Abstract
PCA3 is a prostate-specific non-coding RNA, with utility as urine based early detection biomarker. Here, we report the evaluation of tissue PCA3 expression by RNA in-situ hybridization in a cohort of 41 mapped prostatectomy specimens. We compared tissue PCA3 expression with tissue level ERG expression and matched pre-prostatectomy urine PCA3 and TMPRSS2-ERG levels. Across 136 slides containing 138 foci of prostate cancer, PCA3 was expressed in 55% of cancer foci and 71% of high grade prostatic intraepithelial neoplasia foci. Overall, the specificity of tissue PCA3 was >90% for prostate cancer and high grade prostatic intraepithelial neoplasia combined. Tissue PCA3 cancer expression was not significantly associated with urine PCA3 expression. PCA3 and ERG positivity in cancer foci were positively associated (p<0.01). We report the first comprehensive assessment of PCA3 expression in prostatectomy specimens, and find limited correlation between tissue PCA3 and matched urine in prostate cancer.
Keywords: PCA3, RNA in-situ hybridization, Prostate cancer
INTRODUCTION
PCA3 is a noncoding RNA specifically overexpressed in >90% of prostate cancers(1, 2). The urine assay for PCA3 (PROGENSA, Gen-Probe, San Diego, CA) has recently been approved by the Food and Drug Administration for predicting prostate cancer on rebiopsy. As PCA3 is a noncoding RNA, immunohistochemical-based detection is not feasible. Only a single study evaluating tissue PCA3 RNA expression by in-situ hybridization has been reported, which showed PCA3 expression in the majority of prostate cancer foci and high grade prostatic intraepithelial neoplasia foci, as well as a substantial subset (~30%) of benign glands(3).
In 2005, our group discovered chromosomal rearrangements in prostate cancer resulting in the fusion of the 5’ untranslated region of the androgen-regulated gene TMPRSS2 with members of the E26 transformation-specific family of transcription factors(4, 5). Fusions involving ERG represent 90% of all E26 transformation-specific fusions, and fluorescence in-situ hybridization for the detection of ERG rearrangements has been shown to be highly specific for prostate cancer and high grade prostatic intraepithelial neoplasia adjacent to prostate cancer(6–9). Monoclonal antibodies against ERG have been used as a surrogate for ERG rearrangement status, and have demonstrated diagnostic utility with high sensitivity and specificity for prostate cancer with TMPRSS2-ERG rearrangements(10, 11).
Our group recently evaluated a clinical grade transcription mediated amplification assay that quantifies TMPRSS2-ERG mRNA in post-digital rectal examination urine. We have shown that urine TMPRSS22-ERG in combination with urine PCA3 increases the utility of serum PSA to predict the presence of prostate cancer and significant prostate cancer (per Epstein criteria) upon rebiopsy and correlates with total linear dimension of ERG positive prostate cancer foci on prostatectomy specimens(12).
Unlike ERG, tissue level expression of PCA3 and its correlation with urine expression is not well understood. Therefore, to assess the tissue level expression pattern of PCA3, we used a novel RNA in situ hybridization assay for the detection of PCA3 and evaluated its expression in prostate cancer and high grade prostatic intraepithelial neoplasia. We also compared tissue PCA3 expression with tissue ERG expression by immunohistochemistry, and urine expression of PCA3 and TMPRSS2-ERG.
MATERIALS and METHODS
Study Cohort
The Institutional Review Board approved prostatectomy cohort studied (men who underwent prostatectomy at our institution between 2008 and 2011) was identified from a previously reported cohort of 301 men referred for prostate needle biopsy at the University of Michigan Health System. All cases were assessed for urine TMPRSS2-ERG and urine PCA3 scores by transcription mediated amplification as previously described(12, 13). None of these patients received preoperative radiation or androgen deprivation therapy.
Urine TMPRSS2-ERG and PCA3 assays
Urine TMPRSS2-ERG and PCA3 scores were determined as described previously(12, 13). In short, urine specimens were obtained immediately after attentive digital rectal examination. Expression levels of TMPRSS2-ERG mRNA and PSA mRNA were determined by a third generation, final clinical transcription mediated amplification assay (12–14, 15). The PROGENSA PCA3 assay (Gen-Probe Inc, San Diego, CA, USA) was used to determine PCA3 RNA and PSA mRNA urine levels. PCA3 score was determined as a ratio of PCA3 RNA to PSA mRNA.
Prostatectomy Evaluation
Fresh prostates removed after surgery were weighed, measured, inked, and fixed in 10% neutral formalin. Seminal vesicles, apex, and base were amputated and the remaining prostate was serially sectioned at 4 to 5 mm intervals perpendicular to the long axis of the gland from the base to apex and quartered. All prostatectomy specimens were reviewed by the study pathologists LPK and JIW. Tumor maps were generated by tracking each section and reconstructing them as a whole-mount section. A cancer focus was considered spatially separate or multifocal if it was 3 mm or more from the closest cancer in any single section or 4mm or more from the closest cancer on the adjacent section above or below, as described previously(17). The largest tumor focus was designated as the index nodule and additional smaller tumors were labeled as multifocal tumors. The index nodule showed the highest Gleason score in the majority of cases. In rare cases where a smaller multifocal tumor had higher Gleason scores compared to the index tumor, the smaller multifocal tumor focus with the highest Gleason scores was considered as the index nodule. For each prostatectomy, the total number of tumor foci, linear dimension of index nodule, total linear tumor dimension, and Gleason scores of all tumor foci were documented. At the University of Michigan Health System the greatest linear dimension of the index tumor rather than index tumor volume is reported clinically. Hence, we used the summed greatest linear dimensions of all tumor foci (total tumor linear dimension) as measurement of cancer volume. Adjacent high grade prostatic intraepithelial neoplasia was defined as high grade prostatic intraepithelial neoplasia ≤4 mm from a focus of prostate cancer; the presence of adjacent high grade prostatic intraepithelial neoplasia was noted for all cancer foci. Isolated high grade prostatic intraepithelial neoplasia was defined as high grade prostatic intraepithelial neoplasia >4mm from a focus of prostate cancer and was assessed in 15 randomly selected cases.
Immunohistochemistry for ERG (see below) was previously performed on sections representing all index and multifocal tumor foci from each case. All tumor foci were assigned as as ERG positive or ERG negative. The total linear dimension of ERG positive cancer summed the total linear size of the largest dimension of all ERG positive tumor foci, including the index tumor when ERG positive(12).
PCA3 RNA in situ hybridization was performed on all sections representing all index and multifocal tumor foci from each case. PCA3 score (see below) was assigned based on area of strongest staining in a given tumor focus. PCA3 expression was considered focal if <50% of tumor cells in a given tumor focus were PCA3 positive. The total PCA3 positive linear tumor dimension summed the total linear size of the largest dimension of all PCA3 positive tumor foci in each case. Total PCA3 intensity was assigned as the sum of PCA3 intensity scores of all tumor foci per case. Because PCA3 expression is scored based on a ranking system ranging from 0–4, PCA3 positive tumor volume was additionally scored using PCA3 product score (summed linear dimension of each tumor focus multiplied by its PCA3 intensity score)
ERG Immunohistochemistry
Immunohistochemistry on unstained formalin fixed, paraffin-embedded levels of all tumor foci from the prostatectomy specimen blocks was performed using a monoclonal antibody against ERG, clone EPR 3864 (Epitomics, Burlingame, CA), using the automated Discovery XT staining platform (Ventana Medical Systems-A Roche group, Tucson, AZ) and evaluated by the study pathologists JIW and LPK (12). Staining of vessels was used as a positive control and slides without staining of vessels were excluded from further analysis.
