Abstract
CD44 is a cell-surface glycoprotein involved in many cellular functions including lymphocyte activation, recirculation and homing, hematopoiesis and tumor metastasis, suggesting that CD44 may play an important role in breast cancer development. In this study, we examined if CD44 exon2 polymorphisms are associated with increased susceptibility to breast cancer. Direct nucleotide sequencing analysis showed that multiple single nucleotide polymorphisms (SNPs) were present in the CD44 exon2 coding region in female breast cancer patients. There was no significant difference in the frequency of any single SNP in the CD44 exon2 coding region between breast cancer patients and normal donors. However, CD44 polymorphisms in the CD44 exon2 coding region were identified in approximately 40% of breast cancer patients, which were significantly higher than those seen in normal donors (odds ratio, 9.34; 95% confidence interval = 2.58–33.82; p < 0.0001). Wilcoxon-Mann-Whitney test analysis showed that the patients with the CD44 polymorphisms in CD44 exon2 coding sequence had breast cancer at earlier ages, 49±3 vs. 62±2 (p < 0.0005) and larger tumor burdens (4.9±1.22mm vs. 1.6±0.15mm, p < 0.01) at the time of diagnosis. Interestingly, African American female patients having the CD44 polymorphisms in CD44 exon2 coding sequence were diagnosed with breast cancer at very young age (41±2). Our results demonstrate that CD44 exon2 polymorphisms are associated with breast cancer development, and such analysis may be effectively used in the evaluation of risk, prediction of cancer, prevention, diagnosis and epidemiological studies of breast cancer.
Keywords: Gene polymorphism, breast cancer, CD44, risk, patient
Introduction
Breast cancer has been shown to cause significant cancer-related deaths among women in the United States and globally. Genetic factors play a pivotal role in susceptibility to breast cancer. Women with a family history of breast cancer, especially a first-degree relative, such as mother, sister or daughter, have an increased risk of developing breast cancer (1). The risk is higher if more than one first-degree relative has developed breast cancer and this increases further, the younger the relative was at the time of diagnosis (2). Thus, genetic makeup may be an important determinant of breast cancer risk. A number of breast cancer susceptibility genes have been characterized, notably BRCA1 and BRCA2, which are responsible for approximately 15% of breast cancer cases due to inherited mutations (3, 4). In addition, human epidermal growth factor receptor 2 (HER2) polymorphism, glutathione S-transferase O1 polymorphisms and polymorphic CAG repeats in androgen receptor (AR) gene may confer an increased risk of breast cancer in women (5–7). The current estimates are that all the known breast cancer susceptibility genes account for less than 25% of the familial aggregation of breast cancer (8). This points to the possibility that majority of the familial clustering of breast cancer cannot be explained and thus further studies are needed to identify other breast cancer susceptibility genes. The identification of new genes could make a major impact in risk prediction.
Genome-wide association studies (GWAS) provide an effective approach to identify the functional gene variations that increase or decrease the risk of cancer and thus give us new insights into risk prediction as well as preventive and therapeutic interventions. Studies of the “dark matter” in the human genome that are not captured by the single nucleotide polymorphism (SNP)-based GWAS such as structural and rare gene variants, micro-RNAs, and epigenetics are needed to fully understand the inherited component of cancer. Thus, in the current study, direct nucleotide sequencing strategy was used to perform a fine analysis of CD44 gene polymorphisms in the CD44 exon2 coding region in female breast cancer patients.
CD44 is a cell surface transmembrane glycoprotein, encoded by a single gene. Human CD44 gene is located at the short arm of chromosome 11 and consists of at least 20 exons spanning about 50 kilobases of DNA. The gene is composed of two groups of exons. One group, comprising exons 1–5 and 16–20, are expressed together as the standard form. The 10 variable exons (exons 6–15) can be alternatively spliced and included within the standard exons at an insertion site between exons 5 and 16. CD44 is expressed in a variety of cells including hematopoietic, epithelial, endothelial, and mesodermal origin (9, 10). CD44 plays a role in tumor metastasis (11, 12). CD44s (the standard form) is known to be important in T-cell signaling and a variety of immune cell functions. In addition, CD44s plays a role in T-and B-cell adhesion, cell aggregation, proliferation and cell migration (9). CD44s is also an important cytotoxic triggering molecule on cytotoxic T lymphocytes (CTL), double-negative (DN) T cells and natural killer (NK) cells in mice (13). CD44 has also been shown to interact with hyaluronan in regulation of breast cancer cell proliferation, migration, and invasion, as well as tumor-associated angiogenesis that correlates with patient survival (10, 14). We hypothesize that CD44 exon2 polymorphisms may play an important role in breast cancer development.
Role of CD44 polymorphisms in breast cancer development has not been fully understood. Our previous studies indicated that a unique single nucleotide polymorphism (designated as CD44 Ex2+14 A>G) in the CD44 intron 1 region was identified in 84% of breast cancer patients, which was significantly higher than that seen in normal donors (15). Moreover, the breast cancer patients with homozygous unique SNP in CD44 intron 1 had breast cancer at earlier ages, larger tumor burden, more regional lymph node metastases at the time of diagnosis, and higher cancer recurrence rate (15). In the current study, we examined CD44 polymorphisms in the CD44 exon2 coding region, an important site for CD44 ligand binding (16), in breast cancer patients. The results demonstrated that multiple single nucleotide polymorphisms in the CD44 exon2 coding region were present in approximately 40% of breast cancer patients, which was significantly higher than that in normal donors. Furthermore, the breast cancer patients having the CD44 polymorphisms in the CD44 exon2 coding sequence had breast cancer at an earlier age and significantly larger tumor burden when the patients were diagnosed with breast cancer. The results suggest that CD44 polymorphisms are associated with breast cancer. CD44 polymorphism analysis may be effectively used in evaluation of cancer risk, prediction, prevention, diagnosis and genetic studies of breast cancer.
