Summary
The development of any cell and/or tissue is dependent upon interconnections between several signaling pathways and myriad transcription factors. It is becoming more apparent that these inputs are best studied, not as individual components, but rather as elements of a gene regulatory network. Over the last decade several networks governing the specification of single cells, individual organs and entire stages of development have been described. The current incarnations of these networks are the products of the continual addition of newly discovered genetic, molecular and biochemical interactions. However, as currently envisaged, network diagrams may not sufficiently describe the spatial and temporal dynamics that underlie developmental processes. We have conducted a developmental analysis of a sub circuit of the Drosophila retinal determination network. This sub circuit is comprised of three genes, two (sine oculis and dachshund) of which code for DNA binding proteins and one (eyes absent) that encodes a transcriptional co-activator. We demonstrate here that the nature of the regulatory relationships that exist between these three genes changes as retinal development progresses. We also demonstrate that the response of the tissue to the loss of any of these three RD genes is dependent upon the position of the mutant cells within the eye field. Depending upon its location, mutant tissue will either overproliferate itself or will signal to surrounding cells instructing them to propagate and compensate for the eventual loss through apoptosis of the mutant clone. Taken together these results suggest that the complexities of development are best appreciated when spatial and temporal information is incorporated when describing gene regulatory networks.
Keywords: retinal determination, sine oculis, eyes absent, dachshund, groucho, Notch signaling, Drosophila, compensatory proliferation, gene regulatory networks, GRN
Introduction
The last decade has played witness to the revelation that the specification of tissues and organs, are regulated, not by simple linear cascades, rather by complicated interconnected gene regulatory networks (GRNs). The influence of such networks can be limited to a single context or can extend to multiple developing tissues. Such is the case for the retinal determination (RD) network, which, in addition to the eye, regulates the fate of a number of tissues in both insect and vertebrate systems. First identified in flies, this network also controls the development of learning and memory centers of the brain, several mesodermal derivatives, the gonads and select cells within the central nervous system (Bai and Montell, 2002; Bonini et al., 1998; Callaerts et al., 2001; Chang et al., 2003; Fabrizio et al., 2003; Kammermeier et al., 2001; Kurusu et al., 2000; Mardon et al., 1994; Niimi et al., 1999; Noveen et al., 2000). In addition to its role in vertebrate eye development, the RD network regulates ear, nose, kidney and muscle specification (Brodbeck and Englert, 2004; Gong et al., 2007; Hammond et al., 1998; Hanson, 2001; Heanue et al., 1999; Kalatzis et al., 1998; Laclef et al., 2003; Relaix and Buckingham, 1999; Simpson and Price, 2002; Xu et al., 2003). Over the years members of seven gene families have been identified to function within the RD network. In Drosophila these include the Pax6 genes eyeless (ey) and twin of eyeless (toy), the Pax6(5a) genes eyegone (eyg) and twin of eyegone (toe), the Six family members sine oculis (so) and optix, the founding member of the Eya family of transcriptional co-activators eyes absent (eya), a distant relative of the Ski/Sno family of proto-oncogenes dachshund (dac), the Meis1 homolog homothorax (hth) and the zinc finger transcription factor teashirt (tsh) (reviewed in (Kumar and Moses, 2001b; Treisman, 1999; Treisman and Heberlein, 1998; Weasner et al., 2004).
The evidence that prompted the placement of these genes into a functional network is principally drawn from loss-of-function mutant phenotypes (Bonini et al., 1993; Cheyette et al., 1994; Jang et al., 2003; Mardon et al., 1994; Quiring et al., 1994; Serikaku and O'Tousa, 1994), overlapping expression patterns (Bessa et al., 2002), direct transcriptional activation of one gene by another (Czerny et al., 1999; Niimi et al., 1999; Ostrin et al., 2006; Pauli et al., 2005), protein-protein interactions amongst selected network members (Chen et al., 1997; Pignoni et al., 1997) and the unique ability of these genes to induce ectopic eyes in non-retinal tissues (Bonini et al., 1997; Czerny et al., 1999; Halder et al., 1995; Pan and Rubin, 1998; Seimiya and Gehring, 2000; Shen and Mardon, 1997; Weasner et al., 2007). As additional experimental evidence is gathered, new positive or inhibitory arrows are added resulting in a network with ever increasing complexity. Similar GRNs with equal or greater complexity have been identified in a number of systems including the fly wing and ventral furrow (Aracena et al., 2006; Guss et al., 2001); mouse stem cell, B lymphocyte and brain (Li et al., 2007; Medina et al., 2004; Wang et al., 2007; Zhou et al., 2007); Xenopus mesoendoderm (Loose and Patient, 2004); vertebrate neural crest (Sauka-Spengler and Bronner-Fraser, 2008a; Sauka-Spengler and Bronner-Fraser, 2008b; Sauka-Spengler et al., 2007); Arabidopsis flower development (Espinosa-Soto et al., 2004) and sea urchin embryogenesis (Davidson et al., 2002; Oliveri et al., 2002; Oliveri and Davidson, 2004a; Oliveri and Davidson, 2004b) just to name a few. However, as is the case with any complex system, no single regulatory model can fully describe all of the spatial and temporal events that occur during develo pment (Flores et al., 2000) to produce the final adult tissue.
Eye specification in Drosophila begins during embryogenesis when a small group of cells are set aside to give rise to the future compound eye (Cohen, 1993). Upon emerging as a larva, these cells become organized into a monolayer epithelium called the eye-antennal imaginal disc. During the first two larval instars the eye disc undergoes massive proliferation to generate the large numbers of cells that are required to produce the approximately 800 unit eyes or ommatidia that comprise the adult compound eye. At the start of the third and final instar, pattern formation is initiated at the posterior margin of the epithelium. The wave of morphogenesis can be visualized by a dorso-ventral groove in the epithelium referred to as the morphogenetic furrow (Ready et al., 1976). As the furrow passes, the pool of undifferentiated cells are organized into periodic clusters of developing ommatidia (Ready et al., 1976; Wolff and Ready, 1991). Within each cluster are approximately twenty cells that adopt either photoreceptor or non-neuronal accessory cell fates (Cagan and Ready, 1989; Tomlinson and Ready, 1987a; Tomlinson and Ready, 1987b). These decisions involve complex, stereotyped patterns of gene expression (Dickson and Hafen, 1993; Doroquez and Rebay, 2006; Flores et al., 2000; Kumar and Moses, 1997; Nagaraj and Banerjee, 2007; Voas and Rebay, 2004). Ultimately, the several hundred ommatidia are organized into a precise hexagonal array characteristic of the adult retina.
In the developing fly retina ey is one of the first RD genes to be expressed. Along with toy, ey directly activates the transcription of several downstream targets including itself and three other network genes: so, optix and eya (Halder et al., 1998; Niimi et al., 1999; Ostrin et al., 2006). So and Eya proteins form a composite transcription factor with So contributing a DNA binding domain and Eya providing an activation domain (Pignoni et al., 1997). The So-Eya complex, in turn, activates a number of target genes that play crucial roles in cell proliferation (string, (Jemc and Rebay, 2007), pattern formation (hedgehog, (Pauli et al., 2005) and cell fate specification (lozenge, (Yan et al., 2003). Additionally, So-Eya feeds back to regulate the transcription of the upstream gene ey (Pauli et al., 2005) and the downstream target dac (Pappu et al., 2005). It is this last interaction that is the central focus of this report, as it highlights an instance in which the totality of experimental evidence is not represented by the most current network models.
Consistent with their roles as obligate partners, So and Eya proteins are distributed in completely overlapping expression patterns in the developing eye. Both are expressed in a swathe of undifferentiated cells ahead of the advancing morphogenetic furrow and in all cells posterior to the furrow (Bonini et al., 1993; Cheyette et al., 1994; Serikaku and O'Tousa, 1994). Dac protein distribution ahead of the furrow overlaps that of So and Eya. However, posterior to the furrow dac expression is maintained for approximately eight rows where it is restricted to only a subset of photoreceptors and then quickly tapers off (Mardon et al., 1994). Two enhancers responsible for the activation of dac expression in the retina are under the partial control of both so and eya (Pappu et al., 2005). As the So-Eya complex is still present and functioning in the more posterior cells it is intriguing that dac expression ceases. The seminal experiments that established the regulatory relationships among the RD genes were based in large measure on immunohistochemical assays completed in entirely mutant eye discs in which a furrow failed to initiate (Anderson et al., 2006; Chen et al., 1997; Halder et al., 1998; Pappu et al., 2005) and in ectopic eye assays in which the distribution of RD proteins were measured in response forced expression of either individual or combinations of genes (Bonini et al., 1997; Chen et al., 1999; Czerny et al., 1999; Halder et al., 1995; Shen and Mardon, 1997; Weasner et al., 2007). These experiments have been critical to our understanding of the regulatory interactions that take place during nascent phases of eye development and within the anterior compartment of the developing retina. Several regulatory relationships, first established genetically, have been supported by evidence of protein-protein interactions and direct transcriptional regulatory relationships (Chen et al., 1999; Czerny et al., 1999; Michaut et al., 2003; Niimi et al., 1999; Ostrin et al., 2006; Pauli et al., 2005; Pignoni et al., 1997).
A distinct disadvantage to this historical approach is that interactions taking place along the margins, at the D/V and A/P boundaries, and in cells posterior to the furrow cannot be assessed and thus have largely been neglected. This is particularly true of so, eya and dac, which are the only three RD genes to be expressed posterior the furrow (Bonini et al., 1993; Cheyette et al., 1994; Mardon et al., 1994; Serikaku and O'Tousa, 1994). All three genes are required for furrow initiation and the So-Eya complex is required in the R1, R6 and R7 photoreceptors (Mardon et al., 1994; Pignoni et al., 1997). However it is unclear if the regulatory relationships existing among the three genes in anterior regions of the eye also exist along the posterior regions where pattern formation initiates and in differentiating photoreceptor neurons. In order to verify existing interactions or identify new regulatory relationships among so, eya and dac, we generated randomly distributed retinal mosaic clones for each gene and determined the effect that loss of each gene had on the expression of the other two factors.
