Abstract
Hepatocyte nuclear factor-4 (HNF4), a member of the nuclear receptor superfamily, plays an important role in tissue-specific gene expression, including genes involved in hepatic glucose metabolism. In this study, we show that SRC-1 and GRIP1, which act as coactivators for various nuclear receptors, associate with HNF4 in vivo and enhance its transactivation potential. The AF-2 domain of HNF4 is required for this interaction and for the potentiation of transcriptional activity by these co-activators. p300 can also serve as a coactivator with HNF4, and it synergizes with SRC-1 to further augment the activity of HNF4. HNF4 is also a key regulator of the expression of hepatocyte nuclear factor-1 (HNF1). The overexpression of SRC-1 or GRIP1 enhances expression from a HNF1 gene promoter-reporter in HepG2 hepatoma cells, and this requires an intact HNF4-binding site in the HNF1 gene promoter. Type 1 maturity onset diabetes of young (MODY), which is characterized by abnormal glucose-mediated insulin secretion, is caused by mutations of the HNF4 gene. A mutation of the HNF4-binding site in the HNF1 gene promoter has also been associated with MODY. Thus, HNF4 is involved in the regulation of glucose homeostasis at several levels and along with the SRC-1, GRIP1, and p300 may play an important role in the pathophysiology of non-insulin-dependent diabetes mellitus.
Hepatocyte nuclear factor-41 (HNF4),2 a member of the nuclear receptor superfamily, is primarily expressed in liver, gut, kidney, and pancreas (1). Fatty acyl-CoA thioesters have recently been identified as ligands of HNF4 (2), a finding of considerable interest in view of the critical role HNF4 plays in the expression of genes important in a number of metabolic pathways (1). For example, HNF4 serves as an accessory factor for the glucocorticoid-mediated induction of the phosphoenolpyruvate carboxykinase gene and is necessary for the glucose-mediated increase of liver-type pyruvate kinase gene transcription (3–5). HNF4 also plays an essential role in embryonic development. Disruption of the HNF4 gene in mice results in an embryonic lethal phenotype characterized by a failure of the visceral endoderm to differentiate (6). Furthermore, HNF4 plays a critical role in the regulation of several liver genes. It also stimulates the expression of another liver-enriched transcription factor, HNF1, which itself is required for the expression of several liver genes (7, 8).
Maturity onset diabetes of young (MODY) is a rare form of non-insulin-dependent diabetes mellitus (NIDDM) characterized by defective secretion of insulin. It is inherited in an autosomal dominant pattern. Mutations of the HNF4 and HNF1 genes are directly associated with MODY types 1 and 3, respectively (9, 10), and a mutation that disrupts the HNF4-binding site on the HNF1 gene promoter has been reported in an Italian MODY family (11). Thus, through their effects in liver and beta cells, HNF4 and HNF1 have central roles in glucose homeostasis.
In this study we tested whether the transcriptional activity of HNF4 requires an interaction with a coregulator molecule. HNF4 contains two transactivation domains, designated AF-1 and AF-2. AF-1 is located in the first 24 amino acids of the N-terminal region of the protein, whereas AF-2 extends from amino acids 128 to 370 (12). Several coactivators, including CBP/p300, SRC-1, and members of the SRC-1-related family such as GRIP1/TIF2 or ACTR/RAC3/P/CIP interact with the AF-2 domains of other members of the nuclear receptor super-family and augment their transactivation potential (13–20). A physical and functional interaction between CBP and HNF4 has been recently reported (21). We show that SRC-1 and GRIP1 directly associate with the AF-2 domain of HNF4 in vivo and that these coactivators potentiate the transcriptional activity of HNF4. Furthermore, overexpression of SRC-1 or GRIP1 in HepG2 hepatoma cells enhances HNF1 gene expression, and this effect is dependent on the presence of an HNF4-binding site in the promoter. We propose that HNF4-coactivator interactions play a critical role in the regulation of glucose homeostasis and thus could be of importance in the pathophysiology of NIDDM.
