Abstract
Background:
Mycobacterium tuberculosis complex (MTBC) and non-tuberculous mycobacteria (NTM) may or may not have same clinical presentations, but the treatment regimens are always different. Laboratory differentiation between MTBC and NTM by routine methods are time consuming and cumbersome to perform. We have evaluated the role of GenoType® Mycobacterium common mycobacteria/additional species (CM/AS) assay for differentiation between MTBC and different species of NTM in clinical isolates from tuberculosis (TB) cases.
Materials and Methods:
A total of 1080 clinical specimens were collected from January 2010 to June 2012. Diagnosis was performed by Ziehl-Neelsen staining followed by culture in BacT/ALERT 3D system (bioMerieux, France). A total of 219 culture positive clinical isolates (BacT/ALERT® MP cultures) were selected for differentiation by p-nitrobenzoic acid (PNB) sensitivity test as and BIO-LINE SD Ag MPT64 TB test considering as the gold standard test. Final identification and differentiation between MTBC and different species of NTM were further confirmed by GenoType® Mycobacterium CM/AS assay (Hain Lifescience, Nehren, Germany).
Results:
Out of 219 BacT/ALERT® MP culture positive isolates tested by PNB as 153 MTBC (69.9%) and by GenoType® Mycobacterium CM/AS assay as 159 (72.6%) MTBC and remaining 60 (27.4%) were considered as NTM species. The GenoType® Mycobacterium CM/AS assay was proved 99.3% sensitive and 98.3% specific for rapid differentiation of MTBC and NTM. The most common NTM species were; Mycobacterium fortuitum 20 (33.3%) among rapid growing mycobacteria and Mycobacterium intracellulare 11 (18.3%) among slow growing mycobacteria.
Conclusion:
The GenoType® Mycobacterium assay makes rapid and accurate identification of NTM species as compared with different phenotypic and molecular diagnostic tool and helps in management of infections caused by different mycobacteria.
Keywords: GenoType® Mycobacterium common Mycobacteria/additional species assay, M. tuberculosis complex, non-tuberculous Mycobacteria
INTRODUCTION
Tuberculosis (TB) is a highly infectious disease caused by Mycobacterium tuberculosis complex (MTBC) and potentially fatal disease for human. India is the major TB endemic country and about 1.8 million new cases of TB has been reported annually that accounts for one fifth of new cases in the world – a greater number than in any other country.[1] Management of patients with MTBC and non-tuberculous mycobacteria (NTM) are entirely different, therefore prompt isolation, detection and differentiation is necessary for suitable management.[2,3] Conventional biochemical tests used to identify different mycobacterial species are complex and time consuming.[4] Hence, there was a need for rapid differentiation and identification of different Mycobacterium species for effective treatment of the disease. Recently, various genetic probes and amplification systems for diagnosis of TB have been developed and several of them are available as commercial kits for direct detection and identification of MTBC and NTM in clinical specimens.[5,6] Newer techniques such as high-performance liquid chromatography, chemiluminescent deoxyribonucleic acid (DNA) probes, nucleic acid amplification and sequencing of 16S ribosomal ribonucleic acid (rRNA) genes are more sophisticated, highly expensive and require expensive equipment.[7,8,9] As per recent, World Health Organization objectives to reduce the time for culture, identification and drug resistance detection to as short as 2 days by employing line probe assays (LPA).[10] The GenoType® Mycobacterium common mycobacteria/additional species (CM/AS) assay (Hain Lifescience, Nehren Germany) is a new commercial kit developed to differentiate and identify different species of NTM from cultures. It involves DNA amplification targeting the 23S rRNA gene region, followed by reverse hybridization to specific oligonucleotide probes immobilized on membrane strips. There are two kits – the CM, which identifies 15 Mycobacterium species, including M. tuberculosis complex while the AS kit aims to differentiate 16 additional less common NTM species available for differentiation.[11] The GenoType® Mycobacterium CM/AS assay is very accurate and reliable test for the identification of mycobacterial species, which require a specialized set up and trained laboratory personnel.[12] The aim of this study was to evaluate the role of GenoType® Mycobacterium CM/AS assay based on multiplex polymerase chain reaction (PCR) and reverse hybridization for differentiation between M. tuberculosis complex and different species of NTM in clinical isolates from TB cases.
