Abstract
Recently, a different class of collagen-like molecules has been identified in numerous bacteria. Initial studies have shown that these collagens are readily produced in E. coli and they have been isolated and purified by various small scale chromatography approaches. These collagens are non-cytotoxic, non immunogenic and can be produced in much higher yields than mammalian collagens, making them potential new collagens for biomedical materials. One of the major drawbacks with large scale fermentation of collagens has been appropriate scalable down-steam processing technologies. Like other collagens, the triple helical-domains of bacterial collagens are particularly resistant to proteolysis. The present study describes the development and optimisation of a simple, scalable procedure using a combination of acid precipitation of the E. coli host proteins, followed by proteolysis of residual host proteins to produce purified collagens in large scale without the use of chromatographic methods.
Keywords: Non-animal collagen, precipitation, proteolysis, purification, triple-helix
Introduction
Collagens are the major structural proteins in the extracellular matrix of animals. They are defined by a characteristic triple-helix structural motif that has a (Gly-Xaa-Yaa)n repeating sequence (Brodsky and Ramshaw 1995). In mammalian collagens, the Xaa and Yaa positions are frequently proline, where Pro in the Yaa position is post-translationally modified to hydroxyproline (Hyp) which is essential in enhancing triple-helical stability (Rosenbloom et al. 1973; Brodsky and Ramshaw 1995). In humans, a family of at least 28 different collagen types is present, with each collagen type having a specific biological and structural function (Ricard-Blum 2011).
Collagens are found in a wide variety of commercial products, of which collagen-based biomedical products have received increased attention and have proved largely safe and efficacious in clinical applications (Ramshaw et al. 1996; Ramshaw et al. 2009). The majority of collagen for biomedical products is obtained from animals, especially from bovines. This has lead to an on-going concern regarding the transmission of diseases, especially spongiform encephalopathies (‘Mad cow disease’).
A potential disease free source of collagen is via recombinant production systems. Significant progress has been made in producing human collagens in yeast and plant systems. These systems can be complex as the enzyme prolyl 4-hydroxylase must be introduced into the recombinant organism to enable proline hydroxylation and product stability (Werkmeister and Ramshaw 2012).
An alternative system has recently emerged, based on the large number (more than 100) of (Gly-Xaa-Yaa)n repeating sequences that have been identified in bacterial species (Rasmussen et al. 2003). In a limited number of cases the proposed triple-helical protein has been isolated as a recombinant product and each has been shown to form a stable triple-helix (Xu et al. 2002; Sylvestre et al., 2002; Boydson et al., 2005; Xu et al. 2010; Vandersmissen et al 2010, Ghosh et al., 2012; Pizzaro-Guajardo et al., 2013). Surprisingly, although these sequences do not contain any hydroxyproline, they are nevertheless stable at around body temperature, showing melting temperatures in the 35 – 38 °C range (Xu et al. 2002; Xu et al. 2010).
The best studied of the bacterial, collagen-like triple-helical domains is the Scl2 gene from Streptococcus pyogenes (Xu et al. 2002; Mohs et al. 2007; Yoshizumi et al. 2009; Cosgriff-Hernandez et al. 2010; Peng et al. 2010; Peng et al. 2012). It is non-cytotoxic and non-immunogenic and can be fabricated and stabilised in various formats (Peng et al. 2010). Biologically, the Scl2-derived protein behaves like a ‘blank slate’ in that it shows few, if any interactions with mammalian cells (Cosgriff-Hernandez et al. 2010). However, this is an advantage as it allows design and production of various modified forms where specific, triple-helical functional domains can be introduced by substitution or addition of sequences based on known mammalian sites (Cosgriff-Hernandez et al. 2010; Peng et al. 2012; An et al. 2013).
Non-animal, bacterial collagens, therefore, provide an opportunity for the development of new biomedical products. Previously, we have shown that it is possible to obtain triple-helical protein expression through fermentation at high cell density in very good yields, up to 19 g/L (Peng et al. 2012). To date, however, laboratory protocols for purification have typically used metal affinity resin, ion exchange and gel permeation chromatography (Mohs et al. 2007; Yoshizumi et al. 2009). These approaches are excellent for small scale laboratory work where only milligrams of protein are required, but are unsuitable for larger scale commercial production where many grams to kilograms of purified protein are required. In the present study, we describe a scalable process that achieves >95% pure protein without the need for chromatography columns during the high volume steps, prior to final ‘polishing’ of the preparation.
Material and Methods
Gene constructs
The DNA sequence for the Streptococcus pyogenes protein was based on a fragment of the scl2.28 allele (Q8RLX7) of S. pyogenes encoding the combined globular and collagen-like portions of the Scl2.28 protein, but lacking the C-terminal attachment domain (Peng et al. 2010). It included the addition of a sequence for a His6-tag at the N-terminal of the sequence and a thrombin/trypsin cleavage and spacer sequence LVPRGSP between the N-terminal globular domain (V) and the following (Gly-Xaa-Yaa)n collagen-like domain (CL) sequence. The DNA for this design was synthesised commercially with codon optimisation for expression in E. coli (GeneArt® Gene Synthesis, Regensburg, Germany). The Accession number of the construct used is given below.
The DNA sequence for a tandem construct, comprising two contiguous CL-domains from S. pyogenes, with the two CL-domains linked through a GAAGVM linker sequence, was constructed as described below. The protein also contained a His6-tag and an enzyme cleavage site in the same positions. To produce the tandem construct, the initial monomer construct (above) was modified to introduce a GAAGVM sequence using site directed mutagenesis prior to the start of the CL domain using the following oligonucleotides: 5′ ACGCGGTAGTCCCGGGGCAGCGGGTGTTATGGGGCCCAGAGG 3′ Forward and 3′ CCTCTGGGCCCCATAACACCCGCTGCCCCGGGACTACCGCGT 5′ Reverse
This new, second construct, containing the GAAGVM insert, was then digested with 5′ SmaI (Blunt) and 3′ SspI (Blunt). This digested insert, of the GAAGVM N-terminal modified CL domain, was then subcloned back into the original Scl2 gene at the SmaI site at the end of the original construct. Colonies were grown in 1 × YT media (8g tryptone, 5g yeast extract and 2.5g NaCl per litre) with ampicillin (100 μg/ml) and plasmid DNA extracted to select for clones that contained the additional sequence in the correct orientation. The Accession number of the construct used is given below.
