Abstract
Cancer stem cells (CSCs) or tumor-initiating cells, similar to normal tissue stem cells, rely on developmental pathways, such as the Notch pathway, to maintain their stem cell state. One of the regulators of the Notch pathway is Musashi-1, a mRNA-binding protein. Musashi-1 promotes Notch signaling by binding to the mRNA of Numb, the negative regulator of Notch signaling, thus preventing its translation. Cancer stem cells have also been shown to down-regulate their 26S proteasome activity in several types of solid tumors, thus making them resistant to proteasome-inhibitors used as anti-cancer agents in the clinic. Interestingly, the Notch pathway can be inhibited by proteasomal degradation of the Notch intracellular domain (Notch-ICD), therefore down-regulation of the 26S proteasome activity can lead to stabilization of Notch-ICD. Here we present evidence that the down-regulation of the 26S proteasome in CSCs constitutes another level of control by which Musashi-1 promotes signaling through the Notch pathway and maintenance of the stem cell phenotype of this subpopulation of cancer cells. We demonstrate that Musashi-1 mediates the down-regulation of the 26S proteasome by binding to the mRNA of NF-YA, the transcriptional factor regulating 26S proteasome subunit expression, thus providing an additional route by which the degradation of Notch-ICD is prevented, and Notch signaling is sustained.
Keywords: Breast cancer stem cells, glioma stem cells, radiation, Notch, proteasome, Musashi, NF-Y
Introduction
Cancer cells in solid tumors are heterogeneous with respect to their ability to initiate or regrow tumors and their capability to produce progeny that reflects all cancer cell types found within the tumor. Cells with higher tumorigenicity are often called tumor-initiating cells or cancer stem cells (CSCs) 1. It is important to note that the term CSC does not necessarily imply that CSCs derive from normal tissue stem cells. In breast cancer and glioma, these cells can be prospectively identified based on cell surface marker expression 2, 3, ALDH1 activity 4, or lack of 26S proteasome function 5. For the latter, we have developed an imaging system that allows for prospective identification and tracking of CSCs/tumor-initiating cells. It is based on the stable expression of a fusion between a green fluorescent protein, ZsGreen, and the C-terminal degron of ornithine decarboxylase. In cells with intact proteasome activity the fusion protein is degraded immediately after translation. In CSCs/tumor-initiating cells, the lack of proteasome activity results in the accumulation of the fluorescent fusion protein, and thus in the identification of CSCs/tumor-initiating cells without further manipulation 5.
CSCs/Tumor-initiating cells in solid tumors are thought to be mostly quiescent 5, 6 and in a less energetic metabolic state than their non-tumorigenic progeny 7. Consequently protein turnover in slow cycling, or quiescent CSCs/tumor-initiating cells is expected to be low 8, 9. Therefore, CSCs/tumor-initiating cells are not required to maintain high activities of the 26S proteasome, a multicatalytic protease that requires large amounts of ATP for its assembly and function 10, which is responsible for the targeted degradation of proteins involved in cell signaling and serves as a key protease in protein quality control 11.
Interestingly, key stem cell factors like BMI-1, Oct-4, Sox-2, Nanog and Klf4 12–15, as well as effector proteins in Wnt 16, Notch 17, and Hedgehog 18 signaling are substrates of the 26S proteasome and consequently, low proteasome activities in CSCs/tumor-initiating cells will stabilize these proteins and thus enable sustaining a stem cell state.
Among others, the Notch pathway regulates self-renewal in breast cancer and glioma stem cells 19–21. Activation of the Notch pathway relies on cell-cell interaction, which ultimately leads to nuclear translocation of the intracellular domain of the Notch receptor (Notch-ICD) where it binds to CBF-1 and turns the latter from a transcriptional repressor into a transcriptional activator 22. Notch-ICD can be inhibited by Numb, which in turn is regulated by binding of Musashi-1 to a conserved motif in the 3’-UTR of Numb mRNA, thereby preventing its translation.
In search for a link between low-proteasome activity in CSCs/tumor-initiating cells and the CSC phenotype, we hypothesized that developmental pathways, such as the Notch pathway, down-regulate proteasome activity in order to maintain the stem cell phenotype through stabilization of stem cell factors. We report here, that in CSCs/tumor-initiating cells the RNA-binding protein Musashi-1 binds mRNA of NF-YA (Nuclear transcription factor Y subunit alpha), a subunit of the trimeric master regulatory transcription factor of proteasome subunit expression 23, thereby decreasing NF-YA protein levels and NF-YA DNA-binding activity. As a consequence 26S proteasome subunit expression is down-regulated, thus linking Notch signaling and the CSC state with low proteasome activity.
Methods
Cell culture
Human SUM159PT breast cancer cell line was purchased from Asterand (Detroit, MI). Human MCF-7 and T47D breast cancer cell lines were purchased from American Type Culture Collection (Manassas, VA). GBM146, GBM176, and GBM189 cells were obtained from the UCLA Intellectual and Developmental Disability Research Center Human Cell Core. The U87MG glioma cell line was a kind gift from Dr. P. Michel (Department of Pathology, UCLA, CA). SUM159PT-ZsGreen-cODC, MCF-7-ZsGreen-cODC, T47D-ZsGreen-cODC, U87MG-ZsGeen-cODC and GBM146-ZsGreen-cODC, GBM176-ZsGreen-cODC, and GBM189-ZsGreen-cODC were obtained as described in Vlashi et al. 5. SUM159PT was cultured in log-growth phase in F12 Medium (Invitrogen, Carlsbad, CA) (supplemented with 5% fetal bovine serum [Sigma Aldrich, St Louis, MO] and penicillin (100 units/ml) and streptomycin (100 μg/ml) cocktail [Invitrogen], insulin (5µg/mL) and hydrocortisone (1 μg/ml)), MCF-7 and T47D were cultured in log-growth phase in Dulbecco's Modified Eagle Medium (DMEM) (Invitrogen) (supplemented with 10% fetal bovine serum and penicillin and streptomycin cocktail), and glioma cells were cultured in DMEM/F12 (1:1) (supplemented with 10% fetal bovine serum and penicillin and streptomycin cocktail). All cells were grown in a humidified incubator at 37°C with 5% CO2.
shRNA against Musashi-1
Cells were plated in 6-well plates and incubated (37°C with 5% CO2) until cells reached 70% confluence. Cells were then transfected with shRNA vectors with Lipofectamine 2000® (Invitrogen) according to the manufacturer’s instructions. Briefly, 1 µg of plasmids (shRNAs targeting Msi-1 or scramble shRNA control, Origene) were diluted in 250 µl of OptiMEM (Invitrogen). Lipofectamine 2000® (5 µl) was also diluted in 250 µl of OptiMEM (no serum, no antibiotic), incubated for 5 min at room temperature and then mixed with the plasmid solution. The mixture was incubated for 20 min at room temperature. Cells were rinsed twice with PBS and 500 µl of OptiMEM (no serum, no antibiotic) were added. The plasmid-lipofectamine 2000® mix was added to the cells, and incubated for 5h at 37°C with 5% CO2. Two milliliters of FBS/antibiotic-containing media were added. Cells were incubated for 24 to 48h before experiments.
Sphere-forming capacity
SUM159PT, GBM146, GBM176, or GBM189 cells were transfected with shRNA against Musashi1 or scrambled control shRNA. After 48 hours, cells were plated at a density of 50 cells/ml into ultra-low adhesion 96-well plates in 200µl DMEM/F12, supplemented with 0.4% BSA (Sigma), 10 ml/500ml B27 (Invitrogen) 5 µg/ml bovine insulin (Sigma), 4 µg/ml heparin (Sigma), 20 ng/ml fibroblast growth factor 2 (bFGF, Sigma) and 20 ng/ml epidermal growth factor (EGF, Sigma)). After 15 days in culture mammospheres or neurospheres with diameters larger that 100µm were counted. In parallel, the same cells were seeded into 15 cm suspension dishes at 1,000 cells/ml into the same media to generate enough primary spheres for secondary sphere formation. Primary spheres were mechanically dissociated and counted and used for secondary sphere formation. Tertiary spheres were obtained using the same protocol using cells derived from secondary spheres.
Flow cytometry
We had previously shown that breast cancer and glioma stem cells/tumor-initiating cells could be identified via their low proteasome activity 5,6, which can be assessed by analyzing ZsGreen-cODC protein accumulation by flow cytometry. Cells were defined as "ZsGreen-cODC positive" if the fluorescence in the FL-1H channel exceeded the fluorescence level of 99.9% of the empty vector-transfected control cells.