Materials and Methods
Patient samples
All breast cancer patients in this study signed an institutional review board-approved consent form. The blood samples, breast cancer specimens and adjacent normal breast tissues deposited in the Tissue Bank at South Carolina Cancer Center (Columbia, SC, USA) from 260 breast cancer patients during 2001 and 2008 were used in this research. Among breast cancer patients, 118 were Caucasian American (CA) women, 115 African American (AA) women and 27 unknown ethnic women. The patients had breast cancers from stage I to stage IIIC. The patients were between ages 23 and 90, and had an average age of 55 years at the time of diagnosis with breast cancer.
Normal donors
All normal female donors signed an institutional review board-approved consent form and were recruited in South Carolina Women’s Care Study (Columbia, SC, USA) where they donated blood samples for our research. Among normal donors, 96 were Caucasian American women and 96 African American women. In addition, genomic DNA samples from 40 healthy Caucasian American female normal donors provided by BioChain Institute, Inc. (Hayward, CA, USA) were also used as normal controls.
Genomic DNA isolation
The genomic DNA was isolated from peripheral blood samples derived from female normal donors and breast cancer patients using the AGENCOURT® GENFIND™ v2 Blood & Serum Genomic DNA Isolation kit (Agencourt, Beverly, MA, USA) according to the manufacture’s manual. The DNA concentration of genomic DNA samples was determined by spectrophotometer and the quality of genomic DNA samples was analyzed by agarose gel electrophoresis.
Analysis of CD44 polymorphisms
Direct DNA sequencing method was used to determine the polymorphisms in the CD44 exon2 region including the CD44 exon2 sequence and the boundary sequences between exon2 and upstream intron, and between exon2 and downstream intron. Briefly, 5ng of genomic DNA from each sample was added into 50µl of PCR reaction mixture. M13R-tagged forward primer (5’-CCGGCCTTATTTGACTTTTTAAGGAGTCTG-3’) and M13F-tagged reverse primer (5’-CTCCAGTTGTCATACAGGTTGCAGATTGAC-3’) were used to amplify the CD44 exon2 region from genomic DNA samples by PCR using high fidelity DNA polymerase (Invitrogen, Carlsbad, CA, USA). The PCR products were sent out to MCLAB (South San Francisco, CA, USA) for cleanup and direct sequencing using primers M13R and M13F. At the same time, the PCR products were cloned into pCR2.1 TOPO vector (Invitrogen, Carlsbad, CA, USA), transformed into One Short® TOP10F’ cells (Invitrogen, Carlsbad, CA), and sent out to MCLAB (South San Francisco, CA, USA) for colony sequencing. After cloning and transformation, plasmid DNA samples were also prepared from certain number of samples (including 60 breast cancer patient samples and 40 normal donors) using QIAprep® Spin Miniprep kit (Qiagen, Valencia, CA, USA), and sent out to SeqWright (Houston, TX, USA) for sequencing confirmation. Sequencing analysis using BLAST searching for SNPs was carried out to determine CD44 polymorphisms in the CD44 exon2 region.
Statistical analysis
Odds Ratio Generator Version 1.0.0 (Devilly G.J., 2005, Centre for Neuropsychology, Swinburne University, Australia) was used to calculate odds ratio (OR) and 95% confidence interval (CI) to analyze the statistical differences of CD44 polymorphisms between two groups. The odds ratio ranges from 0 to positive infinity, with 1.0 indicating that the condition or event under study is equally likely in both groups. An odds ratio greater than 1 indicates that the condition or event is more likely in the second group. 2-sample proportion test was used to confirm the statistical differences. Proportion trend test was used to correct the statistics for multiple hypothesis testing. Fisher Exact test and Wilcoxon-Mann-Whitney test were used to determine the association between CD44 polymorphism and breast cancer development, and the association was considered significant at P < 0.05.
Hardy-Weinberg equilibrium (HWE) test of SNP was performed using Michael H. Court’s (2005–2008) online calculator (http://www.tufts.edu/~mcourt01/Documents/Court%20lab%20-%20HW%20calculator.xls). Tests in breast cancer patients (P = 0.4962) and female normal donors (P = 0.7222) did not show any significant deviation from HWE for any of the SNPs.
The Kaplan Meier approach was employed to generate both probability curves and cumulative risk curves for breast cancer between the female breast cancer patients who did not have CD44 polymorphisms in CD44 exon2 coding region and those who had CD44 polymorphisms in CD44 exon2 coding region, which was confirmed by the “survival” package in R statistical software (http://www.r-project.org) using command “survfit”. The Gehan rank test was used to determine the statistical differences, and the difference was considered significant at P < 0.05.
Results
Identification of CD44 polymorphisms in CD44 exon2 coding region
The human CD44 gene has been mapped to the chromosomal locus 11p13 and is composed of two groups of exons. One group, comprising exons 1–5 and 16–20, are spliced together to form a transcript that encodes the ubiquitously expressed standard isoform (abbreviated to CD44s). The 10 variable exons 6–15 (also known as v1–10) can be alternatively spliced and included within the standard exons at an insertion site between exons 5 and 16 (17). CD44 exon2 is critical for CD44 binding to its ligand, hyaluronan (18). Therefore, at first, we were interested in analyzing the polymorphisms in the CD44 exon2 region and their role in breast cancer development. The direct nucleotide sequencing analysis indicated that CD44 polymorphism in the CD44 coding region was present in breast cancer patients (Fig. 1). Thus, in addition to the unique CD44 Ex2+14 A>G polymorphism in the upstream intron region (intron1) of CD44 exon2 (15), one or more additional polymorphisms in the CD44 exon2 coding sequence were also present in breast cancer patients.
