Abstract
Homologous recombination is an important pathway for error-free repair of DNA lesions, such as single-and double-strand breaks, and for rescue of collapsed replication forks. Here, we describe protocols for live cell imaging of single-lesion recombination events in the yeast Saccharomyces cerevisiae using fluorescence microscopy.
Keywords: Homologous recombination, fluorescence microscopy, DNA damage, DNA double-strand break repair
1. Introduction
In the budding yeast Saccharomyces cerevisiae, homologous recombination (HR) is catalyzed by proteins encoded by the RAD52 epistasis group of genes including RAD50-59, XRS2, MRE11, and RFA1-3 (1). However, many additional proteins regulate and coordinate HR according to the molecular nature of the DNA lesion, cell cycle, and developmental phase. Most of these proteins are expressed constitutively, but their copy number per cell varies greatly from <50 molecules of Tel1 to >5,000 molecules of Rfa1. During HR, these proteins are assembled in a coordinated manner into dynamic giga-Dalton complexes at the site of the DNA lesion (2, 3) (Fig. 30.1a). These assemblies of high local concentration of HR and other DNA damage response proteins appear as cytological foci (Fig. 30.1b). Remarkably, the appearance of DNA damage-induced foci is highly conserved from yeast to human (4). Therefore, insight into the molecular principles that govern these DNA repair factories in yeast is likely to be extendible to higher eukaryotes. For single-lesion analysis, yeast has the advantage over higher eukaryotes that its relatively small genome experiences fewer spontaneous DNA lesions per cell. As a consequence, it is easier to discern the signal from a single specific lesion from the background of spontaneous random lesions.
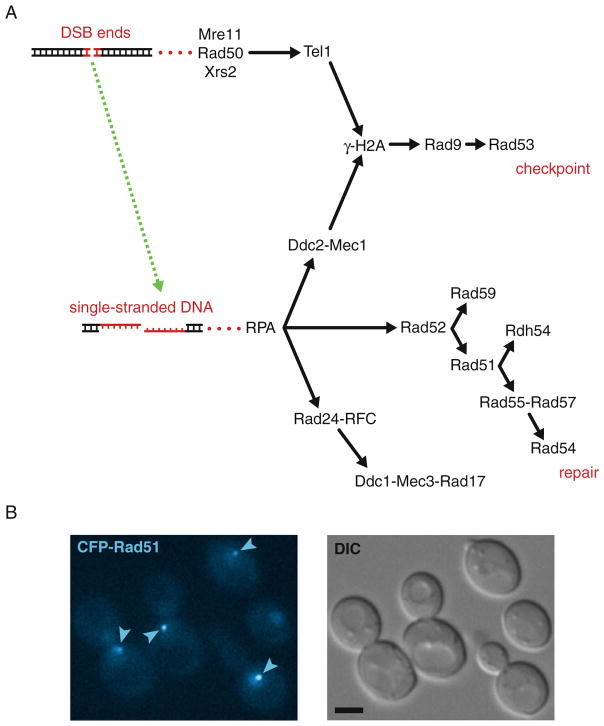
Fig. 30.1.
Assembly of checkpoint and recombination proteins in response to DNA double-strand breaks. (a) Sequential assembly of cytological foci. Arrows indicate the sequential assembly of DNA repair proteins at a DNA double-strand break as described (20). DSB, double-strand break. (b) Formation of Rad51 foci in response to DNA damage. Exponentially growing cells (strain ML494-15C) expressing CFP-Rad51 from the endogenous locus were examined for Rad51 foci. In brief, cells were grown by shaking in liquid SC medium containing 100 μg/ml adenine at 25°C to an OD600 of 0.2–0.3 before addition of 200 μg/ml zeocin. After continued shaking for 1 h, cells were harvested by centrifugation at 3,000 rpm and processed for fluorescence microscopy as described (Section 3). The CFP fluorophore was visualized on a Zeiss AxioImager Z1 wide-field microscope using a Zeiss Plan-Apo 100×/1.40 objective (Carl Zeiss, Jena, Germany), a band-pass CFP filter set from Chroma (Brattleboro, VT), and an ORCA C4742-95-12ER CCD camera (Hamamatsu, Japan). Images were acquired using Volocity software (Improvision, Coventry, UK). DIC, differential interference contrast. Arrows indicate Rad51 foci. Scale bar, 3 μm.