RNA in situ Hybridization
Formalin fixed paraffin embedded sections were baked at 60°C for one hour. RNA in situ hybridization was performed using PCA3 RNA probe (NR_015342 region 1683–2816- http://www.ncbi.nlm.nih.gov/nuccore/nr_015342) and RNAscope FFPE Reagent Kit 2.0 (Advanced Cell Diagnostics, Hayward, CA) according to manufacturer’s instructions. Briefly, tissues were deparaffinized by immersing in xylene twice for 15 minutes each with periodic agitation. The slides were then immersed in 100% ethanol twice for 3 minutes each with periodic agitation then air-dried for 5 minutes. Tissues were circled using a pap pen (Vector, #H-4000), allowed to dry and treated with Pretreatment 1 buffer for 10 minutes. Slides were rinsed in deionized water and then boiled in 1× Pretreatment 2 buffer for 15 minutes. Slides were rinsed again in deionized water and then treated with Pretreatment 3 buffer for 30 minutes at 40°C in a humidity chamber. Slides were rinsed twice in deionized water and then incubated with target probes for PCA3 and DapB, a bacterial gene used as negative control, for 2 hours at 40°C in a humidity chamber. Slides were then washed in 1× Wash Buffer twice for 2 minutes. Slides were then treated with Amp 1 solution for 30 minutes, Amp 2 solution for 15 minutes, Amp 3 solution for 30 minutes and Amp 4 solution for 15 minutes, all at 40°C in a humidity chamber with 2 washes in 1× Wash Buffer for 2 minutes after each step. Slides were then treated with Amp 5 solution for 30 minutes and Amp 6 solution for 15 minutes at room temperature in a humidity chamber with 2 washes in 1× Wash Buffer for 2 minutes after each step. Color was developed by adding a 1:60 solution of Fast Red B: Fast Red A to each slide and incubated for 10 minutes. Slides were washed twice in deionized water and then immersed in a 50% hematoxylin (Fisher, #SH26-4D) solution for 2 minutes. Slides were rinsed several times in deionized water and then immersed in a 0.01% ammonium hydroxide solution. Slides were rinsed in deionized water then dehydrated by immersing 5 times in 70% ethanol twice, immersing 5 times in 95% ethanol twice, immersing twice in 100% ethanol for 5 seconds each and in xylene for 5 seconds. The slides were mounted in Cytoseal XYL (Thermo Scientific, #8312-4) for viewing under bright-field microscope. Positive controls were performed for all runs using a POLR2A gene-specific RNA probe. PCA3 RNA-ISH was not repeated in any tumor focus showing a negative result in order to avoid discrepant analysis.
RNA in situ hybridization evaluation criteria
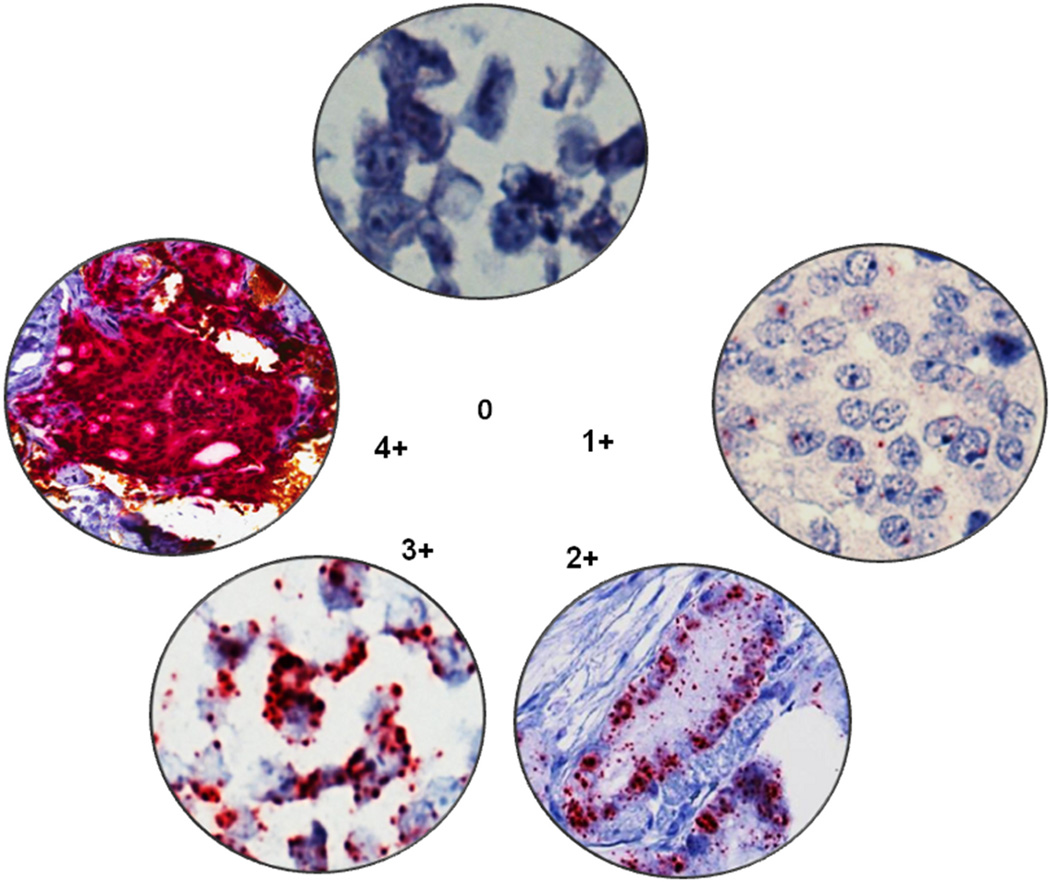
RNA in situ hybridization expression intensity scoring guidelines were established to classify tumor foci as PCA3-positive or PCA3-negative. PCA3 expression by RNA in situ hybridization appeared as distinct cytoplasmic punctate dots. All tumor foci were evaluated and scanned at 20× magnification. Scoring for an entire tumor focus was based on the highest PCA3 intensity using these criteria. Based on the number of dots/cell, we established five grading levels ranging from 0–4: tumor foci with no staining or less than 1 dot/cell under 20× magnification were scored as zero; foci with1–3 dots/cell in more than 5% of the tumor were scored as 1; tumor foci with 4–10 dots/cell with no or very few dot clusters (fused overlapping dots) in more than 5% of tumor were scored as 2; tumor foci with more than 10 dots/cell with less than 10% of positive cells having dot clusters in more than 5% of tumor were scored as 3; tumor foci with more than 10 dots/cell with more than 10% of positive cells having dot clusters in more than 5% of tumor were scored as 4 (Figure 1). Expression was considered focal when dots were seen in <50% of cells in a tumor focus. For the purposes of this study, tumor foci showing scores 2–4 were considered PCA3-positive, while tumor foci showing scores 0–1 were considered PCA3-negative. All PCA3 slides were reviewed by study pathologists LPK and JIW.
Figure 1.
PCA3 intensity scores; tumor foci with no staining or less than 1 dot/cell under 40× magnification were scored as zero; foci with1–3 dots/cell in more than 5% of the tumor were scored as 1; tumor foci with 4–10 dots/cell with no or very few dot clusters (fused overlapping dots) in more than 5% of the tumor were scored as 2; tumor foci with more than 10 dots/cell with less than 10% of positive cells having dot clusters in more than 5% of the tumor were scored as 3; tumor foci with more than 10 dots/cell with more than 10% of positive cells have dot clusters in more than 5% of the tumor were scored as 4.
Tissue PCA3 expression by in situ hybridization vs transcription mediated amplification
Because urine PCA expression by transcription mediated amplification was compared to tissue PCA3 expression by in situ hybridization, tissue PCA3 expression values by in situ hybridization were compared to tissue values by transcription mediated amplification. Nine tissue blocks were selected, on which PCA3 RNA in situ hybridization had been performed - 4 blocks demonstrating prostate cancer with expression intensity of 4 (diffuse), and 5 blocks with expression intensity of 0. Two 10 micron sections were taken from each block, changing the blade between blocks to prevent cross-contamination. Both sections from each block were placed in specimen transport medium, heated in a 60° C waterbath for 30 minutes with occasional swirling, then placed on ice for 5 minutes. Separate sterile swabs were used to remove solidified and floating paraffin. Specimen tubes were then capped, mixed by inverting 5 times, and stored at −70° C until testing. Prior to analysis, samples were warmed to 60° C in a water bath for <20 minutes. PCA3 scores were determined by transcription mediated amplification, as previously described(13).
