Abstract
Studies in Drosophila and mammals have made it clear that genetic mutations that arise in somatic tissues are rapidly recognized and eliminated, suggesting that cellular fitness is tightly monitored. During development, damaged, mutant, or otherwise unfit cells are prevented from contributing to the tissue and are instructed to die, whereas healthy cells benefit and populate the animal. This cell selection process, known as cell competition, eliminates somatic genetic heterogeneity and promotes tissue fitness during development. Yet cell competition also has a dark side. Super competition can be exploited by incipient cancers to subvert cellular cooperation and promote selfish behavior. Evidence is accumulating that MYC plays a key role in regulation of social behavior within tissues. Given the high number of tumors with deregulated MYC, studies of cell competition promise to yield insight into how the local environment yields to and participates in the early stages of tumor formation.
In developing tissues, cell competition encourages healthy cells to grow and kills weaker cells. But exploitation of competitive mechanisms by MYC “super competitors” can kill healthy cells and promote cancer progression.
In multicellular animals, cells reside within structured communities—tissues and organs—and rely on signals and interactions between them to communally oversee each cell’s identity, survival, and rate of growth. Tissues and organs can therefore be considered social groups that are governed by societal rules. According to kin-selection theory the identical genetic relatedness of somatic cells fosters cooperative behavior (Hamilton 1964; West et al. 2002), which will promote development of organs as functional units and thereby the reproductive success of the animal as a whole. However, somatic mutations that allow cells to cheat or ignore communal rules can lead to pathologies such as cancer. Evolution has provided a variety of mechanisms of enforcing cooperation including those that safeguard the genome, ensure normal tissue architecture, and uphold developmental boundaries. In this review I discuss a process known as “cell competition” that is receiving new attention because of its relevance to development and cancer. Cell competition is initiated upon the recognition of cells perceived as weak by their more robust neighbors. The recognition elicits interactions that prevent the weaker cells from contributing to the animal. However, competitive behavior is also exploited by cells with deregulated oncogenes or tumor suppressors, to expand their territory at the expense of—and with assistance from—their cooperating, wild-type neighbors. A recent surge of interest in understanding competitive interactions between cells has fueled work to research to identify the genes and pathways involved. In this review, my primary aim is to discuss what is known about competitive interactions that are regulated by cell-to-cell differences in MYC activity.
COMPETITION: A MECHANISM OF ELIMINATING GENETIC HETEROGENEITY
Although organ development demands cooperation between cells, the introduction of a population of genetically dissimilar cells can promote interactions that are competitive.
This first caught the eye of Drosophila researchers during the generation of mosaics in imaginal discs, the primordial cells of the adult appendages. Mosaic discs composed of wild-type cells and cells with haploinsufficient mutations in genes that encode ribosomal proteins (called Minutes [M]) led to strong competitive interactions. On their own, M/+ cells are viable but slow growing because their ribosome deficiency makes them inherently weaker, but they produce phenotypically normal flies. However, when surrounded by wild-type cells, M/+ cells are actively eliminated and the wild-type cells overtake the tissue (Morata and Ripoll 1975; Simpson 1979; Simpson and Morata 1981). It was later shown that the elimination of the M/+ cells are eliminated by apoptosis, which requires the presence of nearby wild-type neighbors (Moreno et al. 2002). Thus, interactions between the weaker M/+ cells and relatively stronger wild-type cells cause loss of the M/+ cells. The cellular interactions that mediate cell competition are local, and competition between wild-type and M/+ cells is most intense at clone borders. The sharp increase in cell death at the clone edges has allowed competition between wild-type and M/+ cells to be followed by staining for the apoptosis marker, activated caspase-3 (Simpson and Morata 1981; Li and Baker 2007; Li et al. 2009). Cell competition is thus a mechanism that promotes cooperation among cells by eliminating genetic heterogeneity in tissues.
Social interactions of any kind have consequences for the actor and the recipient, and in cell competition the M/+ cells die and the wild-type cells proliferate to compensate for their loss. There is debate over whether the compensatory proliferation of “winner” cells is accelerated over normal rates in M/+ backgrounds (Martin et al. 2009; de la Cova et al. 2014), but it is well documented in other genetic backgrounds (Neto-Silva et al. 2010; Wu and Johnston 2010). The consequence of the additional growth is that the tissue has a larger than normal fraction of the “winner” cells at the expense of the weaker cells (in the vernacular of cell competition, the “losers”); however, normal overall tissue size is maintained. Defining characteristics of cell competition are listed in Table 1.
Table 1.
Hallmarks of cell competition
| Context dependent: occurs only in mixed cell populations |
| Triggered by local differences in growth or cellular metabolism |
| Proliferation of winner cells is stimulated by loss of loser cells |
| Short range effect, stronger at the interface between loser and winner cells |
| Restricted by boundaries of developmental compartments |
| Tissue maintains appropriate size and pattern |
CELL COMPETITION IS CONSERVED AND COMES IN DIFFERENT FLAVORS
Since the discovery of cell competition using Minutes, a handful of other genes has been shown to induce cell competition when activated, and mutations in several signaling pathways have been shown to be subject to cell competition in wild-type mosaics (Tables 2 and 3). The first documentation of competition in a mouse Minute mutant came when the Belly Spot and Tail (Bst) mutant was found to carry a mutation in RpL24 (Oliver et al. 2004). Linkage of the mutation to coat color showed a clear competitive underrepresentation of RpL24/+ cells, indicating loser status, in chimeras. In Drosophila the outcome of the loser population is apoptosis, but this may not be a general rule. When mildly stressed mouse hematopoietic stem cell precursors (HSCPs) compete with nonstressed HSPCs in mixed bone marrow repopulation experiments, the loser cells initiate a program of senescence rather than apoptosis (Bondar and Medzhitov 2010). Here, the relative cellular level of the tumor suppressor p53 dictates the direction of competition, and this is dependent on the presence of both HSPC populations. Cells with higher p53 activity induce the senescence program only when in mixed company with low-p53 cells (Bondar and Medzhitov 2010).
Table 2.
Mutations that lead to cell competition in mosaics
| Gene name | Function | Allele type | Species | Outcome | Involve Myc? | Phenotype |
|---|---|---|---|---|---|---|
| RpL/+, RpS/+ | Ribosome biogenesis | Loss of function (LOF) | Drosophila melanogaster, Mus musculus | Loser | N | Apoptosis |
| myc−/− | Growth regulation | LOF | D. melanogaster, M. musculus | Loser | Y | Apoptosis |
| stat−/− | Growth, immune regulator | LOF | D. melanogaster | Loser | N | Apoptosis |
| yki−/− | Hippo signaling | LOF | D. melanogaster | Loser | Y | Apoptosis |
| tkv/Bmpr1a−/− | Dpp/BMP signaling | LOF | D. melanogaster, M. musculus | Loser | Y | Extrusion, apoptosis |
| scrib−/− | Epithelial polarity/integrity | LOF | D. melanogaster | Loser | Y | Apoptosis |
| Igl−/− | Epithelial polarity/integrity | LOF | D. melanogaster | Loser | Y | Apoptosis |
| mahj−/− | Epithelial polarity/integrity | LOF | D. melanogaster | Loser | ND | Apoptosis |
| csk−/− | Src inhibitory kinase | LOF | D. melanogaster, MDCK cells | Loser | ND | Extrusion, apoptosis |
| p53 | Transcription factor | Gain of function (GOF) | M. musculus | Loser | ND | Sensescence |
Y, yes; N, no; ND, no data.
