Abstract
Introduction
microRNA (miRNA) are small non-coding RNA species that are transcriptionally processed in the host cell and released extracellularly into the bloodstream. Normally involved in post-transcriptional gene silencing, the deregulation of miRNA has been shown to influence pathogenesis of a number of diseases.
Background
Next-generation deep sequencing (NGS) has provided the ability to profile miRNA in biological fluids making this approach a viable screening tool to detect miRNA biomarkers. However, collection and handling procedures of blood needs to be greatly improved for miRNA analysis in order to reliably detect differences between healthy and disease patients. Furthermore, ribonucleases present in blood can degrade RNA upon collection rendering extracellular miRNA at risk of degradation. These factors have consequently decreased sensitivity and specificity of miRNA biomarker assays.
Methods
Here, we use NGS to profile miRNA in various blood components and identify differences in profiles within peripheral blood compared to cell-free plasma or serum and extracellular vesicles known as exosomes. We also analyse and compare the miRNA content in exosomes prepared by ultracentrifugation methods and commercial exosome isolation kits including treating samples with RNaseA.
Conclusion
This study demonstrates that exosomal RNA is protected by RNaseA treatment and that exosomes provide a consistent source of miRNA for disease biomarker detection.
Keywords: microRNA, exosomes, serum, plasma, deep sequencing
There has been significant interest in using microRNA (miRNA) as biomarkers for diagnosis and therapeutic monitoring of diseases such as cancer, neurodegenerative disorders, heart disease and infection. Currently, there are more than 2,000 known human miRNA that have been found to influence gene regulation of essential biological pathways such as cellular development, proliferation, apoptosis and cellular signalling (1,2). From its intracellular origin, miRNA can be secreted extracellularly bound to lipoproteins (3) or secreted in cell-derived extracellular vesicles such as exosomes as a method of cell-to-cell communication (4). miRNA have been implicated in many diseases and have been shown to be taken up by distant cells as cargo in exosomes to influence disease pathogenesis and progression. Together, the pathogenic nature of miRNAs and their ability to be secreted extracellularly into biological fluids, where they remain relatively stable in exosomes, present miRNA as a promising biomarker.
Expression profiling of miRNA associated with disease has been explored where the majority of researchers have used peripheral blood (5,6) or cell-free serum or plasma (7,8). Although these sources are rich in miRNA, it can be difficult to differentiate disease-specific miRNA biomarkers from those expressed both in healthy and diseased patients. Recently, the development of using exosomes as a source of miRNA biomarkers has led to standardized protocols for isolating and analysing exosomes from cell lines which can now be applied to biological fluids (9,10). Exosomes can be isolated from peripheral blood using modified differential ultracentrifugation (UC) and, more recently, using exosomal isolation kits now available in the market. Early investigations into miRNA involved detection by qRT-PCR and microarrays; however, these methods limit the researcher to analysing a selected array of known miRNA using specific primers.
With the current advances in next-generation deep sequencing (NGS), the entire spectrum of known and novel miRNA can be profiled with minimal RNA input (11,12). Thus, the analysis of exosomal miRNA from biological samples such as blood is more feasible than ever before. Furthermore, the cargo contained in exosomes may provide an enriched population of miRNA free of endogenous RNA contaminants such as ribosomal RNA (rRNA). Therefore, these exosomes may house a disease-specific miRNA signature (13), which could be useful for diagnostic purposes. Using NGS, we set out to profile whole blood collected in PAXgene tubes (intracellular RNA), cell-free plasma and serum in comparison to exosomes isolated by UC and by a commercial kit with the aim to determine the optimal sample for miRNA biomarker discovery.
Materials and methods
Study sample
Samples used for this study were obtained from 3 healthy control patients comprising 2 males and 1 female. The mean age of the participants was 28.6±2.08. Whole blood (overnight fasting) was collected by venepuncture into Sarstedt S-Monovette serum-gel 7.5 ml tubes 01.1602.001 (Sarstedt, Germany), Sarstedt S-Monovette EDTA 7.5 ml tubes (Sarstedt, Germany) and PAXgene Blood RNA Tubes (PreAnalytiX, Australia). Collection into Sarstedt S-Monovette serum-gel 7.5 ml tubes for serum was left at RT for 30 minutes for coagulation before centrifugation at 1,800 g for 15 minutes. Collection into Sarstedt S-Monovette EDTA 7.5 ml tubes for plasma was maintained at 4°C for 10–15 minutes before centrifugation at 3,000 g for 10 minutes. The resulting plasma and serum was transferred into 1.5 ml Lo-bind DNA tubes (Eppendorf) and snap frozen immediately. Whole blood collected in PAXgene tubes were kept at RT for 2 hours before transferring to −20°C and then −80°C as per manufacturer's instructions. Informed consent was obtained from all participants and ethics were approved by the appropriate committees.
Exosome isolation by UC
Serum and plasma were diluted 1:2 with cold PBS before low-centrifugation at 500 g for 20 minutes to remove cellular debris. The supernatant was further diluted 1:5 and transferred to polycarbonate Type 70.1 Ti Beckman and Coulter tubes followed by UC at 11,000 g for 45 minutes to remove large membrane vesicles. The supernatant was then transferred to new tubes and ultracentrifuged at 100,000 g for 90 minutes. The resulting exosome pellet was resuspended in 250 µl of cold PBS for RNA extraction. Exosomes prepared for Western blotting and transmission electron microscopy (TEM) were resuspended in 150 µl of cold PBS. Samples were stored at −80°C until required. Exosomes isolated via the ultracentrifuge are denoted as plasma UC and serum UC throughout the manuscript.
Total RNA isolation
RNA extracted from PAXgene tubes were processed using the PAXgene Blood miRNA Kit (Qiagen, Australia) according to manufacturers’ protocol. RNA from cell-free plasma and serum including exosome samples isolated via the UC method were extracted using the miRNeasy Mini Kit (Qiagen, Australia). The manufacturers’ protocol was followed with a slight modification of the protocol involving the use of Trizol LS (Life Technologies, Australia) instead of QIAzol solution (Qiagen, Australia). Briefly, the exosome pellet resuspended in 250 µl of PBS was lysed with 750 µl of Trizol LS and 200 µl of chloroform. After centrifugation, the aqueous phase was removed and the miRNeasy protocol was followed. In addition, a plasma/serum exosomal RNA isolation kit (Norgen Biotek (NG), Canada) was used to isolate exosomes and extract total exosomal RNA according to the manufacturers’ protocol. Exosomal RNA isolated via NG Kit are denoted as plasma NG and serum NG throughout the manuscript. RNaseA treatment (100 ng/ml, Qiagen, Australia) where indicated involved incubation of samples at 37°C for 10 minutes. The total RNA yield (comprising of mostly small RNA), composition and quality was analysed using the Agilent 2,100 Bioanalyser for small RNA profiles with the Small RNA kit (Agilent, Tokyo).
