Abstract
NEDD8, in plants and yeasts also known as RELATED TO UBIQUITIN (RUB), is an evolutionarily conserved 76 amino acid protein highly related to ubiquitin. Like ubiquitin, NEDD8 can be conjugated to and deconjugated from target proteins, but unlike ubiquitin, NEDD8 has not been reported to form chains similar to the different polymeric ubiquitin chains that have a role in a diverse set of cellular processes. NEDD8-modification is best known as a post-translational modification of the cullin subunits of cullin-RING E3 ubiquitin ligases. In this context, structural analyses have revealed that neddylation induces a conformation change of the cullin that brings the ubiquitylation substrates into proximity of the interacting E2 conjugating enzyme. In turn, NEDD8 deconjugation destabilizes the cullin RING ligase complex allowing for the exchange of substrate recognition subunits via the exchange factor CAND1. In plants, components of the neddylation and deneddylation pathway were identified based on mutants with defects in auxin and light responses and the characterization of these mutants has been instrumental for the elucidation of the neddylation pathway. More recently, there has been evidence from animal and plant systems that NEDD8 conjugation may also regulate the behavior or fate of non-cullin substrates in a number of ways. Here, the current knowledge on NEDD8 processing, conjugation and deconjugation is presented, where applicable, in the context of specific signaling pathways from plants.
Keywords: CAND1, COP9 signalosome (CSN), cullin, E3 ubiquitin ligase, F-BOX PROTEIN (FBP), NEDD8, RELATED TO UBIQUITIN (RUB), ubiquitin
NEDD8 IS AN EVOLUTIONARILY CONSERVED REGULATOR
NEDD8 (neural precursor cell expressed, developmentally down-regulated8), in plants and yeasts also known as RELATED TO UBIQUITIN (RUB, hitherto referred to as NEDD8), is a 76 amino acid protein that was originally identified as a highly expressed gene from embryonic mouse brains (Kumar et al., 1993). Amongst all ubiquitin-like modifiers (UBLs), NEDD8 and ubiquitin are most closely related to each other and NEDD8 proteins, like other UBLs, display remarkable sequence conservation across species (Vierstra, 2012). Like ubiquitin, NEDD8 is conjugated to its substrate protein through the formation of an isopeptide bond between its C-terminal glycine and a lysine residue of the target protein (neddylation) but there is no known biological function for free NEDD8.
NEDD8 orthologs can be identified in all eukaryotic species that have sequenced genomes. While NEDD8 is a single gene in humans, mouse, and fruit fly, several copies of NEDD8 are encoded by the genomes of the plant species Arabidopsis (Arabidopsis thaliana; Rao-Naik et al., 1998), rice (Oryza sativa), Brachypodium (Brachypodium distachyon), and the moss Physcomitrella patens (Figure 1). All NEDD8 proteins require proteolytic processing of their C-termini to generate mature NEDD8 with a C-terminal glycine required for NEDD8 conjugation (Figure 1). An additional unique feature of plant NEDD8 is the existence of ubiquitin-NEDD8 gene fusions. While gene fusions of ubiquitin to ubiquitin itself or other genes have been reported in other species, NEDD8 is an unfused gene in animals and yeasts but not in plants. This ubiquitin-NEDD8 fusion structure is found in Arabidopsis RUB1 and RUB2 and seems to be conserved among plants, mosses and algae (Figure 1; Rao-Naik et al., 1998; Vierstra and Callis, 1999; Shin et al., 2011). In RUB1 and RUB2, a single ubiquitin is fused head-to-tail to the N-terminus of NEDD8 and both ubiquitin-NEDD8 fusions then require post-translational processing to release monomeric ubiquitin and NEDD8 (Figure 1). Furthermore, plant genomes contain an unfused monomeric form of NEDD8, RUB3 in Arabidopsis, that can additionally be distinguished from the other RUB genes because it lacks an intron that is present at a conserved position in other RUBs, e.g., in Arabidopsis RUB1 and RUB2 (Figure 1). The absence of an intron suggests that this less complex RUB3 may be more ancient than the intron-containing RUB1 or RUB2 or that RUB3 originated from an mRNA intermediate and a retrotransposition event (Huang et al., 2012).
FIGURE 1.
Protein domain organization of NEDD8 proteins from several representative species. Gene identification numbers (Gene IDs) are as listed in www.ensemblgenomes.org. Specifically indicated are the last five amino acids of the respective proteins before the proteolytic cleave sites, the first amino acid of NEDD8 and the proteins’ C-terminal amino acids. Proteolytic processing occurs after the C-terminal RGG residues and is indicated by an underscore. Positions of introns in the respective genes are indicated by a triangle. The light gray area in BRADI4G28550 highlights an apparent 22 amino acid deletion in the ubiquitin part of the protein. A. thaliana (Arabidopsis thaliana), O. sativa (Oryza sativa, rice), B. distachyon (Brachypodium distachyon), P. patens (Physcomitrella patens, moss), C. reinhardtii (Chlamydomonas reinhardtii, algae), D. melanogaster (Drosophila melanogaster, fruit fly), S. cerevisiae (Saccharomyces cerevisiae, baker’s yeast), S. pombe (Schizosaccharomyces pombe, fission yeast).
Similarly to the high sequence conservation observed between human and Arabidopsis ubiquitin (96% amino acid sequence identity), also NEDD8 proteins are highly conserved between species (83% identity between human and Arabidopsis). This high level conservation is suggestive for an important function of NEDD8 conjugation (neddylation) in eukaryotic cells and a highly conserved neddylation and deneddylation machinery. Indeed, loss-of-NEDD8 function causes lethality at an early developmental stage in most model organisms and also in plants, with the notable exception of Saccharomyces cerevisiae (Lammer et al., 1998; Liakopoulos et al., 1999; Jones and Candido, 2000; Osaka et al., 2000; Tateishi et al., 2001; Ou et al., 2002; Dharmasiri et al., 2003; Maytal-Kivity et al., 2003; Bostick et al., 2004). In Arabidopsis, not the single but the combined knockout of the genes RUB1 and RUB2 leads to a developmental arrest at the embryonic two-cell stage (Bostick et al., 2004). Thus, NEDD8 genes and neddylation are essential for growth and development in plants. Plants with reduced NEDD8 gene expression are dwarfed, partially insensitive to root growth inhibitory concentrations of the plant hormone auxin and also partially defective in auxin-induced lateral root formation (Bostick et al., 2004). As will be outlined below, auxin insensitivity phenotypes are reliable and at the same time the most obvious readouts of neddylation pathway mutants.
NEDD8 PROCESSING
NEDD8 is conjugated to the protein substrates via an isopeptide bond between its C-terminal glycine and a lysine of the target protein (Figure 2). NEDD8, like ubiquitin and most UBLs, is expressed as an inactive precursor with a short C-terminal extension that consists of one or several amino acids that need to be cleaved off to allow for NEDD8 conjugation (Figures 1 and 2; Jentsch and Pyrowolakis, 2000). It has been proposed that the C-terminal extension of ubiquitin, NEDD8, and other UBLs serves to prevent unprocessed proteins to enter into the conjugation pathway but there is, in fact, no experimental evidence supporting this hypothesis (Callis et al., 1995; Rao-Naik et al., 1998). The plant ubiquitin-NEDD8 fusion proteins additionally require removal of the N-terminal ubiquitin by proteolytic cleavage.
FIGURE 2.
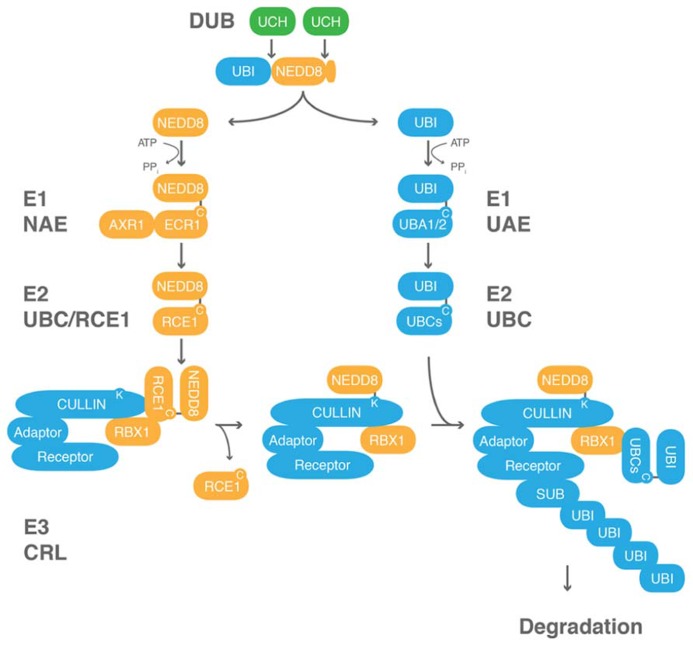
Neddylation and ubiquitin modification are biochemically related processes. Ubiquitin c-terminal hydrolases (UCHs) belong to the family of DUBs that process ubiquitin (UBI)-NEDD8 fusion proteins. UBI and NEDD8 are activated by their conjugation to E1 ubiquitin/NAEs. UBI or NEDD8 from the E1 are then passed via a transthiolation reaction to a protein of the family of E2 ubiquitin-conjugating enzymes, RUB1 CONJUGATING ENZYME1 (RCE1) in Arabidopsis. The ubiquitin-charged E2 can then form a complex with an E3 ubiquitin ligase, and ultimately, ubiquitin and NEDD8 are transferred to a lysine residue on the cullin of the E3 and the substrate, respectively. UBI but not NEDD8 can form chains. RBX1 and other proteins not shown here are the proposed NEDD8 ligases. Cullin-RING ligases (CRLs) are the superfamily of E3 ubiquitin ligases that are regulated by neddylation. CRLs are generally composed of a cullin subunit, RBX1, as well as a substrate (SUB) recognition module composed of an adaptor and a substrate receptor protein. C, cysteine; K, lysine.
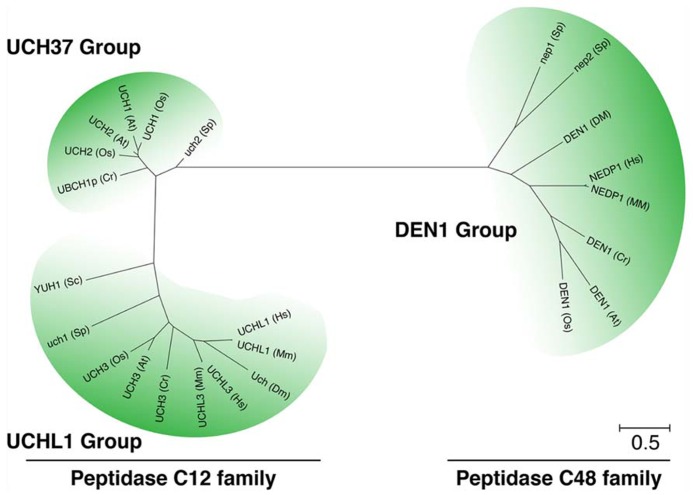
NEDD8 processing is carried out by ubiquitin C-terminal hydrolases (UCH) from the family of deubiquitinating enzymes (DUBs). In S. cerevisiae and humans, NEDD8 precursor C-terminal processing is facilitated by a dual specificity UCH of the C12 family peptidases, Yuh1 (yeast) or UCHL3 (human, mouse), which also processes the C-terminal extensions of ubiquitin (Figure 3; Wada et al., 1998; Johnston et al., 1999; Linghu et al., 2002; Hemelaar et al., 2004; Frickel et al., 2007; Yu et al., 2007). To date, the only isopeptidase known to function exclusively in NEDD8 processing and deconjugation is the C48 family peptidase DEN1/NEDP1/SENP8 from Drosophila, and human (Figure 3; Gan-Erdene et al., 2003; Mendoza et al., 2003; Wu et al., 2003; Shen et al., 2005; Chan et al., 2008; Shin et al., 2011). However, mouse knockouts of UCHL3 or Drosophila and Aspergillus knockouts of DEN1 are viable although NEDD8 and neddylation are essential in the respective organisms (Kurihara et al., 2000; Chan et al., 2008; Christmann et al., 2013). These findings suggest that mutants of these processing enzymes cannot be fully impaired in NEDD8 processing.
FIGURE 3.