Co-Immunoprecipitation of Musashi-1 and NF-YA mRNA
mRNAs bound by Musashi-1 were isolated and detected following an adapted protocol described by Peritz 22. Cells from 7-days old spheres were washed two times with PBS (1X) and lysed with polysome lysis buffer (100 mM KCl, 5 mM MgCl2, 10 mM HEPES, pH 7.0, 0.5% Nonidet P-40, 1 mM DTT, 100 U/ml RNAse OUT (Invitrogen), 2 mM vanadyl ribonucleaside complexes solution (Sigma-Aldrich), 25 μl/ml protease inhibitor cocktail for mammalian tissues (Sigma-Aldrich). Lysates were transferred to a tube on ice and centrifuged at 16,000 × g for 15 min at 4°C.
150 μl of protein A-agarose beads (Invitrogen) per 1 ml of lysate were equilibrated by two washes with 0.5 ml of polysome lysis buffer and the initial volume of beads was restored with polysome lysis buffer. The lysate was pre-cleaned with 50 μl of protein A-agarose beads by incubating for 1h at 4°C, and then centrifuged to pull down the beads. The pre-clearing step was repeated a second time with the supernatant. Cleaned supernatant was divided into 1 ml aliquots, 3 aliquots per sample (Musashi-1 or isotype control [Rabbit Polyclonal IgG], IP-control without antibody; all antibodies were from Abcam). Antibodies were added to the lysates and incubated overnight at 4°C with gentle agitation. 50 μl of protein A-agarose beads were added to each aliquot, the samples rotated at 4°C for 4 hours, and collected by brief centrifuging. Beads were washed 4 times with 0.5 ml of polysome lysis buffer by rotating for 5 min at 4°C, and then washed four more times with polysome lysis buffer including 1 M urea. Beads were re-suspended in 100 μl of polysome lysis buffer with 0.1% SDS and 30 μg proteinase K, and incubated for 30 min at 50°C in a heating block. 0.5 ml of Tri-Reagent (Sigma-Aldrich) were added and incubated for 5 min at room temperature. 0.2 ml of chloroform was added, shaken vigorously, and incubated for 10 min at room temperature. Aliquots were centrifuged for 15 min at 12,000 × g. Aqueous phases were recovered and the pellets (beads) were re-suspended in 100 μl of polysome lysis buffer with 0.1% SDS and 30 μg proteinase K for a second RNA extraction. RNA extraction steps and reverse transcription (RT) with Superscript III were performed according to the manufacturer’s instructions. 1 μl of RT was used to perform a first Real-Time PCR for 35 cycles using specific primers for NF-YA. Specific primers for Numb were used as a positive control for Msi-1 immunoprecipitation. A second Real-Time PCR was performed for 40 cycles using 1 μl of the first PCR product.
Quantitative Reverse Transcription-PCR
Quantitative PCR was performed in the My iQ thermal cycler (Bio-Rad, Hercules, CA) using the 2× iQ SYBR Green Supermix (Bio-Rad). Ct for each gene was determined and ∆∆Ct was calculated relative to the designated reference sample. Gene expression values were then set equal to 2−∆∆Ct as described by the manufacturer of the kit (Applied Biosystems). PCR primers were synthesized by Invitrogen and designed for the human sequence of human of NF-YA (Forward primer: 5’-GAGTCTCGGCACCGTCAT-3’, and Reverse primer: 5’-TGCTTCTTCATCGGCTTG-3’) and the human proteasome subunits X (Forward primer: 5’-ACGTGGACAGTGAAGGGAAC-3’, and Reverse primer: 5’-CTGCATCCACCCTCTTTCAG-3’), Y (Forward primer: 5’-AGGAGAAAGATGGCGGCTAC-3’, and Reverse primer: 5’- AGGACCCAGTGGTTGTTCTG-3’), Z (Forward primer: 5’-GGAGGAGGAAGCCAAGAATC-3’, and Reverse primer: 5’-CTTCCAGCACCTCAATCTCC-3’), PA28a (Forward primer: 5’-AGACAAAGGTCCTCCCTGTG-3’, and Reverse primer: 5’-CTGGACAGCCACTCCAAAAT-3’), PA28b (Forward primer: 5’-ACTCCCTCAATGTGGCTGAC-3’, and Reverse primer: 5’-GCAGGGACAGGACTTTCTCA-3’), LMP2 (Forward primer: 5’-GCAATAGCGTCTGTGGTGAA-3’, and Reverse primer: 5’-ATGCTGACTCGACAGCCTTT-3’), LMP7 (Forward primer: 5’-ATGTCAGTGACCTGCTGCAC-3’, and Reverse primer: 5’-CTGCTGAGCCCGTACTCTCT-3’), PSMC1 (Forward primer: 5’-GACCCCGATGTCAGTAGGAA-3’, and Reverse primer: 5’-GGATCCGTGTCATCCATCAG-3’), PSMD4 (Forward primer: 5’-CAACGTGGGCCTTATCACAC-3’, and Reverse primer: 5’-ATTGTCCTCCACTGGGCTTC-3’), and MECL1 (Forward primer: 5’-GTGCTAGAAGACCGGTTCCA-3’, and Reverse primer: 5’-CTCTGTGGGTGAGCTCAGTG-3’). GAPDH (Forward primer: 5’-CGATCAACTCACCGCCAACA-3’, and Reverse primer: 5’-GTGGCTTCATCTGCTTCCTGTC-3’) or RPLP0 (Forward primer: 5’-GTGATGTGCAGCTGATCAAGACT-3’, and Reverse primer: 5’-GATGACCAGCCCAAAGGAGA-3’) were used as loading control.
Proteasome Function Assays
Chymotryptic, tryptic, and caspase-like proteasome activities were measured as described previously 23 with a few minor modifications. Cells were washed with PBS, and pelleted by centrifugation. Glass beads and homogenization buffer (25 mM Tris, pH 7.5, 100 mM NaCl, 5 mM ATP, 0.2% (v/v) Nonidet P-40 and 20% glycerol) were added and tubes were vortexed for 1 minute. Beads and cell debris were removed by centrifugation at 4°C. Protein concentration was determined by the Micro BCA protocol (Pierce, Rockford, IL) with BSA (Sigma) as standard. To measure 26S proteasome activity, 100 µg of protein from crude cellular extracts of each sample was diluted with buffer I (50 mM Tris, pH 7.4, 2 mM DTT, 5 mM MgCl2, 2 mM ATP) to a final volume of 1 mL in quadruplicates. The fluorogenic proteasome substrates Suc-LLVY-AMC (chymotryptic substrate, Biomol, Plymouth Meeting, PA), Z-ARR-AMC (tryptic substrate, Calbiochem, San Diego, CA), and Z-LLE-AMC (caspase-like substrate, Biomol), were dissolved in DMSO and added to a final concentration of 80 µM in 1% DMSO. Proteolytic activities were continuously monitored by measuring the release of the fluorescent group, 7-amido-4-methylcoumarin (AMC), in a fluorescence plate reader (Spectramax M5, Molecular Devices, Sunnyvale, CA) at 37° C, at excitation and emission wavelengths of 380 and 460 nm, respectively.
NF-YA Transcription factor DNA-binding activity assays
ZsGreen-cODC-positive CSCs/tumor-initiating cell and ZsGreen-cODC-negative non-tumor-initiating cells were sorted from SUM159PT-ZsGreen-cODC and U87MG-ZsGreen-cODC monolayer and sphere cultures, respectively. Total nuclear protein extracts were obtained using the Nuclear Extraction Kit (ActiveMotif, Carlsbad, CA) and protein content was measured using the ProStain™ Protein Quantification Kit (ActiveMotif). DNA-binding activity of NF-YA was assessed using the TransAM® Transcription Factor ELISA (ActiveMotif) following the manufacturer’s instructions. The assay measures the binding of active NF-YA transcription factor to its consensus binding sequence on a short DNA oligonucleotide. The latter is immobilized onto an ELISA plate. After incubation and washing, NF-YA bound to its consensus binding motif can be detected with a specific antibody against NF-YA and quantified using a spectrophotometer. The specificity of the binding reactions is shown in competition experiments using excess non-immobilized double-stranded DNA containing the wild-type or a mutated binding sequence for NF-YA.
