Abstract
Adipose tissue macrophages (ATMs) undergo a phenotypic switch from alternatively activated antiinflammatory M2 macrophages in lean individuals to classically activated proinflammatory M1 macrophages in obese subjects. However, the molecular mechanism underlying this process remains unclear. In this study we aim to determine whether DNA methyltransferase 3b (DNMT3b) regulates macrophage polarization and inflammation. We found that the expression of DNMT3b was significantly induced in macrophages exposed to the saturated fatty acid stearate, was higher in ATMs isolated from obese mice, but was significantly lower in alternatively activated M2 vs classically activated M1 ATMs, suggesting a role for DNMT3b in regulation of macrophage polarization and inflammation in obesity. DNMT3b knockdown promoted macrophage polarization to alternatively activated M2 phenotype and suppressed macrophage inflammation, whereas overexpressing DNMT3b did the opposite. Importantly, in a macrophage-adipocyte coculture system, we found that DNMT3b knockdown significantly improved adipocyte insulin signaling. The promoter of peroxisome proliferator activated receptor (PPAR)γ1, a key transcriptional factor that regulates macrophage polarization, is enriched with CpG sites. Chromatin immunoprecipitation assays showed that DNMT3b bound to the methylation region at PPARγ1 promoter, which was further enhanced by stearate. Moreover, pyrosequencing analysis revealed that stearate increased DNA methylation at PPARγ1, which was prevented by DNMT3b deficiency. Therefore, our data demonstrate that DNMT3b plays an important role in regulating macrophage polarization through epigenetic mechanisms. In obesity, elevated saturated fatty acids enhance DNMT3b expression, leading to DNA methylation at the PPARγ1 promoter, which may contribute to deregulated adipose tissue macrophage polarization, inflammation, and insulin resistance.
Chronic inflammation is a key link between obesity and insulin resistance/type 2 diabetes (1, 2). An important feature of obesity-induced inflammation is the infiltration of macrophages into adipose tissue (3, 4). Adipose tissue macrophages (ATMs) play an important role in obesity-induced inflammation and insulin resistance (3, 4). ATMs can be distinguished into M1 and M2 macrophages by their differential expression of surface markers F4/80, CD11c, and CD206/mannose receptor C type 1 (Mrc1) (5–8). ATMs from lean animals show an alternatively activated, M2 phenotype (9, 10). These M2 macrophages are normally induced by Th2 cytokines IL-4 and IL-13, and typically have down-regulated inducible nitric oxide synthase and up-regulated arginase 1 (Arg1) expression and elevated expression of antiinflammatory factors (IL-10 and IL-1 receptor antagonist [IL-1RN]), therefore attenuating inflammation (11, 12). The cellular signal mediating the activation of M2 macrophage polarization has been investigated. IL-4 and IL-13 induce the tyrosyl phosphorylation and activation of signal transducer and activator of transcription 6 (STAT6). STAT6, in turn, regulates gene expression characteristic of M2 activation (2). Recently, it was reported that peroxisome proliferator activated receptor (PPAR)γ and PPARδ also play important roles in this process (13–18). Obesity is associated with deregulated ATM polarization, characterized by an increase in classically activated M1 ATMs (F4/80+CD11c+CD206−) and a reduction in alternatively activated M2 ATMs (F4/80+CD11c−CD206+) (5–8), which eventually tips the balance of ATMs toward a more proinflammatory phenotype and contributes to obesity-induced inflammation and insulin resistance. A critical question is how obesity causes defective ATM alternative polarization and tips the balance of ATMs toward more proinflammatory phenotypes.
Whereas numerous studies have been devoted to the evaluation of genetic factors related to obesity and its associated complications, much less is known about epigenetic changes that occur without alterations in the DNA sequence. Epigenetic regulation, including DNA methylation, is a molecular link between environmental factors (eg, diets) and complex diseases, including obesity and diabetes. DNA methylation of cytosines at primarily CpG dinucleotides is the most common epigenetic modification. CpGs are often enriched in the promoter and the first exon/5′-untranslated region of genes (19). De novo methylation is mediated by DNA methyltransferase (DNMT) 3a and 3b. Once established, DNA methylation is then maintained through mitosis primarily by the maintenance enzyme DNMT1 (20). Promoters of transcriptionally active genes are typically hypomethylated (21), whereas DNA hypermethylation can result in gene silencing by affecting the binding of methylation-sensitive DNA binding proteins and/or by interacting with various histone modifications and corepressors that alter DNA accessibility to transcriptional factors (20, 22). Alterations of DNA methylation have also been implicated in chronic inflammation-related diseases (23, 24). However, little is known about the role of epigenetics in obesity-induced inflammation. Recently, histone methylation has been implicated in the regulation of macrophage alternative activation (25, 26). However, it is not known whether changes in DNA methylation are involved in this process.
In the present study, we tested the hypothesis that DNMT3b mediates epigenetic regulation of macrophage polarization and inflammation, alteration of which may contribute to deregulated adipose tissue macrophage polarization, inflammation, and insulin resistance in obesity. To test this hypothesis, we examined the expression of DNMT3b in macrophages exposed to the saturated fatty acids (SFAs), the circulating levels of which are commonly increased in obesity. We also examined the DNMT3b expression in ATMs isolated from obese vs lean mice, and in alternatively activated M2 vs classically activated M1 ATMs. Using gain or loss of function of DNMT3b, we examined the M2 macrophage alternative activation and macrophage inflammation in response to lipopolysaccharide and the SFAs. To determine whether alterations of macrophage polarization and inflammation by DNMT3b affect adipocyte insulin sensitivity, we established a macrophage-adipocyte coculture system and examined adipocyte insulin signaling. Finally, we investigated the mechanism underlying DNMT3b regulation of macrophage polarization and determined whether DNMT3b binds to PPARγ1 promoter and alters its DNA methylation, leading to regulation of macrophage polarization and inflammation.
Materials and Methods
Animals
For diet-induced obesity studies, 5-week-old male C57BL/6J mice were purchased from The Jackson Laboratory and were put on either a low-fat (catalog no. D12450B; 10% calories from fat; Research Diets, Inc) or a high-fat diet (catalog no. D12492; 60% calories from fat; Research Diets, Inc) for 24 weeks. ob/ob mice 8 weeks of age and lean control (+/?) were also purchased from The Jackson Laboratory and were fed a regular chow diet (Lab diet5P00; fat content 14% by calorie). All animals were housed with a 12-hour light/12-hour dark cycle in a temperature-controlled facility and had free access to water and food. All animal studies were approved by the Institutional Animal Care and Use Committee at Wake Forest School of Medicine.