NEDD8 processing is mediated by at least three different classes of peptidases. Unrooted phylogenetic tree of representative members of the UCH and DEN1/NEDP1/SENP8 protein families from Arabidopsis thaliana (At), rice (Oryza sativa, Os), algae (Chlamydomonas reinhardtii, Cr), baker’s yeast (Saccharomyces cerevisiae, Sc), fission yeast (Schizosaccharomyces pombe, Sp), fruit fly (Drosophila melanogaster, Dm), mouse (Mus musculus, Mm), and human (Homo sapiens, Hs). Molecular phylogenetic analysis was performed based on the Maximum Likelihood method and the JTT matrix-based model (Jones et al., 1992) using CLUSTALW algorithm at the EMBL-EBI website (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The tree with the highest log likelihood (-5916.8263) is shown. Initial tree(s) for the heuristic search were obtained automatically by applying the Maximum Parsimony method. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 24 protein sequences. All positions containing gaps and missing data were eliminated. There were a total of 145 positions in the final dataset. Evolutionary analyses were conducted in MEGA5 (Tamura et al., 2011).
In non-plant species where NEDD8 is expressed as an unfused gene, examining NEDD8 precursor processing in vivo is not trivial because NEDD8 has only a short C-terminal extension. Therefore, conjugation of processed NEDD8 to cullins is used as an indirect indication for proper processing. Since cullin neddylation was not impaired in any of the UCH gene mutants examined to date, it can be inferred that also NEDD8 processing and conjugation are at least partially functional in these mutants (Kurihara et al., 2000; Chan et al., 2008; Christmann et al., 2013). In the fission yeast Schizosaccharomyces pombe, even a Δnep1 Δnep2 Δuch1 Δuch2 quadruple mutant lacking the two C48 peptidases orthologous to DEN1 (NEP1 and NEP2) as well as the two C12 peptidases orthologous to yeast Yuh1 (UCH1 and UCH2) shows efficient cullin neddylation (O’Donoghue et al., 2013). Only the additional knockout of the cullin deneddylating enzyme COP9 SIGNALOSOME SUBUNIT5 (CSN5) hinted to a reduced efficiency of NEDD8 precursor processing in this complex mutant because it revealed the absence of cullin1 hyperneddylation that can be observed in the Δcsn5 deletion strain (O’Donoghue et al., 2013). Taken together, these various findings suggest that there is a functionally redundant family of NEDD8 processing enzymes and that there may be other as yet unknown peptidases that also participate in NEDD8 processing. Furthermore, it is possible that there is no strict specificity among the ubiquitin and NEDD8 processing enzymes in vivo.
NEDD8 PROCESSING IN PLANTS
NEDD8 processing hydrolases from plants remain to be identified. In Arabidopsis, several peptidases can be classified as C12 and C48 family peptidases based on their homology to UCHs from other species (Figure 3). To date, only the C12 peptidases UCH1 and UCH2 have been analyzed at the biological level (Yang et al., 2007). Based on their homology to C12 peptidases, these UCHs would be predicted to have a role in ubiquitin processing. However, although both UCH proteins showed the predicted ubiquitin processing activity in vitro, neither the uch1 uch2 double mutant nor UCH1 overexpressing lines had apparent changes in the pattern of ubiquitin conjugate formation or in the abundance of free monomeric ubiquitin. At the phenotypic level, mutants or overexpression lines display impaired shoot and flower development and changes in the rate of leaf formation. Altering the abundance of the two UCH genes also affects auxin and cytokinin responsiveness. These phenotypes may be explained by defects in selective rather than general protein degradation and this hypothesis is supported by the observation that the degradation of the auxin-labile AUX/IAA AUXIN RESISTANT3 (AXR3) but not that of the light-labile phytochrome A or ELONGATED HYPOCOTYL5 (HY5) proteins appeared to be affected in the uch mutants. In summary, these findings suggest that UCH1 and UCH2 may not only act at the level of ubiquitin processing but may also act by selectively regulating the proteasomal degradation of proteins by antagonizing substrate ubiquitylation. Whether UCH1 and UCH2 have a role in NEDD8 processing remains to be examined but the comparatively weak morphological phenotype as well as the absence of an apparent cullin neddylation phenotype already suggests that the two UCH proteins may not have a major function in this process.
NEDDYLATION
NEDD8 is conjugated to target proteins in a manner that is highly similar to ubiquitin conjugation (Figure 2). NEDD8 is activated by an E1 NEDD8 activating enzyme (NAE) and then passed on to an E2 NEDD8 conjugating enzyme of the ubiquitin-conjugating (UBC) enzyme family from where the protein is ultimately transferred to its substrate protein. The best-studied NEDD8 conjugates are the cullin subunits of cullin-RING-type E3 ubiquitin ligases (CRLs; Hua and Vierstra, 2011). CRLs are a family of evolutionarily conserved E3 ligases that are composed of a core complex, comprised of a cullin subunit and the RING BOX PROTEIN1 (RBX1), as well as a ubiquitylation substrate recognition module. In plants, three different types of CRL complexes can be distinguished based on the identity of the cullin subunits CULLIN1, CULLIN3, or CULLIN4 and the identity of their respective substrate recognition module (Lammer et al., 1998; Ruegger et al., 1998; Dieterle et al., 2005; Figueroa et al., 2005; Gingerich et al., 2005; Bernhardt et al., 2006; Chen et al., 2006). The CRL subunit RBX1 is common to all CRLs and serves to promote NEDD8 conjugation.
Structural analyses of cullin neddylation revealed that NEDD8 conjugation causes a conformational change in subdomains of the cullin and RBX1 subunits (Duda et al., 2008; Boh et al., 2011). Neddylation also eliminates the binding of the exchange factor CULLIN-ASSOCIATED-NEDD8-DISSOCIATED1 (CAND1) and locks the CRL in an active state. Thus, neddylation controls CRL activity by promoting conformational changes that favor substrate ubiquitylation. CRL neddylation can then also lead to the recruitment of additional regulatory factors (den Besten et al., 2012).
THE NEDDYLATION PATHWAY AND AUXIN INSENSITIVITY
As will be discussed in more detail below, loss of cullin neddylation or cullin deneddylation affect CRL function by promoting or, respectively, preventing interactions with the substrate receptor exchange factor CAND1. In the plant and neddylation biology context, the Arabidopsis CULLIN1-containing E3 ligase SCFTIR1 with the substrate recognition module composed of the F-box protein (FBP) TRANSPORT INHIBITOR RESISTANT1 (TIR1) and its adaptor subunit ARABIDOPSIS SKP1 (ASK) is highly relevant (Ruegger et al., 1998; del Pozo and Estelle, 1999). TIR1, functioning at the same time also as an auxin receptor, binds AUX/IAA transcriptional repressors in an auxin-dependent manner and targets AUX/IAAs for ubiquitylation and degradation by the 26S proteasome (Gray et al., 2001; Tan et al., 2007). AUX/IAAs and auxin-induced AUX/IAA degradation regulate a number of important developmental and morphological processes throughout plant development: bodenlos (bdl) mutants expressing a stabilized (non-degradable) variant of the AUX/IAA protein bodenlos/iaa14 (bdl/iaa14) are deficient in embryonic root differentiation and are consequently rootless (Hamann et al., 1999; Weijers et al., 2005). axr3 mutants express the stabilized axr3/iaa17 protein and this mutation allows for root elongation in the presence of root growth-inhibiting auxin concentrations (Gray et al., 2001). The link between auxin insensitivity (auxin resistance), AUX/IAA degradation and CULLIN1 could be established because the CULLIN1 alleles axr6-1 and axr6-2 were identified based on their auxin insensitivity (Hobbie et al., 2000; Shen et al., 2002; Hellmann et al., 2003; Quint et al., 2005; Esteve-Bruna et al., 2013). While homozygous axr6-1 and axr6-2 loss-of-function mutants arrest development during embryogenesis, the heterozygousmutants display the auxin-insensitive root growth elongation phenotype. Furthermore, double mutants of axr6-1 or axr6-2 with other mutants of the auxin and neddylation pathway are defective in root differentiation and thereby mimic the characteristic phenotype of the bdl mutant (Hobbie et al., 2000; Hellmann et al., 2003; Quint et al., 2005; Esteve-Bruna et al., 2013).
NEDD8 ACTIVATION
Both, defects in root differentiation as well as auxin-insensitive root elongation have been used extensively as phenotypes for the identification and characterization of Arabidopsis neddylation mutants. auxin resistant1 (axr1) is a mutant of the NAE enzyme AXR1 and was identified due to its defects in auxin response that could later be explained by impairment in the degradation of the AUX/IAA protein AXR3 (Lincoln et al., 1990; Leyser et al., 1993; del Pozo et al., 1998). In Arabidopsis, axr1 mutants display an auxin-insensitive root growth phenotype but to fully impair NEDD8 conjugation the function of the AXR1-paralog AXR1-LIKE (AXL) also needs to be deleted (Dharmasiri et al., 2007). axr1 axl1 double mutants have a more severe phenotype than axr1 mutants in that they are defective in embryonic root differentiation and mimic the bdl mutant phenotype (Dharmasiri et al., 2007). Arabidopsis AXR1 and AXL proteins appear to be equivalent at the biochemical level but interestingly have a differential ability to complement the axr1 mutant phenotype when expressed from the AXR1 promoter (Dharmasiri et al., 2007; Hotton et al., 2011). While NEDD8 activation is carried out by a single protein in animals and yeasts, NAE is a heterodimer in plants of AXR1/AXL and E1 C-TERMINAL RELATED1 (ECR1) corresponding to the protein’s N- and C-termini, respectively (Figure 2; del Pozo et al., 1998; Hotton et al., 2011). An ecr1-1 mutant was identified in a screen for mutants with differential auxin sensitivity and axr1/axl mutants as well as ecr1 mutants are defective in cullin neddylation (Woodward et al., 2007).
NEDD8 CONJUGATION
RUB1 CONJUGATING ENZYME1 (RCE1) was identified in Arabidopsis based on its homology to human UBC12 (del Pozo and Estelle, 1999). An rce1-1 insertion mutant with significantly reduced RCE1 expression levels was subsequently isolated and found to be strongly impaired in cullin neddylation (Dharmasiri et al., 2003). rce1-1 mutants display auxin insensitive root growth phenotypes and fail to differentiate a primary root when combined with axr1. Two additional rce1 alleles were recently found in a suppressor screen of the auxin overproducing sur2 mutant (Pacurar et al., 2012). Interestingly, both of these rce1 alleles would be expected to interfere significantly with the biochemical activity of RCE1 since they carry a nonsense and splice site mutation in exon 4, respectively. Analyzing the extent to which RCE1 function is affected in these alleles is certainly interesting because the unexpectedly weak phenotype of these supposedly strong alleles could be considered indicative for the existence of functionally redundant NEDD8 conjugating enzymes.
NEDD8 LIGATION
The CRL core subunit RING BOX1 (RBX1) is one candidate for an E3 NEDD8 ligase (Morimoto et al., 2003). RBX1 is encoded by two genes in Arabidopsis and its function as CRL subunit and as NEDD8 ligase was addressed in mutants, antisense and overexpression lines (Gray et al., 2002; Lechner et al., 2002; Schwechheimer et al., 2002). RBX1 interacts with RCE1 and while cullin neddylation is decreased in the absence of RBX1 it is increased when RBX1 is overexpressed (Gray et al., 2002).
The protein DEFECTIVE IN CULLIN NEDDYLATION1 (DCN1) has also been described as an E3 NEDD8 ligase (Kurz et al., 2005; Kurz et al., 2008; Meyer-Schaller et al., 2009). Based on yeast studies, it has recently been proposed that DCN1 increases the substrate specificity of RBX1 by directing the RBX1-bound NEDD8-E2 toward the cullin (Scott et al., 2010). Additionally, it was shown that the interaction between DCN1 and UBC12 is regulated by the N-terminal acetylation of the UBC12 E2 enzyme (Scott et al., 2011). The analysis of DCN1-LIKE proteins, of which there are five in humans, has also revealed that at least one member of the protein family, DCNL3, is bound to the plasma membrane (Meyer-Schaller et al., 2009). Studies of a mammalian CULLIN2-containing CRL further revealed that DCN1-LIKE1 can engage in interactions between the cullin and the respective substrate receptor subunit, and more importantly, that this interaction is strengthened when the substrate receptor is loaded with cargo (Heir et al., 2013). At least in this case, DCN1-LIKE1 may function as a sensor for degradation substrate availability and consequently promote neddylation. Thus, DCN1 proteins may contribute to the regulation of E3 ligase activity by targeting E3 ligases to or by activating them in specific subcellular locations.