2. Materials
10× Pfu reaction buffer: 200 mM Tris–HCl (pH 8.8 at 25°C), 100 mM (NH4)2SO4, 100 mM KCl, 1% (v/v) Triton X-100, 1 mg/ml BSA, and 20 mM MgSO4.
20× PBS (phosphate buffered saline): 160 g NaCl, 4 g KCl, 28.8 g NA2HPO4, 4.8 g KH2PO4 in 1,000 ml. Adjust pH to 7.4 at 25°C with either HCl or NaOH as appropriate.
YPD (yeast extract peptone dextrose) liquid medium: 10 g yeast extract, 20 g peptone, 20 g glucose in 1,000 ml. Autoclave to sterilize.
SC (synthetic complete) powder: 100 g yeast nitrogen base without amino acids and without NH4SO4 (MP Biomedicals, Solon, OH), 293.11 g NH4SO4, 1.2 g adenine sulfate, 1.2 g L-arginine sulfate, 1.2 g L-histidine-HCl, 1.8 g L-isoleucine, 3.6 g L-leucine, 1.8 g L-lysine-HCl, 1.2 g L-methionine, 3 g L-phenylalanine, 1.2 g L-tryptophan, 1.8 g L-tyrosine, 1.2 g uracil, 9 g L-valine (5). Grind in ball mill overnight.
SC liquid medium: 7.25 g SC powder and 20 g glucose in 1,000 ml. Adjust pH to 5.8 with NaOH/HCl.
5-FOA (5-fluoroorotic acid) solid medium: Autoclave solution I (12 g agar in 255 ml H2O). Filter sterilize solution II (4.35 g SC powder, 18 mg uracil, 450 mg 5-FOA, and 12 g glucose in 345 ml). Cool solution I to 60°C after autoclaving and heat solution II to 60°C. Gently mix the two solutions and cool to 45°C before pouring into the plates.
All PCR fragments are agarose gel purified using GeneJET™ Gel Extraction Kit (Fermentas, Germany).
The following chemicals were used: bleomycin (Sigma-Aldrich) (6), zeocin (Invitrogen), camptothecin (Sigma-Aldrich), methyl methanesulfonate (Sigma-Aldrich), raffinose (Sigma-Aldrich), galactose (Sigma-Aldrich), petroleum jelly (Vaseline; VWR Scientific Products), beeswax (Sigma-Aldrich), lanolin (Sigma-Aldrich), NuSieve® GTG® low-melt agarose (Lonza Rockland, Inc., Rockland, ME), and 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; Sigma-Aldrich).
Yeast strains ML193-3B (MATa ADE2 RAD5 TetI-mRFP1::iYGL119W) and ML494-15C (MATa ADE2 RAD5 YFP-RAD55 CFP-RAD51) are derivatives of W303-1A (7, 8).
The sequences of plasmids pWJ716, pWJ1108, pWJ1162, pWJ1163, pWJ1164, pWJ1165, pWJ1350, pWJ1351, and pWJ1379 are available upon request (9–12). Plasmid pJH132 is described previously (13).
Microscope hardware. For microscopy, yeast cells are relatively small and the number of proteins per cell is low compared to vertebrate systems. For this reason, the essential elements of the microscope hardware are a high-sensitivity cooled CCD or EM-CCD camera and a high-magnification objective (100×) with high numerical aperture (≥1.4).