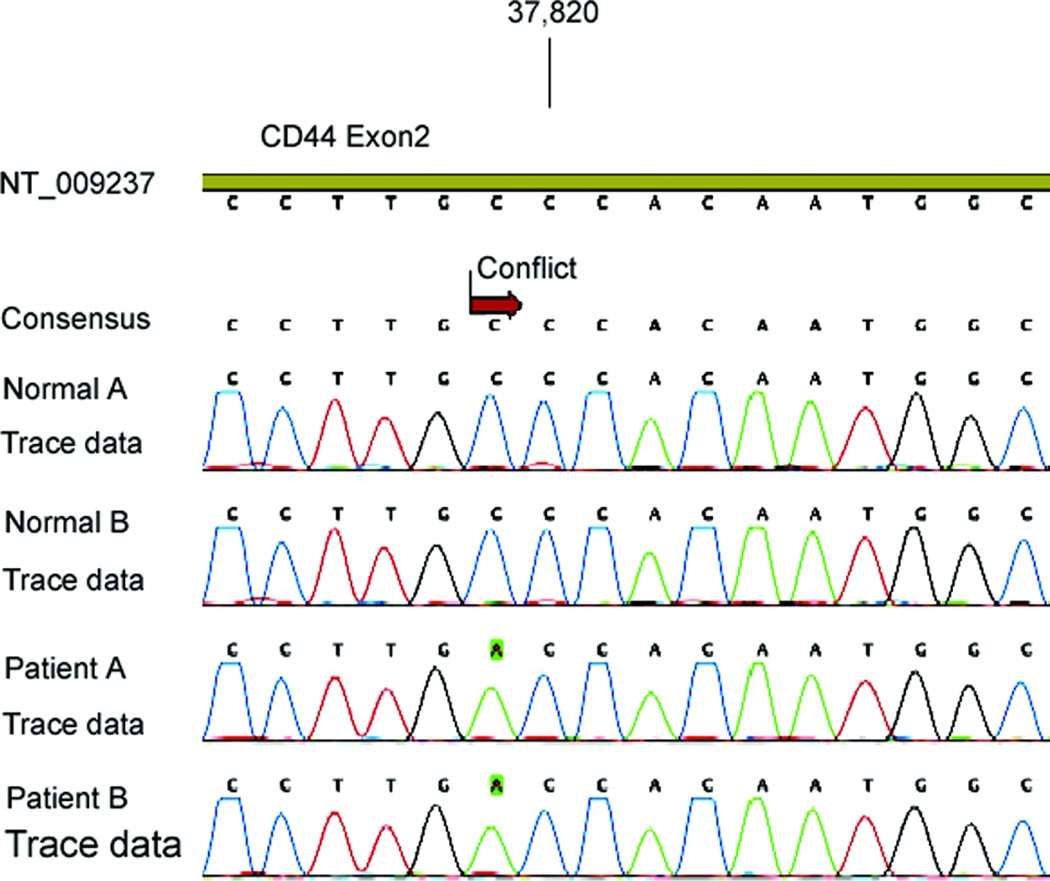
Figure 1.
Representative chromatogram of a single nucleotide polymorphism in CD44 exon2 coding region identified in female breast cancer patients. NT_009237 was the access number of Homo sapiens chromosome 11 genomic contig containing CD44 gene sequence in GenBank. The CD44 polymorphic change from C to A was highlighted in green at mRNA nucleotide position 615 in CD44 exon2 coding sequence, leading to amino acid substitution of proline by threonine at amino acid position 61. A, T, G and C nucleotide were denoted in green, red, black and blue curve, respectively.
Comparison of CD44 polymorphisms in CD44 exon2 coding region between female breast cancer patients and normal donors
The direct nucleotide sequencing analysis revealed 23 additional polymorphic loci within the CD44 exon2 coding region in breast cancer patients (Fig. 2A). We observed 10 synonymous changes and 13 non-synonymous changes, one of which introduced a stop codon at amino acid position 67. The nonsynonymous changes all resulted in nonconservative amino acid substitutions, suggesting that CD44 polymorphisms in the CD44 exon2 coding region may affect CD44 expression and functions.
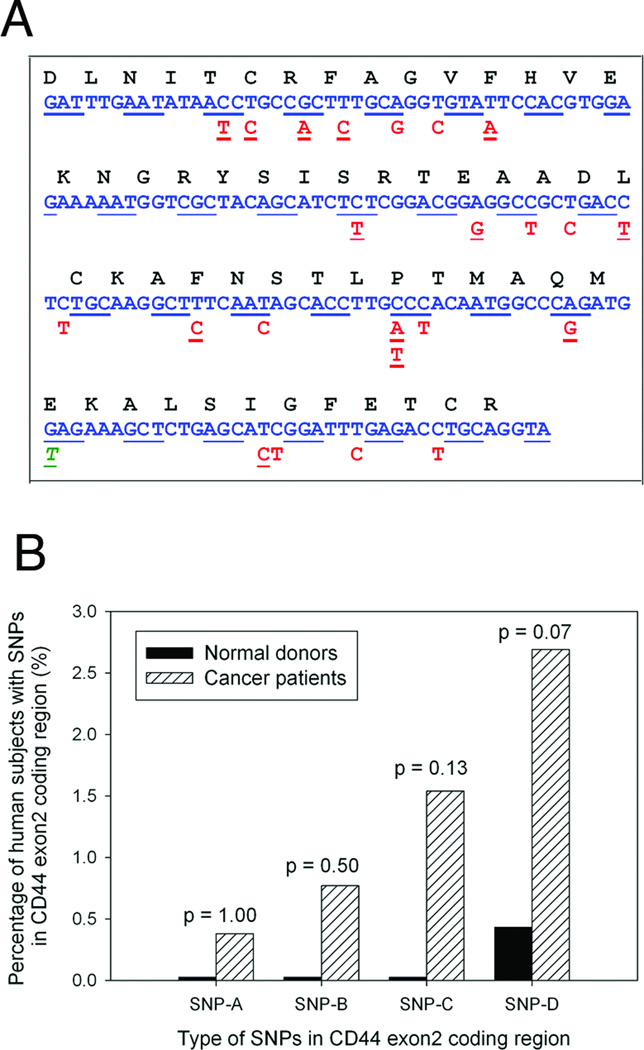
Figure 2.
