Abstract
Understanding genetic influences on both healthy and disordered brain function is a major focus in psychiatric neuroimaging. We utilized task-related imaging findings from an fMRI auditory oddball task known to be robustly associated with abnormal activation in schizophrenia, to investigate genomic factors derived from multiple single nucleotide polymorphisms (SNP’s) from genes previously shown to be associated with schizophrenia. Our major aim was to investigate the relationship of these genomic factors to normal/abnormal brain functionality between controls and schizophrenia patients. We studied a Caucasian-only sample of 35 healthy controls and 31 schizophrenia patients. All subjects performed an auditory oddball task, which consists of detecting an infrequent sound within a series of frequent sounds. Each subject was characterized on 24 different SNP markers spanning multiple risk genes previously associated with schizophrenia. We used a recently developed technique named parallel independent component analysis (para-ICA) to analyze this multimodal dataset (Liu et al., 2008). The method aims to identify simultaneously independent components of each modality (functional imaging, genetics) and the relationships between them. We detected 3 fMRI components significantly correlated with two distinct gene components. The fMRI components, along with their significant genetic profile (dominant SNP) correlations were as follows: 1) Inferior frontal- anterior/posterior cingulate-thalamus-caudate with SNPs from Brain derived neurotropic factor (BDNF) & Dopamine Transporter, (DAT) [r=−0.51; p<0.0001], 2) Superior/middle temporal gyrus-Cingulate-premotor with SLC6A4_PR and SLC6A4_PR_AG (Serotonin transporter promoter; 5HTTLPR) [r=0.27; p=0.03], 3) Default Mode-fronto-temporal gyrus with Brain derived neurotropic factor & Dopamine Transporter (BDNF, DAT) [r=−0.25; p=0.04]. Functional components comprised taskrelevant regions (including PFC, ACC, STG and MTG) frequently identified as abnormal in schizophrenia. Further, gene-fMRI combinations 1 (Z=1.75; p=0.03), 2 (Z=1.84;p=0.03) and 3 (Z=1.67; p=0.04) listed above showed significant differences between controls and patients, based on their correlated loading coefficients. We demonstrate a framework to identify interactions between “clusters” of brain function and of genetic information. Our results reveal the effect/influence of specific interactions, (perhaps epistastatic in nature), between schizophrenia risk genes on imaging endophenotypes representing attention/working memory and goal directed related brain function, thus establishing a useful methodology to probe multivariate genotype-phenotype relationships.
Keywords: auditory oddball, DAT, BDNF, fMRI, gene, parallel ICA, multivariate, 5HTTLPR
Introduction
Schizophrenia is a debilitating psychological condition, whose etiology remains unclear, but which has a demonstrated genetic component (Cardno and Gottesman, 2000; Cardno et al., 1999; Farmer et al., 1987) with biological accompaniments (Allen et al., 2009; Laurent et al., 1999). Polygenic models suggests that interaction and convergence of either multiple common risk alleles, or rare copy number variants along with a variety of environmental factors, can result in schizophrenia (Alaerts and Del-Favero, 2009; Harrison and Weinberger, 2005; Pearlson and Folley, 2008; St Clair, 2009). This model hypothesizes that what is inherited is not the disease per se, but combinations of individually small, unfavorable genetic variants influencing biological processes within the normal range, but increasing disease liability significantly in epistatic combination when all contribute adversely to molecular interactions at a crucial bottleneck, (Pearlson and Folley, 2008). While neuroimaging can contribute detailed information regarding biological relationships between genetic variation and clinical phenotypes (Glahn et al., 2008), to date, potential interactions between multiple liability genes underlying functional brain abnormalities have only been studied in simple combinations, most usually two alleles and one brain region at a time, (Prata et al., 2009). In part this is due to the complexities of studying multiple genetic variants simultaneously in terms of their influence on neuroimaging parameters. Understanding both the normal function and disease-related dysfunction related to identified risk alleles may help clarify the etiopathology of the disorder.
The current search for endophenotypes (quantitative biological disease propensity markers) employs several different strategies (Allen et al., 2009). With respect to schizophrenia research, one of the oldest and most robust functional electrophysiological endophenotypes is an abnormal P300 (P3) response in schizophrenia to an infrequent stimulus type, such as in oddball detection tasks. The P3 in general is thought to reflect cognitive processes necessary for updating working memory representation and attention of task-related stimuli (Coles et al., 1988; Polich and Donchin, 1988; Polich and Kok, 1995). However, because of the poor spatial specificity of ERP neural generators in general (Baillet and Garnero, 1997; Pascual-Marqui et al., 1994) and the P3 in particular (Halgren et al., 1995a; Halgren et al., 1995b; Halgren et al., 1998), it is not clear which brain structures might be modulated by the oddball task. The measurement of brain hemodynamics using fMRI provides a means to identify structures influenced by oddball target interval manipulations. One of the largest fMRI auditory oddball detection (AOD) studies to date (Kiehl et al., 2005) replicated and extended more than a dozen previous fMRI oddball target detection studies, showing that hemodynamic activity is elicited in numerous, widespread cortical and subcortical brain structures during target detection and novelty processing. The fMRI AOD task has thus been increasingly studied as a schizophrenia endophenotype.
The AOD task used in this study was designed to test attention orientation to task-relevant (salient) infrequent stimuli. It has previously been used by us to assess ADHD (Attention Deficit Hyperactivity Disorder) and schizophrenia, among other conditions (Garrity et al., 2007; Stevens et al., 2007). Response to the AOD task in schizophrenia has been associated with impaired ability to attend to novel stimuli, and with a diverse range of brain areas associated with attention, executive function, and working memory, although patients behavioral response to the task is relatively unimpaired.
Research on the genetic component of schizophrenia has led to the identification of a number of common risk variants that could play a part in the expression of the disorder, most of which have been identified using univariate methods (e.g. candidate gene approaches). For schizophrenia several such risk genes have been identified through such methods, including catechol-O-methyl transferase (COMT) (Harrison and Weinberger, 2005) where a specific polymorphism influences prefrontal cortical and executive function (Avramopoulos et al., 2002; Egan et al., 2001; Goldberg and Weinberger, 2004). A study examining epistasis between the a 3’ untranslated region (UTR) variable number tandem repeat in the dopamine transporter (DAT) and the above COMT polymorphism found that these alleles interact nonadditively to modulate cortical function during executive processing, which is significantly altered in schizophrenia (Prata et al., 2009). Other recently identified potential risk genes for schizophrenia include Disrupted in Schizophrenia 1 (DISC1), Brain-derived neurotrophic factor (BDNF), and Dysbindin (Callicott et al., 2005; Craddock et al., 2006a; Craddock et al., 2006b; Harrison and Weinberger, 2005).
In the current study, to better capture the effect of genetic polymorphism on cognitive function, genetically characterized subjects performed an AOD task during functional magnetic resonance imaging (fMRI). We then used parallel independent component analysis (para-ICA; (Liu et al., 2008)), a recently developed technique, to perform multivariate analysis of combined imaging and genomic data. This method utilizes blind source separation to separate high dimensional data to discover patterns associated with, for example, clusters (components) of linked fMRI regions derived from quantitative brain measures, and/or components of associated SNPs derived from a gene array. The technique can identify and quantify associations between these two sets of components and determine significant differences in a patient-versus-control context embedded in the components (Liu et al., 2009a; Liu et al., 2009b). Para-ICA is a variant of ICA designed for multimodality processing, that extracts components using an entropy term based on information theory to maximize independence (Bell and Sejnowski, 1995) and enhances the interconnection by maximizing the linkage function in a joint estimation process (Liu et al., 2008). Thus this method can extract the intrinsic relationship between the identified independent components from both functional imaging and genetic modalities based on higher order statistics. Prior knowledge of neither specified genes nor fMRI patterns is needed, making this a hypothesis-free approach constituting an unsupervised algorithm, analogous to those used in discovering novel genes for revealing regulatory networks through analyzing large datasets (Lee and Batzoglou, 2003). The technique has been validated by examining multivariate relationships between SNP’s and fMRI-measured activity (Calhoun et al., 2006). In the context of gene-brain exploration, parallel ICA has been previously applied to find simultaneously independent components from a large (>300 SNP) gene array and, in one case a functional MRI AOD task in schizophrenia and controls and in a second from EEG data and a SNP array data set in healthy controls (Liu et al., 2009a; Liu et al., 2009b). The method has the advantage that it is able to detect significant associations in modest-sized data sets (Liu et al., 2008; Liu et al., 2009a; Liu et al., 2009b).
