Abstract
Objective
Multiple studies have demonstrated that single-nucleotide polymorphisms (SNPs) in the ITGAM locus (including the non-synonymous SNPs rs1143679, rs1143678, rs1143683) are associated with SLE. ITGAM encodes the protein CD11b, a subunit of the β2 integrin Mac-1. The purpose of this study was to determine the effects of ITGAM genetic variation on the biological functions of neutrophil Mac-1.
Methods
Neutrophils from ITGAM genotyped and sequenced healthy donors were isolated for functional studies. The phagocytic capacity of neutrophil ITGAM variants was probed with complement coated erythrocytes, serum treated zymosan, heat treated zymosan and IgG coated erythrocytes. The adhesion capacity of ITGAM variants, in adhering to either purified intercellular adhesion molecule 1 or tumor necrosis factor α-stimulated endothelial cells was assessed in a flow chamber. Expression levels of total CD11b and activation of CD11b were assessed by flow cytometry.
Results
Mac-1–mediated neutrophil phagocytosis, determined in cultures with 2 different complement-coated particles, was significantly reduced in individuals with nonsynonymous variant alleles of ITGAM. This reduction in phagocytosis was related to variation at either rs1143679 (in the β-propeller region) or rs1143678/rs1143683 (highly linked SNPs in the cytoplasmic/calf-1 regions). Phagocytosis mediated by Fcγ receptors was also significantly reduced in donors with variant ITGAM alleles. Similarly, firm adhesion of neutrophils was significantly reduced in individuals with variant ITGAM alleles. These functional alterations were not attributable to differences in total receptor expression or activation.
Conclusion
The nonsynonymous ITGAM variants rs1143679 and rs1143678/rs113683 contribute to altered Mac-1 function on neutrophils. These results underscore the need to consider multiple nonsynonymous SNPs when assessing the functional consequences of ITGAM variation on immune cell processes and the risk of SLE.
Introduction
Recent genome-wide association studies (GWAS) of human systemic lupus erythematous (SLE) have revealed strong association between single nucleotide polymorphisms (SNPs) in the ITGAM locus and susceptibility to SLE (1, 2). Following the initial reports of ITGAM SNP association with SLE (1, 2), this observation has been replicated in many independent genetic studies across different ethnic groups (3–5). The ITGAM gene encodes the α subunit (known as CD11b) of the β2 integrin Mac-1 (also called CR3) (6). Notably, even before these results in prior genetic studies had implicated ITGAM as a major susceptibility locus in SLE, a study using an experimental mouse model demonstrated that lupus-prone MRL/MpJ-Faslpr mice rendered deficient in CD11b had an exaggerated autoimmune phenotype (7).
Mac-1 is broadly expressed on cells of the myeloid lineage and on a subset of lymphocytes (8–11). Mac-1 is a surface receptor involved in numerous cellular functions. On neutrophils for example, Mac-1 is constitutively expressed, can be rapidly up-regulated upon cell activation, and is important for promoting firm adhesion to endothelial cells and subsequent transendothelial migration (via Mac-1 binding to ligands such as intercellular adhesion molecule 1 (ICAM-1), ICAM-2, among others) (12, 13). Mac-1 also mediates neutrophil phagocytosis of both complement opsonized and unopsonized particles (14–17). Furthermore, Mac-1 can modify the functions of other co-expressed receptors, such as Fc receptors and Toll-like receptors (18–20).
Based on the results of genetic studies, it has been suggested that the observed association of ITGAM with SLE in Caucasian and African American populations is attributable to the variation at the non-synonymous SNP rs1143679 (4), which encodes an amino acid change from Arg to His at amino acid position 77 in the extracellular domain of CD11b. Since then, studies of the impact of ITGAM genetic variation on Mac-1–mediated biologic processes have almost exclusively focused on the influence of the rs1143679 SNP on the functions of Mac-1 in transduced cell lines and primary human monocytes (21–23). While these studies have variously reported that rs1143679 affects cell adhesion, phagocytosis and cytokine production, it has also been observed that this SNP can occur in conjunction with other nonsynonymous ITGAM SNPs, which are in high linkage disequilibrium (LD) in this locus (4). The potential impact of these linked non-synonymous SNPs on Mac-1-mediated functions not been addressed in previous studies. Indeed, analyses in different ethnicities have indicated a more complex association pattern between ITGAM variation and SLE susceptibility (5). Therefore, multiple ITGAM SNPs, in addition to rs1143679, could be contributing to the genetic risk of SLE development.
In the present study, using a cohort of 1,815 healthy donors, we confirmed that multiple ITGAM nonsynonymous SNPs exist, including SNP rs1143679, rs1143678 (Pro to Ser at amino acid position 1146) and rs1143683 (Ala to Val at amino acid position 858) and that these SNPs show strong LD. Furthermore, we provide the first experimental evidence that these multiple SLE associated non-synonymous ITGAM SNPs independently alter Mac-1-mediated neutrophil functions. The alterations in neutrophil Mac-1 functions associated with the variant ITGAM alleles, including decreased firm adhesion of neutrophils and reduced phagocytosis, could not be attributed to changes in the expression or activation of Mac-1, but could be linked to altered Fc receptor–mediated functions. The results of our study highlight the need for caution when interpreting the potential contribution to SLE of any single variant in the ITGAM gene, as any functional differences observed between the common and variant forms of Mac-1 could be due to one or more highly linked variants.
Material and Methods
Reagents
RPMI medium, fetal bovine serum (FBS), phosphate-buffered saline (PBS), tumor necrosis factor-α( TNF-α) and the SuperScript First-Strand Synthesis System for RT-PCR kit were all from Invitrogen. Antibodies against CD11b (ICRF44 and CBRM1/5) and isotype matched immunoglobulins were from eBioscience. Rabbit anti-sheep erythrocyte IgM antibody was from Fitzgerald Industries International. Purified ICAM-1/Fc and P-selectin/Fc chimeras were from R & D systems. Heavy ficoll (Histopaque-1119), fMLP, Protein A, gelatin, fibronectin, zymosan, human AB serum, rabbit anti-sheep erythrocyte IgG antibody and human C5 deficient serum were all from Sigma-Aldrich. Lymphocyte separation medium (light ficoll) was from Mediatech. HUVEC cells and culture media were from Lonza. Sheep erythrocytes were from Colorado Serum.
Human participants and genotyping
Healthy human donors with no chronic autoimmune diseases (n=1,815) were recruited for our genotyping and functional assays. Healthy subjects were chosen in order to avoid the confounding effects of inflammation or medications on cellular functions. All donors recruited for these studies gave informed consent to participate, and the study was approved by the Institutional Review Board.