Identification of SNPs in CD44 exon2 coding region in female breast cancer patients. A, CD44 polymorphisms in CD44 exon2 coding region. Multiple different CD44 polymorphic changes were identified in female breast cancer patients. Synonymous changes were noted bold in red, non-synonymous missense mutations were underlined in red, and the stop codon mutation was underlined and italicized in green. B, Comparison of CD44 polymorphisms in CD44 exon2 coding region between female breast cancer patients and normal donors. Based on the frequency of single nucleotide polymorphisms in CD44 exon2 coding region identified in breast cancer patients, four types of SNPs, SNP-A, SNP-B, SNP-C and SNP-D (from the lowest frequency through the highest frequency), were distinguished (Table 1). Fisher Exact test was used to determine the statistical difference of SNP frequencies between two groups. No significant difference of any SNPs in CD44 exon2 coding region between breast cancer patients and normal donors were found.
The frequency of any single SNP in the CD44 exon2 coding region in breast cancer patients was very low (Table 1). Based on the frequency of single nucleotide polymorphisms in CD44 exon2 coding region identified in breast cancer patients, four types of SNPs, SNP-A, SNP-B, SNP-C and SNP-D at a frequency of 0.38%, 0.77%, 1.54% and 2.69% in breast cancer patients respectively, were distinguished. There was no significant difference in frequency of any single SNPs in the CD44 exon2 coding region between breast cancer patients and normal donors (Fig. 2B). However, CD44 polymorphisms in the CD44 exon2 coding region were identified in approximately 40% of all breast cancer patients under investigation, which were significantly higher than those seen in normal donors (odds ratio, 9.34; 95% confidence interval = 2.58–33.82; p < 0.0001) (Fig. 3). Furthermore, when compared with female healthy normal donors, both Caucasian and African Americans breast cancer patients had a significantly increased frequency of CD44 polymorphisms in the CD44 exon2 coding region. There was also a significant difference of CD44 polymorphism frequencies between Caucasian and African American patients (p < 0.001; Fig. 3). The results indicated that CD44 polymorphisms in the CD44 exon2 coding sequence may also play a role in breast cancer development.
Table 1.
Frequency of CD44 polymorphisms in CD44 exon2 coding region identified in breast cancer patients
| Type of SNP | Frequency (%) | SNP Examples |
|---|---|---|
| SNP-A | 0.38 | 516T→C, 568C→T, 662C→T |
| SNP-B | 0.77 | 577A→G, 649T→C, 650C→T |
| SNP-C | 1.54 | 520G→A, 584T→C, 656T→C |
| SNP-D | 2.69 | 514C→T, 615C→A, 633G→T |
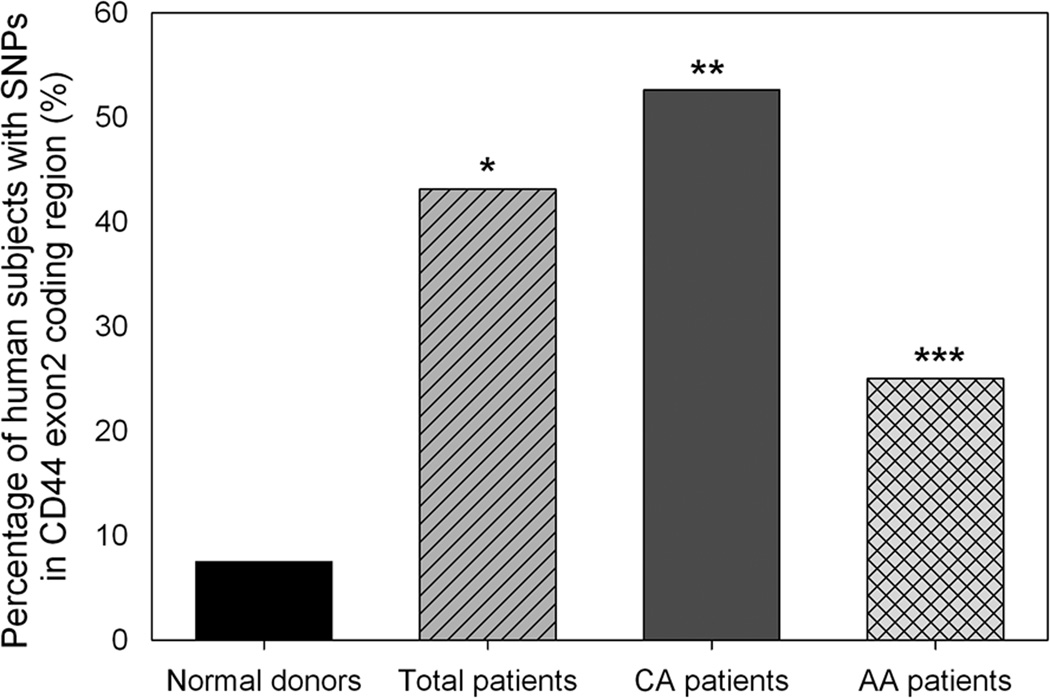
Figure 3.