Table 3.
Super competitors
| Gene name | Function | Species | Outcome | Involve Myc? | Phenotype |
|---|---|---|---|---|---|
| Myc GOF | Growth regulation | D. mel, M. musculus | Winner | Y | Proliferation |
| Stat GOF | Growth, immune regulator | D. melanogaster | Winner | N | Proliferation |
| APC−/− | Wnt signaling | D. melanogaster | Winner | N | Proliferation |
| axin−/− | Wnt signaling | D. melanogaster | Winner | N | Proliferation |
| hippo−/− | Hippo signaling | D. melanogaster | Winner | Y | Proliferation |
| warts−/− | Hippo signaling | D. melanogaster | Winner | Y | Proliferation |
| Yki GOF | Hippo signaling | D. melanogaster | Winner | Y | Proliferation |
| RasV12 GOF | GTPase | MDCK cells | Loser | Y | Extrusion |
| Mahj GOF | Epithelial integrity | D. melanogaster, MDCK cells | Winner | ND |
Y, yes; N, no; ND, no data.
Repopulation experiments provide good examples of competitive behavior, and such experiments have shown that competition for antigen contributes to homeostatic size regulation of the immune system in vertebrates (Freitas et al. 1995). In these experiments irradiated mice were repopulated with equal mixtures of B lymphocyte precursors of different antibody diversity (McLean et al. 1997). The cell populations expanded equally well when seeded in hosts on their own but when mixed the cells with limited antibody diversity contributed significantly less to the final population size. Interestingly, the populations initially grew at the same rate, but competition altered their growth dynamics as the tissue reached its appropriate size (McLean et al. 1997). Whether this process is the same as cell competition is not clear, but other suggestive cases in vertebrates include the reconstitution of diseased rat liver with healthy fetal liver progenitor cells, where repopulation was competitive and was accompanied by increased apoptosis of host cells adjacent to donor cells (Oertel et al. 2006). Some of the first evidence for competitive interactions between genetically different cell populations was actually provided by mammalian embryologists using the creation of chimeric or mosaic embryos to generate cellular heterogeneity in studies of developmental potential (McLaren 1976; Rossant and Spence 1998; Tarkowski 1998).
An important caveat to studies of competition is that although an inability of mutant cells to populate an organ is often observed in mosaics, it may not always result from competitive interactions. Mutations in genes involved in regulation of growth, pattern formation, and cell fate specification can weaken cells or make them subviable. Such mutations may prevent proliferation through defects such as cell lethality, a block in the cell cycle or growth machinery, or loss of integrity leading to delamination. Although these cells are disadvantaged they are not “competed” from the tissue, rather they have an inherent defect that prevents their survival or proliferation. Competition can be distinguished from an inherent defect through a variety of experiments. Commonly, mutant cells are introduced into a genetic background, such as a strong M/+, that gives them a relative advantage. Viability and proliferation of mutant cells in the context of surrounding M/+ cells indicates that their demise in wild-type mosaics is owing to competitive interactions; if the mutant cells still die (or cannot proliferate) in a M/+ background, it is assumed that the mutation confers an inherent inability to populate the tissue.
Of all genes found to be involved in cell competition (Table 2), Drosophila MYC is the most extensively studied. MYC is encoded by the diminutive (dm) gene and is the sole homolog of c-MYC (see Gallant 2013). It was noticed that hypomorphic alleles of MYC led to flies that were smaller than normal with thin thoracic bristles, a phenotype usually associated with severe Minute mutations (Johnston et al. 1999). This raised the possibility that dm mutants would be subject to cell competition like M/+ cells. Clonal analysis confirmed that dm mutant cells were rarely recovered in mosaic tissues, and it was determined that these cells were eliminated through apoptosis (Johnston et al. 1999). Subsequent studies have shown that the intensity of competition by wild-type cells is determined by the level of MYC in the mutant cells: Competition is strongest against cells carrying the dm4 null allele, whereas for cells with the hypomorphic dmP0 allele, in which MYC is still expressed at approximately 15% of wild type, competition is less severe (Wu and Johnston 2010; de la Cova et al. 2014). Very recently, two groups reported convincing evidence for MYC-regulated cell competition in mouse embryos (discussed below) (Clavería et al. 2013; Sancho et al. 2013).
MYC AND SUPER COMPETITION
In 2004 two groups reported that elevating the cellular level of Drosophila MYC less than twofold above normal in mosaic tissues had the striking effect of causing the death of nearby wild-type cells (de la Cova et al. 2004; Moreno and Basler 2004). Both groups engineered systems to heritably express MYC in marked clones of cells and followed the fate of the MYC-expressing cells and that of their neighbors. As reported previously (Johnston et al. 1999), MYC-expressing clones grew larger than controls; however, the surprise was that marked sister clones composed of wild-type cells actually grew less than controls (de la Cova et al. 2004). These experiments defined the phenomenon of “super competition” (Abrams 2002) as robustly growing cells that are able to not only outgrow but also actively trigger the elimination of nearby wild-type cells from the tissue.
Super competition by MYC obeys the rules of cell competition defined in the original M/+ experiments (Table 1) (Morata and Ripoll 1975; Simpson 1979; Simpson and Morata 1981). There are some interesting differences, however. For example, apoptosis of wild-type loser cells competing with high-MYC cells occurs diffusely over short range in vivo (de la Cova et al. 2004), whereas in M/+ loser cells apoptosis is detected specifically at the boundary of the cell populations (Li and Baker 2007; Li et al. 2009). The range over which wild-type loser cells die in response to MYC super-competitor cells suggested that cell–cell contact was not necessary, and this was confirmed in cell culture experiments. Cocultures of wild-type Drosophila S2 cells and S2 cells that expressed MYC led to competitive interactions identical to the experiments in vivo (Senoo-Matsuda and Johnston 2007). By using a transwell assay, it was shown that physical contact between the two cell populations was not necessary. Furthermore, conditioned medium (CM) from cocultures of these cells was sufficient to cause naive S2 cells to behave as if in competition: Apoptosis was induced in wild-type S2 cells when cultured in the CM, whereas the S2 cells that expressed extra MYC proliferated at an accelerated rate (Senoo-Matsuda and Johnston 2007). These results indicate that the competitive interactions are mediated by secreted factors, which appear to be produced by both cell populations (Senoo-Matsuda and Johnston 2007).
