Abstract
Ovarian follicles form through a process in which somatic pregranulosa cells encapsulate individual germ cells from germ cell syncytia. Complementary expression of the Notch ligand, Jagged1, in germ cells and the Notch receptor, Notch2, in pregranulosa cells suggests a role for Notch signaling in mediating cellular interactions during follicle assembly. Using a Notch reporter mouse, we demonstrate that Notch signaling is active within somatic cells of the embryonic ovary, and these cells undergo dramatic reorganization during follicle histogenesis. This coincides with a significant increase in the expression of the ligands, Jagged1 and Jagged2; the receptor, Notch2; and the target genes, Hes1 and Hey2. Histological examination of ovaries from mice with conditional deletion of Jagged1 within germ cells (J1 knockout [J1KO]) or Notch2 within granulosa cells (N2 knockout [N2KO]) reveals changes in follicle dynamics, including perturbations in the primordial follicle pool and antral follicle development. J1KO and N2KO ovaries also contain multi-oocytic follicles, which represent a failure to resolve germ cell syncytia, and follicles with enlarged oocytes but lacking somatic cell growth, signifying a potential role of Notch signaling in follicle activation and the coordination of follicle development. We also observed decreased cell proliferation and increased apoptosis in the somatic cells of both conditional knockout lines. As a consequence of these defects, J1KO female mice are subfertile; however, N2KO female mice remain fertile. This study demonstrates important functions for Jagged1 and Notch2 in the resolution of germ cell syncytia and the coordination of somatic and germ cell growth within follicles of the mouse ovary.
Oocytes are derived from specialized cells known as primordial germ cells that arise during embryogenesis at the base of the incipient allantois and migrate to and colonize the genital ridge of the bipotential gonad (1–3). Primordial germ cells undergo several rounds of mitosis within the gonad without complete cytokinesis (4–7). Consequently, these cells remain connected by cytoplasmic bridges (7, 8) to produce clusters known as germ cell syncytia or germ cell nests that aggregate to form the ovigerous cords (9, 10). Within syncytia, germ cells initiate meiosis anteriorly to posteriorly across the ovary and enter dictyate arrest (11). Although the function of germ cell syncytia has not been established, there is likely utility from a common local environment, eg, to share macromolecular resources, to respond simultaneously to environmental stimuli, or to develop in a coordinated manner (12).
Following meiotic arrest, germ cells within partially fragmented syncytia (9) undergo follicle histogenesis in which individual diplotene-arrested germ cells, now called oocytes, are encapsulated by a single layer of squamous pregranulosa cells to form primordial follicles (13). This process is critical for the formation of the pool of follicles available throughout the reproductive lifetime of a female (14). Because follicles are required to support oocyte development, the establishment of these structures is essential for fertility. Interestingly, by the completion of follicle histogenesis in both rodents and humans, the number of germ cells that remain is significantly reduced, with estimates of germ cell loss ranging from 30% to as much as 70% (7, 15, 16). Various hypotheses have been proposed to account for this dramatic loss, including cell apoptosis, necrosis, autophagy, and germ cell extrusion (13, 17).
A number of factors including estrogens (18–20), progesterone (21, 22), extracellular matrix-related factors (23, 24), neurotrophins (25, 26), KITL/KIT signaling (27, 28), and members of the TGFβ superfamily (29–32) have been shown to influence follicle assembly. Aberrant levels or altered signaling of many of these factors lead to defects in follicle histogenesis, which often causes the generation of follicles containing multiple oocytes, called multioocytic follicles (MOFs), that result from the incomplete fragmentation of syncytia. The fate of oocytes that develop within MOFs in vivo remains unclear, although it is likely that oocytes from these follicles have a significantly reduced fertilization capacity (33).
Once formed, primordial follicles remain quiescent until select cohorts are recruited at various times, through mechanisms that remain unclear, and transition from primordial to primary follicles (34–36). This process, termed follicle activation, results in morphological and physiological changes in both the germ and somatic cells. Oocytes within activated follicles begin to grow, whereas squamous pregranulosa cells transition into cuboidal granulosa cells, become proliferative and later steroidogenic and responsive to pituitary gonadotropins (34–36). A number of factors have been implicated in follicular activation, including members of the TGFβ superfamily (37–39) and the phosphatidylinositol 3-kinase (PI3K)/phosphatase and tensin analog (PTEN)/AKT signaling pathway (40–42).
Notch signaling (43–46) is a widely used pathway for a number of cellular processes including cell-fate specification, cellular migration, mesenchymal/epithelial transition, cell survival/death, cell division, and cell adhesion. In mammals, Notch signaling involves the interaction of one of 4 Notch receptors (Notch1, Notch2, Notch3, and Notch4) with one of 5 Notch ligands (Jagged1, Jagged2, delta-like1, delta-like3, and delta-like4). Notch-mediated juxtacrine signaling is transmitted by binding of the extracellular domain of Notch ligands to the extracellular domain of Notch receptors. Receptor-ligand interactions are further modulated by posttranslational modification of specific receptor epidermal growth factor repeats by fringe proteins (Rfng, Lfng, and Mfng). Notably, transmission of Notch signaling does not involve classical secondary messengers; rather, binding of the Notch ligand causes the Notch receptor to undergo a conformational change that initiates sequential proteolytic cleavages at the receptor juxtamembrane region that results in the release of the Notch intracellular domain from the plasma membrane and allows it to translocate to the nucleus. Within the nucleus, the Notch intracellular domain interacts with the DNA-binding transcriptional repressor, known as RBP-Jκ in the mouse, to displace corepressors and recruit transcriptional coactivators to promote the transcription of Notch target genes.
Investigation of the Drosophila ovariole has suggested that the Notch signaling pathway mediates important interactions between germ line cyst cells and surrounding somatic epithelial cells (47–49). In addition, Notch signaling has an important role in the gonad of Caenorhabditis elegans, where it functions in the mitotic to meiotic transition of germ cells (50). Previous studies have also indicated substantial conservation in the expression of Notch signaling components within the mammalian ovary (51). In the neonatal murine ovary, the Notch ligands, Jagged1 and Jagged2, are expressed in germ cells (52, 53), whereas the Notch receptors, Notch1 and Notch2, are expressed in pregranulosa cells (51, 52). Deletion of Lfng in mice results in female subfertility as a result of defects in meiotic maturation (54). Disruption of various Notch ligands, receptors, and target genes has also been shown to impact ovarian function (53, 55–58). Pharmacological inhibition of Notch signaling using the γ-secretase inhibitors N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester and L-685,458, which act to block receptor proteolysis, results in disrupted follicle histogenesis (52) and altered granulosa cell proliferation and survival (59).
In this study, we detail the expression of Notch ligands, receptors, and target genes during ovigerous cord fragmentation and follicle histogenesis. We used a Notch-responsive reporter mouse to describe the initiation of Notch activation within the mouse ovary and illustrate the organization of Notch active somatic cells immediately prior to follicle histogenesis. In addition, we generated 2 conditional knockout mouse models to study the complementary functions of Jagged1 in germ cells and Notch2 in granulosa cells. These experiments show that Notch signaling between germ and somatic cells is important for follicle histogenesis, the proliferation and survival of granulosa cells, the coordination of germ and somatic cell growth within follicles, and fertility.
Materials and Methods
Mice
Floxed Jagged1 mice (60) were generously provided by Julian Lewis (Cancer Research UK London Research Institute, London, England). Mice with a floxed Notch2 allele (61, 62) were obtained from Ursula Zimber-Strobl and Lothar Strobl (Institute of Clinical Molecular Biology and Tumor Genetics Helmholtz Zentrum, Munich, Germany). A transgenic Cre recombinase line under the control of the mouse Vasa homolog (Ddx4) proximal promoter (63) was kindly provided by Diego Castrillon (University of Texas Southwestern Medical Center, Dallas, Texas) to mediate deletion specifically within germ cells. Mice with Cre recombinase knocked into the Antimullerian hormone receptor 2 (Amhr2) gene (64) were generously provided by Richard Behringer (The University of Texas M.D. Anderson Cancer Center, Houston, Texas) to target tubular and follicular structures of the female reproductive tract, which includes granulosa cells. All conditional knockout lines were maintained on a C57BL/6J (C57BL6) background (The Jackson Laboratory). The transgenic Notch responsive (TNR) enhanced green fluorescent protein (EGFP) reporter mice (65) were a generous gift from Nicholas Gaiano (Johns Hopkins University, Baltimore, MD) and were maintained on a CD1 background (Charles River Laboratories). The TNR line contains 4 tandem CBF1/RBP-Jκ-binding sites and the basal simian virus 40 promoter that drives expression of EGFP in Notch active cells. Rosa26 reporter (Rosa26R) (FVB.129S4(B6)-Gt(ROSA)26Sortm1Sor/J) and conditional tdTomato (B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J) mice were purchased from The Jackson Laboratory. Mice were housed in controlled environmental conditions with access to water and food ad libitum on a 12-hour light, 12-hour dark cycle. Mice were fed a diet free of alfalfa and soybean meal to minimize levels of naturally occurring phytoestrogens and reduce autofluorescence in tissue samples used for ex vivo imaging (2919 Teklad Diets, for breeding; and 2916 Teklad Diets, for maintenance; Harlan). When timed matings were used, embryonic day 0.5 (E0.5) was designated as 12:00 pm on the day of vaginal plug detection. Postnatal day (PND) 0 was designated as the first 24 hours after birth. All procedures were approved by the Northwestern University Institutional Animal Care and Use Committee.