Western immunoblotting
Cell-free (1 μl) and isolated exosomes from plasma and serum (1 μl) were resuspended in 19 µl lysis buffer (50.0 mM Tris pH 7 .4, 150 mM sodium chloride, 1% TritonX-100, 1% sodium-deoxycholic acid and complete ULTRA protease inhibitors (Roche)) on ice for 15 minutes. Samples were resuspended with 4×SDS loading buffer, heated at 95°C for 5 minutes and resolved on 4–12% Bis-Tris NUPAGE SDS-PAGE gels (Life Technologies, Mulgrave, VIC, Australia) and transferred onto Immobilon-P PVDF membrane (Millipore). Membranes were blocked with 3% skimmed milk buffer in PBS-T followed by incubation with primary antibodies to anti-Flotillin, anti-Hsp70, anti-CD63, anti-transferrin and anti-PrP (109–112). Immunoreactive bands were visualized using enhanced Chemiluminescence kit (Amersham Biosciences, Rydelmere, NSW, Australia), exposed to Hyperfilm (Amersham) and developed by the Exomat automated developing system.
Transmission electron microscopy
Isolated exosomes were fixed with 2% glutaraldehyde for 1 hour at room temperature and 6 µl absorbed onto glow discharged carbon-Formvar coated 200-mesh Cu grids (ProSciTech, Kirwan, QLD, Australia) for 5 minutes. Grids were washed twice with MilliQ water and stained with 1.5% uranyl acetate. TEM was performed at ×15,000 to ×36,000 magnification on a Tecnai G2 F30 (FEI, Eindhoven, NL) TEM operating at 300 kV. Electron micrographs were captured with a Gatan UltraScan® 1,000 2 k×2 k CCD camera (Gatan, Pleasanton, CA).
Size distribution analysis
Scanning ion occlusion sensing analysis was performed using the qNano system (Izon, Christchurch, New Zealand). Plasma and serum samples were diluted (1:4 in PBS) and passed through a 0.22 µm filter (Sartorius Australia, Dandenong South, Victoria, Australia). Samples were measured using an NP100A pore with an applied pressure of 10 cm.H2O. The current was recorded for a minimum of 60 s and 800 blockade events. Sample size distributions were calibrated by Izon Control Suite 2.2 using beads of known size, diluted in PBS and measured at identical settings.
Small RNA deep sequencing and bioinformatics analysis
PAXgene, cell-free and exosomal RNA was converted into cDNA libraries using the Ion Total RNA-Seq Kit V2 (Life Technologies, Australia) and prepared for deep sequencing as described previously (12). Pooled libraries with unique barcodes were loaded on 318 sequencing chips and run on the Ion Torrent Personal Genome Machine (Life Technologies, Australia). The Torrent Suite 3.4.1 was used to manage the Ion Torrent PGM to process raw signals and perform base calling. The sequences are then assessed for quality and primer-adapter sequences are trimmed by the Torrent Suite software, followed by alignment to the human reference genome (HG19). The trimmed and aligned data was transferred to Partek Genomics Suite and mapped to known miRNA using miRBase V.20 and Ensembl Release 74. The number of reads for each miRNA were normalized to reads per million (RPM) across all samples. Samples containing less than 5 RPM were removed. Partek Genomics suite and statistical package was used to perform statistical analysis, hierarchical clustering and to identify unique miRNA in each sample type. Data have been uploaded to Vesiclepedia (http://microvesicles.org/).
Results
Exosomal small RNA including miRNA are protected against RNaseA treatment
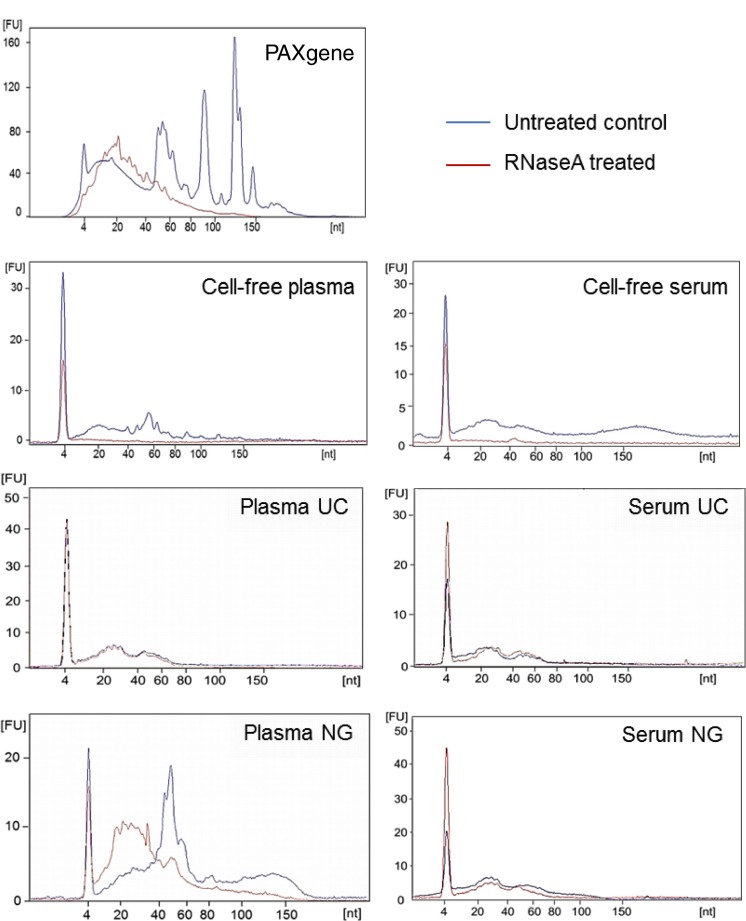
To compare the profiles of miRNA found in different blood components, whole blood, cell-free blood (serum and plasma) and exosomes were collected and prepared from 3 age-matched healthy individuals according to the study design depicted in Fig. 1 for NGS. One PAXgene tube was collected per individual to isolate intracellular RNA and matched whole blood was separated for cell-free plasma and serum. One aliquot remained cell-free whereas the remaining was further processed for exosomal isolation via differential UC or exosomal RNA isolation by the Plasma/Serum exosomal RNA kit from NG. The yield of small RNA obtained upon RNA extraction is displayed in Table I. To investigate whether circulating miRNAs are degraded or resistant to the presence of RNase, blood cell pellets and cell-free blood were treated with RNaseA (100 ng/ml) before RNA extraction or exosomal isolation as indicated in Fig. 1. Whole blood collected by PAXgene tubes resulted in high-quality RNA isolations (RNA integrity number of 9, data not shown) and a small RNA profile containing abundant peaks indicative of cellular RNA containing miRNA, rRNA and transfer RNA (tRNA) (Fig. 2). PAXgene tubes contain additives that stabilize RNA upon collection thus, preventing RNA degradation. As expected, RNaseA treatment of the blood cell pellet resulted in the degradation of RNA (Fig. 2). RNA isolated from cell-free plasma and serum did not contain rRNA or tRNA but consisted of RNA fragments up to 200 nt. The percentage of miRNA in cell-free plasma and serum was 53.9±8.0% and 53.3±10.3%, respectively (Table I). In support of the literature, circulating miRNA in cell-free plasma and serum are not protected against the presence of RNAse (Fig. 2) (14). The concentration of RNase in the circulatory system has not been defined and it would be difficult to determine the degree of RNA degradation biologically. Deep sequencing would provide a better insight into the identity of these small RNA fragments (intact or degraded) through sequence alignment to the human genome.