Statistical analysis
All statistical analyses were performed using the Stats package in the R programming language. The two-tailed t-test was used to analyze tumor focus size vs. PCA3 status, and PCA3-positive tumor size in urine PCA3-high vs. PCA3-low groups. Fisher’s exact test was used to analyze ERG status vs. PCA3 status of tumor foci and, PCA3 status of tumor foci vs. PCA3 status of adjacent high grade prostatic intraepithelial neoplasia. Spearman ρ (rs) was used to test associations between urine PCA3 and total tumor linear dimension, total number of tumor foci, total PCA3 intensity, PCA3-positive tumor linear dimension, and index nodule PCA3 score. Spearman ρ (rs) was also used to evaluate correlations between urine TMPRSS2-ERG and number of PCA3-positive tumor foci, urine TMPRSS2-ERG, number of ERG positive tumor foci, number of PCA3-positive tumor foci, and number of ERG positive tumor foci. Wilcox rank sum test was used to analyze total PCA3 intensity, and PCA3 product score, in urine PCA3-high vs. urine PCA3-low groups.
RESULTS
Prostatectomy cohort
As previously reported, the 41 prostatectomy specimens in this study had a median of 3 cancer foci (range 1–11) and median total linear dimension of 2.5 cm (range 0.4–5.5 cm)(12). The majority of cases were confined to the prostate (pT2, 37/41, 90%) and had index tumor Gleason score of 7 (31/41, 76%).
PCA3 expression by ISH and correlation with ERG expression by IHC
A total of 138 tumor foci were evaluated with PCA3 RNA in situ hybridization. Of the 159 tumor foci identified in our previous study(12), 21 small tumor foci were lost on deeper sectioning. Of the 138 tumor foci evaluated, 77 (56%) were PCA3-positive. PCA3 intensities for all tumor foci were: 4 (43 foci, 31%), 3 (29 foci, 21%), 2 (5 foci, 4%), 1 (20 foci, 14%), and 0 (41 foci, 30%). The index tumor was PCA3-positive in 25 cases (61%). PCA3 intensities for index tumor nodules were: 4 (16 tumor nodules, 39%), 3 (9 nodules, 22%), 2 (1 nodule, 2%), 1 (6 nodules, 15%), and 0 (9 nodules, 22%). Of all PCA3-positive cancer foci, 29 (37%) showed focal PCA3 staining. PCA3 expression was noted in at least one tumor focus in 36 cases (88%). PCA3 status of index tumor foci did not associate with Gleason score (p=0.34, Spearman correlation).The pathologic data, PCA3 status, and urine PCA3 scores are summarized in Table 1. Microphotographs demonstrating diffuse and focal PCA3 expression are present in Figure 2.
Table 1.
Pathologic data and PCA3 scores for all case.
| Case number | Summed tumor dimension (cm) |
Summed PCA3+ tumor dimension (cm) |
Index nodule dimension (cm) |
Index nodule PCA3 score (0- 4) |
Index nodule (ERG status) |
Index nodule Gleason score |
Total number cancer foci |
Total number PCA3+ cancer foci |
Urine PCA3 score |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2.2 | 0 | 1.2 | 0 | negative | 3+3 | 2 | 0 | 34.8 |
| 2 | 1.6 | 0.1 | 1.4 | 0 | negative | 3+4 | 3 | 1 | 27 |
| 3 | 3.5 | 3.5 | 1.8 | 4 | negative | 4+3 | 2 | 2 | 2.6 |
| 4 | 1.2 | 0.1 | 0.8 | 0 | negative | 3+3 | 3 | 1 | 66.3 |
| 5 | 2 | 2 | 1 | 3 | negative | 3+4 (tertiary 5) | 2 | 2 | 59.6 |
| 6 | 2.8 | 2.3 | 1.5 | 4 | negative | 4+3 | 3 | 2 | 19.1 |
| 7 | 1.7 | 1 | 0.7 | 3 | negative | 3+4 | 3 | 2 | 73.9 |
| 8 | 1 | 0 | 0.6 | 1 | negative | 3+4 | 2 | 0 | 49.7 |
| 9 | 0.4 | 0.2 | 0.2 | 1 | negative | 3+3 | 2 | 1 | 12.6 |
| 10 | 3.4 | 0.6 | 2.8 | 1 | negative | 4+3 (tertiary 5) | 3 | 2 | 74 |
