Abstract
Sudden cardiac death (SCD), which is primarily caused by lethal heart disorders resulting in structural and arrhythmogenic abnormalities, is one of the prevalent modes of death in most developed countries. Myocardial ischemia, mainly due to coronary artery disease, is the most common type of heart disease leading to SCD. However, postmortem diagnosis of SCD is frequently complicated by obscure histological evidence. Here, we show that certain mRNA species, namely those encoding hemoglobin A1/2 and B (Hba1/2 and Hbb, respectively) as well as pyruvate dehydrogenase kinase 4 (Pdk4), exhibit distinct postmortem expression patterns in the left ventricular free wall of SCD subjects when compared with their expression patterns in the corresponding tissues from control subjects with non-cardiac causes of death. Hba1/2 and Hbb mRNA expression levels were higher in ischemic SCD cases with acute myocardial infarction or ischemic heart disease without recent infarction, and even in cardiac death subjects without apparent pathological signs of heart injuries, than control subjects. By contrast, Pdk4 mRNA was expressed at lower levels in SCD subjects. In conclusion, we found that altered myocardial Hba1/2, Hbb, and Pdk4 mRNA expression patterns can be employed as molecular signatures of fatal cardiac dysfunction to forensically implicate SCD as the primary cause of death.
Keywords: heart failure, hemoglobin, postmortem RNA expression, pyruvate dehydrogenase kinase, sudden cardiac death
INTRODUCTION
Sudden cardiac death (SCD) is a prevalent mode of death in most developed countries, with an estimated overall incidence of 1 to 2 per 1000 population per year. Over the last decade, significant advances have been made in understanding the clinical and genetic basis of SCD, which is primarily caused by heart disorders with structural and arrhythmogenic abnormalities. In forensic cases, ischemic heart disease, caused mainly by coronary artery disease, is the leading cause of SCD, accounting for more than half of all cases. Both acute myocardial infarction and ischemic coronary disease without recent infarction are major causes of ischemic SCD. Dilated and hypertrophic cardiomyopathies as well as congenital heart diseases such as congenital long QT syndrome, catecholaminergic polymorphic ventricular tachycardia, and Brugada syndrome can also result in fatal arrhythmia and cardiac death. However, forensic diagnosis of cardiac dysfunction associated with the sudden onset of arrhythmia or heart failure in autopsy samples is frequently complicated because it is difficult to interpret the severity of or determine the difference between preexisting and terminal dysfunctions based on conventional morphological findings (Oliva et al., 2011).
Postmortem RNA expression profiling, which is an emerging field of forensic medical science, may provide alternative tools for forensic diagnosis of SCD in autopsy cases. Indeed, extensive studies are currently been carried out to address key issues in forensic science regarding the cause and process of death, postmortem interval, and pathophysiological conditions of diseases and injuries (Maeda et al., 2010; Vennemann and Koppelkamm, 2010a). Distinct RNA compositions in autopsy materials have been proposed to imply certain causes and circumstances of death (Chen et al., 2012; Chung et al., 2012; Ikematsu et al., 2005; Miyazato et al., 2012; Takahashi et al., 2009; Zhao et al., 2006; 2009; Zhu et al., 2008). Notably, Maeda and colleagues proposed that left ventricular (LV) expression profiles of several mRNA species encoding hypoxia-inducible factor 1α and natriuretic peptides were relatively higher in autopsy materials of cardiac death subjects, particularly those from subjects with ischemic heart injuries (Chen et al., 2012; Zhu et al., 2008). However, the application of molecular pathology to diagnose unexpected cardiac death is still challenging because of the lack of well-established forensic RNA biomarkers of fatal cardiac dysfunction.
Our goal in this study was to identify novel molecular signatures of cardiac dysfunction after death for the forensic diagnosis of SCD. For this purpose, we explored the pattern of expression of differentially expressed genes (DEGs) associated with both ischemic and non-ischemic SCD and compared it with patterns found in non-cardiac death control subjects by examining genome-wide postmortem RNA expression in autopsy materials from the LV free wall.
MATERIALS AND METHODS
Subjects and tissue preparation
Non-infarcted myocardial tissue from the LV free wall was collected from selected medicolegal autopsy cases within 5 days postmortem with the help of the National Forensic Service of Korea. The causes of death examined in the present study were traumatic death without injuries to the heart (TD), ischemic heart disease (IH) with or without old infarction, acute myocardial infarction (MI), and non-ischemic SCD (NI), as classified by routine morphological, biochemical, and toxicological findings (Table 1). Dissected tissues were treated immediately with RNAlater™ solution (QIAGEN GmbH, Germany) according to the manufacturer’s instructions and stored at −70°C until use.
Table 1.
Subject information
| Group | Size/Sex | Age (years)
|
BMI (mean ± SEM) | Note |
|---|---|---|---|---|
| Range (mean ± SEM) | ||||
| TD | n = 16/M | 25–73 (45.47 ± 2.84) | 23.69 ± 0.85 | No injuries in cardiac tissues |
| IH | n = 19/M | 40–69 (52.68 ± 1.44) | 26.91 ± 2.49 | Old infarction in some cases (10/19) |
| MI | n = 15/M | 40–63 (54.60 ± 2.00) | 25.22 ± 0.53 | Calcification/thrombosis (15/15) |
| NI | n = 20/M | 22–72 (47.10 ± 3.23) | 23.27 ± 0.86 | No ischemic pathology |
TD, Traumatic death; IH, Ischemic heart disease without recent infarction; MI, Acute myocardial infarction; NI, Non-ischemic sudden cardiac death
RNA isolation and microarray analyses
Total RNA was isolated from 20–50 mg tissue using the RNeasy mini kit (QIAGEN) and then quantified by spectrophotometry. Gene expression profiling by microarray analysis was performed to explore SCD-associated human RNA transcripts as described previously with minor modifications (Chung et al., 2012). Group-pooled cardiac RNA samples representing TD, CC, MI, and NI cases were subjected to microarray analysis. Two hundred nanogram aliquots of total RNA were amplified and labeled. Five micrograms of labeled cDNA was then hybridized to GeneChip Human Gene 1.0 ST arrays (Affymetrix Inc., USA), and the arrays were then washed, stained, and scanned according to the protocol described in the Affymetrix GeneChip Expression Analysis Manual (Affymetrix Inc.). Signal intensities of each gene of interest were then normalized to those of glyceraldehyde 3-phosphate dehydrogenase (Gapdh).
