The protein kinase TORC1 regulates cell growth in response to nutrients. This study demonstrates that phosphatidylinositol 3,5-bisphosphate (PI(3,5)P2) is a critical upstream modulator of TORC1 activity in yeast. In this capacity, PI(3,5)P2 is required for TORC1-dependent regulation of autophagy and nutrient-dependent endocytosis.
Abstract
TORC1, a conserved protein kinase, regulates cell growth in response to nutrients. Localization of mammalian TORC1 to lysosomes is essential for TORC1 activation. Phosphatidylinositol 3,5-bisphosphate (PI(3,5)P2), an endosomal signaling lipid, is implicated in insulin-dependent stimulation of TORC1 activity in adipocytes. This raises the question of whether PI(3,5)P2 is an essential general regulator of TORC1. Moreover, the subcellular location where PI(3,5)P2 regulates TORC1 was not known. Here we report that PI(3,5)P2 is required for TORC1 activity in yeast and regulates TORC1 on the vacuole (lysosome). Furthermore, we show that the TORC1 substrate, Sch9 (a homologue of mammalian S6K), is recruited to the vacuole by direct interaction with PI(3,5)P2, where it is phosphorylated by TORC1. Of importance, we find that PI(3,5)P2 is required for multiple downstream pathways via TORC1-dependent phosphorylation of additional targets, including Atg13, the modification of which inhibits autophagy, and phosphorylation of Npr1, which releases its inhibitory function and allows nutrient-dependent endocytosis. These findings reveal PI(3,5)P2 as a general regulator of TORC1 and suggest that PI(3,5)P2 provides a platform for TORC1 signaling from lysosomes.
INTRODUCTION
The mechanistic target of rapamycin (mTOR) is an evolutionarily conserved protein kinase that is critical for homeostatic control of cell growth and metabolism (reviewed in Kim and Guan, 2009; Laplante and Sabatini, 2009; Loewith and Hall, 2011). mTOR functions within two distinct complexes, TORC1 and TORC2 (Loewith et al., 2002). Although most organisms have a single TOR gene, some yeast, including Saccharomyces cerevisiae, have two, TOR1 and TOR2. In yeast, TORC1 consists of four proteins: Tor1 or Tor2, Kog1 (homologue of Raptor), Lst8 (homologue of mLST8/GβL), and Tco89. TORC1 responds to nutritional status and positively regulates protein synthesis and cell growth while negatively regulating autophagy (reviewed in Loewith and Hall, 2011).
TORC1 resides predominantly on the vacuole membrane in yeast (Reinke et al., 2004; Araki et al., 2005; Urban et al., 2007; Sturgill et al., 2008; Binda et al., 2009). The localization and activity of TORC1 on the vacuole is critical for yeast viability. Moreover, several vacuole-related mutants are synthetically lethal with the tor1Δ mutant, which retains some TORC1 function (Zurita-Martinez et al., 2007). Mammalian TORC1 is recruited to late endosomes and lysosomes in response to the presence of amino acids (Sancak et al., 2008). This localization is critical for TORC1 function (Sancak et al., 2010; Zoncu et al., 2011). However, the mechanisms that localize TORC1 to the yeast vacuole and the mammalian lysosome are not known.
Some key regulators of TORC1 are also localized on the yeast vacuole and mammalian lysosome. These include the yeast EGO complex—in mammalian cells termed Ragulator (Dubouloz et al., 2005; Gao and Kaiser, 2006; Binda et al., 2009; Kogan et al., 2010; Sancak et al., 2010)—and the mammalian lysosomal V-ATPase (Zoncu et al., 2011).
Phosphatidylinositol 3,5-bisphosphate (PI(3,5)P2), a crucial, low-abundance signaling phospholipid found in the endomembrane system, is generated from phosphatidylinositol 3-phosphate (PI3P) by the lipid kinase Fab1/PIKfyve (reviewed in Dove et al., 2009; McCartney et al., 2014). Fab1/PIKfyve is the sole enzyme that generates PI(3,5)P2 (Gary et al., 1998; Zolov et al., 2012). In yeast, Fab1 is positively regulated by Vac7 (Bonangelino et al., 2002; Gary et al., 2002), which has no known mammalian homologue. In addition, Fab1 activity requires the conserved scaffolding protein Vac14 (Botelho et al., 2008; Jin et al., 2008; Sbrissa et al., 2008). PI(3,5)P2 is converted to PI3P by the conserved lipid phosphatase Fig4 (Gary et al., 2002; Rudge et al., 2004), which paradoxically also serves as a positive regulator of Fab1 (Duex et al., 2006b).
Mutations predicted to cause only minor changes in PI(3,5)P2 levels are associated with severe neurological diseases (Chow et al., 2007, 2009). Moreover, loss of human Fig4 results in a multisystem congenital disorder that leads to infant mortality (Campeau et al., 2013). Yet, despite its importance in human physiology, only a fraction of the processes regulated by PI(3,5)P2 have been identified. Moreover, few proteins that bind directly to PI(3,5)P2 have been identified, and there are no known motifs that predict an interaction with PI(3,5)P2.
Recent studies revealed that the WD-40 domain of mammalian Raptor binds PI(3,5)P2 in vitro (Bridges et al., 2012), which raised the intriguing possibility that PI(3,5)P2 is an upstream regulator of TORC1. Consistent with this hypothesis, knockdown of PIKfyve, Vac14, or Fig4 in adipocytes resulted in a loss of the ability of insulin to stimulate TORC1 phosphorylation of S6 kinase (S6K). However, in these prior studies the conditions used to knock down Fig4 and Vac14 did not have a significant effect on PI(3,5)P2 levels, and knockdown of PIKfyve had a modest effect. Thus how the short hairpin RNA experiments affected TORC1 function is unclear. Furthermore, in stimulated adipocytes, TORC1 was found at the plasma membrane, raising questions about the subcellular location where PI(3,5)P2 activates TORC1. In addition, the effect of PI(3,5)P2 on general pathways regulated by TORC1 was not assessed.
Here we report that PI(3,5)P2 is a positive regulator of TORC1 activity on the yeast vacuole. We find that the TORC1 substrate Sch9, a yeast homologue of S6K, localizes on the vacuole via direct binding to PI(3,5)P2 and that this localization is required for TORC1-dependent phosphorylation of Sch9. Moreover, we find that PI(3,5)P2 regulates TORC1 phosphorylation of multiple targets and regulates multiple TORC1 functions. We show that PI(3,5)P2 promotes nutrient-regulated endocytosis from the plasma membrane and inhibits autophagy via a requirement for PI(3,5)P2 in TORC1-dependent phosphorylation of Npr1 and Atg13, respectively. Moreover, the requirement of PI(3,5)P2 in TORC1 inhibition of autophagy is exacerbated by a TORC1-independent role for PI(3,5)P2 in the degradation of autophagic bodies. Thus, under nutrient-rich conditions, mutants with defects in PI(3,5)P2 carry a load of autophagic bodies in the vacuole. Together these findings show that PI(3,5)P2 is a conserved upstream regulator of TORC1 and suggest that PI(3,5)P2 provides a platform for TORC1 signaling from the vacuole/lysosome. Moreover, these studies show that PI(3,5)P2 is a key regulator of cellular response to nutrient status via modulation of TORC1.
RESULTS
PI(3,5)P2 is required for TORC1 activity in vivo
Inhibition of PIKfyve causes a defect in insulin-dependent activation of mTORC1 in L1-3T3 adipocytes (Bridges et al., 2012). To determine whether PI(3,5)P2 is a conserved, general regulator of TORC1, we tested whether PI(3,5)P2 plays a role in TORC1 activation in yeast. Yeast mutants with defects in TORC1 activity are hypersensitive to the TORC1 inhibitor rapamycin (Heitman et al., 1991; Loewith et al., 2002). Thus we tested growth on 1 ng/ml rapamycin, which is sublethal for wild-type yeast. Mutants with impaired TORC1 function, tor1Δ and gtr1Δ, exhibit hypersensitivity to rapamycin (Figure 1A; Binda et al., 2009). Similarly, vac14Δ, vac7Δ, and fab1Δ mutants, which contain little or no detectable PI(3,5)P2, respectively (Bonangelino et al., 2002; Gary et al., 2002; Duex et al., 2006a), also exhibited hypersensitivity to rapamycin (Figure 1A). To further address the hypothesis that PI(3,5)P2 regulates TORC1 activity, we tested whether the hyperactive, dominant FAB1VLA mutant suppressed the rapamycin hypersensitivity of the vac7Δ and vac14Δ mutants. Expression of an additional copy of Fab1 from a low-copy plasmid in the vac7Δ mutant had little effect on the levels of PI(3,5)P2 (Duex et al., 2006b) or on the suppression of the sensitivity of vac7Δ to rapamycin (Figure 1B). In contrast, expression of the hyperactive FAB1VLA mutant in vac7Δ or vac14Δ yeast, which raises the levels of PI(3,5)P2 above levels found in wild-type cells by ∼1.5- and 3-fold, respectively (Duex et al., 2006b), suppressed the sensitivity of the vac7Δ and vac14Δ mutants to rapamycin (Figure 1B). This finding strongly suggests that PI(3,5)P2 levels rather than the Vac7 or Vac14 protein per se are required for TORC1 function. Moreover, in the vac7Δ mutant, whereas PI(3,5)P2 levels are deeply lowered, PI3P levels are twofold higher, yet this elevation in PI3P does not rescue TORC1 function; furthermore, both phosphatidylinositol 4-phosphate (PI4P) and phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) levels are similar to wild-type levels (Gary et al., 2002). Similarly, in the vac14Δ mutant, only PI(3,5)P2 levels are lowered. In this mutant, PI3P levels are elevated above wild-type levels by 20%, whereas PI4P and PI(4,5P)2 levels are similar to wild-type levels (Dove et al., 2002). Together these studies strongly suggest that PI(3,5)P2 modulates TORC1 function.
FIGURE 1:
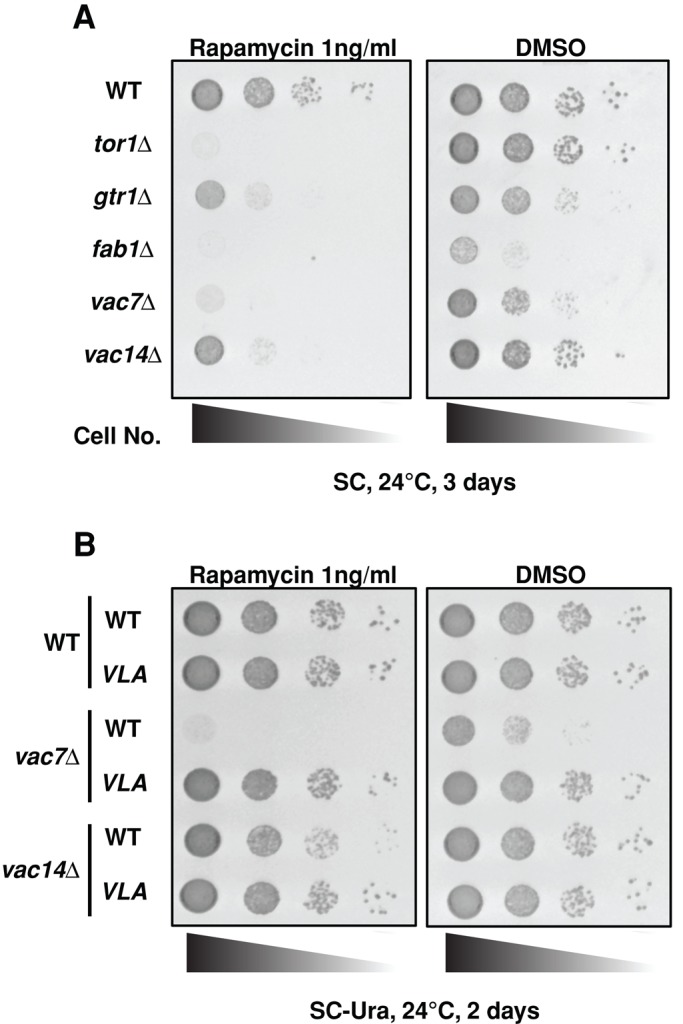
PI(3,5)P2 is required for TORC1 activity. (A) The fab1Δ, vac7Δ, and vac14Δ mutants are hypersensitive to rapamycin. The indicated strains were grown to mid log phase in YEPD medium and then diluted onto SC plates containing DMSO (control) or 1 ng/ml rapamycin, a sublethal dose for wild-type yeast. tor1Δ and gtr1Δ mutants have known defects in TORC1 function. Correlation between the ability to grow in the presence of 1 ng/ml rapamycin and intracellular PI(3,5)P2 levels was indicated by fab1Δ mutants, which have no detectable PI(3,5)P2, vac7Δ cells with very low levels of PI(3,5)P2, and vac14Δ cells with more PI(3,5)P2 than vac7Δ. (B) Increasing PI(3,5)P2 in vac7Δ or vac14Δ mutants restores TORC1 activity. Sensitivity of vac7Δ or vac14Δ mutants to 1 ng/ml rapamycin is rescued by elevation of PI(3,5)P2 levels. Wild-type yeast and vac7Δ or vac14Δ mutants with plasmids expressing Fab1 or dominant active FAB1VLA were grown to mid log phase in SC-Ura medium and then diluted onto SC-Ura plates containing DMSO (control) or 1 ng/ml rapamycin.