MATERIALS AND METHODS
A prospective observational study was conducted at Mycobacteriology Laboratory, Department of Microbiology, Sanjay Gandhi Postgraduate Institute of Medical Sciences (SGPGIMS) Lucknow, India. Specimens were collected from Kasturaba Chest Hospital, Department of Pulmonary Medicine, King George Medical University, Uttar Pradesh, Lucknow, India and from various out-patient department and wards of SGPGIMS, Lucknow, India. A total number of 1,080 clinical specimens were collected from both suspected cases of pulmonary tuberculosis (PTB) and extra-pulmonary tuberculosis (EPTB) from January 2010 to June 2012. Approximately, 2-10 ml of non-repeated specimen was collected from 1080 patients both from PTB and EPTB cases. The specimens were included as bronchoalveolar lavage, bronchial lavage, lung biopsies and sputum from suspected cases of PTB and ascitic fluid, biopsy materials, bone marrow aspirates, cerebrospinal fluid, cold abscesses, gastric aspirate, lymph node aspirate, peritoneal fluid, pleural fluid, pus, synovial fluid, urine and wound swabs from suspected cases of EPTB. Patients from all ages were considered for the study and included both male and female population from urban and rural settings. The study was approved by the Institutional Ethical Committee of King George Medical University Uttar Pradesh, Lucknow, India (Approval No: XXVIII extracellular matrix-B/P2). A written informed consent was obtained from all subjects before enrollment into the study.
Processing and microbiological analysis of specimens
All the clinical specimens were grouped into two types based on presence and absence of normal commensal bacterial flora as non-sterile and sterile specimens. Non-sterile specimens (respiratory) were processed as per standard N-acetyl-L-cysteine-NaOH method[13] and sterile specimens were processed as per standard laboratory protocol (quantity >0.5 ml were centrifuged and <0.5 ml were directly inoculated) in Class II Biosafety cabinet. After specimens processing, centrifuged pellets were subjected to smear microscopy by Ziehl-Neelsen (ZN) staining method and 0.5 ml were inoculated into BacT/ALERT® MP vials in the BacT/ALERT 3D system (bioMerieux, France) containing modified Middlebrook 7H9 with an antibiotic supplement (amphotericin B [0.018%, wt/vol], azlocillin [0.0034%, wt/vol], nalidixic acid [0.04%, wt/vol], trimethoprim [0.00105%, wt/vol], polymyxin B [10,000 U] and vancomycin [0.0005%, wt/vol]).[14] The reading of BacT/ALERT® MP vials were monitored continuously by the BacT/ALERT 3D system.[15] Positive vials were subjected to smear microscopy for the presence of acid fast bacilli (AFB). No growth after 6 weeks of incubation was treated as negative for mycobacteria. Growth of different Mycobacterium species were differentiated by based on battery of tests which includes niacin production, catalase activity at 68°C at pH 7 and p-nitrobenzoic acid (PNB).[16]
Niacin production test
BBL Taxo TB Niacin test strips (Becton and Dickinson, USA), absorbent paper strips and TB Niacin positive test control paper discs were used according to the manufacturer's instruction. In brief, with the help of sterile transfer pipette, approximately 0.6 ml of the positive culture broth extract was transferred to the bottom of 20 mm × 125 mm screw cap test tube. Negative control was also prepared. The strips were dropped with arrow downward into the tubes: Positive and negative controls, test culture and stopper immediately. The colors of the extracts were then compared after 15 min. Niacin accumulation was indicated by vivid appearance of a yellow color in the extract.[17]
Catalase test
The catalase assay for mycobacteria was performed using 30% H2O2(Superoxol) in 10% Tween-80 and the test performed both at 22-25°C and 68°C. MTBC has catalase activity at 22-25°C, but not at 68°C (i.e., heat-labile catalase). In addition to determining catalase activity at 68°C, the strength of the catalase reaction was also evaluated to differentiate MTBC from other mycobacteria. This test was performed by adding Superoxol to a positive culture of mycobacteria at 37°C. 5 min after addition of the Superoxol, the strength of the catalase reaction was determined by measuring the height of the bubbles, characteristic of the catalase reaction, in the tube. MTBC has low catalase activity.[18]
BIO-LINE SD Ag MPT64 TB test
A total volume of 100 μl of broth from BacT/ALERT culture was added to the well of BIO-LINE SD Ag MPT64 TB rapid kit cassette. Inoculated immunochromatographic cassettes were kept at room temperature for 20 min in Class II Biosafety cabinet and examined for the presence of control and test band. Appearance of the band in the “C” region confirmed the validity of the test. Additional appearance of the band in the “T” region was interpreted as positive for the presence of MPT64 Ag antigen in the test broth culture [Figure 1]. No band in the “C” region was interpreted as invalid test.[3,19] Standard reference strain M. tuberculosis complex H37Rv ATCC 27294 was used as a positive control.