The current study differs from our previously published results in that we proposed to use a smaller number of alleles, chosen from among those previously identified as contributing toward schizophrenia risk, in a more hypothesis-driven manner. For example, we conjectured that dopamine-related genes such as COMT, DAT, DRD2 and DBH might interact epistatically. This strategy was also chosen to increase validity of the results of our current study while simultaneously focusing on schizophrenia. Using a known set of schizophrenia genes helps reduce statistical noise (by reducing interactions with other genes not known to be related to the disease) and increases the power of finding robust relationships, in particular interactions among the putative risk genes for the disorder.
In regard to methodology, a number of recent studies have used the combination of ICA (or related variants such as canonical correlation analysis) and the AOD task primarily to capture or build better algorithms to better discriminate brain patterns of schizophrenia from those of healthy controls (Correa et al., 2008; Demirci et al., 2008; Kim et al., 2008; Kim et al., 2009; Sui et al., 2009). However it is important to reiterate that the aim of the current study was slightly different from the above studies, with the main purpose of investigating meaningful relationships between brain activity (during AOD target detection) and clusters of risk SNP’s while simultaneously looking for differences in connections between the above modalities across groups.
Given that risk for a complex disorder such as schizophrenia is influenced or conferred by multiple risk genes acting together, it becomes very important to understand the product of this putatively interactive process. Therefore, the purpose of this study was to simultaneously identify the association patterns of several schizophrenia risk SNP’s or allelic variations with brain functionality representing performance on an attention related task (AOD) in healthy controls and schizophrenia patients using a previously published, recently developed multivariate parallel ICA technique that has validity in studies with small sample sizes. In addition, we also sought to identify and characterize differering gene-fMRI association patterns between controls and schizophrenia and to detect possible epistatic interactions among known schizophrenia risk genes.
Material and Methods
We used data from two different modalities, functional MRI (fMRI) and genotype (SNP) to reveal relationships between brain function from an AOD task and SNPs that also differed between schizophrenia-diagnosed and healthy control groups.
Participants
Sixty-six Caucasian subjects (35 healthy controls (29 right handed/6 left handed), 31 schizophrenia patients (25 right handed/3 left handed/3 ambidextrous)) who had volunteered for functional imaging and genetic testing were included from studies at the Olin Neuropsychiatry Research Center. Groups were matched on age, gender and handedness. All participants were assessed for DSM-IV-TR Axis I disorders using the SCID-IV (First, 2002). Medication information was available for twenty-two schizophrenia patients. Medications included; Second-generation antipsychotics: aripiprazole (5), ziprasidone (3), rispridone (3), quetiapine (2), olanzapine (5); First-generation antipsychotics: haloperidiol (3), fluphenazine (2), perphenazine (1); Mood stabilizers: divalpoex (3), lithium (7), oxcarbazepine (1), gabapentin (1); SSRI/SNRIs: venlafaxine (2), paroxetine (2), citalopram (1), fluoxetine (1), sertraline (1); tricyclics: amitriptyline (5); benzodiazepines: lorazepam (2), diazepam (1) and anticholinergics: benztropine (7). Exclusion criteria for controls included any present or past Axis I disorder or family history of psychotic disorder and for all participants, any significant history of medical or neurological, head injury, or substance abuse within 6 months prior to participation. Participants gave written informed consent using procedures approved by the Yale and Harford Hospital institutional review boards.
Genetic Analysis
Saliva samples were taken from subjects by trained personnel prior to the functional imaging task, and sent to the laboratory of Dr. Joel Gelernter at Yale University (New Haven, Connecticut, USA) for allelic determination. Single nucleotide polymorphism (SNP) analysis for 24 SNP’s spanning 14 schizophrenia putative risk genes was carried out using a fluorogenic 5' nuclease (TaqMan) assay (Shi et al., 1999), with the ABI PRISM 7900 Sequence Detection System (ABI, Foster City, California, USA). All samples were genotyped in duplicate for quality control, with no discrepancies. The 24 SNPs genotyped included: COMT (rs4680); BDNF (rs6265); solute carrier family 6 (neurotransmitter transporter, serotonin member 4; SLC6A4) (rs25531); dopamine receptor D2 (DRD2) (rs6277, rs1799732); GABA-A receptor (GABRA) (rs279837, rs567926, hCV8262927) alpha-7 nicotinic cholinergic receptor, (CHRNA7) (rs868437, rs2337506); alpha 5 muscarinic cholinergic receptor, (CHRM5) (rs16969968); cannabinoid receptor 1 (CB1) (rs1049353); doublecortin domain containing 2 (DCDC2) (dbSTS BV677278); solute carrier family 24, member 5 (rs1426654); dopa decarboxylase (DDC) (aromatic L-amino acid decarboxylase) (rs11238214); dopamine beta-hydroxylase (DBH) (dopamine beta-mono-oxygenase) (rs1611115); Dopamine transporter/Solute carrier family 6 (DAT or SLC6A3) (neurotransmitter transporter, dopamine), member 3; solute carrier family 6 (5HTTR) (neurotransmitter transporter, serotonin member 4) (rs25531); solute carrier family 6 (5HTR; SLC6A2) (neurotransmitter transporter, serotonin) member 2 and Disrupted-in-Schizophrenia 1 (DISC 1) (rs751229, rs3738401, rs980989, rs821616, rs1411771).
Auditory Oddball Task (fMRI)
In the AOD task subjects are presented with a series of novel and standard sounds, and required to detect a random, infrequent target sound within the series. The standard stimulus was a 500-Hz tone, with an 80% occurrence probability. The target stimulus was a 1000-Hz tone, and the novel stimuli were random, non-repeating digital noises (e.g. tone sweep, whistles). Both the novel and the target stimuli had an occurrence probability of 10%.
The stimulus durations were 200 ms with a 2000 ms stimulus onset asynchrony (SOA). Target (or Novel) stimuli were preceded by 3–5 standard stimuli. Stimuli were presented with a pseudorandom distribution of interstimulus interval duration (8–12 seconds), so that each type of stimulus (standard, target, and novel) appeared with equal probability throughout the task. Subjects were instructed to press a button with the right index finger whenever they heard the target stimulus, and not to respond to the standard or novel stimuli. Since the MRI sequence noise is high, verbal confirmation was made as to whether the subjects heard the stimuli proper. In addition, high end noise cancelling headphones were used to minimize external acoustic noise interactions with the in-scanner auditory paradigm. Behavioral responses for the task including mean reaction time and accuracy rate to the target events were recorded in real-time. The AOD paradigm and methods of data acquisition have been described previously in more detail (Kiehl et al., 2005; Stevens et al., 2007)).
Image Acquisition
Blood oxygenation level-dependent (BOLD) functional magnetic resonance (fMRI) images were recorded while subjects performed the AOD task. Subjects were scanned at the Olin Neuropsychiatry Research Center at the Institute of Living, using a Siemens Allegra 3T dedicated head scanner (Erlangen, Germany) equipped with 40mT/m gradients and a standard quadrature head coil. We used a conventional gradient-echo echo-planar imaging with the following imaging parameters: TR= 1.5 seconds, TE= 27msec, field of view= 24cm, acquisition matrix= 64 × 64, flip angle= 70°, voxel size= 3.75 × 3.75 × 4mm, gap= 1mm, 29 slices, ascending axial acquisition. To compensate for longitudinal equilibrium, the first six images were discarded.
