Abstract
Phosphoenolpyruvate carboxylase (PEPC) is known to play a key role in the initial fixation of CO2 in C4 photosynthesis. The PEPC gene from sugarcane (a C4 plant) was introduced into indica rice (Hang2), a process mediated by Agrobacterium tumefaciens. Integration patterns and copy numbers of the gene was confirmed by DNA blot analysis. RT-PCR and western blotting results showed that the PEPC gene was expressed at both the mRNA and protein levels in the transgenic lines. Real-time PCR results indicated that expression of the sugarcane PEPC gene occurred mostly in green tissues and changed under high temperature and drought stress. All transgenic lines showed higher PEPC enzyme activities compared to the untransformed controls, with the highest activity (11.1 times higher than the controls) being observed in the transgenic line, T34. The transgenic lines also exhibited higher photosynthetic rates. The highest photosynthetic rate was observed in the transgenic line, T54 (22.3 μmol m−2 s−1; 24.6 % higher than that in non-transgenic plants) under high-temperature conditions. Furthermore, the filled grain and total grain numbers for transgenic lines were higher than those for non-transgenic plants, but the grain filling (%) and 1,000-grain weights of all transgenic lines remained unchanged. We concluded that over-expression of the PEPC gene from sugarcane in indica rice (Hang2) resulted in higher PEPC enzyme activities and higher photosynthesis rates under high-temperature conditions.
Electronic supplementary material
The online version of this article (doi:10.1007/s11033-014-3070-4) contains supplementary material, which is available to authorized users.
Keywords: Phosphoenolpyruvate carboxylase gene (PEPC gene), C4 photosynthesis, Gene expression, Transgenic rice
Introduction
Photosynthesis is one of the major factors that affect rice yield. In total, 90 % of the crop yield directly comes from organic compounds created by photosynthesis. The yield per unit can increase significantly if the photosynthetic rate of the crop is increased to its full capacity [1–5]. The C4 photosynthesis pathway was discovered in the 1960s [6]. Since then, a number of studies have indicated that the C4 photosynthetic system is superior to the C3 system [7]. This photosynthetic superiority may result from increased CO2 concentration around Rubisco and the repression of photorespiration may increase the net photosynthetic rate. In addition, the efficiency of C4 Rubisco is higher than C3 catalysts. The C4 photosynthetic pathway also has an efficient photosynthetic transformation capacity. Therefore, the transformation of some C4 photosynthetic advantages, such as repressed photorespiration, increased utilization rates of water and nitrogen and improved photosynthetic rates, etc., into C3 crops may result in several plant improvements [8–10].
Many genes related to C4 photosynthesis (e.g., NAD-ME, PEPC, PPDK and NADP-MDH) have been successfully transformed into C3 genomes [11, 12]. The NADP-ME gene from sorghum was introduced into Nongken 58, a rice variety, using an Agrobacterium-mediated system, resulting in 1–7-fold elevation of NADP-ME activity and increased photoinhibition under high light intensity [13]. The PEPC gene from Zea mays was recently cloned and transformed into the rice genome using Agrobacterium tumefaciens [14]. This resulted in an increase in the density and area of stoma in the leaves and in the accumulation of dry matter in the transgenic rice. Oat (Avena sativa; C3) × maize (Zea mays; C4) addition lines (OMAs) allow the effects of individual maize chromosomes to be investigated in oat. When genes encoding phosphoenolpyruvate carboxylase (PEPC), orthophosphate dikinase, pyruvate and the 29-oxoglutarate/malate transporter in maize were expressed in oat, larger bundle sheath cells with increased cell wall lipid deposition were observed in oat leaves [15].
Phosphoenolpyruvate carboxylase, a key enzyme in photosynthesis, is an important carboxylase for plant cell metabolism. PEPC, which appears to fix CO2 in mesophyll cells to form C4 dicarboxylic acids, is more specific for inorganic carbon and improves the photosynthetic rate of C4 plants. The C4-type PEPC gene from sugarcane was transformed into a high-quality indica restorer line, “Hang2”, using A. tumefaciens, which has been extensively used to create super hybrid rice combinations in China. Here, we describe the expression pattern of the PEPC gene, PEPC gene expression under stress treatment in transgenic rice, PEPC enzyme activity and total nitrogen content in the transgenic plants; In addition, we report the physiological photosynthetic characteristics and yield characteristics of the transgenic rice.
Materials and methods
Transformation of indica rice (Hang2)
The plasmid pc1380, which contained the intact sugarcane PEPC gene driven by its endogenous promoter and hph as a marker, was used for the transformation via A. tumefaciens. The embryogenic calli of mature embryos of the rice cultivar indica restorer line, Hang2, were obtained and transformed using A. tumefaciens, as described by Datta [16]. The embryogenic calli were cultured in NB medium [17], containing 50 mg L−1 hygromycin, for three to four selection cycles with a 14-day duration per cycle. Embryogenic calli were selected for regeneration and the resulting plantlets were first grown in the culture solution before being transferred to pots. Putative primary transformants and subsequent seed progenies were grown in the transgenic network house.
Polymerase chain reaction (PCR) assay and southern blot analysis
Genomic DNA was isolated from 1 month old plants by CTAB, and 25–50 ng of the DNA template was used for PCR analysis using PEPC gene specific primers (PEPC 1F and PEPC 1R). The reaction mixture contained 1 μL 10× buffer, 0.8 μL dNTP (2.5 mmol L−1), 0.4 μL PEPC 1F (10 μmol L−1), 0.4 μL PEPC 1R (10 μmol L−1), 0.1 μL rTaq, 25–30 ng DNA and enough water to bring the final volume to 10 μL. The PCR program was as follows: 94 °C denaturation for 5 min, 36 cycles of 94 °C for 30 s, 60 °C for 30 s and 72 °C for 1 min, extension for 10 min at 72 °C and final holding at 4 °C.
The plant genomic DNA was extracted from freshly harvested leaves of transgenic and non-transgenic control plants for southern blot analysis, following a modified CTAB method [18]. A total of 100 μg of DNA per sample was digested with HindIII at 37 °C overnight. The digested DNA was fractionated in 1 % (w/v) TAE agarose gels and then transferred to a Hybond-N nylon membrane (Amersham, Arlington Heights, IL, USA), according to the manufacturer’s instructions. The Hygromycin coding sequence was isolated from the plasmid and labeled using an AlkPhos direct labeling kit (Amersham, Arlington Heights, IL, USA) in order to make the hybridization probes.