Here we show that the response of the eye to discontinuities in the retinal determination network is not static across the eye field but rather is dynamic and position dependent. In particular we demonstrate that, unlike regions anterior the furrow, removal of so and eya in posterior positions of the eye lead to an attempt by these cells to reinitiate the retinal determination program by expressing RD genes that are normally found exclusively in the anterior compartment. This attempt fails and is then followed by cell suicide via programmed cell death but not before the so and eya mutant cells non-autonomously signal through the Notch pathway to adjacent undifferentiated cells instructing them to compensate for their loss by activating dac expression and proliferating. These surrounding cells, which are not competent to properly execute the RD program neither adopt a retinal fate nor die, therefore they assume a default head cuticle fate. We also demonstrate that the loss of either so or eya at the margins of the eye epithelium results in a different developmental path. In these cases, the mutant cells themselves will autonomously overproliferate thereby bypassing any requirement for communication with adjacent cell populations. Consistent with this, the adjacent undifferentiated cells do not activate Notch signaling, express dac or proliferate. The conclusion that we draw from these observations is that the gene regulatory networks governing early specification and patterning decisions are not static sets of connections but rather are temporally and spatially dynamic.
Materials and Methods
Fly Stocks
The following stocks were used to generate retinal mosaic clones: w; FRT40A dacE462 (gift from Graeme Mardon), w; FRT42D so3 (gift from Francesca Pignoni), w; FRT42D eya2, and w;; FRT82B groE48 (gift from Janice Fischer) with the following FRT lines: w; FRT40A Ubi-GFP, w; FRT 40A Pw+, w; FRT42D Ubi-GFP, w; FRT42D Pw+, w;; FRT82B Ubi-GFP RpS3 and yweyflp. The following stocks were used for generalized and flpout over-expression assays; ey-G4, GMR-G4, ywhsflp22, act5C>yellow>GAL4, in conjunction with UAS-so/CyO, UAS-gro (gift from Albert Courey), UAS-dac (gift from Graeme Mardon), UAS-hid (gift from Andreas Bergmann), and UAS-NICD (gift from Sujin Bao). Unless noted otherwise, the above stocks are available from the Bloomington Drosophila Stock Center.
Reagents
The following primary antibodies were used in this study: mouse anti-Dac (1:5), mouse anti-Eya (1:5), rat anti-Elav (1:10), mouse anti-2B10 (1:100), mouse anti-Delta (1:10), and mouse anti-NotchICD (1:4) all of which are available from the Developmental Studies Hybridoma Bank. The guinea pig anti-So (1:200), rabbit anti-Hth (1:500) and rabbit anti-Tsh (1:3000) antibodies are kind gifts from Ilaria Rebay, Richard Mann and Steve Cohen, respectively. The rabbit anti-Cleaved Caspase-3 (1:100) antibody is from Cell Signaling Technology and Phalloidin-Cy5 (1:1000) is from Molecular probes. The following secondary antibodies from Jackson Laboratories were used in this study at 1:100 dilutions: goat anti-mouse TRITC, donkey anti-mouse TRITC, goat anti-rat TRITC, donkey anti-rat TRITC, donkey anti-rat CY5, goat anti-rabbit TRITC, goat anti-guinea pig TRITC.
Microscopy
Imaginal discs were dissected in phosphate buffer, fixed in 4% paraformaldehyde, washed in wash buffer (0.2% Triton), and then incubated in primary antibody overnight. Secondary antibody incubations lasted 2–3 hours after which tissues were further dissected in wash buffer then mounted on slides in Vectashield (Vector Laboratories). Tissues were examined using a Zeiss Axioplan2 with Apotome and imaged using a Zeiss Axiocam MRm camera. Adult flies with clones were either imaged live using a Nikon SMZ1500 microscope equipped for fluorescence, frozen at −80°C for 20 minutes then imaged using Zeiss Discovery scope with color camera, or prepared for SEM by drying through a series of ethanol dilutions and HMDS treatments.
Results
Dac is Repressed by So-Eya in the Posteror Eye
Based on the expression profiles of the eye specification genes, the developing eye can be divided into five zones (Fig. 1A, (Bessa et al., 2002; Bonini et al., 1993; Cheyette et al., 1994; Czerny et al., 1999; Mardon et al., 1994; Pan and Rubin, 1998; Quiring et al., 1994; Seimiya and Gehring, 2000; Serikaku and O'Tousa, 1994). The cells that constitute zone 1 reside at the most anterior regions of the eye and express the two eye specification genes, tsh and hth, in addition to cut, which antagonizes the retinal determination pathway. Zone 2 is located adjacent to zone 1 and extends to the morphogenetic furrow. Every eye specification gene, with the exception of hth, is expressed within this region. Zone 3 lies posterior to the furrow and essentially is defined by the expression pattern of dac. Additionally, so and eya, are expressed in these cells. Zone 4 begins where dac expressions ceases and extends to the posterior edge of the eye field. Finally, the posterior-lateral margins of the eye field constitute zone 5. Two eye specific enhancers within dac are under the partial control of the so and eya (Fig. 1B; Ostrin et al., 2006), however, dac expression is activated in only a subset of cells that contain the So-Eya transcription factor (Fig. 1C–E; (Bonini et al., 1993; Cheyette et al., 1994; Mardon et al., 1994; Serikaku and O'Tousa, 1994). We generated retinal mosaic clones mutant for either so, eya or dac with the intent of understanding the complexity of their regulatory relationship throughout the five defined expression zones of the developing eye.
Figure 1. Spatial and Temporal Expression of the RD Network in the Fly Eye.
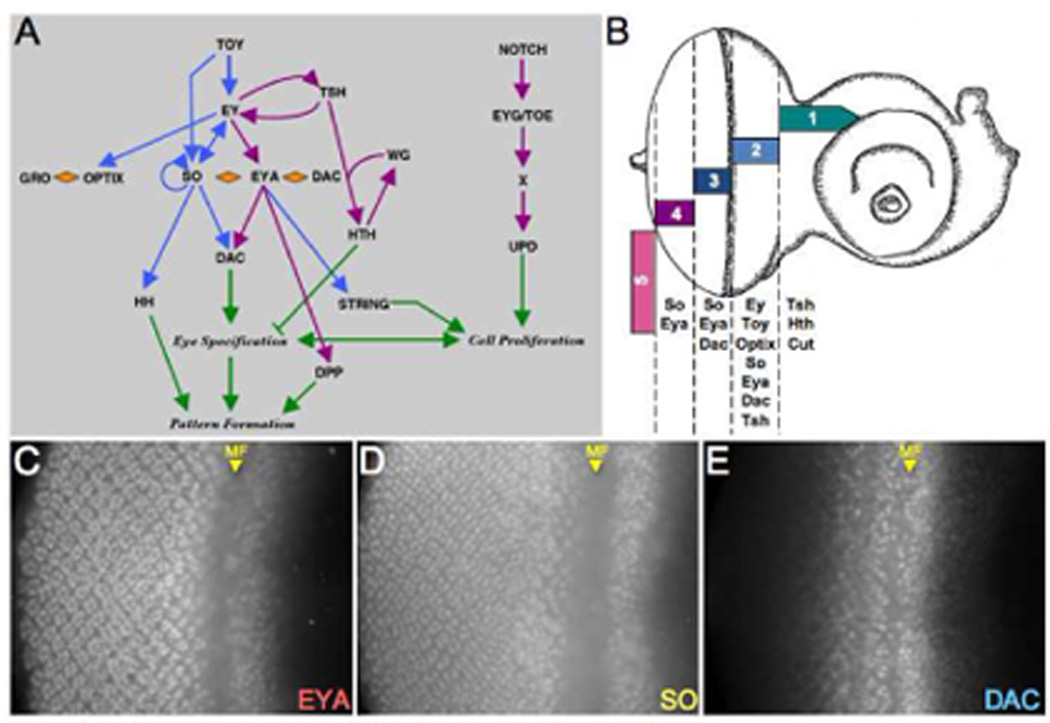
(A) Schematic diagram of the retinal determination network. (B) Schematic drawing of a third instar eye-antennal imaginal disc. The expression zones are modeled after the data presented in Bessa et al., 2002. (C–E) Immunofluorescence images of wild type eye discs. Immunostained proteins are at listed at the bottom right of each panel. MF = morphogenetic furrow. Anterior is to the right.
A prediction of the current RD network model is that removal of either so or eya should result in the cessation of dac expression. As expected, in regions of the eye where Dac protein is normally distributed (zones 2 and 3) removal of either so and eya leads to a halt in dac expression (Fig. 2A,B, blue arrow). This is consistent with earlier reports on whole mutant discs in which dac expression is drastically reduced but not eliminated from so1 and eya2 mutants (Anderson et al., 2006; Pappu et al., 2005). However, removal of either gene within zone 4 has the effect of non-autonomously activating dac expression in cells surrounding the mutant patch (Fig. 2A,B, purple arrow). This is surprising as our a priori expectation was that the loss of either so or eya in non-dac expressing cells would have no appreciable effect on Dac transcription. Our results indicate that, contrary to such expectations, so and eya function early to first activate dac expression but then later reverse course to suppress it (see model in Fig. 8C). We also find that cells along the margin react differently to the loss of So-Eya activity than the nearby cells that populate the interior of the retina. Mutant clones at the margins (zone 5) are unable to induce dac expression in neighboring cells (Fig. 2A,B, pink arrow).
Figure 2. Position Dependency of the So-Eya-Dac Regulatory Relationship.