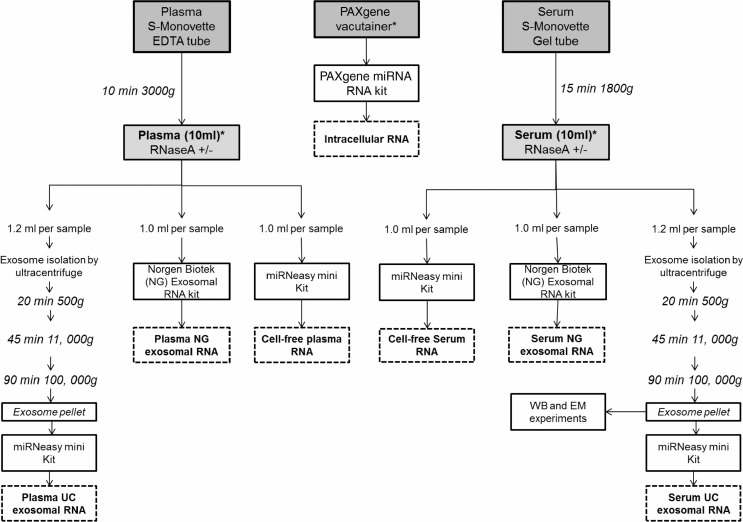
Fig. 1.
Work flow of study design and sample processing. Whole blood from 3 different individuals was collected by venepuncture into each tube using a Multi-fly and processed to analyse intracellular, cell-free and exosomal miRNA. Asterisks indicate the point of RNaseA treatment (100 ng/ml, 37°C for 10 minutes) to investigate RNA degradation in these samples. The workflow outlines the sample collection and preparation from 1 individual. The number of tubes collected from each volunteer was: 2×PAXgene 2.5 ml tubes, 3×Sarstedt S-Monovette serum-gel 7.5 ml tubes and 3×Sarstedt S-Monovette EDTA 7.5 ml tubes. Upon centrifugation of the Sarstedt S-Monovette EDTA tubes, approximately 10 ml of plasma was obtained across 3 Sarstedt S-Monovette tubes which are then separately aliquoted into Lo-Bind DNA tubes (4×1 ml, 2×1.2 ml tubes) for RNA analysis and deep sequencing. The remaining plasma was aliquoted for Western immunoblotting (WB, 1.2 ml), transmission electron microscopy (EM, 1.2 ml) and qNano (1 ml) analysis. For the RNA work involving RNaseA treatment, samples were allocated for an untreated control and RNaseA treatment: 2×1.2 ml for the ultracentrifugation exosomal RNA isolation (Plasma UC), 2×1 ml for the Norgen Biotek exosomal RNA isolation (Plasma NG), and 2×1 ml aliquot was reserved for cell-free plasma RNA isolation. The collection process and sample allocation are repeated for serum collection. Exosomes isolated from serum via the ultracentrifuge are denoted as Serum UC. Exosomal RNA isolated by the Norgen Biotek Kit are denoted as Serum NG. As for the 2×PAXgene tubes, RNA is isolated from 2.5 ml of whole blood per tube and isolated as recommended by the manufacturers protocol. One tube was treated with RNaseA and one was left untreated.
Table I.
Yield and percentage of miRNA extracted from intracellular, cell-free and exosomal samples prepared from plasma and serum
| PAXgene | Plasma cell-free | Serum cell-free | Plasma UC exosomes | Serum UC exosomes | Plasma NG exosomes | Serum NG exosomes | |
|---|---|---|---|---|---|---|---|
| [small RNA] ng | 196.7±46.3 | 4.1±0.08 | 6.3±1.4 | 6.6±1.6 | 8.2±1.0 | 16.8±1 | 8.3±0.9 |
| [miRNA] ng | 50.9±61.4 | 2.2±0.5 | 3.4±1.0 | 4.7±2.1 | 5.0±1.6 | 4.9±1.8 | 3.7±0.7 |
| % miRNA | 30.4±40.4 | 53.9±8.0 | 53.3±10.3 | 67.9±17.0 | 60.9±17.4 | 29.2±11.3 | 44.8±4.5 |
Values are mean±SD, n=3. Small RNA and miRNA yield was determined by the Small RNA Bioanalyser assay. The concentration of small RNA is measured between 6 and 200 nt and the concentration of miRNA is measured between 10 and 40 nt.
Fig. 2.
Small RNA profiles extracted from intracellular, cell-free and exosomal isolation from blood before and after RNaseA treatment. RNA was extracted from samples and run on a Small RNA Bioanalyser assay. Experiments shown here are representative of samples collected from 1 volunteer.
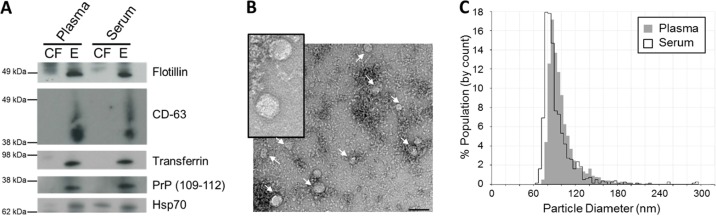
Exosomes were then isolated to determine whether they provided a protective vesicle against degradation of transport miRNA by RNase. Upon differential UC, exosome pellets from both cell-free plasma and serum were positive for exosomal markers, flotillin and CD-63 (Fig. 3A). Exosomes were also found to be enriched with transferrin receptors which are largely responsible for the import of iron into cells via endosomes. Endosomes, similar to exosomes, are multi-vesicular bodies that are produced from the endosomal protein sorting system. Furthermore, prion protein (PrP) was detected in plasma and serum exosomes. PrP has been previously described to be enriched in highly purified exosomes isolated by sucrose gradients (15). As expected, the constitutively expressed Hsp70 protein was detected in both cell-free and exosome pellets. The morphology of vesicles isolated by UC were found to be consistent with previous reports, measuring 70–110 nm in size as observed by electron microscopy (Fig. 3B) and the qNano instrument (Fig. 3C). Upon RNA extraction, exosomes isolated from cell-free plasma and serum using differential UC contained small RNA of up to 80 nt in length which were protected upon RNaseA treatment (Fig. 2). This suggests that exosomes provide an RNase free vesicle for transport within the RNase rich environment of the circulatory system. Approximately 6–9 ng of small RNA (60–80% miRNA) was obtained from exosomes isolated from 1.2 ml of plasma or serum (Table I).
Fig. 3.