Western Blotting
Cells were lysed on ice using RIPA buffer. The lysate was centrifuged at 10,000 × g for 10 min, and the protein concentration in the supernatant was determined using the microBCA protocol (Pierce, Thermo Scientific, Rockford, IL). The protein lysate was separated by SDS-PAGE and then transferred to PVDF membranes (BioRad). The membranes were blocked with 5% BSA/TBS-0.1% Tween20 and then probed with rabbit anti-Musashi1 primary antibodies (Abcam ab52865, Cambridge, MA, 1/2,000) or rabbit anti-NF-YA primary antibodies (Abcam ab6558, Cambridge, MA, 1/1,000) for 1 h at room temperature. After washing, the blots were incubated with an anti-rabbit horseradish peroxidase-conjugated secondary antibody (SigmaAldrich, 1/50,000) and visualized by ECL plus Membrane Blotting Detection System (GE Healthcare) using a laser scanner (Typhoon 9410, GE Healthcare).
Statistics
Unless reported otherwise all results were derived from 3 biologically independent experiments. A p-value equal to or smaller than 0.05 in a Student’s t-test was considered statistically significant.
Results
The RNA-binding protein Musashi-1 binds the regulatory transcription factor of proteasome subunit expression, NF-YA mRNA
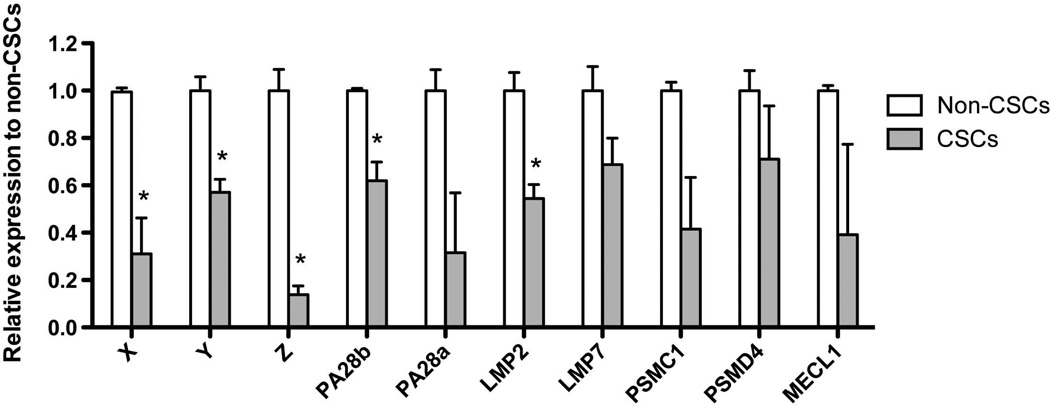
We had previously reported that breast cancer and glioma stem cells/tumor-initiating cells have low proteasome activity 5, 6, 24 and that proteasome subunit mRNAs were down-regulated in glioma CSCs/tumor-initiating cells 5. In order to analyze if the proteasome subunits are also down-regulated at the mRNA level in breast cancer, we sorted SUM159PT-ZsGreen-cODC+ CSCs/tumor-initiating cells and SUM159PT-ZsGreen-cODC− non-tumorigenic cells, and performed qRT-PCR for the proteasome subunits X, Y, Z, PA28a, PA28b, LMP2, LMP7, PSMC1, PSMD4, and MECL1. When we compared proteasome subunit mRNA expression levels in non-tumorigenic cells with high proteasome activity and CSCs/tumor-initiating cells with low proteasome activity, mRNA expression of all subunits tested was significantly down-regulated in CSCs/tumor-initiating cells (Figure 1).
Figure 1. Expression analysis of 26S proteasome and immunoproteasome subunits by real-time PCR.
mRNA were extracted from sorted SUM159PT non-CSCs (ZsGreen-cODC−) and CSCs (ZsGreen-cODC+). 1 µg of total RNA for each condition was treated with DNAse 1 and then reverse-transcribed into cDNA. 1µl of cDNA was used for each reaction of the semi-quantitative PCR. In CSCs mRNA expression levels for proteasome subunits were consistantly down-regulated. Normalized means ± SEM from 3 independent experiments are shown.
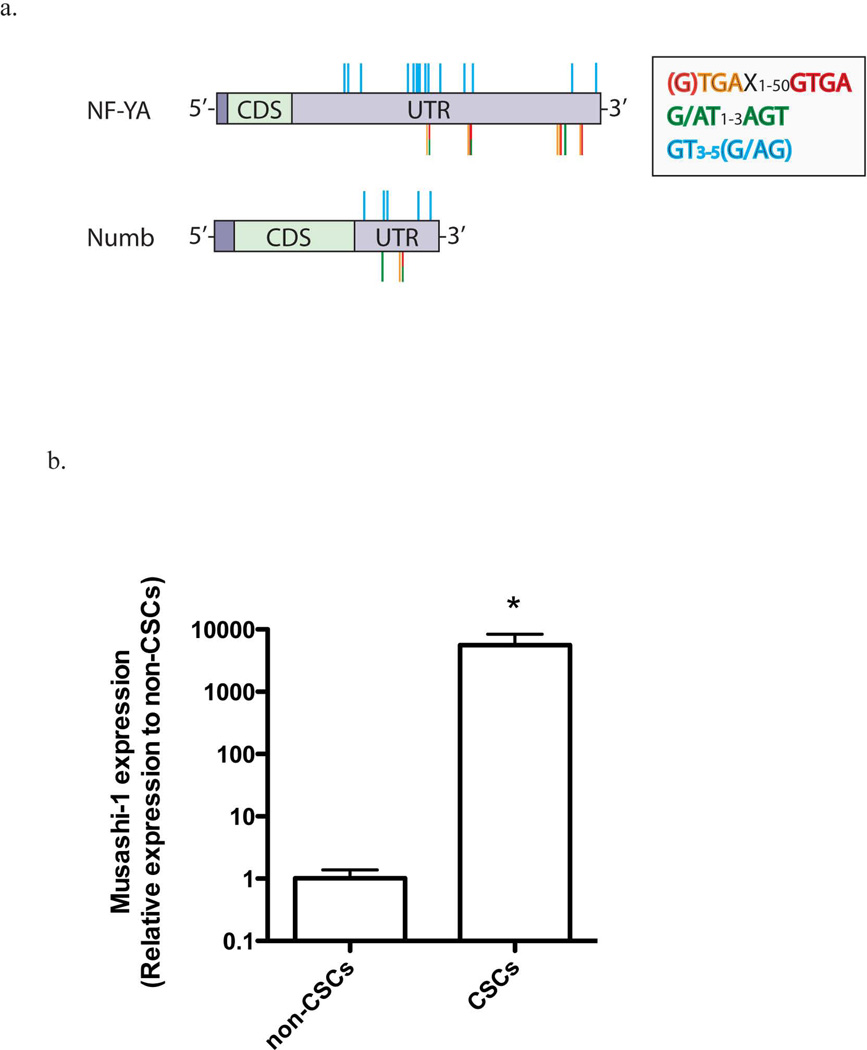
In search for a link between Notch signaling and down-regulation of proteasome subunit mRNA expression, we performed an in-silico analysis to test if the 3’-UTR of NF-YA mRNA, the regulatory transcription factor of proteasome subunit expression, contains binding motifs for Musashi-1 comparable to the sequence of the known Musashi-1 target numb (Fig. 2a and Suppl. Fig. 1). We found several of the putative binding motifs currently discussed for Musashi-1: 3 repeats of the G/AU1–3AGU motif 25, 15 repeats of the GU3–5(AG/G) motif 26, and 5 repeats of the recently proposed minimal binding motif UAGX/GUAGX1–50GUAG 27 (Figure 2a, Suppl. Figure 2 and 3). This suggested that post-transcriptional regulation of NF-YA could be linked to Notch signaling.
Figure 2. Musashi-1 binding sites on NF-YA and Numb mRNA, and Musashi-1 expression.
(a) Schematic representation of the NF-YA (P23511) and Numb mRNA (P47757) sequences. Both mRNAs contain putative binding sequences for Musashi-1 in their 3’-UTR. (b) Realtime RT-PCR for Musashi-1 mRNA in SUM159PT non-tumorigenic cells and CSCs. The expression of Musashi-1 mRNA is significantly up-regulated in CSCs (5590-fold ± 2820, p<0.05, two-sided Student’s t-test). Normalized means ± SEM from 3 independent experiments are shown.
We addressed this possibility experimentally by first analyzing if breast CSCs/tumor-initiating cells with low proteasome activity expressed Musashi-1. When we sorted ZsGreen-cODC− non-tumorigenic cells and ZsGreen-cODC+ CSCs/tumor-initiating cells from SUM159PT breast cancer cells, we found that Musashi-1 mRNA was up-regulated 5590-fold (± 2820, p<0.05, two-sided Student’s t-test) in ZsGreen-cODC+ CSCs/tumor-initiating cells (Figure 2b).
