1. INTRODUCTION
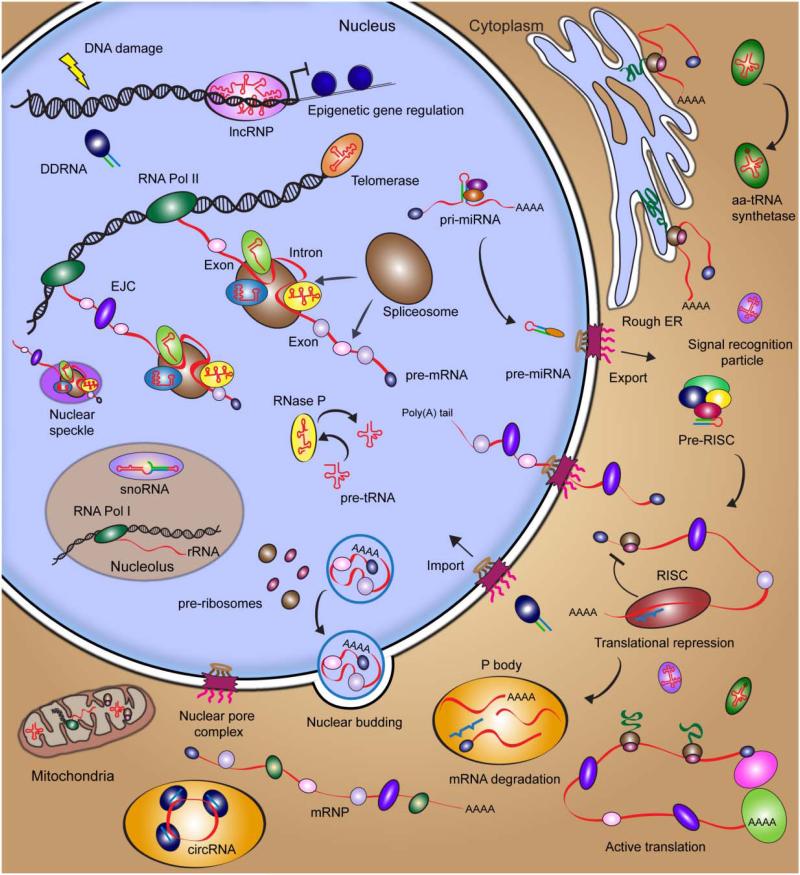
The eukaryotic cell is highly complex. Ever since Robert Hooke discovered “cells” in 1665 when training his comparably primitive microscope on a sliver of cork, scientists have aimed to identify and characterize all functional components of the cell. Around the turn of the millennium, the Human Genome Project laid open our entire cellular catalogue, but shockingly discovered that less than 21,000 protein-coding genes – just ~5-times the number of a bacterium such as Escherichia coli – span only ~1.2% of the over 3 billion base pairs of the human genome.1-4 This lack of proteomic inventory initially perplexed the scientific community, but then spurred debates of possible underlying RNA contributions to cellular complexity.5,6 The Encyclopedia Of DNA Elements (ENCODE) project, an international collaborative research effort, was initiated to provide a comprehensive picture of all functional elements within the human genome through unbiased, transcriptome-wide coverage by RNA deep-sequencing (RNA-seq).7 Particularly striking are the discoveries that at least 75% of the genome is transcribed and that by far most of these transcripts do not code for proteins, but rather “non-coding” RNAs (ncRNAs), many of which are still uncharacterized in terms of their structure and function.7,8 Currently, more than 80,000 distinct ncRNAs have been identified in human cells, which reveals an unexpected and exciting RNA landscape in our body (with excerpts highlighted in Figure 1).9 Many RNA elements have been found to originate from overlapping loci, suggesting that similar RNA sequences can be distinctly generated or processed to perform different biological functions.10,11 In an effort to understand the complex functional networks these RNAs are involved in, systems biology approaches are beginning to be implemented. Abetting such holistic approaches are single molecule methods that promise to provide quantitative mechanistic details for individual biomolecules within living cells.
Figure 1. Survey of the RNA biology in a eukaryotic cell.
Detailed descriptions of RNA and RNP complexes are provided in Section 2.
While RNA-seq has proven powerful for discovering novel cellular RNAs, the approach is limited by the ensemble averaging and loss of spatiotemporal information caused by the isolation of cellular RNA. It thus remains unclear whether, for example, functionally important ncRNAs are expressed in low quantities across all cells of a sample or selectively expressed only in a few cells, which feigns low expression by dilution within the averaged measurement. Single molecule approaches have emerged as an unparalleled means to resolve complex cellular processes that are otherwise masked by such ensemble averaging. The recent implementation of single molecule fluorescence tools to characterize of mRNA expression rates and levels, mRNA and microRNA localization, and ribonucleoprotein complex (RNP) association in living cells, together with the emergence of super-resolution imaging techniques such as PALM and STORM,12 endows single molecule techniques with the potential to broadly dissect the functions and mechanisms of ncRNAs.
In this review, we begin with an overview of the different classes of RNAs in eukaryotic cells, in terms of their biogenesis, function and localization (Figure 1). Given the extraordinary amount of literature on these subjects, where appropriate we guide the reader to pertinent reviews for further detail. Next, we summarize recent technical achievements of single molecule fluorescence microscopy in visualizing RNA and RNA-protein complexes in vivo. Finally, we highlight some applications of single molecule tools over the last 15 years that investigate RNA function within cells. Throughout the text, we will promote a vision of uniquely resolving the still shrouded multitude of functional mechanisms of RNAs, especially ncRNAs, through single molecule approaches.
2. CELL BIOLOGY OF RNA
2.1. Life Cycle of mRNA
2.1.1 Transcription and Splicing of Pre-mRNA
The best-characterized RNAs of the cell are protein-coding messenger RNAs (mRNAs) and the ncRNAs involved in their processing. Over the last 50 years, biochemical, structural and biophysical studies have provided a wealth of information on mRNA biogenesis, function and localization. It is well known that mRNA does not function as a naked biomolecule, but rather as part of larger RNP complexes.13-15 RNA-seq technologies coupled with RNA-protein crosslinking have been successful in mapping RNA target binding sites of RNA-binding proteins on a genomic scale.16-19 These data have revealed extensive, sometimes unexpected RNP networks within the cell that are summarized in a recent review.20 Not surprisingly, single molecule studies have been employed most extensively to study mRNA transcriptional kinetics, expression levels, processing and localization (see section 4), motivated by the stochasticity and cell-to-cell variability associated with such processes.21 Here, we survey the numerous mRNA-protein (mRNP) complexes formed during biogenesis and processing of precursor-mRNAs (pre-mRNAs) into mature transcripts (Figure 1) and what role each processing event plays in the ultimate fate of an mRNA. Within this section, we also provide descriptions of the housekeeping ncRNAs that are involved in each step of mRNA maturation.
Pre-mRNAs are predominantly transcribed by RNA Polymerase (Pol) II and typically contain three distinguishable elements: protein-coding exons, flanking untranslated regions (5’- and 3’-UTRs), and (long) non-coding introns.22 By the act of splicing, introns are excised from the pre-mRNA, ultimately resulting in a processed mRNA with joined, contiguous exons.23,24 This process is catalyzed by the spliceosome, an RNP of large size, based on certain features on the pre-mRNA splice site: usually an intronic GU 5'-end splice site, an internal A-branch site, and AG 3'-end splice site. In humans and other complex metazoans, pre-mRNA is co-transcriptionally bound by several proteins that play a role in splicing, 5’-end capping and 3’-end polyadenylation. Ubiquitously expressed alternative splicing factors, such as heterogeneous nuclear ribonucleoproteins (hnRNPs) and serine-arginine rich-domain containing proteins (SR proteins), function to silence or activate splicing, respectively, and impact polyadenylation and mRNA export.25-30 Sequence-dependent binding of these proteins, as well as other tissue- and developmental- stage specific alternative splicing factors, to the pre-mRNA affects its structure and, consequently, its interactions with additional RNA-binding proteins. The ensuing sequence of hierarchical binding events ultimately determines the splicing potential of any given pre-mRNA splice site.31,32 Another mechanism of alternative splicing involves riboswitches, RNA structural motifs embedded in intergenic regions and 3’-UTRs that bind small metabolites, which in turn induce RNA conformational changes.33 Unlike in bacteria, where transcription and translation are coupled and hence regulation of gene expression by ubiquitous 5’-UTR-encoded riboswitches generally involves direct transcription termination or inhibition of translation initiation,34,35 in eukaryotes riboswitches are typically embedded next to splice sites that they obscure through formation of secondary structure. Once the riboswitch (or, more precisely, its “aptamer” motif) binds the cognate metabolite, the ensuing conformational change makes the splice site accessible, leading to changes in splicing pattern. The end result is an alternatively spliced mRNA that may, for example, contain internal stop codons that cause translation of aberrant peptides, premature translation termination, or destabilization of the transcript.33
While some splicing events are constitutive, high-throughput sequencing studies have revealed that nearly all multi-exon gene transcripts can be alternatively spliced, thus promoting transcriptomic and proteomic diversity in eukaryotic cells.36,37 One of the most profound examples of alternative splicing occurs in the DSCAM (Down Syndrome Cell Adhesion Molecule) gene in D. melanogaster that codes for 38,016 protein isoforms,38 We note that this extreme example is most likely an exception, at least in mammals, as it has recently been shown that most mammalian genes code for one dominant transcript.39 However, given the vast number of possible exon combinations and the challenge to maintain single-nucleotide splicing accuracy to avoid loss of the codon reading frame, it is not surprising that aberrant alternative splicing can result in the malfunction of proteins and ultimately disease.40,41 In fact, it has been suggested that 60% of all human disease causing genetic mutations act through altering the splicing code.42
The spliceosome itself is a dynamic macromolecular RNP machine, containing five small nuclear RNAs (snRNAs) termed U1, U2, U4, U5 and U6 that function in concert with cognate proteins to form snRNPs.43 The snRNAs function as structural scaffolds and mediators of splice site selection.44 Most snRNAs are transcribed by RNA Pol II, with the exception of U6, which is transcribed by RNA Pol III.45 In total, over 200 individual RNA and protein components are assembled and disassembled during spliceosomal mediated excision of an intron, ultimately linking together two exons via two transesterification reactions.23 In human cell lines, approximately 80% of splicing occurs co-transcriptionally, while it has been proposed that post-transcriptional splicing occurs within interchromatin foci termed nuclear speckles.46 Nuclear speckles consist of active, highly dynamic spliceosomal protein components, yet their direct role in post-transcriptional splicing remains debated.47 Once an intron is excised from the pre-mRNA, a multi-protein exon-junction complex (EJC) is deposited ~20 nucleotides (nt) upstream of the adjoined exon-exon boundary, and in turn affects mRNA transport, translation and stability.48,49
In contrast to RNP-mediated splicing, self-catalyzed RNA splicing occurs in Group I and Group II introns, largely based on structural rearrangements of the RNA.50-52 In most cases, it has been shown that high salt (and Mg2+ in particular) promotes RNA catalysis of these introns in vitro, proving that they are RNA-based enzymes or “ribozymes”, yet some proteins are necessary in vivo. In addition to self-splicing introns, numerous other naturally occurring ribozymes have been characterized, including the hairpin, hammerhead, hepatitis delta virus (HDV), Varkud satellite (VS), and glmS ribozymes, in some cases using single molecule fluorescence tools in vitro.53-66 Interestingly, structural motif searches, in vitro selections, and biochemical validations of ribozyme catalytic activity have led to the discovery that the hammerhead and HDV ribozymes in particular exist as ncRNA elements within the genomes of diverse organisms, including humans.67-71 The finding that RNA can catalyze enzymatic reactions supported the RNA World hypothesis, wherein RNA spawned life as we know it by both self-replicating and catalyzing the metabolic reactions necessary to sustain life independent of proteins.72-75
2.1.2. Capping and Polyadenylation of Pre-mRNA
In addition to intron removal, pre-mRNA is modified within the nucleus with a 5’-end 7-methylguanosine cap (5’-cap) and a 3’-end poly(A) tail. The 5’-cap protects the mRNA from nucleolytic cleavage, serves as signal for the ribosome to start translation, and has been shown to have roles in mRNA splicing, nuclear export, stability, and translation.76 A 3’-end canonical hexanucleotide polyadenylation signal, AAUAAA, is found 10-30 bases upstream of the polyadenylation site. The length and location of poly(A) tails can vary, both of which can affect mRNA stability, translational efficiency and transport from the nucleus to the cytoplasm.77 The resulting mature mRNA typically contains a 5’-cap, a 5’-UTR, protein coding exons, a 3’-UTR, and a poly(A) tail. UTRs, just like introns, are cis-acting regulatory ncRNA elements, whose primary sequence and secondary structure directly affect protein and RNA binding and ultimately play critical roles in the regulation of gene expression.22,78 Interestingly, the length of UTRs and the fraction of alternatively spliced genes scale with the developmental complexity in animals, indicative of the greater sophistication of mRNA regulation in higher organisms.22
2.1.3. Nuclear Export of mRNA
Processed, mature mRNAs remain coated with RNA-binding proteins, including the EJC,TREX complex, Aly, Nxfl and SR proteins, that serve to package and compact the mRNA during transport across the nuclear envelope (from the nucleus into the cytoplasm) through the nuclear pore complex (NPC)79-83 or through the recently discovered nuclear envelope budding.48 Such transport processes, especially via the nuclear pore, have been extensively investigated using microscopy techniques, to unravel structural and mechanistic details.48,81,84-88 Classically, the NPC is considered the prevalent mode of RNP shuttling between the nucleus and cytoplasm. The nuclear pore is an almost cylindrical macromolecular complex comprised of nucleoporin protein building blocks.82 Recently, it was found that RNPs can also be transported from the nucleus into the cytoplasm by nuclear envelope budding using mechanism similar to the release of herpes virus capsids.87 Single molecule microscopy presents an exciting avenue to study these yet-to-be characterized RNP transport processes.
2.1.4. Translation of mRNA
Once in the cytoplasm, mRNAs contain numerous signals that are recognized by the cytoplasmic processing machinery that ultimately determines the individual fate of each mRNA. Some mRNAs will be destined to be translated by the ribosome, while others will be targeted for translational repression and decay by miRNAs or siRNAs (see section 2.2). As transcription and mRNA maturation are not fully accurate, some transcripts will contain premature stop codons and are destroyed by the cell via nonsense-mediated mRNA decay (NMD). Each of these processes occurs in sub-compartments of the cytoplasm and has been the focus of numerous studies that are nicely summarized by, for example, Martin and Ephrussi.89
To be efficiently translated, mRNAs must contain a 5’-cap, appropriately positioned EJC, and a poly(A) tail greater than 50 nt with a poly(A) binding protein (PABP)90,91 bound. The translating ribosome in eukaryotes is comprised of each a small (40S) and a large (60S) subunit, together referred to as the 80S ribosome. The 40S subunit is comprised of one ribosomal RNA (18S rRNA) and 33 proteins, while the 60S subunit is composed of three RNAs (5S rRNA, 5.8S rRNA and 28S rRNA) and 46 proteins. Most rRNAs are transcribed in the nucleolus by RNA Pol I, with the exception of 5S RNA, which is transcribed by RNA Pol III. rRNAs are chemically modified by small nucleolar RNA(snoRNA)-directed methylation and pseudouridylation.92 The individual rRNA and ribosomal protein components assemble in a hierarchical manner and form pre-ribosomal components in the nucleus that are exported into the cytoplasm where assembly is completed.93
snoRNAs represent one of the best characterized classes of non-coding RNAs.94-96 Localized to the nucleolus, snoRNAs are often transcribed from intronic regions of the genes they modify. The two major classes of snoRNAs are distinguished by the type of modification they mediate on rRNAs, snRNAs, and tRNAs: C/D box snoRNAs define the target sites for 2′-O-ribose methylation, whereas H/ACA box snoRNAs define the target sites for pseudouridylation. The RNA structure varies between these classes and likely mediates the binding between a snoRNA and its cognate modifying protein to produce a mature snoRNP.97 Recent data have linked snoRNAs to cancer and as precursors to miRNAs, suggesting that these RNAs will need to be examined in new contexts.98,99
Once eukaryotic initiation factors (eIFs) bind distinct segments of the 5‘UTR, such as eIF4E the mRNA cap, translation is primed. The full 80S ribosome is then assembled and the ribosome begins to translocate along the mRNA to synthesize proteins via the sequence specific recognition of three nucleotide codons by aminoacyl-tRNAs. tRNAs are transcribed by RNA Pol III (similar to 5S rRNA) and are heavily site-specifically modified guided by snoRNAs.100 tRNAs are evolutionarily ancient and characterized by a compact L-shaped tertiary structure, in aggregate carrying over 100 types of modifications, discovered by the first ever RNA crystallization experiment.101 In many organisms, multiple copies of tRNA genes give rise to distinct levels of any given tRNA species, which may affect translation rates.102-104 Maturing tRNAs are processed by endonucleolytic 5’-end cleavage by RNase P, an evolutionarily conserved RNP found in all three kingdoms of life and one of the first catalytic RNAs to be discovered.105,106
Nascent polypeptides sequester another RNP highly conserved in all three kingdoms of life, termed signal recognition particle (SRP), which in eukaryotes contains one conserved RNA and at least six proteins, that direct the nascent peptide to the endoplasmic reticulum (ER) or plasma membrane.104,107,108 The RNA component serves both as a scaffold and mediates global rearrangements of the SRP in response to binding its polypeptide cargo. The SRP directs the translocation of the growing polypeptide into the lumen of the ER, where the protein is then folded into its native form.109
2.1.5. Nonsense-Mediated Decay and mRNA Turnover
Nonsense-mediated decay (NMD) is a mechanism by which the cell eliminates mRNAs that contain premature stop codons, many of which result from alternative or aberrant splicing. Numerous RNA-binding proteins, including UPF1, UPF2, and UPF3 (the latter two are components of the EJC) mediate NMD and are associated with the mRNA, at least transiently, within cytoplasmic processing bodies (P-bodies),110 cellular foci that are enriched in RNA processing and degrading enzymes.111 One proofreading round of translation is sufficient to target the mRNA for NMD. We direct the reader to some reviews for further mechanistic details of NMD.112-114
Protein expression is highly correlated with the amount of its mRNA available. To be able to modulate the expression pattern of a cell over time, it is advantageous for aging mRNAs to be degraded.115 Degradation occurs via two pathways, the first involving shortening of the poly(A) tail by a deadenylase followed by decapping of the 5’-cap by Dcp1p and Dcp2p, which exposes the RNA to digestion by 5’-to-3’-exonucleases. The second mechanism requires mRNA deadenylation, followed by digestion by the cytoplasmic exosome.
Certain disease-related proteins have been shown to affect mRNA localization and gene expression. For example, fragile X syndrome-associated fragile X mental retardation protein (FMRP) has been shown to bind mRNAs to direct their localization within the cell and ultimately affect protein expression of target mRNAs.116,117 In addition, it was shown that fragile-X-mental-retardation-related protein 1 (FXR1) and Argonaute 2 (AGO2) bind AU-rich elements (AREs) in a microRNA dependent manner within the 3’-UTR of mRNAs to activate translation during cellular quiescence, thereby providing mechanistic evidence of the importance of cis-activating regulatory elements in 3’-UTRs.118 In addition to FMRP, several other RNA binding proteins (RBPs), such as Staufen and zip-code binding proteins (ZBP), bind specific sequences within UTRs to localize a large fraction of transcripts to distinct sub-cellular domains.89 In the following sections, we will discuss more broadly the mechanisms by which small and long ncRNAs control gene regulation.
2.2. SMALL NON-CODING RNA
2.2.1. Types and Functions of Small ncRNAs
RNA silencing is an evolutionarily conserved mechanism of gene silencing involving three main classes of small ncRNAs, including microRNAs (miRNAs), small interfering RNAs (siRNAs) and PIWI-interacting RNAs (piRNAs).119 The classical biogenesis and cytoplasmic mechanisms of miRNA- and siRNA-mediated gene silencing are similar, as both types of ncRNAs are processed from a relatively longer RNA duplex into an ~22-nt short single strand that engages an Argonaute-containing protein complex to bind and silence target mRNAs. However, miRNAs, siRNAs and piRNAs differ in origin, structure, and their detailed mechanism of silencing. miRNAs are endogenously expressed (genome-encoded), highly sequence-conserved, small ncRNAs that are only imperfectly complementary to typically the 3’-UTRs of mRNAs and mediate translational repression and mRNA decay. By contrast, siRNAs are found either endogenously or administered exogenously and bind to mRNAs by perfect sequence complementarity to mediate site-specific mRNA cleavage.120 Since target destruction is more immediate and absolute, siRNA mediated repression tends to be stronger than that achieved by miRNAs. Finally, piRNAs are ~26-30 nt in length, engage PIWI proteins, and function to silence transposons in the animal germline.121 In addition to their canonical functions, the last several years have revealed important roles of these and similar small ncRNAs in epigenetic gene regulation122 and the DNA damage response.123,124
In 1993, Victor Ambros and colleagues described the first miRNA, lin-4, as a protein expression regulator during normal larval development of the nematode worm Caenorhabditis elegans, although the mechanism remained somewhat elusive.125 In 1998, Andrew Fire and Craig Mello laid the foundation for RNA interference (RNAi), a tool that exploits the introduction of exogenous siRNAs into the cellular RNA silencing pathway to mediate mRNA cleavage, for which they shared the 2006 Nobel Prize in Physiology or Medicine.126 Since these initial reports, small ncRNAs have been identified in plants, animals, and even bacteria (although these sRNAs are often processed from protein-coding transcripts),127 and have been found to be a predominant mechanism for regulating gene expression in eukaryotes.128,129 On the one hand, it is now estimated that at least 60% of protein coding genes are regulated by at least one miRNA.130 On the other hand, siRNAs are routinely exploited in functional genomics, and their therapeutic implications are slowly being realized, although off-target effects and cell-specific delivery remain challenging.131 There are also numerous emerging classes of small and mid-sized ncRNAs that will not be discussed here for brevity, but are summarized in a recent review.132 Given the relatively recent discovery of small ncRNAs and their expanding repertoire of types and functions, we will discuss, where appropriate, outstanding questions and the potential of single molecule microscopy to address them. We will specifically focus on the biogenesis, localization and function of siRNA and miRNAs because of their pervasive functions and the emergence of reports that use single molecule microscopy for functional and mechanistic probing.133-136
2.2.2. Biogenesis of Small ncRNAs
miRNAs are the most ubiquitous small ncRNA in humans, with over 1,500 different mammalian miRNA sequences discovered to date that represent more than 1% of the entire genome and thus the largest gene family.137-139 These RNAs are usually transcribed by RNA Polymerase II as long primary miRNA (pri-miRNA) transcripts.140,141 pri-miRNAs adopt hairpin structures with numerous bulges that are recognized and cleaved by the nuclear endonucleolytic microprocessor complex, mainly comprised of the RNase III enzyme Drosha and its cofactor DGCR8 (Pasha in invertebrates).142 The resulting pre-miRNA hairpin, ~65- to 70-nts in length and containing a 2-nt 3’-overhang, is then bound by Exporin-5 and RanGTP for export from the nucleus to the cytoplasm through the NPC.
Once in the cytoplasm, pre-miRNAs as well as long double-stranded RNAs are recognized and cleaved by the RNase III enzyme Dicer and its cofactor TRBP into short, 20-24 nt duplexes, with characteristic 2 nt 3’-overhangs bearing 3’-OH groups and 5’-phosphates.139 The mature miRNA duplex is loaded into the multiprotein RNA-induced silencing complex (RISC) loading complex (RLC) that includes an Argonaute (AGO) protein.143 Strand selection, thought to be dependent on the thermodynamic stability of the duplex and/or presence of RISC-associated protein components,144 occurs within the RLC, wherein one strand of the duplex (the passenger strand) is cleaved and/or dissociates from the complex. The mature RISC complex contains the miRNA guide strand bound by Argonaute and can now seek out its complementary RNA sequence. Of note, the cytoplasmic portion of the siRNA biogenesis pathway in mammals is very similar.119
In addition to this canonical miRNA biogenesis pathway, whose multi-step nature allows for tight regulation,145 miRNAs are also generated using the mirtron pathway146,147 wherein short hairpins derived from excised introns serve as Dicer substrates to generate miRNAs. Since mirtons are initially processed by the spliceosome, they bypass regulation of the nuclear Drosha processing step and merge with the canonical cytoplasmic miRNA biogenesis pathway only upon nuclear export.
2.2.3. Spatial and Functional Requirements of Small ncRNAs in the Cytoplasm
In the cytoplasm, miRNA-loaded RISC (miRISC) binds mRNA targets to repress translation and then promote mRNA decay, possibly within P-bodies,135,148-150 through specific sequence requirements. Nucleotides 2-7 located on the 5’-end of the miRNA guide strand comprise the seed sequence that is the primary determinant for stable binding to the 3’-UTR of miRNA targets.130 Additional structural elements in the 5’-UTR of targeted mRNAs have been recently shown to elicit synergistic effects with miRNA binding sites in the 3’-UTR to enhance RNA silencing.151 Although several bioinformatic portals are available to predict putative miRNA targets (TargetScan©, PicTar©, and miRanda©, to name a few), few have been experimentally validated and their accuracy is still poor, largely owing to their reliance on relatively short seed sequences whose frequency of occurrence is high despite a requirement for phylogenic conservation.152,153 In light of recent reports that have underscored the importance of target site accessibility154 and seed-independent miRNA binding,155 these target prediction algorithms warrant an overhaul. Furthermore, apart from specific seed matches miRNA-mediated decay requires the recruitment of additional protein components, in particular the Argonaute-associated proteins GW182, CCR4-NOT and RNA helicase eIF4A2.151,156 One such protein is also thought to be responsible for the spatial organization of RISC assembly and miRNA mediated target repression. Li et al. described the altered meristem program1 (AMP1) protein dependent localization of miRNA-loaded AGO1 to the ER, proposing the ER as the main sub-cellular site of repression in Arabidopsis.157 Stalder et al. further reported evidence that the rough ER is the nucleation site for RNA silencing where both miRISC assembly and target repression occur,158 and hypothesized that ER localization is mediated by TRBP and PACT. Regardless of spatial and sequence constraints, it is still unclear whether reduced protein output is achieved by a miRNA efficiently repressing only a subset of molecules of a given type of mRNA or less efficiently repressing a large number of the same mRNA molecules. Moreover, the binding stoichiometries of miRNAs to mRNAs are yet to be fully determined. Such questions are only accessible via single molecule microscopy.
A novel class of non-coding circular RNAs (circRNAs) was recently identified and characterized in mammals.159,160 These RNAs are processed by the spliceosome in an unusual head-to-tail fashion, resulting in circular transcripts that contain multiple miRNA binding sites and act as miRNA sponges or decoys to deplete the cell of specific miRNAs, essentially alleviating repression of the mRNAs they target.161 Single molecule fluorescence in situ hybridization (FISH) studies have shown that circRNA-miRNA complexes localize to P-bodies,160 although the reasons are unknown. Further functional characterizations of this abundant class of ncRNAs will be necessary to determine how universal this mechanism is for sequestering miRNAs inside cells.
2.2.4. Nuclear Localization and Function of Small ncRNAs
Recent reports, summarized here, have provided support for novel roles of miRNAs and siRNAs within the nucleus,128 including their canonical function of post-transcriptional gene regulation. How do ncRNAs that are processed and function in the cytoplasm localize to the nucleus? In human cancer cell lines, a 3’-end hexanucleotide nuclear localization signal, AGUGUU, has been shown to regulate the import of miR-29b from the cytoplasm into the nucleus where it may function to bind a unique set of targets.162 However, it has also been shown that miRNAs that lack such canonical import sequences are also imported into the nucleus, but indirectly.163,164 Nucleocytoplasmic shuttling proteins like Importin8 or TNRC6A (a GW182 isoform) associate with miRISC for transport into the nucleus, possibly triggered by specific cellular cues.165
Numerous research groups, working predominantly with C. elegans, have shown that small ncRNAs and the RNAi machinery have critical roles in epigenetic DNA modification and heterochromatin formation.166,167 For example, exposing the nematode worm to double-stranded RNA results in heritable expression of siRNAs and the heritable epigenetic modification of DNA in the form of histone 3 lysine 9 methylation (H3K9).122,168 Certain miRNAs also influence DNA methylation and histone modification of protein-coding and ncRNA genes, thereby affecting gene expression.169 In another report, it was shown that Argonaute CSR-1 associates with small RNAs (termed 22G-RNAs) and other cofactors to target and efficiently segregate chromosomes during cell division.170 Many 22G-RNAs are antisense to germline-specific protein coding genes, suggesting this mechanism as a potentially common mode of regulation. Another abundant small RNA in C. elegans is 21U-RNA,171 which was found to associate with PIWI-like proteins, and thus to have germline-related functions.172,173 In fission yeast, small RNAs termed pri-RNAs were shown to be Dicer-independent mediators of RNAi involved in heterochromatin formation.174
Small ncRNAs have also been found to associate with pre-mRNAs in the nucleus. For example, it was shown in C. elegans that Argonaute-associated siRNAs are able to inhibit RNA polymerase II and silence pre-mRNAs co-transcriptionally in a process termed RNA induced transcriptional silencing (RITS).175 Another report suggested that human AGO1 and AGO2, which are generally associated with their RNAi functions, can also be involved in alternative splicing of pre-mRNA, a process that is possibly mediated by a small ncRNA component.176 Small ncRNAs have also been shown to autoregulate their own biogenesis, as shown with Argonaute-associated mature let-7 miRNA binding and cleaving the 3’-end of let-7 pri-miRNAs to promote let-7 maturation.129
Finally, a novel class of DNA damage response associated RNAs (DDRNAs) has recently been identified and characterized in mammals, zebrafish and plants.123,124,177 Similar to miRNAs, these RNAs are processed by Dicer and Drosha to generate short, 20-35 nt products. Yet, unlike miRNAs, DDRNAs are not further processed and instead localize to specific DNA damage sites in the nucleus where they may function to recruit proteins involved in DNA damage repair. With the sophisticated high-throughput sequencing and screening tools available today, we will likely discover many more yet unknown small ncRNA-mediated pathways, all of which can in principle be probed by the single molecule techniques highlighted here.
2.3. LONG NON-CODING RNA
2.3.1. Discovery of Long ncRNAs
Long non-coding RNAs (lncRNAs) or long intergenic non-coding RNAs (lincRNAs) are an abundant class of non-coding RNAs that have recently emerged from deep-sequencing data as ubiquitous cellular transcripts of high structural and functional diversity.178-180 Unlike the “housekeeping” small ncRNAs that display clear evolutionary conservation in terms of sequence and structure, lncRNAs are more difficult to classify due to a lack of evolutionary conservation based on primary sequence so that they have remained somewhat of an enigma, despite often exhibiting functional conservation.32
lncRNAs are greater than 200 nt in length with little or no protein-coding capacity. This diverse group of RNAs is expressed tissue-specifically and is classically defined by their function in epigenetics to condense chromatin and regulate DNA methylation and histone modification, thereby positively or negatively affecting the expression of nearby genes.181 Genetic studies from the early 1990's revealed the first lncRNA, Xist, as an ~17,000 nt long RNA that coats and inactivates one X chromosome during dosage compensation in sex determination of mammals. Other lncRNAs, such as H19 and Air, are also involved in genetic imprinting by silencing adjacent alleles through DNA methylation and histone modifications.182-184 Many novel lncRNAs have been identified by high-throughput sequencing of cell type-specific transcriptomes, and subsequent characterization has only begun to illuminate the functional nuclear and cytoplasmic niches of lncRNAs. The biogenesis, cognate protein partners, and functions of lncRNAs remain the most elusive of all ncRNAs so that we discuss only a subset of the best characterized lncRNAs. For further detail, we refer the reader to several recent reviews on specific lncRNAs, including promoter-associated RNAs (PARs).179,185-187
2.3.2. Biogenesis of Long ncRNAs
lncRNAs are found throughout the genome, including intergenic regions (lincRNAs), in antisense, overlapping, intronic, and bidirectional regions relative to protein-coding genes, as well as in UTRs, promoters and enhancers.8,188,189 The biogenesis of lncRNAs is quite similar to that of mRNAs, in that they are typically transcribed by RNA Polymerase II, spliced and further processed to contain a 5’-cap and polyA tail. In fact, some mRNAs have been shown to function as lncRNAs.190 In addition, recent studies suggest that lncRNAs can be chemically modified, a feature classically associated with rRNAs and tRNAs.179 It is possible that chemical modifications are present to stabilize lncRNA secondary and tertiary structures, or that they have evolved to preclude activation of the innate immune response. For example, it was recently shown that modified, but not unmodified, tRNAs avert activation of the innate immune response protein dsRNA-activated protein kinase R (PKR).191 Yet another layer of lncRNA complexity pertains to the presence of adjacent snoRNAs (sno-lncRNAs) loci.192 Thus, this novel class of ncRNAs harbors many surprises, leading to many functionally interesting questions that can be addressed using single molecule approaches.
