The yeast cytochrome oxidase Cox3p assembly module is shown to consist of Cox3p, Cox4p, Cox7p, Cox13p, and accessory factor Rcf1p. The results support an assembly model in which three modules, each containing one of the three core subunits and a unique subset of nuclear-derived subunits, interact to form the holoenzyme.
Abstract
Yeast cytochrome oxidase (COX) was previously inferred to assemble from three modules, each containing one of the three mitochondrially encoded subunits and a different subset of the eight nuclear gene products that make up this respiratory complex. Pull-down assays of pulse-labeled mitochondria enabled us to characterize Cox3p subassemblies that behave as COX precursors and contain Cox4p, Cox7p, and Cox13p. Surprisingly, Cox4p is a constituent of two other complexes, one of which was previously proposed to be an intermediate of Cox1p biogenesis. This suggests that Cox4p, which contacts Cox1p and Cox3p in the holoenzyme, can be incorporated into COX by two alternative pathways. In addition to subunits of COX, some Cox3p intermediates contain Rcf1p, a protein associated with the supercomplex that stabilizes the interaction of COX with the bc1 (ubiquinol-cytochrome c reductase) complex. Finally, our results indicate that although assembly of the Cox1p module is not contingent on the presence of Cox3p, the converse is not true, as none of the Cox3p subassemblies were detected in a mutant blocked in translation of Cox1p. These studies support our proposal that Cox3p and Cox1p are separate assembly modules with unique compositions of ancillary factors and subunits derived from the nuclear genome.
INTRODUCTION
Yeast cytochrome oxidase (COX) is a genetic hybrid consisting of a three-subunit catalytic core encoded by mitochondrial genes and eight other subunits that are all products of nuclear genes. The balanced output of COX proteins expressed in two separate subcellular compartments is achieved through regulation of mitochondrial translation of Cox1p, one of the core subunits (Perez-Martinez et al., 2003, 2009; Barrientos et al., 2004; Pierrel et al., 2007; Mick et al., 2010; Fontanesi et al., 2010, 2011). Mss51p, a translation activator of the COX1 mRNA, plays a central role in this mechanism by virtue of its ability to bind to the product Cox1p (Perez-Martinez et al., 2003). When bound to Cox1p, Mss51p is prevented from exercising its function in translation until it is released at a downstream assembly step involving the dissolution of the Cox1p-Mss51p complex (Barrientos et al., 2004; Mick et al., 2011). This complex has been shown to also contain subunits Cox5ap, Cox6p, and Cox8p of COX (Mick et al., 2007; Fontanesi et al., 2010; McStay et al., 2013c) and the accessory factors Cox14p, Coa3p, and Coa1p that are believed to stabilize the complex (Barrientos et al., 2004; Pierrel et al., 2007; Mick et al., 2010; Fontanesi et al., 2011) and prevent Cox1p from forming unproductive aggregates (McStay et al., 2013b). We recently reported the existence in yeast mitochondria of five Cox1p complexes, designated as D1–D5, that differ in their compositions of COX subunits and accessory factors and in pulse-chase experiments behave as precursors of COX (McStay et al., 2013c). The finding that none of the intermediates contained the core subunits Cox2p and Cox3p led us to propose that COX is assembled from three different modules, each consisting of a mitochondrially translated core subunit and a unique set of nuclear-encoded subunits (McStay et al., 2013c).
The present study was undertaken to further verify the modular model of COX assembly. Using the same approach that enabled us to identify Cox1p intermediates, we present evidence that Cox3p transitions through a series of intermediates that contain a subset of nuclear gene products distinct from those present in the Cox1p module. The subunits associated with the Cox3p module make direct contact with Cox3p in the holoenzyme, lending support to the notion that the primary interactions between the core subunits and their immediate partners are established before formation of the enzyme core.
RESULTS
Expression of tagged Cox3p
The mitochondrial COX3 gene was modified at the 3′ and separately at the 5′ end to express Cox3p with a C-terminal polyhistidine tag or an N-terminal double tag consisting of the protein C and the hemagglutinin (HA) epitopes (HAC). The hybrid genes were introduced by recombination into a mutant in which COX3 had been replaced with ARG8m (Steele et al., 1996). Growth tests indicated that whereas the strain with the C-terminal polyhistidine tag on Cox3p grew as well as wild type on rich glycerol/ethanol, the N-terminal HAC tag on Cox3p elicited a partial growth defect (Supplemental Figure S1A). The slower growth was paralleled by approximately twofold reduction in mitochondrial cytochrome oxidase, as evidenced by the spectrum of cytochromes a and a3 (Supplemental Figure S1B) and the reduced level of Cox3p-HAC in mitochondria (Supplemental Figure S1C). Cox3p with either the C-terminal polyhistidine or N-terminal HAC tag could be purified on nickel-nitriloacetic acid (unpublished data) and protein C antibody beads, respectively (Supplemental Figure S1D). Cox3p obtained after solubilization of mitochondria in lauryl maltoside and fractionation of the extract on protein C antibody beads copurified with the other subunits of COX, but this preparation was less pure (Supplemental Figure S1E) than the one obtained by the same procedure from mitochondria expressing Cox1p-HAC (McStay et al., 2013c).
Notwithstanding the lower efficiency of COX assembly or instability of Cox3p with the N-terminal HAC tag, this strain proved to be experimentally more suitable for the present studies because of the low background of nonspecific Cox3p adsorption to protein C antibody beads.
Detection of Cox3p intermediates in pulse- and pulse-chase-labeled mitochondria
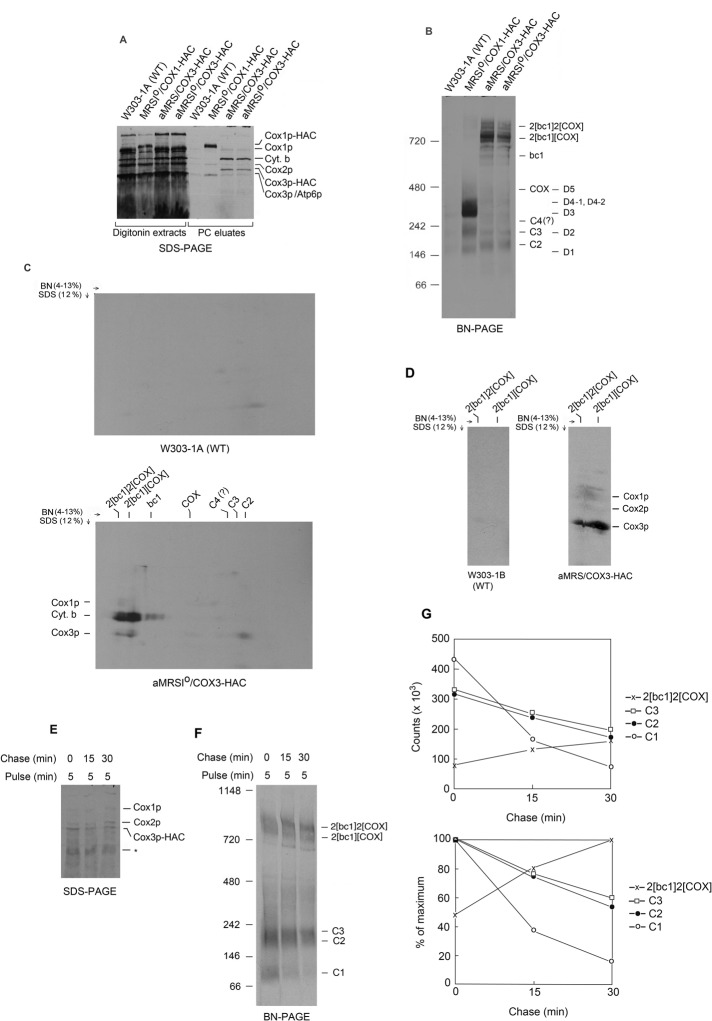
To identify Cox3p assembly intermediates, we labeled mitochondria of MRS/COX3-HAC and MRSIo/COX3-HAC, each containing the COX3-HAC fusion gene, in vitro with a mixture of [35S]methionine/cysteine. Both strains had been allowed to incubate for 2 h in chloramphenicol to increase mitochondrial pools of nuclear-encoded COX subunits. In subsequent experiments we found that the chloramphenicol incubation did not appreciably enhance either labeling or incorporation of Cox3p into COX, and we used mitochondria from cells that had been grown in rich galactose medium without the chloramphenicol incubation. Mitochondria from the haploid strain W303-1A containing a wild-type mitochondrial genome and from MRSIo/COX1-HAC expressing Cox1p-HAC were also labeled as controls. Mitochondria were extracted with digitonin and Cox3p-HAC or Cox1p-HAC purified on protein C antibody beads. The digitonin extracts and the eluates from the antibody beads were separated by SDS–PAGE and blue native (BN)-PAGE. In agreement with earlier studies, separation of the eluate from the control strain MRSIo/COX1-HAC by SDS–PAGE showed enrichment of radiolabeled Cox1p-HAC, Cox2p, and Cox3p (Figure 1A) and of the five previously reported Cox1p intermediates, D1–D5 (Figure 1B). The antibody eluates from MRSIo/COX3-HAC and MRS/COX3-HAC, each expressing Cox3p-HAC, were enriched for Cox1p and Cox3p-HAC, which migrated more slowly than the wild-type protein in SDS–PAGE due to the presence of the tag. Although a faint Cox2p band was also present in this fraction, it was comparable to the nonspecific background binding seen in the eluate of W303-1A. The presence of a strong radiolabeled band at the position of cytochrome b indicated that some of this newly translated protein had been incorporated into the supercomplexes and was coimmunoadsorbed with Cox3p-HAC on the antibody beads. In Saccharomyces cerevisiae the major supercomplex (respirasome) consists of the bc1 complex (ubiquinol-cytochrome c reductase) and COX in a 2:2 ratio (Heinemeyer et al., 2007; Mileykovskaya et al., 2012). A second, less abundant supercomplex is also observed on native gels with a 2:1 ratio of bc1 to COX. The incorporation of cytochrome b into the supercomplexes in mitochondria of cells that are incubated in the presence of chloramphenicol was also reported in coimmunoprecipitates of strains expressing Cox1p-HAC (McStay et al., 2013c).