Generation of Jagged1 and Notch2 conditional knockout mice
Vasa-Cre mice were crossed with Jagged1fl/fl mice to produce Vasa-Cre male breeders with a single recombined Jagged1 allele (Jagged1+/−;Vasa-Cre). Male breeders with specific and robust Cre expression were used to improve the overall efficiency of obtaining the desired conditional knockouts. The paternal line was maintained by mating with C57BL6 females. To generate conditional Jagged1 knockouts, Jagged1+/−;Vasa-Cre males were backcrossed to Jagged1fl/fl females to produce Jagged1fl/−;Vasa-Cre (J1 knockout [J1KO]). Jagged1fl/fl mice were maintained by breeding Jagged1fl/fl male and female mice.
We used Vasa-Cre mice to delete a single floxed Notch2 allele within the germ line. This was followed by a subsequent cross to Amhr2-Cre mice, in order to replace Vasa-Cre with Amhr2-Cre to generate Notch2+/−;Amhr2-Cre males for breeding. Males with specific and robust Cre expression were then crossed with Notch2fl/fl females to generate conditional Notch2 knockout mice (Notch2fl/−;Amhr2-Cre or N2 knockout [N2KO]). The paternal line was later maintained by crossing with C57BL6 females. Notch2fl/fl mice were maintained by breeding Notch2fl/fl male and female mice.
Genotyping was performed by PCR using primer sets provided in Supplemental Table 1 published on the Endocrine Society's Journals Online web site at http://mend.endojournals.org. Heterozygous and Jagged1fl/+;Vasa-Cre (J1het) and normal littermate Jagged1fl/+ animals were used as littermate controls for J1KO samples, whereas Notch2fl/+;Amhr2-Cre (N2het) and Notch2fl/+ (normal littermate) samples were used as littermate controls for N2KO samples. C57BL6 samples were used as additional controls for both conditional knockout lines.
Multiphoton microscopy of ovarian tissues
Ovaries were dissected from embryonic and neonatal TNR mice and placed in a culture dish of PBS for imaging. Two-photon excitation microscopy (66) of whole ovaries was performed using an Olympus BX-51WIF with Coherent Chameleon Ultra II tunable laser or a Nikon A1R-MP+ multiphoton microscope equipped with a Coherent Ti:S Chameleon Vision S laser. Images were processed using FIJI/ImageJ (NIH).
Histological examination and immunologic detection methods
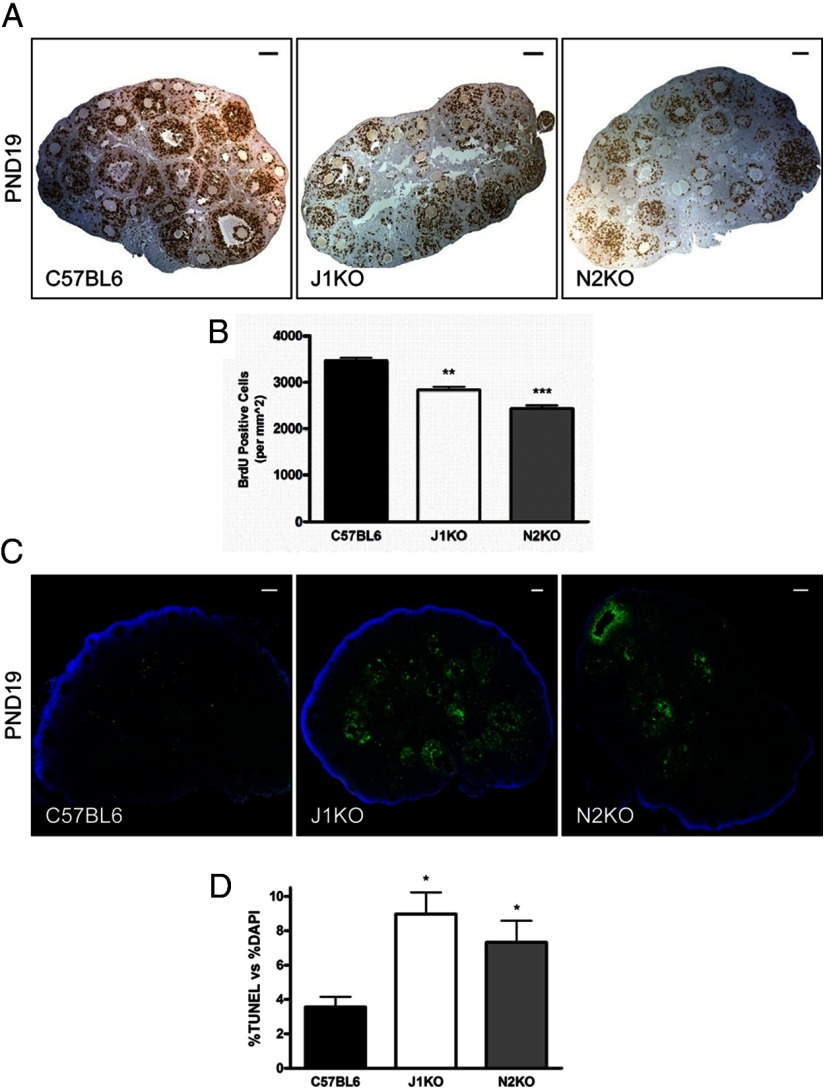
Ovarian tissue samples used for histological examination were dissected with intact bursa and fixed overnight in 4% paraformaldehyde in PBS at 4°C and dehydrated in grades of ethanol for short-term storage at 4°C. Samples were embedded in paraffin and sectioned at 5 μm for histological analysis. Hematoxylin and eosin staining, immunohistochemistry (IHC), and immunofluorescence (IF), were performed as previously described (52). To examine cell proliferation in the ovaries of conditional knockout lines, a single injection of bromodeoxyuridine (BrdU) (100 mg/kg) was administered by ip injection. Six hours postinjection, ovaries from these mice were harvested, fixed, sectioned, and immunohistochemically stained (see below). Cells that were positive for BrdU incorporation were counted using FIJI/ImageJ (NIH). To assess cellular apoptosis, terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate nick end labeling (TUNEL) was performed using the DeadEnd Fluorometric TUNEL System Kit (Promega Corp).
Primary antibodies used include a goat polyclonal JAGGED1 antibody (SC-6011) from Santa Cruz Biotechnology, a rat monoclonal antibody against NOTCH2 (C651.6DbHN) from the Developmental Studies Hybridoma Bank (Iowa City, IA), a rat anti-BrdU antibody (ab6326) from Abcam, and a rabbit polyclonal Phospho-S6 Ribosomal Protein (Ser240/244) antibody (catalog no. 2215) from Cell Signaling Technology. Biotinylated conjugated secondary antibodies against goat (BA-5000), rabbit (BA-1000), and rat (BA-9401) were purchased from Vector Laboratories and used for IF with the Tyramide Signal Amplification kit (NEL701A001KT, Perkin Elmer) or for IHC using the VECTASTAIN Elite ABC Kit (PK-6200) from Vector Laboratories. A horseradish peroxidase-conjugated secondary antibody against rat IgG (catalog no. A0545, Sigma-Aldrich) was used for Western blotting.
RNA isolation and quantitative real-time PCR
Ovaries were dissected and the bursae were removed. Samples were immediately preserved in RNAlater reagent (Life Technologies) overnight at 4°C and then stored at −80°C prior to RNA extraction. Total RNA was extracted using an RNeasy Plus Mini Kit (QIAGEN). RNA concentration and quality were assessed using a NanoDrop spectrophotometer (Thermo Scientific). RNA was reverse transcribed to cDNA using Superscript VILO master mix (Life Technologies). Quantitative real-time PCR (RT-qPCR) assays were performed using SYBR Green PCR Master Mix (Life Technologies) using either an Applied Biosystems 7300 (Life Technologies) or Bio-Rad CFX384 (Bio-Rad Laboratories) thermocycler. The comparative cycle threshold (ΔΔCt) method (67) was implemented for relative quantification using 18SrRNA or Rpl19 as an internal control (68). The sequences of all primers used for RT-qPCR are provided in Supplemental Table 1.
Morphometric analysis of ovarian follicles
Histological sections were stained with hematoxylin and eosin and examined for various follicle populations using a classification system based upon morphological criteria. Primordial and primary follicles were scored as follicles containing an oocyte surrounded by a single layer of squamous pregranulosa cells or cuboidal granulosa cells, respectively. Secondary follicles were classified as follicles containing an oocyte with 2 or more layers of granulosa cells. Antral follicles were scored as follicles containing an oocyte surrounded by multiple layers of granulosa cells with a visible antrum. In addition, the number of corpora lutea and atretic follicles (ie, follicle structures containing only the remnant of a zona pellucida or where the oocyte is detached from the granulosa cell layer) were scored in ovarian sections from samples collected following the completion (PND225) of a fertility study (see below). Aberrant follicle types were also classified histologically. Multioocytic follicles were defined as follicles containing more than a single oocyte. Follicles with a single layer of squamous or cuboidal somatic cells with an enlarged oocyte were designated as prematurely activated. Oocytes from follicles with a single somatic layer were considered enlarged when the oocyte diameter was greater than 30 μm, as oocytes from primordial (12.6 μm) and primary (28.8 μm) follicles generally do not exceed this diameter (69). Oocyte diameters were determined by averaging 2 perpendicular measurements (major/minor axes) across each oocyte. Analysis of follicle populations was performed by scoring follicles that contained an oocyte with a visible nucleolus on the medial section from a minimum of 6 different ovarian samples for each genotype. Detailed analysis of abnormal follicles was conducted using counts and measurements obtained using every 15th section across the entire ovary from a minimum of 6 different ovarian samples for each genotype. Scoring was performed using FIJI/ImageJ (NIH).
