This study isolated many peptide aptamers containing the SxIP motif that binds to Drosophila EB1 and human EB1 and EB3. Interaction sequences are similar to but distinct from each other. Aptamers can competitively displace endogenous EB1-interacting proteins from microtubule plus ends, and their expression in developing flies alters microtubule dynamics.
Abstract
EB1 is a conserved protein that plays a central role in regulating microtubule dynamics and organization. It binds directly to microtubule plus ends and recruits other plus end–localizing proteins. Most EB1-binding proteins contain a Ser–any residue–Ile-Pro (SxIP) motif. Here we describe the isolation of peptide aptamers with optimized versions of this motif by screening for interaction with the Drosophila EB1 protein. The use of small peptide aptamers to competitively inhibit protein interaction and function is becoming increasingly recognized as a powerful technique. We show that SxIP aptamers can bind microtubule plus ends in cells and functionally act to displace interacting proteins by competitive binding. Their expression in developing flies can interfere with microtubules, altering their dynamics. We also identify aptamers binding to human EB1 and EB3, which have sequence requirements similar to but distinct from each other and from Drosophila EB1. This suggests that EB1 paralogues within one species may interact with overlapping but distinct sets of proteins in cells.
INTRODUCTION
Microtubules are a major constituent of the cytoskeleton in all eukaryotic cells. They are essential for cell morphogenesis and motility and form the spindle to segregate chromosomes during mitosis. Microtubules are polar filaments with two differentially regulated ends—a plus end and a minus end. Whereas minus ends are often anchored to subcellular structures, plus ends constantly switch between phases of growth and shrinkage and also interact with subcellular structures (Howard and Hyman, 2003). Therefore precise spatial and temporal regulation of microtubule plus ends is crucial for microtubule organization and function.
A growing number of proteins are known to localize to the polymerizing microtubule plus ends and are collectively called microtubule plus end–tracking proteins (also known as +TIPs; Akhmanova and Steinmetz, 2008). These proteins may regulate dynamics of microtubule ends, anchor them to subcellular structures, or be transported as a cargo. Among them, EB1 is considered to play a central role. It is one of a few proteins that directly bind growing microtubule plus ends and is responsible for recruiting most microtubule-tracking proteins through a direct interaction (Busch and Brunner 2004; Dzhindzhev et al., 2005; Akhmanova and Steinmetz, 2008).
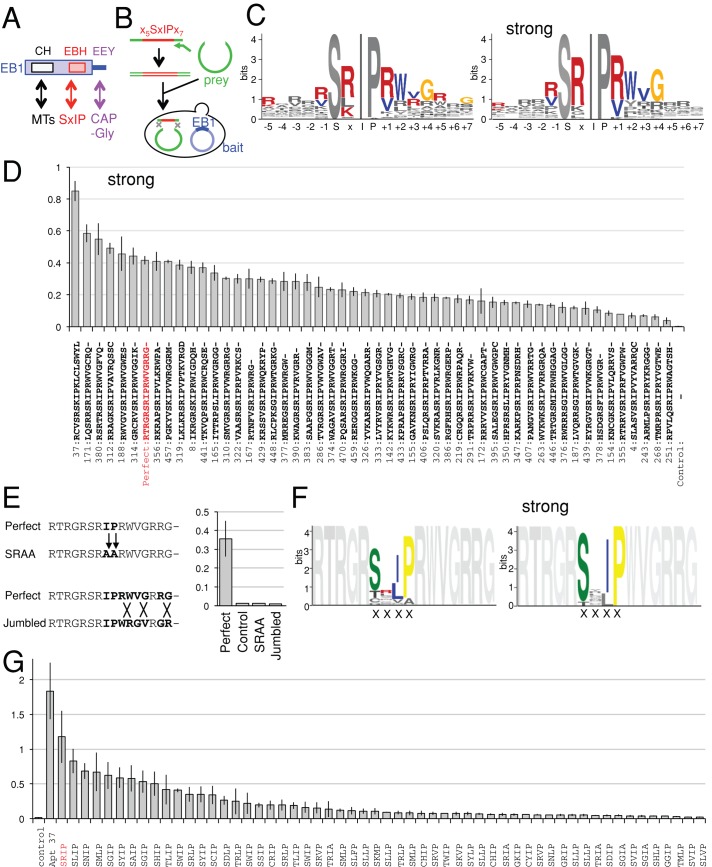
EB1 is a highly conserved eukaryotic protein (Slep et al., 2005). Although yeasts have only one gene, higher eukaryotes have multiple homologues in their genomes—for example, EB1, EB2, and EB3 in humans (Tirnauer and Bierer, 2000). EB1 and its homologues share an overall structure with a calponin homology (CH) domain in the N-terminal region, a central linker region, the EB1 homology domain (EBH), and a disordered tail containing EEY/F in the C-terminal region (Slep, 2010; Figure 1A). The CH domain is essential for association with microtubule plus ends (Zimniak et al. 2009). The EBH domain and the C-terminal EEY/F motif are the binding sites for EB1-interacting proteins (Honnappa et al., 2006, 2009).
FIGURE 1:
Aptamers revealed amino acid sequences surrounding SxIP that promote interaction with Drosophila EB1. (A) Diagram of the EB1 domain structure. The EBH domain of EB1 interacts with SxIP motifs of many microtubule plus end–binding proteins. (B) Isolation of Drosophila EB1 aptamers by yeast two-hybrid screening. Yeast (Y190) containing the EB1 bait plasmid was cotransformed with a linearized prey plasmid and DNA encoding xxxxxSxIPxxxxxxx flanked by sequences corresponding to a prey plasmid for gap repair. (C) Amino acids surrounding SxIP motif overrepresented among EB1 aptamers, and EB1 aptamers with strong interaction. The total height of each stack represents the “information content” in bits at each position and is divided by the frequency of each residue (Bailey et al., 2006). Colored residues indicate that they are more frequently found in aptamers (p < 0.01) than in nonselected peptides and random peptides expected from codon usages. (D) Strength of two-hybrid interactions of the 51 strongest aptamers or aptamer Perfect (marked in red) with EB1. The expression of the reporter gene LacZ was measured by quantitative assay for β-galactosidase activity and normalized for cell density (A420/A600). The empty bait plasmid was used as control. Bars, SEM (n = 3). Full sequence of Aptamer 37, RCVSRSKIPKLCLSWYLIRAREIYES. (E) Two controls (SRAA, Jumbled) generated by mutating aptamer Perfect. Strength of two-hybrid interactions were compared between aptamers Perfect, SRAA, and Jumbled. Bars, SEM (n = 3). (F) Residues within the SxIP motif overrepresented among 56 interactors showing any interaction with EB1 and among the 15 strongest EB1 interactors selected from a library based on the aptamer Perfect sequence in which SRIP was replaced with four random residues (XXXX library). Colored residues indicate that they are more frequently found in aptamers (p < 0.01) than in nonselected peptides and random peptides expected from codon usages. (G) Strength of two-hybrid interactions between EB1 and aptamers from a screen of peptide sequences in which SRIP of aptamer Perfect was replaced with four random residues. It is measured by a quantitative assay for β-galactosidase activity and normalized for cell density (A420/A600).
These two binding sites mediate two distinct modes of interaction through two types of EB1-interacting motif (Figure 1A). One is the CAP-Gly domain, which is present in the dynactin subunit p150Glued and the CLIP-170 family of proteins (Steinmetz and Akhmanova, 2008) and binds the C-terminal EEY/F motif of EB1 (Honnappa et al., 2006). The second motif is the Ser–any residue–Ile-Pro (SxIP) motif, which has been identified in a large number of diverse EB1-interacting proteins (Honnappa et al., 2009; Jiang et al., 2012). This motif binds the EBH domain of EB1 (Honnappa et al., 2009). The SxIP motif was originally characterized as capable of mediating protein interactions with EB1, and an SxIP-containing, 30–amino acid peptide derived from MACF2 has been cocrystallized with EB1 (Honnappa et al., 2009). Although many proteins contain sequences potentially matching the SxIP motifs, only some of these actually bind EB1 and are recruited to microtubule plus ends. Residues comprising and surrounding the SxIP motif of MACF2 were systematically replaced one by one and tested for EB1 binding (Buey et al., 2012), highlighting the contribution of surrounding sequences to EB1 interaction.
EB1 influences the microtubule plus end behavior not only by recruiting other proteins, but also by a direct effect on microtubule dynamics (Akhmanova and Steinmetz, 2008). It has been shown that EB1 has important roles in cell division and microtubule organization and dynamics in various cell types. In cultured Drosophila cells, knockdown of EB1 dramatically reduced the dynamicity of microtubules and increased microtubule pausing in interphase. It also resulted in abnormal organization and positioning of the spindle and reduced astral microtubules in mitosis (Rogers et al., 2002). In cultured mouse fibroblasts, EB1 is important for primary cilia assembly (Schroder et al., 2007). At the whole-animal level, mutations in Drosophila EB1 compromise neuromuscular function, especially function of the chordotonal sensory organs (Elliott et al., 2005). The roles of mouse EB1 and EB3 in myogenesis were studied using a cell line in which differentiation can be induced. It was found that knockdown of EB1 or EB3 prevents elongation and fusion of myoblasts into myotubes (Straube and Merdes, 2007; Zhang et al., 2009). Overexpression of EB3 can restore cell elongation but not cell fusion (Zhang et al., 2009). This suggests that EB1 and EB3 have overlapping but distinct functions during myogenesis in mouse. EB1 homologues are likely to play crucial roles in cell morphogenesis during differentiation in many cell types. It is a challenge to investigate roles of proteins essential for mitosis, such as EB1, in differentiated cells, as their removal disrupts cell divisions that form precursors of the differentiated cells.