Preprocessing
Each participant’s fMRI regressors were determined by extracting the time of stimulus onset for correct target, novel, and standard stimuli. Modeling was carried out using a synthetic hemodynamic response function and temporal derivative. Data from each participant’s fMRI performance were preprocessed using the statistical parametric mapping (SPM2) software (http://www.fil.ion.ucl.ac.uk/spm/software/spm2/). A motion correcting algorithm, INRI-align, was used to realign the images (Freire and Mangin, 2001). Image data were spatially normalized into standard Montreal Neurological Institute space (Friston et al., 1995). Following normalization, data were spatially smoothed with a 10×10×10 mm3 full width at half-maximum Gaussian kernel.
Post-Processing (Statistical Analysis)
As part of the first level or subject level analysis, planned comparisons for the primary condition of interest (target vs baseline) was computed for each subject using SPM2, in which each voxel represented the estimated amplitude of hemodynamic response. More accurate hemodynamic response estimates were provided by a latency variation amplitude-correction method. To remove low-frequency artifacts, a high-pass filter (cutoff period = 128 sec) was included in the model. To statistically control motion-related signal change, six motion-correction parameter estimates (x, y, and z displacement and roll, pitch, and yaw rotations) were included as covariates of no interest. The resulting contrast images for the target v baseline condition were then carried forward as the input to the parallel ICA algorithm to compute IC networks.
Parallel ICA (Liu et al., 2009a; Liu et al., 2009b) was applied using FIT (Fusion ICA Toolbox) in MATLAB 6.5 on the fMRI and SNP data described above to identify the independent functional networks, SNP associations and their interrelationships. In the current manuscript we provide a brief introduction to the para-ICA methods. The mathematical details of the algorithm and methodology of parallel ICA is described in detail in (Liu et al., 2009b). Similar to a conventional ICA analysis, extracted fMRI components are maximally spatially independent and measure the localized task related hemodynamic changes and their variation among the individuals. Components extracted from SNP data are distinct, independent, linear combinations of SNPs that are most likely highly associated with network functionality, or related phenotypes (Dawy et al., 2008). As part of the ICA algorithm loading parameters (Liu et al., 2008) are computed, which express the weights of every component on each subject. A correlation value is calculated between the fMRI component and the genetic component across subjects, based on their loading parameters, to identify the association between the fMRI and SNP components.
The parallel ICA method aims to solve three issues simultaneously. Two of them relate to maximizing independence between the different modalities. The third is determining the relationship between modalities. As seen from Figure 1, A is a linear mixing matrix, relating the sources to the contamination measurements. W is an unmixing matrix and Z is the estimated component matrix. Therefore, the essence of ICA is to find W so that so that Z is as close as possible to the true independent components contained in S. Para-ICA is conceptually similar, but the above is done on two different modalities concurrently. In addition a term linking the two modalities that optimally assesses the relationship between the two is also estimated. Each unmixing matrix for both modalities is thus computed using information derived from both modalities. Figure 1 depicts the para-ICA process in the form of a flow diagram.
Figure 1.
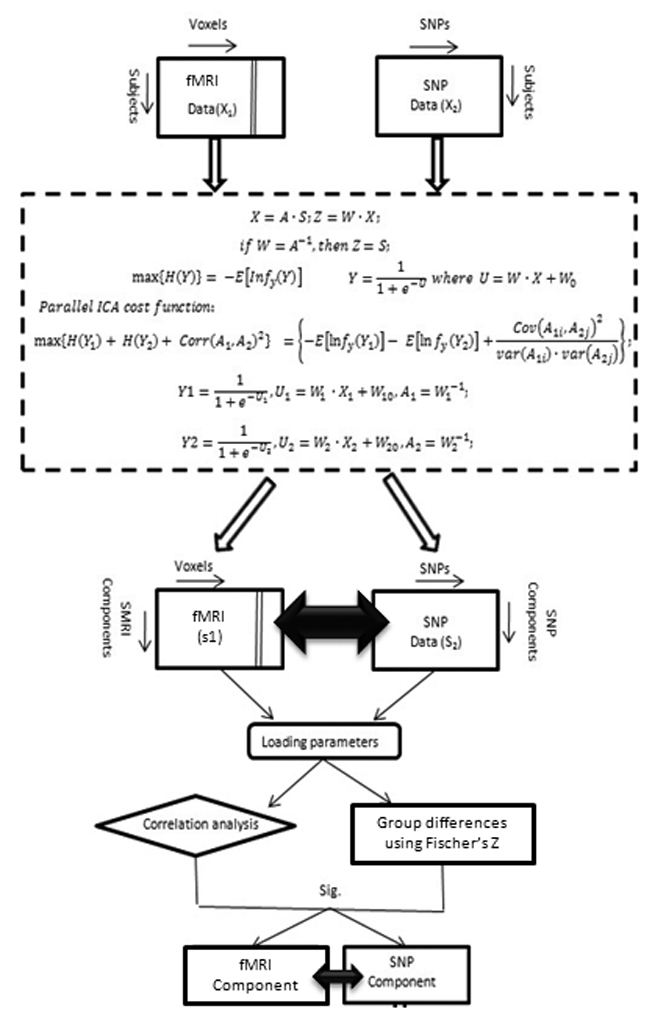
Detailed flow diagram depicting the parallel ICA procedure.
fMRI data including both control and patients group were constructed as a matrix of subjects by utilizing their first level contrast images from the target vs baseline condition, represented as a set of spatially independent voxels that are linearly mixed (Calhoun et al., 2001a). SNP data from both groups were organized as matrix of subjects by SNPs. These two data matrices are the input to the parallel ICA. Group spatial ICA (Calhoun et al., 2001b) in the GIFT software toolbox (http://icatb.sourceforge.net/) was used to identify the number of components. The component set size was estimated using both Akaike information criterion (AIC) and minimum description length (MDL) criteria, which is the standard method for estimating the components from the aggregated data set (Calhoun et al., 2001a, b). The number of estimated components using the above algorithms in our data set was 5 for feature 1 (fMRI) and 4 for feature 2 (gene).
Parallel ICA identified five paired independent components from fMRI and SNP data. The validity of the findings was further evaluated using a leave-one-out crossevaluation. In this method, one subject at a time is randomly omitted and 65 of 66 subjects analyzed repeatedly with the same specification and tested for consistency of the components and connections. All the component maps were thresholded at a |Z|>2.0 for visualization purposes.
Correlation values were calculated for all the fMRI-SNP component pairs, where the value represents the relationship between the fMRI network and the SNPs. Statistically significant correlations were extracted for all component pairs corrected for multiple comparison using a false discovery rate (FDR) with significance level by q<0.05 (Genovese et al., 2002). Further, within group correlation between fMRI and SNP components were computed and the resulting correlation values were compared using a Fisher’s Z-transform (Steel et al., 1997) to reveal group differences between normal controls and schizophrenia patients. Finally, we also computed pearson correlations between fMRI loading factors and behavior to examine any meaningful relationship between the two.
Results
Parallel ICA identified significant relationships between three independent fMRI networks (fMRI-a, b and c) and two gene components (gene-a, b). The fMRI networks identified comprised of the following regions 1) fMRI-a : Inferior frontal gryus, anterior cingulate, posterior cingulate, thalamus, caudate, lingual gyrus, precuneus and cerebellum, 2) fMRI-b : Superior temporal gyrus, middle/medial temporal gyrus, cingulate and premotor regions, 3) fMRI-c : medial frontal gyrus, middle temporal gyrus, posterior parietal (constituents of default mode network) superior temporal gyrus, orbito frontal cortex, prefrontal cortex and motor regions. Identified fMRI-SNP correlations (between loading parameters of each modality) were as follows 1) fMRI-a and gene-a (DAT and BDNF; r=−0.51; p<0.0001), 2) fMRI-b and gene-b (SLC6A4 5HTTLPR and 5HTTLPR_AG; r=0.27; p=0.03) and 3) fMRI-c and gene-a (DAT and BDNF; r=−0.25; p=0.04). Spatial regions of the three fMRI networks thresholded at Z > 2 are shown in Figure 2. Similarly, the genetic components containing SNP’s with loading weights were thresholded at Z > 2. Additionally, other non-significant component combinations (fMRI-d & DAT, BDNF; fMRI-e & DAT) are illustrated in supplementary Table 1.