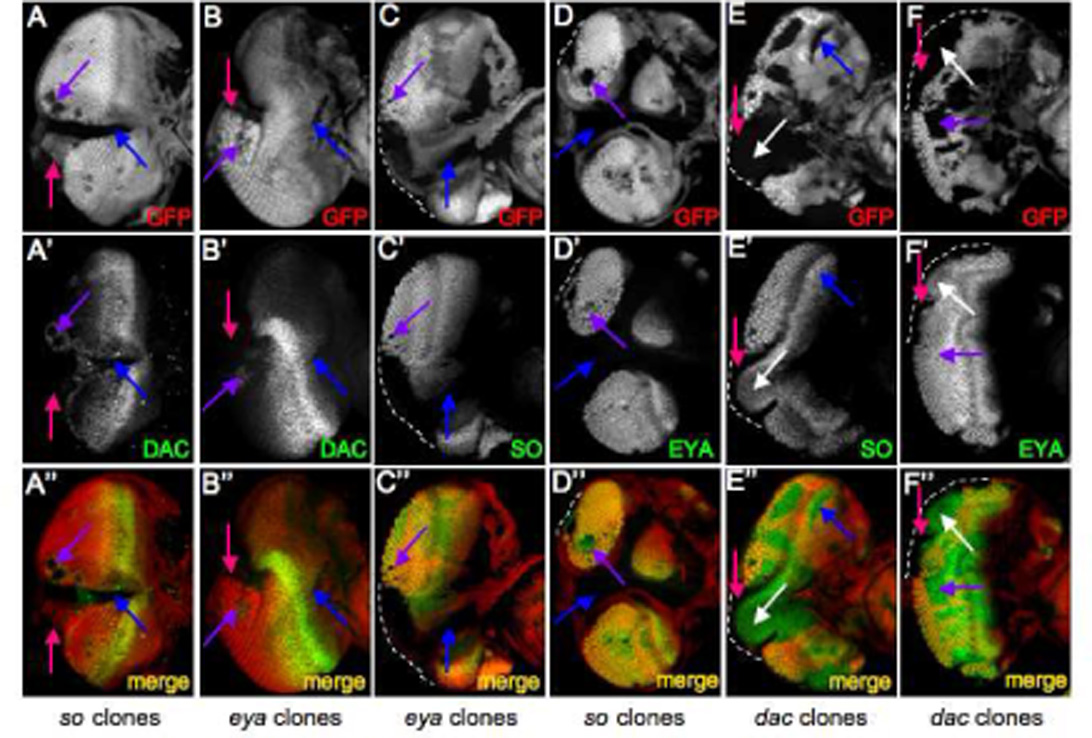
(A–F) Immunofluorescence images of eye discs containing either so3, eya2 or dac4 mutant clones. Immunostained proteins are at listed at the bottom right of each panel. GFP is labeled red and RD proteins are labeled green. Genotypes are listed at the bottom of each column. Anterior is to the right.
As dac expression is under the control of both so and eya (Anderson et al., 2006; Pappu et al., 2005), we set out to determine if the de-repression of dac in zone 4 is primarily due to either so, eya, or both genes equally. We generated loss of function clones mutant for either so or eya and examined the distribution pattern of the other protein. We observe that loss of eya always results in the elimination of so expression, irrespective of geographical location within the eye field (Fig. 2C, blue, purple arrows). In contrast, there is only a reciprocal requirement of so for eya expression within zone 2 and zone 5 (Fig. 2D, blue arrow). It should be noted that when large clones span the furrow, the cells respond as zone 2 cells. In so mutant cells located strictly posterior to the furrow (zones 3 and 4), Eya protein levels appear to be unaffected (Fig. 2D, purple arrow). Thus it appears that in zone 4 the ectopic activation of dac results primarily from the loss of so.
dac has been shown to be regulated by dpp prior to the initiation of the morphogenetic furrow (Curtiss and Mlodzik, 2000). Loss of either dpp or dac at the posterior margin (zone 5) inhibits the initiation of the morphogenetic furrow (Chanut and Heberlein, 1997; Mardon et al., 1994). In addition, loss of either so or eya in zone 5 also leads to an arrest in pattern formation (Pignoni and Zipursky, 1997). We sought to determine if so, eya and dac function at the same level or if a feedback step from dac to so and eya exists during furrow initiation. We generated dac mutant clones and observed that both so and eya are absent in a narrow strip of cells with the mutant patches that contact zone 5 (Figs, 2E,F, pink arrow) indicating that at least at the point of initiation dac does, in fact, feedback and regulate the expression of both so and eya. Interestingly, both So and Eya proteins are still present within the center of the mutant patches suggesting that these cells may still retain their anterior fate (Fig. 2E,F, white arrow). We also observed that removal of dac in cells completely within the eye disc proper appears to have no detrimental effect on the expression of either so or eya (Fig. 2E,F, blue, purple arrows). From these results we draw the conclusion that the regulatory relationships that exist among the so, eya and dac genes is dynamic across the developing eye field. We also propose that the differential requirement for dac in furrow initiation and progression (Mardon et al., 1994) may be the result of dac regulating so and eya at the margin but not within the eye field.
So-Eya Mutant Cells Reinitiate the RD program
As the So-Eya complex regulates dac expression we turned our attention to zone 4 where the loss of either gene unexpectedly leads to the non-autonomous de-repression of dac in cells bordering the mutant patch (Fig. 3A). We stained clones with antibodies against a number of molecular markers in an attempt to determine the molecular identity of the dac expressing cells bordering the clones and the mutant cells themselves. The cells experiencing the non-autonomous de-repression of dac, express neither the neuron specific protein ELAV nor the cone cell marker Cut (Fig B,C). These cells are also unlikely to be pigment cells, as this cellular fate is not assigned until the pupal stage (Cagan and Ready, 1989; Wolff and Ready, 1993). Therefore, these cells most probably belong to the undifferentiated pool of cells that are present within the developing eye disc at this stage. Consistent with earlier reports on the role that so and eya play in cell fate decisions (Pignoni et al., 1997), cells mutant for so fail to express neuronal and cone cell specific markers (Fig 3B,C). We wanted to determine the developmental paths that these two cell populations follow.
Figure 3. Molecular Signature of so Mutant Retinal Clones.
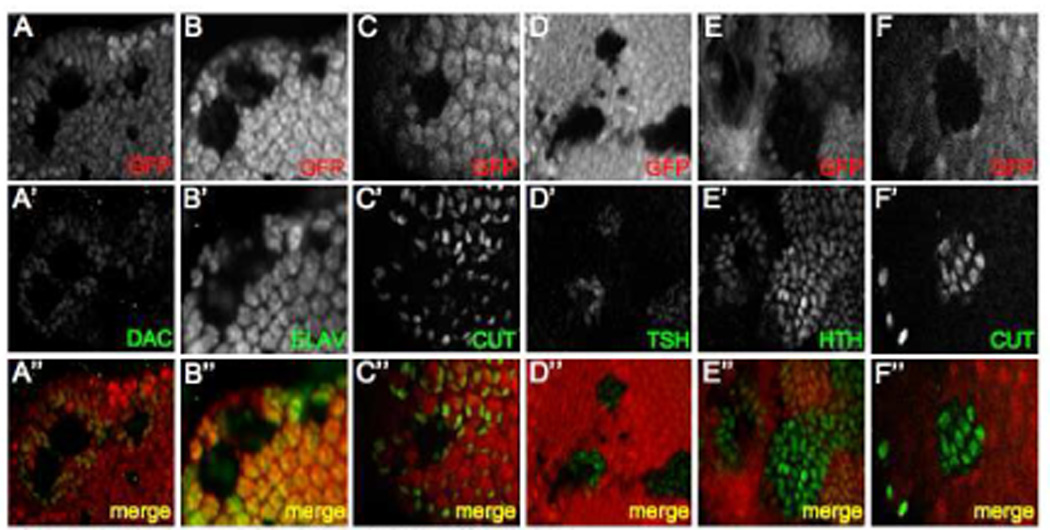
(A–F) Immunofluorescence images of so3 mutant clones residing within zone 4. Immunostained proteins are at listed at the bottom right of each panel. GFP is labeled red and indicated RD proteins are labeled green. Anterior is to the right.
In order to determine the developmental path of so mutant cells and neighboring dac expressing cells, we used antibodies against several proteins that are expressed in zones 1 and 2 (Fig. 1B). Our analysis of Tsh, Hth and Cut distribution is that so and eya mutant cells express each of these genes while the surrounding cells do not (Fig. 3D–F). It should be noted here that the Cut protein that is shown within Fig. 3C is in cells that are not within the cone cell layer. Rather, the cells are at a more basal position, consistent with the position of the cut positive cells located ahead of the furrow and within the antenna in zone 1. As Tsh is normally present in zone 2 and both Hth and Cut are distributed in zone 1, it appears that cells mutant for either RD gene adopt an anterior fate that approximates but does not exactly recreate the molecular environment ahead of the furrow. Interestingly, while Dac, So and Eya are present within the undifferentiated cells surrounding the clones (Fig. 3A, data not shown) they do not express other anterior genes. We conclude that the two cell populations are molecularly distinct with the mutant cells adopting a hybrid zone 1–2 identity and the surrounding cells assuming a fate that is most closely associated with zone 3. What is the ultimate fate of these two cell populations in the eye?