Isolation of adipose macrophages and M1 and M2 macrophages
Isolation of adipose tissue stromal vascular cells was performed as we described previously (27). Stromal vascular cells were then incubated with rat antimouse F4/80 polyclonal antibodies (AbD Serotec), followed by a pull down of F4/80-positive cells with sheep antirat microbeads using a magnetic-activated cell sorting system according to manufacturer's instructions (Miltenyi Biotec). Isolated F4/80+ adipose tissue macrophages (ATMs) were used for gene expression analysis. In another experiment, stromal vascular cells were labeled with APC-F4/80, PE-Cy7-CD11c, and PE-CD206, and M1/M2 macrophage subsets were isolated using BD FACSAria Cell Sorting machine (Becton Dickinson).
Cell culture
Raw264.7 macrophages were cultured in DMEM containing 10% heat-inactivated fetal bovine serum. Fatty acids (stearate, Sigma-Aldrich) were conjugated with BSA at a 4:1 M ratio before treatment. Fatty acids were dissolved in 95% ethanol at 60°C and then were mixed with prewarmed BSA (10%) to yield a stock concentration of 5 mM. Bone marrow-derived macrophages (BMDMs) were cultured in 10% fetal bovine serum and 30% L929 for 8 days.
Macrophage migration assay was performed using a CytoSelect 96-Well Cell Migration Assay kit (Cell Biolabs) according to the manufacturer's instruction. Arg 1 activity was performed as previously described (7). Cells were plated in the membrane chamber that was placed on top of the feeder tray and then incubated with 10 ng/mL MCP-1 at 37°C for 20 hours. Cells that migrated were lyzed and CyQuant GR dye was measured fluorometrically. TNFα protein secretion into medium was measured using the mouse TNFα ELISA kit (R&D Systems).
The macrophage-adipocyte coculture experiment was conducted as we previously described (28). Briefly, macrophages were plated in a transwell insert (0.4-μm porous membrane) and placed into a well containing 3T3-L1 adipocytes at day 8 after the induction of differentiation (29). Four days after coculture, adipocytes were stimulated with 100 nM insulin for 5 minutes. Cells were harvested and homogenized in a modified radioimmunoprecipitation assay lysis buffer.
Immunoblotting
Cells were harvested and homogenized in a modified radioimmunoprecipitation assay buffer containing 50 mM Tris (pH 7.4), 1 mM EDTA (pH 8.0), 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 1 mM each of NaF, NaVO3, and phenylmethylsulfonyl fluoride, 1% of protease inhibitor cocktail (Sigma-Aldrich), and 1% each of serine and tyrosine phosphatase inhibitor cocktail (Sigma-Aldrich). Immunoblotting was conducted as we previously described (28). Rabbit polyclonal antibodies against insulin receptor (Tyr-1162/1163) and insulin receptor substrate 1 (Tyr-612) phosphorylation were obtained from Upstate Biotechnology, Inc.
Small interfering RNA (siRNA) knockdown
Raw264.7 macrophages were reversely transfected with DNMT3b and or PPARγ1 siRNA (SMART pool; Dharmacon; Thermo Scientific) using Lipofectamine RNAiMAX (Invitrogen). Briefly, siRNA was premixed and incubated with the RNAiMAX reagents. The siRNA-RNAiMAX complexes were then mixed with cell suspension at 1 × 105cells/mL before being plated into 24-well plates (10 nM siRNA at final concentration). BMDMs were electroporated with DNMT3b siRNA using Nucleofector II (Lonza) with the Mouse Macrophage Nucleofector kit (Lonza).
Lentiviral constructs and infection
The DNMT3b cDNA vector was purchased from Open Biosystems and was further subcloned into pLVX lentiviral vectors (CLONTECH). The pLVX DNMT3b lentivirus was generated according to the instruction from CLONTECH. Medium containing the lentivirus was harvested and filtered and was used to infect Raw264.7 cells. The infected cells were selected with puromycin (8 μg/mL) for 10 days, and the surviving cells were pooled for further experiments.
Total RNA extraction and quantitative RT-PCR
Macrophage total RNA was extracted using the Tri Reagent kit (Molecular Research Center), according to the manufacturer's protocol. The expression of genes of interest was assessed by quantitative RT-PCR (ABI Universal PCR Master Mix; Applied Biosystems) using a Stratagene Mx3005p thermocycler (Stratagene), as we previously described (28, 30). The primer and probe pairs used in the assays were purchased from Applied Biosystems.
Chromatin immunoprecipitation (ChIP) assay
ChIP was conducted using a ChIP assay kit (Upstate Biotechnology) as we previously described (28). Briefly, cells were fixed with 1% of formaldehyde and then harvested in cell lysis buffer (5 mM 1,4-piperazinediethane sulfonic acid, 85 mM KCl, and 0.5% Nonidet P-40, supplemented with protease inhibitors, pH 8.0). The lysates were sonicated to shear genomic DNA to an average fragment length of 200–1000 bp. Lysates were centrifuged, and the supernatants were collected. The supernatants underwent overnight immunoprecipitation, elution, reverse cross-link, and protease K digestion. A mock immunoprecipitation with normal serum IgG was also included as a negative control for each sample. The DNAs recovered from phenol-chloroform extraction were used for PCR, the primer sequences of which are: PPARγ1 forward, 5′-GGCTGTGAGGAGCAAGGCGG-3′; reverse, 5′-CCGGGGCGACTCTGACCTGA-3′.
Bisulfite conversion and pyrosequencing
Genomic DNA will be prepared by phenol-chloroform extraction. Bisulfite conversion will be performed using EpiTech Bisulfite Kit (QIAGEN). The primers that were used to amplify PPARγ1 promoter/5′-untranslated region covering CpG sites were commercially designed by EpiGenDx. Bisulfite-converted DNA (1 μg) was amplified by PCR, and pyrosequencing was performed by EpiGenDx.
PPARγ1 promoter cloning and luciferase reporter assays
A 1.5-kb fragment covering PPARγ1 proximal promoter and part of 5-untranslated region was PCR amplified from PPARγ1 bacterial artificial chromosome with primers as follows. PPARγ1 forward-1(F1), 5′-GTCTGGTACCTCTGGTGAGGATGGTTTGTA-3′; PPARγ1 reverse-1 (R1), 5′-CCGGGGCGACTCTGACCTGA-3′; PPARγ1 forward-2 (F2), 5′-GGCTGTGAGGAGCAAGGCGG-3′; PPARγ1 reverse-2 (R2), 5′-CCCTAGATCTTTGTCTGTCACACAGTCCTG-3′. The primer sets F1/R1 and F2/R2 amplify 2 overlapping fragments on the PPARγ1 promoter (see Figure 5A). The two PCR fragments were digested with KpnI/ApalI and ApalI/BglII, respectively. The digested fragments were ligated to pGL3-Basic at KpnI/BglII sites to generate pGL3-PPARγ1. The constructs were confirmed by sequencing. To obtain unmethylated promoter, the reporter constructs were transformed into the dam-/dcm-Escherichia coli strain (New England Biolabs). To obtain fully methylated reporter, constructs were incubated with 3 U/μg SssI methylase (New England Biolabs) in the presence of 160 μM of S-adenosylmethionine at 37°C for 3 hours (31, 32). Methylation was confirmed by checking the resistance of reporter constructs to HpyCH4IV digestion (31). The unmethylated or methylated PPARγ1 reporter constructs were transfected into RAW cells using a SuperFect Transfection Reagent kit (QIAGEN), and luciferase activity was measured using a Dual-Luciferase Reporter Assay kit (Promega Corp) as we described (28).