Statistical comparison of CD44 polymorphisms in CD44 exon2 coding region between female breast cancer patients and normal donors. When compared to female normal donors, * indicated that the percentage of female breast cancer patients having CD44 polymorphisms in CD44 exon2 coding region was significantly higher (odds ratio = 9.34; 95% confidence interval = 2.58–33.82; p < 0.0001), ** indicated that the percentage of Caucasian American female breast cancer patients having CD44 polymorphisms was significantly higher (odds ratio = 13.70; 95% confidence interval = 3.60–52.22; p < 0.0001), and *** indicated that the percentage of African American female breast cancer patients with CD44 polymorphisms was also significantly higher (odds ratio = 4.11; 95% confidence interval = 0.87–19.41; p < 0.05). CA, Caucasian American. AA, African American. Fisher Exact test was used to determine the statistical difference of SNP frequencies between two groups.
Relationship between CD44 Polymorphisms and breast cancer development
Considering that female breast cancer patients had significantly increased frequencies of CD44 polymorphisms in the CD44 exon2 region than female normal donors (Fig. 3), we were interested in understanding the clinical significance of CD44 polymorphisms in breast cancer development. Because not all of breast cancer patients exhibited CD44 polymorphisms in the CD44 exon2 coding region (Figs. 2 and 3), we divided the breast cancer patients into two groups: (1) Group I patients are those that did not have the CD44 polymorphisms in the CD44 exon2 coding region, and (2) Group II patients are those that had one or more CD44 polymorphisms in the CD44 exon2 coding sequence. Wilcoxon-Mann-Whitney test demonstrated that the Group II female breast cancer patients having the CD44 polymorphisms in the CD44 exon2 coding sequence had breast cancer at an earlier age and larger tumor sizes when they were diagnosed with breast cancer, which were statistically significant (Fig. 4). Interestingly, as compared with Caucasian American females, African American females having the CD44 polymorphisms in CD44 exon2 coding sequence were diagnosed with breast cancer at a relatively younger age (Fig. 4e). These data indicated that CD44 polymorphisms in the CD44 exon2 coding region may play an important role in breast cancer development.
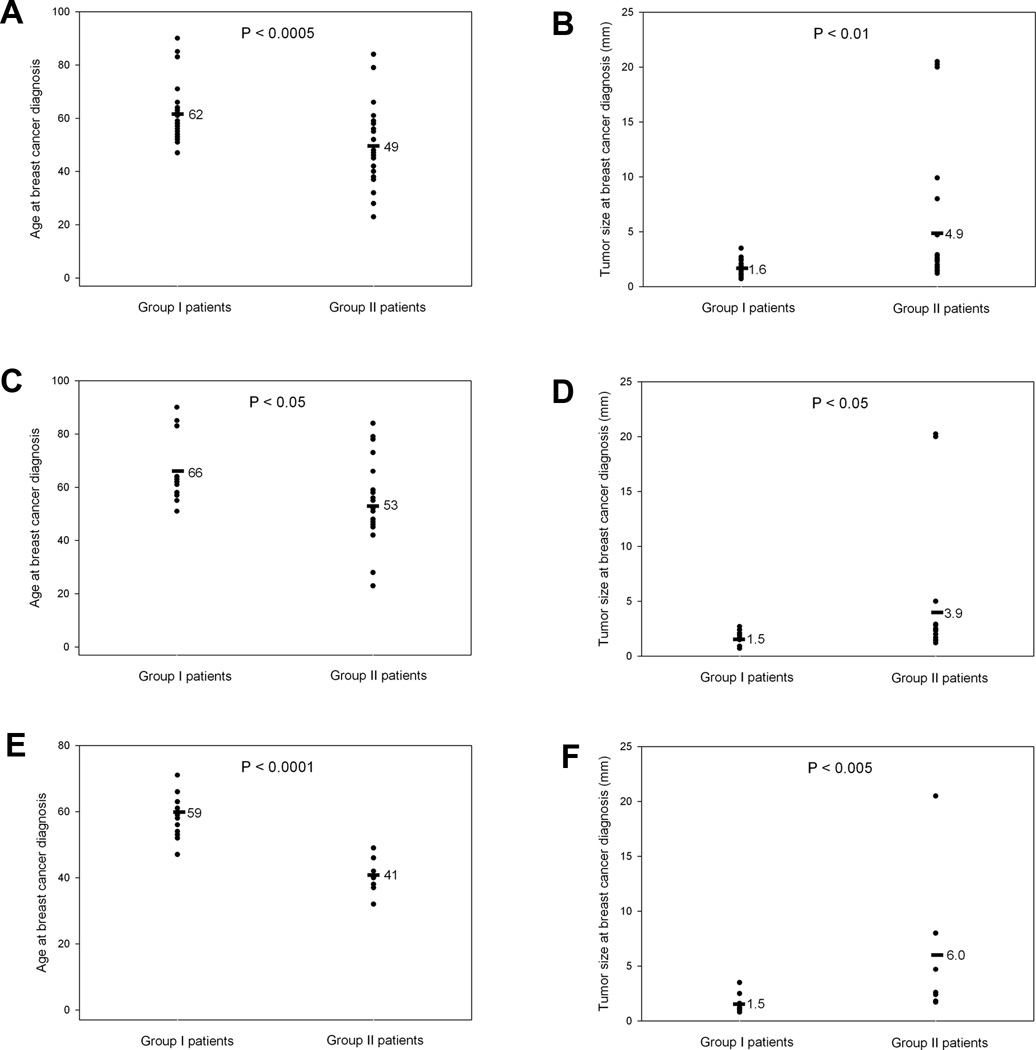
Figure 4.
Association of CD44 polymorphisms with patient age and tumor size at breast cancer diagnosis. The ages were compared at breast cancer diagnosis between Group I and Group II patients from all (A), Caucasian American (C) and African American (E) female breast cancer patients as well as the tumor sizes at breast cancer diagnosis between Group I and Group II patients from all (B), Caucasian American (D) and African American (F) female breast cancer patients. Group I patients are those female breast cancer patients who did not have the CD44 polymorphisms whereas Group II patients are those with CD44 polymorphisms in CD44 exon2 coding sequence as shown in Fig. 2A. Wilcoxon-Mann-Whitney test was used to determine the statistical differences in the age of patients and tumor sizes between two groups.