As yet, DCN1 or DCN1-LIKE proteins have not been analyzed in plants but AT3G12760 is a candidate for a direct DCN1 ortholog from Arabidopsis. A second DCN1-LIKE protein, less closely related to DCN1 than AT3G12760, was identified as anti-auxin resistant3 (AAR3) in a screen for mutants that showed resistance to the anti-auxin p-chlorophenoxyisobutylic acid. The same screen also identified mutant alleles of TIR1 and CULLIN1 and, based on the shared phenotype of these mutants, AAR3/DNC1-LIKE would qualify as a candidate regulator of NEDD8 ligation and SCFTIR1 function (Biswas et al., 2007). Unfortunately, the biochemical function of AAR3/DNC1-LIKE in the context of cullin neddylation has not been examined as yet. There is also one further Arabidopsis gene that may encode for an additional DCN1-LIKE protein. Thus, the biochemical and biological functions of DCN1-LIKE genes from Arabidopsis remain to be investigated.
In yeast, also TFB3, a RING domain subunit of the general transcription factor TFIIH was found to promote neddylation in addition to RBX1 and DCN1 (Rabut et al., 2011). The identification of TFB3 was based on initial observations that CULLIN4 neddylation in yeast was independent from RBX1 and DCN1. The analysis of RING domain protein mutants from yeast then led to the subsequent discovery of Tfb3 as a RING domain protein responsible for the neddylation of CULLIN3 and CULLIN4. A clear homolog of TFB3 is not easily discernable in the plant genomes but the identification of TFB3 from yeast per se indicates that it cannot be ruled out that besides RBX1 and DCN1 also other NEDD8 ligases exist in plants.
MLN4924 – A NEDDYLATION INHIBITOR
The importance of the NEDD8-modification pathway in the control of plant development has recently been elucidated in a study with the neddylation inhibitor MLN4924 (Hakenjos et al., 2011). MLN4924 was initially described as an inhibitor of the human NAE E1 enzyme but was subsequently found to also inhibit the NAE E1 subunit ECR1 from predictably all plant species (Soucy et al., 2009; Brownell et al., 2010; Hakenjos et al., 2011). MLN4924 inhibits neddylation in plants and the impairment of CRL function results in the degradation of a number of CRL substrates such as the AUX/IAAs of the auxin pathway, DELLA proteins of the gibberellin pathway and the cell cycle regulator KRP1 (Hakenjos et al., 2011). While the severe phenotypes of strong NEDD8 pathway mutants in Arabidopsis and the absence of neddylation mutants in other plant species has as yet hampered studying the role of neddylation in all stages of plant development or in non-Arabidopsis species, the availability of MLN4924 now overcomes this limitation (Hakenjos et al., 2011).
NEDDYLATION MUTANTS ARE IMPAIRED IN MANY DIFFERENT CRL FUNCTIONS
As outlined above, AUX/IAA degradation is partially or fully impaired in all mutants of the NEDD8 conjugation pathway and auxin responses are partially or fully blocked in these mutant backgrounds. However, the phenotype of the NEDD8 conjugation mutants is much more complex and not only the consequence of defects in the auxin response pathway. In this regard, it is important to realize that plants predictably have many hundreds of CRLs and that all these CRLs should be impaired in neddylation mutants (Xu et al., 2009). Among these CRLs, SCFTIR1 and closely related complexes implicated in auxin responses have a very prominent role because defects in the auxin-regulatory CRLs lead to morphological defects that can easily be examined (Dharmasiri et al., 2005a,b). However, while malfunction of SCFTIR1 and closely related complexes is the most visible phenotype of neddylation mutants, all CRL functions should be affected in axr1 mutants in a manner similar to the defects observed in the auxin pathway. This fact is sometimes overlooked and particularly axr1 mutants that were amongst the first auxin response mutants to be identified (Lincoln et al., 1990; Leyser et al., 1993) are often being used as auxin pathway-specific mutants. The knowledge about the existence of many other CRL-dependent pathways, also CRL pathways that affect plant growth and morphology clearly argue against using axr1 mutants or other neddylation mutants as auxin pathway-specific mutations for morphological analyses or genetic interaction studies (Dill et al., 2004; Shen et al., 2007; Stirnberg et al., 2007; Nelson et al., 2011; Waters and Smith, 2013).
CSN PROMOTES CULLIN DENEDDYLATION
NEDD8 can be deconjugated from CRLs through the activity of the COP9 signalosome (CSN; Figure 4). CSN is evolutionarily conserved and in most species including plant and mammalian species composed of eight subunits (Chamovitz et al., 1996; Seeger et al., 1998; Wei and Deng, 1998). CSN was originally identified in plants based on mutants that display a constitutively photomorphogenic (cop) phenotype and named following the identification of the causative mutation in the cop9 mutant (Wei and Deng, 1992; Wei et al., 1994). Similarly to light-grown seedlings, cop mutants have a short hypocotyl, open cotyledons, and express light-regulated genes when grown in the dark.
FIGURE 4.

CAND1 regulates the cells CRL repertoire by promoting the exchange of substrate receptor subunits. Schematic representation of the exchange of a hypothetical F-BOX PROTEIN1 (FBP1) of the CRL SCFFBP1 against FBP2 following CULLIN1 deneddylation. ARABIDOPSIS SKP1 (ASK) proteins are adaptor subunits that link FBPs with CULLIN1. CULLIN1, ASK, RBX1 and FBP form an SCF-type CRL. COP9 SIGNALOSOME (CSN) promotes CULLIN deneddylation. CULLIN-ASSOCIATED-NEDD8-DISSOCIATED1 (CAND1) is an exchange factor that can only associate with deneddylated cullins and promotes substrate receptor exchange. C, cysteine; K, lysine.
CSN REPRESSES PHOTOMORPHOGENESIS IN ARABIDOPSIS
In Arabidopsis, loss-of-function of the eight CSN subunits results in most cases in the destabilization of the entire CSN complex and in a phenotypically indistinguishable cop phenotype, marking constitutive photomorphogenesis as a hallmark phenotype for loss of CSN function (Serino et al., 1999, 2003; Dohmann et al., 2005; Gusmaroli et al., 2007). The cop phenotype of csn mutants can be explained by their inability to degrade photomorphogenesis regulatory transcription factors such as HY5 in dark-grown seedlings through the activity of the E3 ligase COP1 (Osterlund et al., 2000; Chen et al., 2006; Lau and Deng, 2012). COP1 function is impaired in csn mutants leading to a stabilization of the COP1 targets also in the dark. In the wildtype, photomorphogenic development during germination is seemingly controlled by the light-controlled nucleocytoplasmic shuttling of COP1 (von Arnim and Deng, 1994; Osterlund et al., 2000; Pacin et al., 2013). In contrast to the strong photomorphogenesis phenotype of csn loss-of-function mutants, mutants with partially impaired CSN function display a number of phenotypes, also including auxin insensitive root elongation (Schwechheimer et al., 2001, 2002; Dohmann et al., 2005, 2008b; Stuttmann et al., 2009; Huang et al., 2013b). This phenotypic similarity was indicative for a connection between CSN, the neddylation pathway, and SCFTIR1-dependent plant growth regulation when knowledge about the biochemical interplay of these components was still unclear (Schwechheimer et al., 2001).
CULLIN DENEDDYLATION IS A FUNCTION OF THE MPN+ DOMAIN SUBUNIT CSN5
COP9 signalosome is closely related to the “lid” of the 26S proteasome. In plants and animals, both protein complexes are composed of six so-called PCI domain subunits and two MPN-domain subunits and they share a set of subunit–subunit interactions within the respective complexes (Glickman et al., 1998; Wei et al., 1998; Fu et al., 2001; Enchev et al., 2010; Kotiguda et al., 2012). The relatedness of the two complexes and their in part shared biochemical function are nicely reflected by the fact that a proteasomal “lid” subunit functionally replaces a “missing” CSN subunit in Saccharomyces cerevisiae (Yu et al., 2011). CSN as well as the “lid” have two MPN-domain proteins, which can be further subdivided into an MPN+ domain protein with a catalytically active metalloprotease site, and a catalytically inactive MPN-domain protein that must be derived from the MPN+-domain counterpart (Maytal-Kivity et al., 2002). The MPN+ domain subunits CSN5 and RPN11 confer deneddylation and deubiquitylation activity to the CSN and proteasome “lid” complexes, respectively (Cope et al., 2002; Ambroggio et al., 2004). csn mutants from Arabidopsis are fully impaired in cullin deneddylation and only traces of, presumably de novo synthesized, unneddylated cullin can be detected in csn mutants. Interestingly, CSN5 is only functional as a cullin deneddylase when associated with CSN. CSN physically interacts with the cullin and RBX1 subunits of CRLs through its subunits CSN2 and CSN6 and it is thought that these interactions provide CSN with the affinity for its CRL targets (Schwechheimer et al., 2001). Interesting is also the recently identified csn3-3 allele, which carries a missense mutation in the CSN3 gene. This mutation strongly impairs auxin responses in the mutant but does neither obviously affect cullin deneddylation nor CSN protein complex integrity (Huang et al., 2013a). Thus, the affected domain of CSN3 may be required for an as yet unknown essential CSN function such as CRL subunit interactions or for the ability of the protein or protein complex to engage in other interactions required for normal auxin responses. This is supported by biochemical analyses combined with structural electron microscopy that suggest that CSN2 and CSN5 interact with the cullin E3 ligase subunit whereas F-box substrate receptors interact with CSN1 and CSN3 (Enchev et al., 2012).
Since csn mutants are impaired in the function of presumably hundreds of E3 ligases, it is not surprising that additional physiological defects have been identified in these mutants that can be explained by defects in other CRLs and include defects in SCFCOI1-mediated jasmonate signaling (Schwechheimer et al., 2002; Hind et al., 2011), SCFSLY1-mediated gibberellin signaling (Dohmann et al., 2010) as well as defects in cold response (Schwechheimer et al., 2002), cell cycle progression (Dohmann et al., 2008a), and the control of ascorbic acid synthesis (Wang et al., 2013).
CSN REGULATION AND CSN REGULATORS
In view of the large number of CRLs that exist in eukaryotic cells and the importance of CSN-dependent cullin deneddylation for CRL function, it has to be asked how CRL neddylation and CSN-dependent deneddylation are regulated. In this context, it was shown for different CRLs that the availability of ubiquitylation substrate receptor and ubiquitylation substrate promotes CRL formation and cullin neddylation (Bornstein et al., 2006; Emberley et al., 2012). The association of FBP-SKP1 dimers can inhibit CSN function on selected SCF-complexes several fold (Emberley et al., 2012). Thus, in the case of SCF-complexes cullin neddylation and deneddylation are regulated by the presence of FBP-SKP1 dimers and particularly by the presence of a given FBP, assuming that there is no regulation on the level of the SKP1 adaptor subunit, which does not confer substrate specificity to the SCF complexes. CSN is even more strongly inhibited in the presence of degradation substrate and thus degradation substrate and degradation substrate receptor availability negatively regulate CSN activity (Emberley et al., 2012). Furthermore, CSN associates tightly with deneddylated SCF and CSN thereby keeps the CRL complex in a state of low activity after substrate degradation (Emberley et al., 2012). Through this interaction, CSN also prevents cullin neddylation, unless binding of a ubiquitylation substrate triggers its dissociation and allows for cullin neddylation (Enchev et al., 2012).
In addition to the regulation of CSN by CRLs, their subunits and their substrates, there are also other candidate regulators whose function remains to be determined. The 7 kDa protein SMALL ACIDIC PROTEIN1 (SMAP1) is an interesting CSN-interaction partner (Nakasone et al., 2012). smap1 mutants were identified as anti-auxin resistant1 (aar1) in a mutant screen that also identified mutants of TIR1, CULLIN1, and DCN1-LIKE. Thus the aar1/smap1 phenotype may well be explained by a defect in the E3 ligase SCFTIR1 or its neddylation or deneddylation. Importantly, immunoprecipitates of SMAP1 are very strongly enriched in at least six CSN subunits, indicating that SMAP1 is a CSN interactor and may regulate CSN function. As yet, the analysis of CULLIN neddylation patterns did not reveal any apparent defects in its neddylation or deneddylation but the aar3/smap1 mutant phenotype together with the SMAP1-CSN interaction strongly suggests that SMAP1 is linked to CSN function. Since there is not apparent homolog of SMAP1 outside of the plant kingdom, this function should be plant specific.
The analysis of the CSN-interacting Rig-G protein, a protein related to the Arabidopsis protein SPINDLY, provided some insights into how interaction partners could interfere with CSN activity. In the case of Rig-G it is proposed that the protein recruits CSN subunits to the cytoplasm and thereby interferes with CSN assembly in the nucleus (Xu et al., 2013).