Total RNA isolation and reverse transcription polymerase chain reaction (RT-PCR) analysis of the transgenic rice
The total RNA was isolated from mature green leaves of the transgenic and control plants using Trizol reagent (Invitrogen, USA). RT-PCR was performed on 2 μg of the total RNA using specific primers for pepc (PEPC 2F and PEPC 2R) and actin (Actin 1F and Actin 1R), following a previously described method [19] (Table 1). The RT-PCR products were resolved on 1 % TAE-agarose gel.
Table 1.
Primers used in this study
| Primer name | Primer sequence (5′ → 3′) |
|---|---|
| PEPC 1F | ATCAAGGAGAAACTGGATG |
| PEPC 1R | TCAGGAAAGAACTAGACTGC |
| PEPC 2F | AGTCGGACATCGAGGAGACG |
| PEPC 2R | ACCACCCATCCAAGAACAGA |
| Actin 1F | CAT CTT GGC ATC TCT CAG CAC |
| Actin 1R | AAC TTT GTC CAC GCT AAT GAA |
| PEPC 3F | CCATCTGCTGGCTTCTGGAGTTTC |
| PEPC 3R | TTGTCGCCGCAGTCACACAGTG |
| Actin 2F | AGTGTCTGGATTGGAGGAT |
| Actin 2R | TCTTGGCTTAGCATTCTTG |
Quantitative reverse transcription PCR (qRT-PCR) analysis of PEPC gene expression
To analyze PEPC gene expression in different tissues of transgenic lines, green leaves, stems and roots of 3 weeks old transgenic seedlings were harvested. Expression of the PEPC gene under abiotic stress was also analyzed. The transgenic seedlings were placed onto filter papers to induce drought stress. Samples were taken after 0, 2, 4, 8, 12 and 24 h treatment. The transgenic seedlings were then placed in a 42 °C chamber and sampling took place after 0, 1, 3, 6, 12, and 24 h of treatment.
Total RNA was isolated from the samples using Trizol (Invitrogen, USA) and reverse transcription was performed on 2 μg of the total RNA using the RevertAid™ First Strand cDNA Synthesis Kit (Fermentas, LTU). The qRT-PCR was performed on a 7500 real-time PCR system (Applied Biosystems) with the Faststart Universal SYBR Green Master system (Roche, USA), following the manufacturer’s protocol. PEPC 3F and PEPC 3R were used as primers. Transcripts of the Rice actin2 gene were amplified using Actin 2F and Actin 2R primers and used as the endogenous control. The relative quantitative method (△△CT) was used to evaluate the quantitative variation in the examined replicates.
Extraction of resolved protein, western blotting and PEPC enzyme activity analysis
Green leaf segments, of about 5 cm in length, were harvested from the midsection of the uppermost fully expanded leaf and immediately frozen in liquid nitrogen until needed. Unless stated otherwise, the leaf samples were harvested at 11:00 a.m. on sunny days. To extract soluble proteins from the leaf segment, the samples were ground in extraction buffer containing 50 mmol L−1 Tris–HCl (pH 7.5), 1 mmol L−1 MgCl2, 5 mmol L−1 DDT, 1 mmol L−1 EDTA and 5 % (w/v) glycerol with a small amount of sea sand (Shanghai Sangon Biotech Co., Ltd., P.R. China). The homogenate was centrifuged at 13,000×g for 10 min at 4 °C and the resulting supernatant was collected as soluble protein extract. Protein concentration was determined using the method described by [20]. Bovine serum albumin was used as the standard. The precipitated samples were separated by SDS-PAGE and subjected to immune blot analysis using the PEPC antibody (antibodies for research, Agrisera). The assay mixture contained 50 mmol L−1 HEPES–KOH (pH 8.0), 5 mmol L−1 MgCl2, 2 mmol L−1 PEP, 0.2 mmol L−1 nicotinamide-adenine dinucleotide, 10 mmol L−1 NaHCO3 and 1.5 U of malate dehydrogenase (from pig heart; Roche Diagnostics, Basel, Switzerland) and the reaction was started by adding 20 μL of the enzyme extract.
Measurement of photosynthetic characteristics
The flag leaf CO2 assimilation rate, stomatal conductance and internal CO2 concentration (Ci) were measured using a Li-6400 portable photosynthesis system (Li-Cor Inc., Lincoln, NE) and the chlorophyll relative content was measured using a SPAD-502 chlorophyll meter (Beijing Hezhong Bopu Technology Develop Co., Ltd.) between 10:00 a.m. and 12:00 a.m. under saturated photosynthetic photon flux density conditions (1,200 μmol m−2 s−1). The air temperature (T-air) in the greenhouse, as measured by the LI-6400 system, was 30 °C on average. Three fully expended flag leaves from each transgenic line and from the control plants were selected and measured at the beginning of the heading stage. The flow rate was maintained at 200–250 μmol s−1 so that the relative humidity inside the chamber was similar to ambient conditions. The measurements were made for 20 s immediately after a stable decrease in CO2 concentration was achieved inside the chamber. Statistical analyses were undertaken using SPSS and MS Excel 2010.
Total nitrogen evaluation in the flag leaves of transgenic lines
The flag leaves of transgenic lines and the control plants were harvested and the dry weight was determined after oven-drying to a constant weight at 70 °C. A total of 0.1 g of the sample was used to determine the total nitrogen content by the micro-Kjeldhal method [21]. Statistical analyses were undertaken using SPSS and MS Excel 2010.
Results
Identification of transgenic plants and PEPC gene expression in rice
The sugarcane PEPC gene construct (Fig. 1) was introduced into the indica restorer line, Hang2. A total of 600 putative hygromycin-resistant transgenic plants (T0) were obtained. The PCR results indicated that there was a 384 bp amplicon for 132 putative transgenic plants (Fig. 2). In order to analyze copy number and integration patterns in the PCR positive transgenic plants, genomic DNA (gDNA) was digested with the HindIII enzyme and hybridized with the Hph-specific probe. Plants with a single copy of the PEPC gene were used for further research (Fig. 3). Expression analysis was carried out using RT-PCR (Fig. 4) in order to verify expression of the sugarcane PEPC gene in the transgenic lines. The results indicated that sugarcane PEPC was expressed at the mRNA level in transgenic plants, but not in wild-type plants. These transgenic plants exhibited normal phenotypes with a normal life cycle, grew to maturity, flowered and set seed. Transgenic plants were grown up to the T4 generation for further research.