Figure 2.
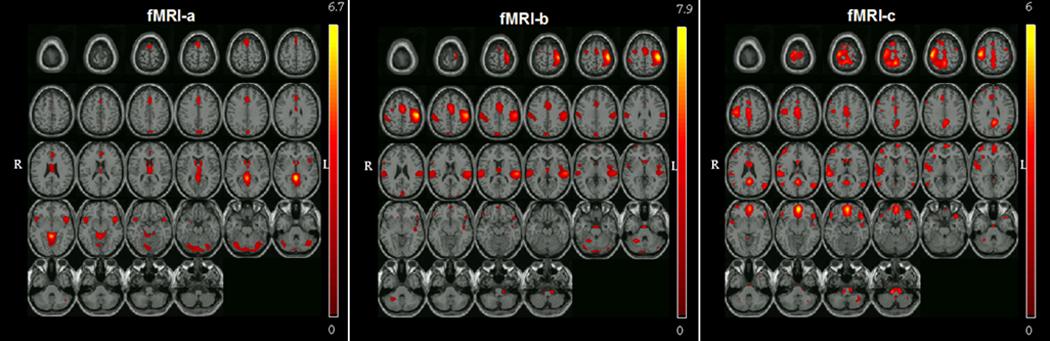
Spatial representation of all three significant brain networks (Total N = 66; 35 Controls & 31 Schizophrenia patients) derived from parallel ICA thresholded at |Z| > 2. The fMRI networks identified comprised of the following regions fMRI-a : Inferior frontal gryus, anterior cingulate, posterior cingulate, thalamus, caudate, lingual gyrus, precuneus and cerebellum, fMRI-b : Superior temporal gyrus, middle/medial temporal gyrus, cingulate and premotor regions, fMRI-c : medial prefrontal gyrus, middle temporal gyrus, posterior parietal and precentral gyrus (constituents of default mode network) superior temporal gyrus, orbito frontal cortex.
The within-group correlation values were compared using a Fisher’s Z-transform on all the above three fMRI-SNP combinations to assess control-patient relationship differences. All the above combinations demonstrated a significant group difference. The individual within-group correlation values along with their significant p-values for the group comparisons are provided in Table 1.
Table 1.
Overall and within-group correlation values between fMRI-SNP combinations along with significance values for the difference between controls and schizophrenia.
| fMRI component |
Linked gene component (Dominant SNP’s of the gene component thresholded at Z=2) |
Overall correlation coefficient between fMRI and Gene components |
Within-group correlation coefficient between fMRI and Gene components |
Comparison of correlation values in Controls versus Schizophrenia (Fisher’s Z and p- values) |
|
|---|---|---|---|---|---|
| Controls | Schizophrenia | ||||
| fMRI-a | gene-a (DAT & BDNF rs6265) |
−0.51 (p<0.0001) | −0.33 (p=0.05) |
−0.66 (p<0.0001) |
Z=1.75; p=0.03 |
| fMRI-b | gene-b (SLC6A4_PR , SLC6A4_Pr_ AG rs25531) |
0.27 (p=0.03) | −0.04 (p=NS) |
0.40 (p=0.02) | Z=1.84;p=0.03 |
| fMRI-c | gene-a (DAT & BDNF rs6265) |
−0.25 (p=0.04) | −0.001 (p=NS) |
−0.40 (p=0.02) | Z=1.67; p=0.04 |
Behaviorally, groups differed significantly only on mean response time (RT) (F=13.48; p<0.001) to the target stimuli. Overall as expected, schizophrenia patients (Mean RT±SD: 504.5±113.4) took longer to respond to target events that their healthy counterparts (Mean RT±SD: 411.3 ± 82.9 msec). Although the control group (Mean accuracy percent±SD: 98 ± 9) performed the task with greater accuracy than patients with schizophrenia (Mean accuracy ± SD: 91 ± 13), groups did not significantly differ on this behavioral measure. In addition, our analysis revealed a single significant positive correlation between fMRI-a and task accuracy (r=0.3; p=0.02).
Discussion
This study is one of the few investigations that has taken a multivariate approach to examine correlations between a presumed fMRI endophenotype and various schizophrenia risk genes while simultaneously probing for differences in relationship patterns between controls and schizophrenia. As we articulated earlier, the parallel ICA methodology used here provides a means for identifying larger-scale clusters of SNPs that are related to functional neuroimaging patterns, and that might theoretically identify epistatic combinations.
A number of recent studies have used techniques similar to para-ICA to evaluate data in a multivariate fashion primarily to develop imaging biomarkers for various disease models (Correa et al., 2008; Demirci et al., 2008; Kim et al., 2008; Kim et al., 2009; Sui et al., 2009). A recent report by Sui et al (2009) utilized a combination of coefficient constrained ICA and principal component analysis to extract optimal fMRI “features” simultaneously from two different fMRI tasks (Sternberg working memory task and AOD) to serve as biomarkers for schizophrenia. Another study by Franco et al (2008) used joint ICA (a technique that uses the same correlation profile of the two modalities as opposed to para-ICA which simultaneously uses two correlated profiles) to relate connectivity measures between diffusion tensor imaging and fMRI. In direct contrast to the above methodologies, para-ICA is considered to contain a more flexible statistical/mathematical model which makes it more appropriate when applied to fusing multimodal data that includes genetic information.
The two dozen SNPs chosen for the current study are in many cases already also known to be related together in a variety of contexts associated with neurotransmitter or cell signaling pathways. The current analysis utilized a Caucasian-only sample to maximize uniformity with respect to ethnicity and thereby avoid any possible population stratification effects. Our results identified three significant correlations between functional and genetic networks. From these, two different functional networks (fMRI-a and fMRI-c) were influenced by or related to the same genetic component (gene-a) which was primarily driven by change in signaling of the risk SNP’s from the DAT and BDNF genes, both of which have been independently proven to be risk markers for schizophrenia (Golimbet et al., 2008a; Golimbet et al., 2008b; Jeong et al., 2004; Talkowski et al., 2008; Wonodi et al., 2009). Both the above relationships indicated that the genetic component were negatively correlated to the functional components (r=−0.51 and −0.25 respectively), suggesting that differences in genetic signaling were associated with lower functional response in these networks. The most significant relationship was found between the combination of components fMRI-a and gene-a, the fMRI component by itself comprised regions including the inferior frontal cortex, parahippocampus, anterior/posterior cingulate, thalamus and striatum (caudate and putamen). Prior research using the oddball paradigm suggests that this network supports the goal-directed and attention processing of task-relevant target events. (Hampshire et al., 2009; Laurens et al., 2005; Yoshiura et al., 1999). The second functional component (fMRI-c) that was associated with this genetic component mostly represented the classic default mode network (DMN; Fox and Raichle, 2007) along with various right temporal and frontal areas. This network is believed to represent a baseline or “idling” brain condition. It has been functionally implicated in attending to external and internal stimuli (Gusnard et al., 2001; Gusnard and Raichle, 2001; Raichle et al., 2001). The DMN has also been shown to be disrupted in several mental disorders including schizophrenia (Broyd et al., 2009; Garrity et al., 2007), however its genetic underpinnings have not been studied, although this topic is important (Meyer-Lindenberg, 2009; Whitfield-Gabrieli et al., 2009). However, our results are partly consistent with the current literature that supports association of the BDNF polymorphism with psychiatric illnesses including depression that is associated with altered resting state EEG activity (Gatt et al., 2008). In addition current evidence suggests that dopamine plays an important role in modulating the DMN (Gatt et al., 2008; Nagano-Saito et al., 2009; Tomasi et al., 2009; van Eimeren et al., 2009). Interestingly a recent study by Tomasi et al 2009, shows evidence that dopamine transporter measures in striatum correlate with the default mode network during a visuospatial attention task mediated through the precuneus. This overlaps partially with the current study, where we observe that both fMRI-a (that includes the precuneus) and fMRI-c (that included the default mode) networks are mediated or associated with differences in genetic signaling from the dopamine transporter and BDNF genes.
