Abstract
Excessive bone resorption is the cause of several metabolic bone diseases including osteoporosis. Thus, identifying factors that can inhibit osteoclast formation and/or activity may define new drug targets that can be used to develop novel therapies for these conditions. Emerging evidence demonstrates that the master regulator of hematopoiesis, Runx1, is expressed in preosteoclasts and may influence skeletal health. To examine the potential role of Runx1 in osteoclast formation and function, we deleted its expression in myeloid osteoclast precursors by crossing Runx1 floxed mice (Runx1F/F) with CD11b-Cre transgenic mice. Mice lacking Runx1 in preosteoclasts (CD11b-Cre;Runx1F/F) exhibited significant loss of femoral trabecular and cortical bone mass compared with that in Cre-negative mice. In addition, serum levels of collagen type 1 cross-linked C-telopeptide, a biomarker of osteoclast-mediated bone resorption, were significantly elevated in CD11b-Cre;Runx1F/F mice compared with those in Runx1F/F mice. Tartrate-resistant acid phosphatase–positive osteoclasts that differentiated from bone marrow cells of CD11b-Cre;Runx1F/F mice in vitro were larger, were found in greater numbers, and had increased bone resorbing activity than similarly cultured cells from Runx1F/F mice. CD11b-Cre;Runx1F/F bone marrow cells that were differentiated into osteoclasts in vitro also had elevated mRNA levels of osteoclast-related genes including vacuolar ATPase D2, cathepsin K, matrix metalloproteinase 9, calcitonin receptor, osteoclast-associated receptor, nuclear factor of activated T cells cytoplasmic 1, and cFos. These data indicate that Runx1 expression in preosteoclasts negatively regulates osteoclast formation and activity and contributes to overall bone mass.
Mature osteoclasts, which originate from hematopoietic stem cells, are multinuclear giant cells that efficiently resorb bone (1). It is well established that myeloid progenitor cells differentiate into preosteoclasts after exposure to macrophage colony-stimulating factors (M-CSF) (also known as CSF-1) (2). Early osteoclast precursors progress through a stage of being CD11b+ during their differentiation into mature osteoclasts (3, 4). Hence, expression of CD11b can be considered a marker of late preosteoclasts. Differentiation of CD11b+ cells into mature osteoclasts occurs in response to exposure to M-CSF and receptor activator of nuclear factor-κB ligand (RANKL) (5).
Excessive bone resorption is observed in pathological conditions such as osteoporosis, tumor-induced osteolysis, or wear debris osteolysis (6). In the past 2 decades, antiresorptive drugs such as aminobisphosphonates and, more recently, anti-RANKL antibodies were developed to inhibit bone resorption and decrease the incidence of fractures (7). However, increasing evidence has linked their use to a number of significant side effects, including atypical subtrochanteric femoral fractures (8) and osteonecrosis of the jaw (9). Therefore, there is a critical need to identify new drug targets that can be used to develop novel, safer, and more effective therapies for metabolic bone diseases that result from enhanced bone resorption.
Multiple signaling pathways, activating key transcription factors, have been shown to regulate various genes that are necessary for normal osteoclast differentiation and function. Transcriptional activators such as nuclear factor-κB, nuclear factor of activated T cells, cFos, microphthalmia-associated transcription factor, and PU.1 have been well established as inducers of osteoclastogenesis and osteoclast activity (10). Recent studies also showed that RANKL signaling was necessary to inhibit transcriptional repressors of osteoclastogenesis such as Id (11), MafB (12), IRF-8 (13), BCL6 (14), and RBP-J (15) to initiate osteoclast formation and function.
Genetic evidence has demonstrated that Runx1 is required for the development of hematopoiesis in mice (16, 17). In addition, deletion of Runx1 broadly in hematopoietic cells in adult mice produced a phenotype that was characterized by more myeloid cells (18), which include osteoclast precursor (3). Similar to other previously identified repressors of osteoclast differentiation, Runx1, which is widely expressed in hematopoietic progenitors within the bone marrow (19), was shown to be inhibited by RANKL in osteoclasts (20). Bai et al (21) previously reported that TNF receptor–associated factor 6, which is a downstream mediator of RANKL signaling, binds a scaffolding protein, four and a half LIM domain 2 (FHL2), to recruit Runx1 into a transcriptional complex capable of transactivating the fhl2 promoter and thereby increasing FHL2 expression. They also found that FHL2 subsequently blunts RANKL-mediated osteoclast formation and function (22). Taken together, these data suggest a potential function for Runx1 in bone resorption. However, whether Runx1 plays a direct role in regulating osteoclastic activity in vivo remains unknown. To examine this question, we performed in vitro and in vivo experiments that demonstrate for the first time that Runx1 is an inhibitor of osteoclast differentiation and function.