Fig. 1.
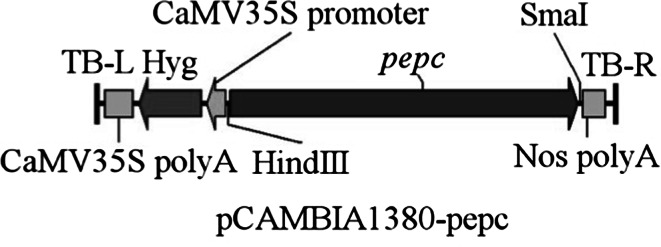
Schematic diagram of the intact PEPC gene from sugarcane, the PEPC gene from sugarcane is an 8.084 kb fragment containing 10 exons, 9 introns, and the promoter (from −1,300) and terminator (2.5 kb) sequences. Black rectangles exons; lines introns and the selective antibiotic resistant gene hygromycin phosphotransferase (hph) used for rice transformation
Fig. 2.
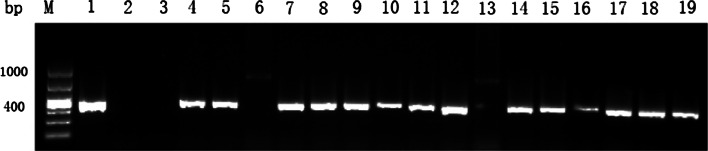
PCR analysis for putative transgenic plants with PEPC gene of sugarcane; M 1,000 bp DNA marker; 1 blank control (H2O); 2 negative control (untransformed plant); 3 positive control (Plasmid); 4–19 putative transgenic plants (6 and 13 were PCR negative)
Fig. 3.
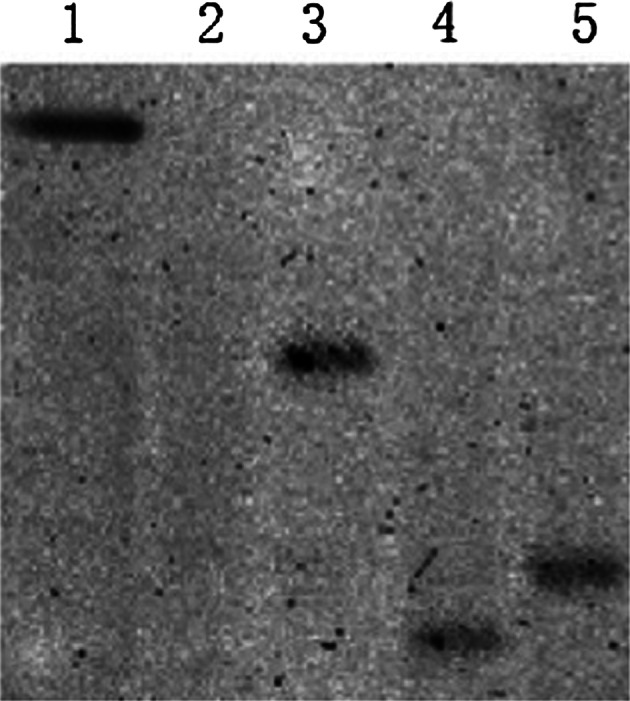
Southern blot analysis of PCR positive transgenic plants with hph gene probe; 1 positive control (plasmid); 2 negative control (untransformed plant); 3–5 transgenic plants
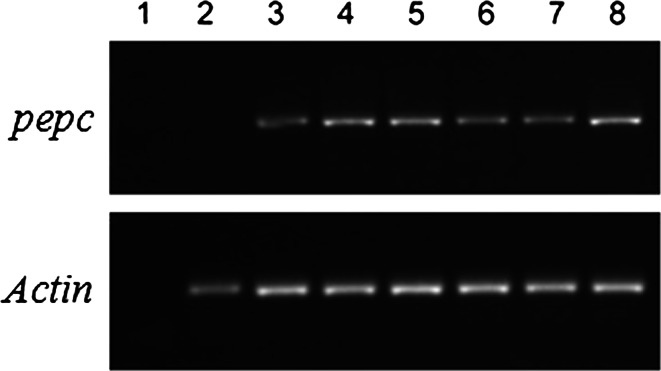
Fig. 4.
RT-PCR analysis of transgenic plants with sugarcane PEPC gene and actin (control); 1 blank control; 2 untransformed plant; 3–8 transgenic plants
Western blotting was performed on four selected transgenic lines, named: T34, T51, T53 and T54, using non-transgenic rice plants as controls. The internal control gene, HSP82, was found to be expressed in both the transgenic lines and the control plants. Strong hybridization signals for PEPC were observed in the transgenic lines, but not in the non-transgenic rice (Fig. 5). This suggested that PEPC protein expression levels in transgenic lines with the sugarcane PEPC gene were higher than in the non-transgenic rice.
Fig. 5.
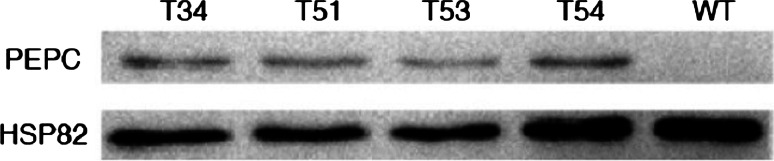
Western blotting analysis of PEPC protein expression of transgenic lines with sugarcane PEPC gene. T34, T51, T53, T54 were transgenic lines; WT was untransformed plant. HSP82 was shown as loading control
Analysis of PEPC gene expression in different tissues of the transgenic lines
The quantitative reverse transcription PCR (qRT-PCR) data for the transgenic line, T34, indicated that PEPC gene expression occurred mostly in the leaves (about ten times higher than that in stems) of the transgenic lines. Expression of the PEPC gene was the weakest in the roots of the transgenic lines (Fig. 6), indicating that PEPC gene expression is found mostly in the green tissues of transgenic lines and demonstrating that PEPC is an enzyme that is involved in photosynthesis.
