This paper introduces a new, easy-to-use method of fluorescence single-molecule speckle microscopy for actin with nanometer-scale accuracy. This new method reveals that actin flows in front of mature focal adhesions (FAs) are fast and biased toward FAs, suggesting that mature FAs are actively engaged in pulling and remodeling the local actin network.
Abstract
Speckle microscopy directly visualizes the retrograde actin flow, which is believed to promote cell-edge protrusion when linked to focal adhesions (FAs). However, it has been argued that, due to rapid actin turnover, the use of green fluorescent protein–actin, the lack of appropriate analysis algorithms, and technical difficulties, speckle microscopy does not necessarily report the flow velocities of entire actin populations. In this study, we developed a new, user-friendly single-molecule speckle (SiMS) microscopy using DyLight dye-labeled actin. Our new SiMS method enables in vivo nanometer-scale displacement analysis with a low localization error of ±8–8.5 nm, allowing accurate flow-velocity measurement for actin speckles with lifetime <5 s. In lamellipodia, both short- and long-lived F-actin molecules flow with the same speed, indicating they are part of a single actin network. These results do not support coexistence of F-actin populations with different flow speeds, which is referred to as the lamella hypothesis. Mature FAs, but not nascent adhesions, locally obstruct the retrograde flow. Interestingly, the actin flow in front of mature FAs is fast and biased toward FAs, suggesting that mature FAs attract the flow in front and actively remodel the local actin network.
INTRODUCTION
Cell migration is a dynamic, actin-based cellular process that is important for many phenomena in multicellular organisms. It involves coordination of actin-based protrusion at the cell front, adhesion of the newly protruded domains to the substrate, and actomyosin-mediated contraction at the cell rear (Mitchison and Cramer, 1996; Pollard and Cooper, 2009). A lamellipodium is a thin, sheet-like pseudopodium and contains a dense actin filament network. Actin polymerization in the lamellipodium generates a forward protrusion force at the cell membrane. At the same time, the entire actin network moves toward the cell center; this is called the retrograde actin flow (Wang, 1985). The interaction between the actin flows and the focal adhesions (FAs) has been proposed to enhance the membrane protrusion (Mitchison and Kirschner, 1988; Jay, 2000). However, how FAs influence local retrograde flows is not fully understood.
There are technical difficulties in measuring the velocities of actin flows accurately. First, filament turnover in the lamellipodial actin network is very rapid, as nearly one-third of filaments have short lifetimes of <10 s in certain types of cells (Watanabe and Mitchison, 2002). Such ephemeral filaments move only short distances (<100–300 nm), and it therefore requires an exceedingly high spatiotemporal resolution to track the filaments. Second, if movements of actin filaments are heterogeneous, individual filaments must be tracked to define the flow. Approaches that monitor a mass of actin filaments, such as photoactivation of fluorescence (Theriot and Mitchison, 1991, 1992), fluorescence recovery after photobleaching (FRAP; Star et al., 2002), and polarization microcopy (Oldenbourg et al., 2000) are not suited for measuring heterogeneous actin movements because of their limited spatial resolutions.
Fluorescent speckle microscopy (FSM) enables higher-resolution analysis of actin assembly/disassembly and movement than the above methods (Waterman-Storer et al., 1998). FSM has been developed in two different directions: one is quantitative FSM (qFSM; Danuser and Waterman-Storer, 2006), and the other is fluorescence single-molecule speckle (SiMS) microscopy (Watanabe and Mitchison, 2002; Watanabe, 2012). In qFSM, resolvable fluorescent speckles containing multiple (2–10) fluorophore probes provide quantitative information on appearance, disappearance, and trajectories through automatic computational analysis (Danuser and Waterman-Storer, 2006). qFSM is a potent method with high statistical power, because it extracts information from thousands of actin speckles within one image, yielding a heat-map representation of actin assembly/disassembly rates in subcellular areas. However, the output of qFSM may contain intrinsic errors, as has been recently argued (Danuser, 2009; Vallotton and Small, 2009). Additionally, it can be difficult to accurately measure the actin retrograde flow velocity by qFSM for the following reasons: 1) automated object tracking may misidentify individual speckles when they are densely packed and 2) a partial exchange of molecules in a multiple-fluorophore speckle may alter the speckle's shape, leading to an imperfect velocity measurement. These problems would complicate measurements of the actin flow rate in lamellipodia (LP).
SiMS microscopy is the ideal approach to disclose the velocities of local actin flows. In SiMS analysis, individual enhanced green fluorescent protein (EGFP)-actin molecules are tracked in cells expressing a very low level of EGFP-actin under the control of the defective CMV promoter (delCMV; Watanabe and Mitchison, 2002; Watanabe, 2012). However, the previous SiMS method has four potential problems. First, there have been warnings against the use of EGFP-actin. Overexpressed EGFP-actin might influence actin dynamics (Aizawa et al., 1997; Deibler et al., 2011; Feng et al., 2005). Moreover, EGFP-actin might be excluded from formin-assembled actin filaments, as GFP-actin expressed in fission yeast does not incorporate into the contractile ring assembled primarily by formin homology protein Cdc12p (Chen et al., 2012). Formins play critical roles in contractile ring formation (Severson et al., 2002; Watanabe et al., 2008), migration of neurons (Shinohara et al., 2012), thin filament assembly in myofibrils (Kan-o et al., 2012; Mi-Mi et al., 2012), and mechanosensing (Higashida et al., 2013). Although GFP-actin incorporates into contractile rings in cultured mammalian cells (Murthy and Wadsworth, 2005; Zhou and Wang, 2008) and has widely been used for live-cell imaging in various model systems (Fischer et al., 1998; Honkura et al., 2008; Schneider et al., 2002), the use of GFP-actin may partially hinder observation of formin-based actin functions (see also Supplemental Figure S1).
Second, the spatiotemporal resolution of SiMS microscopy has been insufficient to measure the velocities of the short-lived actin filaments. For example, our recent analysis of single-molecule EGFP-actin speckles reported uniform flow rates in LP of XTC cells (Ryan et al., 2012). However, we had to limit the analysis to the actin speckles with lifetimes greater than 20 s to accurately measure speckle displacement.
Third, this approach demands some experience to find cells containing fluorescent probes at optimal levels. The low expression levels achieved by delCMV are desirable for most protein localization studies, as exemplified by the recent demonstration of paxillin-labeled FA dynamics in cells migrating through a three-dimensional matrix (Kubow and Horwitz, 2011). However, even with this defective promoter, only a minor population of cells expresses a sufficiently low level of EGFP-actin, which makes SiMS microscopy challenging for inexperienced researchers.
Fourth, Lai et al. (2008) called for revision of our previous SiMS study, because their conclusion based on the FRAP experiments is not consistent with our early SiMS study (Watanabe and Mitchison, 2002). To elucidate whether FRAP and SiMS microscopy contradict each other or not, we used mathematical modeling to compare SiMS and FRAP data on the same cell types and found that there is no fundamental disagreement between the two types of experiments (Smith et al., 2013). In this study, we reexamine actin turnover in LP with a new fluorescent actin probe, the biochemical properties of which are well characterized in vitro, and confirm the conclusion of the early SiMS study (Watanabe and Mitchison, 2002).
In this study, we developed an improved method of SiMS microscopy for measuring actin dynamics with high resolution and elucidated the velocities of local actin flows near FAs in LP. We used fluorescent actin labeled with DyLight dye on lysine, which partially retains the ability to collaborate with formins and profilin in vitro. In cells, DyLight (DL)-actin labels actin networks with improved photostability and brightness compared with GFP-actin. These favorable properties of DL-actin in combination with semiautomatic speckle tracking by Speckle TrackerJ software (Smith et al., 2011) enables in vivo nanometer-scale displacement analysis of actin flow. Using the new method, we readdressed the lamella hypothesis by measuring the actin flow rates, including those of short-lived actin speckles. Furthermore, the new SiMS microscopy revealed the complex interactions between FAs and the local actin flow.
RESULTS
Actin labeled on lysine marks mDia-assembled F-actin effectively but is eliminated from FMNL2-associated actin assembly in vitro
The FH1-FH2 domain of mDia1 moves directionally over tens of microns by processively associating with the growing F-actin barbed end in the cell (Higashida et al., 2004). We noticed that coexpression of red fluorescent protein–tagged actin (mRFP1-actin) impaired processive actin elongation by an FH1-FH2 domain mutant of mDia1 (aa 543–1192), EGFP-mDia1∆N3 (Higashida et al., 2004). In the control cells expressing no mRFP-actin, 73% (30 of 41 speckles, n = 2 cells) of processive mDia1∆N3 SiMS maintained constant speed for more than 2.5 s (Figure S1, A and B). Similarly, in the cell expressing mRFP1-actin at low level, 70% (55/79, n = 3 cells) of processive mDia1∆N3 SiMS maintained constant speed for more than 2.5 s, but once the movement stopped, mDia1∆N3 rarely restarted the motion in a few seconds (Figure S1, C–E). Strikingly, only 38% (11 of 29 speckles) and 13% (2/15) of mDia1∆N3 speckles maintained processive movement for more than 2.5 s in the cells expressing a high level and an excessively high level of mRFP1-actin, respectively (Figure S1, G and H). In these cells (Figure S1, G and H), the speed of mDia1∆N3 was variable compared with that in control cells (Figure S1, A and B), presumably because mDia1∆N3 speckles frequently stopped in the cells expressing mRFP1-actin at high levels. These results indicate that mRFP1-actin interferes with processive actin elongation by mDia1∆N3. Therefore fluorescent protein–tagged actin might not be suitable to monitor formin-based actin structures in vertebrate cells.
To overcome the above problem, we tested fluorescent DyLight-labeled (DL-labeled; Sarkar et al., 2010) rabbit muscle actin on lysines. Mass spectrometry showed that one to three lysines were labeled with DL, but gel filtration using a Superdex 200 enabled the enrichment of single-labeled actin (Figure S2). The labeled actin was eluted in one peak (Figure S2A). Double- or triple-labeled actin was detected mainly in the front part of the peak, whereas the majority of actin in the back part of the peak was unlabeled and single-labeled actin (Figure S2B). We used DL-actin in the back part of the peak in the following experiments.
Previous studies have demonstrated that fluorescent actin labeled on lysine incorporated effectively into mDia1-, Bni1p-, and Cdc12p-elongated actin filaments (Kovar and Pollard, 2004; Kovar et al., 2006; Michelot et al., 2007). We examined incorporation of DL-actin into F-actin that associated with mDia1∆N3, mDia2 FH1-FH2 (aa 610–1077), and FMNL2 FH1-FH2 (aa 566–1092) in vitro by using total internal reflection fluorescence (TIRF) microscopy (Kovar et al., 2006). In the absence of profilin, DL549-actin incorporated into either mDia1- or FMNL2-assembled actin filaments with high efficiency (Figure S3, C–E).