FIGURE 1:
Pull-down assays of Cox3p-HAC. (A) W303-1A (WT), MRSIo/COX1-HAC, and MRSIo/COX3-HAC (intronless mtDNA, expressing tagged Cox1p and Cox3p, respectively), and MRS/COX3-HAC (normal mtDNA and tagged Cox3p) were grown in YPGal to early stationary phase. All of the strains except MRSIo/COX1-HAC were allowed to incubate for an additional 2 h in fresh YPGal containing 2 mg/ml chloramphenicol before preparation of mitochondria. Mitochondria (250 μg protein) were labeled with [35S]methionine/cysteine for 20 min as described in Materials and Methods. Digitonin extracts (8% of total) and eluates (17% of total) from the protein C antibody beads were separated by SDS–PAGE on a 17% polyacrylamide gel. Proteins were transferred to nitrocellulose and exposed to x-ray film. (B) The eluates (36% of total) obtained in A from the protein C antibody beads were separated by BN-PAGE on a 4–13% polyacrylamide gel. C2–C4 refers to complexes containing Cox3p-HAC. D1–D5 correspond to previously identified Cox1p intermediates. Proteins were transferred to a polyvinylidene fluoride (PVDF) membrane and exposed to x-ray film. (C) The eluates (36% of total sample) obtained in A from the protein C antibody beads were separated in the first dimension by BN-PAGE on a 4–13% polyacrylamide gel and in the second dimension by SDS–PAGE on a 12% polyacrylamide gel. Proteins were transferred to nitrocellulose and exposed to x-ray film. (D) The wild-type strain W303-1B and aMRS/COX3-HAC were grown to late log phase in YPGal without additional growth in chloramphenicol. Mitochondria were labeled as in A and the eluates from the protein C antibody beads separated in two dimensions as in C. Proteins were transferred to a nitrocellulose membrane and exposed to x-ray film for 4 d. Only the regions with the supercomplexes are shown. The identity of the band above Cox1p in the panel with aMRS/COX3-HAC was not determined. (E, F) Mitochondria (4 mg) from aMRS/COX3-HAC grown in YPGal without subsequent growth in chloramphenicol medium were labeled in a final volume of 1.25 ml with [35S]methionine/cysteine for 5 min and translation terminated by addition of 24 μg of puromycin and 0.24 μg of methionine plus cysteine. Samples representing 33% of the starting translation mixture were withdrawn at 0, 15, and 30 min after addition of puromycin and cold amino acids. The samples were extracted with digitonin and adsorbed to protein C antibody beads as in A, except that the volumes were increased proportionately to the volume of the extract. The eluates from the antibody beads were separated by SDS–PAGE on a 17% polyacrylamide gel (20% of total) and by BN-PAGE on a 4–13% polyacrylamide gel and radiolabeled protein as in A and B. (G) The C1–C3 and 2[bc1]2[COX] supercomplex bands in E were quantified with a phosphorimager. The radioactivity is expressed as total counts per minute (top) and percentage of the 0- or 30-min chase (bottom).
Cox3p-HAC eluted from the protein C antibody beads contained two radiolabeled bands of ∼200 and 230 kDa referred to as C2 and C3, respectively. A less-well-labeled third band of 300 kDa, C4, was also observed in some experiments. In addition, the mitochondria incorporated the radiolabel into COX and the supercomplexes (Figure 1B). The radioactive band between C4 and COX, also seen in the W303-1A control, is probably nonspecifically adsorbed Cox1p intermediate D3, which is much more abundant and highly labeled than any of the Cox3p complexes. The smaller supercomplex with only one COX was more heavily labeled in mitochondria with Cox3p-HAC, probably as a result of an insufficiency of COX in this strain (Figure 1B). This is also supported by the presence of free bc1 complex in the fraction eluted from the beads. The two-dimensional (2D) gel of the eluate with Cox3p-HAC confirmed that most of the radiolabeled Cox3p-HAC was present in C2 and the supercomplexes, but some also migrated in the regions corresponding to C3, C4, and COX. Although newly translated Cox1p and Cox3p were detected in the supercomplexes, most of the radioactivity in this region was contributed by cytochrome b (Figure 1C). To reduce the amount of labeled cytochrome b, which interferes with detection of Cox2p, we isolated mitochondria containing Cox3p-HAC from a strain that was grown without the additional chloramphenicol treatment. SDS–PAGE separation of the fraction eluted from the antibody beads indicated only background levels of cytochrome b, comparable to that seen in the wild-type control (unpublished data). The 2D gel of this fraction confirmed the presence of all three newly translated COX subunits in the supercomplexes. In agreement with previous results with mitochondria expressing Cox1p-HAC (McStay et al., 2013c), Cox3p was the most highly labeled COX subunit in the supercomplexes (Figure 1D).
The precursor–product relationship of C2–C4 to COX was examined by pulse-chase labeling of mitochondria. Because of the very limited incorporation of the radioactive precursors at shorter pulse times, mitochondria were labeled for 5 min followed by 15 and 30 min of chase in the presence of puromycin and excess unlabeled methionine plus cysteine. The eluate from the 5-min pulse revealed, in addition to C2 and C3, a new prominent radiolabeled product (C1) of ∼100 kDa that was only seen as a very faint band with longer pulse times. The chase resulted in reduction of C1–C3 and increase in radiolabel in the two supercomplexes (Figure 1, E– G).
Based on the radioactivity of the bands seen on native gels, the intermediates with Cox1p-HAC are much more abundant than those with Cox3p-HAC (Figures 1B and 2A). The higher steady-state pools of Cox1p intermediates, particularly D3 and D4, act to dilute newly translated and radiolabeled Cox1p. This accounts for what appears to be a less efficient incorporation of Cox1p in COX (supercomplexes) compared with Cox3p (see also McStay et al., 2013c). Our evidence indicates that the steady-state concentrations of Cox2p intermediates are also considerable higher than those of Cox3p, which can also explain the less than stoichiometric incorporation of this subunit. It is significant that pull-down assays with HAC tagged Cox2p show a more stoichiometric coenrichment of Cox1p, Cox2p, and Cox3p in the supercomplexes (Supplemental Figure S2).
FIGURE 2:
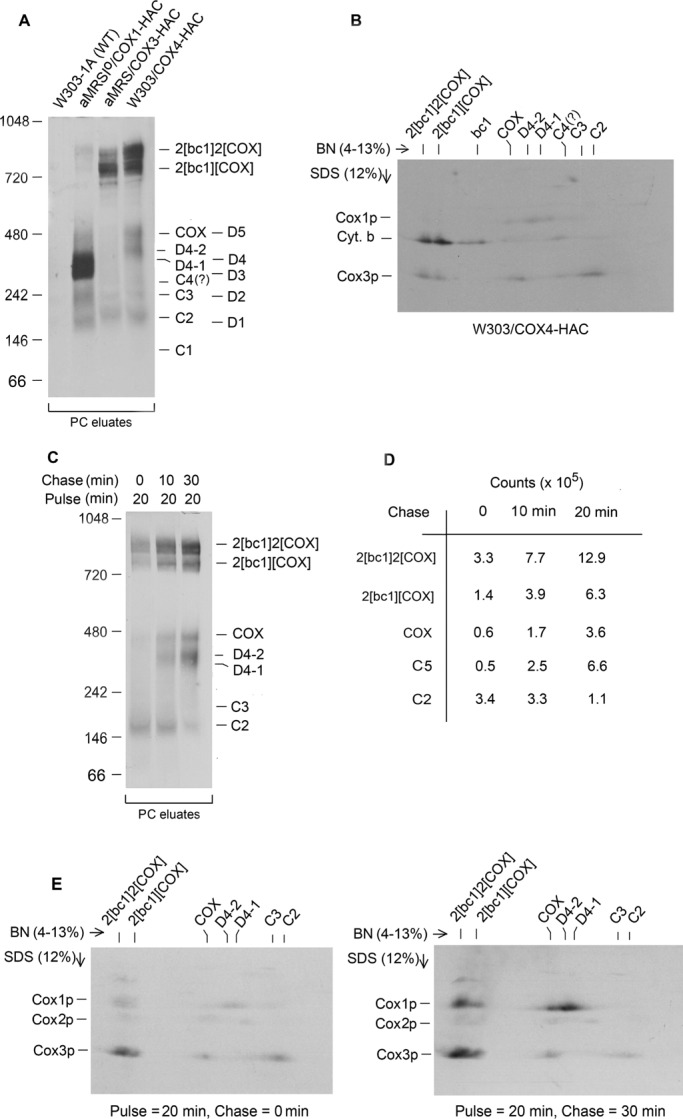
Cox4p is associated with Cox3p in C1-C4. (A) W303-1A (wild type) and aMRSIo/COX1-HAC (intronless DNA, expressing Cox1p-HAC) were grown in YPGal without additional growth in chloramphenicol. Both aMRS/COX3-HAC (Cox3p-HAC expresser) and W303/COX4-HAC (Cox4p-HAC expresser) were grown in YPGal with additional 2 h of growth in chloramphenicol medium. Mitochondria (250 μg of protein) from each strain were labeled, extracted, and reacted with protein C antibody beads as in Figure 1A. Half of the eluate from the antibody beads was separated by BN-PAGE on a 4–13% polyacrylamide gel, transferred to a PVDF membrane, and exposed to x-ray film. (B) Half of the eluate obtained from W303/COX4-HAC was separated by BN-PAGE in the first dimension and by SDS–PAGE on a 12% polyacrylamide gel in the second dimension. Proteins were transferred to nitrocellulose and exposed to x-ray film. (C) W303/COX4-HAC was grown in YPGal without further chloramphenicol treatment. Mitochondria (750 μg of protein) were labeled for 20 min as in Figure 1A and chased for the indicated times after addition of puromycin and cold amino acids. The samples were extracted with digitonin and Cox4p-containing complexes enriched on protein C antibody beads as in Figure 1A. The eluates from the beads were separated by BN-PAGE on a 4–13% polyacrylamide gel, transferred to a PVDF membrane, and exposed to x-ray film. (D) The indicated bands in the pulse-chase experiment of C were quantified with a phosphorimager. (E) Mitochondria of W303/COX4-HAC, were labeled by pulse chase as in D, extracted with digitonin, and purified on protein C antibody beads. The eluates of the samples from the 0- and 30-min chase were separated by BN-PAGE on a 4–13% polyacrylamide gel in the first dimension and by SDS–PAGE on a 12% polyacrylamide gel in the second. Proteins were transferred to nitrocellulose and exposed to x-ray film.