Blood samples were obtained from all donors, and genomic DNA was isolated from EDTA-treated whole blood using the PureGene DNA Isolation Kit (Qiagen). SNP rs1143679 was genotyped via TaqMan assay (Applied Biosystems), using an ABI7900HT instrument. SNP rs1143678 was genotyped with the Pyrosequencing method, as we have previously described (24), using the sense primer (5’-Biotin–AGC TCG GCT TCT TCA AGC GGC A-3’), antisense primer (5’-CAC CGA GAG GCA GCT CTG-3’), and the pyrosequencing primer (5’-GGG TTC GGC CCC CGG-3’). In blood samples from a subset of 31 healthy donors whose neutrophils were used in the functional studies, the full-length ITGAM complementary DNA (cDNA) was directly sequenced using the Sanger sequencing strategy. The full-length ITGAM cDNA was then amplified with RT-PCR, using total RNA isolated from human leukocytes. A series of overlapping sequencing primers was used to sequence the whole open-reading frame of the ITGAM cDNA (details available from the corresponding author upon request).
Neutrophil isolation
Fresh anticoagulated blood was collected from participants by phlebotomy and neutrophils were separated on a discontinuous gradient as previous described (24). The neutrophils were then washed and resuspended in complete RPMI medium. All functional assays were performed in duplicates using neutrophils from different pairs of overlapping genotyped donors.
Phagocytosis assays
Sheep erythrocytes coated with rabbit anti-sheep RBC IgG antibody (referred to as EA), sheep erythrocytes coated with rabbit anti-sheep RBC IgM antibody plus complement (referred to as EAC), STZ (serum treated zymosan) and HTZ (heat treated zymosan) were prepared fresh each day as previous described (25). Neutrophils from healthy donors were suspended in complete RPMI medium at a density of 5x106 cells per ml. Neutrophils (with or without priming for 10 min with 10−8 M fMLP) were incubated with EA or EAC (1:20 ratios) for 30 min, or incubated with STZ or HTZ (1:10 ratios) for 15 min, both at 37 °C. Particles engulfed by neutrophils were counted by light microscopy with oil immersion. For each condition, at least 200 neutrophils were inspected, and the phagocytic index (number of ingested particles per 100 neutrophils) was calculated. The genotype of each neutrophil donor was unknown to the observers at the time of data acquisition.
Flow chamber adhesion assay
Neutrophil adhesion to the purified ICAM-1 or to the activated HUVEC was performed under flow conditions as previously described (26). Briefly, a mixture of 25 mg/ml recombinant human ICAM-1/Fc chimera and 0.5 mg/ml recombinant human P-selectin/Fc chimera (to initiate cell capture, facilitate cell rolling and firm adhesion to ICAM-1) were used as the purified substrates. HUVECs were cultured to full confluence (in 25-mm Corning culture dishes) and primed with 20 ng/ml human TNFα for 6 hours prior to each experiment.
Neutrophils (5x105cells/ml in complete RPMI medium) were injected into the flow chamber at a shear stress of 1.5 dyne/cm2 via a programmable syringe pump, and cell movement was viewed via microscopy and recorded with a CCD camera (30 images per second). All experiments were done at 37 °C. The number of firmly adherent neutrophils was determined by two independent scorers (YZ and NBW), without knowledge of participant genotype. Four-minute long digital movies of each adhesion experiment were viewed, and cells that were captured and adhered in the field of view were determined (expressed as adherent cells/min) (results available from the corresponding author upon request). A cell that moved a distance of < 1 cell diameter in 5 seconds was considered to be firmly adhered. For each donor, a minimum of 3 separate experiments were performed.
Mac-1 expression analyses
Freshly obtained blood (2 ml) was centrifuged, and peripheral blood cells were washed and suspended in PBS. Aliquots of 50ul blood were prepared and fMLP was added to each (final concentration of 5x10−9M). Thereafter, the cells were incubated at 37 °C for 15 min. After washing, fluorochrome-labeled anti-human CD11b antibodies (ICRF44 and CBRM1/5) were added to measure total and activated CD11b (27), respectively. The cells were incubated on ice for 30 min. Erythrocytes were lysed with BD lysis buffer, and flow cytometry was done on a FACSscan Instrument. The captured flow cytometry results were analyzed using the FlowJo software package (Tree Star). Neutrophils were gated based on forward and side scatter patterns.
Statistical analysis
All data are represented as the mean ± SEM. Because the phagocytic probes and adhesion substrates were freshly prepared daily, all studies assessing differences in phagocytosis and adhesion, using cells from homozygous donors, were performed using a paired experimental design, and results were analyzed using Student’s paired t-test (Graphpad Prism, version5). For analysis of donors heterozygous for variation at the ITGAM locus, analysis of variance models were built to test whether the mean level of phagocytosis varied between genotypes. Least squares means were calculated and utilized for pairwise comparisons.
Results
Characterization of non-synonymous variants in the human ITGAM locus
Variants in the human ITGAM locus strongly and reproducibly associate with SLE susceptibility and one non-synonymous SNP, rs1143679, has been purported to be causal for this genetic association (4). However, several other non-synonymous ITGAM SNPs are in strong LD with rs1143679 and the impact of these additional SNPs is seldom considered. Indeed, based on currently available data and our own genotyping results, in addition to rs1143679 there are three other non-synonymous SNPs in the ITGAM locus with a minor allele frequency (MAF) >2% (Supplement Table 1). SNP rs1143679, rs1143683 and rs1143678 have all been shown to have strong association with SLE in Caucasian populations. Attributing disease causality solely to the ITGAM SNP rs1143679 may, therefore, be premature. To examine the possible biological consequences of non-synonymous variants in high LD with rs1143679, we characterized ITGAM SNPs and their impact on neutrophil Mac-1 function, using a large population of genotyped healthy donors.
SNP rs1143679 encodes a c.328G>A, p.Arg77His change in the exon 3 of ITGAM, which encodes part of the extracellular β-propeller domain of CD11b (Figure 1). It has been hypothesized that this change is sufficient to alter CD11b protein’s overall structure and, thereby, alter its activity (3). Based on the presence of conserved amino acids between CD11a and CD11b that have been mapped to the calf-1 region of CD11a (28), SNP rs1143683 (c.2671C>T) encodes a conservative amino acid change (p.Ala858Val) in or near the extracellular calf-1 region of CD11b while rs1143678 (c.3436C>T) encodes a non-conservative amino acid change (p.Pro1146Ser) in the cytoplasmic tail of CD11b (Figure 1).