Figure 5.
DNMT3b regulates PPARγ1 expression. A and B, DNMT3b regulates PPARγ1 mRNA. C and D, DNMT3b regulates PPARγ1 protein. DNMT3b siRNA knockdown or overexpression in Raw264.7 macrophages was conducted as described in Figure 2. PPARγ1 mRNA was measured by real-time RT-PCR and normalized to cyclophilin and protein was measured by immunoblotting. All data are expressed as mean ± SEM (n = 6/group); *, P < .05. 3bKD, DNMT3b knockdown; Dnmt3b, pLVX DNMT3b.
Statistics
All data are expressed as mean ± SEM. Data were evaluated for statistical significance by one-way ANOVA, and statistical significance for comparison of means of different groups was calculated by the least-significant-difference test using the SPSS software package version 11.5. P < .05 was considered significant.
Results
DNMT3b expression is associated with macrophage polarization in obesity
We first determined the physiological relevance of DNMTs in obesity-induced macrophage phenotypic switch. We isolated M1 and M2 ATM populations using fluorescent-activated cell sorting according to the surface expression of F4/80, CD11c (M1), and Mrc1/CD206 (M2) (5, 6, 9, 10) (Supplemental Figure 1). We found that DNMT3b expression was much lower in F4/80+CD11c−CD206+ M2 ATMs than that in F4/80+CD11c+CD206− M1 ATMs (Figure 1A), whereas there was no difference in the expression of DNMT3a between these 2 populations (Figure 1A) (we will report the DNMT1 data elsewhere). We further determined the expression of DNMT3a and 3b in ATMs isolated from lean and obese mice. Interestingly, DNMT3b expression was also significantly elevated in ATMs isolated from ob/ob mice compared with that of lean controls, whereas no difference in DNMT3a expression was observed (Figure 1B). Similar results, to a lesser extent, were observed in ATMs isolated from diet-induced obese mice (Figure 1C). Dietary SFAs, levels of which are elevated in obesity, have been implicated in promoting the metabolic disorders, including obesity and insulin resistance/type 2 diabetes, partly by increased recruitment of proinflammatory M1 macrophages into inflamed adipose tissue (33). We determined whether these obesity-associated factor SFAs would affect DNMT3b expression. We found that the SFA stearate stimulated DNMT3b mRNA and protein levels in macrophages (Figure 1, D and E). In contrast, DHA, an antiinflammatory polyunsaturated fatty acid, suppressed DNMT3b expression (Figure 1F). Interestingly, IL-4, a Th2 cytokine that induces M2 macrophage polarization, also inhibited DNMT3b mRNA and protein levels (Figure 1, G and H). These data suggest that DNMT3b may be an important determinant of the deregulated ATM polarization in the obese adipose tissue.
Figure 1.
DNMT3b expression is associated with macrophage polarization in obesity. A, The expression of DNMT3b, but not DNMT3a, is decreased in F4/80+CD11c−CD206+ M2 ATMs more than that in F4/80+CD11c+CD206− M1 ATMs. B and C, The expression of DNMT3b, but not DNMT3a, is elevated in ATMs isolated from ob/ob (B) and diet-induced obese mice (C). Isolation of M1/M2 ATMs and total ATMs was conducted as described in Materials and Methods. LF: low fat diet, HF: high fat diet (n = 4/group). D–H, Stearate stimulates DNMT3b mRNA (D) and protein (E) levels, whereas docosahexaenoic acid (DHA) (F) inhibits DNMT3b mRNA and IL-4 inhibits both DNMT3b mRNA (G) and protein (H) levels in macrophages. Raw264.7 macrophages were treated with BSA, stearate (C:18, 200 μM), DHA (50 μM), or IL-4 (10 ng/mL) for 48 hours (n = 6/group). Gene expression was measured by real-time RT-PCR and normalized to cyclophilin. Protein levels were measured by immunoblotting. All data are expressed as mean ± SEM; *, P < .05.
DNMT3b regulates macrophage polarization and inflammation
To determine the role of DNMT3b in regulation of macrophage polarization, we used gain- or loss-of-function approaches to either knock down or overexpress DNMT3b in Raw264.7 macrophages and then determined the expression of M2 macrophage markers. Raw264.7 macrophages were transfected with DNMT3b siRNA using a reverse transfection approach. The DNMT3b mRNA was suppressed by 73% in knockdown macrophages (Supplemental Figure 2A), which was consistent with a significant decrease at DNMT3b protein levels (Supplemental Figure 2B). We found that DNMT3b knockdown was sufficient to induce M2 macrophage polarization, evident by increased expression of M2 macrophage markers, such as arginase 1 (Arg1), mannose receptor C type 1 (Mrc1), and macrophage galactose-type c-type lectin (Mgl1) (Figure 2, A–C). Likewise, DNMT3b knockdown also increased Arg1 activity (Supplemental Figure 3). Moreover, DNMT3b deficiency further promoted IL-4-induced expression of M2 macrophage markers (Figure 2, A–C). We also performed a gain of function study by overexpressing DNMT3b in macrophages. The DNMT3b protein level was elevated by overexpression (Supplemental Figure 4). In contrast to the loss of function data shown above, overexpressing DNMT3b inhibited IL-4-induced Arg1 expression in macrophages (Figure 2D). Similar results were observed in BMDMs with DNMT3b knockdown, indicated by increased expression of M2 macrophage markers (Supplemental Figure 5). These data suggest that DNMT3b is an important determinant of macrophage polarization.