Figure 1.
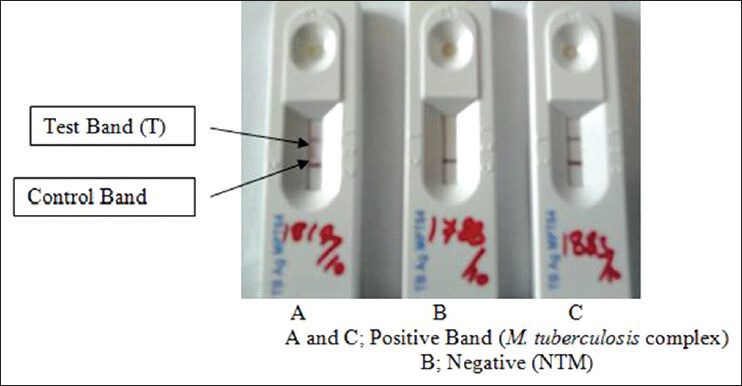
Differentiation of M. tuberculosis complex and non-tuberculous mycobacteria by BIO-LINE SD Ag MPT64 tuberculosis test
GenoType® Mycobacterium CM/AS assay
All isolates were identified using conventional biochemical techniques[20] before molecular identification by the GenoType® Mycobacterium CM/AS assay. The final confirmation of MTBC and NTM species were done by the GenoType® Mycobacterium CM/AS assay of BacT/ALERT® MP culture positive mycobacterial isolates.[11,21] DNA extraction was performed by sonication. In brief, 1 ml positive culture broth were centrifuged at 13,400 g for 2 min and discard supernatant and resuspend bacteria in 100-300 μl of water, then followed by heating to 95°C for 20 min and incubated for 15 min in an ultrasonic bath, then spin at 13,400 g for 2 min and directly used 5 μl of supernatant for the assay.[11] Identification of MTBC and NTM species were carried out by using specific sets of primers designed to amplify a species specific 23S rRNA gene sequence of Mycobacterium species. Amplification mixture (45 μl) was prepared in DNA free room, including 5 μl extracted DNA (20-100 ng DNA) in the reaction mixture contained 35 μl primer nucleotide mix, 5 μl 10 × polymerase incubation buffer for HotStar Taq (QIAGEN, Hilden, Germany), 2 μl 25 mM MgCl2 solution, 0.2 μl HotStar Taq and 3 μl water (biology grade water). Amplification was carried out in a thermal cycler (MJ Research, PTC-100 Thermal Cycler, GMI, Inc, USA), which involved 01 cycles of denaturation solution (DEN) at 95°C for 15 min, annealing of primers at 95°C for 30 s, 2 min at 58°C for 10 cycles, then 20 cycles at 95°C for 25 s, 53°C for 40 s and 70°C for 40 s and final primer extension at 70°C, 8 min for 01 cycle. The amplified products were stored at +8 to −20°C until hybridization was done in hybridization machine (Profiblot; Tekan, Maennedorf, Switzerland).
This was followed by hybridization; in which 20 μl of DEN (blue) was dispensed in the corner of each well and then 20 μl of amplified product was added and incubated for 5 min at room temperature. Then 1 ml of pre-warmed hybridization buffer (HYB, Green) was added, followed by gentle shaking until a homogenous color was developed. Now strip was placed in a manner to make sure complete flooding of solution over strips. Then tray was placed in TwinCubator and was incubated for 30 min at 45°C, followed by complete aspiration of HYB. Washing was done by 1 ml of stringent wash solution to each strips and incubated for 15 min at 45°C in TwinCubator. Again strips were washed once with 1 ml of ringer solution for 1 min on TwinCubator. Then 1 ml of diluted conjugate was added to each strips and incubated for 30 min on TwinCubator. Strips were washed again with 1 ml of ringer solution for 1 min, after that 1 ml of diluted substrate were added to each strips and incubated for 3-20 min in the absence of light without shaking. Ringing was done twice with distilled water to stop the reaction. Strips were removed and dried between two layers of absorbent paper. Evaluation and interpretation of results were done based on the presence and absence of different bands and compared with reference band as provided in the kit.[11] The presence of distinct band at position 10, 16 indicated a positive test for M. tuberculosis complex as shown in Figure 2. Standard strain of M. tuberculosis complex H37 Rv ATCC 27294, Mycobacterium fortuitum ATCC 6841, Mycobacterium intracellulare ATCC 13950 and M. abscessus ATCC 19977 were used as control in this study.
Figure 2.
