Abstract
Purpose of review
In recent years, the emerging role of p53 in metabolic regulation has been a topic of great interest. While apoptotic and growth arrest functions of p53 remain as important mechanisms for preserving genomic stability, metabolic functions of p53 show increasing potential in contributing to p53-mediated tumor suppression. Numerous studies in the past year provided further insights on the metabolic functions of p53 and their implications in tumorigenesis. These findings illuminate the significance of p53 metabolic regulation in tumor biology.
Recent findings
Several novel p53 metabolic targets have been identified that are involved in various aspects of metabolism. Furthermore, while some studies demonstrate the potential tumor suppressive function of p53 metabolic genes, others reveal the pro-survival role of those targets in both tumor and normal cells. Specifically, TIGAR has been thought to promote tumor suppression through metabolic fine-tuning, yet TIGAR-deficient mice, in fact, display reduction in tumorigenesis. Finally, characterization of the 3KR mouse model underscored the significance of p53 metabolic regulation in tumor suppression, while also alluding to the potential mechanism for selective regulation of p53 metabolic targets.
Summary
Recent evidence highlighted the ever-growing complexity of p53 metabolic functions. Expression of many p53 metabolic genes elicits both anti-tumor and tumorigenic effects, suggesting that p53 may in fact contribute to cellular protection as well as tumor suppression. Future studies must carefully dissect the duality of p53 metabolic function, and greater understanding of how metabolic targets are regulated by p53 may prove useful in designing cancer therapies.
Keywords: p53, metabolism, ROS, tumorigenesis
Introduction
p53 is one of the most important tumor suppressors that prevents malignant transformation in mammalian cells [1–4]. Classically, p53 functions as a transcription factor that regulates downstream targets responsible for cell fate control. In response to cellular stresses such as hypoxia, genotoxic and oncogenic stress, p53 activity and level is increased to induce growth arrest or apoptosis [5,6]. Under mild stress, p53 induces cell cycle arrest to allow cells to repair damage or recover from stress before resuming normal replication. Alternatively, upon severe stress that causes irreparable damage, p53 activates a number of pro-apoptotic genes to terminate the cell, and thus, preventing potential oncogenic mutations to prevail [3]. Not until recent discoveries that link p53 to cellular metabolism, tumor suppressing functions of p53 have long been attributed to its ability to regulate cell cycle through apoptosis, growth arrest, and senescence [7,8]. However, characterizations of mice deficient in p21 and Puma, key p53 targets in growth arrest and apoptosis, respectively, showed no increase tendency towards tumor development in these mice, suggesting that other non-canonical p53 functions might be at play in tumor suppression [9–11]. As tumorigenesis is increasingly viewed as a process that involves complex metabolic perturbations, the role of p53 in suppressing tumor formation is also increasingly seen as a metabolic one.
Cancer cells acquire numerous metabolic alterations to enable rapid growth and proliferation, including aerobic glycolysis (the Warburg effect) and enhanced biosynthesis of macromolecules [12,13]. Aerobic glycolysis allows cancer cells to rapidly generate ATP at the cost of efficiency while providing intermediates for de novo synthesis and oxidative balance. Processes such as aerobic glycolysis and glutaminolysis promote biosynthesis of protein, lipids and nucleic acids, which promote cancer cells to grow and proliferate more rapidly, and in some cases, confer survival advantages of tumor cells [14,15]. Reversing the Warburg effect compromises tumorigenicity and survival of cancer cells, while inhibiting glutaminolysis hinders oncogenic transformation of normal cells [16–18].
Interestingly, many p53 metabolic targets counteract the effects of the metabolic alterations observed in tumor cells, which include inhibition of aerobic glycolysis, upregulation of mitochondrial OXPHOS, and promotion of fatty acid oxidation [7]. Additionally, several of these targets aim to increase antioxidant production and to lower intracellular ROS, a protective mechanism to thwart ROS-induced DNA damage and subsequent malignant transformation [19,20]. Most recently, a study done by Li et al. characterized mice carrying mutant p53 that contain lysine-to-arginine mutations at three p53 acetylation sites (p533KR/3KR; K117R + K161R + K162R) [21**]. The 3KR mutant p53, although impaired in its ability to induce apoptosis or growth arrest, retains the ability to prevent spontaneous tumor formation in p533KR/3KR mice. Surprisingly, regulation of many p53-dependent metabolic targets remains intact in p533KR/3KR mice, which sheds light upon the significance of p53 metabolic functions on tumor suppression [21**].