CAND1 – A SUBSTRATE RECEPTOR EXCHANGE FACTOR FOR CRLs
Important progress has been achieved in the understanding of the role of neddylation and deneddylation of cullins in the context of CRL assembly and function through analysis of the protein Cullin-associated-Nedd8-dissociated-1 (CAND1). As its name already reveals, CAND1 was identified as an interactor of non-neddylated cullins (Liu et al., 2002; Zheng et al., 2002; Oshikawa et al., 2003). Through a series of elegant experiments from at least three independent laboratories it was recently shown that CAND1 functions as a novel type of exchange factor for CRLs (Pierce et al., 2013; Wu et al., 2013; Zemla et al., 2013). In a highly quantitative and not only therefore remarkable analysis of the diverse protein–protein interactions that can take place between the subunits of SCF-type CRL complexes and CAND1, it could be shown that CAND1 can promote the disassembly of SCF complexes and that FBPs can remove CAND1 from CULLIN1 (Pierce et al., 2013; Figure 4). When testing 21 different FBPs it was found that 20 of these could be exchanged using CAND1 as an exchange factor. Thus, in the case of SCF-type CRLs and most likely also in the case of CRLs that are formed with the other cullins, CAND1 can modulate the CRL-complex repertoire of the cell.
CAND1 is unable to interact with neddylated cullins and cullin neddylation stabilizes specific CRLs to prevent substrate receptor exchange (Emberley et al., 2012). Upon cullin deneddylation, CAND1 can become active and modulate the CRL repertoire to optimally match substrate receptor demand. Thus, there must be mechanisms to control CRL deneddylation. Indeed, cullin neddylation and deneddylation are controlled by the presence and absence of degradation substrates or their interactions with substrate receptors (Bornstein et al., 2006; Chew and Hagen, 2007; Emberley et al., 2012). Furthermore, CSN binds preferentially to neddylated CRLs, which may also recruit CSN-associated proteins important for CRL regulation (den Besten et al., 2012). CSN can bind deneddylated cullins but is dissociated in the presence of degradation substrate receptors and degradation substrates (Choo et al., 2011). Consequently, it can be inferred that csn as well as cand1 mutants are deficient in releasing specific substrate receptors. This hypothesis could be experimentally confirmed and it could be shown that substrate receptor activation and substrate degradation are delayed in such mutant backgrounds (Zemla et al., 2013).
CAND1 IN PLANTS
In plants, cand1 mutants were identified and analyzed in reverse and forward genetic screens. Mutants deficient in CAND1 were isolated as auxin-resistant mutants and the mutant spectrum of cand1 mutants was recognized as being highly similar to, but also to exceed that of axr1 mutants (Cheng et al., 2004; Chuang et al., 2004; Feng et al., 2004). Three further cand1 alleles were identified based on mutants with severe defects in leaf vein patterning (Alonso-Peral et al., 2006). In rice, CAND1 is required for the formation of crown roots and defects in crown root formation are associated with a cessation in the G2/M phase progression in these mutants (Wang et al., 2011). In this context it is interesting to note that also Arabidopsis csn mutants are defective in G2/M phase progression (Dohmann et al., 2008a).
Arabidopsis CAND1 also preferentially binds to non-neddylated cullins (Feng et al., 2004). Importantly, two sets of weak Arabidopsis mutants exist, the semi-dominant axr6-1 and axr6-2 on the one side and the recessive cul1-6 on the other, carrying missense mutations in almost adjacent positions of CULLIN1. Interestingly, the respective mutant proteins interact differentially with CAND1. While the cul1-6 protein is deficient in CAND1 interaction, axr6-1 and axr6-2 bind more strongly to CAND1 (Feng et al., 2004; Moon et al., 2007). The availability of mutants with weak and strong defects in cullin function, cullin deneddylation, and CAND1 interaction has already permitted to assay the biochemical interactions of the various components at the genetic level (Zhang et al., 2008).
NEDDYLATION SUBSTRATES
EVIDENCE FOR NON-CULLIN NEDDYLATION SUBSTRATES
Despite extensive research, the role and importance of neddylation in cellular processes besides the regulation of CRL activity remains poorly understood. Contrary to the expanding knowledge about ubiquitylated proteins in eukaryotes including plants (Kim et al., 2013) similar studies for NEDD8 have so far not succeeded in consistently identifying non-cullin neddylated proteins (Li et al., 2006; Norman and Shiekhattar, 2006; Jones et al., 2008; Xirodimas, 2008; Bennett et al., 2010; Hakenjos et al., 2011; Hotton et al., 2012). However, there is evidence for the existence of a broad range of neddylated proteins and several non-cullin neddylated proteins have already been identified as summarized in Table 1. Loss of function mutants of DEN1/NEDP1/SENP8 from three different species, namely fruit fly, Schizosaccharomyces pombe and Aspergillus nidulans accumulate neddylated proteins over a broad range of molecular weights. At the same time, these mutants do generally not accumulate neddylated cullins suggesting that DEN1/NEDP1/SENP8 is an important deneddylating enzyme of these non-cullin neddylation substrates (Zhou and Watts, 2005; Chan et al., 2008; Christmann et al., 2013). Also overexpression of NEDD8 leads to the apparent enrichment of many neddylated proteins and this neddylation can be blocked with the inhibitor MLN4924 (Hakenjos et al., 2011). Thus, there is evidence that NEDD8-modified proteins other than the cullins can exist but may be low in abundance or only transiently modified under normal conditions.
Table 1.
Neddylation substrates.
| Neddylated protein | Proposed function of neddylation | Species | Reference |
|---|---|---|---|
| E3 ubiquitin ligases | |||
| Cullins, Cul7, and PARC | Increases activity | Eukaryotes | Hori et al. (1999), Sarikas et al. (2011), Calabrese et al. (2011) |
| Mdm2 | Decreases activity | Human | Xirodimas et al. (2004) |
| Parkin | increases activity | Human | Um et al. (2012), Choo et al. (2012) |
| BRAP2 | – | Human | Takashima et al. (2013) |
| pVHL | Changes pVHL protein interaction | Human | Stickle et al. (2004), Russell and Ohh (2008) |
| DIAP1/XIAP | - | Fruit fly/human | Broemer et al. (2010) |
| DDB1 | - | Arabidopsis | Hotton et al. (2012) |
| Transcription factors | |||
| p53 | Inhibits transcriptional activity | Human | Xirodimas et al. (2004), Abida et al. (2007) |
| p73 | Inhibits transcriptional activity by sequestering Tap73 to the cytoplasm | Human | Watson et al. (2006) |
| AICD | Inhibits transcriptional activity | Human | Lee et al. (2008) |
| E2F1 | Inhibits transcriptional activity by blocking protein interaction | Human | Aoki et al. (2012), Loftus et al. (2012) |
| HIF1α | Stabilizes protein | Human | Ryu et al. (2011) |
| Transcriptional inhibitors | |||
| BCA3 | Activates by promoting protein interaction | Human | Gao et al. (2006) |
| RCAN1 | Stabilizes by inhibiting proteasomal degradation | Human | Noh et al. (2012) |
| Receptors | |||
| EGFR | Promotes receptor ubiquitylation and ligand induced degradation | Mammals | (Oved et al., 2006) |
| TβRII | Stabilizes protein | Human | (Zuo et al., 2013) |
| Kinases | |||
| PINK1 | Stabilizes the cytosolic protein form | Human | Choo et al. (2012) |
| CK1α | – | Human | Huart et al. (2012) |
| Other | |||
| L11, S14, and other ribosomal proteins | Stabilizes the protein | Human | Xirodimas et al. (2008), Zhang et al. (2012) |
| SHC | – | Human | Jin et al. (2013) |
| HUR | Stabilizes the protein | Human | Embade et al. (2012) |
| Histone H4 | Induces complex formation and amplifies Ubi cascade | Human | Ma et al. (2013) |
| drICE/caspase 7 | Reduces catalytic activity | Fruit fly/human | Broemer et al. (2010) |
| Lag2 | – | Yeast | Siergiejuk et al. (2009) |
| ML3 | – | Arabidopsis | Hakenjos et al. (2013) |
PARK, Parkin-like cytoplasmic protein; Mdm2, murine double minute 2; Parkin, Parkinson juvenile disease protein 2; BRAP2, BRCA1-associated protein; DIAP1, Drosophila inhibitor of apoptosis 1; XIAP, X-linked inhibitor of apoptosis protein; DDB1, damaged DNA binding protein1; VHL, Von Hippel–Lindau disease tumor suppressor; p53, cellular tumor antigen p53; p73, tumor protein p73; AICD, amyloid beta A4 protein; E2F1, transcription factor E2F1; HIF1α, hypoxia-inducible factor 3-alpha; BCA3, breast cancer-associated gene 3; RCAN1, regulator of calcineurin 1; EGFR, epidermal growth factor receptor; TβRII, transforming growth factor-beta receptor type II; PINK1, PTEN-induced putative kinase protein 1; CK1α, casein kinase 1 alpha; L11, ribosomal protein L11; S14, ribosomal protein S14; SHC, Src homology 2 domain-containing-transforming protein C1; HUR, Hu-antigen R; DrICE, Drosophila interleukin-1β-converting enzyme; Lag2, longevity-assurance protein 2; ML3, myeloid differentiation factor-2-related lipid-recognition domain protein 3.
At the functional level, genetic experiments in Schizosaccharomyces pombe showed that the introduction of a specific cullin1 mutant, which constitutively activates CRLs and therefore renders these CRLs independent from the neddylation machinery, was unable to rescue the phenotype of NEDD8 conjugation mutant uba3-10 (Girdwood et al., 2011, 2012). This finding may suggest additional biological functions for neddylation that are impaired in uba3-10 besides the neddylation defect of cullins.
IDENTIFICATION OF NEDDYLATION SUBSTRATES - A DIFFICULT ISSUE
A major problem for the identification of new neddylation substrates is the high sequence similarity between NEDD8 and ubiquitin. Both proteins are sequence identical at their C-termini just downstream of a trypsin cleavage site resulting in an identical di-glycine footprint on a modified protein after trypsin digestion for proteomic analyses (Figure 1). Therefore, a di-glycine modification on a lysine of a given peptide cannot unanimously be attributed to either ubiquitin or NEDD8 conjugation.
Ubiquitin and NEDD8 also show a remarkable similarity in their three-dimensional structure and key residues are conserved between the two proteins, most prominently three amino acids (L8, I44, and V70) in the hydrophobic patch that are involved in mediating ubiquitin-protein interactions (Rao-Naik et al., 1998; Whitby et al., 1998; Choi et al., 2009; Girdwood et al., 2011). Thus, a high substrate specificity is required to avoid leakage of ubiquitin or NEDD8 into the respective other modification pathway. Indeed, it has become apparent in the last years that there is a crosstalk between the NEDD8 and ubiquitin conjugation machineries (Hjerpe et al., 2012b; Leidecker et al., 2012; Singh et al., 2012). NEDD8 can be activated by the ubiquitin E1 UBA1 and once activated is conjugated to substrates in a manner similar to ubiquitin (Singh et al., 2012). Inversely, however, NAE specifically activates NEDD8 and does not use ubiquitin as a substrate (Singh et al., 2012). NEDD8 is incorporated in the ubiquitin pathway by UBA1 when the ratio of free NEDD8 to free ubiquitin, which under normal conditions is close to one, shifts toward NEDD8 (Hjerpe et al., 2012a; Leidecker et al., 2012). The leakage of NEDD8 into the ubiquitin pathway leads to the formation of mixed chains with NEDD8 possibly functioning as chain terminator. This mechanism is likely the explanation for the identification of NEDD8 chains in a proteomic study by Jones et al. (2008) using overexpression of a tagged NEDD8 construct. The ability of NEDD8 to form chain linkages is not essential in vivo as demonstrated by the viability of a Schizosaccharomyces pombe strain carrying a Ned8p mutant construct where all lysine residues that could potentially engage in chain formation were mutated to alanine (Girdwood et al., 2011). Depletion of cellular ubiquitin levels can be caused by knockdown or inhibition of the 26S proteasome but can also have physiological causes such as temperature or oxidative stress (Hjerpe et al., 2012a; Leidecker et al., 2012). While this atypical neddylation has been proposed to act as a stress response to ubiquitin depletion, it is still unclear whether this type of atypical neddylation is biologically relevant in vivo (Hjerpe et al., 2012a; Leidecker et al., 2012).