Figure 1. Schematic diagram of the CD11b protein domains and the location of 3 nonsynonymous SNPs.
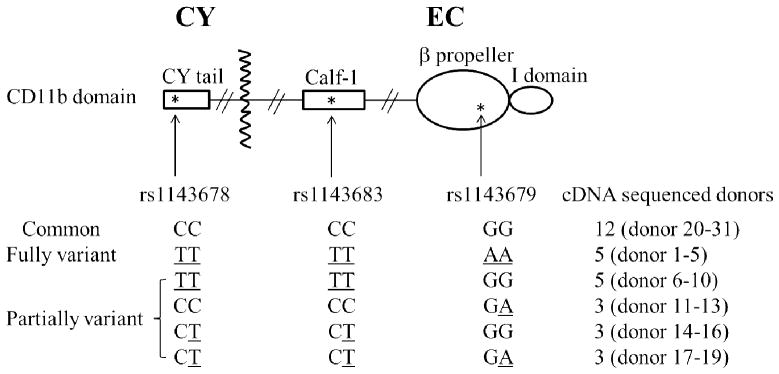
Variant alleles of SNP rs1143678, rs1143683, and rs1143679 are underlined. The codes of each donor with the indicated genotypes are shown. CY = cytoplasmic domain; EC = extracellular domain.
Existing HapMap data, the results of prior published (5) and the ITGAM cDNA sequencing results from all donors used in our functional studies demonstrated that rs1143683 and rs1143678 are in perfect LD. In addition, it was found that strong LD exists between the SNP rs1143678 and the SNP rs1143679 (D’=0.97, r=0.82, n=1815) (Supplement Table 2 and ref 4). Notably, in present study, 86% of donors carrying the rs1143679 variant also carried the non-synonymous rs1143678 variant (Supplement Table 2). Strong LD between rs1143678 and rs1143679 was also found in patients with SLE (3, 4 and Edberg JC: unpublished results). Remarkably, of the 21 donors who were homozygous for the variant (A) allele of SNP rs1143679 (Supplement Table 2, leftmost column), all carried at least one variant (T) allele of SNP rs1143678. Variation at rs1143680 was not observed in any of the donors used in our functional studies. These results underscore that any functional effects attributed to SNP rs1143679 must take into account the potential role of SNP rs1143678 (and rs1143683, which is in perfect LD in our donors).
Association of ITGAM variant alleles with differing capacity of neutrophils to phagocytose opsonized particles
To reveal the possible functional consequences of SLE associated ITGAM variants, we examined Mac-1 mediated phagocytosis by neutrophils harvested from genotyped donors. Full length ITGAM cDNAs from all 31 donors used for functional studies were prepared and sequenced, to confirm the allelic status at the trio of SNPs rs1143678/rs1143683/rs1143679 (Figure 1). In these 31 individuals, we did not detect any other non-synonymous ITGAM genetic variation.
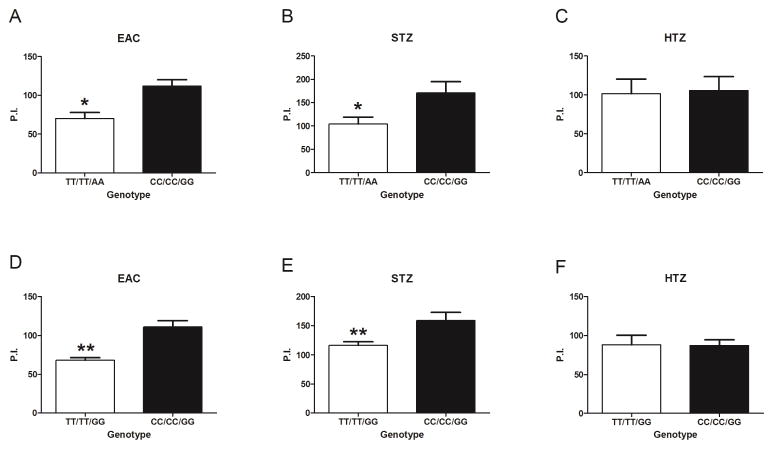
In culture of fMLP primed neutrophils from donors homozygous for the common alleles at rs1143678/rs1143683/rs1143679 (hereafter the common CC/CC/GG; see Figure 1) and from those donors homozygous for the respective minor alleles (hereafter the fully variant TT/TT/AA; See Figure 1), we observed a consistent and statistically significant decrease in phagocytosis of EAC and STZ (complement opsonized particles that are known to engage the I-domain of Mac-1 (14–17)) by neutrophils from donors with the TT/TT/AA ITGAM genotype (Figure 2, A and B). In contrast using HTZ, which can bind to Mac-1 via a lectin domain different from the complement binding site (17) and other cell surface receptors such as dectin-1 (29), phagocytosis was not significantly different between the fully common and fully variant genotypes (Figure 2C).
Figure 2. Non-synonymous ITGAM variants alter complement-mediated neutrophil phagocytosis.
Quantitative phagocytosis by neutrophils from genotyped (rs1143678/rs1143683/rs1143679) healthy donors was assessed. Neutrophils were pretreated with 10−8 M fMLP for 10 minutes, and then incubated with sheep erythrocyte antigen coated with complement (EAC) (A, D), serum-treated zymosan (STZ) (B, E) or heat-treated zymosan (HTZ) (C, F). All experiments were performed in a paired manner. The TT/TT/AA genotype was compared with the CC/CC/GG genotype (A–C) using 3 pairs of donors (donors 1–3 and donors 20–22). The TT/TT/GG genotype was compared with the CC/CC/GG genotype (D–F) using 5 pairs of donors (donors 6–10 and donors 24–28). Results are the mean ± SEM phagocytic index (P. I.) (the number of internalized probes per 100 neutrophils). * = P < 0.05; ** = P < 0.01 by Student's paired t-test.
As noted earlier, in our cohort of 1815 healthy donors, none were homozygous for the variant allele only at rs1143679 (i.e. none had the genotype CC/CC/AA) (Supplement Table 2). Interrogating the influence of the extracellular SNP rs1143679 in isolation was therefore not possible. However, compared to neutrophils from the common CC/CC/GG donors, neutrophils from donors common at rs1143679 but variant at rs1143678/rs1143683 (i.e. partially variant TT/TT/GG genotype, see in Figure 1) still showed a statistically significant defect in phagocytosis of EAC and STZ (Figure 2, D and E). Again, phagocytosis of HTZ was not affected (Figure 2F). These results show that ITGAM variation in the cytoplasmic/calf-1 domain is sufficient to alter neutrophil biology even when there is no SNP variation in the β-propeller domain.