Materials and Methods
Animals
CD11b-Cre;Runx1F/F mice were generated by breeding Runx1F/F mice (18) with CD11b-Cre transgenic mice (23). Only male mice were examined because the CD11b-Cre transgene that was used in these studies was inserted into the Y chromosome (23). Mice were housed at the University of Connecticut Health Center animal facilities according to state and federal law requirements. The use of animals in all experiments was approved by the institutional animal care and use committee of University of Connecticut Health Center.
Flow cytometry
Bone marrow cells, isolated from murine long bones, were used to determine the distribution of osteoclast precursors. Cells were stained with antibodies conjugated to fluorochromes and analyzed by FlowJo software (Tree Star, Inc) using an LSR II flow cytometer (BD Biosciences) as described previously (3).
Fluorescence-activated cell sorting (FACS)
Flow cytometry was performed as described previously (3) to isolate CD11b+ cells. Sorting was performed on a FACSAria cell sorter (BD Biosciences) equipped with 5 lasers and 18 fluorescence detectors.
Micro-computed tomography (CT)
Femoral trabecular bone morphometry was measured using cone-beam microfocus X-ray CT (Scanco Medical AG) as described previously (24, 25). A 0.96-mm segment of trabecular bone was quantitatively analyzed at a 0.96-mm distance from the growth plate toward the femoral midshaft. In addition, cortical thickness was measured within a 0.6-mm segment of cortical bone, located 4.5 mm from the growth plate.
Serum C-terminal telopeptide of type 1 collagen (CTX)
Serum samples obtained from both Runx1F/F and CD11b-Cre;Runx1F/F mice were used to measure CTX with a RatLaps enzyme immunoassay as described previously (25, 26). For these studies, mice were fasted overnight with free access to water before killing.
Static histomorphometric analyses
Femurs fixed with 10% neutral buffered formalin were decalcified in 10% EDTA and then embedded in paraffin. Five-micron-thick serial sections were stained using a tartrate-resistant acid phosphatase (TRAP) staining kit (Kamiya Biomedical Company) and then were counterstained with hematoxylin. The same regions of interest for micro-CT were analyzed for static histomorphometry via OsteoMeasure software (OsteoMetrics, Inc), as described previously (26).
In vitro assays
Bone marrow macrophage cells were isolated from the long bones of either Runx1F/F or CD11b-Cre;Runx1F/F mice as described previously (27). Cells were induced to become osteoclasts in vitro by treatment with 30 ng/mL M-CSF and various doses of RANKL (0, 10, or 30 ng/mL). At the termination of experiments, assays were performed for osteoclastogenesis, pit formation, and gene expression.
Osteoclast formation assays
The ability of progenitor cells to form mature osteoclasts was evaluated via cell counting of multinucleated (>3 nuclei) TRAP+ cells as described previously (27).
Pit formation
Bone marrow macrophage cells placed on bovine cortical bone slices were incubated with 30 ng/mL M-CSF and 30 ng/mL RANKL at the bottom of a 96-well plate for 2 weeks as detailed previously (3). Cells were removed from the bone slices using a sonicator. The bone slices were then stained with toluidine blue and used to quantitatively measure numbers of pits and area using cellSens dimension software (Olympus America Inc).
Gene expression
Total RNA was extracted using TRIzol reagent (Life Technologies). RNA was converted to cDNA by a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) as described previously (25). cDNA from each sample was mixed with gene-specific PCR primers and TaqMan probes (Applied Biosystems) to assay for levels of gene expression by real-time PCR amplification using an ABI PRISM 7500 sequence detection system (Applied Biosystems). Relative gene expression for each specific target was normalized to the expression of glyceraldehyde 3-phosphate dehydrogenase.
Statistical analysis
Values in all in vivo studies are presented as means ± SE (n = 5–8/group). An unpaired t test was performed to determine which means of groups were significantly different (P < .05) using GraphPad Prism 4 (GraphPad Software, Inc). For in vitro studies, data are means ± SE (n = 3) and are representative of 3 replicate experiments. The unpaired t test was used to determine which means were significantly different (P < .05) or had a trend to be different (P < .1).