PI(3,5)P2 activates TORC1 on the vacuole
Mammalian Raptor binds PI(3,5)P2, in vitro (Bridges et al., 2012), and thus we tested whether Kog1, the yeast homologue of Raptor, binds to PI(3,5)P2. A construct encoding glutathione S-transferase (GST) fused to amino acids 1162–1557 of Kog1, which contains the WD-40 domain, was used for expression and purification of the protein from Escherichia coli. We found that Kog1(1162–1557) bound PI(3,5)P2 with a dissociation constant of 19 ± 6 μM (Supplemental Figure S1). These data suggested the possibility that PI(3,5)P2 plays a role in the activation of TORC1 via its association with Kog1. Note that this affinity is substantially lower than that observed for Atg18, which exhibits a dissociation constant of <0.2 μM (Table 1; Dove et al., 2004). However, the affinity of Atg18 for PI(3,5)P2 is exceptionally strong, likely due to two adjacent phosphoinositide-binding sites (Baskaran et al., 2012; Krick et al., 2012; Watanabe et al., 2012). Of importance, dissociation constants of 20 μM are commonly observed in biologically significant interactions.
TABLE 1:
Surface plasmon resonance analysis of Kog1- or Sch9-binding affinity for PI(3,5)P2.
| Kd (μM) | SEM (Kd) | Rmax | SEM (Rmax) | R2 | |
|---|---|---|---|---|---|
| A. Kog1- and Sch9-binding affinity for PI(3,5)P2 | |||||
| Kog1 | 19 | 6 | 1627 | 206 | 0.8 |
| Sch9 | 11 | 2 | 2743 | 154 | 0.96 |
| B. Atg18-binding affinity for PI(3,5)P2 | |||||
| i | 0.046 | 0.010 | 5053 | 332 | 0.8 |
| ii | 0.076 | 0.015 | 4341 | 217 | 0.89 |
Phosphorylated phosphoinositide lipids often function by recruiting downstream effectors to specific membrane subdomains. The translocation of mTORC1 onto the lysosome is important for mTORC1 activation under amino acid stimulation in mammalian cells (Sancak et al., 2010). This raised the possibility that PI(3,5)P2 regulates yeast TORC1 function in part by recruiting TORC1, especially Kog1, to the vacuole or a subdomain on the vacuole.
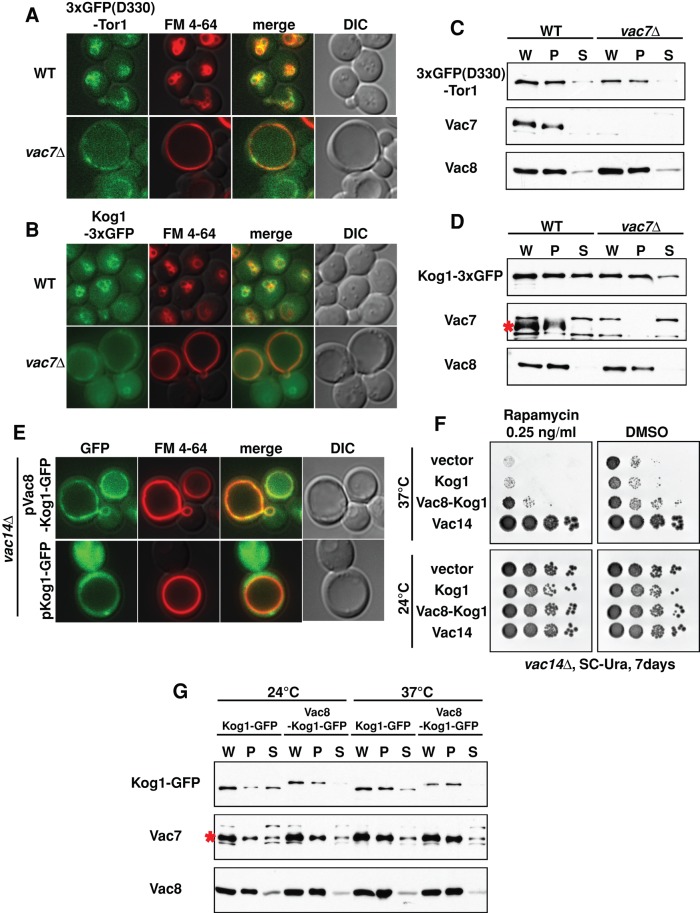
Using fluorescence microscopy (Figure 2, A and B), as well as cell fractionation (Figure 2, C and D), we probed the effect of PI(3,5)P2 on the localization of Tor1–3x green fluorescent protein (GFP) and Kog1-3xGFP. However, by cell fractionation, we did not detect large differences in the localization of Tor1-3xGFP or Kog1-3xGFP in the vac7∆ strain. Note that these tagged proteins were functional as measured by rapamycin sensitivity (Supplemental Figure S2, A and B). Similarly, the localization of Tor1-3xGFP(D330) and Kog1-3xGFP was largely unaffected by expression of the hyperactive FAB1VLA mutant (Supplemental Figure S2, C–G). In addition, the association of Kog1 with Tor1 was similar in wild type and the vac7Δ mutant (Supplemental Figure S2H). Nonetheless, we hypothesized that a small difference in localization of Kog1, and hence TORC1 to the vacuole, might result in a significant defect in TORC1 activity. Accordingly, we undertook a second approach to monitor the effect of PI(3,5)P2 on Kog1 localization.
FIGURE 2:
PI(3,5)P2 is required for TORC1 activity on the vacuole, but TORC1 remains on the vacuole in mutants with low levels of PI(3,5)P2. (A) 3xGFP(D330)-Tor1 (green) and (B) Kog1-3xGFP (green) localizes on the vacuole membrane (red) in wild-type and vac7Δ mutant yeast. Vacuole membranes were visualized with FM 4-64. (C, D) In either the presence or absence of PI(3,5)P2, 3xGFP(D330)-Tor1 or Kog1-3xGFP is in the pellet (P) fraction, which includes vacuole membranes. Cell lysates of the indicated strains were centrifuged at 13,000 × g for 10 min at 4°C. Supernatant (S) fractions and whole-cell lysates (W) were suspended in the same volume of sample buffer, and pellet (P) fractions were suspended in twice the volume of sample buffer. Loading control for cell fractionation: Vac7 and Vac8, which tightly localize on the vacuole membrane. Red asterisk, Vac7 protein. (E) More Vac8-Kog1-GFP (green) localizes on the vacuole membrane (red, FM 4-64) than Kog1-GFP. Note that both Vac8-Kog1-GFP and Kog1-GFP are overexpressed, which likely accounts for the partial cytoplasmic localization of Kog1-GFP. (F) Expression of the Vac8-Kog1 fusion protein partially suppressed the rapamycin sensitivity of vac14Δ mutants at 37°C. The vac14Δ mutants with plasmids expressing empty vector (mock), Kog1, Vac8-Kog1, or Vac14 were grown to mid log phase in SC-URA medium and then diluted onto SC-URA plates containing DMSO (control) or 0.25 ng/ml rapamycin. Note that this level of rapamycin does not interfere with growth of the vac14Δ mutant at 24°C for 7 d. (G) In the vac14Δ mutant, Vac8-Kog1 cofractionates with the P13 fraction at either 24 or 37°C (30 min), which includes the vacuole. Cell lysates were centrifuged at 13,000 × g for 10 min at 4°C. Supernatant (S) fractions and whole-cell lysates (W) were resuspended in the same volume of sample buffer, and the pellet (P) fractions were resuspended in twice the volume of sample buffer. Loading controls for cellular fractionation: Vac7 and Vac8, which tightly localize on the vacuole membrane. Red asterisk, Vac7 protein.
We tested and found that artificially tethering Kog1 to the vacuole partially suppresses the rapamycin hypersensitivity of the vac14Δ mutant. Vac8, a vacuolar membrane protein (Wang et al., 1998), does not require PI(3,5)P2 for its localization to the vacuole. Thus we used Vac8 as a PI(3,5)P2-independent tether. In contrast to Kog1-GFP, which displayed some diffuse cytosolic staining pattern in the vac14∆ strain, we found that the Vac8-Kog1 fusion protein showed better colocalization on the vacuole membrane in the vac14Δ mutant (Figure 2E). Vac8-Kog1 partially suppressed the rapamycin hypersensitivity of the vac14Δ mutant (Figure 2F). Note that the partial rescue by Vac8-Kog1 was not due to changes in the amount of Kog1, as the levels of the two proteins were similar at both 24 and 37°C (Figure 2G). Similarly, although Kog1 was distributed between the pellet and supernatant fractions of cell lysates at 24 and 37°C, Vac8-Kog1 was found primarily in the P13 membrane fraction, which includes the vacuole (Figure 2G); the integral membrane proteins Vac7 and Vac8 were also localized to the P13 fraction. These data strongly support the hypothesis that at least one role of PI(3,5)P2 in TORC1 function occurs on the vacuole membrane. Furthermore, these data link the rapamycin hypersensitivity of the vac14Δ mutant to defects in both TORC1 localization and function.
The TORC1 substrate Sch9 localizes on the vacuole via PI(3,5)P2
To test for additional roles for PI(3,5)P2, we investigated whether PI(3,5)P2 recruits a subset of TORC1 substrates onto the vacuole membrane. Accordingly, we examined a well-characterized direct target of TORC1, Sch9, which is partially localized on the vacuole (Urban et al., 2007). Yeast Sch9 shares similar functions with mammalian S6 kinase, a downstream target and effector of mTORC1.
We first tested whether TORC1-dependent phosphorylation of Sch9 requires PI(3,5)P2. We used Sch9T570A, which contains five sites in its C-terminus that are directly phosphorylated by TORC1 and is missing a TORC1-independent phosphorylation site (Urban et al., 2007). On 6% SDS–PAGE gel, Sch9T570A-HA5 migrates as multiple bands. The upper bands are due to phosphorylation; treatment with λ-phosphatase selectively removed the upper-migrating species (Supplemental Figure S3A). Examination of these proteins on 15% SDS–PAGE gel indicated that there was no apparent difference in stability of the phosphorylated versus nonphosphorylated forms of the protein (Supplemental Figure S3A).