NOVEL NEDDYLATION SUBSTRATES
As outlined above, various studies have led to the identification of novel NEDD8-modified proteins. At the biochemical level, these proteins belong to different protein families including E3 ubiquitin ligases or transcription factors and neddylation has been shown to positively or negatively interfere with their activity (Table 1). Neddylation changes the biochemical properties of its target proteins by inducing conformational changes as well as allowing or precluding protein-protein interactions. Apart from its pleiotropic effects on protein degradation through CRL neddylation, neddylation has a prominent role in cell cycle regulation and cellular stress response pathways in human cells and has also been linked to Alzheimer and Parkinson disease (Table 1). In plants, the only known non-cullin neddylation substrates are DAMAGED DNA BINDING PROTEIN1 (DDB1) and ML3 from Arabidopsis. DDB1 is a subunit of a cullin4 E3 ubiquitin ligase and therefore biochemically close to the cells neddylation machinery (Hotton et al., 2012). ML3 is a protein with intriguing cell biological features that plays a role in pathogen responses (Hakenjos et al., 2013). Interestingly, there is also evidence that ML3 has the ability to bind neddylated proteins in a non-covalent manner. Since the precise biochemical function of ML3 remains to be determined, it is at present unclear what role neddylation has in the control of ML3 function.
Given the very recently acquired understanding of the close interplay between ubiquitylation and neddylation, new and old neddylation targets should undergo close scrutiny to ensure that they are genuine targets for NEDD8 modification (Hjerpe et al., 2012b). A catalog of appropriate characterization criteria has been published (Rabut and Peter, 2008).
CONCLUSION
In this review, we have summarized the current knowledge of the neddylation pathway in eukaryotes with an emphasis on the role of neddylation in plants. While the enzyme pathway for the conjugation and deconjugation of NEDD8 has been elucidated in plants, findings from other eukaryotic model organisms suggest that there are more, unknown players in this pathway that need to be identified to gain a full understanding of the process and its regulation. Particularly the areas of NEDD8 processing, NEDD8 ligases and the identification of non-cullin NEDD8 substrates will require further detailed investigations in the future. Also the presence of ubiquitin-NEDD8 fusion proteins is unique to plants. The analysis of their processing could bear information that may allow understanding how the highly homologous ubiquitin and NEDD8 proteins were derived from each other during evolution.
AUTHOR CONTRIBUTIONS
The authors have contributed in equal parts to the preparation of this manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Work on NEDD8 in the authors’ laboratory is funded by the Priority Program SPP 1365 of the Deutsche Forschungsgemeinschaft.
REFERENCES
- Abida W. M., Nikolaev A., Zhao W., Zhang W., Gu W. (2007). FBXO11 promotes the Neddylation of p53 and inhibits its transcriptional activity. J. Biol. Chem. 282 1797–1804 10.1074/jbc.M609001200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Peral M. M., Candela H., Del Pozo J. C., Martinez-Laborda A., Ponce M. R., Micol J. L. (2006). The HVE/CAND1 gene is required for the early patterning of leaf venation in Arabidopsis. Development 133 3755–3766 10.1242/dev.02554 [DOI] [PubMed] [Google Scholar]
- Ambroggio X. I., Rees D. C., Deshaies R. J. (2004). JAMM: a metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol. 2:e2 10.1371/journal.pbio.0020002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki I., Higuchi M., Gotoh Y. (2012). NEDDylation controls the target specificity of E2F1 and apoptosis induction. Oncogene 32 3954–3964 10.1038/onc.2012.428 [DOI] [PubMed] [Google Scholar]
- Bennett E. J., Rush J., Gygi S. P., Harper J. W. (2010). Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell 143 951–965 10.1016/j.cell.2010.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt A., Lechner E., Hano P., Schade V., Dieterle M., Anders M., et al. (2006). CUL4 associates with DDB1 and DET1 and its downregulation affects diverse aspects of development in Arabidopsis thaliana. Plant J. 47 591–603 10.1111/j.1365-313X.2006.02810.x [DOI] [PubMed] [Google Scholar]
- Biswas K. K., Ooura C., Higuchi K., Miyazaki Y., Van Nguyen V., Rahman A., et al. (2007). Genetic characterization of mutants resistant to the antiauxin p-chlorophenoxyisobutyric acid reveals that AAR3, a gene encoding a DCN1-like protein, regulates responses to the synthetic auxin 2,4-dichlorophenoxyacetic acid in Arabidopsis roots. Plant Physiol. 145 773–785 10.1104/pp.107.104844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boh B. K., Smith P. G., Hagen T. (2011). Neddylation-induced conformational control regulates cullin RING ligase activity in vivo. J. Mol. Biol. 409 136–145 10.1016/j.jmb.2011.03.023 [DOI] [PubMed] [Google Scholar]
- Bornstein G., Ganoth D., Hershko A. (2006). Regulation of neddylation and deneddylation of cullin1 in SCFSkp2 ubiquitin ligase by F-box protein and substrate. Proc. Natl. Acad. Sci. U.S.A. 103 11515–11520 10.1073/pnas.0603921103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick M., Lochhead S. R., Honda A., Palmer S., Callis J. (2004). Related to ubiquitin 1 and 2 are redundant and essential and regulate vegetative growth, auxin signaling, and ethylene production in Arabidopsis. Plant Cell 16 2418–2432 10.1105/tpc.104.024943tpc.104.024943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broemer M., Tenev T., Rigbolt K. T., Hempel S., Blagoev B., Silke J., et al. (2010). Systematic in vivo RNAi analysis identifies IAPs as NEDD8-E3 ligases. Mol. Cell. 40 810–822 10.1016/j.molcel.2010.11.011 [DOI] [PubMed] [Google Scholar]
- Brownell J. E., Sintchak M. D., Gavin J. M., Liao H., Bruzzese F. J., Bump N. J., et al. (2010). Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol. Cell. 37 102–111 10.1016/j.molcel.2009.12.024 [DOI] [PubMed] [Google Scholar]
- Calabrese M. F., Scott D. C., Duda D. M., Grace C. R., Kurinov I., Kriwacki R. W., et al. (2011). A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases. Nat. Struct. Mol. Biol. 18 947–949 10.1038/nsmb.2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis J., Carpenter T., Sun C. W., Vierstra R. D. (1995). Structure and evolution of genes encoding polyubiquitin and ubiquitin-like proteins in Arabidopsis thaliana ecotype Columbia. Genetics 139 921–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamovitz D. A., Wei N., Osterlund M. T., Von Arnim A. G., Staub J. M., Matsui M., et al. (1996). The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell 86 115–121 10.1016/S0092-8674(00)80082-3 [DOI] [PubMed] [Google Scholar]
- Chan Y., Yoon J., Wu J. T., Kim H. J., Pan K. T., Yim J., et al. (2008). DEN1 deneddylates non-cullin proteins in vivo. J. Cell Sci. 121 3218–3223 10.1242/jcs.030445 [DOI] [PubMed] [Google Scholar]
- Chen H., Shen Y., Tang X., Yu L., Wang J., Guo L., et al. (2006). Arabidopsis CULLIN4 forms an E3 ubiquitin ligase with RBX1 and the CDD complex in mediating light control of development. Plant Cell 18 1991–2004 10.1105/tpc.106.043224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Dai X., Zhao Y. (2004). AtCAND1, a HEAT-repeat protein that participates in auxin signaling in Arabidopsis. Plant Physiol. 135 1020–1026 10.1104/pp.104.044495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew E. H., Hagen T. (2007). Substrate-mediated regulation of cullin neddylation. J. Biol. Chem. 282 17032–17040 10.1074/jbc.M701153200 [DOI] [PubMed] [Google Scholar]
- Choi Y. S., Jeon Y. H., Ryu K. S., Cheong C. (2009). 60th residues of ubiquitin and Nedd8 are located out of E2-binding surfaces, but are important for K48 ubiquitin-linkage. FEBS Lett. 583 3323–3328 10.1016/j.febslet.2009.09.034 [DOI] [PubMed] [Google Scholar]
- Choo Y. S., Vogler G., Wang D., Kalvakuri S., Iliuk A., Tao W. A., et al. (2012). Regulation of parkin and PINK1 by neddylation. Hum. Mol. Genet. 21 2514–2523 10.1093/hmg/dds070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo Y. Y., Boh B. K., Lou J. J., Eng J., Leck Y. C., Anders B., et al. (2011). Characterization of the role of COP9 signalosome in regulating cullin E3 ubiquitin ligase activity. Mol. Biol. Cell 22 4706–4715 10.1091/mbc.E11-03-0251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christmann M., Schmaler T., Gordon C., Huang X., Bayram O., Schinke J., et al. (2013). Control of multicellular development by the physically interacting deneddylases DEN1/DenA and COP9 signalosome. PLoS Genet. 9:e1003275 10.1371/journal.pgen.1003275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang H. W., Zhang W., Gray W. M. (2004). Arabidopsis ETA2, an apparent ortholog of the human cullin-interacting protein CAND1, is required for auxin responses mediated by the SCF(TIR1) ubiquitin ligase. Plant Cell 16 1883–1897 10.1105/tpc.021923tpc.021923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope G. A., Suh G. S., Aravind L., Schwarz S. E., Zipursky S. L., Koonin E. V., et al. (2002). Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 298 608–611 10.1126/science.1075901 [DOI] [PubMed] [Google Scholar]
- del Pozo J. C., Estelle M. (1999). The Arabidopsis cullin AtCUL1 is modified by the ubiquitin-related protein RUB1. Proc. Natl. Acad. Sci. U.S.A. 96 15342–15347 10.1073/pnas.96.26.15342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo J. C., Timpte C., Tan S., Callis J., Estelle M. (1998). The ubiquitin-related protein RUB1 and auxin response in Arabidopsis. Science 280 1760–1763 10.1126/science.280.5370.1760 [DOI] [PubMed] [Google Scholar]
- den Besten W., Verma R., Kleiger G., Oania R. S., Deshaies R. J. (2012). NEDD8 links cullin-RING ubiquitin ligase function to the p97 pathway. Nat. Struct. Mol. Biol. 19 511–516 S511 10.1038/nsmb.2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Estelle M. (2005a). The F-box protein TIR1 is an auxin receptor. Nature 435 441–445 10.1038/nature03543 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Weijers D., Lechner E., Yamada M., Hobbie L., et al. (2005b). Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 9 109–119 10.1016/j.devcel.2005.05.014 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Weijers D., Karunarathna N., Jurgens G., Estelle M. (2007). AXL and AXR1 have redundant functions in RUB conjugation and growth and development in Arabidopsis. Plant J. 52 114–123 10.1111/j.1365-313X.2007.03211.x [DOI] [PubMed] [Google Scholar]
- Dharmasiri S., Dharmasiri N., Hellmann H., Estelle M. (2003). The RUB/Nedd8 conjugation pathway is required for early development in Arabidopsis. EMBO J. 22 1762–1770 10.1093/emboj/cdg190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterle M., Thomann A., Renou J. P., Parmentier Y., Cognat V., Lemonnier G., et al. (2005). Molecular and functional characterization of Arabidopsis Cullin 3A. Plant J. 41 386–399 10.1111/j.1365-313X.2004.02302.x [DOI] [PubMed] [Google Scholar]
- Dill A., Thomas S. G., Hu J., Steber C. M., Sun T. P. (2004). The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmann E. M., Kuhnle C., Schwechheimer C. (2005). Loss of the CONSTITUTIVE PHOTOMORPHOGENIC9 signalosome subunit 5 is sufficient to cause the cop/det/fus mutant phenotype in Arabidopsis. Plant Cell 17 1967–1978 10.1105/tpc.105.032870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmann E. M., Levesque M. P., De Veylder L., Reichardt I., Jurgens G., Schmid M., et al. (2008a). The Arabidopsis COP9 signalosome is essential for G2 phase progression and genomic stability. Development 135 2013–2022 10.1242/dev.020743 [DOI] [PubMed] [Google Scholar]