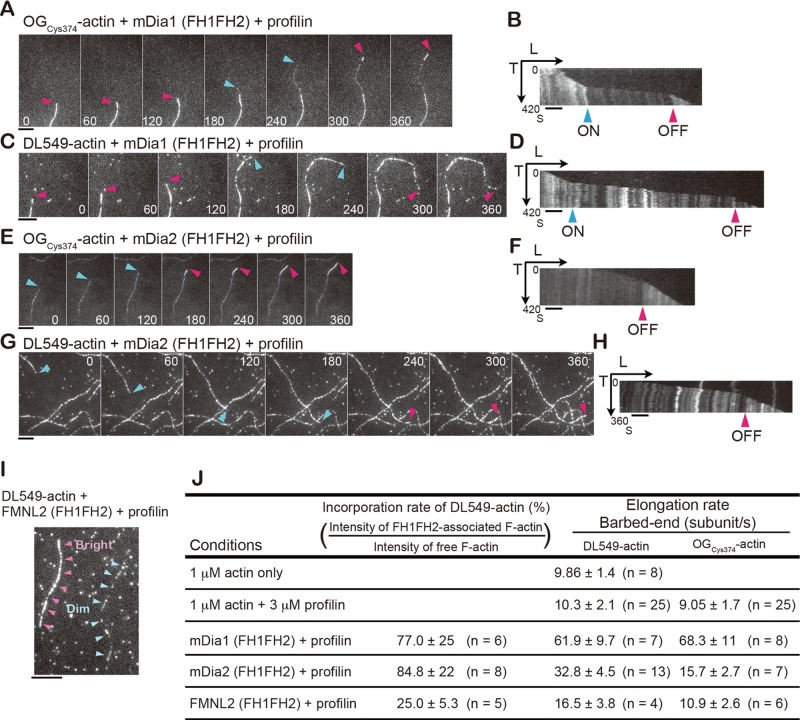
In the presence of profilin, DL549-actin marked mDia1- and mDia2-associated filaments, but it poorly incorporated into FMNL2-associated filaments. We observed growing actin filaments attached to the glass surface by N-ethyl-maleimide (NEM)-inactivated myosin II. Under this condition, it was possible to distinguish formin-bound fast-growing filaments from spontaneously elongating filaments by the filament growth rate. First, we tested mDia1 and mDia2. We confirmed the previous finding (Kovar et al., 2006) that actin labeled on Cys-374 with Oregon Green (OGCys374-actin), which binds profilin weakly (Malm, 1984), incorporates into formin and profilin-assembled F-actin less densely than spontaneously elongating F-actin. Fast-growing dim OGCys374-actin filaments (Figure 1, A and E, blue arrows) occasionally switched to slowly growing bright segments (Figure 1, A and E, red arrows; Supplemental Movies S1 and S2). Kymographs demonstrate the coincidence of the growth rate decrease with the increase in filament fluorescence, which represents the mDia FH1-FH2 dissociation event (Figure 1, B and F, red arrows).
FIGURE 1:
Incorporation of DL549-actin into formin-associated actin filaments in vitro. Actin assembly of 1 μM actin containing OGCys374-actin (10%) or DL549-actin (4%) in the presence of 3 μM profilin and 1 nM GST-mDia1∆N3, 7.5 nM GST-mDia2 (FH1-FH2), or 7.5 nM GST-FMNL2 (FH1-FH2) on the glass coverslip coated with NEM-myosin II. (A, C, E, and G) Time-lapse images of actin growth of OGCys374-actin (A and E) or DL549-actin (C and G) with time in seconds indicated in images. Red arrows indicate the formin-free barbed ends growing at slow rates, and blue arrows indicate the barbed ends undergoing fast mDia1∆N3-associated (A and C) or mDia2 (FH1-FH2)-associated (E and G) growth. The background spots in DL549-actin images represent nonspecifically bound single DL549-actin molecules on the glass surface. (B, D, F, and H) Kymographs show the length (x-axis) of OGCys374-actin filaments (B and F) or DL549-actin filaments (D and H) in (A, C, E, and G) vs. time (y-axis). Blue arrows indicate association (ON) of GST-mDia1∆N3 or mDia2 (FH1FH2), and red arrows indicate dissociation (OFF) of formins. (I) Actin filaments assembled under the condition of 1 μM actin containing DL549-actin, 3 μM profilin, and 7.5 nM GST-FMNL2 (FH1-FH2) on a glass coverslip coated with NEM-myosin II. Pink arrows indicate a bright actin filament, and blue arrows indicate a dim filament. (A–I) Scale bars: 5 μm. (J) Summary of incorporation rates of DL-549-actin to formin-associated filaments and elongation rates at barbed ends under indicated conditions. Rates are represented as mean ± SD.
Similarly, DL549-actin assembled into filaments with two distinct speeds in the presence of either mDia1 or mDia2 (Figure 1, C, D, G, and H, and Movies S1 and S2). The intensity of DL549-actin fluorescence in the fast-growing mDia-associated segments (Figure 1, C and G, blue arrows) was 77 ± 25% of that in the slow-growing mDia-free segments (Figure 1, C and G, red arrows) in individual filaments (n = 6) for mDia1 and 84.8 ± 22% (n = 8) for mDia2 (Figure 1J). The elongation rate of mDia1-enhanced filament growth with DL549-actin was 61.9 ± 9.7 subunits s–1 (n = 7; Figure 1J), which is comparable with that of OGCys374-actin, 68.3 ± 11 subunits s–1 (n = 8; Figure 1J). On the other hand, the elongation rate of mDia2-assembled filaments with DL549-actin was 32.8 ± 4.5 subunits s–1 (n = 13), which is higher than that of OGCys374-actin, 15.7 ± 2.7 subunits s–1 (n = 7; Figure 1J). These results suggest OGCys374-actin might interfere with profilin-mediated actin polymerization of mDia2.
In contrast, a dim portion appeared in the DL549-actin filaments assembled with FMNL2 and profilin (Figure 1I). The intensity of DL549-actin fluorescence in dim segments was 25 ± 5.6% of that in the bright segments (n = 5; Figure 1J). These dim DL549-actin filaments assembled with a slightly enhanced elongation rate, 16.5 ± 3.8 subunits s–1 (n = 4) (Figure 1J). In the same assay with OGCys374-actin, dim actin segments assembled from FMNL2-associated barbed ends at 10.9 ± 2.6 subunits s–1 (n = 6; Figures 1J and S3, A and B). These results indicate that FMNL2 weakly accelerates actin elongation at barbed ends. We further confirmed low incorporation of DL549-actin to FMNL2-assembled filaments by monitoring processive actin elongation at the glass-bound FMNL2 (Movie S3). Strikingly, poorly labeled filaments assembled at fixed points, which represent processively elongating filaments from glass-bound FMNL2, whereas bright segments appeared in the same filaments when they detached from the glass (Movie S3).
Taken together, the new DL-actin probe in the SiMS analysis is anticipated to report the dynamics of 77, 85, and 25% of mDia1-, mDia2-, and FMNL2-assembled F-actin, respectively. Although the filament incorporation rate of DL-actin may vary with different formins, knowledge on the above biochemical properties of DL-actin would be subservient to evaluating the data of the SiMS analysis.
In the following SiMS analysis, we estimate that the molar ratio of DL549-actin to endogenous actin is 1:200,000 by assuming 1000 μM actin in LP (Abraham et al., 1999). Thus the frequency of interaction between Arp2/3 and DL549-actin is expected to be extremely low.
DyLight549-actin is efficiently delivered into the cytoplasm by electroporation and shows improved photostability and brightness
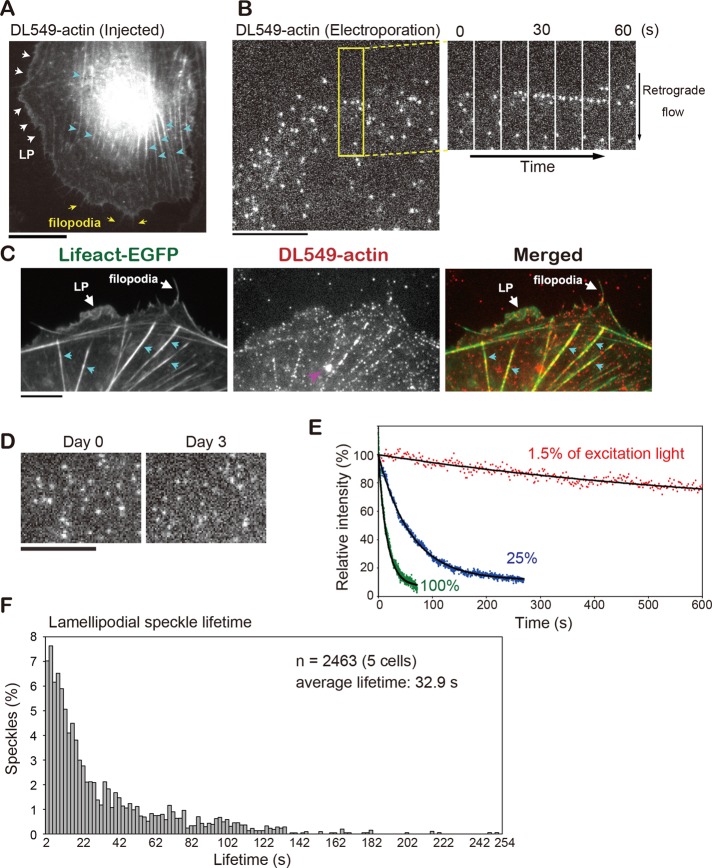
We verified that DL-actin coassembled with cellular actin structures. Microinjected DL549-actin incorporated into actin structures, including stress fibers, LP, and filopodia in XTC cells (Figure 2A and Movie S4).
FIGURE 2:
Characterization of DL549-actin in vivo. (A) Live image of XTC cells microinjected with DL549-actin. White arrows, yellow arrows, and blue arrows indicate LP, filopodia, and stress fibers, respectively. (B) Images of DL549-actin speckles in LP of XTC cells loaded with DL549-actin by electroporation. Time-lapse images paneled at 10-s intervals in the area (rectangle) are shown on the right. (C) DL549-actin coassembles to actin structures visualized by Lifeact-EGFP. White arrows show LP and filopodia as indicated. Blue arrows indicate stress fibers. A pink arrow indicates a fluorescent organelle-like structure, which is apparently an artifact of electroporation. (D) Images of LP DL549-actin speckles that were delivered into cells by electroporation on day 0 and day 3. XTC cells were cultured on poly-l-lysine– and fibronectin-coated coverslips (A and C) or on poly-l-lysine–coated coverslips (B and D). (E) Photostability of DL549-actin in vivo. XTC cells loaded with DL549-actin by electroporation were continuously exposed to excitation light using the 100-W mercury illumination system and acquired without interval. The excitation light used was full (green dots), attenuated to 25% (blue dots), or attenuated to 1.5% (red dots). Black lines show the single-exponential curve fitted to the data. Relative intensity (y-axis) expressed percentages derived from the average fluorescence intensity of the peripheral region of the XTC cell in each frame divided by the value at y-intercept of the fitted curve. (F) Lifetime distribution of DL549-actin speckles in LP. Scale bars: 10 μm.