In this study, we use peptide aptamers to further understand the interaction of EB1 with other proteins. Peptide aptamers are small proteins containing a peptide region typically 10–25 amino acids long, which are capable of binding target molecules with high affinity and specificity (Colas et al., 1996; Seigneuric et al., 2011). High-level expression of an aptamer can competitively disrupt a specific protein–protein interaction and thus inhibit a specific protein function (Cohen et al., 1998). Peptide aptamers can define residues involved in protein–protein interactions and so can provide the basis for small-molecule design (Fabbrizio et al., 1999; Butz et al., 2000; Baines and Colas, 2006). Aptamers can be used in designing small-molecule screens in the search for compounds that can bind to the target protein and thus displace the aptamer. Aptamers have advantages over RNA interference (RNAi) or genetic methods for drug target discovery or validation, as they can affect specific interactions rather than lower global levels of a target (Crawford et al., 2003). An ability to potentially inhibit specific protein interactions at desired times and in desired cell types in a whole organism (Kolonin and Finley, 1998, 2000; Yeh et al., 2013) makes them powerful tools to dissect protein functions and molecular pathways in cells and living organisms. Here we describe the isolation of a large number of aptamers that bind Drosophila EB1, human EB1, and human EB3. We use them to identify SxIP-containing peptides that show an increased affinity to EB1 and to identify residues within the motif that determine EB1 homologue binding specificity. We also explore various methods to isolate high-affinity EB1 aptamers that can successfully compete with natural EB1-interacting proteins to displace them from microtubule ends and alter microtubule dynamics.
RESULTS
Isolation of EB1 aptamers using yeast two-hybrid screening
Peptide aptamers are useful tools for identifying interaction motifs and manipulating protein–protein interactions in vitro and in vivo (Fabbrizio et al., 1999; Wickramasinghe et al., 2010; Yeh et al., 2013; Supplemental Figure S1). We sought to systematically identify EB1 aptamers, peptides that bind to EB1. We used yeast two-hybrid screening to screen a large number of semirandom peptides for interaction with full-length Drosophila EB1 (Figure 1B). The peptide prey library used for screening was designed to express peptides with core motif SxIP preceded by five and followed by seven random residues, as this region was shown to be sufficient for interaction with EB1 (Honnappa et al., 2009; Buey et al., 2012). Oligonucleotides encoding these SxIP-containing peptides were synthesized with flanking sequences corresponding to the ends of a linearized prey vector to facilitate library construction by gap repair in yeast. Five million transformants were screened, of which ∼500 showed activation of both reporter genes. Because a large majority of clones in the library contained SxIP (Supplemental Figure S2) but did not interact with EB1, SxIP alone is not sufficient for the interaction.
EB1 aptamers reveal SxIP-flanking sequences that promote EB1 binding
To determine which sequences were capable of interacting with EB1, we sequenced the plasmid insert from 45 positive transformants from the two-hybrid screen. As a control, we also sequenced 39 random transformants not selected for activation of reporter genes, which reflect the composition of a library used for screening. A large majority of the sequences from these control nonselected transformants encoded peptides with SxIP and an unbiased mixture of residues at other positions (Supplemental Figure S2).
Sequencing of the 45 EB1 aptamers revealed that they have sequences significantly different from those of control nonselected peptides and also from theoretical random peptides (Figure 1C and Supplemental Figure S3). Detailed analysis showed that some residues are significantly overrepresented at certain positions. Of note, we saw preferences of arginine or lysine at the x position of the SxIP motif, arginine or valine at the +1 position (the next residue after SxIP), tryptophan at the +2 position, and glycine at the +4 position. This gives a consensus of (R/K)Tx(R/F)(R/V)S(R/K)IP(R/V)WVGRxG, which represents the most preferred residue at each position (x representing no significant preference).
Selection of strong interactors
Our goal was to isolate strong aptamers that tightly associate with EB1 and could potentially compete with endogenous proteins for EB1 interaction. We also hoped that analysis of the sequence of aptamers that showed a particularly high affinity for EB1 would inform us about the residues that were essential for this strong interaction. It is known that activation of two hybrid reporter genes can be roughly correlated with the affinity of a protein–protein interaction (Estojak et al., 1995). To identify the aptamers that potentially had the highest affinity among those identified in our screen, we measured the expression of a reporter, LacZ, in 281 transformants from the screen in a quantitative assay for β-galactosidase activity.
To identify aptamers with very strong two-hybrid interactions, we retested 51 transformants with the highest LacZ activation together in triplicate (Figure 1D). Aptamer 37 consistently gave the strongest two-hybrid interaction with EB1. Sequence analysis of these strong aptamers showed that the preferred residues among strong EB1 aptamers were similar to those among EB1 aptamers with any strength of interaction, but these residues appeared more frequently among strong aptamers (Figure 1C).
Creation of designer aptamers
To identify the highest-affinity EB1-binding aptamers, we decided to test the hypothesis that we could use semirational design to devise an aptamer sequence based on the consensus information available from the original screen. We generated a potential aptamer (“aptamer Perfect”) that consists of the most frequently presented amino acid at each position, RTRGRSRIPRWVGRRG. The interaction of aptamer Perfect was compared with that of the strongest aptamers found in the screens, using a quantitative two-hybrid assay. Aptamer Perfect gave strong reporter expression, which was comparable to the 10 strongest aptamers, although it was not the strongest (marked in red in Figure 1D). Because the preferred amino acids were chosen from aptamers, most of which have weak interactions, this demonstrated a novel, simple way to design very strong aptamers by examining the consensus without measuring and comparing the interaction of many aptamers.
To test the possibility that the strong interaction found for aptamer Perfect may simply reflect overall amino acid composition rather than a sequence specific interaction, we generated two control aptamers. In the first control (SRAA), the SRIP motif was mutated to SRAA (Figure 1E), and in the second (Jumbled) the order of the residues downstream of SxIP was changed without changing the overall composition (Figure 1E). Quantitative two-hybrid assays showed that these control mutant aptamers do not interact with EB1 (Figure 1E). This demonstrates that the two-hybrid interactions between this aptamer and EB1 are sequence-specific interactions.
Importance of the SxIP motif for EB1 binding
We then examined the involvement of the conserved residues within the SxIP motif in the EB1 interaction to test the possibility that other residues within this motif might result in a higher affinity. We performed a yeast two-hybrid screen for EB1 interaction using a library based on the aptamer Perfect sequence in which SxIP was replaced with four random residues (Supplemental Figure S4). In total, 57 positive transformants were isolated from ∼1 million screened. Sequence analysis showed a strong and significant preference for serine at the first position, isoleucine or leucine at the third position, and proline at the fourth position (Figure 1F). Of interest, we also found a low frequency of other amino acids among weak interactors, but in such cases this unusual amino acid was limited to only one of the positions (e.g., CRIP, SRVP or SRIA, but not CRVP; Figure 1, F and G). This suggests that some degree of flexibility is allowed within this motif, but only if the surrounding sequence is optimal for interaction with EB1.
Although arginine is strongly preferred at the second position for aptamers selected from SxIP-containing peptides, no such strong preference is observed when the SxIP residues are randomized but the flanking regions are kept the same. This suggests that if the surrounding sequence is optimal for interaction with EB1, more flexibility is allowed for the X position.
These results underline the importance of SxIP motifs. Change of these conserved residues within the motif can be tolerated to a limited degree but could not further increase the strength of the interaction.
Tandem repeats of aptamers
Endogenous EB1-interacting proteins often contain multiple SxIP motifs, and it was previously shown that tandem repeats of SxIP-containing regions increased the robustness of the localization at microtubule plus ends (Honnappa et al. 2009). We therefore generated constructs expressing tandem repeats of aptamer 37 or aptamer Perfect.
DNA encoding tandem repeats of these aptamers with flanking sequences for gap repair were synthesized commercially and tested in a yeast two-hybrid assay. The four-residue spacer SGSG was included between each repeat. Several transformants of each construct were checked for the integrity of the plasmid. From four repeats of aptamer 37, all transformants tested contained the plasmid as designed. In contrast, from seven repeats of aptamer Perfect, the number of the repeats varied between transformants, but within each transformant the number appeared stable. Although we scrambled codon usage as much as possible, it seems that during gap repair, recombination reduced the number of tandem repeats. We tested two, four, and seven repeats of aptamer Perfect and four repeats of aptamer 37 for two-hybrid interaction with EB1 and also tested two repeats of aptamer 37 that we later generated. A quantitative assay for reporter expression showed that whereas only two repeats of aptamer Perfect indeed increased the interaction, all other repeats of aptamers reduced the interaction, and four or seven repeats nearly abolished the interaction (Figure 2A).