2.3.3. Epigenetic Gene Regulation and Other Functions of Long ncRNAs
lncRNAs were first characterized for their nuclear functions related to epigenetic gene regulation by DNA methylation and chromatin remodeling. These functions require lncRNAs to associate with proteins such as polycomb complexes or histone modifying proteins.188,193,194 The lncRNAs guide modifying proteins to specific DNA sites to repress gene expression though histone methylation of H3K9 and trimethylation of H3K27. Within the last five to ten years, however, numerous reports have expanded the functional roles of lncRNAs to the cytoplasm where they have been shown to associate with importin-β proteins to prevent nuclear import of a transcription factor (i.e., NRON lncRNA); bind an antisense mRNA to increase protein synthesis in response to stress (i.e., UCHL2 lncRNA); and bind the 3’UTRs of mRNAs to induce decay by dsRNA-recognition protein Staufen1.195-197 Recent studies have also revealed that pseudogenes can act as ncRNAs to regulate gene expression of their protein-coding counterparts.198 Further characterization of the elusive class of lncRNAs will be necessary to determine the full extent of their cellular functions.
2.4. TELOMERASE RNA
Telomerase RNA is a specialized type of lncRNA that, similar to other lncRNAs, acts on DNA in the nucleus. DNA telomere sequences are located at the ends of chromosomes and function to delay cellular senescence.199,200 These sequences are maintained by telomerase, a large ~1,000 kDa (in vertebrates) RNP, whose function was discovered by Blackburn, Greider and Szostak, for which they received the Nobel Prize in Physiology or Medicine in 2009. Telomerase is comprised of an RNA component containing a template sequence, a reverse transcriptase protein component that extends the telomere as guided by the template, and numerous accessory proteins. Aberrant telomerase activity has profound cellular consequences, where telomerase up-regulation in most immortalized cancer cell lines is thought to prevent cellular senescence whereas its inactivity expedites cell death in some diseases.201,202 Telomerase recruitment to chromosome ends has been investigated using single molecule fluorescence approaches, as described in section 4.11.
3. PRINCIPLES OF INTRACELLULAR SINGLE MOLECULE FLUORESCENCE MICROSCOPY OF RNA
As surveyed above, our appreciation for the diversity of cellular RNAs has exponentially increased over the last decade. With the rapid advancement of deep-sequencing and bioinformatics technologies, we are likely to unearth still other classes of RNAs, a further increased functional diversity, as well as novel RNA-protein interactions. The current ensemble-averaged approaches clearly will continue to provide a wealth of information on RNA biology. However, biology is fundamentally stochastic in nature, leading to diverse, spatiotemporally inhomogeneous distributions of molecules within cells as well as across individual cells, even within a clonal cell line or (tumor) tissue. The resulting heterogeneities, short-lived and/or rare pathway and reaction intermediates, dispersed cellular localization and time evolution, multitude of parallel mechanisms of action and non-linear responses from complex, multi-hub networks together form the very foundation of biomolecular function. The omnipresence of such molecular dispersions warrants the development of ultra-sensitive, non-invasive techniques that expose them, leading to the application of emergent single molecule microscopy techniques to biological samples. Some of the earliest implementations of single molecule microscopy have been used to characterize biological processes in unprecedented detail – as exemplified by the observation of single β-galactosidase molecules trapped in microdroplets in the presence of a fluorogenic substrate,203 tracking of single (oftentimes tethered) beads or particles in vitro or in cellulo,204-208 recording of the absorption or fluorescence of single pentacene molecules in p-terphenyl crystalline matrices at liquid-helium temperature,209,210 and measurement of single enzyme turnovers.211
Single molecule microscopy (SMM) can broadly be divided into two categories, optical observation and mechanical manipulation tools. In this review, we will focus on optical methods that employ single molecule fluorescence microscopy hereon referred to as SMM) to probe the intracellular function of RNA. Imaging tools such as atomic force microscopy (AFM) and methods that apply mechanical manipulation to single molecules such as optical and magnetic tweezers are beyond the scope of this article, but a broad overview of such techniques can be found in several reviews.212,213
It turns out that SMM is primed to break the classical optical diffraction limit. According to Abbe's law or Rayleigh's resolution limit,212 diffraction limits our ability to distinguish two features located closer (on the lateral plane) than half the wavelength of the illuminating or emitted light, thereby imposing a theoretical limit on the resolution of fluorescence microscopy of 200-300 nm (using visible, ~500-nm illumination light). Consequently, the image of a single fluorescent probe, typically a few nanometer in diameter, is spread after passing the microscope optics over a few 100 nm on the detector. The intensity distribution of such a diffraction-limited spot can be mathematically described by a point spread function (PSF) and approximated as a simple two-dimensional (2D) Gaussian function. The center of the Gaussian curve, which coincides with the intensity maximum of the diffraction limited spot, can be localized with accuracy similar to the size of the fluorescent emitter, effectively breaking the diffraction barrier. Recent advancements in instrumentation have thus facilitated our ability to visualize single molecules under ambient conditions in situ at nanometer spatial resolution,212,214,215 previously accessible only to biologically invasive techniques such as electron microscopy.
However, the application of intracellular SMM presents a unique pair of challenges: (i) The need to reach an appropriately low sample concentration to delineate individual molecules within the dense and complex milieu of a cell; and (ii) the requirement to detect photons (signal) from individual molecules within the uneven background (noise), contributed mostly by both autofluorescence and signal from out-of-focus molecules, with minimal phototoxic effects on the cell. The former is specifically difficult to control when probing endogenous biomolecules, especially RNA, whose intracellular abundance can vary from a few to several (tens of) thousand(s of) molecules per cell. The latter, especially autofluorescence that is primarily contributed by fluorescent intracellular metabolites, cofactors and pigments, is omnipresent. Put together, these obstacles render the successful implementation of intracellular SMM non-trivial. Nevertheless, a careful choice of labeling strategies and imaging conditions can make this seemingly daunting task relatively seamless. For instance, titratable reporters,216 controlled delivery of labeled probes135,217,218 and ultra-high resolution microscopy methods that systematically probe only a subset of all labeled probes at any given time212,215,219 have judiciously tackled the concentration challenge, whereas improved optical configurations (illumination sources, strategies and detectors) and fluorescent probes have successfully dealt with the latter. In this section, we will review and present a “panorama” of the fluorescent probes, labeling strategies and imaging schemes that have been employed to achieve in cellulo single RNA/RNP molecule detection, along with their respective advantages and disadvantages.
3.1. Fluorescent Probes
During the early stages of intracellular single particle tracking (SPT) and single molecule microscopy large (0.25-2 μm), either fluorescent or non-fluorescent beads were popular as reporters.204,220 Their large size enabled convenient high-precision imaging without the risk of undesired signal photobleaching, even when using microscopes with unsophisticated optics. However, conjugating biomolecules to large beads comes with the caveat that the attachment of a bulky load may skew the molecule's function, localization, and/or diffusion, or introduce other artifacts.220 Moreover, limited options for multiplexing means that non-fluorescent beads cannot be used to probe multiple types of biomolecules simultaneously. Thus, small fluorescent probes, available in various colors, soon superseded beads as the primary choice of visual reporters in SMFM.
Upon their discovery in the 1960's221 and cloning in the 1990's,222 fluorescent proteins (FPs) quickly became a mainstay of intracellular fluorescence microscopy.223 The ease with which FP genes can be expressed as fusions with cellular protein targets and the availability of a broad FP “color-palette” that spans the entire visible and near-IR part of the spectrum224 make them attractive probes. One of the main caveats of this labeling method is that protein fusions are often expressed exogenously, thus resulting in overexpression compared to endogenous levels, which jeopardizes physiological relevance. Additionally, overexpression typically increases intracellular particle density to an extent that it becomes refractory to single molecule visualization. Using weak promoters, inducible expression systems, controllable viral transduction or creating/selecting for stable cell lines with low expression are a few adaptations that can be employed to mitigate the effects of overexpression.225,226 In addition, recent genome editing technologies using Zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and clustered regulatory interspaced short palindromic repeat (CRISPR)/Cas-based methods have emerged as powerful tools that can function to regulate endogenous expression of FP fusions and thus, future implementation of these technologies may help circumvent the above hurdles.227 Despite suffering from frequent intensity fluctuations (blinking) and limited photostability,228 FPs are still preponderant in intracellular single molecule microscopy of RNA due to the ease of creating and delivering them as genetically encoded fluorescence markers. Even otherwise deleterious blinking properties have found compelling applications in super-resolution imaging,219,229,230 enhancing our ability to image samples of high probe density. The emergence of photoactivatable, photoconvertible and fluorescent dimer FPs224,231 has further improved super-resolution imaging schemes219 and tremendously aided in photosynchronization experiments.232 Moreover, a majority of current RNA labeling schemes invoke the binding of multiple FPs per RNA,233 wherein a few well-folded FPs compensate for the blinking or photobleaching of a subset of others within the complex.
Organic fluorophores (of the rhodamine, cyanine, oxazine, bodipy, perylene and other structural scaffolds) are typically preferred over FPs in intracellular SMM for their small size and superior photophysical properties, i.e., they don't blink as often, typically emit more fluorescence photons prior to photobleaching, and their undesired photophysical properties can be suppressed using several additives (as discussed in 3.1.1). In further contrast to FPs, organic dyes are predominantly conjugated to biomolecules ex vivo, via several well standardized conjugation chemistries.234-237 This labeling scheme often mandates the careful purification of probe labeled molecules from unlabeled molecules and unbound dye impurities, and for intracellular imaging, the specific delivery of tagged molecules to cells. Such hurdles are potentially overcome by the use of various genetically encodable tags that form a covalent adduct with organic dye substrates, such as SNAP® tags (NEB), Halo® tags (Promega) and tetracysteine motif bearing peptides (Invitrogen). These labeling strategies effectively combine the elegance of intracellular labeling via genetic engineering with tagging photophysically superior organic dyes. The development of bioorthogonal labeling strategies238,239 and fluorogenic photoaffinity probes240,241 has further broadened the scope of in cellulo labeling methods.
Fluorescent beads and quantum dots (QDs) have several favorable photophysical properties, as they are typically brighter, more photostable and the latter specifically have narrower emission spectra than organic fluorophores.242 However, akin to non-fluorescent beads, these probes are typically large (similar in size to a protein or small RNA) and have a high propensity to affect the intracellular physicochemical characteristics and function of the conjugated biomolecule. Their large size additionally inhibits efficient intracellular delivery and imposes steric constraints during target binding of QD/fluorescent bead labeled probes. Additional limitations of QDs include the potential for cytotoxicity of the composite transition metal ions and tendency for frequent blinking,243 where the latter is a bane for both single molecule counting (as it confounds intensity values that are used to measure copy number) and particle tracking (as it introduces difficulty in assigning contiguous tracks when particles temporarily vanish from observation).
3.1.1. Photophysical Properties Required for Detecting Single Fluorescent Probes
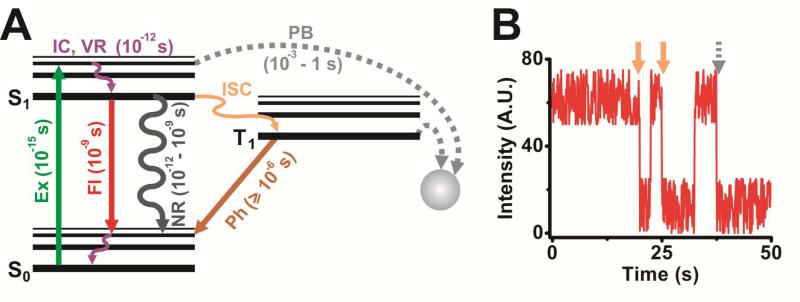
A fluorophore suitable for intracellular SMM should have high quantum yield (i.e., ratio of the rate of fluorescence to the sum of all relaxation rates; reflects the net efficiency of fluorescence), high brightness (i.e., measure of photon output calculated as the product of a fluorophore's extinction coefficient and quantum yield), favorable photophysical properties and sufficient inertness so that the label does not interfere with the function of the molecule to be tagged. Among these, brightness and inertness are inherent characteristics of the probe's chemical nature, and the brightness in particular is significantly influenced by the immediate chemical environment of the probe. It is thus critical to evaluate several probes and choose an appropriately bright fluorophore, such that the fluorescent signal is significantly more intense than the cellular autofluorescence. This cellular background is caused by naturally fluorescent molecules present inside cells, such as NADH, FADH and heme. Two ways to circumvent such background is to use: (i) cell culture media devoid of any naturally fluorescent molecules, especially vitamins;244 and (ii) fluorophores that absorb light and fluoresce in the far-red visible or NIR part of the electromagnetic spectrum where cellular components show minimal emission.228,245,246 Another dye and environment dependent, important photophysical property of fluorophores is their fluorescence lifetimes, i.e., the time taken for an excited singlet electron to transition back to the ground state and concomitantly release a photon (Figure 2). As fluorophore excitation (at femtoseconds, or fs) occurs much faster than photon emission (at nanoseconds, or ns) (Figure 2), excited singlet states have a propensity for electronic saturation, limiting the maximally possible yield of photons.
Figure 2. Photophysical properties of fluorophores.
(A) Simplified Jablonski diagram representing excitation (Ex), fluorescence (Fl) emission, internal conversion (IC), vibrational relaxation (VR), non-radiative decay (NR), intersystem crossing (ISC), phosphorescence (Ph) and photobleaching (PB), and the respective timescales at which these processes occur. S0, singlet ground state; S1, singlet excited state; T1, triplet state. (B) A simulated intensity trajectory of a single molecule with two blinking events (orange arrow) and a single photobleaching step (dotted grey arrow).
In contrast to its intrinsic brightness, undesirable photophysical processes affecting a given fluorophore, such as large intensity fluctuations and photobleaching, to an extent can be controlled extrinsically. Intensity fluctuations are typically characterized by reversible changes of the fluorophore between bright and dark states (i.e., blinking), whereas photobleaching signifies an irreversible switch to a dark state (Figure 2). Both processes markedly affect the quality and length of single molecule recordings. Blinking is predominantly induced by intersystem crossing (ISC, Figure 2), wherein fluorophore excitation populates electronic triplet states instead of singlet states. Relaxation back to the ground state from triplet states, which is a prerequisite for further cycles of electronic excitation and subsequent fluorescence, is quantum mechanically forbidden and takes ~1,000-fold longer than relaxation from singlet states so that probes are temporarily rendered dark. Blinking and photobleaching may also occur due to the chemical reaction of excited state molecules with radical species, induced by the excitation light or provided by the chemical environment of the dye. In certain cases, the excitation light may itself suffice to transform a fluorescent dye into its dark state in a phenomenon termed photochromism. However, several chemical agents such as cyclooctatetraene (COT), trolox, ascorbic acid, mercaptoethylamine (MEA), 4-nitobenzyl alcohol, 1,4-diazabicyclo[2.2.2.]octane (DABCO) and n-propyl gallate can be used to quench triplet states and radical species and, thus, reduce blinking and increase fluorophore longevity.228,247-249 It is noteworthy that the quenching action of some chemical agents is dye specific, for instance, MEA has proven to effectively quench triplet states of Rhodamine 6G250 but increase blinking in cyanine dyes like Cy5.247 Although it is still unclear, this detrimental action of MEA is attributed to its function as a reducing agent, especially considering that other reducing agents such as dithiothreitol (DTT), β-mercaptoethanol (BME) and tris(2-carboxyethyl)phosphine (TCEP) also induce such deleterious effects. This effect, termed redox blinking, can be induced by oxidants such as methyl viologen as well. Regardless of whether they enhance photophysical characteristics, the addition or removal of any of these chemical agents should be contingent upon their tolerability by and the viability of cells, especially in live cell imaging, whereas such stringency is not required for imaging fixed cells, where the choice of reagents can be purely dye-based. Another chemical that has been widely attributed as the cause of photobleaching, presumably via photooxidation of the fluorophore, is molecular oxygen and related species. Especially during intracellular imaging, excited fluorophores can react with molecular oxygen within cells, resulting in the accumulation of phototoxic free radicals that can compromise sub-cellular compartments or even the entire cell's livelihood.251 Enzymatic oxygen scavenging systems (OSS), such as those containing glucoseoxidase and catalase (GODCAT),252 protocatechiuc acid and protocatechuate-3,4-dioxygenase (PCA/PCD)247 or oxyfluor©253 utilize molecular oxygen as a substrate in enzymatic reactions, thereby effectively depleting it. These OSS prevent fast photobleaching and oxygen induced free radical production, resulting in increased signal longevity. However, it is critical to reduce the exposure of cells to OSS as they may lead to hypoxic shock,254 as well as to include good buffering agents in the imaging solution to overcome harmful effects of pH changes induced by certain OSS.255 As molecular oxygen is also an efficient quencher of triplet states, addition of OSS may increase fluorophore lifetimes at the cost of increased blinking rates.256 Thus, it is mandatory to include triplet state quenchers in imaging solutions that also contain OSS. Other deleterious photophysical effects, such as light induced free radical production and phototoxicity, are reduced by striking a balance between the excitation laser power (and wavelength) and the time over which the sample is illuminated while maintaining single molecule sensitivity.
3.2. Labeling Strategies of RNA for Intracellular Single Molecule Fluorescence Microscopy
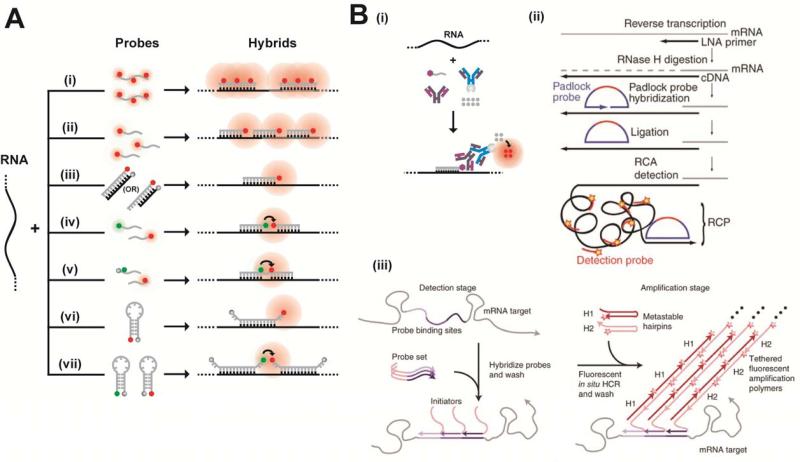
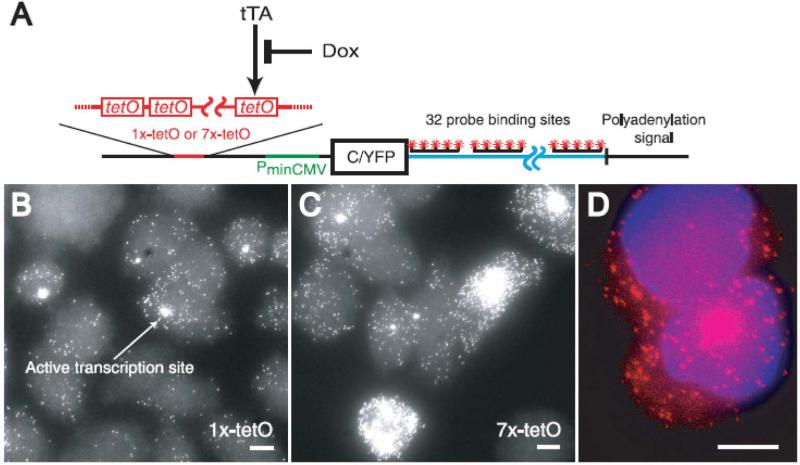
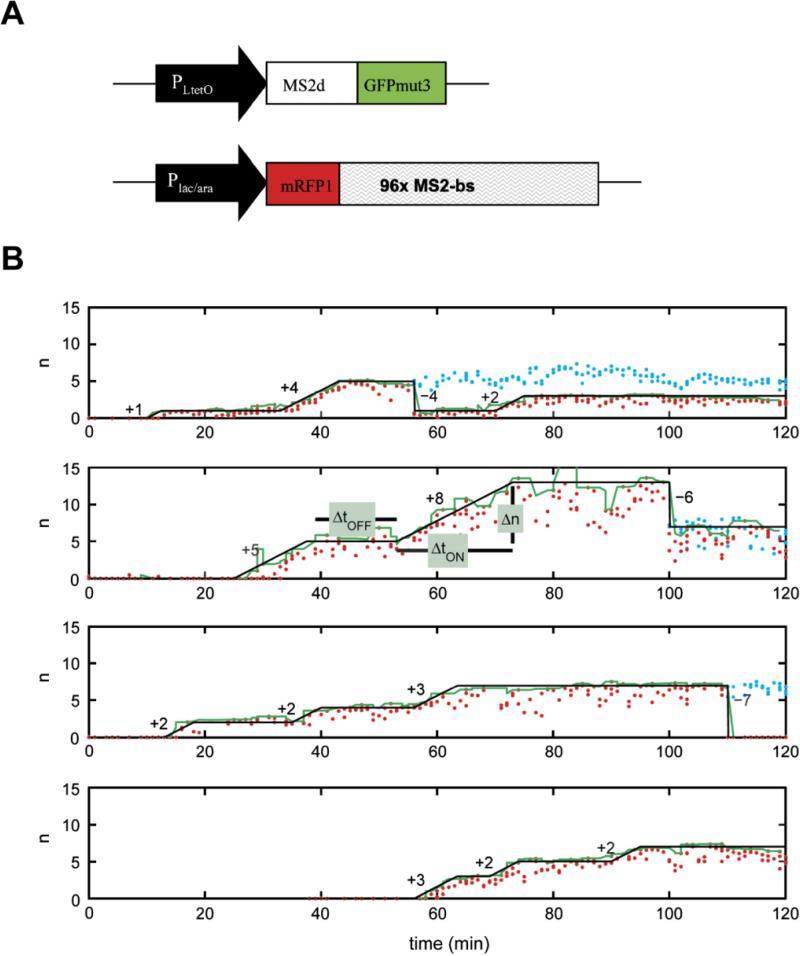
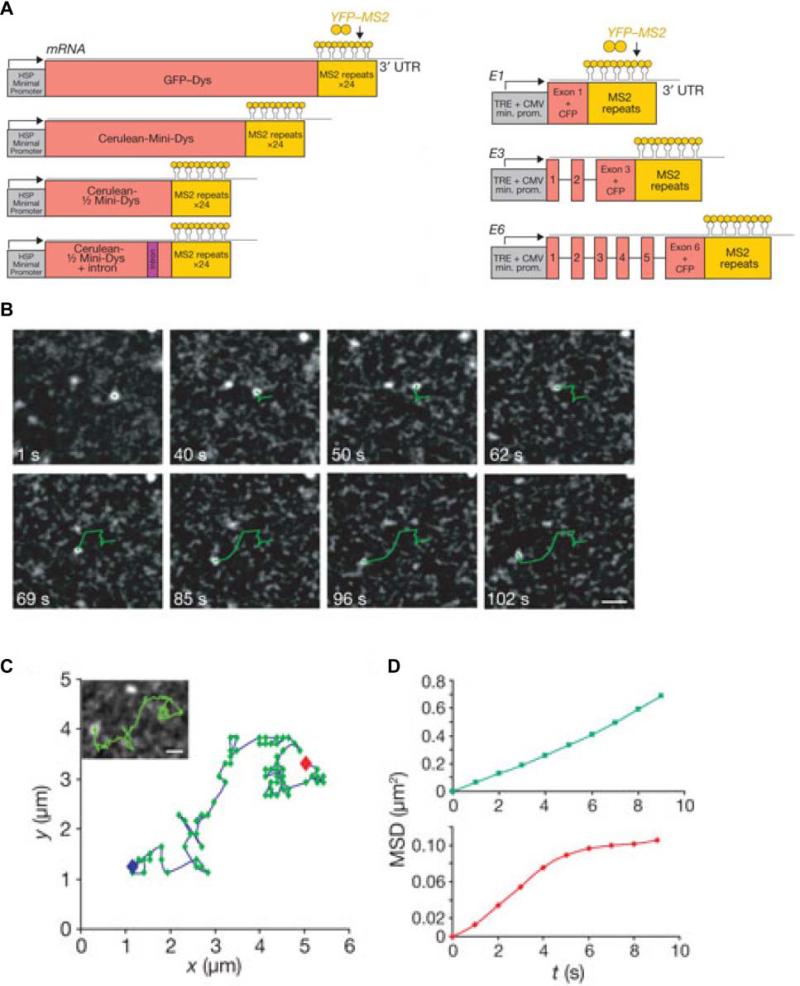
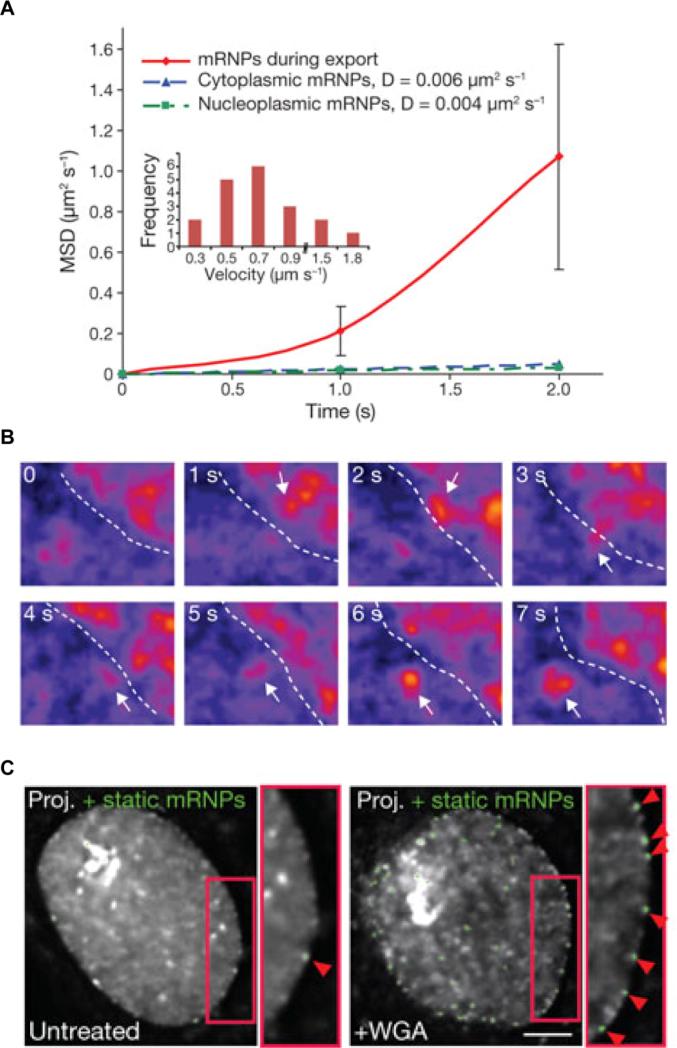
RNA labeling strategies may be crudely divided into two main categories, indirect and direct labeling schemes. The former employs sequence-complementary oligonucleotides (Figures 3 and 4) or fluorophore labeled RNA binding probes, such as RNA binding proteins, RBPs (Figure 5), which associate with appropriate RNA motifs to (indirectly) tag RNA with fluorophores. Conversely, direct labeling schemes exploit chemically reactive functional groups or structural motifs within the RNA, naturally present or introduced by chemical synthesis or RNA modifying proteins, for fluorophore conjugation (Figure 6). Currently, indirect labeling schemes are more predominant in intracellular SMM of RNA as they have the capability to probe endogenous targets, in addition to exogenous constructs, thereby finding widespread application in in situ gene expression profiling with single molecule sensitivity.21 In this review, we will focus on well-established RNA labeling schemes used in intracellular SMM, but also describe a few methods that have strong potential. A majority of these labeling strategies has been optimized to probe mRNAs; however, applications to the world of ncRNAs are slowly emerging.
Figure 3. Fluorescently labeling RNA by hybridization of labeled probes.
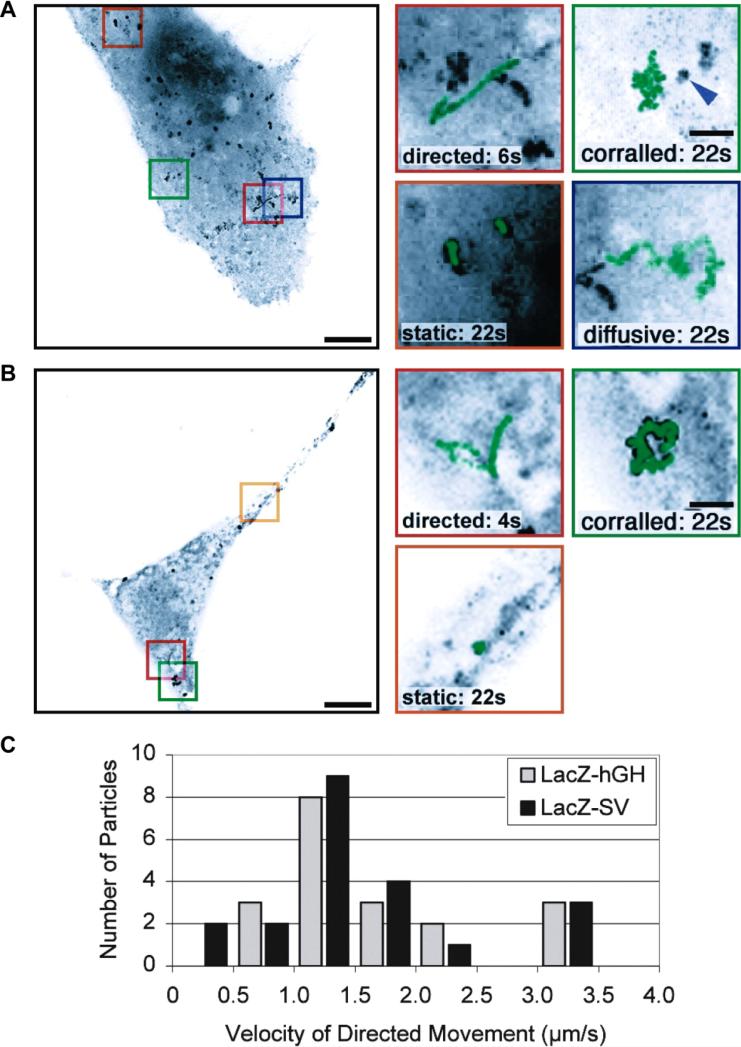
(A) Hybridization probes that do not employ signal amplification strategies. Represented here are schematics of the Singer approach using few multiply labeled probes (i), the Tyagi method of many singly labeled probes (ii), competitive hybridization (iii), and inherently quenched molecular beacons (iv). Green and red circles are spectrally distinct dye molecules and shaded grey circles are quenchers. Schematics iv–v and vii represent hybridization methods that have been widely used in ensemble imaging of intracellular RNA, with immense potential in single molecule imaging. Side-by-side probes493 (iv) are designed to bind target RNAs at adjacent positions, such that the binding configuration brings fluorophores on the two probes into close proximity to enable FRET. In a variant of this scheme, called quenched-autoligation494,495 (v), the probe containing the FRET donor also contains a quencher to suppress the fluorescence from unbound oligonucleotides. Once the functionalized probes bind side-by-side, they self-ligate, removing the quencher from the vicinity of the donor probe, thereby resulting in unquenched FRET. Another variant of the side-by-side scheme consists of dual molecular beacon FRET probes436,496 (vii); here signal specificity is enhanced by two beacons, one containing the FRET donor and another containing the FRET acceptor, which bind at adjacent locations to generate a FRET signal. Each probe contains a quencher to reduce fluorescent background from unbound probes. (B) Signal amplification in hybridization probes. Schematics representing the ELF approach (i), padlock probes (ii) (Reprinted with permission from ref. 273. Copyright 2010 Nature Publishing Group.) , and the HCR approach (iii). Reprinted with permission from ref. 290. Copyright 2010 Nature Publishing Group. RCP, rolling circle product.
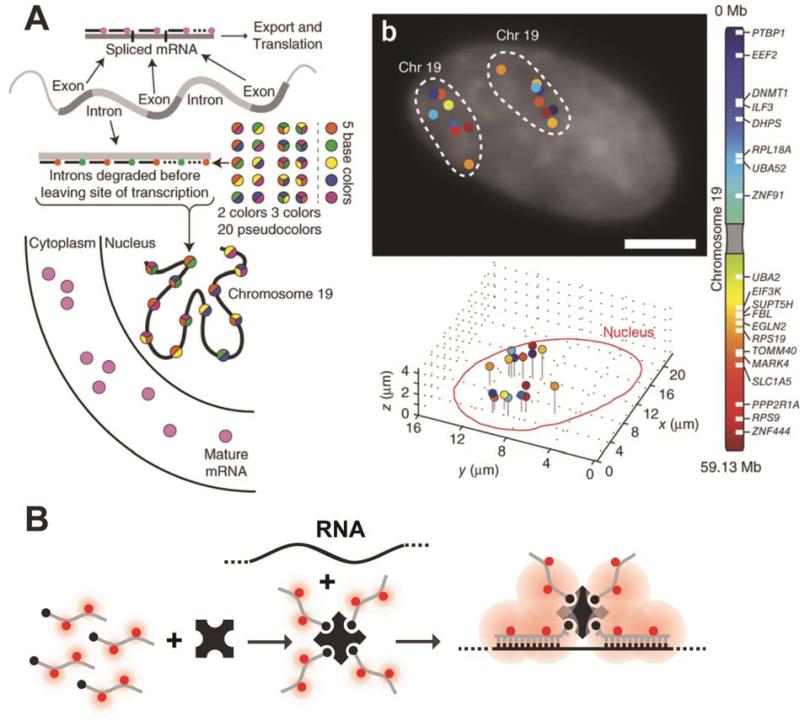
Figure 4. Recent techniques for fluorescently labeling RNA by hybridization of labeled probes.