All of the above identified regions have also been shown to be dysfunctional or abnormally structured in schizophrenia (Jeong et al., 2009; Kiehl et al., 2005; Kim et al., 2009; Meda et al., 2008; Wolf et al., 2007). A recent ICA study by (Kim et al., 2009) identified the above three functional networks as part of a larger network set in their study that was abnormal in schizophrenia subjects performing an auditory oddball task. (Correa et al., 2008) used a novel canonical correlation analysis method to find joint relationships between structural and fMRI (oddball task) in schizophrenia, reporting that less activity in temporal and motor areas was associated with less gray matter in the patient population. Both prior functional and structural studies show that the above regions found in our study may be separately modulated by polymorphisms in both the DAT and BDNF genes, both in controls and in schizophrenia (Bertolino et al., 2008; Chang et al., 2009; Franklin et al., 2009; Rybakowski, 2008), consistent with our most significant result. Prior studies also indicate that significant inverse correlations exist between DAT binding potential in the striatum and the severity of negative symptom scores along with cognitive and depression/anxiety scores on the PANSS (Schmitt et al., 2006). Schofield et al., (2009) demonstrated that the BDNF polymorphism examined in the current study was involved with disturbances in selective information procession as evidenced by a modulated P300 ERP response during the oddball task and corresponding alterations in gray matter in the fronto-hippocampal system.
The other fMRI-gene combination that significantly correlated and differentiated schizophrenia and controls contained spatial regions that encompassed primarily the bilateral superior/middle temporal gyrus and the cingulate regions, which represents a classic network driven by the functional auditory oddball task. In the present study we show the above network was significantly associated with the gene-b component whose genetic signaling was mainly influenced by SNP’s of the SLC6A4_promoter gene. This gene plays an important role in synaptic modulation of serotonin and acts in neuronal division, differentiation and adult neurogenesis (Bayle et al., 2003; Hallmayer, 2004). This gene has also been shown to be individually associated with risk for schizophrenia and other major psychiatric illnesses in previous studies (Liu et al., 1999; Luddington et al., 2009; Mendes de Oliveira et al., 1998; Vijayan et al., 2009). Our results are consistent with previous fMRI and ERP studies that support a role for serotonin in oddball response (Ehlers et al., 1991; Golimbet et al., 2006; Hughes et al., 1999; Wang et al., 2009)
Prior evidence also indicates that the above gene products interact in the central nervous system. (Bertolino et al., 2008) studied the epistatic effect between dopamine-regulating genes such as COMT and DAT and identified a modulated nonlinear response of the hippocampus during working memory tasks. (Pezawas et al., 2008) reported a biologic epistatsis between SLC6A4 and BDNF in the human brain by identifying a neural mechanism linking serotonergic and neurotropic signaling at the neural systems level.
Dopamine (DA) and BDNF are interlinked at many levels; for example, as reviewed by (Hunnerkopf et al., 2007), BDNF is expressed by DA neurons, helps induce the DA phenotype in fetal human cerebral cortex culture; DA stimulation increases BDNF mRNA in neuronal cultures, BDNF infusion into the substantia nigra increases striatal DA turnover, while loss of BDNF expression leads to DA downregulation and DA-ergic neuronal death. Some of the identified genes in the current study have also been shown to specifically interact in the context of psychiatric illnesses (Hunnerkopf et al., 2007; Murphy et al., 2003). Interestingly, (Murphy et al., 2003) show evidence of significant interactions between the Serotonin transporter (SERT) gene with BDNF and DAT in a mouse model that might lead to various psychiatric diseases. The results from our study might therefore be uncovering an interesting epistatic effect between BDNF, DAT and the SLC6A4_PR (5HTTLPR) genes in turn influencing attention-based functional brain networks.
As seen in table 1, even though all three combinations served to discriminate significantly between groups, for combinations 2 and 3, the within-group fMRI-SNP relationship was only significant for the schizophrenia group and not for controls, unlike combination 1 which was significant for both the healthy control and schizophrenia groups. This observation adds to the validity of the results by confirming strong associations of these risk genes and fMRI components especially within the schizophrenia population.
Results from this study also partly confirm similar fMRI-gene combinations to a prior study from our group (Liu et al., 2009b). Specifically we saw that most of the posterior visual association areas identified in our previous study remained consistent with our current results. However, this component (fMRI-a) also included additional fronto-temporal regions that were not identified in our prior study. However the gene associations with this imaging component did not overlap with our previous results. In addition we found two separate fMRI-gene relationships in the current study that we did not observe previously. These differences could be due to two reasons 1) the samples used in our study were not the same as those studied previously and 2) the set of genetic markers was very different between the two studies. The (Liu et al., 2009b) study examined >300 SNPs from a custom SNP chip that, overlapped to a very limited extent with the hypothesis-guided nature of the current study. A potential drawback of our study was the small number of SNP’s included and the relatively low subject numbers. Even though (as previously mentioned) the Parallel ICA algorithm is robust with smaller datasets (Liu et al., 2008; Liu et al., 2009a; Liu et al., 2009b), it will be important to measure such relationships in a larger sample with SNPs derived from the entire genome to increase statistical power and effect size of associations. Another issue that we could not account for in the present study was the influence of medication on fMRI patterns and in turn their relationship with gene components, primarily due to the lack of complete pertinent information. Also, future studies might be needed to investigate relationships between similar risk genes and other aspects of functionality (e.g. attention, resting state etc.) that are known to be compromised in schizophrenia as well as studying non-Caucasian populations.
In conclusion, we present a focused multimodal study to reveal relationships between AOD functionality and specific risk genes for schizophrenia. Our data thus validate previously found associations found via univariate methods and extends our knowledge to the inter-relationships between different risk genotypes to attention processing in schizophrenia. In addition, we also show new genetic correlations with a “default mode” network that successfully discriminated schizophrenia patients from a control population. The findings of the study thus indicate that heritable endophenotypes such as the fMRI-AOD have value in elucidating genotype-phenotype relationships in a multivariate fashion.