| 11 | 2.1 | 2 | 2 | 4 | negative | 3+4 | 2 | 1 | 42.4 |
| 12 | 2.3 | 2.1 | 1.9 | 4 | negative | 4+3 | 3 | 2 | 86.3 |
| 13 | 1.7 | 0.1 | 0.7 | 0 | negative | 3+3 | 6 | 1 | 22 |
| 14 | 0.6 | 0 | 0.6 | 0 | positive | 4+4 | 1 | 0 | 3.2 |
| 15 | 0.9 | 0 | 0.6 | 0 | positive | 3+3 | 3 | 0 | 82.9 |
| 16 | 1.8 | 0.9 | 0.9 | 1 | negative | 3+4 | 2 | 1 | 15.3 |
| 17 | 2.3 | 0.6 | 0.7 | 0 | negative | 3+4 | 6 | 2 | 71.8 |
| 18 | 4.5 | 1.6 | 1.6 | 3 | negative | 3+4 | 5 | 1 | 54.9 |
| 19 | 2.2 | 1.7 | 1.1 | 4 | positive | 3+4 | 3 | 2 | 6.3 |
| 20 | 1.1 | 1.1 | 1.1 | 4 | positive | 3+4 | 1 | 1 | 7.6 |
| 21 | 1.3 | 1.2 | 1.1 | 4 | positive | 3+4 | 3 | 2 | 38.3 |
| 22 | 1.3 | 1.2 | 1.2 | 3 | positive | 3+4 | 2 | 1 | 12.4 |
| 23 | 1.3 | 0 | 1.3 | 0 | positive | 3+4 | 1 | 0 | 31.2 |
| 24 | 3.4 | 3.4 | 1.6 | 4 | negative | 3+4 | 4 | 4 | 16.9 |
| 25 | 3 | 1.5 | 0.8 | 3 | positive | 3+4 | 4 | 2 | 104.5 |
| 26 | 2.3 | 1.9 | 1 | 4 | positive | 3+3 | 6 | 3 | 95.2 |
| 27 | 3.2 | 2.1 | 2.1 | 4 | positive | 4+3 | 2 | 1 | 35.8 |
| 28 | 2.9 | 2.6 | 1 | 3 | positive | 3+3 | 4 | 3 | 37 |
| 29 | 2.7 | 1.1 | 1.6 | 0 | negative | 3+4 | 3 | 2 | 125.9 |
| 30 | 3.1 | 2.6 | 1.9 | 4 | positive | 3+4 | 3 | 2 | 32 |
| 31 | 5.1 | 5.1 | 2 | 4 | positive | 3+4 | 6 | 6 | 74.5 |
| 32 | 2.7 | 2.1 | 2.1 | 4 | positive | 3+4 | 2 | 1 | 10.5 |
| 33 | 5.5 | 4.2 | 1.5 | 3 | positive | 3+4 | 4 | 3 | 46.9 |
| 34 | 2.6 | 1.6 | 1 | 4 | positive | 3+4 | 5 | 3 | 32.9 |
| 35 | 4.9 | 2.8 | 1.6 | 3 | positive | 3+4 | 5 | 2 | 68.9 |
| 36 | 3.5 | 3.5 | 2.4 | 4 | positive | 3+4 | 3 | 3 | 41.6 |
| 37 | 3.4 | 1.4 | 2 | 1 | positive | 3+4 | 3 | 2 | 10.6 |
| 38 | 4.9 | 4.9 | 1.4 | 3 | positive | 3+3 | 7 | 7 | 43.2 |
| 39 | 3.8 | 3.8 | 1.9 | 4 | positive | 3+4 | 3 | 3 | 105.1 |
| 40 | 5.3 | 1.6 | 0.8 | 1 | positive | 3+4 | 11 | 3 | 186.3 |
| 41 | 5.1 | 2.1 | 2.1 | 2 | positive | 3+4 | 3 | 1 | 39.6 |
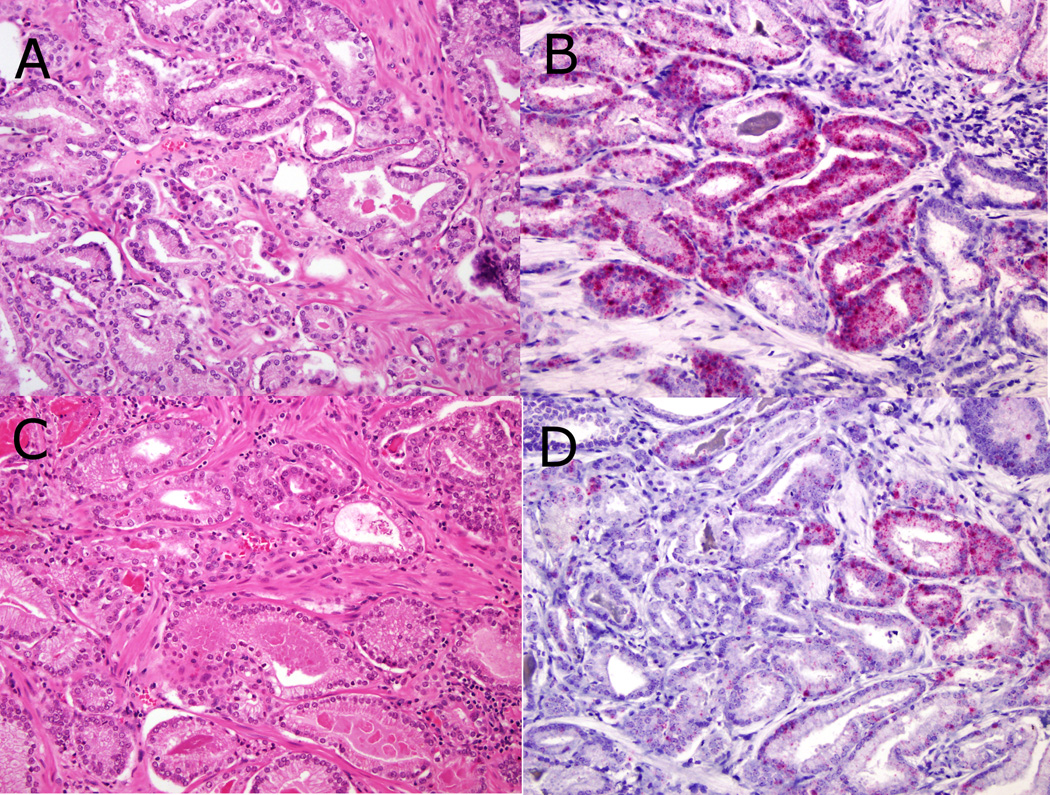
Figure 2.
PCA3 (RNA in-situ hydridization) expression with corresponding H&E stained sections. Diffuse expression with maximum intensity score 4 (2A H&E, 2B PCA3 RNA in-situ hybridization; both 200× magnification). Focal expression with intensity score 4 at right side of image and 0 at left side of image (2C H&E, 2D PCA3 RNA in-situ hybridization; both 200× magnification).
The median total PCA3-positive linear tumor dimension in the 41 patients was 1.6 cm (range 0–5.1 cm). The median number of PCA3-positive tumor foci per case was 2 (range 0–7). The median total PCA3 intensity across all 41 cases was 7 (range 0–26). The median PCA3 product score was 6 (range 0–20). PCA3-positive tumor foci were overall larger than PCA3-negative tumor foci (median 0.9 cm vs. 0.6 cm respectively; two-tailed t-test, p<0.01).
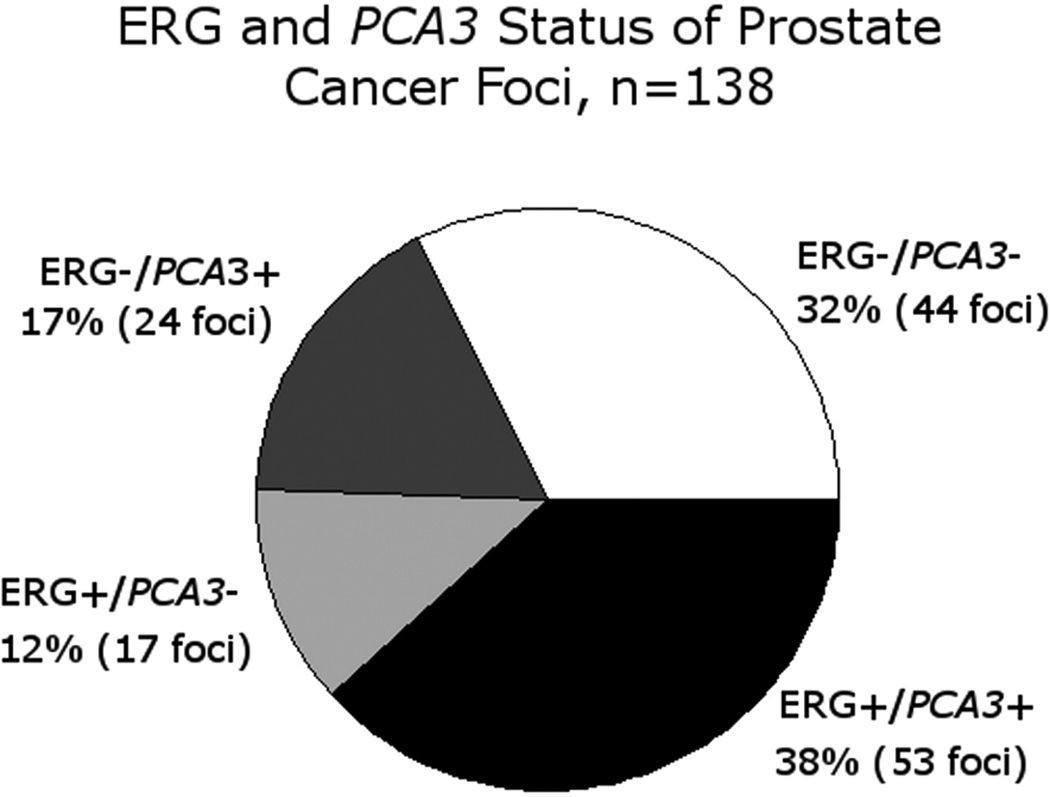
ERG expression was identified in 70 tumor foci (50%), and 53 tumor foci (38%) were positive for both ERG and PCA3. Overall, across all tumor foci, 94 foci (68%) were positive for ERG, PCA3, or both (Figure 3). There was a significant positive association between PCA3 status by RNA in situ hybridization and ERG status by immunohistochemistry in tumor foci (p<0.01, Fisher’s Exact test). The median PCA3 intensity was 3+ in ERG-positive nodules and 1+ in ERG-negative nodules. Of the 41 cases, 95% (39/41) had at least one tumor focus positive for either PCA3 by RNA in situ hybridization and/or ERG expression by immunohistochemistry.
Figure 3.
PCA3 RNA in-situ hybridization and ERG immunohistochemistry status of all tumor foci.