So-Eya Mutant Cells Commit Suicide
We noticed that so clones residing in zone 4 are typically smaller than those that are located in more anterior zones. Since recombination is induced early in eye development, the difference in clone size, several days later, could be attributed to programmed cell death. We stained so and eya mutant discs with an antibody that recognizes activated cleaved Caspase-3 (Cas-3), a marker for cells that are undergoing apoptosis. We observe that cells mutant for so are, in fact, undergoing cell death (Fig. 4A). This was an unexpected finding as we often observe cuticular outgrowths emanating from the adult compound eye (Fig. 4B). Our initial assumption was that these outgrowths were descended from the so and eya mutant clones. However, the elevated levels of Cas-3 in the clone coupled with its absence in the surrounding cells suggests that the outgrowths are derived from another pool of cells. We attempted to determine which cell population gives rise to the cuticular outgrowths. Normally, when clones are generated and assayed in the adult retina, the mutant tissue is identified by the absence of red pigment, which results from the loss of a mini-white construct during recombination. However, since the cells of interest are comprised of head cuticle and not retinal tissue, eye pigmentation is not a useful marker. Instead we made use of a GFP reporter that is under the control of the ubiquitously activated Ubi-63E promoter, thus, as in the eye disc, the presence of GFP can be used to mark wild type tissue in the adult (compare Fig. 4C–F). The cuticular outgrowths in adult flies express GFP indicating that they are wild type. As the undifferentiated, Dac expressing cells surrounding so clones are the only cells in the tissue that have not yet adopted a retinal fate, we believe that it is this population that gives rise to the cuticular outgrowths (Fig. 4G,H). This is consistent with the elevated levels of Cas-3 in mutant clones and also consistent with the fact that the dac positive cells are as yet undifferentiated. We next sought to determine whether decisions to proliferate and express dac are a response to transmitted signals from the mutant tissue to the surrounding cell population.
Figure 4. Fate of so and eya Mutant Tissue in the Developing and Adult Retina.
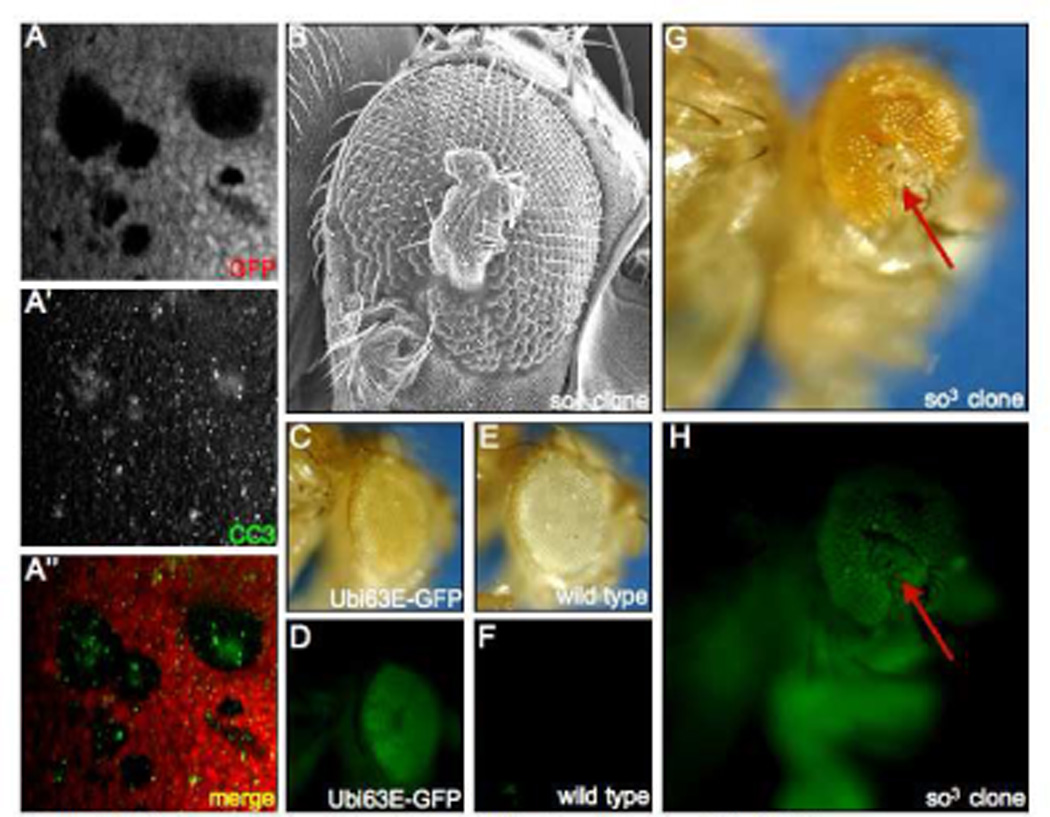
(A) Immunofluorescence images of a so3 mutant clone. Immunostained cleaved Caspase-3 is labeled green and GFP is labeled red. (B). Scanning electron micrograph of a cuticular outgrowth emanating from the compound eye. (C,D) Light and fluorescent images of an adult fly harboring a GFP reporter under the control of the Ubi63E promoter. Note that GFP is expressed in both the retina and the surrounding cuticle. (E,F) Light and fluorescent images of a fly lacking the Ubi63E-GFP transgene. Note that GFP expression is not observed. (G, H) Light and fluorescent images of an adult fly harboring so3 mutant clones. Red arrows point to cuticular outgrowth from the eye. Note the presence of GFP in the cuticular outgrowth. Genotypes are listed at the bottom right of each panel. Anterior is to the right.
So-Eya Mutant Tissue Instruct Surrounding Cells Via Delta-Notch
Our expectation is that the two populations of cells do, indeed, communicate with each other. As the Notch signaling pathway plays roles in both eye specification (Kumar and Moses, 2001a; Kurata et al., 2000) and cell proliferation (Baonza and Freeman, 2005; Chao et al., 2004; Dominguez et al., 2004; Ferres-Marco et al., 2006; Reynolds-Kenneally and Mlodzik, 2005; Singh et al., 2006) we stained discs containing either so mutant clones with antibodies against the intracellular domain of the Notch receptor (NICD) and the ligand, Delta (Dl). We observe an up-regulation of Dl within the mutant tissue (Fig. 5A) and activated Notch in the surrounding, undifferentiated dac positive cells (Fig. 5B). These results suggest that cells mutant for the So-Eya complex, prior to their ultimate demise, signal to the surrounding undifferentiated cells via Notch signaling. This signaling has the effect of inducing compensatory proliferation and dac expression in surrounding cells. As Notch signaling has already been shown to induce cell proliferation when ectopically expressed (Reynolds-Kenneally and Mlodzik, 2005) we set out to determine if ectopic Notch signaling posterior to the furrow was sufficient, on its own to induce dac expression. We used the "flp-out" technique to over-express NICD in clones within zone 4. We observe the expression of NICD was indeed sufficient to induce dac expression both autonomous and non-autonomously (Fig. 5C). We then looked to see if the compensatory proliferation that we observe is mediated by dac or if it is a distinct effect of Notch signaling. Expression of dac either in flp-out clones or using a GMR-GAL4 driver in zones 3 and 4 failed to induce cell proliferation (data not shown). We conclude from these results that the Delta-Notch signal from cells mutant for either so or eya is sufficient to induce both dac expression and cell proliferation but that these two outputs are separable and thus distinct from each other.
Figure 5. so Mutant Tissue Signals to Surrounding Cells via the Notch Pathway.
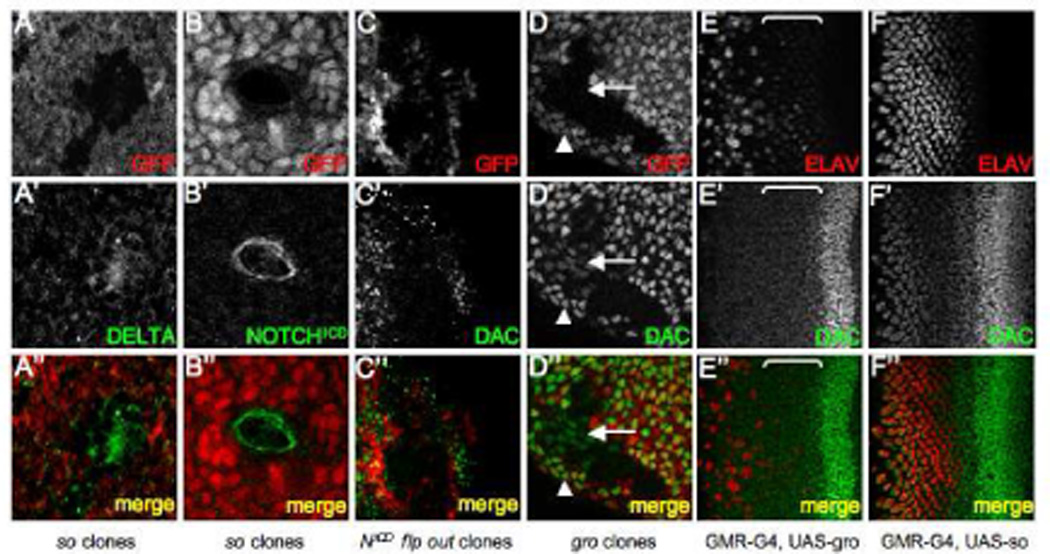
(A–D) Immunofluorescence images of eye discs containing so3 mutant, NICD flp-out, or gro mutant clones. All clones are positioned in zone 4. GFP is labeled in red. Immunostained proteins are in green. (E–F) Immunofluorescence images of eye discs in which GRO or SO is expressed behind the morphogenetic furrow via a GMR-GAL4 driver. ELAV is labeled red and DAC is green. The brackets in panels E indicate zone 3. Genotypes are listed at the bottom of each column. Anterior is to the right.