Supplementary Material
Acknowledgements
We thank the research staff at the Olin Neuropsychiatry Research Center who helped to collect and process the data. This research was supported by the National Institutes of Health, under grants R01 EB005846 & 1 R01 EB006841 (to VDC), and 2 RO1 MH43775, 5 RO1 MH52886 (to GP) and a grant from the MIND Institute (NPB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alaerts M, Del-Favero J. Searching genetic risk factors for schizophrenia and bipolar disorder: learn from the past and back to the future. Hum Mutat. 2009;30:1139–1152. doi: 10.1002/humu.21042. [DOI] [PubMed] [Google Scholar]
- Allen AJ, Griss ME, Folley BS, Hawkins KA, Pearlson GD. Endophenotypes in schizophrenia: a selective review. Schizophr Res. 2009;109:24–37. doi: 10.1016/j.schres.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avramopoulos D, Stefanis NC, Hantoumi I, Smyrnis N, Evdokimidis I, Stefanis CN. Higher scores of self reported schizotypy in healthy young males carrying the COMT high activity allele. Mol Psychiatry. 2002;7:706–711. doi: 10.1038/sj.mp.4001070. [DOI] [PubMed] [Google Scholar]
- Baillet S, Garnero L. A Bayesian approach to introducing anatomo-functional priors in the EEG/MEG inverse problem. IEEE Trans Biomed Eng. 1997;44:374–385. doi: 10.1109/10.568913. [DOI] [PubMed] [Google Scholar]
- Bayle FJ, Leroy S, Gourion D, Millet B, Olie JP, Poirier MF, Krebs MO. 5HTTLPR polymorphism in schizophrenic patients: further support for association with violent suicide attempts. Am J Med Genet B Neuropsychiatr Genet. 2003;119B:13–17. doi: 10.1002/ajmg.b.10037. [DOI] [PubMed] [Google Scholar]
- Bell AJ, Sejnowski TJ. An information-maximization approach to blind separation and blind deconvolution. Neural Comput. 1995;7:1129–1159. doi: 10.1162/neco.1995.7.6.1129. [DOI] [PubMed] [Google Scholar]
- Bertolino A, Di Giorgio A, Blasi G, Sambataro F, Caforio G, Sinibaldi L, Latorre V, Rampino A, Taurisano P, Fazio L, Romano R, Douzgou S, Popolizio T, Kolachana B, Nardini M, Weinberger DR, Dallapiccola B. Epistasis between dopamine regulating genes identifies a nonlinear response of the human hippocampus during memory tasks. Biol Psychiatry. 2008;64:226–234. doi: 10.1016/j.biopsych.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Broyd SJ, Demanuele C, Debener S, Helps SK, James CJ, Sonuga-Barke EJ. Default-mode brain dysfunction in mental disorders: a systematic review. Neurosci Biobehav Rev. 2009;33:279–296. doi: 10.1016/j.neubiorev.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Giuliani NR, Pekar JJ, Kiehl KA, Pearlson GD. Method for multimodal analysis of independent source differences in schizophrenia: combining gray matter structural and auditory oddball functional data. Hum Brain Mapp. 2006;27:47–62. doi: 10.1002/hbm.20166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pearlson GD, Pekar JJ. A method for making group inferences from functional MRI data using independent component analysis. Hum Brain Mapp. 2001a;14:140–151. doi: 10.1002/hbm.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pearlson GD, Pekar JJ. Spatial and temporal independent component analysis of functional MRI data containing a pair of task-related waveforms. Hum Brain Mapp. 2001b;13:43–53. doi: 10.1002/hbm.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callicott JH, Straub RE, Pezawas L, Egan MF, Mattay VS, Hariri AR, Verchinski BA, Meyer-Lindenberg A, Balkissoon R, Kolachana B, Goldberg TE, Weinberger DR. Variation in DISC1 affects hippocampal structure and function and increases risk for schizophrenia. Proc Natl Acad Sci U S A. 2005;102:8627–8632. doi: 10.1073/pnas.0500515102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardno AG, Gottesman II. Twin studies of schizophrenia: from bow-and-arrow concordances to star wars Mx and functional genomics. Am J Med Genet. 2000;97:12–17. [PubMed] [Google Scholar]
- Cardno AG, Jones LA, Murphy KC, Sanders RD, Asherson P, Owen MJ, McGuffin P. Dimensions of psychosis in affected sibling pairs. Schizophr Bull. 1999;25:841–850. doi: 10.1093/oxfordjournals.schbul.a033423. [DOI] [PubMed] [Google Scholar]
- Chang HA, Lu RB, Shy MJ, Chang CC, Lee MS, Huang SY. Brain-derived neurotrophic factor Val66Met polymorphism: association with psychopathological symptoms of schizophrenia? J Neuropsychiatry Clin Neurosci. 2009;21:30–37. doi: 10.1176/jnp.2009.21.1.30. [DOI] [PubMed] [Google Scholar]
- Coles MG, Gratton G, Donchin E. Detecting early communication: using measures of movement-related potentials to illuminate human information processing. Biol Psychol. 1988;26:69–89. doi: 10.1016/0301-0511(88)90014-2. [DOI] [PubMed] [Google Scholar]
- Correa NM, Li YO, Adali T, Calhoun VD. Canonical Correlation Analysis for Feature-Based Fusion of Biomedical Imaging Modalities and Its Application to Detection of Associative Networks in Schizophrenia. IEEE J Sel Top Signal Process. 2008;2:998–1007. doi: 10.1109/JSTSP.2008.2008265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craddock N, O'Donovan MC, Owen MJ. Genes for schizophrenia and bipolar disorder? Implications for psychiatric nosology. Schizophr Bull. 2006a;32:9–16. doi: 10.1093/schbul/sbj033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craddock N, Owen MJ, O'Donovan MC. The catechol-O-methyl transferase (COMT) gene as a candidate for psychiatric phenotypes: evidence and lessons. Mol Psychiatry. 2006b;11:446–458. doi: 10.1038/sj.mp.4001808. [DOI] [PubMed] [Google Scholar]
- Dawy Z, Sarkis M, Hagenauer J, Mueller JC. Fine-scale genetic mapping using independent component analysis. IEEE/ACM Trans Comput Biol Bioinform. 2008;5:448–460. doi: 10.1109/TCBB.2007.1072. [DOI] [PubMed] [Google Scholar]
- Demirci O, Clark VP, Calhoun VD. A projection pursuit algorithm to classify individuals using fMRI data: Application to schizophrenia. Neuroimage. 2008;39:1774–1782. doi: 10.1016/j.neuroimage.2007.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MF, Goldberg TE, Kolachana BS, Callicott JH, Mazzanti CM, Straub RE, Goldman D, Weinberger DR. Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc Natl Acad Sci U S A. 2001;98:6917–6922. doi: 10.1073/pnas.111134598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Wall TL, Chaplin RI. Long latency event-related potentials in rats: effects of dopaminergic and serotonergic depletions. Pharmacol Biochem Behav. 1991;38:789–793. doi: 10.1016/0091-3057(91)90243-u. [DOI] [PubMed] [Google Scholar]
- Farmer AE, McGuffin P, Gottesman II. Twin concordance for DSM-III schizophrenia. Scrutinizing the validity of the definition. Arch Gen Psychiatry. 1987;44:634–641. doi: 10.1001/archpsyc.1987.01800190054009. [DOI] [PubMed] [Google Scholar]
- First MB. The DSM series and experience with DSM-IV. Psychopathology. 2002;35:67–71. doi: 10.1159/000065121. [DOI] [PubMed] [Google Scholar]
- Fox MD, Raichle ME. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat Rev Neurosci. 2007;8:700–711. doi: 10.1038/nrn2201. [DOI] [PubMed] [Google Scholar]
- Franklin TR, Lohoff FW, Wang Z, Sciortino N, Harper D, Li Y, Jens W, Cruz J, Kampman K, Ehrman R, Berrettini W, Detre JA, O'Brien CP, Childress AR. DAT genotype modulates brain and behavioral responses elicited by cigarette cues. Neuropsychopharmacology. 2009;34:717–728. doi: 10.1038/npp.2008.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco AR, Ling J, Caprihan A, Calhoun VD, Jung RE, Heileman GL, Mayer AR. Multimodal and Multi-tissue measures of connectivity revealed by joint independent component analysis. IEEE J Sel Top Signal Process. 2008 Dec 1;2(6):986–997. doi: 10.1109/JSTSP.2008.2006718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire L, Mangin JF. Motion correction algorithms may create spurious brain activations in the absence of subject motion. Neuroimage. 2001;14:709–722. doi: 10.1006/nimg.2001.0869. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Holmes AP, Poline JB, Grasby PJ, Williams SC, Frackowiak RS, Turner R. Analysis of fMRI time-series revisited. Neuroimage. 1995;2:45–53. doi: 10.1006/nimg.1995.1007. [DOI] [PubMed] [Google Scholar]