(A) A pseudocolored scheme for spectral barcoding in iceFISH to simultaneously detect 20 transcript (left panel). The right panel contains a representative cell with the transcriptional activity of 20 genes spatially annotated. Reprinted with permission from ref. 292. Copyright 2013 Nature Publishing Group. (B) Schematic of the MTRIPs labeling method.
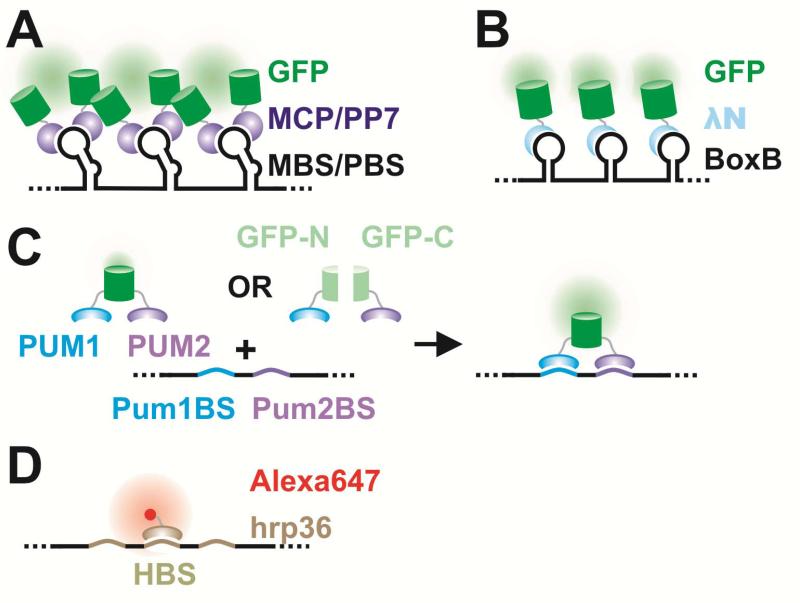
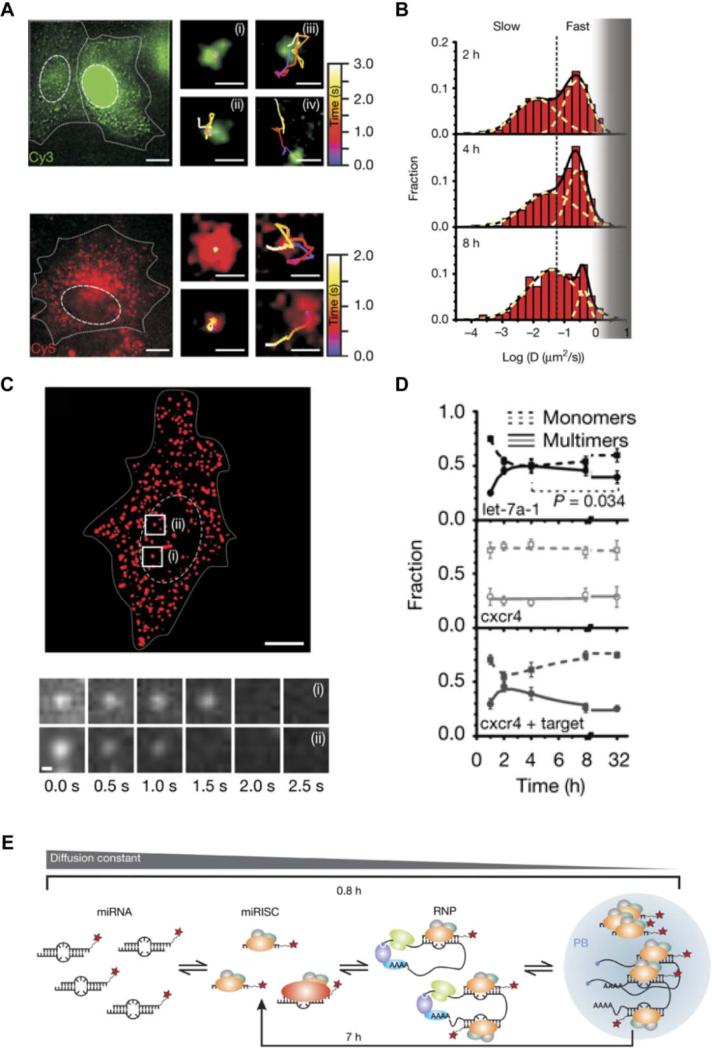
Figure 5. RNA labeling by various protein-RNA tethering approaches.
A detailed description is provided in section 3.
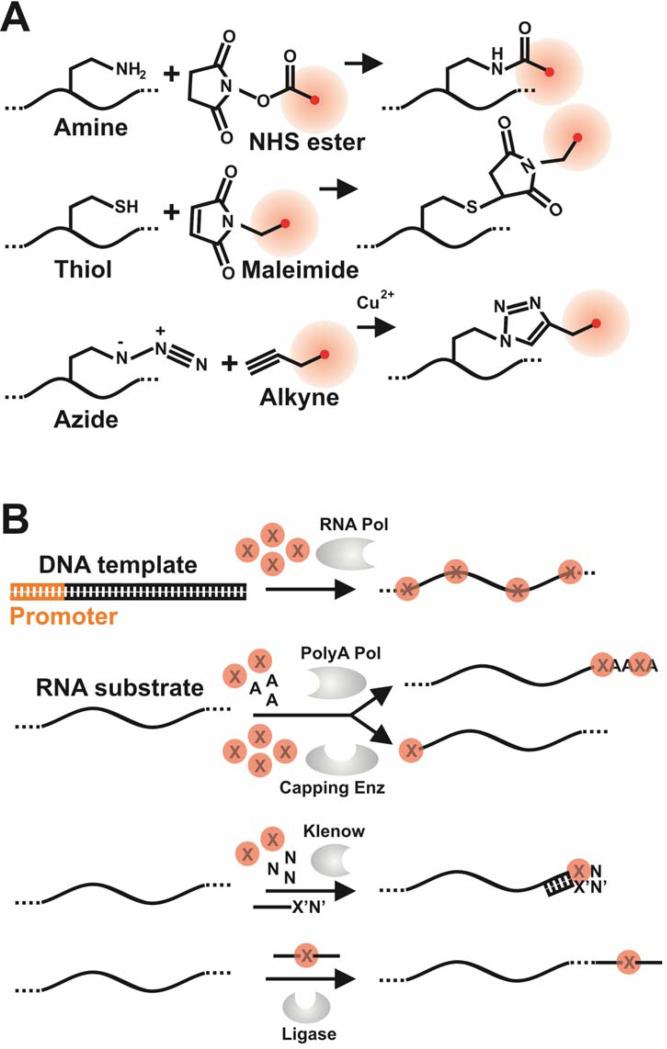
Figure 6. Chemical (A) and enzymatic (B) methods for direct fluorophore labeling RNA.
(A) Direct labeling of RNA by chemical methods. Red circles are dye molecules. (B) Direct labeling of RNA by enzymatic methods. X represents appropriately modified NTPs that are either directly conjugated to fluorophores or contain functional groups for subsequent chemical conjugation of dyes. RNA pol, RNA polymerase; polyA pol, polyA polymerase; Capping Enz, 5‘ end capping enzyme; Klenow, Klenow fragment of DNA polymerase I from E. coli; Ligase, typically one of the two T4 RNA ligases.24
3.2.1. Labeling by Fluorescence in situ Hybridization (FISH): An Early Glimpse at the Power of Intracellular SMM
Labeling target RNAs by hybridizing sequence complementary oligonucleotides in situ upon fixing and permeabilizing a cell was one of the earliest strategies to reach single molecule sensitivity. The method quickly gained widespread use because of its ability to probe the sub-cellular distribution and abundance of endogenous RNA and led to the inception of single cell gene expression analysis, the importance of which is underscored by the ubiquitous occurrence of cell-to-cell variations in gene expression.21,257,258 For instance, tumors that are often considered as a single lump of cells are comprised of many distinct cell types, each bearing distinct gene expression programs.259 Furthermore, the microenvironment of such tumors influences gene expression;259 for example, a cell in the center of a tumor or tissue expresses a different set of transcripts than one in the periphery. As an additional layer of complexity, gene expression is spatially organized even within individual cells.260-262 Such heterogeneities are often hidden within the averaged measurement or statistical error of an ensemble method (such as Northern blotting, quantitative reverse transcription PCR (qRT-PCR), microarray or deep-sequencing), traditionally used to quantify gene expression at high-throughput on a genomic scale. Techniques to access these important heterogeneities, such as single-cell RNA sequencing, microfluidics aided single-cell qRT-PCR, microdissection, fluorescence activated cell sorting and sub-cellular fractionation, are slowly emerging as attractive technologies for single cell transcriptome analysis,263,264 but are still not very efficient and/or introduce quantification or sequence biases through amplification steps in the protocol. Moreover, these ensemble methods still do not provide critical information on the spatiotemporal distribution of transcripts within tissues or individual cells. Therefore, it is becoming increasingly important to complement bulk measurements with techniques that characterize gene expression within individual cells in situ to decipher the stochastically driven, essential dispersities of gene expression within complex genetic networks. Progress in solid state synthesis of fluorophore labeled oligonucleotides and imaging/image analysis technology coupled with incessant advances of sequencing and bioinformatics analysis are now culminated in our ability to probe – in principle – any transcript, coding or non-coding, within the entire transcriptome at single molecule resolution in cellulo.
Traditionally used for DNA profiling and later modified for RNA detection,265in situ hybridization (ISH) protocols generally entail a sequence of fixation, permeabilization, hybridization of long (>100 nt) oligonucleotide probes to their corresponding complementary sequences, thorough washing to remove unbound probes, and image acquisition. Oligonucleotides are either directly labeled with fluorophores (fluorescence in situ hybridization, or FISH) in a stochastic fashion via enzymatic reactions (for example, transcription, 3’-end extension, nick translation and ligation; Figure 6B) or coupled to haptens, such as biotin or digoxigenin. In the latter case, the sample is then treated with avidin or antibody to digoxigenin, which are either directly labeled with fluorophores or coupled to chromogenic enzymes like alkaline phosphatase (AP) or horseradish peroxidase (HRP) whose enzymatic products yield an amplified light signal. Alternatively, secondary antibodies specific to avidin or primary antibody to digoxigenin are fluorophore or enzyme labeled to further amplify the signal from a single hybridization event. Even though these protocols are extremely useful in providing qualitative information on gene expression and localization patterns, they lack quantitative detail due to three main reasons: (i) Random distribution of fluorophores within oligonucleotides often results in a heterogeneously labeled population of probes and sometimes even localizes fluorophores close enough to mutually quench each other, both of which shroud intensity measurements that are critical for calculating the molecule copy number; (ii) long probes are poorly cell-permeable and thus result in incomplete labeling of RNA in situ; and (iii) this original protocol suffered from low sensitivity due to high background caused by unbound and non-specifically bound probes not removed by the washing. To overcome these caveats, multiple short oligonucleotide probes complementary to adjacent sequences within an RNA of interest effectively have now replaced long probes when performing FISH at the single molecule level266,267 (Figures 3 and 4). Each of these short oligonucleotide probes, small enough to surpass the permeability issue, are labeled with multiple266 or single fluorophores267 and designed such that the distance between fluorophores within individual hybridization probes and between different probes minimizes proximity mediated fluorescence self-quenching. Moreover, the fluorophore is attached to a specific nucleotide within the probe, resulting in more homogeneous labeling. The collective fluorescence arising from the binding of multiple such probes to a single RNA molecule is much higher than the fluorescence from a single labeled oligonucleotide, effectively delineating specific signal from unbound, non-specifically or sub-optimally bound oligonucleotides and cellular autofluorescence. In an alternative experimental scheme, endogenous transcripts containing multiple repeats of a specific sequence268 or exogenous transcripts tagged with such a repeat sequence array,269,270 are labeled with multiple copies of a single fluorophore tagged oligonucleotide sequence, essentially mitigating oligonucleotide synthesis costs. Following the basic principle of signal amplification, other modifications to the ISH procedure include the use of molecular beacons,269,270 modified nucleic acid backbone,271,272 padlock probes,273 branched DNA oligonucleotides274 or multivalent RNA hybridization probes.217 With the appropriate calibration controls, instrumentation and image analysis methods (described below) such modifications to the traditional ISH protocol result in single RNA molecule sensitivity.
Singer and coworkers spearheaded single molecule FISH (smFISH) methods by using five or more short (~50 nt) oligonucleotide probes that bound complementary sequences within an RNA of interest266 (Figure 3A, (i)). Each probe was labeled with 3 or 5 fluorophores at predefined positions and had a GC content of ~50%, suitable for optimal hybridization at relatively low temperatures (37-47 °C). Probes were then independently imaged in vitro at different concentrations to derive a calibration curve, which was consequently used to confirm the identity of individual fluorescent particles as single RNA molecules. To this end, various dilutions of the fluorophore labeled oligonucleotide were imaged in vitro in a sample chamber of known volume, using the same microscope settings as during intracellular imaging. A calibration curve of fluorescence signal versus number of oligonucleotide molecules (calculated from the concentration and sample holder volume) per voxel (a 3D pixel element) was plotted and the intensity of individual oligonucleotides was extrapolated from this curve. The authors found that the number of dye labeled oligonucleotide probes within individual fluorescent particles in a deconvolved image, as computed by dividing the particle signal by the signal of a single oligonucleotide, coincided with that expected to bind to a single mRNA.266,275 This method has been employed by several groups for spatial annotation of transcripts and counting;276-279 however, it suffers from one major drawback – high variability in the number of oligonucleotide probes bound per target.266,267,275 More specifically, >50% of all fluorescent spots contain only one or two of the possible five or more oligonucleotide probes, which complicates the reliable distinction of specific over non-specific binding. As each oligonucleotide probe has 3-5 dye labeling sites, incomplete labeling and inefficient separation of fully from partially labeled probes may result in the false annotation of probe density per transcript and thus have an impact on the quantification accuracy.
Tyagi, van Oudenaarden and coworkers modified Singer's protocol by targeting a single transcript with 48-96 oligonucleotide probes, each spanning ~17-22 nt and labeled at the 3‘end with just one fluorophore to allow for the efficient purification of labeled from unlabeled oligonucleotides, thereby improving labeling homogeneity of the target (Figure 3A, (ii)).267 Such short probes also require less stringent conditions for hybridization and washing: compare 28-37°C and 10% formamide267,280 to 37-47°C and 50% formamide266,275 in the Singer protocol or 65°C and 50% formamide281 in traditional FISH. Less harsh conditions allow for combining FISH with immunofluorescence or immunohistochemistry to probe both RNAs and (associated) proteins, frequently referred to as immunoFISH.282 Probes are designed to bind adjacent sequences on a single transcript such that the minimum spacing between them is 3 nt, thus minimizing self-quenching. Compared to the Singer approach, this strategy results in an increased fluorescence enhancement from individual transcripts, to an extent that even transcripts bound by endogenous RBPs or partially degraded are more efficiently detected. By contrast, signal arising from the non-specific binding of just one or two probes is typically insignificant enough to avoid false positives. The single molecule sensitivity of this method was validated by multiple complementary approaches.267,280,283 The method's sensitivity and inherent simplicity have led to its rapid commercialization (Biosearch Technologies ©) and to the availability of intuitive websites for probe design (http://www.singlemoleculefish.com) for any RNA target. However, the approach cannot be employed to detect short transcripts and small ncRNAs. In an effort to overcome this caveat, Shepherd et al. developed a competitive hybridization based approach (Figure 3A, iii).284,285 Herein, double-stranded probes contain a fluorophore on the 5’-end of the strand complementary to target and a quencher on the 3’-end of the other probe strand such that the former probe strand's fluorescence is quenched as long as the two oligonucleotides remain in the duplex.286 The target gradually replaces the quencher strand to bind the fluorophore labeled probe strand, leading to loss of quenching. Shepherd et al. exploited this property to reduce background fluorescence from free probe. To probe smaller RNAs, they additionally reduced the number of probes (5-10 compared to 48-96) and relaxed several probe design criteria, including requirements for ~50% GC content and large separation between probes. However, this method also suffers from variability in the number of probes bound per target, largely due to reduced stringency in probe design. Inefficient labeling due to poor kinetics of probe strand separation is another possible drawback. One solution is to make the fluorophore labeled, target-binding strand longer than the quencher strand, such that the overhang of their duplex is complementary to the target RNA. This allows for a more rapid removal of the quencher by strand displacement.285 Sunney Xie's group has reportedly overcome these drawbacks by probing single mRNA molecules with a single fluorophore labeled oligonucleotide in E. coli.287
The use of probes bearing a modified oligonucleotide backbone that allows them to hybridize more stably to RNA has enabled the detection of short transcripts with high specificity. Hybridization is sensitive enough to distinguish single nucleotide differences and detect single RNA molecules with just a single probe. These properties were exploited by Lu and Tsourkas272 to detect miRNAs in situ at single molecule sensitivity using locked nucleic acid (LNA or 2′-O, 4′-C-methylene-linked ribonucleotide) probes aided by enzyme labeled fluorescence (ELF) based signal amplification (Figure 3B, i). The LNA oligonucleotide probe is labeled with digoxigenin at its 3’-end to be recognized by an anti-DIG-AP chimeric antibody. ELF is achieved by the cleavage of a pro-luminescent substrate by AP (or HRP). Precipitation of the product and multiple turnover by the enzyme result in a fluorescent spot at the site of enzyme activity that is 20- to 40-fold brighter than a single fluorophore. The authors confirmed single molecule sensitivity based on the similarity in copy number distribution of ectopically expressed control transcripts that were detected by either standard smFISH or LNA-ELF-FISH.272 Probes with other backbone modifications, such as peptide nucleic acids (PNA), have been used to detect telomeres and assess their length in situ.271
The possibility of fluorescent ELF amplification products diffusing away during washing or detection has spurred the development of other signal amplification methods. Initially standardized for DNA, Larsson et al.273,288 developed “padlock” probes to detect single nucleotide polymorphisms (SNPs) in RNA, i.e., distinguishing transcripts that differ only by a single nucleo base, via enzyme independent signal amplification. They first reverse transcribed the RNA to cDNA using LNA primers, RNase H treated to degrade any portion of the RNA complementary to the cDNA, hybridized linear padlock probes to the target such that the 5’- and 3’-ends are juxtaposed, enzymatically ligated the ends and used them as templates (and the cDNA as the primer) for rolling circle amplification by Phi29 DNA polymerase (Figure 3B, ii).273 A single-stranded DNA containing tandem repeats of the padlock probe was thus created at the mRNA localization site, to which fluorophore labeled detection oligonucleotides were hybridized to yield a bright fluorescent spot. The specificity of LNA hybridization contributes to the initial specificity in targeting transcripts and tethering the cDNA to the intracellular transcript location, whereas the target dependent padlock probe ligation aided SNP detection. Another signal amplification approach uses branched DNA hybridization.274,289 Here, a single gene specific probe contains flanking sequences that hybridize to a pre-amplifier probe, which in turn binds multiple amplifier oligonucleotides. Each amplifier oligonucleotide binds multiple detection oligonucleotides, thereby resulting in bright fluorescent spots, especially when multiple gene specific probes target a single transcript. This technology has been commercialized as QuantiGene ViewRNA (Affymetrix ©), with the advantage of using universal pre-amplifier, amplifier and detection oligonucleotides for any gene specific set of probes. A related system, named hybridization chain reaction (HCR290), uses flanking sequences on gene specific probes (initiator oligonucleotide) to initiate self-assembly of metastable fluorescent RNA hairpins into large amplification polymers (Figure 3B, iii). All of these protocols improve signal quality and quantity, yet they have a major drawback of amplifying false positive signals as well, necessitating stringent probe design criteria.
Although multiplexing has been achieved with many of these methods by using distinct fluorophore colors, conventional optics and broad emission spectra typically limit the number of simultaneously detectable transcripts to three, beyond which non-specific excitation and spectral bleed-through confound signal identification. To overcome this limitation, Singer and colleagues developed an approach they termed “spectral barcoding”291 wherein gene specific probes are divided into groups labeled with spectrally distinct fluorophores. Hybridization of these groups of probes with their cognate transcripts and careful registration of multiple fluorescent channels results in fluorescent spots that are multi-colored. With each gene designed to bind probes with a specific color combination, one can, in theory, have 2n – 1 color combinations of n spectrally resolvable fluorophores, effectively multiplexing and increasing throughput. Recently, Levesque and Raj292 doubled the number of simultaneously detected transcripts (from 10 to 20) in an adaptation of Singer's method they called intron chromosomal expression FISH (iceFISH). The authors took advantage of the fact that most introns are unstable upon their removal from a pre-mRNA by splicing near their site of transcription and labeled introns to thus probe chromosomal structure (Figure 4A).
A majority of smFISH approaches are not extendible to living cells because it is difficult to deliver such a large number of probes into cells by methods other than irreversible membrane permeablization. Moreover, FISH protocols rely on hybridization under (mildly) denaturing conditions and multiple wash steps to remove unbound probes, both of which are difficult to accomplish in living cells. Tyagi and coworkers269 addressed these drawbacks by microinjecting molecular beacon probes (Figure 3A, vi) into live cells. Molecular beacons are designed to have small complementary sequences on either end such that they adopt a (weak) hairpin structure (with a small stem and a large loop), bringing a fluorophore at the 5’-end close to a quencher at the 3’-end for effective quenching of fluorescence from unbound probes. Upon target hybridization, the hairpin stem is disrupted and the fluorophore becomes unquenched. 2‘-O-methyl (2OMe) oligonucleotides were used instead of DNA probes to alleviate RNase H mediated cleavage of RNA within DNA-RNA hybrids and to prevent probe degradation by cellular nucleases. For fluorescence enhancement from individual transcripts, the authors created an exogenous transcript that binds 96 copies of the probe, thereby requiring only a single probe sequence for target hybridization. To demonstrate that each fluorescent spot contained a single RNA, the group prepared in vitro transcribed RNA containing 16, 32, 64 or 96 probe binding sites, pre-hybridized the probes in solution and microinjected them into cells. As expected, the intensity of particles was proportional to the number of binding sites, the intensity distribution of particles was well represented by a single Gaussian, and the average particle counts per cell coincided with qRT-PCR results. The establishment of live cell single RNA detection allowed the group to understand nuclear trafficking of RNPs. A similar approach was used by Kubitscheck and colleagues to probe endogenous Balbiani ring (BR) 1 and 2 mRNPs. The group used a fluorophore labeled oligonucleotide that sub-stoichiometrically targeted a stretch of repeat sequences (~80 repeats) within the mRNA.268 Ishihama and Funatsu similarly used microinjection to track the diffusive behavior of polyA-tailed ftz mRNA, pre-hybridized with QD labeled oligonucleotide U(22), within the nucleus of mammalian cells.293
As microinjection may lead to the passive transport of probes into the nucleus and consequently hamper cytoplasmic RNA labeling, Santangelo and coworkers217 used a combination of reversible permeabilization by streptolysin O (SLO) for intracellular delivery and a unique set of hybridization probes called multiply labeled tetravalent RNA imaging probes (MTRIPs) (Figure 4B). Individual detection probes were created by binding streptavidin to 2’-O-methyl RNA-DNA chimera that contained a 5’-biotin and 3-5 well spaced internal fluorophores. As streptavidin contains four biotin binding sites, a tetravalent probe forms with a four-fold fluorescence enhancement over a single probe. SLO allowed for the delivery of such large probes into the cells. Single molecule sensitivity was supported by assessing intensity distributions of single probes immobilized to glass and those in cells and further confirmed by comparisons of the signal from monovalent with that of tetravalent probes and the intracellular distribution of scrambled with that of specific probes. Major advantages of this method include the need for fewer tetravalent probes (2 - 3) per RNA and the ability to visualize the dynamics of endogenous (as opposed to engineered) RNA. Recently, the same group replaced streptavidin with multi-armed PEG294 covalently attached to fluorescent oligonucleotides to reduce toxicity from spurious binding of streptavidin to endogenous biotin, and to increase the number of oligonucleotides, and thus fluorophores, per MTRIPs.
RNA labeling via hybridization, although popular, still suffers from several drawbacks. The cross-linking of RBPs to RNA during fixation and tightly formed secondary structures severely affect probe accessibility and binding,295 which may compromise imaging sensitivity. Additionally, accurate RNA counting within large, intracellular aggregates, such as transcription sites or RNA-protein aggregates is seldom straightforward and often error-prone. Another concern pertains to RNA probing in live cells, wherein hybridization of oligonucleotides, especially a large number of them, may compete with endogenous RBP binding or regulatory RNA elements vital to RNA function or trigger antisense or RNA silencing responses that degrade the RNA. Moreover, the translation machinery and other RNA helicases may denature probe-RNA hybrids, resulting in reduced sensitivity. Probes should therefore be designed within the UTRs of RNA, specifically selecting binding sites that do not affect RNA function. Finally, the abundance of unbound probes (that cannot be washed away) can contribute to high background and false positive signal in live cells.
3.2.2. Labeling with RNA Binding Proteins
Since proteins can be easily appended with FPs, labeling RNAs using FP tagged proteins that bind them, rather than using oligonucleotide probes, is a logical extension. This method has allowed for the real-time detection of RNA localization and trafficking, and has provided valuable information on the temporal signature of gene expression in living cells, complementary to transcript counting in fixed cells.296 As a result, it promises the possibility of investigating the entire life cycle of an RNA, from transcription, transport and translation to degradation, in a single experiment, a feat that is difficult to achieve with smFISH. The labeling strategy entails the intracellular expression of the RNA of interest as a fusion with multiple copies of an RNA motif that binds a specific protein. RBP-FP chimeras are simultaneously expressed and the binding of many copies of these fusion proteins to the RNA labels the target above background (Figure 5). Both the target RNA and the RBP, or just the RBP, are genetically engineered into plasmids that are either transiently transfected or stably integrated into the cellular genome for intracellular synthesis. The binding of multiple FP-tagged-RBPs (RBP-FP) through a repetitive RBP binding sequence (RBS) renders a single RNA molecule much brighter than a single FP molecule so that it becomes well distinguishable from unbound RBP-FP molecules and cellular autofluorescence.233 Additionally, certain schemes favor fluorescence enhancement by confinement of individual nucleic acid bound FPs.297 Here, the experiment is designed such that unbound RBP-FPs are highly expressed and diffuse much faster than the time resolution of image acquisition, constituting a fluorescent blur spread over the entire cell. Once RBP-FPs associate with slow moving or immobile RNAs via their cognate RBS, they are confined to the extent that acquisition time is not a limiting factor. Fluorescence from such slowly diffusing complexes supersedes that of the unbound probes and can be detected as distinct diffraction limited spots, effectively enhancing the signal-to-background ratio for single molecule detection. However, in either case unbound RBP-FPs still contribute significant background fluorescence, especially when over-expressed. To mitigate this issue, a nuclear localization signal (NLS) needs to be tagged to the RBP-FP gene as a means to concentrate unbound RBP-FPs in the nucleus and improve the sensitivity of (m)RNA detection in the cytoplasm.233 Alternatively, fluorescence recovery by reconstitution of split GFP fragments can be employed.298 Here, two distinct RBPs are fused to the non-fluorescent N-terminal and C-terminal halves of GFP. Binding of both RBPs to their respective, adjacently located RBS's brings the two fragments in close proximity to promote association, resulting in a fluorescent protein. In this fashion, unbound and singularly bound RBP probes are effectively rendered non-fluorescent. To robustly label the RNA, it is essential that the RBP binds the RBS with great specificity and affinity (preferably with a KD of <10 nM), and that fluorescence recovery from the assembled fragments is reasonably efficient.
One archetypical high-affinity protein-RNA tethering system, pioneered by Singer and coworkers, consists of the MS2 coat protein (MCP) and matching MCP binding sequence (MBS, Figure 5A).233 Derived from the MS2 bacteriophage (or its closely related R17 bacteriophage), the MCP is an ~13.7 kDa regulatory RBP that readily dimerizes in solution, whereas the MBS is an ~21 nt RNA fragment that spontaneously adopts a stem-loop structure.299 Other viral protein-RNA tethering systems that have been derivatized for tagging RNA with FPs include a system derived from the PP7 bacteriophage, an evolutionary cousin of the MS2 bacteriophage, which infects P. aeruginosa.300,301 As in the MS2 system, two copies of the PP7 coat protein (PCP) bind one copy of the stem-loop structure of the PP7 RNA binding site (PBS) with high affinity (KD = ~1 nM).300 Despite the functional similarity, the PCP bears only ~15% sequence identity to the MCP and the PBS differs from the MBS in both size and nucleotide composition. This feature results in orthogonality between the PP7 and MS2 systems, i.e., the PCP does not recognize the MBS and vice versa.300 Another orthogonal system comprises the λN peptide, which spans ~22 amino acid (aa), and its corresponding ~15-nt long RNA binding motif, box B,299,302,303 derived from lambda bacteriophage (Figure 5B). In the virus, the λN protein (12.2 kDa and 107 aa), which contains the λN peptide at its N-terminus, binds box B stem-loop motifs in a 1:1 stoichiometry and with equal specificity and affinity (KD = ~1.5 nM) as the full-length version.299 These three orthogonal protein-RNA tethering systems provide the opportunity to simultaneously image up to three different mRNAs303 or probe three discrete segments of a single mRNA. Furthermore, they have evolved in nature to minimize non-specific interactions with other proteins or RNAs so that they can be expected to serve as “inert” tags for probing the intracellular localization and dynamics of target RNAs.
In its earliest manifestation, 24 copies (24x) of the viral RBS were appended to an RNA, resulting in the binding of up to 48 molecules of FP labeled RBPs,233 although versions that utilize lower304 or significantly higher305,306 number of RBS repeats have also been reported. mRNA labeling via these large appendages is typically achieved by incorporating the RBS into the 5’- or 3’-UTR such that the tag does not affect the transcript's translation or UTR-mediated regulation.233 Nevertheless, control experiments need to be performed to test whether a tagged RNA retains its biological function. Additional control experiments should be performed to verify that individual intracellular fluorescent particles represent single RNA molecules.233,307 To this end, Singer and coworkers used a combination of FP titration and cross-validation by smFISH.233,307 The FP titration aided calibration curve was constructed similar to that discussed in segment 3.2.1, with purified FPs replacing fluorophore labeled oligonucleotides. This measurement was complemented with smFISH, performed on the same set of RBP-FP expressing cells, to exclude the possibility that clusters of sub-optimally labeled mRNAs were misidentified as single mRNAs. Colocalization of smFISH probes with FP labeled particles and calibrations curves that identified the number of oligonucleotide probes bound per RNA (as also described in segment 3.2.1) further supported the FP titration data.
Both the MS2 and PP7 systems have been well characterized, yet similar characterizations of the λN:boxB pair have so far been lacking. Notably, such characterizations have shed light on surprising anomalies and led to the development of improved MCP and PCP probes.308 Anomalies included incomplete and heterogeneous occupancy of MBS by MCP,233,307,308 i.e., the observations that only a subset of the 24 or so MBS is bound by FP tagged MCPs and that MBS or PBS tagged mRNAs were not uniformly labeled across different cells or sometimes within the same cell. Especially the latter undermines the ability of this labeling method to quantify gene expression at single molecule sensitivity. For instance, a single RNA-containing particle that is half as bright as another owing to non-uniform labeling may be scored as harboring half the number of RNA molecules even though the two are stoichiometrically identical. These limitations were attributed to the expression-level dependent dimerization of MCP or PCP.308 At low RBP abundance, dimerization as a prerequisite to RBS binding is compromised and results in incomplete RBS occupancy. As the RBP expression increases, extent of dimerization and hence occupancy increases, but saturates based on the number of RBS repeats and concomitantly increases background fluorescence from unbound RBP-FPs. In an alternative strategy, Singer and coworkers created tandem dimers of MCP (tdMCP) or PCP (tdPCP) conjugated to an FP, wherein two copies of the RBP are engineered in tandem with a flexible linker in the middle.308 This scheme promotes concentration independent intramolecular dimerization, thus effectively resolving issues of expression level mediated non-uniform labeling. While maximum occupancy is achieved with the PCP-PBS system (~48 copies of the PCP or 24 copies of tdPCP bound to 24x PBS), the MCP-MBS pair always shows sub-stoichiometric labeling (only ~26 MCPs or 13 tdMCPs associated with 24x MBS), for reasons still unknown.308
Regardless of their widespread use and efficacy, the protein-RNA tether labeling strategies discussed so far are only useful to probe genetically engineered constructs. Recently, two groups independently developed distinct methods capable of labeling both engineered and endogenous RNAs with single molecule sensitivity.213,268,309,310 Both methods exploit the binding of fluorophore labeled RBPs to specific RNA sequences, rather than structured motifs. In the first technique, Ozawa and coworkers213,309,310 utilized the RNA binding properties of PUMILIO, a protein that mediates eukaryotic posttranscriptional gene regulation (Figure 5C).311 This protein binds RNA via the Pumilio homology domain (PUM-HD), which contains an array of eight modular elements that specifically recognize the RNA sequence UGUANAUA312,313 (where N is any nucleotide), also termed PumBS. As each module recognizes a single RNA base,312 the binding specificity of PUM-HD can be altered by simply changing the base recognizing amino acid residues within each element such that is matches a specific sequence within an RNA of interest. Notably, just a single base mutation within the first half of the consensus sequence results in an ~100- to 3,000-fold loss in affinity for the wild-type PUM-HD that can be rescued by appropriate PUM-HD mutants.312,313 This modular RNA binding feature was exploited to create FP labeled tandem PUM-HD probes, wherein each engineered PUM-HD recognizes a distinct, yet closely located sequence within an endogenous target213,309,310 (Figure 5C). This tandem binding approach utilizes two different 8-nt PumBS's located adjacent to each other and can theoretically distinguish one among 416 (~4.3 × 109) transcripts, unique enough to identify a specific target RNA within a single cell. In one version, endogenous mitochondrial ND6309 or β-actin mRNA314 were visualized by the split-GFP approach. Here, the two mutants of PUM-HD (mPUM1 and mPUM2) were fused to the non-fluorescent N-terminal and C-terminal segments of GFP, respectively; binding of both PUM-HDs to their respective PumBS reconstituted GFP. A variant of this strategy was employed to image β-actin mRNA,213 in which a single RBP-FP fusion was constructed by bridging GFP between two PUM-HDs. In contrast to the split-GFP approach, this version necessitates the construction and intracellular delivery of only one RBP-FP probe (as opposed to two) and overcomes limitations of slow GFP reconstitution. The loss of the background reduction through the split GFP, however, had to be compensated by targeting unbound RBP-FP to the nucleus, and imaging sensitivity was further improved by fluorescence enhancement via confinement. Both versions of this technique utilize tags that are significantly smaller than viral RBS-RBPs, a preferable trait, and can use single-step photobleaching of FPs to confirm detection of single RNAs. The system, however, suffers from diminished fluorescence enhancement and short observation windows, as the RNA is labeled with just one fluorescent RBP that binds its cognate RBS only for a limited amount of time. Disappearance of signal in a single step, often assumed to be photobleaching, can also occur by dissociation of the RBP from the RBS or from the abrupt diffusion of the RNA out of the focal plane. Additional limitations are related to the ‘metastability’ of the reconstituted GFP, wherein the reconstructed probe persists beyond the dissociation or degradation of the RNA that nucleated assembly of the split GFP, often leading to false-positive signal.