Real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
qRT-PCR reactions were performed as described previously with minor modifications (Chung et al., 2012). Relative mRNA levels were deduced according to a previous report (Pfaffl et al., 2002). Briefly, 1 μg of each total RNA sample was reverse-transcribed using MMLV reverse transcriptase (Promega, USA) by the random priming method. Then, aliquots of cDNA were subjected to quantitative real-time PCR in the presence of SYBR Green I (Life Technologies Corp., USA). Reference cDNA was prepared by pooling aliquots of each sample and its three-fold serial dilutions were utilized to calibrate PCR reactions as well as to construct a regression curve for calculating PCR efficiencies of each cycle. Hemoglobin α1/α2 (Hba1/2), hemoglobin β (Hbb), and pyruvate dehydrogenase kinase 4 (Pdk4) mRNA expression levels were normalized to those of Gapdh. Because Hba1 and Hba2 mRNA share identical coding sequences, these two isoforms were not distinguished in the present study. All PCR products were cloned into the pGEM-T vector (Promega), and their sequence identities were confirmed by chain termination sequencing. Primer sequences used for real-time qRT-PCR were as follows: Hba1/2 up, 5′- TTA AGG GCC ACG GCA AGA AG-3′; Hba1/2 dn, 5′- GCA GTG GCT TAG GAG CTT GA-3′; Hbb up, 5′- GTG AAC GTG GAT GAA GTT GG-3′; Hbb dn, 5′- ACT TTC TTG CCA TGA GCC TT-3′; Pdk4 up, 5′-GCT GAT GAA CCA GCA CAT TC-3′; Pdk4 dn, 5′- CAC TCA AAG GCA TCT TGG AC-3′; Gapdh up, 5′- ATG TTC GTC ATG GGT GTG AA-3′; Gapdh dn, 5′- GGT GCT AAG CAG TTG GTG GT-3′. PCR efficiencies (E) for each set of primers from three independent measurements were as follows (in mean ± SEM): EHba1/2 = 2.0526 ± 0.0012; EHbb = 2.0349 ± 0.0101; EPdk4 = 2.0827 ± 0.0053; EGapdh = 2.0557 ± 0.0069
Statistical analysis
Data were statistically evaluated by Student’s t-test or one-way analysis of variance (ANOVA) followed by post-hoc comparison by the Newman-Keuls test. Statistical significance was set at p < 0.05.
RESULTS
Cardiac and coronary pathology
We first examined pathological features of the subjects used in the present study to justify our medicolegal classification of the cases. In comparison with TD as controls, SCD cases showed a significant increment in heart weights (Fig. 1A; F(3, 61) = 9.21, p < 0.01 by one-way ANOVA; p < 0.01 for IH and MI, and p < 0.05 for NI vs. TD by post-hoc comparison) as well as LV wall thickening (Fig. 1B; F(3, 51) = 5.39, p < 0.01 by ANOVA; p < 0.01 for IH, and p < 0.05 for MI and NI vs. TD). SCD subjects with ischemic heart injuries (IH and MI) exhibited more severe cardiac hypertrophy features than NI cases. Three SCD groups were clearly distinguishable based on the extent of main occlusions in the left anterior descending artery (LAD; Fig. 1C) and right coronary artery (RCA; Fig. 1D) (for LAD, F(3, 63) = 61.07, p < 0.01 by ANOVA and p < 0.01 for IH/MI vs. TD/NI by post-hoc comparison; for RCA, F(3, 63) = 27.66, p < 0.01 by ANOVA and p < 0.01 for IH/MI vs. TD/NI). Moderate coronary stenosis was found in IH subjects (60.26 ± 7.14% for LAD; 52.11 ± 5.55% for RCA), while severe or complete blockade was observed in the MI cases (93.33 ± 1.52% for LAD, p < 0.01 for IH vs. MI; 81.67 ± 7.77% for RCA, p < 0.01 for IH vs. MI). In contrast to SCD subjects with ischemic heart injuries, the extent of LAD and RCA occlusion in NI subjects was essentially the same as that observed in controls (usually less than 20%).
Fig. 1.