Estimation of breast cancer probability and risk
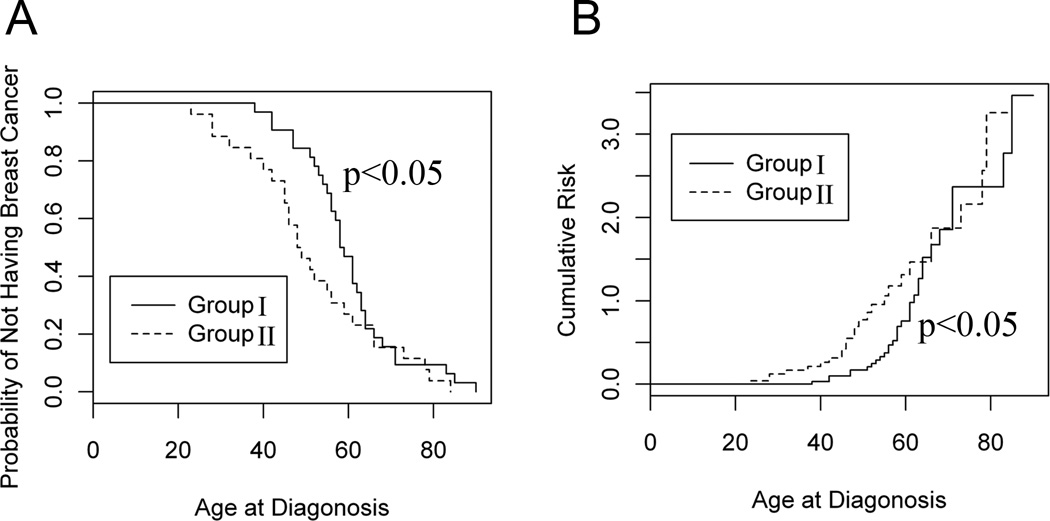
Statistical analysis demonstrated that the patients having the CD44 polymorphisms in CD44 exon2 coding sequence had significantly higher probability and higher cumulative risk for breast cancer (Fig. 5). For instance, a patient with a CD44 polymorphism in CD44 exon2 coding sequence had 0.23 (1–0.77 = 0.23, or 23%) probability for breast cancer at age 40 and 0.73 (1–0.27 = 0.73 or 73%) probability at age 60, whereas the patients without any CD44 polymorphisms in CD44 exon2 coding sequence had 0.03 probability for breast cancer at age 40 and 0.53 probability at age 60 (Fig. 5). Thus, based on the breast cancer probability curve and cumulative risk curve, we may establish a prediction model for breast cancer risk.
Figure 5.
Assessment of breast cancer risk in patient population. A, Comparison of breast cancer probability between the patients who did not have CD44 polymorphisms in CD44 exon2 coding region and those who had CD44 polymorphisms in CD44 exon2 coding sequence. Y axis was the probability of NOT having breast cancer. B, Comparison of cumulative risk for breast cancer between the patients having no CD44 polymorphisms and those having CD44 polymorphisms in CD44 exon2 coding region. The Kaplan Meier approach was used to generate both probability and cumulative risk curves. The Gehan rank test was used to determine the statistical difference.
Discussion
CD44 is a multi-functional transmembrane protein involved in cell proliferation, angiogenesis, invasion and metastasis (19). CD44 and its interaction with hyaluronan may regulate breast cancer cell proliferation, migration, and invasion, as well as tumor-associated angiogenesis and are correlated with patient survival (10, 14). Thus, CD44 may play a role in breast cancer development. Our previous studies demonstrated that the unique single nucleotide polymorphism (CD44 Ex2+14 A>G) in CD44 intron 1 is associated with breast cancer development (15). In the current study, we further examined the gene polymorphisms in CD44 exon2 coding sequence in breast cancer patients. Our results showed that CD44 gene polymorphisms were present in the CD44 exon2 coding region in breast cancer patients (Figs. 1 and 2A). As compared with normal donors, the frequencies of single nucleotide polymorphisms in the CD44 exon2 coding region in breast cancer patients are not higher (Fig. 2B and Tab. 1), however, the combined frequency of all SNPs in breast cancer patients was significantly higher than that in normal donors (Fig. 3), suggesting that CD44 polymorphisms in CD44 exon2 coding sequence play a role in breast cancer development. Recent report also indicated that germline polymorphisms in CD44 gene are associated with clinical outcome in localized gastric adenocarcinoma (20) and sarcoma incidence and survival (21). Thus, CD44 gene variations may be a risk factor for cancers.
Multiple SNPs in the CD44 exon2 coding region were identified in breast cancer patients (Fig. 2). Some of these polymorphic changes were nonsense mutations, however, it is possible that these nonsense mutations may somehow influence RNA stability and thus decrease CD44 expression. Some of them were sense mutations, resulting in the mutated CD44 proteins, and one of them was Amber stop codon mutations, resulting in the truncated CD44 proteins. Therefore, the CD44 polymorphisms in CD44 exon2 coding region may affect CD44 expression and functions. Further studies will be needed to determine the relationship of CD44 polymorphisms with CD44 expression and functions. Nonetheless, it is clear that the female breast cancer patients having the CD44 polymorphisms in CD44 exon2 coding sequence had breast cancer disease at earlier ages and larger tumor burdens when they were diagnosed with breast cancer, which were statistically significant (Fig. 4), suggesting that CD44 polymorphisms in the CD44 exon2 coding region play an important role in breast cancer development.