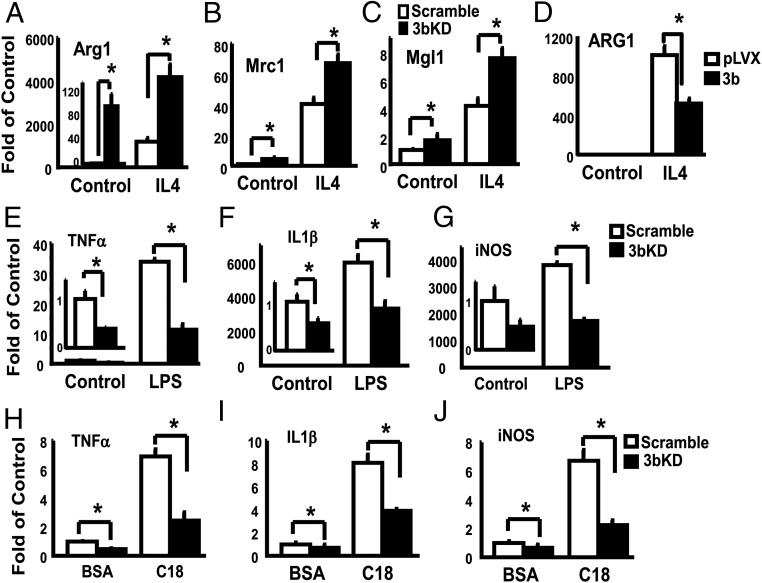
Figure 2.
DNMT3b regulates M2 macrophage polarization and inflammation. A–D, DNMT3b knockdown induces M2 macrophage polarization. Raw264.7 macrophages were reversely transfected with DNMT3b siRNA and then treated with IL-4 (10 ng/mL) for 24 hours. Arg1 (A), Mrc1 (B), and Mgl1 (C) mRNA were measured by real-time RT-PCR and normalized to cyclophilin. All data are expressed as mean ± SEM (n = 6/group); *, P < .05. D, Overexpressing DNMT3b inhibits IL-4-induced Arg1 expression. Raw264.7 macrophages were infected with pLVX DNMT3b lentivirus and selected with puromycin. Cells were then treated with IL-4 (10 ng/mL) for 24 hours. Arg1 mRNA was measured by real-time RT-PCR and normalized to cyclophilin. All data are expressed as mean ± SEM (n = 4/group); *, P < .05.3b. KD, DNMT3b knockdown; Dnmt3b: pLVX DNMT3b. E–J, DNMT3b knockdown inhibits macrophage inflammation. E–G, DNMT3b knockdown inhibits lipopolysaccharide (LPS)-stimulated expression of proinflammatory genes. H–J, DNMT3b knockdown inhibits stearate-stimulated expression of proinflammatory genes. Raw264.7 macrophages were reversely transfected with DNMT3b siRNA and then treated with LPS (100 ng/mL) or stearate (C18, 500 μM) for 4 hours. TNFα, IL-1β, and inducible nitric oxide synthase (iNOS) mRNA were measured by real-time RT-PCR and normalized to cyclophilin. All data are expressed as mean ± SEM (n = 6/group); *, P < .05. 3bKD, DNMT3b knockdown.
To determine whether regulation of macrophage polarization by DNMT3b further affects macrophage inflammation, we measured inflammatory gene expression in Raw264.7 macrophages with DNMT3b knockdown. We found that macrophages with DNMT3b knockdown displayed decreased expression of inflammatory genes such as TNFα, IL-1β, and inducible nitric oxide synthase (Figure 2, E–J). Further, DNMT3b knockdown substantially blocked lipopolysaccharide- and stearate-induced expression of inflammatory genes (Figure 2, E–J). Likewise, DNMT3b knockdown also largely suppressed TNFα secretion into the medium at both basal and lipopolysacchride-stimulated levels (Supplemental Figure 6). We also determined whether DNMT3b deficiency affects macrophage functionality and performed macrophage chemotatic assays. We found that DNMT3b knockdown decreased the ability of macrophage to migrate toward a chemotatic gradient of MCP-1 (Supplemental Figure 7). These data suggest that endogenous DNMT3b is a key regulator of macrophage polarization and inflammation.
Macrophage DNMT3b deficiency improves adipocyte insulin signaling
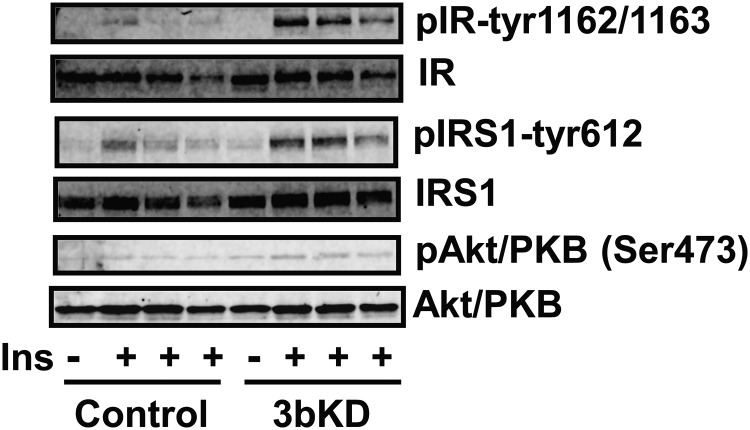
To mimic the physiological condition of adipose tissue in which ATMs constantly interact with adipocytes in a paracrine fashion, we established a coculture system in which DNMT3b-kockdown macrophages were cocultured with differentiated 3T3-L1 adipocytes in transwells. After 2-day coculture, adipocytes were stimulated with 100 nM insulin. We found that insulin signaling events, including insulin-stimulated tyrosyl phosphorylation of insulin receptor, insulin receptor substrate 1 and serine phosphorylation of Akt/PKB, were markedly improved in adipocytes cocultured with DNMT3b-knockdown macrophages compared with those of adipocytes cocultured with control macrophages (Figure 3). These data suggest that inactivation of macrophage DNMT3b likely down-regulates macrophage inflammation and cytokine expression, which may lead to improved insulin sensitivity in adipocytes in a paracrine effect.
Figure 3.
Knockdown of macrophage DNMT3b improves adipocyte insulin (Ins) signaling in a coculture system. Raw264.7 macrophages were reversely transfected with DNMT3b siRNA and then cocultured with differentiated 3T3–L1 adipocytes for 2 days. Adipocytes were stimulated with 100 nM insulin for 5 minutes, and insulin signaling was measured as described in Materials and Methods. 3bKD, DNMT3b knockdown; IR, insulin receptor; IRS, insulin receptor substrate.