FIGURE 2:
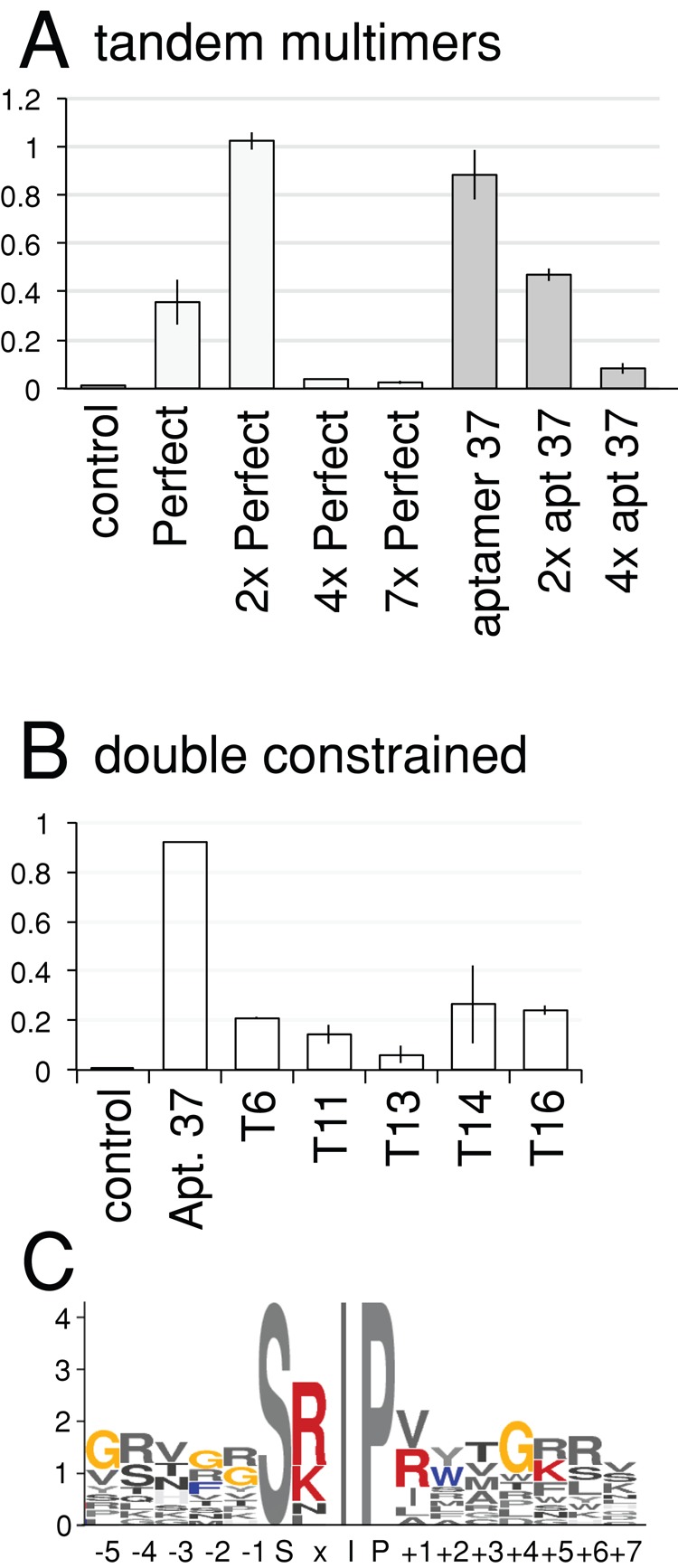
Tandem repeats of aptamers and double-constrained aptamers. (A) Strength of two-hybrid interactions of tandem repeats of aptamers Perfect and 37 with EB1. It is measured by a quantitative assay for β-galactosidase activity and normalized for cell density (A420/A600). Bars, SEM (n = 3). (B) Strength of two-hybrid interactions of double-constrained aptamers embedded in thioredoxin with EB1. Bars, SEM (n = 3; except aptamer 37, n = 1). (C) Residues overrepresented among double-constrained EB1 aptamers.
Isolation of double-constrained aptamers
Aptamers embedded in a scaffold protein (“double-constrained” aptamers) are often used, as they potentially have a higher affinity for their target, and more resistance to proteolysis, than single-constrained (or free) aptamers (Geyer and Brent, 2000; Supplemental Figure S1). To isolate double-constrained aptamers, we used random SxIP-containing peptides of 16 residues expressed as a constrained loop in the active site of thioredoxin (Supplemental Figure S4). These were screened for EB1 binding by the yeast two-hybrid system, and 18 positive transformants (prefixed with T in figures) were identified from 1 million screened. This was a significantly lower frequency than that seen with singly constrained aptamers (∼100/million), which was not unexpected, as both primary and secondary structures are significant in constrained aptamers. Sequencing showed that the preference of amino acids at each position in double-constrained aptamers was not significantly different from that seen in single-constrained aptamers (p > 0.08; Figure 2C and Supplemental Figure S5). Quantitative assays of reporter gene expression (LacZ) indicated that the double-constrained aptamers we isolated did not have a stronger interaction than the strongest single-constrained aptamers (Figure 2B). However, it is still possible that double-constrained aptamers embedded in a scaffold protein may be more active in cells because of increased stability.
Peptide aptamers tightly bind to EB1 in vitro
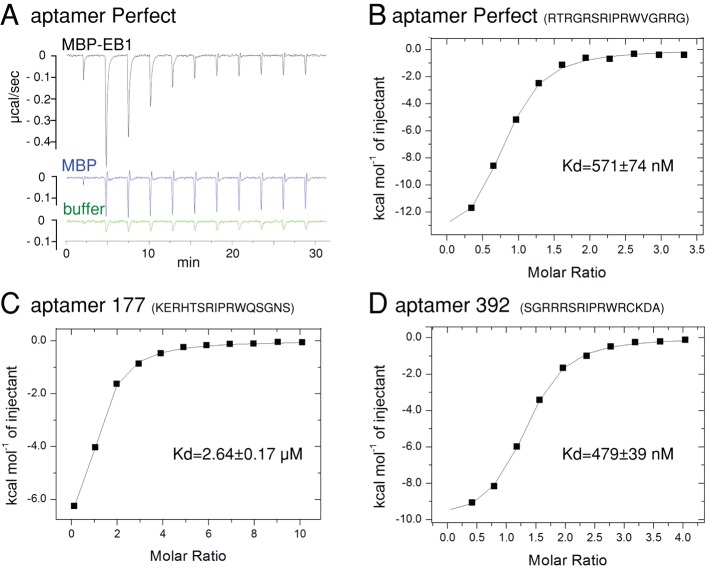
To estimate the affinity of aptamers to EB1 in vitro, we chemically synthesized peptides corresponding to aptamers and tested their interactions with recombinant maltose-binding protein (MBP) and MBP-EB1 by isothermal titration calorimetry (ITC). ITC measures heat released or absorbed when a protein–peptide interaction occurs, from which interaction parameters, including the dissociation constant (Kd), can be calculated. Peptides corresponding to the two aptamers with strong two-hybrid interactions (Perfect and 37) and two aptamers with weaker interactions (392 and 177) were synthesized. The peptide corresponding to aptamer 37 was not sufficiently soluble for ITC. The solubility of other synthetic peptides was also limited but sufficient to estimate rough Kd values. Recombinant MBP-EB1 and MBP were produced in Escherichia coli and purified. In an isothermal calorimeter, MBP-EB1, MBP, and buffer alone were titrated with aptamer Perfect peptide. The specific heat above the buffer alone was plotted against the molar ratio of the aptamer peptide over MBP-EB1 or MBP (Figure 3, A and B). MBP and the peptide did not show significant interaction. We estimated that the Kd between EB1 and each peptide is roughly 570 nM, 2.6 μM, and 480 nM, and for aptamers Perfect, 177, and 392, respectively (Figure 3, B–D). The Kd measurements by ITC in vitro did not perfectly correlate with the affinity estimates based on yeast two-hybrid data. The Kd between human EB1 and adenomatous polyposis coli protein was reported to be ∼5 μM (Honnappa et al. 2005), whereas more systematic analysis showed that the Kd between human EB1 and various native EB1-interacting proteins range between 1.5 and 140 μM (Buey et al., 2012). Therefore we isolated peptides that appear to have higher affinity for Drosophila EB1 than endogenous interacting proteins for human EB1.