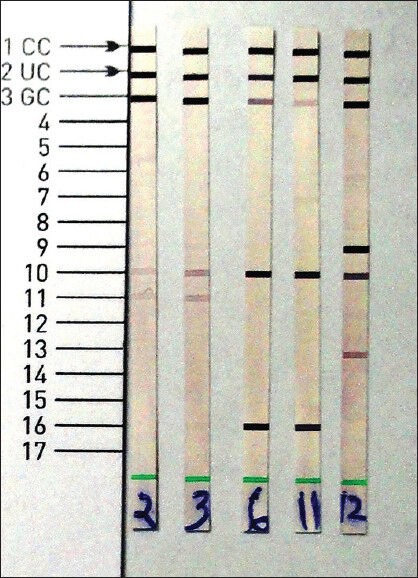
GenoType® mycobacterium common mycobacteria/additional species (CM/AS) assay for rapid differentiation of different species of Mycobacteria. Strain number 2, 3, 12 were identified as non-tuberculous mycobacteria and 6, 11 as Mycobacterium tuberculosis complex by GenoType® Mycobacterium CM/AS
Statistical analysis
Data were analyzed using the statistical package for the social sciences 15.0 (Chicago, IL, USA) for Windows. The sensitivity of different tests that were used to differentiate MTBC and NTM was calculated as sensitivity (Tp/[Tp + Fn]) ×100; specificity (Tn/[Tn + Fp]) ×100; Tp = total number of positives; Tn = total number of negatives; Fp = total number of false positive, Fn = total number of false negative; respectively.
RESULTS
A total number of 1080 clinical specimens from suspected cases of pulmonary and EPTB were screened for the presence of Mycobacterium species. In of 1,080 specimens, 723 (66.9%) specimens pulmonary and remaining 357 (33.1%) were from different extra-pulmonary sources. Smear positivity was 195/1080 (18.1%) for AFB by ZN microscopy. Among which only 219/1080 (20.3%) cases were BacT/ALERT® MP culture positive. The contamination rate of BacT/ALERT® MP culture vials was 4.6% after proper decontamination and digestion of clinical specimens. The distribution of different clinical specimens with respect to smears and culture positivity are shown in Table 1. Niacin production and catalase activity were positive in 163/219 (74.4%) strains and it was phenotypically indicative of MTBC. All the 219 BacT/ALERT® MP culture positive isolates were tested by conventional PNB test using BacT/ALERT® MP vial, rapid BIO-LINE SD Ag MPT64 TB test for rapid differentiation between MTBC and NTM. Final differentiation of MTBC and NTM in BacT/ALERT® MP positive isolates was done by LPA using GenoType® Mycobacterial CM/AS assay. PNB and rapid BIO-LINE SD Ag MPT64 TB test was performed over a total of 219 BacT/ALERT® MP positive mycobacterial isolates irrespective of their day of culture positivity. In of 219 culture positive isolates, 153 (69.9%) were sensitive to PNB (considered as MTBC) and remaining isolates were resistant to PNB (considered as NTM) as shown in Table 2. By using rapid BIO-LINE SD Ag MPT64 TB test, 158 (72.1%) isolates showed the band in the test region (“T”) confirming the presence of MPT 64 antigen expression and thus presence of MTBC in these culture and remaining isolates were identified as NTM as shown in Table 2. By using confirmatory (gold standard) test by GenoType® Mycobacterium CM/AS assay, 159 (72.6%) isolates showed the presence of distinct band at position 10, 16 indicated a positive test for M. tuberculosis complex and remaining 60 (27.4%) isolates were identified as different species of NTM.
Table 1.
Distribution of different specimens among smears and culture positivity
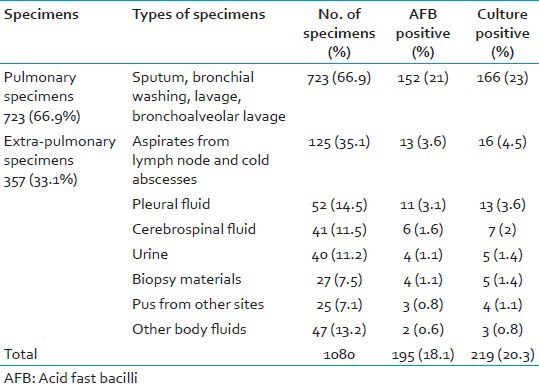
Table 2.