Results
Runx1 gene is deleted by CD11b-driven Cre recombinase in vivo
We first examined the ability of RANKL to suppress Runx1 expression in maturing osteoclasts in vitro. Mouse bone marrow macrophage (BMM) cultures were incubated with either M-CSF or M-CSF plus RANKL for 3 days, and then RNA was extracted to measure Runx1 mRNA levels (Supplemental Figure 1). We found that RANKL decreased Runx1 mRNA by >80% in cultures from both male and female mice. This result confirmed a previous study (20), which demonstrated that RANKL dramatically decreased Runx1 gene expression in osteoclastogenic cultures.
To examine the effect of Runx1 on the formation and activity of osteoclasts in vivo, we used a conditional knockout approach. CD11b-Cre;Runx1F/F mice were generated by crossing Runx1F/F mice (18) with CD11b-Cre transgenic mice (23), which express Cre recombinase under the control of the 5′ regulatory region of CD11b, a marker of osteoclast precursors (4). To measure the degree of rearrangement of the Runx1 gene by CD11b-mediated Cre recombinase, we isolated DNA from CD11b+ bone marrow cells of CD11b-Cre;Runx1F/F mice by FACS to high purity (>99%) as described previously (3). Controls were FACS-isolated CD11b+ cells from Runx1F/F mice. As expected, there was no rearrangement of the Runx1 gene in cells of Runx1F/F mice (Figure 1). In contrast, Cre recombinase–induced deletion of the Runx1 gene (∼60%) was observed in CD11b+ bone marrow cells of CD11b-Cre;Runx1F/F mice.
Figure 1.
Runx1 gene was deleted by Cre recombinase in vivo. DNA isolated from CD11b+ bone marrow cells of 8-week-old Runx1F/F and CD11b-Cre;Runx1F/F mice was used to measure CD11b-Cre–mediated Runx1 gene deletion. After PCR for the Runx1 loci using a 3-primer set (18), PCR products from CD11b+ cells of both Runx1F/F and CD11b-Cre;Runx1F/F mice were run on ethidium bromide–stained 2% agarose gels. F, Runx1 floxed allele; Δ, deleted allele. The percentage of Cre recombination was quantitatively measured using ImageJ software (National Institutes of Health).
Deletion of Runx1 gene in CD11b+ bone marrow cells reduced bone mass and enhanced resorption in vivo
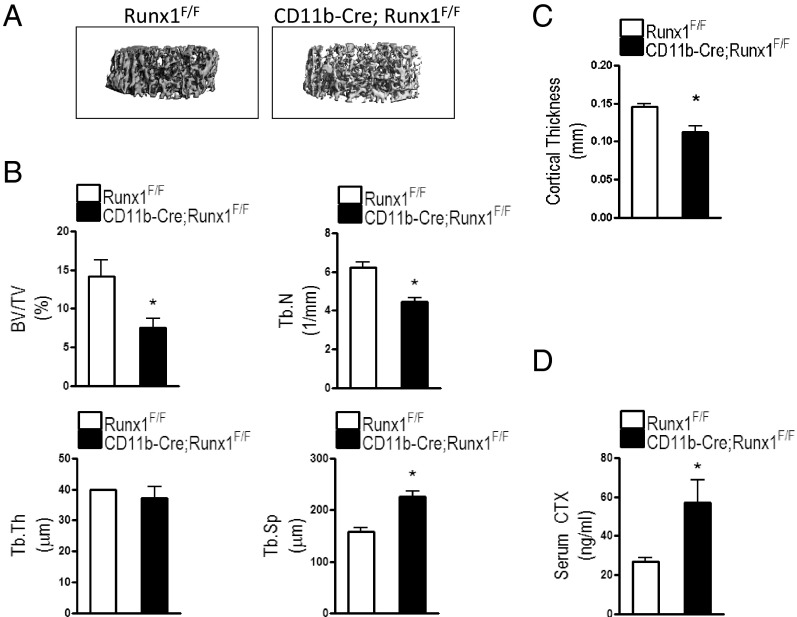
Micro-CT imaging showed that femoral trabecular bone in CD11b-Cre;Runx1F/F mice was microstructurally more disconnected with a larger vacant gap between trabeculae than that in Runx1F/F mice (Figure 2A). Micro-CT quantitatively revealed that mice conditionally lacking Runx1 in preosteoclasts exhibited a 47% decrease in femoral trabecular bone mass (BV/TV) compared with that of Runx1F/F mice (Figure 2B). CD11b-Cre;Runx1F/F mice had a 28% decrease in trabecular number (Tb.N) and a 43% increase in trabecular spacing (Tb.Sp) compared with that in Runx1F/F mice. However, trabecular thickness (Tb.Th) was comparable between the 2 genotypes. In addition, cortical thickness in the femurs of CD11b-Cre;Runx1F/F mice was decreased by 23% compared with that in femurs of Runx1F/F mice (Figure 2C). Hence, mice lacking Runx1 exhibited decreased bone mass.