A number of prediction models such as Gail Model (22) and Clause Model (23) for breast cancer risk have been developed (24, 25). Parl et al. proposed a mathematical model that forecasts breast cancer risk for a woman based on three factors: (a) estimated estrogen exposure, (b) kinetic analysis of the oxidative estrogen metabolism pathway in the breast, and (c) enzyme genotypes responsible for inherited differences in the production of carcinogenic metabolites (26, 27). Fasching et al. evaluated MENDEL, BRCAPRO(Claus), BRCAPRO(Ford) as well as the Tyrer-Cuzick mathematical models in the assessment of breast cancer risk (28). Our statistical analysis demonstrated that the patients having the CD44 polymorphisms in CD44 exon2 coding sequence had significantly higher probability and higher cumulative risk for breast cancer (Fig. 5). Thus, based on the breast cancer probability curve and cumulative risk curve of CD44 polymorphisms, we may establish a prediction model for breast cancer risk.
CD44 is an adhesion molecule of the hyaluronate receptor family. CD44 function depends on its binding to its ligands such as hyaluronan. It has been reported that the function of CD44-hyaluronan binding is responsible for cell-to-cell and cell-to-extracellular matrix interactions (29), apoptosis inhibition (30), lymphocyte stimulation (31) and augmentation of tumor cell motility and metastasis (32). CD44 has two binding domains for hyaluronan, one in exon2 region and another in exon5 region (16). Therefore, we are interested in understanding gene polymorphisms in CD44 exon2 and exon5 in breast cancer. In the current study, multiple CD44 polymorphisms in the CD44 exon2 coding region were identified in breast cancer patients. However, we cannot rule out that CD44 polymorphisms in other exons may also play a critical role in breast cancer development. Furthermore, analysis of CD44 polymorphisms in a larger sample size is necessary to confirm the association between CD44 polymorphisms and breast cancer risk in order to establish a prediction mode for breast cancer risk assessment.
Acknowledgments
We thank Dr. Phillip J. Buckhaults and Ms. Ella S. Weinkle for assistance in obtaining genomic DNA samples, blood samples, breast cancer specimens and adjacent normal breast tissues from breast cancer patients in the Tissue Bank at South Carolina Cancer Center. We also wish to thank Dr. Kim Creek for providing genomic DNA samples from female normal donors.
Grant support: This study was supported by the Innovative and Exploratory Grant Program (IEGP) of University of South Carolina School of Medicine (J. Zhou), NIH grants R01ES09098, R01DA016545 and P01AT003961 (P. Nagarkatti) and NIH grants R01AI053703, R01AI058300, and R01HL058641 (M. Nagarkatti).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Loman N, Johannsson O, Kristoffersson U, Olsson H, Borg A. Family history of breast and ovarian cancers and BRCA1 and BRCA2 mutations in a population-based series of early-onset breast cancer. J Natl Cancer Inst. 2001;93:1215–1223. doi: 10.1093/jnci/93.16.1215. [DOI] [PubMed] [Google Scholar]
- 2.Familial breast cancer: collaborative reanalysis of individual data from 52 epidemiological studies including 58,209 women with breast cancer and 101,986 women without the disease. Lancet. 2001;358:1389–1399. doi: 10.1016/S0140-6736(01)06524-2. [DOI] [PubMed] [Google Scholar]
- 3.Peto J, Collins N, Barfoot R, Seal S, Warren W, Rahman N, et al. Prevalence of BRCA1 and BRCA2 gene mutations in patients with early-onset breast cancer. J Natl Cancer Inst. 1999;91:943–949. doi: 10.1093/jnci/91.11.943. [DOI] [PubMed] [Google Scholar]
- 4.Anglian Breast Cancer Study Group. Prevalence and penetrance of BRCA1 and BRCA2 mutations in a population-based series of breast cancer cases. Br J Cancer. 2000;83:1301–1308. doi: 10.1054/bjoc.2000.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pinto D, Vasconcelos A, Costa S, Pereira D, Rodrigues H, Lopes C, et al. HER2 polymorphism and breast cancer risk in Portugal. Eur J Cancer Prev. 2004;13:177–181. doi: 10.1097/01.cej.0000130015.91525.c7. [DOI] [PubMed] [Google Scholar]
- 6.Olsen A, Autrup H, Sorensen M, Overvad K, Tjonneland A. Polymorphisms of glutathione S-transferase A1 and O1 and breast cancer among postmenopausal Danish women. Eur J Cancer Prev. 2008;17:225–229. doi: 10.1097/CEJ.0b013e3282b6fe1e. [DOI] [PubMed] [Google Scholar]
- 7.Wu MH, Chou YC, Yu CP, Yang T, You SL, Chen CJ, et al. Androgen receptor gene CAG repeats, estrogen exposure status, and breast cancer susceptibility. Eur J Cancer Prev. 2008;17:317–322. doi: 10.1097/CEJ.0b013e3282f75e7f. [DOI] [PubMed] [Google Scholar]
- 8.Thompson D, Easton D. The genetic epidemiology of breast cancer genes. J Mammary Gland Biol Neoplasia. 2004;9:221–236. doi: 10.1023/B:JOMG.0000048770.90334.3b. [DOI] [PubMed] [Google Scholar]
- 9.Bourguignon LY, Zhu D, Zhu H. CD44 isoform-cytoskeleton interaction in oncogenic signaling and tumor progression. Front Biosci. 1998;3:d637–d649. doi: 10.2741/a308. [DOI] [PubMed] [Google Scholar]