The DNA sequence for the triple helix repeat-containing collagen from Methylobacterium sp 4−46 (ACA18713) was obtained from the National Center for Biotechnology Information database (NIH, USA). A trypsin cleavage sequence, Arg-Ala, was introduced between the V and CL domains. The resulting gene construct was synthesised commercially with codon optimisation for expression in E. coli (GeneArt® Gene Synthesis). This construct did not include a His6-tag, which can be provided by the vector if needed. The Accession number of the construct used is given below.
The DNA sequence for the triple helix repeat-containing collagen from Solibacter usitatus Ellin6076 was obtained from the data provided in the National Center for Biotechnology Information database. This sequence was combined with the N-terminal V-domain sequence from S. pyogenes, as above, and the composite construct was synthesised commercially with codon optimisation for expression in E. coli (GeneArt® Gene Synthesis). This construct did not include a His6-tag, which can be provided by the vector, nor an additional enzyme cleavage site. The Accession number of the construct used is given below.
The DNA sequence for the S. usitatus collagen segment was combined with the sequence for the C-terminal V-domain from Rhodopseudomonas palustris, obtained (ABJ82342) from the National Center for Biotechnology Information database. The protein sequences were translated into nominal DNA sequences and a composite gene was designed with a Met initiation signal. A C-terminal His6-tag and a termination codon were also added. This composite construct was synthesised (GeneArt® Gene Synthesis) with a DNA sequence that retained the original amino acid sequences while optimising for expression in E coli. The Accession number of the construct used is given below.
The sequences of all constructs were confirmed by sequencing prior to transformation and protein expression.
Protein expression
DNA sequences were sub-cloned into the pColdI (which included a His6-tag) or pColdIII (Takara Bio, Shiga, Japan) or into the pET vector system (Invitrogen) for expression in E. coli, using 5′ NdeI and 3′ BamHI restriction sites. PCR colony screening was used to detect positive clones. To expand the vector quantity, the clones were grown in 100 ml culture medium and plasmid purified using Qiagen (Germantown, US) kits.
For expression using pCold vectors, a selected positive clone was transformed into the E. coli BL21-DE3. Cells were grown with shaking at 200 rpm in 2 × YT Media (16g tryptone, 10g yeast extract and 5g NaCl per litre) with ampicillin (50 μg/ml) at 37 °C for 24 h until the optical density A600 reached around 3-6. Cultures were then cooled to 25 °C and 1 mM isopropyl beta-D-thiogalactopyranoside (IPTG) added to induce protein expression. After 10 h incubation at 25 °C, the temperature was decreased to 15 °C for a further 14 h after which cells were harvested by centrifugation. Expression using a pET vector was also completed using E. coli BL21 DE3. The cells were grown at 37 °C until A600 reached around 0.6 then protein expression was induced by addition of 1 mM IPTG; the cells were incubated at 37 °C for a further 3 h prior to harvest by centrifugation.
For expression in stirred tanks (using pCold vector systems), a 2L stirred tank bioreactor connected to a Biostat B (Sartorius Stedim, Germany) control system was used, as previously described (Peng et al. 2012). Briefly, the initial volume of medium in the fermenter was 1.6 L and glucose as used as the carbon source. Seed culture was added to the bioreactor to attain an A600 of 0.25. A high cell density fed-batch process was used, with a feed solution of 400 mL of 660 g/L glucose solution to which 40 mL of 1 M MgSO4.7H2O was added. The feed flow rate was 15 mL/h and the feed was initiated 8.5 h after inoculation. The culture was cooled (over a 20 minute period) to 25 °C 24 h after inoculation to activate the cold shock component of the vector and protein expression was induced by addition of 1mM IPTG. After 10 h incubation at 25 °C, the temperature was decreased to 15 °C for a further 14 h incubation after which the cells were collected by centrifugation (Peng et al. 2012).
Selection of extraction conditions for expressed proteins
For extraction, each 1 gram of cell paste was resuspended in 20 ml of 50 mM acetate buffer at selected pH values between pH 2 and pH 7 and the cells ruptured by sonication, using a Misonix S4000 instrument, with a Enhance Booster #1 probe, at 30 A (instrument scale) for 5 min. The cell lysate was clarified by centrifugation (15,000 × g for 30 min, 4 °C) and the clear supernatant containing the triple helical protein was retained. The clarified lysate was held at 4 °C for 16 h and any precipitate that had formed was removed by centrifugation (15,000 × g for 30 min, 4 °C) and the total protein content of the supernatant estimated by measuring absorbance at 280 nm. The retention of soluble triple-helical protein was confirmed by SDS-PAGE (Laemmli 1970). In the present work, with E. coli as the expression host, an extraction pH of 2.2 ± 0.2 in 50 mM acetic acid/HCl was selected for further studies (see below).
Digestion of soluble host cell proteins
The supernatant obtained after removal of acid precipitated proteins was treated by proteolysis. For an extract at pH of 2.2 ± 0.2 in 50 mM acetic acid/HCl, pepsin (0.01 mg/ml) was added and the mixture incubated for 16 h at 4 °C. The reaction was stopped by adjusting the pH of the digest to pH 7. For papain digestion, the extract at pH of 2.2 ± 0.2 was adjusted with dilute NaOH to pH 6.5 and 50 mM Na EDTA and 50 mM cysteine added. Proteolysis by papain (0.01 mg/ml) was completed at 4 °C for 24 h. For trypsin or chymotrypsin digestion, the extract at pH of 2.2 ± 0.2 was adjusted by addition of 50 mM Na2HPO4 and the pH adjusted with dilute NaOH to pH 8.0 and proteolysis by trypsin or chymotrypsin (0.01 mg/ml) completed at 4 °C for 24 h.
The location of the cleavage site N-terminal to the collagen domain was examined in certain cases by automated N-terminal Edman sequencing (Australian Proteome Analysis Facility, Macquarie University).