Previous studies using a chemical fragmentation assay showed that phosphorylation of Sch9T570A-HA5 is dependent on TORC1 activity, as measured by loss of phosphorylation in cells treated with rapamycin (Urban et al., 2007). Furthermore, phosphorylation of Sch9T570A-HA5 is virtually absent in a gtr1Δ mutant (Binda et al., 2009). Consistent with these studies, the gel shift assay also indicated that phosphorylation of Sch9T570A-HA5 was blocked by addition of rapamycin and required the presence of Gtr1 (Supplemental Figure S3A). Of note, Sch9T570A-HA5 was significantly less phosphorylated in the fab1Δ and vac7Δ mutants (Figure 3A). These findings demonstrate that mutants that do not maintain normal levels of PI(3,5)P2 are defective in TORC1-dependent phosphorylation of Sch9.
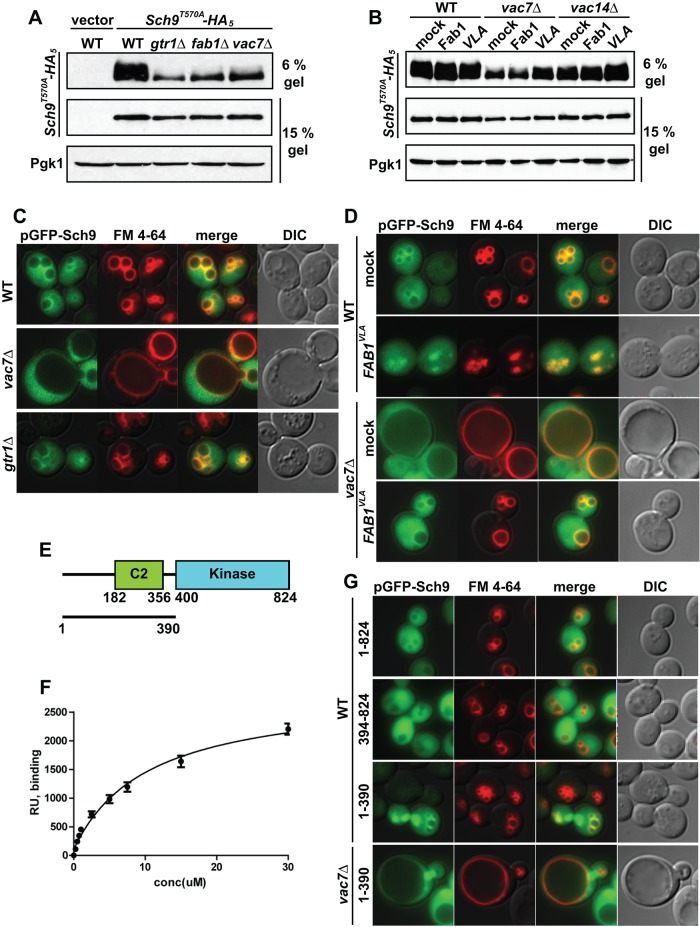
FIGURE 3:
PI(3,5)P2 is required for the localization of Sch9 on the vacuole. (A) Phosphatase treatment indicates that phosphorylation of Sch9-T570A-HA can be assayed by altered mobility on 6% SDS–PAGE. Lysates from 5 × 106 cells from a wild-type strain transformed with a plasmid expressing Sch9-T570A-HA5 were incubated at 30°C for 1 h with (+) or without (–) 6000 U of λ-phosphatase (New England Biolabs, Ipswich, MA). As assessed by mobility on 6% SDS–PAGE, the degree of TORC1-dependent phosphorylation of Sch9-T570A-HA5 is greatly diminished in the gtr1Δ mutant. The degree of TORC1-dependent phosphorylation of Sch9-T570A-HA5 is also greatly diminished in fab1Δ and vac7Δ mutants. (B) TORC1-dependent phosphorylation of Sch9-T570A-HA5 in vac7Δ and vac14Δ mutants is rescued by elevation of PI(3,5)P2 levels. TORC1-dependent phosphorylation of Sch9-T570A-HA5 assessed by mobility on 6% SDS–PAGE in wild-type, vac7Δ, or vac14Δ cells with a vector control or plasmids expressing Fab1 or FAB1VLA. In A and B, loading controls were Sch9-T570A-HA5 and Pgk1 run on 15% SDS–PAGE. (C) GFP-Sch9 (green) partially colocalizes with the vacuole membrane (red) in wild-type and gtr1Δ yeast but not in the vac7Δ mutant. Vacuole membranes were visualized with FM 4-64. (D) Increasing PI(3,5)P2 restores the localization of GFP-Sch9 in vac7Δ mutants. Mock or FAB1VLA plasmids were expressed in wild-type or vac7Δ cells. Analysis of degree of colocalization of GFP-Sch9 and FM 4-64 is shown in Supplemental Figure S3, B and C. (E) Schematic of Sch9 indicating the peptide (aa 1–400) tested for binding to PI(3,5)P2, the predicted calcium/lipid-binding C2 domain (aa 182–356), and the kinase domain (aa 400–824). (F) Recombinant GST-Sch9 fusion protein was tested for its ability to bind PI(3,5)P2, using surface plasmon resonance. The peptide including the C2 domain of Sch9 binds PI(3,5)P2 with a dissociation constant of 11 ± 2 μM (Table 1). Equilibrium-state response levels (RU) for each protein concentration were assessed. Data from three independent experiments. (G) GFP fused to the N-terminal fragment of Sch9 (aa 1–390) localizes on the vacuole in wild-type cells but not in vac7Δ mutants. Vacuole membranes were visualized with FM 4-64. In wild-type or vac7Δ mutants, GFP fused to the full length (aa 1–824), N-terminus fragment (aa 1–390), or C-terminus fragment (aa 391–824) of Sch9 (green) was overexpressed. Vacuole membranes were visualized with FM 4-64.
To extend our analysis, we examined the complementary situation, using the FAB1VLA mutant to generate elevated PI(3,5)P2. We found that concomitant with suppression of rapamycin sensitivity (Figure 1B), expression of the hyperactive mutant in vac7Δ or vac14Δ yeast resulted in increased phosphorylation of Sch9T570A-HA5 (Figure 3B; see 6% gel). In contrast, expression of an additional one or two copies of wild-type Fab1 had little effect on Sch9 phosphorylation in the vac7∆ strain and showed partial suppression in vac14∆ cells, in agreement with lack of suppression of rapamycin sensitivity. In addition, the degree of FAB1VLA-dependent phosphorylation of Sch9T570A-HA5 was lower in vac7Δ compared with the vac14Δ mutant. This parallels the levels of PI(3,5)P2, which are lower in the former mutant compared with the latter (Duex et al., 2006a). Together these observations provide strong support for the hypothesis that PI(3,5)P2 is required for TORC1-dependent phosphorylation of Sch9.
In wild-type yeast, Sch9 is found both on the vacuole membrane and in the cytoplasm (Urban et al., 2007; Figure 3C). Thus we tested whether PI(3,5)P2 is required for Sch9 recruitment to the vacuole. Note that the localization of GFP-Sch9 to the vacuole did not require active TORC1 (Figure 3C); GFP-Sch9 localization in a gtr1Δ mutant was similar to that observed in the wild-type strain. However, the vacuolar localization of GFP-Sch9 was compromised in a vac7Δ mutant. Moreover, elevation of PI(3,5)P2 in the vac7Δ mutant by expression of hyperactive FAB1VLA restored the localization of Sch9 to the vacuole membrane (Figure 3D and Supplemental Figure S3, B and C). These findings show that PI(3,5)P2 recruits the TORC1 substrate Sch9 to the vacuole and further suggest that Sch9 might directly bind this lipid.
To test this hypothesis, we generated a GST fusion protein of the first 400 amino acids of Sch9, which includes a C2 domain (Figure 3E). C2 domains often bind specific phosphoinositide lipids (Lemmon, 2008). The Sch9 C2 domain–containing peptide bound PI(3,5)P2 with a dissociation constant of 11 ± 2 μM (Figure 3, E and F, and Table 1). It is possible that Sch9 associates with other phosphoinositide lipids in vivo, including PI3P, which is also synthesized on endosomes. However, PI3P cannot recruit Sch9 in vivo: PI3P levels in a wild-type strain are 30-fold higher than PI(3,5)P2 levels (Duex et al., 2006a), and in the vac7Δ mutant, the levels of PI3P are twofold higher than levels in a wild-type strain (Gary et al., 2002). Yet despite this high level of PI3P, Sch9 is not recruited to the vacuole. In addition, PI4P and PII4,5)P2 are not responsible for the recruitment of Sch9: in wild-type cells, PI4P and PI(4,5)P2 levels are ∼20-fold higher than that of PI(3,5)P2, and in the vac7Δ mutant the levels of PI4P and PI(4,5P2) remain unchanged (Gary et al., 2002). Together these data indicate that the association of Sch9 with the vacuole requires PI(3,5)P2 and that PI3P, PI4P, and PI(4,5)P2 cannot substitute for PI(3,5)P2.
We further tested and found that the N-terminal region of Sch9 (amino acids [aa] 1–390) is sufficient for the localization of Sch9 on the vacuole in vivo. Full-length Sch9-GFP (1–824), as well as the C2 domain–containing fragment, Sch9-GFP (1–390), localized on the vacuole membrane in the cytoplasm and the nucleus (Figure 3G). However, in the vac7Δ mutant with low levels of PI(3,5)P2, the localization of Sch9-GFP (1–390) to the vacuole was defective (Figure 3G). Together these data strongly suggest that association of Sch9 with the vacuole requires PI(3,5)P2, and that the N-terminal region of Sch9 contains the PI(3,5)P2-binding site.
PI(3,5)P2 is required for TORC1-dependent phosphorylation of Sch9 on the vacuole membrane
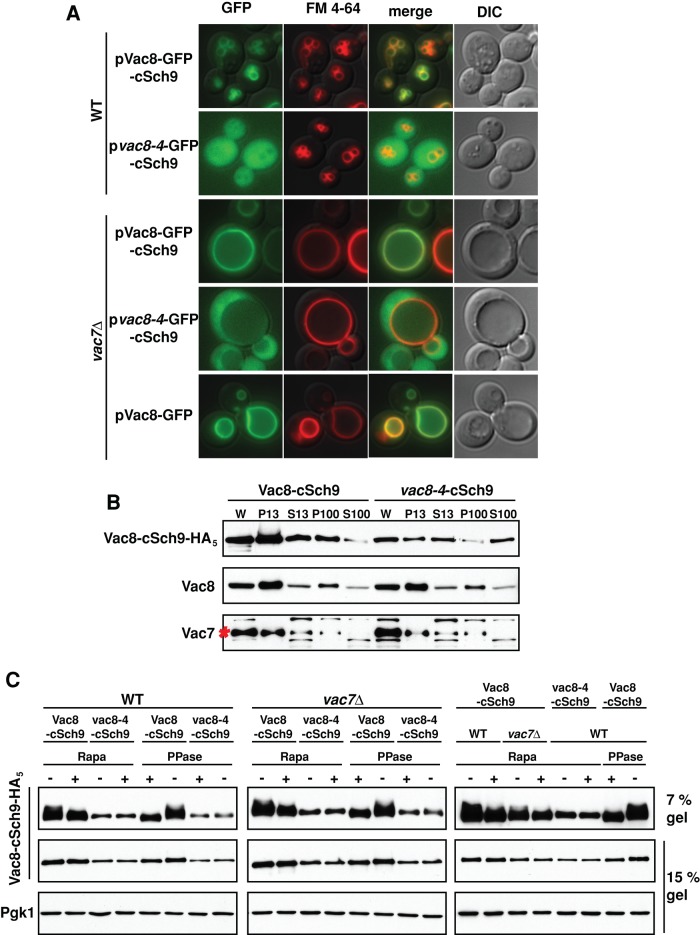
That Sch9 associates with the vacuole membrane via PI(3,5)P2 suggests that PI(3,5)P2 serves as a platform for TORC1 and its target, Sch9. To address this hypothesis, we used an Sch9 C-terminal fragment (aa 709–824; cSch9), which contains all the TORC1 phosphorylation sites, S711, T723, S726, T737, S758, and S765 (Urban et al., 2007). We fused this fragment to either wild-type Vac8 or a mutant version, vac8-4; the vac8-4 protein is mutated at its myristoylation and palmitoylation sites (G2A, C4G, C5T, and C7S) and does not localize on the vacuole membrane (Wang et al., 1998). Accordingly, in wild-type yeast, Vac8-GFP-cSch9 localized on the vacuole membrane, whereas vac8-4–cSch9 was found in the cytoplasm (Figure 4A). Similarly, in a membrane fractionation assay, Vac8 and Vac8-cSch9 were distributed in the P13 and P100 membrane fractions (Figure 4B). In contrast, a large amount of vac8-4–cSch9 was also found in S13 and S100 cytosolic fractions (Figure 4B). Of note, Vac8-cSch9 but not vac8-4–cSch9 was phosphorylated by TORC1 (Figure 4C). These data suggest that the localization of Sch9 on the vacuole is required for its TORC1-dependent phosphorylation.