The second endogenous RNA probing technique exploits the binding of hrp36, an hnRNPA1-like protein found in C. tetans, to hrp36 binding sites in BR2 mRNA (Figure 5D, shown is a general labeling scheme).268,315 Although the hrp36 binding site is found in other mRNAs, its copy number in BR2 mRNA is significantly higher. Thus, at any given moment a larger number of hrp36 molecules bind BR2 mRNA than any other mRNA, allowing for selective fluorescence enhancement.268,315 In these studies, organic dye labeled hrp36 was used instead of FP for three reasons: (i) It was difficult to genetically manipulate primary cells extracted from C. tetans salivary glands; b) overexpression of FP labeled hrp36 was expected to result in high background of hrp36 free or bound to other mRNAs; and c) intracellular delivery and density of labeled hrp36 could be controlled by microinjection. Orthogonal in its approach, multiple repeats of the hrp36 binding site can be tethered to any exogenous RNA, although the system has limited application in probing other endogenous mRNAs. Additional protein-RNA tethering system labeling approaches include the use of PABP,316 two split eIF4A domains,317 and MCP-zipcode binding protein (ZBP),318 none of which have yet been utilized for intracellular SMM.
Although a majority of these FP-based labeling schemes has the foremost advantage of genetic tractability, they suffer from a few drawbacks. First, the relatively high molecular weight of protein-RNA tethering systems limits their application to large RNAs. mRNAs in particular are bound by endogenous RBPs and the translation machinery, and therefore exist as bulky messenger ribonucleoprotein complexes (mRNPs); thus, the molecular weight of large mRNAs does not appreciably change upon introduction of a few tethered RBPs, but these can adversely affect the mobility of smaller mRNAs. Second, the inclusion of the NLS to the RBP-FPs may affect the labeled RNA's function and localization by increasing the propensity of NLS-RBP-FP bound cytoplasmic RNAs to localize within the nucleus. Third, the presence of unbound RBP-FPs in the cytoplasm, mediated by their overexpression and/or their limited binding affinity and resulting significant dissociation from the target RNA, results in significant background noise the removal of which often requires image processing algorithms such as deconvolution.233 Fourth, single molecule counting is not straightforward and entails the painstaking construction of fluorescence calibration curves,233,307 as well as requires the implementation of other complementary methods like fluorescence in situ hybridization (FISH).233,307 Fifth, non-uniformity of labeling further confounds counting (discussed above), but strategies to overcome this issue are slowly emerging.308 Sixth, RBPs – especially PumHDs – are not strictly specific to their corresponding RBS and can generate spurious signal by non-specifically binding to other RNAs. Nonetheless, the reliability of these well-standardized labeling schemes is evidenced by the availability of general methods to tag and image any mRNA with the MCP-MBS system in both lower eukaryotes, such as yeast,304 and complex mammals like mice.226 The transgenic mouse model226 in particular allows for probing the intracellular distribution of any mRNA over different cell types, tissues and even whole animals.
Another approach to overcome some of the drawbacks of FPs is to reduce the protein in size to its chromophore and bind it to a specifically in vitro selected aptamer incorporated into the target RNA. Early versions of this approach used malachite green as the fluorophore with an aptamer that modulates the dye conformation and increases its fluorescence.319,320 A more recent incarnation of the approach uses a GFP-derived group of fluorophores bound by an RNA aptamer termed “spinach” that again enhances their fluorescence.321,322 Because of their relatively low binding affinity and rapid exchange with free dye, the GFP-derived chromophores appear to photobleach only slowly321, but whether such comparably weak binding of small-molecule fluorophores will allow for single molecule detection of spinach-tagged RNA remains to be seen.
3.2.3. Direct Labeling of RNA
The direct, covalent coupling of fluorescent probes, typically organic dyes, to an RNA of interest can be achieved by several chemical and enzymatic methods234-237,323-330 (Figure 6). Generally, small RNAs (<100 nt) are chemically synthesized to contain fluorophore labels or reactive functional groups, which in turn are conjugated to fluorophores (Figure 6A), at predefined positions. Larger RNAs are either labeled at random positions using modified NTPs during transcription or specifically labeled, mostly at the 5’- or 3’-end or internally through ligation, by chemical or enzymatic modification (Figure 6B). Even here, modified NTPs directly coupled to fluorophores or containing reactive functional groups can be used, however, the larger size of the former may inhibit labeling.331 In any case, two critical steps in this labeling strategy are the removal of free dye from the sample and separation of labeled from unlabeled RNA. As the molecular weight of the free dye is much smaller than that of the RNA, the first purification step is easily and efficiently achieved by, for example, ultrafiltration, gel filtration, or gel electrophoresis. Size cannot be used, however, for the second purification step, as the difference in molecular weight between labeled and unlabeled RNA is often negligible. Small RNAs are efficiently purified by reversed-phase HPLC since labeled RNAs are more hydrophobic than their unlabeled counterparts;328 however, this difference becomes less for large RNAs. The ligation of unique sequences to large RNAs can aid purification of the product via beads coupled to complementary hybridization probes, whereas other enzymatic reactions require thorough and time-consuming standardization to achieve high labeling efficiencies.237 Moreover, the incorporation of modified NTPs during transcription results in a heterogeneous population of labeled RNAs, with both varying location and number of modifications. Nevertheless, non-specific RNA labeling via transcription332 or alkylating agents333 has been used to incorporate multiple fluorophores within individual RNA molecules as a fluorescence enhancement strategy.
Although intracellular delivery of directly labeled RNA has challenges, it also offers three main advantages over other labeling strategies. First, RNA counting by stochastic photobleaching of fluorophores, wherein the stepwise reduction in intensity of labeled particles is a proxy of molecular count, is more accurate and straightforward.135,334,335 Other methods require calibration curves and read out the intensity with little ability to consider the broad distribution of intensity fluorophores typically exhibit.336 Second, this method does not require any elaborate probe design or attachment of significant RNA extensions, only that fluorophores are appropriately positioned to not affect RNA function. Finally, particularly small ncRNAs such as siRNAs and miRNAs, which cannot be probed via conventional hybridization or protein-RNA tethering strategies, can be directly labeled and visualized in both live and fixed cells at single molecule resolution.135
3.3. Intracellular Delivery of Fluorophore Labeled RNA
The choice of labeling strategy directly influences that of the intracellular delivery method. For instance, a majority of RBS-RBP labeling strategies largely employs chemical transfection213,233,308-310 or viral transduction226,307,308,337. Plasmids encoding both the RBS and RBP are transiently expressed or stably integrated into the cellular genome using either of these delivery methods. Numerous transfection reagents are available in various chemical formulations – cationic lipids, polyethyleneimine, DEAE dextran, calcium phosphate and cationic or neutral dendrimers to name a few – and serve to neutralize the innately negatively charged DNA (or RNA), which allows for the efficient uptake of the resulting self-assembled particles through negatively charged cell membranes, presumably via endocytosis. As transfection reagent-plasmid complexes are heterogeneous in size and distribution, each cell receives a different amount of the plasmid (or RNA). Thus, transient transfection leads to heterogeneous (over)expression of both the RBS and RBP, a major obstacle for uniform RNA labeling.308 Creation of stably transfected cell lines may mitigate non-uniform expression. Transduction with a lenti- or adenovirus that harbors only a single copy of the DNA offers better control over intracellular delivery, but is often a laborious process. Furthermore, direct synchronization of experiments using any of these methods is difficult because self-assembly, endocytosis, exit from endocytosed vesicles, migration into the nucleus, and subsequent gene expression each take significant amounts of time that have a wide distribution across cells. In addition to these steps, viral transduction also requires virus production, which further adds to the lead time prior to RNA visualization. However, the presence of inducible expression systems225,269,270,303,304,317,338-340 for plasmids can aid in defining a more precise experimental start time, albeit often accompanied by a basal level of “leaky” expression.
RNAs labeled via other strategies are predominantly delivered by physical force133-135,268-270,293,315,333,341-345 or via cell membrane permeabilization.217,225,226,267,270,272,273,278,282,292,294,303,307,337,340,346-348 The former includes electroporation and microinjection, whereas the latter utilizes pore forming peptides or detergents and organic solvents that partially dissolve the cell membrane. More specifically, pore forming peptides function by reversible permeabilization,217,294,346,347 making them popular for intracellular SMM of hybridization-labeled RNA in live cells. SLO is a classic example of a pore forming peptide, which is produced by gram-positive bacteria as a cytolytic toxin.349 The addition of SLO, typically under serum free conditions, results in its binding to cholesterol on the cell membrane and subsequent oligomerization to form pores of ~25–30 nm diameter that permits the influx of hybridization probes or directly labeled RNA.350 Once the SLO containing medium is removed and replaced with fresh growth medium, SLO dissociates and the membrane reseals. However, based on a cell type's cholesterol composition, the incubation temperature, time, cell number, and SLO concentration needs to be optimized and, as is the case for transfection, non-uniform delivery of probe and/or RNA across the cell population is commonplace, and SLO cannot per se be used for intranuclear delivery. Permeabilization of the cell membrane by detergents (Triton-X100, NP40, deoxycholate, etc.) or organic solvents (acetone, ethanol, and methanol) is irreversible and is only used to mediate the delivery of hybridization probes into fixed cells.
In contrast to membrane permeabilization methods, delivery methods employing physical force are amenable for the intracellular delivery of both plasmids and RNA without any additives and are often the method of choice in hard-to-transfect cells. Among them, microinjection offers maximal control over delivered amount and flexibility in experimental design, especially for RNA that is directly labeled.135,343,345 The process is conceptually simple and uses a micromanipulator, which can traverse distances as low as ~10 nm, and an injector pump. First, injection samples are filled into glass capillaries whose openings are typically ~0.5-1 μm wide, small enough not to puncture cells. Microinjection is then achieved via three precisely timed steps that occur within a second: (i) assisted by the micromanipulator the capillary pierces the cell; (ii) the pump applies a pre-set pressure (~10-150 hPa) to the capillary that allows for sample ejection; and (iii) the capillary is removed from the cell. In this fashion, cells are sequentially and uniformly microinjected with a few femtoliters of RNA solution, and controlled delivery is achieved by modulating the sample concentration and injection pressure. Furthermore, the ability to inject RNA selectively into either the cytosol or nucleus provides for the opportunity to evaluate RNA function in distinct cellular compartments and to study nucleocytoplasmic transport processes.351 Disadvantages of this technique include the requirement for expensive instrumentation and its applicability only to relatively large cell types (i.e., not bacteria), and that only a limited number of cells can be manipulated in a given time period. However, the latter has the advantage that different RNA types or concentrations, as necessary for titrating and measuring reaction kinetics, can be injected into small, spatially separated groups of cells, maximizing experimental throughput within a single culture dish. Compared to SLO and transfection mediated intracellular delivery that take ~30 min and >6 h, respectively, fluorophore labeled RNA can be visualized immediately after microinjection, allowing for precise temporal control. Additionally, transfection often leads to the entrapment of probes within endocytic vesicles and their degradation via the endosomal/lysosomal pathway.158 Emerging alternatives employ CPPs,294,352 such as HIV-Tat, that are transported across the cell membrane via cell surface receptors. Here, RNA is directly tagged with the CPP and the addition of this fusion molecule to cell culture medium results in close to quantitative intracellular delivery.
3.4. Instrumentation
As with any ultrasensitive technique, intracellular SMM requires sophisticated instrumentation. However, the availability of a plethora of fluorescence enhancement strategies has made it possible to visualize single bright RNA molecules even with simple microscopes. In addition to the labeling strategy, the choice of excitation source, optics, illumination scheme and detector contribute towards single molecule sensitivity. In the following we provide a broad overview of available instrumentation options and, in Figure 7, describe a typical single molecule fluorescence microscope.
Figure 7. Schematic of our home-built single molecule microscope.
Our Olympus IX81 microscope is equipped with two high NA, 60x and 100x, oil-immersion objectives. The microscope also has low NA, 10x and 20x, objectives. It features an internal 1.6x magnification and is additionally equipped with a 1-4x magnification changer (Olympus). Thus, a maximum magnification of 740x (100x1.6x4) can be achieved. Samples are positioned on a nanometer-precision piezo-controlled stage. The microscope also contains an infrared laser based zero-drift control module (Olympus) to correct for focal drift. Solid state lasers with wavelengths of 405 nm, 488 nm, 532 nm and 640 nm are directed through an acousto-optical tunable filter (AOTF) and split into separate fiber-optic cables within a laser launch system. The AOTF allows for computer-guided selection of the appropriate laser wavelength for illumination and modulation of laser intensity. AOTF coupling also enables sub-millisecond switching between multiple laser wavelengths, forming the basis for alternating or interlaced excitation schemes. Alternatively, a broadband super-continuum laser capable of emitting multiple wavelengths of excitation light in a pulsed fashion (ns-ps) can replace the multi-laser system. The fiber-coupled laser beams are directed into a cell-TIRF laser-combining module (Olympus). All laser beams are focused on the back-focal plane of the objective and aligned to travel parallel to the optical axis such that the incident angle of illumination at the dish-medium interface can be controlled electronically by changing the distance of the beam from the optical axis at ~0.01° angular resolution using the cell-TIRF module. Fluorescence from the sample is detected typically via an EMCCD. For multi-color imaging, the emitted light is split onto two different detectors using a single beamsplitter or onto two regions of the same detector. The former strategy can be used even with a point detector such as an APD or PMT, whereas the latter requires a large detector area and can be implemented only with a CCD. Appropriate mirrors, filters and dichroic beamsplitters are used to guide and spectrally filter the light source and emitted light. QB, Quadband. Cells grown on dishes (Bioptechs) are maintained at 37 °C on the microscope stage while imaging using a stage incubator. The micromanipulator of a microinjection system (Eppendorf Femtojet) is attached to the microscope for intracellular probe delivery.
3.4.1. Light Source
Lamps were initially popular light sources in both ensemble and single molecule microscopy. Due to their polychromatic light, a single lamp could be used to excite distinctly colored fluorophores by spectrally refining the excitation light with relatively inexpensive narrow-bandwidth filters. However, inefficient filtering is typical and leads to the leakage of other excitation light colors that either spuriously excites the sample or adds to the noise of the detector and severely compromises sensitivity. Additionally, most of these arc-based lamps suffer from limited lifetime and non-uniform illumination intensity over their spectral range of operation. Since lasers emit monochromatic, coherent and collimated light, they now have effectively replaced lamps as the primary light source in intracellular SMM. To minimize phototoxicity, especially during live cell imaging, and to reduce photobleaching, lasers operating at 1 μW -100 mW power, with a power density (i.e., the light flux or number of photons per unit area) of 1 W/cm2 to 100 kW/cm2 is typical in intracellular SMM applications.353,354 The choice of illumination wavelength depends on the spectral properties of the fluorescent probe, but typically spans the visible part of the electromagnetic spectrum (~400-700 nm) because transmission properties of available optics are best in the visible range and the best-documented fluorophores emit within this regime. Imaging at wavelengths greater than ~500 nm is preferred to overcome cellular autofluorescence and to increase sample penetration depth via reduced scattering so that far-red and near-infrared (NIR) probes and lasers are slowly gaining popularity. Ultraviolet (UV) light is not suitable for imaging living cells as it induces DNA damage and apoptosis.355
3.4.2. Optics
At the heart of a microscope lies the objective lens. It transmits light from the illumination source and collects (fluorescent) light from the sample to create a focused image on the detector. Two important parameters, the numerical aperture (NA), an indirect measure of photon collection ability, and the magnification together define objectives. High NA (1.25 – 1.65) and optimal magnification (60x – 150x) are preferred to collect as many photons as possible, especially under the low light conditions of single molecule experiments. Notably, a further increase in magnification without a corresponding increase in the brightness of the light source reduces the number of photons collected per unit square area of the detector, thoroughly compromising the visibility of single molecules. A good objective additionally minimizes chromatic aberrations (distortions in an image caused by differential focusing of different wavelengths of light). Thus, achromat or apochromat versions of objectives are mandatory for multicolor imaging. Downstream of the objective lens are other optical elements that mediate spectral selection/filtering, important aspects of intracellular SMM and multicolor imaging. These include dichroic beamsplitters and emission filters, whose choice is based on the spectral characteristics of the light source and the fluorescent probe. The former is oriented at an angle of 45° with respect to the illumination or emission light to spectrally separate them, typically reflecting the light source and transmitting fluorescence from the sample (in which case it is called as a long-pass dichroic). This property is also used in multicolor imaging to separate colors above and below the dichroic's wavelength cut-off. Emission filters placed further downstream are oriented orthogonal to the incoming light to selectively transmit only the fluorescent light and effectively block other stray radiation, including a significant amount of excitation light that leaks past the dichroic.
Although the basic optical elements (objective, other focusing lenses, beamsplitters, filters, etc.) suffice for intracellular SMM of RNA over the lateral (x,y) plane of the sample, 3-dimensional (3D) motion inherent to all biomolecules including RNA is not captured. To extract such axial (z) information, certain optical accessories, such as astigmatic lenses can be used. First introduced by Kao and Verkman,356 and later modified by Zhuang and coworkers,357 the method uses a cylindrical lens in the optical path to adjudicate axial localization based on the intensity profile of the sample under investigation – particles in focus have evenly distributed radial intensity profiles whereas particles just above or below the focal plane have elliptical/ellipsoidal intensity profiles elongated along different major axes. Recently, Moerner and coworkers used the combination of achromatic lenses and a spatial light modulator in a so-called 4f imaging system to extract 3D diffusional information of mRNPs in yeast358,359 at 25 nm lateral and 50 nm axial accuracy. In this case, the PSF of each single molecule is bi-lobed, whose relative position varies with the particle's axial position, essentially carving out the backbone of a DNA double helix through the axial movement of the particle; hence the eponym double helical PSF (DH-PSF). Other accessories such as a defocusing lens360 and wedge prisms361 have also been used to resolve 3D diffusion of particles and hold promise in 3D SPT of RNA.
3.4.3. Illumination Geometry
In intracellular SMM, far-field illumination geometries are typically used, wherein the distance between the objective lens and sample is at least an order of magnitude greater than the wavelength of illumination light (Figure 8). There are primarily two types of far-field illumination: wide-field (WF) and narrow-field (NF) illumination. The main difference between the two types of illumination geometry arises in the sample area (the field of view, or FoV) they excite: >100 μm2 of the sample is illuminated in WF illumination, whereas it is <1 μm2 in NF illumination. Examples of WF illumination include epi-, total internal reflection (TIR), highly inclined and laminated optical sheet (HILO, also termed near-TIR), and various other forms of selective plane illumination (SPI), whereas examples of NF illumination include confocal and single point edge excitation sub-diffraction (SPEED) schemes (Figure 8).
Figure 8. Various types of illumination geometries.
(A) Wide-field, (B) TIRF, (C) HILO/VAEM with a field diaphragm to reduce illumination light diameter, (D) SPI with LSFM (left) and RLSM (right) with a magnified view of the illumination plane. Although the geometry is similar for the two approaches, the dimensions of the illumination plane are very different. (E) Standard narrow-field illumination, with a blow-up of the illumination spot. Pinholes for the excitation (Ex.) light and emitted (Em.) light are used to reduce excitation volume and decrease background from out-of-focus fluorescence, respectively.
Simplest to implement, WF epi-illumination (Figure 8A) is widely used in intracellular SMM of RNA, largely due to several fluorescence enhancement strategies that render single RNA molecules significantly brighter than the cellular background. However, excitation light passes through the entire depth of the sample and out-of-focus fluorescent molecules contribute towards significant background noise, limiting the method to samples with molecules sparsely distributed over all three spatial dimensions. In contrast, only a thin lamina of ~150 nm at the sample surface is illuminated using TIR fluorescence microscopy (TIRF, Figure 8B), effectively reducing background fluorescence from outside regions.362 This scheme is based on Snell's law, wherein light that is transmitted through a medium of higher refractive index (ni; e.g., coverglass) into one of lower refractive index (nr; e.g., water), is totally reflected within the first medium at incident angles (θi) greater than the critical angle given by θc = sin−1(nr/ni). Because of the wave properties of light, TIR creates a thin lamina of “evanescent” wave within the aqueous phase, only exciting molecules that are within ~150 nm of the coverglass-sample interface. TIR is rarely used in intracellular SMM of RNA because it only excites the basal plasma membrane and ~100 nm of the adjacent cytoplasm, whereas most RNAs are present also throughout the cell and in cellular compartments (such as the nucleus) that the evanescent wave cannot penetrate. To achieve greater penetration into the sample without significantly compromising the signal-to-noise ratio, HILO microscopy (HILO, Figure 8C), also termed variable angle epi-fluorescence microscopy (VAEM) or near-TIR fluorescence microscopy (near-TIRF), was developed.363,364 Here, θi < θc so that the light is refracted into the sample at high inclination from the optical axis, thus illuminating an angled layer within the sample and reducing illumination of molecules not in focus. A field diaphragm that maintains the laminar width of the beam controls the width of this illuminated layer within the sample. The excitation beam path for epi-illumination, through-objective (or objective-type) TIRF and HILO are so similar that a single microscope can be used to implement all three schemes (Figure 7).
Several alternative SPI methods have been developed to further increase the penetration depth of illumination and restrict sample excitation to the focal plane, such that single molecules can be visualized within tissues or even whole organs, a realm that is not accessible to WF-epi, TIRF or HILO. One such advancement is called light sheet based fluorescence microscopy (LSFM, Figure 8D).268,315,365-367 In its most recent embodiment,315 the method uses two perpendicular objectives, one with low NA (~0.3) and the other with higher NA (>1) for illumination and detection, respectively. Such a configuration allowed focusing the illumination light as an ~3 μm thick, elliptical sheet, which can be moved across the z-axis of the specimen for optical sectioning. Depth of imaging is only limited by the working distance of the detection objective. A possible alternative to LSFM is reflected light sheet microscopy (RLSM, Figure 8D).354 In this method a small disposable mirror, such as an aluminum coated tipless AFM cantilever, is attached at an angle of ~45° to a high (0.8) NA illumination objective, positioned to face the detection objective with a slight lateral offset. The mirror reflects the light sheet from the illumination objective by 90° and projects it horizontally across the sample, thus attaining optical sectioning capabilities. The upright geometry allowed for imaging samples mounted on standard sample holders and results in a light sheet thickness of ~1 μm (FWHM). However, the positioning of the mirror and the shape of the light sheet introduce a gap between the surface and light sheet that cannot be illuminated. The RLSM illumination geometry holds promise for optical sectioning of thick specimens, but so far has been primarily used to image cultured cells.354
A majority of NF illumination schemes aims to increase spatial resolution by physically reducing the focal volume to a small, typically ellipsoidal or spherical, spot rather than a large plane (Figure 8E). In this fashion, only a few molecules are excited within this small volume element, typically ~500 nm wide and ~1 μm deep. Conventional confocal microscopes are augmented by more sophisticated optical configurations, such as spot-scanning 4Pi microscopy (4PiM)368 and other reversible saturable optical fluorescence transitions (RESOLFT) techniques,369 which achieve significantly higher spatial resolution by further reducing the illumination volume by at least an order of magnitude. Unlike WF illumination, throughput is minimal with NF illumination, i.e., the sample is illuminated only one volume element at a time and has to be scanned, but NF illumination has advantages of superior spatial (axial) resolution. Modifications to the standard confocal setup, such as the spinning-disc or Nipkow-disc confocal microscopy,370 significantly improve scanning rates and throughput by simultaneously illuminating distinct regions of the sample. Recently, Ma and Yang371 increased the FoV of NF illumination by incorporating oblique angle illumination (projected onto the sample at an angle of 45° with respect to the optical axis) and an ~400 μm wide pinhole in the excitation path within a standard WF microscope, in what they termed SPEED microscopy. This modification results in an inclined ellipsoid excitation volume, tilted at an angle of 45° with respect to the optical axis, that selectively illuminates ~320 nm of the sample in all three dimensions, leading to ~6-fold larger lateral FoV and ~3-fold smaller axial FoV than standard confocal microscopy. By restricting the acquisition area on an EMCCD camera, the time resolution of acquisition was improved to ~1 ms,371 which is ~20-to 100-fold better than standard WF imaging using the entire CCD chip.
3.4.4. Detectors
Illumination geometry dictates the choice of detector. Intensified charged coupled devices (ICCDs) or electron multiplying CCDs (EMCCDs), which feature a relatively large detection area (~8x8 mm2), are the detectors of choice in WF illumination. For NF illumination, where photons are collected from only a small region of the entire sample, point detectors like avalanche photodiodes (APDs) or photomultiplier tubes (PMTs) are used. Almost unlimited in time resolution, APDs and PMTs are sensitive enough to detect and accurately count single photons in the sub-millisecond regime, showcasing the superior sensitivity and better time resolution of NF imaging (with the caveats of low throughput and the need for scanning a larger sample). Although current CCDs also have photon counting capabilities, they still suffer from limited time resolution (millisecond time regime) and spectral range of detection. The complementary metal oxide semiconductor (CMOS) is an emerging technology that promises faster acquisition rates for WF imaging, but suffers from lower quantum efficiency (the ratio of detected photons over total photons that reach the detector) than CCDs.
3.4.5. Specialized Illumination Schemes
Once a specific illumination geometry is chosen, several illumination schemes can be employed to enhance sensitivity and spatiotemporal resolution. For instance, alternating excitation (ALEX) schemes372 have been routinely used in multicolor imaging to spectrally and temporally separate photon acquisition from multiple fluorophores by sequential imaging. Other multicolor acquisition schemes rely on spatially separating fluorescence signals from distinct fluorophores onto different regions of the same detector or onto multiple detectors, adding to the cost of instrumentation, whereas ALEX is useful in multicolor imaging on a single detector. ALEX is especially useful in reducing background associated with simultaneous multicolor illumination and in fluorescence resonance energy transfer (FRET) applications to confirm the presence of both the donor and acceptor fluorophores through the course of image acquisition. Another illumination scheme, termed stroboscopic illumination,220,373 can be employed to detect rapidly moving single particles that typically look blurry under slow CCD frame rates. Here, high-powered laser pulses, much shorter than the camera integration time, are used to briefly excite the sample. During this excitation period, single particles do not move over large distances and appear as distinct PSFs. In this fashion, the temporal resolution of acquisition is controlled by the duration of the pulse, however, at the cost of very high laser intensity that may be phototoxic to cells. The method also relies on relatively slow moving particles distributed at a very low density, such that it is easy to track the position of a single particle over time despite the pulse train's dark periods and the presence of other proximal particles, which can obscure particle identity. Finally, when photoactivatable or photoconvertible probes are used, tandem excitation schemes of photoactivation followed by sample excitation can be used to execute stochastic optical reconstruction microscopy (STORM) or photoactivation localization microscopy (PALM) and attain ~10 nm-scale spatial resolution, even within densely populated samples.212,219
3.5. Single Molecule Observables
A single intracellular SMM experiment allows the user to monitor a multitude of observables, each of which provides information about the molecule under study and its immediate surroundings.212 Experiments are typically performed on one of two distinct sample types, live cells or fixed cells, where each provides mutually complementary information. Among the two, live-cell intracellular SMM or SPT experiments shed light on the dynamic aspects of cellular function, such as the mobility of individual molecules and the extent of interaction between multiple distinct sets of molecules. Although fluorescence recovery after photobleaching (FRAP)374 and fluorescence correlation spectroscopy (FCS)133,134,344,375 have been traditionally used to extract such dynamic information, they only measure ensemble-averaged properties and probe a small sub-cellular section at any given time, frequently missing variations in molecular motion that occur over the entirety of the cell. SPT microscopy by WF illumination overcomes this caveat by accounting for almost all fluorophore labeled molecules within a cell in a sequence of images (or video). Individual particles are localized in each frame of the video at nanometer-scale accuracy and this position is linked between frames to construct particle trajectories. Several well-established methods are available to execute both of these steps205,376-379 whose choice is dependent on the signal-to-noise ratio for the former and the ratio of particle density to interframe displacement for the latter. Mobility of RNA is then expressed in the form of velocity and a 1D, 2D or 3D diffusion coefficient (the average spatial unit a molecule traverses per unit time given any constraints on its movement). Particle velocity and displacement are physical properties that provide valuable insight into the processivity and overall directionality of motion, which are important, for example, in the characterization of motor protein-mediated RNA transport.380 Diffusion coefficients encode information about the molecular mass of the mRNPs or the nature of its surrounding milieu, in terms of sub-cellular viscosity and the presence of potential interaction partners.381,382 Furthermore, the diffusive pattern (Brownian, biased, corralled, or stationary) can be used to predict the role of motor proteins or the nature and amount of obstacles in the path of a diffusing molecule.233 For instance, prolonged observation and analytical methods like velocity autocorrelation359 have shown that the diffusive pattern of a single RNP often varies even within an individual trajectory,233,359 highlighting the different intracellular interactions a molecule experiences through its lifetime. More specific interactions are revealed by simultaneous tracking and colocalization dwell times of molecules that are labeled with spectrally distinct fluorophores307,347 in two-color imaging schemes. Concordant motion adds another layer of confidence to traditional colocalization studies that often obfuscates non-interacting particles bearing similar x,y coordinates but distinct z-positions with physical association (complex formation). The protein-RNA,233,307 hybridization by molecular beacons269 or MTRIPs,217,346,347 and direct labeling systems135 have all been used in SPT of RNA in living cells; however, the protein-RNA tethering systems are particularly popular among cell biologists because of their arguably easier and gentler conditions for delivering probes, which may better preserve cell integrity.
Sub-cellular localization and counting of individual mRNA molecules yields single cell gene expression data and thus has revolutionized our understanding of cellular homeostasis and the effects of deviations from it.21,135,383-385 Stoichiometry measurements by stepwise photobleaching, which assess the number of molecules within individual, not further resolved fluorescent particles, increase the accuracy of counting and are useful in discerning the ubiquitous assembly/disassembly of molecular complexes135,334 and in functionally annotating sub-cellular localizations.282,347 However, the abrupt change in intensity by the diffusion of particles out of the focal plane in conventional 2D imaging confounds single molecule counting and stoichiometry measurements in live cells. FCS is useful in such cases, wherein the diffusion of molecules through a well-calibrated focal volume results in intensity bursts whose frequency is used to measure intracellular concentration. This method, however, has the inherent disadvantage of having to assume a homogeneous distribution of the molecule throughout the cell or, conversely, requires the slow, sequential probing of different regions within the cell to correct for heterogeneity. Molecule counting and stoichiometry analysis are more reliable in fixed cells, where particles are easily tractable and spatially confined. A majority of single molecule counting experiments of RNAs, especially mRNAs, has been achieved by smFISH. 3D image stacks of probe-treated cells are acquired and individual particles are detected by image processing steps that include image enhancement and intensity thresholding.267,283 Gaussian fitting,205 deblurring by deconvolution217,266,275 and image convolution by high-pass filtering,267,280 such as a Laplacian of Gaussian (LoG) filter, are a few image enhancement strategies that have been applied to particle detection via smFISH. Molecular stoichiometry in fixed cells can be measured either by particle counting in smFISH images or by stochastic photobleaching of fluorophores.135,334 The latter is more effective when single RNA molecules are labeled with relatively few fluorophores, typically achieved by direct labeling schemes. Photobleaching of RNPs decorated with a large number of fluorophores, as in the case for most protein-RNA tethering and hybridization labeling schemes, yields non-discernible and frequently overlapping photobleaching steps that yield incorrect molecular count.335 In such cases, calibration curves are constructed by imaging (free) fluorescent probes266,275 at various concentrations, from which the intensity of a single probe is extrapolated. The number of probe repeats per RNA is considered in calculating the approximate intensity of a single RNA molecule, which is then used to compute the molecular copy number within individual particles. Alternatively, the mean intensity value of smFISH-detected particles is used as a measure of molecular count.267,283 Although stoichiometries extracted from these curves are often either error-prone or approximate, they are useful in observing trends. Overall, both live and fixed cell imaging are powerful in discerning the spatiotemporal organization of intracellular RNAs.