In this review, we will highlight many of the novel metabolic regulation by p53 and discuss how metabolic and ROS regulations by p53 are implicated in tumor biology. We will also address current clues and evidence on how p53 might differentially regulate metabolic genes among all of its downstream targets.
p53 metabolic regulation
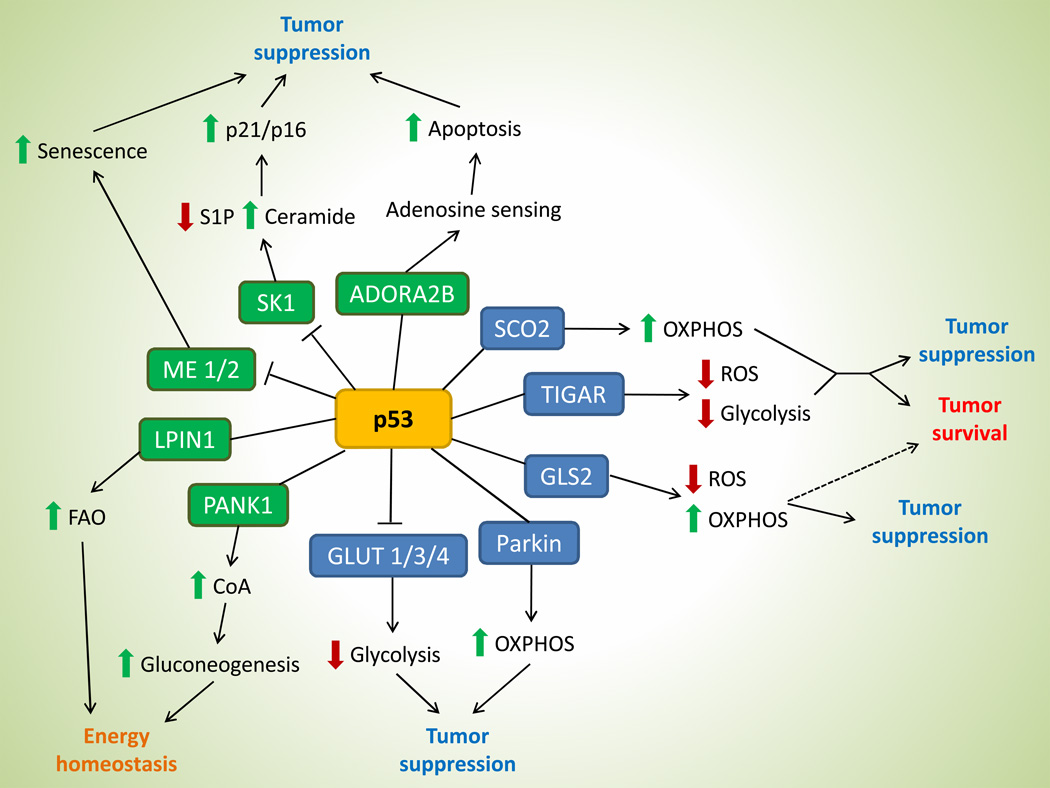
The Warburg Effect, a phenomenon of high glycolytic rate in the presence of oxygen, has long been characterized as the quintessential metabolic perturbation observed in tumor cells [12]. Emerging evidence, however, suggest that metabolic alterations in cancer cells go beyond glucose metabolism and now also include amino acid and lipid metabolism, nucleotide biosynthesis, and antioxidant response [13,15]. Surprisingly, p53 regulates all of these metabolic pathways through its downstream targets (Figure 1).
Figure 1. p53 exerts a diverse array of metabolic functions.
A broad spectrum of metabolic targets are regulated by p53, leading to various functional outcomes in tumor and normal physiology. Genes in blue boxes are previously-identified p53 targets, while those in green boxes are most recently discovered. Dashed line in the GLS2 axis illustrates the potential of this target in enhancing tumor survival based on its function. Abbr. OXPHOS, oxidative phosphorylation; S1P, sphingosine-1-phosphate; FAO, fatty acid oxidation; CoA, coenzyme A.
As a tumor suppressor, p53 functions to bolster mitochondrial oxidative phosphorylation (through activating targets such as SCO2/GLS2/Parkin) and inhibits aerobic glycolysis (via expression of TIGAR and repression of GLUT1/3/4), which antagonizes the predominant utilization of glycolysis in cancer cells [22–28]. Sco2 regulates the cytochrome C oxidase complex of the electron transport chain and increases mitochondrial respiration [22], while Gls2 converts glutamine to glutamate, which can either enter the TCA cycle or be used as a substrate for glutathione synthesis [23,24]. Expression of Parkin also promotes mitochondrial oxidative phosphorylation by increasing the cellular level of acetyl-CoA through the upregulation of pyruvate dehydrogenase E1α1 [25]. On the other hand, TIGAR inhibits glycolysis and shunts glycolytic intermediates into the pentose phosphate pathway for NADPH production and glutathione recycling [26], while repression of glucose transporters GLUT1/3/4 prevents the uptake of glucose [27,28].
More recently, p53 has been implicated in lipid metabolism as well. Indeed, many tumor cells also exhibit high rates of de novo lipid synthesis, and several p53 metabolic targets may possibly counteract such phenomenon via promoting fatty acid oxidation. Jiang et al. identified two repression targets of p53, malic enzyme isoforms 1 and 2 (ME1 and ME2) [29*]. ME1 and ME2 catalyze the oxidative decarboxylation of malate in the TCA cycle to pyruvate and NADH/NADPH. Increase in lipid production was observed in p53-deficient cells due to abundance of NADPH, while silencing of malic enzymes decreases glutamine consumption and glutaminolysis. Interestingly, reduction of ME1/2 expressions can reciprocally activate p53, which led to increased cellular senescence and decreased tumorigenicity of cancer cells, likely through metabolic perturbations that are unfavorable for cancerous growth [29*].