3. Methods
3.1. Fluorescence Tagging of Endogenously Expressed HR Proteins
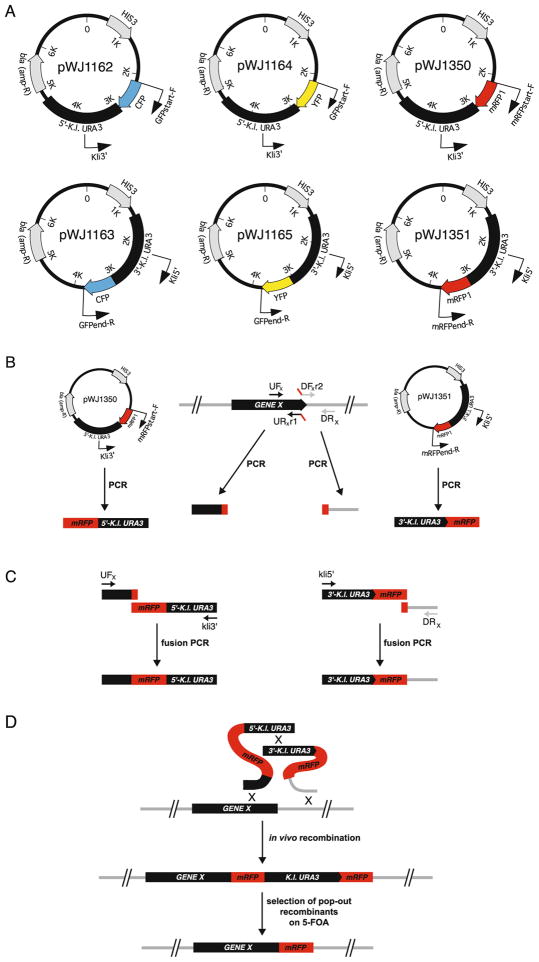
We have previously developed a PCR-based method for fluorescence tagging of endogenously expressed HR proteins with cyan, yellow, or red fluorescent protein (CFP, YFP, and RFP, respectively, or XFP, collectively) (11). In brief, approximately 300 bp on either side of the genomic integration site is amplified by PCR and fused in a second PCR to a cassette containing the gene encoding a fluorescent protein and two-thirds of either the 5′-or 3′-end of the Kluyveromyces lactis URA3 marker (Fig. 30.2). The sequence of the K. lactis URA3 gene is sufficiently divergent from the URA3 gene of S. cerevisiae to prevent targeting to this locus and thus preventing a potential source of false-positive Ura+ transformants. The split marker approach may ease the fusion PCR by allowing for shorter PCR fragments compared to generating one fusion PCR with the full URA3 marker and both sequences of homology. The resulting PCR fragments are co-transformed into the target strain and transformants selected on synthetic complete medium lacking uracil (SC-Ura). The integration generates a direct repeat of the gene encoding the fluorescent protein, which allows for subsequent popout of the URA3 marker by genetic recombination. The protocol for monomeric red fluorescent protein (mRFP) tagging is shown here:
Fig. 30.2.
Cloning-free fluorescence tagging of endogenous genes. (a) Vectors pWJ1162, pWJ1163, pWJ1164, pWJ1165, pWJ1350, and pWJ1350 harboring XFP-K.l.URA3 cassettes for CFP, YFP, or mRFP1 tagging (21–23). (b) PCR amplification of targeting sequences. (c) Adaptamer-mediated fusion PCR. A sequence overlap of 18–22 nucleotides between the two PCR products facilitates their fusion in a second PCR. (d) Gene targeting and marker elimination by popout recombination.
Template genomic DNA is isolated from the target strain as described (14).
The mRFP-5′-K.l.URA3 fragment is amplified by PCR from pWJ1350 using a high-fidelity polymerase such as Pfu and primers Kli3′ and mRFPstart-F. The 3′-K.l.URA3-mRFP fragment is amplified by PCR from pWJ1351 using primers Kli5′ and mRFPend-R (Fig. 30.2a). PCR conditions: 95°C for 2 min, 30 cycles of 95°C for 30 s, 52°C for 30 s, and 72°C for 4 min, followed by 72°C for 1 min and subsequent cooling to 4°C. The PCR fragments are agarose gel purified.
A region of approximately 300 bp immediately upstream of the mRFP integration site is amplified from the genomic DNA using a high-fidelity polymerase such as Pfu and primers UFx and URxr1 (Table 30.1 and Fig. 30.2b). Similarly, a region of approximately 300 bp immediately downstream of the mRFP integration site is amplified using primers DFxr2 and DRx. See Note 1 for primer design. The PCR fragments are agarose gel purified.
Approximately 200 ng of the mRFP-5′-K.l.URA3 fragment is fused by PCR to an equimolar amount of the gene-specific upstream fragment using a high-fidelity polymerase such as Pfu and primers Kli3′ and UFx. Similarly, 200 ng of the 3′-K.l.URA3-mRFP fragment is fused by PCR to an equimolar amount of the gene-specific downstream fragment using primers Kli5′ and DRx. PCR conditions: 95°C for 2 min, 30 cycles of 95°C for 30 s, 52°C for 30 s, and 72°C for 4.5 min, followed by 72°C for 2 min and subsequent cooling to 4°C. The PCR fragments are gel purified.