PCA3 expression in high grade prostatic intraepithelial neoplasia
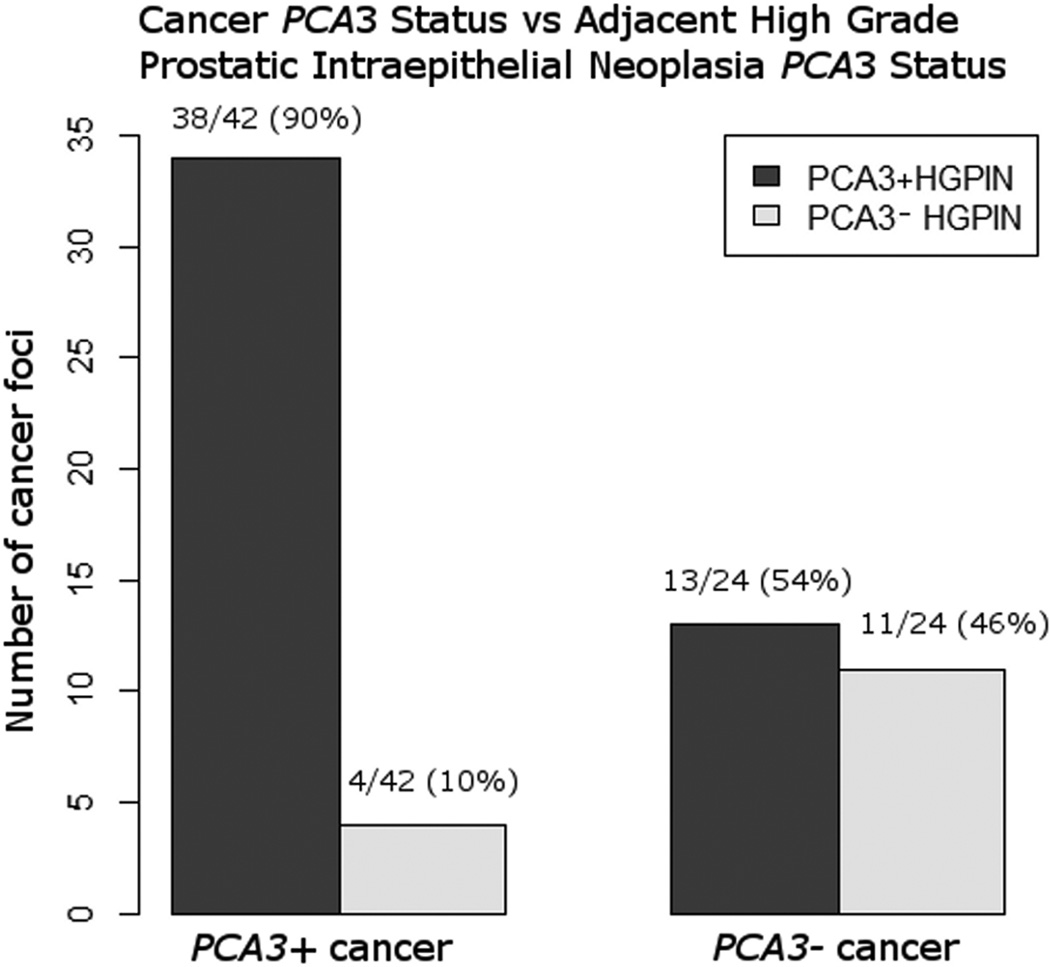
We identified 66 foci of high grade prostatic intraepithelial neoplasia adjacent to cancer, of which 51 (77%) were PCA3-positive. Tissue PCA3 intensities for adjacent high grade prostatic intraepithelial neoplasia were: 4 (29, 44%), 3 (19, 29%), 2 (3, 5%), 1(4, 6%) and 0 (11, 17%). High grade prostatic intraepithelial neoplasia adjacent to PCA3-positive cancer tended to be PCA3-positive (90%, 38/42 foci). In contrast, high grade prostatic intraepithelial neoplasia adjacent to PCA3-negative cancer was nearly equally divided into PCA3-positive (54%; 13/24 tumor foci) and PCA3-negative (46%; 11/24 tumor foci) (Figures 4 and 5).
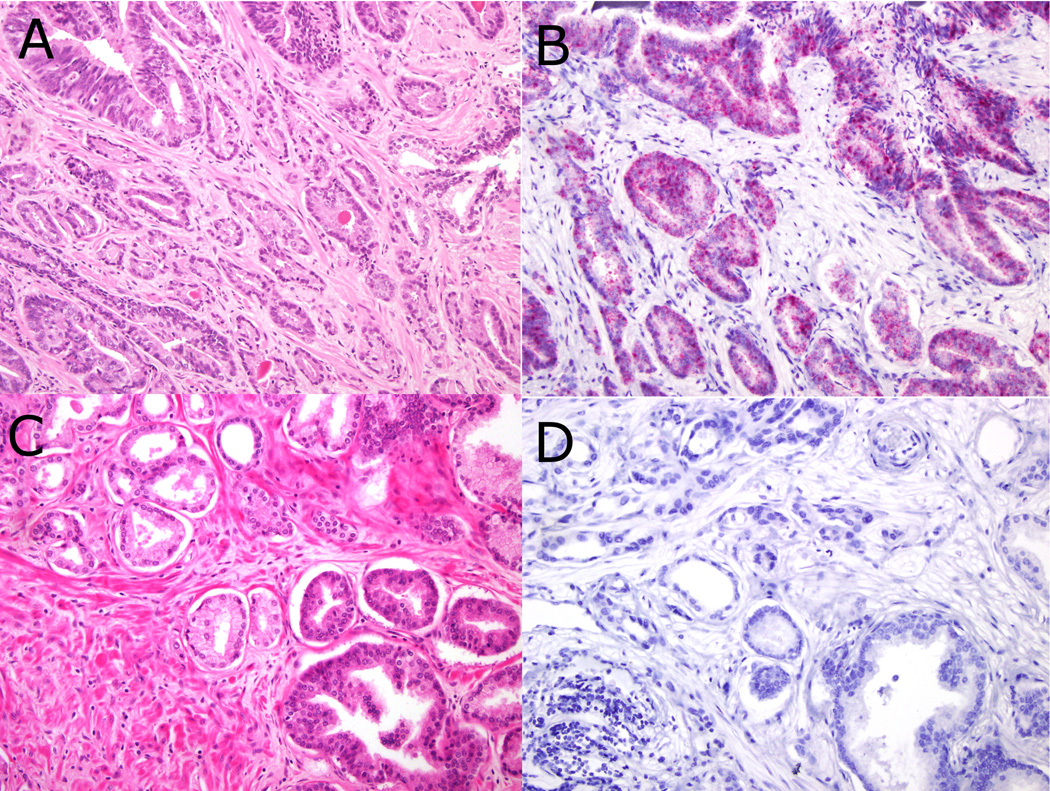
Figure 4.
PCA3 RNA in-situ hybridization in prostate cancer and adjacent high grade high grade prostatic intraepithelial neoplasia showing concordant positive (3A H&E, 3B PCA3 RNA in-situ hybridization; both 200× magnification) and negative (3C H&E, 3D PCA3 RNA in-situ hybridization; both 200× magnification) PCA3 status.
Figure 5.
PCA3 RNA in-situ hybridization status of prostate cancer and adjacent high grade prostatic intraepithelial neoplasia.
We found 16 foci of isolated high grade prostatic intraepithelial neoplasia after careful review of 15 randomly selected cases. Of these, 56% (9/16) were PCA3-positive by RNA-ISH. PCA3 scores for isolated high grade prostatic intraepithelial neoplasia were as follows: 4 (4, 25%), 3 (3, 19%), 2 (2, 13%), 1 (3, 19%) and 0 (4, 25%). In total, 71% (60/84) of all identified high grade prostatic intraepithelial neoplasia foci were PCA3-positive.
All benign glands were examined for PCA3 expression. Of the benign glands, 50–60 acini (on 10 slides from 9 cases; intensity range 2–3) were PCA3-positive. These glands were frequently seen close to (<3 mm from) PCA3-positive tumor foci and the remaining benign prostatic glands, including all foci of atrophy and adenosis, were PCA3-negative. Thus, across 136 tissue sections, positive PCA3 expression by RNA in situ hybridization showed a specificity of >90% for combined prostate cancer and high grade prostatic intraepithelial neoplasia.
Urine PCA3 and urine TMPRSS2-ERG
All 41 patients had sufficient urine RNA for evaluation of PCA3 and TMPRSS2-ERG. Median urine PCA3 score was 40 (SD 38, range 3–186). Of these, 24 (59%) had urine PCA3 scores>35, a proposed optimal cutoff for detection of cancer on biopsy(18), and 29 (71%) had urine PCA3 score>25, the current Food and Drug Administration approved cutoff for predicting prostate cancer after a negative biopsy. Urine PCA3 score correlated well with number of tumor foci (rs=0.51, p<0.01) and correlated weakly with summed total linear tumor dimension (rs=0.29, p=0.06), consistent with our previous study using this cohort(12). Urine PCA3 did not correlate significantly with Gleason score of index tumor nodule (p=0.89, Spearman correlation).