To deliver DL-actin into cells for SiMS microscopy, we applied electroporation. A mixture of 0.5 μM DL549-actin with 1.5 μM profilin, which prevents spontaneous filament nucleation, was loaded into XTC cells using the Neon Transfection System (Invitrogen) with two pulses (1005-V pulse voltage and 35-ms pulse width). DL549-actin was successfully delivered into the cytoplasm at a sufficiently low fluorescent speckle density suitable for SiMS microscopy (Figure 2B and Movie S5). In fixed XTC cells, DL-actin speckles photobleached at a single step, which confirmed that the speckles consisted of single DL fluorophore (Figure S4). All observed cells contained DL549-actin at similar densities (three independent experiments, total n = 72 cells), indicating the electroporation method is highly consistent and would help researchers unfamiliar with single-molecule observation to find the cells labeled at the optimal density. DL549-actin speckles incorporated into actin structures, including LP, stress fibers, and filopodia that are visualized by Lifeact-EGFP in live XTC cells (Figure 2C). XTC cells retained DL549-actin speckles for SiMS microscopy up to 3–4 d after electroporation (Figure 2D). There was no apparent cell damage after electroporation, as the morphology and actin organization were indistinguishable between DL549-actin–loaded cells and control cells without electroporation. Bright vesicle-like structures were often labeled by DL549-actin in electroporated cells (Figures 2C and S5), which were not found in microinjected cells. We speculate that the vesicle-like structure is an artifact arising from endocytosis of DL-actin during the electroporation procedure. Because such vesicle-like structures are markedly larger and brighter than SiMS and they move in stochastically changing directions (Figure S5), they can be easily distinguished from DL549-actin speckles in the cytoplasm and eliminated from the analysis.
Compared with an EGFP tag, the brightness and photostability of DyLight dyes are highly improved, which allows tracking of actin speckle trajectories on extended timescales. Indeed, with a 2-s exposure time, the 100-W mercury excitation could be attenuated to 1.5% using neutral density filters to achieve visualization of single-molecule DL549-actin in the filament (Figure 2, B and D). Importantly, photobleaching of DL549-actin was slow under this condition. Continuous exposure of the whole cell area to 1.5% of the mercury excitation for 600 s reduced DL549-actin fluorescence to only 76% of the original value (Figure 2E). Thus DL549-actin extends the temporal limit of SiMS analysis several-fold. The F-actin lifetime distribution in LP measured by DL549-actin showed a basically identical result to our previous data with EGFP-actin (Watanabe and Mitchison, 2002), and DL549-actin increased the accuracy of detecting long-lived speckles, including speckles with lifetimes longer than 3 min (Figure 2F).
We further verified rapid actin turnover by performing FRAP of DL488-actin speckles in LP. Multiple DL488-actin speckles reappeared in the bleached region within 30 s after bleaching (Figure S6 and Movie S11; 14/17 experiments, nine cells).
DL549-actin enables simultaneous analysis of actin dynamics with diverse timescales
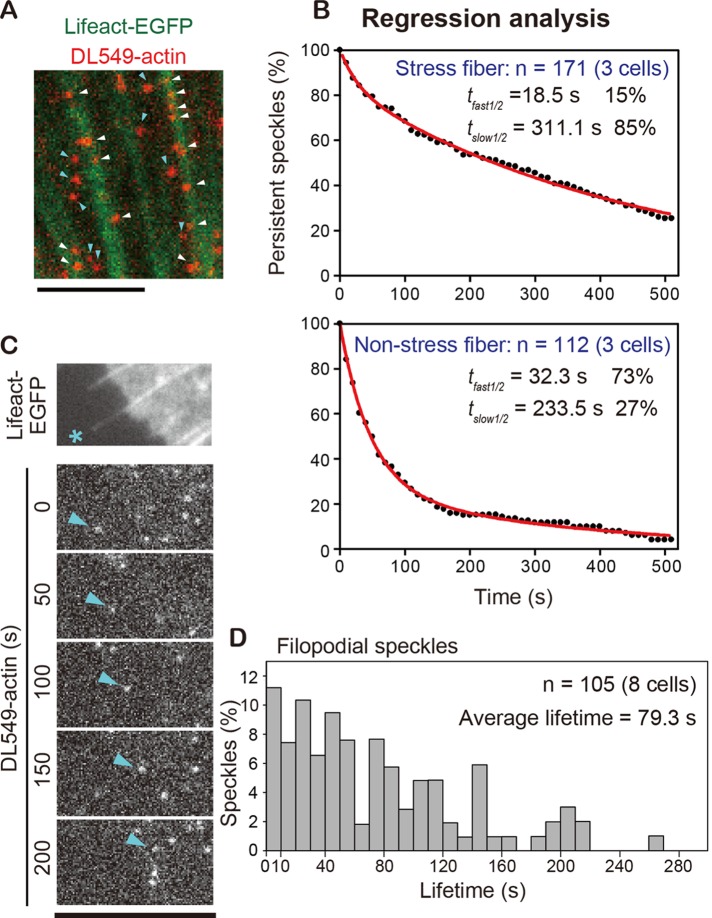
The extended photostability of DL549-actin provides an opportunity to simultaneously monitor actin structures with markedly distinct disassembly kinetics. We compared the lifetime of DL549-actin speckles associating with actin stress fibers (Figure 3A, white arrows) with that of speckles outside stress fibers (Figure 3A, blue arrows). We carried out regression analysis to follow the surviving fraction of preexisting actin speckles, which reports actin disassembly (Watanabe and Mitchison, 2002). The data revealed that DL549-actin speckles outside stress fibers (non–stress fiber) disassembled much faster than those associated with stress fibers (Figure 3B). A double-exponential fit of the data suggested that speckles in both the stress fiber-associating and non–stress fiber filaments comprised two populations with distinct disassembly rates. Stress fiber-associated filaments disassembled at t1/2 of 18.5 s (15%) and t1/2 of 311.1 s (85%). Non–stress fiber filaments disassembled at t1/2 of 32.3 s (73%) and t1/2 of 233.5 s (27%). Thus the majority of actin stress fibers disassembled with a half-life of 5 min, which is in agreement with a previous analysis of photoactivation of fluorescence (Theriot and Mitchison, 1991), whereas a large fraction of non–stress fiber actin filaments disassembled within 1 min. In addition, we analyzed the lifetime distribution of filopodia actin speckles (Figure 3, C and D). The average lifetime of actin filaments in filopodia was 79.3 s, indicating that filopodial actin filaments are more stable than LP filaments, whose average lifetime was 32.9 s (Figure 2F).
FIGURE 3:
Simultaneous regression analyses of rapid and stable actin dynamics within and outside of stress fibers. (A) Merged image of DL-549 actin speckles (red) and stress fibers visualized by EGFP-Lifeact (green). White arrows indicate speckles in stress fibers. Blue arrows indicate speckles that were outside stress fibers. (B) Speckle regression analysis of speckles in stress fibers (n = 171) or outside stress fibers (n = 112) in three cells. Red lines: the two-phase exponential curve fitted to the data gives 18.5 s (15%) and 311.1 s (85%) for the half-lives of speckles in stress fibers, whereas it gives 32.3 s (73%) and 233.5 s (27%) for speckles outside stress fibers. (C) Time-lapse images of a DL549-actin speckle (blue arrows) in filopodia (asterisk) paneled at 50-s intervals. (A–C) XTC cells were cultured on poly-l-lysine– and fibronectin-coated coverslips. (D) Lifetime distribution of DL549-actin speckles in filopodia (n = 105). Scale bars: 5 μm
Inhibition of myosin II decreases the retrograde flow rate in lamella but not in LP
SiMS analysis is often capable of revealing the rapid response of molecules in live cells to pharmacological treatments (Higashida et al., 2008; Fujita et al., 2009). We performed a time-resolved analysis of the effect of the myosin II ATPase inhibitor blebbistatin (Straight et al., 2003) on retrograde actin flows in LP and lamella (LM). DL549-actin is well-suited for combination with green-fluorescent blebbistatin, whose activity is lost upon blue-light irradiation (Kolega, 2004; Sakamoto et al., 2005).
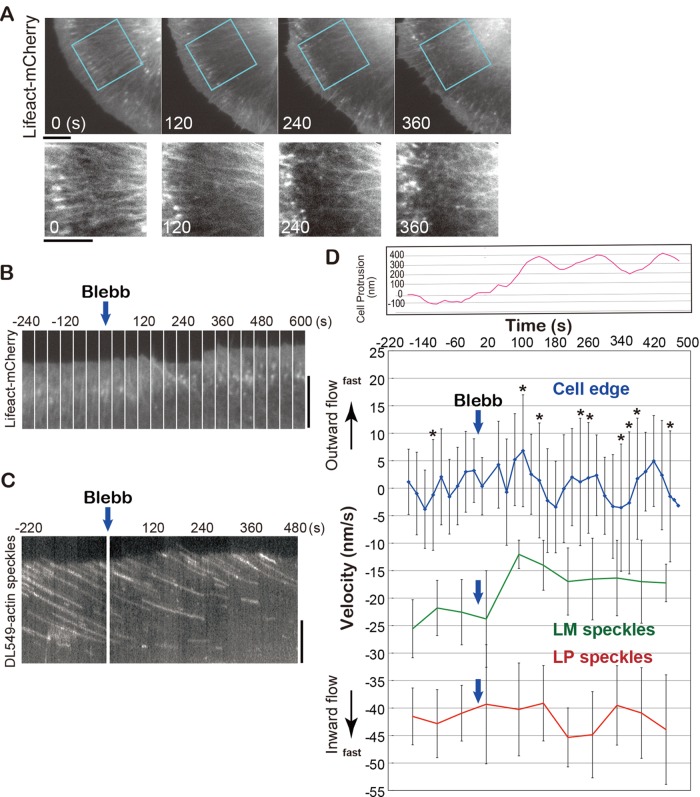
First we monitored the effect of blebbistatin on the whole actin network (n = 7 cells). One to two minutes after perfusion of 50 μM blebbistatin, Lifeact-mCherry fluorescence was reduced in a zone behind LP, and the LM actin bundles became disorganized (Figure 4A and Movie S6). After 2–4 min, the uniform intensity of Lifeact-mCherry in LP became uneven, the width of LP decreased, and cell protrusion and retraction activities gradually increased (Figure 4, A and B, and Movie S6). The LM actin network was transformed into randomly oriented meshwork-like structures in 4 min (Figure 4A).
FIGURE 4:
Effects of blebbistatin on cell peripheral actin networks. (A) Time-lapse images of cell peripheral actin networks visualized by Lifeact-mCherry at 120-s intervals after 50 μM blebbistatin treatment. The indicated areas are enlarged below. LM actin bundles organized in parallel at 0 s, became disorganized at 120 s, and deformed into meshwork-like structures at 240 s. (B) Time-lapse images of LP of a Lifeact-mCherry–expressing cell at 40-s intervals. (C) Kymograph showing the behavior of DL549-actin speckles at the cell periphery before and after 50 μM blebbistatin treatment. (D) Variations in average velocities of retrograde flows of LP speckles (red line) and LM speckles (green) and average velocity of the cellular edge (blue, 1° intervals for 85°) before and after blebbistatin treatment. Bars indicate SD. The asterisks indicate the SDs whose value is more than 10. The velocity is indicated in positive numbers (outward movement) or in negative numbers (inward movement). The retrograde flow velocities at each time point show the average velocities (with SD) of 20 speckles in LP or LM. Top panel shows the relative cell protrusion (pink line) calculated by the average cell-edge velocity. (A–C) XTC cells were cultured on poly-l-lysine–coated coverslips.