Fertility analysis of conditional knockout lines
The fertility of J1KO and N2KO female mice was assessed beginning at 6 weeks of age by continuous mating with a proven C57BL6 male for 6 months. The number of pups born from each female was recorded at the time of birth. Pups from J1KO females were genotyped to assess for the transmission of a recombined Jagged1 allele.
Statistical analysis
Data are presented as means ± SEM. Experiments were performed using replicates and control groups as stated. Differences between groups were calculated with GraphPad Prism (GraphPad Software, Inc) using ANOVA with Bonferroni correction, as appropriate, with differences considered to be significant if P < .05.
Results
Establishment and organization of Notch active cells during ovarian follicle formation
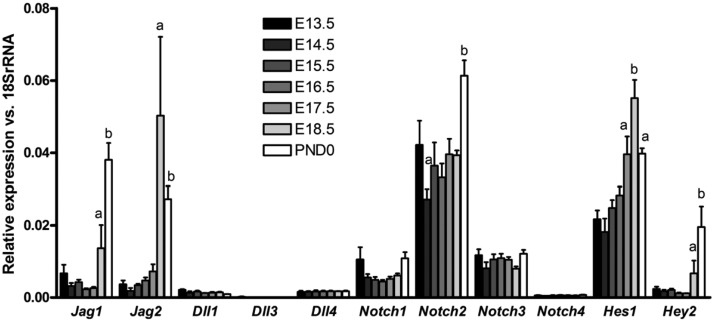
We examined the expression of Notch ligands, receptors, and target genes in the ovaries of CD1 mice from embryonic day 13.5 (E13.5) to PND0 by RT-qPCR (Figure 1) to determine the specific Notch signaling components present during ovigerous cord fragmentation and follicle histogenesis. Expression of the Notch ligands Jagged1 and Jagged2 is strongly up-regulated around E18.5, with Jagged1 being the most highly expressed ligand at PND0. Notch2 is the most abundantly expressed receptor in the embryonic ovary and is the only receptor that is up-regulated during the time frame investigated. The Notch target genes Hes1 and Hey2 are also up-regulated, consistent with elevated activation of the Notch pathway during embryonic ovarian development. The expression of other Notch ligands and receptors is significantly lower and generally constant over the time frame examined. These findings suggest that up-regulation of Jagged1 and Jagged2 may activate the Notch pathway through interaction with NOTCH2 during follicle histogenesis.
Figure 1.
Expression of Notch ligands, receptors, and target genes in the embryonic ovary. Embryonic ovaries from CD1 mice were collected from timed matings at E13.5 to PND0. Multiple ovaries were pooled to create 3 biological replicates that were used for analysis. Levels of each gene are plotted relative to 18S rRNA to show the comparative expression of Notch ligands, receptors, and target genes at each time point. For each gene, letters are used to indicate statistical differences (P < .05) in the level of expression compared with adjacent time points.
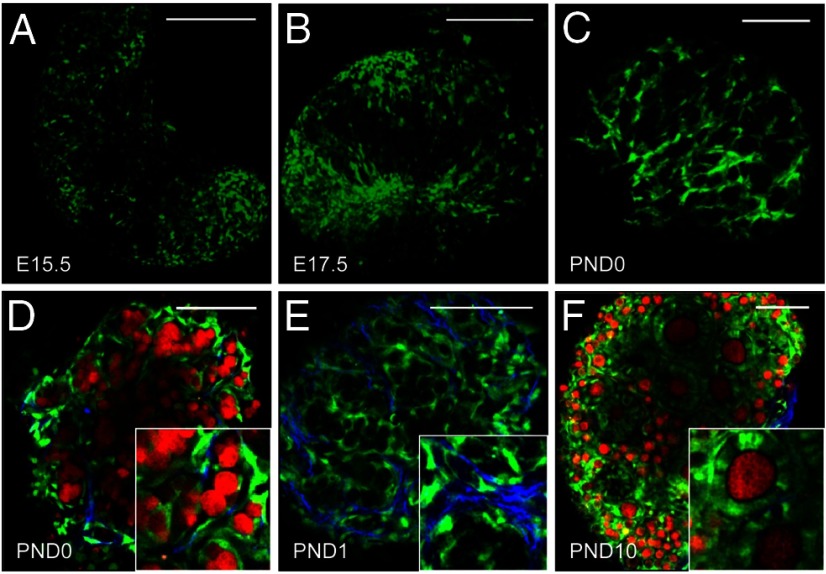
To determine the spatial and temporal pattern of Notch signaling in the developing ovary, we examined ovaries from TNR EGFP reporter mice. The earliest detectable expression of EGFP in TNR ovaries was observed in somatic cells at E15.5 (Figure 2A). At E17.5, Notch active cells are more pronounced and appear dispersed throughout the ovary (Figure 2B). At birth, which coincides with the initiation of follicle histogenesis, Notch active cells are highly organized around germ cell syncytia forming a cage-like pattern (Figure 2C). In addition, Notch active cells can be seen sending projections around individual germ cells within syncytia and enveloping oocytes of primordial follicles (Figure 2D). Some Notch active cells are also associated with collagen fibrils (Figure 2E), which could potentially facilitate pregranulosa cell migration and cellular reorganization during follicle histogenesis (70). At PND10, EGFP is still visible in granulosa cells of developing follicles; however, the levels of expression among cells of the follicle appear more variable than that observed in primordial follicles (Figure 2F).
Figure 2.
Notch active cells undergo extensive reorganization during follicle histogenesis. A, Expression of EGFP (green) from the Notch reporter line was first detected in embryonic ovaries at E15.5, although the levels were low. B, At E17.5, Notch active cells can be seen within the ovary, but are not well organized. C, By PND0, Notch active cells are arranged in a cage-like pattern around germ cell syncytia. D, In addition, Notch active cells (green) can be seen encapsulating individual germ cells (red) within syncytia that are labeled with a conditional allele for tdTomato under the direction of Vasa-Cre (63). E, Notch active cells can also be seen along collagen fibrils (blue), detected by second harmonic generation (79). F, EGFP continues to be detected in granulosa cells of primary follicles, although there is variability in the levels detected between neighboring cells. Scale bars, 100 μm.
Generation of mice with germ cell-specific deletion of Jagged1 and granulosa cell-specific deletion of Notch2
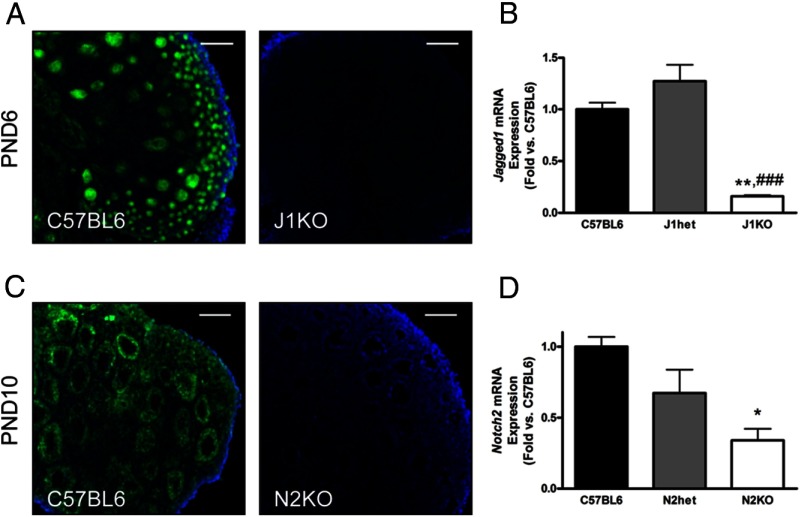
Given that JAGGED1 is known to interact with NOTCH2 (71), and based on the expression patterns and abundance of other Notch ligands and receptors, we hypothesized that Jagged1 expressed in germ cells signals through Notch2 expressed in granulosa cells to facilitate follicle assembly. Because global deletion of either Jagged1 or Notch2 results in embryonic lethality (72, 73), we used conditional approaches to investigate the role of Jagged1 and Notch2 during follicle histogenesis. Analysis of J1KO ovarian sections by IHC for JAGGED1 demonstrated that conditional deletion of Jagged1 using Vasa-Cre leads to uniform deletion across germ cells at PND6 (Figure 3A), although recombination (Supplemental Figure 1, A and B) was not always fully complete at birth. Analysis of RNA from PND19 J1KO ovaries demonstrated that Jagged1 expression was significantly reduced (84% decrease vs C57BL6) (Figure 3B). Conditional deletion of Notch2 was confirmed by decreased NOTCH2 in granulosa cells, as shown by IHC (Figure 3C). Notably, we find that recombination by Amhr2-Cre occurs in the gonads by E12.5 and was specific to somatic cells at PND0 (Supplemental Figure 1, C and D); however, it was often variable between animals and heterogeneous among the cells of individual follicles (Supplemental Figure 1, E and F). Despite this variability, Notch2 mRNA was significantly reduced (66% decrease vs C57BL6) in PND19 N2KO ovaries, as demonstrated by RT-qPCR (Figure 3D).
