Dear Editor,
In the past few years, the use of sequence-specific nucleases for efficient targeted mutagenesis has provided plant biologists with a powerful new approach for understanding gene function and developing new traits. These nucleases create DNA double-strand breaks at chromosomal targeted sites that are primarily repaired by the non-homologous end joining (NHEJ) or homologous recombination (HR) pathways. NHEJ is often imprecise and can introduce mutations at target sites resulting in the loss of gene function. In contrast, HR uses a homologous DNA template for repair and can be employed to create site-specific sequence modifications or targeted insertions (Moynahan and Jasin, 2010).
In spite of being developed only 3 years ago, TALENs have become the reagent of choice for efficiently modifying eukaryotic genomes in a targeted fashion (Baker, 2012; Liu et al., 2012; Tong et al., 2012). Like ZFNs, TALENs are composed of an engineered array of DNA-binding domains fused with a non-specific FokI nuclease domain (Christian et al., 2010). Two TALEN monomers bind target DNA sequences allowing the FokI domains to dimerize and cleave the sequence between the two recognition sites (Supplemental Figure 1A). Each repeat in the DNA-binding domain of a TALEN recognizes one nucleotide in the target in a largely context-independent fashion, making the engineering of nucleases with new DNA sequence specificities easier and more reliable than ZFNs and meganucleases (Reyon et al., 2012).
Although TALENs have been demonstrated to function at high efficiency in many animal species and in human cell lines, the use of TALENs for plant genome modification has only been demonstrated for three species—Arabidopsis, tobacco, and rice. Furthermore, most studies have used TALENs to create mutations by NHEJ (Cermak et al., 2011; Li et al., 2012; Zhang et al., 2012). In this study, we report highly efficient genome modification with TALENs in two model monocot species—rice and Brachypodium—at a total of 12 genetic loci.
The experimental protocol for our TALEN-mediated genomic modification strategy is shown in Figure 1A. Four rice genes, including OsDEP1 (LOC_Os09g26999), OsBADH2 (LOC_Os08g32870), OsCKX2 (LOC_Os01g10110), and OsSD1 (LOC_Os01g40720), and eight Brachypodium genes, including BdABA1 (Bradi5g11750), BdCKX2 (Bradi2g06030), BdSMC6 (Bradi4g08527), BdSPL (Bradi2g03740), BdSBP (Bradi4g33770), BdCOI1 (Bradi2g23730), BdRHT (Bradi1g11090), and BdHTA1 (Bradi1g25390), were targeted to generate knockout mutations. A total of 13 TALEN sites were identified in the coding sequences of these genes using the TALE-NT program (Doyle et al., 2012). TALEN repeat arrays recognizing all the target sites (Supplemental Table 1) were constructed using the Golden Gate assembly method (Cermak et al., 2011; Zhang et al., 2012). For ease of analysis, each TALEN recognition sequence contained a restriction enzyme site within the spacer region (Supplemental Figure 1B and Supplemental Table 1).
Figure 1.
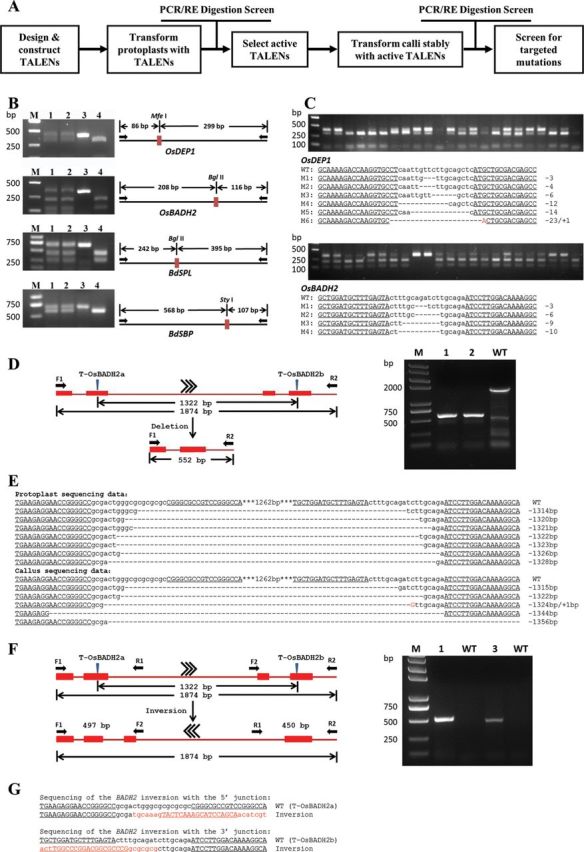
High-Efficiency Gene Targeting Using TALENs in Rice and Brachypodium.