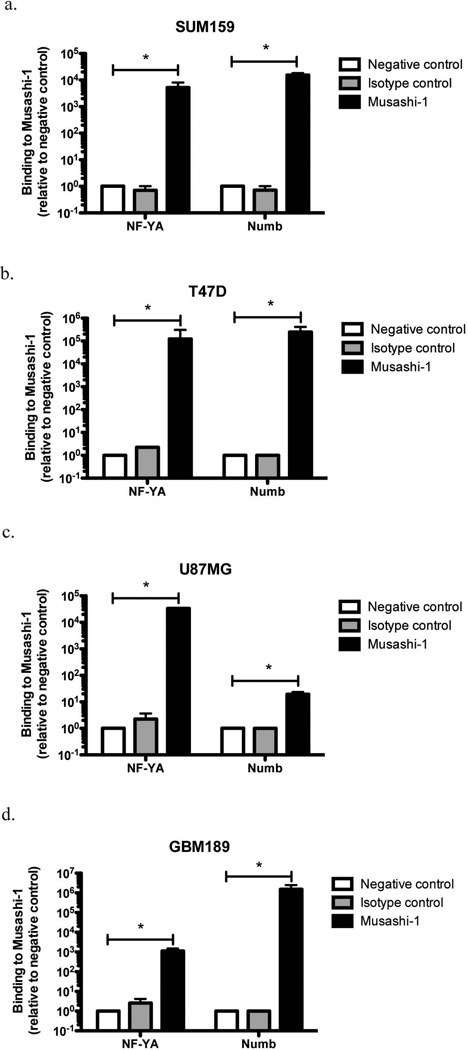
To test if Musashi-1 actually binds NF-YA mRNAs in cells, we immunoprecipitated Musashi-1 from breast cancer and glioma cell lysates using established breast cancer (SUM159PT, T47D) and U87MG glioma cell lines and the primary glioma patient line GBM189 and subjected the co-precipitated mRNA to two rounds of qRT-PCR using specific primers for NF-YA mRNA. We used specific primers for Numb as a positive control and an isotype control antibody and a precipitation without antibodies as negative controls. Consistent with our in-silico analysis, mRNA of NF-YA co-precipitated with Musashi-1 and binding levels were comparable to binding levels of Numb mRNA (Figure 3).
Figure 3. Identification of NF-YA mRNA as a specific target for Musashi1.
Immunoprecitation of total cellular lysates from (a) SUM159PT, (b) T47D, (c) U87MG, and (d) GBM189 cells. Co-precipitated mRNA was extracted, reverse transcribed and amplified using specific primers for human NF-YA (black bars). Numb mRNA amplification was used as a positive control. Immuoprecipitations without antibody (white bars) and isotype control antibody (grey bars) were used as negative controls. NF-YA mRNA could be consistently co-precipitated with Musashi-1 in breast and glioma samples. Normalized means ± SEM from 3 independent experiments are shown.
NF-YA DNA-binding activity is down-regulated in cancer stem cells
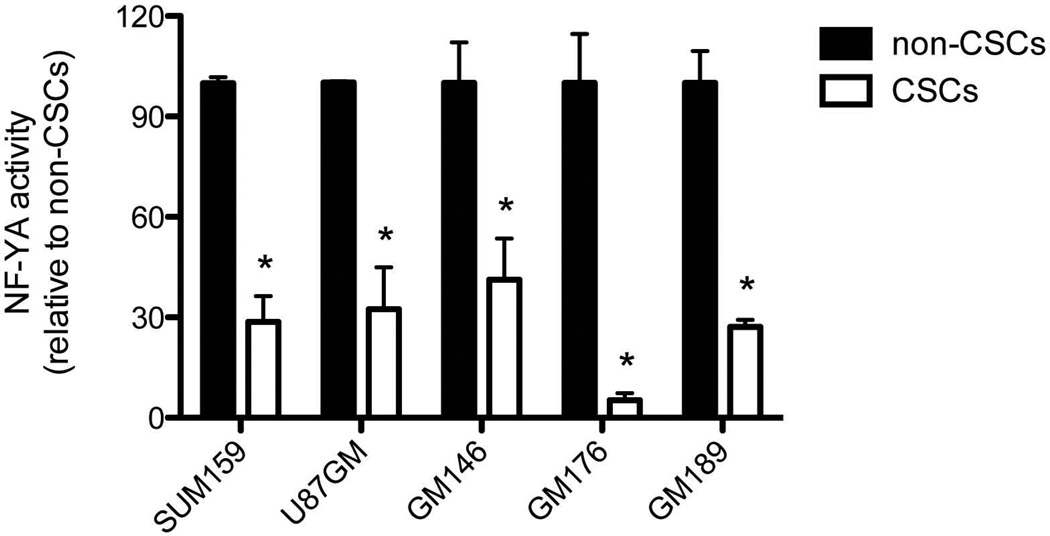
In order to compare the DNA-binding activity of NF-YA in CSCs/tumor-initiating cells and non-tumorigenic cells, we sorted SUM159PT-ZsGreen-cODC and U87MG-ZsGreen-cODC cells as well as cells from three primary glioma patient lines (GBM146-ZsGreen-cODC, GBM176-ZsGreen-cODC, GBM189-ZsGreen-cODC) based on proteasome function. Nuclear protein extracts of CSCs/tumor-initiating cells and non-tumorigenic cells of both lines were analyzed using a NF-YA-specific transcription factor ELISA-assay. The specificity of the assay was confirmed in control experiments in which unbound oligonucleotide containing either the consensus binding motif or a mutated binding motif was added to the reaction in excess (Suppl. Fig 3a).
CSCs/Tumor-initiating cells with low proteasome activity (ZsGreen-cODC+) showed significantly reduced DNA-binding activity of NF-YA (SUM159PT-ZsGreen-cODC+ 28.65 ± 7.7%, n=6, p<0.0001; U87MG-ZGreen-cODC+ 32.5 ± 12.4%, n=4, p=0.0016; GBM146-ZsGreen-cODC+ 41.3 ± 12.3 %, n=6, p=0.0066; GBM176-ZsGreen-cODC+ 5.3 ± 2.1 %, n=6, p<0.0001; GBM-189-ZsGreen-cODC+ 27.1 ± 2.2 %, n=6, p<0.0001; two-sided Student’s t-test (Figure 4). Western blotting using extracts from SUM159PT non-tumorigenic cells and CSCs/tumor-initiating cells confirmed down-regulation of nuclear NF-YA protein in CSCs/tumor-initiating cells (Supplementary Figure 3b).
Figure 4. Activity of the transcription factor NF-YA in non-tumorigenic cells and CSCs.
Nuclear extracts from sorted SUM159PT, U87MG, GBM146, GBM176, and GBM189 non-tumorigenic cells and CSCs were prepared and DNA-binding activity of NF-YA was assessed using the competitive TransAM® Transcription Factor ELISA. DNA-binding activity of NF-YA in CSCs was significantly down-reglated. SUM159PT CSCs 28.65 ± 7.7%, n=6, p<0.0001; U87MG CSCs 32.5 ± 12.4%, n=4, p=0.0016; GBM146 CSCs 41.3 ± 12.3 %, n=6, p=0.0066; GBM176 CSCs 5.3 ± 2.1 %, n=6, p<0.0001; GBM189 CSCs 27.1 ± 2.2 %, n=6, p<0.0001. Normalized means ± SEM from 3 independent experiments are shown.