Methylation at PPARγ1 promoter is a target for DNMT3b
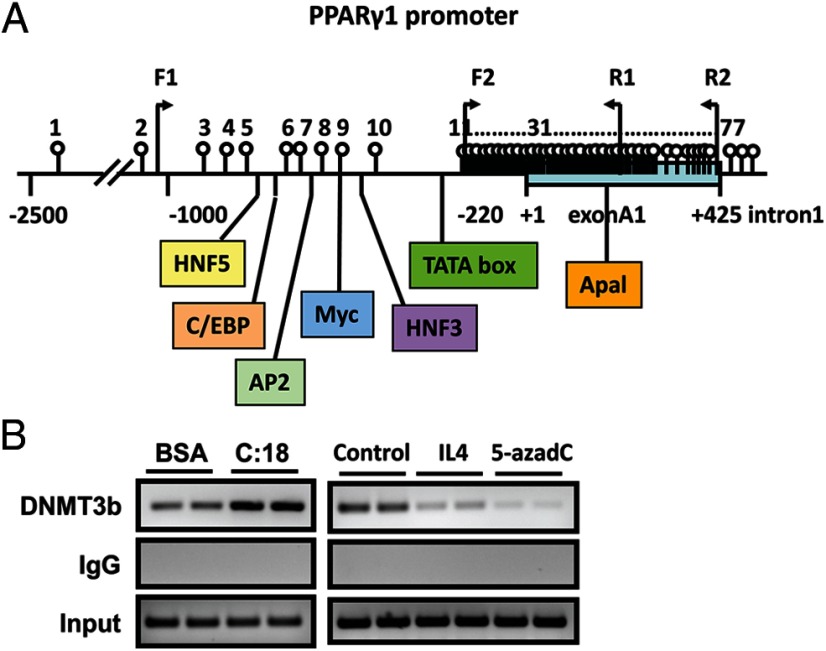
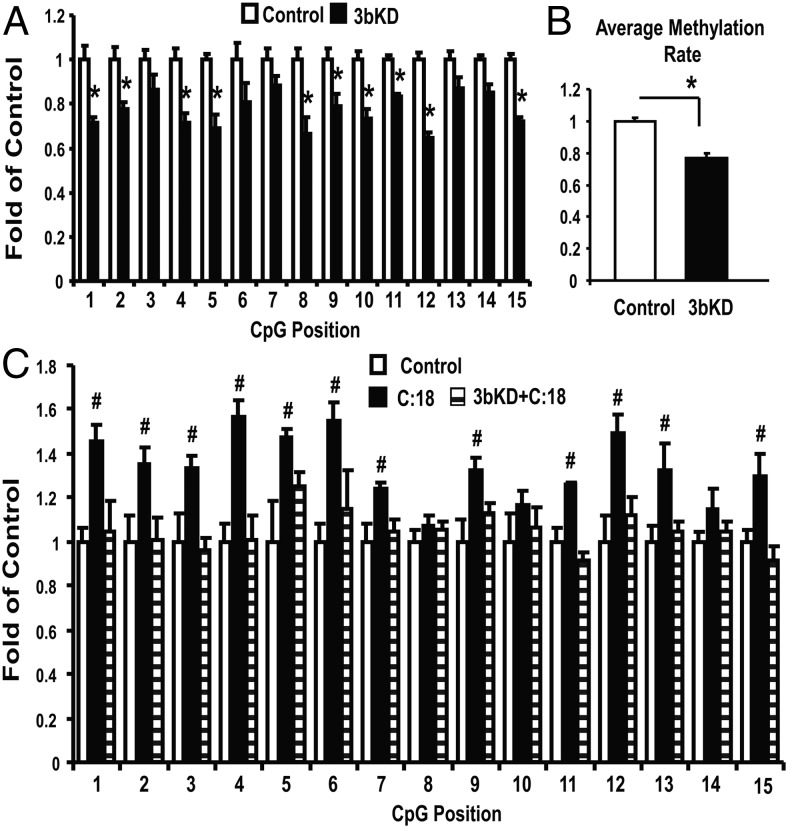
The nuclear receptor PPARγ is a key transcriptional factor that controls macrophage alternative activation (14, 17). We found that the proximal promoter and 5′-untranslated region of PPARγ1 are enriched with CpG sites (Figure 4A). This raised a possibility that PPARγ1 may be subjected to epigenetic regulation. Indeed, our ChIP assays showed that the SFA stearate significantly increased, whereas IL4 reduced, the binding of DNMT3b to PPARγ1 promoter in macrophages (Figure 4B). 5-aza-2′-deoxycytidine, a DNMT inhibitor that can degrade the DNMT protein (34), was used as a positive control in the ChIP assay. We then determined whether the binding of DNMT3b to PPARγ1 promoter regulates PPARγ1 expression. We found that DNMT3b knockdown stimulated basal and IL-4-induced expression of PPARγ1 mRNA (Figure 5A), whereas DNMT3b overexpression did the opposite (Figure 5B). Likewise, DNMT3b knockdown up-regulated PPARγ1 protein levels whereas overexpression decreased its contents (Figure 5, C and D). The data are consistent with the effect of DNMT3b knockdown on promoting M2 macrophage polarization (Figure 2), suggesting that PPARγ1 might be a target for DNMT3b to regulate macrophage polarization. Considering the importance of PPARγ in the regulation of macrophage alternative activation, we determined whether PPARγ1 promoter methylation is a target for DNMT3b to regulate macrophage polarization. Using pyrosequencing, we examined the DNA methylation status at PPARγ1 promoter in macrophages with DNMT3b knockdown. We found that DNMT3b knockdown significantly decreased DNA methylation at PPARγ1 promoter (CpG sites from 15–29 shown in Figure 4A) (Figure 6, A and B). Interestingly, DNA methylation at PPARγ1 promoter can be dynamically regulated, indicated by enhanced methylation by the SFA stearate (Figure 6C). However, DNMT3b knockdown prevented stearate-induced increase on PPARγ1 promoter methylation (Figure 6C). To determine whether the PPARγ1 promoter activity is indeed regulated by methylation, we examined the fully methylated vs unmethylated PPARγ1 promoter activity using luciferase assays. A 6-hour transfection assay (the data on longer transfection times will be reported elsewhere) revealed that the luciferase activity of the unmethylated promoter constructs was higher than that of the fully methylated promoter constructs (Figure 7), suggesting that the PPARγ1 promoter activity is highly regulated by DNA methylation. These data suggest that PPARγ1 may be a target for DNMT3b to regulate macrophage polarization.
Figure 4.
DNMT3b binds to PPARγ1 promoter. A, Schematic illustration of PPARγ1 promoter and 5′-untranslated region. The transcription start site is indicated as +1. TATA box, exon A1, and intron 1 are indicated. The CpG sites are indicated as upward vertical lines with open circles. Positions 1–10 are located upstream of the TATA box, and positions 11–77 are located between TATA box and the end of exon A1. The HNF5, CCAAT enhancer-binding protein (C/EBP), Ap2, Myc, and HNF3 motifs are shown in colored boxes. Arrows indicate positions of forward and reverse primer used in cloning of the 1.5-kb promoter and ChIP assays. B, ChIP assays indicating DNMT3b binding to PPARγ1 promoter. Raw264.7 macrophages were treated with BSA, stearate (C18, 250 μM), IL-4 (10 ng/mL), or 5-aza-2′-deoxycytidine (5-aza-dC) (0.5 μM) for 24 hours. ChIP assays were conducted as described in Materials and Methods.
Figure 6.