Fig. 6.
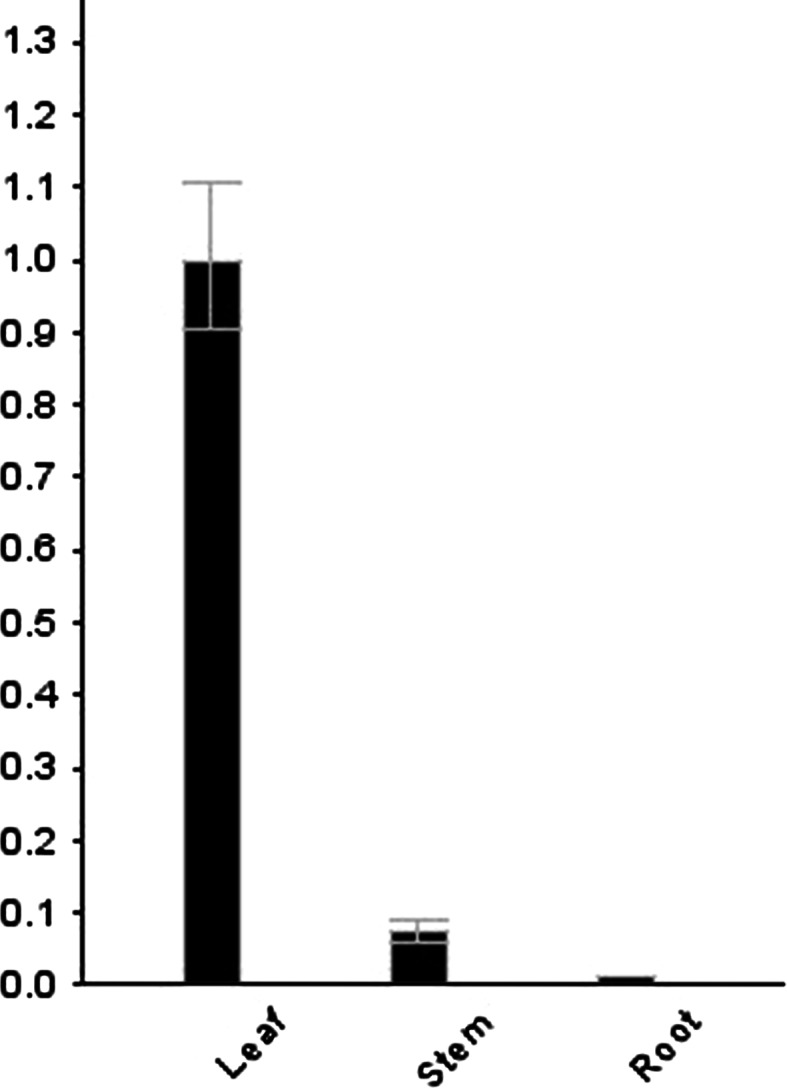
qRT-PCR analysis of PEPC gene expression in different tissues of transgenic line T34
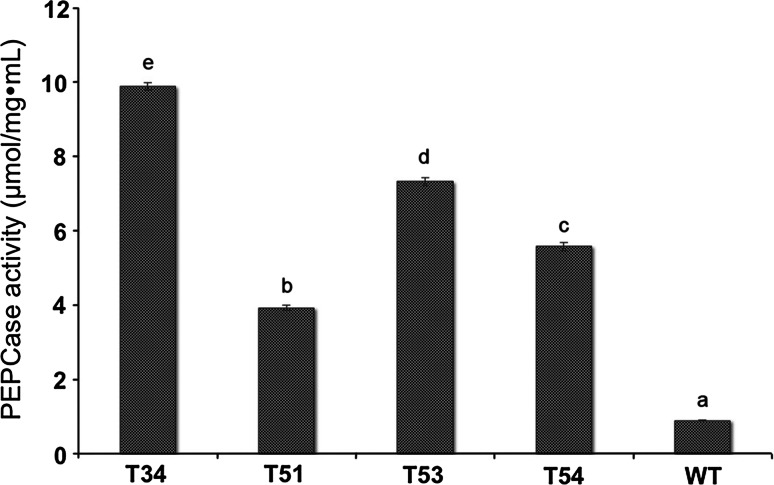
PEPC enzyme activity analysis in transgenic lines
PEPC enzyme activities in the transgenic lines: T34, T51, T53 and T54, were higher than in the non-transformed rice (Fig. 7) at the 5 % significance level. The highest PEPC enzyme activity was observed in the transgenic line, T34, which was 11.1 times higher than in the non-transgenic plants. This was consistent with the higher PEPC protein levels found in the transgenic lines.
Fig. 7.
Analysis of PEPC enzyme activities. T34, T51, T53, T54 were transgenic lines; WT was untransformed plant
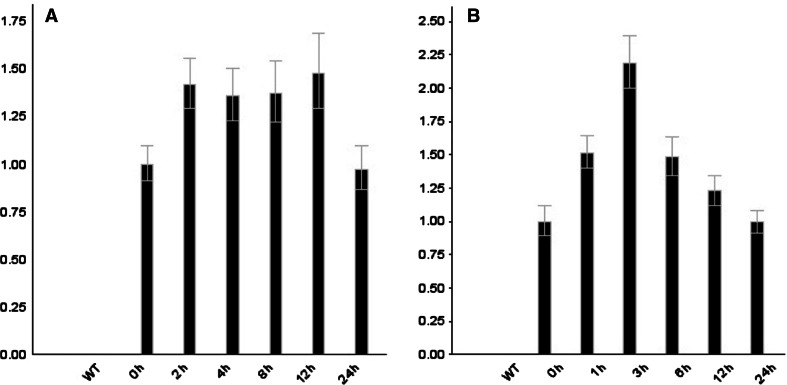
Analysis of PEPC gene expression under stress treatment in transgenic rice
Under abiotic stress, expression of many proteins related to oxidative stress, photosynthesis, carbohydrate metabolism and regulation are altered [22–25]. In order to examine how expression of PEPC was affected by abiotic stress, the T34 transgenic line was subjected to drought and high temperature stress. The qRT-PCR analyses indicated that expression of the sugarcane PEPC gene peaked under drought stress after 12 h and then declined (Fig. 8a), reaching levels equivalent to that seen at 0 h after 24 h under drought stress.