Groucho Represses dac Expression in the Retina
Our results suggest that dac expression is repressed in more posterior regions (zone 4) of the retina by a complex containing So and an unidentified transcriptional co-repressor. One candidate, groucho (gro), is a member of the TLE family of transcriptional repressors (Fisher and Caudy, 1998; Parkhurst, 1998; Paroush et al., 1994; Stifani et al., 1992). Gro functions within the Enhancer of split complex (Delidakis et al., 1991; Ziemer et al., 1988) whose products are downstream components of the Notch signaling cascade (reviewed in (Artavanis-Tsakonas and Simpson, 1991; Campos-Ortega, 1993; Campos-Ortega and Jan, 1991). The main role for gro in the eye appears to be in the specification of photoreceptor cell fates although its loss can also be accompanied by extensive overproliferation of imaginal disc tissue (Chanut et al., 2000; Fischer-Vize et al., 1992). Additionally, So and other members of the SIX family form composite transcriptional repressors with Gro and its vertebrate homologs (Kenyon et al., 2005; Kobayashi et al., 2001; Lopez-Rios et al., 2003; Silver et al., 2003; Zhu et al., 2002). We generated mutant clones of gro and observed that dac expression was ectopically activated in zone 4 cells surrounding the clone as well as in some, but not all, cells within the clone (Fig. 5D arrowhead, arrow). Therefore it is possible that Gro functions both in a cell autonomous and a cell non-autonomous manner in zone 4 to repress dac transcriptional activation in both photoreceptor neurons and undifferentiated cells (Fig. 7B).
Figure 7. Models for So-Eya-Dac Regulation.
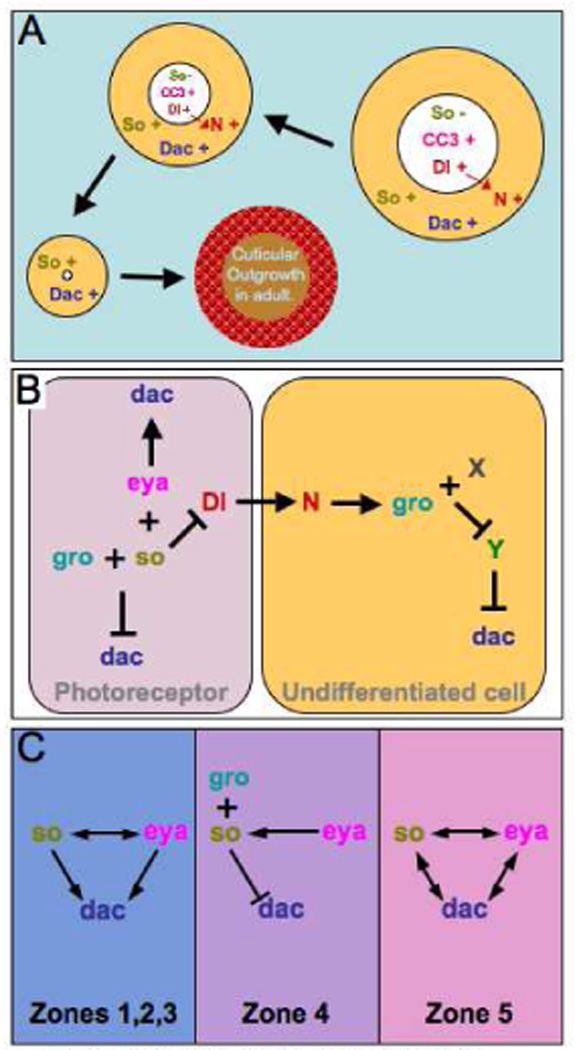
(A). Schematic of the events observed in so or eya clones located in zone 4 of the developing eye disc. Note that the mutant tissue in the disc undergoes programmed cell death and is eliminated by the adult stage and is denoted by the shrinking size of the clone. The cuticular outgrowth in the adult is derived from the dac positive undifferentiated cells that surround the mutant clones in the eye disc. (B) A schematic depicting a potential mechanism for the repression of dac in photoreceptor neurons and undifferentiated cells in zone 4 of the retina. (C) A series of models describing the genetic interactions that we observe between so, eya and dac in the developing retina. Note that the relationships change depending upon the spatial orientation within the eye field.
To test whether gro is sufficient to repress dac transcription we used a GMR-GAL4 driver (Hay et al., 1994) to over-express it in all cells behind the furrow. Our expectation was that if Gro is able to repress dac transcription, we should see Dac protein levels drop immediately behind the furrow zone 3. As expected, we see a reduction within the photoreceptors that normally express dac in this region (Fig. 5E, bracket). However, more strikingly, we observe ectopic Dac expression in all undifferentiated of cells behind the furrow (Fig. 5E Note the absence of ELAV stain in Dac positive nuclei).These data suggest that Gro overexpression influences Dac expression differently in photoreceptors vs. undifferentiated cells. Interestingly, over-expression of so using GMR-GAL4 results in an increase in dac expression predominantly in a different cell population; namely the developing photoreceptor neurons and accessory cone cells (Fig. 5F Note that nuclei ectopically expressing Dac also are ELAV positive). These results suggest that the So-Eya and So-Gro complexes play critical and complex roles in regulating dac expression (Fig. 7A,B).
Distinct Responses to RD Gene Loss at the Eye Field Margin
It had been shown previously that loss of particular RD genes along the margins of the eye field not only resulted in a block in pattern formation but also induced cell proliferation (Mardon et al., 1994). We set out to determine if the mechanisms underlying cell proliferation that accompanies RD gene loss along the margin (zone 5) are similar to or distinct from those governing internal regions (zone 4). Similar to the loss of dac, removal of so and/or eya along the margins leads to a block in the initiation of pattern formation (Fig. 2A,B; (Pignoni et al., 1997) and to an increase in cell proliferation (Fig. 6A). It appears that the mutant cells are themselves proliferating as the cuticular outgrowths that we see along the margins of the adult eye are derived from the mutant tissue as these cells do not express the GFP reporter that we use for marking wild type cells (Fig. 6D,E; compare arrow to arrowhead). This result suggests that, unlike zone 4, mutant cells along the margin (zone 5) proliferate autonomously. Consequently, we do not see upregulation of either Dl within the clone or Notch in adjacent cells or Cas-3 within the clone (data not shown). It should be noted here that the posterior-lateral margin of the retinal field appears to be the only location within the eye disc in which dac feeds back and positively regulates that expression of both so and eya (Fig. 2E,F; 7C).
Figure 6. Autonomous growth in so clones on the margin.
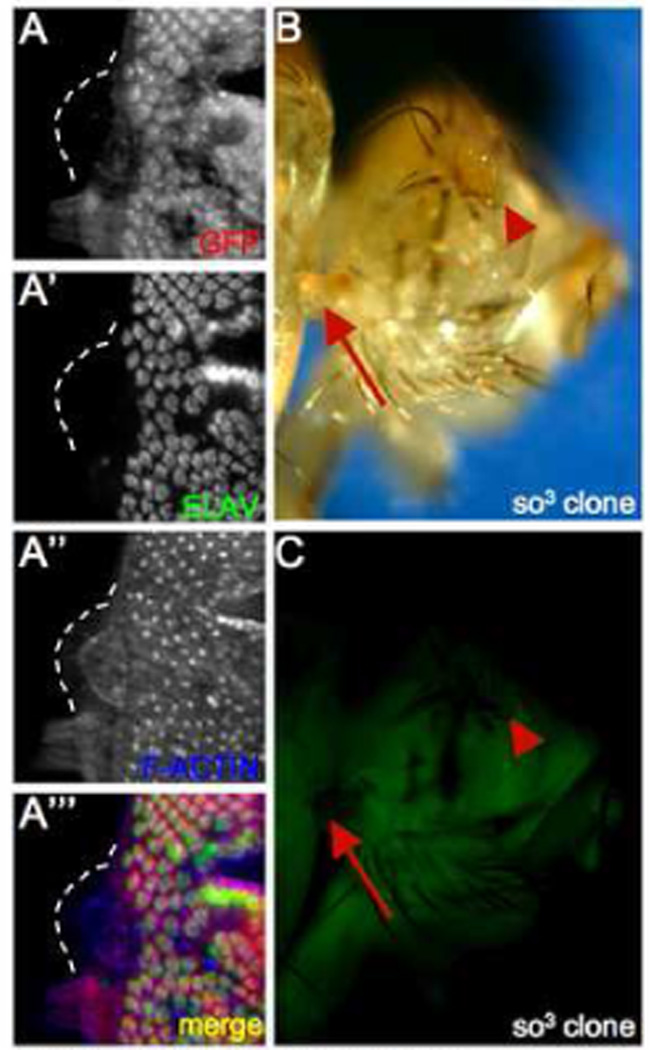
(A) Immunofluorescence images of a so3 mutant clone. GFP is labeled red, immunostained ELAV is labeled green and F-actin is labeled blue. (B, C) Light and fluorescent images of an adult fly with so3 mutant clones. Red arrows point to cuticular outgrowth from the head, red arrowheads point to the eye. Note the absence of GFP in the cuticular outgrowth. Genotypes are listed at the bottom right of each panel. Anterior is to the right.
Discussion
Here we have demonstrated that the so-eya-dac sub circuit of the retinal determination gene regulatory network is dynamically organized across the developing eye field. First, along the margins of the field (Fig. 1B, zone 5), where pattern formation is initiated, positive feedback loops link all three genes to each other (Fig. 7C, right panel). The So-Eya composite transcription factor is thought to partially regulate dac expression through two eye specific enhancers that contain putative So binding sites suggesting that the regulation may be direct (Pappu et al., 2005). Dac has been shown recently to also contact DNA (Kim et al., 2002): however, a consensus binding site has not been identified so it is unclear if the regulation of so and eya by dac is direct or goes though other RD gene intermediates such as ey. As the furrow sweeps across the epithelium the once vast pool of undifferentiated cells are canalized towards a retinal fate: cells in the most anterior regions are the least committed while cells in progressively posterior regions are funneled along a path of terminal differentiation. In regions straddling the advancing morphogenetic furrow (Fig. 1B, zones 2 and 3), where competent, undifferentiated cells are transformed into periodic clusters of photoreceptors, the character of the so-eya-dac sub circuit is altered and becomes unidirectional with So-Eya maintaining its regulation of dac but not visa versa (Fig. 7C, middle panel). In the most posterior portions of the eye (Fig. 1B, zone 4), where retinal cell fate differentiation is complete, the So-Eya complex appears to no longer activate dac transcription. Instead, a complex most likely composed of So and the Gro transcriptional co-repressor (So-Gro) represses dac (Fig. 7C, middle). Our interpretation of the results presented here is that static gene regulatory network maps, in many instances, may not accurately represent the shifting alliances amongst genes during development. It is more likely the case that relationships amongst genes will differ depending upon temporal and/or spatial context.