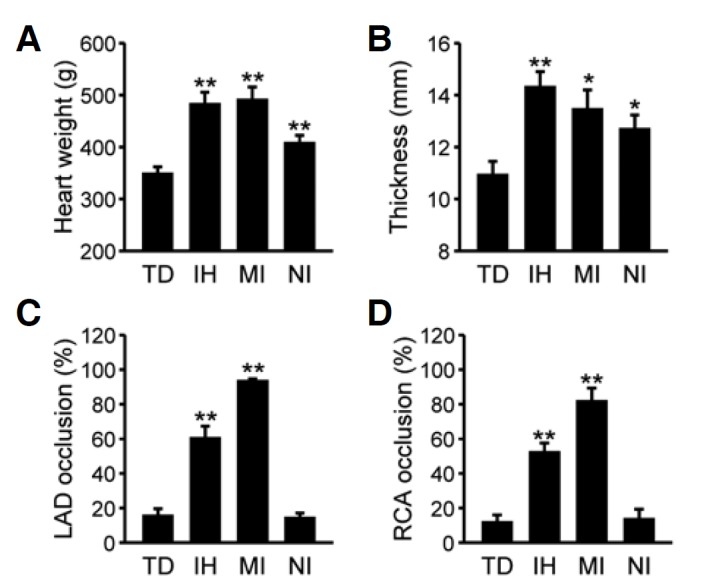
Cardiac and coronary features according to different causes of death. Heart weight (A) and the thickness of the left ventricular free-wall (B) were measured in subjects who died due to traumatic death (TD), ischemic heart disease (IH), acute myocardial infarction (MI), or non-ischemic sudden cardiac death (NI), and are expressed as means ± SEM. Extent of occlusion at the site where a major stenosis was found in the left anterior descending arteries (LCA; C) or the right coronary arteries (RCA; D) is expressed as the mean ± SEM (%). Data were statistically evaluated by one-way ANOVA followed by the Newman-Keuls test (*p < 0.05 and ** p < 0.01 vs. TD).
Identification of SCD-associated differentially expressed genes (DEGs)
To identify mRNA species associated with SCD, we carried out microarray analysis on postmortem RNA samples derived from LV myocardial tissues. In our initial approach, we explored individual genes differentially expressed between controls (TD) and SCD subjects categorized into the three subgroups of IH, MI, and NI. Out of 13,288 RNA species that showed differential expression between control and SCD tissues, 85 (IH), 517 (MI), and 514 (NI) DEGs showed a more than 1.5-fold difference in expression compared with the TD controls. Among these, eight gene transcripts were common to all subgroups of SCD (Table 2). mRNA expression levels of hemoglobin α1/α2 (Hba1/2) and hemoglobin β (Hbb), which are components of the adult hemoglobin tetramer, were higher in all SCD cases than controls, whereas several gene transcripts such as pyruvate dehydrogenase kinase 4 (Pdk4), nuclear receptor 4, subgroup A, member 2 (Nr4a2), and ficolin 3 (Fcn3) showed lower expression in SCD cases. Considering relative expression levels (expressed as % of Gapdh as shown in Table 2) as well as fold-differences between SCD and control cases, we focused mainly on Hba1/2, Hbb, and Pdk4 mRNAs to examine whether alterations in levels of these mRNAs in individual autopsy materials could suggest fatal cardiac dysfunction leading to SCD.
Table 2.
Differentially expressed genes
| Gene symbol | Gene accession | Fold change
|
Expression levels** (% of GAPDH) | ||
|---|---|---|---|---|---|
| IH | MI | NI | |||
| Hba1/2* | NM_000558/NM_000517 | 1.84 | 2.31 | 2.19 | 57.47 |
| Hbb | NM_000518 | 1.86 | 2.25 | 2.24 | 75.53 |
| Areg | NM_001657 | −2.29 | −2.48 | −1.99 | 1.33 |
| B3gnt5 | NM_032047 | −2.94 | −1.68 | −2.09 | 1.72 |
| Fcn3 | NM_003665 | −2.01 | −2.12 | −1.83 | 3.32 |
| Nr4a2 | NM_006186 | −3.01 | −1.64 | −2.72 | 3.51 |
| Pdk4 | NM_002612 | −2.92 | −3.06 | −6.80 | 21.64 |
Hba1 and Hba2 are considered to be as single gene here because they have identical coding sequences and can therefore not be distinguished based on microarray or RT-PCR analyses.
Average expression levels of the four groups for the indicated genes relative to that of Gapdh.
Hba1/2, Hbb, and Pdk4 mRNA profiles in individual subjects with known causes of death
In the next set of experiments, we compared cardiac Hba1/2, Hbb, and Pdk4 mRNA profiles in individual subjects with three different modes of SCD to those in TD cases using qRT-PCR results. Hba1/2 and Hbb mRNA levels tended to be higher in all SCD-related groups than in the TD group by approximately 4-fold (Fig. 2A; for Hba1/2: F(3, 66) = 2.33, p = 0.082 and for Hbb: F(3, 66) = 2.35, p = 0.081 by ANOVA; p < 0.05 vs. TD by Student’s t-test for each SCD-related group). In contrast, Pdk4 mRNA levels were significantly lower in the SCD-related samples (F(3, 66) = 4.60, p < 0.01 by ANOVA; p < 0.05 for TD vs. NI and p < 0.01 vs. TD for IH or MI cases by Neuman-Keuls post-hoc comparison). Although these mRNA species exhibited group differences according to cause of death, large variations within groups were found. Box-and-whisker diagrams for differentially expressed mRNA transcripts show that more than one-half of measured values in a group overlapped with values in other groups, indicating poor discrimination (Fig. 2B). In contrast to myocardial tissues, Hba1/2, Hbb, and Pdk4 mRNA levels in the occipital cortices of the brain were not different between TD and SCD subjects (Fig. 3). Brain tissues are known to be highly vulnerable to hypoxic/anoxic states leading to cellular stress responses and metabolic disturbances during the agonal stage as well as the acute postmortem period (Franz et al., 2005). Therefore, our results strongly suggest that SCD-related alterations in Hba1/2, Hbb, and Pdk4 mRNA expression reflect myocardial dysfunction rather than cellular damage during agonal and/or postmortem periods.