To integrate mRFP into the genome, 0.3–1 μg of each of the fusion fragments is co-transformed into the target strain by the LiAc method (15). Transformants are selected on synthetic complete medium lacking uracil (SC-Ura). Targeting efficiency varies with the genomic locus by at least a factor 10.
The integration generates a direct repeat of the mRFP sequence flanking the K.l.URA3 marker (Fig. 30.2d). This configuration allows for subsequent popout of the URA3 marker. Efficient popout is achieved by growing cells overnight in 2 ml of yeast extract peptone dextrose (YPD) medium before plating 200 μl of the culture on plates containing 5-fluoroorotic acid (5-FOA) (5) (see Note 2).
Table 30.1.
Primers used in these protocols
| Name | Sequence (5′ to 3′) |
|---|---|
| DFxr2 | GCATGGATGAACTATACAAATGAXXXXXXXXXX |
| Down-e | CGATCTTCTACCCAGAATCACXXXXXXXXXXXXXX |
| DRx | XXXXXXXXXXXXXXXXXXX |
| E-Kl | GTGATTCTGGGTAGAAGATCG |
| F-Kl | CGATGATGTAGTTTCTGGTT |
| GFPend-R | TTTGTATAGTTCATCCATGC |
| GFPstart-F | ATGAGTAAAGGAGAAGAAC |
| I-SceI-Down-F | TTACGCTAGGGATAACAGGGTAATATAGCGXXXXXXXX |
| I-SceI-Up-R | CGCTATATTACCCTGTTATCCCTAGCGTAAXXXXXXXXXX |
| Kli3′ | GAGCAATGAACCCAATAACGAAATC |
| Kli5′ | CTTGACGTTCGTTCGACTGATGAGC |
| mRFPend-R | GGCGCCGGTGGAGTGG |
| mRFPstart-F | ATGGCCTCCTCCGAGGAC |
| UFx | XXXXXXXXXXXXXXXXXXX |
| Up-f | AACCAGAAACTACATCATCGXXXXXXXXXXXX |
| URxr1 | GTTCTTCTCCTTTACTCATnnnnnnXXXXXXXXXXX |
Linker sequences are indicated by nnnnnn and gene-specific sequences are indicated by XXXXXX.
3.2. Fluorescence Visualization of an Inducible DSB Site
Specific genomic mega-endonuclease restriction sites such as I-SceI and HO can be marked fluorescently by integration of a tandem array of 100–200 copies of a recognition sequence for a DNA-binding protein fused to XFP. We have good experience with Tet and Lac repressor binding sites (16, 17) (see Note 3). The protocol for genomic integration of the I-SceI cut site and the tetO tandem array is shown here:
Template genomic DNA is isolated from the target strain as described (14).
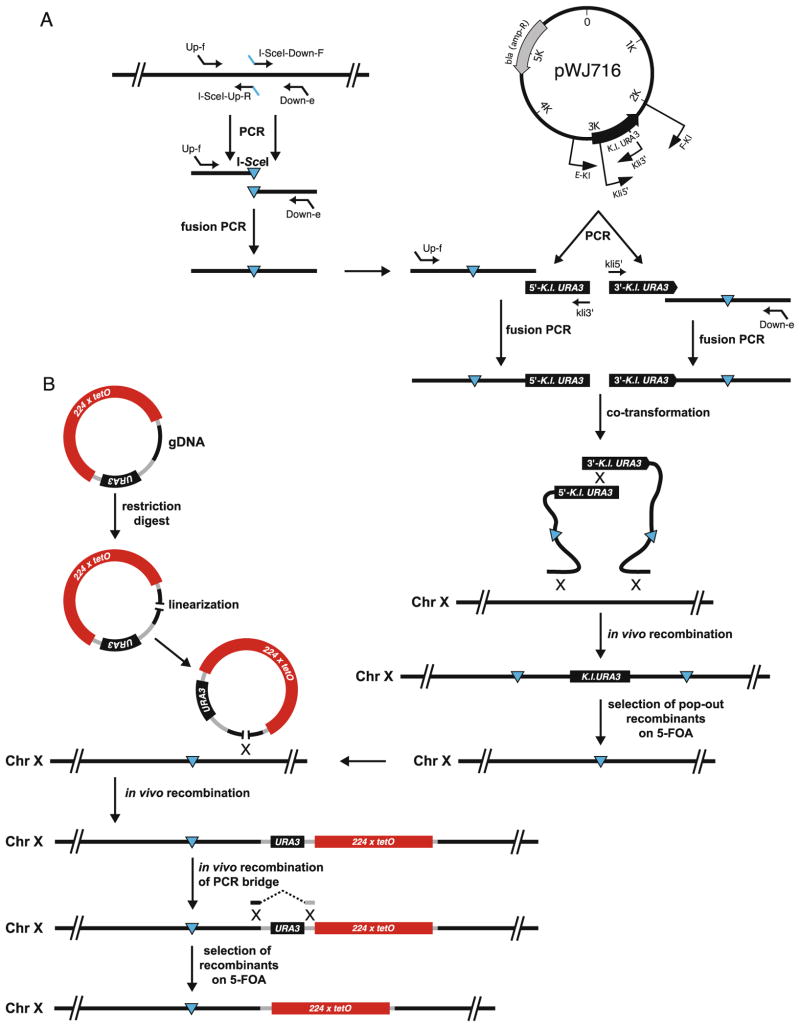
The I-SceI cut site is integrated into the genome essentially as described (9). In brief, a region of approximately 300 bp immediately upstream of the I-SceI integration site is amplified from the genomic DNA using a high-fidelity polymerase such as Pfu and primers Up-f and I-SceI-Up-R (Table 30.1). Similarly, a region of approximately 300 bp immediately downstream of the I-SceI integration site is amplified using primers I-SceI-Down-F and Down-e (Fig. 30.3a). The PCR fragments are agarose gel purified and fused in a second PCR using primers Up-f and Down-e. The PCR fusion is agarose gel purified.