Collection of triple helical protein product by cross-flow filtration
After removal of impurities by acid precipitation followed by proteolysis, fractions containing recombinant triple-helical protein were pooled, concentrated and buffer-exchanged into 20 mM sodium phosphate buffer, pH 8.0 using a 10 kDa cross-flow filtration membrane (Pall Life Sciences). Where required, samples were further dialysed against 20 mM acetic acid prior to freeze drying. All steps were performed at temperatures lower than the melting temperature of the triple-helix, typically at 2-8 °C. The purity of products was assessed by SDS-PAGE (Laemmli 1970).
Accession numbers for cDNA constructs
The cDNA sequences used in this study have been lodged with the European Nucleotide Archive and have the Accession Numbers that are indicated below along with the Accession Numbers for the original sequences on which these new constructs were based.
HG518485. A synthetic construct based on the S. pyogenes bacterial collagen Scl2 fragment (AY069936.1).
HG518483. A synthetic construct based on a tandem repeat of CL domains from S. pyogenes bacterial collagen Scl2 fragment (AY069936.1).
HG518484. A synthetic construct based on the Methylobacterium sp. 4-46 bacterial collagen (ACA18713.1).
HG518482. A synthetic chimeric construct based on the CL domain of Candidatus Solibacter usitatus (ABJ82342) and the N-terminal non-collagenous domain of S. pyogenes bacterial collagen Scl2 fragment (AY069936.1).
HG518486. A synthetic chimeric construct based on the CL domain of Candidatus Solibacter usitatus (ABJ82342) and the C-terminal non-collagenous domain from the Rhodopseudomonas palustris collagen-like protein (YP_001993084).
Results
Construct production
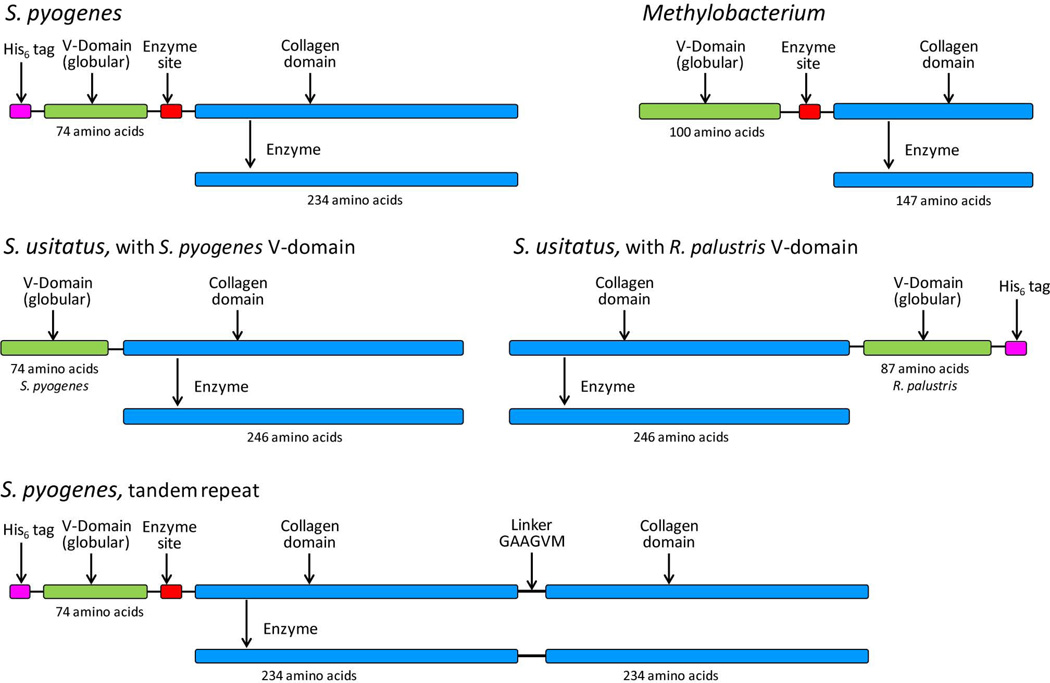
Constructs based on 3 different bacterial collagen-like sequences from S. pyogenes, Methylobacterium sp 4−46 and Solibacter usitatus Ellin6076 were produced, as shown schematically in Fig. 1. The collagen domain from S. pyogenes was 234 amino acids long. The collagen sequence from Methylobacterium was much shorter (147 amino acids). The V domain was slightly larger, at 100 amino acids, than that of S. pyogenes. The S. usitatus sequence length was similar to that from S. pyogenes, being 246 amino acids.
Figure 1.
Schematic representations of the monomer collagen domain constructs that have been expressed and purified using the described method. In some cases a His6 tag was provided through the expression vector.