- Garrity AG, Pearlson GD, McKiernan K, Lloyd D, Kiehl KA, Calhoun VD. Aberrant "default mode" functional connectivity in schizophrenia. Am J Psychiatry. 2007;164:450–457. doi: 10.1176/ajp.2007.164.3.450. [DOI] [PubMed] [Google Scholar]
- Gatt JM, Kuan SA, Dobson-Stone C, Paul RH, Joffe RT, Kemp AH, Gordon E, Schofield PR, Williams LM. Association between BDNF Val66Met polymorphism and trait depression is mediated via resting EEG alpha band activity. Biol Psychol. 2008;79:275–284. doi: 10.1016/j.biopsycho.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Genovese CR, Lazar NA, Nichols T. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. Neuroimage. 2002;15:870–878. doi: 10.1006/nimg.2001.1037. [DOI] [PubMed] [Google Scholar]
- Glahn D, Reichenberg A, Frangou S, Ormel H. Psychiatric neuroimaging: joining forces with epidemiology. Eur Psychiatry. 2008;23:315–319. doi: 10.1016/j.eurpsy.2007.09.014. [DOI] [PubMed] [Google Scholar]
- Goldberg TE, Weinberger DR. Genes and the parsing of cognitive processes. Trends Cogn Sci. 2004;8:325–335. doi: 10.1016/j.tics.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Golimbet VE, Korovaitseva GI, Abramova LI, Kasparov SV, Uvarova LG. [Association between the Val66Met polymorphism of brain-derived neurotrophic factor gene and schizophrenia in Russians] Mol Biol (Mosk) 2008a;42:599–603. [PubMed] [Google Scholar]
- Golimbet VE, Lebedeva IS, Alfimova MV, Korovaitseva GI, Lezheiko TV, Abramova LI, Kaleda VG. [Acoustic evoked potentials, serotonin transporter gene polymorphism and some psychopathological and psychological features in patients with schizophrenia and their relatives] Zh Nevrol Psikhiatr Im S S Korsakova. 2006;106:44–49. [PubMed] [Google Scholar]
- Golimbet VE, Lebedeva IS, Korovaitseva GI, Lezheiko TV, Yumatova PE. Association of 5-HTTLPR serotonin transporter gene polymorphism and Val66Met brain-derived neurotrophic factor gene polymorphism with auditory N100 evoked potential amplitude in patients with endogenous psychoses. Bull Exp Biol Med. 2008b;146:605–608. doi: 10.1007/s10517-009-0348-y. [DOI] [PubMed] [Google Scholar]
- Gusnard DA, Akbudak E, Shulman GL, Raichle ME. Medial prefrontal cortex and self-referential mental activity: relation to a default mode of brain function. Proc Natl Acad Sci U S A. 2001;98:4259–4264. doi: 10.1073/pnas.071043098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusnard DA, Raichle ME. Searching for a baseline: functional imaging and the resting human brain. Nat Rev Neurosci. 2001;2:685–694. doi: 10.1038/35094500. [DOI] [PubMed] [Google Scholar]
- Halgren E, Baudena P, Clarke JM, Heit G, Liegeois C, Chauvel P, Musolino A. Intracerebral potentials to rare target and distractor auditory and visual stimuli. I. Superior temporal plane and parietal lobe. Electroencephalogr Clin Neurophysiol. 1995a;94:191–220. doi: 10.1016/0013-4694(94)00259-n. [DOI] [PubMed] [Google Scholar]
- Halgren E, Baudena P, Clarke JM, Heit G, Marinkovic K, Devaux B, Vignal JP, Biraben A. Intracerebral potentials to rare target and distractor auditory and visual stimuli. II. Medial, lateral and posterior temporal lobe. Electroencephalogr Clin Neurophysiol. 1995b;94:229–250. doi: 10.1016/0013-4694(95)98475-n. [DOI] [PubMed] [Google Scholar]
- Halgren E, Marinkovic K, Chauvel P. Generators of the late cognitive potentials in auditory and visual oddball tasks. Electroencephalogr Clin Neurophysiol. 1998;106:156–164. doi: 10.1016/s0013-4694(97)00119-3. [DOI] [PubMed] [Google Scholar]
- Hallmayer J. Getting our AKT together in schizophrenia? Nat Genet. 2004;36:115–116. doi: 10.1038/ng0204-115. [DOI] [PubMed] [Google Scholar]
- Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry. 2005;10:40–68. doi: 10.1038/sj.mp.4001558. image 45. [DOI] [PubMed] [Google Scholar]
- Hampshire A, Thompson R, Duncan J, Owen AM. Selective tuning of the right inferior frontal gyrus during target detection. Cogn Affect Behav Neurosci. 2009;9:103–112. doi: 10.3758/CABN.9.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AM, Dixon R, Dane A, Kemp J, Cummings L, Yates RA. Effects of zolmitriptan (Zomig) on central serotonergic neurotransmission as assessed by active oddball auditory event-related potentials in volunteers without migraine. Cephalalgia. 1999;19:100–106. doi: 10.1046/j.1468-2982.1999.019002100.x. discussion 173. [DOI] [PubMed] [Google Scholar]
- Hunnerkopf R, Strobel A, Gutknecht L, Brocke B, Lesch KP. Interaction between BDNF Val66Met and dopamine transporter gene variation influences anxietyrelated traits. Neuropsychopharmacology. 2007;32:2552–2560. doi: 10.1038/sj.npp.1301383. [DOI] [PubMed] [Google Scholar]
- Jeong B, Wible CG, Hashimoto RI, Kubicki M. Functional and anatomical connectivity abnormalities in left inferior frontal gyrus in schizophrenia. Hum Brain Mapp. 2009 doi: 10.1002/hbm.20835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong SH, Joo EJ, Ahn YM, Kim YS. Association study of dopamine transporter gene and schizophrenia in Korean population using multiple single nucleotide polymorphism markers. Prog Neuropsychopharmacol Biol Psychiatry. 2004;28:975–983. doi: 10.1016/j.pnpbp.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Kiehl KA, Stevens MC, Celone K, Kurtz M, Krystal JH. Abnormal hemodynamics in schizophrenia during an auditory oddball task. Biol Psychiatry. 2005;57:1029–1040. doi: 10.1016/j.biopsych.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DI, Mathalon DH, Ford JM, Mannell M, Turner JA, Brown GG, Belger A, Gollub R, Lauriello J, Wible C, O'Leary D, Lim K, Toga A, Potkin SG, Birn F, Calhoun VD. Auditory oddball deficits in schizophrenia: an independent component analysis of the fMRI multisite function BIRN study. Schizophr Bull. 2009;35:67–81. doi: 10.1093/schbul/sbn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurens KR, Kiehl KA, Ngan ET, Liddle PF. Attention orienting dysfunction during salient novel stimulus processing in schizophrenia. Schizophr Res. 2005;75:159–171. doi: 10.1016/j.schres.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Laurent A, Moreaud O, Bosson JL, Naegele B, Boucharlat J, Saoud M, Dalery J, D'Amato T. Neuropsychological functioning among non-psychotic siblings and parents of schizophrenic patients. Psychiatry Res. 1999;87:147–157. doi: 10.1016/s0165-1781(99)00064-5. [DOI] [PubMed] [Google Scholar]
- Lee SI, Batzoglou S. Application of independent component analysis to microarrays. Genome Biol. 2003;4:R76. doi: 10.1186/gb-2003-4-11-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Bixler JN, Calhoun VD. A multimodality ICA study - integrating genomic single nucleotide polymorphisms with functional neuroimaging data. IEEE Bioinformatics and Biomedicine. 2008 [Google Scholar]
- Liu J, Kiehl KA, Pearlson G, Perrone-Bizzozero NI, Eichele T, Calhoun VD. Genetic determinants of target and novelty-related event-related potentials in the auditory oddball response. Neuroimage. 2009a;46:809–816. doi: 10.1016/j.neuroimage.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Pearlson G, Windemuth A, Ruano G, Perrone-Bizzozero NI, Calhoun V. Combining fMRI and SNP data to investigate connections between brain function and genetics using parallel ICA. Hum Brain Mapp. 2009b;30:241–255. doi: 10.1002/hbm.20508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Gu N, Feng G, Li S, Bai S, Zhang J, Shen T, Xue H, Breen G, St Clair D, He L. Tentative association of the serotonin transporter with schizophrenia and unipolar depression but not with bipolar disorder in Han Chinese. Pharmacogenetics. 1999;9:491–495. [PubMed] [Google Scholar]