In another study, Heffernan-Stroud et al. reported that p53 negatively regulate sphingosine kinase 1 (SK1) via a proteolytic pathway [30*]. SK1 is a key enzyme in sphingolipid metabolism that maintains the homeostatic balance of ceramide and sphingosine. Specifically, p53-null mice exhibit increase in SK1 levels that leads to an increase in the pro-growth sphingolipid sphingosine-1-phosphate and a decrease in the anti-growth sphingolipid ceramide [30*]. Interestingly, loss of SK1 promotes tumor cell senescence in the thymus of p53-null mice through the elevation of p21 and p16 expressions. Ablation of SK1 reduces tumor formation in p53-null and p53 heterozygous mice, indicating a novel mechanism of p53 tumor suppressing function through sphingolipid regulation [30*].
Aside from its function in tumor suppression, p53 also regulate metabolic targets that serve to maintain homeostasis in normal cells and tissues. A recently identified p53 target, Lipin1 is a nuclear transcriptional co-activator that regulates the expression of genes involved in fatty acid oxidation through peroxisome proliferator-activated receptor alpha (PPAR α) [31]. Under conditions of low glucose, cells upregulate Lipin1 expression through ROS-induced p53 activity and increase fatty acid oxidation to utilize fatty acid as an alternative source of energy [31]. Study by Wang et al. revealed yet another metabolic gene, pantothenate kinase 1 (PANK1), as a p53 target [32*]. PanK1 is an enzyme that catalyzes the rate-limiting step of coenzyme A (CoA) synthesis. Although induced by DNA damage in a p53-dependent manner, PanK1 does not play a role in p53-mediated apoptosis or cell growth arrest. Instead, PanK1 expression is maintained under glucose starvation in the presence of p53 [32*]. Similar to Pank1-null mice, p53-null mice exhibit impairment in gluconeogenesis after starvation compared to wild-type mice, presumably due to lowered CoA levels [32*,33]. Together, Lipin1 and PanK1 link p53 function to maintaining energy and lipid homeostasis in cells and tissues under normal physiological conditions.
In summary, p53 metabolic targets are involved in increasing mitochondrial respiration, decreasing glycolysis, promoting antioxidant defense and regulating lipid metabolism, all of which could contribute to p53-mediated tumor suppression. Moreover, metabolic functions of p53 further extend outside of tumor suppression to regulate cellular homeostasis in normal physiology, underscoring the multitude of p53 regulation.
Functions of p53 metabolic targets in cancer
Many p53 metabolic targets have functions that counteract the metabolic alterations commonly seen in cancer cells, as previously described. Indeed, loss of GLS2 expression correlates with neoplastic transformation in human hepatocellular carcinomas [23,24], while low SCO2 expression is a factor of poor prognosis in patients with breast cancer [34]. The rationale behind this is two-fold: 1) cancer cells display a heightened state of aerobic glycolysis that confers several growth advantages over normal cells, and p53 targets such as SCO2, TIGAR and GLS2 may contribute to p53-mediated tumor suppression by preventing such metabolic transformation; and 2) upregulation of antioxidant production by p53 metabolic targets prevents the increase in oxidative stress associated with malignant transformation. This notion is also supported by the observation that the 3KR mice retain metabolic regulation by p53 and are not prone to spontaneous tumor formation [21**].
However, while aerobic glycolysis promotes rapid growth in tumors, it is not without pitfalls. The inefficiency of glycolysis for energy generation could become perilous for cancer cells that have outgrown the resource capacity of their tumor environments. Similarly, although cancer cells generally have increased levels of ROS, which perpetuates cell proliferation and are involved in tumor initiation and progression, excessive accumulation of ROS is also toxic to cancer cells. Evidence shows that cancer cells require robust antioxidant capacity to offset the intrinsic oxidative stress for survival [35–38]. Given that p53 metabolic targets have the capability of modulating energy homeostasis and decreasing intracellular ROS levels, they could potentially support cancerous growth under certain circumstances. Nevertheless, precisely how these targets affect tumorigenicity or survival of cancer cells remains elusive. Recent studies have attempted to further our understanding of the function of p53 metabolic targets in the context of tumor biology, but the findings only broadened the complexity of tumor metabolism. Current evidence demonstrates that certain cancers gain survival advantages by retaining wild-type p53 and the subsequent activation of p53 metabolic targets. Wanka et al. showed that glioma and colon cancer cells that express wild-type p53 are less sensitive towards hypoxia-induced cell death [39*]. These cancer cells were able to maintain mitochondrial respiration under hypoxic conditions through the activation of SCO2, while cells depleted of p53 or SCO2 undergo necrotic cell death in the presence of low oxygen [39*]. TIGAR is also reported to have tumor-protective functions as well. Cheung et al. reported that TIGAR plays a role in enhancing intestinal adenoma proliferation as well as regeneration of intestinal epithelium [40**]. Through several rescue-experiments, the authors concluded that Tigar expression prevents ROS accumulation through increasing GSH:GSSG ratio and increases nucleotide synthesis that allow intestinal tumor cell to grow more efficiently. Indeed, tumor burden is decreased in TIGAR knock-out mice compared to TIGAR wild-type mice using an intestinal adenoma mouse model [40**]. Similarly, another study by Wanka et al. revealed that Tigar expression protects glioma cells from glucose starvation and hypoxia-induced cell death, which coincides with TIGAR been overexpressed in glioblastomas [41*]. Together, these studies provide evidence that p53 may protect tumor cells from their harsh microenvironment or oxidative stress through the regulation of metabolic targets, which are also suggested by previous studies [42,43].