Figure 3.
Conditional deletion of Jagged1 in germ cells and Notch2 in granulosa cells. The Notch ligand Jagged1 was conditionally deleted in germ cells by crossing Jagged1fl/fl females with Jagged1+/−;Vasa-Cre males. A, IF demonstrates uniform deletion of JAGGED1 in J1KO ovarian sections. B, Disruption of Jagged1 is shown quantitatively by analysis of RNA collected from PND19 J1KO ovaries (n = 3). Granulosa cell-specific deletion of Notch2 was carried out by crossing a Notch2fl/fl female with a Notch2+/−;Amhr2-Cre male. C, Despite heterogeneity in Amhr2-Cre-mediated recombination, some N2KO ovaries show generally uniform deletion of Notch2 as demonstrated by IF. (D) Analysis of whole-ovary RNA collected from PND19 N2KO mice (n = 3) revealed a reduction in Notch2 transcripts, consistent with disruption of Notch2. * or **, P < .05 or P < .01 vs C57BL6; ###, P < .001 vs J1het. Scale bars, 100 μm.
Disruption of Jagged1 or Notch2 results in multioocytic follicles and uncoordinated follicular growth
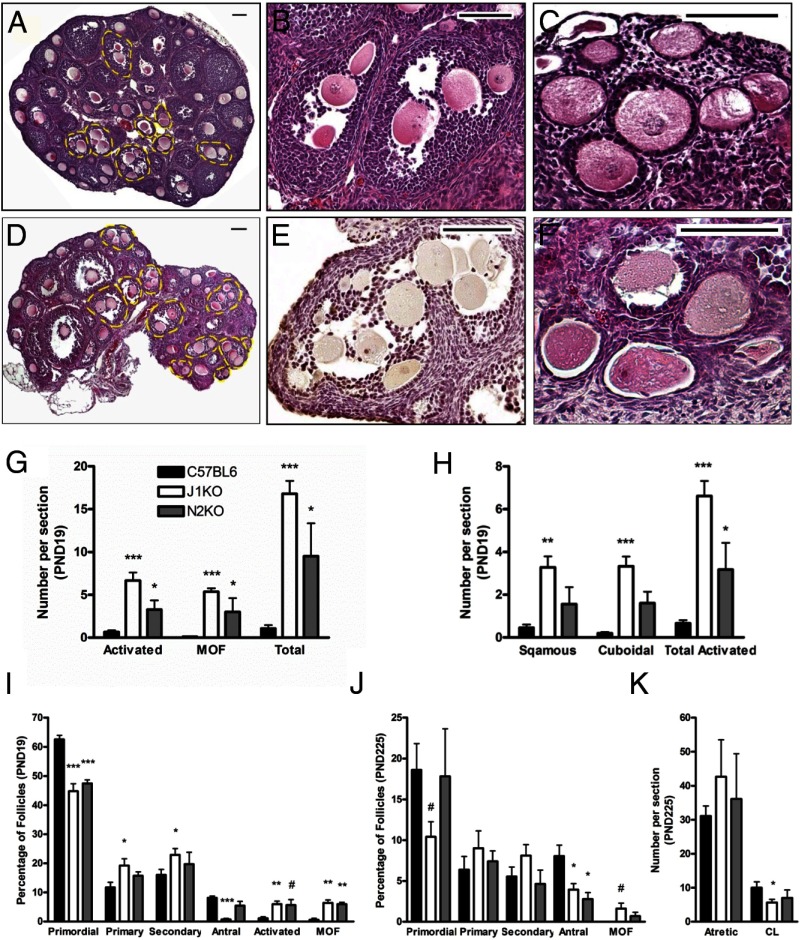
Histological examination of PND19 J1KO (Figure 4, A–C) and N2KO ovaries (Figure 4, D–F) revealed a substantial increase in the prevalence of MOFs (50-fold and 28-fold increase vs C57BL6 for J1KO and N2KO, respectively) (Figure 4, B, E, and G). MOFs observed in conditional knockout ovaries contained as many as 6 (ie, J1KOs) or 15 (ie, N2KOs) oocytes within a single section of a follicle, indicating strong disruption of ovigerous cord fragmentation. In addition, we also observe abnormal follicles with enlarged oocytes but lacking granulosa cell growth in PND19 J1KO and N2KO ovaries (Figure 4, C, F, and G). These follicles are morphologically similar to those previously reported in models in which follicles undergo premature activation (40, 41, 74), and were further categorized as containing a single layer of either squamous or cuboidal granulosa cells (Figure 4H). In addition, we examined phosphorylation of ribosomal protein S6 (rpS6), which is a downstream effector of the PI3K/PTEN/AKT pathway in oocytes. Immunohistochemical detection and subsequent analysis revealed that the percentage of oocytes that stained positive for p-rpS6 in J1KO and N2KO ovary sections was not significantly different from controls (Supplemental Figure 2, A–C).
Figure 4.
Disruption of Jagged1 or Notch2 leads to altered follicular composition and aberrant follicles. Hematoxylin and eosin-stained PND19 J1KO (A–C) and N2KO (D–F) ovarian sections revealed numerous MOFs (outlined in yellow) and abnormal follicles with enlarged oocytes encapsulated by a single squamous or cuboidal granulosa cell layer. Examples of MOFs seen in each conditional knockout are shown (B and E). In one extreme example (E), a MOF from a PND19 N2KO ovary can be seen with 15 oocytes in a single section. Examples of prematurely activated follicles in sections of J1KO (C) and N2KO (F) ovaries are also shown. G, Quantification of MOFs and prematurely activated follicles (n = 6) were scored by examination of every 15th section. H, The prematurely activated follicle phenotype is further characterized by the morphology of the surrounding somatic cells (n = 6). The distribution of follicle populations from the medial section of PND19 and PND225 conditional knockout ovaries (n = 6) is shown (I and J, respectively). K, Quantitative analysis of atretic follicles and corpora lutea observed in the medial section from PND225 ovaries are shown (n = 6). #, P < .07; *, P < .05; **, P < .01; ***, P < 0.001 or vs C57BL6. Scale bar, 100 μm.
The presence of aberrant follicles led us to explore more fully the dynamics of the follicle pool in these Notch pathway-knockout mice. Analysis of PND6 (Supplemental Figure 3A), PND10 (Supplemental Figure 3B), PND19 (Figure 4I), and PND225 (Figure 4J) conditional knockout ovaries revealed alterations in specific follicle populations. Ovaries from PND19 J1KO and N2KO mice have a moderate reduction in the number of primordial follicles, which is associated with an increase in the number of primary and secondary follicles in J1KO ovaries and aberrant follicles in both conditional knockout lines. The proportion of antral follicles present in PND19 J1KO ovaries is also significantly reduced. Antral follicles were reduced in both J1KO and N2KO ovaries at PND225 (Figure 4J), as were the number of corpora lutea observed in PND225 J1KO ovaries (Figure 4K). We did not observe changes in the number of atretic follicles at PND225 (Figure 4K and Supplemental Figure 3C) or total number of follicles at any of the times analyzed (Supplemental Figure 3D).
We also crossed the 2 conditional knockout lines to create a double conditional knockout (dKO) in which Jagged1 and Notch2 are deleted in both germ cells and somatic cells. Generation of dKO mice occurred far below the expected frequency, suggesting embryonic lethality. We speculate that this is perhaps due to ectopic expression of Cre recombinase, which occurs infrequently with each Cre allele but is likely more common with the multiple Cre alleles. Histological examination of PND19 dKO ovaries revealed that the effect of combined deletion of Jagged1 and Notch2 resulted in ovarian phenotypes that were similar to the conditional deletion of either gene individually (Supplemental Figure 3F), consistent with a model in which Jagged1 in germ cells signals through Notch2 in granulosa cells.
Cell proliferation and apoptosis are altered in PND19 J1KO and N2KO ovaries
We next examined conditional knockout ovaries for altered cell proliferation to investigate mechanisms that might explain the presence of aberrant follicles and the alterations in normal follicle populations. Proliferating cells, of which the vast majority (∼80%) were granulosa cells, were labeled through ip injection of BrdU (Figure 5A). Immunohistochemical detection of BrdU and subsequent quantitative analysis revealed that the number of proliferating cells was significantly decreased in J1KO (20% less than C57BL6) and N2KO (30% less than C57BL6) ovaries at PND19 (Figure 5B).
Figure 5.
Cell proliferation and apoptosis in the ovaries of J1KO and N2KO mice. A, Ovaries from J1KO and N2KO mice injected with BrdU were harvested 6 hours postinjection and examined for BrdU incorporation, by IHC, as a measure of cellular proliferation. B, The number of BrdU-labeled cells in PND19 J1KO (n = 3) and N2KO (n = 3) ovaries, normalized to section area, was significantly reduced compared with C57BL6 (n = 3) controls. C, Histologic sections of PND19 J1KO (n = 3) and N2KO (n = 3) ovaries labeled by TUNEL demonstrate (D) increased levels of cell death relative to C57BL6 (n = 3) controls. Cell death is predominantly localized to somatic cells in both J1KO and N2KO ovaries. *, P < .05; **, P < .01; ***, P < .001; or vs C57BL6. Scale bars, 100 μm.