We also describe a second phenomenon in which cells that suffer from interruptions in the RD gene regulatory network appear to actively attempt several corrections. First, mutant cells will attempt to restart the retinal determination program by activating all of the RD genes that are normally expressed in more anterior segments of the eye primordium. As this attempt proves futile, the mutant cells, in a second attempt to correct this deficiency, will then communicate to adjacent undifferentiated cells via Delta-Notch signaling instructing them to both proliferate and attempt an initiation of retinal development. Coupled to this effort is a decision to commit cell suicide via programmed cell death, which clears the defective cells from the developing retina (Fig. 7A). We interpret these findings to suggest that programmed cell death may not, as is often implied, be the first option for cells that are genetically or molecularly compromised. However, it appears that cells mutant for components of gene regulatory networks go to considerable lengths to self-correct prior to their eventual decision to eliminate themselves from the developing tissue. While our analysis is limited to mutations within three different RD network genes, we suggest that this mechanism may not be limited but be a rather common response in other developmental contexts.
And finally our results suggest that within a single cell different transcriptional complexes, involving one common component, in this case a DNA binding protein, and at least two different co-factors, one a transcriptional co-activator and one a transcriptional co-repressor, can both regulate the transcription of a target gene. We have demonstrated that So, Eya and Gro are co-expressed in developing photoreceptor cells. However, in newly differentiated cells just adjacent to the morphogenetic furrow the So-Eya complex activates dac transcription. As these cells mature, a So-Gro repression complex shuts down expression of dac despite the continued presence of the Eya co-activator. We have also shown that hyper-expression of either so or gro affects dac expression in very different cell types although both genes were expressed uniformly. It is likely that So-Eya and So-Gro exist as independent complexes and not as a larger super complex as it has been possible to biochemically isolate just the individual heterodimers (Jemc and Rebay, 2007; Silver et al., 2003). How spatial and temporal information is integrated into the promoter to differentially recruit these differing activation and repression complexes is unclear but is also likely to be important for shaping gene regulatory networks.
Acknowledgements
We would like to thank the following colleagues for their generosity with reagents and equipment: Sujin Bao, Andreas Bergman, Steve Cohen, Albery Courey, Janice Fischer, Thom Kaufman, Richard Mann, Graeme Mardon, Francesca Pignoni, and Ilaria Rebay. We would like to thank Stacy Holtzman for technical assistance with microscopy and Rudy Turner for Scanning Electron Micrograph images. Also, we thank Shera Lesly for discussions on proliferation and shared protocols. Claire L. Salzer is supported through the NIH Molecular Biology and Genetics Training Grant. Justin P. Kumar is supported by a grant from the National Eye Institute (R01 E401463).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson J, Salzer CL, Kumar JP. Regulation of the retinal determination gene dachshund in the embryonic head and developing eye of Drosophila. Dev Biol. 2006;297:536–549. doi: 10.1016/j.ydbio.2006.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aracena J, Gonzalez M, Zuniga A, Mendez MA, Cambiazo V. Regulatory network for cell shape changes during Drosophila ventral furrow formation. J Theor Biol. 2006;239:49–62. doi: 10.1016/j.jtbi.2005.07.011. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Simpson P. Choosing a cell fate: a view from the Notch locus. Trends Genet. 1991;7:403–408. doi: 10.1016/0168-9525(91)90264-q. [DOI] [PubMed] [Google Scholar]
- Bai J, Montell D. Eyes absent, a key repressor of polar cell fate during Drosophila oogenesis. Development. 2002;129:5377–5388. doi: 10.1242/dev.00115. [DOI] [PubMed] [Google Scholar]
- Baonza A, Freeman M. Control of cell proliferation in the Drosophila eye by Notch signaling. Dev Cell. 2005;8:529–539. doi: 10.1016/j.devcel.2005.01.019. [DOI] [PubMed] [Google Scholar]
- Bessa J, Gebelein B, Pichaud F, Casares F, Mann RS. Combinatorial control of Drosophila eye development by eyeless, homothorax, and teashirt. Genes Dev. 2002;16:2415–2427. doi: 10.1101/gad.1009002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonini NM, Bui QL, Gray-Board GL, Warrick JM. The Drosophila eyes absent gene directs ectopic eye formation in a pathway conserved between flies and vertebrates. Development. 1997;124:4819–4826. doi: 10.1242/dev.124.23.4819. [DOI] [PubMed] [Google Scholar]
- Bonini NM, Leiserson WM, Benzer S. The eyes absent gene: genetic control of cell survival and differentiation in the developing Drosophila eye. Cell. 1993;72:379–395. doi: 10.1016/0092-8674(93)90115-7. [DOI] [PubMed] [Google Scholar]
- Bonini NM, Leiserson WM, Benzer S. Multiple roles of the eyes absent gene in Drosophila. Dev. Biol. 1998;196:42–57. doi: 10.1006/dbio.1997.8845. [DOI] [PubMed] [Google Scholar]
- Brodbeck S, Englert C. Genetic determination of nephrogenesis: the Pax/Eya/Six gene network. Pediatr Nephrol. 2004;19:249–255. doi: 10.1007/s00467-003-1374-z. [DOI] [PubMed] [Google Scholar]
- Cagan RL, Ready DF. The emergence of order in the Drosophila pupal retina. Dev. Biol. 1989;136:346–362. doi: 10.1016/0012-1606(89)90261-3. [DOI] [PubMed] [Google Scholar]
- Callaerts P, Leng S, Clements J, Benassayag C, Cribbs D, Kang YY, Walldorf U, Fischbach KF, Strauss R. Drosophila Pax-6/eyeless is essential for normal adult brain structure and function. J Neurobiol. 2001;46:73–88. doi: 10.1002/1097-4695(20010205)46:2<73::aid-neu10>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega JA. Mechanisms of early neurogenesis in Drosophila melanogaster. J Neurobiol. 1993;24:1305–1327. doi: 10.1002/neu.480241005. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega JA, Jan YN. Genetic and molecular bases of neurogenesis in Drosophila melanogaster. Annu Rev Neurosci. 1991;14:399–420. doi: 10.1146/annurev.ne.14.030191.002151. [DOI] [PubMed] [Google Scholar]
- Chang T, Shy D, Hartenstein V. Antagonistic relationship between Dpp and EGFR signaling in Drosophila head patterning. Dev Biol. 2003;263:103–113. doi: 10.1016/s0012-1606(03)00448-2. [DOI] [PubMed] [Google Scholar]
- Chanut F, Heberlein U. Role of decapentaplegic in initiation and progression of the morphogenetic furrow in the developing Drosophila retina. Development. 1997;124:559–567. doi: 10.1242/dev.124.2.559. [DOI] [PubMed] [Google Scholar]
- Chanut F, Luk A, Heberlein U. A screen for dominant modifiers of ro(Dom), a mutation that disrupts morphogenetic furrow progression in Drosophila, identifies groucho and hairless as regulators of atonal expression. Genetics. 2000;156:1203–1217. doi: 10.1093/genetics/156.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao JL, Tsai YC, Chiu SJ, Sun YH. Localized Notch signal acts through eyg and upd to promote global growth in Drosophila eye. Development. 2004;131:3839–3847. doi: 10.1242/dev.01258. [DOI] [PubMed] [Google Scholar]
- Chen R, Amoui M, Zhang Z, Mardon G. Dachshund and Eyes Absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell. 1997;91:893–903. doi: 10.1016/s0092-8674(00)80481-x. [DOI] [PubMed] [Google Scholar]
- Chen R, Halder G, Zhang Z, Mardon G. Signaling by the TGF-b homolog decapentaplegic functions reiteratively within the network of genes controlling retinal cell fate determination in Drosophila. Development. 1999;126:935–943. doi: 10.1242/dev.126.5.935. [DOI] [PubMed] [Google Scholar]
- Cheyette BN, Green PJ, Martin K, Garren H, Hartenstein V, Zipursky SL. The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron. 1994;12:977–996. doi: 10.1016/0896-6273(94)90308-5. [DOI] [PubMed] [Google Scholar]
- Cohen SM. In: Imaginal disc development. Bate M, Martinez Arias A, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 747–841. [Google Scholar]
- Curtiss J, Mlodzik M. Morphogenetic furrow initiation and progression during eye development in Drosophila: the roles of decapentaplegic, hedgehog and eyes absent. Development. 2000;127:1325–1336. doi: 10.1242/dev.127.6.1325. [DOI] [PubMed] [Google Scholar]
- Czerny T, Halder G, Kloter U, Souabni A, Gehring WJ, Busslinger M. twin of eyeless, a second Pax-6 gene of Drosophila, acts upstream of eyeless in the control of eye development. Mol Cell. 1999;3:297–307. doi: 10.1016/s1097-2765(00)80457-8. [DOI] [PubMed] [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, Otim O, Brown CT, Livi CB, Lee PY, Revilla R, Schilstra MJ, Clarke PJ, Rust AG, Pan Z, Arnone MI, Rowen L, Cameron RA, McClay DR, Hood L, Bolouri H. A provisional regulatory gene network for specification of endomesoderm in the sea urchin embryo. Dev Biol. 2002;246:162–190. doi: 10.1006/dbio.2002.0635. [DOI] [PubMed] [Google Scholar]
