Abstract
T lymphocytes responding to microbial infection give rise to effector cells that mediate acute host defense and memory cells that provide long-lived immunity, but the fundamental question of when and how these cells arise remains unresolved. Here we combine single-cell gene expression analyses with machine-learning approaches to trace the transcriptional roadmap of individual CD8+ T lymphocytes throughout the course of an immune response in vivo. Gene expression signatures predictive of eventual fates could be discerned as early as the first T lymphocyte division and may be influenced by asymmetric partitioning of the interleukin-2 receptor during mitosis. These findings underscore the importance of single-cell analyses in understanding fate determination and provide new insights into the specification of divergent lymphocyte fates early during an immune response to microbial infection.
During a microbial infection, responding T lymphocytes give rise to two distinct classes of cellular progeny, effector cells that provide acute host defense and long-lived memory cells that provide durable immunity1. Terminally differentiated, short-lived effector cells (Tsle) can be identified phenotypically by high expression of the lectin-like receptor (KLRG1) and low expression of the interleukin 7 receptor (IL-7R)2. At least two distinct subsets of long-lived memory cells, central memory (Tcm) and effector memory (Tem), have been described and can be distinguished on the basis of their proliferative capacity, cytotoxicity, anatomic localization and expression of certain homing and chemokine receptors, including L-selectin (CD62L) and CCR73,4.
Prior studies using single-cell adoptive transfer and genetic barcoding approaches5,6 have elegantly demonstrated that a single naïve CD8+ T lymphocyte can give rise to more than one fate, and importantly, is capable of generating all of the diverse cellular fates necessary for an immune response. The process by which a single activated T lymphocyte yields effector- and memory-fated progeny and the timing at which these differentiation pathways begin to diverge, however, remain unresolved. One possibility is that the progeny of an activated naïve CD8+ T lymphocyte progress along a linear differentiation path, initially becoming effector cells, with a subset of these cells later acquiring the memory fate1,7,8. An alternative possibility is that the first CD8+ T cell division in vivo is asymmetric9,10, thereby enabling lymphocyte fates to diverge early during an immune response owing to unequal inheritance of certain determinants, such as the interferon γ (IFN-γ) receptor and the T-box transcription factor, T-bet.
Tracing individual lymphocytes sequentially as they differentiate in vivo might distinguish whether lymphocytes progress along a linear differentiation pathway1,7,8 or diverge early during an immune response. While genomic profiling studies have begun to elucidate the transcriptional networks that control lymphocyte fate specification11-13, these studies have been based on analyses of bulk cellular populations, making it impossible to discern cell fate decisions made by individual T cells. Recent technological advances that have coupled microfluidics technologies with high-throughput qRT-PCR analyses have enabled detailed analyses of cell fate decisions in C. elegans development, induced stem cell reprogramming and cancer biology14-17. Here, we applied single-cell gene expression profiling to investigate the ontogeny of effector and memory CD8+ T lymphocytes during a microbial infection in vivo, uncovering evidence for heterogeneity in gene expression within individual lymphocytes early after the initiation of an adaptive immune response.
RESULTS
Single-cell gene expression analyses of CD8+ T lymphocytes in vivo
In order to delineate the hierarchy and mechanism of CD8+ T cell differentiation during an adaptive immune response at the single-cell level, we employed an experimental system that allowed us to interrogate the gene expression of individual CD8+ T lymphocytes throughout the course of a microbial infection in vivo. CD8+ T cells transgenic for the OT-1 T cell receptor that recognize a specific ovalbumin epitope were adoptively transferred into wild-type recipient mice. Mice were infected intravenously 24 hours later with recombinant Listeria monocytogenes bacteria expressing ovalbumin (Lm-OVA) and CD8+ T cells were sorted throughout the course of infection for single-cell analysis (Fig. 1). In addition, we selected for analysis terminally differentiated short-lived effector cells (Tsle, KLRG1hiIL-7Rlo)2, putative memory precursor cells (Tmp, KLRG1loIL-7Rhi)2, and central memory (Tcm, CD44hiCD62Lhi) and effector memory (Tem, CD44hiCD62Llo)3,4 cells (Fig. 1).
Figure 1.
Gating strategy and experimental approach for single-cell gene expression analyses of CD8+ T cell subsets isolated from uninfected (naïve, CD8+CD44loCD62Lhi) or CD45.2 recipient mice infected with Lm-OVA 24h after intravenous adoptive transfer of unlabeled or CFSE-labeled CD45.1+OT-1 CD8+ T cells. CD8+ T cell subsets were isolated at various time points post-infection: division 1 (CD8+CD45.1+CD44hi cells within 2nd brightest CFSE peak); days 3, 5, and 7 post-infection; day 7 short lived effector (Tsle) (CD8+CD45.1+CD44hiKLRG1hiIL-7Rlo), day 7 putative memory precursor (Tmp) (CD8+CD45.1+CD44hiKLRG1loIL-7Rhi), day 45 central memory (Tcm) (CD8+CD45.1+CD44hiCD62Lhi), and day 45 effector memory (Tem) (CD8+CD45.1+CD44hiCD62Llo). Data are representative of three experiments. Data analysis approaches included unsupervised Principal Component analysis (PCA) and Jensen-Shannon Divergence (JSD), and supervised binary classifier and Hidden Markov Model (HMM).
Quantitative real-time PCR analysis was performed using Fluidigm 96.96 Dynamic Arrays, enabling simultaneous measurement of expression for 96 genes in 96 individual cells (Supplementary Fig. 1a). Among the 94 gene targets (Table 1 and Supplementary Table 1) we selected for analysis were transcriptional regulators previously reported to influence CD8+ T lymphocyte differentiation18-25; cytokines, chemokines, and their receptors19; and molecules associated with tissue homing and survival19.
Table 1.
94 selected gene targets grouped according to their function.
| Class | Gene |
|---|---|
| Apoptosis | Bnip2, Bnip3l, Casp3, Casp9, Cflar, Pdcd1 |
| Cytokine/chemokine receptors | Ccr5, Ccr6, Ccr7, Cxcr3, Ifngr1, Ifnar1, Il2ra, Il2rb, Il7r, Klrc1, Klrg1, Tnfrsf1, Tnfrsf9 |
| Cytokines/chemokines/granzyme | Ccl3, Ccl4, Ccl5, Cxcl10, Gzmb, Ifng, Il2, Il3, Lif, Xcl1 |
| Housekeeping | Actb, Gapdh, Rn18s, Rpl35 |
| Polarity/proteasome | Prkcz, Psmb1, Psmb7 |
| Signaling/proliferation/self-renewal | Bag2, Bmi1, Bmp2, Cd28, Cd4, Cd44, Cd69, Cd8a, Grap2, Hk2, Lag3, Lgals1, Mapk3, Mapk8, Mapk14, Mela, Mtor, Myc, Ptprc, RelA, Sema7a, Serpinb6b, Serpinb9, Setd7, Sell, Thy1 |
| Transcription factors | Atf1, Bcl11b, Bcl6, Bhlhe40, Eomes, Foxo1, Foxo3, Foxp1, Foxp3, Gata3, Hopx, Id2, Id3, Irf4, Irf8, Klf2, Lef1, Nfatc1, Nfatc2, Prdm1, Rel, Runx1, Runx2, Stat1, Stat4, Tbx21, Tcf3, Tcf7, Tcf12, Tox, Zeb2, Zfp281 |
After excluding failed reactions, expression data from 1,300 single cells were retained for in-depth analyses (Supplementary Fig. 1b). Because expression of “housekeeping” genes has been shown to vary substantially across cell types and states of differentiation26, the expression of each gene of interest was utilized without normalization for all of the analyses performed herein.