Since the realization of these properties of MYC, the list of super-competitor genes has expanded rapidly. The list includes the genes encoding APC and Axin, regulators of the Wingless/Wnt pathway (Vincent et al. 2011), the signal transducer and activator of transcription (STAT) (Rodrigues et al. 2012), and Yorkie (Yki) (Tyler et al. 2007; Neto-Silva et al. 2010; Ziosi et al. 2010), the Drosophila homolog of the Yes-associated protein (Yap) transcriptional coactivator (Table 3). In addition, certain tumor suppressor genes act as “anti-super-competitors,” as cells carrying mutations in these genes are converted into super competitors. Anti-super-competitors include the Warts/Lats and Hippo/Mst kinases (Tyler et al. 2007; Neto-Silva et al. 2010; Ziosi et al. 2010), the cell polarity determinants Scribbled (Scrib) and Lethal giant large (Lgl) (Froldi et al. 2010; Menendez et al. 2010; Chen et al. 2012; Schroeder et al. 2012), and the Lgl-associated protein Mahjong/VprBP (Tamori et al. 2010), among others (Table 3). Of interest here, the super-competitor function of several of the genes is due partially to increased MYC expression.
ROLES FOR CELLULAR BIOSYNTHESIS AND METABOLISM IN CELL COMPETITION
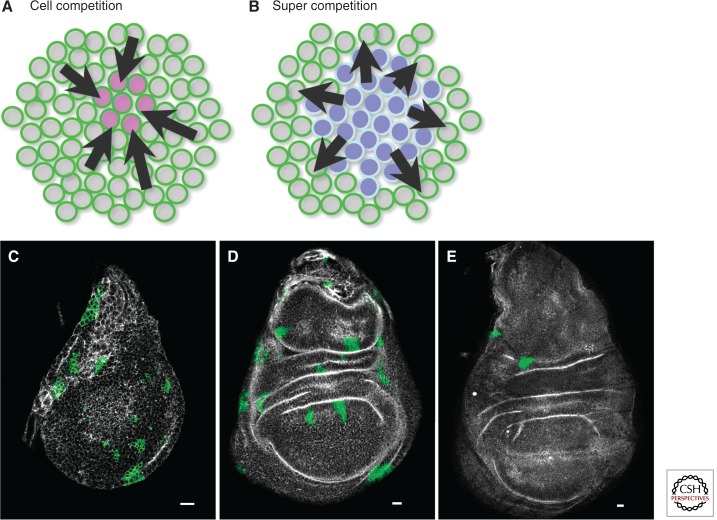
Cell competition requires mechanisms that alert cells to the presence of suboptimal or damaged cells, and implies that cells “know” themselves and can recognize other cells as similar or different. Because somatic cells are essentially all related, genetic differences that arise through mutation are probably immediately recognizable—for example, a mutation that causes differences in cell adhesion. How are more subtle changes like heritable alterations in gene expression recognized? Because cell competition is context dependent (so clearly illustrated by the opposite response of wild-type cells to simple changes in MYC expression) (Fig. 1A,B) alterations that change growth or metabolism are obvious considerations.
Figure 1.
Cell competition and super competition. (A) Wild-type cells (gray and green) become “winners” when a population of relatively less fit cells (pink and green) appears. The wild-type cells instruct (arrows) the less fit cells to induce apoptosis, thereby eliminating them from the tissue. (B) In super-competition, wild-type cells (gray and green) become “losers” and undergo apoptosis owing to signals (arrows) from super-competitor cells (blue). The switch of wild-type cells from “winners” to “losers” illustrates the context-dependent nature of the competitive interactions. (C–E) MYC super-competition assay in Drosophila wing imaginal discs. All cells are wild-type but express a transgene (tub > MYC > Gal4;UAS-GFP, in which “>” denotes Flp recognition target (FRT) sequences) that imparts approximately a 1.5-fold increase in MYC expression above endogenous levels. Excision of the >MYC> cassette by heat-shock-driven Flp recombination yields green fluorescent protein (GFP)-positive clones of cells that no longer express extra MYC. These cells become “losers” owing to a relatively lower level of MYC and are competed away over time by surrounding cells that retain the MYC cassette. (C) Wing imaginal disc with GFP-positive “loser” clones 24 hours after clone induction. (D) Fewer clones remain in the wing disc after 48 hours because of competition from GFP-negative cells. (E) By 72 hours after clone induction, almost all GFP-positive clones are eliminated from the tissue.
Ribosome Biogenesis and Protein Synthesis
The slow growth of M/+ mutants suggested that rates of cell growth or division could be important determinants in competition. MYC’s conserved role in regulation of a number of genes involved in protein synthesis and ribosome biogenesis, including ribosomal protein (Rp) genes, which when mutant are classified as Minutes (Grandori et al. 2005; Grewal et al. 2005; see also Campbell and White 2014), also suggests a role in cell competition. Moreover, cells that overexpress MYC but also carry a heterozygous M/+ mutation no longer function as super competitors (Moreno and Basler 2004), and c-MYC’s oncogenic behavior in a mouse model is abolished in the background of an Rp mutation (Barna et al. 2008).
However, several other reports indicate that local differences in rates of cell division or protein synthesis are not sufficient to induce competitive interactions. Increased activation of Dp110, the Drosophila PI3K and a potent growth regulator that increases protein synthesis does not lead to cell competition (Weinkove et al. 1999; de la Cova et al. 2004). Neither do cell-to-cell differences in the activity of Cyclin D/CDK4, another regulator of growth, cell division, and metabolism (Datar et al. 2000; de la Cova et al. 2004; Frei and Edgar 2004). In addition, an increase of the growth-promoting JAK/STAT pathway in cells in mosaic imaginal discs leads to competition with wild-type cells without increasing ribosome activity or MYC expression (Rodrigues et al. 2012). Similarly, APC or axin mutant cells, which have increased Wnt/Wingless signaling, compete effectively against wild-type cells in the absence of increased MYC or changes in ribosomal activity (Vincent et al. 2011).
These studies argue that although alterations in cell division, cell growth, or ribosome function are commonly associated with cell competition they are not sufficient to induce it. It is possible that these processes act as sensing devices in some cases, however. One way this could occur is if alterations in protein synthesis affected translation of key mRNAs that then modulated a process common to all competitive contexts.