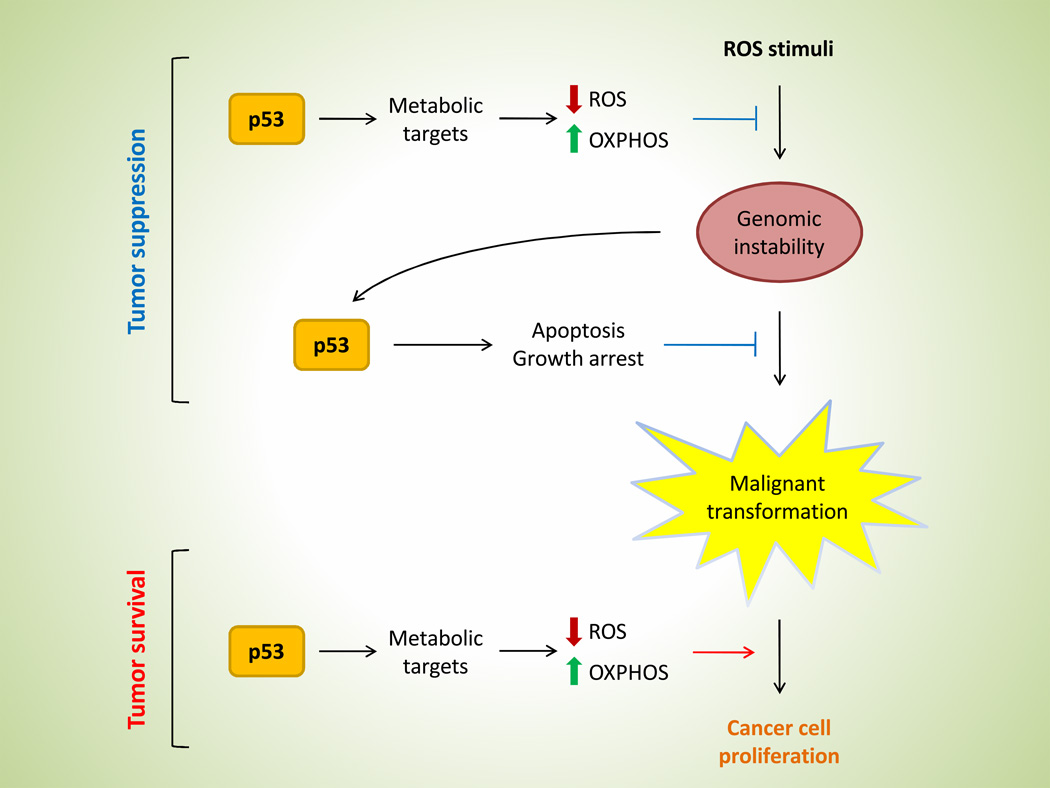
These findings present a rather puzzling question – does p53 suppress tumor formation or does it enhance tumor cell survival? The answer, it seems, is that p53 may act as a double-edged sword (Figure 2). While metabolic functions of p53 do indeed contribute to the overall tumor suppressing activity of p53, they also serve to protect cells, normal or tumor alike, from metabolic stress. By lowering ROS levels and maintaining energy homeostasis, p53 metabolic targets may avert crises that would otherwise result in cell death. For example, while normal healthy cells must maintain oxidative balance for survival, cancer cells have an even lower threshold for oxidative insults [20]. In order for tumor cells to thrive, they must upregulate pathways for antioxidant production, many of which are controlled by p53 targets. Moreover, tumor cells often suffer from hypoxic and nutrient stress due to rapidly growing cells competing for finite amount of resources, especially for cells residing in the center of the tumor bulk where blood supply is limited. Under such conditions, tumor cells will benefit from a shift from aerobic glycolysis to the higher efficient oxidative phosphorylation for energy production, which is also regulated by p53 activity. If indeed p53 function manifests such duality, then when in the evolution of normal to tumor cells does p53 seizes to exist as a tumor suppressor and don the role of enhancing tumorigenicity? Further studies are warranted in order to dissect the intricate balance of cell death and survival in p53 function on tumorigenesis.