EXPERIMENTAL PROCEDURES
Plasmid Construction
The expression plasmids that encode the GAL4 DNA-binding domain-HNF4 fusion proteins were generated by subcloning PCR-amplified DNA fragments into the simian virus 40 (SV40) promoter-enhancer-driven GAL4 expression plasmid, pSG424 (22). The HNF4 cDNA was provided by Dr. Frances Sladek (University of California at Riverside). Briefly, the nucleotide sequences of rat HNF4 corresponding to amino acids 1–374, 1–360, and 128–374 were amplified by the PCR, so that the fragments contained BamHI and KpnI sites at the 59- and 39-ends, respectively. These distinct PCR fragments were then digested and subcloned into the BamHI-KpnI site of pSG424. The reporter plasmid (GAL4)5E1bLuc was provided by Dr. Kazuya Yamada (Fukui Medical University, Japan) and was constructed by taking the BamHI/XhoI fragment from (GAL4)5E1bCAT (23), making it blunt-ended using the Klenow reaction, and subcloning this into the SmaI site of the pGL3 vector. The expression plasmids that encode p300 and GRIP1 have been previously described (24, 25) and were obtained from Dr. David Livingston (Dana Farber Institute, Boston) and Dr. Michael Stallcup (University of Southern California, Los Angeles), respectively. The SRC-1 expression plasmid was provided by Dr. Ming-Jer Tsai (Baylor Collage of Medicine). The HNF1 gene promoter reporter plasmids were provided by Dr. Naoyuki Miura (Akita University, Japan) and have been previously described (8).
The yeast expression plasmids encoding the GAL4-HNF4 chimeric proteins (called pGBT9-HNF4(128–374) and pGBT9-HNF4(128–360)) were generated by the subcloning of EcoRI-BamHI PCR-amplified DNA fragments into the pGBT9 plasmid (CLONTECH). The yeast expression vectors that encode fusion proteins of the GAL4 AD and full-length GRIP1 and SRC-1 (called pGAD424 GRIP1/FL and pGAD424 SRC-1) were described previously (24).
Cell Culture, Transient Transfection, and Luciferase Assays
The transient transfection and maintenance of HepG2 and HeLa cells have been described previously (24, 25). Luciferase activity was measured by the dual luciferase kit (Promega).
Yeast Two-hybrid Assays
Protein-protein interaction assays were performed using the Matchmaker yeast two-hybrid system (CLONTECH). A yeast plasmid, pGBT9-HNF4(128–374) or pGBT9-HNF4(128–360), was transformed into the yeast strain SFY526, and transformants were selected on synthetic dropout medium lacking Trp. Yeast plasmids, pGAD424 GRIP1/FL and pGAD424 SRC-1, were then transformed into the yeast strain SFY526 that contained either pGBT9-HNF4(128–374) or pGBT9-HNF4(128–360). The transformants were selected on synthetic dropout medium lacking Leu and Trp. The colonies that grew in these selection media were subjected to colony filter assays, as described in the technical manual of the Matchmaker yeast two-hybrid system (CLONTECH).