Cox4p association with the Cox3p module
Cox4p, one of the nuclear-encoded subunits of COX, interacts with both Cox1p and Cox3p in the holoenzyme (Tsukihara et al., 1996). Mitochondria from a strain expressing Cox4p-HAC and Cox1p-HAC or Cox3p-HAC as control strains were labeled for 20 min. The antibody eluates purified from mitochondria with either Cox3p-HAC or Cox4p-HAC displayed the same pattern of radiolabeled C2-C4, COX, and supercomplexes (Figure 2A). However, there was substantially more COX in the eluate with Cox4p-HAC, which also had a new, broad band that migrated in the region of the Cox1p intermediate D4 (Figure 2A). The 2D gel of the antibody eluate with Cox4p-HAC confirmed the presence of Cox3p in C2, C4, COX and the supercomplexes but not in the novel band, which was resolved into two bands (D4-1 and D4-2), both of which contained newly translated Cox1p (Figure 2B).
Lauryl maltoside is known to dissociate the two supercomplexes into their component bc1 and COX complexes. Treatment of the eluate with Cox4p-HAC with 1% lauryl maltoside caused most of the radioactivity to collapse into three bands of ∼300, 350, and 150 kDa (Supplemental Figure S3A). The 300- and 350-kDa bands had COX activity and probably consist of the COX monomer complexed to other proteins. The migration of the smaller radiolabeled band differs from both C1 and C2, but because of the presence of lauryl maltoside, the identity of this band with either cannot be deduced.
The precursor–product relationship of the different Cox3p intermediates and COX was studied by pulse-chase labeling of mitochondria expressing Cox4p-HAC. Notwithstanding the relatively long pulse, the chase resulted in a time-dependent loss of radiolabel from C1 and C2 and an increase of label in COX and the two supercomplexes (Figure 2, C and D). Neither C3 nor C4 was sufficiently well labeled and demarcated to be quantified. The eluates obtained after the 0- and 30-min chases were also separated on a 2D gel, which showed loss of radiolabeled Cox3p from C2 and an increase of Cox1p and Cox3p-HAC in COX and the supercomplexes, indicating continued assembly of COX during the 30-min chase (Figure 2E). D4-1 and D4-2 also acquired more newly translated Cox1p but not Cox3p during the chase.
The pulse-chase analysis of the COX4-HAC strain was repeated with a shorter pulse time of 2 min. Under these conditions the C1 band was more prominent and was progressively reduced during the 30-min chase (Figure 3, B and D–F). The chase also led to a reduction in label from C2 and C4 with a concomitant increase in COX and the supercomplexes (Figure 3, B and D–F). It is noteworthy that the short pulse did not elicit incorporation of Cox4p-HAC into either D4-2 or D4-1. The same fractions analyzed by SDS–PAGE revealed that the chase caused an increase of newly translated Cox2p, Cox1p, and cytochrome b and the appearance of a novel band that migrated above Cox1p in the eluates from the PC beads (Figure 3, A and C). The identity of this new band is not known.
FIGURE 3:
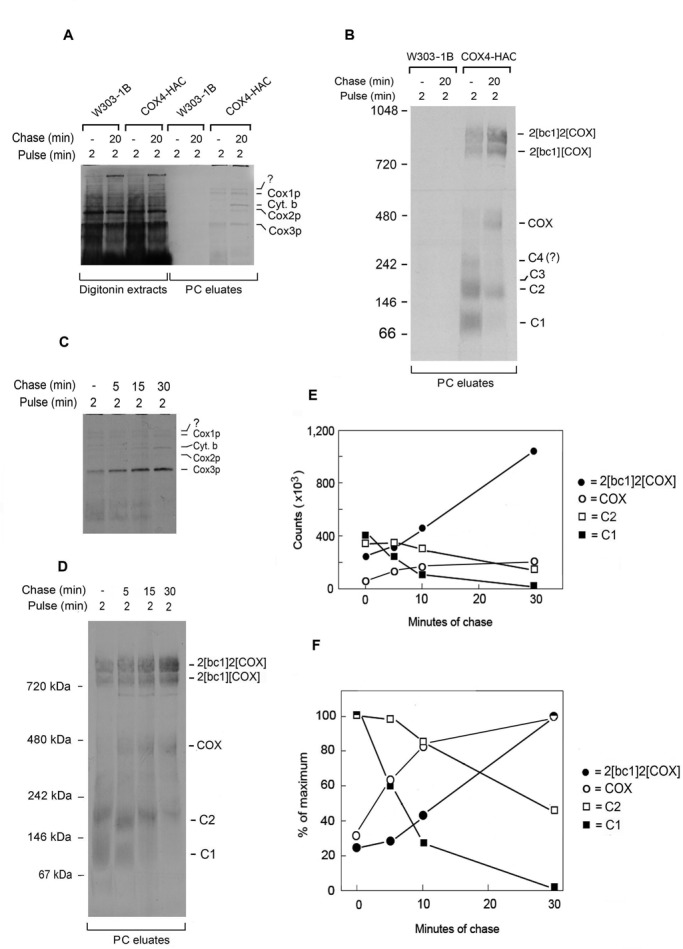
Pulse-chase analysis of Cox4p-HAC incorporation into COX. (A) W303-1B (WT) and W303/COX4-HAC (COX4-HAC) expressing tagged Cox4p were grown to early stationary phase in YPGal with additional 2 h of growth in YPGal containing 2 mg/ml chloramphenicol. Mitochondria (0.5 mg of protein) were labeled for 2 min as in Figure 1A. Half of the translation mixture was extracted with digitonin immediately after addition of puromycin and cold amino acids (0-min chase). The other half was allowed to incubate for an additional 20 min (20-min chase) before extraction with digitonin. The extracts were purified on protein C antibody beads as in Figure 1A, and samples of the extracts (6% of total) and eluates (PC eluates) from the antibody beads (15% of total) were separated by SDS–PAGE on a 17% polyacrylamide gel and radiolabeled proteins visualized as in Figure 1A. The radiolabeled bands in the eluates are identified in the margin. (B) The remainder of the eluates (80% of total) from A was separated by BN-PAGE on a 4–13% polyacrylamide gel and radiolabeled proteins visualized. (C) Mitochondria of W303/COX4-HAC were pulse labeled for 2 min and chased for the indicated times as in A before extraction with digitonin. Digitonin extracts were purified on protein C antibody beads and separated by SDS–PAGE on a 17% polyacrylamide gel. (D) Samples purified on antibody beads were separated by BN-PAGE on 4–13% polyacrylamide gel and radiolabeled proteins visualized. (E, F). The radiolabeled bands in D were quantified in a phosphorimager.
Composition of the Cox3p assembly module
The Cox3p module was also tested for the presence of Cox7p, Cox9p, Cox12p, and Cox13p. Mutations in the genes coding for these COX subunits elicit a range of different growth phenotypes (Patterson and Poyton, 1986; Calder and McEwen, 1991; Taanman and Capaldi, 1993; McStay et al., 2013a, c). The HAC tag at the C-terminus of Cox7p does not affect COX assembly or activity (McStay et al., 2013a). This small subunit, which only contacts Cox3p in the holoenzyme (Tsukihara et al., 1996), was a good candidate to be present in Cox3p intermediates. This was confirmed by the enrichment of Cox3p-HAC in the eluate from the antibody C beads (Figure 4A) and the presence of a radiolabeled band at the position of C2 and of bands more weakly at C1 and C3 (Figure 4B). The 2D gel of the eluate from the antibody beads confirmed the presence of newly translated Cox3p in C2, C3, COX, and the supercomplexes (Figure 4C). The cox7 mutant harboring the COX1-HAC contained the Cox1p intermediates D1–D5 (Figure 4, B and D) and, consistent with previous reports, was defective in assembling COX (Aggeler and Capaldi, 1990; Figure 4E). Even though the cox7-null mutant lacks COX and the supercomplexes, the 2D gel indicated the presence of some radiolabeled Cox3p in the region corresponding to D5, consistent with the presence of Cox3p in this intermediate (Figure 4, B and D).