Cellular Metabolism
A defining aspect of cell competition is the compensatory growth of winner cells. As described above, wild-type cells proliferate faster when mutant loser cells are eliminated (Neto-Silva et al. 2010; Wu and Johnston 2010; de la Cova et al. 2014), and in competitive coculture experiments MYC-expressing cells proliferate faster than they do in monocultures (Senoo-Matsuda and Johnston 2007). These observations indicate that acquiring “winner” status leads to growth stimulation, and implies that metabolism may need to be revved up for this to occur. Much evidence documents the powerful effect of increased MYC expression on cellular metabolism (see Dang 2013). Increased c-MYC stimulates glycolysis and lactate production even in an oxygen-rich environment, a symptom of the “Warburg effect,” that is also observed in Drosophila (de la Cova et al. 2014). Glucose uptake and consumption increases, accompanied by up-regulation of genes such as glucose transporter glut 1 and many of the rate-limiting enzymes of glucose metabolism. Production of lactate increases in MYC-expressing cells because of increased expression of the gene encoding lactic acid dehydrogenase (Ldh), which catalyzes the reversible reaction of pyruvate to lactate. This reaction appears to be critical for MYC to promote growth in flies, because null alleles of ldh reduce the growth of MYC-expressing clones in imaginal discs (M. Ziosi, J. Tennessen, and L. Johnston, unpubl.). LDH-A is also required to promote c-MYC-mediated oncogenicity in a variety of MYC transformed cells (Shim et al. 1997), thus stimulation of glycolysis leading to extra lactate production appears to be an important event downstream from deregulated MYC. Increased MYC activity also stimulates mitochondrial biogenesis in vertebrates and invertebrates (see Morrish et al. 2008; Graves et al. 2012; Morrish and Hockenbery 2014).
Interestingly, acute changes in cellular metabolism appear to be critical for the “winner” phenotype during Myc super competition in Drosophila. New studies indicate that under competitive conditions, MYC-stimulated glycolysis is considerably enhanced, a finding that could be very relevant to progression of incipient tumors (de la Cova et al. 2014). Moreover, this enhancement requires p53, suggesting that p53 and MYC synergize in “winner” cells to promote metabolic regulation over and above what MYC does alone. p53 promotes metabolic homeostasis by regulating expression of genes that dampen glycolysis and increase oxidative phosphorylation, including the synthesis of cytochrome C oxidase (scox) gene in Drosophila (de la Cova et al. 2014) and its homolog Sco2 in the mouse (Matoba et al. 2006). Remarkably, p53 is dispensable for growth and survival of nonmosaic MYC-expressing cells, but when these cells exist in mosaics with wild-type cells p53 becomes essential for their survival, for their enhanced glycolysis, and for their ability to kill off nearby cells (de la Cova et al. 2013)—in other words, for all aspects of the Myc super-competitor phenotype. The mosaic loss of p53 in the absence of added stress does not cause competition in the mouse (Bondar and Medzhitov 2010) or in Drosophila (Wells and Johnston 2012), implying that p53 activity is induced because of the confrontation between the two cell populations. p53 may therefore function as a sensor of cell competition. This idea is supported by the observation that accelerated proliferation of wild-type “winner” cells competing with M/+ cells or dm mutant cells is also reduced in p53 mutants, and suggests that the metabolism-boosting role of p53 could be generally required for the winner phenotype during competition (de la Cova et al. 2014).
SIGNALING PATHWAYS IN SENSING AND RECOGNIZING FITNESS DIFFERENCES
Transforming Growth Factor-β/Bone Morphogenetic Protein/Decapentaplegic Signaling
The conserved transforming growth factor-β/bone morphogenetic protein/decapentaplegic (TGF-β/BMP/Dpp) proteins function in signaling gradients to pattern tissues and control numerous processes during animal development, including cell proliferation, epithelial integrity, and cell fate specification (Perrimon et al. 2012). In Drosophila loss of Dpp activity through removal of the Dpp receptor Thickvein (Tkv) in clones of cells leads to their competitive elimination from the imaginal discs (Burke and Basler 1996). The tkv mutant cells are eliminated because they derepress the Dpp inhibitor Brinker (Brk), and Brk up-regulation in tkv mutant cells activates Jun-amino-terminal kinase activity and leads to apoptosis (Adachi-Yamada and O’Connor 2002; Moreno et al. 2002). Loss of Dpp was subsequently implicated in the competitive elimination of M/+ cells in mosaics by a similar mechanism. M/+ clones survived better in areas of the wing disc where brk expression is at its highest, and removing brk from these M/+ clones prevented their elimination by effectively increasing Dpp activity in the clones (Moreno et al. 2002). The elimination of wild-type cells by super-competitor activity of MYC-expressing cells can also be partially overcome by increasing Dpp activity (Moreno and Basler 2004). The Brk-Dpp regulatory axis regulates MYC expression (Prober and Edgar 2000; Doumpas et al. 2013), and because increased MYC will boost Rp gene expression, the ability of Dpp to rescue loser cells from competition could partially be attributed to up-regulation of MYC.
Recently, two exciting papers revealed a role for c-MYC in cell competition in the mouse embryo, one deriving from analysis of loss of BMP activity and its effect on c-MYC expression. In cultures of mouse embryonic stem (ES) cells, Sancho et al. (Sancho et al. 2013) found that reduced BMP activity in one population of mixed cultures of ES cells led to cell competition. ES cells mutant for the Tkv homolog BMP receptor1a (BMPR1a) are defective in BMP signaling but retain pluripotency and in monoculture will proliferate at the same rate as wild-type ES cells. However, when mixed with wild-type ES cells, the Bmpr1a mutant cells had a significant proliferative disadvantage that was traced to an increase in apoptosis. Quite remarkably, the competition between Bmpr1a mutant and wild-type ES cells in coculture followed the same principles as in Drosophila cells (Senoo-Matsuda and Johnston 2007): Competition was associated with a difference in c-MYC expression between the two cell populations, competition did not require direct cell–cell contact between the populations, and competition was mediated by secreted factors shed into the culture medium (although in wild-type and MYC-expressing ES cell cocultures direct or close proximity between the two populations was required) (Clavería et al. 2013). As in flies, the growth advantage of the wild-type “winner” ES cell population was dependent on the death of the “losers.”
The exit from pluripotency that normally occurs in preimplantation embryos and ES cells is accompanied by a decrease in c-MYC expression and correlates with an increase in apoptosis (Sancho et al. 2013). Although c-MYC down-regulation did not occur in monocultures of Bmpr1a mutant ES cells, it did in cocultures of these cells with wild-type ES cells. This prompted examination of whether competition required a pluripotent state. Strikingly, competition was blocked by conditions that maintained pluripotency, and further experiments showed that competition was specific to acquisition of competence to differentiate. An apoptotic fate associated with c-MYC down-regulation also occurred in tetraploid ES cells and Atg5 mutant ES cells deficient for autophagy that were cocultured with wild-type ES cells, suggesting a common mechanism for cell elimination (Sancho et al. 2013). A second paper (Clavería et al. 2013) observed MYC-driven cell competition in epiblast cells of the mouse embryo. The investigators engineered an inducible recombinase-based system to make heritable, marked patches of cells that expressed different levels of c-MYC in mosaic embryos and found that the cells with higher MYC populated more of the developing embryo at the expense of the cells with lower MYC, which died via apoptosis. The highest level of c-MYC expression allowed otherwise wild-type cells to compete against nonexpressing wild-type cells (as did overexpression of c-MYC in ES cells [Sancho et al. 2013]) documenting c-MYC-mediated super competition in mammals (Clavería et al. 2013; Sancho et al. 2013).