Characterization of plasma and serum exosomes isolated by differential ultracentrifugation. (A) Western immunoblotting of exosomal markers flotillin, CD-63, transferrin, PrP (109–112) and Hsp70 in cell-free (CF) and exosomal (E) samples in plasma and serum. (B) Plasma and serum exosomes were analysed under electron microscopy which displayed the same morphology. Plasma exosomes are shown here. Insert is a larger magnification of the exosomal vesicles. Bar = 100 nm (C) Size distribution of exosomes analysed by the qNano particle counter. Experiments shown here are representative of samples collected from 1 volunteer.
We then evaluated the use of a commercial exosomal RNA isolation kit to determine whether the RNA species extracted were similar to those extracted from exosomes isolated by UC. The plasma/serum exosome RNA Isolation kit from NG is a centrifugation-independent kit which enriches for exosomal RNA using a proprietary column. RNA extracted from plasma using the plasma/serum exosomal RNA kit by NG was found to contain 29.2±11.3% miRNA (Table I). Interestingly, a large presence of tRNA was detected in plasma samples isolated using this commercial kit (Fig. 2) which increased the yield of small RNA extraction to 16.8±1.0 ng (Table I). However, upon RNaseA treatment of the plasma, circulating tRNA and potentially other RNA species were degraded indicative that the kit isolates non-exosomal contaminants in addition to the desired exosomal material. It is possible that the tRNA is a cellular contaminant due to haemolysis occurring during sample collection (16). In comparison, tRNA was not observed in exosomes isolated by UC or in cell-free plasma (Fig. 2) hence, the UC protocol did not pellet non-exosomal RNA or cellular RNA contaminants. This highlights the importance of standardizing sample collection, centrifugation of blood and handling for exosomal RNA work. The commercial kit performed better with serum samples (Fig. 2) which isolated 8.3±0.9 ng of small RNA (44.8±4.5% miRNA, Table I). There was no evidence of non-exosomal or cellular RNA contaminants in the NG serum exosomes, and the RNA profile resembled those observed in serum exosomes isolated by UC (Fig. 2).
Exosomes are enriched with miRNA
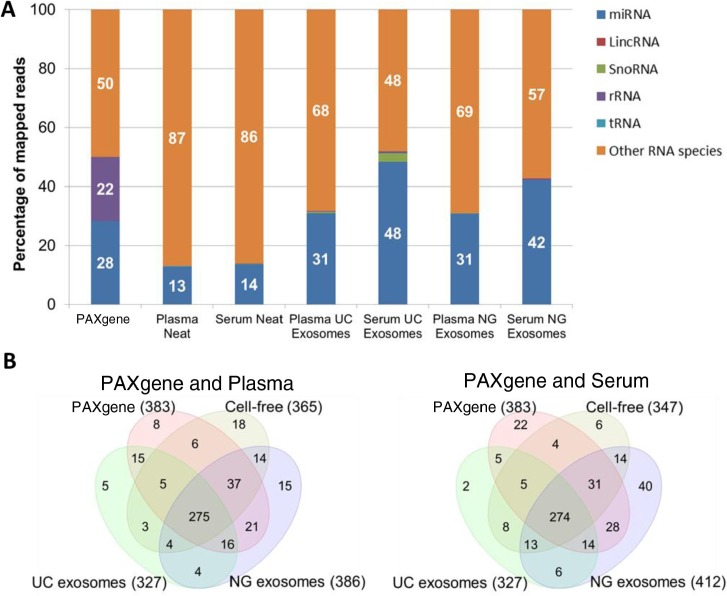
Small RNA libraries of the samples described above were constructed for small RNA-seq by NGS. The number of miRNA identified in this study was 943 of 2,233 known mature miRNA from miRBase V.20. Mapping to Ensembl Release 74 was performed to identify other non-coding RNA such as tRNA, rRNA, small nucleolar RNA (snoRNA) and LincRNA. The average across all 3 samples was calculated to obtain a representative profile of biological diversity between patients (standard deviations of reads in Supplementary file). The PAXgene tube isolated the highest yield of small RNA (Table I), 28% of the small RNA reads were mapped to miRNA (Fig. 4). The remaining reads mapped to other RNA species such as coding mRNA. Size selection to remove the majority of rRNA and tRNA was performed during library construction hence, there is a low percentage of mapping (<1%) to rRNA and tRNA in cell-free and exosomal samples. The high abundance of rRNA and tRNA in blood cells can saturate downstream analysis of miRNA thus ribosomal removal or additional size selection steps are recommended before deep sequencing. As there is a high abundance of rRNA in intracellular RNA, 22% of reads from PAXgene tubes mapped to 5S RNA however, 18S and 28S was successfully removed. Cell-free plasma and serum were found to contain the least amount of reads mapping to miRNA (12–14%). Overall, exosomes isolated from serum contained the highest percentage of miRNA (42–48%) compared to plasma (30%). Serum exosomes isolated by the UC produced the greatest percentage of miRNA (48%) followed closely by the NG kit (42%). The biological enrichment of miRNA in exosomes serves as an advantage when performing deep sequencing as the removal of other RNA contaminants by size selection is not required. An insignificant amount of other non-coding small RNA were detected (<1%) however, serum exosomes isolated by UC were found to contain 2.3% snoRNA. The most abundant snoRNA found in the serum exosomes were SNORD100, SNORD27 and SNORD31 (Supplementary file). The functions of these snoRNA species are largely unknown; however, they have been detected in myeloma (18).
Fig. 4.
Percentage and number of known miRNA species detected in intracellular, cell-free and exosomal samples by NGS. Raw reads were aligned to the human genome (HG19) and mapped to miRBase V.20 and other small RNA from Ensembl Release 74 followed by normalization of raw reads to RPM. The mean of reads per miRNA (n=3) was calculated. A) Percentage of total reads mapped to non-coding small RNA and other RNA species identified by deep sequencing. B) Venn diagrams showing unique and common miRNA detected in different components of blood. miRNA with read counts >5 reads per million were shown for comparison. miRNA identified in this study was uploaded to http://www.microvesicles.org (17).
Upon removing low abundant miRNA with less than 5 normalized reads across all sample types, 488 miRNA of the original 943 miRNA were used for further analysis. Of these, the numbers of miRNA species found in intracellular components of blood (PAXgene) compared to samples isolated from plasma or serum are represented in Fig. 4B. Plasma and serum NG exosomal samples contained the highest number of miRNA species, 386 and 412, respectively. It is highly likely that the NG kit isolates a small percentage of non-exosomal miRNA as there is a large overlap of miRNA found in the PAXgene tube and cell-free samples. Interestingly, the NG exosomal RNA Kit isolated additional miRNA species compared to the intracellular profile of miRNA isolated by the PAXgene tube (383). For example, serum NG exosomal RNA samples were found to have 412 miRNA species including 40 different miRNA species not present in PAXgene tubes, cell-free and UC exosome samples. Similarly, the NG kit isolated an additional 15 different miRNA in plasma. These differences could be due to the binding capacity of the proprietary column depending on the diversity and level of contaminants from haemolysis, thus affecting the efficiency of the NG Kit. Although the UC exosomal samples were found to contain the least number of miRNA species (327), the number of reads per miRNA were greater compared to cell-free samples for most miRNA species (Table II and Fig. 4A).