Fig. 8.
qRT-PCR analysis of PEPC gene expression under drought treatment and under high-temperature. WT was untransformed plant as control; transgenic line T34 was treated by drought for 0, 2, 4, 8, 12, 24 h (a) and treated at 42 °C for 0, 1, 3, 6, 12, 24 h (b)
In addition, the transgenic line, T34, was kept at 42 °C for 0, 1, 3, 6, 12 and 24 h. The qRT-PCR data indicated that PEPC gene expression increased significantly under high temperatures, peaked after 3 h and then declined (Fig. 8b). These results indicated that sugarcane PEPC gene expression in transgenic rice was sensitive to drought stress and high temperatures.
Photosynthetic rate in the transgenic lines
An important consideration for plant breeders is the photosynthetic efficiency of both conventional and transgenic plant varieties [26]. Therefore, the photosynthetic rates of the transgenic plants grown in the transgenic network house were measured at the heading stage. The results showed that the photosynthetic rates of transgenic lines were higher than that of the control plants (Table 2). Moreover, the photosynthetic rates of the transgenic lines: T34, T53 and T54 increased significantly [p = 0.00, 0.00 and 0.00, respectively (p < 0.05), compared with the control], but the photosynthetic rate of the transgenic line, T51, did not increase significantly [p = 0.15, (p > 0.05) compared with the control].
Table 2.
Photosynthetic parameters of the sugarcane pepc gene transgenic lines
| Line | Pn (photosynthesis rate) (μmol m−2 s−1) | SC (stomatal conductance) (cm−1) | Ci (internal CO2 concentration) (μmol mol−1) | Chlorophyll relative content (SPAD) |
|---|---|---|---|---|
| T34 | 19.3 ± 0.2b | 0.074 ± 0.004 | 355.84 ± 0.02 | 31.87 ± 0.92 |
| T51 | 18.3 ± 0.1ab | 0.084 ± 0.001 | 355.8 ± 2.458 | 33.97 ± 1.07 |
| T53 | 21.3 ± 0.1c | 0.075 ± 0.001 | 357.85 ± 0.01 | 31.10 ± 0.35 |
| T54 | 22.3 ± 0.3cd | 0.090 ± 0.010 | 355.15 ± 0.03 | 33.67 ± 0.45 |
| WT | 17.9 ± 0.1a | 0.061 ± 0.005 | 356.56 ± 0.02 | 32.60 ± 0.95 |
Measurement of agronomic data of all the control and transgenic line were recorded based on three replica experiments in identical conditions and the seeds from selected line were randomly chosen. The data was calculated by SPSS, Excel 2010 software package
It has previously been reported that the chlorophyll content of rice decreased slightly when the maize PPDK gene was introduced [27]. The relative chlorophyll content of the sugarcane PEPC gene was measured in the transgenic lines: T34, T51, T53 and T54 using a SPAD-502 chlorophyll meter. The results indicated that the relative chlorophyll content of these transgenic lines did not change significantly compared to non-transformed rice (Table 2).
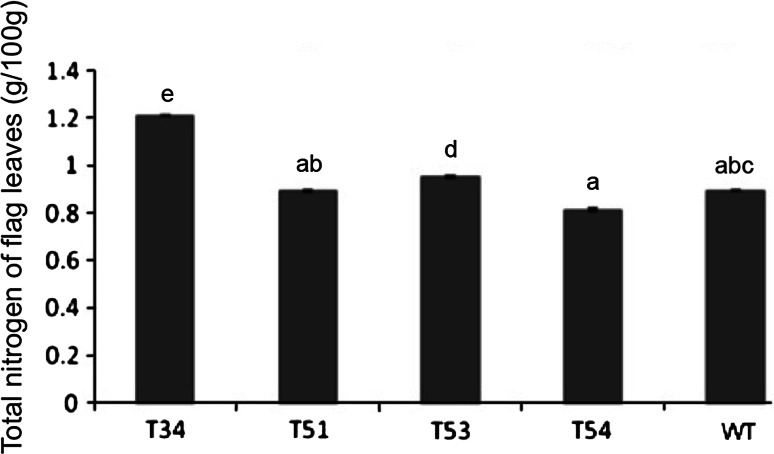
Total nitrogen in the flag leaves of the transgenic lines
Since nitrogen is a limiting factor on the photosynthetic capacity of plants, the total nitrogen content of the flag leaves was measured. The transgenic lines: T34 and T53, had significantly higher nitrogen contents compared to the control (p < 0.05). However, the total nitrogen contents of the transgenic lines: T51 and T54, were not statistically different from that of non-transgenic rice (Fig. 9).
Fig. 9.
Total nitrogen contains of flag leaves. T34, T51, T53, T54 were transgenic lines; WT was untransformed plant
Yield characteristics of transgenic rice
Three mature plants of each transgenic line were harvested in the transgenic network house. The yield characteristics of each transgenic line were observed after drying in a 65 °C oven. With the exception of T54, the biological weights of T34, T51 and T53 increased significantly (p = 0.008, 0.004 and 0.01, respectively) at the 5 % significance level. The panicle lengths of the T51, T53 and T54 lines showed an insignificant trend towards being longer than that of non-transgenic plants (control), while that of the T34 line was significantly longer (p < 0.05). The filled grain and total grain numbers of T34, T51, T53 and T54 were all more than those of non-transgenic plants, but the grain filling (%) and 1,000-grain weights of all the transgenic lines were comparable to those of the non-transgenic plants. In addition, the harvest index increased in some but not all transgenic lines (Table 3). Furthermore, to acquire more detailed description of morphological characteristics in transgenic lines, The T5 generation of transgenic line T34, T51, T53 and T54 were planted in the transgenic network house, and ten mature plants of each transgenic line were harvested to analyze yield characteristics further. The results demonstrated that spike numbers of all transgenic lines were more than that of non-transgenic plants (WT). Meanwhile, total filled grain and average filled grain per spike of transgenic lines were also more than that of non-transgenic plants. The panicle length of T34, T51, T53 and T54 were longer than that of non-transgenic plants, but the panicle length of transgenic line T34, T53 was significantly longer than that of non-transgenic plants (p = 0.044 and 0.025, respectively) (Supplementary Table 4; Fig. 10). According to the data, not all the yield characteristic indexes for the transgenic lines increased significantly, but filled grain and total grain numbers for all transgenic lines did increase significantly. This explains why introducing sugarcane PEPC into rice (C3) results in higher PEPC protein levels and leads to changes in some yield characteristics. It also further illustrates that PEPC plays an important role in rice.