- Dohmann E. M., Levesque M. P., Isono E., Schmid M., Schwechheimer C. (2008b). Auxin responses in mutants of the Arabidopsis CONSTITUTIVE PHOTOMORPHOGENIC9 signalosome. Plant Physiol. 147 1369–1379 10.1104/pp.108.121061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmann E. M., Nill C., Schwechheimer C. (2010). DELLA proteins restrain germination and elongation growth in Arabidopsis thaliana COP9 signalosome mutants. Eur. J. Cell Biol. 89 163–168 10.1016/j.ejcb.2009.12.001 [DOI] [PubMed] [Google Scholar]
- Duda D. M., Borg L. A., Scott D. C., Hunt H. W., Hammel M., Schulman B. A. (2008). Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 134 995–1006 10.1016/j.cell.2008.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embade N., Fernandez-Ramos D., Varela-Rey M., Beraza N., Sini M., Gutierrez De Juan V., et al. (2012). Murine double minute 2 regulates Hu antigen R stability in human liver and colon cancer through NEDDylation. Hepatology 55 1237–1248 10.1002/hep.24795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emberley E. D., Mosadeghi R., Deshaies R. J. (2012). Deconjugation of Nedd8 from Cul1 is directly regulated by Skp1-F-box and substrate, and the COP9 signalosome inhibits deneddylated SCF by a noncatalytic mechanism. J. Biol. Chem. 287 29679–29689 10.1074/jbc.M112.352484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enchev R. I., Schreiber A., Beuron F., Morris E. P. (2010). Structural insights into the COP9 signalosome and its common architecture with the 26S proteasome lid and eIF3. Structure 18 518–527 10.1016/j.str.2010.02.008 [DOI] [PubMed] [Google Scholar]
- Enchev R. I., Scott D. C., Da Fonseca P. C., Schreiber A., Monda J. K., Schulman B. A., et al. (2012). Structural basis for a reciprocal regulation between SCF and CSN. Cell Rep. 2 616–627 10.1016/j.celrep.2012.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve-Bruna D., Perez-Perez J. M., Ponce M. R., Micol J. L. (2013). incurvata13, a novel allele of AUXIN RESISTANT6, reveals a specific role for auxin and the SCF complex in Arabidopsis embryogenesis, vascular specification, and leaf flatness. Plant Physiol. 161 1303–1320 10.1104/pp.112.207779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Shen Y., Sullivan J. A., Rubio V., Xiong Y., Sun T. P., et al. (2004). Arabidopsis CAND1, an unmodified CUL1-interacting protein, is involved in multiple developmental pathways controlled by ubiquitin/proteasome-mediated protein degradation. Plant Cell 16 1870–1882 10.1105/tpc.021949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa P., Gusmaroli G., Serino G., Habashi J., Ma L., Shen Y., et al. (2005). Arabidopsis has two redundant Cullin3 proteins that are essential for embryo development and that interact with RBX1 and BTB proteins to form multisubunit E3 ubiquitin ligase complexes in vivo. Plant Cell 17 1180–1195 10.1105/tpc.105.031989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frickel E. M., Quesada V., Muething L., Gubbels M. J., Spooner E., Ploegh H., et al. (2007). Apicomplexan UCHL3 retains dual specificity for ubiquitin and Nedd8 throughout evolution. Cell Microbiol. 9 1601–1610 10.1111/j.1462-5822.2007.00896.x [DOI] [PubMed] [Google Scholar]
- Fu H., Reis N., Lee Y., Glickman M. H., Vierstra R. D. (2001). Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. EMBO J. 20 7096–7107 10.1093/emboj/20.24.7096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan-Erdene T., Nagamalleswari K., Yin L., Wu K., Pan Z. Q., Wilkinson K. D. (2003). Identification and characterization of DEN1, a deneddylase of the ULP family. J. Biol. Chem. 278 28892–28900 10.1074/jbc.M302890200 [DOI] [PubMed] [Google Scholar]
- Gao F., Cheng J., Shi T., Yeh E. T. (2006). Neddylation of a breast cancer-associated protein recruits a class III histone deacetylase that represses NFkappaB-dependent transcription. Nat. Cell Biol. 8 1171–1177 10.1038/ncb1483. [DOI] [PubMed] [Google Scholar]
- Gingerich D. J., Gagne J. M., Salter D. W., Hellmann H., Estelle M., Ma L., et al. (2005). Cullins 3a and 3b assemble with members of the broad complex/tramtrack/bric-a-brac (BTB) protein family to form essential ubiquitin-protein ligases (E3s) in Arabidopsis. J. Biol. Chem. 280 18810–18821 10.1074/jbc.M413247200 [DOI] [PubMed] [Google Scholar]
- Girdwood D., Robertson M., Gordon C. (2012). Constitutively active Cullin-RING-Ligases fail to rescue loss of NEDD8 conjugation in Schizosaccharomyces pombe. FEBS Lett. 586 1522–1528 10.1016/j.febslet.2012.04.011 [DOI] [PubMed] [Google Scholar]
- Girdwood D., Xirodimas D. P., Gordon C. (2011). The essential functions of NEDD8 are mediated via distinct surface regions, and not by polyneddylation in Schizosaccharomyces pombe. PLoS ONE 6:e20089 10.1371/journal.pone.0020089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman M. H., Rubin D. M., Coux O., Wefes I., Pfeifer G., Cjeka Z., et al. (1998). A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 94 615–623 10.1016/S0092-8674(00)81603-7 [DOI] [PubMed] [Google Scholar]
- Gray W. M., Hellmann H., Dharmasiri S., Estelle M. (2002). Role of the Arabidopsis RING-H2 protein RBX1 in RUB modification and SCF function. Plant Cell 14 2137–2144 10.1105/tpc.003178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray W. M., Kepinski S., Rouse D., Leyser O., Estelle M. (2001). Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414 271–276 10.1038/35104500 [DOI] [PubMed] [Google Scholar]
- Gusmaroli G., Figueroa P., Serino G., Deng X. W. (2007). Role of the MPN subunits in COP9 signalosome assembly and activity, and their regulatory interaction with Arabidopsis Cullin3-based E3 ligases. Plant Cell 19 564–581 10.1105/tpc.106.047571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakenjos J. P., Bejai S., Ranftl Q., Behringer C., Vlot A. C., Absmanner B., et al. (2013). ML3 is a NEDD8- and ubiquitin-modified protein. Plant Physiol. 163 135–149 10.1104/pp.113.221341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakenjos J. P., Richter R., Dohmann E. M., Katsiarimpa A., Isono E., Schwechheimer C. (2011). MLN4924 is an efficient inhibitor of NEDD8 conjugation in plants. Plant Physiol. 156 527–536 10.1104/pp.111.176677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann T., Mayer U., Jurgens G. (1999). The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development 126 1387–1395 [DOI] [PubMed] [Google Scholar]
- Heir P., Sufan R. I., Greer S. N., Poon B. P., Lee J. E., Ohh M. (2013). DCNL1 functions as a substrate sensor and activator of cullin 2-RING ligase. Mol. Cell. Biol. 33 1621–1631 10.1128/MCB.01342-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann H., Hobbie L., Chapman A., Dharmasiri S., Dharmasiri N., Del Pozo C., et al. (2003). Arabidopsis AXR6 encodes CUL1 implicating SCF E3 ligases in auxin regulation of embryogenesis. EMBO J. 22 3314–3325 10.1093/emboj/cdg335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemelaar J., Borodovsky A., Kessler B. M., Reverter D., Cook J., Kolli N., et al. (2004). Specific and covalent targeting of conjugating and deconjugating enzymes of ubiquitin-like proteins. Mol. Cell. Biol. 24 84–95 10.1128/MCB.24.1.84-95.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hind S. R., Pulliam S. E., Veronese P., Shantharaj D., Nazir A., Jacobs N. S., et al. (2011). The COP9 signalosome controls jasmonic acid synthesis and plant responses to herbivory and pathogens. Plant J. 65 480–491 10.1111/j.1365-313X.2010.04437.x [DOI] [PubMed] [Google Scholar]
- Hjerpe R., Thomas Y., Chen J., Zemla A., Curran S., Shpiro N., et al. (2012a). Changes in the ratio of free NEDD8 to ubiquitin triggers NEDDylation by ubiquitin enzymes. Biochem. J. 441 927–936 10.1042/BJ20111671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjerpe R., Thomas Y., Kurz T. (2012b). NEDD8 overexpression results in neddylation of ubiquitin substrates by the ubiquitin pathway. J. Mol. Biol. 421 27–29 10.1016/j.jmb.2012.05.013 [DOI] [PubMed] [Google Scholar]
- Hobbie L., Mcgovern M., Hurwitz L. R., Pierro A., Liu N. Y., Bandyopadhyay A., et al. (2000). The axr6 mutants of Arabidopsis thaliana define a gene involved in auxin response and early development. Development 127 23–32 [DOI] [PubMed] [Google Scholar]
- Hori T., Osaka F., Chiba T., Miyamoto C., Okabayashi K., Shimbara N., et al. (1999). Covalent modification of all members of human cullin family proteins by NEDD8. Oncogene 18 6829–6834 10.1038/sj.onc.1203093 [DOI] [PubMed] [Google Scholar]
- Hotton S. K., Castro M. F., Eigenheer R. A., Callis J. (2012). Recovery of DDB1a (damaged DNA binding protein1a) in a screen to identify novel RUB-modified proteins in Arabidopsis thaliana. Mol. Plant 5 1163–1166 10.1093/mp/sss077 [DOI] [PubMed] [Google Scholar]
- Hotton S. K., Eigenheer R. A., Castro M. F., Bostick M., Callis J. (2011). AXR1-ECR1 and AXL1-ECR1 heterodimeric RUB-activating enzymes diverge in function in Arabidopsis thaliana. Plant Mol. Biol. 75 515–526 10.1007/s11103-011-9750-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z., Vierstra R. D. (2011). The cullin-RING ubiquitin-protein ligases. Annu. Rev. Plant Biol. 62 299–334 10.1146/annurev-arplant-042809-112256 [DOI] [PubMed] [Google Scholar]
- Huang C. R., Burns K. H., Boeke J. D. (2012). Active transposition in genomes. Annu. Rev. Genet. 46 651–675 10.1146/annurev-genet-110711-155616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Quint M., Gray W. M. (2013a). The eta7/csn3-3 auxin response mutant of Arabidopsis defines a novel function for the CSN3 subunit of the COP9 signalosome. PLoS ONE 8:e66578 10.1371/journal.pone.0066578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Ouyang X., Yang P., Lau O. S., Chen L., Wei N., et al. (2013b). Conversion from CUL4-based COP1-SPA E3 apparatus to UVR8-COP1-SPA complexes underlies a distinct biochemical function of COP1 under UV-B. Proc. Natl. Acad. Sci. U.S.A. 110 16669–16674 10.1073/pnas.1316622110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huart A. S., Maclaine N. J., Narayan V., Hupp T. R. (2012). Exploiting the MDM2-CK1alpha protein-protein interface to develop novel biologics that induce UBL-kinase-modification and inhibit cell growth. PLoS ONE 7:e43391 10.1371/journal.pone.0043391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S., Pyrowolakis G. (2000). Ubiquitin and its kin: how close are the family ties? Trends Cell Biol. 10 335–342 10.1016/S0962-8924(00)01785-2 [DOI] [PubMed] [Google Scholar]
- Jin H. S., Liao L., Park Y., Liu Y. C. (2013). Neddylation pathway regulates T-cell function by targeting an adaptor protein Shc and a protein kinase Erk signaling. Proc. Natl. Acad. Sci. U.S.A. 110 624–629 10.1073/pnas.1213819110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston S. C., Riddle S. M., Cohen R. E., Hill C. P. (1999). Structural basis for the specificity of ubiquitin C-terminal hydrolases. EMBO J. 18 3877–3887 10.1093/emboj/18.14.3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D., Candido E. P. (2000). The NED-8 conjugating system in Caenorhabditis elegans is required for embryogenesis and terminal differentiation of the hypodermis. Dev. Biol. 226 152–165 10.1006/dbio.2000.9847 [DOI] [PubMed] [Google Scholar]
- Jones D. T., Taylor W. R., Thornton J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Comp. Appl. Biosci. 8 275–282 [DOI] [PubMed] [Google Scholar]