FIGURE 4:
PI(3,5)P2 is required for TORC1-dependent phosphorylation of Sch9 on the vacuole. (A) Vac8, but not vac8-4, fused to the C-terminal fragment of Sch9 (aa 709–824: cSch9) localizes on the vacuole in both wild-type cells and vac7Δ mutants. Vacuole membranes were visualized with FM 4-64. (B) In wild-type yeast, Vac8-cSch9 cofractionates with the P13 and P100 fractions, which include the vacuole. However, a large amount of vac8-4–cSch9 cofractionates with the S100, which corresponds to the cytosolic fraction. Cell lysates were centrifuged at 13,000 × g for 10 min at 4°C. Supernatant (S13) fractions and whole-cell lysates (W) were resuspended in the same volume of sample buffer, and the pellet (P13) fraction was resuspended in twice the volume of sample buffer. Then, S13 fractions were centrifuged at 100,000 × g for 30 min at 4°C. Supernatant (S100) fractions were resuspended in the same volume of sample buffer, and the pellet (P100) was resuspended in twice the volume of sample buffer. Loading controls for cellular fractionation were Vac7 and Vac8, which tightly localize on the vacuole membrane. Red asterisk, Vac7 protein. (C) Localization of cSh9 on the vacuole via PI(3,5)P2 is required for its TORC1-dependent phosphorylation. TORC1-dependent phosphorylation of Vac8-cSch9 or vac8-4-cSch9 assessed by mobility on 7% SDS–PAGE. Wild-type or vac7Δ yeast, with a plasmid expressing Vac8-cSch9-HA5 or vac8-4-cSch9-HA5, was grown in SC-His medium, and then a final concentration of 200 ng/ml rapamycin (+) was added to the cell culture. Cells were incubated for an additional 1 h. Lysates from 5 × 106 cells with or without rapamycin treatment were incubated at 30°C for 1 h with (+) or without (–) 6000 U λ-phosphatase (New England Biolabs). As assessed by mobility on 7% SDS–PAGE, the degree of TORC1-dependent phosphorylation of Vac8-cSch9-HA5 was greatly diminished in both wild-type and vac7Δ mutant yeast with rapamycin treatment. Loading controls were Vac8- or vac8-4–cSch9-HA5 and Pgk1 run on 15% SDS–PAGE. Right, subset of analyses from left and middle repeated to allow side-by-side comparison of wild-type and vac7Δ strains. Of note, there was less phosphorylation of Vac8-cSch9 in vac7Δ than with a wild-type strain. This provides additional support for the hypothesis that PI(3,5)P2 plays additional roles beyond recruitment of Sch9.
To examine this further, we expressed Vac8-cSch9 or vac8-4-cSch9 in a vac7Δ mutant. Similar to the wild-type strain, in the vac7Δ mutant, Vac8-cSch9 localized on the vacuole, but vac8-4-cSch9 was diffuse in the cytoplasm (Figure 4A). Of importance, Vac8-cSch9 was phosphorylated by TORC1 in the vac7Δ mutant (Figure 4C). These data suggest that PI(3,5)P2-dependent localization of Sch9 on the vacuole is required for its phosphorylation by TORC1. Together these findings strongly support the hypothesis that PI(3,5)P2 is critical for the localization Sch9 on the vacuole, where it is phosphorylated by TORC1.
Of note, when compared side by side, the degree of TORC1-dependent phosphorylation of Vac8-cSch9 in the vac7Δ mutant was less than in a wild-type strain (Figure 4C, rightmost panel). This suggests that in addition to localization of the TORC1 target, PI(3,5)P2 on the vacuole has further roles in the positive regulation of TORC1.
PI(3,5)P2 is required for TORC1 phosphorylation of multiple substrates
TORC1 functions in multiple pathways via phosphorylation of its targets and regulates cell growth. Because yeast mutants with defects in PI(3,5)P2 have pleiotropic defects, we tested whether PI(3,5)P2 is required for multiple TORC1-dependent pathways via regulation of TORC1 phosphorylation of additional targets.
Under nutrient-rich conditions, active TORC1 is required for inhibitory hyperphosphorylation of Npr1 (Schmidt et al., 1998), which allows ubiquitin-mediated endocytosis of selected plasma membrane transporters and their delivery for degradation in the vacuole (MacGurn et al., 2012). Of note, in the vac7Δ mutant, Npr1 was less phosphorylated than in a wild-type strain, and inhibition of TORC1 with rapamycin caused similar loss of phosphorylation of Npr1 in both strains (Figure 5A).
FIGURE 5:
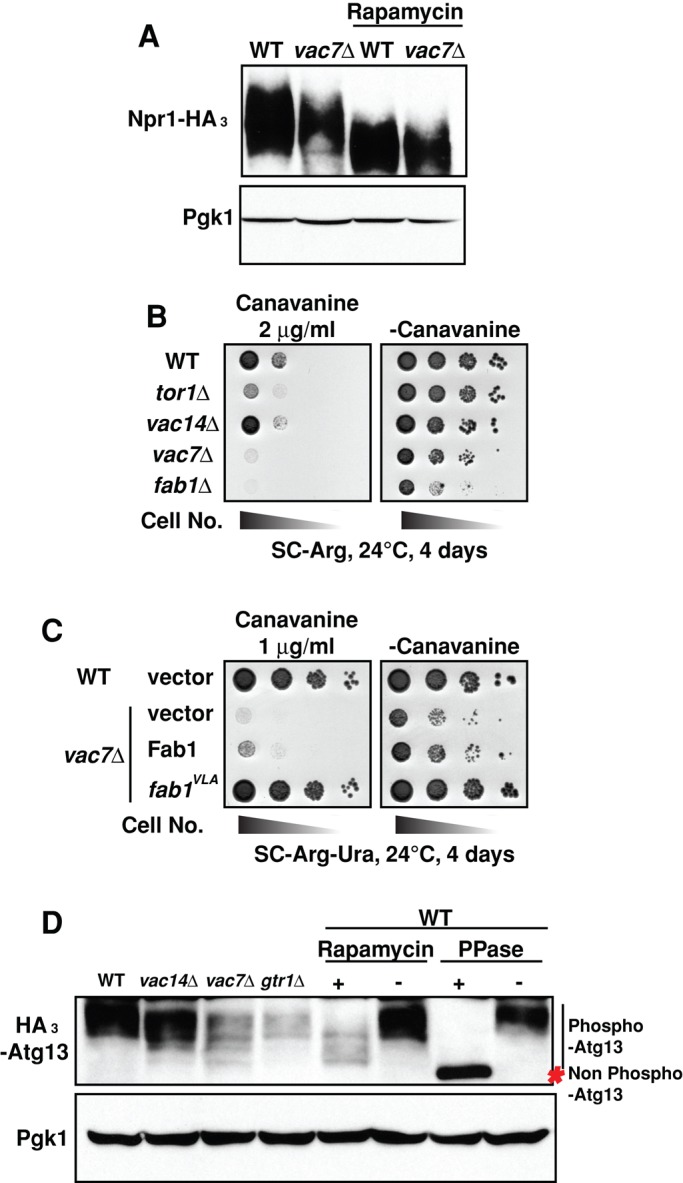
TORC1-dependent regulation of multiple pathways requires PI(3,5)P2. (A) Phosphorylation of Npr1-HA3 is greatly diminished in vac7Δ mutants. Wild-type or vac7Δ strains that express Npr1-HA3 were grown on YEPD; then rapamycin was added to a final concentration of 200 ng/ml. Cells were incubated for an additional 30 min. The degree of phosphorylation of Npr1 was assessed by migration during 6% SDS–PAGE. Loading control: Pgk1 run on 15% SDS–PAGE. (B) The tor1Δ, fab1Δ, and vac7Δ mutants with defects in TORC1 function are hypersensitive to canavanine. The indicated strains were grown to mid log phase in YEPD medium and then diluted onto SC-Arg plates in the presence or absence of 2 μg/ml canavanine, a sublethal dose for wild-type yeast. (C) Sensitivity of vac7Δ mutants to 1 μg/ml canavanine is rescued by elevation of PI(3,5)P2 levels. Wild-type cells with empty vector or the vac7Δ mutant with plasmids expressing mock, Fab1, or FAB1VLA were grown to mid log phase in SC-Ura medium and then diluted onto SC-Arg-Ura plates in the presence or absence of 1 μg/ml canavanine. (D) Phosphorylation of HA3-Atg13 is greatly diminished in vac14Δ, vac7Δ, fab1Δ, and gtr1Δ mutants. Wild-type, vac14Δ, vac7Δ, fab1Δ, or gtr1Δ strains with a plasmid expressing HA3-Atg13 were grown in SC-Leu medium, and then either a final concentration of 200 ng/ml rapamycin (+) or DMSO alone (–) was added to the cell culture. Cells were incubated for an additional 2 h. Lysates from 5 × 106 cells from each strain were incubated at 30°C for 1 h with (+) or without (–) 6000 U of l-phosphatase (New England Biolabs). The degree of phosphorylation of Atg13 was assessed by migration in 6% SDS–PAGE. Loading control: Pgk1 run on 15% SDS–PAGE.
TORC1-dependent phosphorylation of Npr1 releases its inhibitory block on the endocytosis of Can1, an arginine permease (MacGurn et al., 2012). Canavanine is a toxic analogue of arginine. Thus, under nutrient-rich conditions, mutants that do not have normal inhibition of Npr1 have excess Can1 on the plasma membrane and are hypersensitive to canavanine (Lin et al., 2008). The finding that less PI(3,5)P2 resulted in a defect in TORC1-dependent phosphorylation of Npr1 prompted us to test whether PI(3,5)P2 is required for TORC1-regulated inhibition of Npr1 to release its block on the endocytosis of Can1. We therefore analyzed canavanine sensitivity in vac7Δ and fab1Δ mutants. The tor1Δ mutant, as well as the vac7Δ and fab1Δ mutants, were hypersensitive to canavanine (Figure 5B). vac14Δ, which is less defective in PI(3,5)P2 levels, was less sensitive than the vac7Δ mutant. To test further whether this defect was due to low levels of PI(3,5)P2, we expressed the hyperactive FAB1VLA mutant in the vac7Δ mutant and analyzed canavanine sensitivity. The increase in levels of PI(3,5)P2 by FAB1VLA suppressed the sensitivity to canavanine in the vac7Δ mutant (Figure 5C). These data suggest that transport of Can1 from the plasma membrane via TORC1-dependent inhibitory phosphorylation of Npr1 requires PI(3,5)P2.
It is not known whether Npr1 is a direct target of TORC1. Thus, in addition to the direct target Sch9, we chose to examine Atg13, another well-characterized direct target of TORC1. Atg13 functions in the initiation of autophagy (Kamada et al., 2010). Starvation or treatment of cells with rapamycin induces autophagy via inhibition of TORC1-inhibitory phosphorylation of Atg13.