FIGURE 4:
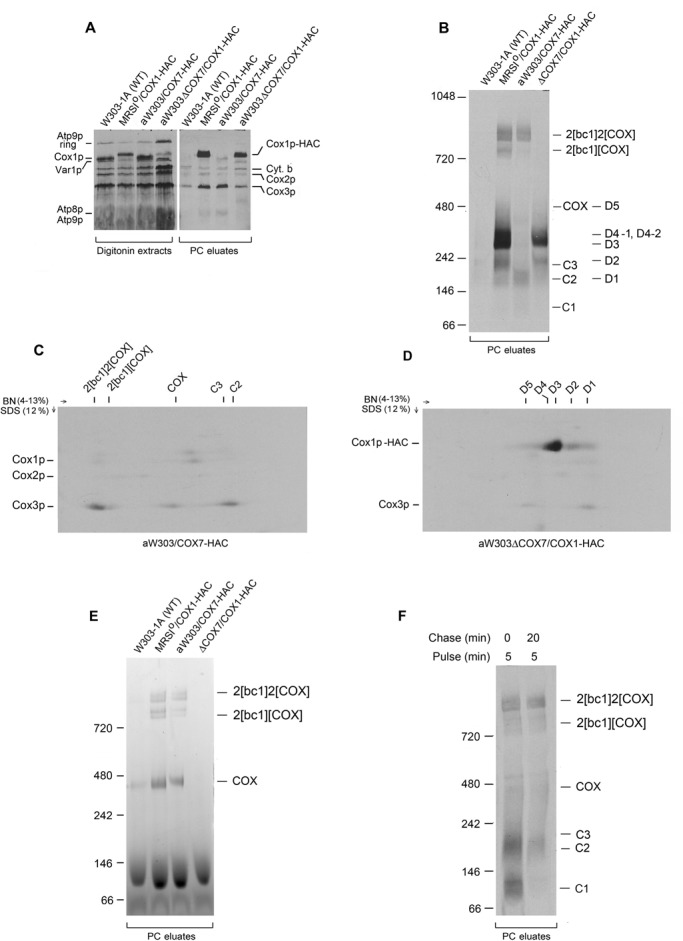
Association of Cox7p with the Cox3p module. W303-1A (WT), MRSIo/COX1-HAC expressing Cox1p with the double HAC tag, W303/COX7-HAC expressing Cox7p with the same tag, and aW303ΔCOX7/COX1-HAC, a respiratory-deficient COX mutant expressing Cox1p with the HAC tag, were grown in YPGal without further growth in chloramphenicol medium. The mitochondria (500 μg of protein) were labeled for 20 min and extracted with digitonin, and the extracts were purified on protein C antibody beads as in Figure 1A. Samples of the digitonin extracts (8% of total) and eluates (17% of total) from the antibody beads (PC eluates) were separated by SDS–PAGE on a 17% polyacrylamide gel (A) and by BN-PAGE on a 4–13% polyacrylamide gel (B) and radiolabeled proteins visualized. (C, D) The eluates from aW303/COX7-HAC and aW303ΔCOX7/COX1-HAC were separated by BN-PAGE on a 4–13% polyacrylamide gel in the first dimension and a 12% polyacrylamide gel in the second dimension. The Cox3p and Cox1p intermediates are identified above each gel and the different COX subunits in the left-hand margins. (E) Mitochondria (500 μg of protein) from the indicated strains were extracted with digitonin, purified on protein C antibody beads, and separated by BN-PAGE on a 4–13% polyacrylamide gel. The gel was stained for COX activity as described previously (McStay et al., 2013c). (F) aW303/COX7-HAC mitochondria were labeled as in Figure 1A for 5 min. Half of the translation mixture (0 time) was extracted with digitonin immediately after addition of puromycin and cold amino acids. The other half was chased for 20 min before extraction. Extracts were purified on protein C antibody beads and separated by BN-PAGE on a 4–13% polyacrylamide gel.
After a 5-min pulse, the eluate from the strain expressing Cox7p-HAC was enriched in radiolabeled C1, C2, and the supercomplexes. C2 separated into two distinct bands, suggesting that this broad band may be a mixture of intermediates with different compositions that are not always resolved by BN-PAGE. The 20-min chase resulted in loss of C1 and C2 but only a small increase in the supercomplexes (Figure 4F). Similar results were obtained when the pulse was increased to 20 min, except that there was much less labeled C1 (Supplemental Figure S4, A and B).
Mitochondria containing C-terminally tagged Cox9p-HAC, Cox12p-HAC, and Cox13p-HAC were analyzed in the same way. Only the eluate obtained from mitochondria with Cox13p-HAC was found to contain radiolabeled C1-C3, COX, and the two supercomplexes. Although the native gel of the eluate with Cox13p-HAC showed the presence of radioactivity in the region of D4-1 and D4-2, it was too low and heterogeneous to conclude that these complexes were present in this strain (Figure 5B). Mitochondria with Cox13p-HAC were pulse labeled for either 5 or 20 min, followed by a chase for up to 30 min. Both visual inspection and quantification of the labeled bands seen with the short pulse time indicated a decrease in C1, C2, and C3 and increase in the larger but not smaller supercomplex (Figure 5, D and E). Like the Cox7p-HAC eluate, the radiolabel in the region of C1 appeared to separate into two different-size bands on the native gel. A decrease of labeled C2 and concomitant increase in the supercomplexes was also observed during a chase of 30 min when mitochondria were pulsed for 20 min (Supplemental Figure S4, B and C).
FIGURE 5:
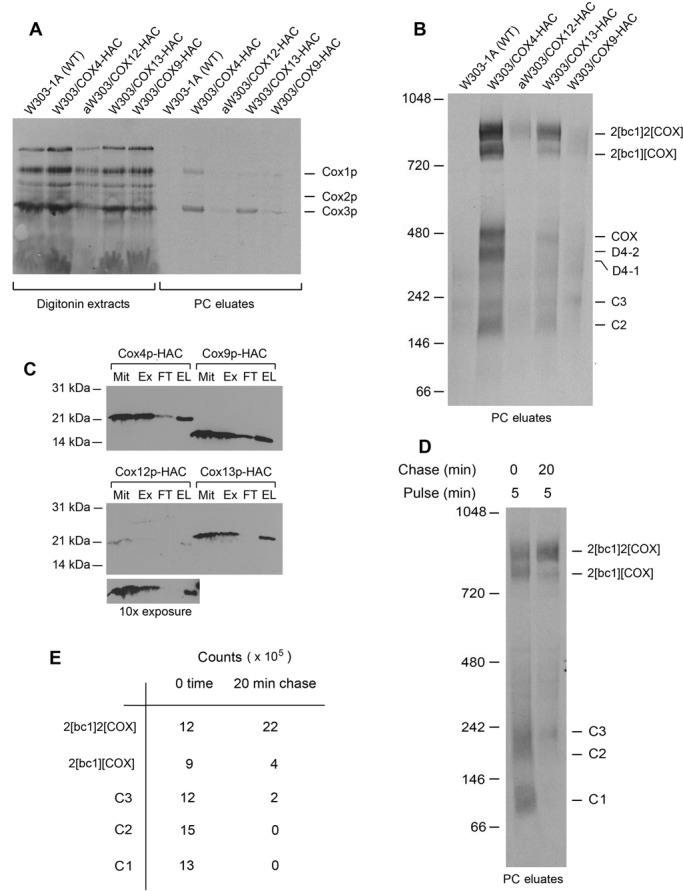
Analysis of Cox9p, Cox12p, and Cox13p in Cox3p intermediates. (A) W303-1A (WT) and strains expressing HAC-tagged Cox4p (W303/COX4-HAC), Cox9p (W303/COX9-HAC), Cox12p (aW303/COX12-HAC), and Cox13p (W303/COX13-HAC) were grown in YPGal to early stationary phase without chloramphenicol. Mitochondria (250 μg of protein) were labeled for 20 min, extracted with digitonin, and purified on protein C antibody beads as in Figure 1A. The digitonin extracts (11% of total) and eluates from the antibody beads (13% of total) were separated by SDS–PAGE on a 17% polyacrylamide gel, transferred to nitrocellulose, and exposed to x-ray film. (B) The PC eluates from A (64% of total) were separated by BN-PAGE on a 4–13% polyacrylamide gel and radiolabeled proteins visualized. (C) Mitochondria with the indicated tagged proteins were extracted with digitonin at a final concentration of 2% and purified on protein C antibody beads. Equivalent samples of (Mit), digitonin extracts (Ex), the protein fraction not adsorbed to the beads (FT), and the eluates from the beads (EL), normalized to 10 μg of the starting mitochondrial protein, were separated by SDS–PAGE on a 15% polyacrylamide gel, transferred to nitrocellulose, and probed with a primary rabbit antibody against the protein C tag (GenScript, Piscataway, NJ). The bottom-most panel with the immunoblot of the Cox12p-HAC samples was exposed for 10 times longer than those shown in the top two panels. (D) W303/COX13-HAC mitochondria were labeled for 5 min as in Figure 1A. Translation was terminated by addition of puromycin and cold amino acids, and a sample representing half of the mixture was immediately removed and extracted with digitonin (0-min chase). The remainder of the translation mixture was incubated for an additional 20 min before extraction with digitonin (20-min chase). The extracts were purified on protein C antibody beads and separated by BN-PAGE on a 4–13% polyacrylamide gel, and radiolabeled proteins were visualized. (E) The radiolabeled bands identified in the native gel of C were quantified with a phosphorimager.
The poor recovery of Cox9p-HAC and Cox12p-HAC from the antibody beads did not permit these subunits to be assigned to or excluded from the Cox3p module (Figure 5, A and B). Unlike the other COX subunits examined in this study, Cox9p was less well adsorbed on the protein C antibody beads (Figure 5C). This, combined with the reduced amount of the 2[bc1]2[COX] supercomplex in mitochondria with Cox9p-HAC (McStay et al., 2013c), could explain the low recovery of this subunit in the antibody eluate. Although most of the Cox12p-HAC in the digitonin extract was adsorbed on the antibody beads (Figure 5C), based on the signal strength on the immunoblot, the mitochondrial concentration of this subunit was considerably lower than of the other subunits (Figure 5C). We do not know whether the tag prevents assembly and/or stability of Cox12p-HAC.
Characterization of D4-1 and D4-2 in Cox1p-, Cox2p-, and Cox3p- deficient mutants
D4-1 and D4-2 are prominent bands in the eluate of the strain expressing Cox4p-HAC but not Cox3p-HAC, Cox7p-HAC, or Cox13p-HAC (Figures 2A, 4B, and 5B). The radioactivity associated with these bands, when resolved by 2D gel electrophoresis, is contributed by Cox1p, as neither contains radiolabeled Cox3p or Cox2p (Figures 2E and 6C). To gain better insight into their compositions, we introduced COX4-HAC into mss51-, pet494-, and pet111-null mutants deficient in translation of Cox1p, Cox3p, and Cox2p, respectively. The mutant strains were pulsed for 20 min and Cox4p-HAC purified on protein C antibody beads. The eluate from the respiratory-competent control strain with Cox4p-HAC was enriched for Cox1p, Cox2p, and Cox3p, whereas the eluate from the pet494 mutant contained radiolabeled Cox2p but no significant radiolabeled Cox1p or Cox3p (Figure 6A). Only background levels of newly translated COX subunits were present in the eluates of the mss51 and pet111 mutants (Figure 6, A and E).