Figure 2.
Conditional deletion of Runx1 in preosteoclasts decreased bone mass and increased osteoclast activity. A, Micro-CT imaging demonstrating the 3-dimensional microstructure of femoral trabecular bones of 8-week-old Runx1F/F and CD11b-Cre;Runx1F/F mice. B, Femoral BV/TV, Tb.N, Tb.Th, and Tb.Sp were measured in Runx1F/F and CD11b-Cre;Runx1F/F mice using micro-T imaging analyses. C, cortical thickness was also measured in the femurs of Runx1F/F and CD11b-Cre;Runx1F/F mice. D, CTX, a bone resorption degradation product, was assayed in serum obtained from 8-week-old Runx1F/F and CD11b-Cre;Runx1F/F mice. Data are means ± SE. n = 5 to 8 mice per group. *, P < .05 vs Runx1F/F mice
To further examine whether the decreased bone mass in CD11b-Cre;Runx1F/F mice was due to increased bone resorption, we measured serum CTX, a bone resorption index, in Runx1F/F and CD11b-Cre;Runx1F/F mice. CTX levels were significantly increased (by 2-fold) in mice with CD11b-mediated Runx1 gene deletion compared with those in Runx1F/F mice (Figure 2D).
CD11b+-mediated deletion of Runx1 gene increases osteoclast surface without alternating osteoblasts in vivo
To complement the micro-CT data, we analyzed the femurs of mice by histomorphometry (Figure 3 and Supplemental Figure 2). As expected, BV/TV was decreased by 39% in CD11b-Cre;Runx1F/F mice compared with that in Runx1F/F mice (Figure 3A). Likewise, Tb.N was significantly decreased and Tb.Sp was significantly increased in CD11b-Cre;Runx1F/F mice compared with those in Runx1F/F mice (Figure 3, B and C). There was no difference in trabecular thickness (Tb.Th) between CD11b-Cre;Runx1F/F and Runx1F/F mice (Figure 3D). CD11b-Cre;Runx1F/F mice also exhibited a 2-fold increase in osteoclast surface over bone surface with compared with that in Runx1F/F mice (Figure 3E). However, osteoblast surface over bone surface in CD11b-Cre;Runx1F/F mice was not significantly different from that of Runx1F/F mice (Figure 3F). In addition, we found no significant difference in mineral apposition or bone formation rates between CD11b-Cre;Runx1F/F and Runx1F/F mice (Supplemental Figure 2). These results imply that the decreased bone mass of CD11b-Cre;Runx1F/F mice was most likely due to increased osteoclast-mediated resorption.
Figure 3.
Loss of Runx1 in preosteoclasts decreased bone mass and increased osteoclasts in vivo without affecting osteoblasts. Femurs of 8-week-old Runx1F/F and CD11b-Cre;Runx1F/F mice were used to measure femoral BV/TV (A), Tb.N (B), Tb.Sp (C), Tb.Th (D), osteoclast surface over bone surface (Oc.S/BS) (E), and osteoclast surface over bone surface (Oc.S/BS) (F) by static histomorphometric analyses. Data are means ± SE. n = 5 to 8 mice per group. *, P < .05 vs Runx1F/F mice
CD11b+-mediated deletion of Runx1 does not affect osteoclast precursors in bone marrow
To examine whether loss of Runx1 in CD11b+ cells affects the distribution of the osteoclast precursor population, we analyzed bone marrow cells from the long bones of both Runx1F/F and CD11b Cre;Runx1F/F mice. The percentage of hematopoietic stem cells (Lin−ckit+Sca1+) in CD11b Cre;Runx1F/F mice was comparable to that of Runx1F/F mice (Figure 4A). Similar results were observed in the distribution of the osteoclast precursor population (Figure 4, B–D). Furthermore, there was no difference in more committed osteoclasts precursors (B220−CD3−CD11b+) between CD11b-Cre;Runx1F/F or Runx1F/F mice (Figure 4, E and F). These results indicate that a decrease of Runx1 in CD11b+ cells does not alter the number of B220−CD3−CD11b+ osteoclast precursors.
Figure 4.