In addition, we analyzed J1KO and N2KO ovaries for changes in cell apoptosis by terminal deoxynucleotide transferase-mediated dUTP nick end labeling (TUNEL) assay (Figure 5C). The results revealed a substantial increase (2.5-fold and 2.1-fold increase vs C57BL6 for J1KOs and N2KOs, respectively) in the percentage of nuclei that were positive for TUNEL at PND19 (Figure 5D). Notably, the cells that were positive for TUNEL were mostly granulosa cells, suggesting that Notch signaling originating from the oocyte serves as an important signal for granulosa cell survival. Interestingly, we did not observe appreciable changes in cell proliferation or cell death in ovaries from PND0 or PND6 knockout mice (data not shown).
Disruption of Jagged1 or Notch2 leads to altered gene expression in the ovary
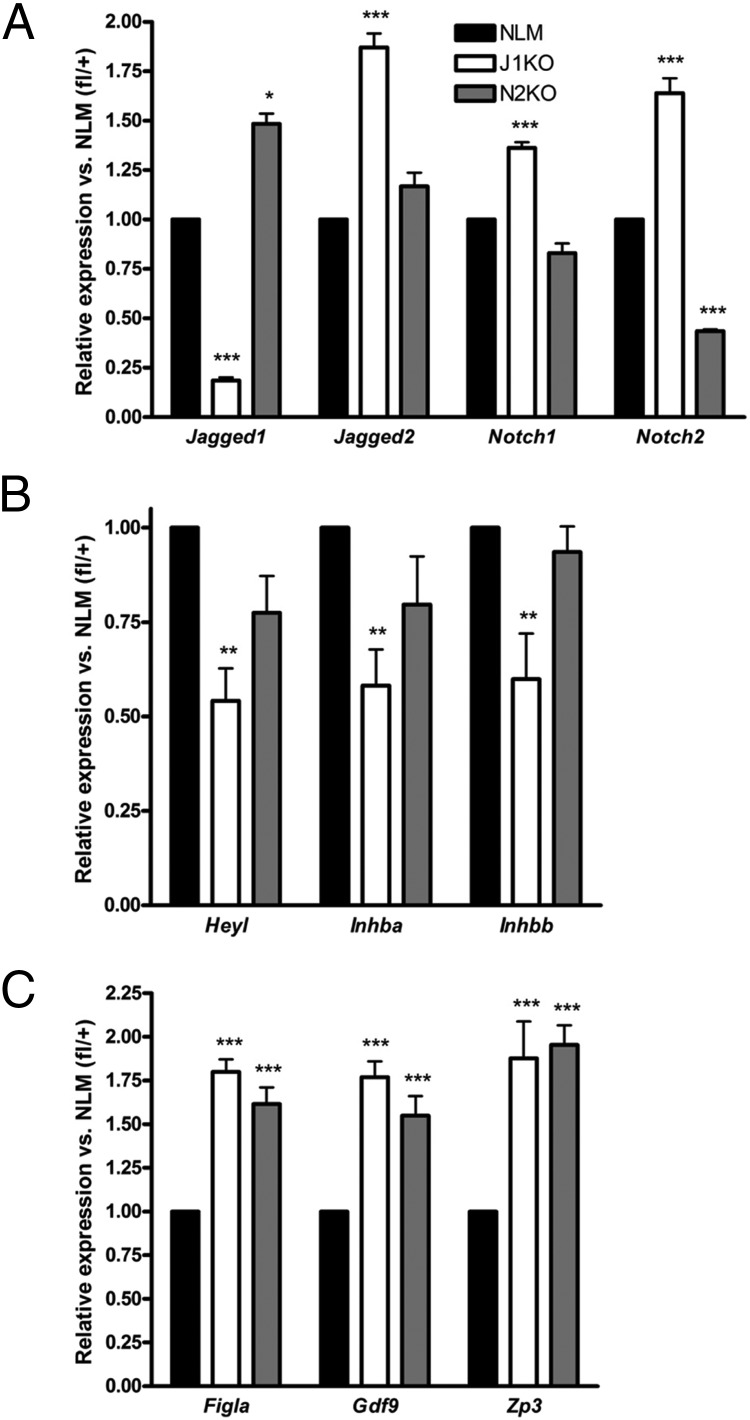
To gain further insight into the phenotypes of these conditional knockout lines, we analyzed RNA extracted from PND19 ovaries by RT-qPCR (Figure 6, A–C and Supplemental Figure 4, A–D) to explore the expression of genes of the Notch pathway, as well as genes involved in follicular development. As expected, the expression of Jagged1 or Notch2 is significantly reduced in each respective model (Figure 6A). Interestingly, the expression of Notch2 is elevated in J1KO ovaries, and conversely, the expression of Jagged1 is elevated in N2KO ovaries (Figure 6A), indicating an underlying mechanism of compensatory action between Jagged1 and Notch2. In addition, there is increased expression of Jagged2 and Notch1 in J1KO ovaries (Figure 6A), suggesting that Jagged1 loss might be compensated by increased expression of alternative Notch ligands and receptors. Despite the potential for compensatory action, the Notch target gene Heyl is significantly reduced in J1KO ovaries (Figure 6B), although the expression of several other target genes examined is unchanged (Supplemental Figure 4C).
Figure 6.
Altered gene expression in PND19 J1KO and N2KO ovaries. The expression of (A) select Notch ligands and receptors, (B) the Notch target gene, Heyl, and the inhibin/activin β-subunit genes Inhba and Inhbb, and (C) oocyte-specific genes was examined from RNA extracted from J1KO (n = 5), N2KO (n = 5), and normal littermates (ie, Jagged1fl/+ or Notch2fl/+) (n = 5) ovaries by RT-qPCR. Statistics: *, P < .05; **, P < .01; or ***, P < .001; or vs normal littermate (NLM).
Several genes associated with granulosa cell function were significantly decreased in J1KO ovaries, including Inhba and Inhbb (Figure 6B). Consistent with the abundance of follicles with uncoordinated growth observed in J1KO and N2KO ovaries, we detected significant increases in the levels of genes that are expressed in the oocytes of activated follicles, including Figla, Gdf9, and Zp3 (Figure 6C) (38, 75, 76). Together, these results demonstrate a critical role for Notch signaling in the coordination of follicle activation and growth.
Mice with germ cell-specific disruption of Jagged1 are subfertile
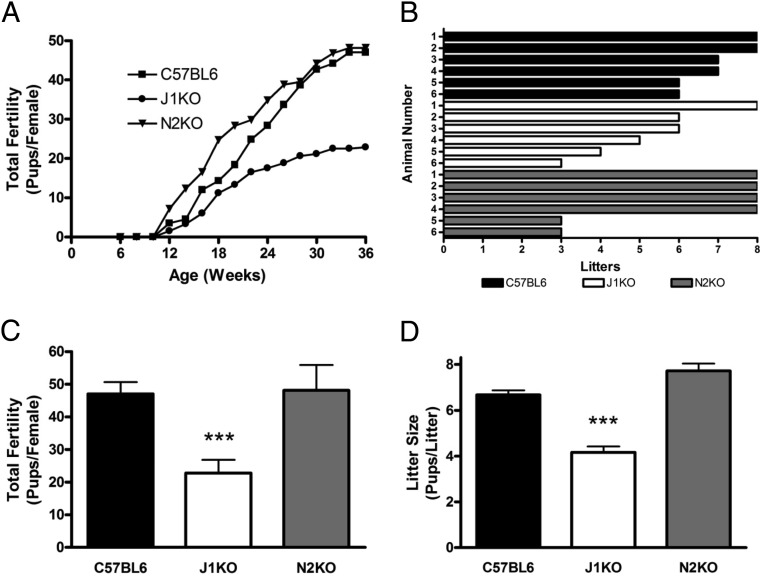
To determine whether the altered ovarian phenotypes of J1KO and N2KO mice would impact their fecundity, we conducted a fertility study for both conditional knockout lines (Figure 7, A–D). J1KO female mice were found to be subfertile (C57BL6 vs J1KO), as measured by a reduction in the total fertility (47.0 vs 22.8 pups/female; Figure 7, A and C) and average litter size (6.7 vs 4.2 pups/litter; Figure 7, B and D). Some of the J1KO females gave birth intermittently throughout the 6-month study, suggesting that some pregnancies might have been aborted. Notably, all of the offspring of J1KO females that were screened by PCR showed transmission of a recombined Jagged1 allele, suggesting that recombination was completely penetrant at the time of fertilization. In addition, we examined the fertility of J1KO males through continuous mating with a C57BL6 female and found no detectable defect in fecundity, and histological sections of J1KO testes appeared grossly normal (data not shown).
Figure 7.
Fertility assessment of J1KO and N2KO female mice. The fertility of J1KO (n = 6) and N2KO (n = 6) females was assessed in comparison with C57BL6 (n = 6) females beginning at 6 weeks of age by continuous mating with a proven C57BL6 male for 6 months. A, The number of pups in each litter was recorded, averaged, and binned in 2-week increments. J1KO females were subfertile, whereas N2KO mice remained fertile. B, The total number of litters from each individual female is represented. One of 6 J1KO and 2 of 6 N2KO females delivered 3 consecutive litters before undergoing premature reproductive senescence. The average total reproductive output (C) and average litter size (D) of J1KO and N2KO females is shown in comparison with C57BL6 female controls. ***, P < .001 vs C57BL6.