RESULTS
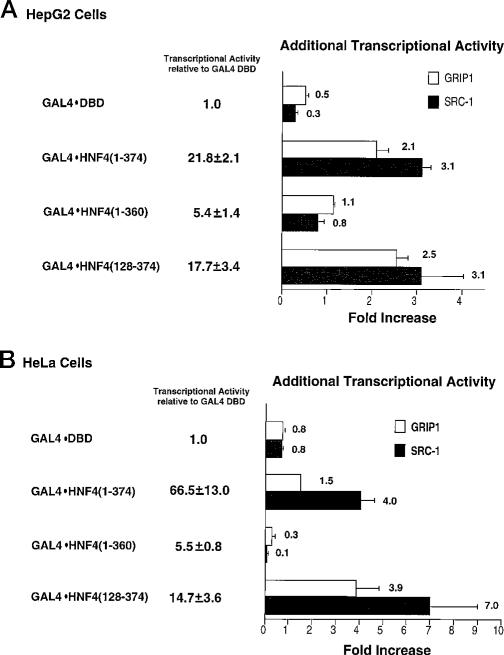
The GAL4 system, previously used to identify the transactivation domains of HNF4 (12), was used to determine whether SRC-1 and GRIP1 function as coactivators with HNF4. Expression plasmids that encode either SRC-1 or GRIP1 were cotransfected into HepG2 hepatoma cells with plasmids that encode the GAL4-HNF4 fusion proteins described under “Experimental Procedures” and with a reporter gene, (GAL4)5E1bLuc, that contains five copies of the GAL4-binding site. The activity of each GAL4-HNF4 chimeric protein was measured relative to that of the GAL4-DBD. Overexpression of SRC-1 and GRIP1 did not augment the transcriptional activity provided by GAL4-DBD in this system (Fig. 1, A and B). As noted previously, GAL4-HNF4(1–374) and GAL4-HNF4(128–374) both possess a strong transactivation function, whereas GAL4-HNF4(1–360), because of the absence of the AF-2 domain, is a weaker trans-activator (Fig. 1A and Ref. 12). The transcriptional activity of GAL4-HNF4(1–374), which contains both the AF-1 and AF-2 activation domains but lacks the C-terminal repression domain, is increased about 3-fold by overexpression of SRC-1 (Fig. 1A). The transcriptional activity of GAL4-HNF4(128–374), which contains only the AF-2 activation domain, was also augmented about 3-fold by overexpression of SRC-1 (Fig. 1A). In contrast, the transcriptional activity of GAL4-HNF4(1–360), which contains the AF-1 domain but lacks the AF-2 domain, was not influenced by the overexpression of SRC-1 (Fig. 1A). Overexpression of GRIP1 also increased the transcriptional activity of GAL4-HNF4(1–374) and GAL4-HNF4(128–374) but had no effect on that of GAL4-HNF4(1–360) (Fig. 1A). The effect of SRC-1 is consistently greater than that of GRIP-1, although significantly so only in the case of GAL4-HNF4(1– 374). These results show that both SRC-1 and GRIP1 can act as coactivators with HNF4 and that these effects are dependent on the presence of the AF-2 domain in HNF4.
Fig. 1. SRC-1 and GRIP1 potentiate the activity of GAL4-HNF4 fusion proteins.
A, expression plasmids that encode SRC-1 or GRIP1 (5 μg) and GAL4-HNF4 fusion proteins (0.5 μg) were cotransfected into HepG2 cells along with the reporter construct (GAL4)5E1bLuc (2.5 μg). The transcriptional activity of each GAL4-HNF4 fusion protein is expressed as the -fold increase relative to the activity of the GAL4-DBD. The additional increase of activity provided by SRC-1 or GRIP1 is presented relative to the activity of the GAL4-HNF4 fusion proteins obtained when equal amounts of DNA from the control vectors were transfected. These results represent the mean ± S.E. of at least three experiments. B, expression plasmids that encode SRC-1 or GRIP1 (10 μg) and the GAL4-HNF4 fusion proteins (2 μg) were cotransfected into HeLa cells with (GAL4)5E1bLuc (2 μg). The data are presented as described above.
We next tested the ability of SRC-1 and GRIP1 to potentiate the transcriptional activities of these three GAL4-HNF4 fusion proteins in HeLa cells to determine whether this phenomenon is cell type-specific. SRC-1 and GRIP1 both increased the trans-activation function of GAL4-HNF4(1–374) and GAL4-HNF4(128–374) but had no effect on that of GAL4-HNF4(1–360) (Fig. 1B). Thus, the effect of SRC-1 and GRIP1 on the transcriptional activity of HNF4 is not cell type-specific. SRC-1 was again a more potent coactivator than GRIP-1. These differences could be because of the different expression levels of these two coactivators in the cells or could reflect a difference in the strength of the interactions.