Fig. 2.
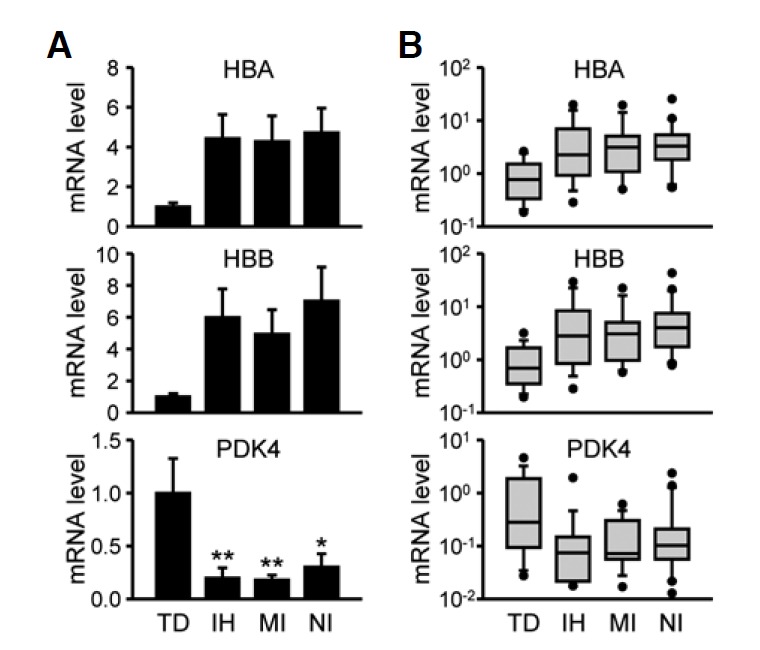
Hba1/2, Hbb, and Pdk4 mRNA expression in the left ventricular free wall in association with different causes of death. Hba1/2, Hbb, and Pdk4 mRNA expression profiles were quantitatively analyzed by real-time qRT-PCR in subjects who died due to traumatic death (TD), ischemic heart disease (IH), acute myocardial infarction (MI), or non-ischemic sudden cardiac death (NI). (A) Data were normalized to Gapdh mRNA levels and expressed as the mean ± SEM in arbitrary units (A.U.), where the mean values of the TD group were set to 1. Data were statistically evaluated by one-way ANOVA followed by the Newman-Keuls test (*p < 0.05 and **p < 0.01 vs. TD). (B) Relative mRNA levels were expressed by box-and-whisker diagrams with log scales. Median values are indicated by the lines in the middle of the boxes.
Fig. 3.
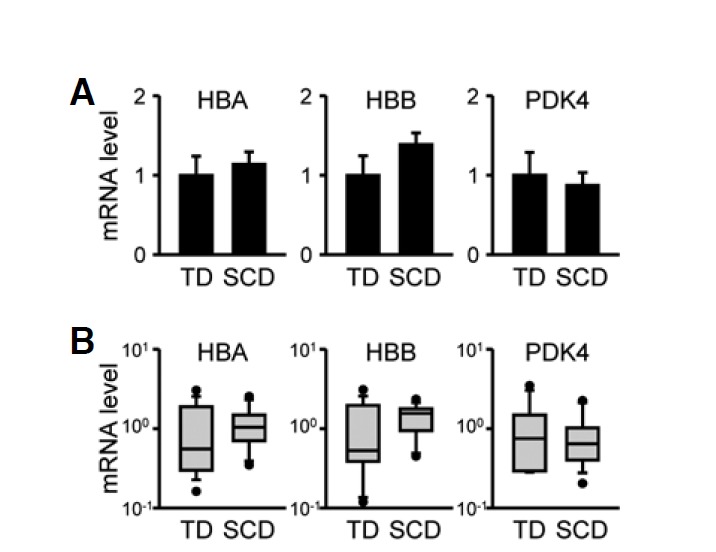
Hba1/2, Hbb, and Pdk4 mRNA expression in the occipital lobes of cerebral cortices. Hba1/2, Hbb, and Pdk4 mRNA expression profiles were analyzed quantitatively by real-time qRT-PCR in subjects who died due to traumatic death (TD) or sudden cardiac death (SCD) including both ischemic and non-ischemic heart injuries. (A) Data were normalized to Gapdh mRNA levels and expressed as the mean ± SEM in arbitrary units (A.U.), where the mean values of the TD group were set to 1. (B) Relative mRNA levels are shown in box-and-whisker diagrams with log scales. Median values are indicated by the lines in the middle of the boxes.
Ratios of Hba1/2 or Hbb mRNA levels relative to those of Pdk4 as potential forensic biomarkers
Despite the group differences in Hba1/2, Hbb, and Pdk4 mRNA expression depending on cause of death, large variations within the group were found in all selected genes as shown in Fig. 2B. These features suggest that Hba1/2, Hbb, or Pdk4 mRNA expression relative to that of a housekeeping gene, such as Gapdh employed in the present study, may not singly provide practical information by itself. However, concurrent alterations in the relative composition of the marker mRNA species could be used as a valuable index to better discriminate among causes of death, as we demonstrated in our previous study of mRNA transcripts encoding heat shock proteins (Chung et al., 2012). To test this idea, we bi-directionally plotted relative expression levels of Hba1/2 or Hbb mRNA normalized by Gapdh in relation to those of Pdk4. All SCD-related groups occupied positions distinct from those of TD subjects in the plot (Fig. 4A). We therefore compared ratios of Hba1/2 or Hbb mRNA abundance to those of Pdk4. As shown in Fig. 4B, the relative ratio of Hba1/2- or Hbb-to-Pdk4 expression levels was higher in all SCD-related groups, including NI subjects, compared to TD controls (for the Hba1/2-to-Pdk4 ratio, F(3, 66) = 4.3180, p < 0.01 by one-way ANOVA; for Hbb-to-Pdk4 ratio, F(3, 66) = 2.8520, p < 0.05). In box-and-whisker diagrams of these ratios, overlapping sections were reduced by approximately ∼25%, indicating better resolution between TD and SCD subjects (Fig. 4C). Taken together, our findings suggest that concurrent alterations in Hba1/2, Hbb, and Pdk4 expression may be good molecular indices for postmortem diagnosis of sudden unexpected cardiac death due to a wide range of causes.