Competition occurred early in development in a restricted window around embryonic day (E) 6.5, coinciding with the increase in endogenous apoptosis also noticed by Sancho et al. (Sancho et al. 2013). In addition, both groups noticed that c-MYC is expressed in the epiblast in a periodic manner (Clavería et al. 2013; Sancho et al. 2013), similar to MYC expression in the developing Drosophila wing disc (Johnston et al. 1999; Neto-Silva et al. 2010; Wu and Johnston 2010). In mouse epiblasts low-MYC expression correlated with cell death, potentially reflecting competitive interactions. Clavería and colleagues tested for endogenous cell competition by blocking competition-driven death in one population of mosaic embryos. Consistent with competition, most cells now survived and contributed to the embryo. However, not all cell death was prevented, suggesting that heterogeneity in MYC expression may rid the embryo of weak (but viable) cells, whereas competition-independent mechanisms eliminate nonviable cells that are unable to participate in embryogenesis (Clavería et al. 2013). Thus these two reports provocatively suggest that at early stages of mammalian embryogenesis cells survey their fitness relative to their neighbors and weed out those that are deemed unfit.
MYC and the Hippo Tumor Suppressor Pathway
Recent work in Drosophila indicates that MYC and the conserved Hippo growth inhibitory pathway engage in a codependent relationship with implications for growth regulation, cell competition, and neoplasia. In both mammals and in Drosophila, the activity of the Hippo signaling pathway regulates the activity of Yki/Yap in control of developmental growth and in neoplastic growth by preventing it from entering the nucleus and activating target genes (Pan 2010). Interestingly, Yki requires MYC function to drive cell proliferation in imaginal discs (Neto-Silva et al. 2010; Ziosi et al. 2010). Furthermore, Yki binds to the dm gene in wing discs and regulates its transcription. These proteins are codependent, because in the absence of Yki, expression of MYC is also unable to sustain proliferation. However, MYC remains able to promote cellular biosynthesis in the absence of Yki, suggesting that MYC’s primary contribution to Yki-dependent growth is enhancement of cellular biosynthesis (Neto-Silva et al. 2010). Intriguingly, MYC indirectly regulates its own expression by controlling the expression of Yki, through both transcriptional and posttranscriptional mechanisms (Neto-Silva et al. 2010). This finding provides a mechanism for balancing MYC-regulated growth-promoting activity with Hippo pathway-mediated growth-suppressing activity. It also suggests that MYC expression is modulated on a cell-by-cell basis, and perhaps provides a mechanism for the heterogeneity in MYC expression that occurs in wing discs and in mouse embryos, mentioned above.
As would be predicted from the activation of MYC, clones of cells that express Yki acquire super-competitive properties and kill nearby wild-type cells, just as high-MYC cells do (Neto-Silva et al. 2010; Ziosi et al. 2010). Their super-competitive capability is owing to induction of MYC expression, because if the dose of MYC is genetically reduced the competitive advantage of Yki-expressing cells is abolished (Ziosi et al. 2010). Interestingly, coexpression of MYC and Yki leads to significantly more growth than expression of either factor alone, consistent with findings from mouse models that Yap and MYC synergize to accelerate tumor formation (Zender et al. 2006). Amplification of 11q22, the chromosomal region that contains Yap, is observed in several human cancers (Overholtzer et al. 2006), suggesting that deregulation of MYC by amplified Yap could contribute to tumorigenesis. Because c-MYC expression is increased by Yap activity in some mouse tissues, the combination of Yap amplification with the super-competitive properties of MYC could lead to particularly aggressive tumors.
Indeed, the relationship of Yki and MYC acts as a tumor accelerant after cells mutant for the cell polarity determinants Lgl, Dlg, or Scrib gain ground within a tissue. These genes are required to maintain epithelial cell integrity, and their absence in the whole tissue eventually leads to disorganized, rampant growth and loss of normal tissue structure. However, within mosaics, cells that lack one of these genes are eliminated by nearby wild-type cells via cell competition (Agrawal et al. 1995; Brumby and Richardson 2003). Recent work sheds some light on this difference in behavior. Clones of cells that lack lgl express MYC at low levels, which helps explain their competitive elimination by wild-type cells. The elimination of lgl or scrib mutant cell clones can be rescued by expression of MYC, activated Ras (RasV12) or Yki (both of which cause up-regulation of endogenous MYC), but also results in neoplastic transformation of the cells (Froldi et al. 2010; Menendez et al. 2010; Chen et al. 2012). RasV12 stimulates nuclear localization of Yki and activates Yki target genes in lgl mutant cells (Menendez et al. 2010); with time, these cells take on super-competitor status and expand their territory, lose normal polarity, and can acquire metastatic ability (Brumby and Richardson 2003; Pagliarini and Xu 2003; Igaki et al. 2006; Menendez et al. 2010).
Remarkably, lgl mutant cells can up-regulate MYC and become super competitors simply by being surrounded by a relatively weaker cell population such as M/+ cells (Froldi et al. 2010). The context-dependent behavior of polarity mutant cells thus appears to be completely determined by the fitness status of their neighbors, rather than the lgl mutation itself. The cells are competitively eliminated when surrounded by more fit cells, but acquire super-competitor aggressiveness when surrounded by relatively weaker cells (Froldi et al. 2010; Chen et al. 2012).
Other factors also play important roles in the competition of cells deficient for polarity genes in Drosophila, including signaling by the c-Jun amino-terminal kinase (JNK) pathway and the tumor necrosis factor Eiger (reviewed in de Beco et al. 2012), and by the JAK/STAT pathway (Schroeder et al. 2012). In terms of MYC, however, an interesting insight into the aggressiveness of polarity mutant neoplastic tumors is that the negative-feedback relationship between MYC and Yki described above is lost in scrib mutant tissue (Chen et al. 2012), implying that MYC’s ability to down-regulate Yki (Neto-Silva et al. 2010) is critical for restraining the tumorigenic potential of polarity mutant cells. This raises the very interesting prospect that polarity determinants play a role in the feedback mechanism.
The Code of Flower, a Putative Calcium Channel
An integral transmembrane protein thought to be a calcium channel has also been identified as a mediator of cell competition. Flower (Fwe) was first characterized in the nerve terminals of Drosophila photoreceptors as an integral membrane protein that associates with and regulates synaptic vesicles (Yao et al. 2009). Subsequently, it was identified in microarrays as a gene up-regulated during competition between wild-type and Myc-expressing wing imaginal disc cells (Portela et al. 2010; Rhiner et al. 2010). Fwe is normally expressed as three different isoforms with one, FweUbi, predominating (Yao et al. 2009; Rhiner et al. 2010). During competition FweUbi is down-regulated in the loser cells, whereas expression of two other isoforms, FweLoseA and FweLoseB, is increased. Because FweUbi remains expressed in the winner cells, a differential is thus set up (Rhiner et al. 2010). Expression of either FweLose isoform is sufficient to induce apoptosis in cells as long as they reside next to cells expressing FweUbi (Rhiner et al. 2010). This suggests that the different Fwe isoforms serve as flags that communicate cell identity or fitness. How the FweLose forms function mechanistically in the loser cells is not known, but presumably they have a role in regulating hid and/or reaper, the proapoptotic genes responsible for killing loser cells. Interestingly, another gene found in the microarray, the extracellular matrix protein Sparc, has a temporary protective effect on the loser cells, which might allow some designated loser cells a chance to recover and survive (Portela et al. 2010). Whether the FweLose isoforms have a role in inducing (or repressing) Sparc is not clear, because initial results suggest that expression or inhibition of either one does not affect the other (Portela et al. 2010).