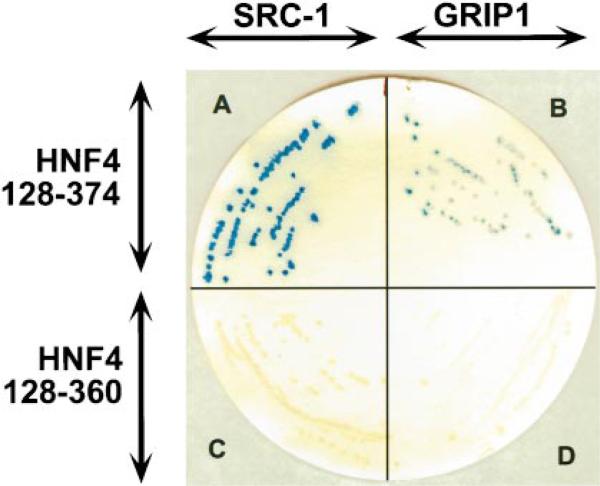
The yeast two-hybrid assay was used to test for an in vivo interaction between HNF4 and either SRC-1 or GRIP1. The expression of the GAL4-HNF4(128–374) or the GAL4-HNF4(128–360) fusion proteins in yeast cells failed to activate reporter gene activity (β-galactosidase, data not shown). Thus, neither of these chimeric proteins possesses transcriptional activity in yeast. β-Galactosidase activity was also not observed when a plasmid (pGAD424) that encodes the GAL4 AD was coexpressed with either of the GAL4-HNF4 fusion proteins (data not shown). However, β-galactosidase activity was strongly induced when a chimeric protein that contained the GAL4 AD and SRC-1 or GRIP1 was coexpressed with GAL4-HNF4(128–374) (Fig. 2). In colony filter assays, yeast that contained the GAL4 AD and SRC-1 fusion protein and GAL4-HNF4(128–374) turned blue 30 min after initiation of the reaction (Fig. 2A). In contrast, yeast that contained the GAL4 AD-GRIP1 fusion protein and GAL4-HNF4(128–374) turned blue 4 h after initiation of the assay (Fig. 2B). Thus, the interaction between SRC-1 and HNF4 is stronger than between GRIP1 and HNF4. Coexpression of the GAL4 AD-SRC-1 or GAL4 AD-GRIP1 fusion proteins with GAL4-HNF4(128–360) did not induce β-galactosidase activity (Fig. 2, C and D). These results suggest that SRC-1 and GRIP1 both interact with HNF4 and that these interactions are dependent on the presence of the AF-2 domain of HNF4. These results are consonant with the functional results shown in Fig. 1.
Fig. 2. In vivo protein-protein interactions between GAL4-HNF4 chimeric proteins and SRC-1 or GRIP1.
Yeast cells were co-transformed with combinations of the yeast plasmids, pGBT9-HNF4(128–374), pGBT9-HNF4(128–360), pGAD424 GRIP1/FL, or pGAD424 SRC-1, as indicated, and were grown on synthetic dropout plates lacking Leu and Trp. Yeast colonies were then subjected to a colony filter assay for β-galactosidase activity. Yeast cells that contained pGBT9-HNF4(128–374) and pGAD424-SRC-1 turned dark blue 30 min after the reaction was initiated (A). Yeast cells that contained pGBT9-HNF4(128–374) and pGAD424-GRIP1 turned blue 4 h after the reaction was initiated (B). In liquid β-galactosidase activity assays, yeast that contain the GAL4 AD-SRC-1 fusion protein and GAL4-HNF4(128–374) showed stronger activity than yeast that contain the GAL4 AD-GRIP1 fusion protein and GAL4-HNF4(128–374) (data not shown).
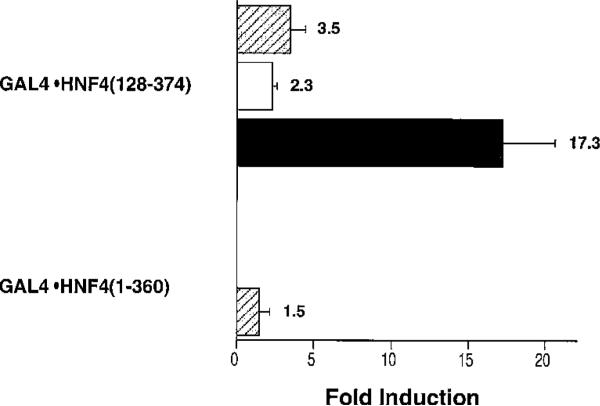
CBP is known to associate with HNF4, and it potentiates the activity of a reporter construct that contains multiple HNF4-binding sites (21). We therefore tested whether p300, another member of the CBP coactivator family, potentiates the transcriptional activity of GAL4-HNF4 chimeric proteins. Overexpression of p300 in HeLa cells does enhance the activity of GAL4-HNF4(128–374) but not that of GAL4-HNF4(1–360) (Fig. 3). Thus, coactivation of HNF4 by p300 also requires an intact AF-2 domain. The transcriptional activity of GAL4-HNF4(128–374) was further enhanced when both p300 and SRC-1 were overexpressed in HeLa cells (Fig. 3), as has been reported in the analysis of other nuclear receptors (20, 28).