In the next section, specific recent examples of characterizing the biological function of RNAs and the mechanisms of their biogenesis, transport, regulation and turnover through single molecule observation are presented.
4. RECENT APPLICATIONS OF SINGLE MOLECULE APPROACHES TO RNA IN CELLULO
Earlier in this review, RNA was introduced in its various forms as a highly versatile molecule, coding for proteins, serving as structural scaffold, performing enzymatic functions, and regulating gene expression. To perform such widespread functions, RNA production, maturation, interactions, and spatial distributions require tight regulation, which in turn is the foundation of cellular homeostasis and survival. Intuitively, the bulk properties observed of such processes and others are potentially the sum of all single molecule events. For example, ensemble experiments have convincingly shown that a significant fraction of β-actin mRNA localizes to the leading edge of a cell,260,386 while a majority of transcripts still seems to be spread throughout the cytoplasm. Yet only with the aid of intracellular SMM it could be resolved that their diffuse cytoplasmic distribution is a cumulative result of individual transcripts either remaining stationary or undergoing: (i) Brownian diffusion; (ii) corralled motions; and (iii) active transport.233,346 It was additionally found that transcripts are not in a constant diffusive state, but rather switch between multiple modes of movement.233 Despite the fact that only ~9-20% of all transcripts exhibit biased motion, interrupting active transport significantly alters intracellular distribution and almost abolishes localization at the leading edge.233,260,346,386 Thus, intracellular SMM has been indispensable for understanding mechanisms of cellular RNA localization and the underlying kinetic parameters controlling them. To fully appreciate these findings, this section will detail the results of select studies performed over the last fifteen years and how they have contributed to our understanding of cellular biology. By necessity, this selection will be incomplete, due to the rapid and accelerating progress in intracellular single molecule fluorescence studies of RNA.
4.1. Stochasticity of Transcription
Gene expression is canonically, and minimally, perceived to result from the transcription of genetic information into mRNAs (i.e., mRNA synthesis), followed by transcript decoding and protein synthesis (translation).387,388 The intracellular copy number of a given mRNA is quite low (~1-500 copies is typical),389 yet each mRNA molecule can be translated tens to hundreds of times. Thus, small fluctuations in mRNA production or degradation will potentially result in large changes in protein concentration257,390,391 and potentially alter the cellular phenotype.392-395 As transcriptional fluctuations are ubiquitous,21,395 it is critical to understand the causes and impacts of such bursts with special attention to perturbation of gene regulation programs. A variety of single molecule techniques have been applied over the years to probe the extent of transcription variability and its impact on gene expression. Below is a comprehensive discussion of their findings.
In first approximation, a stochastic model of gene expression can be assumed to follow a Poisson process,396 wherein the synthesis and degradation of transcripts occur at random, but the probability of transcript production is time-invariant, i.e., the rate of transcript production is constant. Consequently, the mean expression level of the transcript for a single cell and all cells in a clonal population should coincide with the distribution's variance, or the Fano factor (variance/mean) should equal unity. Conversely, describing gene expression patterns using Poisson distributions assumes that they are completely random. Deviations of the Fano factor from unity entail either transcriptional bursting (super-Poissonian or Fano factor >1) or synchronized and homogeneous transcript production over the cell population (sub-Poissonian or Fano factor <1), implying highly regulated transcription.267,397
Transcriptional bursting is a term that is used to describe genes that are synthesized discontinuously, typically with a temporally varying rate of transcript synthesis.398,399 Tyagi and coworkers used smFISH to count individual transcripts and determine that the mean cellular production of an inducible exogenous mRNA in mammalian cells is significantly less than the variance, indicating super-Poissonian trends400 or bursting. They determined that the extent of intercellular gene variability is not heavily weighted on transcription initiation events, as genes bearing multiple promoter sites failed to alter the noisiness in mRNA number. Additionally, modeling the distributions of transcriptional states, cellular locations of single molecule mRNA (nuclear, cytoplasmic, or both), and number of mRNAs produced within the assayed time period for a colony of cells lent additional evidence to transcriptional bursting (Figure 9).400 Transcription was found to be intrinsically random and dependent on single gene activation and deactivation processes, as transcription factor concentrations have little impact on the extent of randomness. Furthermore, genes within a similar DNA locus have synergistic bursting kinetics, unlike instances where they are located within dissimilar loci. One can argue that smFISH only provides a snapshot of events within a fixed cell that cannot account for the temporally changing contributions of mRNA degradation, cell division, and RNA aggregates on the calculated super-Poissonian trends. In a complementary line of experiments, Golding et al. used the MS2 labeling system to detect transcriptional behaviors in live E. coli cells.306 Despite the change in techniques and species, transcriptional bursting was equally seen within single cells, whereby RNA is created by pulses of transcription that are interspersed by longer periods of inactivity (Figure 10).306 Unlike in mammalian cells, the randomness of bacterial transcription appears dependent on extrinsic factors, such as transcription factor or inducer abundance.306,401 Recently, Golding and coworkers used smFISH to probe the transcription of another artificial mRNA construct mRNA when driven by one of 20 different promoter sequences in E. coli and unveiled a correlation between promoter activity and increased ‘burstiness’ during transcription.340 Their work suggests that only the duration of gene activity dictates the oscillatory behavior of transcription, regardless of the stimulus that initiates transcription or the specific promoter sequence controlling it. These and other data suggest that bursting results from topological DNA changes, RNA polymerase dynamics, or some effects of broad-target DNA binding proteins.287,402,403 Transcriptional bursting has also been demonstrated using single molecule techniques in social amoeba,404 humans,291,403,405 and yeast.278
Figure 9. Transcriptional bursting measured by smFISH in mammalian cells.
(A) Schematic of the doxycycline controlled yellow fluorescent protein gene containing 32x smFISH probe binding sites in the transcript's 3’-UTR used in this study. (B, C) Representative images of cells with active or inactive transcription sites. (D) A representative image of two sister cells with variable mRNA localization. The upper cell displays primarily cytoplasmic localization of mRNA (shown in red), while the bottom cell displays both nuclear (stained blue with DAPI) and cytoplasmic mRNA distributions. The scale bar represents 5 μm. Reprinted with permission from ref. 400 Copyright 2006 Public Library of Science.
Figure 10. Transcriptional bursting as measured by mRNAs bound by MCP-GFP in individual E. coli cells.
(A) Schematic of MCP-GFP and mRFP constructs with 96xMS2 binding sites used in this study. (B) Plots from four cells depicting typical estimated numbers of transcripts (n) as a function of time. Red dots indicate raw data, the green line represents smoothed fit, and the black line depicts a linear function that shows periods of transcriptional activity and inactivity. Cyan dots represent the number of transcripts in the daughter cell after cell division. The numbers represent the change in the number of transcripts. Reprinted with permission from ref. 306. Copyright 2005 Elsevier.
Contrary to the work by Golding, Cox and coworkers, Muthukrishnan et al. demonstrated that select genes in E. coli exhibit sub-Poissonian behavior.401 While they too used the MS2 labeling system, these authors elected to monitor gene transcription at the active site, rather than counting mRNA numbers. This group relied on the monotonic increase in signal of the MCP-FP once it is bound to the MBS at the transcription site, so that a stepwise increase in signal (per unit time) reflects the appearance of a novel transcript. This increase persists until the eventual release of the RNA from the transcription site, at which point the relatively low time resolution of data acquisition blurs the signal from fast moving transcripts. Plotting the various lag times between transcriptional events for these mRNA resulted in a Fano factor <1, or sub-Poissonian behavior, which was further corroborated by the low intercellular mRNA variability. Based on these results, the authors argued that transcription cannot be dictated solely by the randomness of binding and release of transcription factors from the DNA surface. Similar observations were made in an earlier study by Kandhavelu et al.406
Not all mRNA production is dependent on an external stimulus or the stage of the cell cycle, as some mRNAs are constitutively (constantly) expressed. For constitutively transcribed genes in yeast cells, specifically housekeeping genes, the variance in intercellular protein expression is remarkably small,407 despite the low transcript numbers. Zenklusen et al.278 concluded that these low variances are related to production of translated mRNA molecules. Intracellular mRNA copy number, as detected by smFISH, is best fit with a Poisson distribution, which along with computational modeling suggests that the transcription of constitutively expressed genes is dependent on the probability of forming the transcription initiation complex.278 The authors discuss these effects as contributed by transcription initiation events that are digitally ‘separated in time’ instead of ‘bursts’.278
Thus far, we have discussed the variability that surrounds gene transcription in a cell culture context. However, cell culture conditions of bacterial or immortalized mammalian cells might not represent those that transpire in tissue or primary eukaryotic cells. For example, in aggregative slime mold development, each endogenous gene exhibits its own unique transcriptional profile, with increased RNA production correlating with increased ‘bursting’ behavior.402 It was further found that transcriptional variability is minimized for transcripts produced in low quantities and for proteins that require tight regulation. Similar effects were observed in developing C. elegans embryos, where intestinal specification during the initial stages of organogenesis is initiated by various environmental cues that induce the transcription of select developmental genes.283 These genes then initiate a signal cascade that instigates heterochronic gene expression and exhibits sub-Poissonian transcription properties enabling their tight regulation in the cell.280 To understand the effects of ‘bursting’ on non-bursting genes, the authors introduced a mutation into an upstream developmental gene that resulted in highly variable mRNA expression profiles of the remaining downstream genes.283 Increasing variability in the transcription of these genes results in a defective intestinal tract. Such strict developmental control is also observed during the induction of pluripotent stem cells.408
The possibility that stochastic transcription may be associated with chromosome structure spurred Levesque and Raj to adopt a spectral barcoding approach that simultaneously visualizes transcript production and relative chromosomal localization.292 Using the iceFISH method, the authors simultaneously visualized 20 different transcripts spread over individual chromosomes. They found that genes on the t(13:19) chromosome of HeLa cells were up to five-fold more active than those on chromosome 19. Although a majority of genes within a given chromosome were transcribed independently, one specific pair of transcripts had anti-correlated expression, even if several megabases apart on the chromosome. Moreover, such an effect was not found on the other allele, suggesting that the effect is mediated by cis- and not trans-acting elements.
These single molecule experiments have demonstrated that transcription is controlled by a wide variety of mechanisms that can vary in a gene- and species-dependent manner. The sensitivity associated with intracellular SMM has revealed that transcription can produce RNA in a highly controlled fashion with low variability (Sub-Poisson),306,401,406,409 in a completely random fashion (Poisson),278,340,402 or in a highly fluctuating, burst-like fashion (Super-Poisson).278,291,306,400,404 Based on these data, a great deal of effort is currently focused on modeling the transcription process as a predictive tool for gene expression.410-413 However, more work in this field is necessary to tease out mechanistic details that control the fate of gene production and the variability surrounding it.
4.2. Transcription Inhibition and Silencing by Long ncRNA
Cell survival, maintenance and self-destruction are all dependent on the well-timed transcription of select genes and the silencing of others. To keep the activity of RNA transcribing polymerase molecules in check, the cell has adopted numerous mechanisms for the short- or long-term silencing of select genes. Important regulators and, at times, targets of such silencing activities are lncRNAs.414,415 smFISH studies have demonstrated that lncRNAs have the ability to inhibit transcription of proximal or overlapping genes in cis, at the site of synthesis,416,417 and in trans, distal to site of synthesis,188,416,418 such as the silencing mediated by Xist lncRNA that inactivates one of the two X chromosomes in female mammals.
In yeast, many genes give rise to antisense transcripts. Some of these transcripts are stable while others, termed cryptic unstable transcripts (CUTs), are rapidly degraded by the nuclear exosome.419 For example, PHO84 AS is an antisense transcript that is involved in the transcriptional silencing of the protein coding gene PHO84 from the same genomic locus and was hypothesized to accumulate at the locus to sustain repression.420 Contradicting this hypothesis, Castelnuovo et al. found using smFISH421 that PHO84 AS is always weakly expressed in wild-type yeast cells, and in fact, a majority of transcripts are localized in the cytoplasm. The latter observation suggested that the antisense transcript essentially behaves like a protein coding mRNA as it is transcribed, polyadenylated and transported to the cytoplasm. However, the expression modes of the sense and antisense transcripts were found to be distinct, with the sense RNA abundantly transcribed in bursts and the antisense synthesized constantly at very low numbers. Although PHO84 and PHO84 AS were expressed within a clonal population of cells, individual cells seldom co-expressed both, an observation often masked in ensemble experiments. Put together, the authors hypothesized that regulation occurs via a switch-like mechanism instead of one where the slow accumulation of one silences the other RNA. Moreover, low expression of the antisense transcript is sufficient for silencing and ensures that sense expression is only activated in the presence of a stronger opposing signal.
4.3. Splicing of Pre-mRNA
Nuclear speckles are membraneless protein aggregates that are enriched in pre-mRNA splicing factors and are located in the interchromatin region of the nucleoplasm.47 Their specific function is still debated as they are not always found near sites of active transcription, but they have been shown to contain pre-mRNAs and thus presumably are sites of post-transcriptional splicing.46 Co-transcriptional splicing is thought to be advantageous as it ensures the fidelity of exon-intron recognition in the RNA molecule before it is released into the nucleoplasm. However, during alternative splicing, post-transcriptional splicing is arguably more important as the complete synthesis of all exons and introns provides greater options for exon selection. To test the frequency of co- and post-transcriptional splicing, Vargas et al. performed smFISH and molecular beacon-mediated live cell imaging of a set of GFP reporter genes, bearing 12 distinct arrays of intronic oligonucleotide binding sites.270 The 3’-UTR or GFP coding sequence (CDS) was simultaneously labeled to annotate spliced and unspliced transcripts – fluorescent spots only containing signal for the 3’-UTR (or GFP CDS) were considered spliced, whereas those that showed colocalization with the intronic array signal were annotated as unspliced transcripts. Using this system, the authors found that the nucleoplasmic distribution of pre-mRNAs varied based on the identity of their intronic array, wherein pre-mRNAs with accessible spicing recognition sites were found almost exclusively at the transcription site (TS), while ones with recognition sites obscured by RNA secondary structure were found diffusely distributed throughout the nucleoplasm. The latter type of pre-mRNA was observed to interact with, but not enter, speckle sites as marked by immunofluorescence.270 Secondary structure analysis predicted that the post-transcriptional splicing phenotype was well correlated with the accessibility of particularly the intron's polypyrimidine tract. Burying this tract in introns known to splice near the TS within strong secondary structure predictably recapitulated the distal splicing phenotype and thus the uncoupling of transcription and splicing.270
Recently, real-time imaging of co-transcriptional splicing has been used to characterize the kinetics of intron excision in live cells.422 In this report, single-molecule sensitivity was achieved using protein-RNA tethering systems. This study revealed the rate of transcription by RNA polymerase II (3-6 kb min−1) is the rate-limiting step in splicing, thus providing mechanistic insight into genetic regulation.
Most mRNAs within the human genome are known to undergo alternative splicing36,37 to generate protein isoforms in the various tissues of an organism. Vargas et al. suggested that the relative abundances of alternative splicing factors dictate the relative quantities of transcript isoforms.270 Waks et al. set out to use smFISH to probe the extent of isoform heterogeneity of a clonal population for non-cancerous and cancerous cells.423 Using bioinformatics approaches, they found CAPRIN1 and MKNK2 to each express two isoforms that differ in either their terminal exon sequence or their 3’-end length. By designing smFISH probes that bind sequences unique to each isoform, the authors were able to use a two-color approach to quantify the copy number of each isoform. MKNK2 pre-mRNA is located near the TS and is thought to be spliced constitutively, whereas as an alternatively spliced pre-mRNA CAPRIN1 is found distal from the TS. In non-cancerous Rpe1 cells, mRNA isoform ratios varied as minimally as could be expected given the stochastic nature of transcription, whereas HeLa cells demonstrated significantly more cell-to-cell variability in the isoform ratios in a manner driven by alternative splicing factor activity. Taken together, these data suggest that the splicing pathway has acquired control mechanisms to reduce isoform fluctuations in non-cancerous cells.423
4.4. Nuclear Diffusion of RNA
Regardless of the relative distances between the transcription and pre-mRNA processing sites, RNA must eventually traverse the nucleoplasm to reach the nuclear envelope for exportation. Active transcription sites are not always located at peripheral regions of the nucleus, but can occur in chromatin dense regions.424,425 Transcribed RNA molecules in these regions traverse chromatin free tracks towards the nuclear envelope,426 sparking the hypothesis that RNA may be actively transported via motor proteins along the nucleoskeleton as a more efficient means of RNA delivery to processing centers and the NPC. An electron microscopy study was the first to demonstrate that BR mRNAs in C. tentans salivary glands are randomly distributed throughout the nucleoplasmic space, suggesting that they move primarily via simple diffusion and not active transport.427 Siebrasse et al. labeled and tracked the movements of single BR mRNA molecules, labeled with fluorophore-conjugated DNA or 2’-O-methyl-RNA oligonucleotides, or with recombinant fluorescent hrp36 (an hnRNP protein), in living C. tentans salivary gland cells268 to corroborate the EM findings. Mean square displacement (MSD) analyses of the SPT data demonstrated deviations from linearity, and thus from Brownian diffusion behavior, with increased lag time, suggesting corralled motions.268,428 This behavior is potentially mediated by the binding of BR transcripts to cellular factors or by general crowding effects that occur in the dense milieu of the nucleus. Nucleoplasmic diffusion coefficients of BR mRNA were significantly slower than those of similarly sized dextran molecules (D = 2.2 μm2/s). These data coupled with Monte Carlo simulations suggested that BR mRNA may be slowed as the result of a series of transient interactions with nuclear components. These results effectively strengthened claims from prior ensemble microscopy experiments performed in chromatin bearing mammalian cells, where mobility of mRNPs was found to involve passive diffusion and ATP-dependent processes.316,429
Shav-Tal et al. employed the MCP-MBS labeling system to selectively label stably integrated and inducible exogenous genes in mammalian cells and, using SPT, studied a significant part of their lifecycle, from transcription to nuclear export.430 Labeled RNA diffused throughout the nucleoplasm avoiding regions occupied by large nuclear structures (chromatin, nucleolus, etc.), with 58% of the tracks Brownian in nature and 42% exhibiting corralled motion (Figure 11).225,430 Motions were most confined or immobile when in close proximity to the TS.269,430 Outside the transcription site, RNA molecules were not observed to stall or stop, implying that they do not make stable interactions with sub-nuclear compartments. Under hyper-osmolar conditions, corralled motions of mRNPs were enhanced due to the entrapment of transcripts within dense DNA regions and their sequestration into speckle domains, a process that is reversible upon returning the cells to isotonic conditions. Vargas et al. obtained similar results with temperature reduction or ATP depletion,269 strongly suggesting a (perhaps indirect) role for DNA dynamics in modulating RNP diffusion.
Figure 11. Single particle tracking of mRNPs in mammalian cells.
(A) Schematic of the mRNA constructs with 24xMS2 binding sites used in this study. Nuclear diffusion data for each construct are summarized in Table 2. (B) Deconvolved time-series images of nuclear Cerulean-½ Mini-Dys + intron mRNP diffusing in the nucleoplasm (green tracks). (C) Complete track of the nuclear Cerulean-½ Mini-Dys + intron mRNP. (D) MSD analysis of single nuclear mRNPs displaying Brownian (green) or corralled (red) motions in the nucleoplasm. Corralled motions deviate from linearity at longer lag times. Scale bar represents 1 μm. Reprinted with permission from ref. 225. Copyright 2010 Nature Publishing Group.
The diffusion coefficients of MCP-MBS labeled mRNPs are much smaller than those reported in ensemble mRNA experiments.428,242 To test if this effect is due to particle size or transient RNP interaction with other proteins, Mor et al. designed seven constructs of increasing size or bearing the same size but different numbers of introns (Table 2).225 2D and 3D SPT of these transcripts showed trends of diffusion coefficients decreasing with RNA size, eventually reaching a saturation point for the three largest mRNAs tested (Table 2). Cytoplasmic diffusion coefficients of the smaller transcripts correlated well with their nuclear diffusion coefficients, however, the larger transcripts diffused more slowly in the nucleoplasm. 3D tracking of mRNPs in cells with Hoechst 33342-stained DNA suggested that the larger RNAs exhibit more corralled motions in the narrow inter-chromatin spaces than the smaller RNA, contributing to their small diffusion coefficients. Thus, the three largest mRNAs have equal nuclear diffusion coefficients but divergent, faster cytoplasmic diffusion coefficients simply due to the strong (and non-discriminating) sieving effects of the comparably narrow inter-chromatin channels they traverse in the nucleus.225
Table 2.
Nuclear diffusion characteristics of RNAs of varying length. Data taken from ref. 225.
| Gene Name | mRNA length (kb) | Diffusion (μm2/sec) | Percent Corralled | Percent Brownian |
|---|---|---|---|---|
| Full-Dys | 14 | 0.005 | 60% | 40% |
| Mini-Dys | 8 | 0.005 | 75% | 25% |
| ½-mini-Dys | 4.8 | 0.005 | 50% | 50% |
| ½-mini-Dys + 1 intron | 4.8 | 0.004 | 45% | 55% |
| E1 | 1.7 | 0.009 | NA | NA |
| E3 | 2.1 | 0.010 | NA | NA |
| E6 | 2.3 | 0.023 | NA | NA |
Labeling of particles with spectrally distinct fluorophores facilitates colocalization and co-tracking during the assembly of biomolecular complexes. A two-color approach was used to study the interaction of snRNPs with nuclear speckles, in an effort to understand the interaction of splicing factors with these largely elusive nuclear foci. To this end, Grunwald et al. microinjected non-specifically labeled U1 snRNP-Cy5 conjugates into the cytoplasm of HeLa cells expressing ASF/SF2-GFP, a protein that concentrates in nuclear speckles.343 Upon nuclear import, the snRNPs distributed into three sub-populations: immobile (77.5%), slowly diffusing (15%, with average diffusion coefficients of 0.51 ± 0.05 μm2/s), and fast diffusing (7.3%, with 8.2 ± 3 μm2/s). Interestingly, not all immobile snRNPs were observed to colocalize with fluorescent markers of nuclear speckles343 where splicing occurs and most spliceosomal snRNPs are thought to reside. Notably, those snRNPs that did colocalize remained in speckles over periods ranging from milliseconds to seconds with a double-exponential time distribution, suggesting that their binding mechanisms are complex.343 In another study, it was observed that spliceosomal components are not stored in nuclear speckles, but rather splicing proteins are found diffusing throughout the nucleus and collide randomly and transiently with pre-mRNAs.431
4.5. Nuclear Export of RNA
The nuclear pore complex, or NPC, is a large protein complex that creates, in first approximation, size-limited perforations on the nuclear envelope to mediate nucleocytoplasmic transport. NPC dependent transport is the primary pathway to deliver RNAs into the cytoplasm. The rate and mechanism of these transport processes are critical to understanding spatiotemporal regulation of gene expression. Due to the stochasticity and rapidity of RNA transport across the NPC, intracellular SMM is an ideal tool to probe its kinetics and mechanism.
As discussed, RNA transcribed in the nucleus traverses chromatin-free channels for the vectorial diffusion to select regions of the nuclear envelope. Regardless of the position of the transcription site or the channel, however, RNA is uniformly distributed throughout the nucleoplasm. Therefore, it seems unlikely that transcribed RNA is automatically targeted for export through the NPC. Supporting this notion, several groups have observed that the anchoring of mRNPs to NPCs does not necessitate transport, and that a single mRNP can interact multiple times with a single or multiple NPCs prior to transport (Figure 12).88,225,307 Additionally, the extent of transport, derived by transcript counting in the nuclear and cytoplasmic compartments over time, also depends on the presence of introns, thereby alluding to a splicing mediated binding of appropriate nuclear export factors.88,225
Figure 12. Nuclear export of MS2 labeled mRNA in mammalian cells.
(A) MSD plot comparison of the nuclear export (red) relative to cytoplasmic (blue) and nucleoplasmic (green) diffusion. (B) Pseudo-colored time-series of an mRNP (red), marked with a white arrow, during nucleocytoplasmic transport across the nuclear envelope (dotted white line). (C) Stalled (red arrows) MS2 labeled RNA (green) at the nuclear envelope before (left image) and after (right image) ATP depletion. Reprinted with permission from ref. 225. Copyright 2010 Nature Publishing Group.
mRNP transport through the NPC is more rapid than nucleoplasmic diffusion (Figure 12A).427 The distribution of translocation times for β-actin mRNP particles across the NPC is, however, very wide, stretching from milliseconds to seconds and fitting a double-exponential time distribution.307 The slowest 10% of mRNPs, predominantly resulting from direction change in the pore, exhibited monophasic transport kinetics, while the faster population exhibited biphasic kinetics, indicating two rate-limiting steps. Additionally, the slowest transported β-actin particles appeared to be positioned most centrally to the NPC, while the fastest population aligned themselves more proximal with the edges of the NPC. β-actin RNPs were also observed to stall at the nuclear and cytoplasmic sides of the NPC, but not in the channel. Depleting cellular ATP levels did not affect binding of β-actin RNA with the NPC, but no fluorescent signal was discovered inside the channels, again suggesting active transport across the pore, which is possibly required to linearize the RNP to fit the pore of the NPC. Based on these data, the authors proposed a three-step mechanism for mRNP translocation across the NPC: a relatively long mRNP docking phase on the nucleoplasmic side for ~80 ms, rapid translocation through the channel within 5-20 ms, and release on the cytoplasmic side within the next ~80 ms.307
One potential caveat when using the MS2 system for studying rates of nuclear export may be the loss of the (relatively weakly bound) MS2 coat protein during translocation through the NPC. Consistent with this notion, Mor et al. reported that the MS2 labeled RNA dimmed during transport, compromising the accuracy of kinetic measurements.225 Furthermore, the addition of the MS2 stem loops and binding of MS2 coat proteins may have deleterious effects on the rates of NPC transport by adding size to the mRNP or preventing the binding of key proteins necessary for NPC interaction. However, it is known that numerous endogenous nuclear RNA binding proteins, like hrp36, will bind to transcribed mRNA in the nucleus and remain bound even during transport through the NPC. Exploiting this phenomenon, Siebrasse et al. were able to track individual BR mRNPs simply by microinjecting fluorescently labeled, recombinant hrp36, a BR mRNA binding protein, into C. tentans salivary gland cells.315 Trajectories of the various moving particles displayed mRNPs that collided with the nuclear envelope and returned to the nucleoplasm (nuclear probing) or returned into the cytoplasm (cytoplasmic probing); showed active export, as well as nucleoplasmic and cytoplasmic particles binding to the NPC with no further interaction; and indicated particles that started and ended at the NPC for the duration of the observation window. Nuclear and cytoplasmic probing events, especially, were thought to be instances where collisions with the nuclear envelope or with the NPC resulted in unsuccessful docking and were the predominant phenomena in hrp36 (nuclear export protein) labeled mRNPs and Dbp5 (cytoplasmic helicase responsible for mRNA remodeling), respectively. Only 25% of all trajectories corresponded to export with time constants of 65 ± 5 ms (for 87% of that population) and 350 ± 25 ms (the remaining 13%). Taken together, the authors hypothesized that a rate-limiting step for export occurs at the nuclear basket, which may represent proper RNA orientation to penetrate the NPC.
In a recent study, SPEED microscopy88,371 was utilized to map the 3D pathway taken by mRNPs as they traverse through the NPC of eukaryotic cells.88 At an unprecedented spatiotemporal accuracy of 8 nm and 2 ms, the authors determined that MCP-FP labeled mRNPs decelerated at the center of the NPC during nuclear export, adopting a fast-slow-fast diffusion pattern during their brief (~12 ms) interaction with the NPC. This contrasts with an earlier report attained at a lower spatiotemporal resolution of 26 nm and 20 ms that predicted a slow-fast-slow model wherein mRNP diffusion was proposed to be attenuated at the NPC periphery.307 Based on the faster SPEED microscopy data88,371, 3D reconstruction of both mRNP and small (protein) molecule export routes was possible and revealed that mRNPs primarily interact with the periphery on the nucleoplasmic side and in the center of the NPC, where they are selected for export, whereas small molecules passively diffuse via the central axial conduit. These results are consistent with a multilane traffic model wherein the transport receptor facilitated diffusion of large mRNP complexes and the passive diffusion of small molecules follow distinct paths through the NPC88,371,432. These observations also demonstrate how improvements in instrumentation often lead to refined biological insights.
4.6. Cytoplasmic Localization of RNA
Much like proteins, the sub-cellular localization of RNAs is critical to their function and regulation.433 Localization can be diffuse, wherein RNA molecules move in a manner that allows equal sampling of the cytoplasmic space, or polarized, where the RNA is directed to and localized in or near unique sub-cellular features. For instance, certain neuronal mRNAs are enriched in select dendritic regions for their local and rapid production of proteins that control potentiation, a process necessary for cellular memory. These asymmetric RNA distributions were alternatively hypothesized to result from: (i) continuous active transport; (ii) active transport and anchoring; (iii) diffusion and anchoring; and/or (iv) increased stability in sub-cellular distributions. However, it is still unclear if these processes simply occur to varying degrees or if different RNAs follow distinct localization pathways. Live cell, single molecule experiments are an ideal tool to provide answers to these questions.
The first recorded experiment utilizing the MS2 RNA labeling system was designed to probe the mechanism of mother-to-daughter (bud) Ash1p mRNA delivery and sequestration in yeast.434 SPT of Ash1p mRNA revealed directional movements at velocities concordant with that of myosin-mediated transport (320 nm/s), strongly suggesting active transport of these mRNPs on the actin cytoskeleton. Ash1p mRNA stably resides at the bud cortex for periods of >1 min, suggesting an anchoring mechanism. It was later found that similar transport mechanisms mediate the asymmetric distribution of other yeast RNAs.303,435 RNA transport mechanisms were found to be widespread in higher eukaryotes such as Drosophila,436-439C. elegans,440 chicken fibroblasts,441 and human cells.213,345,346,442,443
Although individual actively transported mRNPs are large and typically contain the transport and translation machinery among others, they primarily entail only single molecules of mRNA.345,442-445 In lieu of single RNP transport, the packaging and co-transport of multiple RNPs as large granules have also been observed, in both individual cells and multicellular embryos.303,439 In either case, RNA in route to its destination can oscillate between anterograde and retrograde transport,436,444,446 and the extent of bidirectionality can be stimulus dependent. For instance, Rook et al. used the MS2 system to demonstrate that the dendritic localization of CaMKIIα mRNA is increased by cell depolarization.446
In cellulo single molecule imaging has also shed light on novel RNA transport mechanisms. Specifically, Fusco et al.233 used the MS2 labeling strategy to demonstrate the heterogeneous and probabilistic movement of individual mRNPs in living COS cells.233 They found that mRNPs exhibit either a multitude of diffusive behaviors (biased, corralled and Brownian) or remain stationary and frequently switch between different diffusive patterns (Figure 13A and B), a phenomenon observed by other groups as well.346,447 Importantly, the authors demonstrated a sequence specific bias of mRNPs towards select motion types by being able to increase the propensity for directed motion in RNPs tagged with a zipcode sequence contained in the 3’-UTR of β-actin mRNA (Figure 13C).233 Other cytoskeleton independent RNP transport processes were also found in complex multicellular systems, wherein transport is mediated by fluid flow within the organism and localization can either be actin dependent or independent438,448.
Figure 13. Cytoplasmic mRNP dynamics.
(A) Movements of an MCP-FP labeled LacZ-hGH mRNA. Overlay of a 200-frame video (left image). The scale bar located in the bottom right corner represents 10 μm. Magnifications of select regions of the projection image are shown in the right images. The scale bar here represents 2 μm. Green lines represent the tracks the particles traverse. (B) Movements of an MCP-FP labeled Lacz-SV mRNA, imaged similar to panel A. (C) Distributions of LacZ-SV mRNP motions with and without a β-actin 3’-UTR. Reprinted with permission from ref. 233. Copyright 2003 Elsevier.