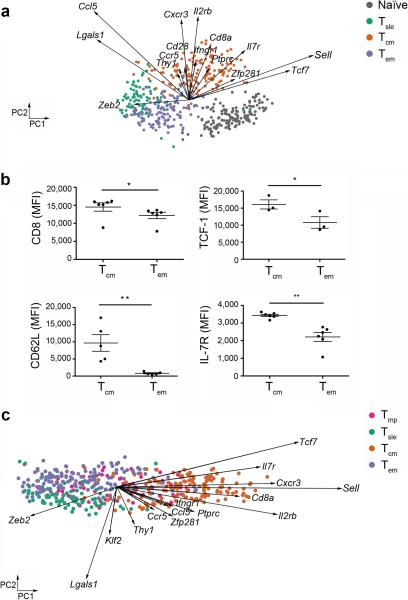
We used principal component analysis (PCA) to visualize the expression data globally. PCA is an unsupervised dimensionality reduction method that we used to project the data into 2 dimensions by its coordinates in the first two principal components (PC1 and PC2) that account for the largest variations in the data. These PCs are linear combinations of the 94 original genes. PCA revealed that naïve, Tsle, Tem, and Tcm cells are clustered distinctly (Fig. 2a). Expression of Sell and Tcf7, which encode the trafficking molecule CD62L and the transcription factor TCF-1, distinguished naïve from Tsle cells, consistent with previous findings2,4. Though Tsle cells formed a distinct cluster, these cells were projected closest to Tem cells (Fig. 2a), suggesting related gene expression profiles that may underlie some of their functional similarities, such as cytotoxicity and secretion of pro-inflammatory cytokines27. This clustering was driven by Zeb2, a transcription factor expressed in Tsle cells12. In addition, Tem and Tcm cells occupied distinct clusters, with higher expression of Tcf7, Il2rb, Il7r, Cxcr3 and Sell mRNA in Tcm cells and higher expression of Zeb2 mRNA in Tem cells accounting for the variance between these memory cell populations. Some of the disparities observed at the transcriptional level were confirmed at the protein level (Fig. 2b), supporting the finding that Tcm and Tem cells are molecularly distinct. The higher expression of Il7r and Tcf7, regulators of T lymphocyte survival and longevity25,28, that we observed in Tcm cells may underlie the superior capacity of these cells to persist in vivo29. Putative memory precursor (Tmp) cells did not form a distinct cluster but overlapped with Tsle, Tem and Tcm cells (Fig. 2c). These results suggested that putative Tmp cells are molecularly heterogeneous, raising the possibility that this population may not represent memory precursor cells, but instead may be comprised of “mature” memory and terminally differentiated effector cells. Together these findings suggest that Tsle, Tcm, and Tem cells, but not putative Tmp cells, exhibit similar gene expression profiles at the single-cell level.
Figure 2.
Effector and memory CD8+ T lymphocyte subsets are molecularly distinct on a single-cell level. (a) Principal component (PC) projections (PC1, horizontal axis; PC2, vertical axis) of single-cell gene expression data derived from individual lymphocytes from the indicated populations. Each circle represents an individual cell of the indicated population: naïve (gray), Tsle (green), Tcm (orange), and Tem (purple) cells. Each vector emanating from the origin represents an individual gene. PC1 and PC2 account for 11% and 9% of the variance, respectively. (b) Mean fluorescence intensity (MFI) of CD8 (Cd8a), TCF-1 (Tcf7), CD62L (Sell), and IL-7R (Il7r) protein expression in Tcm and Tem cells, assessed by flow cytometry. * P < 0.05, ** P < 0.01 (Kolmogorov-Smirnov test). Data are representative of two experiments with at least 3 mice in each experiment (error bars, s.e.m.). (c) PC projections of single-cell gene expression data derived from individual lymphocytes from the indicated populations: Tmp (pink), Tsle (green), Tcm (orange), and Tem (purple) cells. Each vector emanating from the origin represents an individual gene. PC1 and PC2 account for 11% and 6% of the variance, respectively.
Molecular heterogeneity at the single-cell level early after infection
To assess whether single responding CD8+ T cells comprised distinct clusters early after infection, we analyzed the gene expression profiles of individual CD8+ T cells isolated throughout the course of infection (Fig. 3a). PCA revealed substantial heterogeneity among cells isolated early after infection (division 1 and day 3) compared to cells isolated at later timepoints (day 5 and day 7, Tsle, Tcm, and Tem). The first two principal components captured 17% of the variance in our dataset, slightly lower than that previously observed15, likely a reflection of a higher degree of heterogeneity in lymphocytes during differentiation and the greater number of genes analyzed in our study. In agreement with our findings using PCA, an alternative unsupervised method, t-distributed Stochastic Neighbor Embedding analysis30, was also performed and showed similar results (Supplementary Fig. 2). To test whether the heterogeneity observed using data from single cells could be recapitulated using data from bulk cells, we formally compared the analyses using data derived from single versus bulk populations (Fig. 3a). We found that the heterogeneity we observed at the single-cell level within putative Tmp cells and cells isolated early after infection was not apparent in the bulk analysis (Fig. 3a), thus illustrating the power and necessity of using a single-cell approach.
Figure 3.
Early heterogeneity of gene expression exhibited by individual CD8+ T lymphocytes during an immune response. (a) Projections of single-cell gene expression data derived from individual lymphocytes from the indicated populations (top). Each circle represents an individual cell of the indicated population representing: naïve (gray), division 1 (brown), day 3 (yellow), day 5 (light green), day 7 (green), Tmp (pink), Tsle (teal), Tcm (orange), and Tem (purple) cells. PC1 and PC2 account for 10% and 7% of the variance, respectively. Analysis derived from pooled “bulk” samples from each experimental condition, shown as colored stars (bottom). Stars filled with each color represent “bulk” naïve (gray), division 1 (brown), day 3 (yellow), day 5 (light green), day 7 (green), Tmp (pink), Tsle (teal), Tcm (orange), and Tem (purple) cells with grayed-out single-cell data points in the background for clarity. (c) Intra- (left) and inter-population (right) Jensen-Shannon Divergence (JSD) of mean gene expression within and between the indicated CD8+ T cell populations is shown.
To further evaluate the degree of heterogeneity within and between cell populations at each time point, which was not previously possible using bulk analysis, we applied the Jensen-Shannon Divergence (JSD) metric, a non-parametric, model-free measure of similarity between two empirical probability distributions. In general, the intra-population JSD was lowest in naïve cells and highest in cells isolated early after infection (Fig. 3b and Supplementary Fig. 3a). We observed that the intra-population JSD decreased as a function of time following infection, with the notable exception of putative Tmp cells (Fig. 3b). These cells exhibited a high degree of intra-population divergence, consistent with the apparent heterogeneity of these cells by PCA (Fig. 3a). Comparing JSD pair-wise between all cell populations (naïve, division 1, days 3, 5, and 7 post-infection, Tmp, Tsle, Tcm, and Tem) yielded similar observations, with the greatest divergence found between cells isolated early versus late after infection (Fig. 3b and Supplementary Fig. 3a). Importantly, the inter-population JSD metric was not affected by group size (Supplementary Fig. 3b). Together these results demonstrate that CD8+ T lymphocytes responding to microbe exhibit substantial molecular heterogeneity at the single-cell level early after infection that diminishes with time.