Table II.
Normalized reads of the top 25 miRNA detected in PAXgene intracellular samples compared to cell-free and exosomal samples prepared from plasma and serum
| Plasma | Serum | ||||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| miRNA | PAXgene | NG exosomes | Cell-free | UC exosomes | NG exosomes | Cell-free | UC exosomes |
| hsa-miR-451a | 124,894 | 19,012 | 27,779 | 141,194 | 109,409 | 33,598 | 42,650 |
| hsa-miR-191-5p | 25,960 | 11,712 | 9,011 | 12,422 | 7,711 | 4,376 | 2,861 |
| hsa-miR-486-5p | 23,982 | 2,118 | 4,461 | 10,788 | 12,477 | 11,102 | 4,838 |
| hsa-miR-223-3p | 20,034 | 42,972 | 30,254 | 16,438 | 40,852 | 14,803 | 12,628 |
| hsa-miR-484 | 13,061 | 3,987 | 3,225 | 6,957 | 3,312 | 3,077 | 1,357 |
| hsa-miR-16-5p | 7,463 | 3,958 | 1,355 | 7,070 | 14,891 | 3,816 | 8,673 |
| hsa-miR-126-3p | 4,729 | 12,298 | 3,711 | 1,674 | 7,025 | 1,431 | 2,019 |
| hsa-miR-425-5p | 4,009 | 712 | 726 | 3,116 | 731 | 547 | 260 |
| hsa-miR-26a-5p | 3,103 | 6,764 | 533 | 730 | 2,598 | 736 | 804 |
| hsa-miR-486-3p | 2,821 | 432 | 714 | 1,013 | 1,599 | 1,393 | 419 |
| hsa-miR-185-5p | 2,771 | 2,215 | 1,413 | 1,222 | 2,273 | 1,841 | 996 |
| hsa-miR-126-5p | 2,665 | 6,506 | 859 | 412 | 4,271 | 900 | 1,006 |
| hsa-miR-19b-3p | 2,345 | 1,342 | 1,081 | 3,268 | 1,657 | 569 | 518 |
| hsa-miR-103a-3p | 1,938 | 2,207 | 986 | 695 | 1,294 | 506 | 419 |
| hsa-miR-103b | 1,938 | 2,207 | 986 | 695 | 1,294 | 506 | 419 |
| hsa-miR-93-5p | 1,922 | 1,172 | 514 | 936 | 1,754 | 831 | 558 |
| hsa-miR-17-5p | 1,585 | 1,635 | 960 | 650 | 3,472 | 949 | 972 |
| hsa-miR-15b-5p | 1,395 | 1,397 | 173 | 2,003 | 943 | 250 | 242 |
| hsa-miR-30d-5p | 1,350 | 125 | 249 | 957 | 226 | 113 | 72 |
| hsa-miR-342-3p | 1,264 | 810 | 929 | 799 | 1,714 | 965 | 640 |
| hsa-miR-92a-3p | 1,170 | 729 | 241 | 884 | 1,416 | 577 | 581 |
| hsa-let-7g-5p | 1,063 | 1,434 | 328 | 847 | 2,578 | 551 | 812 |
| hsa-miR-320a | 1,015 | 1,333 | 2,895 | 300 | 1,764 | 2,089 | 579 |
| hsa-miR-150-5p | 1,015 | 254 | 353 | 751 | 1,306 | 572 | 923 |
| hsa-miR-18a-5p | 925 | 1,850 | 280 | 164 | 1,592 | 399 | 194 |
Reads are normalized to reads per million. Standard deviation and other miRNA can be found in supplementary data.
The mean (n=3) of normalized reads from the top 25 as ordered by PAXgene tubes in comparison to plasma and serum samples can be seen in Table II. The reads of the remaining miRNA can be found in Supplementary file. Within the 25 most abundant miRNA, not surprisingly, blood cells obtained the highest amount of reads owing to the presence of WBCs which are the richest source of RNA. A significant amount of reads was observed for hsa-miR-451a which was also heavily present in the other samples. Other highly abundant miRNA found in all samples were hsa-miR-191-5p, hsa-miR-486-5p, hsa-miR-223-3p and hsa-miR-484.
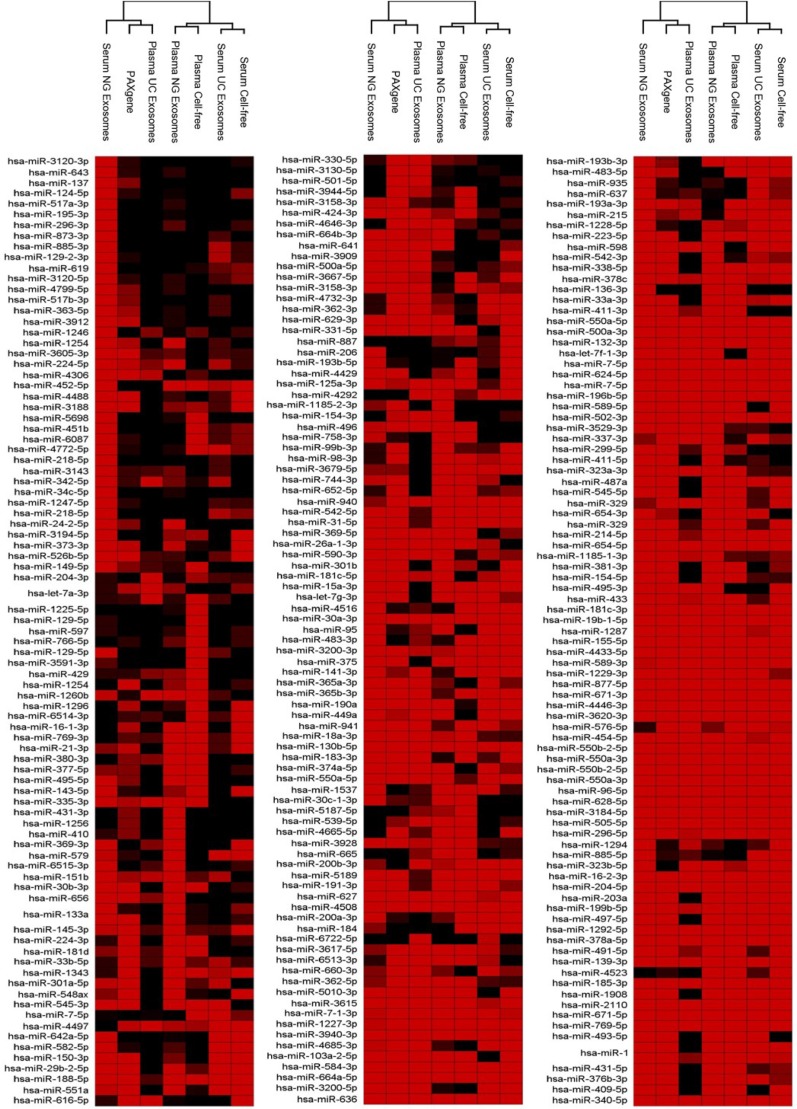
Upon unsupervised hierarchical clustering of deep sequencing data, 2 major clusters across all samples were identified (Fig. 5). Cluster 1 comprised serum NG exosomes, PAXgene and plasma UC exosomes. The PAXgene profile and plasma UC exosomes were found to show more similarity compared to the serum NG exosomes. Cluster 2 comprised plasma NG exosomal RNA, cell-free plasma, serum UC exosomes and cell-free serum. As observed in Fig. 4B, plasma NG exosomal samples share more similarity to cell-free plasma. Serum UC exosomes displayed the most similarity to cell-free serum owing to the NG kit isolating 40 different miRNA not present in other samples (Fig. 5). Figure 5 also illustrates the presence and absence of all 488 abundant miRNA detected across all sample types.