4.7. Translation of mRNA
As discussed earlier, transcription can be a highly variable process that will lead to changes in cellular mRNA levels. Correlating transcript number to protein output further defines the stochastic nature of translation and its overall contribution to noise in gene expression.399 Notably, spatiotemporally controlled, localized mRNA translation is a ubiquitous phenomenon, where proteins are synthesized at their site of activity, thus reducing the need for macromolecular transport - a prototypical example is that of actin mRNA translation at the leading edge of cells.449 Tatavarty et al. probed the translational output of individual mRNAs in neuronal cell dendrites by taking advantage of the facts that mRNPs are typically transported within relatively slow moving granules along dendrites and that translation is primarily localized to the dendritic spine.345 In essence, the authors had a priori knowledge of the location of translation and could visualize both the mRNA and translated protein at the temporal resolution of their microscope. To this end, they non-specifically labeled the mRNA coding for the Venus FP with Cy5 by incorporating Cy5-UTP during in vitro transcription and microinjected the labeled mRNA into the perikaryon of cultured hippocampal cells. They used an appropriate concentration of labeled RNA such that a single granule had at most a single RNA. Correlating mRNA signal with that of translated Venus FP presented evidence for Poissonian (sporadic) or super-Poissonian (Bursting) translation, where bursting was hypothesized to result from polysome formation (mRNA bearing >1 ribosome).345 The authors demonstrated that polysome formation occurs less frequently than monosome formation and that its extent is controlled by receptor mediated elongation or initiation inhibition and excitation.345 Intriguingly, there was no correlation between mRNA number and fluorescent protein output in E. coli,287 which the authors attributed to factors extrinsic to translation, like mRNA degradation, or to the static nature of the smFISH technique.287
Stress conditions can also alter the extent of translation of a given gene.450 During bacterial stress, a protein called RelA synthesizes alarmones (signaling molecules synthesized solely under harsh conditions), such as ppGPP and pppGppp to suppresses translation, among other cellular processes. English et al. discovered that both RelA and the ribosomal 70S subunit have the same diffusion coefficient (~0.5 μm2/s) at normal growth conditions, possibly due to their association.450 Upon heat shock or starvation, RelA's diffusion coefficient increases 25-fold, suggesting its release and diffusion throughout the cytoplasm, in a reversible fashion. Taken together, the authors were able to demonstrate that RelA will ‘hop off’ the ribosome under stress conditions in order to induce the cellular adaptation response pathway, consequently inhibiting bacterial translation.450
4.8. Post-Transcriptional Control of Gene Expression
The proper development, growth, and regeneration of multicellular organisms require tight spatiotemporal control of gene expression. Any deviation from the norm, in terms of the number and spatiotemporal localizations of specific RNAs, can contribute to phenotypic changes that are either necessary or deleterious to the organism. Defining these RNA parameters provides mechanistic insight into functional and dysfunctional differentiation processes. Single molecule, particularly smFISH, tools have been used extensively to characterize RNA expression profiles in multicellular, asynchronous and heterogeneous cell populations, such as tissues and developing embryos.273,451,452 For example, Saffer et al. used smFISH to demonstrate the impacts of genetic mutations in synMuv genes on cellular lin-3 RNA distribution and the consequent influence on vulva development in C. elegans.453 A three-color smFISH study by Itzkovitz et al. discovered the location of specific stem cells in mouse intestines by probing for cell specific genes.384 Their findings revealed that two distinct populations of stem cells are harbored in intestinal crypts, each exhibiting a unique expression profile for stem-cell specific genes that changes as a function of age and active regeneration.384 Lionnet et al. developed a transgenic mouse that co-expresses an endogenous MCP-FP fusion protein together with a β-actin mRNA carrying MBS repeats that can be used to study mRNA expression in any tissue.226 In addition to coding mRNAs, smFISH techniques have been utilized to probe miRNA expression patterns.454,455 Neely et al. utilized the Direct miRNA assay to characterize the expression patterns of 45 miRNAs in as many as 16 different tissue types.454 Finally, Li et al. utilized miRNA In Situ Hybridization (MISH) coupled with enzyme-labeled fluorescence (ELF) to show reduced quantities of miRNA-375 expression in esophageal squamous cell carcinoma cell lines relative to normal tissue.455 Collectively, these single molecule RNA counting assays have and will continue to provide much needed spatial and temporal information of gene regulation in asynchronous and heterogeneous cellular systems.
The cell has developed a multitude of mechanisms to control gene expression and prevent infection through the sequestration and degradation of both endogenous and exogenous parasitic RNAs. Mechanistic and kinetic analyses of RNA decay processes reveal important parameters to understand and predict their cytoplasmic spatial distributions and lifetimes. In cellulo single molecule counting is ideal when aiming to quantify changes in RNA levels within a single cell or a heterogeneous sample. Trcek et al. counted cytoplasmic mRNA using smFISH and uncovered a cell cycle specific mRNA decay mechanism in yeast.456 They deduced that these mRNA are stable from the S to the G2 phase (halftime, t½ > 66 min), but are more rapidly degraded during mitosis (t½ < 2 min). Additionally, these effects were found to be dependent on the promoter sequence and nuclear protein factors that bind them (Dbf2p and Dbf20p).456 Together, this data present a model of degradation wherein co-transcriptional events dictate an mRNA's lifetime.
Single particle tracking is ideal for measuring kinetic parameters of rapid processes and measuring temporal changes in diffusion patterns. Pitchiaya et al. used a unique blend of SPT, photobleaching analysis and microinjection (in a technique termed intracellular single-molecule, high resolution localization and counting, or iSHiRLoC) to elucidate the spatiotemporal behaviors of microinjected, cyanine dye labeled miRNAs in HeLa cells (Figure 14).135 The authors found that a majority of miRNAs begins to diffuse slowly enough for observation with a 100-ms camera integration time about an hour after microinjection, suggesting that association with target mRNAs occurs on this timescale. Analysis of stepwise photobleaching trajectories in fixed cells indicated little multimer assembly (more than one miRNA per focus) up to this time (Figure 14C and D), supporting the conclusion from live cell imaging.135 Two hours after microinjection and beyond, miRNAs are visible as individual particles with a fluorescence signal enhanced by confinement or binding within comparatively large endogenous mRNPs. These particles exhibit four different types of diffusive motion, Brownian, corralled, directional, and slow Brownian/immobile, and distribute into (at least) two Gaussian distributions of diffusion constants that mimic the diffusion observed for mRNPs (0.26 μm2/s) and the mRNA degrading P-bodies (0.034 μm2/s; Figure 14B). Photobleaching analysis revealed time dependent formation of miRNA multimers (foci containing >1 labeled miRNA), whose temporal evolution fits with a double-exponential function (Figure 14D) and correlates well with the changes in miRNA diffusion coefficients.135 An artificial miRNA with much fewer endogenous targets does not show any temporal change in assembly, unless endogenous mRNA target is co-microinjected (Figure 14D). Taken together, the authors concluded that miRNA bind and sequester their targets into mRNA degrading P-bodies within the first 2 h (k1 = 1.2 ± 0.2 h−1), which are slowly (k2 = 0.14 ± 0.08 h−1) released back to the cytoplasmic pool, likely after target degradation, over the remaining 32 h period. These results correlate with the timing of colocalization between smFISH probed RNA and various immunostained P-body components282 and support a unifying two-stage RNA silencing mechanism where translational repression is kinetically followed by mRNA degradation.135
Figure 14. iSHiRLoC of miRNAs.
(A) Representative pseudocolored images of live HeLa cells injected with Cy3 (green) or Cy5 (red) labeled let-7-a1 miRNAs and representative single particle tracks showcasing the multitude of diffusive patterns these miRNAs exhibit. Scale bars represent 10 μm. (B) Distribution of diffusion coefficients of miRNPs at various time points. (C) Representative pseudocolored and background subtracted image of a fixed HeLa cell microinjected with Cy5 labeled let-7-a1 miRNA. Below, two representative photobleaching trajectories of particles in the image. (D) Fraction of monomers and multimers as a function of time. (E) Unifying model of the RNA silencing pathway consistent with the experimental data. Reprinted with permission from ref. 135. Copyright 2012 Nature Publishing Group.
Stress granules (SGs) are membrane-less cytoplasmic foci that form when cells are exposed to environmental stress, and their formation is accompanied by global translation arrest of housekeeping transcripts.457 These foci are composed of the non-canonical, translationally silent 48S pre-initiation complex (containing the small ribosomal subunit-associated early initiation factors eIF4E, eIF3, eIF4A, eIFG as well as PABP), several mRNA binding proteins that regulate mRNA translation and decay and proteins involved in RNA metabolism.457 P-bodies, by contrast, are cytoplasmic foci predominantly enriched in RNA processing and degradation enzymes, which are present in healthy cells as well. Although SGs and P-bodies are often seen in proximity to each other during stress and both have common protein components, they are distinct cellular granules, with the former functioning to store and aggregate translationally inactive mRNAs and the latter representing RNA decay sites.457 While the two types of foci have been well characterized, their interactions with native mRNAs are still poorly understood. To probe this interaction, Zurla et al. used hybridization based MTRIPs to visualize the interaction of actin and other polyA sequence containing mRNAs with immunostained SGs and P-bodies in fixed cells.347 They observed that under stressed conditions <5% of all transcripts colocalized with SGs, whereas <1% colocalized with P-bodies, and that the extent of P-body localization was similar between stressed and unstressed conditions. Stress induced specifically by translation inhibition changed the localization pattern of the transcripts and moved them closer to the nucleus. By co-imaging the mRNA and tubulin, and by disrupting the cytoskeletal network, the group found that microtubule integrity was necessary for actin mRNA localization to both these foci and the exosome.347
4.9. Retroviral Life Cycle
Retroviruses are unique entities in that they carry their genetic information in the form of RNA, rather than DNA. Upon host infection, their genetic information is reverse-transcribed into DNA and inserted into the host's genome. Through hijacking the cellular transcription and translation machineries, new viruses are created.458 A significant body of work has contributed to our overall understanding of the viral life cycle,458 however, resolving key aspects of the sub-cellular localization of viral assembly intermediates and viral trafficking requires in cellulo single molecule tools. For example, influenza virus A contains eight unique RNA segments that comprise its genetic information. Using smFISH, it was discovered that all eight RNA segments remain associated with one another even after their release from the viral capsid, This association is retained until their entry into the nucleus where they disassemble and are subsequently reverse-transcribed and integrated into the genome.459 Once transcribed, each of the eight influenza A RNAs is exported from the nucleus individually, but then they all assemble into a cytoplasmic RNP in a Rab11 (an endosomal binding protein) dependent process.459 RNP formation prior to capsid assembly provides a checkpoint to ensure accurate packaging of the viral genome. Similarly, HIV-2 RNA was observed to dimerize in the cytoplasm prior to capsid assembly.460 Boireau et al. discovered via FRAP and smFISH techniques that MCP-FP labeled HIV-1 RNA is rapidly (at ~1.9 kb/min) transcribed in U2OS cells.461 Basyuk et al. used SPT of similarly labeled viral progeny RNA molecules to discover that they are actively transported to their host's plasma membrane.462 Actively transported viral RNA was observed to co-track with lysosomes and transferrin-positive endosomes in a Gag and Env protein dependent process. Vesicle transport could represent an efficient means to deliver the budding virus to the plasma membrane for its secretion, as corroborated by later studies.463,464 Jouvenet et al.465 and Ivanchenko et al.466 discovered that HIV-1 RNA viral assembly occurs at the plasma membrane and not within the cytoplasm, squelching the hypothesis that viral particles are co-translationally assembled during capsid protein synthesis. Once transported to the budding site, viral RNA particles are rapidly (at ~5 × 10−3 s−1) assembled for release.464,466
A host that has been infected with two parent viruses gives rise to progeny virions that either contain genetic information from a single provirus (homogeneous) or from two different proviruses (heterogeneous). Chen et al. utilized the MCP-FP system and another protein-RNA tethering system comprising of the E. coli RNA binding protein BglG and its cognate binding sequence to selectively label two different HIV-1 RNA molecules in vivo.467 In this study, the authors determined that viral particles are assembled with high fidelity, wherein >90% of particles contain the appropriate number of HIV-1 genomic RNA molecules. The extent of virion heterogeneity depends on the dimer initiation sequence (DIS, a palindromic dimerization sequence in the 5’ UTR of the viral RNA) on each proviral RNA molecule.467 If both proviral RNAs contain identical DIS sequences, the extent of viron heterogeneity will be random. By contrast, unique DIS sequences will induce bias towards the most thermodynamically stable pairing.467,468 Additionally, HIV-1 amd 2, which share very little sequence homology, can package into similar progeny viruses in a DIS dependent fashion.460,468
4.10. Telomerase
Telomeric DNA is a specialized, repetitive sequence on the ends of chromosomes designed to protect chromosomes from progressive shortening and fusion. Telomerase RNA is a specialized lncRNA that helps maintain the proper length of the chromosomal telomeres.199,200 To better understand the mechanism by which the telomerase RNP is recruited to chromosome ends, Gallardo et al. designed stably integrated telomerase RNA using the in vivo labeled MCP-FP system and performed SPT at various stages of the yeast cell cycle.469 Diffusion and co-tracking indicated that telomerase RNA is associated with the telomere and telomere interacting protein Rap1 only during S phase (when telomeres are supposed to be elongated), yet the telomerase retains enzymatic activity throughout the cell cycle. Thus, active telomerase is recruited to the shortest telomeres only in the late S phase of the cell cycle.469
5. CONCLUSIONS
In this review, we have described numerous RNAs that are found in particularly the eukaryotic cell and have provided a detailed summary of the single molecule tools available to study RNAs intracellularly. A large fraction of single molecules studies published so far have been used to characterize protein-coding mRNAs. However, as we have highlighted in Section 2, the majority of RNAs found in the eukaryotic cell do not code for protein, but rather regulate gene expression via post-transcriptional gene silencing or epigenetic gene regulation. Intracellular single molecule techniques offer an unparalleled means to investigate the behavior of these emerging classes of ncRNAs.
Rapid recent advances in single molecule tool development are likely to further facilitate analysis of the multitude of cellular RNA functions. For example, more pervasive implementation of super-resolution techniques may be necessary to dissect the functions and mechanisms of ncRNAs within densely populated samples.212 In addition, enhanced spatiotemporal resolution using, for example, SPEED microscopy,88 more observables, and an increased field of view will likely facilitate studies of intracellular single molecule RNA dynamics. We anticipate a bright future for discoveries using intracellular single molecule fluorescence microscopy as applied to RNA.
Table 1.
Representative classes of RNAs found in a eukaryotic cell
| Category | Name | RNA Length (nts) | Function | References† | Databases | Single molecule References† |
|---|---|---|---|---|---|---|
| Coding RNA | Messenger RNA (mRNA) | variable | Protein coding | 15,41 | 470,471 | 225,233,268,270,278,292,307,315,345,400,423,430,434 |
| Housekeeping non-coding RNA | Ribosomal RNA (rRNA) | ~120-5,000 | Translation | 472-474 | 475 | 345 |
| Signal Recognition particle RNA | 300 | Translation | 107 | 476 | ||
| Transfer RNA (tRNA) | 70-100 | Translation | 100 | 477,478 | ||
| Small nuclear RNA (snRNA) | 60-450 | Splicing | 43 | 343 | ||
| Small nucleolar (snoRNA) | 60-300 | RNA editing | 92 | 479 | ||
| RNase P RNA | ~500 | tRNA maturation | 105 | 480 | ||
| Telomerase RNA | ~400-1,300 | chromosome extension | 199,200 | 481 | ||
| Small non-coding RNA (< 200-nts) | Hepatitis Delta Virus (HDV) ribozyme | ~84 | Unknown | 69,71,482 | ||
| Hammerhead ribozyme | ~100 | Unknown | 67,68 | |||
| MicroRNA (miRNA) | 20-24 | Post-transcriptional gene silencing | 139,143,148,149 | 483-485 | 135,454,455 | |
| Small interfering RNA (siRNA) | 20-24 | Post-transcriptional gene silencing | 143 | 481,485 | ||
| DNA-damage response RNA (DDRNA) | 20-35 | DNA repair | 123,124,177 | |||
| PIWI-interacting RNA (piRNA) | 24-31 | Germline gene silencing | 121,486 | 487 | ||
| Circular RNA (circRNA) | variable | miRNA sponge | 159,160 | |||
| Primal RNA (priRNA) | 22-23 | heterochromatin assembly | 174 | |||
| Mid-sized ncRNA <1 kb | Promoter-associated RNAs (PARs) | variable | Epigenetic gene regulation | 187 | 481 | |
| Long non-coding NA (> 200-nts) | Long non-coding RNA (lncRNA) | >200 | Epigenetic gene regulation | 179 | 484,488,489 | 188,416-418 |
| Cis-acting ncRNA | 5′-UTR | ~150 | mRNA regulatory element | 78 | 490 | |
| 3′-UTR | ~300 | mRNA regulatory element | 78,118 | 490 | ||
| Introns | Variable (up to thousand) | mRNA regulatory element | 491 | 492 |
Representative references. Please refer to the text for a comprehensive list.
ACKNOWLEDGMENTS
This work was supported by NIH grants GM081025 and GM098023 to N.G.W.
Biography
 Sethuramasundaram Pitchiaya received his bachelor's degree in Industrial
Biotechnology from the Center for Biotechnology at Anna University in Chennai,
India, in 2006. In 2011, he received a Ph.D. degree in Chemistry from the University
of Michigan in Ann Arbor, Michigan, while developing iSHiRLoC under the guidance of
Dr. Nils Walter. Until recently, he served as a postdoctoral fellow in the Single
Molecule Analysis in Real-Time (SMART) center, is now back full-time in the Walter
group, and is broadly interested in understanding the intracellular organization,
trafficking and mechanism of action of eukaryotic non-coding RNAs (ncRNAs) and
ribonucleoprotein granules. Apart from assembling, maintaining and training users on
a high-resolution objective-TIRF type single molecule microscope and an atomic force
microscope at the SMART center, he has been working towards unraveling the
intracellular localization, assembly and function of microRNPs and DNA damage
response associated ncRNAs at single molecule resolution.
Sethuramasundaram Pitchiaya received his bachelor's degree in Industrial
Biotechnology from the Center for Biotechnology at Anna University in Chennai,
India, in 2006. In 2011, he received a Ph.D. degree in Chemistry from the University
of Michigan in Ann Arbor, Michigan, while developing iSHiRLoC under the guidance of
Dr. Nils Walter. Until recently, he served as a postdoctoral fellow in the Single
Molecule Analysis in Real-Time (SMART) center, is now back full-time in the Walter
group, and is broadly interested in understanding the intracellular organization,
trafficking and mechanism of action of eukaryotic non-coding RNAs (ncRNAs) and
ribonucleoprotein granules. Apart from assembling, maintaining and training users on
a high-resolution objective-TIRF type single molecule microscope and an atomic force
microscope at the SMART center, he has been working towards unraveling the
intracellular localization, assembly and function of microRNPs and DNA damage
response associated ncRNAs at single molecule resolution.
 Laurie Heinicke was born in 1981 in St. Paul, Minnesota. She received her B.S.
degree in Biochemistry from the University of Minnesota-Twin Cities. In 2010, she
received her Ph.D. in Chemistry at The Pennsylvania State University under the
direction of Dr. Philip Bevilacqua. She spent three years as a postdoctoral fellow
at The University of Pennsylvania (advisors Drs. Alan Gewirtz and Russ Carstens) and
was supported by an NIH Hematopoiesis training grant. She is currently a
postdoctoral fellow in Dr. Nils Walter's laboratory at the University of Michigan in
Ann Arbor, Michigan. Her previous experience working with in vitro
RNA folding intermediates, RNA-protein interactions and cell culture has facilitated
a smooth transition into using single molecule fluorescence microscopy to examine
the behavior of miRNAs and other ncRNAs in living cells.
Laurie Heinicke was born in 1981 in St. Paul, Minnesota. She received her B.S.
degree in Biochemistry from the University of Minnesota-Twin Cities. In 2010, she
received her Ph.D. in Chemistry at The Pennsylvania State University under the
direction of Dr. Philip Bevilacqua. She spent three years as a postdoctoral fellow
at The University of Pennsylvania (advisors Drs. Alan Gewirtz and Russ Carstens) and
was supported by an NIH Hematopoiesis training grant. She is currently a
postdoctoral fellow in Dr. Nils Walter's laboratory at the University of Michigan in
Ann Arbor, Michigan. Her previous experience working with in vitro
RNA folding intermediates, RNA-protein interactions and cell culture has facilitated
a smooth transition into using single molecule fluorescence microscopy to examine
the behavior of miRNAs and other ncRNAs in living cells.
 Thomas Corey Custer received his Bachelor's degree in Chemistry from Monmouth
College in Monmouth, Illinois, in 2002. He completed his Master's degree in
Chemistry from the University of Colorado - Denver, Colorado, in 2006. While a
Master's candidate, he spent 3 years working as a Drug Metabolism scientist for
Array BioPharma in Boulder, Colorado. He continued his efforts as a Drug Metabolism
scientist with Abbott Laboratories in Abbott Park, Illinois, for five years, four of
which he also spent as an adjunct faculty member for the Chemistry Department at the
College of Lake County in Grayslake, Illinois. He is currently a Ph.D. candidate at
the University of Michigan in Ann Arbor, Michigan, in the Program in Chemical
Biology. As a member of the Walter laboratory, he uses live-cell RNA detection
approaches to study the kinetics and mechanisms of microRNA interactions with RNA
silencing components using single molecule techniques such as iSHiRLoC.
Thomas Corey Custer received his Bachelor's degree in Chemistry from Monmouth
College in Monmouth, Illinois, in 2002. He completed his Master's degree in
Chemistry from the University of Colorado - Denver, Colorado, in 2006. While a
Master's candidate, he spent 3 years working as a Drug Metabolism scientist for
Array BioPharma in Boulder, Colorado. He continued his efforts as a Drug Metabolism
scientist with Abbott Laboratories in Abbott Park, Illinois, for five years, four of
which he also spent as an adjunct faculty member for the Chemistry Department at the
College of Lake County in Grayslake, Illinois. He is currently a Ph.D. candidate at
the University of Michigan in Ann Arbor, Michigan, in the Program in Chemical
Biology. As a member of the Walter laboratory, he uses live-cell RNA detection
approaches to study the kinetics and mechanisms of microRNA interactions with RNA
silencing components using single molecule techniques such as iSHiRLoC.
 Nils G. Walter was born in 1966 in Frankfurt am Main, Germany. He received his
“Vordiplom” (B.S.) and “Diploma” (Masters) from the
Technical University of Darmstadt after performing research with Hans-Günther
Gassen on the physiochemical characterization of a protein dehyrogenase enzyme. He
earned his Dr. rer. nat. while studying molecular in vitro
evolution of DNA and RNA using fluorescence techniques with Nobel laureate Manfred
Eigen at the Max-Planck-Institute for Biophysical Chemistry, Göttingen. For
his postdoctoral studies, he turned to RNA enzymes under the guidance of John M.
Burke at the University of Vermont in Burlington, Vermont. He is currently a
Professor of Chemistry at the University of Michigan in Ann Arbor, Michigan. His
research interests focus on non-coding RNA through the lens of single molecule
techniques. He currently directs the unique Single Molecule Analysis in Real-Time
(SMART) Center at Michigan.
Nils G. Walter was born in 1966 in Frankfurt am Main, Germany. He received his
“Vordiplom” (B.S.) and “Diploma” (Masters) from the
Technical University of Darmstadt after performing research with Hans-Günther
Gassen on the physiochemical characterization of a protein dehyrogenase enzyme. He
earned his Dr. rer. nat. while studying molecular in vitro
evolution of DNA and RNA using fluorescence techniques with Nobel laureate Manfred
Eigen at the Max-Planck-Institute for Biophysical Chemistry, Göttingen. For
his postdoctoral studies, he turned to RNA enzymes under the guidance of John M.
Burke at the University of Vermont in Burlington, Vermont. He is currently a
Professor of Chemistry at the University of Michigan in Ann Arbor, Michigan. His
research interests focus on non-coding RNA through the lens of single molecule
techniques. He currently directs the unique Single Molecule Analysis in Real-Time
(SMART) Center at Michigan.
Footnotes
The authors declare no competing financial interest.
REFERENCES
- 1.Consortium EP, Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P, Boyle PJ, Cao H, Carter NP, Clelland GK, Davis S, Day N, Dhami P, Dillon SC, Dorschner MO, Fiegler H, Giresi PG, Goldy J, Hawrylycz M, Haydock A, Humbert R, James KD, Johnson BE, Johnson EM, Frum TT, Rosenzweig ER, Karnani N, Lee K, Lefebvre GC, Navas PA, Neri F, Parker SC, Sabo PJ, Sandstrom R, Shafer A, Vetrie D, Weaver M, Wilcox S, Yu M, Collins FS, Dekker J, Lieb JD, Tullius TD, Crawford GE, Sunyaev S, Noble WS, Dunham I, Denoeud F, Reymond A, Kapranov P, Rozowsky J, Zheng D, Castelo R, Frankish A, Harrow J, Ghosh S, Sandelin A, Hofacker IL, Baertsch R, Keefe D, Dike S, Cheng J, Hirsch HA, Sekinger EA, Lagarde J, Abril JF, Shahab A, Flamm C, Fried C, Hackermuller J, Hertel J, Lindemeyer M, Missal K, Tanzer A, Washietl S, Korbel J, Emanuelsson O, Pedersen JS, Holroyd N, Taylor R, Swarbreck D, Matthews N, Dickson MC, Thomas DJ, Weirauch MT. Nature. 2007;447:799. [Google Scholar]
- 2.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della Gatta G, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii-Hemming P, Gingeras TR, Gojobori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SP, Kruger A, Kummerfeld SK, Kurochkin IV, Lareau LF, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Madan Babu M, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S. Science. 2005;309:1559. [Google Scholar]
- 3.Carninci P, Hayashizaki Y. Curr. Opin. Genet. Dev. 2007;17:139. doi: 10.1016/j.gde.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, Boychenko V, Hunt T, Kay M, Mukherjee G, Rajan J, Despacio-Reyes G, Saunders G, Steward C, Harte R, Lin M, Howald C, Tanzer A, Derrien T, Chrast J, Walters N, Balasubramanian S, Pei B, Tress M, Rodriguez JM, Ezkurdia I, van Baren J, Brent M, Haussler D, Kellis M, Valencia A, Reymond A, Gerstein M, Guigo R, Hubbard TJ. Genome Res. 2012;22:1760. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eddy SR. Nat. Rev. Genet. 2001;2:919. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- 6.Mattick JS. Nat. Rev. Genet. 2004;5:316. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- 7.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Roder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigo R, Gingeras TR. Nature. 2012;489:101. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Nature. 2009;458:223. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belinky F, Bahir I, Stelzer G, Zimmerman S, Rosen N, Nativ N, Dalah I, Iny Stein T, Rappaport N, Mituyama T, Safran M, Lancet D. Bioinformatics. 2013;29:255. doi: 10.1093/bioinformatics/bts676. [DOI] [PubMed] [Google Scholar]
- 10.Tuck AC, Tollervey D. Trends Genet. 2011;27:422. doi: 10.1016/j.tig.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 11.Martens-Uzunova ES, Olvedy M, Jenster G. Cancer Lett. 2013;340:201. doi: 10.1016/j.canlet.2012.11.058. [DOI] [PubMed] [Google Scholar]
- 12.Henriques R, Griffiths C, Hesper Rego E, Mhlanga MM. Biopolymers. 2011;95:322. doi: 10.1002/bip.21586. [DOI] [PubMed] [Google Scholar]
- 13.Dreyfuss G, Kim VN, Kataoka N. Nat. Rev. Mol. Cell. Biol. 2002;3:195. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 14.Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, Krijgsveld J, Hentze MW. Cell. 2012;149:1393. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 15.Huang Y, Steitz JA. Mol. Cell. 2005;17:613. doi: 10.1016/j.molcel.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 16.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr., Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. Cell. 2010;141:129. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. Nat. Struct. Mol. Biol. 2009;16:130. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang C, Darnell RB. Nat. Biotechnol. 2011;29:607. doi: 10.1038/nbt.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugimoto Y, Konig J, Hussain S, Zupan B, Curk T, Frye M, Ule J. Genome Biol. 2012;13:67. doi: 10.1186/gb-2012-13-8-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muller-McNicoll M, Neugebauer KM. Nat. Rev. Genet. 2013;14:275. doi: 10.1038/nrg3434. [DOI] [PubMed] [Google Scholar]
- 21.Raj A, van Oudenaarden A. Annu. Rev. Biophys. 2009;38:255. doi: 10.1146/annurev.biophys.37.032807.125928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mattick JS, Makunin IV. Hum. Mol. Genet. 2006:15, R17. doi: 10.1093/hmg/ddl046. Spec No 1. [DOI] [PubMed] [Google Scholar]
- 23.Wahl MC, Will CL, Luhrmann R. Cell. 2009;136:701. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 24.Abelson J, Blanco M, Ditzler MA, Fuller F, Aravamudhan P, Wood M, Villa T, Ryan DE, Pleiss JA, Maeder C, Guthrie C, Walter NG. Nat. Struct. Mol. Biol. 2010;17:504. doi: 10.1038/nsmb.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nilsen TW, Graveley BR. Nature. 2010;463:457. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaudhury A, Chander P, Howe PH. RNA. 2010;16:1449. doi: 10.1261/rna.2254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dreyfuss G, Matunis MJ, Pinol-Roma S, Burd CG. Annu. Rev. Biochem. 1993;62:289. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 28.Long JC, Caceres JF. Biochem. J. 2009;417:15. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 29.Lou H, Neugebauer KM, Gagel RF, Berget SM. Mol. Cell Biol. 1998;18:4977. doi: 10.1128/mcb.18.9.4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Danckwardt S, Kaufmann I, Gentzel M, Foerstner KU, Gantzert AS, Gehring NH, Neu-Yilik G, Bork P, Keller W, Wilm M, Hentze MW, Kulozik AE. EMBO J. 2007;26:2658. doi: 10.1038/sj.emboj.7601699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McManus CJ, Graveley BR. Curr. Opin. Genet. Dev. 2011;21:373. doi: 10.1016/j.gde.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mauger DM, Siegfried NA, Weeks KM. FEBS Lett. 2013;587:1180. doi: 10.1016/j.febslet.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Serganov A, Nudler E. Cell. 2013;152:17. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Breaker RR. Mol. Cell. 2011;43:867. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suddala KC, Rinaldi AJ, Feng J, Mustoe AM, Eichhorn CD, Liberman JA, Wedekind JE, Al-Hashimi HM, Brooks III CL, Walter NG. Nucleic Acids Res. 2013;41:10462. doi: 10.1093/nar/gkt798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Nat. Genet. 2008;40:1413. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 37.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Nature. 2008;456:470. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. Cell. 2000;101:671. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- 39.Gonzalez-Porta M, Frankish A, Rung J, Harrow J, Brazma A. Genome Biol. 2013;14:R70. doi: 10.1186/gb-2013-14-7-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tazi J, Bakkour N, Stamm S. Biochim. Biophys. Acta. 2009;1792:14. doi: 10.1016/j.bbadis.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cooper TA, Wan L, Dreyfuss G. Cell. 2009;136:777. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lopez-Bigas N, Audit B, Ouzounis C, Parra G, Guigo R. FEBS Lett. 2005;579:1900. doi: 10.1016/j.febslet.2005.02.047. [DOI] [PubMed] [Google Scholar]