Figure 2. Opposing functions of p53 metabolic regulation.
Metabolic regulation by p53 in tumorigenesis is a double-edged sword: maintaining oxidative balance and energy homeostasis could prevent cancer development in the pre-cancerous stage, but favors tumor survival after malignant transformation.
p53 response to metabolic stress
In order for p53 to activate its downstream targets appropriately in a given context, p53 must be able to detect exogenous and endogenous stress signals. Depending on the severity and the type of stress, p53 can either trigger programmed cell death or initiate adaptive responses to ensure survival [3].
Previous studies have shown that p53 is induced under energy and nutrient deprivation by energy-sensing AMP-activated protein kinase (AMPK) through p53 phosphorylation, which triggers downstream activation of growth arrest pathways [44,45]. Most recently, a number of studies revealed various forms of metabolic stress that can activate p53 via different mechanisms. Lee et al. reported that p53 regulates the decision-making between cell death and survival under starvation via interaction with Atg7, a regulator of autophagy [46*].
Presence of Atg7 is required for p53-mediated p21 induction upon metabolic stress. In contrast, the absence of Atg7 led to increasing levels of mitochondrial ROS and DNA damage, resulting in the preferential expression of apoptotic genes by p53 [46*]. Maddocks et al. also demonstrated the pro-survival function of p53 during metabolic stress. The study showed that p53 promotes cell survival during serine starvation in both tissue culture and xenografts [47*]. Serine is required for purine-nucleotide and glutathione synthesis. During serine starvation, PKM2 is inhibited to divert glycolytic intermediates into serine synthesis pathway for de novo serine synthesis. In p53 +/+ cells, serine depletion induced p21 expression, which led to growth arrest and the preferential synthesis of glutathione over purine-nucleotides. However, in p53 −/− cells, glutathione synthesis is significantly lowered in the absence of p21 induction, leading to elevated ROS and subsequent cell death [47*].
While oftentimes metabolic sensing by p53 trigger adaptive responses, Long et al. described a novel p53 target, ADORA2B (A2B), which senses the level of extracellular adenosine and primes the cell for apoptosis upon adenosine accumulation [48*]. Extracellular adenosine is elevated upon metabolic stress such as nutrient deprivation and oxygen depletion; such conditions are seen in disease pathology (ie. ischemia) as well as in tumor growth [49,50]. The authors showed that A2B can induce cell death upon overexpression or by adenosine-sensing via down-regulation of anti-apoptotic Bcl-2 and Bcl-XL, a novel mechanism that contributes to p53-mediated tumor suppression [48*].
Cumulatively, the findings in these studies implicate p53 as a downstream effector of various metabolic stresses. As a master-regulator that controls the flux of many metabolic pathways, it is conceivable that p53 can detect and respond to metabolic stress in order to 1) meet metabolic demands accordingly to ensure cell survival, or 2) terminate the cell if circumstances are deemed irreparable. These functions of p53 underscore both the pro-survival and prodeath components of the p53 pathway, which are evident in the context of both tumor and normal physiology.
Differential regulation of p53 metabolic functions
With the ever-expanding number of p53 targets involved in a diverse array of cellular processes, it is crucial for p53 to selectively activate its downstream targets to achieve the desired outcome depending on the cellular context. Currently, the mechanism underlying the preferential expression of p53 targets remains largely unknown. Previous evidence, however, points towards the possibility that the acetylation of specific lysine residues of p53 can differentially regulate the expression of pro-apoptotic and growth arrest genes. Acetylation of p53 at the K120 lysine residue by acetyltransferases Tip60/hMOF is crucial for p53-mediated apoptosis but not for cell cycle arrest [51,52]. On the other hand, p53 acetylation at K164 is acetylated by CBP/p300, and loss of acetylation at both K120 and K164 abolishes the ability of p53 to induce both apoptosis and growth arrest [53]. This notion is also supported by the 3KR mouse model. While p53-mediated apoptotic and growth arrest gene expressions are abrogated in the 3KR mice, expression of Mdm2 and many of the metabolic targets are still retained [21**]. These findings suggest that additional post-translational modifications of p53 may be required for the preferential expression of p53 metabolic genes.