(A) Flow diagram of recovery of targeted modification by TALENs.
(B) Four representative gels for analyzing the PCR products from protoplast samples treated with the respective TALENs (Lane 1 and Lane 2). Lane 3 and Lane 4 represent undigested and digested wild-type control, respectively.
(C) Molecular evidence and sequences of TALENs (T-OsDEP1 and T-OsBADH2b) induced mutations in rice plants. Two DNA samples were extracted from two different leaves of each plant and PCR-amplified across the TALEN recognition sites. Each PCR amplicon was analyzed for mutations using the same restriction enzyme digestion method as described above for the protoplast assay. The TALEN target sites are underlined. Deletion and insertions are indicated by dashes and red letters, respectively. The numbers to the side indicate the type of mutation and how many nucleotides were involved.
(D) TALEN-induced OsBADH2 deletions. PCR with primers (black Arrows F1 and R2) flanking the presumptive deletion yield a 552-bp product when both T-OsBADH2a and T-OsBADH2b were introduced into rice cells simultaneously. Molecular evidence of genome deletions were detected in the transformed protoplasts. Lane 1 and Lane 2 represent protoplasts which were co-transformed with T-OsBADH2a and T-OsBADH2b. Lane 3 represents wild-type control.
(E) OsBADH2 deletion sequences. The wild-type sequence is shown at the top with the full T-OsBADH2a and T-OsBADH2b target sites (underlined) and spacers (lowercase letters). The TALEN recognition sites are underlined and deletions are indicated by dashed lines.
(F) TALEN-induced OsBADH2 inversion. The outcome of an inversion event of sequence between the TALEN target sites was predicted. Primers flanking the presumptive flanking sites at the 5′- and 3′-ends of the inversion locus are shown (black arrows) along with predicted product size. PCR products were observed at both the 5’- and 3’-junctions only when both TALENs were introduced into resistant callus lines simultaneously. Lane 1 and Lane 3 represent the expected PCR products with 497 bp (5’ junction) and 450 bp (3’ junction), respectively.
(G) OsBADH2 inversion sequences. The OsBADH2 inversion allele is shown with 5’ and 3’ junctions that were analyzed by sequencing. The TALEN recognition sites are underlined; spacers are indicated by lower-case letters; and inversion sequences are in red.
TALEN repeat arrays were first cloned into expression vector pZHY051 for assessment of nuclease activity in protoplasts (Zhang et al., 2012). A protoplast transient assay was developed in both rice and Brachypodium. TALEN-encoding constructs were introduced into protoplasts using polyethylene glycol (PEG). After a 48-h incubation, genomic DNA was prepared from each sample, and DNA fragments encompassing each target site were amplified by PCR. The PCR products were digested by restriction enzymes and visualized by agarose gel electrophoresis (Figure 1B). The PCR amplicons were then cloned into a T–A cloning vector, and approximately 30–50 individual clones were analyzed for mutations by DNA sequencing (Supplemental Figure 2). Frequencies of TALEN-induced mutations at each endogenous target site in protoplasts were estimated. Seven out of 11 TALENs showed mutagenesis frequencies ranging from 4% to 14% (Supplemental Table 2). For the three TALENs (T-OsSD1, T-BdABA1, and T-BdCKX2), mutations could only be detected by an enrichment PCR assay, in which genomic DNA was digested by the restriction enzymes prior to PCR amplification; another round of restriction enzyme digestion was performed after amplification (Supplemental Table 2) (Zhang et al., 2010). Positive correlations of nuclease activities in protoplasts and transformed calli in tobacco (Zhang et al., 2012) have been observed and it has been demonstrated that the protoplast transient assay is a rapid, accurate, and reliable method to assess nuclease activity in Arabidopsis and tobacco (Zhang et al., 2010, 2012). The similar observations were also obtained on rice in our study (Supplemental Tables 2 and 3). Thus, protoplast assay has proven a valuable tool to quickly screen active TALENs before engaging in the laborious plant transformation process.