Protein expression
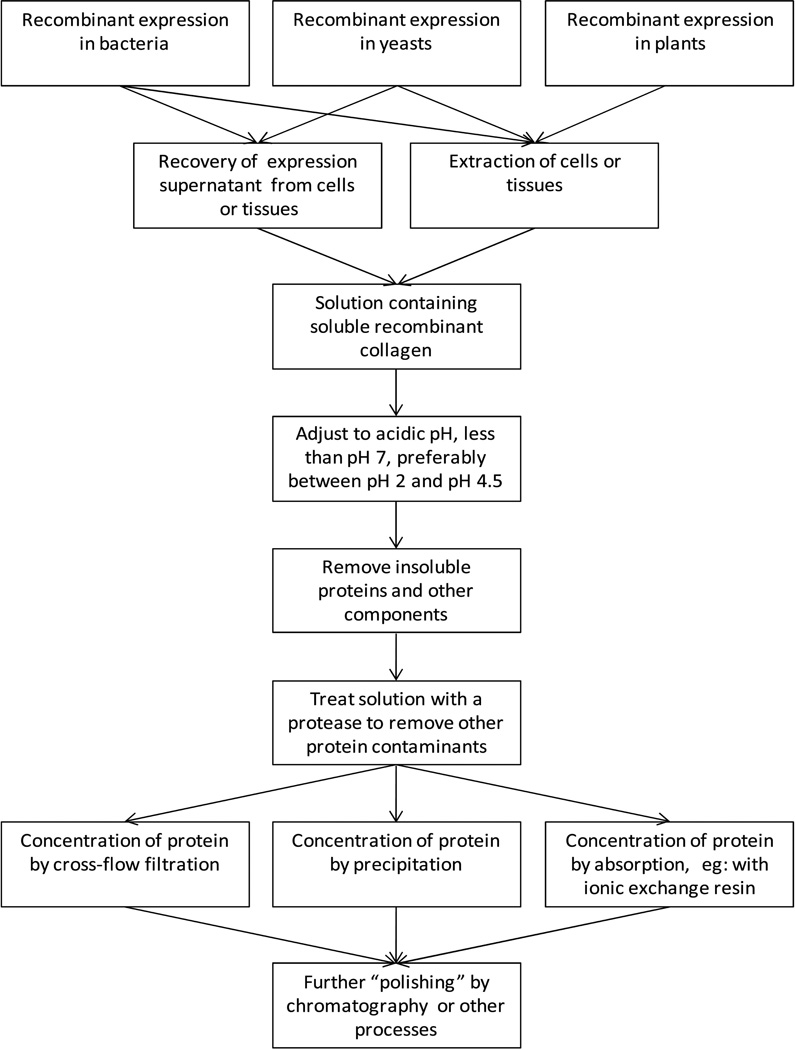
All 5 constructs were found to express recombinant protein well in shake flask cultures, as judged by the yields observed using SDS-PAGE. S. pyogenes V-CL and V-CL-CL, Methylbacterium and S. usitatus CL-domain with the N-terminal S. pyogenes V-domain, were prepared using 2 L bioreactors to enable purification. In the present report, all the expression was completed using E. coli as the expression host, although other bacteria, fungal and plant systems may also suitable (Fig. 2).
Figure 2.
A flow-path for the recombinant collagen purification process.
Protein precipitation
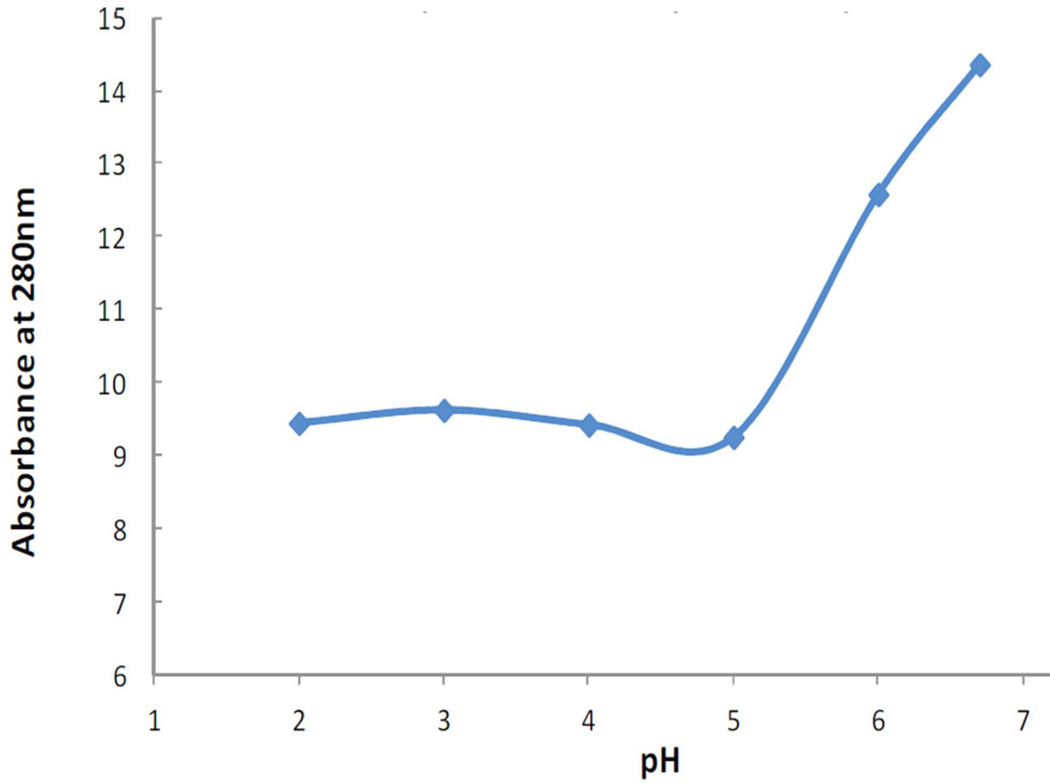
The first step required for a simple collagen purification protocol, was to indentify an efficient means of removing a substantial amount of the contaminating bacterial host proteins. E. coli cells were sonicated at various acidic pH values using an acetate-based buffering system ranging from pH 2.0 t pH 7.0 (Fig. 3). The host proteins were least soluble at a pH less than 5. It was not determined if the acid precipitation was due to denaturation or isoelectric precipitation or a combination of these processes. As can be seen in Figures 4 (A) for S pyogenes V-CL, and Figure 5 for Methylbacterium V-CL, S. usitatus V-CL and S.pyogenes V-CLCL, the respective soluble collagen-containing protein bands are strikingly abundant (labelled as “Ex”). After review of these data, pH 2.2 was selected for further extraction studies.
Figure 3.
The effect of pH using 50 mM acetate buffers on the solubility of E. coli host cell proteins.