Down-regulation of Musashi-1 increases 26S proteasome activity and decreases self-renewal capacity
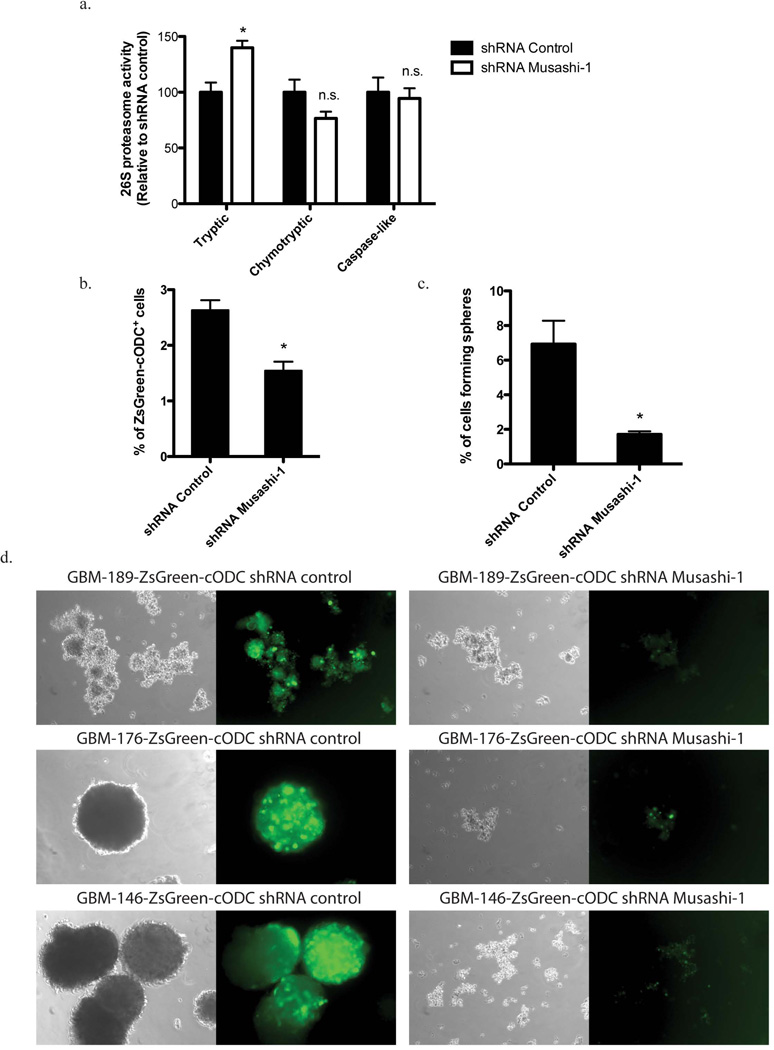
Next we sought to study if down-regulation of Musashi-1 expression would alter the cleavage activity of the 26S proteasome. SUM159PT breast cancer cells were stably transfected with expression constructs for shRNA against Musashi-1 or a scrambled control shRNA, and cultured as monolayer cultures or enriched for breast CSCs/tumor-initiating cells by culturing them as mammospheres. Down-regulation of Msuashi-1 protein shRNA-Musashi-1 was verified by Western blot (Supplementary Figure 4). For assessment of proteasome function, cells were harvested, lysed, and all three cleavage activities of the 26S proteasome were measured using a fluorogenic peptide assay. Suppression of Musashi-1 expression significantly increased the tryptic activity of the 26S proteasome in monolayers and mammospheres (Figure 5a and Suppl. Figure 5) while the chymotryptic and caspase-like activities were not affected. (Tryptic activity shRNA-Msi1: 139.9 +/-8.677%, p=0.0027; Chymotrypic activity shRNA-Msi1: 76.54 +/-11.34%, p=0.0809; Capase-like activity shRNA-Msi1: 94.48 +/-9.072%, p=0.7331).
Figure 5. Influence of Musashi1 expression on proteasome activity and the CSCs phenotype.
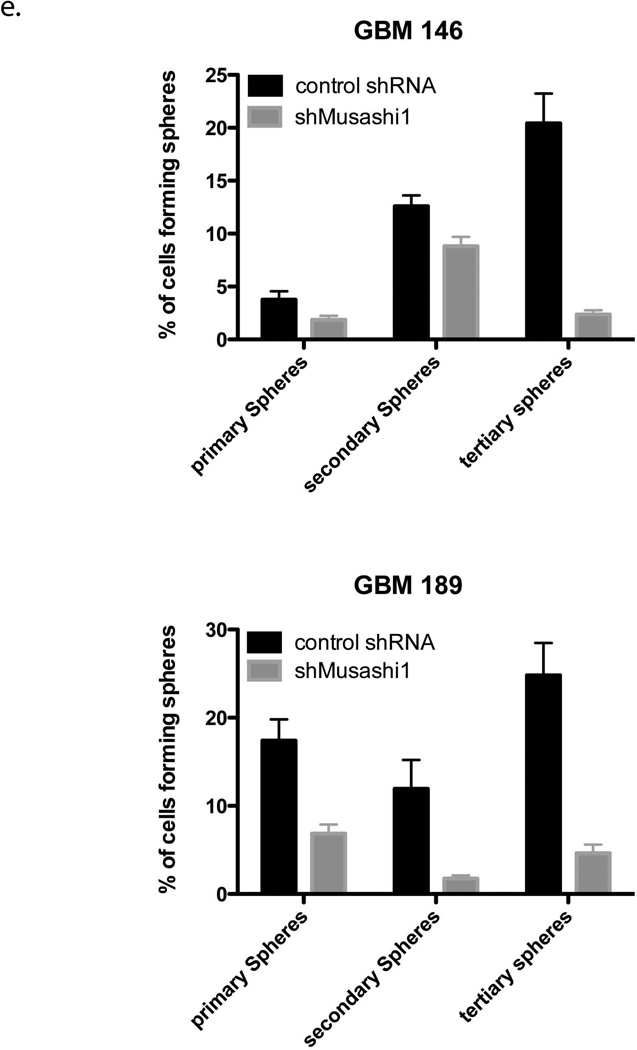
(a) 26S Proteasome function was assessed in total cellular lysates from SUM159PT cells transfected with control shRNA or shRNA targeteing Musashi-1. Suppression of Musashi-1 expression using shRNA significantly increased the tryptic cleavage activity of the 26S proteasome. Tryptic activity shRNA-Msi1: 139.9 ± 8.677%, p=0.0027; Chymotrypic activity shRNA-Msi1: 76.54 ± 11.34%, p=0.0809; Caspase-like activity shRNA-Msi1: 94.48 ± 9.072%, p=0.7331. (b) Flow cytometry analysis of SUM159PT cells. Suppression of Musashi-1 expression using shRNA decreases the number ZsGreen-cODC+ cells. (c) Primary, secondary, and tertiary sphere formation in SUM159PT cells. Suppression of Musashi-1 expression using shRNA significantly reduces self-renewal capacity in primary (170.2 ± 20.67 [n=5] vs. 37 ± 3.27 [n=4], p<0.0001, mean ± SD) and secondary spheres (368 ± 16.02 [n=4] vs. 206.25 ± 23.34, p<0.0001, mean ± SD). (d) Sphere formation assay in primary patient-derived GBM lines. Suppression of Musashi-1 expression reduces the number of ZsGreen-cODC+ cells and abrogates self-renewal capacity. (e) Primary, secondary, and tertiary sphere formation in GBM146 and GBM189 patient-derived specimen after transfection with shRNA against Musashi-1. Los of Musashi-1 expression reduced the sphere-forming capacity. Normalized means ± SEM (a), means ± SEM (b) from 3 independent experiments are shown. (c and e) means ± SD from 5 independent experiments are shown.