- 10.Nebe B, Peters A, Duske K, Richter DU, Briese V. Influence of phytoestrogens on the proliferation and expression of adhesion receptors in human mammary epithelial cells in vitro. Eur J Cancer Prev. 2006;15:405–415. doi: 10.1097/00008469-200610000-00005. [DOI] [PubMed] [Google Scholar]
- 11.Marhaba R, Zoller M. CD44 in cancer progression: adhesion, migration and growth regulation. J Mol Histol. 2004;35:211–231. doi: 10.1023/b:hijo.0000032354.94213.69. [DOI] [PubMed] [Google Scholar]
- 12.Hill A, McFarlane S, Johnston PG, Waugh DJ. The emerging role of CD44 in regulating skeletal micrometastasis. Cancer Lett. 2006;237:1–9. doi: 10.1016/j.canlet.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 13.Rafi-Janajreh AQ, Nagarkatti PS, Nagarkatti M. Role of CD44 in CTL and NK cell activity. Front Biosci. 1998;3:d665–d671. doi: 10.2741/a311. [DOI] [PubMed] [Google Scholar]
- 14.Gotte M, Yip GW. Heparanase, hyaluronan, and CD44 in cancers: a breast carcinoma perspective. Cancer Res. 2006;66:10233–10237. doi: 10.1158/0008-5472.CAN-06-1464. [DOI] [PubMed] [Google Scholar]
- 15.Zhou J, Nagarkatti PS, Zhong Y, Creek K, Zhang J, Nagarkatti M. Unique SNP in CD44 intron 1 and its role in breast cancer development. Anticancer Res. 2010;30:1263–1272. [PMC free article] [PubMed] [Google Scholar]
- 16.Telen MJ, Udani M, Washington MK, Levesque MC, Lloyd E, Rao N. A blood group-related polymorphism of CD44 abolishes a hyaluronan-binding consensus sequence without preventing hyaluronan binding. J Biol Chem. 1996;271:7147–7153. doi: 10.1074/jbc.271.12.7147. [DOI] [PubMed] [Google Scholar]
- 17.Goodison S, Urquidi V, Tarin D. CD44 cell adhesion molecules. Mol Pathol. 1999;52:189–196. doi: 10.1136/mp.52.4.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peach RJ, Hollenbaugh D, Stamenkovic I, Aruffo A. Identification of hyaluronic acid binding sites in the extracellular domain of CD44. J Cell Biol. 1993;122:257–264. doi: 10.1083/jcb.122.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.So JY, Lee HJ, Smolarek AK, Paul S, Wang CX, Maehr H, et al. A Novel Gemini Vitamin D Analog Represses the Expression of a Stem Cell Marker CD44 in Breast Cancer. Mol Pharmacol. 2011;79:360–367. doi: 10.1124/mol.110.068403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winder T, Ning Y, Yang D, Zhang W, Power DG, Bohanes P, et al. Germline polymorphisms in genes involved in the CD44 signaling pathway are associated with clinical outcome in localized gastric adenocarcinoma (GA) Int J Cancer. 2010 doi: 10.1002/ijc.25787. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vazquez A, Grochola LF, Bond EE, Levine AJ, Taubert H, Muller TH, et al. Chemosensitivity profiles identify polymorphisms in the p53 network genes 14-3-3tau and CD44 that affect sarcoma incidence and survival. Cancer Res. 2010;70:172–180. doi: 10.1158/0008-5472.CAN-09-2218. [DOI] [PubMed] [Google Scholar]
- 22.Gail MH, Brinton LA, Byar DP, Corle DK, Green SB, Schairer C, et al. Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. J Natl Cancer Inst. 1989;81:1879–1886. doi: 10.1093/jnci/81.24.1879. [DOI] [PubMed] [Google Scholar]
- 23.Claus EB, Risch N, Thompson WD. Genetic analysis of breast cancer in the cancer and steroid hormone study. Am J Hum Genet. 1991;48:232–242. [PMC free article] [PubMed] [Google Scholar]
- 24.Ready K, Litton JK, Arun BK. Clinical application of breast cancer risk assessment models. Future Oncol. 2010;6:355–365. doi: 10.2217/fon.10.5. [DOI] [PubMed] [Google Scholar]
- 25.Amir E, Freedman OC, Seruga B, Evans DG. Assessing women at high risk of breast cancer: a review of risk assessment models. J Natl Cancer Inst. 2010;102:680–691. doi: 10.1093/jnci/djq088. [DOI] [PubMed] [Google Scholar]
- 26.Parl FF, Egan KM, Li C, Crooke PS. Estrogen exposure, metabolism, and enzyme variants in a model for breast cancer risk prediction. Cancer Inform. 2009;7:109–121. doi: 10.4137/cin.s2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parl FF, Dawling S, Roodi N, Crooke PS. Estrogen metabolism and breast cancer: a risk model. Ann N Y Acad Sci. 2009;1155:68–75. doi: 10.1111/j.1749-6632.2008.03676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fasching PA, Bani MR, Nestle-Kramling C, Goecke TO, Niederacher D, Beckmann MW, et al. Evaluation of mathematical models for breast cancer risk assessment in routine clinical use. Eur J Cancer Prev. 2007;16:216–224. doi: 10.1097/CEJ.0b013e32801023b3. [DOI] [PubMed] [Google Scholar]
- 29.Aruffo A. CD44: one ligand, two functions. J Clin Invest. 1996;98:2191–2192. doi: 10.1172/JCI119026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaneko T, Saito H, Toya M, Satio T, Nakahara K, Hiroi M. Hyaluronic acid inhibits apoptosis in granulosa cells via CD44. J Assist Reprod Genet. 2000;17:162–167. doi: 10.1023/A:1009470206468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lesley J, Hyman R, English N, Catterall JB, Turner GA. CD44 in inflammation and metastasis. Glycoconj J. 1997;14:611–622. doi: 10.1023/a:1018540610858. [DOI] [PubMed] [Google Scholar]
- 32.Thomas GJ, Speight PM. Cell adhesion molecules and oral cancer. Crit Rev Oral Biol Med. 2001;12:479–498. doi: 10.1177/10454411010120060301. [DOI] [PubMed] [Google Scholar]