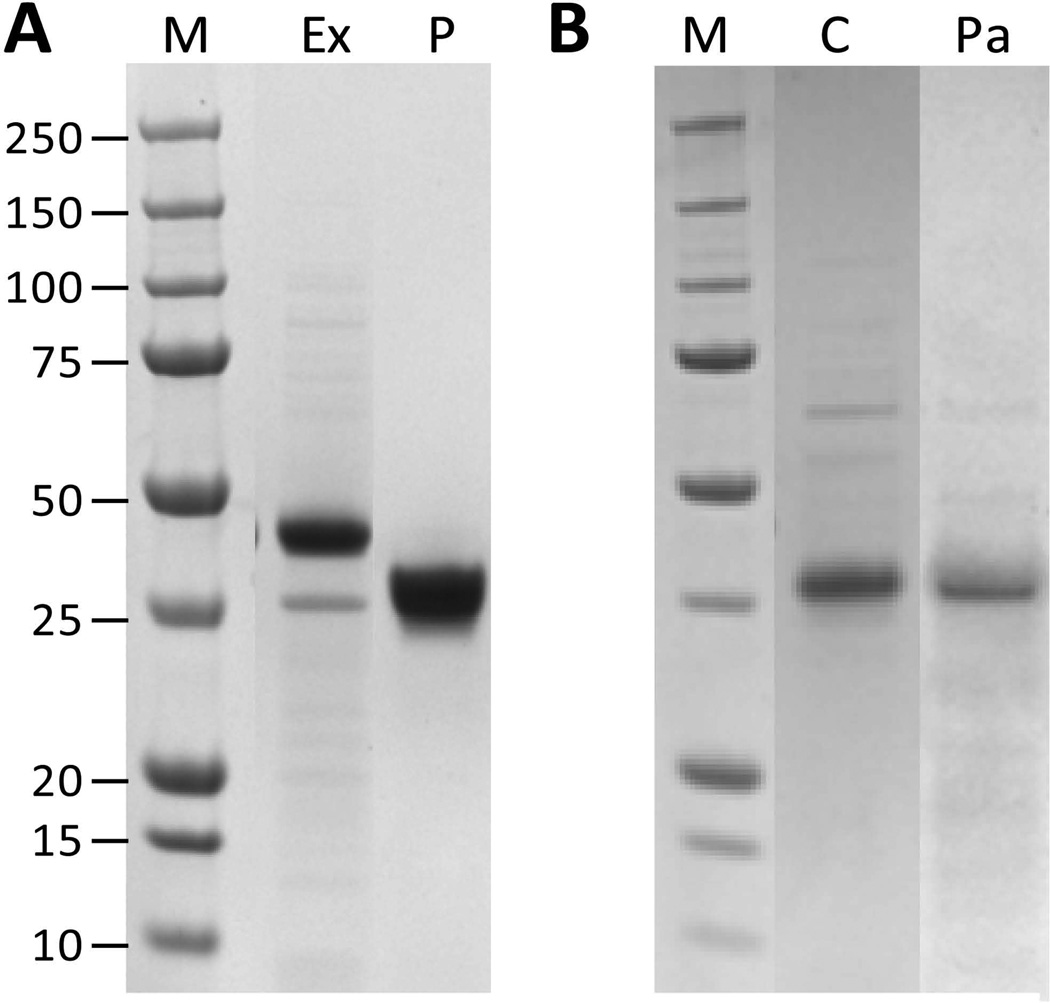
Figure 4.
Collages of SDS-PAGE gels showing the effects of different enzymes in the proteolysis processing step of the purification protocol for S. pyogenes V-CL product. A: After extraction and acid precipitation (Ex) followed by pepsin (P) digestion. B: As A, with chymotrypsin (C), and papain (Pa) digestion. M indicates protein molecular weight standards (kDa).
Figure 5.
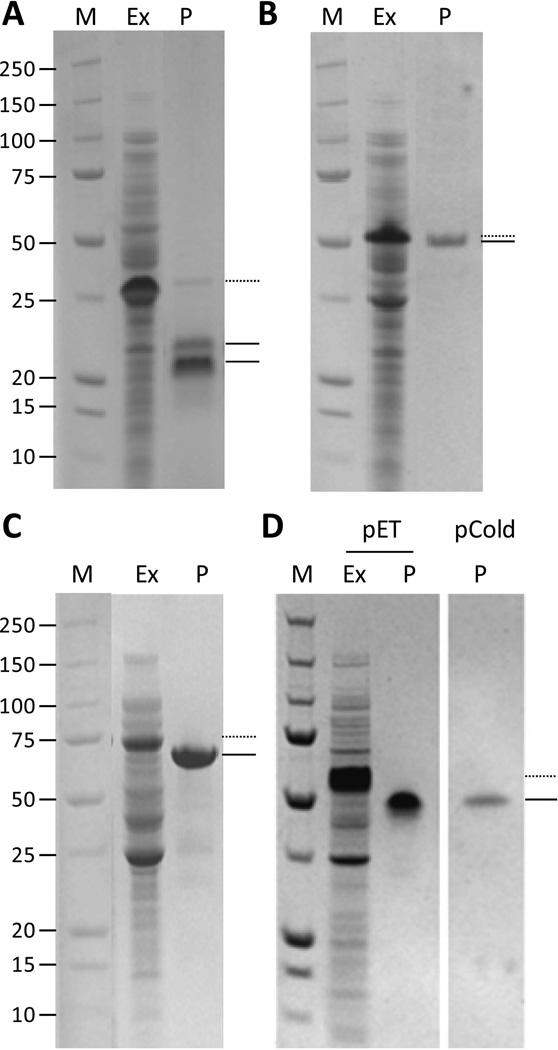
Collages of SDS-PAGE gels showing the effects of pepsin proteolysis (P) in processing step of the purification protocol for various collagen products. A: Methylbacterium, B: S. usitatus CL-domain with the N terminal S. pyogenes V-domain C: S. pyogenes V-CLCL product, D: S. usitatus CL-domain followed by the R. palustris V-domain after expression using pET and pCold vectors. M indicates protein molecular weight standards (kDa). Ex, indicates the extract after expression and extraction, P, indicates the product after purification by precipitation and proteolysis steps. Starting material, including the V-domain and collagen domain is indicated by a dotted line (•••••), while product (collagen domain only) is indicated with a line ( )
)
Proteolysis
Pepsin was very effective in digesting the residual E. coli proteins (Fig. 4A). In the acid precipitated and centrifuged extract from S .pyogenes, the V-CL protein was the major component. The other component seen in extract on the SDS gel is an E. coli protein, which was identified by mass spectroscopy of tryptic peptides as a member of the outer membrane porin family (cl17244: OM_channels) (data not shown). Use of pepsin results in loss of this E. coli protein and detection of a large band comprising the purified CL protein. Other enzymes were also tested, including chymotrypsin (Fig. 4B), trypsin (data not shown), papain (Fig. 4B), ficin and bromelain (data not shown). In the present study pepsin was the preferred enzyme for removal of residual host proteins. Neither trypsin nor chymotrypsin was as effective. Papain, however, was also found to be a suitable enzyme, but may benefit from further optimisation studies. Papain has the advantage that it is a non-animal protease, allowing development of a fully non-animal production (Peng et al. 2012) and purification system.