Distinct transcriptional signatures in CD8+ T cells early after infection
We hypothesized that the heterogeneity observed within lymphocytes early post-infection might reflect distinct gene expression patterns that are predictive of more differentiated cells. We reasoned that supervised classifiers trained on relatively well-defined, differentiated cellular fates, such as sorted Tcm and Tsle cells, could be utilized to assess whether cells isolated early post-infection might be fated towards specific CD8+ T lymphocyte subsets. Boosted decision trees31 were chosen over other classification frameworks with similar performance characteristics because the learned trees are easily interpretable. A decision tree that was built from the data consisted of several predictive rules that compare the expression of Ptprc, Sell, and Ccl5 to thresholds learned from that data to decide whether a cell is more Tcm- or Tsle-like (Supplementary Fig. 4a). Ensembles of decision trees were trained with RobustBoost32 to generate a binary classifier that achieved misclassification error of approximately 4% in leave-one-out cross validation which was split evenly when distinguishing between Tcm versus Tsle cells (Fig. 4a and Supplementary Fig. 4b). The classifier revealed that Sel1 and Il7r were among the most predictive genes whose high expression accurately described Tcm cells, whereas the lack of their expression, along with high expression of Zeb2, defined Tsle cells (Fig. 4b). Application of the classifier to cells isolated at days 5 and 7 post-infection revealed that 49% and 57% of total CD8+ T cells at these timepoints were more like Tsle than Tcm cells (Fig. 4c), consistent with the expected percentages of Tsle cells at days 5 and 7 post-infection2.
Figure 4.
Classifier analysis predicts eventual fates of individual CD8+ T lymphocytes. (a) Predictions by the classifier on sort-purified Tcm and Tsle cells that were cross-validated during training. Horizontal red lines indicate the voting margin for each individual cell and internal confidence of the classifier’s prediction for that cell; percentages indicate rate of misclassification of a Tsle as Tcm and of a Tcm as Tsle. (b) Binary classifier trained to distinguish between a pair of differentiated cell fates (Tcm vs. Tsle). Single vertical lines along the x-axis represent each individual sort-purified Tcm or Tsle cell and its expression of each gene. (c) Individual CD8+ T cells (horizontal blue lines) from the indicated populations (cells isolated at day 5 or 7 post-infection; proximal (“prox”) or distal daughter (“distal”) cells at the first division) were interrogated by the classifier and predictions were sorted by confidence from the most Tcm-like to most Tsle-like cells. Percentages indicate proportion of cells predicted to be more Tcm-like (left) or Tsle-like (right) within each cell population. (d) Violin plots showing expression levels of the indicated genes by first division proximal (blue) and distal daughter (red) cells. Black crosses and squares represent mean and mode values, respectively.
We next asked whether the classifier could discern the fates of responding lymphocytes isolated early during an immune response. It has been previously suggested that asymmetric CD8+ T lymphocyte division yields immune synapse-proximal (“proximal”) and synapse-distal (“distal”) daughter cells that are differentially fated6, raising the possibility that these cells might already exhibit distinct gene expression patterns that are predictive of their eventual fates as early as the first cell division. To test this possibility, putative proximal and distal daughter cells, which can be distinguished by their relative abundance of CD8 and CD11a9, were sorted and analyzed. The classifier revealed that most proximal daughter cells more closely resembled Tsle cells, while most distal daughter cells more closely resembled Tcm cells (Fig. 4c), suggesting that these cells may indeed be differentially fated.
As further evidence that proximal and distal daughter cells display unique molecular patterns that might drive their distinct fates, we observed that these cells exhibited a pronounced disparity in the expression of genes associated with the effector or memory fates (Fig. 4d). Certain genes associated with the memory fate in CD8+ T cells, including Eomes, Sell, Il7r, Il2rb, Tcf7, Id3, and Bcl618,19,21,24,25, were more highly expressed in distal daughter cells. Conversely, certain genes associated with terminally differentiated effector cells, such as Tbx21, Prdm1, and Grzmb19,20,22, were only detected in proximal, but not distal daughter cells. While it remains possible that the gene expression patterns of early lymphocytes might change as the cells continue to differentiate, together these results are indicative of distinct molecular patterns, suggestive of a possible predisposition towards different fates, within cells that may have undergone an asymmetric division in vivo.
Predicting temporal expression of key orchestrators of CD8+ T cell fates
Having determined that the gene expression patterns of less differentiated cells could be utilized to predict their eventual fates, we next sought to develop a simple generative model of CD8+ T lymphocyte fate specification that would capture key genes involved in each step of the differentiation pathway of an individual naïve cell. In contrast to the classifiers we trained on sort-purified cells to discriminate between differentiated cellular fates (Tcm vs. Tsle), we used a Hidden Markov Model (HMM) trained on lymphocytes representing intermediate states of differentiation (division 1, day 3, day 5) between the naïve state and the differentiated fates (Fig. 5a). HMMs have been applied to sequential and time-series analyses in diverse fields and have been particularly useful for modeling “hidden”, unobserved states during biological processes33,34. HMMs not only capture static expression profiles between subpopulations at a particular stage, but can also detect dynamic expression changes responsible for the transitions between them. To construct a temporal paradigm of T lymphocyte fate specification in vivo, we first defined 6 linear and 12 divergent (Supplementary Fig. 4c) HMMs representing possible hypothetical states (pre-Tsle, pre-memory) into which an individual naïve T lymphocyte could transition through prior to differentiating into three observed fates (Tsle, Tcm, and Tem). To evaluate each HMM, all possible paths were analyzed for each individual cell (Supplementary Fig. 4c). Incorporating the single-cell measurements obtained serially within CD8+ T lymphocytes differentiating in vivo, we calculated the likelihood of each of the possible differentiation paths for each defined linear or divergent HMM (Supplementary Fig. 4c). To determine both the significance and robustness of each HMM model, we randomly varied the initial values of the transition matrices by 10% and computed the log likelihood for each iteration. Our results showed that the divergent models generally outperformed the linear models, and an early divergent model was identified as the most likely pathway (Fig. 5b, Supplementary Fig. 4d). The performance of this final model was further evaluated by random ordering of the population labels of the cells as well as the associated expression values. Importantly, the likelihood of the best model was significantly (p=0.00034) higher than the likelihood for shuffled data, showing that the model robustly indicated that an activated CD8+ T lymphocyte gives rise to cells that transition through either a hypothetical pre-Tsle or pre-memory state. Pre-Tsle cells can undergo further differentiation to acquire the Tsle fate, whereas pre-memory cells can further diverge to give rise to Tcm or Tem cells. Together these findings suggest that an early divergent model may be the most likely pathway underlying lymphocyte fate specification in vivo.
Figure 5.