Fig. 5.
Presence and absence of the highly abundant miRNAs identified across intracellular blood, cell-free samples and exosomes samples. Raw reads were aligned to the human genome (HG19) and mapped to miRBase V.20 followed by normalization of raw reads to RPM. The mean was calculated across 3 volunteer samples. miRNA with read counts >5 reads per million were shown for comparison. Hierarchical clustering was performed across samples and miRNA. Data has been uploaded to http://microvesicles.org/.
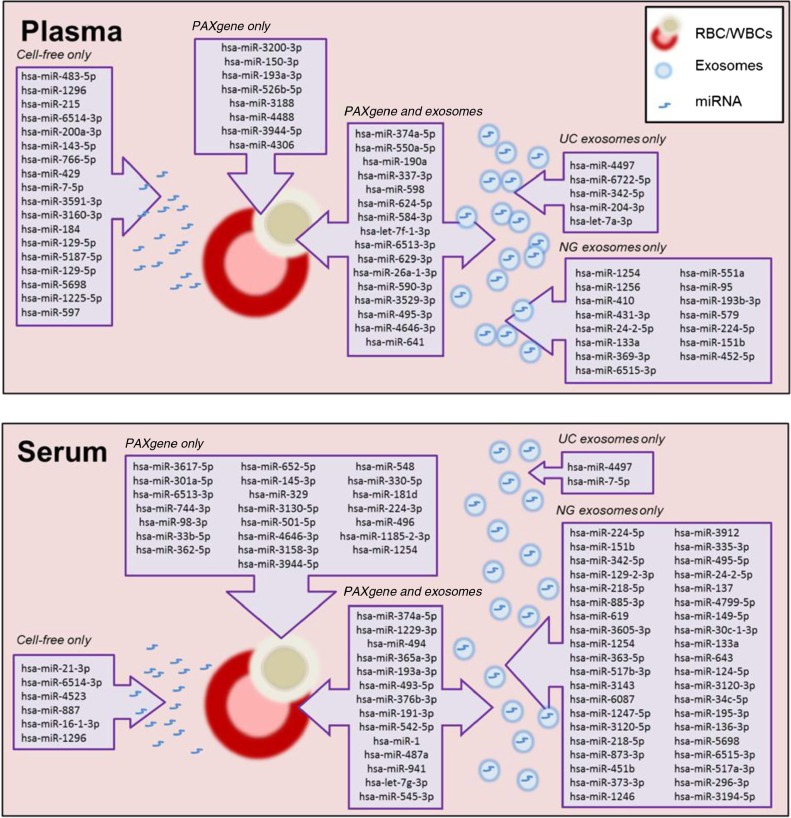
We next investigated the predicted function of the uniquely expressed miRNA found in PAXgene tubes, cell-free samples, exosomal samples, and those found both in PAXgene and exosomes (Fig. 6). Using TargetScan 6.2, the 30 miRNA uniquely found in PAXgene tubes were found to target 4,469 gene targets (Figure 4B, Figure 6 and Supplementary file). These combined targets were then subjected to pathway prediction software collected by the Kyoto Encyclopedia of Genes and Genome (KEGG) to predict the pathways in which these targeted genes collectively regulate. The most targeted pathways found in intracellular blood are Wnt, focal adhesion, glioma, phosphatidylinositol and melanogenesis signalling pathways (Table III). The cell-free samples contained miRNA that targets neuron development and survival such as axon guidance and neurotrophin signalling pathways. Unique miRNA packaged into exosomes targeted neuronal signalling through GABAergic, axon guidance, endocytosis and glutamatergic synapse signalling pathways.miRNA found in both intracellular blood and exosomes targeted TGF signalling and pathways in cancer. The increased enrichment scores seen in exosomes compared to other blood components support the role of exosomes in cell-to-cell communication by targeting these neuronal signalling pathways through specific packaging of miRNA.
Fig. 6.
Schematic summary of unique miRNA detected in intracellular, cell-free and exosomal samples prepared from plasma and serum.
Table III.
Predicted pathways targeted by miRNAs expressed in intracellular, cell-free and exosomal samples
| Enrichment score | P | % of genes in pathway that are present | |
|---|---|---|---|
| PAXgene intracellular miRNA | |||
| Wnt signalling pathway | 17.7 | 1.96E-08 | 10.2 |
| Focal adhesion | 11.5 | 1.01E-05 | 7.8 |
| Glioma | 11.2 | 1.38E-05 | 10.0 |
| Phosphatidylinositol signalling system | 10.4 | 3.12E-05 | 10.3 |
| Melanogenesis | 10.4 | 3.15E-05 | 9.4 |
| Cell-free miRNA | |||
| Neurotrophin signalling pathway | 14.2 | 6.92E-07 | 7.2 |
| Focal adhesion | 11.5 | 1.01E-05 | 6.0 |
| Oocyte meiosis | 11.1 | 1.45E-05 | 7.3 |
| Wnt signalling pathway | 11.1 | 1.45E-05 | 6.8 |
| Axon guidance | 10.8 | 1.98E-05 | 6.6 |
| Exosomal miRNA | |||
| Wnt signalling pathway | 23.8 | 4.67E-11 | 15.9 |
| GABAergic synapse | 17.2 | 3.28E-08 | 17.0 |
| Axon guidance | 16.1 | 9.94E-08 | 13.7 |
| Endocytosis | 15.5 | 1.88E-07 | 12.3 |
| Glutamatergic synapse | 14.3 | 6.10E-07 | 14.1 |
| PAXgene and exosomes | |||
| Wnt signalling pathway | 24.1 | 3.54E-11 | 11.2 |
| Melanogenesis | 12.0 | 6.38E-06 | 9.6 |
| Glioma | 10.0 | 4.56E-05 | 9.4 |
| TGF-beta signalling pathway | 9.5 | 7.66E-05 | 8.9 |
| Pathways in cancer | 8.9 | 1.41E-04 | 6.2 |
miRNA targets were analysed by TargetScan 6.0 and pathway prediction was analysed using KEGG pathways.