The 5′-K.l.URA3 fragment is amplified by PCR from pWJ716 using primers Kli3′ and E-Kl. The 3′-K.l.URA3 fragment is amplified by PCR from pWJ716 using primers Kli5′ and F-Kl. PCR conditions: 95°C for 2 min, 30 cycles of 95°C for 30 s, 52°C for 30 s, and 72°C for 2 min, followed by 72°C for 1 min and subsequent cooling to 4°C. The PCR fragments are agarose gel purified.
Approximately 200 ng of the 5′-K.l.URA3 fragment is fused by PCR to an equimolar amount of the fusion fragment containing the I-SceI cut site (step 2) using primers Kli3′ and Up-f. Similarly, 200 ng of the 3′-K.l.URA3 fragment is fused by PCR to an equimolar amount of the fusion fragment containing the I-SceI cut site using primers Kli5′ and Down-e. PCR conditions: 95°C for 2 min, 30 cycles of 95°C for 30 s, 52°C for 30 s, and 72°C for 4.5 min, followed by 72°C for 2 min and subsequent cooling to 4°C. The PCR fragments are agarose gel purified.
To integrate the I-SceI cut site into the genome, 0.3–1 μg of the fusion fragment is co-transformed into the target strain by the LiAc method (15). Transformants are selected on synthetic complete medium lacking uracil (SC-Ura).
The integration generates a direct repeat of the I-SceI cut site containing sequence flanking the K.l. URA3 marker (Fig. 30.3a). This configuration allows for popout of the URA3 marker. Efficient popout is achieved by growing cells overnight in 2 ml of YPD medium before plating 200 μl of the culture on plates containing 5-FOA (5).
To insert an array of Tet repressor binding sites adjacent to the I-SceI cut site, a 1-kb fragment of genomic target DNA (gDNA) is cloned into an integrative plasmid pWJ1379 containing the tetO array and the URA3 selectable marker (Fig. 30.3b). The gDNA is selected to contain a unique centrally located restriction site for subsequent linearization of the plasmid before transformation into yeast (see Note 4 for plasmid construction).
To integrate the tetO array into the genome, 1 μg of linearized plasmid is transformed into a target strain ML193-3B expressing TetR-mRFP by the LiAc method (15). Transformants are selected on synthetic complete medium lacking uracil (SC-Ura). Initially, transformants are screened visually by fluorescence microscopy to select the clones that harbor the tetO array as determined by the appearance of a red dot in each cell. Subsequently, correct genomic integration is verified by PCR or Southern blot analysis.