Temporal model predicts the differentiation paths of individual CD8+ T lymphocytes. (a) Cells in early states of differentiation (division 1, day 3, day 5) were ranked by their Tsle- or memory-like expression profiles (green to purple gradient in middle row). Cells were then linked to sorted naïve (black top row) and sorted Tsle, Tem and Tcm cells (green to purple to orange gradient in bottom row) in a random fashion, forming hypothetical differentiation paths (black lines) that were analyzed with a Hidden Markov Model. (b) Most likely model of CD8+ T lymphocyte differentiation with key gene expression changes associated with each of 5 unique transitions: naïve to pre-Tsle, naïve to pre-memory, pre-Tsle to Tsle, pre-memory to Tcm, and pre-memory to Tem. Colored circles represent each cell state or fate. (c) Summary of key changes in gene expression during each transition phase predicted by temporal model of CD8+ T lymphocyte differentiation.
We analyzed the changes in expression of all 94 genes during each of these five unique transitions: naïve to pre-Tsle, naïve to pre-memory, pre-Tsle to Tsle, pre-memory to Tcm, and pre-memory to Tem (Fig. 5b, c, and Supplementary Fig. 5). This analysis revealed both shared and unique molecular features of each transition. The naïve to pre-Tsle and naïve to pre-memory transitions, for example, were both associated with increased expression of Lgals1. Notably, however, the naïve to pre-Tsle transition was associated with higher Il2ra and lower Cxcr3, Sell, and Tcf7 expression than the naïve to pre-memory transition, raising the possibility that these genes might influence whether a cell proceeds along the pathway towards terminal differentiation or self-renewal. Like the early transitions from the naïve state, the pre-memory to Tcm and pre-memory to Tem transitions exhibited certain shared molecular regulators, including increased expression of Ccl5 and decreased expression of Foxo1 and Cxcr3. However, the pre-memory to Tcm transition was uniquely associated with increased expression of Tcf7, Il7r and Sell. By contrast, the pre-Tsle to Tsle transition was associated with increased Ccl5 and decreased Il2ra, Il2rb, and Foxo1. Together these results characterize the temporal expression patterns of key genes that influence the fates of CD8+ T lymphocytes responding to microbial infection in vivo.
Asymmetric partitioning of IL-2Rα is associated with distinct cellular fates
The prediction, raised by our temporal model, that Ilr2a might represent an early molecular switch promoting the pathway towards terminal differentiation was intriguing in light of recent work suggesting a role for IL-2 signaling in CD8+ T lymphocyte differentiation35-39. To determine how early in effector- versus memory-fated lineages a possible disparity in IL2ra could be detected, we used flow cytometry to examine the expression of IL-2Rα in CD8+ T cells that had undergone their first division in vivo in response to microbial infection. We observed that differential abundance of IL-2Rα on the cell surface distinguished two populations of 1st daughter cells (Fig. 6a) and that IL-2Rα abundance was inversely correlated with CD62L expression (Fig. 6a), which is highly expressed in Tcm cells. Furthermore, cells with higher expression of IL-2Rα also exhibited an increased capacity for IFN-γ and granzyme B production, characteristic of effector cells (Fig. 6b).
Figure 6.
Asymmetric segregation of IL-2Rα during T lymphocyte division influences the eventual fates of the daughter cells. (a) IL-2Rα and CD62L expression (left) by OT-1 CD8+ T cells undergoing their first division following adoptive transfer into recipients and subsequent infection with Lm-OVA 24h later. Frequencies of IL-2RαloCD62Lhi and IL-2RαhiCD62Llo cells (right); each circle represents an individual mouse and lines indicate the mean. (b) Frequencies of IFN-γ and granzyme B expression by IL-2RαloCD62Lhi and IL-2RαhiCD62Llo cells as in (a). (c) CD62L expression (left) on d49 post-infection by CD45.1+CD8+ T cells in CD45.2+ mice (n=13) that had been previously challenged with Lm-OVA and injected with sort-purified 1st division IL-2RαloCD62Lhi or IL-2RαhiCD62Llo cells 48h later. Frequencies of CD62L+ cells (right); each circle represents an individual mouse and lines indicate the mean. (d) Expansion of CD45.1+CD8+ T cells, assessed by serial bleeding, in mice depicted in (c) that were subsequently re-challenged with Lm-OVA at d50 post-primary infection. (e) Morphology of IL-2Rα (green, top panel) or CD62L (green, bottom panel), β-tubulin (red), and DNA (blue), assessed by confocal microscopy, in sorted OT-1 CD8+ T cells undergoing their first division following adoptive transfer into LM-OVA-infected recipients. Asymmetric segregation of IL-2Rα and CD62L was observed in 60% (n=96) and 62% (n=74) of cells, respectively. Data are representative of 2 (c, d) or 3 experiments (a, b, e); error bars represent s.e.m. * P < 0.05, ** P < 0.01, *** P = 0.0002 (Kolmogorov-Smirnov test).
To test the hypothesis that the amount of IL-2Rα expression conferred a distinct predisposition towards the effector or memory lineages, we sorted IL-2RαhiCD62Llo or IL-2RαloCD62Lhi cells that had undergone their first division in vivo. Cells were then adoptively transferred into recipient wild-type mice that had been infected 48 hours previously with Lm-OVA. We tracked the progeny of adoptively transferred cells at multiple time points throughout the course of the primary response and found that the progeny of both IL-2RαhiCD62Llo and IL-2RαloCD62Lhi cells were detectable following infection (Supplementary Fig. 6). Notably, however, the progeny of the transferred IL-2Rαlo cells exhibited a 4-fold increased capacity to give rise to CD62Lhi central memory cells, compared to the progeny of transferred IL-2Rαhi cells (Fig. 6c). To confirm functionally that these cells were indeed memory lymphocytes, we tested their ability to respond to microbial re-challenge. Recipient mice were re-challenged with Lm-OVA at day 50 after primary infection. We observed a 10-fold increased expansion by the progeny of transferred CD8+ T cells in recipient mice that had received IL-2RαloCD62Lhi cells compared to mice that received IL-2RαhiCD62Llo cells (Fig. 6d), suggesting that these cells exhibit a differential capacity to give rise to memory lymphocytes.
Because certain cytokine and immune receptors can undergo unequal partitioning during cell division9, we hypothesized that asymmetric segregation of IL-2Rα and CD62L during mitosis might provide a mechanism underlying their differential abundance on daughter cells that had undergone their first division in vivo. We used an experimental system that has previously allowed us to examine T cells preparing for their first division in response to a microbe9. OT-1 CD8+ T cells were labeled with CFSE and adoptively transferred into recipient mice that were infected 24 hours previously with Lm-OVA. Undivided donor CD8+ T cells were isolated by flow cytometry at 36 hours after transfer and examined by confocal microscopy. We observed that IL-2Rα and CD62L exhibited a pronounced asymmetric distribution in cells that were preparing for division (Fig. 6e). Taken together, these results suggest that the asymmetric segregation of IL-2Rα and CD62L during the first CD8+ T lymphocyte division in vivo may influence the transcriptional profiles of the nascent daughter cells and their eventual fates.
DISCUSSION
Recent advances in high-throughput single-cell gene expression profiling have enabled their utilization in such diverse fields as embryonic development, hematopoiesis, stem cell reprogramming and cancer biology14-17. These advances, coupled with computational modeling approaches, enabled us to investigate, on a level of molecular detail not previously possible, the ontogeny of effector and memory lymphocytes during a microbial infection in vivo. We find evidence for considerable heterogeneity in gene expression within individual CD8+ T lymphocytes early after the initiation of a microbial infection. Importantly, we demonstrate that this heterogeneity cannot be revealed using traditional bulk population analyses and that many of the computational analyses performed herein, including JSD, classifier and HMM, are possible only with data derived from single cells. These observations provide a compelling argument for the integration of single-cell approaches into future studies of immune cell fate specification.