Urine PCA3 score did not correlate well with any independent measure of PCA3- positive tumor volume, including index nodule PCA3 score (rs= −0.1, p=0.52), total PCA3 positive linear tumor dimension (rs=0.09, p=0.56), or PCA3 product score (rs=0.04, p=0.78). Although urine PCA3 did correlate with the number of PCA3-positive tumor foci per case (rs=0.34, p=0.03), this correlation became non-significant on multivariate linear regression including total number of tumor foci (outcome variable urine PCA3 score; total number tumor foci p<0.01, total number PCA3-positive tumor foci p=0.88). Similarly, although total PCA3 intensity showed correlation with urine PCA3 that approached statistical significance (rs=0.29, p=0.06), this too was statistically non-significant on multivariate analysis (outcome variable urine PCA3 score; total number tumor foci p<0.01, total PCA3 intensity p=0.38). Using both 35 and 25 as cutoff values for urine PCA3, urine PCA3-high and PCA3-low groups showed no statistically significant difference in PCA3-positive tumor linear dimension (p=0.09 for cutoff 35, p=0.46 for cutoff 25; two-tailed t-test) or PCA3 product score (p=0.21 for cutoff 35, p=0.99 for cutoff 25; Wilcox rank sum test) (Table 2).
Table 2.
Fraction of cases positive for urine PCA3 at cutoffs >25 and >35, by total tumor linear dimension (cm) and total PCA3+ tumor linear dimension (cm).
| Summed linear tumor dimension (cm) | Summed PCA3+ linear tumor dimension (cm) | |||||||
|---|---|---|---|---|---|---|---|---|
| 0–1 | 1.1–2.5 | 2.6–3.5 | >3.5 | 0–1 | 1.1–2.5 | 2.6–3.5 | >3.5 | |
| Urine PCA3 score>35 | 0/4 (0%) | 8/16(50%) | 6/8(75%) | 8/13(62%) | 6/13(46%) | 11/18(61%) | 3/6(50%) | 4/4(100%) |
| Urine PCA3 score>25 | 2/4(50%) | 11/16(69%) | 8/8(100%) | 8/13(62%) | 9/13(69%) | 12/18(67%) | 4/6(67%) | 4/4(100%) |
| Urine PCA3 score<25 | 2/4 (50%) | 5/16(31%) | 0/8(0%) | 5/13(38%) | 4/13(31%) | 6/18(33%) | 2/6(33%) | 0/4(0%) |
In our cohort, 5 patients (12%) had no PCA3-positive tumor focus by RNA in situ hybridization (total 9 tumor foci). The median urine PCA3 score in these patients was 34.8 (range 3.2–82.9). Similarly, 2 patients (5%) showed no PCA3-positive focus by RNA in situ hybridization or ERG-positive focus by immunohistochemistry. Urine PCA3 scores in these patients were 34.8 and 49.7.
Of the 41 patients, 12 (29%) had urine PCA3<25. Of these, 92% (11/12) had at least one PCA3-positive tumor focus. Median number of PCA3-positive tumor foci was 1 (range 0–4). Median PCA3 positive linear tumor dimension was 1.3 cm (range 0–3.5). PCA3 intensities of index tumor nodules in these patients were as follows: 4 (6 nodules, 50%), 3 (1 nodule, 8%), 1 (3 nodules, 25%), and 0 (2 nodules, 17%). Of the 7 index nodules with PCA3 intensity 3 or 4, three of them demonstrated diffuse expression within the tumor focus. Median urine PCA3 score in these seven patients with index nodule showing PCA3 intensity 3 or 4 was 10.5 (range 2.3– 19.1). Five patients with urine PCA3 values <25 had total PCA3-positive linear dimension >1.6 cm (the median value).
Urine TMPRSS2-ERG score correlated with number of PCA3-positive tumor foci (rs=0.34, p=0.03). However, number of ERG-positive tumor foci correlated much more strongly with urine TMPRSS2:ERG score (rs=0.64, p<0.01), as shown in our previous study(12), and number of ERG-positive tumor foci correlated strongly with number of PCA3-positive tumor foci (rs=0.63, p<0.01).
Tissue PCA3 expression by in situ hybridization vs transcription mediated amplification
The four tumor foci that were strongly PCA3-positive by RNA in situ hybridization (4/4 intensity) had a median tissue PCA3 score of 10 (range 10–19) by transcription mediated amplification performed on tissue. Conversely, the five tumors that were PCA3-negative by RNA in situ hybridization (0/4 intensity) showed a median tissue PCA3 score of 3 (range 2–4) by transcription mediated amplification. These differences in PCA3 score were statistically significant (Wilcox rank sum test, p=0.02). See Table 3 for details.
Table 3.
PCA3 and PSA RNA values in copies/mL, as measured by transcription mediated amplification
| Case number# | PCA3 RNA value | PSA mRNA value | PCA3:PSA ratio* | PCA3 In situ hybridization intensity |
|---|---|---|---|---|
| 12 - index tumor | 18,458 | 1,870,578 | 10 | 4 |
| 30 - index tumor | 32,681 | 1,694,140 | 19 | 4 |
| 32 - index tumor | 22,178 | 2,199,705 | 10 | 4 |
| 36 - index tumor | 20,544 | 2,093,452 | 10 | 4 |
| 1 - index tumor | 2,360 | 556,125 | 4 | 0 |
| 17 - index tumor | 5,932 | 2,539,530 | 2 | 0 |
| 30 - non-index tumor | 2,364 | 920,239 | 3 | 0 |
| 41 - non-index tumor | 4,282 | 1,542,647 | 3 | 0 |
| 41 - non-index tumor | 3,977 | 1,167,430 | 3 | 0 |
PCA3:PSA ratio calculated as 1,000 × (PCA3 copies/mL) / (PSA copies/mL).
Case number refers to numbering in Table 1.
DISCUSSION
PCA3, a non-coding RNA, was originally reported as specifically overexpressed in prostate cancer by Bussemaker et al(1). In the present study, using a novel RNA in situ hybridization technique to evaluate PCA3 expression in prostate cancer, we have shown that PCA3 RNA is significantly elevated in prostate cancer tissue relative to benign prostatic tissue. This is the first time that PCA3 has been shown to display a strong specificity (>90%) for cancer and high grade prostatic intraepithelial neoplasia in formalin fixed paraffin embedded radical prostatectomy specimens. In this study we systematically mapped all the tumor foci, and found 88% of all cases in our cohort showed at least one PCA3-positive tumor focus. The results of the current study are consistent with previous tissue studies using PCR methods, which showed that although benign prostatic tissue displays low-level PCA3 expression, expression in prostate cancer tissue is considerably higher in comparison to benign tissue(1).
Due to the inability to develop an immunohistochemistry based detection platform for PCA3, there have been very few studies of PCA3 expression in tissue. To our knowledge, a study by Popa et al.(3) is the only other study evaluating PCA3 expression in prostate cancer tissue using in situ hybridization. In a series of 24 and 26 prostate cancers evaluated with chromogenic and radioactive in situ hybridization respectively, their group demonstrated PCA3 expression in the majority of prostate cancer (92%2013;96%) and at least focal cytoplasmic expression in the majority of high grade prostatic intraepithelial neoplasia (71%–96%). PCA3 expression was also noted in almost a third of benign glands (29%–33%), showing at least focal staining, and indicating poor specificity for prostate cancer. Some differences between our study and that of Popa et al. bear mentioning. In the Popa et al. study, the percentage of cells expressing PCA3 was evaluated, and PCA3 expression was reported as positive or negative. The presence of a single positive cell was defined as positive. In contrast, we used a commercially available well standardized RNA in situ detection procedure for bright field application. In this method, oligonucleotide based RNA probes are designed to yield punctate dots for each RNA transcript for a semi quantitative evaluation of tissue level expression of PCA3 RNA. Based on this, we were able to compare differences in the level of PCA3 RNA expression among tumor foci. Therefore, we are the first to introduce the development of evaluation criteria for PCA3 in formalin fixed and paraffin embedded tissue. The present study is also considerably larger, encompassing a well characterized cohort of 138 tumor foci in radical prostatectomy specimens, in contrast to the 48 tissue blocks (28 prostatic cancers and 20 benign prostatic tissues) utilized in the previous study. Also, in contrast to the study by Popa et al., the present study used a non-radioactive method, thereby proposing a more clinically realistic assay.