Fig. 3. SRC-1 and p300 act synergistically to increase the activity of GAL4-HNF4(128–374).
Expression plasmids that encode p300 (10 μg), SRC-1 (10 μg), and GAL4-HNF4 fusion proteins (4 μg) were cotransfected into HeLa cells with the (GAL4)5E1bLuc reporter construct (2 μg). The data are expressed as the -fold induction relative to the activity of the GAL4-HNF4 fusion proteins in cells in which equal amounts of control vectors were transfected. These results represent the mean ± S.E. of at least three experiments. Hatched bars, p300; white bars, SRC-1; black bsrs, p300 + SRC-1.
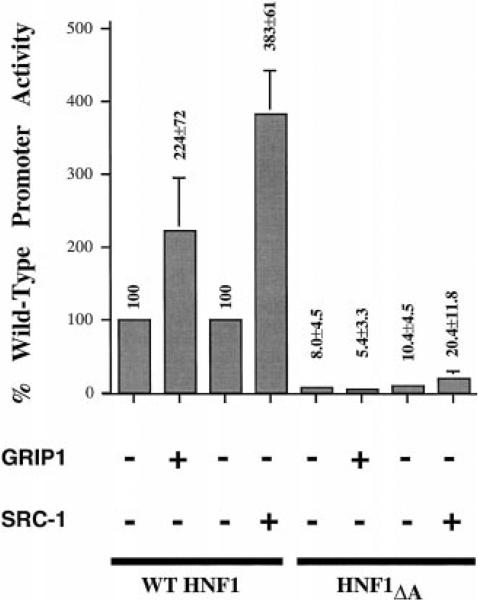
HNF4 binds to its cognate DNA element in the HNF1 gene promoter and enhances transcription through this interaction (7, 8). The coexpression of SRC-1 or GRIP1 should enhance expression from the HNF1 promoter if the HNF4-coregulator interaction is functionally important. In fact, the overexpression of SRC-1 in HepG2 hepatoma cells increased HNF1 gene promoter activity by about 3-fold (Fig. 4). Overexpression of GRIP1 was slightly less effective (Fig. 4). Deletion of the HNF4-binding site in the HNF1 promoter (HNF4ΔA) resulted in a 90% reduction of expression, as expected (8), and the overexpression of either SRC-1 or GRIP1 was without effect when a construct containing this element was used as the promoter in the reporter system (Fig. 4).
Fig. 4. SRC-1 and GRIP1 potentiate transactivation through the HNF1 gene promoter.
Expression plasmids that encode SRC-1 (20 μg) or GRIP1 (5 μg) and a reporter construct that contains either the wild type HNF1 gene promoter (2.5 μg) or the promoter that lacks a functional HNF4-binding site (ΔA) (8) were cotransfected into HepG2 cells. Results are presented relative to wild type HNF1 promoter activity in the absence of cotransfection of either SRC-1 or GRIP1 (100%). These results represent the mean ± S.E. of at least three experiments.