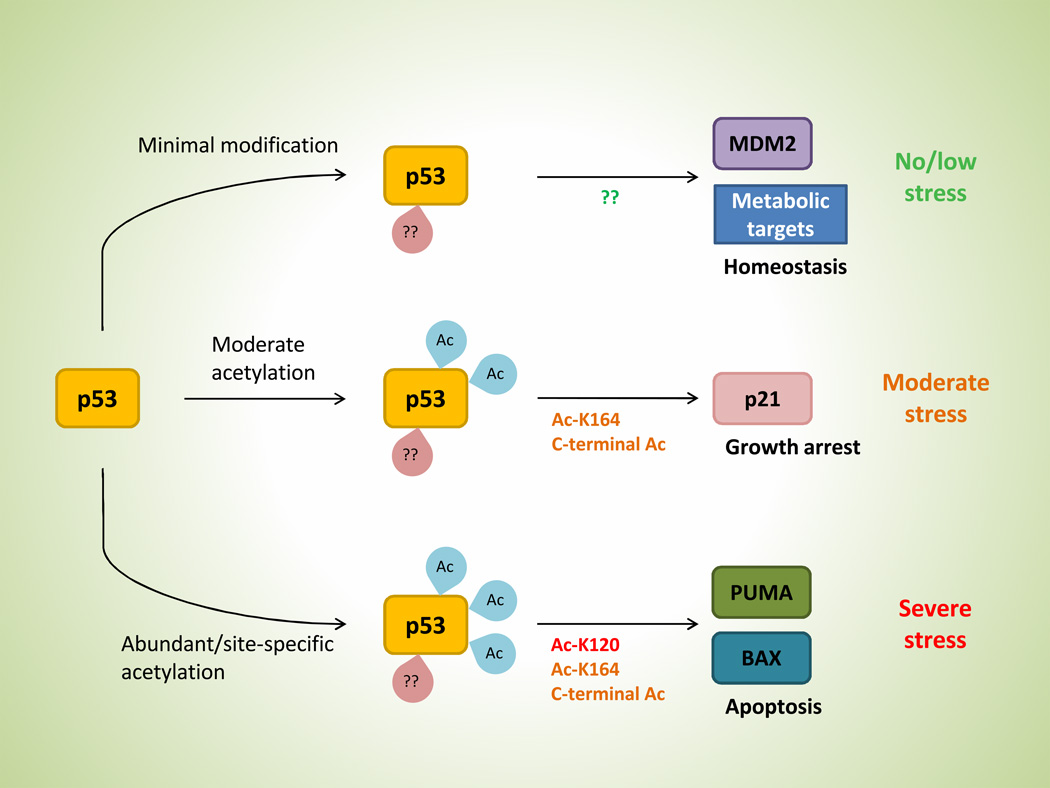
Given our current knowledge, we hypothesize that the regulation of downstream targets by p53 may be hierarchical in nature (Figure 3). Depending on the severity and the type of stress in question, specific sites and number of p53 modifications may vary in which to dictate the appropriate p53 response. Under no or low stress conditions, minimal post-translational modifications of p53 may be sufficient to activate targets such as Mdm2 and various metabolic genes. In the absence of stress, expression of Mdm2 is required to negatively regulate p53 activity in order to avoid p53-induced cell death. Meanwhile, under low stress conditions such as starvation and hypoxia, activation of metabolic targets could enhance cell adaptation and survival. Upon facing moderate stress, however, p53 acetylation increases at K164 and C-terminal lysine residues that lead to growth arrest via p21 induction, which allow cells to recover and repair cellular damage. Ultimately, if the damage from stress becomes irreparable, acetylation at the K120 site would trigger an apoptotic response to circumvent the perpetuation of possible genetic lesions.
Figure 3. A hypothetical model of differential regulation by p53.
p53 is subjected to a hierarchical sequence of post-translational modifications as stress level increases, which induces various functional outcome depending on the combination of modifications (see text for complete description).
The rationale underlying our hypothesis is that p53 metabolic targets, unlike its canonical counterparts involved in apoptosis and growth arrest, are relatively harmless in nature and serve to improve cellular homeostasis, and therefore, are subjected to a hierarchical regulation. This would allow the selective expression of metabolic genes and Mdm2 in the presence of basal p53 expression under normal growth without activating other p53 functions. Since mounting evidence suggest that metabolic regulation by p53 is critical in tumor suppression and/or cell survival and homeostasis, we believe that elucidating the minimal modifications for activating p53 metabolic targets could be the key to understanding the mechanism of how p53 participate in cellular processes such as tumorigenesis and adaptation.
Conclusion
Tumor suppressing abilities of p53 were long been attributed to its canonical regulation on cell fate, but the growing list of p53 metabolic targets shed light on additional mechanisms. These downstream metabolic targets not only contribute to p53-mediated tumor suppression, but also enhance pro-survival adaptations in normal and tumor cells, which further complicated the grand picture of p53 involvement in carcinogenesis. Yet another complexity in p53 biology is the question of how p53 can selectively activate targets from a broad range of its downstream candidates in a context-appropriate manner. Through further understanding of how p53 regulates its metabolic targets, the bipolar attribute in p53 function would potentially allow targeted therapies in both p53 +/+ and p53 −/− tumors.