We analyzed the degree of phosphorylation of Atg13 using mutants that have low levels of PI(3,5)P2. Atg13 is highly phosphorylated by TORC1 (Kamada et al., 2010), and this phosphorylation is largely reversed by treatment with rapamycin or phosphatase (Figure 5D). In the gtr1∆ mutant with reduced TORC1 activity, Atg13 phosphorylation was decreased. Similarly, in vac14Δ or vac7Δ mutants with reduced levels of PI(3,5)P2, Atg13 was less phosphorylated (Figure 5D).
Together these data suggest that PI(3,5)P2 is required for TORC1-dependent phosphorylation of multiple substrates and furthermore that through positive regulation of TORC1, PI(3,5)P2 regulates several pathways, including autophagy and nutrient-regulated endocytosis of Can1.
Autophagy is negatively regulated by PI(3,5)P2
A previous study suggested that PI(3,5)P2 does not play a role in autophagy in yeast (Dove et al., 2004). However TORC1 phosphorylation of Atg13 is required to inhibit autophagy in nutrient-rich conditions. Moreover, TORC1-dependent activity of Sch9 also contributes to the inhibition of autophagy, independent of phosphorylation of Atg13 (Yorimitsu et al., 2007). Our findings that less PI(3,5)P2 caused defects in TORC1 phosphorylation of Atg13 and Sch9 led us to test whether lower levels of PI(3,5)P2 also resulted in a corresponding induction of autophagy under nutrient-rich conditions.
The autophagy pathway involves the sequestration of cytoplasm and/or organelles into autophagosomes, which fuse with the vacuole/lysosome. The breakdown of proteins and membranes in the vacuole provides new building blocks and energy for cellular renovation and homeostasis. We tested whether PI(3,5)P2 inhibits autophagy, using two common assays that monitor Atg8 behavior.
Atg8 is required for both autophagy and the cytoplasm-to-vacuole targeting (Cvt) pathway (Kirisako et al., 1999; Huang et al., 2000). Atg8 localizes on autophagosomes via its conjugation with phosphatidylethanolamine (Mizushima et al., 1998). Atg8 on the inner membrane of the autophagosome is delivered to the vacuole and is degraded there (Kim et al., 2001). In the case of GFP-Atg8, the GFP moiety is resistant to degradation and accumulates in the vacuole lumen. Thus production of free GFP from GFP-Atg8 can be used to monitor autophagy and the Cvt pathway either by microscopy or via Western blot analysis by probing for released GFP.
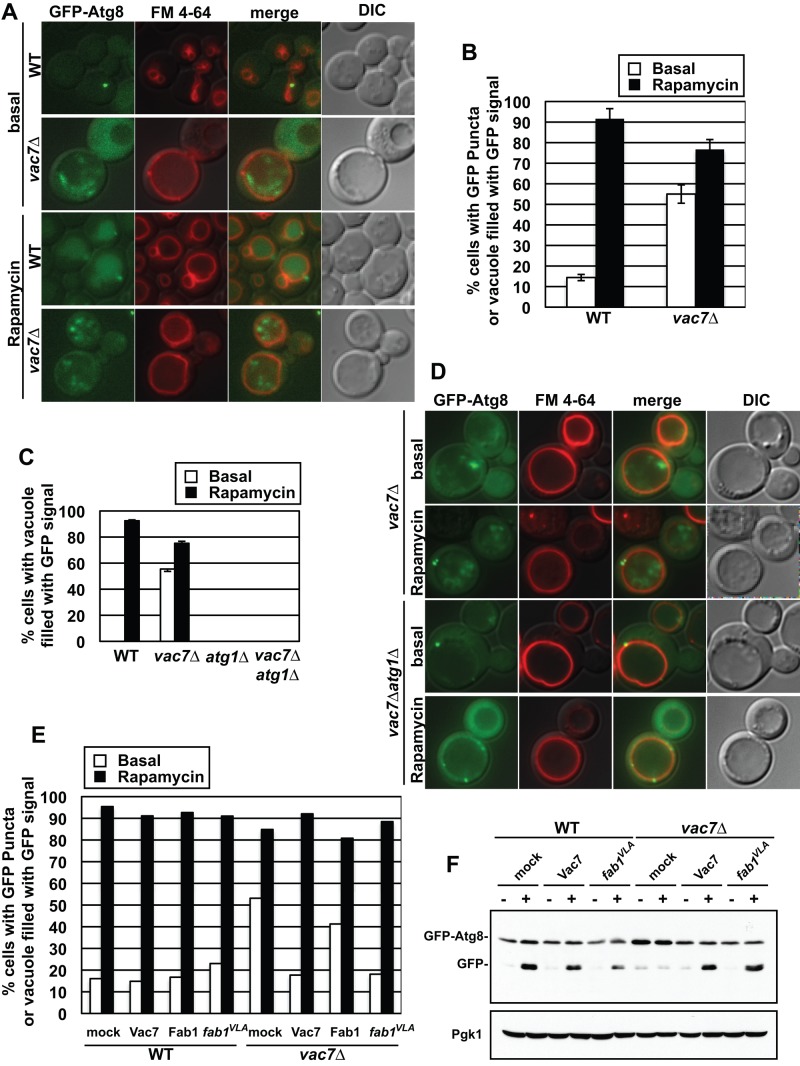
We expressed GFP-Atg8 in wild type or the vac7Δ mutant and quantified the induction of autophagy by microscopy. Wild-type yeast cells contained only single GFP-Atg8–positive puncta near the vacuole membrane under normal conditions, and the population of cells containing GFP puncta was <20% (Figure 6, A and B). In addition, there was no detectable fluorescence signal within the vacuole lumen. On induction of autophagy using rapamycin treatment, GFP-Atg8 was targeted to the vacuole, free GFP was released, and the entire vacuole lumen was filled with GFP (Figure 6A). During rapamycin treatment, the population of cells with GFP puncta or GFP-filled vacuoles increased to >90% (Figure 6B). In contrast, in the vac7Δ mutant, GFP puncta accumulated in the vacuole lumen even under basal conditions, and the population of cells with GFP puncta was >70% (Figure 6, A and B). Note that the GFP-Atg8 in the vacuole lumen of vac7∆ cells was present as puncta compared with the diffuse distribution seen in the lumen of wild-type cells. Of importance, the GFP puncta in the vacuole lumen were a result of the inappropriate induction of autophagy under nutrient-rich conditions. Atg1 plays a key role in the initiation and continuation of autophagy. When an atg1Δ mutant was treated with rapamycin, GFP-Atg8 was not transported to the vacuole lumen because autophagy was blocked (Supplemental Figure S4, A and B; Cheong et al., 2008). Similarly, in a vac7Δ atg1Δ double mutant, whereas GFP-Atg8–positive puncta were observed in the cytoplasm near the vacuole membrane, these were not delivered to the vacuole either in basal conditions or during rapamycin treatment (Figure 6, C and D, and Supplemental Figure S4).
FIGURE 6:
PI(3,5)P2 regulates autophagy. PI(3,5)P2 is required both for inhibition of autophagy during vegetative growth and degradation of autophagic bodies. (A) Wild-type or vac7Δ strains with a plasmid expressing GFP-Atg8 were grown in SC-Ura medium. Rapamycin at a final concentration of 200 ng/ml was added, and cells were incubated for an additional 2 h. Vacuole membranes were visualized with FM 4-64. (B) Quantification of cells with GFP-Atg8 puncta or vacuoles filled with GFP or GFP-Atg8 from A. Three independent trials with >100 cells counted/trial. Error bars in B represent SD of triplicate experiments. (C, D) vac7Δ or atg1Δ vac7Δ strains with a plasmid expressing GFP-Atg8 was grown in SC-Ura medium. Rapamycin at a final concentration of 200 ng/ml was added, and cells were incubated for an additional 2 h. Vacuole membranes were visualized with FM 4-64. (C) Quantification of cells with vacuoles filled with GFP or GFP-Atg8 from D. The data represent three independent trials with >100 cells counted/trial. Error bars in C represent SD of triplicate experiments. (E). Quantification of the number of cells with Atg8 puncta or vacuoles filled with GFP or GFP-Atg8. Wild-type or vac7Δ strain with plasmids expressing GFP-Atg8 and mock, Vac7, Fab1, or FAB1VLA was grown in SC-Ura-Trp medium, and then a final concentration of 200 ng/ml rapamycin was added and cells were incubated for an additional 2 h. The data are an average from two trials with >100 cells/trial. (F) Increasing PI(3,5)P2 by expression of the FAB1VLA mutant restores the defect in the degradation of GFP-Atg8. Wild-type or vac7Δ strain with plasmids expressing GFP-Atg8 and mock, Vac7, or FAB1VLA was grown in SC-Ura-Trp, and then a final concentration of 200 ng/ml rapamycin was added. Cells were incubated for an additional 4 h. Western blot was performed using anti-GFP antibody. Loading control: Pgk1.
To further address whether PI(3,5)P2 is required for the inhibition of autophagy in nutrient-rich conditions, we expressed the hyperactive FAB1VLA mutant in vac7Δ cells and quantified the induction of autophagy by microscopy. Increasing the levels of PI(3,5)P2 by the FAB1VLA mutant or by expression of wild-type VAC7 suppressed the accumulation of GFP puncta in the vacuole under basal conditions (Supplemental Figure S4C) and restored the number of cells without GFP puncta to wild-types levels in basal conditions (Figure 6E). However, expression of Fab1 from a low-copy plasmid did not restore PI(3,5)P2 levels or suppress the inappropriate induction of autophagy under nutrient-rich conditions. Together these data suggest that PI(3,5)P2 is required for inhibition of autophagy under basal conditions.
In an independent test, we performed Western blot analysis for free GFP using whole yeast lysates from the cultures visualized in Figure 6. In the vac7Δ mutant the degradation of GFP-Atg8 was inhibited under both nutrient-rich conditions and rapamycin treatment (Supplemental Figure S4D), consistent with the presence of GFP puncta in the vacuole lumen in the vac7Δ mutant. The presence of GFP puncta in the vacuole suggested that autophagosomes are transported into the vacuole but not degraded, resulting in accumulation of autophagic bodies. Consistent with this hypothesis, increasing the levels of PI(3,5)P2 by expressing the FAB1VLA mutant in the vac7Δ strain resulted in correct degradation of GFP-Atg8 in the vacuole lumen during the induction of autophagy (Figure 6F).
The yeast vacuole is a weakly acidic compartment that contains a wide variety of hydrolases that are only active in this acidic environment. Furthermore, the pH gradient across the vacuole membrane establishes an electrochemical potential that is needed for the transport of various metabolites, including calcium and phosphate ions, establishing a unique milieu that is needed for optimal hydrolase activity. Accordingly, proper acidification is required for the degradation of autophagic bodies in the vacuole lumen but not for the formation or delivery of autophagosomes to the vacuole (Nakamura et al., 1997). Of importance, mutants with low or no PI(3,5)P2 do not have fully acidified vacuoles (Bonangelino et al., 1997, 2002; Dove et al., 2002). Together our studies indicate that the inhibition of autophagy by TORC1 is defective in mutants with low levels of PI(3,5)P2. In addition, we found that PI(3,5)P2 is required for the completion of autophagy. In the absence of PI(3,5)P2, autophagic bodies are not degraded in the vacuole lumen. Thus PI(3,5)P2 negatively regulates the induction of autophagy via positive regulation of TORC1 activity and positively functions in the resolution of autophagy through the degradation of autophagic bodies.