- Delidakis C, Preiss A, Hartley DA, Artavanis-Tsakonas S. Two genetically and molecularly distinct functions involved in early neurogenesis reside within the Enhancer of split locus of Drosophila melanogaster. Genetics. 1991;129:803–823. doi: 10.1093/genetics/129.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson B, Hafen E. Genetic dissection of eye development in Drosophila. In: Bate M, Martinez Arias A, editors. The Development of Drosophila melanogaster. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 1327–1361. [Google Scholar]
- Dominguez M, Ferres-Marco D, Gutierrez-Avino FJ, Speicher SA, Beneyto M. Growth and specification of the eye are controlled independently by Eyegone and Eyeless in Drosophila melanogaster. Nat Genet. 2004;36:31–39. doi: 10.1038/ng1281. [DOI] [PubMed] [Google Scholar]
- Doroquez DB, Rebay I. Signal integration during development: mechanisms of EGFR and Notch pathway function and cross-talk. Crit Rev Biochem Mol Biol. 2006;41:339–385. doi: 10.1080/10409230600914344. [DOI] [PubMed] [Google Scholar]
- Espinosa-Soto C, Padilla-Longoria P, Alvarez-Buylla ER. A gene regulatory network model for cell-fate determination during Arabidopsis thaliana flower development that is robust and recovers experimental gene expression profiles. Plant Cell. 2004;16:2923–2939. doi: 10.1105/tpc.104.021725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio JJ, Boyle M, DiNardo S. A somatic role for eyes absent (eya) and sine oculis (so) in Drosophila spermatocyte development. Dev Biol. 2003;258:117–128. doi: 10.1016/s0012-1606(03)00127-1. [DOI] [PubMed] [Google Scholar]
- Ferres-Marco D, Gutierrez-Garcia I, Vallejo DM, Bolivar J, Gutierrez-Avino FJ, Dominguez M. Epigenetic silencers and Notch collaborate to promote malignant tumours by Rb silencing. Nature. 2006;439:430–436. doi: 10.1038/nature04376. [DOI] [PubMed] [Google Scholar]
- Fischer-Vize JA, Vize PD, Rubin GM. A unique mutation in the Enhancer of split gene complex affects the fates of the mystery cells in the developing Drosophila eye. Development. 1992;115:89–101. doi: 10.1242/dev.115.1.89. [DOI] [PubMed] [Google Scholar]
- Fisher AL, Caudy M. Groucho proteins: transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes Dev. 1998;12:1931–1940. doi: 10.1101/gad.12.13.1931. [DOI] [PubMed] [Google Scholar]
- Flores GV, Duan H, Yan H, Nagaraj R, Fu W, Zou Y, Noll M, Banerjee U. Combinatorial signaling in the specification of unique cell fates. Cell. 2000;103:75–85. doi: 10.1016/s0092-8674(00)00106-9. [DOI] [PubMed] [Google Scholar]
- Gong KQ, Yallowitz AR, Sun H, Dressler GR, Wellik DM. A Hox-Eya-Pax complex regulates early kidney developmental gene expression. Mol Cell Biol. 2007;27:7661–7668. doi: 10.1128/MCB.00465-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guss KA, Nelson CE, Hudson A, Kraus ME, Carroll SB. Control of a genetic regulatory network by a selector gene. Science. 2001;292:1164–1167. doi: 10.1126/science.1058312. [DOI] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Flister S, Walldorf U, Kloter U, Gegring WJ. Eyeless initiates the expression of both sine oculis and eyes absent during drosophila compound eye development. Development. 1998;125:2181–2191. doi: 10.1242/dev.125.12.2181. [DOI] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by target expression of the eyeless gene in Drosophila. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- Hammond KL, Hanson IM, Brown AG, Lettice LA, Hill RE. Mammalian and Drosophila dachshund genes are related to the Ski proto-oncogene and are expressed in eye and limb. Mech Dev. 1998;74:121–131. doi: 10.1016/s0925-4773(98)00071-9. [DOI] [PubMed] [Google Scholar]
- Hanson IM. Mammalian homologues of the Drosophila eye specification genes. Semin Cell Dev Biol. 2001;12:475–484. doi: 10.1006/scdb.2001.0271. [DOI] [PubMed] [Google Scholar]
- Heanue TA, Reshef R, Davis RJ, Mardon G, Oliver G, Tomarev S, Lassar AB, Tabin CJ. Synergistic regulation of vertebrate muscle development by Dach2, Eya2, and Six1, homologs of genes required for Drosophila eye formation. Genes Dev. 1999;13:3231–3243. doi: 10.1101/gad.13.24.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang CC, Chao JL, Jones N, Yao LC, Bessarab DA, Kuo YM, Jun S, Desplan C, Beckendorf SK, Sun YH. Two Pax genes, eye gone and eyeless, act cooperatively in promoting Drosophila eye development. Development. 2003;130:2939–2951. doi: 10.1242/dev.00522. [DOI] [PubMed] [Google Scholar]
- Jemc J, Rebay I. Identification of transcriptional targets of the dual-function transcription factor/phosphatase eyes absent. Dev Biol. 2007;310:416–429. doi: 10.1016/j.ydbio.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalatzis V, Sahly I, El-Amraoui A, Petit C. Eya1 expression in the developing ear and kidney: towards the understanding of the pathogenesis of Branchio-Oto-Renal (BOR) syndrome. Dev Dyn. 1998;213:486–499. doi: 10.1002/(SICI)1097-0177(199812)213:4<486::AID-AJA13>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Kammermeier L, Leemans R, Hirth F, Flister S, Wenger U, Walldorf U, Gehring WJ, Reichert H. Differential expression and function of the Drosophila Pax6 genes eyeless and twin of eyeless in embryonic central nervous system development. Mech Dev. 2001;103:71–78. doi: 10.1016/s0925-4773(01)00328-8. [DOI] [PubMed] [Google Scholar]
- Kenyon KL, Li DJ, Clouser C, Tran S, Pignoni F. Fly SIX-type homeodomain proteins Sine oculis and Optix partner with different cofactors during eye development. Dev Dyn. 2005;234:497–504. doi: 10.1002/dvdy.20442. [DOI] [PubMed] [Google Scholar]
- Kim SS, Zhang RG, Braunstein SE, Joachimiak A, Cvekl A, Hegde RS. Structure of the retinal determination protein Dachshund reveals a DNA binding motif. Structure (Camb) 2002;10:787–795. doi: 10.1016/s0969-2126(02)00769-4. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Nishikawa K, Suzuki T, Yamamoto M. The homeobox protein Six3 interacts with the Groucho corepressor and acts as a transcriptional repressor in eye and forebrain formation. Dev Biol. 2001;232:315–326. doi: 10.1006/dbio.2001.0185. [DOI] [PubMed] [Google Scholar]
- Kumar JP, Moses K. Transcription factors in eye development: a georgeous mosaic? Genes & Dev. 1997;11:2023–2028. doi: 10.1101/gad.11.16.2023. [DOI] [PubMed] [Google Scholar]
- Kumar JP, Moses K. EGF receptor and Notch signaling act upstream of Eyeless/Pax6 to control eye specification. Cell. 2001a;104:687–697. doi: 10.1016/s0092-8674(01)00265-3. [DOI] [PubMed] [Google Scholar]
- Kumar JP, Moses K. Eye specification in Drosophila: perspectives and implications. Semin Cell Dev Biol. 2001b;12:469–474. doi: 10.1006/scdb.2001.0270. [DOI] [PubMed] [Google Scholar]
- Kurata S, Go MJ, Artavanis-Tsakonas S, Gehring WJ. Notch signaling and the determination of appendage identity. Proc Natl Acad Sci U S A. 2000;97:2117–2122. doi: 10.1073/pnas.040556497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurusu M, Nagao T, Walldorf U, Flister S, Gehring WJ, Furukubo- Tokunaga K. Genetic control of development of the mushroom bodies, the associative learning centers in the Drosophila brain, by the eyeless, twin of eyeless, and Dachshund genes. Proc Natl Acad Sci U S A. 2000;97:2140–2144. doi: 10.1073/pnas.040564497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laclef C, Souil E, Demignon J, Maire P. Thymus, kidney and craniofacial abnormalities in Six 1 deficient mice. Mech Dev. 2003;120:669–679. doi: 10.1016/s0925-4773(03)00065-0. [DOI] [PubMed] [Google Scholar]
- Li J, Liu ZJ, Pan YC, Liu Q, Fu X, Cooper NG, Li Y, Qiu M, Shi T. Regulatory module network of basic/helix-loop-helix transcription factors in mouse brain. Genome Biol. 2007;8:R244. doi: 10.1186/gb-2007-8-11-r244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loose M, Patient R. A genetic regulatory network for Xenopus mesendoderm formation. Dev Biol. 2004;271:467–478. doi: 10.1016/j.ydbio.2004.04.014. [DOI] [PubMed] [Google Scholar]
- Lopez-Rios J, Tessmar K, Loosli F, Wittbrodt J, Bovolenta P. Six3 and Six6 activity is modulated by members of the groucho family. Development. 2003;130:185–195. doi: 10.1242/dev.00185. [DOI] [PubMed] [Google Scholar]
- Mardon G, Solomon NM, Rubin GM. dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development. 1994;120:3473–3486. doi: 10.1242/dev.120.12.3473. [DOI] [PubMed] [Google Scholar]
- Medina KL, Pongubala JM, Reddy KL, Lancki DW, Dekoter R, Kieslinger M, Grosschedl R, Singh H. Assembling a gene regulatory network for specification of the B cell fate. Dev Cell. 2004;7:607–617. doi: 10.1016/j.devcel.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Michaut L, Flister S, Neeb M, White KP, Certa U, Gehring WJ. Analysis of the eye developmental pathway in Drosophila using DNA microarrays. Proc Natl Acad Sci U S A. 2003;100:4024–4029. doi: 10.1073/pnas.0630561100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj R, Banerjee U. Combinatorial signaling in the specification of primary pigment cells in the Drosophila eye. Development. 2007;134:825–831. doi: 10.1242/dev.02788. [DOI] [PubMed] [Google Scholar]