Key points.
p53-mediated tumor suppression involves several metabolic pathways
Recent findings implicate p53 metabolic targets in both tumor suppression and tumor survival
p53 metabolic genes may be differentially regulated via post-translational modifications, and elucidating the mechanism of such regulation is potentially critical for therapeutic purposes
Acknowledgement
This work was supported by the National Cancer Institute of the National Institutes of Health under Award 2P01 CA080058-12 and 5RO1 CA172023 to W. Gu. S.-J. Wang is supported by NIH cancer biology training grant T32-CA09503. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 3.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 4.Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 6.Vousden KH, Lu X. Live or let die: the cell's response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 7.Maddocks OD, Vousden KH. Metabolic regulation by p53. J Mol Med (Berl) 2011;89:237–245. doi: 10.1007/s00109-011-0735-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zilfou JT, Lowe SW. Tumor suppressive functions of p53. Cold Spring Harb Perspect Biol. 2009;1:a001883. doi: 10.1101/cshperspect.a001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michalak EM, Villunger A, Adams JM, Strasser A. In several cell types tumour suppressor p53 induces apoptosis largely via Puma but Noxa can contribute. Cell Death Differ. 2008;15:1019–1029. doi: 10.1038/cdd.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choudhury AR, Ju Z, Djojosubroto MW, et al. Cdkn1a deletion improves stem cell function and lifespan of mice with dysfunctional telomeres without accelerating cancer formation. Nat Genet. 2007;39:99–105. doi: 10.1038/ng1937. [DOI] [PubMed] [Google Scholar]
- 11.Valente LJ, Gray DH, Michalak EM, et al. p53 efficiently suppresses tumor development in the complete absence of its cell-cycle inhibitory and proapoptotic effectors p21, Puma, and Noxa. Cell Rep. 2013;3:1339–1345. doi: 10.1016/j.celrep.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 12.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 13.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 14.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 16.Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 17.Christofk HR, Vander Heiden MG, Harris MH, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 18.Wang JB, Erickson JW, Fuji R, et al. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell. 2010;18:207–219. doi: 10.1016/j.ccr.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maillet A, Pervaiz S. Redox regulation of p53, redox effectors regulated by p53: a subtle balance. Antioxid Redox Signal. 2012;16:1285–1294. doi: 10.1089/ars.2011.4434. [DOI] [PubMed] [Google Scholar]
- 20.Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov. 2009;8:579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 21. Li T, Kon N, Jiang L, et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012;149:1269–1283. doi: 10.1016/j.cell.2012.04.026. This study demonstrates that p53 is capable of conferring tumor suppression in the absence of its canonical regulation on cell fate. Moreover, the fact that the 3KR mutant retains metabolic regulation underscores the importance of metabolic targets in preventing malignant transformation.
- 22.Matoba S, Kang JG, Patino WD, et al. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki S, Tanaka T, Poyurovsky MV, et al. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci U S A. 2010;107:7461–7466. doi: 10.1073/pnas.1002459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu W, Zhang C, Wu R, et al. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci U S A. 2010;107:7455–7460. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang C, Lin M, Wu R, et al. Parkin, a p53 target gene, mediates the role of p53 in glucose metabolism and the Warburg effect. Proc Natl Acad Sci U S A. 2011;108:16259–16264. doi: 10.1073/pnas.1113884108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bensaad K, Tsuruta A, Selak MA, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 27.Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 2004;64:2627–2633. doi: 10.1158/0008-5472.can-03-0846. [DOI] [PubMed] [Google Scholar]
- 28.Kawauchi K, Araki K, Tobiume K, Tanaka N. p53 regulates glucose metabolism through an IKK-NF-kappaB pathway and inhibits cell transformation. Nat Cell Biol. 2008;10:611–618. doi: 10.1038/ncb1724. [DOI] [PubMed] [Google Scholar]
- 29. Jiang P, Du W, Mancuso A, et al. Reciprocal regulation of p53 and malic enzymes modulates metabolism and senescence. Nature. 2013;493:689–693. doi: 10.1038/nature11776. This study presents a novel metabolic regulation by p53, and ties metabolic pathways with senescence and tumor suppression.
- 30. Heffernan-Stroud LA, Helke KL, Jenkins RW, et al. Defining a role for sphingosine kinase 1 in p53-dependent tumors. Oncogene. 2012;31:1166–1175. doi: 10.1038/onc.2011.302. This study highlights a novel form of p53 metabolic regulation through sphingolipid metabolism.
- 31.Assaily W, Rubinger DA, Wheaton K, et al. ROS-mediated p53 induction of Lpin1 regulates fatty acid oxidation in response to nutritional stress. Mol Cell. 2011;44:491–501. doi: 10.1016/j.molcel.2011.08.038. [DOI] [PubMed] [Google Scholar]
- 32. Wang SJ, Yu G, Jiang L, et al. p53-dependent regulation of metabolic function through transcriptional activation of pantothenate kinase-1 gene. Cell Cycle. 2013;12:753–761. doi: 10.4161/cc.23597. This study links p53 function to energy homeostasis via CoA synthesis.