Importantly, while phagocytosis of EAC required neutrophil priming with fMLP, STZ and HTZ were internalized by unprimed neutrophils, albeit less efficiently. This allowed us to investigate the influence of ITGAM variation on phagocytosis using unprimed neutrophils. Again, we found that neutrophil phagocytosis of STZ, but not HTZ, was significantly reduced for both the fully variant TT/TT/AA and partially variant TT/TT/GG genotypes (results available from the corresponding author upon request). In summary, these results demonstrate that ITGAM variation affects phagocytosis of complement opsonized particles such as EAC and STZ, but not the non-opsonized particle HTZ. Furthermore, these data are the first to show that non-synonymous SLE-associated SNPs in ITGAM other than rs1143679 can alter the biology of Mac-1.
Lack of effect of ITGAM variation on expression and activation of Mac-1
Similar to other β2 integrins, the conformation of Mac-1 is normally very dynamic, with different conformations associated with different states of activation and affinity (11, 30). When neutrophils are at rest, most cell surface expressed Mac-1 is in a constrained conformation with the major ligand binding domain, the I domain (binding site for ICAM-1, iC3b, etc), buried within the protein’s complex three dimensioned structure. When neutrophils are activated by fMLP or phorbol myristate acetate (PMA) however, Mac-1 undergoes a conformational change that exposes the I domain and thus increases the receptor’s affinity for its ligands (31, 32). Accordingly, it is possible that Mac-1 expression or activation of its I domain is altered by the ITGAM variation. If so, this could explain the observed changes in neutrophil phagocytosis.
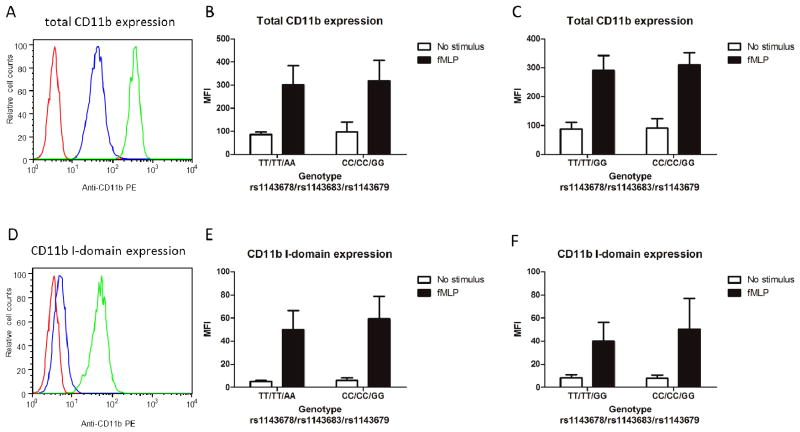
However, our results showed that the expression of total CD11b (assessed using the monoclonal antibody ICFR44), was not different in neutrophils from donors with variant ITGAM alleles compared to those from donors with common ITGAM alleles (Figure 3, A–C). Moreover, we also found that the activation of CD11b (assessed with the I-domain–specific monoclonal antibody CBRM1/5) did not differ by genotype (Figure 3, D–F). Based on these findings, we conclude that the observed differences in neutrophil phagocytosis associated with ITGAM variation (Figure 2) could not be attributed to altered expression of Mac-1 or to changes in its activation state.
Figure 3. CD11b expression and activation on neutrophils are not affected by ITGAM variation.
Total CD11b expression and I-domain activation were measured on neutrophils from genotyped (rs1143678/rs1143683/rs1143679) healthy donors, using flow cytometry. A and D, Representative histograms of total CD11b expression (A) and CD11b I-domain expression (D) are shown. Red line represents neutrophils incubated with isotype control antibody, blue line represents neutrophils incubated with CD11b antibody without stimulus, and green line represents neutrophils incubated with CD11b antibody after 15 minutes of stimulation with 5 × 10 9M fMLP. B, C, E, and F, Anti-CD11b monoclonal antibodies ICRF44 and CBRM1/5 were used to measure total CD11b expression (B and C) and CD11b I-domain expression (E and F), respectively, in fMLP-stimulated and unstimulated cells. All experiments were performed in a paired manner, with 4 different pairs of donors (donors 1–4 and donors 20–23 in B and E; donors 6–9 and donors 24–27 in C and F). Results are the mean ± SEM values of mean fluorescence intensity (MFI) in donors with the TT/TT/AA genotype (B and E) or the TT/TT/GG genotype (C and F) compared to donors with the CC/CC/GG genotype. PE = phycoerythrin.
Alteration of Fc receptor function by ITGAM variation
Previous studies using cells from patients with leukocyte adhesion deficiency (LAD; wherein CD18 is not expressed leading to no β2 integrin expression) or using transduced cells expressing various Fc receptors have shown that full Fc-receptor mediated phagocytosis requires Mac-1 (33, 34). Interestingly, SLE patients also have been reported to have reduced Fc receptor functions (35). Therefore, we investigated whether ITGAM variation might affect Fc receptor mediated phagocytosis of IgG antibody opsonized erythrocytes (EA).
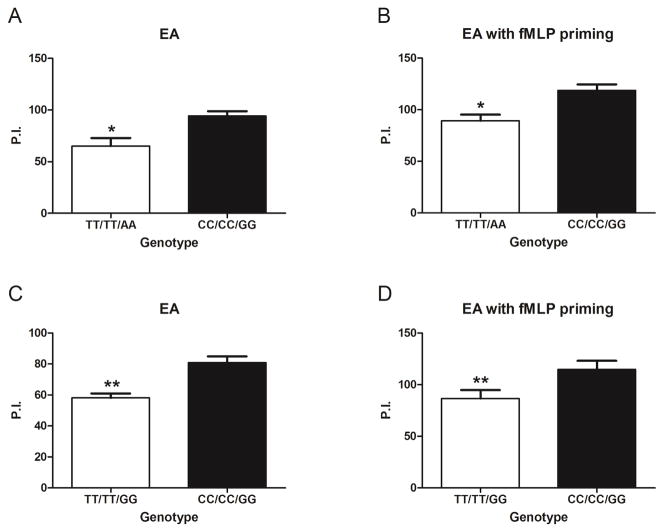
Surprisingly, we found that neutrophils with both the fully variant TT/TT/AA and partially variant TT/TT/GG ITGAM genotypes had significantly reduced EA phagocytosis compared to common CC/CC/GG controls, under both fMLP primed and unprimed conditions (Figure 4). These results are the first to show that SNP variation in ITGAM render neutrophils less able to phagocytose IgG coated particles via Fc receptors.