- 43.Fischer U, Englbrecht C, Chari A. Wiley Interdiscip. Rev. RNA. 2011;2:718. doi: 10.1002/wrna.87. [DOI] [PubMed] [Google Scholar]
- 44.Roca X, Krainer AR, Eperon IC. Genes Dev. 2013;27:129. doi: 10.1101/gad.209759.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matera AG, Terns RM, Terns MP. Nat. Rev. Mol. Cell Biol. 2007;8:209. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 46.Girard C, Will CL, Peng J, Makarov EM, Kastner B, Lemm I, Urlaub H, Hartmuth K, Luhrmann R. Nat. Commun. 2012;3:994. doi: 10.1038/ncomms1998. [DOI] [PubMed] [Google Scholar]
- 47.Spector DL, Lamond AI. Cold Spring Harbor Perspect. Biol. 2011;3:1. doi: 10.1101/cshperspect.a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, Weng Z, Moore MJ. Cell. 2012;151:750. doi: 10.1016/j.cell.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moore MJ, Proudfoot NJ. Cell. 2009;136:688. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 50.Cech TR. Annu. Rev. Biochem. 1990;59:543. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- 51.Saldanha R, Mohr G, Belfort M, Lambowitz AM. FASEB J. 1993;7:15. doi: 10.1096/fasebj.7.1.8422962. [DOI] [PubMed] [Google Scholar]
- 52.Bonen L, Vogel J. Trends Genet. 2001;17:322. doi: 10.1016/s0168-9525(01)02324-1. [DOI] [PubMed] [Google Scholar]
- 53.Rueda D, Bokinsky G, Rhodes MM, Rust MJ, Zhuang X, Walter NG. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10066. doi: 10.1073/pnas.0403575101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McDowell SE, Jun JM, Walter NG. RNA. 2010;16:2414. doi: 10.1261/rna.1829110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Doherty EA, Doudna JA. Annu. Biophys. Biomol. Struct. 2001;30:457. doi: 10.1146/annurev.biophys.30.1.457. [DOI] [PubMed] [Google Scholar]
- 56.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Nature. 2004;428:281. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 57.Lilley DM. Curr. Opin. Struct. Biol. 2005;15:313. doi: 10.1016/j.sbi.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 58.Pereira MJ, Nikolova EN, Hiley SL, Jaikaran D, Collins RA, Walter NG. J. Mol. Biol. 2008;382:496. doi: 10.1016/j.jmb.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rhodes MM, Reblova K, Sponer J, Walter NG. Proc. Natl. Acad. Sci. U.S.A. 2006;103:13380. doi: 10.1073/pnas.0605090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ditzler MA, Aleman EA, Rueda D, Walter NG. Biopolymers. 2007;87:302. doi: 10.1002/bip.20819. [DOI] [PubMed] [Google Scholar]
- 61.McDowell SE, Spackova N, Sponer J, Walter NG. Biopolymers. 2007;85:169. doi: 10.1002/bip.20620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Walter NG, Perumal S. Springer Ser. Biophys. 2009;13:103. doi: 10.1007/978-3-540-70840-7_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhuang X, Kim H, Pereira MJ, Babcock HP, Walter NG, Chu S. Science. 2002;296:1473. doi: 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]
- 64.Bokinsky G, Rueda D, Misra VK, Rhodes MM, Gordus A, Babcock HP, Walter NG, Zhuang X. Proc. Natl. Acad. Sci. U.S.A. 2003;100:9302. doi: 10.1073/pnas.1133280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ditzler MA, Rueda D, Mo J, Hakansson K, Walter NG. Nucleic Acids Res. 2008;36:7088. doi: 10.1093/nar/gkn871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.de Silva C, Walter NG. RNA. 2009;15:76. doi: 10.1261/rna.1346609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seehafer C, Kalweit A, Steger G, Graf S, Hammann C. RNA. 2011;17:21. doi: 10.1261/rna.2429911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.de la Pena M, Garcia-Robles I. EMBO Rep. 2010;11:711. doi: 10.1038/embor.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chadalavada DM, Gratton EA, Bevilacqua PC. Biochemistry. 2010;49:5321. doi: 10.1021/bi100434c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Webb CH, Luptak A. RNA Biol. 2011;8:719. doi: 10.4161/rna.8.5.16226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Salehi-Ashtiani K, Luptak A, Litovchick A, Szostak JW. Science. 2006;313:1788. doi: 10.1126/science.1129308. [DOI] [PubMed] [Google Scholar]
- 72.Crick FH. J. Mol. Biol. 1968;38:367. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- 73.Orgel LE. J. Mol. Biol. 1968;38:381. doi: 10.1016/0022-2836(68)90393-8. [DOI] [PubMed] [Google Scholar]
- 74.Woese CR. The Genetic Code: The Molecular Basis for Genetic Expression. Harper & Row; New York: 1967. [Google Scholar]
- 75.Gesteland RF, Cech TR, Atkins JF. The RNA World, Third Edition. Vol. 43. CSHL Press; Cold Spring Harbor: 2006. [Google Scholar]
- 76.Cowling VH. Biochem. J. 2010;425:295. doi: 10.1042/BJ20091352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Proudfoot NJ. Genes Dev. 2011;25:1770. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mignone F, Gissi C, Liuni S, Pesole G. Genome Biol. 2002;3:1. doi: 10.1186/gb-2002-3-3-reviews0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kohler A, Hurt E. Nat. Rev. Mol. Cell Biol. 2007;8:761. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- 80.Hoelz A, Debler EW, Blobel G. Annu. Rev. Biochem. 2011;80:613. doi: 10.1146/annurev-biochem-060109-151030. [DOI] [PubMed] [Google Scholar]
- 81.Grunwald D, Singer RH, Rout M. Nature. 2011;475:333. doi: 10.1038/nature10318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wente SR, Rout MP. Cold Spring Harbor Perspect. Biol. 2010;2:a000562. doi: 10.1101/cshperspect.a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Katahira J. Biochim. Biophys. Acta. 2012;1819:507. doi: 10.1016/j.bbagrm.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 84.Mehlin H, Daneholt B, Skoglund U. Cell. 1992;69:605. doi: 10.1016/0092-8674(92)90224-z. [DOI] [PubMed] [Google Scholar]
- 85.Davis LI, Blobel G. Cell. 1986;45:699. doi: 10.1016/0092-8674(86)90784-1. [DOI] [PubMed] [Google Scholar]
- 86.Kim J, Izadyar A, Nioradze N, Amemiya S. J. Am. Chem. Soc. 2013;135:2321. doi: 10.1021/ja311080j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Speese SD, Ashley J, Jokhi V, Nunnari J, Barria R, Li Y, Ataman B, Koon A, Chang YT, Li Q, Moore MJ, Budnik V. Cell. 2012;149:832. doi: 10.1016/j.cell.2012.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ma J, Liu Z, Michelotti N, Pitchiaya S, Veerapaneni R, Androsavich JR, Walter NG, Yang W. Nat. Commun. 2013;4:2414. doi: 10.1038/ncomms3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Martin KC, Ephrussi A. Cell. 2009;136:719. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kahvejian A, Svitkin YV, Sukarieh R, M'Boutchou MN, Sonenberg N. Genes Dev. 2005;19:104. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Diem MD, Chan CC, Younis I, Dreyfuss G. Nat. Struct. Mol. Biol. 2007;14:1173. doi: 10.1038/nsmb1321. [DOI] [PubMed] [Google Scholar]
- 92.Bratkovic T, Rogelj B. Cell. Mol. Life Sci. 2011;68:3843. doi: 10.1007/s00018-011-0762-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kressler D, Hurt E, Bassler J. Biochim. Biophys. Acta. 2010;1803:673. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 94.Kiss-Laszlo Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T. Cell. 1996;85:1077. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 95.Tollervey D, Kiss T. Curr. Opin. Cell Biol. 1997;9:337. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- 96.Weinstein LB, Steitz JA. Curr. Opin. Cell Biol. 1999;11:378. doi: 10.1016/S0955-0674(99)80053-2. [DOI] [PubMed] [Google Scholar]
- 97.Watkins NJ, Bohnsack MT. Wiley Interdiscip. Rev. RNA. 2012;3:397. doi: 10.1002/wrna.117. [DOI] [PubMed] [Google Scholar]
- 98.Williams GT, Farzaneh F. Nat. Rev. Cancer. 2012;12:84. doi: 10.1038/nrc3195. [DOI] [PubMed] [Google Scholar]
- 99.Ono M, Scott MS, Yamada K, Avolio F, Barton GJ, Lamond AI. Nucleic Acids Res. 2011;39:3879. doi: 10.1093/nar/gkq1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.El Yacoubi B, Bailly M, de Crecy-Lagard V. Annu. Rev. Genet. 2012;46:69. doi: 10.1146/annurev-genet-110711-155641. [DOI] [PubMed] [Google Scholar]
- 101.Kim SH, Suddath FL, Quigley GJ, McPherson A, Sussman JL, Wang AH, Seeman NC, Rich A. Science. 1974;185:435. doi: 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]
- 102.Goodenbour JM, Pan T. Nucleic Acids Res. 2006;34:6137. doi: 10.1093/nar/gkl725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shah P, Ding Y, Niemczyk M, Kudla G, Plotkin JB. Cell. 2013;153:1589. doi: 10.1016/j.cell.2013.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dittmar KA, Goodenbour JM, Pan T. PLoS Genet. 2006;2:e221. doi: 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Walker SC, Engelke DR. Crit. Rev. Biochem. Mol. Biol. 2006;41:77. doi: 10.1080/10409230600602634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. Cell. 1983;35:849. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 107.Akopian D, Shen K, Zhang X, Shan SO. Annu. Rev. Biochem. 2013;82:693. doi: 10.1146/annurev-biochem-072711-164732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pool MR. Mol. Membr. Biol. 2005;22:3. doi: 10.1080/09687860400026348. [DOI] [PubMed] [Google Scholar]
- 109.Hebert DN, Molinari M. Physiol. Rev. 2007;87:1377. doi: 10.1152/physrev.00050.2006. [DOI] [PubMed] [Google Scholar]
- 110.Durand S, Cougot N, Mahuteau-Betzer F, Nguyen CH, Grierson DS, Bertrand E, Tazi J, Lejeune F. J. Cell Biol. 2007;178:1145. doi: 10.1083/jcb.200611086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Parker R, Sheth U. Mol. Cell. 2007;25:635. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 112.Baker KE, Parker R. Curr. Opin. Cell Biol. 2004;16:293. doi: 10.1016/j.ceb.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 113.Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE. Nat. Genet. 2004;36:801. doi: 10.1038/ng1403. [DOI] [PubMed] [Google Scholar]
- 114.Rebbapragada I, Lykke-Andersen J. Curr. Opin. Cell Biol. 2009;21:394. doi: 10.1016/j.ceb.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 115.Parker R, Song H. Nat. Struct. Mol. Biol. 2004;11:121. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 116.Dictenberg JB, Swanger SA, Antar LN, Singer RH, Bassell GJ. Dev. Cell. 2008;14:926. doi: 10.1016/j.devcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ascano M, Jr., Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, Munschauer M, Dewell S, Hafner M, Williams Z, Ohler U, Tuschl T. Nature. 2012;492:382. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Vasudevan S, Steitz JA. Cell. 2007;128:1105. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ghildiyal M, Zamore PD. Nat. Rev. Genet. 2009;10:94. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Carthew RW, Sontheimer EJ. Cell. 2009;136:642. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Siomi MC, Sato K, Pezic D, Aravin AA. Nat. Rev. Mol. Cell Biol. 2011;12:246. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- 122.Moazed D. Cell. 2011;146:510. doi: 10.1016/j.cell.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Francia S, Michelini F, Saxena A, Tang D, de Hoon M, Anelli V, Mione M, Carninci P, d'Adda di Fagagna F. Nature. 2012;488:231. doi: 10.1038/nature11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chowdhury D, Choi YE, Brault ME. Nat. Rev. Mol. Cell Biol. 2013;14:181. doi: 10.1038/nrm3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lee RC, Feinbaum RL, Ambros V. Cell. 1993;75:843. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 126.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Nature. 1998;391:806. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 127.Storz G, Vogel J, Wassarman KM. Mol. Cell. 2011;43:880. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Castel SE, Martienssen RA. Nat. Rev. Genet. 2013;14:100. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zisoulis DG, Kai ZS, Chang RK, Pasquinelli AE. Nature. 2012;486:541. doi: 10.1038/nature11134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Friedman RC, Farh KK, Burge CB, Bartel DP. Genome Res. 2009;19:92. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jackson AL, Linsley PS. Nat. Rev. Drug Discov. 2010;9:57. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 132.Aalto AP, Pasquinelli AE. Curr. Opin. Cell Biol. 2012;24:333. doi: 10.1016/j.ceb.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ohrt T, Merkle D, Birkenfeld K, Echeverri CJ, Schwille P. Nucleic Acids Res. 2006;34:1369. doi: 10.1093/nar/gkl001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ohrt T, Staroske W, Mutze J, Crell K, Landthaler M, Schwille P. Biophys. J. 2011;100:2981. doi: 10.1016/j.bpj.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pitchiaya S, Androsavich JR, Walter NG. EMBO Rep. 2012;13:709. doi: 10.1038/embor.2012.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Koh HR, Kidwell MA, Ragunathan K, Doudna JA, Myong S. Proc. Natl. Acad. Sci. U.S.A. 2013;110:151. doi: 10.1073/pnas.1212917110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bartel DP. Cell. 2004;116:281. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 138.Bartel DP. Cell. 2009;136:215. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Krol J, Loedige I, Filipowicz W. Nat. Rev. Genet. 2010;11:597. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 140.Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. Trends Genet. 2007;23:614. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 141.Saini HK, Griffiths-Jones S, Enright AJ. Proc. Natl. Acad. Sci. U.S.A. 2007;104:17719. doi: 10.1073/pnas.0703890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Cell. 2006;125:887. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 143.Czech B, Hannon GJ. Nat. Rev. Genet. 2011;12:19. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Noland CL, Doudna JA. RNA. 2013;19:639. doi: 10.1261/rna.037424.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Finnegan EF, Pasquinelli AE. Crit. Rev. Biochem. Mol. Biol. 2013;48:51. doi: 10.3109/10409238.2012.738643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. Cell. 2007;130:89. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Flynt AS, Greimann JC, Chung WJ, Lima CD, Lai EC. Mol. Cell. 2010;38:900. doi: 10.1016/j.molcel.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Huntzinger E, Izaurralde E. Nat. Rev. Genet. 2011;12:99. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 149.Fabian MR, Sonenberg N. Nat. Struct. Mol. Biol. 2012;19:586. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 150.Fabian MR, Sonenberg N, Filipowicz W. Annu. Rev. Biochem. 2010;79:351. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 151.Meijer HA, Kong YW, Lu WT, Wilczynska A, Spriggs RV, Robinson SW, Godfrey JD, Willis AE, Bushell M. Science. 2013;340:82. doi: 10.1126/science.1231197. [DOI] [PubMed] [Google Scholar]
- 152.Sioud M, Cekaite L. Methods Mol. Biol. 2010;629:257. doi: 10.1007/978-1-60761-657-3_16. [DOI] [PubMed] [Google Scholar]
- 153.Xia W, Cao G, Shao N. Science in China. Series C, Life sciences / Chinese Academy of Sciences. 2009;52:1123. doi: 10.1007/s11427-009-0159-4. [DOI] [PubMed] [Google Scholar]
- 154.Long D, Lee R, Williams P, Chan CY, Ambros V, Ding Y. Nat. Struct. Mol. Biol. 2007;14:287. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- 155.Helwak A, Kudla G, Dudnakova T, Tollervey D. Cell. 2013;153:654. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zekri L, Kuzuoglu-Ozturk D, Izaurralde E. EMBO J. 2013;32:1052. doi: 10.1038/emboj.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Li S, Liu L, Zhuang X, Yu Y, Liu X, Cui X, Ji L, Pan Z, Cao X, Mo B, Zhang F, Raikhel N, Jiang L, Chen X. Cell. 2013;153:562. doi: 10.1016/j.cell.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Stalder L, Heusermann W, Sokol L, Trojer D, Wirz J, Hean J, Fritzsche A, Aeschimann F, Pfanzagl V, Basselet P, Weiler J, Hintersteiner M, Morrissey DV, Meisner-Kober NC. EMBO J. 2013;32:1115. doi: 10.1038/emboj.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Nature. 2013;495:333. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 160.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Nature. 2013;495:384. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 161.Ebert MS, Sharp PA. Curr. Biol. 2010;20:R858. doi: 10.1016/j.cub.2010.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hwang HW, Wentzel EA, Mendell JT. Science. 2007;315:97. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 163.Weinmann L, Hock J, Ivacevic T, Ohrt T, Mutze J, Schwille P, Kremmer E, Benes V, Urlaub H, Meister G. Cell. 2009;136:496. doi: 10.1016/j.cell.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 164.Nishi K, Nishi A, Nagasawa T, Ui-Tei K. RNA. 2013;19:17. doi: 10.1261/rna.034769.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Truesdell SS, Mortensen RD, Seo M, Schroeder JC, Lee JH, LeTonqueze O, Vasudevan S. Sci. Rep. 2012;2:842. doi: 10.1038/srep00842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Grewal SI. Curr. Opin. Genet. Dev. 2010;20:134. doi: 10.1016/j.gde.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Law JA, Jacobsen SE. Nat. Rev. Genet. 2010;11:204. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Burton NO, Burkhart KB, Kennedy S. Proc. Natl. Acad. Sci. U.S.A. 2011;108:19683. doi: 10.1073/pnas.1113310108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wu L, Zhou H, Zhang Q, Zhang J, Ni F, Liu C, Qi Y. Mol. Cell. 2010;38:465. doi: 10.1016/j.molcel.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 170.Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, Conte D, Jr., Mello CC. Cell. 2009;139:123. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Cell. 2006;127:1193. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 172.Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, Conte D, Jr., Luo S, Schroth GP, Carrington JC, Bartel DP, Mello CC. Mol. Cell. 2008;31:67. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Das PP, Bagijn MP, Goldstein LD, Woolford JR, Lehrbach NJ, Sapetschnig A, Buhecha HR, Gilchrist MJ, Howe KL, Stark R, Matthews N, Berezikov E, Ketting RF, Tavare S, Miska EA. Mol. Cell. 2008;31:79. doi: 10.1016/j.molcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Halic M, Moazed D. Cell. 2010;140:504. doi: 10.1016/j.cell.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Guang S, Bochner AF, Burkhart KB, Burton N, Pavelec DM, Kennedy S. Nature. 2010;465:1097. doi: 10.1038/nature09095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ameyar-Zazoua M, Rachez C, Souidi M, Robin P, Fritsch L, Young R, Morozova N, Fenouil R, Descostes N, Andrau JC, Mathieu J, Hamiche A, Ait-Si-Ali S, Muchardt C, Batsche E, Harel-Bellan A. Nat. Struct. Mol. Biol. 2012;19:998. doi: 10.1038/nsmb.2373. [DOI] [PubMed] [Google Scholar]
- 177.Wei W, Ba Z, Gao M, Wu Y, Ma Y, Amiard S, White CI, Rendtlew Danielsen JM, Yang YG, Qi Y. Cell. 2012;149:101. doi: 10.1016/j.cell.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 178.Qureshi IA, Mehler MF. Nat. Rev. Neurosci. 2012;13:528. doi: 10.1038/nrn3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Mercer TR, Mattick JS. Nat. Struct. Mol. Biol. 2013;20:300. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 180.Kaikkonen MU, Lam MTY, Glass CK. Cardiovasc. Res. 2011;90:430. doi: 10.1093/cvr/cvr097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Guttman M, Rinn JL. Nature. 2012;482:339. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bartolomei MS, Zemel S, Tilghman SM. Nature. 1991;351:153. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- 183.Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. Nature. 1991;349:38. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 184.Barlow DP, Stoger R, Herrmann BG, Saito K, Schweifer N. Nature. 1991;349:84. doi: 10.1038/349084a0. [DOI] [PubMed] [Google Scholar]
- 185.Wapinski O, Chang HY. Trends Cell Biol. 2011;21:354. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 186.Kim ED, Sung S. Trends Plant Sci. 2012;17:16. doi: 10.1016/j.tplants.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 187.Taft RJ, Kaplan CD, Simons C, Mattick JS. Cell Cycle. 2009;8:2332. doi: 10.4161/cc.8.15.9154. [DOI] [PubMed] [Google Scholar]
- 188.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Proc. Natl. Acad. Sci. U.S.A. 2009;106:11667. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, Oh S, Kim HS, Glass CK, Rosenfeld MG. Nature. 2013;498:516. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Dinger ME, Gascoigne DK, Mattick JS. Biochimie. 2011;93:2013. doi: 10.1016/j.biochi.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 191.Nallagatla SR, Jones CN, Ghosh SK, Sharma SD, Cameron CE, Spremulli LL, Bevilacqua PC. PLoS ONE. 2013;8:e57905. doi: 10.1371/journal.pone.0057905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Yin QF, Yang L, Zhang Y, Xiang JF, Wu YW, Carmichael GG, Chen LL. Mol. Cell. 2012;48:219. doi: 10.1016/j.molcel.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 193.Koziol MJ, Rinn JL. Curr. Opin. Genet. Dev. 2010;20:142. doi: 10.1016/j.gde.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. Nature. 2011;477:295. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. Science. 2005;309:1570. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 196.Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, Forrest AR, Carninci P, Biffo S, Stupka E, Gustincich S. Nature. 2012;491:454. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- 197.Gong C, Maquat LE. Nature. 2011;470:284. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. Nature. 2010;465:1033. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Theimer CA, Feigon J. Curr. Opin. Struct. Biol. 2006;16:307. doi: 10.1016/j.sbi.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 200.O'Sullivan RJ, Karlseder J. Nat. Rev. Mol. Cell Biol. 2010;11:171. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Gilley D, Tanaka H, Herbert BS. Int. J. Biochem. Cell Biol. 2005;37:1000. doi: 10.1016/j.biocel.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 202.Gunes C, Rudolph KL. Cell. 2013;152:390. doi: 10.1016/j.cell.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 203.Rotman B. Proc. Natl. Acad. Sci. U.S.A. 1961;47:1981. doi: 10.1073/pnas.47.12.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Gelles J, Schnapp BJ, Sheetz MP. Nature. 1988;331:450. doi: 10.1038/331450a0. [DOI] [PubMed] [Google Scholar]
- 205.Thompson RE, Larson DR, Webb WW. Biophys. J. 2002;82:2775. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Hua W, Chung J, Gelles J. Science. 2002;295:844. doi: 10.1126/science.1063089. [DOI] [PubMed] [Google Scholar]
- 207.Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Science. 2003;300:2061. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 208.Kural C, Kim H, Syed S, Goshima G, Gelfand VI, Selvin PR. Science. 2005;308:1469. doi: 10.1126/science.1108408. [DOI] [PubMed] [Google Scholar]
- 209.Moerner WE, Kador L. Phys. Rev. Lett. 1989;62:2535. doi: 10.1103/PhysRevLett.62.2535. [DOI] [PubMed] [Google Scholar]
- 210.Orrit M, Bernard J. Phys. Rev. Lett. 1990;65:2716. doi: 10.1103/PhysRevLett.65.2716. [DOI] [PubMed] [Google Scholar]
- 211.Lu HP, Xun L, Xie XS. Science. 1998;282:1877. doi: 10.1126/science.282.5395.1877. [DOI] [PubMed] [Google Scholar]
- 212.Walter NG, Huang CY, Manzo AJ, Sobhy MA. Nat. Methods. 2008;5:475. doi: 10.1038/nmeth.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Yoshimura H, Inaguma A, Yamada T, Ozawa T. ACS Chem. Biol. 2012;7:999. doi: 10.1021/cb200474a. [DOI] [PubMed] [Google Scholar]
- 214.Ando R, Flors C, Mizuno H, Hofkens J, Miyawaki A. Biophys. J. 2007;92:L97. doi: 10.1529/biophysj.107.105882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Sahl SJ, Moerner W. Curr. Opin. Struct. Biol. 2013 doi: 10.1016/j.sbi.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Gossen M, Bujard H. Proc. Natl. Acad. Sci. U.S.A. 1992;89:5547. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Santangelo PJ, Lifland AW, Curt P, Sasaki Y, Bassell GJ, Lindquist ME, Crowe JE., Jr. Nat. Methods. 2009;6:347. doi: 10.1038/nmeth.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Sakon JJ, Weninger KR. Nat. Methods. 2010;7:203. doi: 10.1038/nmeth.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Huang B, Bates M, Zhuang X. Annu. Rev. Biochem. 2009;78:993. doi: 10.1146/annurev.biochem.77.061906.092014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Newby Lambert M, Vocker E, Blumberg S, Redemann S, Gajraj A, Meiners JC, Walter NG. Biophys. J. 2006;90:3672. doi: 10.1529/biophysj.105.067793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Shimomura O, Johnson FH, Saiga Y. J. Cell Comp. Physiol. 1962;59:223. doi: 10.1002/jcp.1030590302. [DOI] [PubMed] [Google Scholar]
- 222.Prasher DC, Eckenrode VK, Ward WW, Prendergast FG, Cormier MJ. Gene. 1992;111:229. doi: 10.1016/0378-1119(92)90691-h. [DOI] [PubMed] [Google Scholar]
- 223.Tsien RY. Annu. Rev. Biochem. 1998;67:509. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 224.Day RN, Davidson MW. Chem. Soc. Rev. 2009;38:2887. doi: 10.1039/b901966a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Mor A, Suliman S, Ben-Yishay R, Yunger S, Brody Y, Shav-Tal Y. Nat. Cell Biol. 2010;12:543. doi: 10.1038/ncb2056. [DOI] [PubMed] [Google Scholar]
- 226.Lionnet T, Czaplinski K, Darzacq X, Shav-Tal Y, Wells AL, Chao JA, Park HY, de Turris V, Lopez-Jones M, Singer RH. Nat. Methods. 2011;8:165. doi: 10.1038/nmeth.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Farzadfard F, Perli SD, Lu TK. ACS Synth. Biol. 2013 doi: 10.1021/sb400081r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Ha T, Tinnefeld P. Annu. Rev. Phys. Chem. 2012;63:595. doi: 10.1146/annurev-physchem-032210-103340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Biteen JS, Thompson MA, Tselentis NK, Bowman GR, Shapiro L, Moerner WE. Nat. Methods. 2008;5:947. doi: 10.1038/NMETH.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Lew MD, Lee SF, Ptacin JL, Lee MK, Twieg RJ, Shapiro L, Moerner WE. Proc. Natl. Acad. Sci. U.S.A. 2011;108:E1102. doi: 10.1073/pnas.1114444108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Wu B, Piatkevich KD, Lionnet T, Singer RH, Verkhusha VV. Curr. Opin. Cell Biol. 2011;23:310. doi: 10.1016/j.ceb.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Subach FV, Subach OM, Gundorov IS, Morozova KS, Piatkevich KD, Cuervo AM, Verkhusha VV. Nat. Chem. Biol. 2009;5:118. doi: 10.1038/nchembio.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, Singer RH, Bertrand E. Curr. Biol. 2003;13:161. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Kapanidis AN, Weiss S. J. Chem. Phys. 2002;117:10953. [Google Scholar]
- 235.Goncalves MS. Chem. Rev. 2009;109:190. doi: 10.1021/cr0783840. [DOI] [PubMed] [Google Scholar]
- 236.Walter NG, Burke JM. Methods Enzymol. 2000;317:409. doi: 10.1016/s0076-6879(00)17027-2. [DOI] [PubMed] [Google Scholar]
- 237.Rinaldi AJ, Suddala KC, Walter NG. Methods Mol. Biol. 2014 doi: 10.1007/978-1-4939-1896-6_6. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Prescher JA, Bertozzi CR. Nat. Chem. Biol. 2005;1:13. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- 239.Shi X, Jung Y, Lin LJ, Liu C, Wu C, Cann IK, Ha T. Nat. Methods. 2012;9:499. doi: 10.1038/nmeth.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Lord SJ, Conley NR, Lee HL, Samuel R, Liu N, Twieg RJ, Moerner WE. J. Am. Chem. Soc. 2008;130:9204. doi: 10.1021/ja802883k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Lord SJ, Conley NR, Lee HL, Nishimura SY, Pomerantz AK, Willets KA, Lu Z, Wang H, Liu N, Samuel R, Weber R, Semyonov A, He M, Twieg RJ, Moerner WE. Chemphyschem. 2009;10:55. doi: 10.1002/cphc.200800581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Science. 2005;307:538. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Sugisaki M, Ren HW, Nishi K, Masumoto Y. Phys. Rev. Lett. 2001;86:4883. doi: 10.1103/PhysRevLett.86.4883. [DOI] [PubMed] [Google Scholar]
- 244.Bogdanov AM, Bogdanova EA, Chudakov DM, Gorodnicheva TV, Lukyanov S, Lukyanov KA. Nat. Methods. 2009;6:859. doi: 10.1038/nmeth1209-859. [DOI] [PubMed] [Google Scholar]
- 245.Billinton N, Knight AW. Anal. Biochem. 2001;291:175. doi: 10.1006/abio.2000.5006. [DOI] [PubMed] [Google Scholar]
- 246.Kredel S, Nienhaus K, Oswald F, Wolff M, Ivanchenko S, Cymer F, Jeromin A, Michel FJ, Spindler KD, Heilker R, Nienhaus GU, Wiedenmann J. Chem. Biol. 2008;15:224. doi: 10.1016/j.chembiol.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 247.Aitken CE, Marshall RA, Puglisi JD. Biophys. J. 2008;94:1826. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Dave R, Terry DS, Munro JB, Blanchard SC. Biophys. J. 2009;96:2371. doi: 10.1016/j.bpj.2008.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Altman RB, Terry DS, Zhou Z, Zheng Q, Geggier P, Kolster RA, Zhao Y, Javitch JA, Warren JD, Blanchard SC. Nat. Methods. 2012;9:68. doi: 10.1038/nmeth.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Widengren J, Chmyrov A, Eggeling C, Lofdahl PA, Seidel CA. J. Phys. Chem. A. 2007;111:429. doi: 10.1021/jp0646325. [DOI] [PubMed] [Google Scholar]
- 251.Olenych SG, Claxton NS, Ottenberg GK, Davidson MW. Curr. Protoc. Cell Biol. 2007 doi: 10.1002/0471143030.cb2105s36. Chapter 21, Unit 21 5. [DOI] [PubMed] [Google Scholar]
- 252.Roy R, Hohng S, Ha T. Nat. Methods. 2008;5:507. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Waterman-Storer CM, Sanger JW, Sanger JM. Cell Motil. Cytoskel. 1993;26:19. doi: 10.1002/cm.970260104. [DOI] [PubMed] [Google Scholar]
- 254.Koritzinsky M, Magagnin MG, van den Beucken T, Seigneuric R, Savelkouls K, Dostie J, Pyronnet S, Kaufman RJ, Weppler SA, Voncken JW, Lambin P, Koumenis C, Sonenberg N, Wouters BG. EMBO J. 2006;25:1114. doi: 10.1038/sj.emboj.7600998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Shi X, Lim J, Ha T. Anal. Chem. 2010;82:6132. doi: 10.1021/ac1008749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Steinhauer C, Forthmann C, Vogelsang J, Tinnefeld P. J. Am. Chem. Soc. 2008;130:16840. doi: 10.1021/ja806590m. [DOI] [PubMed] [Google Scholar]
- 257.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Science. 2002;297:1183. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 258.Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Nat. Genet. 2002;31:69. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- 259.Hanahan D, Weinberg RA. Cell. 2011;144:646. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 260.Oleynikov Y, Singer RH. Curr. Biol. 2003;13:199. doi: 10.1016/s0960-9822(03)00044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Cell. 2007;131:174. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 262.Montero Llopis P, Jackson AF, Sliusarenko O, Surovtsev I, Heinritz J, Emonet T, Jacobs-Wagner C. Nature. 2010;466:77. doi: 10.1038/nature09152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Xue Z, Huang K, Cai C, Cai L, Jiang CY, Feng Y, Liu Z, Zeng Q, Cheng L, Sun YE, Liu JY, Horvath S, Fan G. Nature. 2013;500:593. doi: 10.1038/nature12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Tischler J, Surani MA. Curr. Opin. Biotechnol. 2013;24:69. doi: 10.1016/j.copbio.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 265.Levsky JM, Singer RH. J. Cell. Sci. 2003;116:2833. doi: 10.1242/jcs.00633. [DOI] [PubMed] [Google Scholar]
- 266.Femino AM, Fay FS, Fogarty K, Singer RH. Science. 1998;280:585. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 267.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Nat. Methods. 2008;5:877. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Siebrasse JP, Veith R, Dobay A, Leonhardt H, Daneholt B, Kubitscheck U. Proc. Natl. Acad. Sci. U.S.A. 2008;105:20291. doi: 10.1073/pnas.0810692105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Vargas DY, Raj A, Marras SA, Kramer FR, Tyagi S. Proc. Natl. Acad. Sci. U.S.A. 2005;102:17008. doi: 10.1073/pnas.0505580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Vargas DY, Shah K, Batish M, Levandoski M, Sinha S, Marras SA, Schedl P, Tyagi S. Cell. 2011;147:1054. doi: 10.1016/j.cell.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Lansdorp PM, Verwoerd NP, van de Rijke FM, Dragowska V, Little MT, Dirks RW, Raap AK, Tanke HJ. Hum. Mol. Genet. 1996;5:685. doi: 10.1093/hmg/5.5.685. [DOI] [PubMed] [Google Scholar]