Kymograph analysis revealed that the speed of the retrograde flow was slower in LM than in LP (n = 8 cells; Figure 4C and Movie S7). The average speed of speckles in LM became slower within 1 min after the blebbistatin treatment (Figure 4D, green line), whereas the speckle speed in LP was not altered (Figure 4D, red line), indicating that myosin II–dependent force drives the actin flow in LM but not in LP in XTC cells. We also tracked the cell edges over time using the JFilament plug-in (Ryan et al., 2012) and measured the cell-edge radial velocity with respect to a fixed point near the center. The SDs of cell-edge radial velocities at 1° intervals for 85° were larger after the blebbistatin treatment than before the treatment (Figure 4D, blue line), indicating that both cell-edge protrusion and retraction occurred after the treatment. On average, the cell edge protruded by 300 nm ∼90 s after the blebbistatin treatment (Figure 4D, pink line). Thus the primary target of blebbistatin appears to be the myosin II contractility that drives the actin flow and organizes the actin bundles in LM. In contrast, extensive cell protrusion and retraction occur later than blebbistatin's inhibition of myosin II activity, suggesting that the drug's effect on the cell-edge dynamics is indirect.
Nanometer-scale displacement measurement defines unified retrograde flow in LP
The above results suggest that the flow rates of two actin networks, LP and LM, are separable by the dependence on myosin II activity. On the other hand, a previous qFSM study proposed that actin filaments with two distinct retrograde flow rates coexist in LP of newt lung epithelial cells (Ponti et al., 2004). This is referred to as the lamella hypothesis because the slower flow speed is equivalent to that in LM. However, another study that used several speckle-tracking approaches failed to detect such slowly migrating fluorescent actin speckles in LP (Vallotton and Small, 2009). The authors of the latter study argued against the fidelity of the qFSM method, although both studies (Danuser, 2009; Vallotton and Small, 2009) agreed that there still remained the possibility that slowly migrating actin speckles with short lifetimes, which are not readily detectable by conventional tracking approaches, might exist in the LP.
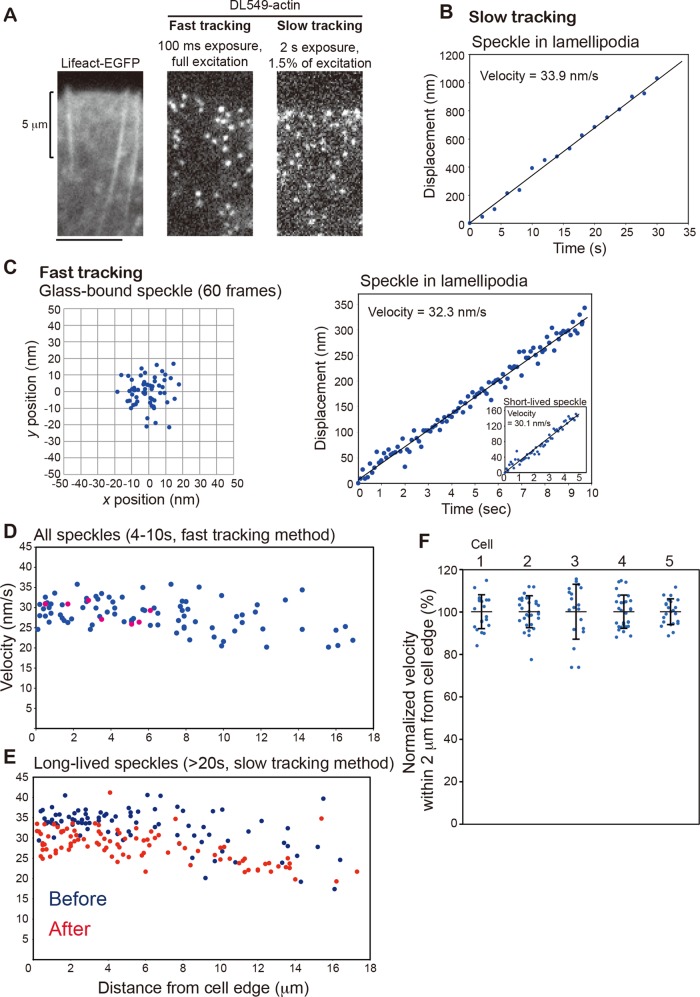
Analysis of DL549-actin speckles in the LP of XTC cells revealed that short-lived actin filaments account for one-third of DL-actin speckles in the LP actin network (745 of 2463 speckles with 2- to 8-s lifetimes, Figure 2F) and could govern a large part of actin flow speed distribution. We increased the accuracy of displacement measurements of short-lived actin speckles by the following two factors: 1) The use of bright DL549-actin illuminated by an unattenuated 100-W mercury-arc lamp with a strong emission peak at 546 nm, enabling monitoring of DL-actin speckles with a high signal-to-noise (S/N) ratio (Figure 5A, middle). We acquired images at 100-ms intervals (fast tracking) for the short duration of 10 s. 2) Subpixel localization of the speckle's centroid using the two-dimensional Gaussian-Fit model of Speckle TrackerJ software (Smith et al., 2011). Under this condition, the centroids of immobile DL549-actin speckles that were immobilized on the glass surface were distributed with an SD of 8.08 and 8.39 nm in the x- and y-axes, respectively (Figure 5C), indicating ∼8-nm accuracy of our position measurement. The localization accuracy of the Gaussian fit model depends on the S/N ratio, as previously demonstrated with simulated images (Cheezum et al., 2001; Sbalzarini and Koumoutsakos, 2005; Smith et al., 2011). From the S/N ratio of the immobile speckle in Figure 5C, the SD of the speckle position in one direction was also estimated to be 8.2 nm. We calculated the speed of individual speckles in LP by the linear fit of centroid displacement (Figure 5C).
FIGURE 5:
Nanometer displacement measurements of DL549-actin speckles in LP. (A) Images of EGFP-Lifeact (left), DL549-actin speckles with fast tracking (middle) or slow tracking (right). One pixel is 100 × 100 nm2. XTC cells were cultured on poly-l-lysine–coated coverslips. (B) Displacement of DL549-actin in LP in the series of slow-tracking images (16 frames). (C) Left, dispersion of the central position of the immobile glass-bound DL549-actin speckle in the series of fast-tracking images (60 frames). The central position was determined by the Gaussian-Fit model of Speckle TrackerJ software. The SDs of the centroid positions in one direction are 8.08 nm for the x-axis and 8.39 nm for the y-axis. Right, displacement plot of the central position of DL549-actin in LP in the series of fast-tracking images (100 frames). The velocity was calculated from a linear fit. Inset shows a displacement plot of a short-lived speckle that appeared and disappeared during fast tracking. (D and E) Retrograde flow velocity plotted against distance from cell edge of all measurable speckles with the fast-tracking method (D) and long-lived speckles (> 20-s lifetime) with the slow-tracking method (E). Pink dots in (D) indicate short-lived speckles that appeared and disappeared in the 10-s acquisition window. The slow-tracking images were acquired before (E, blue dots) or after (E, red dots) taking the fast-tracking image (D). (F) Retrograde flow velocities of actin speckles that flowed within 2 μm from the cell edge (n = 5 cells). The velocities of DL549-actin speckles were calculated with the fast-tracking method, and then were normalized with the average velocity of all counted speckles in each cell. Bars indicate mean ± SD. Cell 1 corresponds to the cell shown in (D); cells 2–5 correspond to cells 2–5 in Figure S7.
The speed distribution of all speckles near the cell edge measured by the fast-tracking method (Figure 5D) was comparable with that of the long-lived speckles (lifetime >20 s) measured by the conventional tracking (slow tracking) method using images acquired at 2-s intervals with attenuated illumination (Figure 5, A, B, and E). The data in Figure 5, D and E, were obtained in three consecutive movies acquired in the order of slow tracking, fast tracking, and slow tracking in the same field of a cell (data from two slow-tracking speckles are indicated as “before” and “after” in Figure 5E). The actin network exhibited typical LP morphology, as indicated by Lifeact-EGFP within 4 μm of the cell edge (Figure 5A, left). In the LP zone, the speed of all speckles measured by the fast-tracking method was 29.7 ± 2.66 nm s−1 (Figure 5D). Those speckles in the fast-tracking movie contained actual short-lived speckles that appeared and disappeared in 10 s (Figure 5D, pink dots), and their speeds were similar to those of the other speckles. The speed of the long-lived speckles in the LP zone was 34.5 ± 2.05 nm s−1 before and 29.7 ± 2.49 nm s−1 after the fast-tracking measurement, which is comparable with the speckles’ speed measured by the fast-tracking method (Figure 5, D and E). The actin flow appears slightly slower after acquisition with the fast-tracking method than before, presumably due to photodamage to the cell (Millius et al., 2012). In the inner LM zone between 10 and 18 μm from the cell edge, the average speeds of speckles were slower, and their speeds were more varied than those in the LP (Figure 5, D and E). The speeds of speckles in the LM were 25.8 ± 4.31 nm s−1 by the fast-tracking method, 28.1 ± 5.77 nm s−1 (before), and 24.2 ± 3.19 nm s−1 (after) by the slow-tracking method. Although the length of LP and the speeds of actin flow varied from one cell to another, the uniform actin flow in LP was observed in all analyzed XTC cells (n = 5 cells; Figures 5F and S7).
These data indicate three points: 1) Bright DL549-actin signals under the unattenuated mercury-arc illumination and the Gaussian fit algorithm enable robust velocity measurement of speckles whose total displacement is only 100–150 nm, that is, within the diffraction limit of a light microscope. 2) The strong illumination used for the fast-tracking method rapidly decreases the speed of actin retrograde flow due to photodamage to the cell (Millius et al., 2012). Special care is required to minimize photodamage by restricting the illuminated area (Watanabe, 2012) and the duration for the nanometer-scale measurement in live cells. 3) In LP of XTC cells, actin filaments undergo retrograde movement with a single flow speed regardless of their lifetimes. Our data with XTC cells spread on a poly-l-lysine–coated surface do not support coexistence of F-actin populations with different flow speeds proposed in the lamella hypothesis.