FIGURE 3:
ITC shows a high affinity of aptamers to Drosophila EB1. (A) Raw ITC data showing that aptamer Perfect interacts with MBP-fused Drosophila EB1, but not MBP, in vitro. Heat released by titrations of 100 μM aptamer Perfect into 5 μM solution of MBP-EB1, MBP, and buffer alone. Each peak corresponds to one injection. An initial smaller injection was followed by 10 injections. For MBP-EB1, the heat became smaller for each injection, as the binding site became saturated. For buffer and MBP, it stayed constant, as heat was released only from dilution of the peptide without specific binding. (B–D) Integrated heat peaks subtracted by the heat of dilution and plotted against the molar ratio of the peptide for aptamer Perfect (B), 177 (C), or 392 (D) to MBP-EB1. The line represents the fit to the single-site binding model by the Origin program.
To gain insight into the high affinity of the aptamers, we carried out structural modeling of Drosophila EB1 complexed with aptamer Perfect or a fragment of endogenous EB1 interactor Sentin (Supplemental Figure S6). This revealed that aptamer Perfect is likely to interact with EB1 more strongly than the fragment of Sentin. The most crucial residues seem to be the ones at the positions +2 and +3 (WV) of aptamer Perfect. This fits nicely with our results that these two residues are significantly overrepresented at these positions among EB1 aptamers (Figure 1C).
EB1 aptamers can outcompete endogenous proteins for microtubule plus end localization in cells
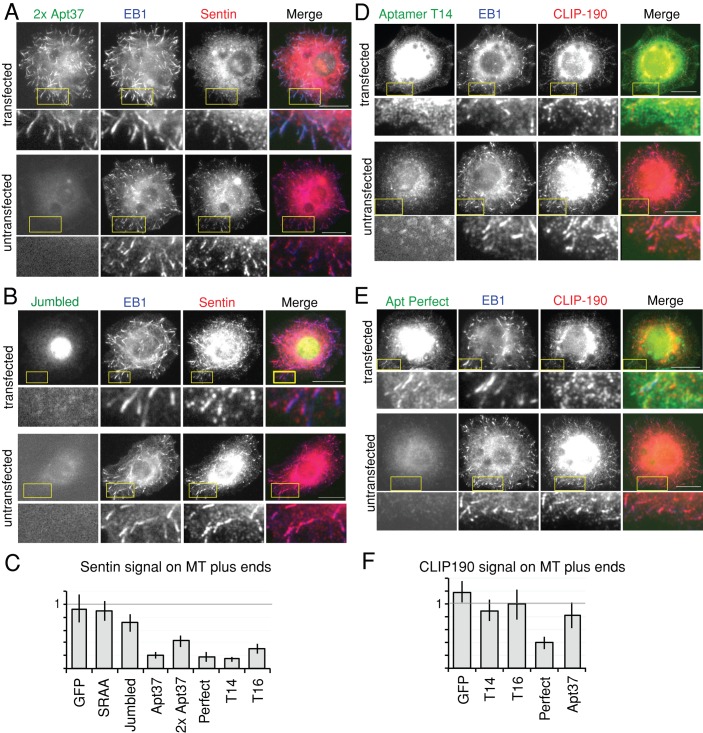
We identified EB1 aptamers with a high affinity of binding in vitro. To test whether these aptamers can bind to EB1 in living cells, we transiently expressed the aptamers as green fluorescent protein (GFP)-fusion proteins under the Actin5C promoter in the Drosophila S2 embryonic cell line. We tested several EB1 aptamers that gave strong two-hybrid interactions in yeast. The fixed cells were immunostained with antibodies against GFP and EB1 or α-tubulin. EB1 localized to microtubule plus ends in a comet-like shape as described (Mimori-Kiyosue et al., 2000). All GFP-aptamer fusion proteins we tested colocalized with EB1 comets at the microtubule plus ends, and signals of aptamers 37, Perfect, T14, and T16 at microtubule ends were particularly strong (Figure 4A). Therefore these aptamers can specifically recognize EB1 in the presence of other proteins in cells.
FIGURE 4:
Aptamers can be recruited to microtubule plus ends and displace the endogenous EB1 interactor Sentin in Drosophila cells. (A) Aptamer 37 dimer colocalizes with EB1 at microtubule ends and competes out Sentin from microtubule plus ends. (B) A mutant aptamer, Jumbled, does not colocalize with EB1 at microtubule plus ends or compete out Sentin from microtubule plus ends effectively. S2 cells were transfected with an expression plasmid expressing aptamer 37 dimer or aptamer Jumbled fused to GFP and immunostained for GFP, EB1, and α-tubulin. A typical transfected and an untransfected cell on the same slide are shown for comparison. Bar, 10 μm. Yellow boxes indicate areas magnified in the images below. (C) The specific signal intensity of Sentin at microtubule plus ends. Sentin intensities in aptamer-expressing cells relative to those in untransfected cells on the same slide are shown with SEM (n = 30). (D) Aptamer T14 colocalizes with EB1 at microtubule plus ends but competes out CLIP-190 from microtubule plus ends. (E) Aptamer Perfect colocalizes with EB1 at microtubule ends and interferes with the localization of CLIP-190 at microtubule plus ends. S2 cells were transfected with an expression plasmid expressing GFP-fused aptamer T14 or aptamer Perfect and immunostained for GFP, EB1, and CLIP-190. A typical transfected and an untransfected cell on the same slide are shown for comparison. Bar, 10 μm. Yellow boxes indicate areas magnified in the images below. (F) The specific signal intensity of CLIP-190 at microtubule plus ends. CLIP-190 intensities in aptamer-expressing cells relative to those in untransfected cells on the same slide are shown with SEM (n = 30).
To test whether aptamers can compete with endogenous EB1-interacting proteins containing SxIP motifs, we determined the localization of one such protein, Sentin. Sentin contains an SxIP motif in the C-terminal region and is considered to be a major cargo of EB1 in S2 cells (Li et al., 2011). S2 cells were transfected with a plasmid expressing a GFP-aptamer fusion protein as described and immunostained with antibodies against GFP, EB1, and Sentin (Figure 4A). In control cells not expressing aptamers, Sentin colocalized with EB1 at microtubule plus ends as expected. In contrast, in cells expressing aptamers, the Sentin signal was greatly reduced or absent from microtubule plus ends. To quantify this reduction, we measured the intensity of the Sentin signal colocalized with EB1 comets at microtubule plus ends in GFP-aptamer–expressing cells and nonexpressing cells on the same slide (Figure 4, A–D). We found that the Sentin signal was greatly reduced in GFP-aptamer–expressing cells relative to nonexpressing cells.
To test whether the effect of the aptamers is due to a sequence-specific interaction with EB1, we tested GFP alone, a mutant aptamer Perfect with SRIP mutated to SRAA (SRAA), and a mutant aptamer Perfect with the residues downstream of SxIP partially jumbled (Jumbled). The quantification of Sentin signals overlapping with EB1 comets showed that expression of GFP alone or of the SRAA mutant had no significant effect on Sentin localization (Figure 4C). However, the Jumbled mutant had a small but significant effect on Sentin localization (Figure 4, B and C). This suggests that the overall amino acid composition may contribute, to a small degree, to the interaction with EB1.
The binding sites of the two known EB1-binding motifs, SxIP motif and CAP-Gly, partially overlap on the C-terminal region of EB1 (Slep, 2010). We investigated whether CLIP-190, which is recruited to microtubules by CAP-Gly but also has a putative SxIP motif, can be outcompeted from the microtubule plus ends by peptide aptamers. Cells transfected with each aptamer and untransfected cells on the same slide were visually examined. CLIP-190 colocalized to EB1 at microtubule plus ends in cells transfected with aptamer 37, T14, or T16 to the same degree as in the untransfected cells (Figure 4, D and F). CLIP-190 colocalization with EB1 was weaker in cells expressing aptamer Perfect (Figure 4, E and F). CLIP-190 is predicted to bind to a site of EB1 distinct from but close to the SxIP-binding region (Honnappa et al., 2006), suggesting that this aptamer caused steric hindrance.
In conclusion, the aptamers we isolated can specifically bind EB1 in cells and displace endogenous EB1-interacting proteins from microtubule plus ends.
Expression of EB1 aptamers in developing flies alters microtubule dynamics
To test whether expression of EB1 aptamers in developing organisms can interfere with vital processes, we made transgenic flies expressing aptamers. Expression of EB1 aptamers fused to GFP controlled by a GAL4-inducible promoter (UASp) was driven by ubiquitously expressed GAL4 under the actin5C promoter. We found that fewer adult flies expressing aptamers were obtained in comparison with the control (Figure 5A). This suggests that aptamers interfere with microtubule function in developing flies.