The top 5 pathways affected are shown here.
Discussion
Diagnostic testing using human blood is considered to be relatively non-invasive and a cost-effective method. Circulating miRNAs in the blood has now offered a new form of biomarker for disease diagnosis. The number of miRNA associated with disease is expanding as a result of new deep sequencing technologies. However, the ability to report a reliable panel of biomarkers has seen challenges with regard to obtaining high sensitivity and specificity upon validation. In addition, consensus has not been reached with regard to sample collection (such as plasma or serum), source of RNA from the sample (for example cell-free or exosomes) and the method used for isolation of exosomes (UC or commercial kits) for analysing miRNA in blood. Reproducibility is also problematic during validation studies largely due to possible changes of miRNA expression in the bloodstream that originate from multiple tissue and organs. Here, we have performed a comprehensive study using a workflow to carry out a complete comparison between samples that are commonly used in miRNA studies. Using this study design, we were also able to investigate whether exosomes indeed contain a source of enriched miRNA compared to non-exosomal samples. For comparison, we isolated RNA from blood samples that are most commonly used in miRNA biomarker studies. This included whole blood collected in PAXgene tubes, cell-free plasma and serum, and exosomes isolated from plasma and serum. Furthermore, we addressed the debate concerning whether to continue the effort with the time consuming UC isolations of exosomes or to use a time-efficient commercial exosomal kit for biomarkers studies. To obtain a comprehensive profile of all these samples, we performed small RNA sequencing using NGS.
Initial studies involving miRNA analysed cellular components of blood (red blood cells, white blood cells and platelets) as they provide a rich source of RNA (5,19). The highest yield of RNA is obtainable from intracellular RNA (PAXgene tubes) which comprise a large variety of coding and non-coding RNA species. There is a possibility that disease related miRNA biomarkers are diluted amongst other RNA species leading to greater signal-to-noise ratio. Analysing miRNA profiles from intracellular fractions for disease biomarkers may not be disease specific as mature RBCs lack a nucleus and therefore DNA transcription or RNA processing does not occur. The majority of RNA extracted from blood is derived from WBCs which encompasses 5 different cell types of the immune system. The study of miRNA in WBCs would be suitable for infectious diseases and immunity. Figure 6 schematically summarizes the various miRNA that were uniquely detected in the various samples analysed in this study. Unique miRNA detected in the cellular fraction were found to be involved in controlling cellular homeostasis through targeting regulatory cell and receptor signalling pathways (Table III).
The least amount of small RNA was obtained in cell-free plasma and serum. The spectra of miRNA were slightly different between cell-free plasma and serum which may be due to possible cellular contamination or the coagulation process of plasma and serum during preparation. This highlights the requirement for standardization of sample preparation for miRNA biomarker studies. A recent study investigated the effect of various plasma processing conditions on miRNA levels and observed high levels of miRNA contamination from platelets within plasma (20). Hence, recommendations for plasma handling include platelet removal, monitoring of RBC contamination or performing platelet counts. Conceivably, those undertaking miRNA biomarker studies would favour serum separator tubes to avoid additional handling during sample preparation. The increased number of detectable miRNA in plasma compared to serum was also observed in another study using Taqman qPCR panels (21). Despite this, there are less reads on average per miRNA in plasma compared to serum. The miRNA found uniquely in cell-free samples were predicted to target neuronal signalling pathways however, the percentage of genes targeted by miRNAs is lower than those collectively found in exosomes (Table III). Presumably, the presence of miRNA in cell-free samples is diluted in the plasma volume. Hence, larger volumes of cell-free plasma or serum would be required to assay miRNA within a detectable range by qRT-PCR.
Exosomes were found to be enriched in miRNA. The top 10 most abundant miRNA found in exosomes were hsa-miR-451a, hsa-miR-223-3p, hsa-miR-16-5p, hsa-miR-191-5p, hsa-miR-486-5p, hsa-miR-126-3p, hsa-miR-484, hsa-miR-126-5p, hsa-miR-26a-5p and hsa-let-7b-5p which displayed more reads compared to most miRNA detected in cell-free samples (Table II). Hsa-miR-451a regulates erythroid development and is expressed under homeostatic conditions (22,23), and hsa-miR-223 is expressed throughout granulocytic differentiation into mature peripheral blood granulocytes (24). In addition, miR-126 (25), miR-191 (26) and miR-26 (27) have been implicated in the mouse brain through regulation of various neuronal proteins such as brain derived neurotropic factor and amyloid precursor protein. Exosomes have recently been successfully isolated from brain tissue (28) and potentially pass through the blood brain barrier (29). The detection of these brain associated miRNA further suggests that exosomes provide a method of communication between the brain and distant organs. Furthermore, miR-16 and let-7b have been highly associated with exosomes in a number of reports (10,30) (31) and have been used as endogenous controls (32). Interestingly, pathway analysis indicated that the unique exosomal miRNA (UC and NG) highly targeted neuronal signalling pathways (Table III).
Although the use of miRNA biomarkers has been widely applied to cancer diagnostics (33–35), it has not been thoroughly investigated in neurodegenerative diseases. We have previously identified a panel of miRNAs that are deregulated in prion diseases which is a neurodegenerative disorder affecting humans (10). The nervous system is a rich source of miRNA expression and a number of deregulated miRNA have been found to contribute to other neurodegenerative disorders such as Alzheimer's disease, including hsa-miR-9, hsa-miR-20a and hsa-miR-132 (36). In this study, we have profiled baseline miRNA expression in blood obtained from 3 age matched healthy volunteers. It would be attractive to profile exosomal miRNA biomarkers and perform differential analysis of miRNA expression between healthy control and a disease condition such as neurodegenerative diseases.
Although there are increasing reports on the application of miRNA as biomarkers for various disease, this study highlights that the profile and concentration of miRNA are different between the intracellular, cell-free and exosomal components of blood. Furthermore, differences in profiles are also observed between plasma and serum samples. Sample type should be deliberated before profiling miRNA for biomarker discovery in blood samples. Biological diversity between patients should also be considered upon discovery studies investigating miRNA biomarkers. Differential expression analysis between healthy and disease samples across a large cohort should ideally be performed on highly abundant miRNA. This ensures success upon biomarker validation across the population and large cohorts. Furthermore, abundantly expressed miRNA will be within detectable levels of molecular technology platforms used for validation such as qRT-PCR.
Acknowledgements
We thank the Advanced Microscopy Facility at Bio21 Molecular Science and Biotechnology Institute, The University of Melbourne, for electron microscopy facilities.
To access the supplementary material to this article, please see Supplementary files under Article Tools online.
Conflict of interest and funding
This work was supported by the Australian Research Council (FT100100560 to AFH) and the National Health and Medical Research Council (628946 to AFH). LC was supported by a University of Melbourne Early Career Researcher Project Grant for the work, BJS is the recipient of an Australian Postgraduate Award, and AFH is an ARC Future Fellow.