Mature FAs but not nascent adhesions attract and influence the actin flow in LP
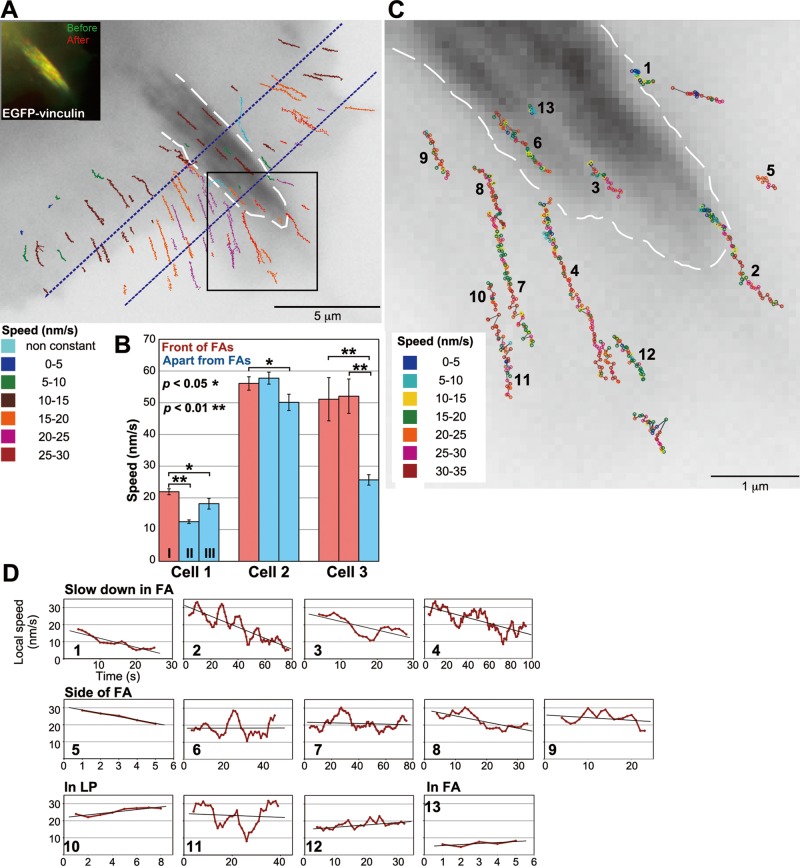
We investigated how FAs locally affect the actin flow in LP. We observed XTC cells that were spread on the glass coverslip doubly coated with poly-l-lysine and laminin (Figures 6, 7, and S8). Under that condition, cells formed nascent adhesions and mature FAs that were marked by EGFP-vinculin in flat LP (Figures 6A and 7A, insets). Strikingly, the trajectories of DL-actin speckles in front and to the side of mature FAs were biased toward the FAs (Figures 6A and S8 and Movie S8), suggesting mature FAs attract surrounding actin filaments.
FIGURE 6:
Effects of mature FAs on local retrograde flow at the cell periphery. (A) A speed and trajectory map of DL549-actin speckles in LP containing a mature FA. A white dotted line outlines the FA. Lines indicating the trajectories of DL549-actin speckles observed within a 100-s time window are shown in an image of EGFP-vinculin. Colors of lines indicate the average speed of speckles. For the speckles that slowed down in the FA area, the speeds before deceleration are shown. In the region between two dotted blue lines, the speed of DL-actin speckles are as follows: speckles flowing near the center of the FA, 9.61 ± 1.10 nm s−1 (n = 10, 2 movies); speckles flowing outside of the FA without changing velocity, 19.2 ± 5.23 nm s−1 (n = 37, 2 movies). The speed of DL-actin speckles in front of the FA was 23.6 ± 4.53 nm s−1 (n = 11, 2 movies). The inset indicates images of EGFP-vinculin before (green) and after (red) acquisition of the SiMS imaging for DL549-actin. (B) Comparison of regional actin flow speeds between the locations in front of FAs and apart from FAs in LP (n = 3 cells). Cell 1 corresponds to the cell in (A); cells 2 and 3 correspond to the cells 2 and 3 in Figure S8. The regions are shown in Figure S9. The speeds of all speckles that flowed inside and crossed the region, except for the speckles in FAs, were measured. The bars show the mean and the SE of speckles’ speeds in each region. For the speckles that slowed down near the FA area, we measured the speeds before speckles encountered FAs. *, p < 0.05, **, p < 0.01, one-tailed Student's t test. (C) A local speed and trajectory map of representative DL549-actin speckles in the boxed region in (A). Circles indicate locations of speckles in each frame. Gray lines link circles in sequences of frames. Local actin flow speeds were measured with a 5-s time window (5 frames), and the circles corresponding to the intermediate time point (the third frame) were colored according to speed. (D) The local speeds of speckles measured with a 10-s time window (10 frames) are plotted against time. The speckles with numbers correspond to the numbered speckles in (C).
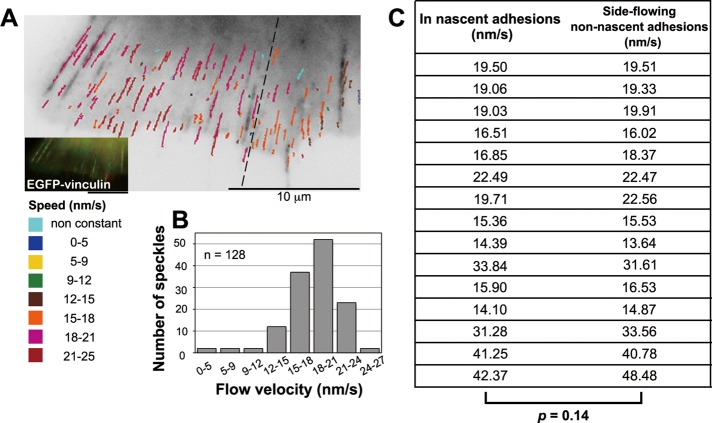
FIGURE 7:
Nascent adhesions have little effect on local retrograde flow at the cell periphery. (A) A speed and trajectory map of the DL549-actin speckles in LP containing nascent adhesions. The inset indicates images of EGFP-vinculin before (green) and after (red) acquisition of the DL549-actin movie (Movie S9). Lines indicating trajectories of DL549-actin speckles as in Figure 6A. (B) The flow speed distribution of DL549-actin speckles shown in (A). n = 128. (C) Comparison of speeds of DL549-actin speckles over nascent adhesions (in adhesion) with those of speckles that flowed near but outside nascent adhesions (side-flowing no adhesion). The speeds for side-flowing no adhesion are averaged from two no adhesion speckles (n = 15, four cells).
DL549-actin speckles in the center of mature FAs moved slower than those flowing in other parts of LP (Figure 6A). Interestingly, the speckles moved at faster actin flow rates in front of mature FAs than those in other areas of LP (Figure 6, A and B). The DL549-actin speckles flowing into the FA area gradually slowed down in front of FA (Figure 6, C and D, speckles 1–4), suggesting that mature FAs locally drag the actin flow. On the other hand, the speckles migrating along the side of FAs moved at a constant speed (Figure 6, C and D, speckles 5–9) that was similar to those flowing near the cell edge (Figure 6, C and D, speckles 10–12). The speckles occasionally moved away from the side of mature FAs, suggesting that they dodged the FAs, presumably because the mature FAs physically impeded the actin flow (Figure 6, C and D, speckle 8). We reproducibly observed these features of local actin flow near mature FAs (Figure S8 and Movie S10). These data indicate that mature FAs attract the flow in front and actively remodel the local actin networks.
In contrast, nascent adhesions had little local effect on the actin retrograde flow. Under that condition, nascent adhesions formed along the direction of the retrograde flow (Figure 7A and Movie S9). The speckles flowing over nascent adhesions moved at similar speeds to those flowing near the nascent adhesions. The distribution of actin flow rates showed one major peak in the LP containing nascent adhesions (Figure 7B). Although speckle speeds were locally similar, they differed between regions that were far apart in a single cell. The speckles moved at the faster speeds of 19.3 ± 2.92 nm s−1 (n = 88) on the left side of the dotted line in Figure 7A and slightly more slowly at 16.3 ± 2.17 nm s−1 (n = 40) on the right side (Figure 7A). In addition, actin speckles in LP of the cells spread on the laminin-coated surface tended to flow more slowly (18.3 ± 3.04 nm s−1; n = 128; Figure 7A) than those in LP of the cells spread on the poly-l-lysine coated surface (29.7 ± 2.66 nm s−1; n = 33; Figure 5D). We further compared the speed of DL549-actin speckles flowing over nascent adhesions to that of speckles that flowed near but outside the adhesions side by side in the same time frames (n = 15; 4 cells; Figure 7C). The flow speeds in and outside nascent adhesions showed no significant difference (p = 0.14), suggesting that nascent adhesions have little effect on the local retrograde actin flow speed (Figure 7C).
DISCUSSION
FSM has been used to measure the retrograde actin flow velocities by direct visualization. However, its limitations and shortcomings have become evident in recent years. It has been debated that speckle microscopy does not always report the flow velocities of the entire actin population (Danuser, 2009; Vallotton and Small, 2009). In this study, we introduce an improved method of SiMS microscopy for measuring actin dynamics. The key advantages of the present method are as follows: 1) A bright and photostable DL-actin enables F-actin lifetime analysis on diverse timescales, from seconds to tens of minutes. 2) In combination with the Gaussian fit of speckle centroids, DL-actin enables measurement of actin speckle displacement below the diffraction limit of light microscopy. 3) This is the easiest method for live-cell single-molecule imaging.
The new method revealed the retrograde flow rates, including those of short-lived actin speckles (lifetime <10 s). Our data indicate that actin filaments move at a uniform speed in LP. On the other hand, mature FAs, but not nascent adhesions, influence the actin flow velocity locally. Actin in front of mature FAs flows fast, and its direction is biased toward FAs, suggesting that mature FAs are actively engaged in pulling and remodeling the local actin network.
The new method extends the performance of SiMS microscopy
DL-actin improved spatial resolution of SiMS and enabled in vivo nanometer-scale movement analysis. In vitro, fluorescence imaging with 1-nm accuracy (FIONA) has been developed to track a single molecule at the nanometer scale (Michelotti et al., 2010) and has been used for studies of molecular motors (Yildiz et al., 2003, 2004) and nucleic acids (Pitchiaya et al., 2012). The step size of chemical fluorophore-labeled myosin V has been measured with 1.5-nm localization accuracy with this technique (Yildiz et al., 2003). With the same basic principles as FIONA, we achieved measurement of DL-actin speckles’ centroids with a low localization error of 8–8.5 nm with 100-ms temporal resolution. In the retrograde flow velocity measurement, speckle displacements of 100–150 nm for 3–4 s were sufficient for reliable measurements. Note that special care must be taken to minimize the photodamage to the cell, as our data indicate that retrograde flow velocity decreased under the unattenuated mercury illumination for 10 s.
DL-actin also extended the temporal resolution and time window of the SiMS analysis. Our data demonstrate that the average actin speckle lifetime was 32.9 s in LP and 79.3 s in filopodia. The majority of actin filaments in stress fibers disassemble with a half-life of 5 min, whereas a large proportion of filaments outside stress fibers disassemble in 1 min. The slow photobleaching of DL-actin would likely enable measurement of actin dynamics on diverse timescales, for instance, the formation and remodeling of thin filaments in myofibrils (Littlefield et al., 2001) and adherens junctions in epithelial cells (Zhang et al., 2005). DL-actin speckles are detectable for several days after electroporation, which is also advantageous for analyzing the dynamics of actin structures during cell differentiation.