The FweLose isoforms were shown to be expressed in loser cells of several different competitive contexts, including scrib mutants, tkv mutants, and M/+ mutant cells (Rhiner et al. 2010), implying that the FweLose forms play an important role in the loser cell fate. This has led to the proposal of a “Flower code” that acts as a downstream signaling event during competition that labels cells as “losers” and leads to their elimination (Rhiner et al. 2010).
CELL COMPETITION IN PREMALIGNANCY AND IN DEVELOPMENT
As is evident from the work described above, the parallels between super competition and the premalignant stages of cancer are striking. In the fly, a simple twofold increase in MYC expression is enough to render cells the ability to kill off their wild-type neighbors. With the demonstration that MYC expression confers super-competitor status in mouse cells (Clavería et al. 2013; Sancho et al. 2013) it seems clear that this non-cell autonomous function of MYC is conserved. This is troubling, as super competition is essentially a mechanism by which cancer cells could propel their own grim momentum. Both precancerous fields and established tumors could use super competition to kill nontransformed neighboring cells to expand their territory. Heritable increases in c-MYC expression occur in numerous cancers of all major types and are often an early event (see Huang and Weiss 2013; Roussel and Robinson 2013; Gabay et al. 2014; Schmitz et al. 2014). It has been proposed (Moreno and Basler 2004) that cell competition could explain “field cancerization,” a clinical finding in which patches of genetically related mutant cells with no overt phenotype slowly expand and acquire additional mutations that lead to frank tumors (Slaughter et al. 1953; Braakhuis et al. 2003). In humans, cells carrying precancerous mutations that are phenotypically silent for many years are not uncommon (Bissell and Hines 2011). Large patches of p53 mutations in skin cells, for example, can exist for years without causing harm (Zlegler et al. 1994) before eventually erupting into full-blown malignancy. What molecular and cellular events determine whether a tissue responds to the appearance of a cell with cancer potential by eliminating it or promoting its growth? The answer remains a mystery, although there are examples in the literature of tumor inhibition by neighboring stromal cells that prevent a precancerous field of cells from expanding (Stoker et al. 1966; Bissell and Hines 2011).
The evidence also argues that cell competition provides a developmental mechanism for culling tissues of cells perceived as dangerous early after their appearance. Cells found to be unfit are killed by apoptosis and thereby prevented from contributing to the animal. A recent demonstration that cell competition can occur in the follicular epithelium of the Drosophila ovary at a stage when the cells are postmitotic suggests that competition can promote homeostasis even in nonproliferating tissues (Tamori and Deng 2013). As pointed out (Clavería et al. 2013), competition may be especially important in animals in which somatic tissues must be functional over long life spans. The mechanisms regulating competitive interactions may have arisen as adaptive traits that promote fitness, first as a developmental cell fitness-monitoring system, and perhaps later as a “gatekeeper” for cancer. In this regard, the identification of an endogenous system of cell competition in mouse embryos and the finding that competition-induced apoptosis contributes to precision in organ size control in Drosophila (de la Cova et al. 2004) suggest that competition enhances tissue flexibility while at the same time setting its limits. In light of the restricted time window of competition in mouse embryos, it is also interesting to consider life strategies for which cell competition may be useful. Epiblast cells contribute to many organs, and competition between them can impact all developmental stages, whereas the contribution of cells of each imaginal disc, which grow during a juvenile (larval) stage are restricted to one organ in the adult Drosophila. Competition at each point has the potential to significantly impact the physiology, survival, and thus reproductive fitness of the animal, and may have evolved as a general mechanism of tissue optimization that is applied at points in development where cooperative interactions are especially important.
CONCLUDING REMARKS
Many of the genes found to be players in cell competition and super competition have vital physiological roles in animal development. The regulatory roles of MYC and p53 in growth, metabolism, and stress sensing are exceptionally well studied, and their freedom from regulation is central to tumorigenesis. The ECM component Sparc has been reported to have both anticancer and metastasis-promoting roles (Chong et al. 2012), and mice deficient in Fwe are less susceptible to skin papillomas, linking it to cancer (Petrova et al. 2011). How these factors and pathways intersect is an important avenue for study. What is the mechanistic contribution of the fundamental cellular processes like ribosome biogenesis or cellular metabolism to sensing and communicating cellular fitness? How does p53 function as a fitness sensor? The explorations into cell competition described here lay the groundwork for new ways of thinking about how a constructive social order is maintained during development and how it is undermined during tumorigenesis.
ACKNOWLEDGMENTS
I thank Cora Bergantinos for providing the photomicrographs in Figure 1, and Paola Bellosta, Cora Bergantinos, Marcello Ziosi, and Lale Elpar for their constructive comments on the manuscript. I thank present and past members of my laboratory and many colleagues whose work has enlightened this review and apologize to those whose work was not cited. Support for work in my laboratory on cell competition has come from the National Institutes of Health and the Rita Allen Foundation.