Fig. 4.
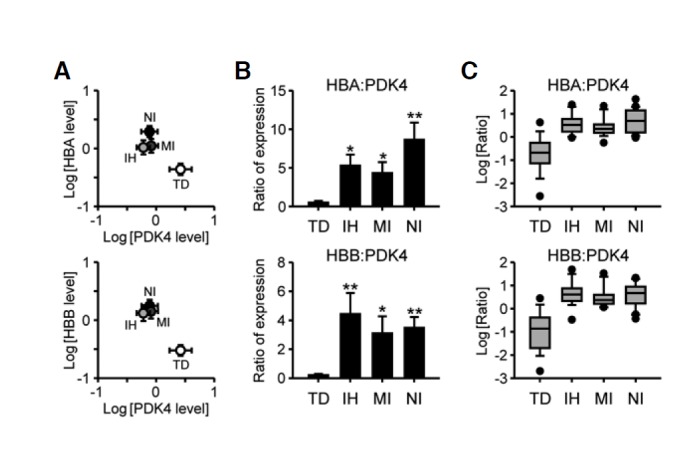
Pairwise analyses of relative mRNA abundances of Hba or Hbb in relation to Pdk4. (A) Pairwise analyses of Hba1/2 or Hbb mRNA levels in comparison with those of Pdk4. Normalized mRNA levels of Hba1/2 (upper) or Hbb (lower) relative to Gapdh were bi-directionally plotted in relation to those of Pdk4 on a logarithmic scale. Each symbol and error bar indicates the mean ± SEM. (B) Ratio of relative Hba1/2 (upper) or Hbb. (lower) mRNA level to that of Pdk4 in each case were calculated and expressed as means ± SEM. Data were statistically evaluated by one-way ANOVA followed by the Newman-Keuls test (*p < 0.05 and **p < 0.01 vs. TD). (C) The log ratios of Hba1/2 or Hbb to Pdk4 were expressed as box-and-whisker diagrams. Median values are indicated by the lines in the middle of the boxes.
DISCUSSION
In the present study, we aimed to identify LV myocardium-enriched mRNA species detectable in postmortem heart tissues with expression profiles associated with fatal cardiac dysfunctions leading to SCD. For this purpose, we compared genome-wide mRNA expression profiles in LV free wall tissues from subjects with three different modes of SCD and compared these profiles with those obtained for TD cases as a control group. We found that patterns of Hba1/2, Hbb, and Pdk4 mRNA expression in SCD cases were distinct from that observed in TD subjects, but common among subgroups of SCD subjects.
It should be noted that the concept of molecular or genetic autopsy was first established in postmortem investigation of SCD. Early molecular studies of SCD cases included detection of viral genomes in inflammatory cardiomyopathies as well as gene mutational analysis of several hereditary heart diseases (Basso et al., 2001; Tester and Ackerman 2006). Along with recent advances in molecular genetics, several genetic defects in cardiac ion channels such as KCNQ1, KCNH2, KCNE1/2, and SCN5A have been shown to be associated with certain cases of heritable and acquired heart malfunctions due to ventricular arrhythmias and sudden death.
However, it is also believed that RNA expression analysis of autopsy tissues from pathologic and non-pathologic hearts might be useful for postmortem investigation of SCD cases. Both qualitative and quantitative features of cardiac mRNA expression have been examined. For example, an increase in non-functional splice variants of the Scn5a gene transcript was shown to be associated with heart failure due to impairment of the cardiac Na+ current along with a reduction in NaV1.5 channel protein levels (Shang et al., 2007). In addition, Maeda and colleagues showed that several hypoxia-responsive gene transcripts encoding hypoxia-inducible factor-1α, erythropoietin, and vascular endothelial growth factor were locally increased in cardiomyocytes from acute myocardial infarction cases compared with asphyxiation cases, but not in recurrent myocardial infarction and uncharacterized SCD cases (Zhu et al., 2008). In a more recent study, these authors also reported that mRNA expression of atrial and brain natriuretic peptides in the LV free wall tended to be higher in various modes of cardiac death including acute ischemic heart disease, chronic congestive heart disease, and right ventricular cardiomyopathy than in LV tissues from acute fatal bleeding and/or mechanical asphyxiation cases (Chen et al., 2012). Therefore, it is crucial as well as practically useful to explore sets of myocardial RNA transcripts closely and commonly related to various modes of heart failure for postmortem diagnoses of SCD cases, even those without apparent anatomical signatures.