DISCUSSION
The endosomal membrane lipid PI(3,5)P2 regulates multiple pathways. Pleiotropic defects observed in yeast, mice, and cultured cells caused by deficiencies in PI(3,5)P2 strongly suggest that many of the pathways regulated by PI(3,5)P2 have not yet been identified. Moreover, these pleiotropic phenotypes make it difficult to determine which individual pathways are directly regulated by this lipid. Identification of effector proteins provides a means to reveal processes that are directly modulated by PI(3,5)P2. Here we show that PI(3,5)P2 regulation of TORC1 is required for at least three TORC1-dependent pathways. Specifically, we show that 1) PI(3,5)P2 is required for the activation of TORC1 on the vacuole membrane; 2) a major substrate of TORC1, Sch9, associates with the vacuole membrane via PI(3,5)P2, and this association is required for TORC1-dependent phosphorylation of Sch9; and 3) PI(3,5)P2 plays a key role in additional TORC1-regulated pathways, including nutrient-dependent endocytosis of Can1 and inhibition of autophagy, via TORC1-dependent phosphorylation of Npr1 and Atg13, respectively. These studies expand our knowledge of pathways regulated by PI(3,5)P2 and show that via modulation of TORC1 function, PI(3,5)P2 is a key regulator of cellular response to nutrient status.
Regulation of TORC1 by PI(3,5)P2 on endosomal compartments
Despite the importance of PI(3,5)P2 for modulation of TORC1, it is likely that PI(3,5)P2 is necessary but not sufficient for TORC1 activation. Note that hyperosmotic stress results in a 20-fold elevation of PI(3,5)P2 (Duex et al., 2006a), yet this same treatment inhibits TORC1 phosphorylation of Sch9 (Urban et al., 2007). The inhibition of TORC1 during hyperosmotic stress occurs via as-yet-unidentified upstream regulators. This divergence in the effects of hyperosmotic shock on PI(3,5)P2 and TORC1 is consistent with the postulate that PI(3,5)P2 is not the sole upstream regulator of TORC1 function.
The precise mechanisms for PI(3,5)P2-dependent TORC1 activation remain to be determined. In yeast, TORC1 localizes on the vacuole with or without amino acid stimulation (Sturgill et al., 2008; Binda et al., 2009). In contrast, mTORC1 translocates onto the lysosome specifically during amino acid stimulation (Sancak et al., 2010). The simple postulate that PI(3,5)P2 acts solely via recruitment of TORC1 to endosomes is unlikely because artificial recruitment of TORC1 to the vacuole in the absence of PI(3,5)P2 only partially restores TORC1 function (Figure 2). Our findings, as well as others, suggest three ways that PI(3,5)P2 may play roles in the activation of TORC1.
First, amino acids play a major role in the regulation of TORC1. Thus PI(3,5)P2 may be involved in sensing amino acids. The vacuole/lysosome is a major reservoir for amino acids (Messenguy et al., 1980), which are released as needed during vegetative growth. In mammals, amino acids stored in the lysosome are detected by the lysosomal V-ATPase, which in turn activates TORC1 (Zoncu et al., 2011). Of note, yeast mutants with defects in PI(3,5)P2 have less-acidified vacuoles and may have defects in V-ATPase activity (Bonangelino et al., 2002). Thus PI(3,5)P2 may indirectly activate TORC1 via direct activation of the V-ATPase. However, this is unlikely to be the sole role of PI(3,5)P2 in TORC1 function. Both Kog1 and Sch9 interact directly with PI(3,5)P2. Moreover, PI(3,5)P2 is required to recruit Sch9 to the vacuole, where it is phosphorylated by TORC1.
Second, PI(3,5)P2 may directly activate TORC1. The essential TORC1 subunit Raptor functions as a scaffold for the TORC1 complex (Hara et al., 2002; Kim et al., 2002; Nojima et al., 2003; Sancak et al., 2008; Yip et al., 2010), and yeast Kog1 likely serves similar roles. Of note, Kog1 (Supplemental Figure S1) and its mammalian homologue, Raptor (Bridges et al., 2012), directly bind PI(3,5)P2 in vitro. Thus PI(3,5)P2 may activate TORC1 through direct interaction with Kog1/Raptor, which may modulate the structure of TORC1. However, integrity of the TORC1 complex does not appear to be grossly affected by loss of PI(3,5)P2; Kog1 interaction with Tor1 still occurs in a vac7Δ mutant (Supplemental Figure S2).
Third, a membrane subdomain that includes PI(3,5)P2 may exist on the vacuole and serve as a platform for TORC1, its substrates, and regulators, including the EGO complex. Recruitment of the TORC1 substrate Sch9 to the vacuole requires PI(3,5)P2. Moreover, in a global study, Ego3, a component of the EGO complex, was shown to bind phosphorylated phosphatidylinositol lipids (Zhu et al., 2001).
TORC1-dependent phosphorylation occurs on vacuole/lysosome membranes
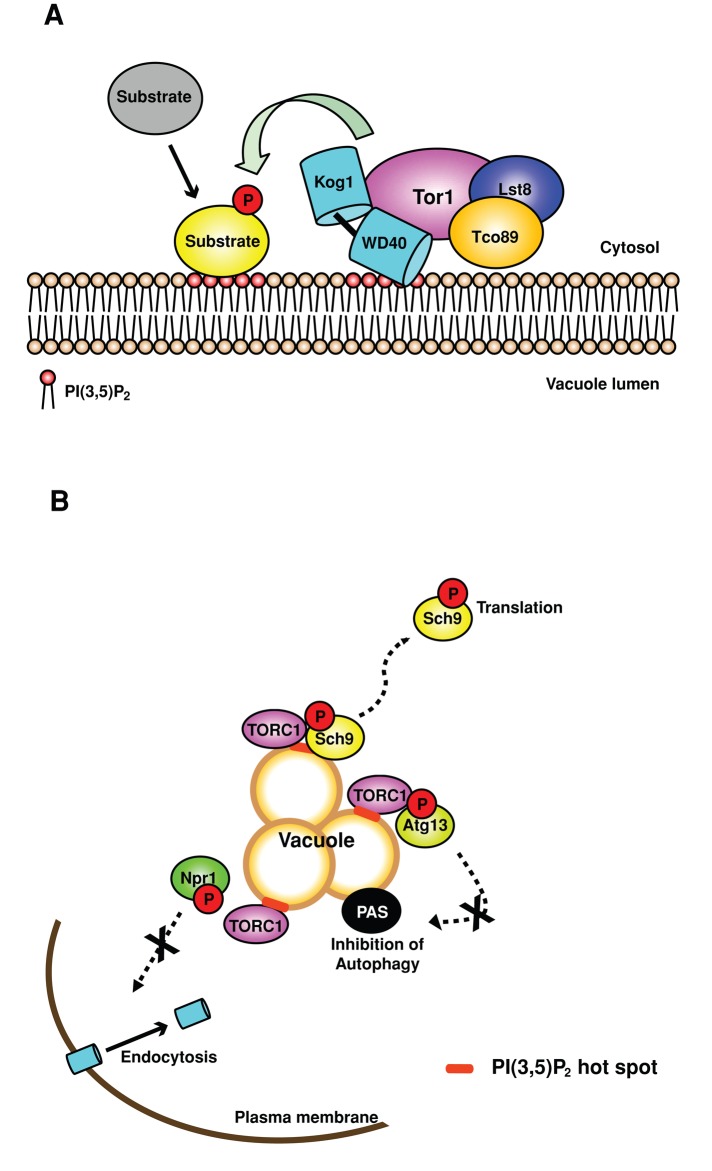
That PI(3,5)P2-dependent localization of Sch9 on the vacuole is required for TORC1-dependent phosphorylation of Sch9 (Figures 3 and 4) makes it tempting to speculate that TORC1 phosphorylation of multiple targets may require their recruitment to the vacuole/lysosome by PI(3,5)P2. In mammals, S6K and eIF4E-binding protein 1 (Brunn et al., 1997; Hara et al., 1997; Burnett et al., 1998; Isotani et al., 1999), the transcription factor EB (TFEB; Settembre et al., 2012), lipin-1 (Peterson et al., 2011), and Atg13 and Ulk1 (Hosokawa et al., 2009), and in yeast, Sch9, Atg13 (Kamada et al., 2010), Sfp1 (Lempiainen et al., 2009), and Gln3 (Bertram et al., 2000), are direct targets of TORC1. With the exception of TFEB and Sch9, the location where these targets are phosphorylated by TORC1 is not known. mTORC1-dependent phosphorylation of TFEB occurs on the lysosome and regulates the translocation of TFEB to the nucleus during lysosomal stress or amino acid stimulation (Pena-Llopis and Brugarolas, 2011; Settembre et al., 2012). The activation of Rag GTPase by the V-ATPase (Settembre et al., 2012) and the association of active Rags with TFEB (Martina and Puertollano, 2013) are required for translocation of TFEB to the lysosome. Whether PI(3,5)P2 is required for TORC1 phosphorylation of TFEB or other targets has not been tested. Perhaps TFEB or other substrates, including S6K, target to the lysosome/vacuole membrane via PI(3,5)P2 and are thereby brought to TORC1 via PI(3,5)P2 signaling (Figure 7).
FIGURE 7:
PI(3,5)P2 is a critical upstream regulator of TORC1. (A) PI(3,5)P2-dependent activation of TORC1 and recruitment of substrates to the vacuole membrane. Kog1, a subunit of TORC1, and the TORC1 substrate, Sch9, are downstream effectors of PI(3,5)P2. PI(3,5)P2 recruits Sch9 and likely additional TORC1 targets to the vacuole, where their TORC1-dependent phosphorylation occurs. (B) PI(3,5)P2 is required for TORC1-dependent phosphorylation of multiple substrates and TORC1-dependent inhibition of autophagy, as well as regulation of nutrient-dependent endocytosis. TORC1-dependent phosphorylation of each substrate tested, Sch9, Npr1, and Atg13, requires PI(3,5)P2, which suggests that phosphorylation of these substrates occurs on the vacuole. In addition, PI(3,5)P2-mediated TORC1-dependent phosphorylation of Npr1 or Atg13 is required to release Npr1-dependent inhibition of Can1 endocytosis and TORC1-dependent inhibition of autophagy, respectively. Note that in addition to regulation of TORC1, PI(3,5)P2 modulates pathways that are independent of TORC1 signaling.
Roles for PI(3,5)P2 in autophagy
Atg13 is a direct target of TORC1 and is hyperphosphorylated under nutrient-rich conditions, which inhibits autophagy (Kamada et al., 2010). Although it had been reported that in yeast PI(3,5)P2 is not involved autophagy (Dove et al., 2004), we found that PI(3,5)P2 is required for TORC1-dependent inhibition of autophagy (Figures 6 and Supplemental Figure S4) via TORC1 phosphorylation of Atg13 (Figure 5D). These findings make it tempting to speculate that Atg13 is an effector of PI(3,5)P2. Atg13, as well as additional proteins, play key roles in the initiation of autophagy and, as part of the initiation, are recruited to the phagophore assembly site (reviewed in Nakatogawa et al., 2009). PI(3,5)P2 may be required for recruitment of Atg13 to the phagophore assembly site via TORC1-dependent phosphorylation of Atg13.
That PI(3,5)P2 is required for TORC1 inhibition of autophagy raises the possibility that Atg18, which plays a role in the formation of autophagosomes (Obara et al., 2008; Nair et al., 2010), may also serve a second role in autophagy. Atg18, independent of its role in forming autophagosomes, is a negative regulator of PI(3,5)P2 synthesis (Dove et al., 2004). Through two adjacent sites, Atg18 tightly binds both PI3P and PI(3,5)P2 (Dove et al., 2004; Baskaran et al., 2012; Krick et al., 2012; Watanabe et al., 2012). These lipid-binding sites on Atg18 are required for Atg18 localization on the vacuole, both for the autophagy pathway (Obara et al., 2008; Nair et al., 2010) and for negative regulation of the synthesis of PI(3,5)P2 (Dove et al., 2004; Efe et al., 2007). Perhaps via inhibition of PI(3,5)P2 synthesis, Atg18 promotes the inhibition of TORC1 and the subsequent activation of autophagy.