FIGURE 6:
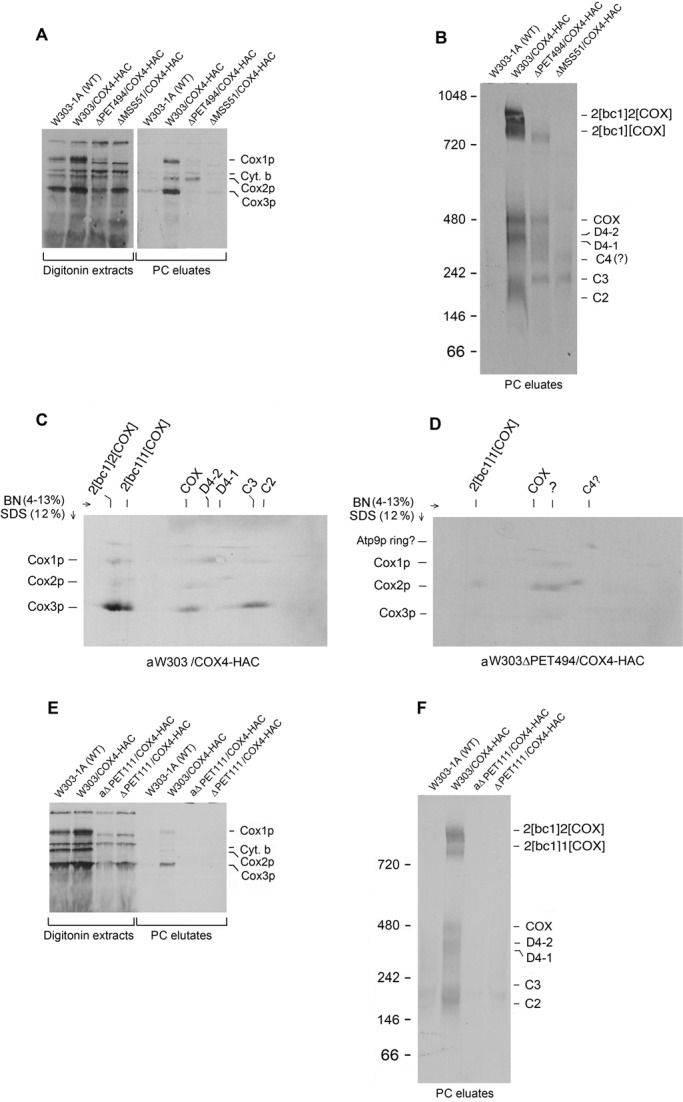
Cox4p containing intermediates in mutants blocked in translation of Cox1p, Cox2p, and Cox3p. (A) W303-1A (WT) and strains expressing tagged Cox4p in a wild-type background (W303/COX4-HAC), in a pet494-null mutant deficient in Cox3p (aW303ΔPET494/COX4-HAC), and in a mss51 mutant deficient in Cox1p (W303ΔMSS51/COX4-HAC) were grown in YPGal to early stationary phase in the absence of chloramphenicol. Mitochondria (500 μg of protein) were labeled for 20 min, extracted with digitonin, and purified on protein C antibody beads as in Figure 1A. Samples of the digitonin extracts (11% of total) and eluates (20% of total) from the antibody beads were separated by SDS–PAGE on a 17% polyacrylamide gel and radiolabeled proteins visualized. (B) The remainder of the eluates from the antibody beads in A was separated by BN-PAGE on a 4–13% polyacrylamide gel and radiolabeled proteins visualized. (C, D) W303/COX4-HAC and aW303ΔPET494/COX4-HAC mitochondria were labeled and purified on protein C beads as in A. The antibody eluates were separated in the first dimension by BN-PAGE on a 4–13% polyacrylamide gel and by SDS–PAGE on a 12% gel in the second dimension. Proteins were transferred to nitrocellulose and exposed to x-ray film. The Cox4p-HAC–containing bands are identified in the upper part of the two gels and the COX subunits in the left-hand margins. The band labeled with a question mark may be related to Cox2p intermediates. (E) The wild-type W303-1A, W303/COX4-HAC, a respiratory-competent strain expressing tagged Cox4p, and W303ΔPET11/COX4-HAC (ΔPET11/COX4-HAC) and aW303ΔPET11/COX4-HAC (aΔPET11/COX4-HAC), two pet111-null mutants expressing tagged Cox4p, were grown to early stationary phase without chloramphenicol. Mitochondria were labeled and extracted, and Cox4p-HAC–containing complexes were purified on protein C antibody beads as in A. The digitonin extracts and eluates from the antibody beads were separated by SDS–PAGE and radiolabeled bands visualized. (F) The eluates from E were separated by BN-PAGE.
BN-PAGE analysis of the antibody eluates from the pet494 mutant disclosed the absence of C2 and the presence of a band at the position of C3 and of broadly distributed radioactivity spanning the region of C4 to D4-2. In addition, some radiolabeled COX and the smaller of the two supercomplexes were also present (Figure 6B). The 2D gel of the eluates confirmed that the newly translated Cox3p in the respiratory-competent strain with Cox4p-HAC was present in C2, C3, COX, and the supercomplexes (Figure 6C). The presence of Cox1p and absence of Cox3p in D4-1 and D4-2 suggests that Cox4p can also interact with late intermediates of Cox1p. The 2D gel of the eluate from the pet494 mutant indicated background amounts of radiolabeled Cox3p, indicating some residual translation of this subunit in the mutant. This is also evident by the very low but detectable COX activity in the mutant (Supplemental Figure S3B). Radiolabeled Cox1p was detected in the region of D4-1 and D4-2 in the 2D gel of the pet494 mutant but only at background levels, most likely because of the severe reduction of Cox1p translation due to translational down-regulation of this subunit in the mutant (Figure 6A). The 2D gel of the pet494 mutant indicated that most of the radiolabeled Cox2p was present in the smaller of the two supercomplexes, in COX, and in the region of D4-1 and D4-2. The last may be complexes of Cox2p and Cox4-HAC that are formed when Cox3p and Cox1p are absent. The native gel of the eluate from the mss51 mutant contained radiolabeled bands of 250 and 300 kDa, which were also present in the pet494 mutant (Figure 6B). These bands are unlikely to be C3 and C4 because neither eluate showed an enrichment of Cox3p, and the 2D gel failed to show the presence of Cox3p in the regions corresponding to these two bands (Figure 6D). The radioactivity at the position of C4 migrated as high–molecular weight protein similar to the Atp9p ring (Figure 6D).
Neither Cox3p nor of any of the radiolabeled complexes seen in the pet494 and mss51 mutants was detected in the antibody eluate of the pet111 mutant (Figure 6, E and F) despite the fact that Cox4p-HAC was expressed in this strain (Supplemental Figure S5). The pet111 mutation blocked translation of Cox2p and in addition completely repressed translation of Cox1p (Figure 6E). Although there was modest reduction in the steady-state concentration of Cox4p-HAC in this strain, unexpectedly, the extractability of the protein was dramatically altered. Most of the protein from the respiratory-competent strain W303/COX4-HAC was extracted by digitonin and recovered in the PC eluate. In contrast, essentially none of the Cox4p-HAC in the pet111 mutant was detected in this fraction (Supplemental Figure S5).
Cox3p complexes with Rcf1p
Rcf1p is an 18-kDa mitochondrial inner membrane protein that interacts with Cox3p and stabilizes the association of COX with bc1 in the supercomplex but is not essential for COX assembly (Chen et al., 2012; Strogolova et al., 2012; Vukotic et al., 2012). A yeast strain expressing Rcf1p with a C-terminal HAC tag was previously used to characterize Rcf1p complexes by BN-PAGE (McStay et al., 2013a). When purified on protein C antibody beads from in vitro–labeled mitochondria, most of the radioactivity was resolved into two bands of 200 and 300 kDa (McStay et al., 2013a; Figure 7, A and D). The broad band centering at 200 kDa spanned the C2–C3 region, and the 300-kDa band migrated like C4 (Figure 7, A and E). On longer exposures to x-ray film, some radioactivity was also detected in the region between C4 and COX and the supercomplexes but not in the C1 region, even when short pulse times were used to label the mitochondrial gene products (Figure 7E). The 2D gel revealed that most of the radioactivity was contributed by Cox3p, although the second-dimension gel revealed the presence of a radiolabeled protein that migrated as Cox1p at the position of C2 and of another protein, which migrated as the Atp9 ring at the position of the 300-kDa complex (Figure 7B). The possible interaction of a small fraction of Rcf1p with Cox1p and Atp9p was not studied further. It is interesting to note, however, that a band corresponding in its migration to C4 in the eluates from the mss51 and pet494 mutants harboring Cox4p-HAC also contained what appeared to be radiolabeled Atp9p ring (Figure 6D).