Table 3.
Yield characteristics of transgenic lines containing the sugarcane pepc gene
| Line | Biological weight (g) | Panicle length (cm) | Filled grain | Total grain | Grain filling (%) | 1,000-grain weight (g) | Harvest index (%) |
|---|---|---|---|---|---|---|---|
| T34 | 128.1 ± 2.1bc | 25.1 ± 1.1 | 1,250.7 ± 148.0b | 2,547.3 ± 231.6b | 47.9 ± 4.7 | 24.7 ± 0.3 | 24.1 ± 2.4a |
| T51 | 135.8 ± 27.6bcd | 23.6 ± 1.2 | 2,496.3 ± 201.8e | 4,261.0 ± 964.9e | 60.2 ± 11.0 | 25.7 ± 0.3 | 48.7 ± 12.1cd |
| T53 | 125.8 ± 28.9b | 22.1 ± 1.9 | 2,289.0 ± 721.1d | 3,997.3 ± 898.9d | 56.5 ± 4.8 | 23.5 ± 1.6 | 42.6 ± 7.8c |
| T54 | 104.2 ± 30.7a | 23.7 ± 3.6 | 2,121.3 ± 2,071.3c | 3,892.0 ± 2,471.1c | 47.3 ± 17.5 | 22.7 ± 0.2 | 41.8 ± 32.2b |
| WT | 67.3 ± 4.1a | 21.0 ± 1.2 | 565.0 ± 163.4a | 1,166.0 ± 182.0a | 47.8 ± 6.3 | 26.2 ± 0.5 | 21.9 ± 5.3a |
Grain filling (%) = filled grain/total grain × 100 %, Harvest index (%) = filled grain weight/biological weight × 100 %. Values shown are averages of three replicates. Statistical analyses were undertaken using SPSS and MS Excel 2010. a, b, c, d, e indicated significant difference
Fig. 10.

The phenotype of spike between transgenic line and untransformed plant. a The phenotype of spike untransformed plant; b The phenotype of spike for transgenic line
Discussion
In order to increase the yield potential of rice, researchers have tried various methods to improve the photosynthetic ability per leaf area, but with limited success [26, 28–32]. Compared to non-transformed, wild-type rice, transgenic rice expressing the maize PEPC gene required a higher light intensity to bring about photosynthesis saturation, had a higher photosynthetic CO2 uptake rate and a higher carboxylation efficiency [33]. The PEPC substrate level (phosphoenolpyruvate) was slightly lower and the reaction product (oxaloacetate) was slightly higher in the transgenic rice, which suggested that the maize PEPC gene was functional in the transgenic rice lines [34]. PEPC from C4 millet (Seteria italica) was introduced into Japonica rice and the transformants only showed improvements in both photosynthesis rates and yields under upland field cultivation conditions [35]. In this study, the photosynthetic rates of Hang2 rice lines were significantly improved by introducing the intact sugarcane PEPC gene.
PEPC enzyme activity has been found to be high in several transgenic plants. It has been reported that a transgenic rice line with the maize PEPC gene expressed large amounts of the maize PEPC protein and exhibits high PEPC activity [36]. PEPC activity was significantly higher in PEPC transgenic rice than in the wild type rice [37, 38]. A 50-fold elevation in PEPC activity, but without a significant effect on overall growth, has been reported in PEPC transgenic rice compared to non-transgenic rice [31]. This study also found that PEPC activity was significantly higher in the transgenic rice lines. There was 11.1 times more activity in the transgenic plants than in the non-transgenic plants.
Nitrogen is required for the biosynthesis of proteins, nucleic acids and chlorophyll. It is unquestionably one of the most important nutrients needed for plant growth. Nitrogen starvation causes lower intracellular concentrations of amino acids and proteins, including those involved in photosynthesis and carbon metabolism [39, 40]. Nitrogen is a limiting factor on the photosynthetic capacity of plants. C4 plants have higher efficiencies than C3 plants when it comes to the utilization of light, water and nitrogen, perhaps owing to the low photorespiration rate of the former, which affects carbon and nitrogen metabolism [41]. Introduction of the intact maize C4-PPDK gene into japonica rice led to significant accumulation of the PPDK protein in the leaf chloroplasts of homozygous lines. Accumulation of PPDK affected the chlorophyll, Rubisco and nitrogen contents in the leaves. The transgenic line, PD25, showed an increase in leaf nitrogen content of approximately 7 %. Rubisco content increased slightly and chlorophyll content decreased slightly on a leaf area basis [27]. In our study, total nitrogen content in most of the transgenic lines was not significantly different to that of the non-transgenic line. Further research is needed to measure the activities of enzymes related to nitrogen metabolism.
Investigation of the main agronomic traits of three types of transgenic rice containing the maize C4 photosynthetic genes: PEPC, PPDK, or PEPC+PPDK, indicated that those containing the maize PEPC and/or PPDK genes had more panicles and higher yields than the untransformed controls [42]. Similarly, PEPC and PPDK transgenic rice have been reported to produce 22–24 % more grains than wild-type plants [33]. PEPC transgenic rice had higher dry weights for leaves, stems, sheaths and panicles than the untransformed wild rice, with the largest increase occurring in the panicles. The soluble sugar and protein contents in the grains of PEPC transgenic rice were also significantly enhanced [38]. The filled grain and total grain numbers of sugarcane PEPC gene transgenic lines were higher than those of untransformed rice. In addition, the biological weights of the transgenic lines were also higher than that of the untransformed rice. The harvest index of most transgenic plants was also higher than that of control plants, which indicated that the yield of transgenic Hang2 plants with the intact sugarcane PEPC gene improved to some degree. Thus, all these studies proved that C4-type genes function in transgenic rice. Recently, the fact that genes in C3 species can be recruited into cell-specific functions in the C4 pathway suggests that the trans-factor(s) responsible for generating cell-specific accumulation of proteins is important when engineering C4 photosynthesis in rice [43]. Further research is needed to reveal the action mechanism of protein encoded by C4-type genes in transgenic rice and what their action pathways are in C3 rice.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported by the National High Technology Research and Development Program of China (863 Program, Grant no. 2006AA100101) and the Fujian provincial technological program (Grant no. F2006AA100101).