Applying electroporation to introduce the fluorescently labeled DL-actin into cells will extend the availability of SiMS microscopy to many users. Electroporation is considerably easier for inexperienced researchers than microinjection. Additionally, electroporation is highly efficient, and the level of the DL-actin protein introduced into the cell can be well controlled, which increases the chance of finding cells suited for SiMS microscopy. In practice, the expression level of GFP-actin is difficult to control, and it is not easy to find cells that express GFP-actin at an optimal level for single-molecule imaging.
Lai and colleagues pointed out the disagreement between FRAP and SiMS studies (Lai et al., 2008). In the SiMS study, a large population of actin was found to be short-lived, and actin turnover was observed throughout LP (Watanabe and Mitchison, 2002). In contrast, the FRAP study suggested that actin assembles at the leading edge and disassembles at the back of LP (Lai et al., 2008). We confirmed that DL-actin reported basically identical results to our early SiMS data with EGFP-actin (Watanabe and Mitchison, 2002). Recently we found that the direct comparison between SiMS and FRAP data on the same cell types showed no large discrepancies between the SiMS and FRAP, and both experiments supported a broad distribution of actin turnover (Smith et al., 2013).
On the other hand, a small discrepancy remained. In combination with mathematical modeling, we proposed that slowly diffusing oligomer-actin might account for the discrepancy by slowing the actin recycling process in LP (Miyoshi et al., 2006; Miyoshi and Watanabe, 2013; Smith et al., 2013). Thus our series of work confirms the validity of our SiMS microscopy.
Actin labeled on lysine with DyLight incorporates effectively into mDia1- and mDia2-assembled filaments but poorly into FMNL2-bound filaments
Our data warn the usage of EGFP-actin might cause one to overlook the formin-based actin dynamics, due to interference between EGFP-actin and formins. We used DL-labeled actin on lysine to avoid the use of EGFP-actin for our SiMS microscopy, because fluorescently labeled actin on lysine has been shown to incorporate into mDia1-, mDia2-, and Bni1-associated filaments with ∼75% efficiency of formin-free filaments in vitro (Kovar et al., 2006). Although we confirmed that DL-actin assembles into mDia1- and mDia2-associated filaments with high efficiencies, DL-actin incorporates into FMNL2-associated filaments much less densely than in formin-free filaments. FMNL2 accumulates at the leading edge and regulates lamellipodial protrusion in B16-F1 melanoma cells (Block et al., 2012). The previous study demonstrated that fission yeast Cdc12p has rigorous structural requirements for actin subunits to incorporate into Cdc12p-mediated actin assembly (Chen et al., 2012). Similarly, FMNL2 and some other vertebrate formins might be intolerant of modification of actin. There are 15 formins in humans (Schonichen and Geyer, 2010). To develop actin probes that are fully compatible with all formins and to reveal the contribution of formins for actin assembly in LP, further studies are required.
Myosin II is dispensable for the retrograde flow in LP
Our data show that blebbistatin decreases the retrograde actin flow rate in LM, but not in LP. We observed newly formed LP in XTC cells 0.5–2 h after spreading on a poly-l-lysine–coated surface. Similarly, blebbistatin has no effect on the retrograde flow in newly formed LP in Jurkat T-cells (Babich et al., 2012). In these fresh LP, a dense actin meshwork most likely neither contains myosin II nor links to myosin II–dependent contractile structures in LM. It is currently unclear what molecular motors drive the retrograde flow in LP. Actin polymerization partly drives the retrograde flow in LP (Henson et al., 1999), but motor proteins other than myosin II might also pull the LP actin network.
Blebbistatin enhances protruding and retracting movements at the cell edge, but this behavior occurs following the deceleration of the actin flow rate in LM. These results suggest the primary target of blebbistatin is myosin II contractility in LM, and the drug's effect on cell-edge protrusion is most likely indirect. Thus time-resolved measurements unambiguously reveal the site of action of the drug.
Revisiting the lamella hypothesis using the in vivo nanometer-scale displacement measurement
Using DL-actin with desirable properties, we reevaluated whether two F-actin populations with distinct retrograde flow velocities coexist in LP, as postulated in the lamella hypothesis (Ponti et al., 2004). We show for the first time that short-lived LP speckles (lifetime <10 s), which move over a few hundred nanometers, flow with speeds similar to the other LP speckles. These results suggest that LP actin filaments form a tightly connected network experiencing a uniform flow rate, regardless of individual filament lifetimes.
In addition, we did not observe the coexistence of two F-actin populations with distinct retrograde flows in LP of XTC cells spread on a laminin-coated surface. In these cells, actin filaments in LP containing nascent adhesions flow at uniform speeds. Mature FAs influence the actin flow rates field across the LP, but actin filaments moving at distinct flow rates exist separately in LP rather than overlappingly. Taking together all of our findings, we conclude that the lamella hypothesis does not apply in LP of XTC cells. We suggest that the SiMS method will be useful for investigating the spatial variance of actin flow in other cell systems. Such studies would test whether the variability in retrograde flow speeds of short- and long-lived speckles reported by Ponti et al. (2004) is intrinsic to the particular cell types or an artifact arising from qFSM particle-tracking errors.
New SiMS microscopy dissects local actin flow near FAs
Previous qFSM studies have shown that the retrograde flow speed of actin speckles decreases when they approach FAs located at the transition zone between LP and LM (Hu et al., 2007; Gardel et al., 2008). In contrast, our present study investigated the interaction of the local actin flow with FAs located in the tip and middle portion of LP. Although nascent adhesions are small, which hinders the accurate measurement of local actin flow near FAs, our SiMS microscopy method enabled us to monitor the fine movement of individual actin filaments.
Our data show that actin filaments in LP flow uniformly even within nascent adhesions, whereas mature FAs locally obstruct actin flow. In addition, mature FAs attract the actin flow in front of FAs toward themselves by increasing the actin flow speed and changing the flow direction. These results suggest that mature FAs significantly influence the local actin flow in LP. In contrast, our study also revealed that individual nascent adhesions have little inhibitory effect on the local retrograde flow. The speed of DL-actin speckles flowing over nascent adhesions was 97.3 ± 0.06% of that of speckles flowing near but outside the adhesions (Figure 7C). However, as a whole, nascent adhesions may decrease the flow speed in LP. We conclude that LP actin filaments form a connected network via cross-linkers and move as a unit.
Mature FAs appear to accelerate and gather actin flows in front of FAs. This suggests that the interaction of mature FAs with the actin network might be more complex than a passive stick-slip interaction as previously suggested (Margadant et al., 2011; Wolgemuth, 2005). Instead, mature FAs may be actively engaged in pulling and remodeling the local actin network. Such actin flows may recruit components of FAs.
The observed modification of actin flow velocity by FAs must involve transformation of the actin network near and within FAs. The fast-moving actin networks flowing toward FAs are presumably engaged with FAs, which transform the fast-moving networks into slow-moving networks. On the other hand, part of the actin network flowing toward the side of FAs seems to dodge the FAs and flow along the sides of FAs at an unaltered speed. The rapid turnover of LP actin networks might indicate a flexible transformation mechanism of the network that allows it to adapt to obstructive FAs.
With the advantages described above, our new actin SiMS microscopy method will provide a powerful tool to unveil actin dynamics of various actin structures, which will provide valuable perspectives on the regulatory mechanisms of actin cytoskeleton functions. The improvements in the usability and success rate of the SiMS method with DL-actin will support the wide application of the method by researchers.
MATERIALS AND METHODS
DyLight-actin
Rabbit skeletal muscle actin was prepared from acetone powder by one cycle of polymerization and pelleting as described previously (Spudich and Watt, 1971). Seven milligrams of actin filaments in 3.5 ml of 50 mM KCl, 50 mM PIPES (pH 6.8), 0.2 mM CaCl2, 0.2 mM ATP was labeled with 0.8 mg of DyLight 488 or 549 NHS-ester (Thermo Fisher Scientific, Lafayette, CO) in 40 μl of N,N-dimethylformamide, which was a 4.9-fold molar excess to actin, for 2 h at 25°C. Nonreacted dye and denatured actin were removed by two cycles of pelleting, depolymerized in G-buffer (2 mM Tris, pH 8.0, 0.2 mM CaCl2, 0.2 mM ATP, 0.2 mM dithiothreitol), and polymerized. Labeled actin was depolymerized in G-buffer and then further purified by gel filtration with HiLoad16/60 Superdex 200 (GE Healthcare, Waukesha, WI). Our mass spectrometry showed that DL-NHS-ester could label one to three lysines of an actin molecule, but we were able to separate a fraction that mainly contained actin labeled at single site with DL by using gel filtration. We used DL-actin in the fraction containing single-DL–labeled G-actin. The concentration of labeled actin was measured by absorbance at 290 nm, 493 nm, and 562 nm using these extinction coefficients: total actin, A290 = 26,600 M−1cm−1; DyLight 488, A493 = 70,000 M−1cm−1; and DyLight 549, A562 = 150,000 M−1cm−1.
Unlabeled actin was purified from rabbit skeletal muscle actin as previously described (Mizuno et al., 2011). OGCys374-actin was prepared as previously described (Kovar et al., 2006). Recombinant GST-mDia1∆N3 (Higashida et al., 2004), GST-mDia2 FH1-FH2 (Block et al., 2012), GST-FMNL2 FH1-FH2 (Block et al., 2012), and human profilin I (Mizuno et al., 2011) were purified as previously described.
Plasmids and reagents
The expression vectors harboring delCMV (Watanabe and Mitchison, 2002) for EGFP-mDia1∆N3, mRFP1-actin, Lifeact-EGFP, and Lifeact-mCherry were as previously described (Higashida et al., 2004; Ryan et al., 2012). Human vinculin cDNA (GenBank accession number: BC039174) was obtained from Open Biosystems and was subcloned into pEGFP-C1 (Clontech, Mountain View, CA). pGEX6P-FMNL2-8P-FH1-FH2-C and pGEX6P-mDia2-FH1-FH2 were gifts from Klemens Rottner (University of Bonn) and Jan Faix (Hannover Medical School), respectively. (S)-(-)-blebbistatin was from Tocris Bioscience (Bristol, UK).
Direct observation of actin elongation with TIRF
Observation of actin elongation from formins bound to the glass surface.
Flow cells were prepared with coverslips as previously described (Mizuno et al., 2011). GST-mDia1∆N3 (5 μM) or GST-FMNL2 FH1-FH2 (1.7 μM) was absorbed on the glass surface of a flow cell for 5 min at room temperature, and then flow cells were washed with 10% (wt/vol) bovine serum albumin (BSA) in Basic buffer (50 mM KCl, 10 mM imidazole-HCl, pH 7.0, 1 mM MgCl2, 1 mM ethylene glycol tetraacetic acid, 0.5% methylcellulose, 100 μg/ml glucose oxidase, 20 μg/ml catalase, 4.5 mg/ml glucose, 2 mM ATP). After 5 min, free BSA was washed out by Basic buffer. Actin polymerization was initiated by mixing 1 μM of DL549-G-actin (4% labeled) or DL488-G-actin (2% labeled) with Basic buffer, and then the mixture was immediately loaded into a flow cell.