FIGURE 7:
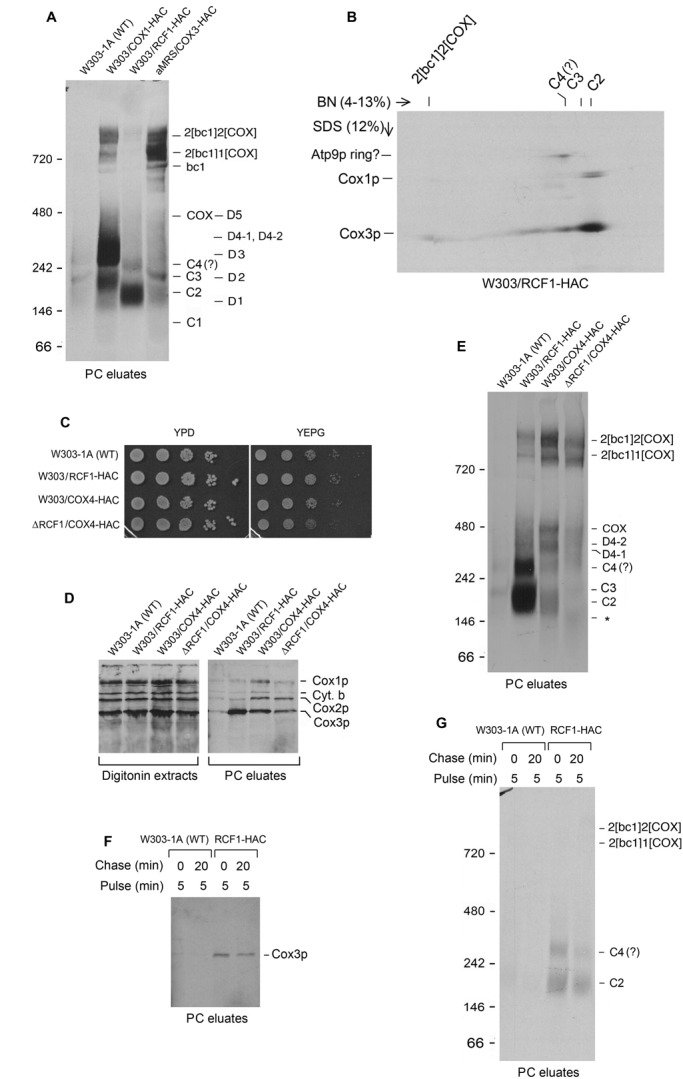
Complexes of Cox3p and Rcf1p. (A) W303-1A (WT) and W303/COX1-HAC, W303/RCF1-HAC, and W303/COX3-HAC expressing tagged Cox1p, Rcf1p, and Cox3p, respectively, were grown to early stationary phase in the absence of chloramphenicol. Mitochondria (500 μg of protein) were labeled for 20 min and extracted with digitonin, and the extracts were purified on protein C antibody beads as in Figure 1A. Half of the eluate was separated by BN-PAGE on a 4–13% polyacrylamide gel. (B) The eluate from W303/RCF1-HAC was separated by BN-PAGE on a 4–13% polyacrylamide gel and by SDS–PAGE on a 12% polyacrylamide gel in the second dimension. (C) The indicated strains were serially diluted and spotted on rich glucose (YPD) and rich glycerol plus ethanol media. The plates were incubated for 28 h at 30°C. (D) W303-1A (WT), W303/RCF1-HAC expressing Rcf1p-HAC, W303/COX4-HAC expressing Cox4p-HAC, and W303ΔRCF1/COX4-HAC (ΔRCF1/COX4-HAC), an rcf1-null mutant expressing Cox4-HAC, were grown in YPGal without chloramphenicol and mitochondria labeled as in Figure 1A. Mitochondria were extracted with digitonin and purified on protein C antibody beads. The extracts (11% of total) and eluates from the antibody beads (20% of total) were separated by SDS–PAGE on a 17% polyacrylamide gel. (E) The eluates from D (60% of total) were separated by BN-PAGE. (F, G) W303-1A (WT) and W303/RCF1-HAC, expressing tagged Rcf1p, were grown to early stationary phase in YPGal without chloramphenicol. Mitochondria (1 mg of protein) were labeled for 5 min as in Figure 1A and chased for 20 min after addition of puromycin and cold amino acids. Mitochondria in the pulsed and chased samples were extracted with digitonin and purified on protein C antibody beads. The eluates (14% of total) from the antibody beads were separated by SDS–PAGE on a 17% polyacrylamide gel and radiolabeled proteins visualized. (G) The eluates (78% of total) from F were separated by BN-PAGE.
The rcf1-null mutant expressing Cox4p-HAC grew more slowly on rich glycerol/ethanol than either the strain expressing Cox4p-HAC or the rcf1 mutant alone (Figure 7C). The decreased growth can be explained by some reduction of COX as a result of the rcf1-null mutation (Supplemental Figure S6). This was also evident from the BN-PAGE of the fraction eluted from the protein C antibody beads of the rcf1-null mutant, which revealed a reduction of radiolabel in the two supercomplexes and the D4-1 and D4-2 region (Figure 7E). The rcf1 mutant lacked C2 and had a faster-migrating band not seen in the parental strain, which presumably corresponds to the Cox3p intermediate lacking Rcf1p (marked with asterisk in Figure 7E).
Mitochondria with Rcf1p-HAC were pulse labeled for 5 min and chased for up to 30 min in the presence of puromycin and excess cold methionine plus cysteine. The 5-min pulse resulted in labeling of the 200- and 300-kDa complexes, both of which underwent a modest decrease after a 20-min chase (Figure 7, F and G). The chase also produced a weak signal in the supercomplexes. The increase of radiolabel in the supercomplexes after the chase was more evident when the pulse was increased to 20 min (Supplemental Figure S4, D–F).
Considerably more of the C2–C4 intermediates were detected in mitochondria with tagged Rcf1p than those with tagged Cox4p, Cox7p, and Cox13p (Figure 7A and Supplemental Figure S4). This suggested that more Cox3p is complexed to Rcf1p than to the other subunits of COX. This was confirmed by comparing the coadsorption to the antibody beads of newly translated Cox3p from mitochondria with Rcf1p-HAC and Cox4p-HAC (Figure 7D and Supplemental Figure S7). It is unlikely that this difference is due to the pool sizes of unassembled Cox4p and of Rcf1p, as only radiochemical amounts of Cox3p are translated in isolated mitochondria. In both cases, however, only a small fraction of newly translated Cox3p is complexed to either Cox4p or Rcf1p.
DISCUSSION
We previously proposed that COX is assembled not by a single linear pathway but by separate pathways culminating in three modules, each consisting of a mitochondrially encoded core subunit and a unique subset of subunits derived from the nucleocytoplasmic genetic system (McStay et al., 2013c). The three modules interact with each other at a late stage of the overall process to form the holoenzyme (McStay et al., 2013a, c). The results of the present study are consistent with this model (Figure 8).
FIGURE 8:
Assembly of COX from Cox1p, Cox2p, and Cox3p modules. In this model each assembly module contains one of the three mitochondrially encoded subunits of COX. The diagram shows the subunits and the regulatory/ancillary factors associated with the different Cox1p and Cox3p subassemblies. The Cox2p module has not yet been analyzed. D3, the most abundant Cox1p intermediate, is indicated to interact with Cox4p to produce D4-1 and D4-2, which were previously identified as D4 (McStay et al., 2013a, c). The order in which the modules interact with each other to form the holoenzyme is not known.
Newly translated Cox3p is detected in COX, the supercomplexes, and three subassemblies ranging from 100 to 200 kDa in mass. These complexes, referred to as C1–C3, are distinct from Cox1p intermediates in their electrophoretic properties. A fourth complex of ∼300 kDa is also observed, although it is not present among the subassemblies pulled down from mitochondrial extracts containing Cox7p-HAC and Cox13-HAC. The assignment of this complex as a Cox3p intermediate is, therefore, questionable at present. A chase in the presence of puromycin and cold amino acids resulted in decrease of radiolabeled C1–C3 and increase in the supercomplexes, suggesting that the subassemblies are precursors of COX. However, unlike the Cox1p intermediates, some of which are present in sufficiently high concentrations for immunological detection, the Cox3p subassemblies are much less abundant and observable only as radiochemical species. The low concentration of Cox3p relative to Cox1p and Cox2p intermediates can explain what appears to be a disproportionately lower incorporation of newly translated Cox1p and Cox2p into COX and the supercomplexes, as the specific radioactivity of the intermediates formed from these subunits would be expected to be diluted by the large preexisting pools of nonradioactive intermediates. In this connection it is significant that a more stoichiometric incorporation of all three subunits into the supercomplexes is seen in pull downs with tagged Cox2p-HAC (Supplemental Figure S2). Although the differences in intermediate pool sizes can help to explain the apparent lower incorporation of Cox1 and Cox2p into the holoenzyme, alternative explanations of subunit exchange, the presence of partially assembled COX in the supercomplexes, or a combination thereof cannot be excluded by our results.
The C1–C3 subassemblies were also immune purified from mitochondria expressing tagged Cox4p, indicating that this nuclear-encoded subunit is part of the Cox3p module. The radiolabel in C1–C3 of mitochondria containing COX4-HAC decreased after the chase, with a concomitant larger increase in COX and the supercomplexes. The 2D gels of the samples that had been labeled for 20 min and analyzed either directly or after a 30-min chase revealed a decrease of radiolabeled Cox3p in C2 and C3 and an increase in the supercomplexes, confirming that these subassemblies are precursors of COX. Although radiolabeled Cox3p increased in COX and the supercomplexes after the chase, there was an even larger increase of Cox1p. These results are consistent with time-dependent assembly of Cox1p and Cox3p modules into COX and of the latter into the supercomplexes. C1, the smallest subassembly, detected in short pulses was also decreased during the chase, suggesting that it too is an assembly intermediate.
Similar results were obtained with mitochondria of strains expressing tagged Cox7p and Cox13p, although the efficiency of recovery of these complexes from the protein C antibody beads was less than with Cox4p-HAC. The enrichment on the antibody beads of radiolabeled C1–C3 that are chased into COX and the supercomplexes indicated that Cox7p and Cox13p are also part of the Cox3p module (Figure 8). Because of the poor recovery of Cox9p-HAC and Cox12p-HAC from antibody beads, the presence or absence of these subunits in the Cox3p module remains to be established. The fact that Cox12p, like Cox4p, Cox7p, and Cox13p, has interfaces with Cox3p suggests that this protein may also be a component of the Cox3p module. The lack of homology between yeast and mammalian Cox9p, which interacts with Cox2p (Tsukihara et al., 1996), precludes an assignment of this subunit to either Cox2p or Cox3p assembly intermediates. The subunit composition of the Cox3p module, combined with the previous assignments of nuclear-encoded subunits to the Cox1p module (McStay et al., 2013a), leaves only two subunits (Cox9p and Cox12p) as possible candidates to be part of the Cox2p module.