PI(3,5)P2 also plays a TORC1-independent role in autophagy. The absence of PI(3,5)P2 causes defects in the degradation of autophagic bodies (Figure 6 and Supplemental Figure S4), which are likely due to defects in the acidification of the vacuole (Bonangelino et al., 1997, 2002; Nakamura et al., 1997). These findings show that there are both TORC1-dependent and TORC1-independent roles for PI(3,5)P2 in autophagy. Moreover, in the absence of PI(3,5)P2, the induction of autophagy during nutrient-rich conditions is likely further exacerbated by PI(3,5)P2-related defects in the degradation of autophagic bodies.
These roles for PI(3,5)P2 in the inhibition of autophagy and the resolution of autophagic bodies are likely conserved. Autophagy is important for neural function and neonatal survival in mice (Kuma et al., 2004; Hara et al., 2006; Efeyan et al., 2013). Similarly, mice deficient in the synthesis of PI(3,5)P2 show defects in the nervous system and early lethality (Zhang et al., 2007; Zolov et al., 2012). These similarities are consistent with a link between PI(3,5)P2 and autophagy in all metazoans. Indeed, defects in Fab1/PIKfyve function cause autophagic dysfunction in mammals (Jefferies et al., 2008; de Lartigue et al., 2009; Ferguson et al., 2009; Martin et al., 2013), Caenorhabditis elegans (Nicot et al., 2006), and Drosophila (Rusten et al., 2007). Similar to yeast, defects in PI(3,5)P2 production in mammalian cells result in the loss of nutrient-dependent inhibition of autophagy (Jefferies et al., 2008; Martin et al., 2013). Thus the requirement for PI(3,5)P2-dependent activation of TORC1 for inhibition of autophagy is likely conserved. The TORC1-dependent and independent roles for PI(3,5)P2 described here join a growing list of pathways modulated by PI(3,5)P2. The pleiotropic defects in yeast, cultured mammalian cells, and mice predict that PI(3,5)P2 likely has multiple additional roles in endosomal physiology.
MATERIALS AND METHODS
Strains and media
Yeast strains and plasmids are listed in the Supplemental Experimental Procedures. Yeast were grown in yeast extract/peptone/dextrose (YEPD) or synthetic minimal medium without selective amino acid(s) at 24°C. Construction of plasmids and strains is described in the Supplemental Experimental Procedures.
Generation of functional Tor1-3xGFP and Kog1-3xGFP
To generate functional Tor1-3xGFP, we expressed Tor1 with internally integrated 3xGFP at amino acid D330, a site that does not impair Tor1 localization or function (Sturgill et al., 2008). This construct was expressed as the sole copy of TOR1 from the TOR1 endogenous locus. For Kog1, the 3xGFP tag was placed at the C-terminus of Kog1. Kog1-3xGFP was expressed as the sole copy of KOG1 from the endogenous locus.
Fluorescence microscopy
Mid–log phase cells were labeled with FM 4-64 (Vida and Emr, 1995). Images were acquired using the DeltaVision RT Restoration Microscopy System (Applied Precision, Issaquah, WA).
Rapamycin sensitivity
Cells were grown as indicated in YEPD or selective media to mid log phase. Cells were collected and resuspended in media, and 10-fold serial dilutions from 0.1 OD/ml were performed. Cells were transferred to YEPD or selective media plates containing dimethyl sulfoxide (DMSO; control) and the indicated concentrations of rapamycin. Cells were incubated at 24°C for 2–7 d or 37°C for 2–7 d. Images were obtained every day starting at day 2.
Protein purification
Proteins were expressed in E. coli BL21 star (DE3) pRARE. Cells were grown in Luria broth with 0.05 mg/ml ampicillin and 0.025 mg/ml spectinomycin at 37°C to an OD600 of 0.3. After 18 h of induction with 0.2 mM isopropyl-β-d-thiogalactoside at 16°C, cells were harvested. Cells were sonicated in lysis buffer (20 mM Tris-HCl, pH 7.4, 200 mM NaCl, 1 mM dithiothreitol [DTT]). Proteins were purified with glutathione Sepharose resin (GE Healthcare, Little Chalfont, Buckinghamshire, England), concentrated by spin filtration using an Ultrafree-4 centrifugal filter 50K or 30K membrane (Millipore, Darmstadt, Germany), and dialyzed in Biacore buffer (25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.4, 150 mM NaCl, 1 mM DTT).
Western blot
Log-phase cells were lysed (Duex et al., 2006b), and the lysates separated by SDS–PAGE and transferred to nitrocellulose. Antibody dilutions were anti-Pgk1 (Invitrogen, Grand Island, NY), 1:10,000; anti-hemagglutinin (Covance, Princeton, NJ), 1:3000; and anti-GFP (Roche, Basel, Switzerland), 1:500.
Surface plasmon resonance analysis
Surface plasmon resonance analysis was performed as described previously (Dove et al., 2004; Narayan and Lemmon, 2006). GST-Atg18 (PI(3,5)P2) served as a positive control (Dove et al., 2004), and GST was a negative control. Proteins were used within 24 h of cell lysis, and lipid surfaces were used within 8 h of generation. Affinity was measured using sensor surfaces containing di-oleyl-phosphatidylcholine liposomes doped with PI(3,5)P2 at a molar ratio of 97:3. In an initial experiment, no binding was detected between GST-Sch9 or GST-Kog1 and the PI(3,5)P2 surface. To increase sensitivity, the molar ratio of PI(3,5)P2 was increased to 22:3. Binding was measured by flowing protein samples over the sensor surface. Lipids (Eschelon) were bound to sensor chips, L1 (Biacore, GE Healthcare). Four sensors were available for each run. The first and second sensors contained only phosphatidylcholine (PC). The subsequent two sensors contained PC:PI(3,5)P2. Two sensors were dedicated to PC:PI(3,5)P2 in order to obtain more accurate binding constants. Phosphoinositides:PC were present at a molar ratio of 22:3. Response to PC alone was subtracted from the responses detected from the phosphoinositide-containing chips. Binding data are presented at the indicated protein concentration injected. The Kd values and maximum binding (Rmax) were calculated by using Prism (GraphPad). Data were fitted to the equation
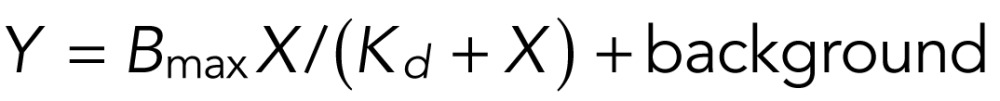 |
where Y is total binding, X is the protein concentration, Bmax is the maximum specific binding in the same units as Y, and Kd is the equilibrium binding constant in the same units as X. Background is the amount of nonspecific binding with no added protein.
Supplementary Material
Acknowledgments
We thank Jason Gestwicki, Andrea Thompson, and Srikanth Patury (University of Michigan) for assistance with surface plasmon resonance. We thank Claudio Virgilio and Scott Powers for strains and plasmids. We thank Mara Duncan, Mark Lemmon, Ken Inoki, Diane Fingar, Michael Hall, Rob Piper, and the Weisman Lab for helpful discussions. This work was supported by National Institutes of Health (NIH) Grants R01-GM50403 (L.S.W.) and GM053396 (D.J.K.), MCubed, University of Michigan (L.S.W., D.J.K.), the Canton of Geneva, the Swiss National Science Foundation, the European Research Council (R.L.), and NIH Grant 5R01DK061618 (A.R.S.). With great sadness we report that Gela Tevzadze passed away in August 2012.
Abbreviation used:
- TORC
target of rapamycin complex
Footnotes
Present addresses: *CHA Cancer Institute, CHA University, Seoul 135-081, South Korea
†Department of Physiology, University of Tennessee Health Sciences Center, and Children's Foundation Research Institute, Le Bonheur Children's Hospital, Memphis, TN 38103.
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-01-0021) on January 29, 2014.
REFERENCES
- Araki T, Uesono Y, Oguchi T, Toh EA. LAS24/KOG1, a component of the TOR complex 1 (TORC1), is needed for resistance to local anesthetic tetracaine and normal distribution of actin cytoskeleton in yeast. Genes Genet Syst. 2005;80:325–343. doi: 10.1266/ggs.80.325. [DOI] [PubMed] [Google Scholar]
- Baskaran S, Ragusa MJ, Boura E, Hurley JH. Two-site recognition of phosphatidylinositol 3-phosphate by PROPPINs in autophagy. Mol Cell. 2012;47:339–348. doi: 10.1016/j.molcel.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram PG, Choi JH, Carvalho J, Ai W, Zeng C, Chan TF, Zheng XF. Tripartite regulation of Gln3p by TOR, Ure2p, and phosphatases. J Biol Chem. 2000;275:35727–35733. doi: 10.1074/jbc.M004235200. [DOI] [PubMed] [Google Scholar]
- Binda M, Peli-Gulli MP, Bonfils G, Panchaud N, Urban J, Sturgill TW, Loewith R, De Virgilio C. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol Cell. 2009;35:563–573. doi: 10.1016/j.molcel.2009.06.033. [DOI] [PubMed] [Google Scholar]
- Bonangelino CJ, Catlett NL, Weisman LS. Vac7p, a novel vacuolar protein, is required for normal vacuole inheritance and morphology. Mol Cell Biol. 1997;17:6847–6858. doi: 10.1128/mcb.17.12.6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonangelino CJ, Nau JJ, Duex JE, Brinkman M, Wurmser AE, Gary JD, Emr SD, Weisman LS. Osmotic stress-induced increase of phosphatidylinositol 3,5-bisphosphate requires Vac14p, an activator of the lipid kinase Fab1p. J Cell Biol. 2002;156:1015–1028. doi: 10.1083/jcb.200201002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botelho RJ, Efe JA, Teis D, Emr SD. Assembly of a Fab1 phosphoinositide kinase signaling complex requires the Fig4 phosphoinositide phosphatase. Mol Biol Cell. 2008;19:4273–4286. doi: 10.1091/mbc.E08-04-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges D, Ma JT, Park S, Inoki K, Weisman LS, Saltiel AR. Phosphatidylinositol 3,5-bisphosphate plays a role in the activation and subcellular localization of mechanistic target of rapamycin 1. Mol Biol Cell. 2012;23:2955–2962. doi: 10.1091/mbc.E11-12-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunn GJ, Hudson CC, Sekulic A, Williams JM, Hosoi H, Houghton PJ, Lawrence JC, Jr, Abraham RT. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci USA. 1998;95:1432–1437. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campeau PM, et al. Yunis-Varon syndrome is caused by mutations in FIG4, encoding a phosphoinositide phosphatase. Am J Hum Genet. 2013;92:781–791. doi: 10.1016/j.ajhg.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H, Nair U, Geng J, Klionsky DJ. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19:668–681. doi: 10.1091/mbc.E07-08-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CY, et al. Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature. 2007;448:68–72. doi: 10.1038/nature05876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CY, et al. Deleterious variants of FIG4, a phosphoinositide phosphatase, in patients with ALS. Am J Hum Genet. 2009;84:85–88. doi: 10.1016/j.ajhg.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lartigue J, Polson H, Feldman M, Shokat K, Tooze SA, Urbe S, Clague MJ. PIKfyve regulation of endosome-linked pathways. Traffic. 2009;10:883–893. doi: 10.1111/j.1600-0854.2009.00915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove SK, et al. Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. EMBO J. 2004;23:1922–1933. doi: 10.1038/sj.emboj.7600203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove SK, Dong K, Kobayashi T, Williams FK, Michell RH. Phosphatidylinositol 3,5-bisphosphate and Fab1p/PIKfyve underPPIn endo-lysosome function. Biochem J. 2009;419:1–13. doi: 10.1042/BJ20081950. [DOI] [PubMed] [Google Scholar]
- Dove SK, McEwen RK, Mayes A, Hughes DC, Beggs JD, Michell RH. Vac14 controls PtdIns(3,5)P(2) synthesis and Fab1-dependent protein trafficking to the multivesicular body. Curr Biol. 2002;12:885–893. doi: 10.1016/s0960-9822(02)00891-6. [DOI] [PubMed] [Google Scholar]