DNMT3b regulates DNA methylation at PPARγ1 promoter. A and B, DNMT3b knockdown decreases DNA methylation at PPARγ1 promoter. A, Methylation levels at CpG sites corresponding to 15–29 on PPARγ1 promoter (see Figure 5A). B, Average methylation levels of all these CpG sites. DNMT3b siRNA knockdown in Raw264.7 macrophages was conducted as described in Figure 2. CpG methylation was measured by pyrosequencing as described in Materials and Methods. All data are expressed as mean ± SEM (n = 4/group0; *, P < .05 vs control. 3bKD, DNMT3b knockdown. C, DNMT3b knockdown prevents stearate-induced increase of PPARγ1 promoter methylation. All data are expressed as mean ± SEM (n = 4/group); #, P < .05 vs control or 3bKD+C:18. 3bKD, DNMT3b knockdown.
Figure 7.
DNA methylation regulates PPARγ1 promoter activity. A 1.5-kb PPARγ1 promoter fragment was cloned into pGL3 luciferase reporter. Unmethylated vs fully methylated PPARγ1 promoter activity was conducted as described in Materials and Methods.
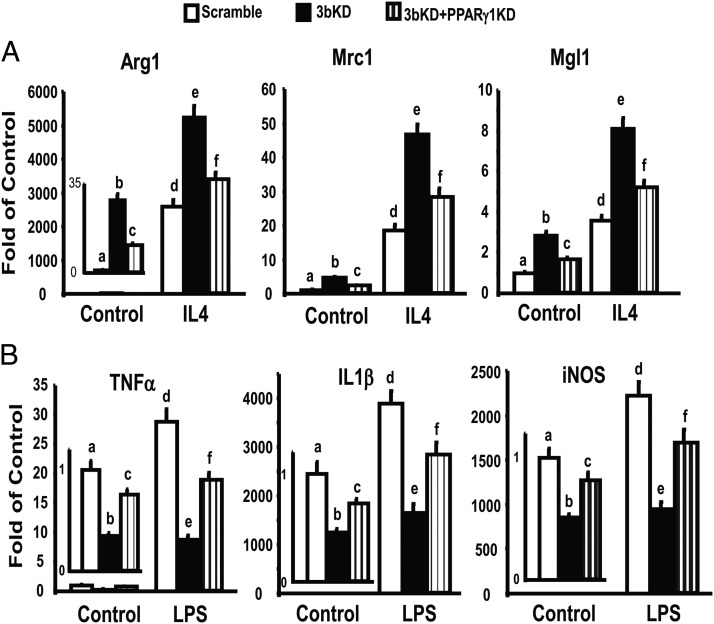
To determine the role of PPARγ1 in mediating M2 macrophage polarization induced by DNMT3b deficiency, we performed double knockdown of both PPARγ1 and DNMT3b in Raw cells. We found that DNMT3b deficiency significantly increased basal and IL-4-stimulated expression of M2 macrophage markers including Arg1, Mrc1, and Mgl1, which was substantially prevented by PPARγ1 knockdown (Figure 8A). Further, PPARγ1 knockdown significantly blocked the ability of DNMT3b deficiency to suppress inflammatory gene expression (Figure 8B). These data suggest that PPARγ1 plays an important role in mediating DNMT3b deficiency-induced M2 macrophage polarization and antiinflammatory phenotypes.
Figure 8.
PPARγ1 knockdown substantially blocks the effects of DNMT3b deficiency on M2 macrophage polarization and inflammation. A, PPARγ1 knockdown substantially blocks the effects of DNMT3b on M2 macrophage polarization. B, PPARγ1 knockdown substantially blocks the effects of DNMT3b on macrophage inflammation. Raw264.7 macrophages were reversely transfected with DNMT3b and PPARγ1 siRNAs and then treated with IL-4 (10 ng/mL) for 24 hours or treated with lipopolysaccharide (LPS) (100 ng/mL) for 4 hours. mRNA was measured by real-time RT-PCR and normalized to cyclophilin. All data are expressed as mean ± SEM (n = 6/group); bars that are labeled with different letters are statistically significant; P < .05. 3bKD, DNMT3b knockdown; PPARγ1KD, PPARγ1 knockdown. INOS, inducible nitric oxide synthase.
Discussion
Our data demonstrate that DNMT3b plays a key role in regulation of macrophage polarization and inflammation, which may involve modulation of DNA methylation at the PPARγ1 promoter. Our study is the first to provide a novel mechanism whereby DNA methylation mediates macrophage alternative activation, which may mediate macrophage phenotypic switch in obesity and, in turn, contribute to obesity-induced inflammation and insulin resistance.
Most complex diseases, including obesity and diabetes, are results of gene and environment interactions. One of the mechanisms by which environmental factors such as diets affect gene expression patterns involves their capacity to reprogram the epigenome (35, 36). To better understand how obesity, a disorder that typically results from environment (eg, high-fat diet) and gene interactions, causes defective ATM alternative polarization and tips the balance of ATMs toward more proinflammatory phenotypes, we focused on epigenetic mechanisms, particularly DNA methylation. We found that DNMT3b expression is significantly induced in macrophages exposed to SFAs and is higher in ATMs isolated from obese mice vs. lean, but is significantly lower in alternatively activated M2 vs classically activated M1 ATMs. The data suggest that the expression of DNMT3b in macrophages is physiologically important and relevant in obesity-associated macrophage phenotypic switch. Our studies with gain or loss of function approaches show that DNMT3b knockdown promoted macrophage polarization to alternatively activated M2 phenotype, leading to attenuation of macrophage inflammation, whereas overexpressing DNMT3b in macrophages prevented M2 macrophage conversion. To test whether DNMT3b regulation of macrophage polarization and inflammation would affect insulin sensitivity in adjacent adipocytes, we established a macrophage-adipocyte coculture system. We found that DNMT3b knockdown significantly improved adipocyte insulin signaling in the coculture system. Our data suggest a scenario in which elevated SFAs in obesity may induce the expression of DNMT3b, which prevents M2 macrophage conversion and tips the balance of ATMs toward more proinflammatory M1 phenotype, leading to exaggerated inflammation in obese adipose tissue. Dietary SFAs, levels of which are elevated in obesity, may therefore contribute to obesity-induced inflammation and insulin resistance via an epigenetic mechanism. Our study is in line with rapidly growing evidence converged to suggest that epigenetic events figure prominently in the development of obesity and diabetes, including epigenetic regulation of insulin resistance in various tissues (37–41).