DISCUSSION
The transcription factor HNF4, a member of the nuclear receptor superfamily, is important for tissue-specific gene expression, including several genes involved in hepatic glucose metabolism (1). We now show that both SRC-1 and GRIP1 interact with HNF4 (Fig. 2) and potentiate its transcriptional activity (Fig. 1). Both are presumably active in vivo, and given the high degree of homology between SRC-1, GRIP1, and other SRC-1-related coactivators such as RAC3, ACTR, and P/CIP, it is possible that these proteins can also function as coactivators for HNF4. It is not known why most tissues express all these coactivators, but they could have selective actions. For example, SRC-1 has been implicated in the action of androgens. The expression of GRIP1 is increased about 2-fold in knockout mice that lack the SRC-1 gene, apparently in compensation for the loss of SRC-1, but these mice still display a partial resistance to the action of androgens (29). These coactivators are thus not completely interchangeable. It is possible that specific coactivators, or different ratios of various of these proteins in cells, result in selective interaction with nuclear receptors, including HNF4.
CBP/p300 serves as a coactivator with most members of the nuclear hormone receptor family (13, 17, 30–32). As shown in this paper, p300 enhances the transactivation function of GAL4-HNF4(128–374). This coactivation effect is dependent on an intact AF-2 domain, which is also required for the interaction of SRC-1 and GRIP1 with HNF4. In addition, p300 acts synergistically with SRC-1 or GRIP1 to further enhance the transcriptional activity of HNF4. A similar phenomenon has been observed with other nuclear receptors (20, 28). Recent studies suggest that transactivation by nuclear receptors requires the formation of a complex with coactivators (32–34). These coactivator complexes can contain SRC-1/GRIP1, p300/CBP, and other coactivators such as P/CAF (34, 35). SRC-1 and GRIP1 have both been shown to directly associate with CBP/p300 (13, 34–36). Moreover, many of these proteins possess histone acetyltransferase activity (34, 37–39). The histone acetylase activity of P/CAF is required for the retinoic acid-mediated induction of gene transcription (34, 40). A two-step model that first requires chromatin remodeling by histone acetylation and then an interaction with the general transcription apparatus has been proposed to explain how this coactivator complex mediates the transactivation of nuclear receptors (32). It is possible that a similar mechanism is required for the action of HNF4, which serves as an accessory factor in the glucocorticoid response unit of the phosphoenolpyruvate carboxykinase gene (5).
HNF4 appears to have a pleiotropic effect on the regulation of glucose homeostasis. The mechanism of HNF4 gene mutations in the pathophysiology of MODY (type 1) has not been established, but it is of interest to note that HNF4 is required for expression of HNF1, another MODY gene (type 3) (7–9). Indeed, at least one MODY pedigree is because of a mutation of the HNF4-binding site in the HNF1 gene promoter. The common denominator of MODY type 1 and 3 may be the loss of HNF1 gene function. In this regard, it may be noteworthy that we found that overexpression of both SRC-1 and GRIP1 increases the activity of the HNF1 gene promoter, and a mutation of the HNF4 DNA-binding site in this promoter disrupts this effect.
In addition to its apparent role in insulin secretion (10) and a demonstrated role in the regulation of hepatic gene expression (1), HNF4 also regulates the expression of other genes (e.g. glucose transporter 2 (Glut 2), aldolase B, and pyruvate kinase that are involved in glucose transport and glucose metabolism in embryonic stem cells and in embryos (41)). The observation that fatty acyl-CoA thioesters are HNF4 ligands (2) provides support for the connection between HNF4 and glucose homeostasis because fatty acids affect the regulation of glucose metabolism by decreasing peripheral glucose utilization, increasing hepatic gluconeogenesis, and influencing insulin secretion and pancreatic β cell hyperplasia (42–44). Ligands affect the binding of nuclear receptors to target DNA sequences but also influence the interactions of these receptors with coactivators or corepressors (34, 45). This phenomenon, along with the potential role of these coactivators in NIDDM, is an interesting area for future investigation.
Acknowledgments
We thank Don Scott and Marc Daniels for a critical reading of this manuscript, Cathy Caldwell for technical assistance, and Deborah C. Brown for helping prepare this manuscript.
Footnotes
This work was supported by National Institutes of Health Grants DK 20593 (the Vanderbilt Diabetes Research and Training Center) and DK 35107. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
HNF4 and HNF1 exist as multiple isoforms. In this paper we are referring to the a isoform of each protein.
The abbreviations used are: HNF, hepatocyte nuclear factor; MODY, maturity onset diabetes of young; NIDDM, non-insulin-dependent diabetes mellitus; PCR, polymerase chain reaction; AD, activation domain; DBD, DNA-binding domain.