Loss of Runx1 does not affect the distribution of osteoclast precursor cells in bone marrow. Cells isolated from murine long bones of either 8-week-old Runx1F/F or CD11b-Cre;Runx1F/F mice were analyzed using flow cytometry for the percentage of hematopoietic stem cells (Lin−ckit+Sca1+) (A) and osteoclast precursors (populations IV, V, and VI) (B–D). The percentage (E) and total number of committed osteoclast precursors (B220−CD3−CD11b+) (F) were also measured. Data are means ± SE. n = 4 to 5 mice per group.
Loss of Runx1 accelerates osteoclastogenesis in vitro
To determine whether Runx1 regulates osteoclastogenesis in vitro, we isolated BMM cells from the long bones of both Runx1F/F and CD11b-Cre;Runx1F/F mice and cultured them in the presence of M-CSF and RANKL. We evaluated the formation of osteoclasts morphologically and numerically after staining differentiated multinucleated cells for TRAP. We also measured resorptive activity by pit formation assays. Marrow cultures from CD11b-Cre;Runx1F/F mice exhibited larger TRAP+ multinuclear cells than Runx1F/F mice at day 5 (Figure 5A). In BMM cultures from either CD11b-Cre;Runx1F/F or Runx1F/F mice, the number of TRAP+ multinucleated cells peaked at day 5 (Figure 5B). However, the number of TRAP+ multinuclear cells was significantly greater in cultures isolated from CD11b-Cre;Runx1F/F mice than in Runx1F/F controls at all time points. In addition, the pit formation assay showed that the area of bone resorbed by TRAP+ multinuclear cells from CD11b-Cre;Runx1F/F mice was significantly increased compared with that in cultures from Runx1F/F mice (Figure 5, C and D). Collectively, these data indicate that loss of Runx1 function in CD11b+ preosteoclasts accelerated the formation and activity of osteoclasts in vitro.
Figure 5.
Loss of Runx1 function accelerated osteoclastogenesis in vitro. Bone marrow cells were isolated from the long bones of either 8-week-old Runx1F/F or CD11b-Cre;Runx1F/F mice. Cells were seeded in 96-well plates at a density of 5000 cells per well and cultured in the presence of 30 ng/mL M-CSF and 30 ng/mL RANKL to induce osteoclast formation. A, Image of multinuclear osteoclasts stained with TRAP at culture day 5 (magnification, ×4). B, TRAP+ multinuclear osteoclasts with greater than 3 nuclei were counted as osteoclasts in bone marrow cells from Runx1F/F and CD11b-Cre;Runx1F/F mice that were cultured for 4, 5, and 6 days. C, Micrographs show osteoclast-mediated resorbing surfaces (pits) formed on bovine bone chips after bone marrow cells from either Runx1F/F or CD11b-Cre;Runx1F/F mice were cultured on bovine cortical bone chips with M-CSF and RANKL (30 ng/mL for each) for 14 days. D, Total pit area per slice was measured in both Runx1F/F and CD11b-Cre;Runx1F/F groups. Data obtained were means ± SE (n = 3) and were representative of 3 replicate experiments. *, P < .05 vs Runx1F/F mice
We also extracted total RNA from bone marrow cells of both CD11b-Cre;Runx1F/F and Runx1F/F mice to assess the expression of genes associated with osteoclast formation and activity including vacuolar ATPase (v-ATPase) D2, cathepsin K (CatK), matrix metalloproteinase 9 (MMP9), calcitonin receptor (CTR), osteoclast-associated receptor (OSCAR), nuclear factor of activated T cells cytoplasmic 1 (NFATc1), and cFos. Figure 6 shows that the RANKL dose dependently enhanced the transcript levels of v-ATPase D2, CatK, MMP9, CTR, OSCAR, NFATc1, and cFos in marrow cultures from both Runx1F/F and CD11b-Cre;Runx1F/F mice after 2 days of culture. Inductions of gene expression for v-ATPase D2 (Figure 6A), CatK (Figure 6B), and MMP9 (Figure 6C) were significantly greater in bone marrow cell cultures from CD11b-Cre;Runx1F/F mice than in those from Runx1F/F mice. There was also a trend for increased expressions of both CTR and OSCAR in cultured cells from CD11b-Cre;Runx1F/F mice compared with those in cells from Runx1F/F mice (Figure 6, D and E). In addition, cells from CD11b-Cre;Runx1F/F mice exhibited higher mRNA levels of NFATc1 and cFos compared with those in cells from Runx1F/F mice for all doses of RANKL (Figure 6, F and G). As expected, treatment with M-CSF alone did not affect the expression levels of any of the osteoclast marker genes in bone marrow cell cultures from either group. However, expressions of osteoclast marker genes in cultured cells from CD11b-Cre;Runx1F/F and Runx1F/F mice at day 4 of culture with M-CSF and RANKL were similar (data not shown). This result implies that Runx1 conditionally deleted preosteoclasts mature more rapidly than wild-type cells.