Despite the presence of numerous abnormal follicles in N2KO ovaries, the fertility of N2KO females (C57BL6 vs N2KO) was unchanged as measured by total fertility (47.0 vs 48.2 pups/female; Figure 7, A and C) and average litter size (6.7 vs 7.7 pups/litter; Figure 7, B and D). However, 2 of the 6 N2KO, and 1 of the 6 J1KO females, produced 3 consecutive litters to begin the study, but failed to produce additional litters thereafter, suggesting that these females underwent premature reproductive senescence (Figure 7B). We also did not identify any defect in the fertility of N2KO males, and histological sections of testes appeared grossly normal (data not shown).
Discussion
The results presented here reveal that activation of the Notch signaling pathway in the mouse ovary occurs in somatic pregranulosa cells between E17.5 and PND0, concomitant with increased expression of Jagged1 and Jagged2, in germ cells, and Notch2, in granulosa cells. Notch active cells within the embryonic mouse ovary undergo extensive reorganization, potentially by movement along collagen fibrils, to form a cage-like pattern surrounding germ cell syncytia. At birth, Notch active cells can be seen invading germ cell syncytia and encapsulating individual germ cells. In addition to this reorganization, the number of Notch active cells appears to increase during follicle histogenesis, indicating that either additional cells are recruited to become Notch active or that Notch active somatic cells proliferate. Recent evidence suggesting that pregranulosa cells are nonproliferative (77) favors a model in which additional pregranulosa cells are recruited to become Notch active following contact with germ cells.
The 2 conditional knockout lines described in this study were used to investigate the role of the Notch-signaling pathway during follicle assembly and development, and they provide intriguing insights into the intercellular signaling between germ cells and pregranulosa cells. Deletion of either Jagged1 or Notch2 leads to changes in follicular dynamics, including a reduction in the primordial follicle pool. In J1KO ovaries, the loss of primordial follicles corresponds with an increase in the levels of primary and secondary follicles; however, the development of antral follicles is significantly reduced, suggesting that disruption of Notch signaling impacts follicular growth of advanced follicles. Consistent with this finding, we observed a decrease in granulosa cell proliferation and an increase in cell apoptosis in the ovaries of both conditional knockouts. This effect was not observed until PND19, however, which is the first time point examined in which antral follicles are present. In addition, both J1KO and N2KO ovaries contain numerous MOFs and follicles with uncoordinated germ and somatic cell growth.
The MOF phenotype is consistent with a model in which interactions between germ cells and pregranulosa cells are disrupted prior to ovigerous cord fragmentation. We previously showed that pharmacologic inhibition of Notch signaling in ex vivo cultured newborn ovaries results in delayed ovigerous cord fragmentation and follicle assembly as seen by the retention of germ cells within syncytia (52). The current study demonstrates that continued disruption of Notch signaling between germ cells and pregranulosa cells results in the formation of MOFs. Thus, disruption of Notch signaling may prevent pregranulosa cells from interacting with germ cells in a manner that allows delineation and encapsulation of individual germ cells. Alternatively, disrupted Notch signaling may decrease the potential of pregranulosa cells to invade germ cell syncytia.
The reduction in primordial follicles and the presence of prematurely activated follicles in the ovaries of J1KO and N2KO mice suggest that Notch signaling also performs a critical role in the maintenance of resting follicles. We previously showed that although the Notch target gene Hes1 is expressed in squamous pregranulosa cells of primordial follicles, it is absent from cuboidal granulosa cells of primary follicles (52). This is despite continued expression of Jagged1 within oocytes and Notch2 within granulosa cells. For this reason, we speculate that Notch signaling between the oocyte and surrounding somatic cells may normally cease at follicle activation, possibly as a result of spatial separation caused by the formation of a zona pellucida. Consequently, the effective loss of Notch signaling between the oocyte and granulosa cells may trigger cellular events that facilitate follicle activation. Consistent with this hypothesis, we find increased expression of genes that are expressed in the oocytes of activated follicles, including Figla, Gdf9, and Zp3 in PND19 J1KO and N2KO ovaries. Interestingly, we did not see an increase in the number of p-rpS6-positive germ cells, a downstream effector of PI3K/PTEN/AKT signaling; however, additional targets or time points might be required to fully examine the impact of disrupted Notch signaling on the PI3K/PTEN/AKT pathway.
As a juxtacrine signaling system, Notch pathway activation requires direct physical contact between a ligand-expressing cell and a receptor-expressing cell. For this reason, a model in which the sole source of Notch ligand in the ovary arises from the oocyte is not consistent with continued EGFP expression in granulosa cells of developing follicles of the Notch reporter line. However, continued EGFP expression in granulosa cells distal to the oocyte of developing follicles could be a consequence of Notch signaling between adjacent granulosa cells or between juxtaposed granulosa and stroma or theca cells. This change in the source of Notch ligand likely occurs at the primary follicle stage when variability in EGFP expression is first observed between adjacent granulosa cells, which indicates that Notch signaling is not activated consistently as it would be from a single uniform source like the oocyte.
Gene expression data from J1KO and N2KO ovaries demonstrate up-regulation of alternative Notch ligands and receptors in both conditional knockout lines, suggesting that the effects of Jagged1 or Notch2 deletion might be masked by compensation or functional complementation by the expression of other Notch ligands or receptors. Interestingly, disruption of either Jagged1 or Notch2 results in up-regulation of the other gene, which supports an important link between the function and regulation of this ligand-receptor pair. Loss of Jagged1 also results in the up-regulation of Jagged2, which is first expressed in the germ cells of neonatal ovaries (53) but later is found in the granulosa cells of developing follicles (51). Consequently, functional complementation of Jagged1 by Jagged2 might be observed until this transition in Jagged2 expression occurs. Additionally, it remains unclear how heterogeneity of Notch2 recombination in the N2KO line fully impacts the ovarian phenotypes. It is possible that incomplete recombination of Notch2 within granulosa cells allows some granulosa cells with functional Notch2 to provide sufficient support for further follicular growth, perhaps explaining, in part, the less severe fertility phenotype in N2KO mice.
The cumulative impact that abnormal follicles have on the fertility of conditional knockout mice remains unclear. We have observed MOFs in the ovaries of J1KO and N2KO females that are more than 1 year old, which suggests that these follicles can persist in the ovary and are not necessarily removed from the follicle pool at an elevated rate. However, it is unlikely that the oocytes from these follicles contribute substantially to the overall fecundity of J1KO and N2KO females (33). In this study, 2 of 6 N2KO and 1 of 6 J1KO females exhibited premature reproductive senescence, which is commonly associated with hyperactivation and exhaustion of the primordial follicle pool (40, 41, 74). Despite a similar fertility defect, we did not observe complete loss of the quiescent follicle pool in the ovaries from these mice, although corpora lutea were either completely lacking or nearly absent. Incomplete disruption of the follicular reserve suggests that other mechanisms may explain the altered fertility of female conditional knockout mice. It is likely that the combined effects of perturbations in follicular composition and growth, altered cell proliferation/apoptosis, changes in gene expression, and the presence of aberrant follicle types all contribute to the overall fertility defects that are observed in female mice in which Notch signaling is disrupted within the ovary.
Recently, a similar genetic approach was reported by Xu and Gridley (57) in which an alternative floxed Notch2 allele was used to disrupt Notch signaling in ovarian somatic cells. The Notch2 allele used in that study targets exon 3, leading to premature truncation of the NOTCH2 protein (73), whereas the allele reported here targets exons 27–29, which encode the transmembrane domain of NOTCH2 (61, 62). There are numerous similarities between the 2 models, including the presence of MOFs, disruption of the resting follicle pool, and alterations in cellular apoptosis. However, Xu and Gridley find that deletion of Notch2 results in subfertility of female mice similar to the reduction we observed in J1KO females. This suggests that differences in the regions of Notch2 targeted for Cre recombination, or the efficacy of recombination, can give rise to substantial phenotypic distinctions. Another recent study by Manosalva et al (56) demonstrated that disruption of the Notch target gene Hes1 or overactivation of the Notch pathway in ovarian somatic cells leads to alterations in the number and size of oocytes and in the number of pregranulosa cells. Both Hes1 disruption and overactivation of the Notch pathway result in decreased female fertility, suggesting that Notch signaling within the ovary must be carefully balanced.
The current study, in conjunction with other recent reports, highlights the importance of the Notch signaling pathway in the development and function of ovarian follicles. We have shown that Notch activation between germ cells and pregranulosa cells is important for the resolution of germ cell syncytia and the formation and maintenance of primordial follicles. Disruption of Notch signaling results in the formation of MOFs and follicles with uncoordinated growth. This is accompanied by decreased cellular proliferation and increased apoptosis of granulosa cells, and a reduction in antral follicles. As a consequence of these defects, J1KO female mice are subfertile, and although N2KO mice remain fertile, they may exhibit premature reproductive senescence. Of particular interest, is the role of Jagged2, which seems to parallel Jagged1 in many respects. Genetic experimentation to investigate the role of Jagged2 in the ovary has not been reported; however, a hypomorphic allele for Jagged2 reportedly has poor female fertility (78). Further exploration of the Notch pathway within the ovary promises to provide new insights into the mechanisms of follicle development and maturation.