Using sequential single-cell gene expression measurements within activated lymphocytes during the course of a microbial infection in vivo, we constructed a temporal model that enables us to predict the timing and changes in the expression of key genes within individual lymphocytes as they transition from the naïve state towards each of several cellular fates. We provide experimental evidence supporting an important prediction of this temporal model-- that differential expression of IL-2Rα may reflect one of the earliest molecular determinants influencing the memory versus effector fate decision. Moreover, we demonstrate that unequal partitioning of IL-2Rα during the first asymmetric division in vivo may result in its disparate abundance in daughter lymphocytes, potentially contributing to their acquisition of distinct gene expression profiles and cellular fates.
Along with prior evidence that other critical signaling molecules, such as IFN-γR, can be unequally partitioned9, these results suggest that asymmetric segregation of cytokine receptors during lymphocyte division may result in increased IL-2 and IFN-γ signaling encountered by proximal daughter cells relative to distal daughters. As IL-2 has previously been shown to induce Prdm1 and repress Bcl6 and Il7ra37,38, while IFN-α is known to induce Tbx2140,41, differential cytokine signaling encountered by proximal and distal daughter cells may initiate a pre-effector or pre-memory gene expression program, respectively, consistent with our experimental observations and with prior work showing that cells that receive prolonged IL-2 signals acquire characteristics of terminally differentiated effector cells37. Continued changes in gene expression patterns, influenced by environmental signals, may enable lymphocytes to continue along distinct pathways towards terminal differentiation or self-renewal.
Recent reports describing additional subsets of memory T lymphocytes, however, raise the possibility that the effector or central memory lineages may not be the exclusive fate choices adopted by the progeny of IL-2RαhiCD62Llo and IL-2Rαlo CD62Lhi cells. Tissue-resident memory T cells42,43 do not circulate and instead remain in the peripheral tissues after pathogen clearance, while so-called “effector-phenotype” memory T cells share certain phenotypic characteristics with terminally differentiated effector cells and mediate robust immune protection in certain infectious settings despite exhibiting poor proliferative recall responses44. Indeed, the progeny of IL-2RαhiCD62Llo cells appear to give rise to a population of lymphocytes that, while poorly proliferative in response to microbial rechallenge, persist in vivo, reminiscent of effector-phenotype memory cells. Thus, it remains possible that the first cellular division, in addition to mediating a divergence of the effector and memory fates, may also facilitate the specification of distinct memory cell subset fates.
Although the generation of long-lived memory lymphocytes is an essential feature of an adaptive immune response, the fundamental question of when and how these cells arise has remained controversial. Resolving whether lymphocytes progress along a linear differentiation pathway, or diverge early during an immune response, owing to asymmetric cell division, necessitated tracing individual lymphocytes as they undergo differentiation in vivo. By interrogating the gene expression patterns of individual lymphocytes during an immune response to microbial pathogen, we have been able to reconstruct the lineage path of single lymphocytes as they differentiate in vivo. This approach has yielded new insights underlying lymphocyte fate specification and provides new evidence supporting an early divergence of lymphocyte fates, via asymmetric division, during an adaptive immune response to a microbial infection. More broadly, we anticipate that single-cell gene expression approaches undertaken by investigators across scientific disciplines, along with ever-improving advances in such technologies as single-cell RNA sequencing45,46 and single-cell mass cytometry47, will continue to provide unprecedented molecular insights into cell fate specification in diverse biological settings, including immunity, development, and cancer.
METHODS
Mice
All animal work was done in accordance with Institutional Animal Care and Use Guidelines of the University of California, San Diego. All mice were housed in specific pathogen-free conditions prior to use. Wild-type C57/BL6J mice were purchased from the Jackson Laboratory and OT-1 TCR transgenic mice recognizing ovalbumin peptide SIINFEKL (residues 257-264)/Kb were used.
Adoptive cell transfers and infections
5 × 103 OT-1 CD45.1+ CD8+ T cells were adoptively transferred into congenic wild-type CD45.2 recipients, followed by infection intravenously one day later with 5 × 103 colony-forming units (CFU) of Listeria monocytogenes expressing full-length chicken ovalbumin (Lm-OVA). Splenocytes were isolated from recipient mice at 5, 7, or 45 days post-infection. To isolate cells at 3 days post-infection, 2 × 104 OT-1 CD8+ T cells were adoptively transferred. To isolate cells that had undergone their first division, 2 × 106 OT-1 CD8+ T cells were first labeled with carboxyfluorescein diacetate succinimidyl ester (CFSE) prior to adoptive transfer and recipient mice were sacrificed at 48 hours post-infection. Cells were stained with fluorochrome-labeled antibodies against CD8, CD44, CD4, CD11b, CD11c, and F4/80, and sorted on a MoFlo (Beckman Coulter) or FACS Aria II (BD Biosciences) flow cytometer.
Microbead-based enrichment
Magnetic bead-based enrichment was performed as previously described48. Single cell suspensions were prepared from infected mice that had received OT-1 CD8+ T cells, stained with PE-conjugated anti-CD45.1 antibody, washed, stained with anti-PE magnetic microbeads (Miltenyi Biotec), and enriched through a magnetic column. Cells were then stained and sorted as described above.
Lymphocyte fate tracking experiments
Splenocytes from infected recipient mice that had received CFSE-labeled OT-1 CD8+ T cells were stained with fluorochromeconjugated antibodies against CD8, CD62L, and IL-2Rα. Cells that had undergone their first division (represented as the second brightest CFSE peak) were electronically gated, and IL-2RαhiCD62Llo or IL-2RαloCD62Lhi cells were sorted. 350 cells of each phenotype were adoptively transferred into separate infection-matched CD45.2+ wild-type recipient mice. The progeny of transferred CD45.1+ T cells were monitored throughout the primary response by serial bleeding. At 50 days post-infection, recipient mice were re-challenged with 5 × 105 CFU of Lm-OVA and expansion of the progeny of donor CD45.1+ T cells tracked in the peripheral blood.
Antibodies and flow cytometry
The following antibodies were used: CD8a (53-6.7), CD45.1 (A20), CD62L (MEL-14), KLRG1 (2F1), IFN-γ (XMG1.2), CD44 (1M7), IL-2Rα (PC61), Vα2 (B20.1), CD4 (RM4-5), B220 (RA3-6B2), CD11b (M1/70), CD11c (N418), F4/80 (BM8), IL-7R (A7R34), and F(ab′)2 anti-rabbit anti-IgG and were obtained from Biolegend or eBioscience. Rabbit anti-TCF-1 (C63D9) antibody was obtained from Cell Signaling Technology. Anti-human PE-conjugated Granzyme B (GB11) was obtained from Life Technologies. For intracellular detection of IFN-γ, CD8+ T cells were stimulated ex vivo with 0.25 ng/ml SIINFEKL in the presence of Brefeldin A (Sigma) for 4 hours at 37°C; cells were fixed in 4% paraformaldehyde (Electron Microscopy Services) and permeabilized prior to staining. All samples were analyzed on an Accuri C6 or FACS Canto (BD Biosciences).