Observation of elongating actin filaments trapped to the glass surface by NEM-myosin II.
We performed these observations following the previous study with slight modifications (Kovar et al., 2006). Briefly, 50 nM NEM-inactivated myosin II in 20 mM HEPES (pH 7.5), 800 mM KCl was absorbed on the glass surface of a flow cell for 5 min. Flow cells were washed with 1% (wt/vol) BSA in Basic buffer. After 5 min, free BSA was washed out by Basic buffer. One micromolar DL549-G-actin (4% labeled) or OGCys374-G-actin (10% labeled) was incubated with 3 μM profilin in G-buffer for 5 min at room temperature, and then 1 nM GST-mDia1∆N3, 7.5 nM GST-mDia2 FH1-FH2, or 7.5 nM GST-FMNL2 FH1-FH2 was added to the mixture. Actin polymerization was initiated by mixing with Basic buffer, and then the mixture was immediately loaded into a flow cell.
Fluorescence images were acquired using a IX71 microscope (Olympus, Tokyo, Japan) equipped with a UAPON 1.49 numerical aperture (NA), 100× TIRFM objective lens (Olympus) and KYMA laser (488 nm, 20 mW; Melles Griot, Carlsbad, CA) and 85YCA laser (561 nm, 25 mW; Melles Griot) excitation using the TIRF condenser (IX2-RFAEVAW, Olympus). Fluorescence was recorded on a CCD camera (UIC-QE; Molecular Devices, Sunnyvale, CA). Elongation rate was measured using the kymograph command in MetaMorph software.
Cell culture, microinjection, and electroporation
XTC cells were maintained as described previously (Watanabe and Mitchison, 2002). For microinjection, 9.2 μM DL549-G-actin (20% labeled) and 25 μM human profilin I were incubated in G-buffer on ice for 30 min. The mixture was centrifuged at 300,000 × g for 30 min, and then half of the supernatant was collected. Injections into XTC cells were performed with an Eppendorf FemtoJet (Eppendorf, Hamburg, Germany) and micromanipulator 5171 attached to an Olympus IX71 microscope.
DL549-actin was electroporated into XTC cells with the Neon transfection system (Invitrogen, Carlsbad, CA). DL549-G-actin (5 μM, 12% labeled) and 15 μM human profilin I were incubated in G-buffer on ice for 30 min, and then the mixture was centrifuged at 300,000 × g for 30 min. Half of the supernatant was collected and diluted to one-tenth its concentration with resuspension buffer R of the Neon transfection system just before electroporation. XTC cells were dissociated by trypsinization, collected by centrifugation, and then washed with serum-free 70% Leibovitz's L15 medium. The XTC cells were collected again as a pellet and then resuspended with the DL549-G-actin (0.5 μM) and profilin I (1.5 μM) mixture in resuspension buffer R at a density of 1.86 × 107 cells/ml. The cells were transferred into a reaction tip, two pulses (1005 V, 35 ms) were applied, and then the cells were washed with serum-free 70% L15 medium.
Live-cell imaging and FSM
Speckle imaging and live-cell imaging were carried out in XTC cells as previously described (Watanabe, 2012). Briefly, XTC cells were allowed to spread on a poly-l-lysine–coated coverslip in 70% L15 Leibovitz medium (Invitrogen) without serum. For observation of stress fibers and FAs, a poly-l-lysine and fibronectin–coated coverslip and a poly-l-lysine and laminin–coated coverslip (both from Sigma-Aldrich) were used, respectively, and the XTC cells were maintained in 70% L15 Leibovitz medium with 10% fetal calf serum. Time-lapse imaging was performed using a microscope (IX71, Olympus) equipped with 100-W mercury illumination, a Plan-Apo 100×, 1.40 NA oil objective (Olympus), and a cooled EMCCD camera (Evolve 512; Photometrics, Tucson, AZ).
We tracked speckles by using the Speckle TrackerJ plug-in (Smith et al., 2011). For the displacement measurement, the lifetime analysis, and the regression analysis, the speckles were tracked using the Constant-Velocity-NCC model, and then each track was carefully checked to exclude tracking errors. The data of lifetime analysis and regression analysis were normalized for photobleaching (Watanabe and Mitchison, 2002; Watanabe, 2012). For the nanometer-scale displacement measurement, we used the Adjustment model to refine the position of speckles to bring the speckle marks close to the center of intensity. We then followed the speckles by used the Gaussian-Fit model to adjust the position of speckles with subpixel accuracy by fitting a two-dimensional Gaussian to the intensity near a speckle mark. To measure cell-edge velocity, we tracked the cell edge over time using the JFilament plug-in (Ryan et al., 2012), and then measured the radial velocity with respect to an arbitrary fixed point around the center of the cell, similar to Ryan et al. (2012)
Supplementary Material
Acknowledgments
We thank Chiharu Higashida for her initial finding of the inhibitory effect of mRFP1-actin on the processive movement of mDia1ΔN3, Nahoko Higashitani and Atsushi Higashitani (Tohoku University) for help with time-of-flight mass spectrometry and data analysis, Klemens Rottner (University of Bonn) and Jan Faix (Hannover Medical School) for pGEX-FLMN2(8P)-C and pGEX-mDia2-FH1FH2, Kazumasa Ohashi (Tohoku University) for help with FRAP of DL549-actin, and Velia M. Fowler (Scripps Research Institute) for critical reading of the manuscript. This work was supported by Human Frontiers Science program grant RGP0061∕2009-C (N.W. and D.V.); NEXT program grant LS013 from the Cabinet Office, Government of Japan (N.W.); a grant from the Takeda Science Foundation (N.W.); a Grant-in-Aid for Young Scientists (S.Y.); and a Grant-in-Aid for Scientific Research on Innovative Areas (S.Y.).
Abbreviations used:
- DL
DyLight
- FA
focal adhesion
- FRAP
fluorescence recovery after photobleaching
- FSM
fluorescent speckle microscopy
- LM
lamella
- LP
lamellipodia
- SiMS
single-molecule speckle
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-03-0162) on February 5, 2014.
REFERENCES
- Abraham VC, Krishnamurthi V, Taylor DL, Lanni F. The actin-based nanomachine at the leading edge of migrating cells. Biophys J. 1999;77:1721–1732. doi: 10.1016/S0006-3495(99)77018-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizawa H, Sameshima M, Yahara I. A green fluorescent protein-actin fusion protein dominantly inhibits cytokinesis, cell spreading, and locomotion in Dictyostelium. Cell Struct Funct. 1997;22:335–345. doi: 10.1247/csf.22.335. [DOI] [PubMed] [Google Scholar]
- Babich A, Li S, O'Connor RS, Milone MC, Freedman BD, Burkhardt JK. F-actin polymerization and retrograde flow drive sustained PLCγ1 signaling during T cell activation. J Cell Biol. 2012;197:775–787. doi: 10.1083/jcb.201201018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block J, et al. FMNL2 drives actin-based protrusion and migration downstream of Cdc42. Curr Biol. 2012;22:1005–1012. doi: 10.1016/j.cub.2012.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheezum MK, Walker WF, Guilford WH. Quantitative comparison of algorithms for tracking single fluorescent particles. Biophys J. 2001;81:2378–2388. doi: 10.1016/S0006-3495(01)75884-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Nag S, Pollard TD. Formins filter modified actin subunits during processive elongation. J Struct Biol. 2012;177:32–39. doi: 10.1016/j.jsb.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danuser G. Testing the lamella hypothesis: the next steps on the agenda. J Cell Sci. 2009;122:1959–1962. doi: 10.1242/jcs.054379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danuser G, Waterman-Storer CM. Quantitative fluorescent speckle microscopy of cytoskeleton dynamics. Annu Rev Biophys Biomol Struct. 2006;35:361–387. doi: 10.1146/annurev.biophys.35.040405.102114. [DOI] [PubMed] [Google Scholar]
- Deibler M, Spatz JP, Kemkemer R. Actin fusion proteins alter the dynamics of mechanically induced cytoskeleton rearrangement. PLoS One. 2011;6:e22941. doi: 10.1371/journal.pone.0022941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Ning Chen W, Vee Sin Lee P, Liao K, Chan V. The influence of GFP-actin expression on the adhesion dynamics of HepG2 cells on a model extracellular matrix. Biomaterials. 2005;26:5348–5358. doi: 10.1016/j.biomaterials.2005.01.069. [DOI] [PubMed] [Google Scholar]
- Fischer M, Kaech S, Knutti D, Matus A. Rapid actin-based plasticity in dendritic spines. Neuron. 1998;20:847–854. doi: 10.1016/s0896-6273(00)80467-5. [DOI] [PubMed] [Google Scholar]
- Fujita A, Shishido T, Yuan Y, Inamoto E, Narumiya S, Watanabe N. Imatinib mesylate (STI571)-induced cell edge translocation of kinase-active and kinase-defective Abelson kinase: requirements of myristoylation and src homology 3 domain. Mol Pharmacol. 2009;75:75–84. doi: 10.1124/mol.108.051706. [DOI] [PubMed] [Google Scholar]
- Gardel ML, Sabass B, Ji L, Danuser G, Schwarz US, Waterman CM. Traction stress in focal adhesions correlates biphasically with actin retrograde flow speed. J Cell Biol. 2008;183:999–1005. doi: 10.1083/jcb.200810060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson JH, Svitkina TM, Burns AR, Hughes HE, MacPartland KJ, Nazarian R, Borisy GG. Two components of actin-based retrograde flow in sea urchin coelomocytes. Mol Biol Cell. 1999;10:4075–4090. doi: 10.1091/mbc.10.12.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashida C, Kiuchi T, Akiba Y, Mizuno H, Maruoka M, Narumiya S, Mizuno K, Watanabe N. F- and G-actin homeostasis regulates mechanosensitive actin nucleation by formins. Nat Cell Biol. 2013;15:395–405. doi: 10.1038/ncb2693. [DOI] [PubMed] [Google Scholar]
- Higashida C, Miyoshi T, Fujita A, Oceguera-Yanez F, Monypenny J, Andou Y, Narumiya S, Watanabe N. Actin polymerization-driven molecular movement of mDia1 in living cells. Science. 2004;303:2007–2010. doi: 10.1126/science.1093923. [DOI] [PubMed] [Google Scholar]
- Higashida C, Suetsugu S, Tsuji T, Monypenny J, Narumiya S, Watanabe N. G-actin regulates rapid induction of actin nucleation by mDia1 to restore cellular actin polymers. J Cell Sci. 2008;121:3403–3412. doi: 10.1242/jcs.030940. [DOI] [PubMed] [Google Scholar]
- Honkura N, Matsuzaki M, Noguchi J, Ellis-Davies GC, Kasai H. The subspine organization of actin fibers regulates the structure and plasticity of dendritic spines. Neuron. 2008;57:719–729. doi: 10.1016/j.neuron.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Hu K, Ji L, Applegate KT, Danuser G, Waterman-Storer CM. Differential transmission of actin motion within focal adhesions. Science. 2007;315:111–115. doi: 10.1126/science.1135085. [DOI] [PubMed] [Google Scholar]