Hba1/2 and Hbb mRNAs have been believed to be predominantly expressed in cells of erythroid lineage. Recent studies, however, reported HBA1/2 and HBB expression in non-erythrocyte cells such as neurons (Richter et al., 2009), alveolar epithelial cells (Newton et al., 2006), endometrial cells (Dassen et al., 2008), hepatocytes (Liu et al., 2011), as well as cardiac cells (Arab et al., 2007; Kittleson et al., 2005; Solskov et al., 2012). In particular, Hare and colleagues showed that Hba1/2 and Hbb mRNA levels were upregulated in myocardial tissues biopsied from both ischemic and non-ischemic cardiomyopathy patients (Kittleson et al., 2005). Although the biological function of cellular hemoglobin in non-erythroid cells requires elucidation, cellular expression of these hemoglobin subunits appears to be induced in response to increased levels of intracellular reactive oxygen species (ROS), and they may play a protective role against oxidative stress (Arab et al., 2007; Li et al., 2013; Liu et al., 2011). In this context, it is noteworthy that oxidative stress, characterized as increased generation of mitochondrial ROS production in cardiac cells, is associated with various modes of heart failure (Belch et al., 1991; Hill and Singal, 1997; Matsushima et al., 2006). Prolonged ROS production can result in a variety of cardiac dysfunctions and damage the failing heart; it can impair cardiac contractility, activate a variety of hypertrophic signaling cascades and transcriptional networks, and directly mediate apoptosis (Tsutsui et al., 2011). It is therefore plausible that Hba1/2 and Hbb mRNA expression might be induced by oxidative stress during myocardial dysfunction leading to cardiac death.
PDK4, a mitochondrial protein, is known to play an important regulatory role in maintaining metabolic flexibility in cardiomyocytes by causing a switch from predominant oxidation of fatty acids to oxidation of glucose by phosphorylating pyruvate dehydrogenase (PDH). Loss of metabolic flexibility, such as that in diabetes mellitus, is closely related to the onset of cardiovascular diseases and heart failure (Alvarez-Guardia et al., 2010; Taegtmeyer et al., 2002; Zhao et al., 2008). It was recently found that a splicing mutant of the Pdk4 gene lacking exon 11, which is a prerequisite for PDK4 protein activity, underlay the pathophysiological phenotypes in a Doberman pinscher model of human dilated cardiomyopathy (Meurs et al., 2012; Wynn et al., 2008). More importantly, a systemic analysis to discover gene networks in the LV myocardium revealed that peroxisome proliferator-activated receptor gamma coactivator-1α (PGC-1α) target gene down-regulation may serve as a key signature of a failing human heart (Sihag et al., 2009). Impaired PGC-1α activity was shown to lead directly to a reduction in PDK4 expression, resulting in an increment in glucose oxidation (Alvarez-Guardia et al., 2010). Therefore, it is tempting to speculate that down-regulation of cardiac Pdk4 mRNA expression, either for genetic or environmental reasons, might be related to susceptibility to cardiac dysfunction. This possibility needs to be further examined.
From a practical viewpoint, routine normalization methods using a housekeeping gene in quantitative analyses of postmortem RNA expression often mislead and may obscure differences between cases and controls due to a variety of ante-mortem and postmortem factors differentially influencing integrity of each RNA species (Gonzalez-Herrera et al., 2013; Preece and Cairns, 2003; Zhang et al., 2013). Rather, pairwise analyses of a set of gene transcripts with concurrently changed, but negatively correlated under certain pathological condition, appeared to be more practical for forensic applications as we previously demonstrated (Chung et al., 2012). As shown in Fig. 4, Hba1/2 or Hbb mRNA expression was negatively correlated with Pdk4 mRNA expression, and paired analyses of two mRNA species (Hba/b and Pdk4) provided better indices implying cardiac death.
In conclusion, we suggest that concurrent alterations in the expression of heart-enriched Hba1/2, Hbb, and Pdk4 mRNAs in the postmortem LV free wall may reflect a wide spectrum of fatal cardiac dysfunctions. Expression profiles of a few mRNA transcripts may not be sufficient for postmortem diagnosis of SCD cases, especially non-ischemic cases; further evaluation of heart failure-related sets/pairs of gene transcripts would therefore be useful to enable practical application of such DEGs to forensically diagnose SCD.
Acknowledgments
This work was supported by the Korean government through the National Forensic Service, Ministry of Security and Public Administration (Grant: 1315000312, NFS-NG-2011-01). G.H.S. was supported by a Korea University Post-doctoral Fellowship. eWorldEditing.com (USA) edited the manuscript by the support of Korea University.