Pepsin digestion was also evaluated on the soluble extracts obtained after acid precipitation of cell lysates derived from each of the other collagen gene constructs, including Methylobacterium (Fig 5A), S. usitatus using the N-terminal S. pyogenes V-domain (Fig 5B), the V-CL-CL construct using the S. pyogenes collagen domain (Fig. 5C), and S. usitatus using the C-terminal R. palustris V-domain (Fig. 5D), where both pET and pCold had been used as expression vectors.
For Methylobacterium (Fig. 5A), digestion was incomplete as a small quantity of starting material remained (dotted line). Also, there were two products after the N-terminal V-domain had been removed. N-terminal sequence analysis (Fig. 6) showed that both these product bands had the same N-terminal sequence, indicating that the smaller of the two proteins had lost a small number of residues from the C-terminal of the CL domain. For S. usitatus using the N-terminal S. pyogenes V-domain (Fig. 5B), which lacked any additional cleavage/spacer insert, the V-domain was not cleaved from the CL-domain by pepsin. So, while the proteolysis removed the residual E. coli host proteins, the V-CL remained intact. The V-CL-CL construct using the S. pyogenes collagen domain (Fig. 5C) was also readily processed by pepsin. In the present example, low levels of host proteins may remain, eg: at about 25 kDa, in some preparations. When the S. usitatus CL-domain followed by the R. palustris V-domain was expressed in either pET and pCold vectors (Fig. 5D), both gave good yields of product, and both extracts processed similarly to give product that was substantially pure and with the V-domain removed.
Figure 6.
N-terminal sequence determination on selected samples after proteolysis with pepsin or papain. Underlined bold residues are the N-terminal sequences that were identified by automated Edman sequencing, while italic bold residues are those in the Gly-Xaa-Yaa repeating collagen sequences. Numbers give the location within the full construct prior to proteolysis.
The SDS-PAGE analyses (Figs. 4 & 5) showed that in most cases the V-domain is removed from the constructs, as the protein in these examples has a lower molecular weight than that initial product in the extracts. However, with the S. pyogenes V-domain linked to the S. usitatus CL domain, which lacked the additional cleavage/spacer sequence, little if any cleavage of the V-domain was found (Fig. 5B). On the other hand, the C-terminal V-domains of the S. usitatus CL domain followed by the R. palustris V-domain, again without a cleavage/spacer insert, was cleaved using pepsin proteolysis (Fig. 5D).
The sites of enzyme cleavage were generally not certain, but are assumed to be close to the triple-helical collagen domain. In certain cases the N-terminal cleavage site preceding the CL domain was established using automated Edman degradation. For S. pyogenes constructs, cleaved with pepsin and papain, the cleavage sties were determined as starting LVP and GSP (underlined in Fig.6), both immediately prior to the start of the CL-domain (italics in Fig.6). For Methylobacterium, the pepsin cleavage site started VAV and was 11 residues prior to the start of the CL-domain (italics in Fig. 6).
In the present study, the post-proteolysis product has been concentrated, dialysed and freeze dried, allowing a dry weight to be determined. In an example, an extract from a production process of the S. pyogenes V-CL construct completed using a 2 L bioreactor that was estimated to contain 19 g/L V-CL based on staining of SDS-PAGE gels, was processed according the present method. Given the removal of the V-domain during proteolysis, the initial yield of 19 g/L equates to an estimated yield of about 14 g/L for an isolated CL domain. After completion of the purification process 20 g of purified bacterial collagen, as the dry weight of the product after freeze drying, was recovered from the 2 L batch indicating a yield of about 71%.
Discussion
All constructs expressed well in shake flask cultures, as judged by SDS-PAGE. They had all been designed to include a V-domain. This domain assists in the registration of the three collagen chains during synthesis and was included to enhance production. For S. usitatus, two different V-domain sequences were examined. Previous data (Xu et al. 2010) had suggested that soluble expression of S. usitatus was low when its own V-domain was used, and a better soluble yield was obtained when replaced with the S. pyogenes V-domain. Another V-domain that improved soluble expression was that from R. palustris (Xu et al. 2010). In contrast to the S. pyogenes V-domain, this R. palustris V-domain is found at the C-terminal in the source sequence and was consequently used in this study as a C-terminal domain in combined construct (Fig. 1) along with the addition of a His6-tag sequence. In some cases (Fig. 1), a His6-tag was included as part of the construct, while in others this was part of the vector sequence, (e.g. pColdI) if needed for small analytical purification protocols. In addition, the His6-tag can be used to quantify collagen expression and yield prior to its removal during the proteolysis step. Similarly, in some cases an additional enzyme site/spacer sequence was present, while others without such a site were available for comparison to assess the need for this insertion. A further construct was also made comprising a collagen sequence that consisted of a tandem construct, being two contiguous collagen domains from the S. pyogenes (Yoshizumi et al. 2009). Similar to the monomeric construct, this construct also had a His6-tag, a single V-domain and an inserted enzyme/spacer site.
The acid purification protocol was an efficient means of removing a substantial amount of the contaminating bacterial host proteins. The host proteins were least soluble at a pH lower than 5. It was not determined if the acid precipitation was due to denaturation or isoelectric precipitation or a combination of these processes. Acid precipitation of contaminating host proteins was very effective for collagen purification. For other proteins, the removal of host protein has been achieved using a simple heat step (Lyons et al. 2007) but this is not appropriate for the current collagens as they denature, and do not readily refold into the triple-helical conformation (Yoshizumi et al. 2009). An important aspect of using acid precipitation of host proteins is that collagens in general are typically soluble under acidic conditions, especially at low pH (Miller and Rhodes 1982). In the present study, the solubility of the various bacterial collagens, which still included attached V-domains, was confirmed by SDS-PAGE.