To further understand the target genes by which DNMT3b regulates DNA methylation, we focused on PPARγ1 the expression of which in macrophages is critical in the regulation of macrophage alternative activation, inflammation, and whole-body glucose metabolism (13, 14, 16–18). Given the fact that the proximal promoter and 5′-untranslated region of PPARγ1 is enriched with CpG sites, PPARγ1 may be subjected to DNA methylation. Using ChIP assays, we found the direct binding of DNMT3b to PPARγ1 promoter/5′-untranslated region in macrophages, suggesting that DNMT3b is the major enzyme responsible for this process. Interestingly, unlike the traditional view of DNA methylation, which was thought to be stable, DNMT3b regulation of PPARγ1 promoter methylation is dynamic, because the SFA stearate regulates DNMT3b binding to PPARγ1 promoter. Indeed, our pyrosequencing assays confirmed that stearate significantly increased DNA methylation at the PPARγ1 promoter, whereas DNMT3b deficiency prevented the SFA's effect on the PPARγ1 methylation. This suggests that DNA methylation at PPARγ1 promoter, which was thought to be a very stable epigenetic marker, is actually dynamic in response to physiological stimuli such as nutritional cues (eg, SFA), and this dynamic methylation change on PPARγ1 promoter requires DNMT3b. In addition, we demonstrate that PPARγ1 promoter activity is highly regulated by DNA methylation. Finally, our loss of function experiment with PPARγ1 knockdown demonstrates that PPARγ1 is required for full capacity of DNMT3b deficiency to induce M2 macrophage polarization and to antagonize inflammation. However, it is noteworthy that PPARγ1 knockdown can not completely prevent the M2 macrophage phenotypes caused by DNMT3b deficiency, suggesting that PPARγ1 may not be the only target gene that DNMT3b acts on to exert its functions. Indeed, other transcriptional factors (eg, PPARδ, CCAAT enhancer-binding protein β, and STAT6, etc,) also play critical roles in regulation of macrophage polarization (2). Therefore, additional studies will be required to determine new downstream signals that mediate DNMT3b's effect on macrophage polarization and inflammation. Nonetheless all these data point to PPARγ1 as an important target, if not the only target, for DNMT3b to regulate macrophage polarization.
Alterations of DNA methylation have also been implicated in chronic inflammation-related diseases (23, 24). However, little is known about the role of DNA methylation in obesity-induced inflammation. It has become apparent that DNA methylation and histone methylation have a close cross talk and DNA methylation can set up a platform for histone modifications (42–45). Evidence has shown that changes in key chromatin histone methylation patterns can affect inflammatory signaling and gene expression in several cell types in diabetic conditions and models (39, 46). Recently, histone methylation has been implicated in the regulation of macrophage alternative activation (25, 26). Therefore, it would be interesting to determine whether altered DNA methylation on the PPARγ1 promoter by SFAs would cause modifications of histone methylation. Interestingly, previous studies show that even a transient hyperglycemic exposure may induce sustained epigenetic changes at histone methylation and subsequent long-lasting expression of inflammatory genes, which has been termed as “hyperglycemic memory” (47, 48). Considering that DNA methylation is a more stable epigenetic marker than histone methylation, future studies are warranted to address whether epigenetic alterations of the DNA methylation at the PPARγ1 promoter also reflect a long-term metabolic memory in response to a transient exposure to SFAs.
In summary, we demonstrate that DNA methylation by DNMT3b plays an important role in the regulation of macrophage polarization and inflammation. Gain and loss of function studies demonstrate an important role for DNMT3b as a negative regulator of M2 macrophage polarization. DNA methylation at the PPARγ1 promoter in macrophages is enhanced by obesity-associated factors SFAs, which is dynamically regulated by DNMT3b. Deregulated DNA methylation at PPARγ1 promoter by DNMT3b, presumably in response to elevated nutritional cues (eg, SFAs) in obesity, may result in deregulated macrophage polarization, thereby contributing to obesity-induced inflammation and insulin resistance. These studies could also help guide the development of epigenetic regulation as new therapeutic targets in the prevention and treatment of obesity-induced insulin resistance/type 2 diabetes.
Additional material
Supplementary data supplied by authors.
Acknowledgments
This work was supported by National Institutes of Health (NIH) Grants R01HL107500 (to B.X.), R01DK084172 (to H.S.), R01DK085176 (to L.Y.), American Heart Association grants 10SDG3900046 (to B.X.) and 11GRNT7370080 (to H.S.), and American Diabetes Association grant 7–13-BS-159 (to H.S.).
Disclosure Summary: The authors have no conflict of interests to disclose
Footnotes
- Arg1
- arginase 1
- ATM
- adipose tissue macrophage
- BMDM
- bone marrow-derived macrophage
- ChIP
- chromatin immunoprecipitation
- DNMT
- DNA methyltransferase
- Mgl1
- macrophage galactose-type c-type lectin
- Mrc1
- mannose receptor C type 1
- PPAR
- peroxisome proliferator-activated receptor
- SFA
- saturated fatty acid
- siRNA
- small interfering RNA
- STAT
- signal transducer and activator of transcription.
References
- 1. Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–867. [DOI] [PubMed] [Google Scholar]
- 2. Olefsky JM, Glass CK. Macrophages, inflammation, and insulin resistance. Annu Rev Physiol. 2010;72:219–246. [DOI] [PubMed] [Google Scholar]
- 3. Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112:1796–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xu H, Barnes GT, Yang Q, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest. 2003;112:1821–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fujisaka S, Usui I, Bukhari A, et al. Regulatory mechanisms for adipose tissue M1 and M2 macrophages in diet-induced obese mice. Diabetes. 2009;58:2574–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li P, Lu M, Nguyen MT, et al. Functional heterogeneity of CD11c-positive adipose tissue macrophages in diet-induced obese mice. J Biol Chem. 2010;285:15333–15345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lumeng CN, Bodzin JL, Saltiel AR. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest. 2007;117:175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu H, Perrard XD, Wang Q, et al. 1c expression in adipose tissue and blood and its role in diet-induced obesity. Arterioscler Thromb Vasc Biol. 2010;30:186–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lumeng CN, DelProposto JB, Westcott DJ, Saltiel AR. Phenotypic switching of adipose tissue macrophages with obesity is generated by spatiotemporal differences in macrophage subtypes. Diabetes. 2008;57:3239–3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Spencer M, Yao-Borengasser A, Unal R, et al. Adipose tissue macrophages in insulin-resistant subjects are associated with collagen VI and fibrosis and demonstrate alternative activation. Am J Physiol Endocrinol Metab. 2010;299:E1016–E1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. [DOI] [PubMed] [Google Scholar]