REFERENCES
- Alvarez-Guardia D., Palomer X., Coll T., Davidson M.M., Chan T.O., Feldman A.M., Laguna J.C., Vázquez-Carrera M. The p65 subunit of NF-kappaB binds to PGC-1alpha, linking inflammation and metabolic disturbances in cardiac cells. Cardiovasc. Res. 2010;87:449–458. doi: 10.1093/cvr/cvq080. [DOI] [PubMed] [Google Scholar]
- Arab S., Konstantinov I.E., Boscarino C., Cukerman E., Mori A., Li J., Liu P.P., Redington A.N., Coles J.G. Early gene expression profiles during intraoperative myocardial ischemia-reperfusion in cardiac surgery. J. Thorac. Cardiovasc. Surg. 2007;134:74–81. doi: 10.1016/j.jtcvs.2007.01.025. [DOI] [PubMed] [Google Scholar]
- Basso C., Calabrese F., Corrado D., Thiene G. Postmortem diagnosis in sudden cardiac death victims: macroscopic, microscopic and molecular findings. Cardiovasc. Res. 2001;50:290–300. doi: 10.1016/S0008-6363(01)00261-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belch J.J., Bridges A.B., Scott N., Chopra M. Oxygen free radicals and congestive heart failure. Br. Heart J. 1991;65:245–248. doi: 10.1136/hrt.65.5.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carturan E., Tester D.J., Brost B.C., Basso C., Thiene G., Ackerman M.J. Postmortem genetic testing for conventional autopsy-negative sudden unexplained death: an evaluation of different DNA extraction protocols and the feasibility of mutational analysis from archival paraffin-embedded heart tissue. Am. J. Clin. Pathol. 2006;129:391–397. doi: 10.1309/VLA7TT9EQ05FFVN4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.H., Michiue T., Ishikawa T., Maeda H. Pathophysiology of sudden cardiac death as demonstrated by molecular pathology of natriuretic peptides in the myocardium. Forensic Sci. Int. 2012;223:342–348. doi: 10.1016/j.forsciint.2012.10.018. [DOI] [PubMed] [Google Scholar]
- Chung U., Seo J.S., Kim Y.H., Son G.H., Hwang J.J. Quantitative analyses of postmortem heat shock protein mRNA profiles in the occipital lobes of human cerebral cortices: implications in cause of death. Mol Cells. 2012;34:473–480. doi: 10.1007/s10059-012-0214-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassen H., Kamps R., Punyadeera C., Dijcks F., de Goeij A., Ederveen A., Dunselman G., Groothuis P. Haemoglobin expression in human endometrium. Hum. Reprod. 2008;23:635–641. doi: 10.1093/humrep/dem430. [DOI] [PubMed] [Google Scholar]
- Franz H., Ullmann C., Becker A., Ryan M., Bahn S., Arendt T., Simon M., Pääbo S., Khaitovich P. Systematic analysis of gene expression in human brains before and after death. Genome Biol. 2005;6:R112. doi: 10.1186/gb-2005-6-13-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Herrera L., Valenzuela A., Marchal J.A., Lorente J.A., Villanueva E. Studies on RNA integrity and gene expression in human myocardial tissue, pericardial fluid and blood, and its postmortem stability. Forensic Sci. Int. 2013;232:218–228. doi: 10.1016/j.forsciint.2013.08.001. [DOI] [PubMed] [Google Scholar]
- Hill M.F., Singal P.K. Right and left myocardial antioxidant responses during heart failure subsequent to myocardial infarction. Circulation. 1997;96:2414–2420. doi: 10.1161/01.cir.96.7.2414. [DOI] [PubMed] [Google Scholar]
- Ikematsu K., Tsuda R., Nakasono I. Gene response of mouse skin to pressure injury in the neck region. Leg. Med (Tokyo) 2005;8:128–131. doi: 10.1016/j.legalmed.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Kittleson M.M., Minhas K.M., Irizarry R.A., Ye S.Q., Edness G., Breton E., Conte J.V., Tomaselli G., Garcia J.G., Hare J.M. Gene expression analysis of ischemic and nonischemic cardiomyopathy: shared and distinct genes in the development of heart failure. Physiol Genomics. 2005;21:299–307. doi: 10.1152/physiolgenomics.00255.2004. [DOI] [PubMed] [Google Scholar]
- Li X., Wu Z., Wang Y., Mei Q., Fu X., Han W. Characterization of adult α- and β-globin elevated by hydrogen peroxide in cervical cancer cells that play a cytoprotective role against oxidative insults. PLoS One. 2013;8:e54342. doi: 10.1371/journal.pone.0054342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W.S., Baker S.S., Baker R.D., Nowak N.J., Zhu L.X. Upregulation of hemoglobin expression by oxidative stress in hepatocytes and its implication in nonalcoholic steatohepatitis. PLoS One. 2011;6:e24363. doi: 10.1371/journal.pone.0024363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H., Zhu B.L., Ishikawa T., Michiue T. Forensic molecular pathology of violent deaths. Forensic Sci. Int. 2010;203:83–92. doi: 10.1016/j.forsciint.2010.07.024. [DOI] [PubMed] [Google Scholar]
- Matsushima S., Kinugawa S., Ide T., Matsusaka H., Inoue N., Ohta Y., Yokota T., Sunagawa K., Tsutsui H. Over-expression of glutathione peroxidase attenuates myocardial remodeling and preserves diastolic function in diabetic heart. Am. J. Physiol. Heart Circ. Physiol. 2006;291:H2237–H2245. doi: 10.1152/ajpheart.00427.2006. [DOI] [PubMed] [Google Scholar]
- Meurs K.M., Lahmers S., Keene B.W., White S.N., Oyama M.A., Mauceli E., Lindblad-Toh K. A splice site mutation in a gene encoding for PDK4, a mitochondrial protein, is associated with the development of dilated cardiomyopathy in the Doberman pinscher. Hum. Genet. 2012;131:1319–1325. doi: 10.1007/s00439-012-1158-2. [DOI] [PubMed] [Google Scholar]