To test whether TALENs can induce high-frequency gene knockouts in rice and Brachypodium, the TALENs targeting the four rice and eight Brachypodium genes were transformed into plants. The constructs were introduced into embryonic cells of rice or Brachypodium using Agrobacterium tumefaciens. After 5–8 weeks of selection, hygromycin-resistant calli appeared, and about 20–50 callus lines from each transformation were sampled. Genomic DNA was extracted from each sample and PCR- amplified across the TALEN recognition sites. Each PCR amplicon was analyzed for mutations using the same restriction enzyme digestion method as described above for the protoplast assay. Undigested DNA fragments were observed in all 13 TALEN-transformed calli, and they were cloned and sequenced. We identified 127 mutant sequences (65 rice mutants and 62 Brachypodium mutants), and each was characterized by small deletions, insertions, and nucleotide substitutions. Most mutations were small deletions ranging from 1 to 20 bp (Supplemental Figure 3). All of the mutations occurred in the spacer region between the TALEN binding sites. In rice, mutations in the OsDEP1 gene were identified in eight out of 23 and 10 out of 36 calli surveyed (mean value 31.3%) from two independent transformation experiments. Mutations at OsCKX2 were recovered from 29 out of 47 calli (61.7%), and mutations at rice OsSD1 were identified in one out of 26 calli (3.8%). In addition, two pairs of TALENs were designed to target OsBADH2. While both TALENs induced mutations, they did so at different frequencies. Mutations disrupting OsBADH2 using T-OsBADH2a occurred in four out of 43 and two out of 22 calli surveyed (mean value 9.2%) in two independent transformation experiments. Mutations disrupting OsBADH2 using T-OsBADH2b were found in eight out of 30, seven out of 30, and 13 out of 44 calli surveyed (mean value 26.9%) in three independent transformations (Supplemental Table 3). Similarly, high-efficiency targeted gene knockouts were also observed in resistant calli resulting from each Brachypodium transformation experiment. Frequencies ranged from 5.9% to 100% (Supplemental Table 3) and most showed >30% mutagenesis as measured both by the restriction enzyme digestion assay and sequencing (Supplemental Figure 3 and Supplemental Table 3). In total, bi-allelic modifications were recovered from calli transformed with five out of 13 TALENs (Supplemental Table 3 and Supplemental Figure 4). Approximately 12.5% (1/8) of T-OsBADH2b-induced mutations, 3.4% (1/29) of T-OsCKX2-induced mutations, 45.5% (5/11) of T-BdCOI1-induced mutations, 22.2% (2/9) of T-BdSBP-induced mutations, and 10.7% (3/28) of T-BdSPL-induced mutations were bi-allelic. The individual transgenic callus lines were further propagated and regenerated into whole plants. Mutations induced by T-OsDEP1 were identified in 10 out of 52 transgenic plants (19.2%) and mutations induced by T-OsBADH2b were recovered from four out of 11 transgenic plants (36.4%). Our results demonstrate that those transformed callus lines can regenerate to whole plants that contain the TALEN-induced genetic changes at the relevant loci (Figure 1C).
In animal systems, it has previously been reported that co-delivery of two TALEN pairs that target the same chromosome can cause large DNA rearrangements, including deletions and inversions (Carlson et al., 2012). In this study, we investigated whether similar DNA arrangements could be recovered with high efficiency in plants. Two rice TALENs, T-OsBADH2a and T-OsBADH2b, separated by 1322 bp in the OsBADH2 gene, were co-introduced into rice protoplasts. We used two PCR primers flanking the targeted region that would amplify a 552-bp fragment if the sequence between the two TALEN sites was deleted (Figure 1D). The appropriately sized PCR products were observed in samples transformed with both TALEN pairs, but they were absent in those transformed only with one TALEN pair as well as in the untransformed samples (Figure 1D). Sequencing of the PCR products confirmed the left side of T-OsBADH2a and the right side of T-OsBADH2b were joined, and the intervening sequence was deleted (Figure 1E). These two TALEN pairs were next co-delivered into rice calli by particle bombardment. Ninety-eight hygromycin-resistant calli were screened after 6 weeks of selection, and three types of modifications were identified—simple modifications (small deletions or insertions) at either target site, large deletions of the sequence between the two target sites, and inversion of the sequence between the two target sites. Five calli (5.1%) contained small indels in the T-OsBADH2a target site, and seven calli (7.1%) showed small indels in the T-OsBADH2b target site. In addition, five calli (5.1%) showed large deletions of the intervening sequence (Figure 1E). These results support the idea that TALEN-induced genomic deletions were mediated via NHEJ. To determine whether the sequence between the two TALEN target sites had been inverted, we designed PCR primers that would amplify the new 5’- and 3’-junctions of the predicted inversion (Figure 1F). PCR amplification of both the 5’- and 3’-junctions identified a putative inversion in one callus (1.0%) (Figure 1F), and DNA sequencing of the PCR product confirmed the inversion and revealed a deletion of 10 nucleotides at the junction (Figure 1G). In general, deletions were recovered at much higher efficiencies than inversions (5.1% and 1.0%, respectively).