Figure 4. Non-synonymous variants in ITGAM alter FcγR mediated neutrophil phagocytosis.
Quantitative phagocytosis by neutrophils from genotyped (rs1143678/rs1143683/rs1143679) healthy donors was assessed using sheep erythrocyte antigen coated with rabbit anti-sheep erythrocyte IgG antibodies (EA). Neutrophils without priming (A and C) or after 10 minutes of priming with 10 8M fMLP (B and D) were used. All experiments were performed in a paired manner. The TT/TT/AA genotype was compared with the CC/CC/GG genotype (A and B) using 3 pairs of donors (donors 1–3 and donors 20–22). The TT/TT/GG genotype was compared with the CC/CC/GG genotype (C and D) using 5 pairs of donors (donors 6–10 and donors 24–28). Results are the mean ± SEM phagocytic index (P. I.) (the number of internalized probes per 100 neutrophils). * = P < 0.05; ** = P < 0.01 by Student's paired t-test
Neutrophil firm adhesion is altered by ITGAM variation
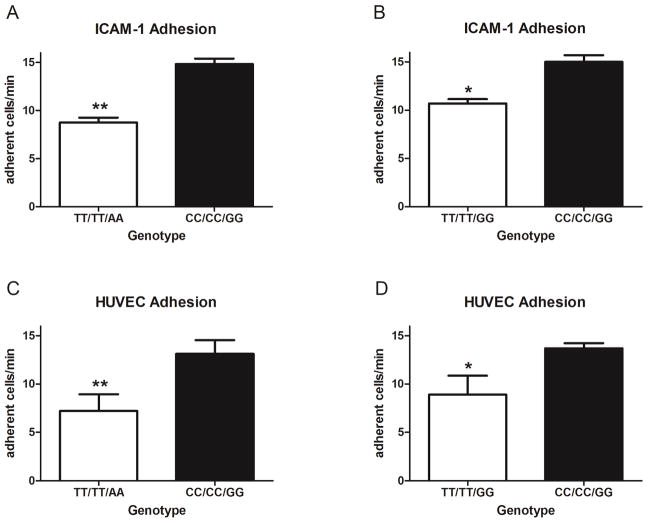
To investigate if ITGAM variation alters additional Mac-1 dependent functions that could increase susceptibility to SLE, we compared firm adhesion of neutrophils isolated from donors with different ITGAM genotypes. In the first series of experiments we used an in vitro flow chamber based assay to analyze neutrophil firm adhesion to the purified Mac-1 ligand ICAM-1. P-selectin was also included on the surface to initiate cell capture and facilitate cell rolling and subsequent firm adhesion to ICAM-1 (26). We found that neutrophil firm adherence was significantly reduced for both the fully variant TT/TT/AA and partially variant TT/TT/GG donors (Figure 5, A and B).
Figure 5. Non-synonymous ITGAM variants alter neutrophil firm adhesion under flow condition.
Firm adhesion under flow conditions was assessed using neutrophils from genotyped (rs1143678/rs1143683/rs1143679) healthy donors after 10 minutes of pretreatment with 10 8M fMLP, followed by incubation with intercellular adhesion molecule 1 (ICAM-1)/P-selectin (A and B) or human umbilical vein endothelial cells (HUVECs) (C and D) coupled in a flow chamber. Firm adhesion was defined as movement of the neutrophil a distance of <1 cell diameter for 5 seconds. The number of adherent cells/minute was calculated from a 4-minute video. All experiments were performed in a paired manner. For ICAM-1 adhesion, 3 different pairs of donors (donors 2, 3, 5, 20, 30, and 31 in A and donors 6–8, 21, 24, and 29 in B) were used. For HUVEC adhesion, 4 different pairs of donors (donors 1–4, 21, 23, 26, and 31 in C and donors 6–9, 23, 25, 26, and 28 in D) were used. Results are the mean ± SEM of adherent cells/minute from a minimum of 3 independent videos recorded for each donor. * = P < 0.05; ** = P < 0.01 by Student's paired t-test.
We next analyzed neutrophil adhesion to TNF-α-stimulated HUVECs. In this series of experiments, we observed the same genotype-associated reduction in neutrophil adhesion (Figure 5, C and D). These results demonstrate ITGAM variation have functional consequences in neutrophils, including the impairment of Mac-1–mediated firm adhesion to both purified and natural ligands under conditions of shear stress.
Independent inhibition of Mac-1 function on neutrophils by SNP rs1143679
Our analysis of donors with homozygous ITGAM variation demonstrates that the rs1143678/rs1143683 variants can independently alter Mac-1 functions in human primary neutrophils. To directly address the ability of rs1143679 variation to alter Mac-1 function, we studied neutrophils from donors heterozygous at rs1143679 but homozygous for the common alleles at rs1143678/rs1143683 (partially variant CC/CC/GA, see Figure 1). Neutrophils from these donors displayed significantly reduced phagocytosis of STZ compared to neutrophils from donors with the common ITGAM genotype (CC/CC/GG) (Figure 6A).
Figure 6. ITGAM SNP rs1143679 variant independently alters neutrophil phagocytosis.
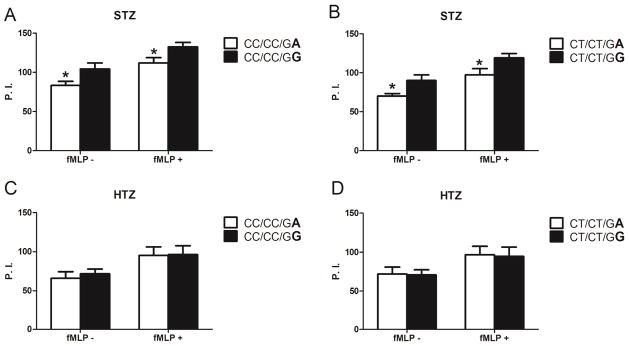
Quantitative phagocytosis by neutrophils from genotyped (rs1143678/rs1143683/rs1143679) healthy donors was assessed using serum-treated zymosan (STZ) (A and B) or heat-treated zymosan (HTZ) (C and D). Neutrophils without priming (fMLP ) or after 10 minutes of priming with 10 8M fMLP (fMLP+) were used. All experiments were performed in a paired manner. For comparisons of donors with the CC/CC/GA genotype and those with the CC/CC/GG genotype (A and C) or donors with the CT/CT/GA genotype and those with the CT/CT/GG genotype (B and D), 3 pairs of donors (donors 11–20, 23, and 26) were assessed. Variations are shown in boldface. Results are the mean ± SEM phagocytic index (P. I.) (number of internalized probes per 100 neutrophils). * = P < 0.005 by analysis of variance.