Footnotes
Editors: Chi V. Dang and Robert N. Eisenman
Additional Perspectives on MYC and the Pathway to Cancer available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Abrams JM 2002. Competition and compensation: Coupled to death in development and cancer. Cell 110: 403–406 [DOI] [PubMed] [Google Scholar]
- Adachi-Yamada T, O’Connor MB 2002. Morphogenetic apoptosis: A mechanism for correcting discontinuities in morphogen gradients. Dev Biol 251: 74–90 [DOI] [PubMed] [Google Scholar]
- Agrawal N, Kango M, Mishra A, Sinha P 1995. Neoplastic transformation and aberrant cell-cell interactions in genetic mosaics of lethal(2)giant larvae (lgl), a tumor suppressor gene of Drosophila. Dev Biol 172: 218–229 [DOI] [PubMed] [Google Scholar]
- Barna M, Pusic A, Zollo O, Cost M, Kondrashov N, Rego E, Rao EH, Ruggero D 2008. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature 456: 971–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell MJ, Hines WC 2011. Why don’t we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med 17: 320–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondar T, Medzhitov R 2010. p53-Mediated hematopoietic stem and progenitor cell competition. Cell Stem Cell 6: 309–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakhuis BJM, Tabor MP, Kummer JA, Leemans CR, Brakenhoff RH 2003. A genetic explanation of slaughter’s concept of field cancerization: Evidence and clinical implications. Cancer Res 63: 1727–1730 [PubMed] [Google Scholar]
- Brumby AM, Richardson HE 2003. Scribble mutants cooperate with oncogenic Ras or Notch to cause neoplastic overgrowth in Drosophila. EMBO J 22: 5769–5779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke R, Basler K 1996. Dpp receptors are autonomously required for cell proliferation in the entire developing Drosophila wing. Development 122: 2261–2269 [DOI] [PubMed] [Google Scholar]
- *.Campbell KJ, White RJ 2014. MYC regulation of cell growth through control of RNA polymerase I and III activities. Cold Spring Harb Perspect Med 10.1101/cshperspect.a018408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Schroeder MC, Kango-Singh M, Tao C, Halder G 2012. Tumor suppression by cell competition through regulation of the Hippo pathway. Proc Natl Acad Sci 109: 484–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong HC, Tan CK, Huang RL, Tan NS 2012. Matricellular proteins: A sticky affair with cancers. J Oncol 2012: 351089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavería C, Giovinazzo G, Sierra R, Torres M 2013. Myc-driven endogenous cell competition in the early mammalian embryo. Nature 500: 39–44 [DOI] [PubMed] [Google Scholar]
- *.Dang CV 2013. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb Perspect Med 3: a014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datar SA, Jacobs HW, de la Cruz AF, Lehner CF, Edgar BA 2000. The Drosophila cyclin D-Cdk4 complex promotes cellular growth. EMBO J 19: 4543–4554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beco S, Ziosi M, Johnston LA 2012. New frontiers in cell competition. Dev Dyn 241: 831–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cova C, Abril M, Bellosta P, Gallant P, Johnston LA 2004. Drosophila myc regulates organ size by inducing cell competition. Cell 117: 107–116 [DOI] [PubMed] [Google Scholar]
- de la Cova C, Senoo-Matsuda N, Wu D-C, Ziosi M, Bellosta P, Quinzii CM, Johnston LA 2014. Super-competitor status of Drosophila Myc cells reprograms metabolism and requires p53 as a fitness sensor. Cell Metab 19: 470–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doumpas N, Ruiz-Romero M, Blanco E, Edgar B, Corominas M, Teleman AA 2013. Brk regulates wing disc growth in part via repression of Myc expression. EMBO Rep 14: 261–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei C, Edgar BA 2004. Drosophila cyclin D/Cdk4 requires Hif-1 prolyl hydroxylase to drive cell growth. Dev Cell 6: 241–251 [DOI] [PubMed] [Google Scholar]
- Freitas AA, Rosado MM, Viale A-C, Grandien A 1995. The role of cellular competition in B cell survival and selection of B cell repertoires. Eur J Immunol 25: 1729–1738 [DOI] [PubMed] [Google Scholar]
- Froldi F, Ziosi M, Garoia F, Pession A, Grzeschik NA, Bellosta P, Strand D, Richardson HE, Pession A, Grifoni D 2010. The lethal giant larvae tumour suppressor mutation requires dMyc oncoprotein to promote clonal malignancy. BMC Biol 8: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Gabay M, Li Y, Felsher DW 2014. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb Perspect Med 10.1101/cshperspect.a014241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Gallant P 2013. Myc function in Drosophila. Cold Spring Harb Perspect Med 3: a014324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ 2005. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol 7: 311–318 [DOI] [PubMed] [Google Scholar]
- Graves JA, Wang Y, Sims-Lucas S, Cherok E, Rothermund K, Branca MF, Elster J, Beer-Stolz D, Van Houten B, Vockley J, et al. 2012. Mitochondrial structure, function and dynamics are temporally controlled by c-Myc. PLoS ONE 7: e37699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SS, Li L, Orian A, Eisenman RN, Edgar BA 2005. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat Cell Biol 7: 295–302 [DOI] [PubMed] [Google Scholar]
- Hamilton WD 1964. The genetical evolution of social behaviour. II. J Theoret Biol 7: 17–52 [DOI] [PubMed] [Google Scholar]
- *.Huang M, Weiss WA 2013. Neuroblastoma and MYCN. Cold Spring Harb Perspect Med 3: a014415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaki T, Pagliarini RA, Xu T 2006. Loss of cell polarity drives tumor growth and invasion through JNK activation in Drosophila. Curr Biol 16: 1139–1146 [DOI] [PubMed] [Google Scholar]
- Johnston LA, Prober DA, Edgar BA, Eisenman RN, Gallant P 1999. Drosophila myc regulates cellular growth during development. Cell 98: 779–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Baker NE 2007. Engulfment is required for cell competition. Cell 129: 1215–1225 [DOI] [PubMed] [Google Scholar]
- Li W, Kale A, Baker NE 2009. Oriented cell division as a response to cell death and cell competition. Curr Biol 19: 1821–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin FA, Herrera SC, Morata G 2009. Cell competition, growth and size control in the Drosophila wing imaginal disc. Development 136: 3747–3756 [DOI] [PubMed] [Google Scholar]
- Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM 2006. p53 regulates mitochondrial respiration. Science 312: 1650–1653 [DOI] [PubMed] [Google Scholar]
- McLaren A 1976. Genetics of the early mouse embryo. Ann Rev Genet 10: 361–388 [DOI] [PubMed] [Google Scholar]
- McLean AR, Rosado MM, Agenes F, Vasconcellos R, Freitas AA 1997. Resource competition as a mechanism for B cell homeostasis. Proc Natl Acad Sci 94: 5792–5797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez J, Perez-Garijo A, Calleja M, Morata G 2010. A tumor-suppressing mechanism in Drosophila involving cell competition and the Hippo pathway. Proc Natl Acad Sci 107: 14651–14656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morata G, Ripoll P 1975. Minutes: Mutants of Drosophila autonomously affecting cell division rate. Dev Biol 42: 211–221 [DOI] [PubMed] [Google Scholar]
- Moreno E, Basler K 2004. dMyc transforms cells into super-competitors. Cell 117: 117–129 [DOI] [PubMed] [Google Scholar]
- Moreno E, Basler K, Morata G 2002. Cells compete for Decapentaplegic survival factor to prevent apoptosis in Drosophila wing development. Nature 416: 755–759 [DOI] [PubMed] [Google Scholar]