Recurrent TMPRSS2-ERG fusions present in approximately 50% of PSA-detected prostate cancers result in overexpression of ERG protein product(4, 8–10, 19). The fusion is reliably detectable using fluorescence in situ hybridization(6, 8, 9), and immunohistochemsitry using monoclonal antibodies directed against ERG has shown high concordance with fluorescence in situ hybridization results(10, 11, 19–21). We and others have shown that ERG IHC is >99% specific for prostate cancer and high grade prostatic intraepithelial neoplasia, showing only rare expression in benign prostatic glands(10, 11). The present study showed 55% of cancer foci were PCA3-positive by RNA in situ hybridization, a fraction slightly higher than that seen for the sensitivity of ERG immunohistochemistry in the present cohort (50%) and previous studies(8, 10). Similarly, while 88% of cases had at least one PCA3-positive cancer focus, slightly fewer (76%) had at least one ERG-positive cancer focus. We found a strong, statistically significant positive association (p<0.01) between ERG status by immunohistochemistry and PCA3 status by RNA in situ hybridization in prostate cancer foci. To the best of our knowledge, this is the first report of such a positive association. A subset of tumor foci (44/138 (32%)) were completely negative for both markers. Despite the strong association between PCA3 and ERG expression in tumor foci, a subset of tumor foci (41, 29%) also showed inverse ERG and PCA3 status. Consistent with these findings, 68% of tumor foci were positive for either PCA3 or ERG.
High grade prostatic intraepithelial neoplasia has displayed molecular abnormalities similar to prostate cancer in numerous studies, including chromosomal losses and gains(22, 23), telomere shortening(24), and AMACR overexpression(25). Consistent with those findings, 71% of high grade prostatic intraepithelial neoplasia foci in the present study were PCA3-positive. Interestingly, while the majority (90%) of high grade prostatic intraepithelial neoplasia foci adjacent to PCA3-positive cancer was also PCA3-positive, slightly over half of isolated high grade prostatic intraepithelial neoplasia (56%) and high grade prostatic intraepithelial neoplasia adjacent to PCA3-negative cancer (44%) were PCA3-positive. The relationship between ERG status of high grade prostatic intraepithelial neoplasia and associated prostate cancer differs from that of PCA3, in that ERG-positive high grade prostatic intraepithelial neoplasia has been shown to be strongly associated with adjacent ERG-positive prostate cancer(13, 17). Furthermore, in contrast to the large fraction of high grade prostatic intraepithelial neoplasia foci showing PCA3 expression (71%) in the present study, several studies have shown that only a small fraction of high grade prostatic intraepithelial neoplasia foci (15–18%) demonstrate ERG expression(10, 21). These findings favor the hypothesis that ERG-positive high grade prostatic intraepithelial neoplasia may indicate unsampled prostate cancer or high grade prostatic intraepithelial neoplasia that may progress to invasive carcinoma, which does not appear to be true of PCA3-positive high grade prostatic intraepithelial neoplasia. In summary, while ERG-positive high grade prostatic intraepithelial neoplasia represents the minority of high grade prostatic intraepithelial neoplasia and may be predictive of adjacent ERG-positive prostate cancer, PCA3-positive high grade prostatic intraepithelial neoplasia appears to represent the majority of high grade prostatic intraepithelial neoplasia and does not appear to be predictive of adjacent prostate cancer.
The great majority of clinical studies of PCA3 have been on urine measurement, which has demonstrated superior sensitivity and specificity to serum PSA for the detection of clinically significant prostate cancer on biopsy(13, 26–28). Similarly, TMPRSS2-ERG mRNA can be detected in the urine of patients with prostate cancer, and correlates with linear extent of ERG-positive cancer by immunohistochemistry(19, 21). In the present study, urine PCA3 score did not correlate with any measure of PCA3-positive tumor burden by RNA in situ hybridization except the number of PCA3-positive tumor foci per case (rs=0.34, p=0.03), which became non-significant on multivariate analysis including total number of tumor foci. In contrast, a strong correlation has been shown between urine TMPRSS2:ERG and both the total ERG-positive tumor dimension (rs=0.68) and the number of ERG positive tumor foci (rs=0.67)(12).
Interestingly, 12% (5/41) of cases in the present study had no PCA3-positive tumor focus. Of these, 80% (4/5) had urine PCA3 values >25, the current Food and Drug Administration approved cutoff value. There are several possible explanations for this lack of correlation between urine and tissue PCA3 expression levels. First, while ERG immunohistochemistry shows strong diffuse expression within positive tumor foci, PCA3 RNA in situ hybridization tended to show focal expression, with variations in intensity within a given tumor focus. Thus, in contrast to ERG, in which quantification in tissue is relatively straight-forward, quantification of PCA3 tissue expression is more challenging. Adding to this difficulty, in contrast to ERG which stains endothelial cells thereby offering a positive internal control, no internal positive control was available for PCA3 RNA in situ hybridization, although successful positive controls(evaluating for the presence of RNA) were run in parallel. Third, PCA3 RNA in situ hybridization was only performed on representative slides containing the majority of the index nodule. Also, although expression of PCA3 was evaluated in high grade prostatic intraepithelial neoplasia that happened to be on cancer-containing slides, the present study was not designed to evaluate PCA3 expression in all high grade prostatic intraepithelial neoplasia within a prostate. It is possible that unmeasured high grade prostatic intraepithelial neoplasia burden contributed to urine PCA3, particularly given the majority of high grade prostatic intraepithelial neoplasia is PCA3-positive. Lastly, a small subset (13%) of small cancer foci was lost on obtaining deeper sections for PCA3 RNA in situ hybridization. These lost foci may contribute to the lack of correlation between urine and tissue PCA3 expression, to a limited extent.
Consistent with this lack of correlation between urine and tissue PCA3 values, the majority of patients with urine PCA3<25 demonstrated at least one PCA3 positive tumor focus, and half demonstrated an index tumor nodule with strong PCA3 expression (4/4 intensity) by RNA in situ hybridization. Insufficient RNA collection is an unlikely explanation for these discrepant cases, because urine prostate specific antigen mRNA was measured in all cases, and those with insufficient values were excluded from the study. Urine PCA3 score may reflect associations between tumor burden and overall prostate volume, with larger prostates contributing more PSA mRNA from benign prostatic tissue, thus lowering the PCA3:PSA ratio. This hypothesis will need to be investigated in future studies. False positive tissue PCA3 RNA in situ hybridization is unlikely, as background benign prostate glands served as a successful negative control. A lack of association between in situ hybridization values and transcription median amplification values also appears to be unlikely, as our data show that the values between these different methodologies appear to correlate. Based on the preliminary observations from an independent and related study of evaluation of tissue PCA3 expression in cases with very high and very low urine PCA3, we found other cases with similar discrepant results (unpublished data from our group, study in progress). It appears that the process by which PCA3 RNA enters the urine from prostate cancer cells may be more complex than we currently realize. Further work on a large cohort of cases is needed to better understand this relationship.
The present study showed that 88% (36/41) and 76% (31/41) of prostatectomy specimens had at least one tumor focus positive for PCA3 RNA in situ hybridization and ERG immunohistochemistry, respectively. Importantly, 95% (39/41) of cases had at least one tumor focus positive for either PCA3 and/or ERG. Both patients with no PCA3-positive or ERG-positive cancer focus had urine PCA3 scores >25. Thus, 100% of patients in this study showed over-expression of ERG or PCA3 in tissue, urine, or both.
The current study has some limitations. This cohort does not reflect the entire spectrum of pathology seen at radical prostatectomy, including cases with high Gleason score and high stage. Hence, our results may not necessarily reflect the performance of PCA3 RNA in situ hybridization in high-grade or late stage cancers. Our cohort also lacks follow-up information precluding the evaluation of associations with outcome, and is relatively small. Hence, our findings regarding the expression of PCA3 by RNA in situ hybridization in prostate cancer need to be validated in a larger series.
Supplementary Material
Acknowledgements
This work was supported in part by the US National Institutes of Health Early Detection Research Network (U01 CA111275 and U01 CA113913), NIH S.P.O.R.E. (P50 CA69568), and R01 CA132874. N.P. and S.A.T. are supported by University of Michigan Prostate SPORE Career Development Awards. A.M.C. is supported by the Howard Hughes Medical Institute, the Doris Duke Foundation and the Prostate Cancer Foundation and is an American Cancer Research Professor and a Taubman Scholar. We would like to thank Angela Wu, Ritu Bhalla and Rohit Mehra for providing help and advice.