- Miyazato T., Ishikawa T., Michiue T., Maeda H. Molecular pathology of pulmonary surfactants and cytokines in drowning compared with other asphyxiation and fatal hypothermia. Int. J. Legal Med. 2012;126:581–587. doi: 10.1007/s00414-012-0698-2. [DOI] [PubMed] [Google Scholar]
- Newton D.A., Rao K.M.K., Dluhy R.A., Baatz J.E. Hemoglobin is expressed by alveolar epithelial cells. J. Biol. Chem. 2006;281:5668–5676. doi: 10.1074/jbc.M509314200. [DOI] [PubMed] [Google Scholar]
- Oliva A., Brugada R., D’Aloja E., Boschi I., Partemi S., Brugada J., Pascali V.L. State of the art in forensic investigation of sudden cardiac death. Am. J. Forensic Med. Pathol. 2011;32:1–16. doi: 10.1097/PAF.0b013e3181c2dc96. [DOI] [PubMed] [Google Scholar]
- Pfaffl M.W., Horgan G.W., Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preece P., Cairns N.J. Quantifying mRNA in postmortem human brain: influence of gender, age at death, postmortem interval, brain pH, agonal state and interlobe mRNA variance. Brain Res. Mol. Brain Res. 2003;118:60–71. doi: 10.1016/s0169-328x(03)00337-1. [DOI] [PubMed] [Google Scholar]
- Richter F., Meurers B.H., Zhu C.N., Medvedeva V.P., Chesselet M.F. Neurons express hemoglobin alpha- and beta-chains in rat and human brains. J. Comp. Neurol. 2009;515:538–547. doi: 10.1002/cne.22062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang L.L., Pfahnl A.E., Sanyal S., Jiao Z., Allen J., Banach K., Fahrenbach J., Weiss D., Taylor W.R., Zafari A.M., et al. Human heart failure is associated with abnormal C-terminal splicing variants in the cardiac sodium channel. Circ. Res. 2007;101:1146–1154. doi: 10.1161/CIRCRESAHA.107.152918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sihag S., Cresci S., Li A.Y., Sucharov C.C., Lehman J.J. PGC-1α and ERRα target gene downregulation is a signature of the failing human heart. J. Mol. Cell. Cardiol. 2009;46:201–212. doi: 10.1016/j.yjmcc.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solskov L., Magnusson N.E., Kristiansen S.B., Jessen N., Nielsen T.T., Schmitz O., Bøtker H.E., Lund S. Microarray expression analysis in delayed cardioprotection: the effect of exercise, AICAR, or metformin and the possible role of AMP-activated protein kinase (AMPK) Mol. Cell. Biochem. 2012;360:353–362. doi: 10.1007/s11010-011-1075-z. [DOI] [PubMed] [Google Scholar]
- Taegtmeyer H., Razeghi P., Young M.E. Mitochondrial proteins in hypertrophy and atrophy: a transcript analysis in rat heart. Clin. Exp. Pharmacol. Physiol. 2002;29:346–350. doi: 10.1046/j.1440-1681.2002.03656.x. [DOI] [PubMed] [Google Scholar]
- Takahashi H., Ikematsu K., Tsuda R., Nakasono I. Increase in dual specificity phosphatase 1, TGF-beta stimulated gene 22, domain family protein 3 and Luc7 homolog (S. cerevisiae)-like messenger RNA after mechanical asphyxiation in the mouse lung. Leg. Med (Tokyo) 2009;11:181–185. doi: 10.1016/j.legalmed.2009.03.008. [DOI] [PubMed] [Google Scholar]
- Tsutsui H., Kinugawa S., Matsushima S. Oxidative stress and heart failure. Am. J. Physiol. Heart Circ. Physiol. 2011;301:H2181–H2190. doi: 10.1152/ajpheart.00554.2011. [DOI] [PubMed] [Google Scholar]
- Vennemann M., Koppelkamm A. mRNA profiling in forensic genetics I: Possibilities and limitations. Forensic Sci. Int. 2010a;203:71–75. doi: 10.1016/j.forsciint.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Wynn R.M., Kato M., Chuang J.L., Tso S.C., Li J., Chuang D.T. Pyruvate dehydrogenase kinase-4 structures reveal a metastable open conformation fostering robust core-free basal activity. J. Biol. Chem. 2008;283:25305–25315. doi: 10.1074/jbc.M802249200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhang P., Ma K.J., Lv Y.H., Li W.C., Luo C.L., Li L.L., Shen Y.W., He M., Jiang J.Q., et al. The selection of endogenous genes in human postmortem tissues. Sci. Justice. 2013;53:115–120. doi: 10.1016/j.scijus.2012.11.005. [DOI] [PubMed] [Google Scholar]
- Zhao D., Zhu B.L., Ishikawa T., Li D.R., Michiue T., Maeda H. Quantitative RT-PCR assays of hypoxia-inducible factor-1alpha, erythropoietin and vascular endothelial growth factor mRNA transcripts in the kidneys with regard to the cause of death in medicolegal autopsy. Leg. Med (Tokyo) 2006;8:258–263. doi: 10.1016/j.legalmed.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Zhao G., Jeoung N.H., Burgess S.C., Rosaaen-Stowe K.A., Inagaki T., Latif S., Shelton J.M., McAnally J., Bassel-Duby R., Harris R.A., et al. Overexpression of pyruvate dehydrogenase kinase 4 in heart perturbs metabolism and exacerbates calcineurin-induced cardiomyopathy. Am. J. Physiol. Heart Circ. Physiol. 2008;294:H936–H943. doi: 10.1152/ajpheart.00870.2007. [DOI] [PubMed] [Google Scholar]
- Zhao D., Ishikawa T., Quan L., Michiue T., Yoshida C., Komatu A., Chen J.H., Wang Q., Zhu B.L., Maeda H. Evaluation of pulmonary GLUT1 and VEGF mRNA levels in relation to lung weight in medicolegal autopsy cases. Leg. Med (Tokyo) Suppl. 2009;1:S290–293. doi: 10.1016/j.legalmed.2009.01.094. [DOI] [PubMed] [Google Scholar]
- Zhu B.L., Tanaka S., Ishikawa T., Zhao D., Li D.R., Michiue T., Quan L., Maeda H. Forensic pathological investigation of myocardial hypoxia-inducible factor-1 alpha, erythropoietin and vascular endothelial growth factor in cardiac death. Leg. Med (Tokyo) 2008;10:11–19. doi: 10.1016/j.legalmed.2007.06.002. [DOI] [PubMed] [Google Scholar]