References
- 1.Ku MS, Agarie S, Nomura M, Fukayama H, Tsuchida H, Ono K, Hirose S, Toki S, Miyao M, Matsuoka M. High-level expression of maize phosphoenolpyruvate carboxylase in transgenic rice plants. Nat Biotechnol. 1999;17:76–80. doi: 10.1038/5256. [DOI] [PubMed] [Google Scholar]
- 2.Leegood RC. C4 photosynthesis: principles of CO2 concentration and prospects for its introduction into C3 plants. J Exp Bot. 2002;53:581–590. doi: 10.1093/jexbot/53.369.581. [DOI] [PubMed] [Google Scholar]
- 3.Parry MA, Reynolds M, Salvucci ME, Raines C, Andralojc PJ, Zhu XG, Price GD, Condon AG, Furbank RT. Raising yield potential of wheat. II. Increasing photosynthetic capacity and efficiency. J Exp Bot. 2011;62:453–467. doi: 10.1093/jxb/erq304. [DOI] [PubMed] [Google Scholar]
- 4.Raines CA. Increasing photosynthetic carbon assimilation in C3 plants to improve crop yield: current and future strategies. Plant Physiol. 2011;155:36–42. doi: 10.1104/pp.110.168559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Von-Caemmerer S, Evans JR. Enhancing C3 photosynthesis. Plant Physiol. 2010;154:589–592. doi: 10.1104/pp.110.160952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voznesenskaya EV, Franceschi VR, Kiirats O, Freitag H, Edwards GE. Kranz anatomy is not essential for terrestrial C4 plant photosynthesis. Nature. 2001;414:543–546. doi: 10.1038/35107073. [DOI] [PubMed] [Google Scholar]
- 7.Long SP, Zhu XG, Naidu SL, Ort DR. Can improvement in photosynthesis increase crop yields? Plant Cell Environ. 2006;29:315–330. doi: 10.1111/j.1365-3040.2005.01493.x. [DOI] [PubMed] [Google Scholar]
- 8.Hibberd JM, Sheehy JE, Langdale JA. Using C4 photosynthesis to increase the yield of rice: rationale and feasibility. Curr Opin Plant Biol. 2008;11:228–231. doi: 10.1016/j.pbi.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Langdale JA. C4 cycles: past, present, and future research on C4 photosynthesis. Plant Cell. 2011;23:3879–3892. doi: 10.1105/tpc.111.092098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sage RF, Zhu XG. Exploiting the engine of C4 photosynthesis. J Exp Bot. 2011;62:2989–3000. doi: 10.1093/jxb/err179. [DOI] [PubMed] [Google Scholar]
- 11.Kajala K, Covshoff S, Karki S, Woodfield H, Tolley BJ, Dionora MJ, Mogul RT, Mabilangan AE, Danila FR, Hibberd JM, Quick WP. Strategies for engineering a two-celled C4 photosynthetic pathway into rice. J Exp Bot. 2011;62:3001–3010. doi: 10.1093/jxb/err022. [DOI] [PubMed] [Google Scholar]
- 12.Miyao M, Masumoto C, Miyazawa S, Fukayama H. Lessons from engineering a single-cell C4 photosynthetic pathway into rice. J Exp Bot. 2011;62:3021–3029. doi: 10.1093/jxb/err023. [DOI] [PubMed] [Google Scholar]
- 13.Chi W, Zhou JS, Zhang F, Wu NH. Photosynthetic features of transgenic rice expressing sorghum C4 type NADP-ME. Acta Bot Sin. 2004;46:873–882. [Google Scholar]
- 14.Li X, Ren CG, Wang M, Gong LJ, Liu W, Cao K, Wang C, Cong W, Zhou YL. Relationship between plant hormone contents and yield in PEPC transgenic rice with high photosynthesis efficiency. Jiangsu J Agric Sci. 2010;26:449–455. [Google Scholar]
- 15.Tolley BJ, Sage TL, Langdale JA, Hibberd JM. Individual maize chromosomes in the C3 plant oat can increase bundle sheath cell size and vein density. Plant Physiol. 2012;159:1418–1427. doi: 10.1104/pp.112.200584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Datta K, Tu J, Oliva N, Ona I, Velazhahan R, Mew TW, Muthukrishnan S, Datta SK. Enhanced resistance to sheath blight by constitutive expression of infection-related rice chitinase in transgenic elite indica rice cultivars. Plant Sci. 2001;160:405–414. doi: 10.1016/S0168-9452(00)00413-1. [DOI] [PubMed] [Google Scholar]
- 17.Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- 18.Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4326. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Datta K, Baisakh N, Thet KM, Tu J, Datta SK. Pyramiding transgenes for multiple resistance in rice against bacterial blight, yellow stem borer and sheath blight. Theor Appl Genet. 2002;106:1–8. doi: 10.1007/s00122-002-1014-1. [DOI] [PubMed] [Google Scholar]
- 20.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 21.Humphries Mineral components and ash analysis. Soil Sci. 1956;100:112–117. [Google Scholar]
- 22.Bie X, Wang K, She M, Du L, Zhang S, Li J, Gao X, Lin Z, Ye X. Combinational transformation of three wheat genes encoding fructan biosynthesis enzymes confers increased fructan content and tolerance to abiotic stresses in tobacco. Plant Cell Rep. 2012;31:2229–2238. doi: 10.1007/s00299-012-1332-y. [DOI] [PubMed] [Google Scholar]
- 23.Huerta C, Freire M, Cardemil L. Expression of hsp70, hsp100 and ubiquitin in Aloe barbadensis Miller under direct heat stress and under temperature acclimation conditions. Plant Cell Rep. 2013;32:293–307. doi: 10.1007/s00299-012-1363-4. [DOI] [PubMed] [Google Scholar]
- 24.Lee DG, Ahsan N, Lee SH, Kang KY, Bahk JD, Lee IJ, Lee BH. A proteomic approach in analyzing heat-responsive proteins in rice leaves. Proteomics. 2007;18:3369–3383. doi: 10.1002/pmic.200700266. [DOI] [PubMed] [Google Scholar]