In addition to C1–C4, pull-down assays of tagged Cox4p uncovered a larger subassembly, which contained newly translated Cox1p instead of Cox3p. When separated on a 2D gel, this complex separated into two close but distinct bands, designated as D4-1 and D4-2. The size and presence in D4-1 of Cox1p but not Cox3p suggest that it probably corresponds to the previously reported Cox1p intermediate D4, which overlaps partially with the more abundant Cox1p intermediate D3 (McStay et al., 2013a, c). In view of the presence of Cox1p in D4-2, it is not clear why this complex was not detected previously on native gels of the antibody eluate purified from mitochondria with tagged Cox1p. The native gel of the eluate with Cox1p-HAC contains radiolabeled material in the D4-2 region, which does not migrate as a distinct band (McStay et al., 2013a; Figures 1B and 2A). A possible explanation is that unlike Cox4p-HAC, tagged Cox1p-HAC in this complex is not available to the antibody.
Pulse-chase labeling experiments revealed an increase of Cox1p in the D4-1 and D4-2 complexes as well as in COX and the supercomplexes during the chase period. This is consistent with previous results indicating that Cox1p-HAC translated during a short pulse is chased from smaller intermediates into D4 (McStay et al., 2013c). The presence of Cox4p in subassemblies containing Cox3p or Cox1p, exclusive of each other, suggests that this structural subunit can be recruited by either module. Whether one pathway is preferred over another cannot be inferred from current data. It is also possible that the C1–C4 or the D4-2 and D4-1 complexes may be heterogeneous with respect to Cox4p. If addition of Cox4p to COX can be mediated by either the Cox3p or the Cox1p module, the more likely scenario is that a module with the Cox4p subunit, whether Cox3p or Cox1p, partners with the complementary module lacking it.
Mutations in either pet494 or pet111, which block translation of Cox2p and Cox3p, respectively, reduce translation of Cox1p. This is even more evident in the pet111 than in the pet494 mutant. Both mutants, however, are able to form all of the Cox1p intermediates in a ratio similar to those present in a respiratory-competent strain (McStay et al., 2013a). The present study shows that the converse is not true. The mss51 mutant lacked the C2 intermediate, and although the blue native gel indicated the presence of radioactive bands with migrations similar to C3 and C4, no Cox3p was detected at those positions in the 2D gel, suggesting that they may be unrelated. The pet111 mutant was deficient in C2–C4 intermediates, even though this mutant is known to translate Cox3p (Poutre and Fox, 1987). These observations raised the interesting possibility that the Cox3p biogenesis pathway may be regulated by Cox1p and Cox2p.
Our evidence indicates that most of Rcf1p is complexed to Cox3p in two complexes that migrate as C2 and C4. Because of the broad nature of the C2 band, we cannot exclude that Rcf1p may also be present in C3. Pull-down assays indicate that some Rcf1p is also present in the supercomplexes and perhaps COX. The absence of radiolabeled C1 in eluates from the antibody beads after short pulse labeling of mitochondria of strains expressing Rcf1p-HAC indicates that this protein is unlikely to be a component of this complex. In agreement with earlier results (Chen et al., 2012; Strogolova et al., 2012; Vukotic et al., 2012), our data indicate that Rcf1p is not essential for COX assembly. Rcf1p, however, does affect the assembly of COX to some degree, as reflected by the slower growth rate on respiratory carbon sources and measurable decrease of COX in the rcf1-null mutant. The rcf1 mutant is completely deficient in C2 and has a novel band of ∼150 kDa, which probably corresponds to this intermediate without Rcf1p. This suggests that C2 lacking Rcf1p is competent in assembling with the other COX modules, albeit not as efficiently as the native complex with Rcf1p.
MATERIALS AND METHODS
Strains and growth media
Supplemental Table S1 lists the genotypes and sources of the S. cerevisiae strains used in this study. The compositions of solid and liquid yeast extract/peptone/dextrose (YPD), yeast extract/peptone/galactose (YPGal), and (yeast extract/peptone/glycerol/ethanol (YEPG) have been described (Myers et al., 1985)
Construction of COX3-deletion allele
The 5′ and 3′ untranslated regions (UTRs) of COX3 were amplified from mitochondrial DNA (mtDNA) of MR6 (Rak et al., 2007) with primer pairs cox3-9/cox3-5n and cox3-7/cox3-10, respectively (Supplemental Table S2). The products were digested with combinations of SacI plus BamH1 and BamH1 plus XbaI. The two fragments were ligated to pJM2 (Mulero and Fox, 1993) digested with SacI plus XbaI. The resultant plasmid (pCOX3/ST4) contained the 5′ and 3′ UTRs of COX3 separated by a BamH1 site. After digestion with BamH1, this plasmid was ligated to a 1.3-kb BamH1 fragment containing recoded ARG8m (Steele et al., 1996) to obtain pCOX3/ST5 with the cox3::ARG8m null allele. The cox3::ARG8m allele in pCOX3/ST5 was first introduced into DFKρo, a kar1 mutant lacking mtDNA, by biolistic transformation (Bonnefoy and Fox, 2007). This ρ− mutant was subsequently used to substitute the mutant for the wild-type COX3 gene by homologous recombination in strain MRS-3A and MRSIo with wild type and an intronless mitochondrial genome, respectively. The resultant respiratory-deficient mutants aMRSΔCOX3 and aMRSIoΔCOX3 were verified genetically to harbor the cox3-null allele.
Construction of COX3-pH and COX3-HAC fusion genes
To obtain the hybrid gene expressing Cox3p with a C-terminal polyhistidine tag (COX3-pH), mtDNA of MR6 was PCR amplified with primer pairs cox3-9/cox3-9n and cox3-10n/cox3-10 and the resultant fragment digested with SacI plus KpnI and KpnI plus XbaI, respectively. The digested fragments were ligated to pJM2 digested with SacI plus XbaI to yield pCOX3/ST6 with the COX3-pH hybrid gene. This plasmid was used to obtain DFK/COX3-pH by biolistic transformation of DFKρo. The COX3-pH gene was recombined with the cox3::ARG8m null allele by crossing the kar1 strain DFK/COX3-pH to aMRSΔCOX3. Recombinants were enriched by first selecting respiratory-competent cells on YEPG. Single colonies were then crossed to an α-mating-type haploid tester with complementary auxotrophic markers and replicated on minimal glucose to distinguish diploid and haploid cells.
The second gene construct was designed to express Cox3p with an N-terminal double HAC tag consisting of the HA followed by the protein C tag. This construct entailed PCR amplification of two fragments. The first, obtained with primers cox3-20 and cox3-21, contained the COX3 5′ UTR plus a methionine codon followed by part of the sequence coding for the protein C tag. The second fragment was amplified with primers cox3-15 and cox3-4. This fragment contained the rest of the sequence for the protein C tag followed by two glycine codons, the HA tag, and the entire COX3 coding sequence minus the initiation codon and terminating with the 3′ UTR of the gene. After digestion of the first and second fragments with combinations of SacI plus ClaI and ClaI plus PstI, respectively, they were ligated to pJM2 (minus the extra PstI site) digested with SacI and PstI. The plasmid pCOX3/ST10 was verified to contain the correct insert and was introduced into the kar1 strain DFKρo by biolistic transformation. The two strains MRS/COX3-HAC and MRSIo/COX3-HAC were obtained by crossing DFK/COX3-HAC containing pCOX3/ST10 to the cox3-null mutants MRSΔCOX3 and MRSIoΔCOX3 and screening recombinants as described for the gene with polyhistidine tag. Both MRS/COX3-HAC and MRSIo/COX3-HAC were verified to express Cox3p with the double tag.
Construction of the Cox2p-HAC allele
The sequence coding for the HAC double tag was ligated to the 3′ end of the COX2 by PCR amplification of the gene by a strategy similar to that used to construct the Cox1p-HAC allele (McStay et al., 2013c). The kar1 strain DFK/COX2-HAC containing the COX2 fusion gene was crossed to a cox2-null mutant kindly provided by Thomas Fox (Cornell University, Ithaca, NY) and recombinants selected for the loss of the ARG8m disruptor of COX2 and the acquisition of growth on nonfermentable carbon sources. Several clones were verified by sequencing to have the COX2-HAC allele in their mtDNA.
Growth of yeast, isolation of mitochondria, and labeling of mitochondrial gene products
Yeast were grown in 2% galactose, 2% peptone, and 1% yeast extract (YPGal) to early stationary phase. In a few experiments, cells were transferred to fresh YPGal containing 2 mg/ml chloramphenicol and grown for an additional 2 h. Mitochondria (250 μg of protein) isolated by the method of Herrmann et al. (1994) were labeled in a final volume of 85 μl for 20 min with a mixture of [35S]methionine/cysteine (3000 Ci/mmol; MP Biochemicals, Solon, OH) at 24°C as described previously (Rak et al., 2011). Translation was stopped by addition of 4 μg of puromycin and 0.04 μmol of methionine and cysteine, and incubation was continued for an additional 10 min. The mitochondria in the translation buffer were mixed with 80 μl of 4% digitonin in extraction buffer (Wittig et al., 2006) and phenylmethanesulfonyl fluoride to a final concentration of 1 mM. After centrifugation at 90,000 × gav for 10 min the supernatant was added to 30 μl of packed protein C antibody beads (Anti–Protein C Affinity Matrix; Roche Applied Science, Indianapolis, IN) that had been washed in PC wash buffer (20 mM Tris-Cl, pH 7.5, 2 mM CaCl2, 0.2 M NaCl) and rotated at 4°C for 90 min. The beads were washed four times with 0.5 ml of wash buffer containing 0.5% digitonin. The protein adsorbed on the beads was eluted at 4°C by rotating with 70 μl of elution buffer (20 mM Tris-Cl, pH 7.5, 7 mM EDTA, 0.2 M NaCl, and 0.5% digitonin). Part of the eluate (20%) was mixed with Laemmli sample buffer and separated by SDS–PAGE on a 17% polyacrylamide gel. The remaining sample was mixed with BN-PAGE sample buffer (Wittig et al., 2006) and separated on a 4–13% gel.