Further supporting a functional role of variation at rs1143679, we also observed significant reduction in neutrophil STZ phagocytosis in donors heterozygous for all three SNPs (partially variant CT/CT/GA, see Figure 1) compared to donors heterozygous at rs1143678/rs1143683 alone (partially variant CT/CT/GG, see Figure 1) (Figure 6B). In contrast, none of the ITGAM variant genotypes affected phagocytosis of HTZ (Figure 6, C and D) which is consistent with our prior results (Figure 2, C and F). These results demonstrate that SNP variation at rs1143679 has an independent impact on Mac-1 function in primary neutrophils.
Discussion
Our study, like others before it (21–23) demonstrates that ITGAM variation alters leukocyte phagocytosis of complement opsonized particles. In addition we have extended the observation that ITGAM variation has impact on other biologically important neutrophil functions, i.e., firm adhesion under shear stress and Fc receptor mediated phagocytosis. Importantly, we have shown that these functional alterations can be the consequence of multiple SLE associated non-synonymous SNPs, not just solely at rs1143679. Indeed, it is likely that these multiple ITGAM SNPs co-segregate as a haplotype and act in concert to affect Mac-1 mediated functions and thereby modify the risk of SLE development.
Among the SLE associated non-synonymous SNPs herein, SNPs rs1143679 and rs114378/rs1143683 independently alter Mac-1 mediated neutrophil functions. Our genotyping results suggest that rs1143679 is in strong LD with rs114378/rs1143683 and that 86% of rs1143679 variant allele carriers also carry an rs1143678 variant allele. This strong LD has been confirmed in another large cohort, which comprised >10,000 SLE cases and controls (Edberg JC: unpublished result). We acknowledge that ITGAM genetic association studies have shown different strengths of association between ITGAM SNPs and SLE in different ethnic groups, perhaps because the LD patterns and MAF values may differ across ethnicities. Nevertheless, our results should serve as a reminder that a full understanding of the reasons for the genetic association between the ITGAM locus and SLE will require studies of multiple ITGAM SNPs, not single ones.
Like others have shown using human monocytes and transfected cell lines (21–23), we demonstrated that in neutrophils neither total Mac-1 receptor expression nor activation of its major ligand binding site I domain is affected by ITGAM variation. Thus, Mac-1 expression or I domain exposure is unlikely to be the mechanism by which neutrophil phagocytosis and adhesion is reduced by ITGAM variation. The caveat is that mAb CBRM1/5 may not detect subtle changes in the I-domain which might nevertheless occur as a consequence of ITGAM variation. This is especially true for rs1143679 SNP, which is located in the extracellular domain of CD11b and proximal to the I domain. If this SNP does alter I-domain structure, it is also likely to affect ligand binding affinity (3).
In contrast, the SNP rs1143683 is not as likely to alter Mac-1 structure, since this SNP introduces a conservative amino acid change (Ala-Val) in the extracellular calf-1 region of CD11b. Furthermore although cation binding sites in the thigh/calf region of integrins are important for its proper activation (28), SNP rs114383 is not within those sites. The association between SNP rs1143683 and SLE thus likely reflects its perfect LD with SNP rs1143678.
Importantly, SNP rs1143678 resides in the cytoplasmic tail of CD11b, which is known to be involved in Mac-1 signaling (36) and is likely to be important in integrin-cytoskeleton interactions and, thus, could play a role in a variety of integrin functions (37–39). The Pro to Ser amino acid change introduced by SNP rs1143678 could potentially affect the structure of the cytoplasmic tail and, thus impact its signaling or interaction with cytoskeleton proteins. Indeed, our results demonstrate that there is a functional consequence of variation at SNP rs1143678 and that this variation alone is sufficient to significantly inhibit neutrophil adhesion, Mac-1 mediated phagocytosis and Fc receptor mediated phagocytosis. This SNP has been overlooked or discounted in previous ITGAM functional studies. The fact that both rs1143679 and rs1143678/rs1143683 appear to independently alter Mac-1 functions suggests multiple mechanisms of functional alterations are involved. All of these hypotheses need further verification.
In this study, we demonstrated that the function of neutrophils from healthy donors carrying SLE associated ITGAM risk alleles is impaired. This finding is consistent with a previous study using CD11b deficient mice, the results of which showed that loss of Mac-1 increased autoimmune disease severity in the MRL/MpJ-Faslpr strain (7). Our data also suggest that the reduction in Fc receptor function reported in SLE patients (35) could be partly due to ITGAM variants, thus extending the importance of these genetic variants to both complement and Fc receptor function. Although our current study does not address the mechanism by which ITGAM variants alter Fc receptor function, prior work has demonstrated physical and functional interactions between FcγRIIIb and Mac-1 (25, 40). Moreover, our study does not address the physiological ligand(s) that interact with Mac-1 to manifest diminished receptor function related to the SLE associated ITGAM variants. Despite these caveats, our results demonstrate that a deficiency in CD11b function is associated with development of an autoimmune phenotype.
The ITGAM variation associated reduction in Mac-1 mediated phagocytosis (Figure 2 and results available from the corresponding author upon request) (21, 22) and Fc receptor mediated phagocytosis (Figure 3) (35) could contribute to altered immune complex clearance and deposition. Indeed, impaired clearance of immune complexes has been observed in patients with SLE (35, 41–43), and it is tempting to speculate that ITGAM variation may provide one mechanism for this deficiency. It is also possible that reduced adhesion resulting from ITGAM variation could affect normal leukocyte trafficking in vivo and thus contribute to SLE pathogenesis. Other Mac-1 functions not probed in our study could also be altered by ITGAM variants and contribute to SLE pathogenesis (20).
To our knowledge our study is the first to focus on the biological impact of multiple disease-associated genetic variants in the ITGAM locus on neutrophils, a cell type that plays an important role in SLE (44). Although prior studies have investigated the impact of ITGAM variation on Mac-1 mediated adhesion, ours is the first to examine this question in terms of the impact on the more physiologically common biologic function of neutrophil adhesion, under conditions of shear stress to both purified ligand (ICAM-1) and endothelial cells. Future functional studies of the ITGAM locus are needed to discern for the potential role(s) of yet other non-synonymous variants at this locus.