Additional material
Supplementary data supplied by authors.
Acknowledgments
We thank Diego Castrillon, MD/PhD, for use of the Vasa-Cre mouse line; Richard Behringer, PhD, for use of the Amhr2-Cre mouse line; Ursula Zimer-Strobl, PhD, and Lothar Strobl, PhD, for use of the floxed Notch2 mouse line; Julian Lewis, PhD, for use of the Jagged1 mouse line; Nicholas Gaiano, PhD, for use of the TNR reporter mouse line; Dan Trombly, PhD, for his initial involvement toward the generation of conditional knockout lines; Liz Gutierrez and Megan Romero within the Ovarian Histology Core for tissue sectioning; David Wokosin, PhD, and Teng-leong Chew, PhD, for invaluable support involving multiphoton microscopy; and colleagues in our laboratory for comments on the manuscript.
This work was supported by a grant from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (P01 HD021921) to K.E.M; D.A.V. and R.D.P. received support from the Cellular and Molecular Basis of Disease Training Program through a grant from the National Institutes of Health, Institute of General Medical Sciences (T32 GM08061); the Northwestern University Multi-Photon Core received support through a grant from the National Institute of Neurological Disorders and Stroke (P30 NS054850).
Disclosure Summary: The authors have nothing to disclose
Footnotes
- BrdU
- bromodeoxyuridine
- dKO
- double conditional knockout
- EGFP
- enhanced green fluorescent protein
- IHC
- immunohistochemistry
- IF
- immunofluorescence
- J1KO
- J1 knockout
- N2KO
- N2 knockout
- PI3K
- phosphatidylinositol 3-kinase
- PND
- postnatal day
- PTEN
- phosphatase and tensin analog
- RT-qPCR
- quantitative real-time PCR
- TNR
- transgenic Notch responsive
- TUNEL
- terminal deoxynucleotide transferase-mediated dUTP nick end labeling.
References
- 1. Saitou M, Yamaji M. Primordial germ cells in mice. Cold Spring Harb Perspect Biol. 2012;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ohinata Y, Payer B, O'Carroll D, et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436:207–213. [DOI] [PubMed] [Google Scholar]
- 3. Molyneaux KA, Stallock J, Schaible K, Wylie C. Time-lapse analysis of living mouse germ cell migration. Dev Biol. 2001;240:488–498. [DOI] [PubMed] [Google Scholar]
- 4. Tam PP, Snow MH. Proliferation and migration of primordial germ cells during compensatory growth in mouse embryos. J Embryol Exp Morphol. 1981;64:133–147. [PubMed] [Google Scholar]
- 5. Anderson EL, Baltus AE, Roepers-Gajadien HL, et al. Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice. Proc Natl Acad Sci USA. 2008;105:14976–14980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pepling ME, Spradling AC. Female mouse germ cells form synchronously dividing cysts. Development. 1998;125:3323–3328. [DOI] [PubMed] [Google Scholar]
- 7. Pepling ME, Spradling AC. Mouse ovarian germ cell cysts undergo programmed breakdown to form primordial follicles. Dev Biol. 2001;234:339–351. [DOI] [PubMed] [Google Scholar]
- 8. Greenbaum MP, Iwamori T, Buchold GM, Matzuk MM. Germ cell intercellular bridges. Cold Spring Harb Perspect Biol. 2011;3:a005850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lei L, Spradling AC. Mouse primordial germ cells produce cysts that partially fragment prior to meiosis. Development. 2013;140:2075–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mork L, Tang H, Batchvarov I, Capel B. Mouse germ cell clusters form by aggregation as well as clonal divisions. Mech Dev. 2012;128:591–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Menke DB, Koubova J, Page DC. Sexual differentiation of germ cells in XX mouse gonads occurs in an anterior-to-posterior wave. Dev Biol. 2003;262:303–312. [DOI] [PubMed] [Google Scholar]
- 12. Mayo K, Jameson L, Woodruff TK. Eggs in the nest. Endocrinology. 2007;148:3577–3579. [DOI] [PubMed] [Google Scholar]
- 13. Pepling ME. Follicular assembly: mechanisms of action. Reproduction. 2012;143:139–149. [DOI] [PubMed] [Google Scholar]
- 14. Tingen C, Kim A, Woodruff TK. The primordial pool of follicles and nest breakdown in mammalian ovaries. Mol Hum Reprod. 2009;15:795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bristol-Gould SK, Kreeger PK, Selkirk CG, et al. Postnatal regulation of germ cells by activin: the establishment of the initial follicle pool. Dev Biol. 2006;298:132–148. [DOI] [PubMed] [Google Scholar]
- 16. Kerr JB, Duckett R, Myers M, Britt KL, Mladenovska T, Findlay JK. Quantification of healthy follicles in the neonatal and adult mouse ovary: evidence for maintenance of primordial follicle supply. Reproduction. 2006;132:95–109. [DOI] [PubMed] [Google Scholar]
- 17. Rodrigues P, Limback D, McGinnis LK, Plancha CE, Albertini DF. Multiple mechanisms of germ cell loss in the perinatal mouse ovary. Reproduction. 2009;137:709–720. [DOI] [PubMed] [Google Scholar]
- 18. Chen Y, Breen K, Pepling ME. Estrogen can signal through multiple pathways to regulate oocyte cyst breakdown and primordial follicle assembly in the neonatal mouse ovary. J Endocrinol. 2009;202:407–417. [DOI] [PubMed] [Google Scholar]
- 19. Chen Y, Jefferson WN, Newbold RR, Padilla-Banks E, Pepling ME. Estradiol, progesterone, and genistein inhibit oocyte nest breakdown and primordial follicle assembly in the neonatal mouse ovary in vitro and in vivo. Endocrinology. 2007;148:3580–3590. [DOI] [PubMed] [Google Scholar]
- 20. Kipp JL, Kilen SM, Bristol-Gould S, Woodruff TK, Mayo KE. Neonatal exposure to estrogens suppresses activin expression and signaling in the mouse ovary. Endocrinology. 2007;148:1968–1976. [DOI] [PubMed] [Google Scholar]
- 21. Nilsson EE, Stanfield J, Skinner MK. Interactions between progesterone and tumor necrosis factor-α in the regulation of primordial follicle assembly. Reproduction. 2006;132:877–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nilsson EE, Skinner MK. Progesterone regulation of primordial follicle assembly in bovine fetal ovaries. Mol Cell Endocrinol. 2009;313:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brown HM, Dunning KR, Robker RL, Pritchard M, Russell DL. Requirement for ADAMTS-1 in extracellular matrix remodeling during ovarian folliculogenesis and lymphangiogenesis. Dev Biol. 2006;300:699–709. [DOI] [PubMed] [Google Scholar]
- 24. Mazaud S, Guyot R, Guigon CJ, Coudouel N, Le Magueresse-Battistoni B, Magre S. Basal membrane remodeling during follicle histogenesis in the rat ovary: contribution of proteinases of the MMP and PA families. Dev Biol. 2005;277:403–416. [DOI] [PubMed] [Google Scholar]
- 25. Kerr B, Garcia-Rudaz C, Dorfman M, Paredes A, Ojeda SR. NTRK1 and NTRK2 receptors facilitate follicle assembly and early follicular development in the mouse ovary. Reproduction. 2009;138:131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nilsson E, Dole G, Skinner MK. Neurotrophin NT3 promotes ovarian primordial to primary follicle transition. Reproduction. 2009;138:697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jones RL, Pepling ME. KIT signaling regulates primordial follicle formation in the neonatal mouse ovary. Dev Biol. 2013;382:186–197 [DOI] [PubMed] [Google Scholar]
- 28. Hutt KJ, McLaughlin EA, Holland MK. KIT/KIT ligand in mammalian oogenesis and folliculogenesis: roles in rabbit and murine ovarian follicle activation and oocyte growth. Biol Reprod. 2006;75:421–433. [DOI] [PubMed] [Google Scholar]
- 29. McMullen ML, Cho BN, Yates CJ, Mayo KE. Gonadal pathologies in transgenic mice expressing the rat inhibin α-subunit. Endocrinology. 2001;142:5005–5014. [DOI] [PubMed] [Google Scholar]
- 30. Nilsson EE, Schindler R, Savenkova MI, Skinner MK. Inhibitory actions of Anti-Mullerian Hormone (AMH) on ovarian primordial follicle assembly. PloS one. 2011;6:e20087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Myers M, Tripurani SK, Middlebrook B, Economides AN, Canalis E, Pangas SA. Loss of gremlin delays primordial follicle assembly but does not affect female fertility in mice. Biol Reprod. 2011;85:1175–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Memon MA, Anway MD, Covert TR, Uzumcu M, Skinner MK. Transforming growth factor β (TGFβ1, TGFβ2 and TGFβ3) null-mutant phenotypes in embryonic gonadal development. Mol Cell Endocrinol. 2008;294:70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Iguchi T, Kamiya K, Uesugi Y, Sayama K, Takasugi N. In vitro fertilization of oocytes from polyovular follicles in mouse ovaries exposed neonatally to diethylstilbestrol. In Vivo. 1991;5:359–363. [PubMed] [Google Scholar]
- 34. Reddy P, Zheng W, Liu K. Mechanisms maintaining the dormancy and survival of mammalian primordial follicles. Trends Endocrinol Metab. 2010;21:96–103. [DOI] [PubMed] [Google Scholar]