- Dubouloz F, Deloche O, Wanke V, Cameroni E, De Virgilio C. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol Cell. 2005;19:15–26. doi: 10.1016/j.molcel.2005.05.020. [DOI] [PubMed] [Google Scholar]
- Duex JE, Nau JJ, Kauffman EJ, Weisman LS. Phosphoinositide 5-phosphatase Fig 4p is required for both acute rise and subsequent fall in stress-induced phosphatidylinositol 3,5-bisphosphate levels. Eukaryotic Cell. 2006a;5:723–731. doi: 10.1128/EC.5.4.723-731.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duex JE, Tang F, Weisman LS. The Vac14p-Fig4p complex acts independently of Vac7p and couples PI(3,5)P2 synthesis and turnover. J Cell Biol. 2006b;172:693–704. doi: 10.1083/jcb.200512105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efe JA, Botelho RJ, Emr SD. Atg18 regulates organelle morphology and Fab1 kinase activity independent of its membrane recruitment by phosphatidylinositol 3,5-bisphosphate. Mol Biol Cell. 2007;18:4232–4244. doi: 10.1091/mbc.E07-04-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeyan A, Zoncu R, Chang S, Gumper I, Snitkin H, Wolfson RL, Kirak O, Sabatini DD, Sabatini DM. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature. 2013;493:679–683. doi: 10.1038/nature11745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson CJ, Lenk GM, Meisler MH. Defective autophagy in neurons and astrocytes from mice deficient in PI(3,5)P2. Hum Mol Genet. 2009;18:4868–4878. doi: 10.1093/hmg/ddp460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Kaiser CA. A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat Cell Biol. 2006;8:657–667. doi: 10.1038/ncb1419. [DOI] [PubMed] [Google Scholar]
- Gary JD, Sato TK, Stefan CJ, Bonangelino CJ, Weisman LS, Emr SD. Regulation of Fab1 phosphatidylinositol 3-phosphate 5-kinase pathway by Vac7 protein and Fig4, a polyphosphoinositide phosphatase family member. Mol Biol Cell. 2002;13:1238–1251. doi: 10.1091/mbc.01-10-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary JD, Wurmser AE, Bonangelino CJ, Weisman LS, Emr SD. Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J Cell Biol. 1998;143:65–79. doi: 10.1083/jcb.143.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- Hara K, Yonezawa K, Kozlowski MT, Sugimoto T, Andrabi K, Weng QP, Kasuga M, Nishimoto I, Avruch J. Regulation of eIF-4E BP1 phosphorylation by mTOR. J Biol Chem. 1997;272:26457–26463. doi: 10.1074/jbc.272.42.26457. [DOI] [PubMed] [Google Scholar]
- Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W-P, Scott SV, Kim J, Klionsky DJ. The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J Biol Chem. 2000;275:5845–5851. doi: 10.1074/jbc.275.8.5845. [DOI] [PubMed] [Google Scholar]
- Isotani S, Hara K, Tokunaga C, Inoue H, Avruch J, Yonezawa K. Immunopurified mammalian target of rapamycin phosphorylates and activates p70 S6 kinase alpha in vitro. J Biol Chem. 1999;274:34493–34498. doi: 10.1074/jbc.274.48.34493. [DOI] [PubMed] [Google Scholar]
- Jefferies HB, et al. A selective PIKfyve inhibitor blocks PtdIns(3,5)P(2) production and disrupts endomembrane transport and retroviral budding. EMBO Rep. 2008;9:164–170. doi: 10.1038/sj.embor.7401155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin N, et al. VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P(2) in yeast and mouse. EMBO J. 2008;27:3221–3234. doi: 10.1038/emboj.2008.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K, Ohsumi Y. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol. 2010;30:1049–1058. doi: 10.1128/MCB.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- Kim E, Guan K-L. RAG GTPases in nutrient-mediated TOR signaling pathway. Cell Cycle. 2009;8:1014–1018. doi: 10.4161/cc.8.7.8124. [DOI] [PubMed] [Google Scholar]
- Kim J, Huang W-P, Klionsky DJ. Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J Cell Biol. 2001;152:51–64. doi: 10.1083/jcb.152.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogan K, Spear ED, Kaiser CA, Fass D. Structural conservation of components in the amino acid sensing branch of the TOR pathway in yeast and mammals. J Mol Biol. 2010;402:388–398. doi: 10.1016/j.jmb.2010.07.034. [DOI] [PubMed] [Google Scholar]
- Krick R, Busse RA, Scacioc A, Stephan M, Janshoff A, Thumm M, Kuhnel K. Structural and functional characterization of the two phosphoinositide binding sites of PROPPINs, a beta-propeller protein family. Proc Natl Acad Sci USA. 2012;109:E2042–E2049. doi: 10.1073/pnas.1205128109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Lempiainen H, Uotila A, Urban J, Dohnal I, Ammerer G, Loewith R, Shore D. Sfp1 interaction with TORC1 and Mrs6 reveals feedback regulation on TOR signaling. Mol Cell. 2009;33:704–716. doi: 10.1016/j.molcel.2009.01.034. [DOI] [PubMed] [Google Scholar]
- Lin CH, MacGurn JA, Chu T, Stefan CJ, Emr SD. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell. 2008;135:714–725. doi: 10.1016/j.cell.2008.09.025. [DOI] [PubMed] [Google Scholar]
- Loewith R, Hall MN. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics. 2011;189:1177–1201. doi: 10.1534/genetics.111.133363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- MacGurn JA, Hsu PC, Smolka MB, Emr SD. TORC1 regulates endocytosis via Npr1-mediated phosphoinhibition of a ubiquitin ligase adaptor. Cell. 2012;147:1104–1117. doi: 10.1016/j.cell.2011.09.054. [DOI] [PubMed] [Google Scholar]
- Martin S, Harper CB, May LM, Coulson EJ, Meunier FA, Osborne SL. Inhibition of PIKfyve by YM-201636 dysregulates autophagy and leads to apoptosis-independent neuronal cell death. PLoS One. 2013;8:e60152. doi: 10.1371/journal.pone.0060152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, Puertollano R. Rag GTPases mediate amino acid-dependent recruitment of TFEB and MITF to lysosomes. J Cell Biol. 2013;200:475–491. doi: 10.1083/jcb.201209135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney AJ, Zhang Y, Weisman LS. Phosphatidylinositol 3,5-bisphosphate: Low abundance, high significance. BioEssays. 2014;36:52–64. doi: 10.1002/bies.201300012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenguy F, Colin D, ten Have JP. Regulation of compartmentation of amino acid pools in Saccharomyces cerevisiae and its effects on metabolic control. Eur J Biochem. 1980;108:439–447. doi: 10.1111/j.1432-1033.1980.tb04740.x. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- Nair U, Cao Y, Xie Z, Klionsky DJ. Roles of the lipid-binding motifs of Atg18 and Atg21 in the cytoplasm to vacuole targeting pathway and autophagy. J Biol Chem. 2010;285:11476–11488. doi: 10.1074/jbc.M109.080374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Matsuura A, Wada Y, Ohsumi Y. Acidification of vacuoles is required for autophagic degradation in the yeast, Saccharomyces cerevisiae. J Biochem. 1997;121:338–344. doi: 10.1093/oxfordjournals.jbchem.a021592. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- Narayan K, Lemmon MA. Determining selectivity of phosphoinositide-binding domains. Methods. 2006;39:122–133. doi: 10.1016/j.ymeth.2006.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicot AS, Fares H, Payrastre B, Chisholm AD, Labouesse M, Laporte J. The phosphoinositide kinase PIKfyve/Fab1p regulates terminal lysosome maturation in Caenorhabditis elegans. Mol Biol Cell. 2006;17:3062–3074. doi: 10.1091/mbc.E05-12-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima H, Tokunaga C, Eguchi S, Oshiro N, Hidayat S, Yoshino K, Hara K, Tanaka N, Avruch J, Yonezawa K. The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J Biol Chem. 2003;278:15461–15464. doi: 10.1074/jbc.C200665200. [DOI] [PubMed] [Google Scholar]
- Obara K, Sekito T, Niimi K, Ohsumi Y. The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. J Biol Chem. 2008;283:23972–23980. doi: 10.1074/jbc.M803180200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena-Llopis S, Brugarolas J. TFEB, a novel mTORC1 effector implicated in lysosome biogenesis, endocytosis and autophagy. Cell Cycle. 2011;10:3987–3988. doi: 10.4161/cc.10.23.18251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson TR, et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke A, Anderson S, McCaffery JM, Yates J, 3rd, Aronova S, Chu S, Fairclough S, Iverson C, Wedaman KP, Powers T. TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J Biol Chem. 2004;279:14752–14762. doi: 10.1074/jbc.M313062200. [DOI] [PubMed] [Google Scholar]
- Rudge SA, Anderson DM, Emr SD. Vacuole size control: regulation of PtdIns(3,5)P2 levels by the vacuole-associated Vac14-Fig4 complex, a PtdIns(3,5)P2-specific phosphatase. Mol Biol Cell. 2004;15:24–36. doi: 10.1091/mbc.E03-05-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusten TE, et al. ESCRTs and Fab1 regulate distinct steps of autophagy. Curr Biol. 2007;17:1817–1825. doi: 10.1016/j.cub.2007.09.032. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbrissa D, Ikonomov OC, Fenner H, Shisheva A. ArPIKfyve homomeric and heteromeric interactions scaffold PIKfyve and Sac3 in a complex to promote PIKfyve activity and functionality. J Mol Biol. 2008;384:766–779. doi: 10.1016/j.jmb.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Beck T, Koller A, Kunz J, Hall MN. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 1998;17:6924–6931. doi: 10.1093/emboj/17.23.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill TW, Cohen A, Diefenbacher M, Trautwein M, Martin DE, Hall MN. TOR1 and TOR2 have distinct locations in live cells. Eukaryotic Cell. 2008;7:1819–1830. doi: 10.1128/EC.00088-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban J, et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell. 2007;26:663–674. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YX, Catlett NL, Weisman LS. Vac8p, a vacuolar protein with armadillo repeats, functions in both vacuole inheritance and protein targeting from the cytoplasm to vacuole. J Cell Biol. 1998;140:1063–1074. doi: 10.1083/jcb.140.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Kobayashi T, Yamamoto H, Hoshida H, Akada R, Inagaki F, Ohsumi Y, Noda NN. Structure-based analyses reveal distinct binding sites for Atg2 and phosphoinositides in Atg18. J Biol Chem. 2012;287:31681–31690. doi: 10.1074/jbc.M112.397570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip CK, Murata K, Walz T, Sabatini DM, Kang SA. Structure of the human mTOR complex I and its implications for rapamycin inhibition. Mol Cell. 2010;38:768–774. doi: 10.1016/j.molcel.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Zaman S, Broach JR, Klionsky DJ. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:4180–4189. doi: 10.1091/mbc.E07-05-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, et al. Loss of Vac14, a regulator of the signaling lipid phosphatidylinositol 3,5-bisphosphate, results in neurodegeneration in mice. Proc Natl Acad Sci USA. 2007;104:17518–17523. doi: 10.1073/pnas.0702275104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- Zolov SN, et al. In vivo, PIKfyve generates PI(3,5)P2, which serves as both a signaling lipid and the major precursor for PI5P. Proc Natl Acad Sci USA. 2012;109:17472–17477. doi: 10.1073/pnas.1203106109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurita-Martinez SA, Puria R, Pan X, Boeke JD, Cardenas ME. Efficient Tor signaling requires a functional class C Vps protein complex in Saccharomyces cerevisiae. Genetics. 2007;176:2139–2150. doi: 10.1534/genetics.107.072835. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.