Footnotes
DISCLOSURE/CONFLICT OF INTEREST
A.M.C. serves on the advisory boards of Gen-Probe, Inc. and Ventana Medical Systems, Inc./Roche. S.A.T. has received honoraria from, and consults for Ventana Medical Systems. S.A.T. and A.M.C. are co-inventors on a patent filed by the University of Michigan covering ETS fusions in prostate cancer. The diagnostic field of use has been licensed to Gen-Probe, Inc., who has sublicensed some rights to Ventana Medical Systems, Inc./Roche. Gen-Probe, Inc. and Ventana/Roche did not play a role in the design and conduct of this study, in the collection, analysis, or interpretation of the data, or in the preparation, review, or approval of the article. NP does receive research funding from Ventana/Roche but this funding was not used for the development of the assay
REFERENCES
- 1.Bussemakers MJ, van Bokhoven A, Verhaegh GW, et al. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res. 1999;59:5975–5979. [PubMed] [Google Scholar]
- 2.Schmidt U, Fuessel S, Koch R, et al. Quantitative multi-gene expression profiling of primary prostate cancer. Prostate. 2006;66:1521–1534. doi: 10.1002/pros.20490. [DOI] [PubMed] [Google Scholar]
- 3.Popa I, Fradet Y, Beaudry G, et al. Identification of PCA3 (DD3) in prostatic carcinoma by in situ hybridization. Mod Pathol. 2007;20:1121–1127. doi: 10.1038/modpathol.3800963. [DOI] [PubMed] [Google Scholar]
- 4.Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 5.Tomlins SA, Laxman B, Dhanasekaran SM, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 6.Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mosquera J-M, Perner S, Genega EM, et al. Characterization of TMPRSS2-ERG fusion high-grade prostatic intraepithelial neoplasia and potential clinical implications. Clin Cancer Res. 2008;14:3380–3385. doi: 10.1158/1078-0432.CCR-07-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rubin MA, Maher CA, Chinnaiyan AM. Common gene rearrangements in prostate cancer. J Clin Oncol. 2011;29:3659–3668. doi: 10.1200/JCO.2011.35.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomlins SA, Bjartell A, Chinnaiyan AM, et al. ETS gene fusions in prostate cancer: from discovery to daily clinical practice. Eur Urol. 2009;56:275–286. doi: 10.1016/j.eururo.2009.04.036. [DOI] [PubMed] [Google Scholar]
- 10.Park K, Tomlins SA, Mudaliar KM, et al. Antibody-based detection of ERG rearrangement-positive prostate cancer. Neoplasia. 2010;12:590–598. doi: 10.1593/neo.10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomlins SA, Palanisamy N, Siddiqui J, et al. Antibody-based detection of ERG rearrangements in prostate core biopsies, including diagnostically challenging cases: ERG staining in prostate core biopsies. Arch Pathol Lab Med. 2012;136:935–946. doi: 10.5858/arpa.2011-0424-OA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Young A, Palanisamy N, Siddiqui J, et al. Correlation of Urine TMPRSS2:ERG and PCA3 to ERG+ and Total Prostate Cancer Burden. Am J Clin Pathol. 2012;138:685–696. doi: 10.1309/AJCPU7PPWUPYG8OH. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomlins SA, Aubin SMJ, Siddiqui J, et al. Urine TMPRSS2:ERG fusion transcript stratifies prostate cancer risk in men with elevated serum PSA. Sci Transl Med. 2011;3 doi: 10.1126/scitranslmed.3001970. 94ra72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cornu J-N, Cancel-Tassin G, Egrot C, et al. Urine TMPRSS2:ERG fusion transcript integrated with PCA3 score, genotyping, and biological features are correlated to the results of prostatic biopsies in men at risk of prostate cancer. The Prostate. 2012;73:242–249. doi: 10.1002/pros.22563. [DOI] [PubMed] [Google Scholar]
- 15.Lin DW, Newcomb LF, Brown EC, et al. Urinary TMPRSS2:ERG and PCA3 in an active surveillance cohort: results from a baseline analysis in the Canary Prostate Active Surveillance Study. Clin Cancer Res. 2013;19:2442–2450. doi: 10.1158/1078-0432.CCR-12-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stephan C, Jung K, Semjonow A, et al. Comparative assessment of urinary prostate cancer antigen 3 and TMPRSS2:ERG gene fusion with the serum prostate-specific antigen-based prostate health index for detection of prostate cancer. Clin Chem. 2013;59:280–288. doi: 10.1373/clinchem.2012.195560. [DOI] [PubMed] [Google Scholar]
- 17.Wise AM, Stamey TA, McNeal JE, et al. Morphologic and clinical significance of multifocal prostate cancers in radical prostatectomy specimens. Urology. 2002;60:264–269. doi: 10.1016/s0090-4295(02)01728-4. [DOI] [PubMed] [Google Scholar]
- 18.Marks LS, Fradet Y, Deras IL, et al. PCA3 molecular urine assay for prostate cancer in men undergoing repeat biopsy. Urology. 2007;69:532–535. doi: 10.1016/j.urology.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 19.Braun M, Goltz D, Shaikhibrahim Z, et al. ERG protein expression and genomic rearrangement status in primary and metastatic prostate cancer--a comparative study of two monoclonal antibodies. Prostate Cancer Prostatic Dis. 2012;15:165–169. doi: 10.1038/pcan.2011.67. [DOI] [PubMed] [Google Scholar]
- 20.Falzarano SM, Zhou M, Carver P, et al. ERG gene rearrangement status in prostate cancer detected by immunohistochemistry. Virchows Arch. 2011;459:441–447. doi: 10.1007/s00428-011-1128-4. [DOI] [PubMed] [Google Scholar]
- 21.He H, Magi-Galluzzi C, Li J, et al. The diagnostic utility of novel immunohistochemical marker ERG in the workup of prostate biopsies with “atypical glands suspicious for cancer.”. Am J Surg Pathol. 2011;35:608–614. doi: 10.1097/PAS.0b013e31820bcd2d. [DOI] [PubMed] [Google Scholar]
- 22.Jenkins RB, Qian J, Lee HK, et al. A molecular cytogenetic analysis of 7q31 in prostate cancer. Cancer Res. 1998;58:759–766. [PubMed] [Google Scholar]
- 23.Qian J, Bostwick DG, Takahashi S, et al. Chromosomal anomalies in prostatic intraepithelial neoplasia and carcinoma detected by fluorescence in situ hybridization. Cancer Res. 1995;55:5408–5414. [PubMed] [Google Scholar]
- 24.Vukovic B, Park PC, Al-Maghrabi J, et al. Evidence of multifocality of telomere erosion in high-grade prostatic intraepithelial neoplasia (HPIN) and concurrent carcinoma. Oncogene. 2003;22:1978–1987. doi: 10.1038/sj.onc.1206227. [DOI] [PubMed] [Google Scholar]
- 25.Kunju LP, Chinnaiyan AM, Shah RB. Comparison of monoclonal antibody (P504S) and polyclonal antibody to alpha methylacyl-CoA racemase (AMACR) in the work-up of prostate cancer. Histopathology. 2005;47:587–596. doi: 10.1111/j.1365-2559.2005.02281.x. [DOI] [PubMed] [Google Scholar]
- 26.Auprich M, Bjartell A, Chun FK-H, et al. Contemporary role of prostate cancer antigen 3 in the management of prostate cancer. Eur Urol. 2011;60:1045–1054. doi: 10.1016/j.eururo.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 27.Groskopf J, Aubin SMJ, Deras IL, et al. APTIMA PCA3 molecular urine test: development of a method to aid in the diagnosis of prostate cancer. Clin Chem. 2006;52:1089–1095. doi: 10.1373/clinchem.2005.063289. [DOI] [PubMed] [Google Scholar]
- 28.Ploussard G, Durand X, Xylinas E, et al. Prostate cancer antigen 3 score accurately predicts tumour volume and might help in selecting prostate cancer patients for active surveillance. Eur Urol. 2011;59:422–429. doi: 10.1016/j.eururo.2010.11.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.