Single-cell gene expression assays
Inventoried TaqMan assays (Life Technologies) were pooled to a final concentration of 0.2X for each of the 94 gene expression assays. Single CD8+ T cells were sorted directly into RT-PreAmp Master Mix (Life Technologies) containing the pooled assays. Cell lysis, sequence-specific RT, and sequence-specific amplification of cDNA were performed as previously described14, and analyzed in 96.96 Dynamic Arrays on a BioMark system (Fluidigm). Ct values were calculated from the BioMark system software. Cells in which both Actb and Rn18s mRNA expression were detected were retained for further analyses.
Statistical analysis
For statistical analysis, the Kolmogorov-Smirnov test was used for model-free comparisons involving two groups (Figs. 2b, 6b-6d). Differences at P < 0.05 were considered significant.
T lymphocyte confocal microscopy
Immunofluorescence of T cells was performed as previously described9 with the following antibodies: anti-β-tubulin (Sigma); anti-IL-2Rα (PC61.5), anti-CD62L (MEL14) (eBioscience); and anti-mouse Alexa Fluor 488 and anti-rat Alexa Fluor 647 (Life Technologies). DAPI (Life Technologies) was used to detect DNA. Cells undergoing cytokinesis were identified by dual nuclei and pronounced cytoplasmic cleft by brightfield. Acquisition of image stacks was performed as previously described9 using a FV1000 laser scanning confocal microscope (Olympus). The volume of 3D pixels (voxels) containing the designated receptor fluorescence was quantified within each nascent daughter in cytokinetic cells as previously described9 using ImageJ software.
Data and pre-processing
The log expression of each gene g was computed as follows: logEg,c = 40-Ctg,c where c is the cell and Ctg,c is the Ct value obtained from the BioMark (Fluidigm). Cells c′ with undefined Ct values (Ctg,c′ = 999) for both g =Rn18s and g=Actb, or cells c″ with at least unexpressed genes were also removed from our analyses. The remaining “good” cells in each population were deemed sufficient for all subsequent analyses since they exceeded the number of free parameters for any supervised model by a factor of at least 5.
Principal component analysis
We used principal component analysis (PCA) to reduce dimensionality of the data with a linear transformation and projected the data X from its original 94 dimensions down to the first two principal components. PCA was performed in Matlab using the function “pca”. In order to visualize the clustering of populations, we projected the cells from their original 94-gene space to the first two principal components of X. Each principal component, also known as eigen-gene, captures some percentage of the total variance in X proportional to its corresponding eigen value in the singular value decomposition of X. The first two eigen-genes have the largest eigen values. In order to visualize the contribution of each original dimension to these eigen-genes, we projected the 94 unit vectors on to the 2D space spanned by the principal components. These projections combine into the scatter and spike plots depicted in Figs. 2a, 2c, and 3a.
T-distributed stochastic neighborhood embedding
To confirm our unsupervised clustering results, we performed t-distributed Stochastic Neighborhood Embedding (tSNE)29, one of the most powerful dimensionality reduction methods, on our dataset. tSNE is specifically designed for visualization of high-dimensional data and has been shown to capture more useful variance and more complex clustering patterns in the data by attempting to preserve the distances between datapoints from high to low dimension without any prior assumptions on the distribution of the data. In contrast, PCA only captures linear relationships between genes and principal components and assumes a single homoscedastic (spherical) Gaussian distribution for the entire dataset.
Jensen-Shannon divergence
To quantify the differences between the populations and heterogeneity within each population, we use the Jensen-Shannon Divergence (JSD), a symmetric version of the Küllback-Liebler (KL) divergence, a parameter- and model-free metric of the distance between empirical distributions. Given two sets of experimental measurements, {x1,x2, ... , xm} and {y1,y2,... ,yn}, such as expression profiles for individual cells from the Tcm vs Tem populations (in this case ), we used the JSD to characterize the distance between the two empirical distributions Px and Py implied by the Tcm and Tem cells, respectively.
| (1) |
| (2) |
M = (Px + Py) / 2 is an equal mixture of the two distributions and the KL divergence can be approximated over discretized histograms of its two input distributions:
This is the common form of JSD, which does not take into account the group sizes m and n. In lieu of using the more general form which allows for arbitrary re-weighting of the contribution from each distribution, we randomly sub-sampled the larger group and concluded that the common form we used was not sensitive to group size differences when those sizes are within a factor of 2, i.e. min(m,n) ≥ max(m,n) /2.
We interpreted each cell's expression profile as a sample from the 94-dimensional empirical distribution of its population. Expression values for each of the 94 genes is discretized in the same bins, so we simply added the single-dimensional JSD between the two populations for each gene. Moreover, we identified the most and least differentially expressed genes between the two populations, which need not match the PCA results exactly since the JSD analysis does not make the simple linear modeling assumption that PCA does. Finally, to quantify the heterogeneity within a single population, we partitioned it in half randomly and measured the JSD between the two halves. Averaging this intra-population JSD for multiple random partitions gave an estimate of the true variation in each population.
This approach is more principled than a previous application15 of JSD to measure single cell diversity, which arbitrarily converted each cell's expression profile into a separate probability distribution over RNA molecules. This was a misrepresentation of the BioMark's output, which does not distribute a fixed budget of expression units over the 94 genes of interest, but rather measures the doubling times for each PCR primer, and can be justified only for single-cell RNAseq experiments in which comparable numbers of reads are produced for each cell.
Rationale for approach to supervised analysis of gene expression data
PCA and other unsupervised dimensionality reduction methods aid in understanding the structure of a cell population. However, these determinations are performed by visual inspection. Suppose we are given a heterogeneous (unsorted) population of cells X'. In order to classify a new cell, i.e., to identify which subpopulation it belongs to, we could co-cluster the new samples with existing labeled data in X. This approach is suboptimal for two reasons: efficiency and accuracy. This co-clustering approach is not efficient because in order to classify even one new cell x′ in X′, we need to re-run PCA on the original data X extended by a single row x′. More importantly, the accuracy of this approach depends not only on the quality of X, but also on that of x′, which we are trying to assess. If some of the new samples in X′ contain bad or noisy readings that are not filtered by our criteria for X, the variance inherent in X′ will eclipse the useful structure observed in X and the co-clustering result will be unrelated to, or even worse, counter to the original clustering of X. To resolve both of these problems, we decided on a supervised analysis, which learns to distinguish between subpopulations of the labeled data X in the form of boosted classifiers and applies the classifiers to the remainder of the cells in X.
Robust boosting
We used RobustBoost32 to train an ensemble of decision trees at depths ≤ 20. We chose boosting over other classification frameworks because the models that are learned are easily interpretable. For example, the Alternating Decision Tree (ADTree)31 for the Tcm vs Tsle classifier (shown in Supplementary Fig. 4a) consisted of simple rules where the expression of Ptprc, Ccl5, and Sell were compared to thresholds learned from the data. The classifier’s confidence was measured by the margin of each prediction (see red bars in Fig. 4a). We evaluated the performance of the classifier by its prediction accuracy in leave-one-out cross-validation, where the m classifiers b1,b2, ... ,bm are each trained on a different subset of m-1 cells. Each classifier bi was tested on cell xc, which corresponded to the cth row of the data matrix X, after being trained on the remaining cells X-c= {x1,x2, ..., xc-1, xc+1, ..., xm}. This cross-validation produced a group of m classifiers that provided an estimate of the generalization error on the validation examples. This also generated an overall margin on the training examples by tallying the predictions of m-1 informed and 1 uninformed classifier for each of the m cells, where γc = ||b1...m(xc)-lc|| and lc is the label of cell c (in this case lc = -1 means Tcm and lc = 1 means Tsle).