The large chromosomal rearrangements obtained through simultaneous expression of two pairs of TALENs could be useful for a variety of purposes. For example, targeted deletions would make possible the selective removal of gene clusters and enables scientists to delete intergenic regions, introns, regulatory elements, and non-coding RNAs. To our knowledge, this is the first study demonstrating highly efficient targeted knockouts in multiple genes in the model monocot species Brachypodium. We anticipate TALEN technology will make targeted gene modification a routine practice not just for these model monocots, but also for economically important crops, such as maize and wheat.
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
The work was supported by the National Natural Science Foundation of China (Grant Nos. 201263, 383601, 31200273, and 31271420) and the National Science Foundation (Grant No. DBI-0923827). No conflict of interest declared.
Supplementary Material
REFERENCES
- Baker M. 2012. Gene-editing nucleases. Nature Methods. 9, 23–26 [DOI] [PubMed] [Google Scholar]
- Carlson D.F., Tan W., Lillico S.G., Stverakova D., Proudfoot C., Christian M., Voytas D.F., Long C.R., Whitelaw C.B., Fahrenkrug S.C. 2012. Efficient TALEN-mediated gene knockout in livestock. Proc. Natl Acad. Sci. U S A. 109, 17382–17387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T., Doyle E.L., Christian M., Wang L., Zhang Y., Schmidt C., Baller J.A., Somia N.V., Bogdanove A.J., Voytas D.F. 2011. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39, e82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M., Cermak T., Doyle E.L., Schmidt C., Zhang F., Hummel A., Bogdanove A.J., Voytas D.F. 2010. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 186, 757–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle E.L., Booher N.J., Standage D.S., Voytas D.F., Brendel V.P., Vandyk J.K., Bogdanove A.J. 2012. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 40, W117–W122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Liu B., Spalding M.H., Weeks D.P., Yang B. 2012. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nature Biotechnol. 30, 390–392 [DOI] [PubMed] [Google Scholar]
- Liu J., Li C., Yu Z., Huang P., Wu H., Wei C., Zhu N., Shen Y., Chen Y., Zhang B., Deng W.-M., Jiao R. 2012. Efficient and specific modifications of the drosophila genome by means of an easy TALEN strategy. J. Genet. Genomics. 39, 209–215 [DOI] [PubMed] [Google Scholar]
- Moynahan M.E., Jasin M. 2010. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nature Reviews Molecular Cell Biology. 11, 196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan M.E., Jasin M. 2010. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nature Reviews Mol. Cell Biol. 11, 196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyon D., Tsai S.Q., Khayter C., Foden J.A., Sander J.D., Joung J.K. 2012. FLASH assembly of TALENs for high-throughput genome editing. Nature Biotechnol. 30, 460–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong C., Huang G., Ashton C., Wu H., Yan H., Ying Q.-L. 2012. Rapid and cost-effective gene targeting in rat embryonic stem cells by TALENs. J. Genet. Genomics. 39, 275–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Maeder M.L., Unger-Wallace E., Hoshaw J.P., Reyon D., Christian M., Li X., Pierick C.J., Dobbs D., Peterson T, et al. 2010. High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc. Natl Acad. Sci. U S A. 107, 12028–12033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang F., Li X., Baller J.A., Qi Y., Starker C.G., Bogdanove A.J., Voytas D.F. 2012. TALENs enable efficient plant genome engineering. Plant Physiol. http://dx.doi.org/10.1104/pp.112.205179 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