Supplementary Material
Acknowledgments
We thank Dr. Robert P. Kimberly for helpful discussions and support. We also thank Dr. Carl Langefeld for advice on statistical analysis and Mark McCrory, Deborrah McDuffie and Ellen Sowell for technical assistance. We also thank Dr. Susan Bellis for helpful discussions.
This work was supported by grants from the NIH (P01-AR49084, 5P30-AR048311, R21-AR058864, R21-DA026956 and UL1TR000165 from the National Center for Advancing Translational Sciences (NCATS)) and the Lupus Research Institute.
References
- 1.Harley JB, Alarcón-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, et al. International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN) Genome–wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–10. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hom G, Graham RR, Modrek B, Taylor KE, Ortmann W, Garnier S, et al. Association of systemic lupus erythematosus with C8orf13–BLK and ITGAM–ITGAX. N Engl J Med. 2008;358:900–9. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 3.Nath SK, Han S, Kim-Howard X, Kelly JA, Viswanathan P, Gilkeson GS, et al. A nonsynonymous functional variant in integrin–alpha (M) (encoded by ITGAM) is associated with systemic lupus erythematosus. Nat Genet. 2008;40:152–4. doi: 10.1038/ng.71. [DOI] [PubMed] [Google Scholar]
- 4.Han S, Kim-Howard X, Deshmukh H, Kamatani Y, Viswanathan P, Guthridge JM, et al. Evaluation of imputation-based association in and around the integrin-alpha-M (ITGAM) gene and replication of robust association between a non-synonymous functional variant within ITGAM and systemic lupus erythematosus (SLE) Hum Mol Genet. 2009;18:1171–80. doi: 10.1093/hmg/ddp007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang W, Zhao M, Hirankarn N, Lau CS, Mok CC, Chan TM, et al. ITGAM is associated with disease susceptibility and renal nephritis of systemic lupus erythematosus in Hong Kong Chinese and Thai. Hum Mol Genet. 2009;18:2063–70. doi: 10.1093/hmg/ddp118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Solovjov DA, Pluskota E, Plow EF. Distinct roles for the alpha and beta subunits in the functions of integrin alphaMbeta2. J Biol Chem. 2005;280:1336–45. doi: 10.1074/jbc.M406968200. [DOI] [PubMed] [Google Scholar]
- 7.Kevil CG, Hicks MJ, He X, Zhang J, Ballantyne CM, Raman C, et al. Loss of LFA-1, but not Mac-1, protects MRL/MpJ-Fas(lpr) mice from autoimmune disease. Am J Pathol. 2004;165:609–16. doi: 10.1016/S0002-9440(10)63325-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harlan JM, Winn RK, Vedder NB, Doerschuk CM, Rice CL. Adhesion: Its Role in Inflammatory Disease. New York, USA: W. H. Freeman and Company; 1992. [Google Scholar]
- 9.Lejtenyi D, Osmond DG, Miller SC. Natural killer cells and B lymphocytes in L-selectin and Mac-1/LFA-1 knockout mice: marker-dependent, but not cell lineage-dependent changes in the spleen and bone marrow. Immunobiology. 2003;207:129–35. doi: 10.1078/0171-2985-00220. [DOI] [PubMed] [Google Scholar]
- 10.Ross GD, Vetvicka V. CR3 (CD11b, CD18): a phagocyte and NK cell membrane receptor with multiple ligand specifi cities and functions. Clin Exp Immunol. 1993;92:181–4. doi: 10.1111/j.1365-2249.1993.tb03377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo BH, Carman CV, Springer TA. Structure Basis of Integrin Regulation and Signaling. Annu Rev Immunol. 2007;25:619–47. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hentzen ER, Neelamegham S, Kansas GS, Benanti JA, McIntire LV, Smith CW, et al. Sequential binding of CD11a/CD18 and CD11b/CD18 defines neutrophil capture and stable adhesion to intercellular adhesion molecule-1. Blood. 2000;95:911–20. [PubMed] [Google Scholar]
- 13.Zen K, Guo YL, Li LM, Bian Z, Zhang CY, Liu Y. Cleavage of the CD11b extracellular domain by the leukocyte serprocidins is critical for neutrophil detachment during chemotaxis. Blood. 2011;117:4885–94. doi: 10.1182/blood-2010-05-287722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayadas TN, Cullere X. Neutrophil beta2 integrins: moderators of life or death decisions. Trends Immunol. 2005;7:388–95. doi: 10.1016/j.it.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 15.Le Cabec V, Carreno S, Moisand A, Bordier C, Maridonneau-Parini I. Complement receptor 3 (CD11b/CD18) mediates type I and type II phagocytosis during nonopsonic and opsonic phagocytosis, respectively. J Immunol. 2002;169:2003–9. doi: 10.4049/jimmunol.169.4.2003. [DOI] [PubMed] [Google Scholar]
- 16.Balsam LB, Liang TW, Parkos CA. Functional mapping of CD11b/CD18 epitopes important in neutrophil-epithelial interactions: a central role of the I domain. J Immunol. 1998;160:5058–65. [PubMed] [Google Scholar]
- 17.Ross GD, Cain JA, Lachmann PJ. Membrane complement receptor type three (CR3) has lectin-like properties analogous to bovine conglutinin and functions as a receptor for zymosan and rabbit erythrocytes as well as a receptor for iC3b. J Immunol. 1985;134:3307–15. [PubMed] [Google Scholar]
- 18.Ortiz-Stern A, Rosales C. Cross-talk between Fc receptors and integrins. Immunol Lett. 2003;90:137–43. doi: 10.1016/j.imlet.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 19.Han C, Jin J, Xu S, Liu H, Li N, Cao X. Integrin CD11b negatively regulates TLR-triggered inflammatory responses by activating Syk and promoting degradation of MyD88 and TRIF via Cbl-b. Nat Immunol. 2010;11:734–42. doi: 10.1038/ni.1908. [DOI] [PubMed] [Google Scholar]