If desired, the co-integrated vector backbone of the tetO plasmid can be deleted by transformation of a PCR fusion bridging the region to be deleted (Fig. 30.3b). The PCR bridge should have >300 bp of homology to the genomic target on each side of the region to be deleted. The PCR bridge is constructed by fusion PCR of regions upstream and downstream of the sequence to be deleted using relevant primers essentially as described in step 2. Since the URA3 marker can also be deleted by loss of the entire tetO-containing region, it advisable to use 1–2 μg of PCR bridge in the LiAc transformation (15). Transformants are incubated for 3 h in YPD in order to allow for the URA3 gene product to disappear before plating on solid medium containing 5-FOA (5) (see Note 5).
Fig. 30.3.
Construction of a fluorescently marked DSB site. (a) PCR-based insertion of a unique site-specific I-SceI site in the genome. (b) Genomic integration of a tetO array in a strain constitutively expressing TetR-mRFP (e.g., strain ML193-3B) gDNA, genomic target DNA.
3.3. Cell Culture for Fluorescence Microscopy
For yeast live cell imaging, the best results are obtained by culturing and mounting cells in identical medium. YPD medium should be avoided for imaging, because it quenches a broad range of wavelengths. By contrast, filter-sterilized minimal medium has excellent optical properties (5). A protocol for imaging exponentially growing cells is shown here:
3.4. Induction of HR Foci by DNA-Damaging Agents
Foci of HR proteins can be induced by a number of agents or enzymes that cause DNA double-strand breaks (DSBs) and/or collapsed replication forks. Examples of agents and doses are 20 Gy of γ-irradiation (1 DSB per haploid cell) (18), 1 μg/ml bleomycin for 1 h (0.5 DSB per haploid cell) (6), 200 μg/ml zeocin for 1 h, 5 μg/ml camptothecin for 1 h, and 0.03% (v/v) methyl methanesulfonate (MMS) for 1 h.
3.5. Induction of HR Foci by Mega-endonucleases
Strains in which a single site-specific endonuclease restriction site such as the HO or the I-SceI site in the genome has been marked fluorescently as described above (Section 3.2) are transformed with a plasmid expressing the appropriate endonuclease from an inducible promoter, e.g., pJH132 (pGAL-HO) or pWJ1108 (pGAL-I-SceI), respectively:
Transform the appropriate plasmid (expressing HO or I-SceI) into the assay strain by the LiAc method (15). Select transformants on the appropriate glucose-containing minimal medium.
Inoculate fresh transformants in 2 ml of the appropriate minimal, raffinose-based medium (2% (w/v) raffinose, autoclaved) (see Note 8).
Grow by shaking at 25°C for 24 h.
Dilute culture to OD600 = 0.2 and grow for one cell cycle (3.5 h for wild type) prior to endonuclease induction.
Add galactose (sterile-filtered, stock solution, 30% (w/v)) to a final concentration of 2% (w/v). This induces expression of the endonuclease.
Grow by shaking at 25°C for another 90 min prior to microscopy.
3.6. Sample Preparation
Live cell imaging is preferred over fixed cells, because fixation may generate artifacts or otherwise decrease the quality of the obtained data. This protocol describes preparation of both types of samples:
Harvest 1.5 ml of cells at OD600 = 0.4–0.6 by centrifugation at 1,500×g.
For live cell imaging, resuspend the pellet of cells in 50 μl of medium by vortexing and proceed to mounting the cells (Section 3.7).
For fixed cell imaging, resuspend the pellet of cells in PBS containing 4% paraformaldehyde (wear gloves).
Incubate shaking for 30 min at room temperature.
Wash twice for 30 min in PBS at room temperature. Store in the dark at 4°C in PBS containing 0.02% sodium azide (optional for long-term storage).