After acid precipitation and centrifugation to remove precipitated proteins, the residual contaminating host proteins in the soluble, collagen-enriched supernatant, were successfully removed by proteolysis. Various enzymes were examined for hydrolysis of the remaining contaminating host proteins and to cleave and digest the V domain away from the CL domain. The preferred enzyme, used in the present study, was pepsin. It had the advantage that there was no requirement for pH adjustment after the precipitation and centrifugation steps.
The use of acidic conditions, while favourable for the precipitation step and retaining the solubility of recombinant collagens, also has potentially adverse effects on the stability of the collagen triple helix in these bacterial proteins. Typically, bacterial collagens are stable at around body temperature, 35 – 40 °C (Xu et al. 2010). However, it has been shown (Xu et al. 2010) that the stability at pH 2.2 of the collagen domains from S. pyogenes was reduced to 25.7 °C, Methylobacterium to 28.3 °C and that of S. usitatus to 27.0 °C. In the bacterial collagens, where there is no hydroxyproline, charged pairing is present and can add to the triple helical stability (Persikov et al. 2005). At lower pH the carboxyl groups become uncharged and their contribution to stabilisation by charge pairs is lost.
The suitability of the present method for purification (Fig. 2) was demonstrated on larger scale preparations, showing in all cases adequate stability of the collagen to acid treatment and the resistance of the collagen domains to proteolysis at low temperature. The collagen triple-helix has long been known to be particularly resistant to proteolysis, and proteolysis was introduced as a step for the solubilisation of animal collagens from tissues (Kuhn et al. 1961). However, there are some exceptions where it is not suitable, typically type III collagens from certain species. For example, kangaroo type III collagen is completely digested by pepsin (Stephens et al. 1991), while from other species a range of susceptibilities are found to pepsin, trypsin and thermolysin (Wang et al., 1978; Birkedal-Hansen et al., 1985). In the present studies, the bacterial collagens are resistant to proteolysis, confirming previous observations (Yoshizumi et al. 2009; Xu et al. 2010), while the need for an added spacer/cleavage sites for removal of the V-domain appears to depend on the specific nature of the individual V- and CL-domains. The removal of the V-domain is advantageous for biomedical products and introduction of an enzyme cleavage site can be used to assist with the proteolysis step. Typically the triple helical domain of collagen is poorly immunogenic; on the other hand, the non-triple helical domains are more immunogenic (Werkmeister and Ramshaw 2000). For S. pyogenes, the collagen domain has been shown to be non-immunogenic (Peng et al. 2010).
The present data has shown that this simple and cost effective method for purification of recombinant proteins give good product yields. However, as the purification process has not been fully optimised, it is possible that other process improvements (e.g. using precipitation rather than cross-flow filtration) may result in better overall yields. If these collagens are to be used as new biomedical materials, then further ‘polishing’ steps may be needed, for example to remove any residual DNA or lipo-polysaccharides originating from the host expression system.
Recombinant bacterial collagens can be produced using straightforward processes and, in contrast to the established animal-derived collagens, have the added advantage that the protein can be designed and modified to enhance or change specific biological properties (Peng et al. 2013). Furthermore, recombinant collagens also lack the composition and batch-to-batch variability that occurs in animal-derived materials due to variations in the age and physiology of the tissue from which the collagen is extracted.
In the present study we have developed a simple purification method that is applicable to a range of different non-animal, bacterial collagens. The method can be adapted to large-scale commercial process as it uses acid precipitation and proteolysis and does not include any potentially expensive chromatographic processes. The approach can be applied to a range of bacterial collagens with different collagen domain sizes (in the present examples ranging from 147 to 474 amino acids), and collagen domains with either acidic or basic isoelectric points. It is suitable for rationally designed constructs, such as tandem constructs, and composite, chimeric constructs where the collagen and V-domains are obtained from different species. Therefore, the combination of effective large-scale production (Peng et al. 2012), and the present simple yet effective purification system, which gives a good overall yield, provides opportunities for larger scale production of recombinant non-animal, bacterial collagens.
Acknowledgments
This work was supported in part through NIH grant #EB011620. This study was facilitated by access to the Australian Proteome Analysis Facility supported under the Australian Government’s National Collaborative Research Infrastructure Strategy (NCRIS).
References
- An B, DesRochers TM, Qin G, Xia X, Thiagarajan G, Brodsky B, Kaplan DL. The influence of specific binding of collagen-silk chimeras to silk biomaterials on hMSC behavior. Biomaterials. 2103;34:402–412. doi: 10.1016/j.biomaterials.2012.09.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkedal-Hansen H, Taylor RE, Bhown AS, Katz J, Lin HY, Wells BR. Cleavage of bovine skin type III collagen by proteolytic enzymes. Relative resistance of the fibrillar form. J Biol Chem. 1985;260:16411–16417. [PubMed] [Google Scholar]