- *.Morrish F, Hockenbery D 2014. Myc and mitochondrial biogenesis. Cold Spring Harb Perspect Med 10.1101/cshperspect.a014225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrish F, Neretti N, Sedivy JM, Hockenbery DM 2008. The oncogene c-Myc coordinates regulation of metabolic networks to enable rapid cell cycle entry. Cell Cycle 7: 1054–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neto-Silva RM, de Beco S, Johnston LA 2010. Evidence for a growth-stabilizing regulatory feedback mechanism between Myc and Yorkie, the Drosophila homolog of Yap. Dev Cell 19: 507–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oertel M, Menthena A, Dabeva MD, Shafritz DA 2006. Cell competition leads to a high level of normal liver reconstitution by transplanted fetal liver stem/progenitor cells. Gastroenterology 130: 507–520 [DOI] [PubMed] [Google Scholar]
- Oliver ER, Saunders TL, Tarle SA, Glaser T 2004. Ribosomal protein L24 defect in belly spot and tail (Bst), a mouse Minute. Development 131: 3907–3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, Deng CX, Brugge JS, Haber DA 2006. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci 103: 12405–12410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini RA, Xu T 2003. A genetic screen in Drosophila for metastatic behavior. Science 302: 1227–1231 [DOI] [PubMed] [Google Scholar]
- Pan D 2010. The hippo signaling pathway in development and cancer. Dev Cell 19: 491–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova E, Soldini D, Moreno E 2011. The expression of SPARC in human tumors is consistent with its role during cell competition. Commun Integr Biol 4: 171–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrimon N, Pitsouli C, Shilo BZ 2012. Signaling mechanisms controlling cell fate and embryonic patterning. Cold Spring Harb Perspect Biol 4: a005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portela M, Casas-Tinto S, Rhiner C, López-Gay JM, Domínguez O, Soldini D, Moreno E 2010. Drosophila SPARC is a self-protective signal expressed by loser cells during cell competition. Dev Cell 19: 562–573 [DOI] [PubMed] [Google Scholar]
- Prober DA, Edgar BA 2000. Ras1 promotes cellular growth in the Drosophila wing. Cell 100: 435–446 [DOI] [PubMed] [Google Scholar]
- Rhiner C, Lopez-Gay JM, Soldini D, Casas-Tinto S, Martin FA, Lombardia L, Moreno E 2010. Flower forms an extracellular code that reveals the fitness of a cell to its neighbors in Drosophila. Dev Cell 18: 985–998 [DOI] [PubMed] [Google Scholar]
- Rodrigues AB, Zoranovic T, Ayala-Camargo A, Grewal S, Reyes-Robles T, Krasny M, Wu DC, Johnston LA, Bach EA 2012. Activated STAT regulates growth and induces competitive interactions independently of Myc, Yorkie, Wingless and ribosome biogenesis. Development 139: 4051–4061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J, Spence A 1998. Chimeras and mosaics in mouse mutant analysis. Trends Genet 14: 358–363 [DOI] [PubMed] [Google Scholar]
- *.Roussel MF, Robinson GW 2013. Role of MYC in medulloblastoma. Cold Spring Harb Perspect Med 3: a014308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho M, di Gregorio A, George N, Pozzi S, Sánchez JM, Pernaute B, Rodríues TA 2013. Competitive interactions eliminate unfit embryonic stem cells at the onset of differentiation. Dev Cell 26: 19–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Schmitz R, Ceribelli M, Pittaluga S, Wright G, Staudt LM 2014. Oncogenic mechanisms in Burkitt lymphoma. Cold Spring Harb Perspect Med 10.1101/cshperspect.a014282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder MC, Chen CL, Gajewski K, Halder G 2012. A non-cell-autonomous tumor suppressor role for Stat in eliminating oncogenic scribble cells. Oncogene 19: 4471–4479 [DOI] [PubMed] [Google Scholar]
- Senoo-Matsuda N, Johnston LA 2007. Soluble factors mediate competitive and cooperative interactions between cells expressing different levels of Drosophila Myc. Proc Natl Acad Sci 104: 18543–18548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, Dalla-Favera R, Dang CV 1997. c-Myc transactivation of LDH-A: Implications for tumor metabolism and growth. Proc Natl Acad Sci 94: 6658–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson P 1979. Parameters of cell competition in the compartments of the wing disc of Drosophila. Dev Biol 69: 182–193 [DOI] [PubMed] [Google Scholar]
- Simpson P, Morata G 1981. Differential mitotic rates and patterns of growth in compartments in the Drosophila wing. Dev Biol 85: 299–308 [DOI] [PubMed] [Google Scholar]
- Slaughter DP, Southwick HW, Smejkal W 1953. “Field cancerization” in oral stratified squamous epithelium. Cancer 6: 963–968 [DOI] [PubMed] [Google Scholar]
- Stoker MGP, Shearer M, O’Neill C 1966. Growth inhibition of polyoma-transformed cells by contact with static normal fibroblasts. J Cell Sci 1: 297–310 [DOI] [PubMed] [Google Scholar]
- Tamori Y, Deng WM 2013. Tissue repair through cell competition and compensatory cellular hypertrophy in postmitotic epithelia. Dev Cell 25: 350–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamori Y, Bialucha CU, Tian AG, Kajita M, Huang YC, Norman M, Harrison N, Poulton J, Ivanovitch K, Disch L, et al. 2010. Involvement of Lgl and Mahjong/VprBP in cell competition. PLoS Biol 8: e1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarkowski AK 1998. Mouse chimaeras revisited: Recollections and reflections. Int J Dev Biol 42: 903–908 [PubMed] [Google Scholar]
- Tyler DM, Li W, Zhuo N, Pellock B, Baker NE 2007. Genes affecting cell competition in Drosophila. Genetics 175: 643–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JP, Kolahgar G, Gagliardi M, Piddini E 2011. Steep differences in wingless signaling trigger myc-independent competitive cell interactions. Dev Cell 21: 366–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinkove D, Neufeld TP, Twardzik T, Waterfield MD, Leevers SJ 1999. Regulation of imaginal disc cell size, cell number and organ size by Drosophila class IA phosphoinositide 3-kinase and its adaptor. Curr Biol 9: 1019–1029 [DOI] [PubMed] [Google Scholar]
- Wells BS, Johnston LA 2012. Maintenance of imaginal disc plasticity and regenerative potential in Drosophila by p53. Dev Biol 361: 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SA, Pen I, Griffin AS 2002. Cooperation and competition between relatives. Science 296: 72–75 [DOI] [PubMed] [Google Scholar]
- Wu DC, Johnston LA 2010. Control of wing size and proportions by Drosophila Myc. Genetics 184: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao CK, Lin YQ, Ly CV, Ohyama T, Haueter CM, Moiseenkova-Bell VY, Wensel TG, Bellen HJ 2009. A synaptic vesicle-associated Ca2+ channel promotes endocytosis and couples exocytosis to endocytosis. Cell 138: 947–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ, et al. 2006. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell 125: 1253–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziosi M, Baena-López LA, Grifoni D, Froldi F, Pession A, Garoia F, Trotta V, Bellosta P, Cavicchi S, Pession A 2010. dMyc functions downstream of Yorkie to promote the supercompetitive behavior of hippo pathway mutant cells. PLoS Genet 6: e1001140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlegler A, Jonason AS, Leffell DJ, Simon JA, Sharma HW, Kimmelman J, Remington L, Jacks T, Brash DE 1994. Sunburn and p53 in the onset of skin cancer. Nature 372: 773–776 [DOI] [PubMed] [Google Scholar]