- 25.Salekdeh GH, Siopongco J, Wade LJ, Ghareyazie B, Bennett J. Proteomic analysis of rice leaves during drought stress and recovery. Proteomics. 2002;2:1131–1145. doi: 10.1002/1615-9861(200209)2:9<1131::AID-PROT1131>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 26.Dunwell JM. Transgenic approaches to crop improvement. J Exp Bot. 2000;51:487–496. doi: 10.1093/jexbot/51.suppl_1.487. [DOI] [PubMed] [Google Scholar]
- 27.Fukayama H, Tsuchida H, Agarie S, Nomura M, Onodera H, Ono K, Lee BH, Hirose S, Toki S, Ku MS, Makino A, Matsuoka M, Miyao M. Significant accumulation of C4-specific pyruvate, orthophosphate dikinase in a C3 plant, rice. Plant Physiol. 2001;127:1136–1146. doi: 10.1104/pp.010641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bandyopadhyay A, Datta K, Zhang J, Yang W, Raychaudhur S, Miyao M, Datta SK. Enhanced photosynthesis rate in genetically engineered indica rice expressing pepc gene cloned from maize. Plant Sci. 2007;172:1204–1209. doi: 10.1016/j.plantsci.2007.02.016. [DOI] [Google Scholar]
- 29.Horton P. Prospects for crop improvement through the genetic manipulation of photosynthesis: morphological and biochemical aspects of light capture. J Exp Bot. 2000;51:475–485. doi: 10.1093/jexbot/51.suppl_1.475. [DOI] [PubMed] [Google Scholar]
- 30.Ishimaru K. Identification of a locus increasing rice yield and physiological analysis of its function. Plant Physiol. 2003;133:1083–1090. doi: 10.1104/pp.103.027607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taniguchi Y, Ohkawa H, Masumoto C, Fukuda T, Tamai T, Lee K, Sudoh S, Tsuchida H, Sasaki H, Fukayama H, Miyao M. Overproduction of C4 photosynthetic enzymes in transgenic rice plants: an approach to introduce the C4-like photosynthetic pathway into rice. J Exp Bot. 2008;59:1799–1809. doi: 10.1093/jxb/ern016. [DOI] [PubMed] [Google Scholar]
- 32.Zhang JF, Bandyopadhyay A, Sellappan K, Wang GY, Xie HA, Datta K, Datta SK. Characterization of a C4 maize pyruvate orthophosphate dikinase expression in C3 transgenic rice plants. Afr J Biotechnol. 2010;9:234–242. [Google Scholar]
- 33.Jiao DM, Huang XQ, Li X, Chi W, Kuang TY, Zhang QD, Ku MS, Cho DH. Photosynthetic characteristics and tolerance to photo-oxidation of transgenic rice expressing C4 photosynthesis enzymes. Photosynth Res. 2002;72:85–93. doi: 10.1023/A:1016062117373. [DOI] [PubMed] [Google Scholar]
- 34.Fukayama H, Hatch MD, Tamai T, Tsuchida H, Sudoh S, Furbank RT, Miyao M. Activity regulation and physiological impacts of maize C4-specific phosphoenolpyruvate carboxylase overproduced in transgenic rice plants. Photosynth Res. 2003;77:227–239. doi: 10.1023/A:1025861431886. [DOI] [PubMed] [Google Scholar]
- 35.Ding ZS, Huang SH, Zhou BY, Sun XF, Zhao M. Over-expression of phosphoenolpyruvate carboxylase cDNA from C4 millet (Seteria italica) increase rice photosynthesis and yield under upland condition but not in wetland fields. Plant Biotechnol Rep. 2013;7:155–163. doi: 10.1007/s11816-012-0244-1. [DOI] [Google Scholar]
- 36.Jiao DM, Li X, Ji BH. Photoprotective effects of high level expression of C4 phosphoenolpyruvate carboxylase in transgenic rice during photoinhibition. Photosynthetica. 2005;43:501–508. doi: 10.1007/s11099-005-0082-2. [DOI] [Google Scholar]
- 37.Ji BH, Zhu SQ, Jiao DM. A limited photosynthetic C4-microcycle and its physiological function in transgenic rice plant expressing the maize PEPC gene. Acta Bot Sin. 2004;46:542–551. [Google Scholar]
- 38.Li X, Wang C. Physiological and metabolic enzymes activity changes in transgenic rice plants with increased phosphoenolpyruvate carboxylase activity during the flowering stage. Acta Physiol Plant. 2013;35:1503–1512. doi: 10.1007/s11738-012-1191-8. [DOI] [Google Scholar]
- 39.Hockin NL, Mock T, Mulholland F, Kopriva S, Malin G. The response of diatom central carbon metabolism to nitrogen starvation is different to that of green algae and higher plants. Plant Physiol. 2012;158:299–312. doi: 10.1104/pp.111.184333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schlüter U, Mascher M, Colmsee C, Scholz U, Bräutigam A, Fahnenstich H, Sonnewald U. Maize source leaf adaptation to nitrogen deficiency effects not only N and C metabolism but also control of P homeostasis. Plant Physiol. 2012;160:1384–3406. doi: 10.1104/pp.112.204420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oaks A. Efficiency of nitrogen utilization in C3 and C4 cereals. Plant Physiol. 1994;106:407–414. doi: 10.1104/pp.106.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang D, Chi W, Wang S, Wang D, Wu S, LI X, LI C, Zhang Y, Luo Y. Characteristics of transgenic rice overexpression maize photosynthetic enzymes for breeding two-line hybrid rice. Acta Agron Sin. 2004;30:248–252. [Google Scholar]
- 43.Brown NJ, Newell CA, Stanley S, Chen JE, Perrin AJ, Kajala K, Hibberd JM. Independent and parallel recruitment of preexisting mechanisms underlying C4 photosynthesis. Science. 2011;331:1436–1439. doi: 10.1126/science.1201248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.