Figure 6.
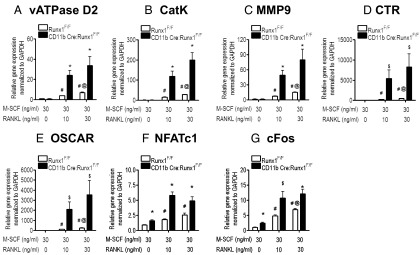
Runx1–regulated genes related to osteoclast differentiation. Bone marrow cells isolated from either 8-week-old Runx1F/F or CD11b-Cre;Runx1F/F mice were cultured in the presence of 30 ng/mL M-CSF with various doses of RANKL (0, 10, and 30 ng/mL) for 2 days. Total RNA, extracted from cells, was used to measure gene expressions of v-ATPase D2 (A), CatK (B), MMP9 (C), CTR (D), OSCAR (E), NFATc1 (F), and cFos (G). The relative quantification of target gene expression was normalized to the expression of a housekeeping gene, glyceraldehyde 3-phosphate dehydrogenase (GAPDH), for each sample using the ΔΔCT method. Results are expressed as means ± SE (n = 3) and are representative of 3 replicate experiments. *, P < .05 vs Runx1F/F mice within the same treatment; $, P < .1 vs Runx1F/F mice within the same treatment; #, P < .05 vs M-CSF alone within Runx1F/F mice; @, P < .05 vs M-CSF plus 10 ng/mL RANKL within Runx1F/F mice.
In summary, our data indicate that loss of Runx1 in preosteoclasts up-regulated RANKL-mediated mature osteoclast-associated gene expression only at an early stage of osteoclast differentiation.
Discussion
Recent evidence demonstrating a strong link between single nucleotide polymorphisms in the Runx1 gene locus and trabecular vertebral bone mineral density in men aged 65 years or older (28) implies that Runx1 is associated with skeletal health. In the current work, we demonstrated that Cre-mediated deletion of the Runx1 gene in preosteoclasts induced bone loss by increasing the number and activity of osteoclasts.
The decrease in trabecular and cortical bone mass, the increase in the osteoclast surface on bone, and the higher levels of circulating CTX in CD11b-Cre;Runx1F/F mice relative to those in the Runx1F/F control mice are evidence that Runx1 represses osteoclastic bone resorption. Our in vitro data further confirm the inhibitory role that Runx1 has on osteoclast activity. This was demonstrated by increased pit formation and enhanced expression of multiple mRNA transcripts involved in osteoclast activity in cultured cells from CD11b-Cre;Runx1F/F mice relative to that in cultured cells from Runx1F/F control mice. Our findings of similar osteoblast number and mineral apposition rates in CD11b-Cre;Runx1F/F and Runx1F/F mice argue that our targeted Runx1 deletion strategy did not significantly affect bone formation in vivo.
We also demonstrated that BMM cells from CD11b-Cre;Runx1F/F mice differentiated in vitro into a greater number of TRAP+ multinuclear osteoclasts in the presence of RANKL relative to that in Runx1F/F control cells. There is a precedent for a role of Runx1 in hematopoietic precursor lineage commitment. Tissue-specific deletion of Runx1 broadly in hematopoietic cells using Mx1-Cre–accelerated conversion of CD11b− cells into CD11b+ cells (18, 29). This result implies that Runx1 may influence osteoclast precursor cell abundance. However, when we examined the percentages and total number of cells in a population of more committed osteoclast precursors (B220−CD−CD11b+) in bone marrow by flow cytometry, using our previously described phenotyping scheme (3), we found no significant differences between CD11b-Cre;Runx1F/F and Runx1F/F mice. This result indicates that deletion of the Runx1 gene in CD11b+ cells, an established pool of late osteoclast precursors (2, 3), did not affect the total number of precursors. Rather, we believe that loss of Runx1 function at the CD11b+ stage of myeloid cell development enhances the rate of commitment toward mature osteoclasts rather than increasing the proliferation of osteoclast precursors. Our finding of increased osteoclast-associated gene expression in M-CSF plus RANKL–treated CD11b-Cre;Runx1F/F cell cultures relative to Runx1F/F cell cultures at day 2 but not at day 4 of culture is consistent with this hypothesis.