- 33.Leonardi R, Rehg JE, Rock CO, Jackowski S. Pantothenate kinase 1 is required to support the metabolic transition from the fed to the fasted state. PLoS One. 2010;5:e11107. doi: 10.1371/journal.pone.0011107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Won KY, Lim SJ, Kim GY, et al. Regulatory role of p53 in cancer metabolism via SCO2 and TIGAR in human breast cancer. Hum Pathol. 2012;43:221–228. doi: 10.1016/j.humpath.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 35.Benassi B, Fanciulli M, Fiorentino F, et al. c-Myc phosphorylation is required for cellular response to oxidative stress. Mol Cell. 2006;21:509–519. doi: 10.1016/j.molcel.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 36.DeNicola GM, Karreth FA, Humpton TJ, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trachootham D, Zhou Y, Zhang H, et al. Selective killing of oncogenically transformed cells through a ROS-mediated mechanism by beta-phenylethyl isothiocyanate. Cancer Cell. 2006;10:241–252. doi: 10.1016/j.ccr.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 38.Young TW, Mei FC, Yang G, et al. Activation of antioxidant pathways in ras-mediated oncogenic transformation of human surface ovarian epithelial cells revealed by functional proteomics and mass spectrometry. Cancer Res. 2004;64:4577–4584. doi: 10.1158/0008-5472.CAN-04-0222. [DOI] [PubMed] [Google Scholar]
- 39. Wanka C, Brucker DP, Bahr O, et al. Synthesis of cytochrome C oxidase 2: a p53-dependent metabolic regulator that promotes respiratory function and protects glioma and colon cancer cells from hypoxia-induced cell death. Oncogene. 2012;31:3764–3776. doi: 10.1038/onc.2011.530. This study shed light on the importance of p53-mediated increase of mitochondrial respiration in cancer cell survival.
- 40. Cheung EC, Athineos D, Lee P, et al. TIGAR is required for efficient intestinal regeneration and tumorigenesis. Dev Cell. 2013;25:463–477. doi: 10.1016/j.devcel.2013.05.001. TIGAR has been thought possess tumor suppressive function by inhibiting glycolysis and lowering intracellular ROS. This study, however, showed that TIGAR expression actually promotes tumorigenicity and tumor survival, suggesting the possibility of p53 having both positive and negative effect on tumorigenesis.
- 41. Wanka C, Steinbach JP, Rieger J. Tp53-induced glycolysis and apoptosis regulator (TIGAR) protects glioma cells from starvation-induced cell death by up-regulating respiration and improving cellular redox homeostasis. J Biol Chem. 2012;287:33436–33446. doi: 10.1074/jbc.M112.384578. This study demonstrates another case of TIGAR manifesting protective role in cancer cells under stress.
- 42.Sinthupibulyakit C, Ittarat W, St Clair WH, St Clair DK. p53 Protects lung cancer cells against metabolic stress. Int J Oncol. 2010;37:1575–1581. doi: 10.3892/ijo_00000811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sung HJ, Ma W, Wang PY, et al. Mitochondrial respiration protects against oxygen-associated DNA damage. Nat Commun. 2010;1:5. doi: 10.1038/ncomms1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones RG, Plas DR, Kubek S, et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 45.Okoshi R, Ozaki T, Yamamoto H, et al. Activation of AMP-activated protein kinase induces p53-dependent apoptotic cell death in response to energetic stress. J Biol Chem. 2008;283:3979–3987. doi: 10.1074/jbc.M705232200. [DOI] [PubMed] [Google Scholar]
- 46. Lee IH, Kawai Y, Fergusson MM, et al. Atg7 modulates p53 activity to regulate cell cycle and survival during metabolic stress. Science. 2012;336:225–228. doi: 10.1126/science.1218395. This study reveals a novel regulation on p53 by autophagy-associated Atg7 in the presence of metabolic stress, highlighting potential mechanism underlying p53-mediated cell fate decision.
- 47. Maddocks OD, Berkers CR, Mason SM, et al. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2013;493:542–546. doi: 10.1038/nature11743. This article characterizes p53 response under serine starvation, emphasizing the importance of p53 in tumor survival under stress.
- 48. Long JS, Crighton D, O'Prey J, et al. Extracellular adenosine sensing-a metabolic cell death priming mechanism downstream of p53. Mol Cell. 2013;50:394–406. doi: 10.1016/j.molcel.2013.03.016. This study presents a novel p53 target and a first example that mediates extracellular sensing and signaling that leads to cell death, which is a unique mechanism of p53-mediated tumor suppression.
- 49.Blay J, White TD, Hoskin DW. The extracellular fluid of solid carcinomas contains immunosuppressive concentrations of adenosine. Cancer Res. 1997;57:2602–2605. [PubMed] [Google Scholar]
- 50.Latini S, Bordoni F, Pedata F, Corradetti R. Extracellular adenosine concentrations during in vitro ischaemia in rat hippocampal slices. Br J Pharmacol. 1999;127:729–739. doi: 10.1038/sj.bjp.0702591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang Y, Luo J, Zhang W, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell. 2006;24:827–839. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 52.Sykes SM, Mellert HS, Holbert MA, et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang Y, Zhao W, Chen Y, et al. Acetylation is indispensable for p53 activation. Cell. 2008;133:612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]