- 35. Adhikari D, Liu K. Molecular mechanisms underlying the activation of mammalian primordial follicles. Endocr Rev. 2009;30:438–464. [DOI] [PubMed] [Google Scholar]
- 36. McLaughlin EA, McIver SC. Awakening the oocyte: controlling primordial follicle development. Reproduction. 2009;137:1–11. [DOI] [PubMed] [Google Scholar]
- 37. Pangas SA. Regulation of the ovarian reserve by members of the transforming growth factor beta family. Mol Reprod Dev. 2012;79:666–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dong J, Albertini DF, Nishimori K, Kumar TR, Lu N, Matzuk MM. Growth differentiation factor-9 is required during early ovarian folliculogenesis. Nature. 1996;383:531–535. [DOI] [PubMed] [Google Scholar]
- 39. Yan C, Wang P, DeMayo J, et al. Synergistic roles of bone morphogenetic protein 15 and growth differentiation factor 9 in ovarian function. Mol Endocrinol. 2001;15:854–866. [DOI] [PubMed] [Google Scholar]
- 40. Reddy P, Liu L, Adhikari D, et al. Oocyte-specific deletion of Pten causes premature activation of the primordial follicle pool. Science. 2008;319:611–613. [DOI] [PubMed] [Google Scholar]
- 41. Adhikari D, Zheng W, Shen Y, et al. Tsc/mTORC1 signaling in oocytes governs the quiescence and activation of primordial follicles. Hum Mol Genet. 2010;19:397–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Reddy P, Adhikari D, Zheng W, et al. PDK1 signaling in oocytes controls reproductive aging and lifespan by manipulating the survival of primordial follicles. Hum Mol Genet. 2009;18:2813–2824. [DOI] [PubMed] [Google Scholar]
- 43. Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. [DOI] [PubMed] [Google Scholar]
- 44. Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Guruharsha KG, Kankel MW, Artavanis-Tsakonas S. The Notch signalling system: recent insights into the complexity of a conserved pathway. Genetics. 2012;13:654–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kopan R. Notch signaling. In: Current Topics in Developmental Biology. 1st ed San Diego, CA: Academic Press; 2010. 1 online resource (xvi, 530 p., 524 p. of plates) [Google Scholar]
- 47. Ruohola H, Bremer KA, Baker D, Swedlow JR, Jan LY, Jan YN. Role of neurogenic genes in establishment of follicle cell fate and oocyte polarity during oogenesis in Drosophila. Cell. 1991;66:433–449. [DOI] [PubMed] [Google Scholar]
- 48. Xu T, Caron LA, Fehon RG, Artavanis-Tsakonas S. The involvement of the Notch locus in Drosophila oogenesis. Development. 1992;115:913–922. [DOI] [PubMed] [Google Scholar]
- 49. López-Schier H, St Johnston D. Delta signaling from the germ line controls the proliferation and differentiation of the somatic follicle cells during Drosophila oogenesis. Genes Dev. 2001;15:1393–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kadyk LC, Lambie EJ, Kimble J. glp-3 is required for mitosis and meiosis in the Caenorhabditis elegans germ line. Genetics. 1997;145:111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Johnson J, Espinoza T, McGaughey RW, Rawls A, Wilson-Rawls J. Notch pathway genes are expressed in mammalian ovarian follicles. Mech Dev. 2001;109:355–361. [DOI] [PubMed] [Google Scholar]
- 52. Trombly DJ, Woodruff TK, Mayo KE. Suppression of Notch signaling in the neonatal mouse ovary decreases primordial follicle formation. Endocrinology. 2009;150:1014–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Guo M, Zhang H, Bian F, et al. P4 down-regulates Jagged2 and Notch1 expression during primordial folliculogenesis. Front Biosci. 2012;4:2731–2744. [DOI] [PubMed] [Google Scholar]
- 54. Hahn KL, Johnson J, Beres BJ, Howard S, Wilson-Rawls J. Lunatic fringe null female mice are infertile due to defects in meiotic maturation. Development. 2005;132:817–828. [DOI] [PubMed] [Google Scholar]
- 55. Dorfman MD, Kerr B, Garcia-Rudaz C, Paredes AH, Dissen GA, Ojeda SR. Neurotrophins acting via TRKB receptors activate the JAGGED1-NOTCH2 cell-cell communication pathway to facilitate early ovarian development. Endocrinology. 2011;152:5005–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Manosalva I, González A, Kageyama R. Hes1 in the somatic cells of the murine ovary is necessary for oocyte survival and maturation. Dev Biol. 2013;375:140–151. [DOI] [PubMed] [Google Scholar]
- 57. Xu J, Gridley T. Notch2 is required in somatic cells for breakdown of ovarian germ-cell nests and formation of primordial follicles. BMC Biol. 2013;11:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hunkapiller NM, Gasperowicz M, Kapidzic M, et al. A role for Notch signaling in trophoblast endovascular invasion and in the pathogenesis of pre-eclampsia. Development. 2011;138:2987–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang CP, Yang JL, Zhang J, et al. Notch signaling is involved in ovarian follicle development by regulating granulosa cell proliferation. Endocrinology. 2011;152:2437–2447. [DOI] [PubMed] [Google Scholar]
- 60. Brooker R, Hozumi K, Lewis J. Notch ligands with contrasting functions: Jagged1 and Delta1 in the mouse inner ear. Development. 2006;133:1277–1286. [DOI] [PubMed] [Google Scholar]
- 61. Saito T, Chiba S, Ichikawa M, et al. Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity. 2003;18:675–685. [DOI] [PubMed] [Google Scholar]
- 62. Schouwey K, Delmas V, Larue L, et al. Notch1 and Notch2 receptors influence progressive hair graying in a dose-dependent manner. Dev Dyn. 2007;236:282–289. [DOI] [PubMed] [Google Scholar]
- 63. Gallardo T, Shirley L, John GB, Castrillon DH. Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis. 2007;45:413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jamin SP, Arango NA, Mishina Y, Hanks MC, Behringer RR. Requirement of Bmpr1a for Müllerian duct regression during male sexual development. Nat Genet. 2002;32:408–410. [DOI] [PubMed] [Google Scholar]
- 65. Duncan AW, Rattis FM, DiMascio LN, et al. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nat Immunol. 2005;6:314–322. [DOI] [PubMed] [Google Scholar]
- 66. Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. [DOI] [PubMed] [Google Scholar]
- 67. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) method. Methods. 2001;25:402–408. [DOI] [PubMed] [Google Scholar]
- 68. Camp TA, Rahal JO, Mayo KE. Cellular localization and hormonal regulation of follicle-stimulating hormone and luteinizing hormone receptor messenger RNAs in the rat ovary. Mol Endocrinol. 1991;5:1405–1417. [DOI] [PubMed] [Google Scholar]
- 69. Griffin J, Emery BR, Huang I, Peterson CM, Carrell DT. Comparative analysis of follicle morphology and oocyte diameter in four mammalian species (mouse, hamster, pig, and human). J Exp Clin Assist Reprod. 2006;3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Berkholtz CB, Lai BE, Woodruff TK, Shea LD. Distribution of extracellular matrix proteins type I collagen, type IV collagen, fibronectin, and laminin in mouse folliculogenesis. Histochem Cell Biol. 2006;126:583–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Shimizu K, Chiba S, Kumano K, et al. Mouse jagged1 physically interacts with notch2 and other notch receptors. Assessment by quantitative methods. J Biol Chem. 1999;274:32961–32969. [DOI] [PubMed] [Google Scholar]
- 72. Xue Y, Gao X, Lindsell CE, et al. Embryonic lethality and vascular defects in mice lacking the Notch ligand Jagged1. Hum Mol Genet. 1999;8:723–730. [DOI] [PubMed] [Google Scholar]
- 73. McCright B, Lozier J, Gridley T. Generation of new Notch2 mutant alleles. Genesis. 2006;44:29–33. [DOI] [PubMed] [Google Scholar]
- 74. Castrillon DH, Miao L, Kollipara R, Horner JW, DePinho RA. Suppression of ovarian follicle activation in mice by the transcription factor Foxo3a. Science. 2003;301:215–218. [DOI] [PubMed] [Google Scholar]
- 75. Soyal SM, Amleh A, Dean J. FIGα, a germ cell-specific transcription factor required for ovarian follicle formation. Development. 2000;127:4645–4654. [DOI] [PubMed] [Google Scholar]
- 76. Rankin T, Familari M, Lee E, et al. Mice homozygous for an insertional mutation in the Zp3 gene lack a zona pellucida and are infertile. Development. 1996;122:2903–2910. [DOI] [PubMed] [Google Scholar]
- 77. Mork L, Maatouk DM, McMahon JA, et al. Temporal differences in granulosa cell specification in the ovary reflect distinct follicle fates in mice. Biol Reprod. 2012;86:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Grüneberg H. 1956 Genetical studies on the skeleton of the mouse. Section XVIII. Three genes for syndactylism. J Genet. 1956;113–145. [Google Scholar]
- 79. Williams RM, Zipfel WR, Webb WW. Interpreting second-harmonic generation images of collagen I fibrils. Biophys J. 2005;88:1377–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.