REFERENCES
- 1.Sladek FM. In: Liver Gene Expression. Tronche F, Yaniv M, editors. Vol. 11. R. G. Landes Company; Austin, TX: 1994. pp. 207–223. [Google Scholar]
- 2.Hertz R, Magenheim J, Berman I, Bar-Tana J. Nature. 1998;392:512–516. doi: 10.1038/33185. [DOI] [PubMed] [Google Scholar]
- 3.Towle HC. J. Biol. Chem. 1995;270:23235–23238. doi: 10.1074/jbc.270.40.23235. [DOI] [PubMed] [Google Scholar]
- 4.Liu Z, Towle HC. Biochem. J. 1995;308:105–111. doi: 10.1042/bj3080105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall RK, Sladek FM, Granner DK. Proc. Natl. Acad. Sci. U. S. A. 1995;92:412–416. doi: 10.1073/pnas.92.2.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen WS, Manova K, Weinstein DC, Duncan SA, Plump AS, Prezioso VR, Bachvarova RF, Darnell JE., Jr. Genes Dev. 1994;8:2466–2477. doi: 10.1101/gad.8.20.2466. [DOI] [PubMed] [Google Scholar]
- 7.Kuo CJ, Conley PB, Chen L, Sladek FM, Darnell JE, Jr., Crabtree GR. Nature. 1992;355:457–461. doi: 10.1038/355457a0. [DOI] [PubMed] [Google Scholar]
- 8.Miura N, Tanaka K. Nucleic Acids Res. 1993;21:3731–3736. doi: 10.1093/nar/21.16.3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamagata K, Oda N, Kaisaki PJ, Menzel S, Furuta H, Vaxillaire M, Southam L, Cox RD, Lathrop GM, Boriraj VV, Chen X, Cox NJ, Oda Y, Yano H, LeBeau MM, Yamada S, Nishigori H, Takeda J, Fajans SS, Hattersley AT, Iwasaki N, Hansen T, Pedersen O, Polonsky KS, Bell GI. Nature. 1996;384:455–458. doi: 10.1038/384455a0. [DOI] [PubMed] [Google Scholar]
- 10.Yamagata K, Furuta H, Oda N, Kaisaki PJ, Menzel S, Cox NJ, Fajans SS, Signorini S, Stoffel M, Bell GI. Nature. 1996;384:458–460. doi: 10.1038/384458a0. [DOI] [PubMed] [Google Scholar]
- 11.Gragnoli C, Lindner T, Cockburn BN, Kaisaki PJ, Gragnoli F, Marozzi G, Bell GI. Diabetes. 1997;46:1648–1651. doi: 10.2337/diacare.46.10.1648. [DOI] [PubMed] [Google Scholar]
- 12.Hadzopoulou-Cladaras M, Kistanova E, Evagelopoulou C, Zeng S, Cladaras C, Ladias JAA. J. Biol. Chem. 1997;272:539–550. doi: 10.1074/jbc.272.1.539. [DOI] [PubMed] [Google Scholar]
- 13.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S-C, Heyman RA, Rose DW, Glass CK, Rosenfeld MG. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 14.Hong H, Kohli K, Garabedian MJ, Stallcup MR. Mol. Cell. Biol. 1997;17:2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Gomes PJ, Chen JD. Proc. Natl. Acad. Sci. U. S. A. 1997;94:8479–8484. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen H, Lin RJ, Schiltz RL, Chakravarti D, Nash A, Nagy L, Privalsky ML, Nakatani Y, Evans RM. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 17.Chakravarti D, LaMorte VJ, Nelson MC, Nakajima T, Schulman IG, Juguilon H, Montminy M, Evans RM. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 18.Onate SA, Tsai SY, Tsai MJ, O'Malley BW. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 19.Voegel JJ, Heine MJ, Zechel C, Chambon P, Gronemeyer H. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 20.Torchia J, Rose DW, Inostroza J, Kamei Y, Westin S, Glass CK, Rosenfeld MG. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 21.Yoshida E, Aratani S, Itou H, Miyagishi M, Takiguchi M, Osumu T, Murakami K, Fukamizu A. Biochem. Biophys. Res. Commun. 1997;241:664–669. doi: 10.1006/bbrc.1997.7871. [DOI] [PubMed] [Google Scholar]