References
- 1.Griffiths-Jones S. The microRNA registry. Nucleic Acids Res. 2004;32:D109–11. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 3.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–8. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 5.Schipper H, Maes O, Chertkow H, Wang E. MicroRNA expression in Alzheimer blood mononuclear cells. Gene Regul Syst Bio. 2007;1:263–74. doi: 10.4137/grsb.s361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaz C, Ahmad HM, Sharma P, Gupta R, Kumar L, Kulshreshtha R, et al. Analysis of microRNA transcriptome by deep sequencing of small RNA libraries of peripheral blood. BMC Genomics. 2010;11:288. doi: 10.1186/1471-2164-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheinerman KS, Tsivinsky VG, Crawford F, Mullan MJ, Abdullah L, Umansky SR. Plasma microRNA biomarkers for detection of mild cognitive impairment. Aging (Albany NY) 2012;4:590–605. doi: 10.18632/aging.100486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsujiura M, Ichikawa D, Komatsu S, Shiozaki A, Takeshita H, Kosuga T, et al. Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer. 2010;102:1174–9. doi: 10.1038/sj.bjc.6605608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coleman BM, Hanssen E, Lawson VA, Hill AF. Prion-infected cells regulate the release of exosomes with distinct ultrastructural features. FASEB J. 2012;26:4160–73. doi: 10.1096/fj.11-202077. [DOI] [PubMed] [Google Scholar]
- 10.Bellingham SA, Coleman BM, Hill AF. Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res. 2012;40:10937–49. doi: 10.1093/nar/gks832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng L, Quek CY, Sun X, Bellingham SA, Hill AF. The detection of microRNA associated with Alzheimer's disease in biological fluids using next-generation sequencing technologies. Front Genet. 2013;4:150. doi: 10.3389/fgene.2013.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng L, Sun X, Scicluna BJ, Coleman BM, Hill AF. Characterization and deep sequencing analysis of exosomal and non-exosomal miRNA in human urine. Kidney Int. 2013 doi: 10.3402/jev.v2i0.19671. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 13.Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One. 2012;7:e30679. doi: 10.1371/journal.pone.0030679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koberle V, Pleli T, Schmithals C, Augusto Alonso E, Haupenthal J, Bonig H, et al. Differential stability of cell-free circulating microRNAs: implications for their utilization as biomarkers. PLoS One. 2013;8:e75184. doi: 10.1371/journal.pone.0075184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vella LJ, Sharples RA, Lawson VA, Masters CL, Cappai R, Hill AF. Packaging of prions into exosomes is associated with a novel pathway of PrP processing. J Pathol. 2007;211:582–90. doi: 10.1002/path.2145. [DOI] [PubMed] [Google Scholar]
- 16.Kirschner MB, Edelman JJ, Kao SC, Vallely MP, van Zandwijk N, Reid G. The impact of hemolysis on cell-free microRNA biomarkers. Front Genet. 2013;4:94. doi: 10.3389/fgene.2013.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalra H, Simpson RJ, Ji H, Aikawa E, Altevogt P, Askenase P, et al. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012;10:e1001450. doi: 10.1371/journal.pbio.1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez-Corral L, Mateos MV, Corchete LA, Sarasquete ME, de la Rubia J, de Arriba F, et al. Genomic analysis of high-risk smoldering multiple myeloma. Haematologica. 2012;97:1439–43. doi: 10.3324/haematol.2011.060780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu S, Yu B, Chertkow H, Wang E, Schipper H. Transcriptional profiling of Alzheimer blood mononuclear cells by microarray. Neurobiol Aging. 2007;28:1795–809. doi: 10.1016/j.neurobiolaging.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 20.Cheng HH, Yi HS, Kim Y, Kroh EM, Chien JW, Eaton KD, et al. Plasma processing conditions substantially influence circulating microRNA biomarker levels. PLoS One. 2013;8:e64795. doi: 10.1371/journal.pone.0064795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang K, Yuan Y, Cho JH, McClarty S, Baxter D, Galas DJ. Comparing the MicroRNA spectrum between serum and plasma. PLoS One. 2012;7:e41561. doi: 10.1371/journal.pone.0041561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu D, dos Santos CO, Zhao G, Jiang J, Amigo JD, Khandros E, et al. miR-451 protects against erythroid oxidant stress by repressing 14-3-3zeta. Genes Dev. 2010;24:1620–33. doi: 10.1101/gad.1942110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rasmussen KD, Simmini S, Abreu-Goodger C, Bartonicek N, Di Giacomo M, Bilbao-Cortes D, et al. The miR-144/451 locus is required for erythroid homeostasis. J Exp Med. 2010;207:1351–8. doi: 10.1084/jem.20100458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–9. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 25.Sonntag KC, Woo TU, Krichevsky AM. Converging miRNA functions in diverse brain disorders: a case for miR-124 and miR-126. Exp Neurol. 2012;235:427–35. doi: 10.1016/j.expneurol.2011.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mellios N, Huang HS, Grigorenko A, Rogaev E, Akbarian S. A set of differentially expressed miRNAs, including miR-30a-5p, act as post-transcriptional inhibitors of BDNF in prefrontal cortex. Hum Mol Genet. 2008;17:3030–42. doi: 10.1093/hmg/ddn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smirnova L, Grafe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG. Regulation of miRNA expression during neural cell specification. Eur J Neurosci. 2005;21:1469–77. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- 28.Haqqani AS, Delaney CE, Tremblay TL, Sodja C, Sandhu JK, Stanimirovic DB. Method for isolation and molecular characterization of extracellular microvesicles released from brain endothelial cells. Fluids Barriers CNS. 2013;10:4. doi: 10.1186/2045-8118-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–5. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 30.Patel RS, Jakymiw A, Yao B, Pauley BA, Carcamo WC, Katz J, et al. High resolution of microRNA signatures in human whole saliva. Arch Oral Biol. 2011;56:1506–13. doi: 10.1016/j.archoralbio.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park NJ, Zhou H, Elashoff D, Henson BS, Kastratovic DA, Abemayor E, et al. Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin Cancer Res. 2009;15:5473–7. doi: 10.1158/1078-0432.CCR-09-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koga Y, Yasunaga M, Moriya Y, Akasu T, Fujita S, Yamamoto S, et al. Exosome can prevent RNase from degrading microRNA in feces. J Gastrointest Oncol. 2011;2:215–22. doi: 10.3978/j.issn.2078-6891.2011.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao X, Zhang R, Qu X, Zhao M, Zhang S, Wu H, et al. MiR-15a, miR-16-1 and miR-17-92 cluster expression are linked to poor prognosis in multiple myeloma. Leuk Res. 2012;36:1505–9. doi: 10.1016/j.leukres.2012.08.021. [DOI] [PubMed] [Google Scholar]
- 35.Si H, Sun X, Chen Y, Cao Y, Chen S, Wang H, et al. Circulating microRNA-92a and microRNA-21 as novel minimally invasive biomarkers for primary breast cancer. J Cancer Res Clin Oncol. 2013;139:223–9. doi: 10.1007/s00432-012-1315-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, et al. Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]