- 20.Reed JH, Jain M, Lee K, Kandimalla ER, Faridi MH, Buyon JP, et al. Complement receptor 3 influences Toll-like receptor 7/8 dependent inflammation: implications for autoimmune diseases characterized by antibody reactivity to ribonucleoproteins. J Biol Chem. 2013 doi: 10.1074/jbc.M112.403303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacPherson M, Lek HS, Prescott A, Fagerholm SC. A systemic lupus erythematosus-associated R77H substitution in the CD11b chain of the Mac-1 integrin compromises leukocyte adhesion and phagocytosis. J Biol Chem. 2011;286:17303–10. doi: 10.1074/jbc.M110.182998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhodes B, Furnrohr BG, Roberts AL, Tzircotis G, Schett G, Spector TD, et al. The rs1143679 (R77H) lupus associated variant of ITGAM (CD11b) impairs complement receptor 3 mediated functions in human monocytes. Ann Rheum Dis. 2012;71:2028–34. doi: 10.1136/annrheumdis-2012-201390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosetti F, Tsuboi N, Chen K, Nishi H, Ernandez T, Sethi S, et al. Human lupus serum induces neutrophil-mediated organ damage in mice that is enabled by Mac-1 deficiency. J Immunol. 2012;189:3714–23. doi: 10.4049/jimmunol.1201594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelley JM, Monach PA, Ji C, Zhou Y, Wu J, Tanaka S, et al. IgA and IgG antineutrophil cytoplasmic antibody engagement of Fc receptor genetic variants influences granulomatosis with polyangiitis. Proc Natl Acad Sci U S A. 2011;108:20736–41. doi: 10.1073/pnas.1109227109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edberg JC, Kimberly RP. Modulation of Fc gamma and complement receptor function by the glycosyl-phosphatidylinositol-anchored form of Fc gamma RIII. J Immunol. 1994;152:5826–35. [PubMed] [Google Scholar]
- 26.Ni N, Kevil CG, Bullard DC, Kucik DF. Avidity modulation activates adhesion under flow and requires cooperativity among adhesion receptors. Biophysical Journal. 2003;85:4122–33. doi: 10.1016/S0006-3495(03)74824-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oxvig C, Lu C, Springer TA. Conformational changes in tertiary structure near the ligand binding site of an integrin I domain. Proc Natl Acad Sci USA. 1999;96:2215–20. doi: 10.1073/pnas.96.5.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie C, Shimaoka M, Xiao T, Schwab P, Klickstein LB, Springer TA. The integrin alpha-subunit leg extends at a Ca2+-dependent epitope in the thigh/genu interface upon activation. Proc Natl Acad Sci U S A. 2004;101:15422–7. doi: 10.1073/pnas.0406680101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor PR, Brown GD, Reid DM, Willment JA, Martinez-Pomares L, Gordon S, et al. The beta-glucan receptor, dectin-1, is predominantly expressed on the surface of cells of the monocyte/macrophage and neutrophil lineages. J Immunol. 2002;169:3876–82. doi: 10.4049/jimmunol.169.7.3876. [DOI] [PubMed] [Google Scholar]
- 30.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2003;110:673–87. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 31.Lefort CT, Hyun YM, Schultz JB, Law FY, Waugh RE, Knauf PA, et al. Outside-In Signal Transmission by Conformational Changes in Integrin Mac-1. J Immunol. 2009;183:6460–8. doi: 10.4049/jimmunol.0900983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimaoka M, Xiao T, Liu JH, Yang Y, Dong Y, Jun CD, et al. Structures of the alpha L I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation. Cell. 2003;112:99–111. doi: 10.1016/s0092-8674(02)01257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krauss JC, Poo H, Xue W, Mayo-Bond L, Todd RF, Petty HR. Reconstitution of antibody-dependent phagocytosis in fibroblasts expressing Fcg receptor IIIb and the complement receptor type 3. J Immunol. 1994;153:1769–77. [PubMed] [Google Scholar]
- 34.Gresham HD, Graham IL, Anderson DC, Brown EJ. Leukocyte adhesion-deficient neutrophils fail to amplify phagocytic function in response to stimulation. J Clin Invest. 1991;88:588–97. doi: 10.1172/JCI115343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelley JM, Edberg JC, Kimberly RP. Pathways: Strategies for susceptibility genes in SLE. Autoimmun Rev. 2010;9:473–6. doi: 10.1016/j.autrev.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fagerholm SC, Varis M, Stefanidakis M, Hilden TJ, Gahmberg CG. Alpha-Chain phosphorylation of the human leukocyte CD11b/CD18 (Mac-1) integrin is pivotal for integrin activation to bind ICAMs and leukocyte extravasation. Blood. 2006;108:3379–86. doi: 10.1182/blood-2006-03-013557. [DOI] [PubMed] [Google Scholar]
- 37.Kucik DF. Rearrangement of integrins in avidity regulation by leukocytes. Immunologic Research. 2002;26:199–206. doi: 10.1385/IR:26:1-3:199. [DOI] [PubMed] [Google Scholar]
- 38.Yu T, Wu X, Gupta KB, Kucik DF. Affinity, lateral mobility, and clustering contribute independently to beta 2-integrin-mediated adhesion. Am J Physiol Cell Physiol. 2010;299:C399–10. doi: 10.1152/ajpcell.00039.2009. [DOI] [PubMed] [Google Scholar]
- 39.Wu X, Yu T, Bullard DC, Kucik DF. SDF-1alpha (CXCL12) regulation of lateral mobility contributes to activation of LFA-1 adhesion. Am J Physiol Cell Physiol. 2012;303:C666–72. doi: 10.1152/ajpcell.00190.2012. [DOI] [PubMed] [Google Scholar]
- 40.Todd RF, 3rd, Petty HR. Beta 2 (CD11/CD18) integrins can serve as signaling partners for other leukocyte receptors. J Lab Clin Med. 1997;129:492–8. doi: 10.1016/s0022-2143(97)90003-2. [DOI] [PubMed] [Google Scholar]
- 41.Hamburger MI, Lawley TJ, Kimberly RP, Plotz PH, Frank MM. A serial study of splenic reticuloendothelial system Fc receptor functional activity in systemic lupus erythematosus. Arthritis Rheum. 1982;25:48–54. doi: 10.1002/art.1780250108. [DOI] [PubMed] [Google Scholar]
- 42.Parris TM, Kimberly RP, Inman RD, McDougal JS, Gibofsky A, Christian CL. Defective Fc receptor-mediated function of the mononuclear phagocyte system in lupus nephritis. Ann Intern Med. 1982;97:526–32. doi: 10.7326/0003-4819-97-4-526. [DOI] [PubMed] [Google Scholar]
- 43.Hebert LA, Cosio G. The erythrocyte-immune complex-glomerulonephritis connection in man. Kidney Int. 1987;31:877–85. doi: 10.1038/ki.1987.81. [DOI] [PubMed] [Google Scholar]
- 44.Warde N. Autoimmunity: The role of neutrophils in SLE: untangling the NET. Nat Rev Rheumatol. 2011;7:252. doi: 10.1038/nrrheum.2011.41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.