The finding of elevated expressions of v-ATPas D2, a key regulator of fusion (30, 31), in cell cultures from Runx1 conditionally deleted mice supports the hypothesis that Runx1 is a negative regulator of osteoclastic fusion, which has effects on cell fusion. Additional evidence arguing for a role of Runx1 in the control of cell fusion was our finding that conditional loss of Runx1 function resulted in increased osteoclast size in cultured CD11b-Cre;Runx1F/F cells relative to Runx1F/F control cells. The size of multinuclear osteoclasts is known to directly influence osteoclast function. Lees et al (32) previously reported that bigger osteoclasts with a greater number of nuclei exhibited higher rates of bone resorption than did smaller osteoclasts.
Collectively, these data imply that Runx1 has temporal functions during osteoclastogenesis and resorption. Future studies will be geared toward dissecting these potential stage-specific roles of Runx1 in osteoclastic bone resorption.
Additional material
Supplementary data supplied by authors.
Acknowledgments
This study was supported in part by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (National Institutes of Health Grants R01 AR052674–01 to H.D., R01 AR048714 to J.L., and R01AR060867 to H.D. and J.L.) and funds from the University of Connecticut Health Center (to H.D.).
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- BMM
- bone marrow macrophage
- BV/TV
- trabecular bone mass
- CatK
- cathepsin K
- CTR
- calcitonin receptor
- CT
- computed tomography
- CTX
- C-terminal telopeptides of type 1 collagen
- FACS
- fluorescence-activated cell sorting
- FHL2
- four and a half LIM domain 2
- M-CSF
- macrophage colony-stimulating factor
- MMP9
- matrix metalloproteinase 9
- NFATc1
- nuclear factor of activated T-cells cytoplasmic 1
- OSCAR
- osteoclast-associated receptor
- RANKL
- receptor activator of nuclear factor-κB ligand
- Tb.N
- trabecular number
- Tb.Sp
- trabecular spacing
- Tb.Th
- trabecular thickness
- TRAP
- tartrate-resistant acid phosphatase
- v-ATPase
- vacuolar ATPase.
References
- 1. Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature. 2003;423:337–342. [DOI] [PubMed] [Google Scholar]
- 2. Arai F, Miyamoto T, Ohneda O, et al. Commitment and differentiation of osteoclast precursor cells by the sequential expression of c-Fms and receptor activator of nuclear factor kappaB (RANK) receptors. J Exp Med. 1999;190:1741–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jacquin C, Gran DE, Lee SK, Lorenzo JA, Aguila HL. Identification of multiple osteoclast precursor populations in murine bone marrow. J Bone Miner Res. 2006;21:67–77. [DOI] [PubMed] [Google Scholar]
- 4. Li P, Schwarz EM, O'Keefe RJ, Ma L, Boyce BF, Xing L. RANK signaling is not required for TNFα-mediated increase in CD11hi osteoclast precursors but is essential for mature osteoclast formation in TNFα-mediated inflammatory arthritis. J Bone Miner Res. 2004;19:207–213. [DOI] [PubMed] [Google Scholar]
- 5. Teitelbaum SL. Bone resorption by osteoclasts. Science. 2000;289:1504–1508. [DOI] [PubMed] [Google Scholar]
- 6. Lorenzo JA, Canalis E, Raisz LG. Metabolic bone disease. In: Kronenberg H, Melmed S, Polonsky KS, Larsen PR, eds. Williams Textbook of Endocrinology. 11 ed Philadelphia, PA: Elsevier Sanders; 2008;11. [Google Scholar]
- 7. Rachner TD, Khosla S, Hofbauer LC. Osteoporosis: now and the future. Lancet. 2011;377:1276–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shane E, Burr D, Ebeling PR, et al. Atypical subtrochanteric and diaphyseal femoral fractures: report of a task force of the American Society for Bone and Mineral Research. J Bone Miner Res. 2010;25:2267–2294. [DOI] [PubMed] [Google Scholar]
- 9. Khosla S, Burr D, Cauley J, et al. Bisphosphonate-associated osteonecrosis of the jaw: report of a task force of the American Society for Bone and Mineral Research. J Bone Miner Res. 2007;22:1479–1491. [DOI] [PubMed] [Google Scholar]
- 10. Takayanagi H. Mechanistic insight into osteoclast differentiation in osteoimmunology. J Mol Med (Berl). 2005;83:170–179. [DOI] [PubMed] [Google Scholar]