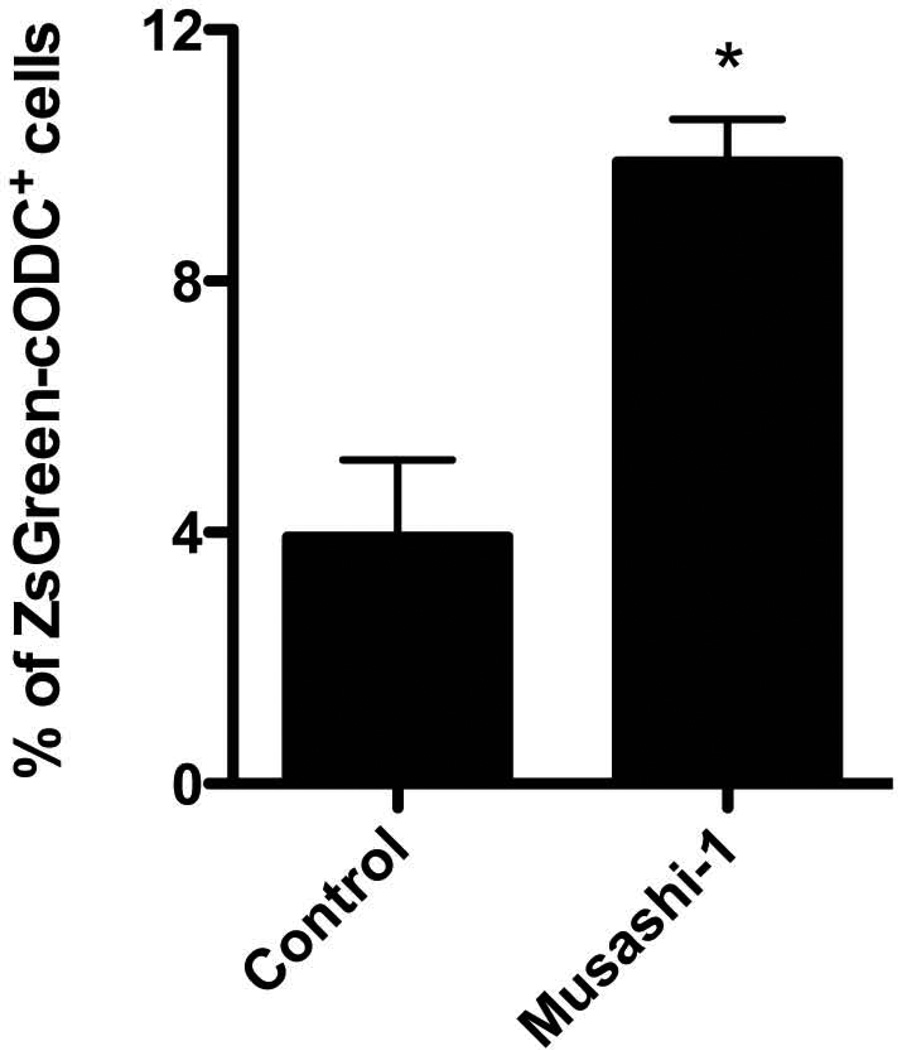
Assessment of mammosphere or neurosphere formation from breast cancer or glioma cells allows for the estimation of the self-renewal capacity of CSCs/tumor-initiating cells. When SUM159PT cells were transfected with shRNA against Musashi-1, the number of ZsGreen-cODC+ CSCs/tumor-initiating cells, as well as the mammosphere forming capacity significantly decreased (Figure 5b and c). To further test the effect of Musashi-1 on the CSCs/tumor-initiating cells population we suppressed Musashi-1 expression in three primary glioma samples derived from 3 different patients, and observed a loss of neurosphere formation and a reduction in the number of ZsGreen-cODC+ CSCs/tumor-initiating cells (Figure 5d). Like in breast cancer, transfection with shRNA against Musashi-1 lead to a loss in primary, secondary and tertiary sphere-forming capacity (Figure 5e). Conversely, when SUM159PT-ZsGren-cODC cells were infected with an expression construct for Musashi-1, the number of CSCs/tumor-initiating cells with low proteasome activity (ZsGreen-cODC+) increased significantly from 3.93 ± 1.23 % to 9.90 ± 0.68% (p=0.013 Student’s t-test, n=3) (Figure 6).
Figure 6. Effect of Musashi1 overexpression on cells with low proteasome activity.
Overexpression of Musashi-1 in SUM159PT cells causes a significant increase in the number of ZsGreen-cODC+ cells from 3.93 ± 1.23 % in control cultures to 9.90 ± 0.68% in Musashi-1 overexpression cells (p=0.013 Student’s t-test, n=3). Means ± SEM from 3 independent experiments are shown.
Discussion
Maintenance of the CSC/tumor-initiating cells state requires activation and maintenance of cellular pathways that differ from those required in transiently amplifying cells or differentiated progeny. CSCs/tumor-initiating cells are thought to be mostly quiescent 6 and thus do not constantly need to double their cellular mass during mitosis. Also, quiescent cells have been shown to have an altered turnover of proteins with lysosomal rather than proteasome-dependent proteolysis as the predominant type of protein degradation 8, 9. While inhibition of proteasome function usually leads to death of tumor cells 28, it seems safe to assume that the 26S proteasome, the cellular machinery responsible for protein degradation in signaling and protein quality control in growing cells 11, can be down-regulated during quiescence without affecting the integrity of the cells. More importantly, down-regulation of the 26S proteasome, which requires considerable amounts of ATP for its assembly, maintenance, and function 29, could increase the metabolic efficiency of quiescent cells. Differences in RNA quality were unlikely the cause for the lower proteasome subunit mRNA expression levels in SUM159PT-ZsGreen-cODC+ CSCs/tumor-initiating cells since expression levels of the ribosomal subunit mRNA RPLP0 did not differ between the two cell populations, RPLP0 expression levels were used to normalized the data, and Musashi-1 mRNA levels were more than 5,000-fold higher in SUM159PT-ZsGreen-cODC+ CSCs/tumor-initiating cells (Fig 2B). Furthermore, we had previously reported that SUM159PT-ZsGreen-cODC+ CSCs/tumor-initiating cells have up-regulated mRNA levels for a large number of stem cell-related genes (Fig. 4 and Suppl. 4 in 30).
We had recently described the lack of 26S proteasome function in breast and glioma CSCs/tumor-initiating cells 5, 6 and others reported similar results in non-small cell lung cancer 31 and pancreatic cancer 32. Furthermore, in breast cancer 33 and glioma patients 7, low levels of proteasome subunit expression in tumors correlates with reduced relapse-free survival and overall survival, respectively.
We, and others have previously proposed that proteasome inhibitors could be a valuable novel class of anticancer drugs. This was based on the observation that drugs, such as MG-132 or bortezomib efficiently eliminated a broad spectrum of solid tumors in vitro and in vivo 28, 34–36 and that established cancer therapies target the proteasome 37–39. Furthermore, proteasome function in malignant cells was found to be up-regulated when compared to normal cells of the same tissue of origin, thus indicating a possible therapeutic window for proteasome inhibitors in the clinic. However, a large number of clinical trials have shown that bortezomib, alone or in combination with radiation or chemotherapy has very little -if any- clinical anticancer effect in solid tumors but instead induces considerable normal tissue toxicity 40–45.
Our observation that low proteasome subunit expression and low proteasome activity are general phenomena in CSCs/tumor-initiating cells from breast cancer and glioma and the apparent resistance of CSCs/tumor-initiating cells to proteasome inhibitors 5 may offer an explanation for these clinical observations. However, it was unclear how the expression of proteasomes and their proteolytic activity were down-regulated in CSCs/tumor-initiating cells and how this related to the CSC phenotype. Therefore, we sought to search for a possible link between developmental pathways activated in CSCs/tumor-initiating cells and the transcriptional regulation of proteasome subunit expression. The regulatory pathways underlying proteasome subunit expression are incompletely understood, but it appears that in mammalian cells proteasome subunit genes are regulated as a gene cluster 46 under the control of the transcription factor NF-YA 23, thereby maintaining the stoichiometry of all the subunits required to assemble a functional 26S proteasome.
Among other developmental signaling pathways the Notch pathway is known to facilitate self-renewal in breast cancer and glioma stem cells 19–21. Upon binding of its ligands Notch receptors undergo three cleavage steps, which finally lead to nuclear translocation of the intracellular domain of the receptor (Notch-ICD) where it binds to CBF-1, thereby turning CBF-1 from a transcriptional repressor into a transcriptional activator. Notch-ICD itself is inhibited by binding to Numb, which leads to ubiquitination of Notch-ICD, and subsequent targeting for proteasomal degradation. Numb expression levels on the other hand are regulated by binding of Musashi-1 to the 3’-UTR of Numb mRNA, thereby preventing its translation 47. The RNA binding protein Musashi-1 was first described in the context of the Notch signaling pathway 47. But subsequently it became clear that Musashi-1 also binds to the 3’-UTR of other mRNAs, including those coding for p21WAF-1 48. Using a systematic search for Musashi-1 targets de Sousa Abreu et al. recently reported 64 additional mRNAs involved in cell proliferation, differentiation, cell cycle control, apoptosis, and protein modification to be controlled by Musashi-1 49. However, de Sousa Abreu et al. did not identify NF-YA mRNA as a Musashi-1 target.
In our study we used a more direct strategy utilizing a monoclonal antibody against Musashi-1 for immunoprecipitation. Subsequent quantitative RT-PCR analysis of precipitates from glioma and breast cancer cells consistently amplified NF-YA mRNA (Figure 3), which codes for the regulatory subunit of the trimeric master regulatory transcription factor of proteasome subunit expression, NF-YA 23. Therefore, its binding to Musashi-1 agrees with our finding that nuclear extracts from ZsGreen-cODC+ breast and glioma CSCs/tumor-initiating cells with low proteasome activity showed reduced DNA-binding activity of NF-YA (Figure 4a).
The role of NF-YA in the context of stem cells has so far mostly been studied in the hematopoietic system. Interestingly, NF-YA was shown to be critical for the expansion of Lin−/Sca+/c-Kit+ hematopoietic stem cells (HSCs) but was dispensable in quiescent HSCs 50. This was consistent with our previous study in which we reported that more than 20% of breast cancer stem cells with low proteasome activity were indeed quiescent 6. Likewise, in glioma, a comparable number of glioma stem cells stained negative for Ki67 5. Similar to HSCs, this subpopulation of cells may not rely on NF-YA activity, however the increase in activity of this transcription factor is required during early differentiation.