- Niimi T, Seimiya M, Kloter U, Flister S, Gehring WJ. Direct regulatory interaction of the eyeless protein with an eye-specific enhancer in the sine oculis gene during eye induction in Drosophila. Development. 1999;126:2253–2260. doi: 10.1242/dev.126.10.2253. [DOI] [PubMed] [Google Scholar]
- Noveen A, Daniel A, Hartenstein V. Early development of the Drosophila mushroom body: the roles of eyeless and dachshund. Development. 2000;127:3475–3488. doi: 10.1242/dev.127.16.3475. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Carrick DM, Davidson EH. A regulatory gene network that directs micromere specification in the sea urchin embryo. Dev Biol. 2002;246:209–228. doi: 10.1006/dbio.2002.0627. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Davidson EH. Gene regulatory network analysis in sea urchin embryos. Methods Cell Biol. 2004a;74:775–794. doi: 10.1016/s0091-679x(04)74032-7. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Davidson EH. Gene regulatory network controlling embryonic specification in the sea urchin. Curr Opin Genet Dev. 2004b;14:351–360. doi: 10.1016/j.gde.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Ostrin EJ, Li Y, Hoffman K, Liu J, Wang K, Zhang L, Mardon G, Chen R. Genome-wide identification of direct targets of the Drosophila retinal determination protein Eyeless. Genome Res. 2006;16:466–476. doi: 10.1101/gr.4673006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Rubin GM. Targeted expression of teashirt induces ectopic eyes in Drosophila. Proc Natl Acad Sci U S A. 1998;95:15508–15512. doi: 10.1073/pnas.95.26.15508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappu KS, Ostrin EJ, Middlebrooks BW, Sili BT, Chen R, Atkins MR, Gibbs R, Mardon G. Dual regulation and redundant function of two eye-specific enhancers of the Drosophila retinal determination gene dachshund. Development. 2005;132:2895–2905. doi: 10.1242/dev.01869. [DOI] [PubMed] [Google Scholar]
- Parkhurst SM. Groucho: making its Marx as a transcriptional co-repressor. Trends Genet. 1998;14:130–132. doi: 10.1016/s0168-9525(98)01407-3. [DOI] [PubMed] [Google Scholar]
- Paroush Z, Finley RL, Jr, Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- Pauli T, Seimiya M, Blanco J, Gehring WJ. Identification of functional sine oculis motifs in the autoregulatory element of its own gene, in the eyeless enhancer and in the signalling gene hedgehog. Development. 2005;132:2771–2782. doi: 10.1242/dev.01841. [DOI] [PubMed] [Google Scholar]
- Pignoni F, Hu B, Kenton HZ, Xiao J, Garrity PA, Zipursky SL. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell. 1997;91:881–891. doi: 10.1016/s0092-8674(00)80480-8. [DOI] [PubMed] [Google Scholar]
- Pignoni F, Zipursky SL. Induction of Drosophila eye development by Decapentaplegic. Development. 1997;124:271–278. doi: 10.1242/dev.124.2.271. [DOI] [PubMed] [Google Scholar]
- Quiring R, Walldorf U, Kloter U, Gehring WJ. Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans [see comments] Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- Ready DF, Hanson TE, Benzer S. Development of the Drosophila retina, a neurocrystalline lattice. Dev. Biol. 1976;53:217–240. doi: 10.1016/0012-1606(76)90225-6. [DOI] [PubMed] [Google Scholar]
- Relaix F, Buckingham M. From insect eye to vertebrate muscle: redeployment of a regulatory network. Genes Dev. 1999;13:3171–3178. doi: 10.1101/gad.13.24.3171. [DOI] [PubMed] [Google Scholar]
- Reynolds-Kenneally J, Mlodzik M. Notch signaling controls proliferation through cell-autonomous and non-autonomous mechanisms in the Drosophila eye. Dev Biol. 2005;285:38–48. doi: 10.1016/j.ydbio.2005.05.038. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell Biol. 2008a;9:557–568. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. Insights from a sea lamprey into the evolution of neural crest gene regulatory network. Biol Bull. 2008b;214:303–314. doi: 10.2307/25470671. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Meulemans D, Jones M, Bronner-Fraser M. Ancient evolutionary origin of the neural crest gene regulatory network. Dev Cell. 2007;13:405–420. doi: 10.1016/j.devcel.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Seimiya M, Gehring WJ. The Drosophila homeobox gene optix is capable of inducing ectopic eyes by an eyeless-independent mechanism. Development. 2000;127:1879–1886. doi: 10.1242/dev.127.9.1879. [DOI] [PubMed] [Google Scholar]
- Serikaku MA, O'Tousa JE. sine oculis is a homeobox gene required for Drosophila visual system development. Genetics. 1994;138:1137–1150. doi: 10.1093/genetics/138.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Mardon G. Ectopic eye development in Drosophila induced by directed dachshund expression. Development. 1997;124:45–52. doi: 10.1242/dev.124.1.45. [DOI] [PubMed] [Google Scholar]
- Silver SJ, Davies EL, Doyon L, Rebay I. Functional dissection of eyes absent reveals new modes of regulation within the retinal determination gene network. Mol Cell Biol. 2003;23:5989–5999. doi: 10.1128/MCB.23.17.5989-5999.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson TI, Price DJ. Pax6; a pleiotropic player in development. Bioessays. 2002;24:1041–1051. doi: 10.1002/bies.10174. [DOI] [PubMed] [Google Scholar]
- Singh A, Shi X, Choi KW. Lobe and Serrate are required for cell survival during early eye development in Drosophila. Development. 2006;133:4771–4781. doi: 10.1242/dev.02686. [DOI] [PubMed] [Google Scholar]
- Stifani S, Blaumueller CM, Redhead NJ, Hill RE, Artavanis-Tsakonas S. Human homologs of a Drosophila Enhancer of split gene product define a novel family of nuclear proteins. Nat Genet. 1992;2:119–127. doi: 10.1038/ng1092-119. [DOI] [PubMed] [Google Scholar]
- Tomlinson A, Ready DF. Cell fate in the Drosophila ommatidium. Dev. Biol. 1987a;123:264–275. doi: 10.1016/0012-1606(87)90448-9. [DOI] [PubMed] [Google Scholar]
- Tomlinson A, Ready DF. Neuronal differentiation in the Drosophila ommatidium. Dev. Biol. 1987b;120:366–376. doi: 10.1016/0012-1606(87)90239-9. [DOI] [PubMed] [Google Scholar]
- Treisman JE. A conserved blueprint for the eye? Bioessays. 1999;21:843–850. doi: 10.1002/(SICI)1521-1878(199910)21:10<843::AID-BIES6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Treisman JE, Heberlein U. Eye development in Drosophila: formation of the eye field and control of differentiation. Curr. Topics in Dev. Biol. 1998;39:119–157. doi: 10.1016/s0070-2153(08)60454-8. [DOI] [PubMed] [Google Scholar]
- Voas MG, Rebay I. Signal integration during development: Insights from the Drosophila eye. Dev Dyn. 2004;229:162–175. doi: 10.1002/dvdy.10449. [DOI] [PubMed] [Google Scholar]
- Wang XP, Suomalainen M, Felszeghy S, Zelarayan LC, Alonso MT, Plikus MV, Maas RL, Chuong CM, Schimmang T, Thesleff I. An integrated gene regulatory network controls stem cell proliferation in teeth. PLoS Biol. 2007;5:e159. doi: 10.1371/journal.pbio.0050159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weasner B, Anderson J, Kumar J. The eye specification network in Drosophila. Proc Ind. Natl Acad Sci. 2004;70:517–530. [PMC free article] [PubMed] [Google Scholar]
- Weasner B, Salzer C, Kumar JP. Sine oculis, a member of the SIX family of transcription factors, directs eye formation. Dev Biol. 2007;303:756–771. doi: 10.1016/j.ydbio.2006.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff T, Ready DF. The beginning of pattern formation in the Drosophila compound eye: the morphogenetic furrow and the second mitotic wave. Development. 1991;113:841–850. doi: 10.1242/dev.113.3.841. [DOI] [PubMed] [Google Scholar]
- Wolff T, Ready DF. Pattern formation in the Drosophila retina. In: Bate M, Martinez Arias A, editors. In The Development of Drosophila melanogaster. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 1277–1326. [Google Scholar]
- Xu PX, Zheng W, Huang L, Maire P, Laclef C, Silvius D. Six1 is required for the early organogenesis of mammalian kidney. Development. 2003;130:3085–3094. doi: 10.1242/dev.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Canon J, Banerjee U. A transcriptional chain linking eye specification to terminal determination of cone cells in the Drosophila eye. Dev Biol. 2003;263:323–329. doi: 10.1016/j.ydbio.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Chipperfield H, Melton DA, Wong WH. A gene regulatory network in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104:16438–16443. doi: 10.1073/pnas.0701014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu CC, Dyer MA, Uchikawa M, Kondoh H, Lagutin OV, Oliver G. Six3-mediated auto repression and eye development requires its interaction with members of the Groucho-related family of co-repressors. Development. 2002;129:2835–2849. doi: 10.1242/dev.129.12.2835. [DOI] [PubMed] [Google Scholar]
- Ziemer A, Tietze K, Knust E, Campos-Ortega JA. Genetic Analysis of Enhancer of Split, a Locus Involved in Neurogenesis in Drosophila Melanogaster. Genetics. 1988;119:63–74. doi: 10.1093/genetics/119.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]