- 272.Lu J, Tsourkas A. Nucleic Acids Res. 2009;37:e100. doi: 10.1093/nar/gkp482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Larsson C, Grundberg I, Soderberg O, Nilsson M. Nat. Methods. 2010;7:395. doi: 10.1038/nmeth.1448. [DOI] [PubMed] [Google Scholar]
- 274.Player AN, Shen LP, Kenny D, Antao VP, Kolberg JA. J. Histochem. Cytochem. 2001;49:603. doi: 10.1177/002215540104900507. [DOI] [PubMed] [Google Scholar]
- 275.Femino AM, Fogarty K, Lifshitz LM, Carrington W, Singer RH. Methods Enzymol. 2003;361:245. doi: 10.1016/s0076-6879(03)61015-3. [DOI] [PubMed] [Google Scholar]
- 276.Hess ST, Girirajan TPK, Mason MD. Biophys. J. 2006;91:4258. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Maamar H, Raj A, Dubnau D. Science. 2007;317:526. doi: 10.1126/science.1140818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Zenklusen D, Larson DR, Singer RH. Nat. Struct. Mol. Biol. 2008;15:1263. doi: 10.1038/nsmb.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Tan RZ, van Oudenaarden A. Mol. Syst. Biol. 2010;6:358. doi: 10.1038/msb.2010.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Raj A, Rifkin SA, Andersen E, van Oudenaarden A. Nature. 2010;463:913. doi: 10.1038/nature08781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Harland RM. Methods Cell Biol. 1991;36:685. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- 282.Shih JD, Waks Z, Kedersha N, Silver PA. Nucleic Acids Res. 2011;39:7740. doi: 10.1093/nar/gkr456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Raj A, Tyagi S. Methods Enzymol. 2010;472:365. doi: 10.1016/S0076-6879(10)72004-8. [DOI] [PubMed] [Google Scholar]
- 284.Morrison LE, Halder TC, Stols LM. Anal. Biochem. 1989;183:231. doi: 10.1016/0003-2697(89)90473-9. [DOI] [PubMed] [Google Scholar]
- 285.Li Q, Luan G, Guo Q, Liang J. Nucleic Acids Res. 2002;30:E5. doi: 10.1093/nar/30.2.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Shepherd DP, Li N, Micheva-Viteva SN, Munsky B, Hong-Geller E, Werner JH. Anal. Chem. 2013;85:4938. doi: 10.1021/ac303792p. [DOI] [PubMed] [Google Scholar]
- 287.Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS. Science. 2010;329:533. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Larsson C, Koch J, Nygren A, Janssen G, Raap AK, Landegren U, Nilsson M. Nat. Methods. 2004;1:227. doi: 10.1038/nmeth723. [DOI] [PubMed] [Google Scholar]
- 289.Battich N, Stoeger T, Pelkmans L. Nat. Methods. 2013;10:1127. doi: 10.1038/nmeth.2657. [DOI] [PubMed] [Google Scholar]
- 290.Choi HM, Chang JY, Trinh le A, Padilla JE, Fraser SE, Pierce NA. Nat. Biotechnol. 2010;28:1208. doi: 10.1038/nbt.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Levsky JM, Shenoy SM, Pezo RC, Singer RH. Science. 2002;297:836. doi: 10.1126/science.1072241. [DOI] [PubMed] [Google Scholar]
- 292.Levesque MJ, Raj A. Nat. Methods. 2013;10:246. doi: 10.1038/nmeth.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Ishihama Y, Funatsu T. Biochem. Biophys. Res. Commun. 2009;381:33. doi: 10.1016/j.bbrc.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 294.Lifland AW, Zurla C, Santangelo PJ. Bioconj. Chem. 2010;21:483. doi: 10.1021/bc9003876. [DOI] [PubMed] [Google Scholar]
- 295.Luebke KJ, Balog RP, Garner HR. Nucleic Acids Res. 2003;31:750. doi: 10.1093/nar/gkg133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Coulon A, Chow CC, Singer RH, Larson DR. Nat. Rev. Genet. 2013;14:572. doi: 10.1038/nrg3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Xie XS, Choi PJ, Li GW, Lee NK, Lia G. Annu. Rev. Biophys. 2008;37:417. doi: 10.1146/annurev.biophys.37.092607.174640. [DOI] [PubMed] [Google Scholar]
- 298.Kerppola TK. Nat. Rev. Mol. Cell Biol. 2006;7:449. doi: 10.1038/nrm1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Keryer-Bibens C, Barreau C, Osborne HB. Biol. Cell. 2008;100:125. doi: 10.1042/BC20070067. [DOI] [PubMed] [Google Scholar]
- 300.Chao JA, Patskovsky Y, Almo SC, Singer RH. Nat. Struct. Mol. Biol. 2008;15:103. doi: 10.1038/nsmb1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Larson DR, Zenklusen D, Wu B, Chao JA, Singer RH. Science. 2011;332:475. doi: 10.1126/science.1202142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Daigle N, Ellenberg J. Nat. Methods. 2007;4:633. doi: 10.1038/nmeth1065. [DOI] [PubMed] [Google Scholar]
- 303.Lange S, Katayama Y, Schmid M, Burkacky O, Brauchle C, Lamb DC, Jansen RP. Traffic. 2008;9:1256. doi: 10.1111/j.1600-0854.2008.00763.x. [DOI] [PubMed] [Google Scholar]
- 304.Haim L, Zipor G, Aronov S, Gerst JE. Nat. Methods. 2007;4:409. doi: 10.1038/nmeth1040. [DOI] [PubMed] [Google Scholar]
- 305.Golding I, Cox EC. Proc. Natl. Acad. Sci. U.S.A. 2004;101:11310. doi: 10.1073/pnas.0404443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Golding I, Paulsson J, Zawilski SM, Cox EC. Cell. 2005;123:1025. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 307.Grunwald D, Singer RH. Nature. 2010;467:604. doi: 10.1038/nature09438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Wu B, Chao JA, Singer RH. Biophys. J. 2012;102:2936. doi: 10.1016/j.bpj.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Ozawa T, Natori Y, Sato M, Umezawa Y. Nat. Methods. 2007;4:413. doi: 10.1038/nmeth1030. [DOI] [PubMed] [Google Scholar]
- 310.Yamada T, Yoshimura H, Inaguma A, Ozawa T. Anal. Chem. 2011;83:5708. doi: 10.1021/ac2009405. [DOI] [PubMed] [Google Scholar]
- 311.Quenault T, Lithgow T, Traven A. Trends Cell Biol. 2011;21:104. doi: 10.1016/j.tcb.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 312.Wang X, McLachlan J, Zamore PD, Hall TM. Cell. 2002;110:501. doi: 10.1016/s0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- 313.Cheong CG, Hall TM. Proc. Natl. Acad. Sci. U.S.A. 2006;103:13635. doi: 10.1073/pnas.0606294103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Katahira J. Biochim. Biophys. Acta. 2012;1819:507. doi: 10.1016/j.bbagrm.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 315.Siebrasse JP, Kaminski T, Kubitscheck U. Proc. Natl. Acad. Sci. U.S.A. 2012;109:9426. doi: 10.1073/pnas.1201781109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Calapez A, Pereira HM, Calado A, Braga J, Rino J, Carvalho C, Tavanez JP, Wahle E, Rosa AC, Carmo-Fonseca M. J. Cell Biol. 2002;159:795. doi: 10.1083/jcb.200203046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Valencia-Burton M, McCullough RM, Cantor CR, Broude NE. Nat. Methods. 2007;4:421. doi: 10.1038/nmeth1023. [DOI] [PubMed] [Google Scholar]
- 318.Rackham O, Brown CM. EMBO J. 2004;23:3346. doi: 10.1038/sj.emboj.7600341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Nguyen DH, DeFina SC, Fink WH, Dieckmann T. J. Am. Chem. Soc. 2002;124:15081. doi: 10.1021/ja027635d. [DOI] [PubMed] [Google Scholar]
- 320.Babendure JR, Adams SR, Tsien RY. J. Am. Chem. Soc. 2003;125:14716. doi: 10.1021/ja037994o. [DOI] [PubMed] [Google Scholar]
- 321.Paige JS, Wu KY, Jaffrey SR. Science. 2011;333:642. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Song W, Strack RL, Jaffrey SR. Nat. Methods. 2013;10:873. doi: 10.1038/nmeth.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Martin G, Keller W. RNA. 1998;4:226. [PMC free article] [PubMed] [Google Scholar]
- 324.Kinoshita Y, Nishigaki K, Husimi Y. Nucleic Acids Res. 1997;25:3747. doi: 10.1093/nar/25.18.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Huang Z, Szostak JW. Nucleic Acids Res. 1996;24:4360. doi: 10.1093/nar/24.21.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Rosemeyer V, Laubrock A, Seibl R. Anal. Biochem. 1995;224:446. doi: 10.1006/abio.1995.1068. [DOI] [PubMed] [Google Scholar]
- 327.Qin PZ, Pyle AM. Methods. 1999;18:60. doi: 10.1006/meth.1999.0757. [DOI] [PubMed] [Google Scholar]
- 328.Walter NG. Curr. Prot. Nucleic Acid Chem. 2003 Chapter 11, Unit 11 10. [Google Scholar]
- 329.Motorin Y, Burhenne J, Teimer R, Koynov K, Willnow S, Weinhold E, Helm M. Nucleic Acids Res. 2011;39:1943. doi: 10.1093/nar/gkq825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Rao H, Tanpure AA, Sawant AA, Srivatsan SG. Nat. Protoc. 2012;7:1097. doi: 10.1038/nprot.2012.046. [DOI] [PubMed] [Google Scholar]
- 331.Winz ML, Samanta A, Benzinger D, Jaschke A. Nucleic Acids Res. 2012;40:e78. doi: 10.1093/nar/gks062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Science. 2005;309:1573. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 333.Tadakuma H, Ishihama Y, Shibuya T, Tani T, Funatsu T. Biochem. Biophys. Res. Commun. 2006;344:772. doi: 10.1016/j.bbrc.2006.03.202. [DOI] [PubMed] [Google Scholar]
- 334.Leake MC, Chandler JH, Wadhams GH, Bai F, Berry RM, Armitage JP. Nature. 2006;443:355. doi: 10.1038/nature05135. [DOI] [PubMed] [Google Scholar]
- 335.Coffman VC, Wu JQ. Trends Biochem. Sci. 2012;37:499. doi: 10.1016/j.tibs.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Mutch SA, Fujimoto BS, Kuyper CL, Kuo JS, Bajjalieh SM, Chiu DT. Biophys. J. 2007;92:2926. doi: 10.1529/biophysj.106.101428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.de Turris V, Nicholson P, Orozco RZ, Singer RH, Muhlemann O. RNA. 2011;17:2094. doi: 10.1261/rna.02918111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Valencia-Burton M, Shah A, Sutin J, Borogovac A, McCullough RM, Cantor CR, Meller A, Broude NE. Proc. Natl. Acad. Sci. U.S.A. 2009;106:16399. doi: 10.1073/pnas.0907495106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Zipor G, Haim-Vilmovsky L, Gelin-Licht R, Gadir N, Brocard C, Gerst JE. Proc. Natl. Acad. Sci. U.S.A. 2009;106:19848. doi: 10.1073/pnas.0910754106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.So LH, Ghosh A, Zong C, Sepulveda LA, Segev R, Golding I. Nat. Genet. 2011;43:554. doi: 10.1038/ng.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Geerts H, De Brabander M, Nuydens R, Geuens S, Moeremans M, De Mey J, Hollenbeck P. Biophys. J. 1987;52:775. doi: 10.1016/S0006-3495(87)83271-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Babcock HP, Chen C, Zhuang X. Biophys. J. 2004;87:2749. doi: 10.1529/biophysj.104.042234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Grunwald D, Spottke B, Buschmann V, Kubitscheck U. Mol. Biol. Cell. 2006;17:5017. doi: 10.1091/mbc.E06-06-0559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Ohrt T, Mutze J, Staroske W, Weinmann L, Hock J, Crell K, Meister G, Schwille P. Nucleic Acids Res. 2008;36:6439. doi: 10.1093/nar/gkn693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Tatavarty V, Ifrim MF, Levin M, Korza G, Barbarese E, Yu J, Carson JH. Mol. Biol. Cell. 2012;23:918. doi: 10.1091/mbc.E11-07-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Lifland AW, Zurla C, Yu J, Santangelo PJ. Traffic. 2011;12:1000. doi: 10.1111/j.1600-0854.2011.01209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Zurla C, Lifland AW, Santangelo PJ. PLoS ONE. 2011;6:e19727. doi: 10.1371/journal.pone.0019727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Trcek T, Chao JA, Larson DR, Park HY, Zenklusen D, Shenoy SM, Singer RH. Nat. Protoc. 2012;7:408. doi: 10.1038/nprot.2011.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Gerlach D, Kohler W, Gunther E, Mann K. Infect. Immun. 1993;61:2727. doi: 10.1128/iai.61.6.2727-2731.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Palmer M, Harris R, Freytag C, Kehoe M, Tranum-Jensen J, Bhakdi S. EMBO J. 1998;17:1598. doi: 10.1093/emboj/17.6.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Dange T, Grunwald D, Grunwald A, Peters R, Kubitscheck U. J. Cell Biol. 2008;183:77. doi: 10.1083/jcb.200806173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Nitin N, Santangelo PJ, Kim G, Nie S, Bao G. Nucleic Acids Res. 2004;32:e58. doi: 10.1093/nar/gnh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Grunwald D, Shenoy SM, Burke S, Singer RH. Nat. Protoc. 2008;3:1809. doi: 10.1038/nprot.2008.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Gebhardt JC, Suter DM, Roy R, Zhao ZW, Chapman AR, Basu S, Maniatis T, Xie XS. Nat. Methods. 2013;10:421. doi: 10.1038/nmeth.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Rehemtulla A, Hamilton CA, Chinnaiyan AM, Dixit VM. J. Biol. Chem. 1997;272:25783. doi: 10.1074/jbc.272.41.25783. [DOI] [PubMed] [Google Scholar]
- 356.Kao HP, Verkman AS. Biophys. J. 1994;67:1291. doi: 10.1016/S0006-3495(94)80601-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Huang B, Wang W, Bates M, Zhuang X. Science. 2008;319:810. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Thompson MA, Lew MD, Badieirostami M, Moerner WE. Nano Lett. 2010;10:211. doi: 10.1021/nl903295p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Thompson MA, Casolari JM, Badieirostami M, Brown PO, Moerner WE. Proc. Natl. Acad. Sci. U.S.A. 2010;107:17864. doi: 10.1073/pnas.1012868107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Toprak E, Balci H, Blehm BH, Selvin PR. Nano Lett. 2007;7:2043. doi: 10.1021/nl0709120. [DOI] [PubMed] [Google Scholar]
- 361.Sun Y, McKenna JD, Murray JM, Ostap EM, Goldman YE. Nano Lett. 2009;9:2676. doi: 10.1021/nl901129j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Axelrod D. J. Cell Biol. 1981;89:141. doi: 10.1083/jcb.89.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Konopka CA, Bednarek SY. Plant J. 2008;53:186. doi: 10.1111/j.1365-313X.2007.03306.x. [DOI] [PubMed] [Google Scholar]
- 364.Tokunaga M, Imamoto N, Sakata-Sogawa K. Nat. Methods. 2008;5:159. doi: 10.1038/nmeth1171. [DOI] [PubMed] [Google Scholar]
- 365.Ritter JG, Veith R, Siebrasse JP, Kubitscheck U. Opt. Express. 2008;16:7142. doi: 10.1364/oe.16.007142. [DOI] [PubMed] [Google Scholar]
- 366.Ritter JG, Veith R, Veenendaal A, Siebrasse JP, Kubitscheck U. PLoS ONE. 2010;5:e11639. doi: 10.1371/journal.pone.0011639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Spille JH, Kaminski T, Konigshoven HP, Kubitscheck U. Opt. Express. 2012;20:19697. doi: 10.1364/OE.20.019697. [DOI] [PubMed] [Google Scholar]
- 368.Lang MC, Engelhardt J, Hell SW. Opt. Lett. 2007;32:259. doi: 10.1364/ol.32.000259. [DOI] [PubMed] [Google Scholar]
- 369.Hell SW. Science. 2007;316:1153. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- 370.Nakano A. Cell Struct. Funct. 2002;27:349. doi: 10.1247/csf.27.349. [DOI] [PubMed] [Google Scholar]
- 371.Ma J, Yang W. Proc. Natl. Acad. Sci. U.S.A. 2010;107:7305. doi: 10.1073/pnas.0908269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Michalet X, Weiss S, Jager M. Chem. Rev. 2006;106:1785. doi: 10.1021/cr0404343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Elf J, Li GW, Xie XS. Science. 2007;316:1191. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Poo M, Cone RA. Nature. 1974;247:438. doi: 10.1038/247438a0. [DOI] [PubMed] [Google Scholar]
- 375.Magde D, Elson E, Webb WW. Phys. Rev. Lett. 1972;29:705. [Google Scholar]
- 376.Cheezum MK, Walker WF, Guilford WH. Biophys. J. 2001;81:2378. doi: 10.1016/S0006-3495(01)75884-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Sbalzarini IF, Koumoutsakos P. J. Struct. Biol. 2005;151:182. doi: 10.1016/j.jsb.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 378.Jaqaman K, Loerke D, Mettlen M, Kuwata H, Grinstein S, Schmid SL, Danuser G. Nat. Methods. 2008;5:695. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Serge A, Bertaux N, Rigneault H, Marguet D. Nat. Methods. 2008;5:687. doi: 10.1038/nmeth.1233. [DOI] [PubMed] [Google Scholar]
- 380.Reck-Peterson SL, Yildiz A, Carter AP, Gennerich A, Zhang N, Vale RD. Cell. 2006;126:335. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Saxton MJ. Biophys. J. 1997;72:1744. doi: 10.1016/S0006-3495(97)78820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Saxton MJ, Jacobson K. Annu. Rev. Biophys. Biomol. Struct. 1997;26:373. doi: 10.1146/annurev.biophys.26.1.373. [DOI] [PubMed] [Google Scholar]
- 383.Itzkovitz S, van Oudenaarden A. Nat. Methods. 2011;8:S12. doi: 10.1038/nmeth.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Itzkovitz S, Lyubimova A, Blat IC, Maynard M, van Es J, Lees J, Jacks T, Clevers H, van Oudenaarden A. Nat. Cell Biol. 2012;14:106. doi: 10.1038/ncb2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Kwon S. BMB Rep. 2013;46:65. doi: 10.5483/BMBRep.2013.46.2.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Shestakova EA, Singer RH, Condeelis J. Proc. Natl. Acad. Sci. U.S.A. 2001;98:7045. doi: 10.1073/pnas.121146098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Shandilya J, Roberts SG. Biochim. Biophys. Acta. 2012;1819:391. doi: 10.1016/j.bbagrm.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 388.Papantonis A, Cook PR. Chem. Rev. 2013;113:8683. doi: 10.1021/cr300513p. [DOI] [PubMed] [Google Scholar]
- 389.Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Cell. 1998;95:717. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 390.Raser JM, O'Shea EK. Science. 2004;304:1811. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Science. 2005;307:1962. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- 392.Thomas MC, Chiang CM. Crit. Rev. Biochem. Mol. Biol. 2006;41:105. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 393.Li B, Carey M, Workman JL. Cell. 2007;128:707. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 394.Saunders A, Core LJ, Lis JT. Nat. Rev. Mol. Cell Biol. 2006;7:557. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 395.Lionnet T, Singer RH. EMBO Rep. 2012;13:313. doi: 10.1038/embor.2012.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Thattai M, van Oudenaarden A. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8614. doi: 10.1073/pnas.151588598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Munsky B, Neuert G, van Oudenaarden A. Science. 2012;336:183. doi: 10.1126/science.1216379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Cai L, Friedman N, Xie XS. Nature. 2006;440:358. doi: 10.1038/nature04599. [DOI] [PubMed] [Google Scholar]
- 399.Li GW, Xie XS. Nature. 2011;475:308. doi: 10.1038/nature10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Muthukrishnan AB, Kandhavelu M, Lloyd-Price J, Kudasov F, Chowdhury S, Yli-Harja O, Ribeiro AS. Nucleic Acids Res. 2012;40:8472. doi: 10.1093/nar/gks583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Muramoto T, Cannon D, Gierlinski M, Corrigan A, Barton GJ, Chubb JR. Proc. Natl. Acad. Sci. U.S.A. 2012;109:7350. doi: 10.1073/pnas.1117603109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Dar RD, Razooky BS, Singh A, Trimeloni TV, McCollum JM, Cox CD, Simpson ML, Weinberger LS. Proc. Natl. Acad. Sci. U.S.A. 2012;109:17454. doi: 10.1073/pnas.1213530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Chubb JR, Trcek T, Shenoy SM, Singer RH. Curr. Biol. 2006;16:1018. doi: 10.1016/j.cub.2006.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Ben-Ari Y, Brody Y, Kinor N, Mor A, Tsukamoto T, Spector DL, Singer RH, Shav-Tal Y. J. Cell Sci. 2010;123:1761. doi: 10.1242/jcs.062638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Kandhavelu M, Mannerstrom H, Gupta A, Hakkinen A, Lloyd-Price J, Yli-Harja O, Ribeiro AS. BMC Syst. Biol. 2011;5:149. doi: 10.1186/1752-0509-5-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS. Nature. 2006;441:840. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 408.Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A, Jaenisch R. Cell. 2012;150:1209. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Makela J, Kandhavelu M, Oliveira SM, Chandraseelan JG, Lloyd-Price J, Peltonen J, Yli-Harja O, Ribeiro AS. Nucleic Acids Res. 2013;41:6544. doi: 10.1093/nar/gkt350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Larson DR, Singer RH, Zenklusen D. Trends Cell Biol. 2009;19:630. doi: 10.1016/j.tcb.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Coppey M, Benichou O, Voituriez R, Moreau M. Biophys. J. 2004;87:1640. doi: 10.1529/biophysj.104.045773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Shahrezaei V, Swain PS. Proc. Natl. Acad. Sci. U.S.A. 2008;105:17256. doi: 10.1073/pnas.0803850105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Iyer-Biswas S, Hayot F, Jayaprakash C. Phys. Rev. E. 2009;79:031911. doi: 10.1103/PhysRevE.79.031911. [DOI] [PubMed] [Google Scholar]
- 414.Maamar H, Cabili MN, Rinn J, Raj A. Genes Dev. 2013;27:1260. doi: 10.1101/gad.217018.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Goodrich JA, Kugel JF. Nat. Rev. Mol. Cell Biol. 2006;7:612. doi: 10.1038/nrm1946. [DOI] [PubMed] [Google Scholar]
- 416.van Werven FJ, Neuert G, Hendrick N, Lardenois A, Buratowski S, van Oudenaarden A, Primig M, Amon A. Cell. 2012;150:1170. doi: 10.1016/j.cell.2012.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Bumgarner SL, Neuert G, Voight BF, Symbor-Nagrabska A, Grisafi P, van Oudenaarden A, Fink GR. Mol. Cell. 2012;45:470. doi: 10.1016/j.molcel.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Ng K, Daigle N, Bancaud A, Ohhata T, Humphreys P, Walker R, Ellenberg J, Wutz A. Mol. Biol. Cell. 2011;22:2634. doi: 10.1091/mbc.E11-02-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Neil H, Malabat C, d'Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. Nature. 2009;457:1038. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 420.Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F. Cell. 2007;131:706. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 421.Castelnuovo M, Rahman S, Guffanti E, Infantino V, Stutz F, Zenklusen D. Nat. Struct. Mol. Biol. 2013;20:851. doi: 10.1038/nsmb.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Martin RM, Rino J, Carvalho C, Kirchhausen T, Carmo-Fonseca M. Cell Rep. 2013;4:1144. doi: 10.1016/j.celrep.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Waks Z, Klein AM, Silver PA. Mol. Syst. Biol. 2011;7:506. doi: 10.1038/msb.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Gibcus JH, Dekker J. Mol. Cell. 2013;49:773. doi: 10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Bickmore WA, van Steensel B. Cell. 2013;152:1270. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 426.Huang S, Deerinck TJ, Ellisman MH, Spector DL. J. Cell Biol. 1994;126:877. doi: 10.1083/jcb.126.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Singh OP, Bjorkroth B, Masich S, Wieslander L, Daneholt B. Exp. Cell Res. 1999;251:135. doi: 10.1006/excr.1999.4490. [DOI] [PubMed] [Google Scholar]
- 428.Veith R, Sorkalla T, Baumgart E, Anzt J, Haberlein H, Tyagi S, Siebrasse JP, Kubitscheck U. Biophys. J. 2010;99:2676. doi: 10.1016/j.bpj.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Politz JC, Tuft RA, Pederson T, Singer RH. Curr. Biol. 1999;9:285. doi: 10.1016/s0960-9822(99)80136-5. [DOI] [PubMed] [Google Scholar]
- 430.Shav-Tal Y, Darzacq X, Shenoy SM, Fusco D, Janicki SM, Spector DL, Singer RH. Science. 2004;304:1797. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Rino J, Carvalho T, Braga J, Desterro JM, Luhrmann R, Carmo-Fonseca M. PLoS Comput. Biol. 2007;3:2019. doi: 10.1371/journal.pcbi.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Fiserova J, Richards SA, Wente SR, Goldberg MW. J. Cell Sci. 2010;123:2773. doi: 10.1242/jcs.070730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Holt CE, Bullock SL. Science. 2009;326:1212. doi: 10.1126/science.1176488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Mol. Cell. 1998;2:437. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 435.Casolari JM, Thompson MA, Salzman J, Champion LM, Moerner WE, Brown PO. PLoS ONE. 2012;7:e31912. doi: 10.1371/journal.pone.0031912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Bratu DP, Cha BJ, Mhlanga MM, Kramer FR, Tyagi S. Proc. Natl. Acad. Sci. U.S.A. 2003;100:13308. doi: 10.1073/pnas.2233244100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Weil TT, Parton R, Davis I, Gavis ER. Curr. Biol. 2008;18:1055. doi: 10.1016/j.cub.2008.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Weil TT, Forrest KM, Gavis ER. Dev. Cell. 2006;11:251. doi: 10.1016/j.devcel.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 439.Wilkie GS, Davis I. Cell. 2001;105:209. doi: 10.1016/s0092-8674(01)00312-9. [DOI] [PubMed] [Google Scholar]
- 440.Middelkoop TC, Williams L, Yang PT, Luchtenberg J, Betist MC, Ji N, van Oudenaarden A, Kenyon C, Korswagen HC. Dev. Biol. 2012;361:338. doi: 10.1016/j.ydbio.2011.10.029. [DOI] [PubMed] [Google Scholar]
- 441.Tyagi S, Alsmadi O. Biophys. J. 2004;87:4153. doi: 10.1529/biophysj.104.045153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Batish M, van den Bogaard P, Kramer FR, Tyagi S. Proc. Natl. Acad. Sci. U.S.A. 2012;109:4645. doi: 10.1073/pnas.1111226109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Mikl M, Vendra G, Kiebler MA. EMBO Rep. 2011;12:1077. doi: 10.1038/embor.2011.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Tubing F, Vendra G, Mikl M, Macchi P, Thomas S, Kiebler MA. J. Neurosci. 2010;30:4160. doi: 10.1523/JNEUROSCI.3537-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Mingle LA, Okuhama NN, Shi J, Singer RH, Condeelis J, Liu G. J. Cell Sci. 2005;118:2425. doi: 10.1242/jcs.02371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Rook MS, Lu M, Kosik KS. J. Neurosci. 2000;20:6385. doi: 10.1523/JNEUROSCI.20-17-06385.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Yamagishi M, Ishihama Y, Shirasaki Y, Kurama H, Funatsu T. Exp. Cell Res. 2009;315:1142. doi: 10.1016/j.yexcr.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 448.Forrest KM, Gavis ER. Curr. Biol. 2003;13:1159. doi: 10.1016/s0960-9822(03)00451-2. [DOI] [PubMed] [Google Scholar]
- 449.Rodriguez AJ, Shenoy SM, Singer RH, Condeelis J. J. Cell Biol. 2006;175:67. doi: 10.1083/jcb.200512137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.English BP, Hauryliuk V, Sanamrad A, Tankov S, Dekker NH, Elf J. Proc. Natl. Acad. Sci. U.S.A. 2011;108:E365. doi: 10.1073/pnas.1102255108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Korzelius J, The I, Ruijtenberg S, Prinsen MB, Portegijs V, Middelkoop TC, Groot Koerkamp MJ, Holstege FC, Boxem M, van den Heuvel S. PLoS Genet. 2011;7:e1002362. doi: 10.1371/journal.pgen.1002362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Harris DT, Horvitz HR. Development. 2011;138:4051. doi: 10.1242/dev.065417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Saffer AM, Kim DH, van Oudenaarden A, Horvitz HR. PLoS Genet. 2011;7:e1002418. doi: 10.1371/journal.pgen.1002418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Neely LA, Patel S, Garver J, Gallo M, Hackett M, McLaughlin S, Nadel M, Harris J, Gullans S, Rooke J. Nat. Methods. 2006;3:41. doi: 10.1038/nmeth825. [DOI] [PubMed] [Google Scholar]
- 455.Li J, Li X, Li Y, Yang H, Wang L, Qin Y, Liu H, Fu L, Guan XY. PLoS ONE. 2013;8:e53582. doi: 10.1371/journal.pone.0053582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Trcek T, Larson DR, Moldon A, Query CC, Singer RH. Cell. 2011;147:1484. doi: 10.1016/j.cell.2011.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Anderson P, Kedersha N. J. Cell Biol. 2006;172:803. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Goff SP. Nat. Rev. Microbiol. 2007;5:253. doi: 10.1038/nrmicro1541. [DOI] [PubMed] [Google Scholar]
- 459.Chou YY, Heaton NS, Gao Q, Palese P, Singer R, Lionnet T. PLoS Pathog. 2013;9:e1003358. doi: 10.1371/journal.ppat.1003358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Ni N, Nikolaitchik OA, Dilley KA, Chen J, Galli A, Fu W, Prasad VV, Ptak RG, Pathak VK, Hu WS. J. Virol. 2011;85:7603. doi: 10.1128/JVI.00562-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Boireau S, Maiuri P, Basyuk E, de la Mata M, Knezevich A, Pradet-Balade B, Backer V, Kornblihtt A, Marcello A, Bertrand E. J. Cell Biol. 2007;179:291. doi: 10.1083/jcb.200706018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Basyuk E, Galli T, Mougel M, Blanchard JM, Sitbon M, Bertrand E. Dev. Cell. 2003;5:161. doi: 10.1016/s1534-5807(03)00188-6. [DOI] [PubMed] [Google Scholar]
- 463.Kemler I, Meehan A, Poeschla EM. J. Virol. 2010;84:6352. doi: 10.1128/JVI.00363-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Jouvenet N, Simon SM, Bieniasz PD. Proc. Natl. Acad. Sci. U.S.A. 2009;106:19114. doi: 10.1073/pnas.0907364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Jouvenet N, Bieniasz PD, Simon SM. Nature. 2008;454:236. doi: 10.1038/nature06998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Ivanchenko S, Godinez WJ, Lampe M, Krausslich HG, Eils R, Rohr K, Brauchle C, Muller B, Lamb DC. PLoS Pathog. 2009;5:e1000652. doi: 10.1371/journal.ppat.1000652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Chen J, Nikolaitchik O, Singh J, Wright A, Bencsics CE, Coffin JM, Ni N, Lockett S, Pathak VK, Hu WS. Proc. Natl. Acad. Sci. U.S.A. 2009;106:13535. doi: 10.1073/pnas.0906822106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Dilley KA, Ni N, Nikolaitchik OA, Chen J, Galli A, Hu WS. J. Virol. 2011;85:10499. doi: 10.1128/JVI.05147-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Gallardo F, Laterreur N, Cusanelli E, Ouenzar F, Querido E, Wellinger RJ, Chartrand P. Mol. Cell. 2011;44:819. doi: 10.1016/j.molcel.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 470.Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, Maglott DR, Searle S, Farrell CM, Loveland JE, Ruef BJ, Hart E, Suner MM, Landrum MJ, Aken B, Ayling S, Baertsch R, Fernandez-Banet J, Cherry JL, Curwen V, Dicuccio M, Kellis M, Lee J, Lin MF, Schuster M, Shkeda A, Amid C, Brown G, Dukhanina O, Frankish A, Hart J, Maidak BL, Mudge J, Murphy MR, Murphy T, Rajan J, Rajput B, Riddick LD, Snow C, Steward C, Webb D, Weber JA, Wilming L, Wu W, Birney E, Haussler D, Hubbard T, Ostell J, Durbin R, Lipman D. Genome Res. 2009;19:1316. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Pruitt KD, Tatusova T, Brown GR, Maglott DR. Nucleic Acids Res. 2012;40:D130. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Lempiainen H, Shore D. Curr. Opin. Cell Biol. 2009;21:855. doi: 10.1016/j.ceb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 473.Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. Cell. Mol. Life Sci. 2008;65:2334. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Tschochner H, Hurt E. Trends Cell Biol. 2003;13:255. doi: 10.1016/s0962-8924(03)00054-0. [DOI] [PubMed] [Google Scholar]
- 475.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. Nucleic Acids Res. 2009;37:D141. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Rosenblad MA, Gorodkin J, Knudsen B, Zwieb C, Samuelsson T. Nucleic Acids Res. 2003;31:363. doi: 10.1093/nar/gkg107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Chan PP, Lowe TM. Nucleic Acids Res. 2009;37:D93. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Juhling F, Morl M, Hartmann RK, Sprinzl M, Stadler PF, Putz J. Nucleic Acids Res. 2009;37:D159. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Ellis JC, Brown DD, Brown JW. RNA. 2010;16:664. doi: 10.1261/rna.1871310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Brown JW. Nucleic Acids Res. 1999;27:314. doi: 10.1093/nar/27.1.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Yang JH, Shao P, Zhou H, Chen YQ, Qu LH. Nucleic Acids Res. 2010;38:D123. doi: 10.1093/nar/gkp943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Webb CH, Riccitelli NJ, Ruminski DJ, Luptak A. Science. 2009;326:953. doi: 10.1126/science.1178084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Kozomara A, Griffiths-Jones S. Nucleic Acids Res. 2011;39:D152. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Paraskevopoulou MD, Georgakilas G, Kostoulas N, Reczko M, Maragkakis M, Dalamagas TM, Hatzigeorgiou AG. Nucleic Acids Res. 2013;41:D239. doi: 10.1093/nar/gks1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Backman TW, Sullivan CM, Cumbie JS, Miller ZA, Chapman EJ, Fahlgren N, Givan SA, Carrington JC, Kasschau KD. Nucleic Acids Res. 2008;36:D982. doi: 10.1093/nar/gkm997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Ishizu H, Siomi H, Siomi MC. Genes Dev. 2012;26:2361. doi: 10.1101/gad.203786.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Sai Lakshmi S, Agrawal S. Nucleic Acids Res. 2008;36:D173. doi: 10.1093/nar/gkm696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS. Nucleic Acids Res. 2011;39:D146. doi: 10.1093/nar/gkq1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Genes Dev. 2011;25:1915. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Grillo G, Turi A, Licciulli F, Mignone F, Liuni S, Banfi S, Gennarino VA, Horner DS, Pavesi G, Picardi E, Pesole G. Nucleic Acids Res. 2010;38:D75. doi: 10.1093/nar/gkp902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Roy SW, Gilbert W. Nat. Rev. Genet. 2006;7:211. doi: 10.1038/nrg1807. [DOI] [PubMed] [Google Scholar]
- 492.Zhou Y, Lu C, Wu QJ, Wang Y, Sun ZT, Deng JC, Zhang Y. Nucleic Acids Res. 2008;36:D31. doi: 10.1093/nar/gkm766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Cardullo RA, Agrawal S, Flores C, Zamecnik PC, Wolf DE. Proc. Natl. Acad. Sci. U.S.A. 1988;85:8790. doi: 10.1073/pnas.85.23.8790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494.Sando S, Kool ET. J. Am Chem. Soc. 2002;124:9686. doi: 10.1021/ja026649g. [DOI] [PubMed] [Google Scholar]
- 495.Abe H, Kool ET. Proc. Natl. Acad. Sci. U.S.A. 2006;103:263. doi: 10.1073/pnas.0509938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Santangelo PJ, Nix B, Tsourkas A, Bao G. Nucleic Acids Res. 2004;32:e57. doi: 10.1093/nar/gnh062. [DOI] [PMC free article] [PubMed] [Google Scholar]