3.7. Mounting of Live Cells
Cells are mounted on standard glass slides and covered by cover glass appropriate to the optics of the microscope. Issues to consider before mounting the cells are the cell density and immobilization. A high cell density is desired to maximize data acquisition. However, at high cell density and growth rate (glucose), the medium quickly becomes saturated with carbon dioxide, which precipitates as gas bubbles that displace cells. Therefore, the optimal cell density must often be determined empirically. For most strains, the cells are immobilized on the slide simply by adjusting the volume applied (usually 2–3 μl) so that the cells settle in a monolayer with the cells touching both the slide and the cover glass. However, for mutant strains that exhibit heterogeneous cell size, it can be necessary to immobilize cells in low-melt agarose. For long-term imaging (>30 min), the edges of the cover glass are sealed to prevent evaporation by a mixture of 1 volume of petroleum jelly (Vaseline), 1 volume of beeswax, and 1 volume of lanolin. The protocol is as follows:
Prepare a solution of 1.2% (w/v) low-melt agarose (gelling at 36°C) in appropriate medium (e.g., SC). Aliquot in microcentrifuge tubes and use each aliquot only once. Melt by boiling and keep at 42°C prior to use. Melt the wax solution (e.g., in a 65°C incubator).
Add 2 μl cell suspension to the slide and mix with 2 μl of agarose solution by pipetting. Apply cover glass as fast as possible.
Seal with melted wax using a flat metal spatula. Heating of the spatula over a gas burner may be necessary to facilitate dispersion of wax along the sides of the cover glass.
3.8. DAPI Staining
Yeast cells can be DAPI stained to visualize DNA content without fixation:
Add DAPI to the liquid medium at 10 μg/ml and grow by shaking for 30 min.
Wash cells in SC without DAPI before microscopy.
Due to the intense DAPI staining of mitochondria, it can be difficult to discern the nuclear compartment. To visualize staining of nuclear DNA, it is often necessary to eliminate mitochondrial DNA. This is achieved by the following protocol:
Inoculate an overnight culture in YPD containing 25 μg/ml ethidium bromide at 30°C.
The next morning, dilute 100-fold into YPD containing 25 μg/ml ethidium bromide. Grow again overnight at 30°C.
Plate for individual colonies on YPD.
To test for loss of mitochondrial DNA, pick five candidates and examine them under the microscope after DAPI staining. Additionally, candidates can be tested for lack of growth on yeast extract peptone medium containing 2% (w/v) galactose due to loss of mitochondria.
3.9. Time-Lapse Microscopy
Imaging of cells over time requires that phototoxicity is minimized and that favorable growth conditions can be maintained. Phototoxicity can be reduced by decreasing the fluorescence exposure time or intensity, and by reducing the number of optical sections acquired. The easiest way to maintain favorable growth conditions is to reduce the cell density of the slide so that only a single cell is present in the field of view, thereby prolonging the time that the cell can grow before nutrients are exhausted locally. For this reason it is not recommended to concentrate the cells by centrifugation before mounting:
Cells are cultured and mounted essentially as described above (Sections 3.3 and 3.7).
The appropriate acquisition parameters for time-lapse microscopy should be determined empirically. In our case, we were able to image Rad52-YFP with minimal phototoxicity for 20 time points over 4 h by reducing the number of optical sections from 11 to 9 (0.5 μm between sections), reducing the neutral density filter for the fluorescence path from 25 to 10% transmission, and by reducing the exposure time from 1 to 0.5 s.
Acknowledgments
The LacI-R197K mutant protein was engineered by Christian Müller. This work was supported by The Danish Agency for Science, Technology and Innovation (ML), the Villum Kann Rasmussen Foundation (ML), GM67055 (RR), and the Lundbeck Foundation (NEB).
Footnotes
Usually, primers are designed to insert a small flexible linker between mRFP and the protein of interest. Obviously the stop codon from the gene to be fused to mRFP should not be included in the primers for tagging at the N terminus. For XFP fused to the N terminus, we have good experience with a Gly2ProGly2 linker and for XFP fused to the C terminus we routinely use an Ala4 linker.
Generally, correct genomic targeting is achieved for >90% of Ura+ colonies. However, if the genomic target sequence contains repetitive or ARS-like sequences, the transformed PCR fragments can circularize to a self-replicating extrachromosomal circle. These incorrect clones are easily identified during the popout step, because they give rise to a confluent layer of Ura− cells on 5-FOA due to the instability of centromere-less extrachromosomal circles.
When using the Lac repressor, a mutant LacI-R197K protein is used, which binds to DNA in the presence of both glucose and galactose.