- 22.Sadowski I, Ptashne M. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lillie JW, Green MR. Nature. 1989;338:39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- 24.Ding XF, Anderson CM, Ma H, Hong H, Uht RM, Kushner PJ, Stallcup MR. Mol. Endocrinol. 1998;12:302–313. doi: 10.1210/mend.12.2.0065. [DOI] [PubMed] [Google Scholar]
- 25.Eckner R, Ewen ME, Newsome D, Gerdes M, DeCaprio JA, Lawrence JB, Livingston DM. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 26.Deleted in proof
- 27.Deleted in proof
- 28.Smith CL, Onate SA, Tsai MJ, O'Malley BW. Proc. Natl. Acad. Sci. U. S. A. 1996;93:8884–8888. doi: 10.1073/pnas.93.17.8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu J, Qiu Y, DeMayo FJ, Tsai SY, Tsai MJ, O'Malley BW. Science. 1998;279:1922–1925. doi: 10.1126/science.279.5358.1922. [DOI] [PubMed] [Google Scholar]
- 30.Dowell P, Ishmael JE, Avram D, Peterson VJ, Neverivy DJ, Leid M. J. Biol. Chem. 1997;272:33435–33443. doi: 10.1074/jbc.272.52.33435. [DOI] [PubMed] [Google Scholar]
- 31.Hanstein B, Eckner R, DiRenzo J, Halachmi S, Liu H, Searcy B, Kurokawa R, Brown M. Proc. Natl. Acad. Sci. U. S. A. 1996;93:11540–11545. doi: 10.1073/pnas.93.21.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenster G, Spencer TE, Burcin MM, Tsai SY, Tsai MJ, O'Malley BW. Proc. Natl. Acad. Sci. U. S. A. 1997;94:7879–7884. doi: 10.1073/pnas.94.15.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurokawa R, Kalafus D, Ogliastro M-H, Kioussi C, Xu L, Torchia J, Rosenfeld MG, Glass CK. Science. 1998;279:700–703. doi: 10.1126/science.279.5351.700. [DOI] [PubMed] [Google Scholar]
- 34.Glass CK, Rose DW, Rosenfeld MG. Curr. Opin. Cell Biol. 1997;9:222–232. doi: 10.1016/s0955-0674(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 35.Yao TP, Ku G, Zhou N, Scully R, Livingston DM. Proc. Natl. Acad. Sci. U. S. A. 1996;93:10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voegel JJ, Heine MJS, Tini M, Vivat V, Chambon P, Grone-meyer H. EMBO J. 1998;17:507–519. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 38.Bannister AJ, Kouzarides T. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 39.Spencer TE, Jenster G, Burcin MM, Allis CD, Zhou J, Mizzen CA, McKenna NJ, Onate SA, Tsai SY, Tsai MJ, O'Malley BW. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 40.Korzus E, Torchia J, Rose DW, Xu L, Kurokawa R, McInerney EM, Mullen T-M, Glass CK, Rosenfeld MG. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 41.Stoffel M, Duncan SA. Proc. Natl. Acad. Sci. U. S. A. 1997;94:13209–13214. doi: 10.1073/pnas.94.24.13209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boden G. Diabetes. 1997;46:3–10. [PubMed] [Google Scholar]
- 43.Newgard CB, McGarry JD. Annu. Rev. Biochem. 1995;64:689–719. doi: 10.1146/annurev.bi.64.070195.003353. [DOI] [PubMed] [Google Scholar]
- 44.Unger RH. Diabetes. 1995;44:863–870. doi: 10.2337/diab.44.8.863. [DOI] [PubMed] [Google Scholar]
- 45.Shibata H, Spencer TE, Onate SA, Jenster G, Tsai SY, Tsai MJ, O'Malley BW. Recent Prog. Horm. Res. 1997;52:141–165. [PubMed] [Google Scholar]