- 11. Lee J, Kim K, Kim JH, et al. Id helix-loop-helix proteins negatively regulate TRANCE-mediated osteoclast differentiation. Blood. 2006;107:2686–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Smink JJ, Bégay V, Schoenmaker T, Sterneck E, de Vries TJ, Leutz A. Transcription factor C/EBPβ isoform ratio regulates osteoclastogenesis through MafB. EMBO J. 2009;28:1769–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhao B, Takami M, Yamada A, et al. Interferon regulatory factor-8 regulates bone metabolism by suppressing osteoclastogenesis. Nat Med. 2009;15:1066–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miyauchi Y, Ninomiya K, Miyamoto H, et al. The Blimp1-Bcl6 axis is critical to regulate osteoclast differentiation and bone homeostasis. J Exp Med. 2010;207:751–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhao B, Grimes SN, Li S, Hu X, Ivashkiv LB. TNF-induced osteoclastogenesis and inflammatory bone resorption are inhibited by transcription factor RBP-J. J Exp Med. 2012;209:319–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Niki M, Okada H, Takano H, et al. Hematopoiesis in the fetal liver is impaired by targeted mutagenesis of a gene encoding a non-DNA binding subunit of the transcription factor, polyomavirus enhancer binding protein 2/core binding factor. Proc Natl Acad Sci USA. 1997;94:5697–5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe AH, Speck NA. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci USA. 1996;93:3444–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Growney JD, Shigematsu H, Li Z, et al. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005;106:494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lorsbach RB, Moore J, Ang SO, Sun W, Lenny N, Downing JR. Role of RUNX1 in adult hematopoiesis: analysis of RUNX1-IRES-GFP knock-in mice reveals differential lineage expression. Blood. 2004;103:2522–2529. [DOI] [PubMed] [Google Scholar]
- 20. Saltman LH, Javed A, Ribadeneyra J, et al. Organization of transcriptional regulatory machinery in osteoclast nuclei: compartmentalization of Runx1. J Cell Physiol. 2005;204:871–880. [DOI] [PubMed] [Google Scholar]
- 21. Bai S, Zha J, Zhao H, Ross FP, Teitelbaum SL. Tumor necrosis factor receptor-associated factor 6 is an intranuclear transcriptional coactivator in osteoclasts. J Biol Chem. 2008;283:30861–30867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bai S, Kitaura H, Zhao H, et al. FHL2 inhibits the activated osteoclast in a TRAF6-dependent manner. J Clin Invest. 2005;115:2742–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ferron M, Vacher J. Targeted expression of Cre recombinase in macrophages and osteoclasts in transgenic mice. Genesis. 2005;41:138–145. [DOI] [PubMed] [Google Scholar]
- 24. Soung DY, Devareddy L, Khalil DA, et al. Soy affects trabecular microarchitecture and favorably alters select bone-specific gene expressions in a male rat model of osteoporosis. Calcif Tissue Int. 2006;78:385–391. [DOI] [PubMed] [Google Scholar]
- 25. Jastrzebski S, Kalinowski J, Stolina M, et al. Changes in bone sclerostin levels in mice after ovariectomy vary independently of changes in serum sclerostin levels. J Bone Miner Res. 2013;28:618–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Soung do Y, Gentile MA, Duong le T, Drissi H. Effects of pharmacological inhibition of cathepsin K on fracture repair in mice. Bone. 2013;55:248–255. [DOI] [PubMed] [Google Scholar]
- 27. Aguila HL, Mun SH, Kalinowski J, Adams DJ, Lorenzo JA, Lee SK. Osteoblast-specific overexpression of human interleukin-7 rescues the bone mass phenotype of interleukin-7-deficient female mice. J Bone Miner Res. 2012;27:1030–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yerges LM, Klei L, Cauley JA, et al. Candidate gene analysis of femoral neck trabecular and cortical volumetric bone mineral density in older men. J Bone Miner Res. 2010;25:330–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang G, Zhang P, Hirai H, et al. PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat Genet. 2008;40:51–60. [DOI] [PubMed] [Google Scholar]
- 30. Lee SH, Rho J, Jeong D, et al. v-ATPase V0 subunit d2-deficient mice exhibit impaired osteoclast fusion and increased bone formation. Nat Med. 2006;12:1403–1409. [DOI] [PubMed] [Google Scholar]
- 31. Kim K, Lee SH, Ha Kim J, Choi Y, Kim N. NFATc1 induces osteoclast fusion via up-regulation of Atp6v0d2 and the dendritic cell-specific transmembrane protein (DC-STAMP). Mol Endocrinol. 2008;22:176–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lees RL, Heersche JN. Macrophage colony stimulating factor increases bone resorption in dispersed osteoclast cultures by increasing osteoclast size. J Bone Miner Res. 1999;14:937–945. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.