Temporal model of CD8+ T cell differentiation
Akin to the Heisenberg uncertainty principle, the problem of observing a cell’s gene expression is that we must modify (i.e. destroy) the cell in order to observe its gene expression. While not a concern in the single-cell analysis of static populations, this is a limitation in capturing the dynamics of tracing the lineage of the cell. We propose a statistical modeling approach to overcome this limitation with approximate single-cell histories sampled from the available time-series gene expression data33. Briefly, we constructed hypothetical differentiation paths and trained a Hidden Markov Model (HMM) on the resulting expression time courses. Starting from each naïve cell, we sampled cells in successively more mature stages whose expression profiles satisfied an ensemble of predictors for one of the terminal fates, matched these samples in the early differentiation stages (division 1 and day 3), connected both ends of each path, and finally estimated the transition and observation parameters of a 6-state HMM in order to learn the state-to-state transition probabilities and in-state mixture components that capture the dynamics of gene expression in the hypothetical histories.
Input data
To capture the temporal structure of T cell differentiation in our time course gene expression data from single cells, we developed a semi-supervised method based on the fate classifier predictions in early heterogeneous populations from Fig. 4d and on the expression profiles of putative pre-memory and pre-effector cells sort-purified from Tmp and Tsle populations on day 5 after infection. Then, we constructed hypothetical differentiation histories of single cells starting from the naïve population, going through an intermediate stage and ending in one of the three terminal fates: Tcm, Tem, or Tsle. To approximate the real distribution of proliferation transitions between these stages, we used 1,000 bootstrap samples from each subpopulation stringed along one of the three main paths according to their classifier scores. This resulted in an empirical distribution over early transitions (naïve → pre-Tsle, naïve → pre-memory) and another distribution over late transitions (pre-memory → Tcm, pre-memory → Tem, pre-Tsle → Tsle). The early transitions were then connected to the late transitions by cells at the intermediate states.
Model structure
Since the differentiation dynamics of individual proliferating T cells are not yet well described, we used an HMM to model the data because of its simple, yet powerful structure which decouples uncertainty in the lineage reconstruction (state transitions) from measurement noise (observations/emissions). We constructed a HMM with 6 states: naïve, pre-memory, Tcm, Tem, pre-Tsle, and Tsle, to capture the signal in each empirical distribution from our temporal approximation input. Each state emits gene expressions from a mixture of two 94-dimensional Gaussians with full covariance matrices.
Due to concerns over our model’s sensitivity to initialization, we constructed 18 biologically plausible differentiation pathways (6 sequential and 12 bifurcating, whose structures are shown in Supplementary Fig. 4c) and fixed the transition parameters to the corresponding adjacency matrix of each structure in turn. Using the learning algorithm described below, we calculated the posterior log likelihood of each pathway. To address any further concerns over the robustness of these results, we reinitialized each structure twice more with 10% random noise drawn from the Uniform[0,1] distribution, which also ensured that there are no zero-probability transitions between any two states.
Transition parameters
For a cell c in state f, the probability of transitioning to state t is . We assumed that other cells whose expression profile in state f is similar to that of cell c will have similar differentiation potential and, in particular, have a similar probability of transitioning to state t. This assumption allowed us to share the parameters Tf,t = P(f→t) which gives the probability of any cell in state f to proliferate to state t.
Observation parameters
Due to the bimodal nature of the violin plots in Fig. 4d, we modeled the observed expression x of cell c in state i as a mixture model of two Gaussians with 94-dimensional means and , 94 × 94 full covariance matrices and . Like the transitions, parameter sharing between cells allowed us to simplify the observation parameters, resulting in the following observation model:
Learning algorithm
First, we initialized the model parameters to their prior distributions. Specifically, the transitions P(f → t) were initialized to the matrix shown in Fig. 5b. The emission parameters for the naïve, Tsle, Tcm, and Tem states were initialized to the maximum likelihood fit for a mixture of two Gaussians to the empirical histograms of gene expression for the respective population. The emission parameters in the intermediate states, pre-memory and pre-Tsle, were fit to the empirical histograms accumulated over all intermediate states. The transition parameters were fixed throughout the duration of each learning run, but were randomized with up to 10% noise as detailed above.
Finally, we optimized the parameters of the HMM using the Expectation Maximization algorithm implemented in pmtk3, the probabilistic modeling toolkit for Matlab/Octave34. The learned emission parameters were used to identify the genes whose relative expression changed the most during each transition, as shown in Fig. 5b-c. While we did not learn the transition probabilities, we did re-sample them from 18 plausible structures and picked the most likely structure whose transition matrix is shown in Supplementary Fig. 4d and whose adjacency graph is on the far bottom right of Supplementary Fig. 4c. To determine the most likely structure, we calculated the posterior likelihoods of each HMM model (starting from 10 random re-initializations) and compared their cumulative distribution functions (CDF) in Supplementary Figure 4c. To further gauge the statistical significance of the best model, we randomly shuffled the input data 100 times for each model and built a background distribution of the resulting log-likelihoods. To determine both the significance and robustness of each HMM model compared to its own shuffled background consisting of 100 random shuffles of the data, we used the non-parametric Kolmogorov-Smirnov method to test if the log-likelihoods of each model are significantly higher on the real data than on the shuffled data and to provide a p-value for the significance of each result. To test the reproducibility of our results with respect to the bootstrap sampling method, we resampled the data and performed another 10 randomly initialized training runs on the “best” model (see bottom right panel of Supplementary Figure 4c). Finally, we performed Kolmogorov-Smirnov tests on each proposed structure to check that the best model’s CDF represented significantly higher log-likelihoods than the other models and its own background.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Hedrick, J. Bui, A. Goldrath, S. Schoenberger and members of the Chang and Yeo labs for helpful discussions and critical reading of the manuscript. This work was supported by US National Institutes of Health (DK080949, OD008469, and AI095277 to J.T.C. and HG004659 and NS075449 to G.W.Y.) and the UCSD Digestive Diseases Research Development Center Grant DK80506; and California Institute for Regenerative Medicine grants (RB1-01413 and RB3-05009 to G.W.Y.). B.K. is a National Science Foundation graduate research fellow. G.W.Y. is a recipient of the Alfred P. Sloan Research Fellowship. J.T.C. is a Howard Hughes Medical Institute Physician-Scientist Early Career Awardee.
Footnotes
AUTHOR CONTRIBUTIONS
J.A. and J.T.C. designed experiments. J.A, P.J.M., and S.H.K. performed experiments. B.K. and G.W.Y. performed data analysis. J.A., B.K., G.W.Y., and J.T.C. wrote the manuscript.