Detection and differentiation between Mycobacterium tuberculosis complex and non-tuberculous mycobacteria by different methods
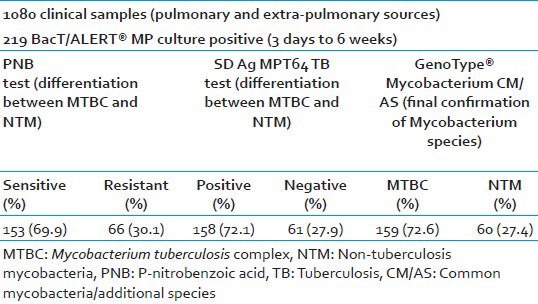
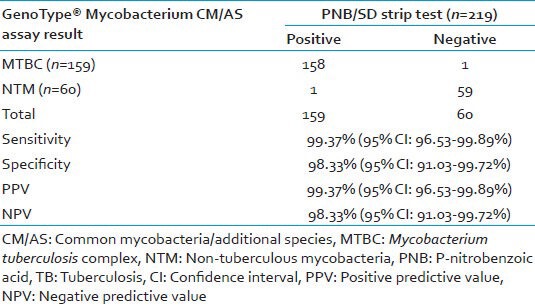
The sensitivity, specificity, positive and negative predictive value of the GenoType® Mycobacterium CM/AS assay was found to be 99.37% (95% CI: 96.53-99.89%), 98.33% (95% CI: 91.03-99.72%), 99.37% (95% CI: 96.53-99.89%) and 98.33% (95% CI: 91.03-99.72%) respectively, when compared with the either PNB or SD strip test positive as the gold standard method shown in Table 3.
Table 3.
Sensitivity of GenoType® Mycobacterium CM/AS assay for differentiation of MTBC and NTM against PNB test, BIO-LINE SD Ag MPT64 TB test as gold standard
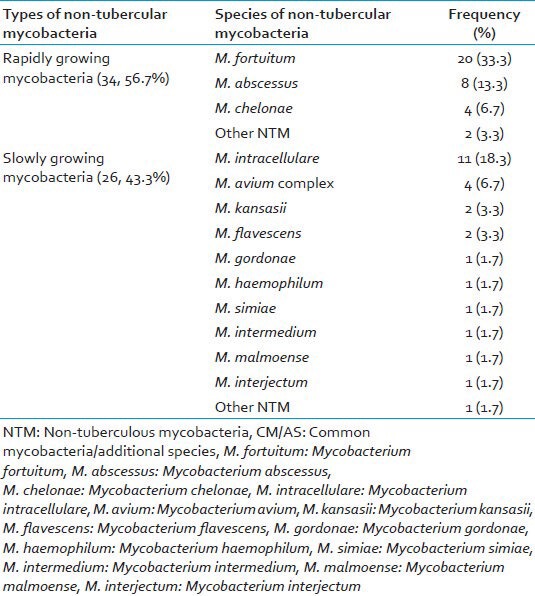
A total of 60/219 (27.4%) were genotypically identified as different species of NTM by GenoType® Mycobacterium CM/AS assay. The distribution of different species of 60 NTM isolates were as; M. fortuitum 20 (33.3%) being the most common NTM species followed by M. intracellulare 11 (18.3%), M. abscessus 8 (13.3%), M. avium complex 4 (6.7%) and M. chelonae 4 (6.7%) as shown in Table 4.
Table 4.
Distribution of different NTM species differentiated by GenoType® Mycobacterium CM/AS assay (n=60)
DISCUSSION
The GenoType® Mycobacterium CM/AS assay is rapid and reliable identification test of mycobacterial species, which may provide patients with an earlier onset of targeted therapy. Smear for AFB is rapid, but does not differentiate between MTBC and NTM.[22] Conventional methods like direct staining of a colony and a panel of different biochemical tests are unable to distinguish more than 100 Mycobacterial species, since they overlap in their biochemical properties.[23,24] A total of 153 Mycobacterial isolates were identified as MTBC and 6 (strains those correctly identified by GenoType® Mycobacterium CM/AS assay as MTBC) were falsely identified as NTM by using PNB test. Previous study reported almost similar result i.e., 98.7% accuracy using MGIT 960 PNB as compared with 96.2% by using BacT/ALERT PNB test in our study.[18] Previous studies reported that 90-100% sensitivity of BIO-LINE SD Ag MPT64 TB kit for differentiation of MTBC and NTM species.[12,25,26] We reported the sensitivity of BIO-LINE SD Ag MPT64 TB, 99.4% for MTBC isolates as compared with this assay. Various other studies that have compared GenoType® Mycobacterium CM/AS assay with different phenotypic and genotypic methods and reported excellent results. Out of a total 60 NTM isolates, 58 (96.6%) NTM strains were correctly identified to species level by using GenoType® Mycobacterium CM/AS assay, which showed similar results (89.3-100%) as obtained in other different studies.[27,28,29,30] Only 3.4% of NTM species were not identifiable by using this assay up to the species level. Because of the different gene probes these two strains were still unidentified to species level. DNA sequencing may be useful for the identification of rare or unidentifiable NTM species.