- Jones J., Wu K., Yang Y., Guerrero C., Nillegoda N., Pan Z. Q., et al. (2008). A targeted proteomic analysis of the ubiquitin-like modifier nedd8 and associated proteins. J. Proteome Res. 7 1274–1287 10.1021/pr700749v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. Y., Scalf M., Smith L. M., Vierstra R. D. (2013). Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell 25 1523–1540 10.1105/tpc.112.108613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotiguda G. G., Weinberg D., Dessau M., Salvi C., Serino G., Chamovitz D. A., et al. (2012). The organization of a CSN5-containing subcomplex of the COP9 signalosome. J. Biol. Chem. 287 42031–42041 10.1074/jbc.M112.387977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Yoshida Y., Noda M. (1993). Cloning of a cDNA which encodes a novel ubiquitin-like protein. Biochem. Biophys. Res. Commun. 195 393–399 10.1006/bbrc.1993.2056 [DOI] [PubMed] [Google Scholar]
- Kurihara L. J., Semenova E., Levorse J. M., Tilghman S. M. (2000). Expression and functional analysis of Uch-L3 during mouse development. Mol. Cell. Biol. 20 2498–2504 10.1128/MCB.20.7.2498-2504.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz T., Chou Y. C., Willems A. R., Meyer-Schaller N., Hecht M. L., Tyers M., et al. (2008). Dcn1 functions as a scaffold-type E3 ligase for cullin neddylation. Mol. Cell. 29 23–35 10.1016/j.molcel.2007.12.012 [DOI] [PubMed] [Google Scholar]
- Kurz T., Ozlu N., Rudolf F., O’Rourke S. M., Luke B., Hofmann K., et al. (2005). The conserved protein DCN-1/Dcn1p is required for cullin neddylation in C. elegans and S. cerevisiae. Nature 435 1257–1261 10.1038/nature03662 [DOI] [PubMed] [Google Scholar]
- Lammer D., Mathias N., Laplaza J. M., Jiang W., Liu Y., Callis J., et al. (1998). Modification of yeast Cdc53p by the ubiquitin-related protein rub1p affects function of the SCFCdc4 complex. Genes Dev. 12 914–926 10.1101/gad.12.7.914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau O. S., Deng X. W. (2012). The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 17 584-593 10.1016/j.tplants.2012.05.004 [DOI] [PubMed] [Google Scholar]
- Lechner E., Xie D., Grava S., Pigaglio E., Planchais S., Murray J. A., et al. (2002). The AtRbx1 protein is part of plant SCF complexes, and its down-regulation causes severe growth and developmental defects. J. Biol. Chem. 277 50069–50080 10.1074/jbc.M204254200 [DOI] [PubMed] [Google Scholar]
- Lee M. R., Lee D., Shin S. K., Kim Y. H., Choi C. Y. (2008). Inhibition of APP intracellular domain (AICD) transcriptional activity via covalent conjugation with Nedd8. Biochem. Biophys. Res. Commun. 366 976–981 10.1016/j.bbrc.2007.12.066 [DOI] [PubMed] [Google Scholar]
- Leidecker O., Matic I., Mahata B., Pion E., Xirodimas D. P. (2012). The ubiquitin E1 enzyme Ube1 mediates NEDD8 activation under diverse stress conditions. Cell Cycle 11 1142–1150 10.4161/cc.11.6.19559 [DOI] [PubMed] [Google Scholar]
- Leyser H. M., Lincoln C. A., Timpte C., Lammer D., Turner J., Estelle M. (1993). Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature 364 161–164 10.1038/364161a0 [DOI] [PubMed] [Google Scholar]
- Li T., Santockyte R., Shen R. F., Tekle E., Wang G., Yang D. C., et al. (2006). A general approach for investigating enzymatic pathways and substrates for ubiquitin-like modifiers. Arch. Biochem. Biophys. 453 70–74 10.1016/j.abb.2006.03.002 [DOI] [PubMed] [Google Scholar]
- Liakopoulos D., Busgen T., Brychzy A., Jentsch S., Pause A. (1999). Conjugation of the ubiquitin-like protein NEDD8 to cullin-2 is linked to von Hippel–Lindau tumor suppressor function. Proc. Natl. Acad. Sci. U.S.A. 96 5510–5515 10.1073/pnas.96.10.5510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln C., Britton J. H., Estelle M. (1990). Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2 1071–1080 10.1105/tpc.2.11.1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linghu B., Callis J., Goebl M. G. (2002). Rub1p processing by Yuh1p is required for wild-type levels of Rub1p conjugation to Cdc53p. Eukaryot. Cell 1 491–494 10.1128/EC.1.3.491-494.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Furukawa M., Matsumoto T., Xiong Y. (2002). NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol. Cell. 10 1511–1518 10.1016/S1097-2765(02)00783-9 [DOI] [PubMed] [Google Scholar]
- Loftus S. J., Liu G., Carr S. M., Munro S, La Thangue N. B. (2012). NEDDylation regulates E2F-1-dependent transcription. EMBO Rep. 13 811–818 10.1038/embor.2012.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T., Chen Y., Zhang F., Yang C. Y., Wang S., Yu X. (2013). RNF111-dependent neddylation activates DNA damage-induced ubiquitination. Mol. Cell. 49 897–907 10.1016/j.molcel.2013.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maytal-Kivity V., Pick E., Piran R., Hofmann K., Glickman M. H. (2003). The COP9 signalosome-like complex in S. cerevisiae and links to other PCI complexes. Int. J. Biochem. Cell Biol. 35 706–715 10.1016/S1357-2725(02)00378-3 [DOI] [PubMed] [Google Scholar]
- Maytal-Kivity V., Reis N., Hofmann K., Glickman M. H. (2002). MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 3:28 10.1186/1471-2091-3-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza H. M., Shen L. N., Botting C., Lewis A., Chen J., Ink B., et al. (2003). NEDP1, a highly conserved cysteine protease that deNEDDylates Cullins. J. Biol. Chem. 278 25637–25643 10.1074/jbc.M212948200 [DOI] [PubMed] [Google Scholar]
- Meyer-Schaller N., Chou Y. C., Sumara I., Martin D. D., Kurz T., Katheder N., et al. (2009). The human Dcn1-like protein DCNL3 promotes Cul3 neddylation at membranes. Proc. Natl. Acad. Sci. U.S.A. 106 12365–12370 10.1073/pnas.0812528106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J., Zhao Y., Dai X., Zhang W., Gray W. M., Huq E., et al. (2007). A new CULLIN 1 mutant has altered responses to hormones and light in Arabidopsis. Plant Physiol. 143 684–696 10.1104/pp.106.091439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto M., Nishida T., Nagayama Y., Yasuda H. (2003). Nedd8-modification of Cul1 is promoted by Roc1 as a Nedd8-E3 ligase and regulates its stability. Biochem. Biophys. Res. Commun. 301 392–398 10.1016/S0006-291X(02)03051-6 [DOI] [PubMed] [Google Scholar]
- Nakasone A., Fujiwara M., Fukao Y., Biswas K. K., Rahman A., Kawai-Yamada M., et al. (2012). SMALL ACIDIC PROTEIN1 acts with RUB modification components, the COP9 signalosome, and AXR1 to regulate growth and development of Arabidopsis. Plant Physiol. 160 93–105 10.1104/pp.111.188409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D. C., Scaffidi A., Dun E. A., Waters M. T., Flematti G. R., Dixon K. W., et al. (2011). F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 108 8897–8902 10.1073/pnas.1100987108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh E. H., Hwang H. S., Hwang H. S., Min B., Im E., Chung K. C. (2012). Covalent NEDD8 conjugation increases RCAN1 protein stability and potentiates its inhibitory action on calcineurin. PLoS ONE 7:e48315 10.1371/journal.pone.0048315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman J. A., Shiekhattar R. (2006). Analysis of Nedd8-associated polypeptides: a model for deciphering the pathway for ubiquitin-like modifications. Biochemistry 45 3014–3019 10.1021/bi052435a [DOI] [PubMed] [Google Scholar]
- O’Donoghue J. E., Bech-Otschir D., Larsen I. B., Wallace M., Hartmann-Petersen R., Gordon C. (2013). Nedd8 processing enzymes in Schizosaccharomyces pombe. BMC Biochem. 14:8 10.1186/1471-2091-14-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaka F., Saeki M., Katayama S., Aida N., Toh E. A., Kominami K., et al. (2000). Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. EMBO J. 19 3475–3484 10.1093/emboj/19.13.3475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshikawa K., Matsumoto M., Yada M., Kamura T., Hatakeyama S., Nakayama K. I. (2003). Preferential interaction of TIP120A with Cul1 that is not modified by NEDD8 and not associated with Skp1. Biochem. Biophys. Res. Commun. 303 1209–1216 10.1016/S0006-291X(03)00501-1 [DOI] [PubMed] [Google Scholar]
- Osterlund M. T., Hardtke C. S., Wei N., Deng X. W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405 462–466 10.1038/35013076 [DOI] [PubMed] [Google Scholar]
- Ou C. Y., Lin Y. F., Chen Y. J., Chien C. T. (2002). Distinct protein degradation mechanisms mediated by Cul1 and Cul3 controlling Ci stability in Drosophila eye development. Genes Dev. 16 2403–2414 10.1101/gad.1011402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oved S., Mosesson Y., Zwang Y., Santonico E., Shtiegman K., Marmor M. D., et al. (2006). Conjugation to Nedd8 instigates ubiquitylation and down-regulation of activated receptor tyrosine kinases. J. Biol. Chem. 281 21640–21651 10.1074/jbc.M513034200 [DOI] [PubMed] [Google Scholar]
- Pacin M., Legris M., Casal J. J. (2013). COP1 re-accumulates in the nucleus under shade. Plant J. 75 631–641 10.1111/tpj.12226 [DOI] [PubMed] [Google Scholar]
- Pacurar D. I., Pacurar M. L., Street N., Bussell J. D., Pop T. I., Gutierrez L., et al. (2012). A collection of INDEL markers for map-based cloning in seven Arabidopsis accessions. J. Exp. Bot. 63 2491–2501 10.1093/jxb/err422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce N. W., Lee J. E., Liu X., Sweredoski M. J., Graham R. L., Larimore E. A., et al. (2013). Cand1 promotes assembly of new SCF complexes through dynamic exchange of F box proteins. Cell 153 206–215 10.1016/j.cell.2013.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M., Ito H., Zhang W., Gray W. M. (2005). Characterization of a novel temperature-sensitive allele of the CUL1/AXR6 subunit of SCF ubiquitin-ligases. Plant J. 43 371–383 10.1111/j.1365-313X.2005.02449.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut G., Le Dez G., Verma R., Makhnevych T., Knebel A., Kurz T., et al. (2011). The TFIIH subunit Tfb3 regulates cullin neddylation. Mol. Cell. 43 488–495 10.1016/j.molcel.2011.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut G., Peter M. (2008). Function and regulation of protein neddylation. `Protein modifications: beyond the usual suspects' review series. EMBO Rep. 9 969–976 10.1038/embor.2008.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao-Naik C., Delacruz W., Laplaza J. M., Tan S., Callis J., Fisher A. J. (1998). The rub family of ubiquitin-like proteins. Crystal structure of Arabidopsis rub1 and expression of multiple rubs in Arabidopsis. J. Biol. Chem. 273 34976–34982 10.1074/jbc.273.52.34976 [DOI] [PubMed] [Google Scholar]
- Ruegger M., Dewey E., Gray W. M., Hobbie L., Turner J., Estelle M. (1998). The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev. 12 198–207 10.1101/gad.12.2.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R. C., Ohh M. (2008). NEDD8 acts as a `molecular switch' defining the functional selectivity of VHL. EMBO Rep. 9 486–491 10.1038/embor.2008.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu J. H., Li S. H., Park H. S., Park J. W., Lee B., Chun Y. S. (2011). Hypoxia-inducible factor alpha subunit stabilization by NEDD8 conjugation is reactive oxygen species-dependent. J. Biol. Chem. 286 6963–6970 10.1074/jbc.M110.188706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarikas A., Hartmann T., Pan Z. Q. (2011). The cullin protein family. Genome Biol. 12 220 10.1186/gb-2011-12-4-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C., Serino G., Callis J., Crosby W. L., Lyapina S., Deshaies R. J., et al. (2001). Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science 292 1379–1382 10.1126/science.1059776 [DOI] [PubMed] [Google Scholar]
- Schwechheimer C., Serino G., Deng X. W. (2002). Multiple ubiquitin ligase-mediated processes require COP9 signalosome and AXR1 function. Plant Cell 14 2553–2563 10.1105/tpc.003434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D. C., Monda J. K., Bennett E. J., Harper J. W., Schulman B. A. (2011). N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science 334 674–678 10.1126/science.1209307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D. C., Monda J. K., Grace C. R., Duda D. M., Kriwacki R. W., Kurz T., et al. (2010). A dual E3 mechanism for Rub1 ligation to Cdc53. Mol. Cell. 39 784–796 10.1016/j.molcel.2010.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M., Kraft R., Ferrell K., Bech-Otschir D., Dumdey R., Schade R., et al. (1998). A novel protein complex involved in signal transduction possessing similarities to 26S proteasome subunits. FASEB J. 12 469–478 [PubMed] [Google Scholar]