- 12. Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677–686. [DOI] [PubMed] [Google Scholar]
- 13. Bouhlel MA, Brozek J, Derudas B, et al. Unlike PPARγ, PPARα or PPARβ/δ activation does not promote human monocyte differentiation toward alternative macrophages. Biochem Biophys Res Commun. 2009;386:459–462. [DOI] [PubMed] [Google Scholar]
- 14. Bouhlel MA, Derudas B, Rigamonti E, et al. PPARγ activation primes human monocytes into alternative M2 macrophages with anti-inflammatory properties. Cell Metab. 2007;6:137–143. [DOI] [PubMed] [Google Scholar]
- 15. Charo IF. Macrophage polarization and insulin resistance: PPARγ in control. Cell Metab. 2007;6:96–98. [DOI] [PubMed] [Google Scholar]
- 16. Kang K, Reilly SM, Karabacak V, et al. Adipocyte-derived Th2 cytokines and myeloid PPARδ regulate macrophage polarization and insulin sensitivity. Cell Metab. 2008;7:485–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Odegaard JI, Ricardo-Gonzalez RR, Goforth MH, et al. Macrophage-specific PPARγ controls alternative activation and improves insulin resistance. Nature. 2007;447:1116–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Odegaard JI, Ricardo-Gonzalez RR, Red Eagle A, et al. Alternative M2 activation of Kupffer cells by PPARδ ameliorates obesity-induced insulin resistance. Cell Metab. 2008;7:496–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Luczak MW, Jagodziński PP. The role of DNA methylation in cancer development. Folia Histochem Cytobiol. 2006;44:143–154. [PubMed] [Google Scholar]
- 20. Maunakea AK, Chepelev I, Zhao K, Bruneau B. Epigenome mapping in normal and disease States. Circ Res. 2010;107:327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. [DOI] [PubMed] [Google Scholar]
- 22. Bäckdahl L, Bushell A, Beck S. Inflammatory signalling as mediator of epigenetic modulation in tissue-specific chronic inflammation. Int J Biochem Cell Biol. 2009;41:176–184. [DOI] [PubMed] [Google Scholar]
- 23. Stenvinkel P, Karimi M, Johansson S, et al. Impact of inflammation on epigenetic DNA methylation—a novel risk factor for cardiovascular disease? J Intern Med. 2007;261:488–499. [DOI] [PubMed] [Google Scholar]
- 24. Kröger H, Dietrich A, Grätz R, Wild A, Ehrlich W. The effect of tryptophan plus methionine, 5-azacytidine, and methotrexate on adjuvant arthritis of rat. Gen Pharmacol. 1999;33:195–201. [DOI] [PubMed] [Google Scholar]
- 25. Ishii M, Wen H, Corsa CA, et al. Epigenetic regulation of the alternatively activated macrophage phenotype. Blood. 2009;114:3244–3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Satoh T, Takeuchi O, Vandenbon A, et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol. 2010;11:936–944. [DOI] [PubMed] [Google Scholar]
- 27. Yang Z, Wang X, He Y, et al. The full capacity of AICAR to reduce obesity-induced inflammation and insulin resistance requires myeloid SIRT1. PLoS One. 2010;7:e49935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang Z, Kahn BB, Shi H, Xue BZ. Macrophage α1 AMP-activated protein kinase (α1AMPK) antagonizes fatty acid-induced inflammation through SIRT1. J Biol Chem. 2010;285:19051–19059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shi H, Tzameli I, Bjørbaek C, Flier JS. Suppressor of cytokine signaling 3 is a physiological regulator of adipocyte insulin signaling. J Biol Chem. 2004;279:34733–34740. [DOI] [PubMed] [Google Scholar]
- 30. Shi H, Kokoeva MV, Inouye K, Tzameli I, Yin H, Flier JS. TLR4 links innate immunity and fatty acid-induced insulin resistance. J Clin Invest. 2006;116:3015–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fujiki K, Kano F, Shiota K, Murata M. Expression of the peroxisome proliferator activated receptor γ gene is repressed by DNA methylation in visceral adipose tissue of mouse models of diabetes. BMC Biol. 2009;7:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shore A, Karamitri A, Kemp P, Speakman JR, Lomax MA. Role of Ucp1 enhancer methylation and chromatin remodelling in the control of Ucp1 expression in murine adipose tissue. Diabetologia. 2010;53:1164–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hotamisligil GS, Erbay E. Nutrient sensing and inflammation in metabolic diseases. Nat Rev Immunol. 2008;8:923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Weisenberger DJ, Velicescu M, Cheng JC, Gonzales FA, Liang G, Jones PA. Role of the DNA methyltransferase variant DNMT3b3 in DNA methylation. Mol Cancer Res. 2004;2:62–72. [PubMed] [Google Scholar]
- 35. Edwards TM, Myers JP. Environmental exposures and gene regulation in disease etiology. Environ Health Perspect. 2007;115:1264–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Skinner MK, Manikkam M, Guerrero-Bosagna C. Epigenetic transgenerational actions of environmental factors in disease etiology. Trends Endocrinol Metab. 2010;21:214–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Campión J, Milagro FI, Martinez JA. Individuality and epigenetics in obesity. Obes Rev. 2009;10:383–392. [DOI] [PubMed] [Google Scholar]
- 38. Holness MJ, Sugden MC. Epigenetic regulation of metabolism in children born small for gestational age. Curr Opin Clin Nutr Metab Care. 2006;9:482–488. [DOI] [PubMed] [Google Scholar]
- 39. Ling C, Groop L. Epigenetics: a molecular link between environmental factors and type 2 diabetes. Diabetes. 2009;58:2718–2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Maier S, Olek A. Diabetes: a candidate disease for efficient DNA methylation profiling. J Nutr. 2002;132:2440S–2443S. [DOI] [PubMed] [Google Scholar]
- 41. Szarc vel Szic K, Ndlovu MN, Haegeman G, Vanden Berghe W. Nature or nurture: let food be your epigenetic medicine in chronic inflammatory disorders. Biochem Pharmacol. 2010;80:1816–1832. [DOI] [PubMed] [Google Scholar]
- 42. Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. [DOI] [PubMed] [Google Scholar]
- 43. Stancheva I. Caught in conspiracy: cooperation between DNA methylation and histone H3K9 methylation in the establishment and maintenance of heterochromatin. Biochem Cell Biol. 2005;83:385–395. [DOI] [PubMed] [Google Scholar]
- 44. Hashimshony T, Zhang J, Keshet I, Bustin M, Cedar H. The role of DNA methylation in setting up chromatin structure during development. Nat Genet. 2003;34:187–192. [DOI] [PubMed] [Google Scholar]
- 45. Lande-Diner L, Zhang J, Ben-Porath I, et al. Role of DNA methylation in stable gene repression. J Biol Chem. 2007;282:12194–12200. [DOI] [PubMed] [Google Scholar]
- 46. Villeneuve LM, Natarajan R. The role of epigenetics in the pathology of diabetic complications. Am J Physiol Renal Physiol. 2010;299:F14–F25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. El-Osta A, Brasacchio D, Yao D, et al. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J Exp Med. 2008;205:2409–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Brasacchio D, Okabe J, Tikellis C, et al. Hyperglycemia induces a dynamic cooperativity of histone methylase and demethylase enzymes associated with gene-activating epigenetic marks that coexist on the lysine tail. Diabetes. 2009;58:1229–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.