Miscellaneous procedures
Standard methods were used for the preparation and ligation of DNA and transformation of Escherichia coli (Sambrook et al., 1989). Yeast was transformed by the lithium acetate method of Schiestl and Gietz (1989). Proteins were separated either by SDS–PAGE on 12 or 17% polyacrylamide gels in Laemmli buffer (Laemmli, 1970) or by BN-PAGE on 4–13% polyacrylamide gels (Wittig et al., 2006). Western blots were treated with monoclonal or polyclonal antibodies followed by a second reaction with anti-mouse or anti-rabbit immunoglobulin G conjugated to horseradish peroxidase (Sigma-Aldrich, St. Louis, MO). Proteins were detected with SuperSignal Chemiluminescent Substrate Kit (Pierce, Rockford, IL). In-gel cytochrome oxidase activity was assayed on digitonin extracts and eluates from protein C antibody beads from 50 μg of starting mitochondrial protein. The gel was immersed in a solution containing 0.5 mg/ml 3,3′-diaminobenzidine and 1 mg/ml horse heart cytochrome c for several hours. Protein concentrations were estimated by the Lowry procedure (Lowry et al., 1951).
Supplementary Material
Acknowledgments
This research was supported by National Institutes of Health Research Grant GM50187.
Abbreviations used:
- BN
blue native
- COX
cytochrome oxidase
- mtDNA
mitochondrial DNA
Footnotes
*Present address: Department of Life Sciences, New York Institute of Technology, Old Westbury, NY 11568.
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-10-0575) on January 29, 2014.
REFERENCES
- Aggeler R, Capaldi RA. Yeast cytochrome c oxidase subunit VII is essential for assembly of an active enzyme. Cloning, sequencing, and characterization of the nuclear-encoded gene. J Biol Chem. 1990;265:16389–16393. [PubMed] [Google Scholar]
- Barrientos A, Zambrano A, Tzagoloff A. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 2004;23:3472–3482. doi: 10.1038/sj.emboj.7600358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnefoy N, Fox TD. Directed alteration of Saccharomyces cerevisiae mitochondrial DNA by biolistic transformation and homologous recombination. Methods Mol Biol. 2007;372:153–166. doi: 10.1007/978-1-59745-365-3_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calder KM, McEwen JE. Deletion of the COX7 gene in Saccharomyces cerevisiae reveals a role for cytochrome c oxidase subunit VII in assembly of remaining subunits. Mol Microbiol. 1991;5:1769–1777. doi: 10.1111/j.1365-2958.1991.tb01926.x. [DOI] [PubMed] [Google Scholar]
- Chen YC, Taylor EB, Dephoure N, Heo JM, Tonhato A, Papandreou I, Nath N, Denko NC, Gygi SP, Rutter J. Identification of a protein mediating respiratory supercomplex stability. Cell Metab. 2012;15:348–360. doi: 10.1016/j.cmet.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanesi F, Clemente P, Barrientos A. Cox25 teams up with Mss51, Ssc1, and Cox14 to regulate mitochondrial cytochrome c oxidase subunit 1 expression and assembly in Saccharomyces cerevisiae. J Biol Chem. 2011;286:555–566. doi: 10.1074/jbc.M110.188805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanesi F, Soto IC, Horn D, Barrientos A. Mss51 and Ssc1 facilitate translational regulation of cytochrome c oxidase biogenesis. Mol Cell Biol. 2010;30:245–259. doi: 10.1128/MCB.00983-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer J, Braun HP, Boekema EJ, Kouril R. A structural model of the cytochrome c reductase/oxidase supercomplex from yeast mitochondria. J Biol Chem. 2007;282:12240–12248. doi: 10.1074/jbc.M610545200. [DOI] [PubMed] [Google Scholar]
- Herrmann JM, Foelsch H, Neupert W, Stuart RA. Isolation of yeast mitochondria and study of mitochondrial protein translation. In: Celis JE, editor. In: Cell Biology: A Laboratory Handbook. Vol. I. San Diego, CA: Academic Press; 1994. pp. 538–544. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- McStay GP, Su CH, Thomas SM, Xu JT, Tzagoloff A. Characterization of assembly intermediates containing subunit 1 of yeast cytochrome oxidase. J Biol Chem. 2013a;288:26546–26556. doi: 10.1074/jbc.M113.498592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay GP, Su CH, Tzagoloff A. Stabilization of Cox1p intermediates by the Cox14p-Coa3p complex. FEBS Lett. 2013b;587:943–949. doi: 10.1016/j.febslet.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay GP, Su CH, Tzagoloff A. Modular assembly of yeast cytochrome oxidase. Mol Biol Cell. 2013c;24:440–452. doi: 10.1091/mbc.E12-10-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mick DU, Fox TD, Rehling P. Inventory control: cytochrome c oxidase assembly regulates mitochondrial translation. Nat Rev Mol Cell Biol. 2011;12:14–20. doi: 10.1038/nrm3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mick DU, Vukotic M, Piechura H, Meyer HE, Warscheid B, Deckers M, Rehling P. Coa3 and Cox14 are essential for negative feedback regulation of COX1 translation in mitochondria. J Cell Biol. 2010;191:141–154. doi: 10.1083/jcb.201007026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mick DU, Wagner K, van der Laan M, Frazier AE, Perschil I, Pawlas M, Meyer HE, Warscheid B, Rehling P. Shy1 couples Cox1 translational regulation to cytochrome c oxidase assembly. EMBO J. 2007;26:4347–4358. doi: 10.1038/sj.emboj.7601862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mileykovskaya E, Penczek PA, Fang J, Mallampalli VK, Sparagna GC, Dowhan W. Arrangement of the respiratory chain complexes in Saccharomyces cerevisiae supercomplex III2IV2 revealed by single particle cryo-electron microscopy. J Biol Chem. 2012;287:23095–23103. doi: 10.1074/jbc.M112.367888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulero JJ, Fox TD. Alteration of the Saccharomyces cerevisiae COX2 mRNA 5’-untranslated leader by mitochondrial gene replacement and functional interaction with the translational activator protein PET111. Mol Biol Cell. 1993;4:1327–1335. doi: 10.1091/mbc.4.12.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers AM, Pape LK, Tzagoloff A. Mitochondrial protein synthesis is required for maintenance of intact mitochondrial genomes in Saccharomyces cerevisiae. EMBO J. 1985;4:2087–2092. doi: 10.1002/j.1460-2075.1985.tb03896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson TE, Poyton RO. COX8, the structural gene for yeast cytochrome c oxidase subunit VIII. DNA sequence and gene disruption indicate that subunit VIII is required for maximal levels of cellular respiration and is derived from a precursor which is extended at both its NH2 and COOH termini. J Biol Chem. 1986;261:17192–17197. [PubMed] [Google Scholar]
- Perez-Martinez X, Broadley SA, Fox TD. Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 2003;22:5951–5961. doi: 10.1093/emboj/cdg566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Martinez X, Butler CA, Shingu-Vazquez M, Fox TD. Dual functions of Mss51 couple synthesis of Cox1 to assembly of cytochrome c oxidase in Saccharomyces cerevisiae mitochondria. Mol Biol Cell. 2009;20:4371–4380. doi: 10.1091/mbc.E09-06-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierrel F, Bestwick ML, Cobine PA, Khalimonchuk O, Cricco JA, Winge DR. Coa1 links the Mss51 post-translational function to Cox1 cofactor insertion in cytochrome c oxidase assembly. EMBO J. 2007;26:4335–4346. doi: 10.1038/sj.emboj.7601861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poutre CG, Fox TD. PET111, a Saccharomyces cerevisiae nuclear gene required for translation of the mitochondrial mRNA encoding cytochrome c oxidase subunit II. Genetics. 1987;115:637–647. doi: 10.1093/genetics/115.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak M, Gokova S, Tzagoloff A. Modular assembly of yeast mitochondrial ATP synthase. EMBO J. 2011;30:920–930. doi: 10.1038/emboj.2010.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak M, Tetaud E, Godard F, Sagot I, Salin B, Duvezin-Caubet S, Slonimski PP, Rytka J, di Rago JP. Yeast cells lacking the mitochondrial gene encoding the ATP synthase subunit 6 exhibit a selective loss of complex IV and unusual mitochondrial morphology. J Biol Chem. 2007;282:10853–10864. doi: 10.1074/jbc.M608692200. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Steele DF, Butler CA, Fox TD. Expression of a recoded nuclear gene inserted into yeast mitochondrial DNA is limited by mRNA-specific translational activation. Proc Natl Acad Sci USA. 1996;93:5253–5257. doi: 10.1073/pnas.93.11.5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strogolova V, Furness A, Robb-McGrath M, Garlich J, Stuart RA. Rcf1 and Rcf2, members of the hypoxia-induced gene 1 protein family, are critical components of the mitochondrial cytochrome bc1-cytochrome c oxidase supercomplex. Mol Cell Biol. 2012;32:1363–1373. doi: 10.1128/MCB.06369-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taanman JW, Capaldi RA. Subunit VIa of yeast cytochrome c oxidase is not necessary for assembly of the enzyme complex but modulates the enzyme activity. Isolation and characterization of the nuclear-coded gene. J Biol Chem. 1993;268:18754–18761. [PubMed] [Google Scholar]
- Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- Vukotic M, et al. Rcf1 mediates cytochrome oxidase assembly and respirasome formation, revealing heterogeneity of the enzyme complex. Cell Metab. 2012;15:336–347. doi: 10.1016/j.cmet.2012.01.016. [DOI] [PubMed] [Google Scholar]
- Wittig I, Braun HP, Schagger H. Blue native PAGE. Nat Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.