FIGURE 6:
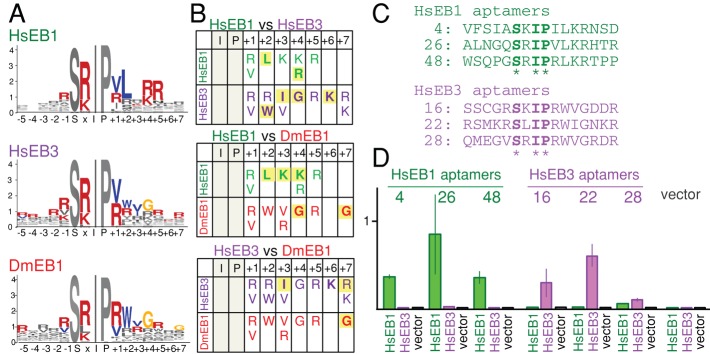
Aptamers for human EB1 and human EB3 have sequence requirements similar to but distinct from each other and aptamers for Drosophila EB1. (A) Residues overrepresented among aptamers to human EB1 (HsEB1), human EB3 (HsEB3), and Drosophila EB1 (DmEB1). Colored residues are more frequently found in aptamers (p < 0.01) than in nonselected peptides and random peptides expected from codon usage. (B) Three-way comparison of amino acid preferences between aptamers for three EB1 homologues. Residues whose frequencies are significantly different from nonselected or random peptides are shown. Yellow background highlights residues whose frequencies are significantly different between a pair of EB1 homologues (p < 0.01). (C) Selected aptamers that are likely to have differential interactions with human EB1 and EB3. (D) The strength of two-hybrid interactions of aptamers with human EB1 or EB3. Error bars, SEM.
To examine whether microtubule dynamics is altered, we used hemocytes from third-instar larvae because isolated cells can be adhered on a coverslip, allowing good resolution of microtubules. We first confirmed that the GFP-fused aptamers were expressed and localized as comets in hemocytes (Supplemental Movie S1). To test whether expression of the aptamers interferes with microtubule dynamics, we used EB1-GFP driven by the mild ubiquitin promoter to mark growing microtubule plus ends (Shimada et al., 2006; Parton et al., 2011). We compared the microtubule growth rate in hemocytes expressing GFP-fused aptamer and EB1-GFP with that in control hemocytes expressing EB1-GFP alone. We found that the microtubule growth was significantly slower in aptamer-expressing hemocytes than in control hemocytes (p < 0.01; Figure 5B and Supplemental Movies S2 and S3). This fits well with previous reports showing reduced growth rate of interphase microtubules in S2 cells depleted of EB1 or Sentin. Our results demonstrate that expression of aptamers can interfere with microtubule dynamics in a developing organism.
Aptamers to human EB1 and EB3 reveal different binding specificity between EB1 homologues
Many metazoans have multiple EB1 paralogues that are homologous within the SxIP-binding region (Tirnauer and Bierer, 2000). However, it is not clear whether these EB1 paralogues in a single species have different functions or interact with different proteins. It is also not clear whether EB1 orthologues in different species have different sequence specificity for interacting proteins. To gain insight into the sequence specificity of different EB1 homologues, we isolated aptamers that bind to human EB1 and human EB3 by yeast two-hybrid screening using the same methodology as for SxIP-containing aptamers of Drosophila EB1. We identified 47 human EB1- and 58 human EB3-positive transformants potentially containing human EB1 and human EB3 aptamers from 2 million SxIP-containing random peptides each.
Sequencing of these aptamers revealed similar but distinct sequence patterns of residues for each of the three EB1 homologues (Figure 6, A and B). Common features among aptamers for all three EB1 homologues include 1) no strong amino acid preferences except within SxIP and up to five downstream positions, 2) a preference for positive residues, especially arginine at the x position, and 3) a preference for valine or arginine at the +1 position. Most distinct features specific to individual EB1 homologues can be found at the positions from +2 to +4. Aptamers binding to human EB1 show a high frequency of leucine at the +2 position, whereas those binding to human EB3 and Drosophila EB1 show a clear preference for tryptophan. At the +3 position, aptamers binding to human EB1 showed a clear preference for lysine, but those binding to human EB3 showed a clear preference of isoleucine and valine. At the +4 position, human EB1 aptamer showed a preference for arginine, but those for human EB3 and Drosophila EB1 showed a preference for glycine. Statistical analysis showed that the frequencies of amino acids are significantly different (p < 0.01) at certain positions between EB1 homologues (marked with yellow background in Figure 6B).
FIGURE 5:
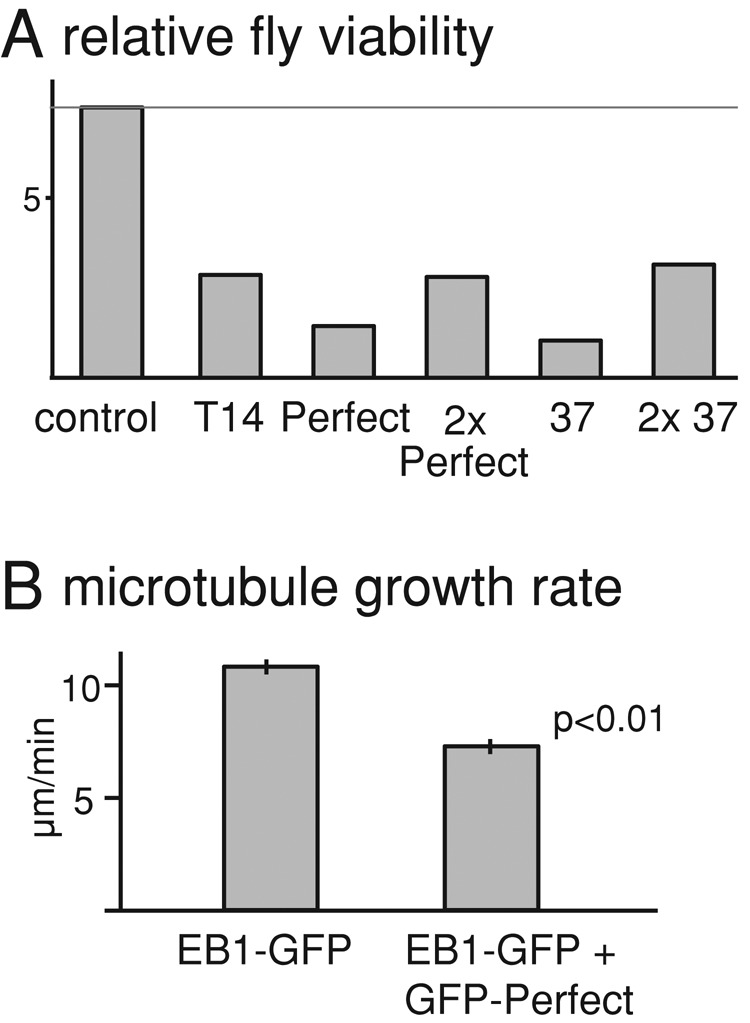
Expression of aptamers alters microtubule dynamics. (A) Number of adult flies expressing GFP-aptamer relative to number of balancer-carrying siblings that do not express GFP-aptamer (relative fly viability). The control has the identical genotype to experimental flies except that it does not carry aptamer transgenes. The relative viabilities of aptamer-expressing flies are significantly different from the control (p < 0.01, except for aptamer Perfect, which gives p = 0.02). (B) Microtubule growth rates at plus ends in hemocytes from third-instar larvae. Sibling larvae with or without expression of GFP-fused aptamer Perfect were compared. EB1-GFP driven by the ubiquitin promoter was used to mark growing microtubule plus ends. Error bars, SEM (n ≥ 146).
On the basis of the sequences, we selected three human EB1 aptamers unlikely to interact with human EB3, and three human EB3 aptamers unlikely to interact with human EB1 (Figure 6C), and tested the interactions with human EB1 and EB3 in a yeast two-hybrid assay. We found that five of six aptamers showed clear differential interactions as predicted. This demonstrates that human EB1 and EB3 can interact with different sequences (Figure 6D).
These results highlight the importance of residues downstream of the SxIP motif for binding specificity and raise the possibility that different EB1 paralogues may have overlapping but different sets of cargoes.
DISCUSSION
In this study we describe the isolation of peptide aptamers that bind to each of the three EB1 homologues, Drosophila EB1, human EB1, and human EB3. EB1 plays a central role in regulating microtubules by recruiting many proteins to growing microtubule plus ends. To improve the efficiency of screening, special biased aptamer libraries were created in which the conserved residues of the SxIP motif were invariant and the other residues randomized. Sequence analysis of these aptamers revealed the common sequence determinants that promote interaction with EB1 homologues and identified residues specific for interaction with each EB1 homologue. Furthermore, we developed novel methods to select and design aptamers that have high affinity to Drosophila EB1. One of the strongest-interacting peptides was estimated to bind to Drosophila EB1 with Kd of ∼570 nM in vitro, higher than known interacting proteins. We successfully demonstrated that the aptamers can competitively displace endogenous EB1-interacting proteins from microtubule ends and alter microtubule dynamics in developing flies.
These aptamers to the EB1 proteins were generated with two initial aims. The first was to gain information on sequences that promote interaction with EB1 and its homologues. The second was to facilitate development of tools that can be used to interfere with EB1 functions at specific times and in specific tissues.