- Boydston JA, Chen P, Steichen CT, Turnbough CL. Orientation within the exosporium and structural stability of the collagen-like glycoprotein BclA of Bacillus anthracis. J Bacteriol. 2005;187:5310–5317. doi: 10.1128/JB.187.15.5310-5317.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky B, Ramshaw JAM. The collagen triple-helix structure. Matrix Biol. 1997;15:545–554. doi: 10.1016/s0945-053x(97)90030-5. [DOI] [PubMed] [Google Scholar]
- Cosgriff-Hernandez E, Hahn MS, Russell B, Wilems T, Munoz-Pinto D, Browning MB, Rivera J, Höök M. Bioactive hydrogels based on designer collagens. Acta Biomater. 2010;6:3969–3977. doi: 10.1016/j.actbio.2010.05.002. [DOI] [PubMed] [Google Scholar]
- Ghosh N, McKillop TJ, Jowitt TA, Howard M, Davies H, Holmes DF, Roberts IS, Bella J. Collagen-like proteins in pathogenic E. coli strains. PLoS One. 2012;7:e37872. doi: 10.1371/journal.pone.0037872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn K, Hannig K, Hörmann H. Die Einwirkung von Trypsin und Pepsin auf natives Kollagen. Das Leder. 1961;10:237–241. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lyons RE, Lesieur E, Kim M, Wong DC, Huson MG, Nairn KM, Brownlee AG, Pearson RD, Elvin CM. Design and facile production of recombinant resilin-like polypeptides: gene construction and a rapid protein purification method. Protein Eng Des Sel. 2007;20:25–32. doi: 10.1093/protein/gzl050. [DOI] [PubMed] [Google Scholar]
- Miller EJ, Rhodes RK. Preparation and characterization of the different types of collagen. Methods Enzymol. 1982;82:33–64. doi: 10.1016/0076-6879(82)82059-4. [DOI] [PubMed] [Google Scholar]
- Mohs A, Silva T, Yoshida T, Amin R, Lukomski S, Inouye M, Brodsky B. Mechanism of stabilization of a bacterial collagen triple helix in the absence of hydroxyproline. J Biol Chem. 2007;282:29757–29765. doi: 10.1074/jbc.M703991200. [DOI] [PubMed] [Google Scholar]
- Peng YY, Yoshizumi A, Danon SJ, Glattauer V, Prokopenko O, Mirochnitchenko O, Yu Z, Inouye M, Werkmeister JA, Brodsky B, Ramshaw JAM. A Streptococcus pyogenes derived collagen-like protein as a non-cytotoxic and non-immunogenic cross-linkable biomaterial. Biomaterials. 2010;31:2755–2761. doi: 10.1016/j.biomaterials.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng YY, Howell L, Stoichevska V, Werkmeister JA, Dumsday GJ, Ramshaw JAM. Pilot production of a collagen-like protein from Streptococcus pyogenes for biomedical applications. Microbial Cell Fact. 2012;11:146. doi: 10.1186/1475-2859-11-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng YY, Stoichevska V, Schacht K, Werkmeister JA, Ramshaw JAM. Engineering multiple biological functional motifs into a blank collagen-like protein template from Streptococcus pyogenes. J. Biomed. Mater. Res. 2013 doi: 10.1002/jbm.a.34898. in press. [DOI] [PubMed] [Google Scholar]
- Persikov AV, Ramshaw JAM, Kirkpatrick A, Brodsky B. Electrostatic interactions involving lysine make major contributions to collagen triple-helix stability. Biochemistry. 2005;44:1414–1422. doi: 10.1021/bi048216r. [DOI] [PubMed] [Google Scholar]
- Ramshaw JAM, Peng YY, Glattauer V, Werkmeister JA. Collagens as biomaterials. J Mat Res Mat Med. 2009;20:S3–S8. doi: 10.1007/s10856-008-3415-4. [DOI] [PubMed] [Google Scholar]
- Ramshaw JAM, Werkmeister JA, Glattauer V. Collagen-based biomaterials. Biotechnol Genet Eng Rev. 1996;13:335–382. doi: 10.1080/02648725.1996.10647934. [DOI] [PubMed] [Google Scholar]
- Rasmussen M, Jacobsson M, Björck L. Genome-based identification and analysis of collagen-related structural motifs in bacterial and viral proteins. J Biol Chem. 2003;278:32313–32316. doi: 10.1074/jbc.M304709200. [DOI] [PubMed] [Google Scholar]
- Ricard-Blum S. The collagen family. Cold Spring Harb Perspect Biol. 2011;3:a004978. doi: 10.1101/cshperspect.a004978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom J, Harsch M, Jimenez S. Hydroxyproline content determines the denaturation temperature of chick tendon collagen. Arch Biochem Biophys. 1973;158:478–484. doi: 10.1016/0003-9861(73)90539-0. [DOI] [PubMed] [Google Scholar]
- Stephens LJ, Werkmeister JA, Tebb TA, Ramshaw JAM. Identification of type III collagen from kangaroo skin. Das Leder. 1991;42:41–44. [Google Scholar]
- Sylvestre P, Couture-Tosi E, Mock M. A collagen-like surface glycoprotein is a structural component of the Bacillus anthracis exosporium. Mol Microbiol. 2002;45:169–178. doi: 10.1046/j.1365-2958.2000.03000.x. [DOI] [PubMed] [Google Scholar]
- Vandersmissen L, De Buck E, Saels V, Coil DA, Anné J. A Legionella pneumophila collagen-like protein encoded by a gene with a variable number of tandem repeats is involved in the adherence and invasion of host cells. FEMS Microbiol Lett. 2010;306:168–176. doi: 10.1111/j.1574-6968.2010.01951.x. [DOI] [PubMed] [Google Scholar]
- Wang HM, Chan J, Pettigrew DW, Sodek J. Cleavage of native type III collagen in the collagenase susceptible region by thermolysin. Biochim Biophys Acta. 1978;533:270–277. doi: 10.1016/0005-2795(78)90571-8. [DOI] [PubMed] [Google Scholar]
- Werkmeister JA, Ramshaw JAM. Immunology of collagen-based biomaterials. In: Wise DL, editor. Handbook of Biomaterials Engineering. New York: Marcel Dekker Inc; 2000. pp. 739–759. [Google Scholar]
- Werkmeister JA, Ramshaw JAM. Recombinant protein scaffolds for tissue engineering. Biomed Mater. 2012;7:012002. doi: 10.1088/1748-6041/7/1/012002. [DOI] [PubMed] [Google Scholar]
- Xu C, Yu Z, Inouye M, Brodsky B, Mirochnitchenko O. Expanding the family of collagen proteins: recombinant bacterial collagens of varying composition form triple-helices of similar stability. Biomacromolecules. 2010;11:348–356. doi: 10.1021/bm900894b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Keene DR, Bujnicki JM, Höök M, Lukomski S. Streptococcal Scl1 and Scl2 proteins form collagen-like triple helices. J Biol Chem. 2002;277:27312–27318. doi: 10.1074/jbc.M201163200. [DOI] [PubMed] [Google Scholar]
- Yoshizumi A, Yu Z, Silva T, Thiagarajan G, Ramshaw JAM, Inouye M, Brodsky B. Self-association of Streptococcus pyogenes collagen-like constructs into higher order structures. Protein Sci. 2009;18:1241–1251. doi: 10.1002/pro.134. [DOI] [PMC free article] [PubMed] [Google Scholar]