- Luddington NS, Mandadapu A, Husk M, El-Mallakh RS. Clinical implications of genetic variation in the serotonin transporter promoter region: a review. Prim Care Companion J Clin Psychiatry. 2009;11:93–102. doi: 10.4088/pcc.08r00656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meda SA, Giuliani NR, Calhoun VD, Jagannathan K, Schretlen DJ, Pulver A, Cascella N, Keshavan M, Kates W, Buchanan R, Sharma T, Pearlson GD. A large scale (N=400) investigation of gray matter differences in schizophrenia using optimized voxel-based morphometry. Schizophr Res. 2008;101:95–105. doi: 10.1016/j.schres.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes de Oliveira JR, Otto PA, Vallada H, Lauriano V, Elkis H, Lafer B, Vasquez L, Gentil V, Passos-Bueno MR, Zatz M. Analysis of a novel functional polymorphism within the promoter region of the serotonin transporter gene (5-HTT) in Brazilian patients affected by bipolar disorder and schizophrenia. Am J Med Genet. 1998;81:225–227. doi: 10.1002/(sici)1096-8628(19980508)81:3<225::aid-ajmg4>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Meyer-Lindenberg A. Neural connectivity as an intermediate phenotype: brain networks under genetic control. Hum Brain Mapp. 2009;30:1938–1946. doi: 10.1002/hbm.20639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DL, Uhl GR, Holmes A, Ren-Patterson R, Hall FS, Sora I, DeteraWadleigh S, Lesch KP. Experimental gene interaction studies with SERT mutant mice as models for human polygenic and epistatic traits and disorders. Genes Brain Behav. 2003;2:350–364. doi: 10.1046/j.1601-1848.2003.00049.x. [DOI] [PubMed] [Google Scholar]
- Nagano-Saito A, Liu J, Doyon J, Dagher A. Dopamine modulates default mode network deactivation in elderly individuals during the Tower of London task. Neurosci Lett. 2009;458:1–5. doi: 10.1016/j.neulet.2009.04.025. [DOI] [PubMed] [Google Scholar]
- Pascual-Marqui RD, Michel CM, Lehmann D. Low resolution electromagnetic tomography: a new method for localizing electrical activity in the brain. Int J Psychophysiol. 1994;18:49–65. doi: 10.1016/0167-8760(84)90014-x. [DOI] [PubMed] [Google Scholar]
- Pearlson GD, Folley BS. Schizophrenia, psychiatric genetics, and Darwinian psychiatry: an evolutionary framework. Schizophr Bull. 2008;34:722–733. doi: 10.1093/schbul/sbm130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezawas L, Meyer-Lindenberg A, Goldman AL, Verchinski BA, Chen G, Kolachana BS, Egan MF, Mattay VS, Hariri AR, Weinberger DR. Evidence of biologic epistasis between BDNF and SLC6A4 and implications for depression. Mol Psychiatry. 2008;13:709–716. doi: 10.1038/mp.2008.32. [DOI] [PubMed] [Google Scholar]
- Polich J, Donchin E. P300 and the word frequency effect. Electroencephalogr Clin Neurophysiol. 1988;70:33–45. doi: 10.1016/0013-4694(88)90192-7. [DOI] [PubMed] [Google Scholar]
- Polich J, Kok A. Cognitive and biological determinants of P300: an integrative review. Biol Psychol. 1995;41:103–146. doi: 10.1016/0301-0511(95)05130-9. [DOI] [PubMed] [Google Scholar]
- Prata DP, Mechelli A, Fu CH, Picchioni M, Toulopoulou T, Bramon E, Walshe M, Murray RM, Collier DA, McGuire P. Epistasis between the DAT 3' UTR VNTR and the COMT Val158Met SNP on cortical function in healthy subjects and patients with schizophrenia. Proc Natl Acad Sci U S A. 2009;106:13600–13605. doi: 10.1073/pnas.0903007106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. A default mode of brain function. Proc Natl Acad Sci U S A. 2001;98:676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybakowski JK. BDNF gene: functional Val66Met polymorphism in mood disorders and schizophrenia. Pharmacogenomics. 2008;9:1589–1593. doi: 10.2217/14622416.9.11.1589. [DOI] [PubMed] [Google Scholar]
- Schmitt GJ, Frodl T, Dresel S, la Fougere C, Bottlender R, Koutsouleris N, Hahn K, Moller HJ, Meisenzahl EM. Striatal dopamine transporter availability is associated with the productive psychotic state in first episode, drug-naive schizophrenic patients. Eur Arch Psychiatry Clin Neurosci. 2006;256:115–121. doi: 10.1007/s00406-005-0618-2. [DOI] [PubMed] [Google Scholar]
- Schofield PR, Williams LM, Paul RH, Gatt JM, Brown K, Luty A, Cooper N, Grieve S, Dobson-Stone C, Morris C, Kuan SA, Gordon E. Disturbances in selective information processing associated with the BDNF Val66Met polymorphism: evidence from cognition, the P300 and fronto-hippocampal systems. Biol Psychol. 2009;80:176–188. doi: 10.1016/j.biopsycho.2008.09.001. [DOI] [PubMed] [Google Scholar]
- St Clair D. Copy number variation and schizophrenia. Schizophr Bull. 2009;35:9–12. doi: 10.1093/schbul/sbn147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel RD, Torrie JH, Dickey TA. Principles and Practice of Statistics: A Biomedical Approach. McGraw Hill; 1997. [Google Scholar]
- Stevens MC, Pearlson GD, Kiehl KA. An FMRI auditory oddball study of combined-subtype attention deficit hyperactivity disorder. Am J Psychiatry. 2007;164:1737–1749. doi: 10.1176/appi.ajp.2007.06050876. [DOI] [PubMed] [Google Scholar]
- Sui J, Adali T, Pearlson GD, Calhoun VD. An ICA-based method for the identification of optimal FMRI features and components using combined group-discriminative techniques. Neuroimage. 2009;46:73–86. doi: 10.1016/j.neuroimage.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkowski ME, Kirov G, Bamne M, Georgieva L, Torres G, Mansour H, Chowdari KV, Milanova V, Wood J, McClain L, Prasad K, Shirts B, Zhang J, O'Donovan MC, Owen MJ, Devlin B, Nimgaonkar VL. A network of dopaminergic gene variations implicated as risk factors for schizophrenia. Hum Mol Genet. 2008;17:747–758. doi: 10.1093/hmg/ddm347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi D, Volkow ND, Wang R, Telang F, Wang GJ, Chang L, Ernst T, Fowler JS. Dopamine transporters in striatum correlate with deactivation in the default mode network during visuospatial attention. PLoS One. 2009;4:e6102. doi: 10.1371/journal.pone.0006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eimeren T, Monchi O, Ballanger B, Strafella AP. Dysfunction of the default mode network in Parkinson disease: a functional magnetic resonance imaging study. Arch Neurol. 2009;66:877–883. doi: 10.1001/archneurol.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayan NN, Iwayama Y, Koshy LV, Natarajan C, Nair C, Allencherry PM, Yoshikawa T, Banerjee M. Evidence of association of serotonin transporter gene polymorphisms with schizophrenia in a South Indian population. J Hum Genet. 2009 doi: 10.1038/jhg.2009.76. [DOI] [PubMed] [Google Scholar]
- Wang L, Mullette-Gillman O, Gadde KM, Kuhn CM, McCarthy G, Huettel SA. The effect of acute tryptophan depletion on emotional distraction and subsequent memory. Soc Cogn Affect Neurosci. 2009 doi: 10.1093/scan/nsp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf DH, Gur RC, Valdez JN, Loughead J, Elliott MA, Gur RE, Ragland JD. Alterations of fronto-temporal connectivity during word encoding in schizophrenia. Psychiatry Res. 2007;154:221–232. doi: 10.1016/j.pscychresns.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wonodi I, Hong LE, Stine OC, Mitchell BD, Elliott A, Roberts RC, Conley RR, McMahon RP, Thaker GK. Dopamine transporter polymorphism modulates oculomotor function and DAT1 mRNA expression in schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:282–289. doi: 10.1002/ajmg.b.30811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield-Gabrieli S, Thermenos HW, Milanovic S, Tsuang MT, Faraone SV, McCarley RW, Shenton ME, Green AI, Nieto-Castanon A, LaViolette P, Wojcik J, Gabrieli JD, Seidman LJ. Hyperactivity and hyperconnectivity of the default network in schizophrenia and in first-degree relatives of persons with schizophrenia. Proc Natl Acad Sci U S A. 2009;106:1279–1284. doi: 10.1073/pnas.0809141106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiura T, Zhong J, Shibata DK, Kwok WE, Shrier DA, Numaguchi Y. Functional MRI study of auditory and visual oddball tasks. Neuroreport. 1999;10:1683–1688. doi: 10.1097/00001756-199906030-00011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


