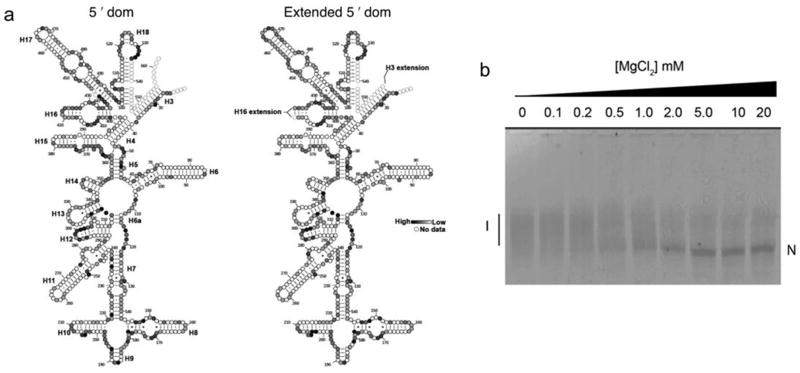
Extended Data Figure 1. Modification of the 5′ domain RNA preserves its structure.
a, The secondary structures of the wild-type and extended 5′ domain RNAs were probed by selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE)60. The 5′ domain RNA (2 pmol) was annealed to unlabeled oligonucleotides and folded in HKM20 buffer (80 mM K-HEPES pH 7.6, 300 mM KCl, 6 mM 2-mercaptoethanol, 20 mM MgCl2) before treatment with 3 mM N-methylisatoic acid (NMIA) at 42 ºC for 26 min. Modifications were detected by primer extension and quantified as previously described19. Results of SHAPE chemical probing of the free RNA structure for the wild-type 5′ domain (left) and the 5′ domain with h16 and h3 extensions after annealing with oligonucleotides (right). Saturation of grey indicates reactivity with NMIA (see Methods). Dashed circles indicate nucleotides that were not detected in our primer extension assay. The results show that the extensions added for the fluorescent labeling of the rRNA do not significantly perturb the rRNA folding. b, Native PAGE folding assay of 5′dom-h3h16. Fluorescently-labeled oligonucleotide (h3P5-Cy5; 25 nM) was annealed to an equimolar concentration of extended 5′ domain RNA in 10 μL CE buffer (20 mM Na-cacodylate, 0.5 mM Na2EDTA) for 5 min at 70 ºC and 5 min at 25 ºC. The RNA-oligonucleotide complex was then folded at 37 ºC for 30 min in varying [MgCl2] (0–20 mM) before electrophoresis on a native 8% polyacrylamide gel containing 10 mM MgCl2. The folding midpoint was 0.5 ± 0.1 mM MgCl2, similar to that of the wild-type 5′ domain RNA (1.4 ± 0.2 mM) reported previously61.