An early gain in 20S proteasome function has been described for embryonic stem cells 51 in which the cells substantially increase the activity of the proteasome core particle upon differentiation to remove oxidized proteins from the cell mass. However, in this study 51 26S proteasome activity was not affected, and ES cells seem to rely on a different isoform of NF-YA 52.
Conclusion and summary
Taken together we propose a circuitry between the already known relationship between the Notch signaling pathway and Musashi-1 on one hand and the NF-YA and the regulation of the proteasome on the other hand that supports maintenance of the cancer stem cell state. In CSCs/tumor-initiating cells, which rely on Notch signaling, the stem cell marker Musashi-1 is expressed at high levels. In addiation to support maintenance of the stem cell phenotype via the Notch pathway, Musashi-1 overexpression in CSCs/tumor-initiating cells also down-regulates proteasome activity, via inhibition of NF-YA, a master regulatory hub for proteasome subunit expression. As a consequence, proteasome levels and activity, which require large amounts of ATP for its assembly and function 29 are low in CSCs/tumor-initiating cells, thus making CSCs/tumor-initiating cells more energy efficient. Finally, our results may explain why proteasome inhibition in solid cancers leads to excellent experimental responses of bulk tumor cell population but spares CSCs/tumor-initiating cells, thus limiting the clinical efficiency of this class of drugs.
Supplementary Material
Supplementary Figure 1. (a) Color-code of the putative binding sequences of Musashi1. (b) Full-length mRNA sequence of Numb annotated for 5’UTR, coding sequence, 3’UTR and localization of Musashi1 binding sites.
Supplementary Figure 2. Full-length mRNA sequence of NF-YA annotated for 5’UTR, coding sequence, 3’UTR and localization of Musashi1 binding sites.
Supplementary Figure 3. (a) DNA-binding competitive activity analysis of NF-YA in SUM159PT, U87MG, GBM146, GBM176 and GBM189. White bar: NF-YA activity control; grey bar: NF-YA activity with mutant competitive NF-YA primer; black bar: NF-YA activity with wild-type competitive NF-YA primer. The specific binding of the wild-type primer on NF-YA binding site significantly prevents NF-YA binding activity. (b) Western Blot analysis of protein expression level of NF-YA in sorted SUM159PT non-CSCs and CSCs. Consistently with Musashi-1 binding NF-YA mRNA and NF-YA activity, the level of NF-YA protein is down-regulated in SUM159PT CSCs.
Supplementary Figure 4. Western Blot analysis Musashi-1 expression in SUM159PT shRNA control and shRNA targeting Musashi-1.
Supplementary Figure 5. Proteolytic activity of the 26S proteasome in SUM159PT cells, cultured as monolayer or mammospheres, before and after transfection with shRNA against Musashi1 or a scrambled control shRNA.
Acknowledgments
Grant Support: FP was supported by a generous gift from Steve and Cathy Fink and grants from the National Cancer Institute (RO1CA137110, 1R01CA161294) and the Army Medical Research & Materiel Command’s Breast Cancer Research Program (W81XWH-11-1-0531).
Footnotes
Authors’ contribution
Chann Lagadec: Collection and assembly of data, Data analysis and interpretation, Manuscript writing
Erina Vlashi: Data analysis and interpretation, Manuscript writing
Patricia Frohnen: Collection and assembly of data
Yazeed Alhiyari: Collection and assembly of data
Mabel Chan: Collection and assembly of data
Frank Pajonk: Conception and design, Data analysis and interpretation, Manuscript writing, Final approval of manuscript
References
- 1.Reya T, Morrison SJ, Clarke MF, et al. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 2.Al-Hajj M, Wicha MS, Benito-Hernandez A, et al. Prospective identification of tumorigenic breast cancer cells. Proc. Natl Acad. Sci. USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hemmati HD, Nakano I, Lazareff JA, et al. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci U S A. 2003;100:15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 Is a Marker of Normal and Malignant Human Mammary Stem Cells and a Predictor of Poor Clinical Outcome. Cell stem cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vlashi E, Kim K, Lagadec C, et al. In vivo imaging, tracking, and targeting of cancer stem cells. J Natl Cancer Inst. 2009;101:350–359. doi: 10.1093/jnci/djn509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lagadec C, Vlashi E, Della Donna L, et al. Survival and self-renewing capacity of breast cancer initiating cells during fractionated radiation treatment. Breast Cancer Res. 2010;12:R13. doi: 10.1186/bcr2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vlashi E, Lagadec C, Vergnes L, et al. Metabolic state of glioma stem cells and nontumorigenic cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:16062–16067. doi: 10.1073/pnas.1106704108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lockwood TD, Minassian IA. Protein turnover and proliferation. Failure of SV-3T3 cells to increase lysosomal proteinases, increase protein degradation and cease net protein accumulation. Biochem J. 1982;206:251–258. doi: 10.1042/bj2060251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lockwood TD, Minassian IA, Roux L. Protein turnover and proliferation. Turnover kinetics associated with the elevation of 3T3-cell acid-proteinase activity and cessation of net protein gain. Biochem J. 1982;206:239–249. doi: 10.1042/bj2060239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Babbitt SE, Kiss A, Deffenbaugh AE, et al. ATP hydrolysis-dependent disassembly of the 26S proteasome is part of the catalytic cycle. Cell. 2005;121:553–565. doi: 10.1016/j.cell.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 11.Pajonk F, McBride WH. The Proteasome in Cancer Biology and Treatment. Radiat Res. 2001;156:447–459. doi: 10.1667/0033-7587(2001)156[0447:tpicba]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 12.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Chen ZY, Wang X, Zhou Y, et al. Destabilization of Kruppel-like factor 4 protein in response to serum stimulation involves the ubiquitin-proteasome pathway. Cancer Res. 2005;65:10394–10400. doi: 10.1158/0008-5472.CAN-05-2059. [DOI] [PubMed] [Google Scholar]
- 14.Baltus GA, Kowalski MP, Zhai H, et al. Acetylation of sox2 induces its nuclear export in embryonic stem cells. Stem Cells. 2009;27:2175–2184. doi: 10.1002/stem.168. [DOI] [PubMed] [Google Scholar]
- 15.Moretto-Zita M, Jin H, Shen Z, et al. Phosphorylation stabilizes Nanog by promoting its interaction with Pin1. Proc Natl Acad Sci U S A. 2010;107:13312–13317. doi: 10.1073/pnas.1005847107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aberle H, Bauer A, Stappert J, et al. beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oberg C, Li J, Pauley A, et al. The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. J Biol Chem. 2001;276:35847–35853. doi: 10.1074/jbc.M103992200. [DOI] [PubMed] [Google Scholar]
- 18.Huntzicker EG, Estay IS, Zhen H, et al. Dual degradation signals control Gli protein stability and tumor formation. Genes Dev. 2006;20:276–281. doi: 10.1101/gad.1380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dontu G, Al-Hajj M, Abdallah WM, et al. Stem cells in normal breast development and breast cancer. Cell Prolif. 2003;36 Suppl 1:59–72. doi: 10.1046/j.1365-2184.36.s.1.6.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu YY, Zheng MH, Cheng G, et al. Notch signaling contributes to the maintenance of both normal neural stem cells and patient-derived glioma stem cells. BMC Cancer. 2011;11:82. doi: 10.1186/1471-2407-11-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dontu G, Jackson KW, McNicholas E, et al. Role of Notch signaling in cell-fate determination of human mammary stem/progenitor cells. Breast Cancer Res. 2004;6:R605–R615. doi: 10.1186/bcr920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weinmaster G. Notch signal transduction: a real rip and more. Curr Opin Genet Dev. 2000;10:363–369. doi: 10.1016/s0959-437x(00)00097-6. [DOI] [PubMed] [Google Scholar]