The isolation of so many EB1 aptamers has provided vital information on the sequences required for EB1 binding. The SxIP motif was previously identified, on the basis that it is overrepresented in known EB1-binding regions and is essential for the interaction with EB1 (Honnappa et al., 2009). However, because many proteins that contain a simple SxIP do not interact with EB1 and do not localize to microtubule plus ends, the surrounding sequence is believed to be important for the EB1 interaction. This sequence requirement was studied by systematically replacing each residue within the SxIP motif and the flanking sequences of human MACF2 and testing the binding to human EB1 in vitro (Buey et al., 2012). We took an opposite approach. We started from random sequences and selected them for EB1 binding in vivo. These two results are largely consistent, but there are some differences in the pattern of sequence requirement. These differences may be due to the assay systems used, as our assays were carried out in the cellular context of yeast, whereas the other studies were carried out on cellulose membranes in vitro. Alternatively, they may be due to different sequences tested for interaction, as we selected interactors from a large combination of random residues at multiple positions, whereas those studies changed one position at a time from a known interacting sequence. Known EB1-binding sequences generally match to the sequences we identified, but none matches perfectly (Supplemental Figure S7A). Very high affinity interactions may be disadvantageous in nature for some reason—for example, because they outcompete other EB1-interacting proteins in cells.
Many higher eukaryotes have multiple EB1 paralogues that share similarity in both the CH domain (interacting with microtubules) and the EBH domain (interacting with cargoes via the SxIP motif; Figure 1A; Slep, 2010). The best-studied examples are human EB1 and EB3. Little is known about whether they have distinct cargoes and, if so, what the determinant is of this specificity. Our comparative study of human EB1, human EB3, and Drosophila EB1 showed that sequence requirements surrounding the SxIP motif for interaction with three EB1 homologues are similar but also have crucial differences at certain positions. Indeed, we were able to predict aptamers that interact differentially with EB1 and EB3 and demonstrated that five of six interact differentially in a yeast two-hybrid assay, suggesting that EB1 orthologues interact with similar but distinct SxIP-containing sequences. Evidence from studies with mammalian EB1 and EB3 also suggests that both have shared functions and cargoes but also distinct functions and cargoes during myogenesis and neurogenesis (Zhang et al., 2009; Geraldo et al., 2008). Although only three proteins are known to interact specifically with human EB3, not EB1 (Geraldo et al., 2008), most SxIP sequences found in these proteins generally match better to the EB3-binding sequences we identified than EB1-binding sequences (Supplemental Figure S7B). Our study provides vital insight into how EB1 paralogues have distinct functions and cargoes at the molecular level.
In this study, we successfully developed novel methods to generate high-affinity aptamers very efficiently. First, we used random peptides with invariant amino acids at specific positions. Although only three positions are fixed, this has the effect of enriching the library ∼8000-fold. We identified ∼500 EB1 aptamers by yeast two-hybrid screening, but without this enrichment, even a 10-fold-larger screen would have identified only 1. This large number of EB1 aptamers allowed us to further select strong aptamers by a quantitative two-hybrid assay. Therefore this is an effective way to identify aptamers when a binding motif is known. Second, we demonstrated that, even without a quantitative assay of individual strength, a high-affinity aptamer can be designed by choosing the best-represented amino acids at each position. The binding affinity of this “designer” aptamer to Drosophila EB1 is estimated to have a higher affinity than that of known endogenous EB1-interacting proteins to human EB1.
We also explored three further approaches to improve the affinity of aptamers. First, we optimized the SxIP motif by screening peptides in which the motif is substituted by random amino acids, with the remainder of the peptide identical to the designed aptamer Perfect. In this case, we found that the residues from the conserved motif gave the strongest interaction. Second, we generated tandem repeats of EB1 aptamers, as it was shown that multiple SxIP motifs increase the microtubule plus end association (Honnappa et al., 2009). However, we found that in the two-hybrid assay, repeats of EB1 aptamers did not increase the interaction. Third, we screened a library of SxIP-containing peptides double constrained using a thioredoxin scaffold. This method has been successfully used in isolating strong aptamers for other proteins (Colas et al., 1996). We successfully used this method to isolate double-constrained aptamers, but none of these showed a higher affinity than the previously identified, singly constrained aptamers. Comparisons between singly and double constrained aptamers have been made by transferring aptamers isolated in one system to another system (Cohen et al., 1998). As far as we are aware, this study is the first example of parallel screens of both singly and doubly constrained aptamers carried out using the same bait protein. Although a yeast two-hybrid assay did not reveal stronger interaction, double-constrained aptamers may be more biologically active due to increased stability in cells (Cohen et al., 1998). Further studies are needed to gain more concrete information on the effectiveness of the double-constrained aptamers identified here.
EB1 has a role in regulating microtubule organization and dynamics in cell division, interphase, and postmitotic cells. Because EB1 is an essential gene, the use of strategies such as knockout and RNAi have limited utility in studying its specific functions, particularly postmitotic ones. Temperature-sensitive mutants or conditional degrons have been used to overcome this type of problem (Lovato et al., 2009; Kanemaki, 2013). However, the ability of peptide aptamers to interfere with specific protein functions means that they can be used as an alternative technique to inactivate EB1 function in specific cells and at specific stages in cell differentiation.
To investigate the functional properties of the high-affinity EB1 aptamers, we expressed EB1-binding sequences as GFP fusions in Drosophila S2 cells. All the strong aptamers tested could outcompete a major EB1 cargo protein, Sentin, for microtubule plus end binding. Of interest, one of the aptamers inhibited plus end localization of CLIP-190 that binds to a site of EB1 distinct from but close to the SxIP-binding region (Honnappa et al., 2006), suggesting steric hindrance. When aptamers are expressed in developing flies, the microtubule growth rate at plus ends is significantly reduced in larval hemocytes. This phenotype is similar to that seen in S2 cells depleted of EB1 or Sentin (Rogers et al., 2002; Li et al., 2011). Furthermore, the expression reduced fly viability. Therefore our study demonstrated that aptamers can be used to interfere with the EB1 function both in cultured cells and developing flies.
MATERIALS AND METHODS
Plasmid expression constructs and yeast two-hybrid methods
The Y190 strain (MATa leu2-3.112 ura3-52 trp1-901 his3-D200 ade2-101 gal4D gal80D cyhR URA3::GAL1-lacZ, LYS2::GAL1-HIS3) was used for all yeast two-hybrid analyses. To make bait plasmids, ∼50 nucleotide fragments complementary to pGBT9 bait vector upstream and downstream of the EcoRI site on pGBT9 were added to Drosophila EB1, human EB1, or human EB3 coding sequences from LD24317 (Drosophila Genomics Resource Center, Bloomington, IN), MHS1010-202693923 (Thermo Scientific, Loughborough, UK), or pJW242 (a kind gift from J. Welburn, Edinburgh University) using PCR, by inclusion of these sequences in primers (oKMT17, 18; 82, 83; 84, 85; Supplemental Figure S7). Y190 was cotransformed with the PCR product and pGBT9 vector linearized with EcoRI for inducing the gap repair. The integrity and the absence of unwanted mutations in bait plasmids were confirmed by restriction digests and sequencing.
To make prey plasmid libraries, two oligonucleotides were synthesized commercially (Eurofins, Billingham, UK). The core region of both sequences encodes Ser-x-Ile-Pro preceded by five and followed by seven random residues (xxxxxSxIPxxxxxxx), where x is encoded by nnk (n = A/C/T/G, k = G/T) and SxIP is encoded by TCCnnkATTCCA. For singly constrained aptamers, this core sequence followed by a stop codon (TGA) was flanked by 48 nucleotides (nt) at the 5′ end and 21 nt at the 3′ end, each of which corresponds to the sequence upstream and downstream of the EcoRI restriction site on the prey vector pACT2 (oKMT46; Supplemental Figure S8). A further 29 nt were added to the 3′ end through inclusion in a primer (oKMT34; Supplemental Figure S8) during synthesis of the complementary strand by PrimeStar polymerase mix (Takara, Saint-Germain-en-Laye, France) for 10 min at 72°C. For selecting aptamers doubly constrained by thioredoxin A (TrxA), the core sequence was directly flanked by 48 and 24 nt corresponding to the sequences upstream and downstream of the RsrII restriction site in the TrxA sequence (oKMT63; Supplemental Figure S5). A further 24 nt were added to the 3′ end using oKMT64 (Supplemental Figure S8) as before.
Y190 carrying a bait plasmid was cotransformed with a DNA fragment as before and a linearized prey vector (using EcoRI for pACT2 or RsrII for pACT2+TrxA). Transformants were selected for HIS3 expression by plating them on yeast minimum media supplemented with 20 mM 3-amino-1,2,4-triazole and incubation for 1 wk. From each transformant, single colonies were isolated and tested for β-galactosidase activity on plates by X-gal overlay. Tests on some selected transformants confirmed that the activation of reporter genes depends on the EB1 bait plasmid. To exclude that activation of the reporter genes was caused by random mutations in the yeast or the plasmid, the regions including the aptamers were amplified from some transformants by PCR. Y190 containing the EB1 bait plasmid was cotransformed with the PCR product and the linear prey plasmid and retested for β-galactosidase activity.