Chihota et al.[12] reported that cost of organism identification per positive culture on BACTEC has been reported as US $35.94 using standard biochemical tests, US $15.49 for anti-MPB64 assay and US $2.28 for cording. However, the cost of the GenoType® Mycobacterium CM/AS assay is around US $50 for rapid identification and differentiation of Mycobacterial species at our setup. The advantage of using GenoType® Mycobacterium CM/AS assay is rapid and accurate identification of a wide range of Mycobacterium species as compared with phenotypic and other molecular techniques, which fails to differentiate different NTM species correctly. The added advantages of this assay are short turnaround time as compared with conventional phenotypic methods and other molecular methods. The limitations of this assay are, it requires extraction, amplification and hybridization, which require more labor, it cannot detect every species of NTM because it has only important probes for most common and clinically important NTM species. We did not evaluate the ability of the GenoType® Mycobacterium CM/AS assay to detect mixed Mycobacterial infection.
Recently, in a multicentric study the frequencies of different NTM species were estimated over a 20 years period in which the NTM species most frequently isolated in 1991 to 1996 were the M. avium - M. intercellulare complex (29.1%), M. gordonae (18.8%), M. xenopi (19%), M. kansasii (10.3%) and M. fortutium (9.8%) accounting for approximately 87% of all NTM species.[21,31] The GenoType® Mycobacterium CM/AS assay was able to identify 96.7% of theses NTM species to species level correctly in our study. We found 27.4% prevalence of NTM was based on genotypic identification by GenoType® Mycobacterium CM/AS assay. However, the isolation rate of NTM from India has been reported ranging from 0.5% to 8.6%.[32] We found M. fortuitum 20 (33.3%) the most common NTM species followed by M. intracellulare 11 (18.3%), M. abscessus 8 (13.3%), M. avium complex 4 (6.7%) and M. chelonae 4 (6.7%). The GenoType® Mycobacterium CM/AS assay, which uses multiplex PCR and the reverse hybridization are used for the robust and reproducible identification test for different Mycobacterial species. The results of this assay are simple, easy to interpret without much expertise such as that required for DNA sequencing. The GenoType® Mycobacterium CM/AS assay can easily be implemented in routine work for Mycobacterial species differentiation in the clinical setting. Rapid identification and differentiation to species level by using GenoType® Mycobacterium CM/AS assay help in targeted therapy and management of infections cause by different Mycobacterial species and indirectly it will help in reducing development of antimicrobial drug resistance in the community.
CONCLUSION
We concluded, the GenoType® Mycobacterium CM/AS assay may be applied to different species of NTM from culture positive media. It can easily differentiate between MTBC and different species of NTM with high sensitivity and high specificity. This assay makes rapid and accurate identification of NTM species as compared with different phenotypic and other molecular diagnostic tools and helps in management of infections caused by different mycobacteria.
ACKNOWLEDGMENT
This work was supported by a grant from Indian Council of Medical Research, New Delhi (Extramural ICMR Project Sanction No. 5/8/5/4/2007-ECD-I). The authors would like to thank technical members of Mycobacteriology Laboratory, Department of Microbiology, Sanjay Gandhi Postgraduate Institute of Medical Science, Lucknow, India for their technical support during the research work.
Footnotes
Source of Support: Indian Council of Medical Research, New Delhi, India.
Conflict of Interest: None declared.