- Jay DG. The clutch hypothesis revisited: ascribing the roles of actin-associated proteins in filopodial protrusion in the nerve growth cone. J Neurobiol. 2000;44:114–125. doi: 10.1002/1097-4695(200008)44:2<114::aid-neu3>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Kan-o M., Takeya R, Taniguchi K, Tanoue Y, Tominaga R, Sumimoto H. Expression and subcellular localization of mammalian formin Fhod3 in the embryonic and adult heart. PLoS One. 2012;7:e34765. doi: 10.1371/journal.pone.0034765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolega J. Phototoxicity and photoinactivation of blebbistatin in UV and visible light. Biochem Biophys Res Commun. 2004;320:1020–1025. doi: 10.1016/j.bbrc.2004.06.045. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124:423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Pollard TD. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci USA. 2004;101:14725–14730. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubow KE, Horwitz AR. Reducing background fluorescence reveals adhesions in 3D matrices. Nat Cell Biol. 2011;13:3–5. doi: 10.1038/ncb0111-3. author reply 5–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai FP, Szczodrak M, Block J, Faix J, Breitsprecher D, Mannherz HG, Stradal TE, Dunn GA, Small JV, Rottner K. Arp2/3 complex interactions and actin network turnover in lamellipodia. EMBO J. 2008;27:982–992. doi: 10.1038/emboj.2008.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlefield R, Almenar-Queralt A, Fowler VM. Actin dynamics at pointed ends regulates thin filament length in striated muscle. Nat Cell Biol. 2001;3:544–551. doi: 10.1038/35078517. [DOI] [PubMed] [Google Scholar]
- Malm B. Chemical modification of Cys-374 of actin interferes with the formation of the profilactin complex. FEBS Lett. 1984;173:399–402. doi: 10.1016/0014-5793(84)80813-3. [DOI] [PubMed] [Google Scholar]
- Margadant F, Chew LL, Hu X, Yu H, Bate N, Zhang X, Sheetz M. Mechanotransduction in vivo by repeated talin stretch-relaxation events depends upon vinculin. PLoS Biol. 2011;9:e1001223. doi: 10.1371/journal.pbio.1001223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelot A, Berro J, Guerin C, Boujemaa-Paterski R, Staiger CJ, Martiel JL, Blanchoin L. Actin-filament stochastic dynamics mediated by ADF/cofilin. Curr Biol. 2007;17:825–833. doi: 10.1016/j.cub.2007.04.037. [DOI] [PubMed] [Google Scholar]
- Michelotti N, de Silva C, Johnson-Buck AE, Manzo AJ, Walter NG. A bird's eye view tracking slow nanometer-scale movements of single molecular nano-assemblies. Methods Enzymol. 2010;475:121–148. doi: 10.1016/S0076-6879(10)75006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millius A, Watanabe N, Weiner OD. Diffusion, capture and recycling of SCAR/WAVE and Arp2/3 complexes observed in cells by single-molecule imaging. J Cell Sci. 2012;125:1165–1176. doi: 10.1242/jcs.091157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi-Mi L, Votra S, Kemphues K, Bretscher A, Pruyne D. Z-line formins promote contractile lattice growth and maintenance in striated muscles of C. elegans. J Cell Biol. 2012;198:87–102. doi: 10.1083/jcb.201202053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Cytoskeletal dynamics and nerve growth. Neuron. 1988;1:761–772. doi: 10.1016/0896-6273(88)90124-9. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ, Cramer LP. Actin-based cell motility and cell locomotion. Cell. 1996;84:371–379. doi: 10.1016/s0092-8674(00)81281-7. [DOI] [PubMed] [Google Scholar]
- Miyoshi T, Tsuji T, Higashida C, Hertzog M, Fujita A, Narumiya S, Scita G, Watanabe N. Actin turnover-dependent fast dissociation of capping protein in the dendritic nucleation actin network: evidence of frequent filament severing. J Cell Biol. 2006;175:947–955. doi: 10.1083/jcb.200604176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Watanabe N. Can filament treadmilling alone account for the F-actin turnover in lamellipodia. Cytoskeleton (Hoboken) 2013;70:179–190. doi: 10.1002/cm.21098. [DOI] [PubMed] [Google Scholar]
- Mizuno H, Higashida C, Yuan Y, Ishizaki T, Narumiya S, Watanabe N. Rotational movement of the formin mDia1 along the double helical strand of an actin filament. Science. 2011;331:80–83. doi: 10.1126/science.1197692. [DOI] [PubMed] [Google Scholar]
- Murthy K, Wadsworth P. Myosin-II-dependent localization and dynamics of F-actin during cytokinesis. Curr Biol. 2005;15:724–731. doi: 10.1016/j.cub.2005.02.055. [DOI] [PubMed] [Google Scholar]
- Oldenbourg R, Katoh K, Danuser G. Mechanism of lateral movement of filopodia and radial actin bundles across neuronal growth cones. Biophys J. 2000;78:1176–1182. doi: 10.1016/S0006-3495(00)76675-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitchiaya S, Androsavich JR, Walter NG. Intracellular single molecule microscopy reveals two kinetically distinct pathways for microRNA assembly. EMBO Rep. 2012;13:709–715. doi: 10.1038/embor.2012.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponti A, Machacek M, Gupton SL, Waterman-Storer CM, Danuser G. Two distinct actin networks drive the protrusion of migrating cells. Science. 2004;305:1782–1786. doi: 10.1126/science.1100533. [DOI] [PubMed] [Google Scholar]
- Ryan GL, Petroccia HM, Watanabe N, Vavylonis D. Excitable actin dynamics in lamellipodial protrusion and retraction. Biophys J. 2012;102:1493–1502. doi: 10.1016/j.bpj.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Limouze J, Combs CA, Straight AF, Sellers JR. Blebbistatin, a myosin II inhibitor, is photoinactivated by blue light. Biochemistry. 2005;44:584–588. doi: 10.1021/bi0483357. [DOI] [PubMed] [Google Scholar]
- Sarkar P, Sridharan S, Luchowski R, Desai S, Dworecki B, Nlend M, Gryczynski Z, Gryczynski I. Photophysical properties of a new DyLight 594 dye. J Photochem Photobiol B. 2010;98:35–39. doi: 10.1016/j.jphotobiol.2009.10.005. [DOI] [PubMed] [Google Scholar]
- Sbalzarini IF, Koumoutsakos P. Feature point tracking and trajectory analysis for video imaging in cell biology. J Struct Biol. 2005;151:182–195. doi: 10.1016/j.jsb.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Schneider ME, Belyantseva IA, Azevedo RB, Kachar B. Rapid renewal of auditory hair bundles. Nature. 2002;418:837–838. doi: 10.1038/418837a. [DOI] [PubMed] [Google Scholar]
- Schonichen A, Geyer M. Fifteen formins for an actin filament: a molecular view on the regulation of human formins. Biochim Biophys Acta. 2010;1803:152–163. doi: 10.1016/j.bbamcr.2010.01.014. [DOI] [PubMed] [Google Scholar]
- Severson AF, Baillie DL, Bowerman B. A formin homology protein and a profilin are required for cytokinesis and Arp2/3-independent assembly of cortical microfilaments in C. elegans. Curr Biol. 2002;12:2066–2075. doi: 10.1016/s0960-9822(02)01355-6. [DOI] [PubMed] [Google Scholar]
- Shinohara R, et al. A role for mDia, a Rho-regulated actin nucleator, in tangential migration of interneuron precursors. Nat Neurosci. 2012;15:373–380. S371–S372. doi: 10.1038/nn.3020. [DOI] [PubMed] [Google Scholar]
- Smith MB, Karatekin E, Gohlke A, Mizuno H, Watanabe N, Vavylonis D. Interactive, computer-assisted tracking of speckle trajectories in fluorescence microscopy: application to actin polymerization and membrane fusion. Biophys J. 2011;101:1794–1804. doi: 10.1016/j.bpj.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MB, Kiuchi T, Watanabe N, Vavylonis D. Distributed actin turnover in the lamellipodium and FRAP kinetics. Biophys J. 2013;104:247–257. doi: 10.1016/j.bpj.2012.11.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- Star EN, Kwiatkowski DJ, Murthy VN. Rapid turnover of actin in dendritic spines and its regulation by activity. Nat Neurosci. 2002;5:239–246. doi: 10.1038/nn811. [DOI] [PubMed] [Google Scholar]
- Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, Mitchison TJ. Dissecting temporal and spatial control of cytokinesis with a myosin II inhibitor. Science. 2003;299:1743–1747. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- Theriot JA, Mitchison TJ. Actin microfilament dynamics in locomoting cells. Nature. 1991;352:126–131. doi: 10.1038/352126a0. [DOI] [PubMed] [Google Scholar]
- Theriot JA, Mitchison TJ. Comparison of actin and cell surface dynamics in motile fibroblasts. J Cell Biol. 1992;119:367–377. doi: 10.1083/jcb.119.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallotton P, Small JV. Shifting views on the leading role of the lamellipodium in cell migration: speckle tracking revisited. J Cell Sci. 2009;122:1955–1958. doi: 10.1242/jcs.042036. [DOI] [PubMed] [Google Scholar]
- Wang YL. Exchange of actin subunits at the leading edge of living fibroblasts: possible role of treadmilling. J Cell Biol. 1985;101:597–602. doi: 10.1083/jcb.101.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N. Fluorescence single-molecule imaging of actin turnover and regulatory mechanisms. Methods Enzymol. 2012;505:219–232. doi: 10.1016/B978-0-12-388448-0.00020-6. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Mitchison TJ. Single-molecule speckle analysis of actin filament turnover in lamellipodia. Science. 2002;295:1083–1086. doi: 10.1126/science.1067470. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Ando Y, Yasuda S, Hosoya H, Watanabe N, Ishizaki T, Narumiya S. mDia2 induces the actin scaffold for the contractile ring and stabilizes its position during cytokinesis in NIH 3T3 cells. Mol Biol Cell. 2008;19:2328–2338. doi: 10.1091/mbc.E07-10-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer CM, Desai A, Bulinski JC, Salmon ED. Fluorescent speckle microscopy, a method to visualize the dynamics of protein assemblies in living cells. Curr Biol. 1998;8:1227–1230. doi: 10.1016/s0960-9822(07)00515-5. [DOI] [PubMed] [Google Scholar]
- Wolgemuth CW. Lamellipodial contractions during crawling and spreading. Biophys J. 2005;89:1643–1649. doi: 10.1529/biophysj.105.066720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- Yildiz A, Tomishige M, Vale RD, Selvin PR. Kinesin walks hand-over-hand. Science. 2004;303:676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- Zhang J, Betson M, Erasmus J, Zeikos K, Bailly M, Cramer LP, Braga VM. Actin at cell-cell junctions is composed of two dynamic and functional populations. J Cell Sci. 2005;118:5549–5562. doi: 10.1242/jcs.02639. [DOI] [PubMed] [Google Scholar]
- Zhou M, Wang YL. Distinct pathways for the early recruitment of myosin II and actin to the cytokinetic furrow. Mol Biol Cell. 2008;19:318–326. doi: 10.1091/mbc.E07-08-0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.