- 23.Xu H, Fu J, Ha SW, et al. The CCAAT box-binding transcription factor NF-Y regulates basal expression of human proteasome genes. Biochim Biophys Acta. 2012;1823:818–825. doi: 10.1016/j.bbamcr.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 24.Lagadec C, Vlashi E, Della Donna L, et al. Radiation-induced Reprograming of Breast Cancer Cells. Stem Cells. 2012 doi: 10.1002/stem.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacNicol MC, Cragle CE, MacNicol AM. Context-dependent regulation of Musashi-mediated mRNA translation and cell cycle regulation. Cell Cycle. 2011;10:39–44. doi: 10.4161/cc.10.1.14388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glazer RI, Wang XY, Yuan H, et al. Musashi1: a stem cell marker no longer in search of a function. Cell Cycle. 2008;7:2635–2639. doi: 10.4161/cc.7.17.6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohyama T, Nagata T, Tsuda K, et al. Structure of Musashi1 in a complex with target RNA: the role of aromatic stacking interactions. Nucleic Acids Res. 2012;40:3218–3231. doi: 10.1093/nar/gkr1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pajonk F, Pajonk K, McBride WH. Apoptosis and radiosensitization of hodgkin cells by proteasome inhibition. Int J Radiat Oncol Biol Phys. 2000;47:1025–1032. doi: 10.1016/s0360-3016(00)00516-2. [DOI] [PubMed] [Google Scholar]
- 29.Liu CW, Li X, Thompson D, et al. ATP binding and ATP hydrolysis play distinct roles in the function of 26S proteasome. Mol Cell. 2006;24:39–50. doi: 10.1016/j.molcel.2006.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lagadec C, Vlashi E, Della Donna L, et al. Radiation-induced reprogramming of breast cancer cells. Stem Cells. 2012;30:833–844. doi: 10.1002/stem.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan J, Zhang Q, Wang Y, et al. 26S proteasome activity is down-regulated in lung cancer stem-like cells propagated in vitro. PLoS One. 2010;5:e13298. doi: 10.1371/journal.pone.0013298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adikrisna R, Tanaka S, Muramatsu S, et al. Identification of pancreatic cancer stem cells and selective toxicity of chemotherapeutic agents. Gastroenterology. 2012;143:234–245. e237. doi: 10.1053/j.gastro.2012.03.054. [DOI] [PubMed] [Google Scholar]
- 33.Elfadl D, Hodgkinson VC, Long ED, et al. A pilot study to investigate the role of the 26S proteasome in radiotherapy resistance and loco-regional recurrence following breast conserving therapy for early breast cancer. Breast. 2011;20:334–337. doi: 10.1016/j.breast.2011.02.017. [DOI] [PubMed] [Google Scholar]
- 34.Pajonk F, van Ophoven A, Weissenberger C, et al. The proteasome inhibitor MG-132 sensitizes PC-3 prostate cancer cells to ionizing radiation by a DNA-PK-independent mechanism. BMC Cancer. 2005;5:76. doi: 10.1186/1471-2407-5-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pervan M, Pajonk F, Sun J-R, et al. The proteasome inhibitor PS-341 is a potential radiosensitizer. Proceedings of the American Association for Cancer Research Annual Meeting. 2001;42:666–667. [Google Scholar]
- 36.Pajonk F, Himmelsbach J, Riess K, et al. The human immunodeficiency virus (HIV)-1 protease inhibitor saquinavir inhibits proteasome function and causes apoptosis and radiosensitization in non-HIV-associated human cancer cells. Cancer Res. 2002;62:5230–5235. [PubMed] [Google Scholar]
- 37.Pajonk F, McBride WH. Ionizing radiation affects 26s proteasome function and associated molecular responses, even at low doses. Radiother Oncol. 2001;59:203–212. doi: 10.1016/s0167-8140(01)00311-5. [DOI] [PubMed] [Google Scholar]
- 38.Pajonk F, van Ophoven A, McBride WH. Hyperthermia-induced proteasome inhibition and loss of androgen receptor expression in human prostate cancer cells. Cancer Res. 2005;65:4836–4843. doi: 10.1158/0008-5472.CAN-03-2749. [DOI] [PubMed] [Google Scholar]
- 39.Fekete MR, McBride WH, Pajonk F. Anthracyclines, proteasome activity and mult-drug-resistance. BMC Cancer. 2005;5:114. doi: 10.1186/1471-2407-5-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trinh XB, Sas L, Van Laere SJ, et al. A phase II study of the combination of endocrine treatment and bortezomib in patients with endocrine-resistant metastatic breast cancer. Oncol Rep. 2012;27:657–663. doi: 10.3892/or.2011.1562. [DOI] [PubMed] [Google Scholar]
- 41.Irvin WJ, Jr, Orlowski RZ, Chiu WK, et al. Phase II study of bortezomib and pegylated liposomal doxorubicin in the treatment of metastatic breast cancer. Clin Breast Cancer. 2010;10:465–470. doi: 10.3816/CBC.2010.n.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lilenbaum R, Wang X, Gu L, et al. Randomized phase II trial of docetaxel plus cetuximab or docetaxel plus bortezomib in patients with advanced non-small-cell lung cancer and a performance status of 2: CALGB 30402. J Clin Oncol. 2009;27:4487–4491. doi: 10.1200/JCO.2009.22.7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Friday BB, Anderson SK, Buckner J, et al. Phase II trial of vorinostat in combination with bortezomib in recurrent glioblastoma: a north central cancer treatment group study. Neuro Oncol. 2012;14:215–221. doi: 10.1093/neuonc/nor198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kraft AS, Garrett-Mayer E, Wahlquist AE, et al. Combination therapy of recurrent prostate cancer with the proteasome inhibitor bortezomib plus hormone blockade. Cancer Biol Ther. 2011;12:119–124. doi: 10.4161/cbt.12.2.15723. [DOI] [PubMed] [Google Scholar]
- 45.O'Neil BH, Raftery L, Calvo BF, et al. A phase I study of bortezomib in combination with standard 5-fluorouracil and external-beam radiation therapy for the treatment of locally advanced or metastatic rectal cancer. Clin Colorectal Cancer. 2010;9:119–125. doi: 10.3816/CCC.2010.n.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sato Y, Sakamoto K, Sei M, et al. Proteasome subunits are regulated and expressed in comparable concentrations as a functional cluster. Biochem Biophys Res Commun. 2009;378:795–798. doi: 10.1016/j.bbrc.2008.11.125. [DOI] [PubMed] [Google Scholar]
- 47.Kurihara Y, Nagata T, Imai T, et al. Structural properties and RNA-binding activities of two RNA recognition motifs of a mouse neural RNA-binding protein, mouse-Musashi-1. Gene. 1997;186:21–27. doi: 10.1016/s0378-1119(96)00673-7. [DOI] [PubMed] [Google Scholar]
- 48.Battelli C, Nikopoulos GN, Mitchell JG, et al. The RNA-binding protein Musashi-1 regulates neural development through the translational repression of p21WAF-1. Mol Cell Neurosci. 2006;31:85–96. doi: 10.1016/j.mcn.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 49.de Sousa Abreu R, Sanchez-Diaz PC, Vogel C, et al. Genomic analyses of musashi1 downstream targets show a strong association with cancer-related processes. The Journal of biological chemistry. 2009;284:12125–12135. doi: 10.1074/jbc.M809605200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bungartz G, Land H, Scadden DT, et al. NF-Y is necessary for hematopoietic stem cell proliferation and survival. Blood. 2012;119:1380–1389. doi: 10.1182/blood-2011-06-359406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hernebring M, Brolen G, Aguilaniu H, et al. Elimination of damaged proteins during differentiation of embryonic stem cells. Proc Natl Acad Sci U S A. 2006;103:7700–7705. doi: 10.1073/pnas.0510944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fluchter P, Muller V, Pajonk FG. [Suicidality: procedure in emergency cases] Medizinische Klinik, Intensivmedizin und Notfallmedizin. 2012;107:469–475. doi: 10.1007/s00063-012-0123-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. (a) Color-code of the putative binding sequences of Musashi1. (b) Full-length mRNA sequence of Numb annotated for 5’UTR, coding sequence, 3’UTR and localization of Musashi1 binding sites.
Supplementary Figure 2. Full-length mRNA sequence of NF-YA annotated for 5’UTR, coding sequence, 3’UTR and localization of Musashi1 binding sites.
Supplementary Figure 3. (a) DNA-binding competitive activity analysis of NF-YA in SUM159PT, U87MG, GBM146, GBM176 and GBM189. White bar: NF-YA activity control; grey bar: NF-YA activity with mutant competitive NF-YA primer; black bar: NF-YA activity with wild-type competitive NF-YA primer. The specific binding of the wild-type primer on NF-YA binding site significantly prevents NF-YA binding activity. (b) Western Blot analysis of protein expression level of NF-YA in sorted SUM159PT non-CSCs and CSCs. Consistently with Musashi-1 binding NF-YA mRNA and NF-YA activity, the level of NF-YA protein is down-regulated in SUM159PT CSCs.
Supplementary Figure 4. Western Blot analysis Musashi-1 expression in SUM159PT shRNA control and shRNA targeting Musashi-1.
Supplementary Figure 5. Proteolytic activity of the 26S proteasome in SUM159PT cells, cultured as monolayer or mammospheres, before and after transfection with shRNA against Musashi1 or a scrambled control shRNA.