Making tandem repeats of peptide aptamer prey plasmids
DNA encoding tandem repeats of aptamer 37 and aptamer Perfect flanked by the sequences corresponding to the prey vector fragments upstream (81 nt) and downstream (74 nt) of the EcoRI site on the prey vector were synthesized commercially (pKMT169, 170; Supplemental Figure S8) and introduced into cloning vectors (Life Technologies, Paisley, UK). DNA encoded seven repeats of aptamer Perfect or four repeats of aptamer 37, which were separated by SGSG linkers in both cases. Various codons with reasonable usages in both yeast and Drosophila were combined to minimize unwanted recombination. The DNA fragment encoding oligomerized peptides was amplified along with the flanking sequences and used for cotransformation of Y190 carrying the EB1 bait plasmid with a linearized prey plasmid to allow gap repair as described earlier. The sequence for two repeats of aptamer 37 was amplified from a plasmid encoding four repeats of aptamer 37 (oKMT31, 69; Supplemental Figure S8), and flanking sequences corresponding to the sequences on the prey vector were added as described to make the prey plasmid. Two repeats of aptamer sequences are unlikely to bind two EB1 molecules simultaneously, but more than two repeats may be able to bind. Expression of aptamers with more than two SxIPs in Drosophila cells altered the localization of EB1 and induced microtubule bundling, which may be caused by cross-linking two EB1 proteins. This may be a reason why these peptides did not work well in our two-hybrid assay.
Measuring the strength of yeast two-hybrid interactions
An overnight yeast culture was diluted into a fresh medium. After culture at 30°C for 3 h, A600 was measured and confirmed to be between 0.5 and 1. The cell culture was centrifuged, and the pellet was sometimes stored at −75°C after being frozen in liquid nitrogen, which did not affect the outcome. Yeast was thawed and incubated with Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM Mg2SO4, 1 mM dithioerythritol, 0.2% [vol/vol] sarcosyl, pH 7) and 4 mg/ml ortho-nitrophenyl-β-galactoside until the color developed (∼15 min). Na2CO3, 1 M, was added, followed by centrifugation. The A420 of the supernatant was measured. The β-galactosidase activity per cell was calculated as A420/A600 ratio. For selection of strong EB1 aptamers, ∼50 different aptamers in each batch were tested simultaneously together with three reference aptamers to facilitate rough comparison between batches.
Generation and expression of peptide aptamer constructs
Expression plasmids for GFP fused aptamer were generated by Gateway cloning (Invitrogen, Paisley, UK). The attB1 and attB2 sites were added to a peptide aptamer coding sequence for cloning into pDONR221 (Invitrogen). The destination vector pAGW was used for the expression of an aptamer fused to a GFP under the Actin5C promoter in S2 cells, and pPGW was used for expression of an aptamers fused to GFP under the UASp promoter in flies. Drosophila Schneider S2 cells were cultured and transfected, and transgenic flies were made by P-element–mediated transformation of w1118 as described previously (Brittle and Ohkura, 2005). To quantify the fly viability, heterozygotes of actin5C-GAL4 over the balancer chromosome TM6C were crossed with flies homozygous for a peptide aptamer gene under the UASp promoter or wild-type flies for control. Relative viability was calculated as the number of aptamer-expressing adults to the number of nonexpressing adults (carrying TM6C).
Immunostaining and image analysis
For immunostaining, S2 cells were adhered to coverslips coated with concanavalin A (ConA) as described previously (Dzhindzhev et al., 2005). Primary antibodies against α-tubulin (dm1A, 1:250; Sigma-Aldrich, Gillingham, UK), Drosophila EB1 (Elliott et al., 2005), GFP (rabbit, mouse anti-GFP, 1:500, A11122, 3E6; Molecular Probes, Paisley, UK), and Sentin (rat anti-Sentin, 1:50) were used. Secondary antibodies conjugated with Alexa Flour 488, Cy3, or Cy5 (Molecular Probes or Jackson Labs) were used. Images were taken with an Axioplan 2 microscope (Zeiss, Cambridge, UK) attached to a charge-coupled device camera (Hamamatsu, Hamamatsu, Japan) controlled by OpenLab 2.2.1 software (ImproVision, Coventry, UK). To measure the Sentin signal at the microtubule plus ends, we used the method described by Dzhindzhev et al. (2005), with the following modifications. The plus end signal was calculated as S − B, where S is the total pixel intensity for a particular plus end signal and B is the total pixel intensity of the local background. The local background signal intensity was measured using the same-sized area beside the plus end signal area. Three plus end signals were measured in at least 10 separate interphase cells using OpenLab 2.2.1.
To assess the growth rate of microtubule plus ends, ubiquitin-EB1-GFP/Y; pUASp-GFP-aptamer/+ males were crossed with females carrying actin5C-GAL4 over a balancer. Hemocytes were isolated from each female larva, plated on ConA-coated MatTek glass-bottom dishes for 2 h, and filmed at room temperature using a microscope (Axiovert; Carl Zeiss) attached to a spinning-disk confocal head (Yokogawa, Tokyo, Japan) controlled by Volocity software (PerkinElmer, Cambridge, UK). Because all female larvae express EB1-GFP but only some express GFP-aptamer, the presence or absence of the pUASp-GFP-aptamer transgenes in each larva was assessed by PCR from purified genomic DNA. From each larva, four or five EB1-GFP comets were measured in each of four or five hemocytes. A total of six or seven larvae were examined for each genotype.
Isothermal titration calorimetry
MBP or MBP fused with Drosophila EB1 was expressed at 37°C in BL21 (DE3) pLysS E. coli strain as described, and all purification steps were performed at 4°C. Cells were disrupted by sonication, and protein extract was loaded onto an MBPTrap HP column (GE Healthcare, Amersham, UK) for affinity purification. The proteins were further purified by size exclusion chromatography using a Superdex 200 10/300 GL column (GE Healthcare). The MicroCal Auto-iTC200 (GE Healthcare, Amersham, UK) system was used for ITC. The aptamer peptides were commercially synthesized (Eurogentec, Southampton, UK) and dissolved in 100% dimethyl sulfoxide (DMSO). We titrated 5 μM MBP or MBP-EB1 at 25°C with 100 μM peptides for aptamer Perfect and 392. For aptamer 177, 5 μM MBP or 4 μM MBP-EB1 was titrated at 25°C with 100 or 200 μM peptide, respectively. MBP-EB1 or peptide was analyzed in the same buffer (120 mM NaCl, 2.7 mM KCl, 30 mM NaH2PO4, 1 nM dithiothreitol, 2% DMSO, pH 7.4). Titrations of peptide to buffer only were performed to allow baseline corrections. The resulting heats were measured, and the parameters were calculated by fitting to a single-site binding model using Origin software (OriginLab, Stoke Mandeville, UK).
Structural modeling
For modeling of Drosophila EB1 with aptamer Perfect and with a Sentin SxIP fragment, a crystal structure (3GJO; Honnappa et al., 2009) of the complex between human EB1cΔC8 and MACFp1 was used as a template to generate a .pdb file by Swiss-Prot. To refine the model, FlexPepDoc was used for energy minimization (London et al., 2011). To calculate buried surface areas, the ArealMol in CCP4 package was used (Lee and Richards, 1971; Winn et al., 2011). The figures were generated by PyMOL (www.pymol.org) and manually labeled.
Supplementary Material
Acknowledgments
We thank members of the Ohkura laboratory for support and discussion, Robin Beaven for critical reading of the manuscript, and Sally Beard, Julie Welburn, and Terrence Murphy for reagents. We especially thank J. P. Arulanandam for help with structural modeling. We also thank the Edinburgh Protein Production Facility and the Bloomington Drosophila Stock Center and Drosophila Genomics Resource Center at Indiana University. This work is funded by a Scottish Universities Life Sciences Alliance/Biotechnology and Biological Sciences Research Council PhD studentship to K.L. and the Wellcome Trust (092076, 098030, 081849).
Abbreviations used:
- GFP
green fluorescent protein
- ITC
isothermal titration calorimetry
- MBP
maltose-binding protein
- SxIP
Ser–any residue–Ile-Pro
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-08-0504) on January 29, 2014.