REFERENCES
- 1.Nirman Bhawan, New Delhi: Central TB Division. Directorate General of Health Services, Ministry of Health and Family welfare; 2010. [Last accessed on 2011 Feb 07]. RNTCP. Status TB Report. Available from: http://www.tbcindia.org . [Google Scholar]
- 2.Hasegawa N, Miura T, Ishii K, Yamaguchi K, Lindner TH, Merritt S, et al. New simple and rapid test for culture confirmation of Mycobacterium tuberculosis complex: A multicenter study. J Clin Microbiol. 2002;40:908–12. doi: 10.1128/JCM.40.3.908-912.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park MY, Kim YJ, Hwang SH, Kim HH, Lee EY, Jeong SH, et al. Evaluation of an immunochromatographic assay kit for rapid identification of Mycobacterium tuberculosis complex in clinical isolates. J Clin Microbiol. 2009;47:481–4. doi: 10.1128/JCM.01253-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marzouk M, Kahla IB, Hannachi N, Ferjeni A, Salma WB, Ghezal S, et al. Evaluation of an immunochromatographic assay for rapid identification of Mycobacterium tuberculosis complex in clinical isolates. Diagn Microbiol Infect Dis. 2011;69:396–9. doi: 10.1016/j.diagmicrobio.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 5.Ting SW, Chia CL, Hsin CL. Current situations on identification of nontuberculous mycobacteria. J Biomed Lab Sci. 2009;21:1–6. [Google Scholar]
- 6.Lee JC, Yu FL, Lin MH, Huang GS, Chang CY, Cheng CL, et al. Utility of immunochromatographic assay for detecting Mycobacterium tuberculosis from positive BACTEC MGIT 960 Cultures. J Biomed Lab Sci. 2010;22:64–8. [Google Scholar]
- 7.French AL, Benator DA, Gordin FM. Nontuberculous mycobacterial infections. Med Clin North Am. 1997;81:361–79. doi: 10.1016/s0025-7125(05)70522-8. [DOI] [PubMed] [Google Scholar]
- 8.Ichiyama S, Iinuma Y, Yamori S, Hasegawa Y, Shimokata K, Nakashima N. Mycobacterium growth indicator tube testing in conjunction with the AccuProbe or the AMPLICOR-PCR assay for detecting and identifying mycobacteria from sputum samples. J Clin Microbiol. 1997;35:2022–5. doi: 10.1128/jcm.35.8.2022-2025.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laughon BE. New tuberculosis drugs in development. Curr Top Med Chem. 2007;7:463–73. doi: 10.2174/156802607780059736. [DOI] [PubMed] [Google Scholar]
- 10.Geneva: 2007. Mar 26, [Last Accessed on 2012 Feb 22]. World Health Orgnization. The Use of Liquid Medium for Culture and Drug Susceptibility Testing (DST) in Low- and Medium-income Settings: Summary of the Expert Group Meeting on the use of liquid culture systems. Available from: http://www.who.int/tb/dots/laboratory/policy/en/index3.html . [Google Scholar]
- 11.Lee AS, Jelfs P, Sintchenko V, Gilbert GL. Identification of non-tuberculous mycobacteria: Utility of the GenoType Mycobacterium CM/AS assay compared with HPLC and 16S rRN A gene sequencing. J Med Microbiol. 2009;58:900–4. doi: 10.1099/jmm.0.007484-0. [DOI] [PubMed] [Google Scholar]
- 12.Chihota VN, Grant AD, Fielding K, Ndibongo B, van Zyl A, Muirhead D, et al. Liquid vs. solid culture for tuberculosis: Performance and cost in a resource-constrained setting. Int J Tuberc Lung Dis. 2010;14:1024–31. [PubMed] [Google Scholar]
- 13.Scott CP, Filho LD, Mello FC, Thornton CG, Bishai WR, Fonseca LS, et al. Comparison of C (18)-carboxypropylbetaine and standard N-acetyl-L-cysteine-NaOH processing of respiratory specimens for increasing tuberculosis smear sensitivity in Brazil. J Clin Microbiol. 2002;40:3219–22. doi: 10.1128/JCM.40.9.3219-3222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carricajo A, Fonsale N, Vautrin AC, Aubert G. Evaluation of BacT/Alert 3D liquid culture system for recovery of mycobacteria from clinical specimens using sodium dodecyl (lauryl) sulfate-NaOH decontamination. J Clin Microbiol. 2001;39:3799–800. doi: 10.1128/JCM.39.10.3799-3800.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Angeby KA, Werngren J, Toro JC, Hedström G, Petrini B, Hoffner SE. Evaluation of the BacT/ALERT 3D system for recovery and drug susceptibility testing of Mycobacterium tuberculosis. Clin Microbiol Infect. 2003;9:1148–52. doi: 10.1046/j.1469-0691.2003.00783.x. [DOI] [PubMed] [Google Scholar]
- 16.Kubica GP. Differential identification of mycobacteria. VII. Key features for identification of clinically significant mycobacteria. Am Rev Respir Dis. 1973;107:9–21. doi: 10.1164/arrd.1973.107.1.9. [DOI] [PubMed] [Google Scholar]