The tetO array should be positioned 3–5 kb away from the I-SceI cut site, in order to prevent or delay its conversion into single-stranded DNA during resection of the DSB generated at I-SceI, which will abolish binding by the Tet repressor. Since the tetO array is unstable in E. coli, it is essential that the tetO plasmid is cloned in a rec− strain at 30°C. For the tetO array, we have good experience with the SURE2 strain (Stratagene, La Jolla, CA), and for the lacO array the STBL2 strain (Invitrogen, Paisley, UK) gives good stability.
Generally, correct deletion of the plasmid sequences by the PCR bridge is achieved for 10–30% in the 5-FOA-resistant colonies.
The extra adenine is necessary only for ade2 mutant strains, which will otherwise accumulate a red pigment that is strongly autofluorescent. However, SC medium containing 100 μg/ml adenine can also be used for ADE2 cells.
Some variants of GFP and RFP mature better at 25°C than at 30 or 37°C (19).
It is important to use fresh transformants in order to avoid cells in which mutations have accumulated in the endonuclease recognition site. Such mutations tend to appear in old transformants due to leakiness of the GAL1-10 promoter.
References
- 1.Krogh B, Symington L. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 2.Lisby M, Rothstein R. DNA damage checkpoint and repair centers. Curr Opin Cell Biol. 2004;16:328–334. doi: 10.1016/j.ceb.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 3.Lisby M, Rothstein R. Localization of checkpoint and repair proteins in eukaryotes. Biochimie. 2005;87:579–589. doi: 10.1016/j.biochi.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 4.Lisby M, Rothstein R. Choreography of recombination proteins during the DNA damage response. DNA Repair (Amst) 2009;8:1068–1076. doi: 10.1016/j.dnarep.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sherman F, Fink GR, Hicks JB. Methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 6.Moore CW, McKoy J, Dardalhon M, Davermann D, Martinez M, Averbeck D. DNA damage-inducible and RAD52-independent repair of DNA double-strand breaks in Saccharomyces cerevisiae. Genetics. 2000;154:1085–1099. doi: 10.1093/genetics/154.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 8.Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- 9.Erdeniz N, Mortensen UH, Roth-stein R. Cloning-free PCR-based allele replacement methods. Genome Res. 1997;7:1174–1183. doi: 10.1101/gr.7.12.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torres-Rosell J, Sunjevaric I, De Piccoli G, Sacher M, Eckert-Boulet N, Reid R, Jentsch S, Rothstein R, Aragon L, Lisby M. The Smc5–Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat Cell Biol. 2007;9:923–931. doi: 10.1038/ncb1619. [DOI] [PubMed] [Google Scholar]
- 11.Reid R, Lisby M, Rothstein R. Cloning-free genome alterations in Saccharomyces cerevisiae using adaptamer-mediated PCR. Methods Enzymol. 2002;350:258–277. doi: 10.1016/s0076-6879(02)50968-x. [DOI] [PubMed] [Google Scholar]
- 12.Lisby M, Mortensen UH, Rothstein R. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat Cell Biol. 2003;5:572–577. doi: 10.1038/ncb997. [DOI] [PubMed] [Google Scholar]
- 13.Jensen RE, Herskowitz I. Directionality and regulation of cassette substitution in yeast. Cold Spring Harb Symp Quant Biol. 1984;49:97–104. doi: 10.1101/sqb.1984.049.01.013. [DOI] [PubMed] [Google Scholar]
- 14.Hoffman CS, Winston F. A ten-minute DNA preparation efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 15.Gietz D, St Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein–protein interactions can mediate sister chromatid cohesion. Curr Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- 17.Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 18.Lisby M, Rothstein R, Mortensen UH. Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci USA. 2001;98:8276–8282. doi: 10.1073/pnas.121006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim CR, Kimata Y, Oka M, Nomaguchi K, Kohno K. Thermosensitivity of green fluorescent protein fluorescence utilized to reveal novel nuclear-like compartments in a mutant nucleoporin NSP1. J Biochem (Tokyo) 1995;118:13–17. doi: 10.1093/oxfordjournals.jbchem.a124868. [DOI] [PubMed] [Google Scholar]
- 20.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response; spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 21.Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 22.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. Proc Natl Acad Sci USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heim R, Tsien RY. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]