REFERENCES
- Akhmanova A, Steinmetz MO. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol. 2008;9:309–322. doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines IC, Colas P. Peptide aptamers as guides for small-molecule drug discovery. Drug Discov Today. 2006;11:334–341. doi: 10.1016/j.drudis.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Brittle A, Ohkura H. Mini spindles, the XMAP215 homologue, suppresses pausing of interphase microtubules in Drosophila. EMBO J. 2005;24:1387–1396. doi: 10.1038/sj.emboj.7600629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buey RM, et al. Sequence determinants of a microtubule tip localization signal (MtLS) J Biol Chem. 2012;287:28227–28242. doi: 10.1074/jbc.M112.373928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch KE, Brunner D. The microtubule plus end-tracking proteins Mal3p and Ttip1p cooperate for cell-end targeting of interphase microtubules. Curr Biol. 2004;14:548–559. doi: 10.1016/j.cub.2004.03.029. [DOI] [PubMed] [Google Scholar]
- Butz K, Denk C, Ullmann A, Scheffner M, Hoppe-Seyler F. Induction of apoptosis in human papillomavirus positive cancer cells by peptide aptamers targeting the viral E6 oncoprotein. Proc Natl Acad Sci USA. 2000;97:6693–6697. doi: 10.1073/pnas.110538897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen BA, Colas P, Brent R. An artificial cell-cycle inhibitor isolated from a combinatorial library. Proc Natl Acad Sci USA. 1998;95:14272–14277. doi: 10.1073/pnas.95.24.14272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas P, Cohen B, Jessen T, Grishina I, McCoy J, Brent R. Genetic selection of peptide aptamers that recognize and inhibit cyclin-dependent kinase 2. Nature. 1996;380:548–550. doi: 10.1038/380548a0. [DOI] [PubMed] [Google Scholar]
- Crawford M, Woodman R, Ko Ferrigno P. Peptide aptamers: tools for biology and drug discovery. Brief Funct Genomic Proteomic. 2003;2:72–79. doi: 10.1093/bfgp/2.1.72. [DOI] [PubMed] [Google Scholar]
- Dzhindzhev NS, Rogers SL, Vale RD, Ohkura H. Distinct mechanisms govern the localisation of Drosophila CLIP-190 to unattached kinetochores and microtubule plus-ends. J Cell Sci. 2005;118:3781–3790. doi: 10.1242/jcs.02504. [DOI] [PubMed] [Google Scholar]
- Elliott SL, Cullen CF, Wrobel N, Kernan MJ, Ohkura H. EB1 is essential during Drosophila development and plays a crucial role in the integrity of chordotonal mechanosensory organs. Mol Biol Cell. 2005;16:891–901. doi: 10.1091/mbc.E04-07-0633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estojak J, Brent R, Golemis EA. Correlation of two-hybrid affinity data with in vitro measurements. Mol Cell Biol. 1995;15:5820–5829. doi: 10.1128/mcb.15.10.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbrizio E, Le Cam L, Polanowska J, Kaczorek M, Lamb N, Brent R, Sardet C. Inhibition of mammalian cell proliferation by genetically selected peptide aptamers that functionally antagonize E2F activity. Oncogene. 1999;18:4357–4363. doi: 10.1038/sj.onc.1202825. [DOI] [PubMed] [Google Scholar]
- Geraldo S, Khanzada UK, Parsons M, Chilton JK, Gordon-Weeks PR. Targeting of the F-actin-binding protein drebrin by the microtubule plus-tip protein EB3 is required for neuritogenesis. Nat Cell Biol. 2008;10:1181–1189. doi: 10.1038/ncb1778. [DOI] [PubMed] [Google Scholar]
- Geyer CR, Brent R. Selection of genetic agents from random peptide aptamer expression libraries. Methods Enzymol. 2000;328:171–208. doi: 10.1016/s0076-6879(00)28398-5. [DOI] [PubMed] [Google Scholar]
- Honnappa S, et al. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 2009;138:366–376. doi: 10.1016/j.cell.2009.04.065. [DOI] [PubMed] [Google Scholar]
- Honnappa S, John CM, Kostrewa D, Winkler FK, Steinmetz MO. Structural insights into the EB1-APC interaction. EMBO J. 2005;24:261–269. doi: 10.1038/sj.emboj.7600529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnappa S, Okhrimenko O, Jaussi R, Jawhari H, Jelesarov I, Winkler FK, Steinmetz MO. Key interaction modes of dynamic +TIP networks. Mol Cell. 2006;23:663–671. doi: 10.1016/j.molcel.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Howard J, Hyman AA. Dynamics and mechanics of the microtubule plus end. Nature. 2003;422:753–758. doi: 10.1038/nature01600. [DOI] [PubMed] [Google Scholar]
- Jiang K, et al. A proteome-wide screen for mammalian SxIP motif-containing microtubule plus-end tracking proteins. Curr Biol. 2012;22:1800–1807. doi: 10.1016/j.cub.2012.07.047. [DOI] [PubMed] [Google Scholar]
- Kanemaki MT. Frontiers of protein expression control with conditional degrons. Pflügers Arch. 2013;465:419–425. doi: 10.1007/s00424-012-1203-y. [DOI] [PubMed] [Google Scholar]
- Kolonin MG, Finley RL. Targeting cyclin-dependent kinases in Drosophila with peptide aptamers. Proc Natl Acad Sci USA. 1998;95:14266–14271. doi: 10.1073/pnas.95.24.14266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolonin MG, Finley RL. A role for cyclin J in the rapid nuclear division cycles of early Drosophila embryogenesis. Dev Biol. 2000;227:661–672. doi: 10.1006/dbio.2000.9916. [DOI] [PubMed] [Google Scholar]
- Lee B, Richards FM. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- Li W, Miki T, Watanabe T, Kakeno M, Sugiyama I, Kaibuchi K, Goshima G. EB1 promotes microtubule dynamics by recruiting sentin in Drosophila cells. J Cell Biol. 2011;193:973–983. doi: 10.1083/jcb.201101108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London N, Raveh B, Cohen E, Fathi G, Schueler-Furman O. Rosetta FlexPepDock web server—high resolution modeling of peptide-protein interactions. Nucleic Acids Res. 2011;39:W249–W253. doi: 10.1093/nar/gkr431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovato TL, Adams MM, Baker PW, Cripps RM. A molecular mechanism of temperature sensitivity for mutations affecting the Drosophila muscle regulator myocyte enhancer factor-2. Genetics. 2009;183:107–117. doi: 10.1534/genetics.109.105056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y, Shiina N, Tsukita S. The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr Biol. 2000;10:865–868. doi: 10.1016/s0960-9822(00)00600-x. [DOI] [PubMed] [Google Scholar]
- Parton R, Hamilton R, Cullen F, Lu W, Ohkura H, Davis I. A PAR-1–dependent orientation gradient of dynamic microtubules directs posterior cargo transport in the Drosophila oocyte. J Cell Biol. 2011;194:121–135. doi: 10.1083/jcb.201103160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SL, Rogers GC, Sharp DJ, Vale RD. Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J Cell Biol. 2002;158:873–884. doi: 10.1083/jcb.200202032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder J, Schneider L, Christensen S, Pedersen L. EB1 is required for primary cilia assembly in fibroblasts. Curr Biol. 2007;17:1134–1139. doi: 10.1016/j.cub.2007.05.055. [DOI] [PubMed] [Google Scholar]
- Seigneuric R, Gobbo J, Colas P, Garrido C. Targeting cancer with peptide aptamers. Oncotarget. 2011;2:557–561. doi: 10.18632/oncotarget.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y, Yonemura S, Ohkura H, Strutt D, Uemura T. Polarized transport of Frizzled along the planar microtubule arrays in Drosophila wing epithelium. Dev Cell. 2006;10:209–222. doi: 10.1016/j.devcel.2005.11.016. [DOI] [PubMed] [Google Scholar]
- Slep KC. Structural and mechanistic insights into microtubule end-binding proteins. Curr Opin Cell Biol. 2010;22:88–95. doi: 10.1016/j.ceb.2009.10.009. [DOI] [PubMed] [Google Scholar]
- Slep KC, Rogers SL, Elliott SL, Ohkura H, Kolodziej PA, Vale RD. Structural determinants for EB1-mediated recruitment of APC and spectraplakins to the microtubule plus end. J Cell Biol. 2005;168:587–598. doi: 10.1083/jcb.200410114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz MO, Akhmanova A. Capturing protein tails by CAP-Gly domains. Trends Biochem Sci. 2008;33:535–545. doi: 10.1016/j.tibs.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Straube A, Merdes A. EB3 regulates microtubule dynamics at the cell cortex and is required for myoblast elongation and fusion. Curr Biol. 2007;17:1318–1325. doi: 10.1016/j.cub.2007.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer JS, Bierer BE. EB1 proteins regulate microtubule dynamics, cell polarity, and chromosome stability. J Cell Biol. 2000;149:761–766. doi: 10.1083/jcb.149.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickramasinghe RD, Ko Ferrigno P, Roghi C. Peptide aptamers as new tools to modulate clathrin-mediated internalisation–inhibition of MT1-MMP internalisation. BMC Cell Biol. 2010;11:58. doi: 10.1186/1471-2121-11-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh JT, Binari R, Gocha T, Dasgupta R, Perrimon N. PAPTi: a peptide aptamer interference toolkit for perturbation of protein-protein interaction networks. Sci Rep. 2013;3:1156. doi: 10.1038/srep01156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Zaal KJ, Sheridan J, Mehta A, Gundersen GG, Ralston E. Microtubule plus-end binding protein EB1 is necessary for muscle cell differentiation, elongation and fusion. J Cell Sci. 2009;122:1401–1409. doi: 10.1242/jcs.039255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimniak T, Stengl K, Mechtler K, Westermann S. Phosphoregulation of the budding yeast EB1 homologue Bim1p by Aurora/Ipl1p. J Cell Biol. 2009;186:379–391. doi: 10.1083/jcb.200901036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.