Accession codes. GEO: gene expression array data, GSE54321.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Ahmed R, Gray D. Immunological memory and protective immunity: understanding their relation. Science. 1996;272:54–60. doi: 10.1126/science.272.5258.54. [DOI] [PubMed] [Google Scholar]
- 2.Joshi NS, et al. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27:281–95. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masopust D, Kaech SM, Wherry EJ, Ahmed R. The role of programming in memory T-cell development. Curr Opin Immunol. 2004;16:217–25. doi: 10.1016/j.coi.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–12. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 5.Stemberger C, et al. A single naive CD8+ T cell precursor can develop into diverse effector and memory subsets. Immunity. 2007;27:985–97. doi: 10.1016/j.immuni.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 6.Gerlach C, et al. One naive T cell, multiple fates in CD8+ T cell differentiation. J Exp Med. 2010;207:1235–46. doi: 10.1084/jem.20091175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchholz VR, et al. Disparate individual fates compose robust CD8+ T cell immunity. Science. 2013;340:630–5. doi: 10.1126/science.1235454. [DOI] [PubMed] [Google Scholar]
- 8.Gerlach C, et al. Heterogeneous differentiation patterns of individual CD8+ T cells. Science. 2013;340:635–9. doi: 10.1126/science.1235487. [DOI] [PubMed] [Google Scholar]
- 9.Chang JT, et al. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science. 2007;315:1687–91. doi: 10.1126/science.1139393. [DOI] [PubMed] [Google Scholar]
- 10.Chang JT, et al. Asymmetric proteasome segregation as a mechanism for unequal partitioning of the transcription factor T-bet during T lymphocyte division. Immunity. 2011;34:492–504. doi: 10.1016/j.immuni.2011.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaech SM, Hemby S, Kersh E, Ahmed R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell. 2002;111:837–51. doi: 10.1016/s0092-8674(02)01139-x. [DOI] [PubMed] [Google Scholar]
- 12.Best JA, et al. Transcriptional insights into the CD8(+) T cell response to infection and memory T cell formation. Nat Immunol. 2013;14:404–12. doi: 10.1038/ni.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarkar S, et al. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J Exp Med. 2008;205:625–40. doi: 10.1084/jem.20071641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo G, et al. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell. 2010;18:675–85. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 15.Buganim Y, et al. Single-Cell Expression Analyses during Cellular Reprogramming Reveal an Early Stochastic and a Late Hierarchic Phase. Cell. 2012;150:1209–22. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalerba P, et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol. 2011;29:1120–7. doi: 10.1038/nbt.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu R, Neff NF, Quake SR, Weissman IL. Tracking single hematopoietic stem cells in vivo using high-throughput sequencing in conjunction with viral genetic barcoding. Nat Biotechnol. 2011;29:928–33. doi: 10.1038/nbt.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ichii H, et al. Role for Bcl-6 in the generation and maintenance of memory CD8+ T cells. Nat Immunol. 2002;3:558–63. doi: 10.1038/ni802. [DOI] [PubMed] [Google Scholar]
- 19.Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol. 2012;12:749–61. doi: 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kallies A, Xin A, Belz GT, Nutt SL. Blimp-1 transcription factor is required for the differentiation of effector CD8(+) T cells and memory responses. Immunity. 2009;31:283–95. doi: 10.1016/j.immuni.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 21.Pearce EL, et al. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302:1041–3. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 22.Rutishauser RL, et al. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 2009;31:296–308. doi: 10.1016/j.immuni.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szabo SJ, et al. Distinct effects of T-bet in TH1 lineage commitment and IFN- gamma production in CD4 and CD8 T cells. Science. 2002;295:338–42. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- 24.Yang CY, et al. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat Immunol. 2011;12:1221–9. doi: 10.1038/ni.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou X, et al. Differentiation and persistence of memory CD8(+) T cells depend on T cell factor 1. Immunity. 2010;33:229–40. doi: 10.1016/j.immuni.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warren L, Bryder D, Weissman IL, Quake SR. Transcription factor profiling in individual hematopoietic progenitors by digital RT-PCR. Proc Natl Acad Sci U S A. 2006;103:17807–12. doi: 10.1073/pnas.0608512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masopust D, Vezys V, Marzo AL, Lefrancois L. Preferential localization of effector memory cells in nonlymphoid tissue. Science. 2001;291:2413–7. doi: 10.1126/science.1058867. [DOI] [PubMed] [Google Scholar]
- 28.Schluns KS, Kieper WC, Jameson SC, Lefrancois L. Interleukin-7 mediates the homeostasis of naive and memory CD8 T cells in vivo. Nat Immunol. 2000;1:426–32. doi: 10.1038/80868. [DOI] [PubMed] [Google Scholar]
- 29.Wherry EJ, et al. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat Immunol. 2003;4:225–34. doi: 10.1038/ni889. [DOI] [PubMed] [Google Scholar]
- 30.van der Maaten LJP, Hinton GE. Visualizing High-Dimensional Data Using t-SNE. Journal of Machine Learning Research. 2008;9:2579–2605. [Google Scholar]
- 31.Freund Y, Mason Llew. The alternating decision tree learning algorithm. Proc. 16th International Conference on Machine Learning. 1999:124–133. [Google Scholar]
- 32.Freund YB, L, Littman M. Linear Separation, Drifting Games and Boosting. 2009 [Google Scholar]
- 33.Beerenwinkel N, Drton M. A mutagenetic tree hidden Markov model for longitudinal clonal HIV sequence data. Biostatistics. 2007;8:53–71. doi: 10.1093/biostatistics/kxj033. [DOI] [PubMed] [Google Scholar]
- 34.Bulla J, B. I. Stylized facts of financial time series and hidden semi-Markov models. Computational Statistics & Data Analysis. 2006;51:2192–2209. [Google Scholar]
- 35.Feau S, Arens R, Togher S, Schoenberger SP. Autocrine IL-2 is required for secondary population expansion of CD8(+) memory T cells. Nat Immunol. 2011;12:908–13. doi: 10.1038/ni.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams MA, Tyznik AJ, Bevan MJ. Interleukin-2 signals during priming are required for secondary expansion of CD8+ memory T cells. Nature. 2006;441:890–3. doi: 10.1038/nature04790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalia V, et al. Prolonged interleukin-2Ralpha expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity. 2010;32:91–103. doi: 10.1016/j.immuni.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 38.Pipkin ME, et al. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity. 2010;32:79–90. doi: 10.1016/j.immuni.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Obar JJ, Lefrancois L. Early signals during CD8 T cell priming regulate the generation of central memory cells. J Immunol. 2010;185:263–72. doi: 10.4049/jimmunol.1000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Afkarian M, et al. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat Immunol. 2002;3:549–57. doi: 10.1038/ni794. [DOI] [PubMed] [Google Scholar]
- 41.Lighvani AA, et al. T-bet is rapidly induced by interferon-gamma in lymphoid and myeloid cells. Proc Natl Acad Sci U S A. 2001;98:15137–42. doi: 10.1073/pnas.261570598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gebhardt T, et al. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat Immunol. 2009;10:524–30. doi: 10.1038/ni.1718. [DOI] [PubMed] [Google Scholar]
- 43.Masopust D, et al. Dynamic T cell migration program provides resident memory within intestinal epithelium. J Exp Med. 2010;207:553–64. doi: 10.1084/jem.20090858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olson JA, McDonald-Hyman C, Jameson SC, Hamilton SE. Effector-like CD8(+) T cells in the memory population mediate potent protective immunity. Immunity. 2013;38:1250–60. doi: 10.1016/j.immuni.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang F, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377–82. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 46.Shalek AK, et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498:236–40. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bendall SC, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–96. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moon JJ, et al. Tracking epitope-specific T cells. Nat Protoc. 2009;4:565–81. doi: 10.1038/nprot.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.