcini ED, McInnerney K, Gerlach R, Livinghouse T, Hilmer JK, Bothner B, Fleckman P, Olerud JE, Stewart PS. Phevalin (aureusimine B) production by Staphylococcus aureus biofilm and impacts on human keratinocyte gene expression. PLoS One. 2012;7:e40973. doi: 10.1371/journal.pone.0040973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. 4. Garland Science; New York, NY: 2002. [Google Scholar]
- 37.Pluskal T, Castillo S, Villar-Briones A, Oresic M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics. 2010;11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katajamaa M, Oresic M. Processing methods for differential analysis of LC/MS profile data. BMC Bioinformatics. 2005;6:179. doi: 10.1186/1471-2105-6-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 40.R.C. Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. [Google Scholar]
- 41.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, Bouatra S, Sinelnikov I, Arndt D, Xia J, Liu P, Yallou F, Bjorndahl T, Perez-Pineiro R, Eisner R, Allen F, Neveu V, Greiner R, Scalbert A. HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41:D801–807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu ZJ, Schultz AW, Wang J, Johnson CH, Yannone SM, Patti GJ, Siuzdak G. Liquid chromatography quadrupole time-of-flight mass spectrometry characterization of metabolites guided by the METLIN database. Nat Protoc. 2013;8:451–460. doi: 10.1038/nprot.2013.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fahy E, Subramaniam S, Murphy RC, Nishijima M, Raetz CR, Shimizu T, Spener F, van Meer G, Wakelam MJ, Dennis EA. Update of the LIPID MAPS comprehensive classification system for lipids. J Lipid Res. 2009;50(Suppl):S9–14. doi: 10.1194/jlr.R800095-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kind T, Fiehn O. Seven Golden Rules for heuristic filtering of molecular formulas obtained by accurate mass spectrometry. BMC Bioinformatics. 2007;8:105. doi: 10.1186/1471-2105-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kanehisa M. Molecular network analysis of diseases and drugs in KEGG. Methods Mol Biol. 2013;939:263–275. doi: 10.1007/978-1-62703-107-3_17. [DOI] [PubMed] [Google Scholar]
- 46.Schmitt C, Gouya L, Malonova E, Lamoril J, Camadro JM, Flamme M, Rose C, Lyoumi S, Da Silva V, Boileau C, Grandchamp B, Beaumont C, Deybach JC, Puy H. Mutations in human CPO gene predict clinical expression of either hepatic hereditary coproporphyria or erythropoietic harderoporphyria. Hum Mol Genet. 2005;14:3089–3098. doi: 10.1093/hmg/ddi342. [DOI] [PubMed] [Google Scholar]
- 47.Rajpurohit R, Koch CJ, Tao Z, Teixeira CM, Shapiro IM. Adaptation of chondrocytes to low oxygen tension: relationship between hypoxia and cellular metabolism. J Cell Physiol. 1996;168:424–432. doi: 10.1002/(SICI)1097-4652(199608)168:2<424::AID-JCP21>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 48.Wang W, Mazurkewich S, Kimber MS, Seah SY. Structural and kinetic characterization of 4-hydroxy-4-methyl-2-oxoglutarate/4-carboxy-4-hydroxy-2-oxoadipate aldolase, a protocatechuate degradation enzyme evolutionarily convergent with the HpaI and DmpG pyruvate aldolases. J Biol Chem. 2010;285:36608–36615. doi: 10.1074/jbc.M110.159509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haudenschild DR, Chen J, Pang N, Lotz MK, D’Lima DD. Rho kinase-dependent activation of SOX9 in chondrocytes. Arthritis Rheum. 2010;62:191–200. doi: 10.1002/art.25051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grodzinsky AJ, Lipshitz H, Glimcher MJ. Electromechanical properties of articular cartilage during compression and stress relaxation. Nature. 1978;275:448–450. doi: 10.1038/275448a0. [DOI] [PubMed] [Google Scholar]
- 51.Mow VC, Kuei SC, Lai WM, Armstrong CG. Biphasic creep and stress relaxation of articular cartilage in compression? Theory and experiments. J Biomech Eng. 1980;102:73–84. doi: 10.1115/1.3138202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.