- Serino G., Su H., Peng Z., Tsuge T., Wei N., Gu H., et al. (2003). Characterization of the last subunit of the Arabidopsis COP9 signalosome: implications for the overall structure and origin of the complex. Plant Cell 15 719–731 10.1105/tpc.009092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serino G., Tsuge T., Kwok S., Matsui M., Wei N., Deng X. W. (1999). Arabidopsis cop8 and fus4 mutations define the same gene that encodes subunit 4 of the COP9 signalosome. Plant Cell 11 1967–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Luong P., Huq E. (2007). The F-box protein MAX2 functions as a positive regulator of photomorphogenesis in Arabidopsis. Plant Physiol. 145 1471–1483 10.1104/pp.107.107227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L. N., Liu H., Dong C., Xirodimas D., Naismith J. H., Hay R. T. (2005). Structural basis of NEDD8 ubiquitin discrimination by the deNEDDylating enzyme NEDP1. EMBO J. 24 1341–1351 10.1038/sj.emboj.7600628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W. H., Parmentier Y., Hellmann H., Lechner E., Dong A., Masson J., et al. (2002). Null mutation of AtCUL1 causes arrest in early embryogenesis in Arabidopsis. Mol. Biol. Cell 13 1916–1928 10.1091/mbc.E02-02-0077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y. C., Tang S. J., Chen J. H., Liao P. H., Chang S. C. (2011). The molecular determinants of NEDD8 specific recognition by human SENP8. PLoS ONE 6:e27742 10.1371/journal.pone.0027742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siergiejuk E., Scott D. C., Schulman B. A., Hofmann K., Kurz T., Peter M. (2009). Cullin neddylation and substrate-adaptors counteract SCF inhibition by the CAND1-like protein Lag2 in Saccharomyces cerevisiae. EMBO J. 28 3845–3856 10.1038/emboj.2009.354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. K., Zerath S., Kleifeld O., Scheffner M., Glickman M. H., Fushman D. (2012). Recognition and cleavage of related to ubiquitin 1 (Rub1) and Rub1-ubiquitin chains by components of the ubiquitin-proteasome system. Mol. Cell. Proteomics 11 1595–1611 10.1074/mcp.M112.022467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucy T. A., Smith P. G., Milhollen M. A., Berger A. J., Gavin J. M., Adhikari S., et al. (2009). An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458 732–736 10.1038/nature07884 [DOI] [PubMed] [Google Scholar]
- Stickle N. H., Chung J., Klco J. M., Hill R. P., Kaelin W. G., Jr., Ohn M. (2004). pVHL modification by NEDD8 is required for fibronectin matrix assembly and suppression of tumor development. Mol. Cell. Biol. 24 3251–3261 10.1128/MCB.24.8.3251-3261.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P., Furner I. J, Ottoline Leyser H. M. (2007). MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J. 50 80–94 10.1111/j.1365-313X.2007.03032.x [DOI] [PubMed] [Google Scholar]
- Stuttmann J., Lechner E., Guerois R., Parker J. E., Nussaume L., Genschik P., et al. (2009). COP9 signalosome- and 26S proteasome-dependent regulation of SCFTIR1 accumulation in Arabidopsis. J. Biol. Chem. 284 7920–7930 10.1074/jbc.M809069200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima O., Tsuruta F., Kigoshi Y., Nakamura S., Kim J., Katoh M. C., et al. (2013). Brap2 regulates temporal control of NF-kappaB localization mediated by inflammatory response. PLoS ONE 8:e58911 10.1371/journal.pone.0058911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28 2731–2739 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X., Calderon-Villalobos L. I., Sharon M., Zheng C., Robinson C. V., Estelle M., et al. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446 640–645 10.1038/nature05731 [DOI] [PubMed] [Google Scholar]
- Tateishi K., Omata M., Tanaka K., Chiba T. (2001). The NEDD8 system is essential for cell cycle progression and morphogenetic pathway in mice. J. Cell Biol. 155 571–579 10.1083/jcb.200104035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um J. W., Han K. A., Im E., Oh Y., Lee K., Chung K. C. (2012). Neddylation positively regulates the ubiquitin E3 ligase activity of parkin. J. Neurosci. Res. 90 1030–1042 10.1002/jnr.22828 [DOI] [PubMed] [Google Scholar]
- Vierstra R. D. (2012). The expanding universe of ubiquitin and ubiquitin-like modifiers. Plant Physiol. 160 2–14 10.1104/pp.112.200667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra R. D., Callis J. (1999). Polypeptide tags, ubiquitous modifiers for plant protein regulation. Plant Mol. Biol. 41 435–442 10.1023/A:1006323317890 [DOI] [PubMed] [Google Scholar]
- von Arnim A. G., Deng X. W. (1994). Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79 1035–1045 10.1016/0092-8674(94)90034-5 [DOI] [PubMed] [Google Scholar]
- Wada H., Kito K., Caskey L. S., Yeh E. T., Kamitani T. (1998). Cleavage of the C-terminus of NEDD8 by UCH-L3. Biochem. Biophys. Res. Commun. 251 688–692 10.1006/bbrc.1998.9532 [DOI] [PubMed] [Google Scholar]
- Wang J., Yu Y., Zhang Z., Quan R., Zhang H., Ma L., et al. (2013). Arabidopsis CSN5B interacts with VTC1 and modulates ascorbic acid synthesis. Plant Cell 25 625–636 10.1105/tpc.112.106880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. F., He F. F., Ma X. X., Mao C. Z., Hodgman C., Lu C. G., et al. (2011). OsCAND1 is required for crown root emergence in rice. Mol. Plant 4 289–299 10.1093/mp/ssq068 [DOI] [PubMed] [Google Scholar]
- Waters M. T., Smith S. M. (2013). KAI2- and MAX2-mediated responses to karrikins and strigolactones are largely independent of HY5 in Arabidopsis seedlings. Mol. Plant 6 63–75 10.1093/mp/sss127 [DOI] [PubMed] [Google Scholar]
- Watson I. R., Blanch A., Lin D. C., Ohh M., Irwin M. S. (2006). Mdm2-mediated NEDD8 modification of TAp73 regulates its transactivation function. J. Biol. Chem. 281 34096–34103 10.1074/jbc.M603654200 [DOI] [PubMed] [Google Scholar]
- Wei N., Chamovitz D. A., Deng X. W. (1994). Arabidopsis COP9 is a component of a novel signaling complex mediating light control of development. Cell 78 117–124 10.1016/0092-8674(94)90578-9 [DOI] [PubMed] [Google Scholar]
- Wei N., Deng X. W. (1992). COP9: a new genetic locus involved in light-regulated development and gene expression in arabidopsis. Plant Cell 4 1507–1518 10.1105/tpc.4.12.1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N., Deng X. W. (1998). Characterization and purification of the mammalian COP9 complex, a conserved nuclear regulator initially identified as a repressor of photomorphogenesis in higher plants. Photochem. Photobiol. 68 237–241 10.1111/j.1751-1097.1998.tb02495.x [DOI] [PubMed] [Google Scholar]
- Wei N., Tsuge T., Serino G., Dohmae N., Takio K., Matsui M., et al. (1998). The COP9 complex is conserved between plants and mammals and is related to the 26S proteasome regulatory complex. Curr. Biol. 8 919–922 10.1016/S0960-9822(07)00372-7 [DOI] [PubMed] [Google Scholar]
- Weijers D., Benkova E., Jager K. E., Schlereth A., Hamann T., Kientz M., et al. (2005). Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J. 24 1874–1885 10.1038/sj.emboj.7600659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby F. G., Xia G., Pickart C. M., Hill C. P. (1998). Crystal structure of the human ubiquitin-like protein NEDD8 and interactions with ubiquitin pathway enzymes. J. Biol. Chem. 273 34983–34991 [DOI] [PubMed] [Google Scholar]
- Woodward A. W., Ratzel S. E., Woodward E. E., Shamoo Y., Bartel B. (2007). Mutation of E1-CONJUGATING ENZYME-RELATED1 decreases RELATED TO UBIQUITIN conjugation and alters auxin response and development. Plant Physiol. 144 976–987 10.1104/pp.107.100404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Yamoah K., Dolios G., Gan-Erdene T., Tan P., Chen A., et al. (2003). DEN1 is a dual function protease capable of processing the C terminus of Nedd8 and deconjugating hyper-neddylated CUL1. J. Biol. Chem. 278 28882–28891 10.1074/jbc.M302888200 [DOI] [PubMed] [Google Scholar]
- Wu S., Zhu W., Nhan T., Toth J. I., Petroski M. D., Wolf D. A. (2013). CAND1 controls in vivo dynamics of the cullin 1-RING ubiquitin ligase repertoire. Nat. Commun. 4 1642 10.1038/ncomms2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xirodimas D. P. (2008). Novel substrates and functions for the ubiquitin-like molecule NEDD8. Biochem. Soc. Trans. 36 802–806 10.1042/BST0360802 [DOI] [PubMed] [Google Scholar]
- Xirodimas D. P., Saville M. K., Bourdon J. C., Hay R. T., Lane D. P. (2004). Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell 118 83–97 10.1016/j.cell.2004.06.016 [DOI] [PubMed] [Google Scholar]
- Xirodimas D. P., Sundqvist A., Nakamura A., Shen L., Botting C., Hay R. T. (2008). Ribosomal proteins are targets for the NEDD8 pathway. EMBO Rep. 9 280–286 10.1038/embor.2008.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Ma H., Nei M., Kong H. (2009). Evolution of F-box genes in plants: different modes of sequence divergence and their relationships with functional diversification. Proc. Natl. Acad. Sci. U.S.A. 106 835–840 10.1073/pnas.0812043106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G. P., Zhang Z. L., Xiao S., Zhuang L. K., Xia D., Zou Q. P., et al. (2013). Rig-G negatively regulates SCF-E3 ligase activities by disrupting the assembly of COP9 signalosome complex. Biochem. Biophys. Res. Commun. 432 425–430 10.1016/j.bbrc.2013.01.132 [DOI] [PubMed] [Google Scholar]
- Yang P., Smalle J., Lee S., Yan N., Emborg T. J., Vierstra R. D. (2007). Ubiquitin C-terminal hydrolases 1 and 2 affect shoot architecture in Arabidopsis. Plant J. 51 441–457 10.1111/j.1365-313X.2007.03154.x [DOI] [PubMed] [Google Scholar]
- Yu H. A., Kim S. G., Kim E. J., Lee W. J., Kim D. O., Park K., et al. (2007). Characterization of ubiquitin C-terminal hydrolase 1 (YUH1) from Saccharomyces cerevisiae expressed in recombinant Escherichia coli. Protein Exp. Purif. 56 20–26 10.1016/j.pep.2007.07.005 [DOI] [PubMed] [Google Scholar]
- Yu Z., Kleifeld O., Lande-Atir A., Bsoul M., Kleiman M., Krutauz D., et al. (2011). Dual function of Rpn5 in two PCI complexes, the 26S proteasome and COP9 signalosome. Mol. Biol. Cell 22 911–920 10.1091/mbc.E10-08-0655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemla A., Thomas Y., Kedziora S., Knebel A., Wood N. T., Rabut G., et al. (2013). CSN- and CAND1-dependent remodelling of the budding yeast SCF complex. Nat. Commun. 4 1641 10.1038/ncomms2628 [DOI] [PubMed] [Google Scholar]
- Zhang J., Bai D., Ma X., Guan J., Zheng X. (2012). hCINAP is a novel regulator of ribosomal protein-HDM2-p53 pathway by controlling NEDDylation of ribosomal protein S14. Oncogene 33 246–254 10.1038/onc.2012.560 [DOI] [PubMed] [Google Scholar]
- Zhang W., Ito H., Quint M., Huang H., Noel L. D., Gray W. M. (2008). Genetic analysis of CAND1-CUL1 interactions in Arabidopsis supports a role for CAND1-mediated cycling of the SCFTIR1 complex. Proc. Natl. Acad. Sci. U.S.A. 105 8470–8475 10.1073/pnas.0804144105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Yang X., Harrell J. M., Ryzhikov S., Shim E. H., Lykke-Andersen K., et al. (2002). CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol. Cell. 10 1519–1526 10.1016/S1097-2765(02)00784-0 [DOI] [PubMed] [Google Scholar]
- Zhou L., Watts F. Z. (2005). Nep1, a Schizosaccharomyces pombe deneddylating enzyme. Biochem. J. 389 307–314 10.1042/BJ20041991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo W., Huang F., Chiang Y. J., Li M., Du J., Ding Y., et al. (2013). c-Cbl-mediated neddylation antagonizes ubiquitination and degradation of the TGF-beta type II receptor. Mol. Cell 49 499–510 10.1016/j.molcel.2012.12.002 [DOI] [PubMed] [Google Scholar]