- 17.Addo K, Owusu-Darko K, Yeboah-Manu D, Caulley P, Minamikawa M, Bonsu F, et al. Mycobacterial species causing pulmonary tuberculosis at the Korle Bu Teaching Hospital, Accra, Ghana. Ghana Med J. 2007;41:52–7. doi: 10.4314/gmj.v41i2.55293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma B, Pal N, Malhotra B, Vyas L. Evaluation of a rapid differentiation test for Mycobacterium tuberculosis from other mycobacteria by selective inhibition with p-nitrobenzoic acid using MGIT 960. J Lab Physicians. 2010;2:89–92. doi: 10.4103/0974-2727.72157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maurya AK, Nag VL, Kant S, Kushwaha RA, Kumar M, Mishra V, et al. Evaluation of an immunochromatographic test for discrimination between Mycobacterium tuberculosis complex and non tuberculous mycobacteria in clinical isolates from extra-pulmonary tuberculosis. Indian J Med Res. 2012;135:901–6. [PMC free article] [PubMed] [Google Scholar]
- 20.O’Hara CM, Weinstein MP, Miller JM. Manual and automated system for detection and identification of microorganisms. In: Murray PR, Baron EJ, Jorgensen JH, Pfaller MA, Yolken RH, editors. Manual of Clinical Microbiology. 8th ed. Vol. 1. Washington DC: ASM Press; 2003. pp. 20036–2904. [Google Scholar]
- 21.Richter E, Rüsch-Gerdes S, Hillemann D. Evaluation of the GenoType Mycobacterium Assay for identification of mycobacterial species from cultures. J Clin Microbiol. 2006;44:1769–75. doi: 10.1128/JCM.44.5.1769-1775.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma SK, Mohan A. Extra pulmonary tuberculosis. Indian J Med Res. 2004;120:316–53. [PubMed] [Google Scholar]
- 23.Somoskovi A, Mester J, Hale YM, Parsons LM, Salfinger M. Laboratory diagnosis of nontuberculous mycobacteria. Clin Chest Med. 2002;23:585–97. doi: 10.1016/s0272-5231(02)00018-7. [DOI] [PubMed] [Google Scholar]
- 24.Tortoli E, Bartoloni A, Böttger EC, Emler S, Garzelli C, Magliano E, et al. Burden of unidentifiable mycobacteria in a reference laboratory. J Clin Microbiol. 2001;39:4058–65. doi: 10.1128/JCM.39.11.4058-4065.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ismail NA, Baba K, Pombo D, Hoosen AA. Use of an immunochromatographic kit for the rapid detection of Mycobacterium tuberculosis from broth cultures. Int J Tuberc Lung Dis. 2009;13:1045–7. [PubMed] [Google Scholar]
- 26.Singh AK, Dhole TN, Maurya AK, Singh AK, Kumar M, Umrao J, et al. Evaluation of rapid TB antigen MPT64 test for identification of Mycobacterium tuberculosis complex in liquid culture isolates at tertiary care center in Northern India. Int J Infect Dis. 2012;16:e294. [Google Scholar]
- 27.Mäkinen J, Marjamäki M, Marttila H, Soini H. Evaluation of a novel strip test, GenoType Mycobacterium CM/AS, for species identification of mycobacterial cultures. Clin Microbiol Infect. 2006;12:481–3. doi: 10.1111/j.1469-0691.2006.01380.x. [DOI] [PubMed] [Google Scholar]
- 28.Padilla E, González V, Manterola JM, Pérez A, Quesada MD, Gordillo S, et al. Comparative evaluation of the new version of the INNO-LiPA mycobacteria and genotype mycobacterium assays for identification of mycobacterium species from MB/BacT liquid cultures artificially inoculated with mycobacterial strains. J Clin Microbiol. 2004;42:3083–8. doi: 10.1128/JCM.42.7.3083-3088.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruiz P, Gutierrez J, Zerolo FJ, Casal M. GenoType mycobacterium assay for identification of mycobacterial species isolated from human clinical samples by using liquid medium. J Clin Microbiol. 2002;40:3076–8. doi: 10.1128/JCM.40.8.3076-3078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russo C, Tortoli E, Menichella D. Evaluation of the new genotype mycobacterium assay for identification of mycobacterial species. J Clin Microbiol. 2006;44:334–9. doi: 10.1128/JCM.44.2.334-339.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martín-Casabona N, Bahrmand AR, Bennedsen J, Thomsen VO, Curcio M, Fauville-Dufaux M, et al. Non-tuberculous mycobacteria: Patterns of isolation. A multi-country retrospective survey. Int J Tuberc Lung Dis. 2004;8:1186–93. [PubMed] [Google Scholar]
- 32.Jani MN, Rodrigues CS, Mehta AP. The neglected and often ignored: Nontuberculous mycobacteria. J Glob Infect Dis. 2011;3:94. doi: 10.4103/0974-777X.77305. [DOI] [PMC free article] [PubMed] [Google Scholar]


